A high-affinity rice potassium transporter plays a major role in K acquisition by roots faced with low external K and in K upward transport from roots to shoots in K-deficient rice plants.
Abstract
In plants, K transporter (KT)/high affinity K transporter (HAK)/K uptake permease (KUP) is the largest potassium (K) transporter family; however, few of the members have had their physiological functions characterized in planta. Here, we studied OsHAK5 of the KT/HAK/KUP family in rice (Oryza sativa). We determined its cellular and tissue localization and analyzed its functions in rice using both OsHAK5 knockout mutants and overexpression lines in three genetic backgrounds. A β-glucuronidase reporter driven by the OsHAK5 native promoter indicated OsHAK5 expression in various tissue organs from root to seed, abundantly in root epidermis and stele, the vascular tissues, and mesophyll cells. Net K influx rate in roots and K transport from roots to aerial parts were severely impaired by OsHAK5 knockout but increased by OsHAK5 overexpression in 0.1 and 0.3 mm K external solution. The contribution of OsHAK5 to K mobilization within the rice plant was confirmed further by the change of K concentration in the xylem sap and K distribution in the transgenic lines when K was removed completely from the external solution. Overexpression of OsHAK5 increased the K-sodium concentration ratio in the shoots and salt stress tolerance (shoot growth), while knockout of OsHAK5 decreased the K-sodium concentration ratio in the shoots, resulting in sensitivity to salt stress. Taken together, these results demonstrate that OsHAK5 plays a major role in K acquisition by roots faced with low external K and in K upward transport from roots to shoots in K-deficient rice plants.
Potassium (K) is one of the three most important macronutrients and the most abundant cation in plants. As a major osmoticum in the vacuole, K drives the generation of turgor pressure, enabling cell expansion. In the vascular tissue, K is an important participant in the generation of root pressure (for review, see Wegner, 2014 [including his new hypothesis]). In the phloem, K is critical for the transport of photoassimilates from source to sink (Marschner, 1996; Deeken et al., 2002; Gajdanowicz et al., 2011). In addition, enhancing K absorption and decreasing sodium (Na) accumulation is a major strategy of glycophytes in salt stress tolerance (Maathuis and Amtmann, 1999; Munns and Tester, 2008; Shabala and Cuin, 2008).
Plants acquire K through K-permeable proteins at the root surface. Since available K concentration in the soil may vary by 100-fold, plants have developed multiple K uptake systems for adapting to this variability (Epstein et al., 1963; Grabov, 2007; Maathuis, 2009). In a classic K uptake experiment in barley (Hordeum vulgare), root K absorption has been described as a high-affinity and low-affinity biphasic transport process (Epstein et al., 1963). It is generally assumed that the low-affinity transport system (LATS) in the roots mediates K uptake in the millimolar range and that the activity of this system is insensitive to external K concentration (Maathuis and Sanders, 1997; Chérel et al., 2014). In contrast, the high-affinity transport system (HATS) was rapidly up-regulated when the supply of exogenous K was halted (Glass, 1976; Glass and Dunlop, 1978).
The membrane transporters for K flux identified in plants are generally classified into three channels and three transporter families based on phylogenetic analysis (Mäser et al., 2001; Véry and Sentenac, 2003; Lebaudy et al., 2007; Alemán et al., 2011). For K uptake, it was predicted that, under most circumstances, K transporters function as HATS, while K-permeable channels mediate LATS (Maathuis and Sanders, 1997). However, a root-expressed K channel in Arabidopsis (Arabidopsis thaliana), Arabidopsis K Transporter1 (AKT1), mediates K absorption over a wide range of external K concentrations (Sentenac et al., 1992; Lagarde et al., 1996; Hirsch et al., 1998; Spalding et al., 1999), while evidence is accumulating that many K transporters, including members of the K transporter (KT)/high affinity K transporter (HAK)/K uptake permease (KUP) family, are low-affinity K transporters (Quintero and Blatt, 1997; Senn et al., 2001), implying that functions of plant K channels and transporters overlap at different K concentration ranges.
Out of the three families of K transporters, cation proton antiporter (CPA), high affinity K/Na transporter (HKT), and KT/HAK/KUP, CPA was characterized as a K+(Na+)/H+ antiporter, HKT may cotransport Na and K or transport Na only (Rubio et al., 1995; Uozumi et al., 2000), while KT/HAK/KUP were predicted to be H+-coupled K+ symporters (Mäser et al., 2001; Lebaudy et al., 2007). KT/HAK/KUP were named by different researchers who first identified and cloned them (Quintero and Blatt, 1997; Santa-María et al., 1997). In plants, the KT/HAK/KUP family is the largest K transporter family, including 13 members in Arabidopsis and 27 members in the rice (Oryza sativa) genome (Rubio et al., 2000; Mäser et al., 2001; Bañuelos et al., 2002; Gupta et al., 2008). Sequence alignments show that genes of this family share relatively low homology to each other. The KT/HAK/KUP family was divided into four major clusters (Rubio et al., 2000; Gupta et al., 2008), and in cluster I and II, they were further separated into A and B groups. Genes of cluster I or II likely exist in all plants, cluster III is composed of genes from both Arabidopsis and rice, while cluster IV includes only four rice genes (Grabov, 2007; Gupta et al., 2008).
The functions of KT/HAK/KUP were studied mostly in heterologous expression systems. Transporters of cluster I, such as AtHAK5, HvHAK1, OsHAK1, and OsHAK5, are localized in the plasma membrane (Kim et al., 1998; Bañuelos et al., 2002; Gierth et al., 2005) and exhibit high-affinity K uptake in the yeast Saccharomyces cerevisiae (Santa-María et al., 1997; Fu and Luan, 1998; Rubio et al., 2000) and in Escherichia coli (Horie et al., 2011). Transporters of cluster II, like AtKUP4 (TINY ROOT HAIRS1, TRH1), HvHAK2, OsHAK2, OsHAK7, and OsHAK10, could not complement the K uptake-deficient yeast (Saccharomyces cerevisiae) but were able to mediate K fluxes in a bacterial mutant; they might be tonoplast transporters (Senn et al., 2001; Bañuelos et al., 2002; Rodríguez-Navarro and Rubio, 2006). The function of transporters in clusters III and IV is even less known (Grabov, 2007).
Existing data suggest that some KT/HAK/KUP transporters also may respond to salinity stress (Maathuis, 2009). The cluster I transporters of HvHAK1 mediate Na influx (Santa-María et al., 1997), while AtHAK5 expression is inhibited by Na (Rubio et al., 2000; Nieves-Cordones et al., 2010). Expression of OsHAK5 in tobacco (Nicotiana tabacum) BY2 cells enhanced the salt tolerance of these cells by accumulating more K without affecting their Na content (Horie et al., 2011).
There are only scarce reports on the physiological function of KT/HAK/KUP in planta. In Arabidopsis, mutation of AtKUP2 (SHORT HYPOCOTYL3) resulted in a short hypocotyl, small leaves, and a short flowering stem (Elumalai et al., 2002), while a loss-of-function mutation of AtKUP4 (TRH1) resulted in short root hairs and a loss of gravity response in the root (Rigas et al., 2001; Desbrosses et al., 2003; Ahn et al., 2004). AtHAK5 is the only system currently known to mediate K uptake at concentrations below 0.01 mm (Rubio et al., 2010) and provides a cesium uptake pathway (Qi et al., 2008). AtHAK5 and AtAKT1 are the two major physiologically relevant molecular entities mediating K uptake into roots in the range between 0.01 and 0.05 mm (Pyo et al., 2010; Rubio et al., 2010). AtAKT1 may contribute to K uptake within the K concentrations that belong to the high-affinity system described by Epstein et al. (1963).
Among all 27 members of the KT/HAK/KUP family in rice, OsHAK1, OsHAK5, OsHAK19, and OsHAK20 were grouped in cluster IB (Gupta et al., 2008). These four rice HAK members share 50.9% to 53.4% amino acid identity with AtHAK5. OsHAK1 was expressed in the whole plant, with maximum expression in roots, and was up-regulated by K deficiency; it mediated high-affinity K uptake in yeast (Bañuelos et al., 2002). In this study, we examined the tissue-specific localization and the physiological functions of OsHAK5 in response to variation in K supply and to salt stress in rice. By comparing K uptake and translocation in OsHAK5 knockout (KO) mutants and in OsHAK5-overexpressing lines with those in their respective wild-type lines supplied with different K concentrations, we found that OsHAK5 not only mediates high-affinity K acquisition but also participates in root-to-shoot K transport as well as in K-regulated salt tolerance.
RESULTS
The Effect of K Supply on OsHAK5 Expression in Rice
It has been reported that the transcripts of OsHAK5 were faint and not significantly affected by prolonged (8–11 d) K starvation in rice roots (Okada et al., 2008), while its transcripts in shoots were enhanced by more than 5-fold (Horie et al., 2011). To resolve the effect of short-term K deprivation on OsHAK5 expression, we detected the change of its transcripts by quantitative reverse transcription (qRT)-PCR between 1 and 72 h after removal of K from culture solution. In roots, OsHAK5 expression increased transiently by about 10-fold at 1 h, 3-fold at 6 h, and 2-fold at 24 h after K removal (Fig. 1A). In basal node and sheath, two major events of transcript increase were detected at 6 and 72 h of K deprivation (Fig. 1B). In leaf blades, the short-term K deprivation increased OsHAK5 expression much less, barely 2-fold (at 48 h; Fig. 1C).
Figure 1.
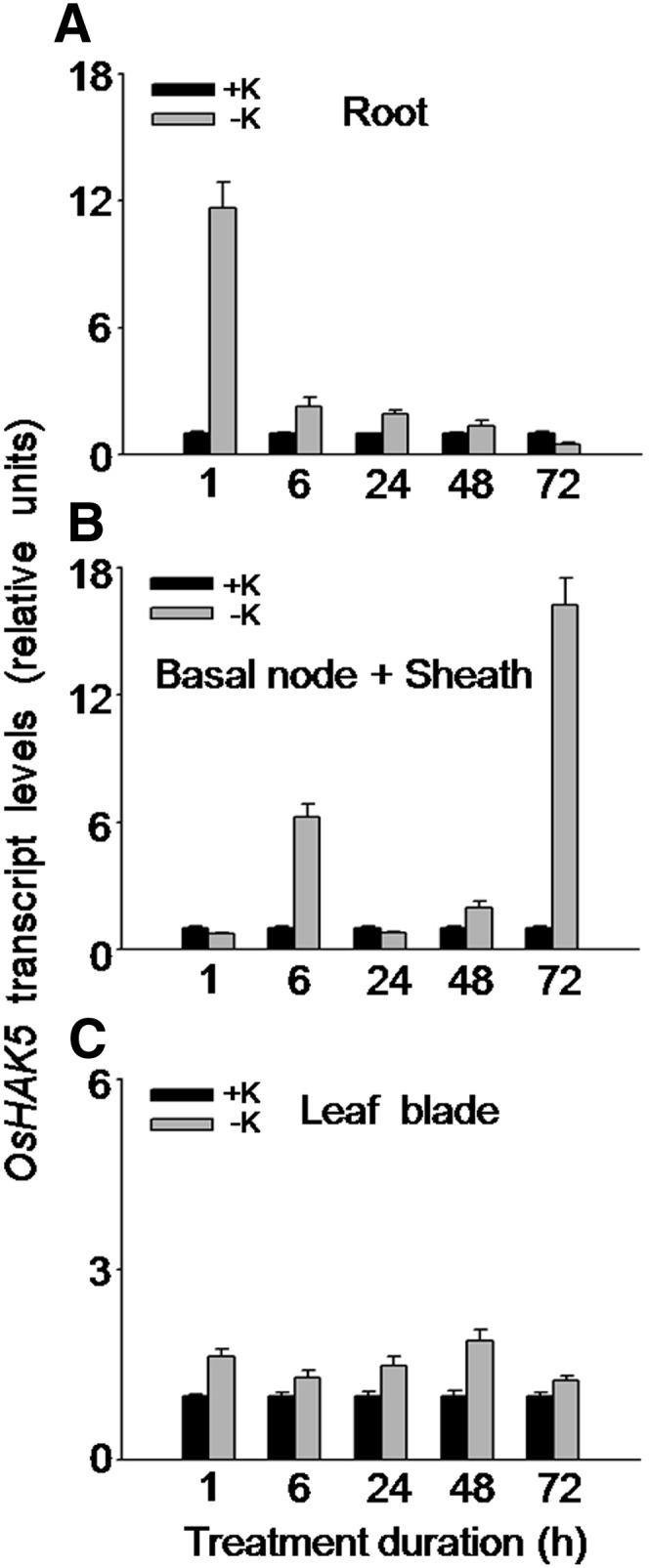
Effect of K deficiency on the transcriptional expression of OsHAK5 in rice. Ten-day-old rice seedlings (cv Nipponbare) were grown in IRRI solution for 2 weeks and then transferred to K-sufficient (1 mm K; +K) and K-deficient (0 mm K; −K) solutions for the indicated durations. Total RNAs were isolated from roots, basal node and sheath, and leaf blades of the seedlings, and OsHAK5 transcript levels were determined by qRT-PCR. A housekeeping gene, ACTIN (OsRac1; accession no. AB047313), was used as an internal standard. Error bars indicate se of three biological replicates (n = 3). All the primers are listed in Supplemental Table S2.
Cellular and Tissue Specificity of OsHAK5 Expression in Rice
Based on the fluorescence signal of GFP fused to the C terminus of OsHAK5, we detected OsHAK5 expression in the plasma membrane in BY2 cells and in rice protoplasts (Supplemental Fig. S1), consistent with its localization in onion (Allium cepa) epithelial cells (Horie et al., 2011).
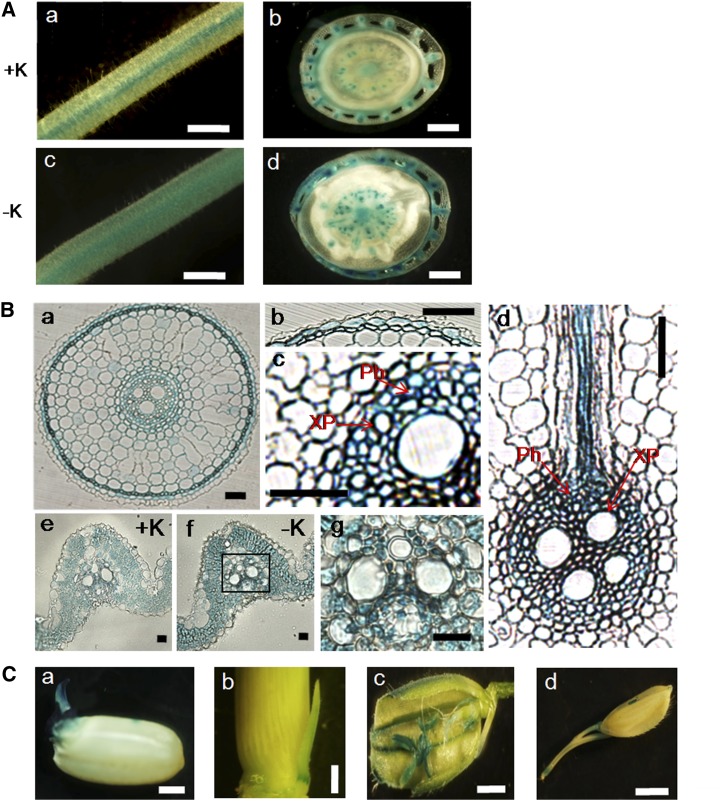
The tissue-specific expression of OsHAK5 in rice was analyzed using the GUS reporter fused to the OsHAK5 promoter. Under normal K supplementation, GUS staining was evident in root (Fig. 2Aa), root-shoot junction (Fig. 2Ab), and leaf sheath (outer layer of Fig. 2Ab). Low-K-enhanced OsHAK5 expression was examined specifically in the root and root-shoot junction (basal node plus leaf sheath; Fig. 2, compare Ac and Ad with Aa and Ab).
Figure 2.
Tissue-specific localization of OsHAK5 in the root and shoot of rice using the GUS reporter driven by the OsHAK5 promoter. Rice was transformed with OsHAK5 promoter:GUS. A, GUS activity was detected in root (a; root hair zone) and root-shoot junction (b; the outer layer is the leaf sheath) of rice grown in solution containing 1 mm K or in root (c) and root-shoot junction (d) after 6 h of K deprivation. Bars = 1 mm. B, GUS staining of the cross sections of root and leaf blade observed by dissection microscopy. a to d, Root cross sections of rice after 6 h of K starvation: a, root cross section (approximately 3.5 cm from root tip); b and c, enlarged epidermis and stele, respectively, of a; and d, cross sections of stele and cortex at the lateral root branching zone. e and f, Cross sections of leaf blades of rice grown in 1 mm K (+K) and in K-depleted solution (−K) for 10 d. g shows a leaf vascular tissue (enlarged part of f). Ph, Phloem; XP, xylem parenchyma. Bars = 100 μm. C, GUS activity was detected in embryonic tissue (a), tiller bud (b), flower (c), and pedicel (d). Bars = 1 mm.
We also examined the cell-specific expression of OsHAK5 in root tissue (Fig. 2Ba). GUS staining was prominent in root epidermis (Fig. 2Bb), parenchyma of stele tissue (Fig. 2Bc), and primordial of the lateral root (Fig. 2Bd). Since epidermal silica bodies interfere with the even staining of GUS in leaf blades, the GUS staining intensity was compared on the cross section. It is clear that OsHAK5 expression was enhanced by K starvation for 10 d (Fig. 2, compare Be with Bf) and predominantly at mesophyll and parenchyma cells of the vascular bundle (Fig. 2, Bf and Bg). In addition, the GUS staining indicated that OsHAK5 was also expressed in geminated embryonic tissue (Fig. 2Ca), young tiller (Fig. 2Cb), flower organs (Fig. 2Cc), and pedicel (Fig. 2Cd).
The Effect of OsHAK5 Expression Level on Direct K Acquisition by Rice Roots
OsHAK5 was reported to complement the growth defects of both the K transport E. coli mutant and the high-affinity K uptake-deficient yeast mutant (Horie et al., 2011). We examined the functional complementation of OsHAK5 in a K uptake-deficient yeast strain, R5421, in a series of K concentrations and found that OsHAK5 improved the yeast growth at the [K] range of 0.05 to 10 mm (Supplemental Fig. S2). To resolve whether OsHAK5 is directly required for K acquisition in rice roots exposed to low external K, we measured the net K fluxes using the high-resolution scanning ion-selective electrode system (SIET) in the primary roots of seedlings. We compared one of the OsHAK5 overexpression (OX) lines (Supplemental Fig. S3) and an OsHAK5 KO mutant line (Supplemental Fig. S4) with the fluxes in their respective wild types.
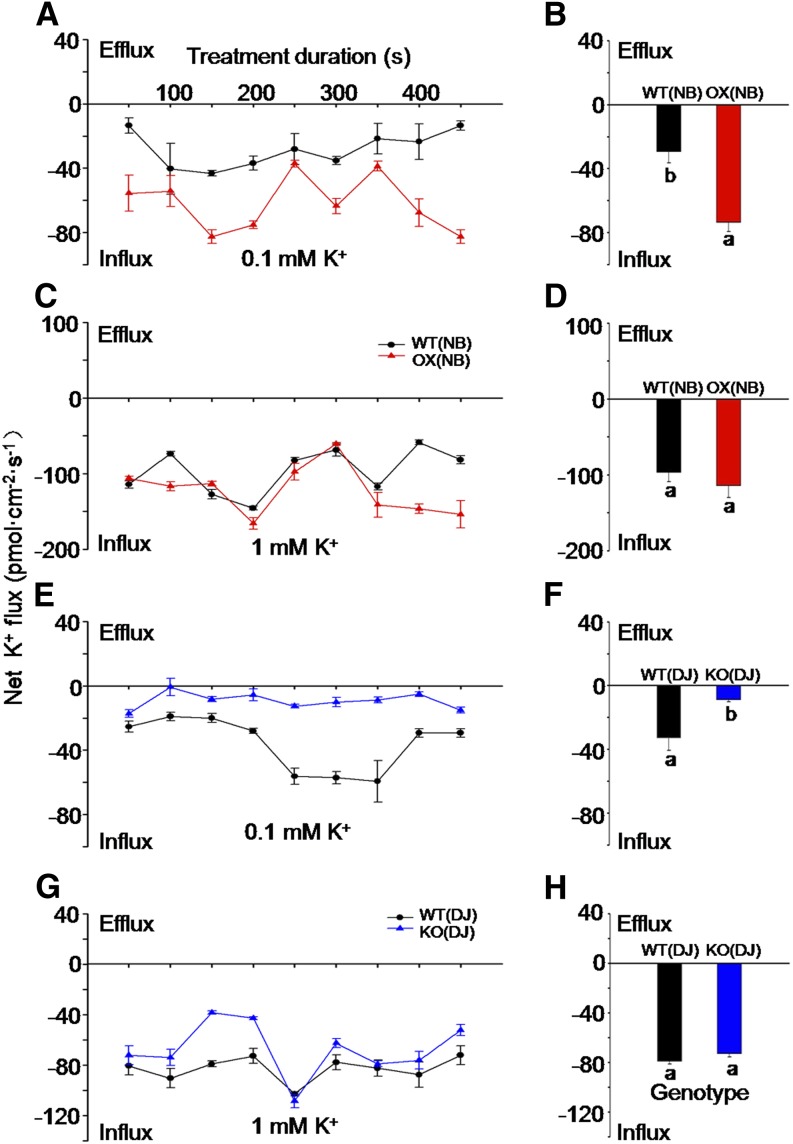
In seedlings supplied with 0.1 mm K, the net K influx in roots of OX lines was considerably larger than that in the wild type throughout the 7.5-min duration of the measurement (Fig. 3A). On average, overexpression of OsHAK5 increased the net K influx rate about 2.6-fold (Fig. 3B). However, when the K concentration was 1 mm, there was no significant difference in the net K influx between OX lines and the wild type during the entire duration of the measurement, except during the last 1.5 min of data sampling (Fig. 3, C and D). In contrast to the OX plants, the OsHAK5 KO line showed much lower net K influx throughout the sampling period (Fig. 3E) and, on average, only about 20% of the flux in the wild type (Fig. 3F). Increasing the K concentration from 0.1 to 1 mm dramatically stimulated the net K influx in both wild-type and oshak5 mutant roots, and K influx in the OsHAK5 KO line largely caught up, becoming only about 15% less than that in the wild type (Fig. 3, G and H). These data suggest that OsHAK5 is directly involved in root K acquisition particularly from low-K medium.
Figure 3.
Effect of OsHAK5 expression level on net plasma membrane K fluxes in the primary root meristem supplied with different concentrations of K. A and C, Net K fluxes (see “Materials and Methods”) in wild-type cv Nipponbare [WT(NB)] and in its OsHAK5-overexpressing lines [OX(NB)] supplied with 0.1 mm K (A) or 1 mm K (C) for 7.5 min. B and D, Mean K fluxes of A and C, respectively, averaged over the entire 7.5 min. E and G, Net K fluxes in wild-type DJ [WT(DJ)] and in its OsHAK5 KO line [KO(DJ)] supplied with 0.1 mm K (E) or 1 mm K (G) for 7.5 min. F and H, Mean K fluxes of E and G, respectively, averaged over the entire 7.5 min. Each symbol and error bar represents, respectively, the mean and se for six to eight individual plants. Significant differences from the wild type in each group (NB or DJ) are indicated by different letters (P < 0.05, one-way ANOVA).
The Effect of OsHAK5 Expression Level on K Uptake and the Growth of Rice Plants
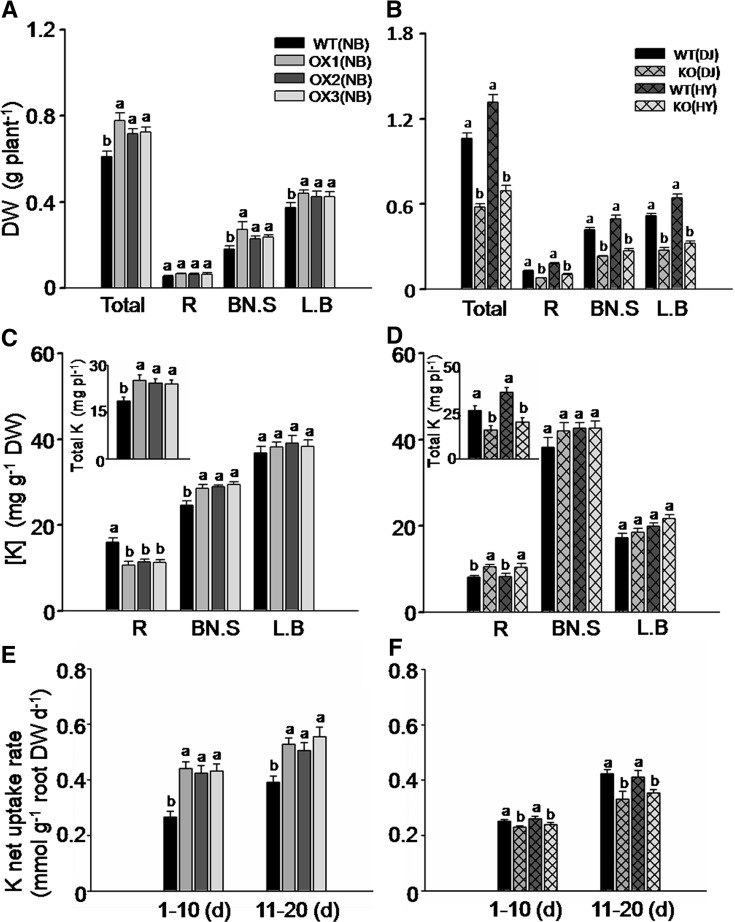
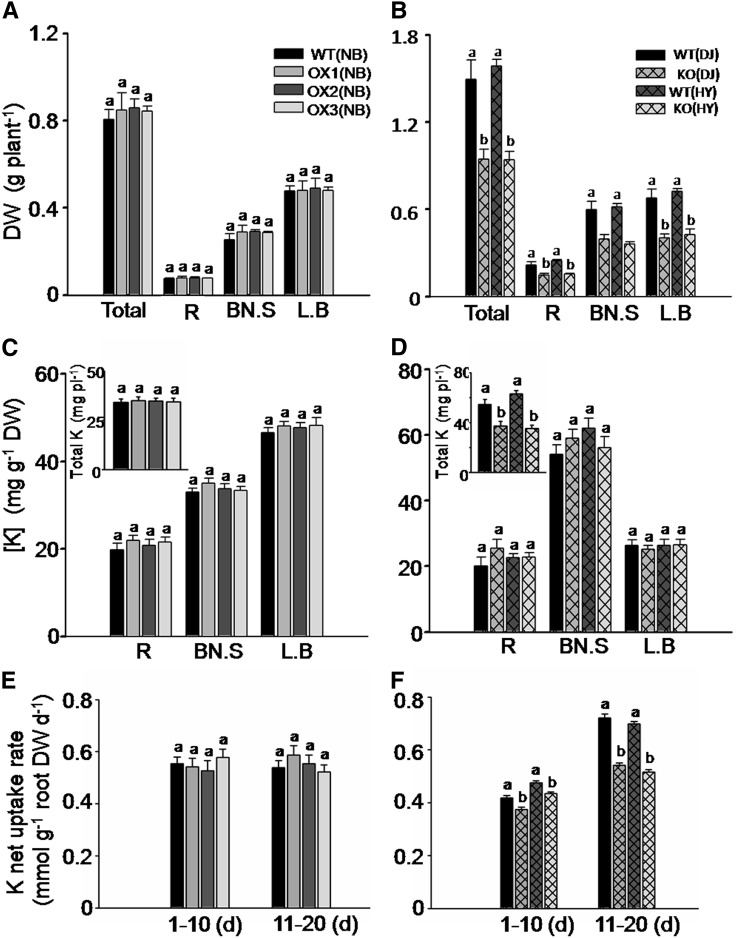
We further evaluated the role of OsHAK5 in K uptake in rice during an extended (2 weeks) growth period in a 0.3 mm K solution. Overexpression of OsHAK5 significantly increased the growth of the aerial parts (Fig. 4A). In contrast, OsHAK5 knockout impaired the growth of both KO lines relative to their respective wild-type cultivars (Fig. 4B; Supplemental Fig. S5). Overexpression of OsHAK5 decreased K concentration in the roots but increased K concentration in the basal node and sheath (Fig. 4C). The amount of the total accumulated K ([K] × biomass) in OsHAK5 OX lines increased by about 25% (Fig. 4C, inset), while in the roots it was about 17% to 21% lower than in the wild type. In contrast, in the roots of the OsHAK5 KO lines, K concentration was higher than in the wild type, but there was no difference of K concentration in the shoots (Fig. 4D). Due to the dramatically impaired biomass accumulation (Fig. 4B), the total K accumulation in both KO lines was only about 56% to 60% relative to their wild types (Fig. 4D, inset). Since these comparisons were based on joint K accumulation before and during the low-K treatment, and the different plant lines likely did not have the same K content at the start of the low-K treatment, we also compared the net K uptake rates occurring during the low-K treatment. The net K uptake rates (total K accumulation per unit of time and unit of root dry weight) were calculated based on sampling the plants three times, at 10-d intervals, starting at 0.3 mm K treatment (Nieves-Cordones et al., 2010; see “Materials and Methods”). Net K uptake rates in the OX lines in the two 10-d sampling periods were significantly higher than those in the wild type; oppositely, the rates in the KO lines were significantly reduced compared with their respective wild type (Fig. 4, E and F). Thus, OsHAK5 overexpression increased rice K acquisition and enhanced upward K transport from roots to shoots at low external K concentration (0.3 mm K), and OsHAK5 knockout impaired this absorption and transport.
Figure 4.
Effect of OsHAK5 expression level on rice growth and K accumulation under conditions of low K supply. Ten-day-old seedlings were grown in IRRI solution containing 1 mm K for 2 weeks, then transferred to a solution containing 0.3 mm K and allowed to grow for 2 weeks (A–D) or 20 d (E and F). A, C, and E, OX lines and their respective wild-type lines. B, D, and F, KO lines and their respective wild-type lines. A and B, Accumulation of dry biomass (dry weight) in root (R), basal node plus sheath (BN.S), and leaf blades (L.B). C and D, Accumulation of K (K concentration and total K content per plant [inset]). E and F, Net K uptake rates during days 1 to 10 and 11 to 20 of growth in solution containing 0.3 mm K. WT(NB), The wild type of cv Nipponbare; OX1 to OX3, three independent lines of OsHAK5-overexpressing plants of cv Nipponbare; WT(DJ), the wild type of the cv DJ; KO(DJ), OsHAK5 knockout mutant line of cv DJ; WT(HY), the wild type of cv HY; KO(HY), OsHAK5 knockout mutant line of cv HY. Error bars indicate se (n = 5 plants); pl, plant. Significant differences from the wild type in each group (NB, DJ, or HY) are indicated by different letters (P < 0.05, one-way ANOVA).
The Effect of OsHAK5 Expression Level on Net K Transport Rate from Root to Shoot
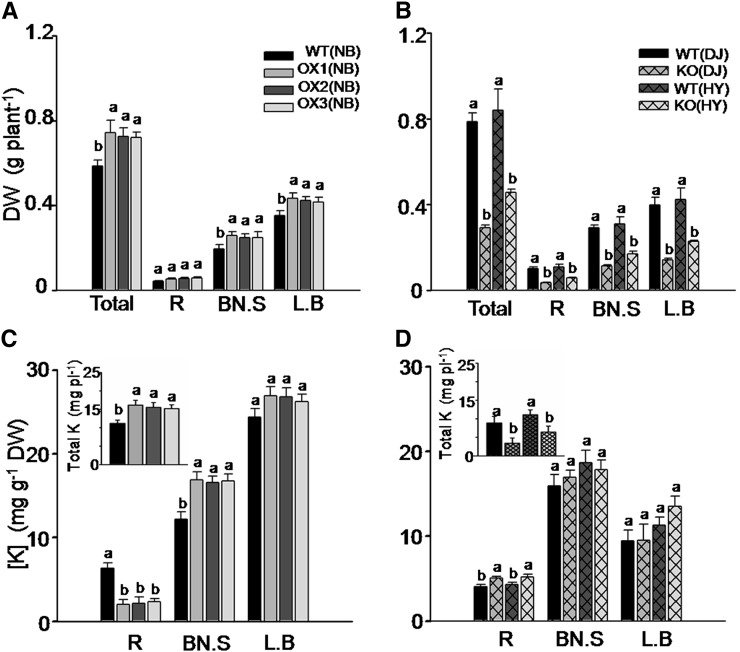
Since the rates of K transport to the shoot also depend on K uptake by the root, to reveal the role of OsHAK5 in the net K transport from root to shoot, we examined this transport in a complete absence of K in the nutrient solution (eliminating K uptake by the root). We compared the growth rate and K distribution between root and shoot by growing rice plants for 2 weeks in nutrient solution completely devoid of K. At the end of this period, the biomass in all three OsHAK5 OX lines, in particular of the basal node and sheath and leaf blade, was significantly higher than that in the wild type (Fig. 5A). In contrast, the biomass in the OsHAK5 KO lines was only about half of that in their respective wild types (Fig. 5B). Although the total K content of OX plants was higher than in the wild type (Fig. 5C, inset), K concentration in the roots of the OX plants decreased to only about 32% to 36% of the wild-type level (Fig. 5C), clearly indicating that K was transported to the shoots of the OX plants. On the other hand, due to the dramatically impaired growth of the KO plants (Fig. 5B), their total K content was much lower than that of their respective wild types, yet K concentration in the roots of KO plants was higher than in the wild types (Fig. 5D), showing a particular impairment in root-to-shoot transfer of K.
Figure 5.
Effect of OsHAK5 expression level on rice growth and K accumulated in the absence of external K in the culture solution. Ten-day-old seedlings were grown in IRRI solution containing 1 mm K for 2 weeks, then transferred to a solution without K and allowed to grow for 2 weeks. A and B, OX lines and their wild types. C and D, KO lines and their wild types. A and C, Accumulation of biomass (dry weight) per plant. B and D, Accumulation of K (K concentration and total K content per plant [insets]). Other details are as in Figure 4.
Could this difference in K transfer from root to shoot be due to the generally poorer thriving status of the KO plants, and in particular the lower biomass and K content of their roots, rather than specifically to the absence of OsHAK5? To answer this, we compared plants that achieved a similar biomass and similar total K content during the assay period: wild-type cv Hwayoung (HY) and cv Dongjin (DJ) plants grown in 1 mm K and KO plants grown in 5 mm K. To facilitate this comparison, we recalculated and replotted the total shoot dry biomass (the merged biomass of leaf sheath and blade) and its K concentration using the data from wild-type plants (cv DJ and cv HY) grown in 1 mm K (from Fig. 7, B and D) and the data from KO plants grown in 5 mm K (Supplemental Fig. S6, A and B). The root biomass values of all these plants were almost the same, and although the total biomass and total K content of the wild types were even higher than in KO lines (Supplemental Fig. S7, A and C), K concentration and K partitioning in the roots of KO lines were still significantly higher (Supplemental Fig. S7, B and D).
Figure 7.
Effect of OsHAK5 expression level on rice growth and on the concentration of accumulated K under continuous supply of normal [K] (1 mm). Ten-day-old seedlings were grown in IRRI solution containing 1 mm K continuously for 4 weeks. A, C, and E, OX lines and their wild types. B, D, and F, KO lines and their wild types. A and B, Accumulation of dry biomass (dry weight) per plant. C and D, Accumulation of K (K concentration and total K content per plant [insets]). E and F, Net K uptake rates in OX and KO lines during days 1 to 10 and 11 to 20 of growth in solution containing 1 mm K. The abbreviated names of wild types and transgenic lines are as in Figure 4. Error bars indicate se (n = 5 plants). Significant differences from the wild type in each group (NB, DJ, or HY) are indicated by different letters (P < 0.05, one-way ANOVA).
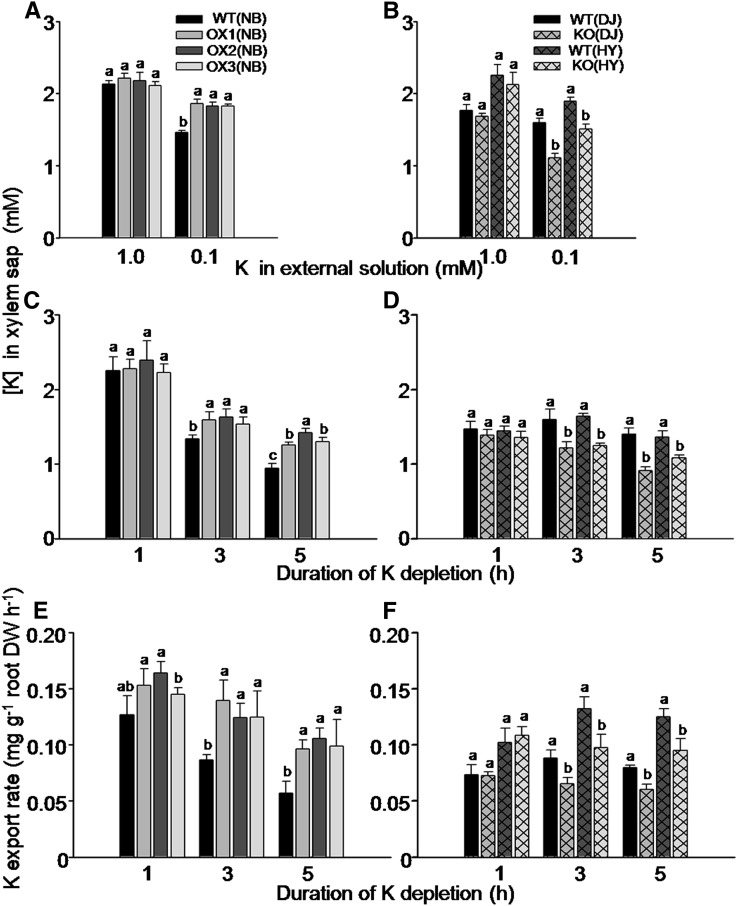
To explore more directly the involvement of OsHAK5 in K transport from the root to the shoot, we collected xylem sap of all these KO mutants, OX lines, and wild types to measure the K concentration. There was no significant difference in K concentration in the xylem sap between these KO mutants or OX lines relative to their respective wild-type lines when they were exposed to 1 mm K solution (Fig. 6, A and B). However, K concentration in the xylem sap was higher by 20% to 25% in the OX lines (Fig. 6A) and was lower by 19% to 24% in the KO mutant lines (Fig. 6B) compared with their respective wild types when they were similarly exposed to 0.1 mm K solution.
Figure 6.
Effect of OsHAK5 expression level on K concentration and transport rate in the xylem sap of rice supplied with different concentrations of K in the culture solution. A, C, and E, OX lines and their wild types. B, D, and F, KO lines and their wild types. A to D, K concentration in the xylem sap. A and B, The seedlings were grown in IRRI solution containing 1 mm K for 8 weeks, then transferred to medium without K for 3 d and further transferred to either 1 mm K or 0.1 mm K solution for collecting xylem sap and K analysis after 1 h of partial shoot excision. C and D, The seedlings were grown in IRRI solution containing 1 mm K for 8 weeks, and then the excision procedure was repeated, but without prestarvation for K. The root with the stump was transferred to 0 mm K solution for up to 5 h. The xylem sap was collected for 1 h in two hourly intervals (i.e. during the first hour, the second to third hour, and the fourth to fifth hour, counting from removal of the K supply). E and F, K export rate in the xylem sap. After the end of xylem sap collection, the entire root system of each plant was excised and its dry weight was determined. The shoot excision, xylem collection, and calculation of K export rate are described in “Materials and Methods.” The abbreviated names of wild types and transgenic lines are as in Figure 4. Error bars indicate se (n = 5 plants). Significant differences from the wild type in each group (NB, DJ, or HY) are indicated by different letters (P < 0.05, one-way ANOVA).
In order to exclude the effect of K acquisition by roots in K export from root to shoot, we collected the xylem sap while the roots bathed in a culture solution without K for 1 h and in two 1-h intervals (i.e. during the first hour, second to third hour, and fourth to fifth hour, counting from removal of the K supply). K concentration in the collected sap samples was determined (Fig. 6, C and D) as well as the sample volumes and the root dry weight. These served to calculate the K export rate from the root (Fig. 6, E and F; see “Materials and Methods”). K concentration in the xylem sap collected during the first hour from different plants did not differ between the transgenic lines and their respective wild-type lines (Fig. 6, C and D), and neither did K export rate (Fig. 6, E and F), but at 3 and 5 h after halting the supply of K to the root, both K concentration and K export rate in the xylem sap were markedly higher in the OX lines (Fig. 6, C and E) and lower in the KO mutant lines (Fig. 6, D and F) compared with their respective wild types.
High-K Supplementation Was Not Fully Able to Rescue the Growth of oshak5 Mutants
In order to test whether OsHAK5 functions in K acquisition and distribution only under conditions of low K supply, we compared K concentration and biomass accumulation in the OX lines and KO mutants with their respective wild types grown in a nutrient solution containing 1 mm K. The total biomass of the OX plants, or the separate biomass values detailed for the root, basal node and sheath, and leaf blade, were no different than in the wild type (Fig. 7A); however, the growth of the OsHAK5 knockout plants was markedly impeded relative to their respective wild types (Fig. 7B). However, when grown in this K-abundant solution, neither the OX lines nor the KO lines showed any differences in K concentration in the various plant organs between the transgenic lines and their respective wild types (Fig. 7, C and D). We then compared the net K uptake rates into these plants in the normal K (1 mm) supply solution by quantifying the difference of total K accumulation at three 10-d-apart time points (see “Materials and Methods”). OsHAK5 overexpression did not improve the rates of net K uptake compared with the wild type (Fig. 7E); however, the KO plants took up K at a considerably lower rate relative to their wild-type counterparts (Fig. 7F). To exclude the possibility that the impairment in biomass accumulation of the KO mutants was due to the deficiency of K in the culture medium, we also compared the mutants and the wild type grown in a solution containing 5 mm K. The supply of 5 mm K appears to have ameliorated considerably the growth of both oshak5 mutants (Fig. 7B; Supplemental Figs. S6A and S7A). Encouraged by this, K concentration in the solution was further increased to 10 or 20 mm in an independent experiment; however, the average total biomass of the mutants was still significantly lower than that of their respective wild types (Supplemental Fig. S6, C and D), indicating that OsHAK5 is also important for the growth of rice under conditions of sufficient K supply.
OsHAK5 Improved the Salt Tolerance of Rice by Enhancing K Uptake and Translocation to the Shoot
OsHAK5 has been demonstrated to play a role in enhancing the salt tolerance of both yeast and BY2 cells in suspension cultures (Horie et al., 2011). To test whether OsHAK5 also plays the same role in enhancing salt tolerance in rice, we first examined the OsHAK5 response to elevated external NaCl at the transcript level (using qRT-PCR). A 100 mm NaCl treatment in the presence of 1 mm K increased the abundance of OsHAK5 transcripts in roots, basal node and sheath, and leaf blade (Fig. 8). There was a prominent transcript peak in all these tissues at 72 h of exposure to NaCl (Fig. 8), but interestingly, already after 1 h of NaCl treatment the transcript level increased close to 2-fold in the roots and more than 3-fold in the basal node and sheath (Fig. 8, A and B).
Figure 8.
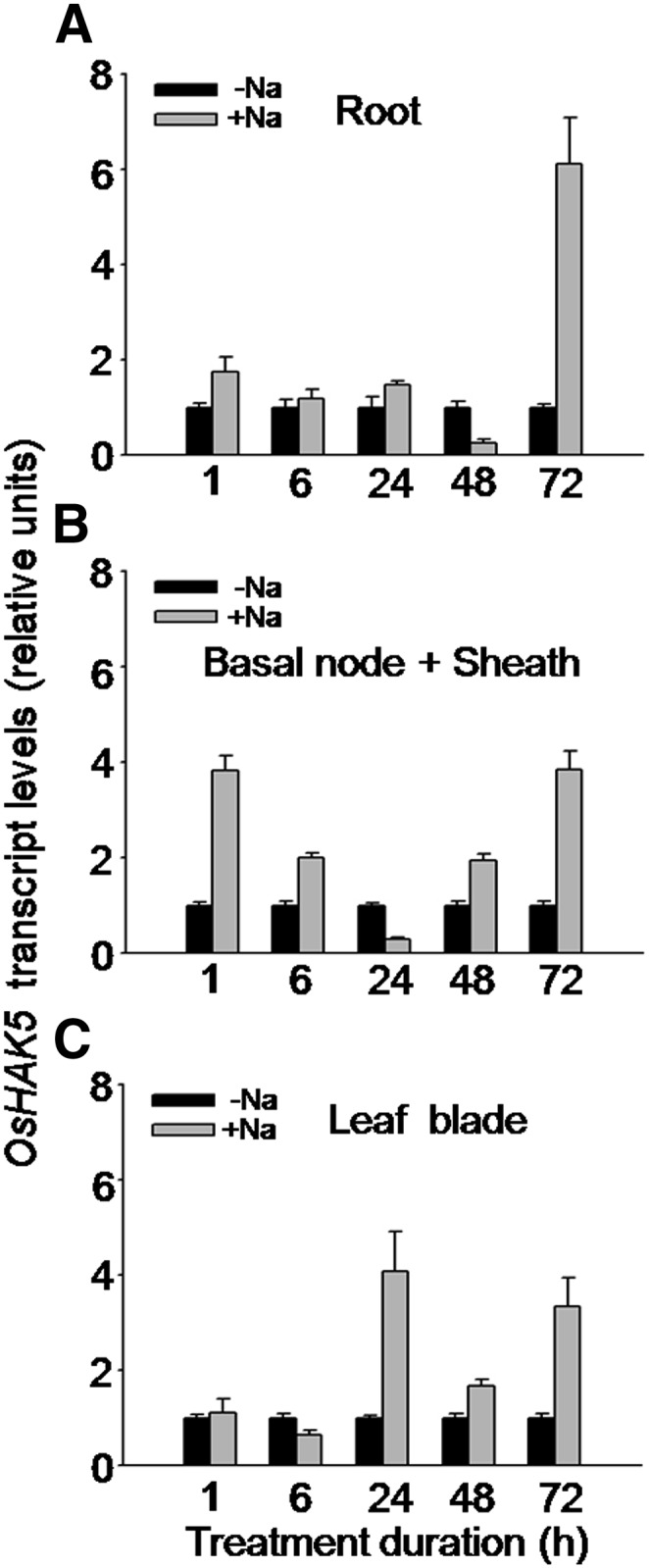
Effect of salt stress on the transcript level of OsHAK5 in rice. Ten-day-old rice seedlings (cv Nipponbare) were grown in IRRI solution for 2 weeks, then transferred to a control solution without salt (0 mm NaCl; −Na) or to a salt-enriched solution (100 mm NaCl; +Na) for the indicated durations. Total RNA was isolated from roots, basal node and sheath, and leaf blades of the seedlings, and OsHAK5 transcript levels were determined by qRT-PCR. A housekeeping gene, ACTIN (OsRac1; accession no. AB047313), was used as an internal standard. Error bars indicate se (n = 3 biological replicates). All primers are listed in Supplemental Table S2.
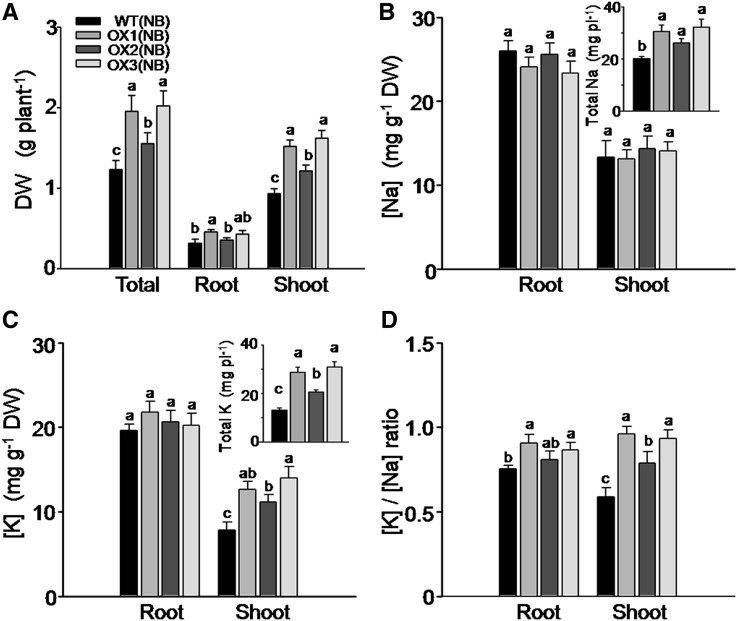
We compared the differences, after 8 d of 100 mm NaCl treatment (in the presence of 1 mm K), in biomass accumulation and in K and Na acquisition in the OX lines versus their respective wild-type lines. While the root biomass accumulation (dry weight) in the OsHAK5 OX lines did not change remarkably, the shoot growth of these plants was improved (Fig. 9A). Na concentration in both roots and shoots remained unaffected, and the total Na accumulated in the OsHAK5 OX plants was increased significantly due to their larger biomass (Fig. 9B). K concentration in the roots of OX plants remained the same as in the wild types, but in the shoots it increased by about 43% to 80% and the total K content of wild-type plants increased by 47% to 115% (Fig. 9C and inset). Consequently, in the OsHAK5 OX lines, the ratio of [K] to [Na] was about 45% higher in the shoot as compared with the wild-type line, while the [K]-to-[Na] ratio in the roots of the OX lines was not consistently higher than in the roots of the wild-type plants (Fig. 9D).
Figure 9.
Effect of OsHAK5 overexpression on rice salt tolerance as indicated by biomass accumulation, K and Na accumulation, and the ratios of their concentrations. Ten-day-old seedlings were grown in IRRI solution containing 1 mm K for 3 weeks, then transferred to a solution containing additionally 100 mm NaCl and allowed to grow for 8 d. WT(NB), The wild type of cv Nipponbare; OX1 to OX3, three independent lines of OsHAK5-overexpressing plants of cv Nipponbare. Error bars indicate se (n = 5 plants). Significant differences from the wild type in each group are indicated with different letters (P < 0.05, one-way ANOVA).
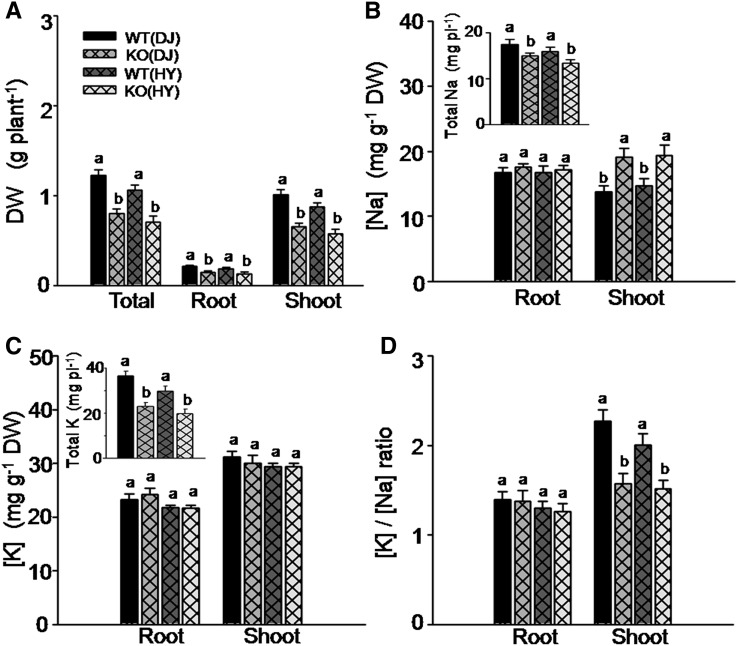
The OsHAK5 knockout mutants were more sensitive to salt stress than their wild-type counterparts, judging by their considerably lower biomass accumulation in the shoot and in the root as compared with their respective wild-type lines (Fig. 10A, Supplemental Fig. S8). Na concentration in the shoot of KO plants was much higher even though total Na accumulation in the KO lines was lower than in the wild types (Fig. 10B). K concentration did not differ from the wild type, either in the shoot or in the root (Fig. 10C); therefore, the [K]-to-[Na] ratio was lower in the shoots of the mutant plants relative to their wild-type counterparts (Fig. 10D). The lower [K]-to-[Na] ratio, concomitant with lower salt tolerance in the OsHAK5 KO lines, suggests that OsHAK5 is required for salt tolerance, probably through enhancing K and/or impeding Na translocation to the shoot.
Figure 10.
Effect of OsHAK5 knockout on salt sensitivity of rice, as indicated by biomass accumulation, K and Na accumulation, and [K]-to-[Na] ratios. Ten-day-old seedlings were grown in IRRI solution containing 1 mm K for 3 weeks, then transferred to a solution containing additionally 100 mm NaCl and allowed to grow for 8 d. WT(DJ), The wild type of cv DJ; KO(DJ), OsHAK5 knockout mutant line of cv DJ; WT(HY), the wild type of cv HY; KO(HY), OsHAK5 knockout mutant line of cv HY. Error bars indicate se (n = 5 plants). Significant differences from the wild type in each group (DJ or HY) are indicated by different letters (P < 0.05, one-way ANOVA).
DISCUSSION
OsHAK5 Contributes to K Acquisition in Roots Supplied with Low K
K concentration in the soil typically ranges between 0.1 and 1 mm (Maathuis, 2009), and the actual concentrations of K at the root surface can be even lower than in bulk soil solution due to the fast K acquisition by the roots (Ashley et al., 2006). K deficiency up-regulated the expression of high-affinity K transporter genes, such as AtHAK5 and OsHAK1 (Gierth et al., 2005; Rubio et al., 2010). In this study, we found that the expression of OsHAK5, another HAK transporter gene in cluster I, in the rice roots was quickly up-regulated by external K depletion (Figs. 1 and 2, Aa, Ab, and Be versus Ac, Ad, and Bf). The GFP reporter and GUS staining showed that OsHAK5 is localized to the plasma membrane (Supplemental Fig. S1) and epidermal cells of root (Fig. 2, Ba and Bb). By comparing the net K influx rate in roots (Fig. 3) and total K accumulation (insets in Figs. 4, 5, and 7) of OsHAK5 OX lines and KO mutants with their respective wild types at different external K concentrations, we provide direct evidence that lowering K in the culture medium (between 0 and 0.3 mm K in this study) enhanced OsHAK5 expression (Figs. 1 and 2), which, in turn, increased K acquisition by the rice plants. The OsHAK5 KO rice lines (in both cv HY and cv DJ genetic backgrounds) could not cope with the lower external K supply; therefore, in the presence of 0.3 mm K, the total K uptake of the mutant lines was only about 60% of their respective wild-type lines (insets in Figs. 4D and 5D). The enhancement in the OX plants did not occur when they were grown in solutions with a normal (1 mm) K concentration (Fig. 7, A, C, and E). Moreover, OsHAK5 enabled K accumulation by a yeast mutant defective in K transport from solutions with a range of K concentrations of 0.05 to 1 mm (Supplemental Fig. S2). All these data strongly support the notion that OsHAK5 contributes to K acquisition by rice. Interestingly, there was a measurable K influx current (Fig. 3, E and F) and a substantial net K uptake rate at external low K concentration not only via roots of the OX plants (Fig. 4E) but also from roots of the KO plants (Fig. 4F). This implies that other K transporter(s) contributed to K uptake in the KO lines. However, the other high-affinity transport(s) could not compensate fully for the loss of function of OsHAK5.
In Arabidopsis, AtHAK5 is the major system for root K uptake at low external K supply (Rubio et al., 2010). Among all 27 HAK members in rice, OsHAK1 and OsHAK5 have the highest identity with AtHAK5 (Gupta et al., 2008). Bañuelos et al. (2002) have shown that OsHAK1 was abundantly expressed in rice roots. They found that the trk1 trk2 yeast mutant expressing a chimeric rice OsHAK1 (the first 48 bp of its open reading frame came from the barley HvHAK1 transporter) showed comparably high affinity values (Km) with rice for K uptake. Therefore, it could be predicted that there are at least two high-affinity K transporters, OsHAK1 and OsHAK5, with distinct functions in rice, for mediating K uptake from low-K solution.
OsHAK5 Functions in Upward Transport of K from Roots to Aerial Parts at Low K Supply Levels
K absorbed at the root surface is transported to the parenchyma cells for xylem loading. Xylem loading has been identified as the main site for the control of K supply to the shoot (Marschner et al., 1996). K is also circulated back to the root via phloem, and a large fraction of it returns to the xylem. We identified that OsHAK5 was strongly expressed at the xylem parenchyma and phloem of root vascular tissues (Fig. 2, Bc and Bd), in particular under K deficiency conditions (Fig. 2, Ba, Bc, and Bd), suggesting that OsHAK5 may be involved in K distribution between roots and shoots.
K transport into the xylem is regulated by the K status of the plant (Véry and Sentenac, 2003). In a K-starved plant, plant growth is aided by the transport to the shoot of the root-stored K (Cai et al., 2012). In comparison with the wild type grown at 0.3 mm K solution, K concentration in the roots of OsHAK5 OX plants was lower by about 30% (Fig. 4C), and it was decreased further to only 30% of the wild-type level in the absence of K (Fig. 5C). In these plants, K was transported upward to the shoots more than in the wild type (Figs. 4C and 5B). Oppositely, when external K was reduced or eliminated, K concentration in the roots of KO plants was higher than in the roots of their respective wild types, but K concentration in their shoot was similar to that in the wild type (Figs. 4D and 5D). Therefore, the K concentration gradient in OsHAK5 KO plants should be more favorable for K transport from root to shoot than that in the wild type. The rate of K transport to the shoot is expected to depend mainly on the activity of K transporters and on the K concentration gradient between the roots and shoot. However, a smaller amount of K was transported to the shoot in KO plants in comparison with that in the wild type, clearly due to the absence of OsHAK5.
Under conditions of normal K supply, K concentrations in roots and shoots of OX plants were similar to those in the wild type (Fig. 7C). Only when external K was low did overexpression of OsHAK5 enhance the rate of net K uptake (compare Figs. 4E and 7E). Thus, in medium supplying normal (1 mm or greater) K, OsHAK5 overexpression did not contribute to K transport from roots to the shoots (Fig. 7C). Interestingly, normal or even high concentrations of K could not compensate for the lack of OsHAK5 and amend the growth (Fig. 7B; Supplemental Fig. S6, A, C, and D) and the K net uptake rate in the KO plants (Fig. 7F). This phenomenon will be discussed later.
We also provided evidence that OsHAK5 contributed to K export in the xylem sap when external K supply was lowered (0.1 mm K) or removed (0 mm K) from the culture medium. In the first 1 h of sap collection after transfer to 0 mm, K concentration in the xylem sap was almost the same in all the mutant plants and their respective wild types (Fig. 6, C and D). In contrast, after 3 to 5 h of exposure to 0 mm, it was 20% to 25% higher in OsHAK5 OX plants than in the wild type (Fig. 6C) and 19% to 23% lower in the oshak5 mutants than in the wild type (Fig. 6D).
Since OsHAK5 functioned in K acquisition in both rice roots and yeast cells (Fig. 3; Supplemental Fig. S2; Horie et al., 2011) and was expressed abundantly in the xylem parenchyma throughout the primary and lateral roots (Fig. 2, Ba, Bc, and Bd), it is likely that OsHAK5 functions in acquiring K into the xylem parenchyma cells. The enriched K in the xylem-adjacent cells may enable K channels to release K efficiently into the xylem sap driven by the K electrochemical gradient (Gaymard et al., 1998). This is particularly important with low intercellular K concentration. When K supply is normal, K diffusion from the intercellular space to the xylem through other transporters or channels, or via the apoplast, may contribute sufficiently to K transport from roots to shoots. Thus, while OsHAK5 mediates root-to-shoot transfer of K at low external K concentrations, like that reported for nitrate distribution in the rice plant by the high-affinity nitrate transporter OsNRT2.3a (Tang et al., 2012), at high external K concentrations a bypass flow-type mechanism may predominate, like that reported for rice during salt stress (Faiyue et al., 2012).
OsHAK5 Enhances Salt Tolerance in Rice by Elevating the K-Na Ratio
Plant roots do not have a specific transporter for Na uptake. The entry of Na into root cells is mediated by nonselective cation facilitators (Malagoli et al., 2008; Munns and Tester, 2008). Eliminating Na uptake and increasing K accumulation in the shoot is an important strategy in plants for salt stress tolerance (Munns and Tester, 2008). Therefore, enhancing the expression of high-affinity HAK transporters is an attractive target for the improvement of K acquisition and plant growth under saline conditions (Nieves-Cordones et al., 2010). In Arabidopsis, the expression of AtHAK5 is greatly up-regulated by K starvation, although this up-regulation was largely counteracted by salt stress; biomass accumulation of the wild type was still significantly higher than in athak5 mutant plants, indicating the role of AtHAK5 in plant salt resistance in low-K conditions (Nieves-Cordones et al., 2010). We report here that the expression of OsHAK5 in roots, basal node, and sheath was significantly up-regulated by high external NaCl even in normal K concentrations (Fig. 8). The up-regulation of OsHAK5 expression was consistent with data showing that OsHAK5 overexpression enhances rice salt tolerance (growth; Fig. 9A). In E. coli and BY2 cells, OsHAK5 has been shown to accumulate K preferably over Na during salt treatment (Horie et al., 2011). Our results from the transgenic rice plants support these findings: in the presence of 100 mm Na, the shoot of all three OsHAK5 OX plants grew much more than that of the wild-type plants (Fig. 9A). Although total Na content was elevated (Fig. 9B, inset), Na concentration in these OX lines was the same as that in the wild type (Fig. 9B). In contrast, not only the total K content but also K concentration increased in the shoots of OX lines, which resulted in a significant increase in the K-Na ratio (Fig. 9, C and D). KO plants were much more sensitive to the salt stress than wild-type plants (Fig. 10A), even though this impairment may not be due necessarily to more severe toxicity of NaCl in the mutants. Na concentration in the shoot of KO plants was significantly higher than in the shoot of wild-type plants (Fig. 10B), with a concomitant significant decrease of the [K]-to-[Na] ratio in the shoot of KO plants (Fig. 10D). Thus, OsHAK5 enhanced rice salt tolerance by elevating the [K]-to-[Na] ratio in the rice shoot.
Although OsHAK5 is a K uptake transporter (Fig. 3; Supplemental Fig. S2; Horie et al., 2011) and is expressed abundantly in mesophyll cells of the shoot (Fig. 2, Be and Bf), currently, its function in these cells is not clear. Activation of OsHAK5 may depolarize the membrane potential of the mesophyll cells, while loss of its function may hyperpolarize the membrane potential and favor Na influx into the cells. This may explain the elevation of Na concentration in the shoot of oshak5 KO plants (Fig. 10B) and the higher sensitivity to high-salt stress compared with the wild type (Fig. 10A). Interestingly, the up-regulation of OsHAK5 expression in the shoot (particularly in the basal node and sheath) occurred much faster than in the root (Fig. 8). It was suggested that the concentration of K in the phloem fluid can act as a signal conveying the shoot demand for K and regulating K uptake from the external medium and K xylem loading (Engels and Marschner, 1992). Taking into consideration also that OsHAK5 was expressed strongly in the phloem tissue (companion cells and sieve elements) and in mesophyll cells (Fig. 2, Be–Bg), it is indeed likely that OsHAK5 is involved in signaling, transducing salt stress onto regulatory pathways in rice.
The OsHAK5 Affect on Rice Growth May Be Other Than Maintaining K Homeostasis
The functional identification of a number of K transporters and channels in different plants has led to some blurring of the distinction between the classical concepts of HATS and LATS for K acquisition and distribution in plants (Alemán et al., 2011; Coskun et al., 2013). Although HAK/KUP/KT family members were considered as members of HATS with regard to K uptake at low K supply levels, some of the group members also played a role in plants under conditions of high external K supply (Fu and Luan, 1998; Kim et al., 1998). In addition, the functional comparison of high-affinity K transporters and K channels from barley and Arabidopsis led to the conclusion that the model of root K acquisition based on the different K transporters from Arabidopsis is not universally applicable (Coskun et al., 2013).
Our data did not fully support the notion that OsHAK5 functions only as a high-affinity K transporter in rice. Unlike the expression pattern of AtHAK5, mainly in the epidermis and vasculature of K-deprived roots (Gierth et al., 2005), OsHAK5 was expressed abundantly in both roots and shoots as well as in reproductive organs of rice (Figs. 1 and 2). High-affinity K transporters mediate K uptake at external K concentrations commonly below 0.5 mm (Epstein et al., 1963; Marschner, 1996), while K concentration in the intercellular or intracellular space is usually in the millimolar range (Coskun et al., 2013). OsHAK5 functioned in K uptake complementation in yeast in K concentrations ranging between 0.05 and 10 mm (Supplemental Fig. S2). The nitrate transporter Chlorate1 (AtNRT1.1) reportedly uses dual-affinity binding and a phosphorylation switch enabling it to sense a wide range of nitrate concentrations in the soil, thereby functioning as an ion sensor in higher plants (Ho et al., 2009). In Arabidopsis, at higher K concentrations, unknown systems can supply sufficient K to promote plant growth even in the absence of AtHAK5 and AtAKT1 (Rubio et al., 2010). If, at low external K, OsHAK5 in rice affected only K uptake, normal or high K supplementation should be able to rescue the K uptake and growth of OsHAK5 KO mutants. However, this rescue was only partially complete when the mutants were grown in normal K supply (1 mm; Fig. 7C), even in 5 to 20 mm K (Supplemental Fig. S6, A, C, and D). The two independent KO mutants from different genetic backgrounds showed almost the same responses to the supply of high K (Fig. 7C; Supplemental Fig. S6). These similarly conflicting results may be reconciled if we assume that OsHAK5 exerts its effect on rice growth under conditions of high K supply by a mechanism other than maintaining K uptake and homeostasis.
There is a link between K transport and auxin transport and distribution in plants. Mutation of TRH1/AtKUP4 in Arabidopsis resulted in tiny root hairs; however, the phenotype was not restored when the mutants were grown at high external K concentrations (Rigas et al., 2001; Desbrosses et al., 2003). Since AtKUP4 affects auxin transport in Arabidopsis roots (Vicente-Agullo et al., 2004; Rigas et al., 2013), the K transport specifically mediated by AtKUP4 might be essential for root hair elongation irrespective of K supply levels. In addition, a point mutation on AtKT2/AtKUP2 of Arabidopsis resulted in a short hypocotyl (short hypocotyl3) when grown in darkness, while this point mutation had no effect on the K transport function of the gene (Elumalai et al., 2002). Rigas et al. (2013) showed that TRH1/AtKUP4 activity is required for the polar localization of PIN1 (an auxin transporter) protein in roots. The activity of PINs is considered rate limiting for auxin polar transport and distribution, which, in turn, patterns the plant architecture (Rigas et al., 2013) and, therefore, growth. A speculation about a seemingly K-unrelated effect of OsHAK5 as a K+-H+ cotransporter on growth could involve a possible effect of its activity on auxin transport and distribution in rice. This speculation should be characterized in the future.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Three cultivars of rice (Oryza sativa subsp. japonica) were used in this work: Nipponbare, DJ, and HY. The plants were grown hydroponically. Ten-day-old rice seedlings of each wild-type and transgenic line, uniform in size, were selected and transferred to International Rice Research Institute (IRRI) nutrient solution (Yoshida et al., 1976) containing the following elements: 1.25 mm NH4NO3, 0.3 mm KH2PO4, 0.35 mm K2SO4, 1 mm CaCl2, 1 mm MgSO4 0.5 mm Na2SiO3, 20 μm NaFeEDTA, 20 μm H3BO3, 9 μm MnCl2, 0.32 μm CuSO4, 0.77 μm ZnSO4, and 0.38 μm Na2MoO4, pH 5.5. The solution was replaced every 2 d. All the plants were grown in a greenhouse with a 16-h-light (30°C)/8-h-dark (22°C) photoperiod, and the relative humidity was controlled at 60% to 70%.
Rice leaf protoplasts were prepared from 2-week-old seedlings grown hydroponically under dark conditions according to Bart et al. (2006) and Miao and Jiang (2007).
For K starvation or depletion experiments, rice seedlings were grown in the IRRI solution containing 1 mm K for 2 weeks and then transferred into a solution in which K2SO4 and/or KH2PO4 was replaced by Na2SO4 and/or NaH2PO4, respectively. At each harvest, the roots of all rice plants grown in the nutrient solution were washed in 0.1 mm CaSO4 for 5 min, separated into roots, basal nodes and sheaths, and leaf blades, before their biomass was recorded and K and Na concentrations were measured as described previously (Ding et al., 2006).
Measurement of Net K Flux Rate in Rice Plants with the SIET
Rice seedlings were grown in the IRRI nutrient solution for 3 weeks and then deprived of K for 7 d. Net K+ flux in the root meristem zone was measured using the noninvasive SIET technique as described previously (Tang et al., 2012). The roots of seedlings were equilibrated in the measuring solution (0.1 mm CaCl2 and 0.3 mm MES, pH 6) for 30 min. The equilibrated seedlings were then transferred to the measuring chamber filled with the solution containing either 0.1 mm K+ or 1 mm K+. Net K+ fluxes were measured under the experimental conditions for 7.5 min to decrease variability due to fluctuations. Prior to the flux measurements, the ion-selective electrodes were calibrated using K+ concentration of 0.05, 0.1, and 0.5 mm. Roots from at least six individual plants were measured in an independent experiment. Each plant was measured once. The measurements were carried out using the SIET system BIO-003A (Younger USA Science and Technology) at Xu-Yue Science and Technology (http://www.xuyue.net).
K Net Uptake Rates
Ten-day-old rice seedlings were grown in IRRI solution for 2 weeks and then transferred to 0.3 or 1 mm K solution for 20 d. Plants were harvested at three time points, on day 0 (just before the treatment) and 10 and 20 d after treatment, for the determination of their dry weight and K concentrations. The K net uptake rates into the plants were calculated according to the following equation (Nieves-Cordones et al., 2010):
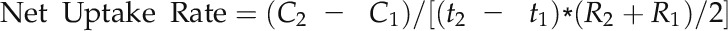 |
where C is the total K content, R is the root dry weight, and t is the time at each of the three harvests; the subscript numbers 1 and 2 indicate the start and end of the period for which the uptake rate is calculated: t2 − t1 = 10 d, and (R2 + R1)/2 = the mean root dry weight.
Detecting K Concentration and Calculation of the Rate of K Export in the Xylem Sap
Shoots were excised from plants grown for 8 weeks in 1 mm K solution or removal of K for 3 d before the treatment at about 3 to 4 cm above the root; then, a preweighed absorbing cotton ball was attached to the cut surface of the stump and covered with plastic film. Immediately afterward, the root with the stump was transferred to 1, 0.1, or 0 mm K solution. The sap was collected for 1 h, during the first hour, and, on the same plants, also during the second to third hour and the fourth to fifth hour, exchanging cotton balls on the same stumps. The cotton ball was weighed to calculate the volume of the exudates from the increase in its weight (assuming a density of 1 g cm−3). The collected xylem sap was squeezed from the cotton ball with a syringe for the determination of K concentration by Inductive Coupled Plasma Emission Spectrometer. After the end of the xylem sap collection, the entire root system of each plant was excised and its dry weight was determined. The collected volume multiplied by K concentration represents the amount of total K (mg) transported in the xylem sap per plant. The total K divided by total root dry weight (g) of each individual plant and the time (h) for the correction represents the rate of K export in the xylem of each plant.
Semiquantitative Reverse Transcription-PCR and Quantitative Real-Time PCR
Total RNA was extracted from different tissues of rice using TRIzol reagent following the manufacturer’s protocol. Reverse transcription (RT)-PCR and qRT-PCR for targeted genes, OsHAK5 (accession no. AK241580) and OsACTIN (accession no. AB047313), were performed according to the protocol described previously (Ai et al., 2009; Tang et al., 2012). All primers for the semiquantitative RT-PCR and quantitative real-time PCR are listed in Supplemental Table S1. All the PCR products were confirmed by sequencing.
Cloning of the OsHAK5 Promoter and Construction of OsHAK5 OX Vectors
A 1,776-bp fragment of the upstream OsHAK5 coding region was amplified from rice genomic DNA (cv Nipponbare) with the specific primers listed in Supplemental Table S2. The fragment was first cloned into the pEasy-Blunt vector and then transferred into the AscI/PacI sites of the binary vector pS1aG3 by replacing the cauliflower mosaic virus 35S promoter (Tang et al., 2012).
The OsHAK5 OX vector was constructed by amplifying the full length (open reading frame of 2,346 bp) of OsHAK5 from complementary DNA of rice shoot with KOD-plus DNA polymerase (Takara Biotechnology). The primers for full-length OsHAK5 are listed in Supplemental Table S3. The product was cloned into pTK303 at KpnI/NheI sites.
To construct enhanced GFP fused with the C terminus of OsHAK5, its PCR-amplified product was under the control of 35S at the HindIII site and in frame with enhanced GFP at the PstI site of pCAMBIA(gfp)1302 vector. The integrity of all these sites in the genes and constructs was confirmed by sequencing the cloned products.
Transient Expression of OsHAK5:GFP and Fluorescence Microscopy Imaging
Transient expression of OsHAK5:GFP in tobacco (Nicotiana tabacum) BY2 cells was conducted by incubation of Agrobacterium tumefaciens harboring the OsHAK5 gene fused with GFP in pCAMBIA(gfp)1302 vector with exponentially growing BY2 cells for 2 to 3 d. Transient expression in rice of OsHAK5:GFP in the pCAMBIA(gfp)1302 vector was conducted using the polyethylene glycol method (Chen et al., 2006). For the plasma membrane localization of OsHAK5, we used a confocal laser scanning microscope (LSM410; Carl Zeiss) as described by Tang et al. (2012).
Generation of OsHAK5 Overexpression Lines, and Detection of Insertional Copy Numbers and GUS Staining
Both vectors harboring OsHAK5 promoter:GUS and 35S:OsHAK5 were transferred into A. tumefaciens strain EHA105 by electroporation, and the agrobacteria were used to infect callus induced from wild-type rice (cv Nipponbare) as described previously (Tang et al., 2012). The transgenic 35S promoter-directed OsHAK5 OX lines were initially screened by hygromycin resistance and GUS staining; a total of 26 transgenic lines were selected after a second round of PCR analysis and Southern-blot verification by following the protocol described by Jia et al. (2011). Among all the lines of the T2 homozygous generation, 15 lines with only one copy of the gene insert and similar intensity of OsHAK5 expression were further sequenced to identify the gene insertion sites. Three independent transgenic rice lines with OsHAK5 integrated at noncoding regions were selected and named OX1, OX2, and OX3 (Supplemental Fig. S3) for detailed physiological and molecular analyses under different K and Na treatments. The histochemical analysis of GUS staining in different tissues of rice was performed as described previously (Ai et al., 2009).
Identification of oshak5 Transfer DNA Insertion Mutant Lines
Two putative transfer DNA (T-DNA) insertion lines of OsHAK5 knockout mutants were identified by searching the SIGNAL database (http://signal.salk.edu/cgi-bin/RiceGE). Their genetic backgrounds were cv HY and cv DJ. The integration positions were verified by sequencing the right border of PCR products with the primers used to identify the T-DNA mutants as informed by the suppliers (Supplemental Table S4). One mutant of cv HY and one mutant of cv DJ were identified with only one copy of the insertion, and the insertional positions were consistent with the supplier’s report. These two OsHAK5 KO lines were further confirmed by qRT-PCR with mRNA from K-starved roots and shoots of the T2 generation. There was no detectable OsHAK5 transcript in both knockout mutants (Supplemental Fig. S4).
Statistical Analysis
Data were analyzed by ANOVA using the SPSS 10 program (SPSS). Different letters above columns in the figures indicate statistical differences (P ≤ 0.05) between the transgenic plants and the respective wild-type plants and/or between different treatments.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Localization of OsHAK5 in the cell plasma membrane.
Supplemental Figure S2. Functional complementation test of OsHAK5 in yeast for absorbing K from culture medium containing different levels of K.
Supplemental Figure S3. Molecular identification of OsHAK5-overexpressing transgenic rice lines in the background of cv Nipponbare.
Supplemental Figure S4. Homozygous T-DNA insertion mutants of the OsHAK5 gene in rice: phenotype and isolation.
Supplemental Figure S5. Effect of OsHAK5 knockout on rice growth in conditions of low K supply.
Supplemental Figure S6. Effect of continuous supply of high K on plant growth and K accumulation of the OsHAK5 KO transgenic rice.
Supplemental Figure S7. Effect of OsHAK5 on K partitioning between root and shoot of the rice plants with similar root size and total K contents.
Supplemental Figure S8. Effect of OsHAK5 knockout on plant growth in the presence of 100 mm NaCl.
Supplemental Table S1. Primers for semiquantitative RT-PCR and real-time quantitative PCR of OsHAK5.
Supplemental Table S2. Primers for the promoter of OsHAK5.
Supplemental Table S3. Primers for OsHAK5 complementary DNA for the construction of overexpression.
Supplemental Table S4. Primers for the identification of two homozygous mutant lines of oshak5.
Supplementary Material
Glossary
- K
potassium
- Na
sodium
- LATS
low-affinity transport system
- HATS
high-affinity transport system
- qRT
quantitative reverse transcription
- SIET
scanning ion-selective electrode system
- OX
overexpression
- KO
knockout
- IRRI
International Rice Research Institute
- RT
reverse transcription
- T-DNA
transfer DNA
- HY
Hwayoung
- DJ
Dongjin
Footnotes
This work was supported by the National Natural Science Foundation (grant no. 31361140357), the National R&D Program for Transgenic Crops, the Research Fund for the Doctoral Program of Higher Education of China (grant no. 20130097110008), the Fundamental Research Funds for the Central Universities (grant no. KYTZ201404), the 111 Project (grant no. 12009), the Innovative Research Team Development Plan of the Ministry of Education of China (grant no. IRT1256), the Priority Academic Program Development of Jiangsu Higher Education Institutions project, and the Legacy Heritage Fund Program of the Israel Science Foundation (grant no. 1842/13 to N.M.).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Ahn SJ, Shin R, Schachtman DP. (2004) Expression of KT/KUP genes in Arabidopsis and the role of root hairs in K+ uptake. Plant Physiol 134: 1135–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai P, Sun S, Zhao J, Fan X, Xin W, Guo Q, Yu L, Shen Q, Wu P, Miller AJ, et al. (2009) Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J 57: 798–809 [DOI] [PubMed] [Google Scholar]
- Alemán F, Nieves-Cordones M, Martínez V, Rubio F. (2011) Root K+ acquisition in plants: the Arabidopsis thaliana model. Plant Cell Physiol 52: 1603–1612 [DOI] [PubMed] [Google Scholar]
- Ashley MK, Grant M, Grabov A. (2006) Plant responses to potassium deficiencies: a role for potassium transport proteins. J Exp Bot 57: 425–436 [DOI] [PubMed] [Google Scholar]
- Bañuelos MA, Garciadeblas B, Cubero B, Rodríguez-Navarro A. (2002) Inventory and functional characterization of the HAK potassium transporters of rice. Plant Physiol 130: 784–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bart R, Chern M, Park CJ, Bartley L, Ronald PC. (2006) A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods 2: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Chen L, Qu H, Lian J, Liu W, Hu YB, Xu G. (2012) Alteration of nutrient allocation and transporter genes expression in rice under N, P, K, and Mg deficiencies. Acta Physiol Plant 34: 939–946 [Google Scholar]
- Chen S, Tao L, Zeng L, Vega-Sanchez ME, Umemura K, Wang GL. (2006) A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol Plant Pathol 7: 417–427 [DOI] [PubMed] [Google Scholar]
- Chérel I, Lefoulon C, Boeglin M, Sentenac H. (2014) Molecular mechanisms involved in plant adaptation to low K+ availability. J Exp Bot 65: 833–848 [DOI] [PubMed] [Google Scholar]
- Coskun D, Britto DT, Li M, Oh S, Kronzucker HJ. (2013) Capacity and plasticity of potassium channels and high-affinity transporters in roots of barley and Arabidopsis. Plant Physiol 162: 496–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeken R, Geiger D, Fromm J, Koroleva O, Ache P, Langenfeld-Heyser R, Sauer N, May ST, Hedrich R. (2002) Loss of the AKT2/3 potassium channel affects sugar loading into the phloem of Arabidopsis. Planta 216: 334–344 [DOI] [PubMed] [Google Scholar]
- Desbrosses G, Josefsson C, Rigas S, Hatzopoulos P, Dolan L. (2003) AKT1 and TRH1 are required during root hair elongation in Arabidopsis. J Exp Bot 54: 781–788 [DOI] [PubMed] [Google Scholar]
- Ding Y, Luo W, Xu G. (2006) Characterisation of magnesium nutrition and interaction of magnesium and potassium in rice. Ann Appl Biol 149: 111–123 [Google Scholar]
- Elumalai RP, Nagpal P, Reed JW. (2002) A mutation in the Arabidopsis KT2/KUP2 potassium transporter gene affects shoot cell expansion. Plant Cell 14: 119–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels C, Marschner H. (1992) Adaptation of potassium translocation into the shoot of maize (Zea mays) to shoot demand: evidence for xylem loading as a regulating step. Physiol Plant 86: 263–268 [Google Scholar]
- Epstein E, Rains DW, Elzam OE. (1963) Resolution of dual mechanisms of potassium absorption by barley roots. Proc Natl Acad Sci USA 49: 684–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faiyue B, Al-Azzawi MJ, Flowers TJ. (2012) A new screening technique for salinity resistance in rice (Oryza sativa L.) seedlings using bypass flow. Plant Cell Environ 35: 1099–1108 [DOI] [PubMed] [Google Scholar]
- Fu HH, Luan S. (1998) AtKuP1: a dual-affinity K+ transporter from Arabidopsis. Plant Cell 10: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajdanowicz P, Michard E, Sandmann M, Rocha M, Corrêa LG, Ramírez-Aguilar SJ, Gomez-Porras JL, González W, Thibaud JB, van Dongen JT, et al. (2011) Potassium (K+) gradients serve as a mobile energy source in plant vascular tissues. Proc Natl Acad Sci USA 108: 864–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaymard F, Pilot G, Lacombe B, Bouchez D, Bruneau D, Boucherez J, Michaux-Ferrière N, Thibaud JB, Sentenac H. (1998) Identification and disruption of a plant shaker-like outward channel involved in K+ release into the xylem sap. Cell 94: 647–655 [DOI] [PubMed] [Google Scholar]
- Gierth M, Mäser P, Schroeder JI. (2005) The potassium transporter AtHAK5 functions in K+ deprivation-induced high-affinity K+ uptake and AKT1 K+ channel contribution to K+ uptake kinetics in Arabidopsis roots. Plant Physiol 137: 1105–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass ADM. (1976) Regulation of potassium absorption in barley roots: an allosteric model. Plant Physiol 58: 33–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass ADM, Dunlop J. (1978) The influence of potassium content on the kinetics of potassium influx into excised ryegrass and barley roots. Planta 141: 117–119 [DOI] [PubMed] [Google Scholar]
- Grabov A. (2007) Plant KT/KUP/HAK potassium transporters: single family-multiple functions. Ann Bot (Lond) 99: 1035–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M, Qiu X, Wang L, Xie W, Zhang C, Xiong L, Lian X, Zhang Q. (2008) KT/HAK/KUP potassium transporters gene family and their whole-life cycle expression profile in rice (Oryza sativa). Mol Genet Genomics 280: 437–452 [DOI] [PubMed] [Google Scholar]
- Hirsch RE, Lewis BD, Spalding EP, Sussman MR. (1998) A role for the AKT1 potassium channel in plant nutrition. Science 280: 918–921 [DOI] [PubMed] [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF. (2009) CHL1 functions as a nitrate sensor in plants. Cell 138: 1184–1194 [DOI] [PubMed] [Google Scholar]
- Horie T, Sugawara M, Okada T, Taira K, Kaothien-Nakayama P, Katsuhara M, Shinmyo A, Nakayama H. (2011) Rice sodium-insensitive potassium transporter, OsHAK5, confers increased salt tolerance in tobacco BY2 cells. J Biosci Bioeng 111: 346–356 [DOI] [PubMed] [Google Scholar]
- Jia H, Ren H, Gu M, Zhao J, Sun S, Zhang X, Chen J, Wu P, Xu G. (2011) The phosphate transporter gene OsPht1;8 is involved in phosphate homeostasis in rice. Plant Physiol 156: 1164–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EJ, Kwak JM, Uozumi N, Schroeder JI. (1998) AtKUP1: an Arabidopsis gene encoding high-affinity potassium transport activity. Plant Cell 10: 51–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagarde D, Basset M, Lepetit M, Conejero G, Gaymard F, Astruc S, Grignon C. (1996) Tissue-specific expression of Arabidopsis AKT1 gene is consistent with a role in K+ nutrition. Plant J 9: 195–203 [DOI] [PubMed] [Google Scholar]
- Lebaudy A, Véry AA, Sentenac H. (2007) K+ channel activity in plants: genes, regulations and functions. FEBS Lett 581: 2357–2366 [DOI] [PubMed] [Google Scholar]
- Maathuis FJM. (2009) Physiological functions of mineral macronutrients. Curr Opin Plant Biol 12: 250–258 [DOI] [PubMed] [Google Scholar]
- Maathuis FJM, Amtmann A. (1999) K+ nutrition and Na+ toxicity: the basis of cellular K+/Na+ ratios. Ann Bot (Lond) 84: 123–133 [Google Scholar]
- Maathuis FJM, Sanders D. (1997) Regulation of K+ absorption in plant root cells by external K+: interplay of different plasma membrane K+ transporters. J Exp Bot 48: 451–458 [DOI] [PubMed] [Google Scholar]
- Malagoli P, Britto DT, Schulze LM, Kronzucker HJ. (2008) Futile Na+ cycling at the root plasma membrane in rice (Oryza sativa L.): kinetics, energetics, and relationship to salinity tolerance. J Exp Bot 59: 4109–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner H (1996) Mineral Nutrition of Higher Plants. Academic Press, London [Google Scholar]
- Marschner H, Kirkby EA, Cakmak I. (1996) Effect of mineral nutritional status on shoot-root partitioning of photoassimilates and cycling of mineral nutrients. J Exp Bot 47: 1255–1263 [DOI] [PubMed] [Google Scholar]
- Mäser P, Thomine S, Schroeder JI, Ward JM, Hirschi K, Sze H, Talke IN, Amtmann A, Maathuis FJ, Sanders D, et al. (2001) Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol 126: 1646–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Jiang L. (2007) Transient expression of fluorescent fusion proteins in protoplasts of suspension cultured cells. Nat Protoc 2: 2348–2353 [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M. (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Nieves-Cordones M, Alemán F, Martínez V, Rubio F. (2010) The Arabidopsis thaliana HAK5 K+ transporter is required for plant growth and K+ acquisition from low K+ solutions under saline conditions. Mol Plant 3: 326–333 [DOI] [PubMed] [Google Scholar]
- Okada T, Nakayama H, Shinmyo A, Yoshida K. (2008) Expression of OsHAK genes encoding potassium ion transporters in rice. Plant Biotechnol J 25: 241–245 [Google Scholar]
- Pyo YJ, Gierth M, Schroeder JI, Cho MH. (2010) High-affinity K+ transport in Arabidopsis: AtHAK5 and AKT1 are vital for seedling establishment and postgermination growth under low-potassium conditions. Plant Physiol 153: 863–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z, Hampton CR, Shin R, Barkla BJ, White PJ, Schachtman DP. (2008) The high affinity K+ transporter AtHAK5 plays a physiological role in planta at very low K+ concentrations and provides a caesium uptake pathway in Arabidopsis. J Exp Bot 59: 595–607 [DOI] [PubMed] [Google Scholar]
- Quintero FJ, Blatt MR. (1997) A new family of K+ transporters from Arabidopsis that are conserved across phyla. FEBS Lett 415: 206–211 [DOI] [PubMed] [Google Scholar]
- Rigas S, Debrosses G, Haralampidis K, Vicente-Agullo F, Feldmann KA, Grabov A, Dolan L, Hatzopoulos P. (2001) TRH1 encodes a potassium transporter required for tip growth in Arabidopsis root hairs. Plant Cell 13: 139–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigas S, Ditengou FA, Ljung K, Daras G, Tietz O, Palme K, Hatzopoulos P. (2013) Root gravitropism and root hair development constitute coupled developmental responses regulated by auxin homeostasis in the Arabidopsis root apex. New Phytol 197: 1130–1141 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Navarro A, Rubio F. (2006) High-affinity potassium and sodium transport systems in plants. J Exp Bot 57: 1149–1160 [DOI] [PubMed] [Google Scholar]
- Rubio F, Alemán F, Nieves-Cordones M, Martínez V. (2010) Studies on Arabidopsis athak5, atakt1 double mutants disclose the range of concentrations at which AtHAK5, AtAKT1 and unknown systems mediate K uptake. Physiol Plant 139: 220–228 [DOI] [PubMed] [Google Scholar]
- Rubio F, Gassmann W, Schroeder JI. (1995) Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science 270: 1660–1663 [DOI] [PubMed] [Google Scholar]
- Rubio F, Santa-Maria GE, Rodriguez-Navarro A. (2000) Cloning of Arabidopsis and barley cDNAs encoding HAK potassium transporters in root and shoot cells. Physiol Plant 109: 34–43 [Google Scholar]
- Santa-María GE, Rubio F, Dubcovsky J, Rodríguez-Navarro A. (1997) The HAK1 gene of barley is a member of a large gene family and encodes a high-affinity potassium transporter. Plant Cell 9: 2281–2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senn ME, Rubio F, Bañuelos MA, Rodríguez-Navarro A. (2001) Comparative functional features of plant potassium HvHAK1 and HvHAK2 transporters. J Biol Chem 276: 44563–44569 [DOI] [PubMed] [Google Scholar]
- Sentenac H, Bonneaud N, Minet M, Lacroute F, Salmon JM, Gaymard F, Grignon C. (1992) Cloning and expression in yeast of a plant potassium ion transport system. Science 256: 663–665 [DOI] [PubMed] [Google Scholar]
- Shabala S, Cuin TA. (2008) Potassium transport and plant salt tolerance. Physiol Plant 133: 651–669 [DOI] [PubMed] [Google Scholar]
- Spalding EP, Hirsch RE, Lewis DR, Qi Z, Sussman MR, Lewis BD. (1999) Potassium uptake supporting plant growth in the absence of AKT1 channel activity: inhibition by ammonium and stimulation by sodium. J Gen Physiol 113: 909–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Fan X, Li Q, Feng H, Miller AJ, Shen Q, Xu G. (2012) Knockdown of a rice stelar nitrate transporter alters long-distance translocation but not root influx. Plant Physiol 160: 2052–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uozumi N, Kim EJ, Rubio F, Yamaguchi T, Muto S, Tsuboi A, Bakker EP, Nakamura T, Schroeder JI. (2000) The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol 122: 1249–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Véry AA, Sentenac H. (2003) Molecular mechanisms and regulation of K+ transport in higher plants. Annu Rev Plant Biol 54: 575–603 [DOI] [PubMed] [Google Scholar]
- Vicente-Agullo F, Rigas S, Desbrosses G, Dolan L, Hatzopoulos P, Grabov A. (2004) Potassium carrier TRH1 is required for auxin transport in Arabidopsis roots. Plant J 40: 523–535 [DOI] [PubMed] [Google Scholar]
- Wegner LH. (2014) Root pressure and beyond: energetically uphill water transport into xylem vessels? J Exp Bot 65: 381–393 [DOI] [PubMed] [Google Scholar]
- Yoshida S, Forno DA, Cock JH, Gomez KA (1976) Routine Procedures for Growing Rice Plants in Culture Solution, Ed 3. International Rice Research Institute, Los Banos, Philippines [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.