Abstract
Accumulations of Tau, a microtubule-associated protein, into neurofibrillary tangles is a hallmark of Alzheimer’s disease and other tauopathies. However, the mechanisms leading to this pathology are still unclear: the aggregates themselves could be toxic or the sequestration of Tau into tangles might prevent Tau from fulfilling its normal functions, thereby inducing a loss of function defect. Surprisingly, the consequences of losing normal Tau expression in vivo are still not well understood, in part due to the fact that Tau knockout mice show only subtle phenotypes, presumably due to the fact that mammals express several MAPs with partially overlapping functions. In contrast, flies express fewer microtubule-associated proteins, with Tau being the only member of the Tau/MAP2/MAP4 family. Therefore, we used Drosophila to address the physiological consequences caused by the loss of Tau. Reducing the levels of fly Tau (dTau) ubiquitously resulted in developmental lethality, whereas deleting Tau specifically in neurons or the eye caused progressive neurodegeneration. Similarly, chromosomal mutations affecting dTau also caused progressive degeneration in both the eye and brain. Although photoreceptor cells initially developed normally in dTau knockdown animals, they subsequently degenerated during late pupal stages whereas weaker dTau alleles caused an age-dependent defect in rhabdomere structure. Expression of wild type human Tau partially rescued the neurodegenerative phenotype caused by the loss of endogenous dTau, suggesting that the functions of Tau proteins are functionally conserved from flies to humans.
INTRODUCTION
The microtubule-associated protein (MAP) Tau was originally identified due to its activity in microtubule assembly and stabilization (Weingarten et al., 1975; Witman et al., 1976). Subsequently, it has gained considerably more attention as the major component of neurofibrillary tangles (Grundke-Iqbal et al., 1986), which together with amyloid plaques, are hallmarks of Alzheimer’s disease (AD). Besides AD, Tau accumulations have also been described in several other neurodegenerative diseases, collectively known as Tauopathies, including Cortico-basal degeneration, Pick’s Disease, Frontotemeporal Dementia and Parkinsonism linked to chromosome-17 (FTDP-17), and Progressive Supranuclear Palsy (Lee et al., 2001). While these Tauopathies all exhibit progressive neurodegeneration they vary in clinical presentation and for many of them the underlying causes are unknown. In the case of FTDP-17, it has been shown that mutations in Tau can actually cause the disease (van Swieten and Spillantini, 2007), confirming the importance of Tau in maintaining neuronal integrity. However, it still remains unclear whether the pathogenesis is due to the loss of functional Tau (Cash et al., 2003), to the potential neurotoxic effects of tangles (Iqbal et al., 2009) or Tau oligomers (Feuillette et al., 2010), to a switch of Tau isoform expression, or a combination of several of these factors. In agreement with a dominant function of Tau, expression of wild type human Tau or Tau variants carrying FTDP-associated mutations in the fly eye causes retinal degeneration, whereby the mutant forms have more severe effects (Wittmann et al., 2001). However, neither construct induced tangle formation. Only after co-expression of Tau kinases, neurofibrillary tangles could be observed (Jackson et al., 2002; Chau et al., 2006). Support for a loss-of-function mechanism comes from the finding that some mutations associated with FTDP-17 are localized in the microtubule-binding sites of Tau and reduce its ability to promote microtubule stabilization (LeBoeuf et al., 2008; Combs and Gamblin, 2012). Likewise, phosphorylation of Tau has been shown to decrease its microtubule binding affinity (Li et al., 2007; Barten et al., 2012), suggesting that disease-related hyperphosphorylation could also result in the loss of normal Tau function (Zhang et al., 2012). Most studies to define Tau activity have been conducted in vitro or have used overexpression assays and therefore authentic physiological functions for Tau are still largely unknown. Even the effects of Tau in regulating microtubule stability have not been confirmed in vivo, as Tau knock-out mice show no overt phenotypes (Ke et al., 2012). In part, this lack of a robust phenotype may be due to redundancy with other MAPs, and indeed MAP1A is upregulated about two-fold in Tau knock-out lines (Harada et al., 1994). Although compensatory mechanism may prevent major problems in brain integrity, it has recently been reported that behavioral changes and motor deficits are detectable when mice lacking Tau were aged (Lei et al., 2012). These results suggest that while the loss of Tau might initially cause relatively subtle effects on the brain, it may lead to more severe deleterious consequences over time.
Drosophila expresses a Tau orthologue (dTau) that is 46% identical and 66% similar to human Tau (Heidary and Fortini, 2001). In comparison to vertebrates, Drosophila expresses fewer MAPs, with dTau being the only member of the Tau/MAP2/MAP4 family (Dehmelt and Halpain, 2005). In addition, the sole MAP1-related protein in flies (Futsch) is more closely related to MAP1B, whereas they lack a MAP1A orthologue (Hummel et al., 2000). With fewer MAPs present in the fly one might expect less redundancy that could prevent the development of more severe phenotypes. Nevertheless, it has previously been reported that even in Drosophila the loss of dTau does not results in morphological or behavioral defects and that these flies are viable and fertile (Doerflinger et al., 2003). However, after re-examination of these flies, we found that they have not completely lost dTau because several transcripts that are made from an internal start site are present. Using a knock-down approach, we now found that dTau is an essential gene in flies and that a reduction in dTau cause defects in rhabdomere formation and progressive degeneration in the eye and central nervous system. The degeneration is enhanced by a mutation in MAP1B, showing that though flies have less MAPs there still is some redundancy in their function. Nevertheless, our results are the first to show that loss of Tau can induce degenerative phenotypes.
METHODS
Drosophila stocks
GMR-GAL4, elav-GAL4, actin-GAL4, tubulin-Gal4, tauHM05101, tubP-GAL80ts, UAS-Dcr-2, tauDf(3R)MR22, Df(3R)BSC499, and UAS-hTau23 (UAS-MAPT.A59A) were obtained from the Bloomington Stock Center. UAS-dTau::GFP and tauEP3597 were a gift of D. St. Johnston (University of Cambridge, Great Britain). Appl-Gal4 was obtained from Laura Torroja (Universidad Autonoma de Madrid, Spain). tauGD25023 was received from the Vienna Drosophila RNAi Center (Vienna, Austria). olk1 mutants are described in (Bettencourt da Cruz et al., 2005). Flies were raised under standard conditions at 25°C unless specifically described. To create the MAP60 and MAP205-expressing flies, we used LD02709 for MAP60 and LD12965, which were cloned into pUAST. Transgenic flies were generated by P-element transformation using Best Gene Inc. (Chino Hills, CA).
RT-PCR
RNA extraction from Drosophila heads and cDNA synthesis was performed as previously described in Bolkan et al. (2011). For quantitative PCR, the SYBR Green PCR Master Mix Kit (Applied Biosystems) was used and samples were analyzed on a Bio-Rad iCycler iQ. We used GGCGGCGAGAAGAAGATAGT and CTTGGCCTTGTCCTTGAAGT as primers for dTau and GCGGCAGCTACAACACAATA and CATCATTACCAGGGCGAAAT for the control reaction using the swiss-cheese gene. Fold changes were calculated using the Pfaffl method (Pfaffl, 2001). For the non-quantitative analysis, cDNA was synthesized in the same way, but the amplification reaction was done using Taq DNA Polymerase (Fermentas, ThermoScience) with 25 cycles. Samples were then run on 1.5% agarose gels and visualized. All reactions employed the same reverse primer as used for quantitative analysis, combined with the forward Primer for exon 1 (GAGGGCTAATCAGGATCAA) and the forward primer for the alternative start site (ATGCCTCCCTCATGTATCTTT).
Tissue sections for light microscopy and transmission electron microscopy
Paraffin sections for light microscopy were prepared as described in (Bettencourt da Cruz et al., 2005). Briefly, whole flies were fixed in Carnoy’s solution and dehydrated in an ethanol series followed by incubation in methyl benzoate before embedding in paraffin. Sections were cut at 7μm and imaged using the auto-fluorescence caused by the dispersed eye pigment. For electron microscopy, ultrathin Epon plastic sections were prepared as described in (Kretzschmar et al., 1997). TEMs were analyzed with a FEI Morgagni 268 electron microscope.
Measurement of the vacuolar pathology
Paraffin sections of heads were imaged and measured in a blinded manner. For an analysis of neurodegeneration in brain sections, the area containing the most severe example of pathology in each sample was photographed for further analysis. Sizes of holes in the central brain were measured in Adobe Photoshop (in pixels), while eye degeneration measurements were done using the threshold discrimination function of ImageJ. For each eye the total was selected and the area within that region that fell below a predetermined fluorescent threshold (corresponding to vacuoles) was measured as a percentage of the total area of the retina. As controls, we used flies from the same cross, that only carried the UAS or GAL4 construct to minimize genetic background effects. Statistics were performed using one-way ANOVA and a Dunett’s post-test in cases where several experimental groups were compared to one control group. Student’s Neuman-Keuls tests were used when only comparing two data sets.
Measurement of microtubules
To determine microtubule density, neurites were selected that contained microtubules cut in cross-sections and were free of mitochondria, as the area occupied by mitochondria may reduce the number of microtubules. We then counted the number of microtubules in a neurite and also determined the the cross-sectional area of the neurite by outlining the perimeter using ImageJ. The number of microtubules per μm2 was then calculated from these measurements. No more than 10 neurites per image were analyzed, and at least 3 animals per genotype were used to determine average microtubule densities. To measure the cross sectional area of individual microtubules, the outer perimeter was circled and the area measured using ImageJ. No more than 30 microtubules per image were analyzed, and at least 3 different animals per genotype were used.
Immunohistochemistry
For vibratome sections, heads were fixed overnight in 4% Paraformaldehyde (PFA), washed, and briefly rinsed in 100% ethanol to enhance binding of the tissue to the 3% agar in which they were embedded. Sections were cut on a Leica VT1000 S Vibratome at a thickness of 50 μm. For detecting the endogenous expression pattern of dTau, we used y1w1118 to minimize interference by autofluorescence from the eye pigment. Sections were stained with a custom anti-dTau antibody at a dilution of 1:2000. Anti-dTau was raised in chickens against a synthetic peptide containing amino acid sequence CZEKKIFDDKDYLKNVEHS by Aves Labs (Tigard, OR). Cy3-labeled anti-chicken secondary antibodies were used at 1:1000 (Jackson ImmunoResearch). All washes were performed with PBS+ 0.1% Triton-X (PBST). Dilutions and blocking steps were done in PBST + 2% goat serum (Jackson ImmunoResearch). Images were taken on an Olympus FW1000 confocal microscope. For immunohistochemical labeling of eye discs and larval brains, brains of 3rd instar larvae were dissected in PBS with the discs attached. They were then fixed for 2 hrs at room temperature in 4% PFA, blocked for 1 hr, and then incubated in primary antibody for 3 days at 4°C; dTau was used at 1:1000 and 24B10 (anti-chaoptin) at 1:300 (Developmental Studies Hybridoma Bank, DSHB). After rinsing, the tissue samples were incubated with corresponding secondary antibodies overnight at 4°C: anti-chicken AF488 (Invitrogen) and anti-mouse-Cy3 (Vector Labs) were both used at 1:1000. Samples were post-fixed in 4% PFA for 15 min, then cleared and mounted in Scale A2 buffer (Hama et al. 2011).
Western Blots
Western Blots were performed as described in (Carmine-Simmen et al., 2009). Lysates of 10 heads were loaded on 8% SDS polyacrylamide gels, blotted onto PVDF membranes (Amersham Biosciences), and labeled with our dTau antisera at 1:20,000. For loading controls, we used nc82 (anti-bruchpilot; DSHB) at 1:250.
RESULTS
Loss of dTau causes retinal degeneration
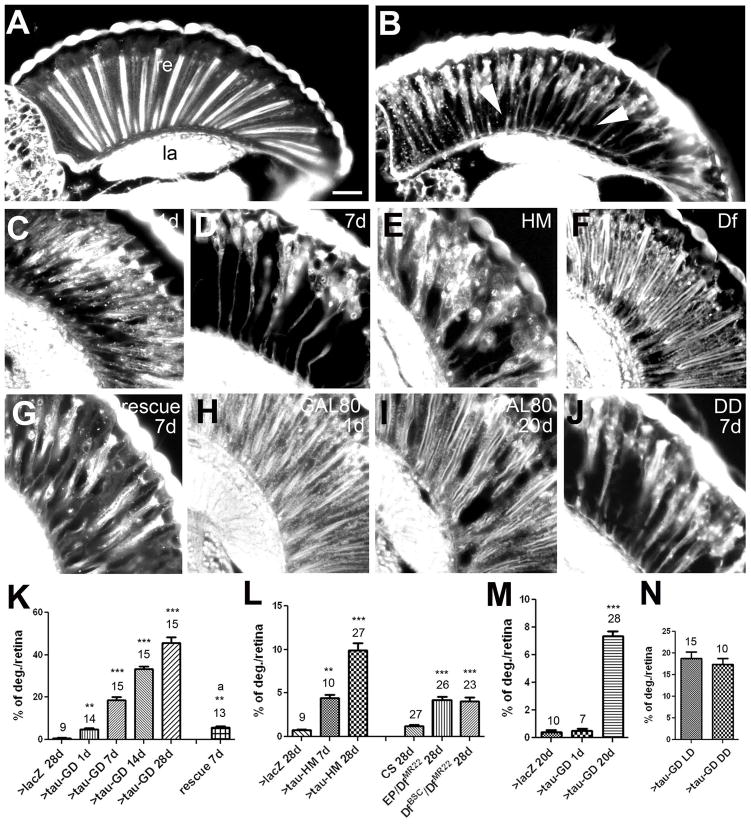
Previous studies reported that dTau is strongly expressed in the fly eye (Heidary and Fortini, 2001). We therefore first investigated whether we could detect a phenotype when dTau expression was reduced in the eye. Indeed, GMR-GAL4; tauGD25023 RNAi flies showed a rapid progressive pattern of retinal degeneration: whereas at 1d post-eclosion (24–36 hours after adult emergence), vacuoles were only found in the most proximal area of the retina (Fig. 1C), the pattern of degeneration had spread about two thirds along the length of the retina after 1 week [Fig. 1(B, D)]. By four weeks, these degenerative effect has progressed even further, encompassing almost the entire retina while leaving only the cornea intact [not shown, for quantification see Fig. 1(K)]. To confirm that this phenotype was specifically due to loss of dTau (and not an artifact of the RNAi construct), we tested another RNAi line (tauHM05101) that, as described below, targets a different region within the dTau mRNA sequence. Inducing this construct with GMR-GAL4 (combined with UAS-Dicer-2) resulted in a similar, although weaker, degeneration that started in the proximal part of the retina and became progressively worse when the flies were aged for 4 weeks [Fig. 1(E), quantification in (L)]. Finally, we analyzed flies that carried an insertion in dTau (tauEP3597) over a deletion that includes dTau (Df(3R)tauMR22), a combination that has been proposed to completely eliminate dTau expression (Doerflinger et al.,2003). In addition, we tested the effects of the Df(3R)tauMR22 deletion over an overlapping deficiency, Df(3R)BSC499 (see fig. 6A), which should also result in a complete loss of dTau. Again these flies showed retinal degeneration [Df(3R)BSC499/Df(3R)tauMR22 in Fig. 1(F)], but this phenotype was weaker than seen in both knockdown lines [Fig. 1 (L)]. However, as described later, these deficiency combinations do not in fact delete all dTau transcripts. Lastly, to verify that the retinal degeneration seen in the knockdown lines was caused by the loss of dTau, we performed a rescue experiment by co-expressing UAS-dTau with tauGD25023. As shown in figure 1G, this resulted in a significant reduction of the retinal degeneration detected at one week of age, compared with age-matched controls co-expressing UAS-LacZ [Fig. 1(K)]. To address whether the retinal degeneration seen in GMR-GAL4; tauGD25023 flies might be due to early developmental defects, we also introduced tubP-GAL80ts into this line. Allowing the flies to develop at the permissive temperature of 18°C did not result in retinal degeneration in 1d old flies [Fig. 1(H)]. In contrast, when these flies were shifted to 29°C after eclosion (to induce expression of the RNAi construct) and aged for 20d, we did again observe retinal degeneration [Fig. 1(I), quantification in (M)]. This experiment revealed that dTau expression is required to maintain retinal integrity during adulthood, although the weaker phenotype seen in these flies (compared to GMR-GAL4; tauGD25023 flies) suggests that dTau might also play a more subtle role in photoreceptor differentiation. Finally, we tested whether rearing the flies in the dark prevented or delayed the degenerative phenotype. However, quantifying the vacuoles in these flies at 7d did not reveal any significant changes compared to GMR-GAL4; tauGD25023 flies kept in a light-dark cycle [Fig. 1(J), quantification in (N)]. This result shows that the retinal degeneration caused by the loss of dTau is not dependent on light and therefore is not affected by phototransduction activity per se.
Figure 1.
Decreasing dTau levels in the eye causes progressive retinal degeneration. A: A four-week-old GMR-Gal4/UAS-LacZ control fly shows an intact retina (re) and underlying lamina (la). B: In contrast, vacuoles (arrowheads) can be detected in the retina of a 7d old GMR-GAL4; tauGD25023 fly. C: This phenotype is already detectable at 1-day-old but increases with age (7d in D). E: Retinal degeneration also occurs in a 4-week-old GMR-GAL4; tau HM05101 fly and a tauDf(3R)MR22/Df(3R)BSC499 fly aged for four weeks (F). G: Co-expression of UAS-dTau with tauGD25023 reduces retinal degeneration when comparing to the age-matched control in (D). H: A GMR-GAL4; tauGD25023; tubP-GAL80ts fly raised at the permissive temperature does not show retinal degeneration when 1d old. I: However, aged for 20d at the restrictive temperature, these flies again show retinal degeneration. J: Aging GMR-GAL4; tauGD25023 flies in constant darkness did not prevent the retinal degeneration observed when 7d old. Quantification of the retinal degeneration observed in GMR-GAL4; tauGD25023 (K), GMR-GAL4; tau HM05101 and tauDf(3R)MR22/Df(3R)BSC499 (L), and in GMR-GAL4; tauGD25023; tubP-GAL80ts (M). N: Quantification of the retinal degeneration in flies raised in a light:dark cycle or in constant darkness. The number of analyzed retinae and the SEMs are indicated for each bar. ** p<0.01, *** p<0.001. Scale bar in A=20μm.
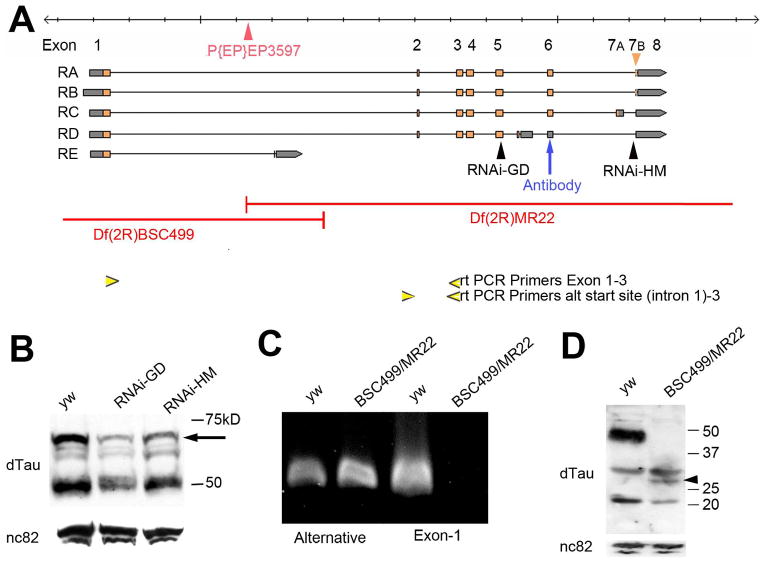
Figure 6.
The dTau locus encodes several alternative transcripts and protein isoforms. A; Map of the dTau locus (modified from flybase.org). The five annotated transcripts are shown and the areas deleted in Df(3R)BSC499 and tauDf(3R)MR22 are indicated by red lines. The small 7B exon only present in the RA and RB transcript is pointed out by an orange arrowhead. Also indicated are the locations of the tauEP3597 insertion (red arrowhead), the regions targeted by the RNAi construct (black arrowheads), and the locations of the antibody epitope (blue arrow). The yellow arrowheads represent the positions of the primers used for the PCR reactions. B: Western blot from head extracts using anti-dTau shows multiple bands corresponding to various dTau isoforms. Whereas induction of tauGD8682 via GMR-GAL4 decreased the levels of all protein isoforms, induction tauHM05101 reduced the levels of the largest isoform (arrow) but did not affect smaller isoforms. C: RT-PCRs reveal products in both y w control flies and tauDf(3R)MR22/Df(3R)BSC499 flies when using primes corresponding to the alternative start site and exon 3 (lane 1 and 2). However, when using primers corresponding to exon 1 and exon 3, a band can only be detected in y w controls (lane 3 and 4). D: A Western blot shows a strong 50kD band and several smaller bands in head extracts from y w controls (lane 1). In head extracts from tauDf(3R)MR22/Df(3R)BSC499 flies the 50kD band is absent whereas the smaller bands are still present and a new band is detectable. B, D: Loading controls using nc-82 (anti-Bruchpilot) are shown below.
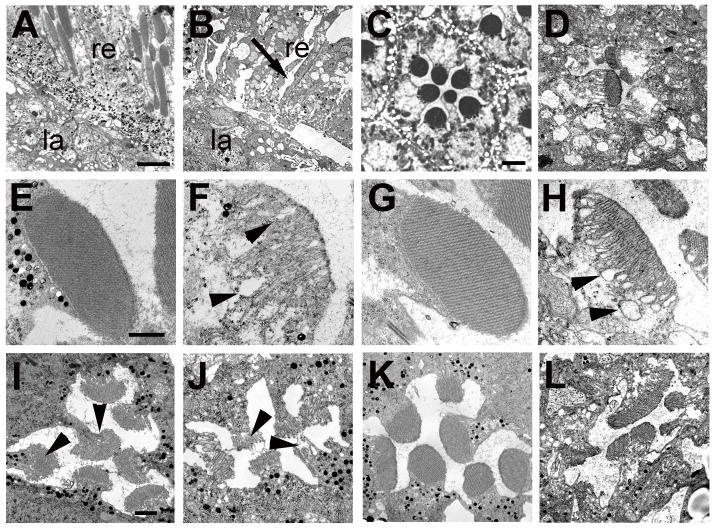
To study the defects on the eye caused by the loss of dTau in more detail, we used transmission electron microscopy. As early as 24–36h after eclosion, the retinas of GMR-GAL4; tauGD25023 flies looked highly disorganized and contained obvious vacuoles [Fig. 2(B), arrow]. Cross sections of these retinas revealed that the ommatidia contained fewer photoreceptor cells than normal [Fig. 2(D)], while the ommatidia that were present had abnormal rhabdomeres with loosened membranes, especially in proximity to the cell body [Fig. 2(F), arrowheads]. In contrast, age-matched control flies showed the highly organized array of 7 photoreceptors with intact rhabdomeres, typical of wild type retinas [Fig. 2 (A, C, E]). The retinas and rhabdomeres of tauDf(3R)MR22/Df(3R)BSC499 flies, which at the light microscopic level showed a much weaker phenotype, were indistinguishable from wild type flies when analyzed 1d after eclosion [Fig. 2(G)], but when aged for 7 days, they also showed a loosening of the microvilli [Fig. 2(H), arrowheads]. In contrast to the knockdown flies, we did not detect photoreceptor loss in these flies at this age.
Figure 2.
Transmission electron microscopy reveals degeneration of rhabdomeres. A: A GMR-GAL4/UAS-LacZ fly at 36 hours post eclosion shows an intact retina (re) and lamina (la). B: In a 36hr old GMR-GAL4; tauGD25023 fly the retina is disorganized and vacuoles have formed in the retina (re, arrow). C: An intact ommatidia in a 36h old GMR-GAL4/UAS-LacZ control with seven photoreceptors present. D: In contrast, a 36h old GMR-GAL4; tauGD25023 fly shows a disrupted ommatidial structure with only a few abnormal looking photoreceptors present. Whereas control flies have highly structured rhabdomeres at this age (E), the rhabdomeres of the photoreceptors that are present in GMR-GAL4; tauGD25023 have an abnormally loose structure with loops forming that are mostly found in proximity to the cell cytoplasm (F, arrowheads). G: In contrast, the rhabdomeres are intact in a 1d old tauDf(3R)MR22/Df(3R)BSC499 fly but become more loose when aged for 7d (H). I: P11 control pupa showing the developing seven rhabdomeres (arrowheads). J: In an age-matched GMR-GAL4; tauGD25023 fly, the rhabdomeres already look abnormal although all photoreceptors are present at this stage. K: Pharate adult control fly. L: A pharate adult GMR-GAL4; tauGD25023 fly has lost several photoreceptors and the remaining photoreceptors are abnormally shaped and have irregular rhabdomeres. Scale bars in A=6μm, in C=2μm, in E=1μm, and in I=1μm.
dTau is required for rhabdomere integrity
As mentioned above, the severe phenotype detectable in GMR-GAL4; tauGD25023 flies by one day after eclosion suggested that dTau could play a role in the development as well as the maintenance of the retina. To determine whether we could detect more subtle defects during photoreceptor and rhabdomere differentiation, we therefore performed sections from pupae at stage P11 (73–78 hours ppf). At this stage, the photoreceptors have started to form rhabdomeres, which are easily detectable in control flies [Fig. 2(I), arrowheads]. Although GMR-GAL4; tauGD25023 flies had also started to form rhabdomeres at this stage, they were noticeably smaller, more disorganized, and less compact [Fig. 2(J), arrowheads]. Surprisingly however, all seven photoreceptors were present at this stage. In contrast, by 12–18 hours later (the pharate adult stage), the ommatidia in knockdown flies had lost several photoreceptor cells and showed severely deformed rhabdomeres [Fig. 2(L)], compared with the well-structured rhabdomeres in age-matched wild type controls [Fig. 2(K)]. Together, these experiments suggest that dTau is not required for the determination and initial development of photoreceptors but does play an essential role in maintaining their structural integrity and survival into adulthood.
dTau is essential for maintenance of the central nervous system
To address whether dTau plays a role in other cells, we first induced the tauGD25023 construct ubiquitously. In contrast to mice, in which the loss of Tau does not cause lethality, the ubiquitous knockdown of dTau via tubulin-GAL4 resulted in complete lethality in flies, with the majority of animals dying as late larvae or pupae. To determine whether this lethality was due to the loss of neuronal dTau, we knocked down dTau pan-neuronally using the Appl-GAL4 and elav-GAL4 driver lines. Expression of tauGD25023 via Appl-GAL4 again resulted in most of the flies dying before adulthood (only 3% survived), suggesting that the developmental lethality observed with tubulin-GAL4 is due to neuronal defects. This lethality could be rescued by co-expression of UAS-dTau, with 80% of the of Appl-GAL4;tauGD25023 flies reaching adulthood, whereas flies that co-expressed the UAS-LacZ control construct still exhibited 88% lethality, confirming that this effect was caused by the loss of dTau. Using elav-GAL4 to knock down dTau had less dramatic consequences than with Appl-GAL4, resulting in 45% of the flies eclosing. Performing head sections from the surviving flies at 1d revealed no obvious abnormalities [Fig 3(A)] however, when aged for 14d, vacuoles could be found scattered throughout the brain [Fig. 3(B)]. We also detected vacuoles in the brain of 28d old Df(3R)BSC499/Df(3R)tauMR22 flies [Fig. 3(C)]. Quantifying this degenerative phenotype by measuring the overall vacuole area in the different lines revealed a mean of 17.1 ± 1.3 μm2 in 14d old dTau knockdown and 19.4 ± 2.1 μm2 in 28d Df(3R)BSC499/Df(3R)tauMR22, compared to 1.4 ± 0.2 μm2 in age-matched control flies (expressing UAS-GFP with elav-GAL4) [p<0.001; Fig 3(D)]. Again, we addressed whether knocking down dTau expression during adulthood was sufficient to induce degeneration by rearing tauGD25023; elav-GAL4/tubP-GAL80ts flies at the permissive temperature of 18°C before shifting them to 30°C after eclosion. Whereas no degeneration was detectable in 1d old flies [Fig. 3(E)], significant vacuolization was detectable at 21d [Fig 3(F, H)] when compared to age-matched controls that were raised and aged under the same paradigm [Fig. 3(G)]. Surprisingly, the 21d old elav-GAL4/tubP-GAL80ts control flies were significantly worse than 1d old experimental flies (8.7 ± 0.8 μm2 versus 3.9 ± 0.5 μm2). We assume that his phenotype is due to the enhanced aging process when raising these flies at 30°C because we previously showed that a sparse number of vacuoles occurs with normal aging (Krishnan et al., 2009).
Figure 3.
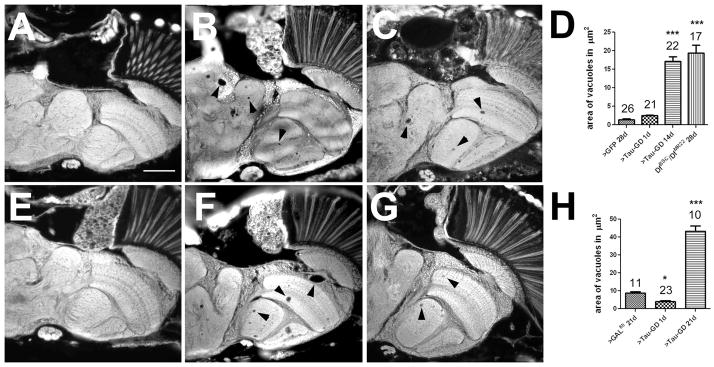
Loss of dTau causes degeneration in the brain. A: No degeneration is detectable in a 1d old tauGD25023; elav-GAL4 fly. However after 14d several vacuoles have formed in the brain (arrowheads, B). C: Vacuoles can also be found in a 28d old tauDf(3R)MR22/Df(3R)BSC499 fly. D: Quantification of the area of vacuoles. E: tauGD25023; elav-GAL4/tubP-GAL80ts flies do not reveal vacuolization at 1d but vacuoles have formed after being aged for 21d at the restrictive temperature (F). G: A elav-GAL4/tubP-GAL80ts aged for 21d at the restrictive temperature also shows a few small vacuoles. H: Quantification of the vacuoles when raised at the restrictive temperature. D, H: The number of analyzed flies and the SEMs are indicated for each bar. * p<0.05, *** p<0.001. Scale bar in A=50μm.
Analyzing electron microscopic sections from these flies revealed that 1d old tauGD25023; elav-GAL4 flies had more rounded axons surrounded by extracellular space [Fig. 4(B), arrowheads], in contrast to the tightly packed, more irregularly shaped axons seen in 1d old wild type flies [Fig 4(A)]. In addition, many of the axons of the tauGD25023;elav-GAL4 flies had intracellular inclusions [Fig. 4(B), arrow]. Although 1d old tauDf(3R)MR22/Df(3R)BSC499 flies were more similar to wild type controls, exhibiting, tighter packed irregularly shaped axons, we also could occasionally detect abnormal gaps between their axons [Fig. 4(C), arrowhead]. However, by 16d, many more and larger extracellular gaps had formed between the axons of these flies [Fig. 4(D), arrowheads], resembling the 1d old tauGD25023; elav-GAL4 flies.
Figure 4.
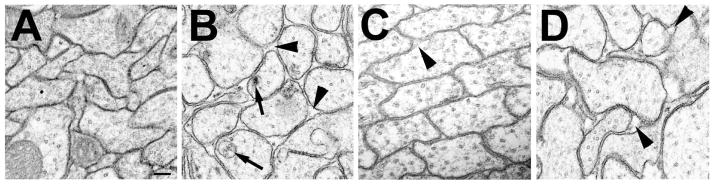
Decreasing the levels of dTau affects axonal morphology. A: Transmission electron microscopy reveals tightly packed axons in a 1d old wild type fly. B: In a 1d old the tauGD25023; elav-GAL4 fly the axons look more rounded and extracellular gaps are visible (arrowheads). In addition, intracellular inclusions and vacuoles can be detected (arrows). C: In tauDf(3R)MR22/Df(3R)BSC499, the axons are tighter packed and only a few extracellular gaps are visible (arrowhead) at 1d of age, however, the number and size of these gaps increases with age as seen in a 16d old fly (D). Scale bar in A=200μm.
Loss of dTau affects microtubule structure and density
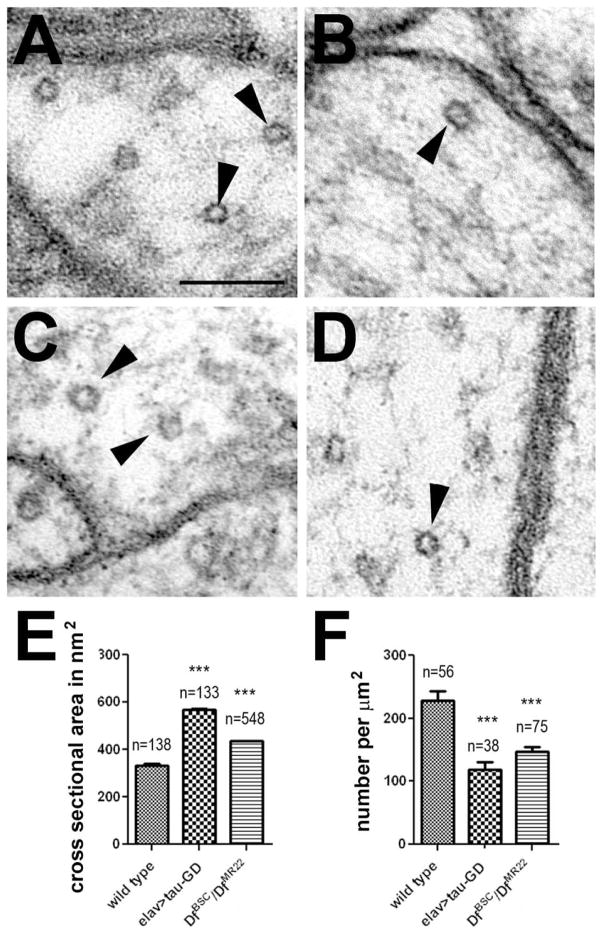
Because Tau has been shown to affect microtubule stability in vitro, we used electron microscopy to detect possible abnormalities in microtubule morphology caused by the loss of dTau. Measuring the area of individual microtubules in cross sections using the outer perimeter, we found an increase in the mean cross sectional area form 335 ± 5.2 nm2 in wild type [Fig. 5(A), quantification in E] to 567 ± 6.1 nm2 in tauGD25023; elav-GAL4 flies [Fig 5(B)]. An increase in the mean cross sectional area of microtubules was also detectable in tauDf(3R)MR22/Df(3R)BSC499 flies [Fig. 5(C)], with 435 ± 1.6 nm2 [Fig. 5(D), quantification in E]. In addition, we measured microtubule density by counting how many microtubules are present in neurites. To account for the different sizes of individual neurites, we measured the area of the neurites and then calculated the number of microtubules per μm2 neurite area. This revealed a significant decrease in microtubule density in the knockdown and the deficiency flies with 119 ± 15 and 148 ± 9 microtubules per μm2 axon area compared to 228 ± 15 per μm2 axon in wild type flies [Fig. 5(F)].
Figure 5.
Decreasing the levels of dTau affects microtubule morphology and density. A: Microtubules (arrowheads) in a wild type fly. B: Less and larger microtubules are found in a tauGD25023;elav-GAL4. C: A 1d old tauDf(3R)MR22/Df(3R)BSC499 fly also has larger microtubules and this in confirmed in a 14d old fly (D). E: Quantification of the microtubule mean cross sectional area in nm2. The number of measured individual microtubules and the SEMs are indicated. F: Number of microtubules found per μm2 axon area. The number of analyzed axons (from at least three animals and at least 6 images) and the SEMs are indicated. All flies were 1d old. *** p<0.001. Scale bar in A=100nm.
The dTau locus encodes multiple alternative transcripts and protein isoforms
Alternative splicing of dTau was first proposed by Doerflinger et al. in 2003, and five alternative transcripts are annotated in Flybase that would encode four different protein isoforms. The tauGD25023 RNAi construct targets exon 4, thereby affecting four out of the five transcripts [Fig. 6(A)]. In contrast, the tauHM05101 RNAi is complementary to a small exon just before the last exon that is only present in the RA and RB transcript [orange arrowhead, Fig 6(A)]. To determine the effects of each RNAi construct on the expression levels of the dTau transcripts, we performed a quantitative RT PCR using primers against a fragment contained in all the transcripts except RE (exons 4–5). This approach revealed a 1.89 fold decrease in dTau mRNA levels in GMR-GAL4;tauHM05101 fly heads and a 4.21 fold decrease in GMR-GAL4;tauGD25023 (note that dTau is still present in the brain), confirming that the tauHM05101 RNAi is less efficient than tauGD25023.
To detect changes in dTau protein levels, we used an antibody that we raised against a peptide within exon 6. Both RNAi constructs reduced the levels of the highest molecular weight band [arrow, Fig. 6(B)], likely corresponding to the RA and RB isoforms, although the effect was weaker with tauHM05101, correlating with the weaker effect of this RNAi construct on mRNA levels. In contrast, lower molecular weight bands (arrowheads) appeared unaffected by inducing expression of the tauHM05101 RNAi construct, whereas they were reduced with tauGD25023. Given the fact that the tauHM05101 sequence is located within an exon that is only contained in the RA and RB isoforms, these results strongly suggest that tauHM05101 RNAi construct does not affect all of the dTau isoforms, therefore causing a much weaker phenotype than tauGD25023.
Similarly, our analysis suggests that flies trans-heterozygous for tauDf(3R)MR22 and Df(3R)BSC499 do not lack all dTau isoforms. Both the the first exon and upstream region of dTau are deleted in tauDf(3R)MR22, whereas Df(3R)BSC499 deletes exons 2–8 [red line, Fig. 6(A)]; therefore, the trans-heterozygotes should lack all annotated transcripts. Nevertheless, these flies are viable and have a relatively mild phenotype. To find an explanation for this conundrum, we investigated whether an internal start site might exist within the dTau coding region, as has been suggested for MAP1B (Liu and Fischer, 1996). Indeed, an analysis of the dTau gene using the McPromoter Prediction Server (Ohler, 2006) hosted by BDGP, fruitfly.org) identified a strongly predicted promoter (probability score 1.0) just upstream of exon 2, with a nearby start site in the correct reading frame. To determine whether mRNAs containing this region are present in fly heads, we designed a corresponding primer and used it in combination with a primer in exon 3 [Fig. 6(A)]. Amplifying cDNAs obtained from y1w1118 control flies, we detected a PCR product of the expected size, demonstrating that an authentic transcript is indeed made from this alternative start site [Fig. 6(C,) lane 1]. This fragment is also detectable in tauDf(3R)MR22/Df(3R)BSC499 flies, with the band appearing even stronger than in the controls (lane 2). In contrast, using primers for a fragment spanning exon 1–3 did not result in a product when using cDNAs from the double deficiency (lane 4), confirming that none of the annotated transcripts that use the original start site is present in these flies. In contrast, this fragment was easily detectable in control flies (lane 3). To determine whether shorter transcripts made from the alternative start site are translated into proteins, we performed Western Blots with our anti-dTau antibody (which should detect the smaller forms), and found that lower molecular weight isoforms could indeed be detected in control flies, although there levels were quite low compared to the major band at 50kD [Fig. 6(D,) lane 1]. In contrast, whereas, the 50kD band was absent in tauDf(3R)MR22/Df(3R)BSC499 head extracts (as expected), the lower molecular forms were readily detectable, along with an additional smaller band (arrowhead). Because the first exon, which is missing in these alternatively transcript variants, does not contain any of the conserved regions identified in Tau (including all the microtubule binding domains), we assume that these dTau isoforms are probably functional. Therefore, we assume that their presence in tauDf(3R)MR22/Df(3R)BSC499 prevents the developmental lethality and more severe degenerative phenotypes seen in our knockdown lines.
dTau is expressed in the eye and central nervous system
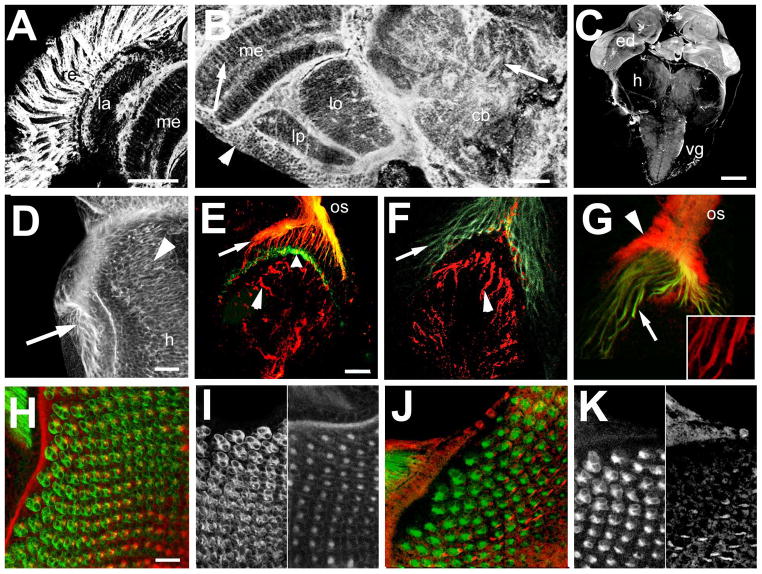
dTau has previously been described as being primarily expressed in the developing and adult eye (Heidary and Fortini, 2001). Using our dTau antiserum, we confirmed strong dTau expression in the retina [Fig. 7(A)] but we also detected significant amounts in all other areas of the adult brain [Fig. 7(B)]. dTau was present in most neuronal cell bodies [arrowhead, Fig. 7(B)] as well as in the neuropils of the optic system and the central brain (arrows). In third instar larvae, dTau expression was most prominent in the eye discs [ed, Fig 7(C)]; however, weaker immmunostaining was also detectable in the ventral ganglia (vg) and in the outer optic anlagen of the brain hemispheres [arrowhead, Fig. 7(D)]. Double staining with the photoreceptor marker 24B10 (chaoptin) revealed that while dTau was present in photoreceptor axons [arrow, Fig. 7(E)], it did not appear to extend into their terminals (broad arrowhead). We also detected dTau in neurons within the optic anlagen that were not stained by 24B10 (arrowhead). In contrast, dTau was hardly detectable in the photoreceptor axons of GMR-GAL4;tauGD25023 larvae [arrow, Fig. 7(F)] whereas, it was still present in neurites in the brain hemispheres (arrowhead), confirming the specific knockdown of dTau in the eye. In agreement with our Western Blot experiments, some dTau could still be detected in tauDf(3R)MR22/Df(3R)BSC499 flies, but the staining pattern was different than in wild type animals. Whereas we could only detect low levels in their photoreceptor axons [arrow, Fig. 7(G), and inset], immunostaining in the optic stalk (os) was much stronger (arrowhead). However, this staining did not colocalize with 24B10, suggesting that it might be due to dTau expression by the glia that ensheath the optic stalk (Hummel et al., 2000). Altered expression of dTau could also be observed in the eye disc. In wild type controls, dTau immunoreactivity was found in a punctuate pattern [Fig. 7(H, I), right panel] that seemed to correspond with developing axons. By comparison, counterstaining with 24B10 outlined the cell bodies and axons [left panel in Fig. 7(I)], consistent with the cell surface localization of chaoptin (Reinke et al., 1988). In contrast, not only was there considerably reduced dTau immunoreactivity in tauDf(3R)MR22/Df(3R)BSC499 flies, it also appeared less punctuate [Fig. 7(J, K), right panel]. The difference in the expression pattern seen in tauDf(3R)MR22/Df(3R)BSC499 flies might be due to the specific localization of isoforms that do not contain exon 1 (and that are still present in this mutant).
Figure 7.
dTau is expressed in the eye and brain. A, B: Vibratome sections stained with anti-dTau show that dTau is strongly expressed in the adult eye (A, confocal stack of 25 0.5μm optical sections) but it can also be found in the brain (B, confocal stack of 20 images). dTau is detectable in cell bodies localized in the cortex (arrowhead) and in neuronal fibers in the neuropils (arrows). C; In the larval brain, strongest expression is found in the eye discs (ed) but lower levels of expression are detectable in the proximal half of the hemispheres (h) and in the ventral ganglia (vg). D: Weak expression can also be detected in the outer optic anlagen (arrowhead, the arrow points to the photoreceptor axons). E: In wild type, dTau (red) co-localizes with chaoptin (24B10, green) in photoreceptor axons projections (arrow) but does not appear to be present in their terminals (broad arrowhead). It can also be seen in fibers in the developing lamina (arrowhead). F: In GMR-GAL4; tauGD25023, dTau is still present in the fibers in the lamina (arrowhead) but is not detectable in photoreceptor axons (arrow). G: In contrast, in tauDf(3R)MR22/Df(3R)BSC499 dTau is still present in photoreceptor axons, though at much lower levels than in wild type (arrows and inset). In addition, there is a strong staining in the optic stalk (os) that does not co-localize with 24B10 (arrowhead). H: dTau is also detectable in the eye disc (stack of 15 optical images of 0.5μm), co-localizing with 24B10. However, the dTau appears restricted to axons (I, right panel), whereas 24B10 also outlines the photoreceptor cell bodies (I, left panel). J, K: In eye discs from tauDf(3R)MR22/Df(3R)BSC499 flies, the pattern again looks different with a less distinct and punctuate dTau staining (K, right panel). Scale bar in A=50μm, in B=25μm, in C=100μm, in D=20μm, in E=25μm, and in H=20μm
Genetic interaction between dTau and other microtubule-associated proteins
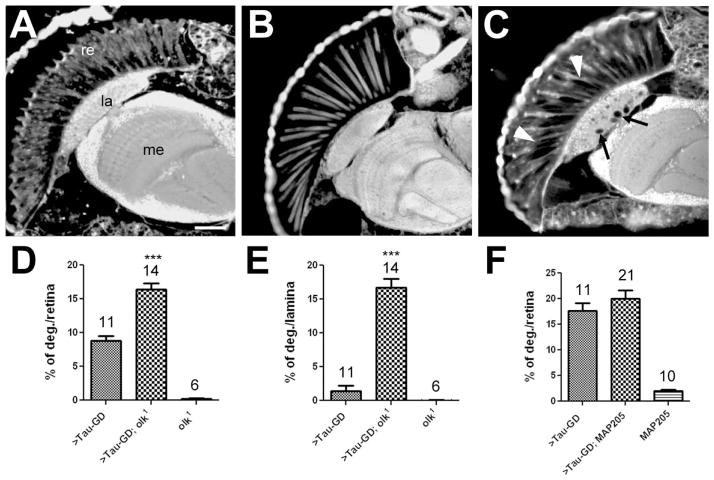
Besides dTau, three other MAPs have been identified in Drosophila. We previously showed that over-expression of dTau can partially rescue the degeneration caused by futscholk1, a hypomorphic mutation of the Drosophila MAP1B orthologue (Bettencourt da Cruz et al., 2005). We therefore tested whether futscholk1 can enhance the degenerative phenotype caused by the loss of dTau, which would provide further evidence that the two proteins have overlapping functions. We first combined futscholk1 with the pan-neuronal knockdown tauGD25023; elav-GAL4; however this did not result in viable flies. As described above, tauGD25023; elav-GAL4 exhibited increased lethality during development, but 45% of the flies normally reach adulthood, showing that the loss of Futsch enhanced the lethality caused by reduced dTau expression. To address whether these flies also have an enhanced degenerative phenotype, we crossed futscholk1 into GMR-GAL4; tauGD25023 knockdown flies. In contrast to the dTau knockdown [Fig. 8(A)], futscholk1 flies did not show any retinal degeneration when aged for three days [Fig. 8(B)]. However, combining futscholk1 with the loss of dTau significantly enhanced the extent of retinal degeneration [Fig. 8(C, D)]. In addition, these flies also exhibited a significant increase in vacuolization of the lamina [arrows, Fig. 8(C, E)], the target field of the outer photoreceptors. We also occasionally found a few vacuoles in the lamina of dTau knockdown flies, but we did not detect lesions of this type in futscholk1 flies at this age [Fig. 8(E)], although they did develop some vacuoles in the lamina when aged for 20d (data not shown). Finally, we analyzed 7d old dTau knockdown flies that overexpressed MAP205 or MAP60 to determine whether either protein could functionally replace dTau, but neither had an effect on the dTau knockdown phenotype [Fig. 8(C) and data not shown]. These results support our previous data that dTau and Futsch have at least partially overlapping functions, whereas the other two MAPs do not appear to be able to functionally replace dTau.
Figure 8.
A mutation in MAP1B/Futsch enhances the dTau phenotype. In contrast to a 3d old GMR-GAL4; tauGD25023 fly (A), a 3d old futscholk1 mutant does not show retinal degeneration (B). C: A 3d old futscholk1; GMR-GAL4; tauGD25023 fly shows a more severe retinal degeneration (arrowheads) and also vacuole formation in the lamina (arrows). Quantification of the degeneration in the retina (D) and lamina (E). F: Co-expressing MAP205 with GMR-GAL4; tauGD25023 had no effect on the retinal degeneration when analyzed at 7d. The number of analyzed retinae and the SEMs are indicated for each bar. *** p<0.001. Scale bar in A=25μm.
Expression of human Tau suppresses the retinal degeneration after loss of dTau
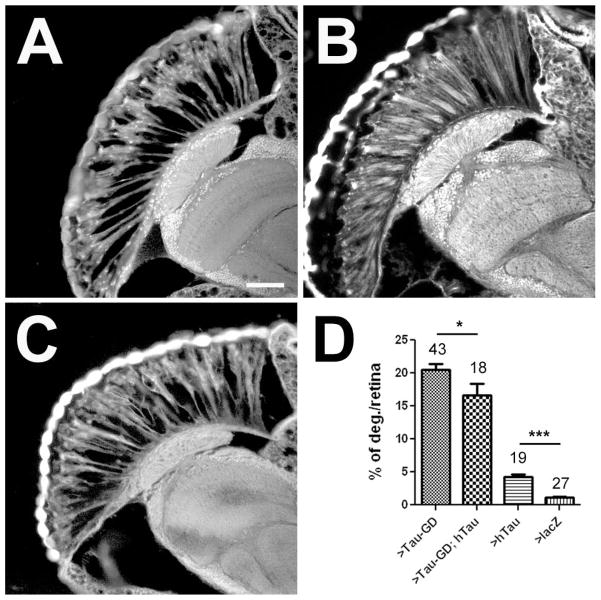
As a final experiment, we investigated whether expression of human Tau could replace Drosophila Tau in our retinal dTau knockdown. As shown by other groups, expressing certain wild type human Tau constructs can induce an external rough eye phenotype (Khurana et al., 2006; Chatterjee et al., 2009). Therefore, we used the Tau23 construct (Williams et al., 2000), for which no such phenotype had been described, and indeed, we found that inducing its expression with GMR-GAL4 did not result in an external phenotype (data not shown). We therefore used the Tau23 construct for our rescue experiments. As shown in figure 9(B), this construct did in fact cause a relatively weak pattern of retinal degeneration, detectable in sections from 7d old GMR-GAL4; UAS-hTau23 flies (compared with controls expressing lacZ) [Fig. 9(D)]. Nevertheless, co-expressing this construct with tauGD25023 in the eye significantly reduced vacuole formation. These results indicate that human Tau can functionally replace dTau, although tight control of its expression level seems to be needed to prevent the induction of an overexpression phenotype.
Figure 9.
Expression of human Tau suppresses the retinal degeneration caused by the loss of dTau. A: 7d old GMR-GAL4; tauGD25023 fly. B: An age-matched GMR-GAL4; UAS-hTau23 fly also shows some vacuoles in the retina. C: Co-expressing hTau23 with GMR-GAL4; tauGD25023 suppresses the retinal degeneration at 7d. D: Quantifying the degeneration in the retina shows a significant decrease when hTau23 is co-expressed with GMR-GAL4; tauGD25023 even though hTau23 expression alone results in a significant retinal degeneration. The number of analyzed retinae and the SEMs are indicated. * p<0.05, *** p<0.001. Scale bar in A=25μm.
DISCUSSION
The initial finding that tubulin cannot assemble into microtubules in vitro in the absence of Tau (Weingarten et al., 1975) and the observation that Tau is highly expressed during neuronal development (Matus, 1990; Tucker, 1990; Gordon-Weeks, 1991), originally suggested that Tau would have a vital function in neuronal growth and function. However, knockout mice lacking Tau were viable and appeared surprisingly normal, with no overt phenotypes (Harada et al., 1994; Dawson et al., 2001). In contrast, our results show that dTau is an essential gene whereby this appears to be due to a function in neurons because only 3% of the flies in which dTau was knocked down specifically in neurons reached adulthood. As mentioned previously, this difference in sensitivity to the loss of Tau is probably due to less redundancy in Drosophila: whereas mice have two other closely related MAPs, MAP2 and MAP4, flies do not express orthologs of these close family members (Dehmelt and Halpain, 2005). Tau additionally seems to overlap in function with MAP1B (DiTella et al., 1996; Takei et al., 2000), while an upregulation of MAP1A was observed in the Tau knockout mice (Harada et al., 1994). Concerning MAP1, flies also have only one family member, Futsch, which is more homologous to MAP1B. However, like in mammals Futsch does seem to have some functional redundancy with dTau, because mutations in Futsch enhance the dTau phenotype. Furthermore, we previously showed that additional expression of dTau partially rescued defects seen in futscholk mutants (Bettencourt da Cruz et al., 2005). Because MAP60 and MAP205 did not affect the dTau phenotype, although they can bind microtubule, dTau and MAP1probably share additional functions besides their microtubule binding activity. Finally, we confirmed a functional conservation of the fly and human Tau proteins, because the expression of human Tau could partially rescue the loss of dTau phenotype. For these experiments, we used flies expressing the hTau23 variant (Mudher et al., 2004) which is the shortest neuronal isoform with only three microtubule binding sites and no N-terminal repeats (Friedhoff et al., 1998). Although this construct had a significant effect on the phenotypes caused by the loss of dTau, it was clearly only a partial rescue. This could be due to a coparatively weak microtubule binding affinity of this construct as it has only three microtubule-binding sites whereas dTau has five (Heidary and Fortini, 2001). Alternatively, the retinal degeneration caused by the expression of this construct alone may partially counteract the rescue ability.
Reducing dTau in neurons resulted in progressive degeneration whereby the effect was more severe for photoreceptor neurons, correlating with the higher expression levels of dTau in the retina. Although weaker, the same phenotypes were detectable with another RNAi construct (tau HM05101) and with chromosomal deletions in the dTau region, both of which only affect some dTau isoforms. Both, the retinal degeneration and degeneration in the brain also occurred when we restricted the knockdown of dTau expression to the adult stage, revealing that dTau is required in maintaining neurons during adulthood. However, we also detected changes in photoreceptor morphology and a loss of photoreceptors in the retinal knockdown in late pupal stages, indicating that dTau is necessary for late stages of photoreceptor development. In contrast, we could not detect defects in neuronal outgrowth, as photoreceptor axons still reached their normal termination fields and the lamina and medulla appear normal in adult flies. Due to the lack of a null allele of dTau, we do currently not know whether the seemingly normal development of these flies in earlier stages of development is due to either an incomplete knockdown or indicates that dTau is not required at those stages, or reflects the fact that the loss of dTau can be compensated by other proteins in earlier stages.
In agreement with the findings by Heidary and Fortini (Heidary and Fortini, 2001), we could readily detect dTau in the nervous system in earlier stages, including a strong expression in the eye disk in 3rd instar larvae. In photoreceptors, dTau was found in axons, whereas it appeared mostly absent in the cell bodies and the photoreceptor terminals. Furthermore, we detect dTau staining in the optic stalk that did not overlap with chaoptin, suggesting that dTau is expressed in glial cells although the levels of dTau in these cells normally appeared to be much lower than in photoreceptors. However, the staining in these cells was much stronger in tauDf(3R)MR22/Df(3R)BSC499 flies than in wild type, suggesting that the glial cells express specific isoforms that are increased in flies carrying this chromosomal aberration. A similar observation has been reported in vertebrates, where some Tau isoforms appear to be mostly expressed in glial cells (Couchie et al., 1988).
In addition to photoreceptor loss, the reduction in dTau caused a marked structural phenotype in the rhabdomeres. Whereas the microvilli were both regularly stacked and tightly packed in control flies, rhabdomeres in dTau knockdowns were more loosely packed, especially in regions close to the cell cytoplasm. Rhabdomeres start forming around 55% of pupal development (approximately P8), and in the mature state, over 90% of the plasma membrane of the photoreceptor is folded into approximately 60,000 microvilli (Leonard et al., 1992). Stabilized and acetylated microtubules are specifically localized to the apical side of the photoreceptor cells that extend these membranes (Chen et al., 2010). In addition, this membrane extension relies on the efficient transport of other proteins along microtubules (League and Nam, 2011). Our observation that the rhabdomeres appeared small and malformed by P11 in dTau knockdown pupae (a time when the photoreceptors were still present) therefore suggests that dTau is required for the stabilization of the microtubule network that in turn is needed for the extension of the rhabdomeric membranes and formation of microvilli. Interestingly, dTau not only seems to be required for both the development of the rhabdomeres and also for maintaining their structure during later stages, as they initially formed normally in tauDf(3R)MR22/Df(3R)BSC499 flies but then became progressively less well organized in 7d old flies. This might indicate that a specific requirement for stabilized microtubules at the apical side of photoreceptor cells, both during development of microvilli and for the subsequent maintenance during adulthood.
That the loss of dTau does indeed affect microtubules was confirmed by our EM analysis, which showed that the density of microtubule is decreased. A similar phenotype was observed in Tau knockout mice, although only in small-caliber axons (Harada et al., 1994). In addition, the mean cross sectional area of microtubules was increased in dTau mutants compared to wild type. This change in size could either be due to differences in proteins that are associated with the microtubules or to less tightly packed protofilaments. We think the latter is more likely because it has been shown in in vitro studies that Tau can regulate the curvature and diameter of microtubules (Choi et al., 2009).
Supplementary Material
Acknowledgments
Many thanks are due Melissa Williams and the OHSU imaging facility, which is supported by the NIH P30NS061800 grant. This work was supported by grants to D.K. from the Medical Research Foundation of Oregon and the Oregon Tax Check-off Program for Alzheimer’s Research. B.B. was supported by training grants from the NIH (5T32AG023477, 5T32HD049309) and the Oregon Tax Check-off Program for Alzheimer’s Research.
Footnotes
The authors declare no conflict of interest.
References
- Barten DM, Fanara P, Andorfer C, Hoque N, Wong PY, Husted KH, Cadelina GW, Decarr LB, Yang L, Liu V, Fessler C, Protassio J, Riff T, Turner H, Janus CG, Sankaranarayanan S, Polson C, Meredith JE, Gray G, Hanna A, Olson RE, Kim SH, Vite GD, Lee FY, Albright CF. Hyperdynamic microtubules, cognitive deficits, and pathology are improved in tau transgenic mice with low doses of the microtubule-stabilizing agent BMS-241027. J Neurosci. 2012;32:7137–7145. doi: 10.1523/JNEUROSCI.0188-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt da Cruz A, Schwarzel M, Schulze S, Niyyati M, Heisenberg M, Kretzschmar D. Disruption of the MAP1B-related protein FUTSCH leads to changes in the neuronal cytoskeleton, axonal transport defects, and progressive neurodegeneration in Drosophila. Mol Biol Cell. 2005;16:2433–2442. doi: 10.1091/mbc.E04-11-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmine-Simmen K, Proctor T, Tschape J, Poeck B, Triphan T, Strauss R, Kretzschmar D. Neurotoxic effects induced by the Drosophila amyloid-beta peptide suggest a conserved toxic function. Neurobiol Dis. 2009;33:274–281. doi: 10.1016/j.nbd.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash AD, Aliev G, Siedlak SL, Nunomura A, Fujioka H, Zhu X, Raina AK, Vinters HV, Tabaton M, Johnson AB, Paula-Barbosa M, Avila J, Jones PK, Castellani RJ, Smith MA, Perry G. Microtubule reduction in Alzheimer’s disease and aging is independent of tau filament formation. Am J Pathol. 2003;162:1623–1627. doi: 10.1016/s0002-9440(10)64296-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Sang TK, Lawless GM, Jackson GR. Dissociation of tau toxicity and phosphorylation: role of GSK-3beta, MARK and Cdk5 in a Drosophila model. Hum Mol Genet. 2009;18:164–177. doi: 10.1093/hmg/ddn326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau KW, Chan WY, Shaw PC, Chan HY. Biochemical investigation of Tau protein phosphorylation status and its solubility properties in Drosophila. Biochem Biophys Res Commun. 2006;346:150–159. doi: 10.1016/j.bbrc.2006.05.112. [DOI] [PubMed] [Google Scholar]
- Chen G, League GP, Nam SC. Role of spastin in apical domain control along the rhabdomere elongation in Drosophila photoreceptor. PLoS One. 2010;5:e9480. doi: 10.1371/journal.pone.0009480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi MC, Raviv U, Miller HP, Gaylord MR, Kiris E, Ventimiglia D, Needleman DJ, Kim MW, Wilson L, Feinstein SC, Safinya CR. Human microtubule-associated-protein tau regulates the number of protofilaments in microtubules: a synchrotron x-ray scattering study. Biophys J. 2009;97:519–527. doi: 10.1016/j.bpj.2009.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combs B, Gamblin TC. FTDP-17 tau mutations induce distinct effects on aggregation and microtubule interactions. Biochemistry. 2012;51:8597–8607. doi: 10.1021/bi3010818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couchie D, Charriere-Bertrand C, Nunez J. Expression of the mRNA for tau proteins during brain development and in cultured neurons and astroglial cells. J Neurochem. 1988;50:1894–1899. doi: 10.1111/j.1471-4159.1988.tb02494.x. [DOI] [PubMed] [Google Scholar]
- Dawson HN, Ferreira A, Eyster MV, Ghoshal N, Binder LI, Vitek MP. Inhibition of neuronal maturation in primary hippocampal neurons from tau deficient mice. J Cell Sci. 2001;114:1179–1187. doi: 10.1242/jcs.114.6.1179. [DOI] [PubMed] [Google Scholar]
- Dehmelt L, Halpain S. The MAP2/Tau family of microtubule-associated proteins. Genome Biol. 2005;6:204. doi: 10.1186/gb-2004-6-1-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiTella MC, Feiguin F, Carri N, Kosik KS, Caceres A. MAP-1B/TAU functional redundancy during laminin-enhanced axonal growth. J Cell Sci. 1996;109 (Pt 2):467–477. doi: 10.1242/jcs.109.2.467. [DOI] [PubMed] [Google Scholar]
- Doerflinger H, Benton R, Shulman JM, St Johnston D. The role of PAR-1 in regulating the polarised microtubule cytoskeleton in the Drosophila follicular epithelium. Development. 2003;130:3965–3975. doi: 10.1242/dev.00616. [DOI] [PubMed] [Google Scholar]
- Feuillette S, Miguel L, Frebourg T, Campion D, Lecourtois M. Drosophila models of human tauopathies indicate that Tau protein toxicity in vivo is mediated by soluble cytosolic phosphorylated forms of the protein. J Neurochem. 2010;113:895–903. doi: 10.1111/j.1471-4159.2010.06663.x. [DOI] [PubMed] [Google Scholar]
- Friedhoff P, von Bergen M, Mandelkow EM, Davies P, Mandelkow E. A nucleated assembly mechanism of Alzheimer paired helical filaments. Proc Natl Acad Sci U S A. 1998;95:15712–15717. doi: 10.1073/pnas.95.26.15712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon-Weeks PR. Growth cones: the mechanism of neurite advance. Bioessays. 1991;13:235–239. doi: 10.1002/bies.950130506. [DOI] [PubMed] [Google Scholar]
- Grundke-Iqbal I, Iqbal K, Quinlan M, Tung YC, Zaidi MS, Wisniewski HM. Microtubule-associated protein tau. A component of Alzheimer paired helical filaments. J Biol Chem. 1986;261:6084–6089. [PubMed] [Google Scholar]
- Harada A, Oguchi K, Okabe S, Kuno J, Terada S, Ohshima T, Sato-Yoshitake R, Takei Y, Noda T, Hirokawa N. Altered microtubule organization in small-calibre axons of mice lacking tau protein. Nature. 1994;369:488–491. doi: 10.1038/369488a0. [DOI] [PubMed] [Google Scholar]
- Heidary G, Fortini ME. Identification and characterization of the Drosophila tau homolog. Mech Dev. 2001;108:171–178. doi: 10.1016/s0925-4773(01)00487-7. [DOI] [PubMed] [Google Scholar]
- Hummel T, Krukkert K, Roos J, Davis G, Klambt C. Drosophila Futsch/22C10 is a MAP1B-like protein required for dendritic and axonal development. Neuron. 2000;26:357–370. doi: 10.1016/s0896-6273(00)81169-1. [DOI] [PubMed] [Google Scholar]
- Iqbal K, Liu F, Gong CX, del Alonso AC, Grundke-Iqbal I. Mechanisms of tau-induced neurodegeneration. Acta Neuropathol. 2009;118:53–69. doi: 10.1007/s00401-009-0486-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson GR, Wiedau-Pazos M, Sang TK, Wagle N, Brown CA, Massachi S, Geschwind DH. Human wild-type tau interacts with wingless pathway components and produces neurofibrillary pathology in Drosophila. Neuron. 2002;34:509–519. doi: 10.1016/s0896-6273(02)00706-7. [DOI] [PubMed] [Google Scholar]
- Ke YD, Suchowerska AK, van der Hoven J, De Silva DM, Wu CW, van Eersel J, Ittner A, Ittner LM. Lessons from tau-deficient mice. Int J Alzheimers Dis. 2012;2012:873270. doi: 10.1155/2012/873270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana V, Lu Y, Steinhilb ML, Oldham S, Shulman JM, Feany MB. TOR-mediated cell-cycle activation causes neurodegeneration in a Drosophila tauopathy model. Curr Biol. 2006;16:230–241. doi: 10.1016/j.cub.2005.12.042. [DOI] [PubMed] [Google Scholar]
- Kretzschmar D, Hasan G, Sharma S, Heisenberg M, Benzer S. The swiss cheese mutant causes glial hyperwrapping and brain degeneration in Drosophila. J Neurosci. 1997;17:7425–7432. doi: 10.1523/JNEUROSCI.17-19-07425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan N, Kretzschmar D, Rakshit K, Chow E, Giebultowicz JM. The circadian clock gene period extends healthspan in aging Drosophila melanogaster. Aging (Albany NY) 2009;1:937–948. doi: 10.18632/aging.100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- League GP, Nam SC. Role of kinesin heavy chain in Crumbs localization along the rhabdomere elongation in Drosophila photoreceptor. PLoS One. 2011;6:e21218. doi: 10.1371/journal.pone.0021218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBoeuf AC, Levy SF, Gaylord M, Bhattacharya A, Singh AK, Jordan MA, Wilson L, Feinstein SC. FTDP-17 mutations in Tau alter the regulation of microtubule dynamics: an “alternative core” model for normal and pathological Tau action. J Biol Chem. 2008;283:36406–36415. doi: 10.1074/jbc.M803519200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee VM, Goedert M, Trojanowski JQ. Neurodegenerative tauopathies. Annu Rev Neurosci. 2001;24:1121–1159. doi: 10.1146/annurev.neuro.24.1.1121. [DOI] [PubMed] [Google Scholar]
- Lei P, Ayton S, Finkelstein DI, Spoerri L, Ciccotosto GD, Wright DK, Wong BX, Adlard PA, Cherny RA, Lam LQ, Roberts BR, Volitakis I, Egan GF, McLean CA, Cappai R, Duce JA, Bush AI. Tau deficiency induces parkinsonism with dementia by impairing APP-mediated iron export. Nat Med. 2012;18:291–295. doi: 10.1038/nm.2613. [DOI] [PubMed] [Google Scholar]
- Leonard DS, Bowman VD, Ready DF, Pak WL. Degeneration of photoreceptors in rhodopsin mutants of Drosophila. J Neurobiol. 1992;23:605–626. doi: 10.1002/neu.480230602. [DOI] [PubMed] [Google Scholar]
- Li B, Chohan MO, Grundke-Iqbal I, Iqbal K. Disruption of microtubule network by Alzheimer abnormally hyperphosphorylated tau. Acta Neuropathol. 2007;113:501–511. doi: 10.1007/s00401-007-0207-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Fischer I. Isolation and sequencing of the 5′ end of the rat microtubule-associated protein (MAP1B)-encoding cDNA. Gene. 1996;171:307–308. doi: 10.1016/0378-1119(95)00061-5. [DOI] [PubMed] [Google Scholar]
- Matus A. Microtubule-associated proteins and the determination of neuronal form. J Physiol (Paris) 1990;84:134–137. [PubMed] [Google Scholar]
- Mudher A, Shepherd D, Newman TA, Mildren P, Jukes JP, Squire A, Mears A, Drummond JA, Berg S, MacKay D, Asuni AA, Bhat R, Lovestone S. GSK-3beta inhibition reverses axonal transport defects and behavioural phenotypes in Drosophila. Mol Psychiatry. 2004;9:522–530. doi: 10.1038/sj.mp.4001483. [DOI] [PubMed] [Google Scholar]
- Ohler U. Identification of core promoter modules in Drosophila and their application in accurate transcription start site prediction. Nucleic Acids Res. 2006;34:5943–5950. doi: 10.1093/nar/gkl608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke R, Krantz DE, Yen D, Zipursky SL. Chaoptin, a cell surface glycoprotein required for Drosophila photoreceptor cell morphogenesis, contains a repeat motif found in yeast and human. Cell. 1988;52:291–301. doi: 10.1016/0092-8674(88)90518-1. [DOI] [PubMed] [Google Scholar]
- Takei Y, Teng J, Harada A, Hirokawa N. Defects in axonal elongation and neuronal migration in mice with disrupted tau and map1b genes. J Cell Biol. 2000;150:989–1000. doi: 10.1083/jcb.150.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker RP. The roles of microtubule-associated proteins in brain morphogenesis: a review. Brain Res Brain Res Rev. 1990;15:101–120. doi: 10.1016/0165-0173(90)90013-e. [DOI] [PubMed] [Google Scholar]
- van Swieten J, Spillantini MG. Hereditary frontotemporal dementia caused by Tau gene mutations. Brain Pathol. 2007;17:63–73. doi: 10.1111/j.1750-3639.2007.00052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingarten MD, Lockwood AH, Hwo SY, Kirschner MW. A protein factor essential for microtubule assembly. Proc Natl Acad Sci U S A. 1975;72:1858–1862. doi: 10.1073/pnas.72.5.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DW, Tyrer M, Shepherd D. Tau and tau reporters disrupt central projections of sensory neurons in Drosophila. J Comp Neurol. 2000;428:630–640. doi: 10.1002/1096-9861(20001225)428:4<630::aid-cne4>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Witman GB, Cleveland DW, Weingarten MD, Kirschner MW. Tubulin requires tau for growth onto microtubule initiating sites. Proc Natl Acad Sci U S A. 1976;73:4070–4074. doi: 10.1073/pnas.73.11.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann CW, Wszolek MF, Shulman JM, Salvaterra PM, Lewis J, Hutton M, Feany MB. Tauopathy in Drosophila: neurodegeneration without neurofibrillary tangles. Science. 2001;293:711–714. doi: 10.1126/science.1062382. [DOI] [PubMed] [Google Scholar]
- Zhang B, Carroll J, Trojanowski JQ, Yao Y, Iba M, Potuzak JS, Hogan AM, Xie SX, Ballatore C, Smith AB, 3rd, Lee VM, Brunden KR. The microtubule-stabilizing agent, epothilone D, reduces axonal dysfunction, neurotoxicity, cognitive deficits, and Alzheimer-like pathology in an interventional study with aged tau transgenic mice. J Neurosci. 2012;32:3601–3611. doi: 10.1523/JNEUROSCI.4922-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.