Testing tomato gene expression with tagged nuclei and ribosomes and CRISPR/Cas9 genome editing shows conservation of SHORT-ROOT gene function.
Abstract
Agrobacterium rhizogenes (or Rhizobium rhizogenes) is able to transform plant genomes and induce the production of hairy roots. We describe the use of A. rhizogenes in tomato (Solanum spp.) to rapidly assess gene expression and function. Gene expression of reporters is indistinguishable in plants transformed by Agrobacterium tumefaciens as compared with A. rhizogenes. A root cell type- and tissue-specific promoter resource has been generated for domesticated and wild tomato (Solanum lycopersicum and Solanum pennellii, respectively) using these approaches. Imaging of tomato roots using A. rhizogenes coupled with laser scanning confocal microscopy is facilitated by the use of a membrane-tagged protein fused to a red fluorescent protein marker present in binary vectors. Tomato-optimized isolation of nuclei tagged in specific cell types and translating ribosome affinity purification binary vectors were generated and used to monitor associated messenger RNA abundance or chromatin modification. Finally, transcriptional reporters, translational reporters, and clustered regularly interspaced short palindromic repeats-associated nuclease9 genome editing demonstrate that SHORT-ROOT and SCARECROW gene function is conserved between Arabidopsis (Arabidopsis thaliana) and tomato.
Agrobacterium rhizogenes (recently revised as Rhizobium rhizogenes; Young et al., 2001) is a soil-borne gram-negative bacterium that induces hairy roots upon wounding and infection of monocot and eudicot plants. Its root-inducing plasmid, containing transfer DNA encoding root locus (rol) gene loci (rolA, rolB, and rolC), is responsible for the stable introduction of genetic material into host cells. This can trigger the profuse production of highly branched hairy roots at the site of infection, usually the hypocotyl or cotyledon. Auxin signaling, but not ethylene signaling, is required for hairy root production (Lima et al., 2009). These root systems can be maintained in culture or hosted by plants with untransformed aerial tissue.
Hairy roots have been used for a variety of purposes over the last 30 years, ranging from recombinant protein production and metabolic engineering to analyses of rhizosphere physiology and biochemistry (for review, see Ono and Tian, 2011). More recently, A. rhizogenes-mediated hairy root production has been utilized as a biotechnology tool in a variety of plant species to discover novel biological insights. Metabolic enzyme function can be determined by gene overexpression or RNA interference approaches using hairy root transformation, for example the elucidation of an enzyme that functions in pyridine alkaloid biosynthesis in Nicotiana glauca (DeBoer et al., 2009; Kajikawa et al., 2009). Stable isotope studies can also be performed on hairy roots to elucidate distinct reactions within a biosynthetic pathway. For instance, studies in hairy roots of Ophiorrhiza pumila determined that camptothecin is derived from the 2C-methyl-d-erythritol 4-phosphate and shikimate pathways (Yamazaki et al., 2004). A. rhizogenes was used to identify plant genes that can suppress programmed cell death triggered in plants by fumonisin B1 by transforming individual members of a complementary DNA (cDNA) library into tomato (Solanum spp.) roots, which were then screened for resistance to fumonisin 1 (Harvey et al., 2008). In addition, A. rhizogenes-transformed tomato roots expressing the baculovirus p35 gene were used to demonstrate the existence in plants of proteases with substrate site specificity that is functionally equivalent to animal caspases (Lincoln et al., 2002). A. rhizogenes can be used as a tool to determine spatial and temporal aspects of gene expression in plants in order to infer signaling pathways associated with pathogen response. A set of candidate genes were tested in tomato and potato (Solanum tuberosum) for their role in the production of a nematode feeding structure upon infection by the cyst nematode Globodera rostochiensis (Wiśniewska et al., 2013). Additional tools to enable the efficiency of hairy root transformation include the pHairy plasmid, which uses the red fluorescent protein DsRed2 under the control of a cauliflower mosaic virus 35S promoter for visual selection of transgenic hairy roots in soybean (Glycine max). This reporter greatly increased the percentage detection of transformants and was used as a tool to determine the function of the soybean Nod factor receptor, GmNFR1α (Lin et al., 2011).
The tomato genome sequence has facilitated the discovery of many genes that regulate tomato growth and development with a particular focus on fruit development. Recent work has characterized tomato root development in the domesticated and wild species Solanum lycopersicum ‘M82’ and Solanum pennellii, respectively (Ron et al., 2013). A wide variety of cellular traits were identified that differ between these two species, including differences in cortex radial patterning, changes in cell number in distinct cell types, and differences in cell type differentiation (Ron et al., 2013). Candidate loci that regulate root length, root growth angle, and cell patterning were inferred using genetic approaches enabled by the sequencing of the tomato genome (Ron et al., 2013). To test the function of these candidate genes, transformation of the appropriate genotype is necessary. Furthermore, given these differences in cellular traits between species, several biological questions were raised as to the function of specific root cell types and their adaptive potential in distinct environments.
Profiling gene expression, chromatin modifications, and other molecular signatures at cell type resolution is of great interest to identify the regulatory events that determine cell type- and species-specific differences. This can be enabled by the use of techniques, including isolation of nuclei tagged in specific cell types (INTACT; Deal and Henikoff, 2010) and translating ribosome affinity purification (TRAP; Zanetti et al., 2005; Mustroph et al., 2009b), which facilitate the interrogation of multiple levels of gene regulation through one-step purification of nuclei and mRNA-ribosome complexes, respectively (Bailey-Serres, 2013). Both INTACT and TRAP can take advantage of promoters that are expressed in specific tissues, regions, or cell types. Once such localized gene regulation is recognized, it becomes important to consider the functional significance of expression in the individual cell type. Recently, RNA-guided genome editing (cluster regularly interspaced short palindromic repeats [CRISPR]-associated nuclease9 [Cas9]) was established as a method for targeted mutation of specific genes in plants (for review, see Belhaj et al., 2013). Testing this technology as a means for genome editing in tomato will enable rapid functional genomics studies in tomato to determine gene function at the cell/tissue-specific level.
Here, we describe the use of A. rhizogenes as a tool for fast and efficient visualization of gene expression at cell type resolution for genes expressed in the root in S. lycopersicum and S. pennellii. We show that the cell type-specific expression of genes is similar in plants transformed by A. rhizogenes as compared with Agrobacterium tumefaciens. Using this methodology, we have generated a root cell type-specific promoter resource for tomato. Tools to capture nuclei using the INTACT method and to immunopurify polyribosomes (polysomes) using the TRAP method in tomato roots were also developed using hairy root transformation. Finally, we show that hairy root transformation could be used as a means to interrogate gene function using CRISPR-mediated gene mutation.
RESULTS
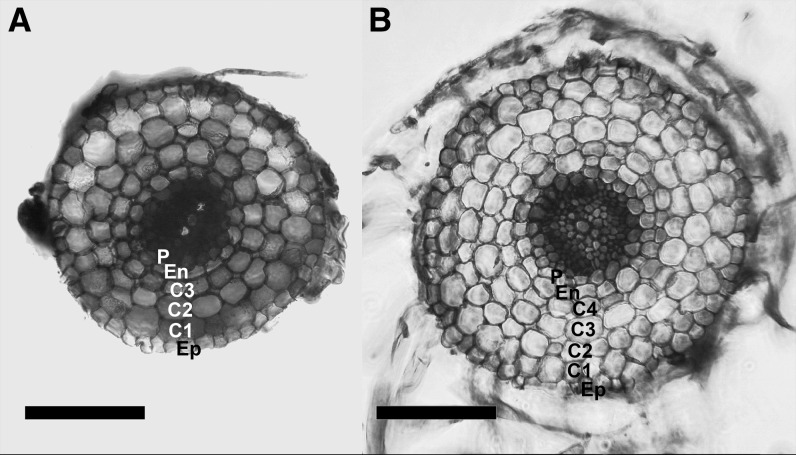
Hairy Roots Are Anatomically Similar to Primary Roots
We carried out hairy root induction in tomato by wounding the cotyledons and coinfiltrating with A. rhizogenes (ATCC15834). Similar to adventitious roots, the resulting roots are derived from shoot tissue, specifically, hypocotyl tissue. Conservation of the expression of key patterning genes in adventitious roots, as compared with primary roots, suggests that adventitious roots are analogous to primary roots, at least in Arabidopsis (Arabidopsis thaliana; Lucas et al., 2011). Since hairy roots might be used as a proxy to explore the expression patterns and functions of cell type-specific genes, we visually characterized the radial patterning of tomato adventitious roots relative to hairy roots in S. lycopersicum cv M82 as well as the anatomy of the hairy roots relative to primary roots. The only difference found was that hairy roots often contain one extra cortex layer (Fig. 1); the overall cellular architecture of the tomato adventitious and hairy roots was otherwise indistinguishable, with radial symmetry of outer cell types and diarch patterning of vascular tissue observed in both root types and primary roots (Ron et al., 2013).
Figure 1.
Cortex layer numbers differ between adventitious roots and hairy roots. Cross sections show an S. lycopersicum cv M82 adventitious root (A) and a hairy root derived from transformation of S. lycopersicum cv M82 with A. rhizogenes (B). C1 to C4, Cortex layers 1 to 4; En, endodermis; Ep, epidermis; P, pericycle. Bars = 100 μm.
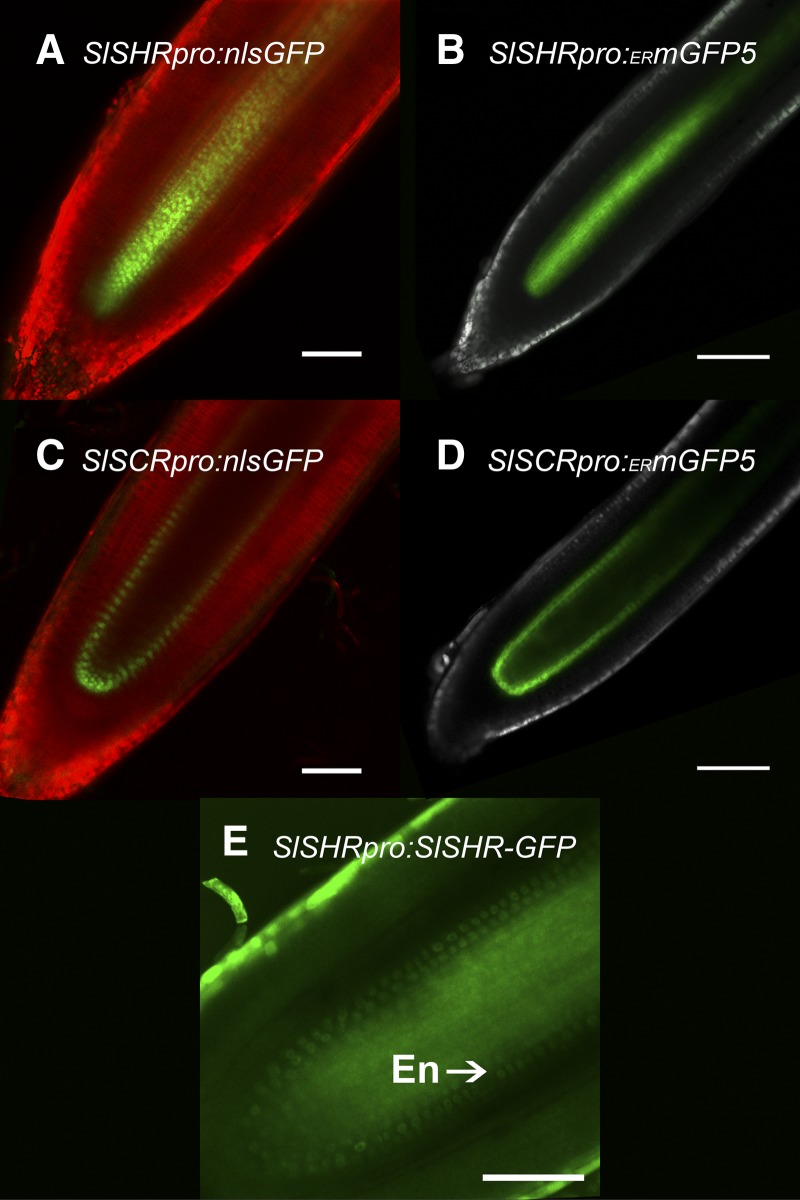
Expression Patterns Driven by the SHORT-ROOT and SCARECROW Promoters Are the Same in A. rhizogenes-Derived Hairy Roots and in Primary Roots of A. tumefaciens Transformants
To explore the molecular signatures of individual cell types, promoters are needed that mark these cell types. However, it was unclear if expression in a hairy root produced by A. rhizogenes would reflect expression as visualized in A. tumefaciens-mediated transformation. The transcripts for the transcription factors SHORT-ROOT (SHR) and SCARECROW (SCR) are expressed in root vascular tissue and in the root endodermis/quiescent center (QC), respectively. These expression patterns are conserved between Arabidopsis and rice (Oryza sativa; Cui et al., 2007), suggesting conserved expression across eudicots, including tomato. Roots transformed using A. tumefaciens and in A. rhizogenes expressing a GFP-GUS transgene driven by the tomato SHR promoter (SlSHR) showed the same pattern of GFP and GUS fluorescence in the root stele of both hairy roots and primary roots (Fig. 2, A and B; Supplemental Fig. S1A). When the SlSCR promoter was used, GFP and GUS were detected in the root endodermis and in the QC of both hairy roots and primary roots (Fig. 2, C and D; Supplemental Fig. S1B). Furthermore, an SlSHR-GFP translational fusion, driven by the SlSHR promoter in hairy roots, was able to expand its domain of abundance (Fig. 2E). These expression and movement patterns are consistent with observations in Arabidopsis and rice and suggest that the role of SHR as a cell nonautonomous regulator may be conserved (Cui et al., 2007).
Figure 2.
Promoter activity is recapitulated in A. rhizogenes-transformed hairy roots relative to A. tumefaciens-transformed primary roots of tomato. A and B, The SlSHR promoter drives expression in the vasculature in both an SlSHRpro-nlsGFP (G10-90pro-TagRFP-LTI6b)-transformed hairy root (A) and an SlSHRpro-ERGFP-transformed primary root (B). C and D, The SlSCR promoter drives expression in the QC and endodermis in both an SlSCRpro-nlsGFP (G10-90pro-TagRFP-LTI6b)-transformed hairy root (C) and an SlSCRpro-ERGFP-transformed primary root (D). In A to D, GFP fluorescence is green, TagRFP fluorescence is red, and the green component of autofluorescence is white. E, SlSHRpro-SHR-GFP protein fusion shows cytosolic subcellular localization of the SHR protein in the vasculature and movement of the protein outside of the vasculature, where it localizes to nuclei. En, Endodermis. This image was taken without linear unmixing, so green represents both GFP and autofluorescence. All images were taken with a 20× objective. Bars = 100 μm.
A Cell Type-Specific Promoter Resource in S. lycopersicum
Promoters driving gene expression in specific root cell types or tissues of Arabidopsis have been utilized to develop profiles of gene expression, chromatin modification, proteins, metabolites, and transcripts targeted for translation at cell type resolution (Birnbaum et al., 2003; Lee et al., 2006; Brady et al., 2007; Mustroph et al., 2009b; Deal and Henikoff, 2010; Petricka et al., 2012; Bailey-Serres, 2013; Moussaieff et al., 2013). Such promoter resources are of key importance to determining novel biological insights in other plant species.
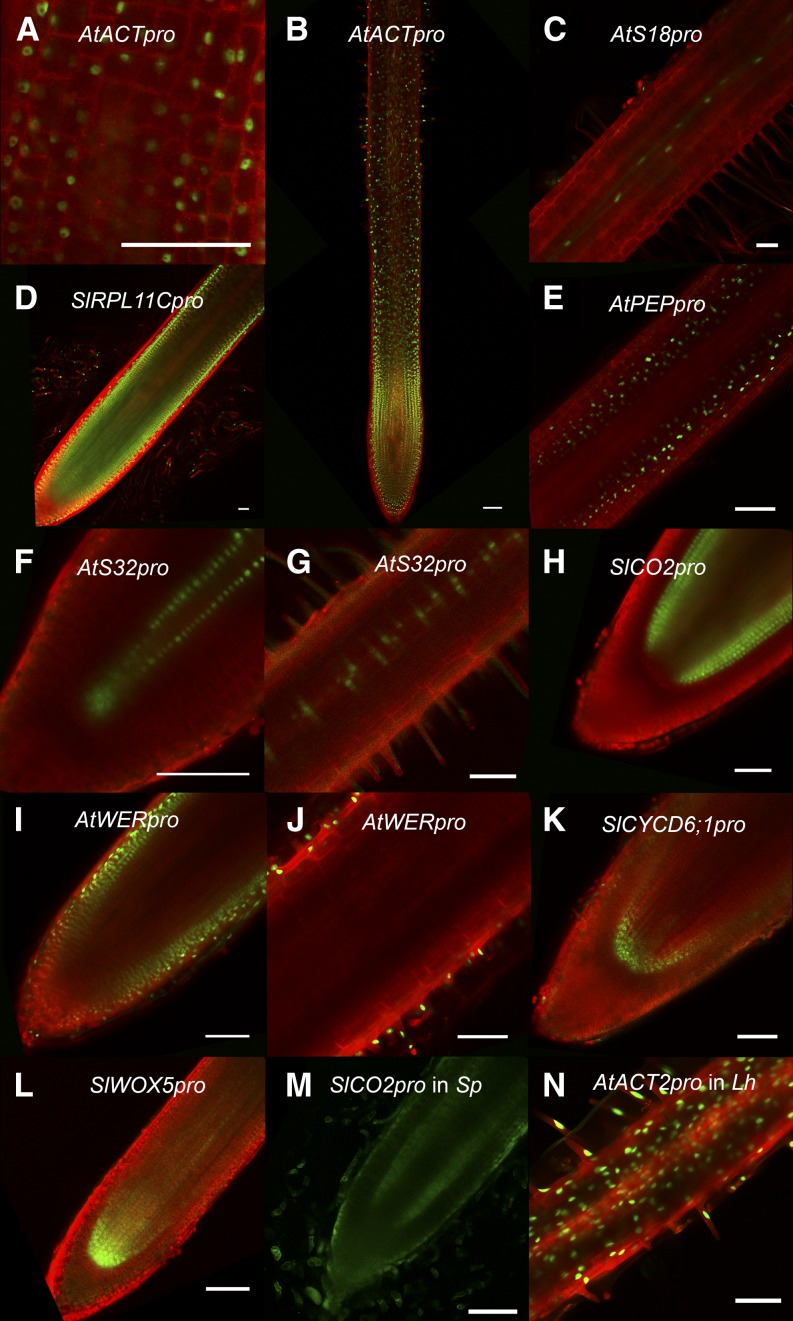
Using A. rhizogenes-mediated transformation, we developed a suite of promoters that mark cell- or tissue-specific expression in S. lycopersicum cv M82. These include promoters described in the previous section that drive expression in the stele (SlSHR), endodermis (SlSCR), QC and initials (S. lycopersicum CYCLIN-D6-1 [SlCYCD6;1] and S. lycopersicum WUSCHEL-related homeobox5 [SlWOX5]; Fig. 3, K and L), phloem (Arabidopsis S32 [AtS32]; Fig. 3, F and G; Supplemental Fig. S2A), maturing xylem (AtS18; Fig. 3C; Supplemental Fig. S2B), meristematic cortex cells (S. lycopersicum Cortex specific transcript [SlCO2]; Fig. 3H; Supplemental Fig. S1C), meristematic, elongating, and mature cortex cells (Arabidopsis endopeptidase [AtPEP]; Fig. 3E), lateral root cap and epidermal cells (Arabidopsis WEREWOLF [AtWER]; Fig. 3, I and J; Supplemental Fig. S2C), as well as a constitutive promoter (Arabidopsis ACTIN2 [AtACT2]; Fig. 3, A and B) and a promoter that drives expression in the tomato root meristem (S. lycopersicum RIBOSOMAL PROTEIN L11C [SlRPL11C]; Fig. 3D). The tomato promoters that we identified had similar (SlSCR, SlSHR, SlCO2, and SlRPL11C) or expanded (SlCYCD6;1 and SlWOX5) expression domains compared with their putative Arabidopsis orthologs (Haecker et al., 2004; Mustroph et al., 2009b; Sozzani et al., 2010). We also tested promoters of genes that drive cell type-specific expression in Arabidopsis for their ability to drive expression in tomato. In at least five cases (AtACT2, AtS32, AtS18, AtPEP, and AtWER), these promoters were able to drive similar expression in tomato (Williams and Sussex, 1995; An et al., 1996; Mace et al., 2006; Brady et al., 2007; Mustroph et al., 2009b). In addition, we were able to use the same protocol to drive the expression of AtPEP in S. pennellii (Fig. 3M) and AtACT2 in Lepidium hyssopifolium (Fig. 3N), demonstrating the utility of this method across different species.
Figure 3.
A cell type- and tissue-specific promoter toolbox. GFP expression patterns driven by a variety of promoters were tested in the hairy root transformation system by cloning them upstream of an nlsGFP-GUS fusion in a vector containing a ubiquitously expressing plasma membrane marker: G10-90pro-TagRFP-LTI6b (A–D, G–I, and K) or 35Spro-TagRFP-LTI6b (E and F). A and B, AtACT2pro drives near constitutive expression (under 63× and 10× magnification, respectively). The nlsGFP (green) shows nuclear localization, and TagRFP (red) shows plasma membrane localization. C, AtS18pro drives expression in the maturing xylem (10×). This image contains an overlay of GFP fluorescence from two different focal planes to show both phloem strands. D, SlRPL11Cpro expression is most pronounced in the meristematic zone (10×). E, AtPEPpro drives expression in the cortex throughout the root, including the elongation zone (20×). F and G, AtS32pro drives expression in the phloem, starting in the meristematic zone (F; 40×) all the way to mature root (G; 20x). H, SlCO2pro drives expression in cortex layers only in the meristematic zone (20×). I and J, AtWERpro drives expression in the lateral root cap (I; 20×) and all epidermal cells throughout the root (I and J; 20×). K, SlCYCD6pro drives expression in the QC and vascular initials (20×). L, SlWOX5pro drives expression in the QC and vascular cells in the proximal meristem (20×). M, SlCO2pro drives expression of AtRPL18CDS-GFP in the meristematic cortex of S. pennellii (20×). This image was taken without linear unmixing, so green represents both GFP and autofluorescence. N, AtACT2pro also drives near constitutive expression in L. hyssopifolium hairy roots (20×). Bars = 100 μm.
Tomato roots pose certain challenges for confocal microscopy. Considerable autofluorescence is observed in the lateral root cap and epidermal cells of both primary roots and hairy roots. This autofluorescence overlaps with the GFP emission spectrum but can be separated using the linear unmixing function in advanced confocal laser scanning microscope technology (Supplemental Fig. S3). In Arabidopsis, it is common to use propidium iodide (PI) staining to visualize the cell wall as a countermark to promoter-driven GFP expression. However, the use of PI in tomato roots to mark cell walls was unsuccessful due to the presence of an exodermis layer that inhibits the entry of this dye (Naseer et al., 2012; Ron et al., 2013). We employed an alternative approach of using a membrane-tagged protein, LTI6b, encoded by At3g05890 (Cutler et al., 2000; Federici et al., 2012) fused to a red fluorescent protein (TagRFP) driven by a constitutive promoter (35S or G10-90) to countermark the promoter-driven GFP expression and identify root cell types (Supplemental Fig. S4). By combining promoters of interest driving GFP and this membrane-bound red fluorescent protein marker in the same backbone, we could quickly and easily visualize cell type-specific expression in tomatoes transformed with A. rhizogenes.
Optimization of INTACT and TRAP for Tomato
In addition to identifying a suite of cell type-specific promoters, we tested molecular tools for organ- to cell type-specific analyses in tomato using hairy root transformation. We generated optimized vectors for studying cell type-specific nuclear transcriptome and chromatin modifications by INTACT (Deal and Henikoff, 2010) and transcripts targeted for TRAP (Zanetti et al., 2005; Mustroph et al., 2009b) in tomato. Binary vectors were constructed to allow Gateway cloning of cell type-specific promoters into the INTACT and TRAP constructs designed for tomato. These constructs are modular with unique restriction sites at junctions of optimizable elements to allow later modifications and application to other species (Supplemental Fig. S5). These unique restriction sites allow for optimization of the plant antibiotic resistance gene as well as the promoter that drives the expression of this gene for both INTACT and TRAP binary vectors. INTACT-specific optimizable elements include the biotin ligase gene, the nuclear tagging protein, and the biotin ligase recognition peptide. TRAP-specific optimizable elements include the tagged ribosomal protein.
INTACT methodology is based on the expression of a cell type-specific nuclear envelope-localized biotin ligase recognition peptide and a constitutive biotin ligase gene, leading to cell type-specific biotin tagging of nuclear envelopes. The biotin-streptavidin interaction is utilized for affinity purification of biotinylated nuclei. These cell type-specific nuclear preparations can be used for global-scale analyses including RNA sequencing and chromatin immunoprecipitation (ChIP) sequencing. To perform INTACT in tomato, we created a binary vector containing both the biotin ligase (BirA) cassette and the nuclear tagging fusion (NTF) cassette. To reduce the possibility of an unstable transcript, we used Arabidopsis codon use-optimized mBirA, as tomato has similar codon usage to Arabidopsis (http://solgenomics.net/misc/codon_usage/codon_histogram.pl). The expression of mBirA was driven by the SlACT2 promoter (Solyc11g005330), which we characterized to be constitutive in tomato roots by GFP fusion and hairy root expression. The NTF cassette is a fusion of the tryptophan-proline-proline (WPP) domain of Arabidopsis RAs-related Nuclear protein GTPase-Activating Protein (GAP)1, enhanced GFP (eGFP), and a biotin ligase recognition peptide (Deal and Henikoff, 2010). This was placed 3′ of a Gateway recombination site that enables the insertion of a cell type-specific promoter to drive the NTF expression (Hartley et al., 2000).
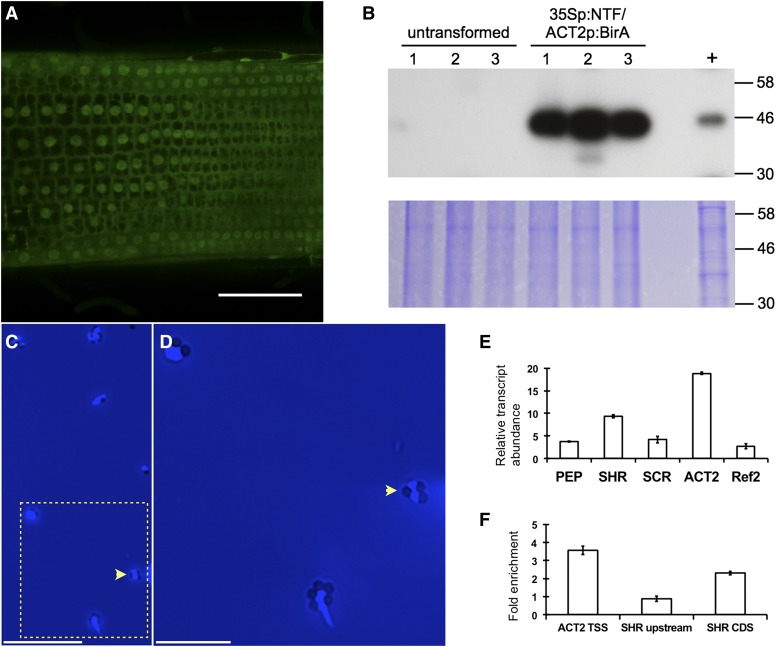
The INTACT vector carrying both the 35Spro:NTF and SlACT2pro:mBirA cassettes was tested by transformation of tomato roots with A. rhizogenes. GFP fluorescence, indicative of NTF expression, was detected in all cells and localized to the nuclear envelope and cytosol (Fig. 4A). Successful biotinylation of the NTF was tested by the fractionation of protein extracts from roots transformed with a transfer DNA encoding the 35Spro:NTF and SlACT2pro:mBirA constructs by detection with streptavidin conjugated to horseradish peroxidase (Fig. 4B). A biotinylated protein of the expected size for NTF (42 kD) was detected in extracts from hairy roots obtained in three independent transformation experiments and absent in untransformed roots (Fig. 4B). We also confirmed that NTF-tagged nuclei could be purified from tomato roots using streptavidin-coated magnetic beads, as was reported previously for Arabidopsis (Deal and Henikoff, 2011). Indeed, using the INTACT purification procedure established for Arabidopsis, we recovered up to 7 × 105 nuclei from 300 mg of transformed roots and found that these nuclei were highly pure and free of cytoplasmic contamination (Fig. 4, C and D; Supplemental Fig. S6). Very few nuclei were obtained from the same amount of untransformed roots in mock purifications.
Figure 4.
Application of INTACT methodology to tomato with the hairy root transformation system. A, 35Spro-driven NTF protein expression in tomato hairy roots shows GFP localization (green) in the nuclear envelope and cytosol. Bar = 100 μm. B, Detection of biotinylated NTF protein in root extracts. Total protein was isolated from tomato roots that were either untransformed or transformed with 35Spro-NTF and ACT2pro-BirA transgenes. Transgenic Arabidopsis roots carrying the ADF8pro-NTF and ACT2pro-BirA transgenes were included as a positive control (+) for biotinylated NTF. Molecular mass markers are shown on the right. The expected size of NTF is 46 kD. C and D, Capture of biotinylated nuclei from transformed tomato roots carrying the 35Spro-NTF and ACT2pro-BirA transgenes with streptavidin-coated magnetic beads. Nuclei were stained with 4′,6-diamino-phenylindole (DAPI) to visualize DNA. C, View with mixed white light and DAPI-channel fluorescence illumination. Seven bead-bound nuclei are visible in the field shown, with a representative nucleus indicated by the yellow arrowhead. The nuclei are DAPI-bright patches surrounded by darker colored spherical magnetic beads. Bar = 50 μm. D, Magnification of the boxed region in C. Bar = 20 μm. E, Quantitative analysis of selected transcripts in INTACT-purified tomato nuclei. qPCR was used to assay the relative abundance of mRNAs. Data represent averages ± sd of two biological replicates of nuclei purification experiments. F, ChIP and qPCR to examine the abundance of H3K4me3 in INTACT-purified tomato nuclei. Relative enrichment of the modification in the promoter of the ACT2 and SHR genes as well as within the SHR gene body is shown relative to input. Data represent average ± sd enrichment from two biological replicate experiments. Solyc01g014230 was used as a reference for normalization.
To confirm that the nuclei purified from 35Spro:NTF/SlACT2pro:mBirA-transformed roots were suitable for molecular analyses, we analyzed the relative abundance of multiple transcripts in the RNA extracted from these nuclei and also performed ChIP for the transcription-associated histone modification, histone H3 Lys-4 trimethylation (H3K4me3). For the transcript abundance analyses, total nuclear RNA was isolated, reverse transcribed, and subjected to quantitative PCR (qPCR) with primers specific to six genes. All transcripts were amplified, and the relative abundance of each was generally reflective of its abundance in total RNA (Fig. 4E; Koenig et al., 2013). No amplification was observed in control RNA samples from which reverse transcriptase was omitted, indicating that the PCR products obtained in experimental samples were derived from cDNA and not residual genomic DNA contamination. For ChIP, the INTACT purification was performed with formaldehyde-treated 35Spro:NTF/SlACT2pro-mBirA-transformed roots. The chromatin was isolated, fragmented by sonication, and immunoprecipitated with an antibody against H3K4me3 or an anti-GFP antibody as a negative control. Immunoprecipitated DNA was subjected to qPCR to measure the enrichment of H3K4me3 at the ACT2 and SHR promoters as well as the SHR gene body relative to input DNA (Fig. 4F). Among these genomic regions, the data show that H3K4me3 was most abundant in the ACT2 promoter, less so within the SHR gene body, and at background level in the SHR promoter. These DNA fragments were undetectable by qPCR in the anti-GFP immunoprecipitated sample. These results demonstrate that the INTACT method can be translated to hairy roots of A. rhizogenes-transformed tomato.
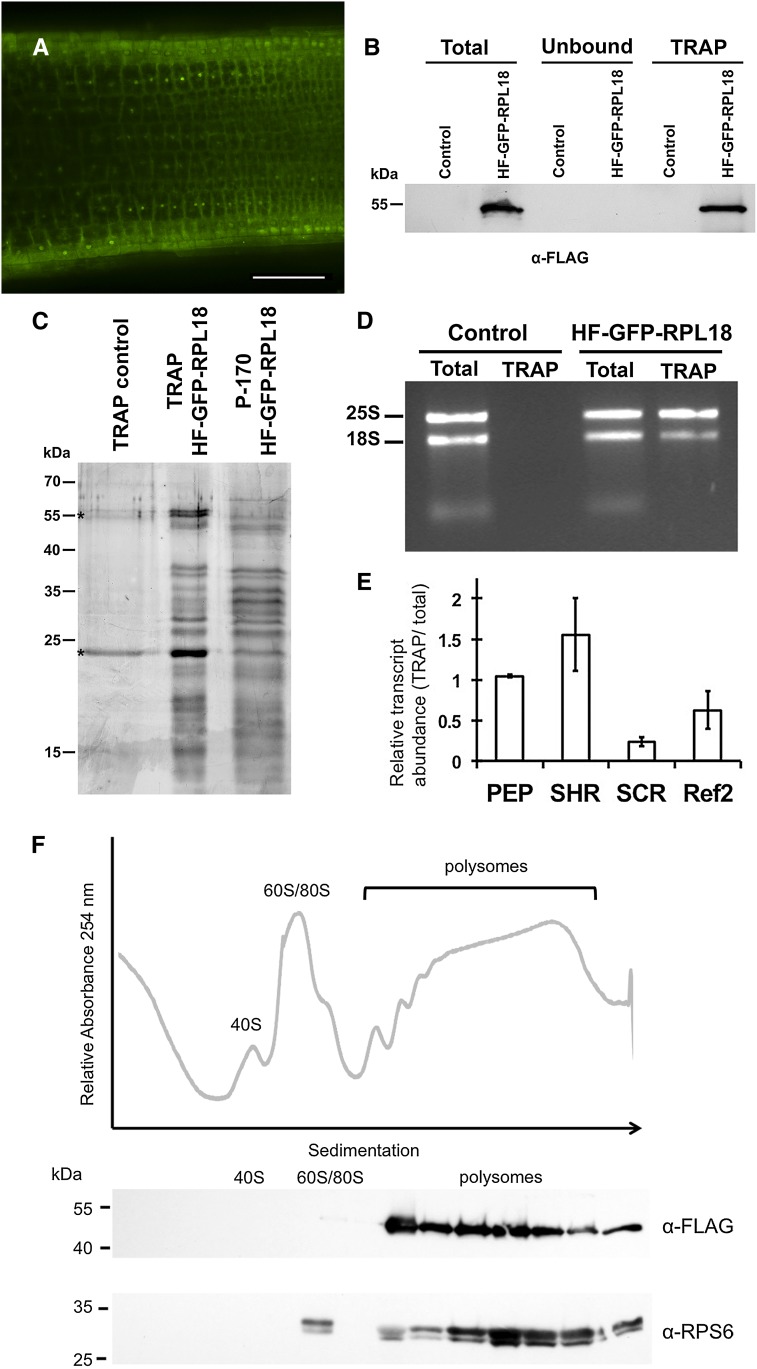
We also explored whether A. rhizogenes root transformation could extend the application of TRAP in tomato, as accomplished for Medicago truncatula (Reynoso et al., 2013). TRAP is based on the expression of an epitope-tagged version of RIBOSOMAL PROTEIN L18b (RPL18b) that is a core component of the large (60S) ribosomal subunit in the cytoplasm (Zanetti et al., 2005; Bailey-Serres 2013). Translation involves the scanning of the 5′ untranslated region of the mRNA by the 40S ribosomal subunit, the recognition of an AUG codon as the start site, and joining of the 60S subunit to form an 80S ribosome-mRNA complex. As this ribosome continues the elongation of translation, additional ribosomes can initiate on the transcript to form a polysome complex. The affinity purification of the FLAG epitope-tagged ribosomes yields mRNA that can be used for profiling of mRNAs or footprinting of individual ribosomes (Zanetti et al., 2005; Juntawong et al., 2014).
For TRAP, we generated a binary vector in which His6-FLAG-GFP-tagged RPL18b (Mustroph et al., 2009b) is driven by a specific promoter inserted with Gateway technology (Hartley et al., 2000). For tomato, we used the RPL18B coding sequence from Arabidopsis due to greater than 82% amino acid sequence identity between this deduced protein and the two RPL18 proteins encoded by S. lycopersicum. The TRAP vector was tested in A. rhizogenes-transformed hairy roots with the 35S promoter. The subcellular localization of GFP-tagged RPL18b was both nucleolar and cytosolic (Fig. 5A), consistent with previous studies (Mustroph et al., 2009b). Successful affinity purification of mRNA-ribosome complexes was achieved with S. lycopersicum extracts from 35Spro:HF-GFP-RPL18 hairy roots using anti-FLAG antibody-conjugated agarose beads, as described previously (Zanetti et al., 2005; Mustroph et al., 2009b). Western-blot analysis detected a His6-FLAG-GFP-RPL18 polypeptide of an estimated mass of 50 kD with antiserum against the FLAG epitope in the crude cell supernatant (total) and immunopurified ribosome (TRAP) fractions (Fig. 5B). This protein was not detectable in the fractions obtained with control nontransformed root tissue processed in parallel (Fig. 5B). To confirm that the immunopurification procedure yielded ribosomes, we compared the polypeptide composition of the TRAP fraction with that of ribosomes isolated by ultracentrifugation (P-170; Fig. 5C). The proteins that coimmunopurified with His6-FLAG-GFP-RPL18 showed a similar electrophoretic mobility and stoichiometric intensity to those present in conventionally purified ribosomes (Fig. 5C, second and third lanes, respectively). The limited number of proteins in the sample from wild-type roots (Fig. 5C, first lane) confirmed the specificity of the immunopurification. The presence of 25S and 18S ribosomal RNAs (rRNAs) in the TRAP fraction from 35Spro-HF-GFP-RPL18 roots and their absence in the TRAP from control wild-type roots further validated the successful purification of ribosomes from hairy roots (Fig. 5D). In addition, the detection of the transcripts of ACT2, SCR, SHR, PEP, and Solyc04g064820 (Fig. 5E) in the TRAP fraction by quantitative reverse transcription-PCR analysis confirmed that mRNA-ribosome complexes were obtained. We noted that SCR mRNA showed lower relative abundance in the TRAP versus total mRNA fractions when compared with PEP and SHR (Fig. 5E). With the exception of PEP, these results are similar to what was observed previously in Arabidopsis (Mustroph et al., 2009b). As an additional control, we evaluated the incorporation of His6-FLAG-GFP-RPL18 into polysome complexes. To do so, ribosomal complexes from 35Spro-HF-GFP-RPL18 hairy roots were fractionated through a 15% to 60% Suc density gradient and the presence of the tagged protein was evaluated by immunodetection. His6-FLAG-GFP-RPL18 was present in fractions containing 80S monosomes and small and large polysomes (Fig. 5F). Ribosomal protein S6 (RPS6), a core component of the small ribosomal subunit, was similarly detected in polysome fractions. This protein was resolved as two distinct bands, most likely reflecting variations in its C-terminal phosphorylation, as shown for Arabidopsis and maize (Zea mays; Williams et al., 2003; Chang et al., 2005). Overall, these data confirm that the expression of His6-FLAG-GFP-RPL18 allowed the efficient immunopurification of mRNA-ribosome complexes from hairy roots of tomato.
Figure 5.
Application of TRAP methodology to tomato with the hairy root transformation system. A, 35Spro-driven His6-FLAG epitope-tagged GFP-RPL18 (HF-GFP-RPL18) fusion protein in the tomato hairy root system shows GFP localization (green) in the nucleolus and cytosol. Bar = 100 μm. B, HF-GFP-RPL18 is efficiently immunopurified by TRAP. Equal fresh weight of tissue from wild-type roots (control) and HF-GFP-RPL18 hairy roots was extracted in a polysome-stabilizing buffer and processed to obtain a clarified cell supernatant (total), which was incubated with anti-FLAG agarose beads to obtain the TRAP fraction containing ribosomes and associated mRNA. Cell components not bound to the matrix remained in the unbound fraction. Molecular mass markers are shown on the left. The expected size of HF-GFP-RPL18 is 51 kD. C, Comparison of immunopurified proteins from untransformed roots or HF-GFP-RPL18-transformed hairy roots by TRAP and ribosomal proteins isolated by conventional ultracentrifugation (P-170). Proteins were visualized by silver staining. Equal proportions of TRAP samples were loaded in lanes 1 and 2. Molecular mass markers are shown on the left. Asterisks indicate bands corresponding to IgG from the affinity matrix. D, Ethidium bromide staining of RNA isolated from the total and TRAP fractions from wild-type (control) and HF-GFP-RPL18 roots. The 25S and 18S rRNA sizes are indicated on the left. E, Quantitative reverse transcription-PCR analysis of selected transcripts in the TRAP fractions. Relative transcript level in the TRAP versus total fraction was determined. Data represent average ± sd level of each transcript in two biological replicates. ACT2 was used as a reference for normalization. F, HF-GFP-RPL18 is incorporated into small to large polysomal complexes. HF-GFP-RPL18 hairy root ribosomal complexes were fractionated by ultracentrifugation through a 15% to 60% Suc density gradient, and the A254 was recorded. The positions of ribosomal subunits 40S and 60S, monosomes (80S), and polysomes are indicated. Fourteen fractions were analyzed by immunoblot with anti-FLAG and anti-RPS6 antisera. Two electrophoretic variants of RPS6 were detected by the antisera. Molecular mass markers are indicated on the left.
The Function of SHR in Tomato as Determined Using Hairy Root Transformation and CRISPR
RNA-guided genome editing using the bacterial type II CRISPR/Cas9 system has emerged as an efficient tool to edit genomes in microbial, animal, and plant systems. Recently CRISPR/Cas9-based genome editing was demonstrated in Arabidopsis, tobacco (Nicotiana tabacum), wheat (Triticum aestivum), rice, and sorghum (Sorghum bicolor; for review, see Belhaj et al., 2013). Two components are needed for CRISPR genome editing: the Cas9 endonuclease and a synthetic RNA chimera (single-guide RNA [sgRNA]; Jinek et al., 2012). The sgRNA contains a guide sequence region of 19 to 22 bp (also known as the protospacer) that matches the target DNA sequence to be mutated, which is fused to the trans-activating CRISPR RNA. An additional requirement for the Cas9 nuclease activity is the presence of the protospacer-associated motif (PAM) NGG trinucleotide downstream of the target site. Thus, the target region follows the consensus (N)19-22NGG.
While the CRISPR/Cas9 system has been tested in several other plant species, it has not been evaluated in tomato, nor has it been evaluated with hairy root transformation. Therefore, we performed a series of experiments to test the potential of the Cas9 system to induce gene knockouts using hairy root transformation in tomato. First, we constructed a binary vector containing both a Nicotiana spp. codon-optimized variant of Cas9 fused to two nuclear localization signals (NLS) driven by the 35S promoter together with an sgRNA cassette driven by the Arabidopsis U6 promoter (Supplemental Fig. S7). Individual target sequences can be cloned into the sgRNA cassette to obtain an all-in-one targeting plasmid for hairy root transformation.
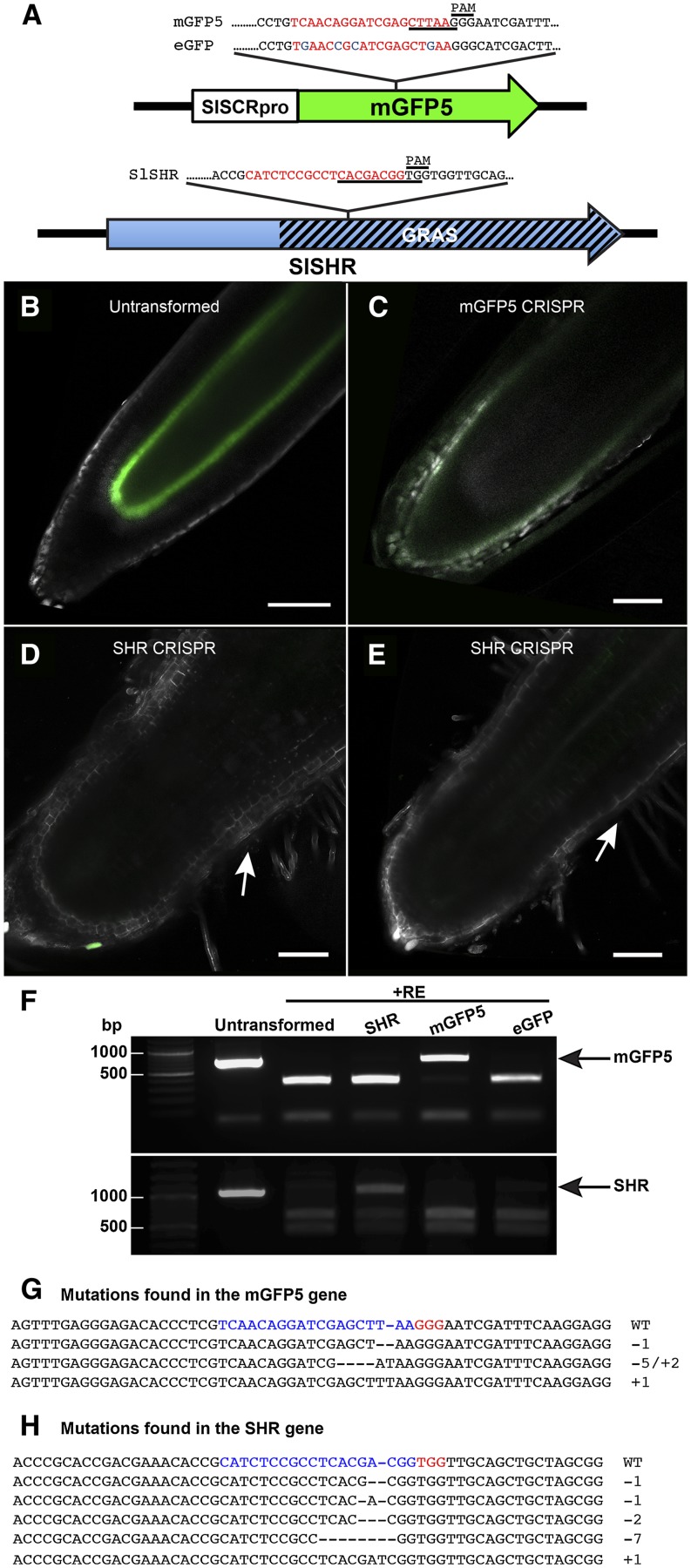
Second, we generated three CRISPR/Cas9 constructs to be transformed into a stable transgenic line expressing SlSCRpro:modified (m) mGFP5. The first construct tested the ability of the CRISPR/Cas9 system to mutate GFP in tomato. The 19-bp seed was designed to be complementary to the mGFP5 coding region. Individual transgenic hairy roots were identified with varying levels of GFP expression upon A. rhizogenes-mediated transformation of this construct into the SlSCRpro:mGFP5 transgenic line (Fig. 6, B and C; Supplemental Fig. S7). To easily identify sgRNA-guided, Cas9-induced mutations in the mGFP5 gene, we used the restriction enzyme digestion/PCR amplification (RE/PCR) method (Nekrasov et al., 2013; Voytas, 2013). The target sequence within mGFP5 contains an AflII restriction site at the predicted cleavage site of the Cas9 endonuclease. Genomic DNA was isolated from independent hairy roots with varying levels of GFP fluorescence, digested with AflII, and PCR was performed with primers flanking the target site (Fig. 6, A and F; Supplemental Table S1). In this manner, we successfully enriched for mGFP5 genes carrying mutations that remove the AflII site. The PCR product was purified and cloned, and individual clones were sequenced. Both deletion and insertion mutants were identified (Fig. 6G).
Figure 6.
The CRISPR/Cas9 system introduces mutations in hairy root transformants and can be used to study gene function in roots. A, Schematic representation of SlSCRpro-mGFP5 and SlSHR gene structures, DNA sequences, and relative locations of the targets designed for use with CRISPR. The restriction site used to screen for mutations in each target is underlined. The target sequence is in red. SNPs between mGFP5 and eGFP are in blue. The PAM follows the consensus sequence NGG. B, Control hairy root (containing no CRISPR binary vector) induced from the SlSCRpro-mGFP5 transgenic line shows strong ERGFP expression in the QC and endodermis. C to E, sgRNAs designed to complement mGFP5 and SlSHR coding sequences were transformed in an SlSCRpro-mGFP5 transgenic background using hairy root transformation. An sgRNA designed to complement mGFP5 led to the reduction or elimination of SlSCRpro-mGFP5 expression (C). Hairy root transformation with an sgRNA designed against the endogenous SHR coding sequence induced short roots with stunted meristematic and elongation zones as well as the reduction or elimination of SlSCRpro-mGFP5 expression (D and E). White arrows point to root hairs and the start of the maturation zone 0.3 mm from the root tip. This domain is outside the field of view in the control (B). B to E were imaged with a 20× objective and constant settings of 488-nm excitation, 70% laser power, 1.87-Airy unit pinhole, and 583 gain. GFP expression is shown in green with both autofluorescence spectra, and residuals are shown in white. Bars = 100 μm. F, Genomic DNA was extracted from roots transformed with Cas9 and sgRNAs against SHR, mGFP5, and eGFP. DNA was predigested with AleI and AflII for SHR and mGFP5/eGFP, respectively, and genes were amplified using primers flanking the target sequence. Amplicons were digested again. Untransformed indicates SlSCRpro-mGFP5 (same as in B) but not transformed with Cas9/CRISPR. Black arrows indicate no digestion. G and H, Alignment of sequences with Cas9-induced mutations obtained from roots transformed with Cas9/sgRNA for mGFP5 (G) and SHR (H). The wild-type (WT) sequence is shown at the top. The sequence targeted by the synthetic sgRNA is shown in red, and mutations are shown in blue. The changes in length relative to the wild type are shown to the right.
Next, to assess the specificity of the sgRNA, an sgRNA was designed to target a homologous 19-bp region from eGFP (Fig. 6A; Mali et al., 2013). This 19-bp region includes four single-nucleotide polymorphisms (SNPs) that distinguish mGFP5 from eGFP, with two of these SNPs included in the critical 3′ region of the seed adjacent to the PAM (Cong et al., 2013; Jiang et al., 2013; Liu et al., 2013). This sgRNA was transformed into SlSCRpro-mGFP5 using A. rhizogenes. Thus, if the sgRNA is nonspecific in recognition of its target site, then mutations in mGFP5 would be observed. If the sgRNA is specific, then no mutation in mGFP would be observed. In this case, no transgenic roots were observed with reduced GFP expression (Supplemental Fig. S4). We performed the RE/PCR assay on genomic DNA extracted from these roots and sequenced individual clones as described previously. No mutations were identified showing that the CRISPR/Cas9 system can be used to mutate genes with specificity (Fig. 6F).
The third construct we generated was designed to test whether the CRISPR/Cas9 system would allow us to determine if SHR function is conserved between tomato and Arabidopsis. SHR regulates SCR expression in Arabidopsis, and mutations in the AtSHR gene result in a loss of SCR expression as well as a short-root phenotype resulting from defects in stem cell division and cell patterning (Helariutta et al., 2000). An sgRNA was designed to target the SlSHR coding region, specifically in the region encoding the GRAS (for GIBBERELLIC-ACID INSENSITIVE [GAI], REPRESSOR of GAI, and SCR) domain. This construct was also transformed into the SlSCRpro-mGFP5 transgenic line using A. rhizogenes. Several transgenic hairy roots were identified with no GFP expression and a short meristematic size, as determined by the premature early presence of root hairs, which mark the maturation zone (Fig. 6, D and E; Supplemental Fig. S8). Using the RE/PCR method, we confirmed that the roots contain a variety of insertion and deletion mutations in the SlSHR coding region (Fig. 6, F and H). These alterations in root phenotype, consistent with the Arabidopsis shr mutants (Benfey et al., 1993), support the conclusion that SHR function is evolutionarily conserved with respect to the regulation of its downstream targets and root length in Arabidopsis and tomato. Together, these data demonstrate that A. rhizogenes and the CRISPR/Cas9 system provide a facile means to test gene function in root development.
DISCUSSION
Root development has been characterized at great spatiotemporal resolution in model plants, including Arabidopsis and rice. With the great advances in short-read sequencing technology, the potential to understand root development at such resolution in other crop species, which had previously been recalcitrant to such approaches, has increased significantly. Despite these considerable technological improvements, functional genomic approaches in crops with a long generation time hit a bottleneck due to difficulties in transformation-based validation using A. tumefaciens. In tomato, hypotheses could be rapidly tested using transient approaches in the related species Nicotiana benthamiana; however, results obtained from such methods need to be interpreted cautiously, as they usually do not take into account cellular context and constructs are usually driven with ubiquitous, nonhost promoters. The experiments outlined here using A. rhizogenes in tomato allow for rapid functional testing of genes at cell type resolution that are highly relevant in planta. Furthermore, these approaches can also be translated to other species, including Arabidopsis (Supplemental Fig. S5K).
Our understanding of Arabidopsis root development has greatly benefited from the ease of imaging using laser scanning confocal microscopy coupled with the use of PI to counterstain cell boundaries. The presence of the lignified, suberinized Casparian strip partially limits study of the root vasculature, as PI diffusion past this cellular boundary is restricted. Most plant species contain an exodermis, which is similarly suberinized, and thus the use of PI to counterstain cell boundaries in such species is not possible. One major advantage of our approach with respect to characterizing root development at cell type resolution was the use of TagRFP in the binary vector to visualize the cell membrane. Use of such a marker will facilitate the elucidation of root cell type development and function in other plant species.
The ancestor of tomato and Arabidopsis diverged approximately 100 to 120 million years ago (Moore et al., 2010; Sato et al., 2012). Despite this evolutionary distance, a small number of Arabidopsis regulatory sequences have been shown to drive similar gene expression patterns in tomato. Here, we transformed multiple Arabidopsis promoter:GFP fusions and tested the cellular specificity of GFP expression in tomato. Of seven cell type-specific promoters from Arabidopsis tested, only four were able to drive similar gene expression patterns in planta in tomato. Interestingly, both AtSCR and AtSHR promoters were not active in tomato, even though the native SlSHR and SlSCR share the same expression patterns as their Arabidopsis orthologs. This suggests that the regulatory sequences or their cognate transcription factors likely diverged significantly during the course of evolution, leading to these two species. Conserved regulatory sites can be identified in those promoters that were able to drive similar gene expression patterns in both species (AtS18, AtS32, AtPEP, AtWER, and AtACT2). Furthermore, expression patterns can demonstrate the conservation of developmental programs between the evolutionarily distant Arabidopsis and tomato. For instance, the AtPEP promoter drives expression in all root cortex layers, suggesting that cortex function as measured by this molecular marker is likely similar between all three layers in tomato, in contrast to Arabidopsis, where only a single cortex layer is present. Furthermore, the AtPEP marker line suggests that there is little functional difference between each of the four cortex layers, although more cortex-specific promoters should be tested to assess the validity of this hypothesis. Additionally, different promoters can help define distinct developmental stages of cell types. For instance, the SlCYCD6 and SlWOX5 promoters show differences in the number of cells marked in the QC/stem cell niche. This suggests that there are two distinct populations of cells in the stem cell niche of tomato.
The INTACT and TRAP technologies have also revolutionized our ability to address the functions of individual cell types by monitoring their molecular signatures (Mustroph et al., 2009b; Deal and Henikoff, 2010; Juntawong et al., 2014). Critical to our approach was testing the different components of both the INTACT and TRAP cassettes in planta using A. rhizogenes. Specifically, we were able to rapidly assess the proper nuclear and ribosome localization of each fusion protein prior to the lengthy process of generating and genotyping stable transgenics using traditional A. tumefaciens transformation. Of particular note, two components differ between the INTACT construct used in Arabidopsis and that used in tomato. A codon-optimized version of BirA was utilized; the version used in Arabidopsis was derived from Escherichia coli (Deal and Henikoff, 2010). Second, we used the tomato ACTIN2 (SlACT2) promoter to drive BirA expression instead of Arabidopsis ACT2 (AtACT2). Our immunoprecipitations and subsequent chromatin or transcript amplification of the SHR, SCR, and PEP DNA or transcripts, respectively, demonstrate that INTACT and TRAP technology can be utilized successfully in tomato. Nuclear immunopurification and mRNA qPCR reflected the promoter:GFP expression patterns observed. Of particular note, the variation in the relative levels of individual transcripts in the immunopurified versus total RNA fractions is consistent with the differential translation of individual mRNAs (Juntawong et al., 2014).
Testing gene function using transformation is essential to functional genomic approaches. CRISPR/Cas9 technology has greatly excited the plant community with its ability to edit genomes, as opposed to transposon-based insertion to disrupt gene function. The use of CRISPR/Cas9 technology in other crop species yielded similar mutation types to those observed here, insertions and deletions, characteristic of the error-prone nonhomologous end-joining DNA repair. Our data suggest that SHR function is conserved between Arabidopsis and tomato with respect to its ability to regulate SCR expression and root length. Further analyses are needed to determine the influence on asymmetric cell division.
Together, the experiments presented here clearly show that A. rhizogenes can be a transformative tool to rapidly test gene expression and function at cell type resolution in the context of root development and even in response to the environment, as has been described previously in M. truncatula (Reynoso et al., 2013). Furthermore, the cell type-specific promoter collection and tomato-optimized INTACT and TRAP vectors will enable high-throughput profiling of chromatin and transcripts undergoing translation in individual cell types. Finally, CRISPR/Cas9 technology can be used in tomato to edit the genome in a targeted, specific manner.
MATERIALS AND METHODS
Rhizogenes Transformation
Tomato (Solanum spp.) and Lepidium hyssopifolium seeds were surface sterilized in 70% (v/v) ethanol for 5 min followed by 50% (v/v) commercial bleach for 20 min and three washes with sterile deionized water. Seeds were plated on Murashige and Skoog (MS) plates containing 4.3 g L−1 MS medium (Caisson; catalog no. MSP01-50LT), 0.5 g L−1 MES, 30 g L−1 Suc, pH 5.8, and 30 g L−1 agar (Difco; catalog no. 214530) in Magenta boxes and placed in a 23°C incubator (16 h of light/8 h of darkness) for 7 to 10 d until cotyledons were fully expanded and the true leaves were just emerged.
Agrobacterium rhizogenes transformation followed the protocol reported previously (Harvey et al., 2008) with modifications as follows. Competent A. rhizogenes was transformed by electroporation with the desired binary vector, plated on custom-made MG/L medium plates (Caisson; catalog no. MQP04-1LT; for composition, see Supplemental Appendix S1) with the appropriate antibiotics, and incubated for 2 to 4 d at 26°C to 28°C. A transformed A. rhizogenes culture was inoculated from plates into Caisson 10 mL of liquid medium (MG/L) with the appropriate antibiotics (100 mg L−1 spectinomycin) and was grown overnight at 26°C to 30°C with shaking at 200 rpm. The A. rhizogenes culture was used to transform 30 to 50 tomato cotyledons. Using a scalpel, the cotyledons were cut from 8- to 12-d-old tomato seedlings and immediately immersed in bacterial suspension at an optical density at 600 nm of 0.3 in MS liquid medium for 20 min, then blotted on sterile Whatman filter paper and transferred (adaxial side down) onto MS agar plates without antibiotics. After 2 to 3 d of incubation at 25°C with shading (Oberpichler et al., 2008), the cotyledons were transferred to MS agar plates with 200 mg L−1 cefotaxime (Caisson; catalog no. C032) and antibiotics (50 mg L−1 kanamycin, 4 mg L−1 glufosinate-ammonium, or 20 mg L−1 hygromycin B) and returned to 25°C. At least three to five independent roots arise from each cotyledon. Antibiotic-resistant roots that emerged were further transferred to new selection plates with the same antibiotic concentration for further analysis. Ten independent roots were subcloned for each construct, and at least five to 10 of these independent roots showed expression of the marker. A complete and detailed protocol can be found in Supplemental Appendix S1.
Plasmid DNA Constructs
Promoter Design
Promoters were cloned from Arabidopsis (Arabidopsis thaliana) Columbia-0 and Solanum lycopersicum ‘M82’ genomic DNA. For Arabidopsis promoters, previously identified promoters were used (An et al., 1996; Lee et al., 2006; Mustroph et al., 2009b). Tomato homologs of previously identified cell type-specific genes in Arabidopsis were found using Reciprocal Best BLAST hits: SlSHR (Solyc02g092370), SlSCR (Solyc10g074680), SlCO2 (Solyc08g078890), SlCYCD6;1 (Solyc07g054950), SlWOX5 (Solyc03g096300), and SlRPL11C (Solyc02g086240). Cloning primers (Supplemental Table S1) were designed to amplify approximately 2,000 to 3,000 bp upstream of the translational start site. The promoters were amplified from genomic DNA using Phusion DNA polymerase (New England Biolabs). All promoters were cloned into pENTR/D-Topo or pENTR5′/Topo (Invitrogen) and confirmed by sequencing. The promoters were then cloned into the binary vectors using LR Clonase II Enzyme mix (Invitrogen).
Transcriptional Reporter Constructs
The binary vectors for transcriptional reporters were recombined into the Multisite Gateway vector pK7m24GW together with endoplasmic reticulum-localized GFP or into pMK7S*NFm14GW backbone (https://gateway.psb.ugent.be/search). 35Spro:TagRFP-LTI6b-NosT was created using Gibson Assembly (Gibson et al., 2009) by replacing GFP in 35Spro:GFP-LTI6b-NosT (Federici et al., 2012) with TagRFP-T (Shaner et al., 2008). Next, the 35Spro:TagRFP-LTI6b-NosT cassette was PCR amplified from and cloned using In-Fusion (Clontech) into HindIII/ApaI-linearized pMK7S*NFm14GW to generate pMR074 (Supplemental Fig. S4). The G10-90 promoter was amplified from pMDC07 (Curtis and Grossniklaus, 2003). The G10-90pro-TagRFP-LTI6b-35STer cassette was cloned as three pieces by In-Fusion (Clontech) into HindIII/ApaI-linearized pMK7S*NFm14GW to generate pMR099 (Supplemental Fig. S4).
Translational Reporter Constructs
The binary vector for the SlSHR translational fusion was recombined into the Multisite Gateway vector pK7m34GW (https://gateway.psb.ugent.be/search) as an SlSHRpro-SlSHR-GFP fusion.
TRAP and INTACT Constructs
The binary vectors for TRAP and INTACT (for vector maps, see Supplemental Fig. S5) were constructed into the pK7WG binary backbone (https://gateway.psb.ugent.be/search). pK7WG was digested with KpnI and partially with EcoRV. TRAP and INTACT cassettes were amplified by PCR using previously published vectors as templates (Mustroph et al., 2009b; Deal and Henikoff, 2010) and primers listed in Supplemental Table S1. The cassettes were amplified in multiple fragments in order to introduce unique restriction sites around optimizable elements. The TRAP cassette was amplified in two pieces: SnaBI-His6-FLAG-GFP-StuI and StuI-AtRPL18-XbaI-Tocs; the INTACT cassette was amplified in three pieces: StuI-WPP-MfeI, MfeI-GFP-BLRP-Tnos, and KpnI-AtACT2p-XhoI-BirA-SnaBI-T35S. These cassettes were introduced into the digested backbone with Gibson Assembly Master Mix (New England Biolabs). Further replacement of the BirA element with codon use-optimized mBirA+3xmyc (a gift of Dolf Weijers and Joakim Palovaara) and AtACT2p with tomato SlACT2p (Solyc11g005330) was carried out using In-Fusion (Clontech) cloning technology using primers listed in Supplemental Table S1. The binary vector backbones were propagated in One Shot ccdB Survival Cells (Invitrogen).
CRISPR Constructs
A plasmid encoding Nicotiana spp. codon-optimized Cas9 was a kind gift from Savithramma Dinesh-Kumar (University of California, Davis). A 3× FLAG tag and an NLS were placed at the N terminus of Cas9, and an additional NLS was placed at the C terminus of Cas9. These three fusion elements were plant codon use optimized using GeneArt (Life Technologies) and synthesized by Life Technologies. The modified Nicotiana spp. codon-optimized Cas9 was placed under the control of the 35S promoter of pPLV26 (De Rybel et al., 2011) using In-Fusion (Clontech). The Cas9 cassette was amplified and cloned into pENTR/D-Topo followed by an LR reaction into pB7WG binary vector (https://gateway.psb.ugent.be/search) to generate plasmid pMR091. The AtU6pro-sgRNA cassette was amplified and cloned into XbaI-linearized pMR091 by In-Fusion (Clontech) to generate plasmid pMR093 (Supplemental Fig. S5). The SlSHR target site of 19 nucleotides was selected manually following the criteria described previously (Mali et al., 2013). The chosen target sequence was BLASTed against the tomato genome (http://solgenomics.net/tools/blast/), and no other potential matches in the genome were identified. The mGFP5 and eGFP target sequences were based on a previously described target sequence for eGFP (Mali et al., 2013). The selected target sequence (19 bp) was incorporated into two 60-bp oligonucleotides (Supplemental Table S1). The two primers (final concentration of 10 µm for each primer) were annealed and extended (one cycle) to make a 100-bp double-stranded DNA fragment using Phusion polymerase (New England Biolabs). Then, the 100-bp double-stranded DNA was incorporated into an NcoI-linearize pMR093 using Gibson Assembly (New England Biolabs).
CRISPR RE/PCR Assay
Emerging transgenic roots were screened using a fluorescence dissecting microscope (Zeiss SteREO Discovery.v12), and the roots with decreased or lack of SlSCRpro-endoplasmic reticulum-localized GFP expression were then confirmed by imaging using laser scanning confocal microscopy. To identify the mutations, genomic DNA was extracted from roots using the DNeasy Plant Mini Kit (Qiagen) and PCR amplified with Phusion DNA polymerase (New England Biolabs) using primers spanning the sgRNA target sites. The amplicons were then digested with a restriction enzyme that recognizes the wild-type target sequences (AflII for mGFP5 and AleI or MslI for SHR). Mutations introduced by nonhomologous end joining were resistant to restriction enzyme digestion due to loss of the restriction site and resulted in undigested bands. Enzyme digestion products were visualized by agarose gel electrophoresis. Alternatively, 25 ng of the genomic DNA was digested by AflII or AleI, and the digested DNA was used as a template in a PCR with the same primers as before. The resulting PCR fragments were purified and cloned into the pJET1.2 plasmid (Thermo Scientific). The mutated sequences were identified by sequencing the PCR product isolated directly from single colonies.
Laser Scanning Confocal Microscopy
Laser scanning confocal microscopy was carried out with tomato hairy roots immersed in perfluorodecalin (Acros Organics; Littlejohn and Love, 2012) using the LSM 710 laser scanning microscope (Carl Zeiss; most figures) or tomato hairy roots immersed in sterile water using the LSM 700 laser scanning microscope (Carl Zeiss; Figs. 3M, 4A, and 5A).
As tomato roots autofluoresce at green wavelengths (490–620 nm; Supplemental Fig. S3J), the linear unmixing capability of the LSM 710 was used to resolve the GFP fluorescence patterns (Supplemental Fig. S3). First, reference spectra for autofluorescence were generated by exciting a nonfluorophore-expressing tomato hairy root with the 488-nm laser, and the emitted light was measured across the visible spectrum in 9.7-nm sections (a spectral scan). Zen 2011 software (Carl Zeiss) was used to autodetect two spectral components of autofluorescence, and these autofluorescence spectra were saved for later use (Supplemental Fig. S3, A–C and J). To image the transgenic roots, GFP was imaged using 488-nm excitation, and a spectral scan was collected (Supplemental Fig. S3C). The spectral scan was used for linear unmixing with the Zen 2011 software to separate tomato root autofluorescence and GFP fluorescence (Supplemental Fig. S3, E–G). In Figures 2, A and C, and 3, A to I and K, the autofluorescence is not shown, and in Figures 2, B and D, and 5, it is shown in white. For imaging TagRFP, the sample was excited with a 561-nm laser, and emitted light was recorded according to the preset DsRed emission spectrum (Supplemental Fig. S3H). Finally, GFP and TagRFP channels were overlaid (Supplemental Fig. S3I), and 100-μm scale bars were added using Zen 2011 software.
All laser scanning confocal microscopy imaging for Figures 2D and 6, B to E, and Supplemental Figure S7, B to E, was carried out with the following settings: 20× objective, 488-nm excitation laser with 70% power, 1.87-Airy unit pinhole, 583 gain, and the linear unmixing was carried out in nonweighted fashion. Imaging with the LSM 700 confocal microscope was carried out with 488-nm excitation and the preset eGFP emission spectrum.
Nuclei Purification
Biotinylated nuclei were purified from approximately 300 mg of 35Spro:NTF/SlACT2pro:mBirA-transformed tomato roots essentially as described previously for Arabidopsis roots (Deal and Henikoff, 2011) with a modification to the magnetic bead-capture procedure. In brief, transformed roots were ground in liquid nitrogen and resuspended in 10 mL of nuclei purification buffer (NPB; 20 mm MOPS, 40 mm NaCl, 90 mm KCl, 2 mm EDTA, 0.5 mm EGTA, 0.5 mm spermidine, and 0.2 mm spermine, pH 7) containing Roche Complete protease inhibitors. These extracts were then filtered with 70-μm nylon mesh and centrifuged at 1,000g for 5 min at 4°C to pellet the nuclei. Nuclei were washed with 1 mL of NPB, pelleted again, and then resuspended in 1 mL of NPB. Twenty-five microliters of M-280 streptavidin-coated Dynabeads (approximately 1.5 × 107 beads; Life Technologies; catalog no. 11205D) was added to the nuclei, and this mixture was rotated at 4°C for 30 min to allow capture of the biotinylated nuclei on beads. The nuclei/bead suspension was then diluted to 10 mL in a 15-mL tube, mixed thoroughly, and placed in a Dynamag 15 magnet (Life Technologies; catalog no. 12301D) at 4°C to capture bead-bound nuclei. The supernatant was carefully decanted, and the beads were resuspended in 10 mL of cold NPB, placed on a rotating mixer for 30 s, and then placed back in the Dynamag 15 magnet to capture the beads and nuclei from solution. This wash step was repeated once more, and bead-bound nuclei and free beads were finally resuspended in 1 mL on NPB for counting and analysis of purity as described previously (Deal and Henikoff, 2011).
For ChIP experiments, nuclei were purified using the same procedure and amount of tissue as described above except that roots were first treated with 1% (v/v) formaldehyde for 15 min in order to cross-link histones to the DNA.
Ribosome Immunopurification
The isolation of ribosomes from 35Spro:HF-GFP-RPL18 transgenic roots or nontransformed control roots was performed by immunopurification or conventional ultracentrifugation, respectively, as described by Mustroph et al. (2009a). Immunopurification used the EZ View Red ANTI-FLAG M2 Affinity Gel (Sigma-Aldrich) as the matrix for the antibodies recognizing the FLAG peptide. Frozen tissue was pulverized in polysome extraction buffer (PEB; 200 mm Tris-HCl, pH 9, 200 mm KCl, 35 mm MgCl2, 25 mm EGTA, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 50 µg mL−1 cycloheximide, 50 µg mL−1 chloramphenicol, 1% [v/v] Triton X-100, 1% [v/v] Brij-35 [polyoxyethylene 23 lauryl ether], 1% [v/v] Tween 20 [polyoxyethylene sorbitan monolaurate 20], 1% [v/v] Igepal CA 630 [octylphenyl-polyethylene glycol], and 1% [v/v] polyoxyethylene 10 tridecyl ether). For TRAP, 0.5-mL packed volume of frozen pulverized root tissue was used. One milliliter of frozen tissue was used to obtain the P-170 pellet fraction enriched in cellular ribosomes by differential centrifugation. Five milliliters of pulverized root tissue was used for Suc density gradient fractionation of polysomes for western-blot analysis of ribosomal proteins. Briefly, tissue was hydrated in 2 volumes of PEB, and a clarified crude cell extract (total fraction) was obtained by centrifugation at 16,000g for 15 min and filtration through Miracloth. A portion of this supernatant was reserved for protein and RNA analyses as the total fraction. The remainder was used for pelleting by centrifugation for 3 h through a 2 m Suc cushion at 170,000g to obtain the P-170 fraction. For fractionation of polysomes, the P-170 fraction was centrifuged for 1.5 h through a 15% to 60% (w/v) Suc gradient at 237,000g. Fractions of 400 µL were collected, and complexes were precipitated by the addition of 2 volumes of 100% (v/v) ethanol, overnight incubation at 4°C, and centrifugation at 16,000g. For TRAP, the clarified cell extract was incubated with 100 µL of agarose beads washed twice with buffer containing 200 mm Tris-HCl, pH 9, 200 mm KCl, 35 mm MgCl2, and 25 mm EGTA. The beads and extract were incubated for 2 h at 4°C with gentle shaking. After this step, the beads were collected by centrifugation at 8,200g for 2 min at 4°C, and the supernatant was reserved as the unbound fraction. The beads were washed four times with wash buffer (PEB without detergents) without being allowed to dry. A total of 200 µL of elution buffer (wash buffer added with 3× FLAG peptide [Sigma-Aldrich] at a final concentration of 200 ng µL−1) was added to the beads and incubated at 4°C with gentle shaking for 30 min. Ten microliters of both total and unbound fractions (1% [v/v] of the clarified extract) and 20 µL of the eluate (10% [v/v] of the clarified extract) were used for the western-blot analysis.
Western Blotting
To detect the biotin-labeled nuclear tagging fusion, protein extracts were made from 35Spro:NTF/SlACT2pro:mBirA-transformed or untransformed tomato hairy roots as well as transgenic Arabidopsis roots carrying both ADF8pro:NTF and ACT2pro:BirA transgenes. Three independent samples of transformed and untransformed tomato roots were analyzed. Tissues were ground in liquid nitrogen and resuspended in 2 volumes of RIPA buffer (50 mm Tris, 150 mm NaCl, 1% [w/v] Nonidet P-40, 0.5% [v/v] sodium deoxycholate, and 0.1% [w/v] SDS, pH 7.5) containing Roche Complete protease inhibitors. This extract was cleared by centrifugation and mixed with 1 volume of 2× SDS loading buffer (100 mm Tris-HCl, pH 7.5, 10% [w/v] SDS, 30% [v/v] glycerol, 1% [v/v] β-mercaptoethanol, and 0.2% [w/v] bromphenol blue), heated to 100°C for 5 min, and equal aliquots of each sample were electrophoresed on two separate 12% (w/v) SDS polyacrylamide gels. One gel was stained with Coomassie Brilliant Blue to visualize protein amounts. Proteins from the other gel were electrophoretically transferred to a nitrocellulose membrane, which was blocked in phosphate-buffered saline with Triton X-100 (PBSt; 11.9 mm sodium phosphate, 137 mm NaCl, 2.7 mm KCl, and 0.1% [v/v] Triton X-100, pH 7.4) with 10% (w/v) milk for 30 min, washed twice for 5 min with PBSt, and incubated with a 1:2,000 dilution of streptavidin-horseradish peroxidase (HRP; GE; catalog no. RPN1231) in PBSt with 1% (w/v) bovine serum albumin for 30 min. The membrane was then washed three times for 5 min with PBSt, and biotinylated proteins were detected using enhanced chemiluminescence reagents (Pierce; catalog no. 34075).
To detect FLAG-tagged RPL18, fractions obtained in the purification procedure were separated by 15% (w/v) SDS-PAGE and detected by silver staining or immunodetection with a FLAG HRP-conjugated monoclonal antibody (1:1,000; Sigma-Aldrich) as described previously by Zanetti et al. (2005). RPS6 was detected using a polyclonal antiserum against maize (Zea mays) RPS6 protein (1:5,000; Williams et al., 2003), and an HRP-conjugated goat anti-rabbit IgG was used as a secondary antibody (1:10,000; Bio-Rad).
For the analysis of cytoplasmic and nuclear protein distribution during INTACT purification (Supplemental Fig. S6), 400 mg of 35Spro:NTF/SlACT2pro:mBirA-transformed tomato roots was ground in liquid nitrogen and resuspended in 1 mL of NPB containing Roche Complete protease inhibitors. These extracts were then filtered through 70-μm nylon mesh to generate the extract fraction containing cytoplasmic components and released nuclei. An aliquot of this initial extract was combined with 1 volume of 2× SDS loading buffer and placed on ice for the remainder of the procedure. The extract fraction was then centrifuged at 1,000g for 5 min at 4°C to pellet the nuclei and other debris. The supernatant from this centrifugation was mixed with 1 volume of 2× SDS loading buffer and kept on ice (supernatant fraction), while the pellet was resuspended in 1 mL of NPB. An aliquot of the resuspended pellet was again centrifuged to pellet the nuclei and debris, and the pelleted material was resuspended in 2× SDS loading buffer and kept on ice (pellet fraction). To the remainder of the resuspended pellet was added 25 μL of M-280 streptavidin-coated Dynabeads, and tagged nuclei were purified as described above. The final purified nuclei were resuspended in 2× SDS loading buffer to generate the purified nuclei fraction, and all fractions were heated to 100°C for 5 min and run on 12% (w/v; for actin) or 16% (w/v; for histone H3) SDS polyacrylamide gels. Separated proteins were transferred to nitrocellulose membranes, and membranes were blocked as described above. Blocked membranes were probed with either a histone H3 antibody (Abcam; catalog no. Ab1791; 1:2,000 dilution) or an ACTIN8 antibody (mouse monoclonal clone 10B3; a gift from Richard Meagher; 1:2,000 dilution). Blots were washed three times for 5 min with PBSt and probed with the appropriate horseradish peroxidase-coupled secondary antibody for chemiluinescent detection.
Transcript Quantification
To detect and quantify transcripts from nuclei, RNA was isolated from approximately 7 × 105 nuclei purified from 35Spro:NTF/SlACT2pro:mBirA-transformed roots using the Qiagen RNeasy Plant Mini Kit. Two biological replicates were performed, starting with separate samples of transformed roots. Each RNA sample was treated with RNase-free DNase I (Ambion; AM1907), and cDNA was prepared using the SuperScript III kit (Invitrogen; 18080-051) with oligo(dT) primers according to the manufacturer’s instructions. Real-time PCR was performed on an Applied Biosystems Step One Plus instrument using SYBR Green detection chemistry. Relative quantities of each transcript were calculated using the 2−ddCt method (Livak and Schmittgen, 2001). 2-ddCt is a method that uses the difference (d) of reference and target gene cycle numbers to reach a set threshold (Ct) and takes their difference (d) between different sample. Transcripts assayed were PEP (Solyc04g076190), SHR (Solyc02g092370), SCR (Solyc10g074680), and ACT2 (Solyc11g005330), with transcript Solyc01g014230 serving as the endogenous control transcript to which the others were normalized. All primer sequences are given in Supplemental Table S1.
Total and TRAP RNA were extracted from 400 µL of clarified extract and from 200 µL of TRAP (ribosome immunopurified) eluate using 800 and 400 µL of Trizol reagent (Invitrogen), respectively, and quantified by the use of a Nanodrop ND-1000 UV-Vis spectrophotometer (Nanodrop Technology). rRNA and tRNA were evaluated by electrophoresis on 1.2% (w/v) agarose gels. To evaluate specific transcripts, 0.5 μg of the extracted RNA was treated with DNase (RNase-Free DNase Set; Qiagen) and used to synthesize the first-strand cDNA using oligo(dT) primer and reverse transcriptase (Thermo Scientific Maxima H minus reverse transcriptase). qPCR was performed using the CFX96 real-time system and the SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) according to the manufacturer’s instructions. Three technical and two biological replicates were performed for each gene. Relative expression levels were calculated as described by Livak and Schmittgen (2001) using ACT2 (Solyc11g005330) levels for normalization.
ChIP and qPCR
Biotinylated nuclei were purified from formaldehyde-treated 35Spro:NTF/SlACT2pro:mBirA-transformed roots as described above. Two biological replicates of these experiments were performed, starting with separate samples of transformed roots. Purified nuclei (approximately 3 × 105 nuclei) were lysed in 120 μL of nuclei lysis buffer (50 mm Tris, 10 mm EDTA, and 1% [w/v] SDS, pH 8) and sonicated using a Diagenode Bioruptor to yield chromatin fragments with an average size of approximately 500 bp. Sonicated chromatin was cleared by centrifugation and diluted to a 1.2-mL final volume with ChIP dilution buffer (16.7 mm Tris, 1.2 mm EDTA, 1.1% [v/v] Triton X-100, and 167 mm NaCl, pH 8). An aliquot of this chromatin was reserved as the input sample, and the remainder was then divided into two aliquots of equal volume and 1 to 3 μg of antibody was added to each aliquot. In these experiments, we used antibodies against H3K4me3 (Abcam; catalog no. Ab8580) and GFP (Santa Cruz Biotechnology; catalog no. sc-8334) as negative controls. Antibodies were incubated with chromatin at 4°C for 2 h on a rocking platform, and then 30 μL of protein A Dynabeads (Life Technologies; 10001D) was added with rocking at 4°C for an additional 1 h. Beads were washed once for 5 min at 4°C in 0.5 mL of each of the following buffers: low-salt wash buffer (20 mm Tris, pH 8, 150 mm NaCl, 0.1% [w/v] SDS, 1% [v/v] Triton X-100, and 2 mm EDTA), high-salt wash buffer (20 mm Tris, pH 8, 500 mm NaCl, 1% [w/v] sodium deoxycholate, 1% [v/v] nonylphenoxypolyethoxylethanol [Nonidet P-40], and 1 mm EDTA), LiCl wash buffer (10 mm Tris, pH 8, 250 mm LiCl, 0.1% [w/v] SDS, 1% [v/v] Triton X-100, and 2 mm EDTA), and Tris-EDTA (10 mm Tris, pH 7.5, and 1 mm EDTA). Chromatin was eluted from the beads in 200 μL of elution buffer (100 mm NaHCO3 and 1% [w/v] SDS) with vortexing for 5 min, NaCl was added to 0.5 m, and eluted chromatin was heated to 100°C for 15 min to reverse cross-links. DNA was isolated by treating the chromatin with RNase A, proteinase K, and purification using the Qiagen MinElute kit. Input chromatin samples were processed in parallel with the ChIP samples.
qPCR was used to assay the relative enrichment of H3K4me3 on the ACT2 (Solyc11g005330) promoter region as well as the SHR (Solyc02g092370) promoter and gene body. PCR was performed on an Applied Biosystems Step One Plus instrument using SYBR Green detection chemistry. Relative enrichment at each genomic location was calculated by the 2−ddCt method (Livak and Schmittgen, 2001) using the 3′ end of the ACT2 gene as the endogenous control region and the input chromatin sample as the calibrator. No detectable amplification was observed in the anti-GFP negative control ChIP samples. All primer sequences are given in Supplemental Table S1.
Tissue Embedding, Sectioning, and GUS Staining
Embedding and sectioning of roots were done as previously described in Ron et al. (2013). Detection of GUS expression in roots was done as previously described in Ron et al. (2010). After GUS staining, roots were embedded and sectioned as above but without the FAA fixing step. Images were captured with a Zeiss Axioplan Imaging 2 microscope with a CCD Zeiss camera using the AxioVision 4.8 software.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Cell type- and tissue-specific expression from SlSHR, SlSCR, and SlCO2 promoters.
Supplemental Figure S2. Cell type- and tissue-specific expression from AtS32, AtS18, and AtWER promoters.
Supplemental Figure S3. Use of spectral scanning and linear unmixing.
Supplemental Figure S4. Maps of vectors for transcriptional fusions.
Supplemental Figure S5. Vector maps of TRAP and INTACT backbones.
Supplemental Figure S6. INTACT-purified nuclei are free of cytoplasmic contamination.
Supplemental Figure S7. High-efficiency targeted gene editing in tomato plants.
Supplemental Figure S8. Targeting the SHR gene using CRISPR/CAS9.
Supplemental Table S1. Complete primer list.
Supplemental Appendix S1. Complete hairy root transformation protocol.
Supplementary Material
Acknowledgments
We thank Savithramma Dinesh-Kumar for the plant codon-optimized Cas9 cassette, Joakim Palovaara and Dolf Weijers for the Arabidopsis codon use-optimized mBirA gene, and Joger Tsien for TagRFP-T.
Glossary
- cDNA
complementary DNA
- INTACT
isolation of nuclei tagged in specific cell types
- TRAP
translating ribosome affinity purification
- CRISPR
cluster regularly interspaced short palindromic repeats
- Cas9
CRISPR-associated nuclease9
- QC
quiescent center
- PI
propidium iodide
- NTF
nuclear tagging fusion
- ChIP
chromatin immunoprecipitation
- H3K4me3
histone H3 Lys-4 trimethylation
- qPCR
quantitative PCR
- rRNA
ribosomal RNA
- sgRNA
single-guide RNA
- PAM
protospacer-associated motif
- RE/PCR
restriction enzyme digestion/PCR amplification
- SNPs
single-nucleotide polymorphisms
- MS
Murashige and Skoog
- NLS
nuclear localization signal
- NPB
nuclei purification buffer
- PEB
polysome extraction buffer
- HRP
horseradish peroxidase
Footnotes
This work was supported by the National Science Foundation (Plant Genome grant no. 1238243 to G.P., M.A.R., K.Z., S.W., N.S., R.B.D., J.B.-S., and S.M.B. and grant no. 1052395 to M.R. and S.M.B.), by a Finnish Cultural Foundation postdoctoral fellowship (to K.K.), by the Engineering and Physical Sciences Research Council and the National Science Foundation (grant no. EP/H019162/1 to F.F.), and by Comisión Nacional de Investigación Científica y Tecnológica-Programa de Atracción e Inserción de Capital Humano Avanzado (grant no. 82130027 to F.F.).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- An YQ, McDowell JM, Huang S, McKinney EC, Chambliss S, Meagher RB. (1996) Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J 10: 107–121 [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J. (2013) Microgenomics: genome-scale, cell-specific monitoring of multiple gene regulation tiers. Annu Rev Plant Biol 64: 293–325 [DOI] [PubMed] [Google Scholar]
- Belhaj K, Chaparro-Garcia A, Kamoun S, Nekrasov V. (2013) Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 9: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey PN, Linstead PJ, Roberts K, Schiefelbein JW, Hauser MT, Aeschbacher RA. (1993) Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development 119: 57–70 [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN. (2003) A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Chang IF, Szick-Miranda K, Pan S, Bailey-Serres J. (2005) Proteomic characterization of evolutionarily conserved and variable proteins of Arabidopsis cytosolic ribosomes. Plant Physiol 137: 848–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Levesque MP, Vernoux T, Jung JW, Paquette AJ, Gallagher KL, Wang JY, Blilou I, Scheres B, Benfey PN. (2007) An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316: 421–425 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR. (2000) Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA 97: 3718–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Henikoff S. (2010) A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev Cell 18: 1030–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Henikoff S. (2011) The INTACT method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana. Nat Protoc 6: 56–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deboer KD, Lye JC, Aitken CD, Su AKK, Hamill JD. (2009) The A622 gene in Nicotiana glauca (tree tobacco): evidence for a functional role in pyridine alkaloid synthesis. Plant Mol Biol 69: 299–312 [DOI] [PubMed] [Google Scholar]
- De Rybel B, van den Berg W, Lokerse A, Liao CY, van Mourik H, Möller B, Peris CL, Weijers D. (2011) A versatile set of ligation-independent cloning vectors for functional studies in plants. Plant Physiol 156: 1292–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federici F, Dupuy L, Laplaze L, Heisler M, Haseloff J. (2012) Integrated genetic and computation methods for in planta cytometry. Nat Methods 9: 483–485 [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, III, Smith HO. (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6: 343–345 [DOI] [PubMed] [Google Scholar]
- Haecker A, Gross-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T. (2004) Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131: 657–668 [DOI] [PubMed] [Google Scholar]
- Hartley JL, Temple GF, Brasch MA. (2000) DNA cloning using in vitro site-specific recombination. Genome Res 10: 1788–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey JJ, Lincoln JE, Gilchrist DG. (2008) Programmed cell death suppression in transformed plant tissue by tomato cDNAs identified from an Agrobacterium rhizogenes-based functional screen. Mol Genet Genomics 279: 509–521 [DOI] [PubMed] [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN. (2000) The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101: 555–567 [DOI] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. (2013) RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31: 233–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntawong P, Girke T, Bazin J, Bailey-Serres J. (2014) Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc Natl Acad Sci USA 111: E203–E212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajikawa M, Hirai N, Hashimoto T. (2009) A PIP-family protein is required for biosynthesis of tobacco alkaloids. Plant Mol Biol 69: 287–298 [DOI] [PubMed] [Google Scholar]
- Koenig D, Jiménez-Gómez JM, Kimura S, Fulop D, Chitwood DH, Headland LR, Kumar R, Covington MF, Devisetty UK, Tat AV, et al. (2013) Comparative transcriptomics reveals patterns of selection in domesticated and wild tomato. Proc Natl Acad Sci USA 110: E2655–E2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Colinas J, Wang JY, Mace D, Ohler U, Benfey PN. (2006) Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc Natl Acad Sci USA 103: 6055–6060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima JE, Benedito VA, Figueira A, Peres LE. (2009) Callus, shoot and hairy root formation in vitro as affected by the sensitivity to auxin and ethylene in tomato mutants. Plant Cell Rep 28: 1169–1177 [DOI] [PubMed] [Google Scholar]
- Lin MH, Gresshoff PM, Indrasumunar A, Ferguson BJ. (2011) pHairyRed: a novel binary vector containing the DsRed2 reporter gene for visual selection of transgenic hairy roots. Mol Plant 4: 537–545 [DOI] [PubMed] [Google Scholar]
- Lincoln JE, Richael C, Overduin B, Smith K, Bostock R, Gilchrist DG. (2002) Expression of the antiapoptotic baculovirus p35 gene in tomato blocks programmed cell death and provides broad-spectrum resistance to disease. Proc Natl Acad Sci USA 99: 15217–15221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlejohn GR, Love J. (2012) A simple method for imaging Arabidopsis leaves using perfluorodecalin as an infiltrative imaging medium. J Vis Exp pII: 3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Wang Z, Xiao A, Zhang Y, Li W, Zu Y, Yao S, Lin S, Zhang B. (2013) Efficient gene targeting in zebrafish mediated by a zebrafish-codon-optimized Cas9 and evaluation of off-targeting effect. J Genet Genomics 41: 43–46 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lucas M, Swarup R, Paponov IA, Swarup K, Casimiro I, Lake D, Peret B, Zappala S, Mairhofer S, Whitworth M, et al. (2011) Short-Root regulates primary, lateral, and adventitious root development in Arabidopsis. Plant Physiol 155: 384–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace DL, Lee JY, Twigg RW, Colinas J, Benfey PN, Ohler U. (2006) Quantification of transcription factor expression from Arabidopsis images. Bioinformatics 22: e323–e331 [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. (2013) RNA-guided human genome engineering via Cas9. Science 339: 823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE. (2010) Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc Natl Acad Sci USA 107: 4623–4628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussaieff A, Rogachev I, Brodsky L, Malitsky S, Toal TW, Belcher H, Yativ M, Brady SM, Benfey PN, Aharoni A. (2013) High-resolution metabolic mapping of cell types in plant roots. Proc Natl Acad Sci USA 110: E1232–E1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A, Juntawong P, Bailey-Serres J. (2009a) Isolation of plant polysomal mRNA by differential centrifugation and ribosome immunopurification methods. Methods Mol Biol 553: 109–126 [DOI] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJH, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J. (2009b) Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA 106: 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseer S, Lee Y, Lapierre C, Franke R, Nawrath C, Geldner N. (2012) Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proc Natl Acad Sci USA 109: 10101–10106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V, Staskawicz B, Weigel D, Jones JD, Kamoun S. (2013) Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol 31: 691–693 [DOI] [PubMed] [Google Scholar]
- Oberpichler I, Rosen R, Rasouly A, Vugman M, Ron EZ, Lamparter T. (2008) Light affects motility and infectivity of Agrobacterium tumefaciens. Environ Microbiol 10: 2020–2029 [DOI] [PubMed] [Google Scholar]
- Ono NN, Tian L. (2011) The multiplicity of hairy root cultures: prolific possibilities. Plant Sci 180: 439–446 [DOI] [PubMed] [Google Scholar]
- Petricka JJ, Schauer MA, Megraw M, Breakfield NW, Thompson JW, Georgiev S, Soderblom EJ, Ohler U, Moseley MA, Grossniklaus U, et al. (2012) The protein expression landscape of the Arabidopsis root. Proc Natl Acad Sci USA 109: 6811–6818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynoso MA, Blanco FA, Bailey-Serres J, Crespi M, Zanetti ME. (2013) Selective recruitment of mRNAs and miRNAs to polyribosomes in response to rhizobia infection in Medicago truncatula. Plant J 73: 289–301 [DOI] [PubMed] [Google Scholar]
- Ron M, Dorrity MW, de Lucas M, Toal T, Hernandez RI, Little SA, Maloof JN, Kliebenstein DJ, Brady SM. (2013) Identification of novel loci regulating interspecific variation in root morphology and cellular development in tomato. Plant Physiol 162: 755–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron M, Saez MA, Williams LE, Fletcher JC, McCormick S. (2010) Proper regulation of a sperm-specific cis-nat-siRNA is essential for double fertilization in Arabidopsis. Genes Dev 24: 1010–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Tabata S, Hirakawa H, Asamizu E, Shirasawa K, Isobe S, Kaneko T, Nakamura Y, Shibata D, Aoki K, et al. (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, Tsien RY. (2008) Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods 5: 545–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzani R, Cui H, Moreno-Risueno MA, Busch W, Van Norman JM, Vernoux T, Brady SM, Dewitte W, Murray JAH, Benfey PN. (2010) Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature 466: 128–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytas DF. (2013) Plant genome engineering with sequence-specific nucleases. Annu Rev Plant Biol 64: 327–350 [DOI] [PubMed] [Google Scholar]
- Williams AJ, Werner-Fraczek J, Chang IF, Bailey-Serres J. (2003) Regulated phosphorylation of 40S ribosomal protein S6 in root tips of maize. Plant Physiol 132: 2086–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams ME, Sussex IM. (1995) Developmental regulation of ribosomal protein L16 genes in Arabidopsis thaliana. Plant J 8: 65–76 [DOI] [PubMed] [Google Scholar]
- Wiśniewska A, Dąbrowska-Bronk J, Szafrański K, Fudali S, Święcicka M, Czarny M, Wilkowska A, Morgiewicz K, Matusiak J, Sobczak M, et al. (2013) Analysis of tomato gene promoters activated in syncytia induced in tomato and potato hairy roots by Globodera rostochiensis. Transgenic Res 22: 557–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki Y, Kitajima M, Arita M, Takayama H, Sudo H, Yamazaki M, Aimi N, Saito K. (2004) Biosynthesis of camptothecin: in silico and in vivo tracer study from [1-13C]glucose. Plant Physiol 134: 161–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JM, Kuykendall LD, Martínez-Romero E, Kerr A, Sawada H. (2001) A revision of Rhizobium Frank 1889, with an emended description of the genus, and the inclusion of all species of Agrobacterium Conn 1942 and Allorhizobium undicola de Lajudie et al. 1998 as new combinations: Rhizobium radiobacter, R. rhizogenes, R. rubi, R. undicola and R. vitis. Int J Syst Evol Microbiol 51: 89–103 [DOI] [PubMed] [Google Scholar]
- Zanetti ME, Chang IF, Gong F, Galbraith DW, Bailey-Serres J. (2005) Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression. Plant Physiol 138: 624–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.