Absence of the RNAse H2 complex, mediating the excision of ribonucleotides from DNA, was found to overcome the growth inhibitory phenotype of plants lacking a functional replication checkpoint because of tolerating the substitution of deoxynucleotides with ribonucleotides. This substitution, however, results in replication errors, demonstrating the need of RNase H2 activity for genome stability.
Abstract
The WEE1 kinase is an essential cell cycle checkpoint regulator in Arabidopsis thaliana plants experiencing replication defects. Whereas under non-stress conditions WEE1-deficient plants develop normally, they fail to adapt to replication inhibitory conditions, resulting in the accumulation of DNA damage and loss of cell division competence. We identified mutant alleles of the genes encoding subunits of the ribonuclease H2 (RNase H2) complex, known for its role in removing ribonucleotides from DNA-RNA duplexes, as suppressor mutants of WEE1 knockout plants. RNase H2 deficiency triggered an increase in homologous recombination (HR), correlated with the accumulation of γ-H2AX foci. However, as HR negatively impacts the growth of WEE1-deficient plants under replication stress, it cannot account for the rescue of the replication defects of the WEE1 knockout plants. Rather, the observed increase in ribonucleotide incorporation in DNA indicates that the substitution of deoxynucleotide with ribonucleotide abolishes the need for WEE1 under replication stress. Strikingly, increased ribonucleotide incorporation in DNA correlated with the occurrence of small base pair deletions, identifying the RNase H2 complex as an important suppressor of genome instability.
INTRODUCTION
Faithful duplication of the genome is important for error-free transmission of the genetic information from one generation to the next. Because during growth and development the DNA is prone to damage induced by environmental stress and endogenous factors, DNA synthesis is accompanied by several quality checks, called checkpoints, which arrest the cell cycle for DNA repair upon damage. Similar to other organisms, in plants, two central players of this DNA damage response are the Ataxia Telangiectasia Mutated (ATM) and ATM- and RAD3-related (ATR) protein kinases. In general, ATR is triggered by stalled replication forks and single-stranded DNA (ssDNA), whereas ATM is activated by double-strand breaks (DSBs) (Garcia et al., 2003; Culligan et al., 2004, 2006; Ricaud et al., 2007; Ciccia and Elledge, 2010). In animals, DNA stress checkpoint activation by ATM and ATR triggers eventually a transient or permanent cell cycle arrest through the phosphorylation of a number of downstream proteins, including the CDC25 phosphatase and WEE1 kinase, which operate as the on and off switches of cyclin-dependent kinase (CDK) activity (Harper and Elledge, 2007).
Plants lack an orthologous CDC25 gene (Boudolf et al., 2006) but possess a homolog of the WEE1 protein kinase (Sun et al., 1999; Sorrell et al., 2002; Gonzalez et al., 2004). Arabidopsis thaliana WEE1 transcript levels are strongly induced upon treatment with replication-inhibitory drugs in an ATR-dependent manner. Moreover, whereas WEE1 knockout (WEE1KO) mutants show a wild-type phenotype under normal growth conditions, they are hypersensitive to drugs inducing replication stress (De Schutter et al., 2007). Molecular analysis revealed that absence of WEE1 results in altered S-phase kinetics, suggesting a role in the adaptation of the DNA replication rate in response to replication defects. The inability to do so eventually triggers premature differentiation and a cell death phenotype within the root meristem (Cools et al., 2011).
Replication stress can be triggered by the application of hydroxyurea (HU), inhibiting the activity of the ribonucleotide reductase (RNR) enzyme that is responsible for the reduction of ribonucleotides (rNTPs) to deoxyribonucleotides (dNTPs) and consequently limiting dNTP availability for DNA polymerases. These polymerases have evolved mechanisms to efficiently recognize and incorporate dNTPs, but not rNTPs, into the genome. This is achieved through sugar-type discrimination, as dNTPs possess deoxyribose, whereas ribose is the sugar backbone of rNTPs (Brown and Suo, 2011). However, because rNTP levels are 10- to 2000-fold higher than dNTP levels (Ferraro et al., 2010; Nick McElhinny et al., 2010b), DNA polymerases may misincorporate rNTPs into genomic DNA every 10,000th to 100,000th nucleotide, making ribonucleoside monophosphate (rNMP) the most prevalent aberrant nucleotide occurring in DNA (Nick McElhinny et al., 2010a, 2010b; Reijns et al., 2012; Clausen et al., 2013). Ribonucleotides have a reactive 2’OH on the sugar part and this makes them more sensitive to strand cleavage, resulting in genome instability (Nick McElhinny et al., 2010a). Therefore, organisms evolved a repair pathway, called ribonucleotide excision repair, which is initiated by ribonucleases H (RNase H) that specifically catalyzes the cleavage of RNA in DNA-RNA duplexes (Stein and Hausen, 1969). There are two main types of RNase H. Type 1 RNase H (RNase H1) needs at least four sequential rNMPs for recognition and cleavage to occur, while type 2 (RNase H2) is able to cut even single rNMPs and is the only enzyme known to hydrolyze ribonucleotides misincorporated during genomic replication (Cerritelli and Crouch, 2009; Bubeck et al., 2011). When a single rNTP is incorporated in DNA, RNase H2 incises the DNA 5′ of the ribonucleotide, which produces DNA containing 3′ hydroxyl and 5′ phospho-ribonucleotide ends. Upon DNA replication by the POL δ and/or POL ε polymerases, the ribonucleotide is displaced and the resulting flap is excised by the FEN1 and/or EXO1 exonucleases, followed by ligation of the nick through the LIG1 ligase (Rydberg and Game, 2002; Sparks et al., 2012; Williams et al., 2012).
Here, we report that a mutation within the catalytic subunit of the RNase H2 complex was found to partially overcome the replication phenotype of WEE1KO plants. Similarly, the mutation of the regulatory subunits of the RNase H2 complex rescued WEE1-deficient plants under replication stress. Rather than the observed increase in homologous recombination, the ability to overcome replication stress was found to correlate with an increased incorporation of rNMPs in DNA. This substitution of dNTPs with rNTPs restored replication kinetics of the WEE1KO plants but resulted in replication errors, highlighting the need for RNase H2 activity to maintain genome stability.
RESULTS
A Mutation in the RNase H2 Subunit A Gene Rescues HU Hypersensitivity of WEE1KO Plants
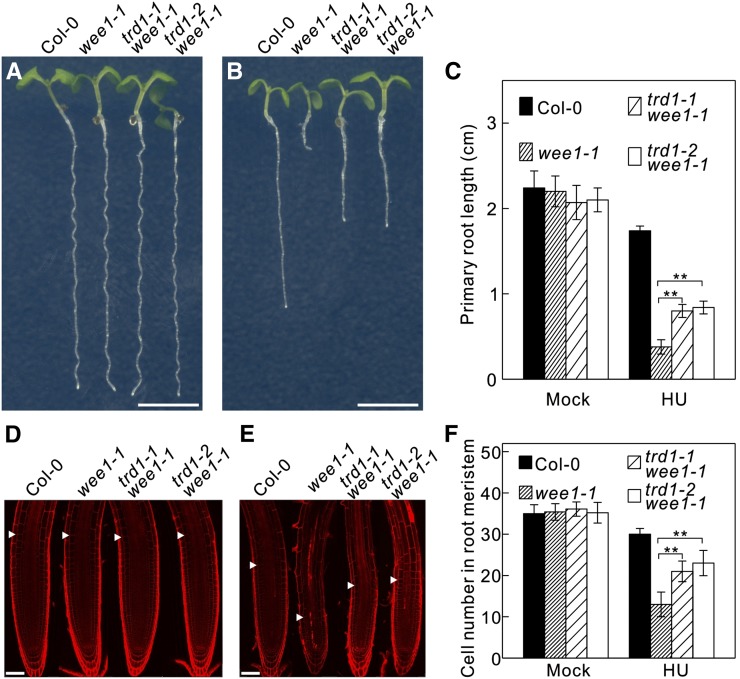
WEE1KO seedlings (wee1-1) show a strong inhibition of root growth when grown on HU-containing medium (Figures 1A to 1C). To identify complementing mutations, an ethyl methanesulfonate-mutagenized wee1-1 seed stock was screened in the M2 generation for restored root growth in the presence of 0.75 mM HU. Among the identified mutants, the triffid1-1 (trd1-1) wee1-1 mutant resulted in a partial recovery of root growth (Figures 1A to 1C) and an almost complete inhibition of the cell death phenotype (Figures 1D and 1E), as observed by the decrease in number of fluorescent cells upon treatment with propidium iodide, a fluorescent dye that outlines the walls of living cells but also penetrates through the plasma membrane of dead cells. The root meristem size of the different genotypes was measured by counting the meristematic cortex cells, which showed that under control growth conditions, there was no statistically significant difference between the wild-type (Columbia-0 [Col-0]) and the wee1-1 and wee1-1 trd1-1 mutant plants (Figures 1D and 1F). In the presence of HU, the number of dividing cortex cells was clearly reduced in wee1-1 plants compared with wild-type seedlings (Figures 1E and 1F), whereas the meristem size of wee1-1 trd1-1 mutants did differ significantly from that of wild-type plants.
Figure 1.
A Mutation in the Catalytic Subunit of the RNase H2 Complex Partially Rescues HU Hypersensitivity of WEE1KO Plants.
(A) and (B) Root growth of 7-d-old wild-type (Col-0) and wee1-1, trd1-1 wee1-1, and trd1-2 wee1-1 mutant plants grown on control medium (A) or medium supplemented with 0.75 mM HU (B). Bar = 0.5 cm.
(C) Quantification of the root length of plants shown in (A) and (B). Data represent mean ± sd (n > 10, **P value < 0.01, two-sided Student’s t test).
(D) and (E) Representative confocal microscopy images of plants shown in (A) and (B) stained with propidium iodide. Arrowheads indicate the meristem size based on the cortical cell length. Bar = 50 μm.
(F) Number of meristematic cortex cells. Data represent mean ± sd (n > 10, **P value < 0.01, two-sided Student’s t test).
[See online article for color version of this figure.]
Through next-generation sequencing-based gene mapping, the underlying mutation in the trd1-1 mutant was pinpointed to a base pair change at codon position 584 of the At2g25100 gene, resulting in a glycine (GGA) to glutamic acid (GAA) substitution (Figure 2A). This base pair change was confirmed by direct sequencing of the original mutant. The At2g25100 gene is annotated as the catalytic subunit A of the RNase H2 protein complex. The amino acid change is located in a conserved domain (Figure 2B), directly next to a tyrosine finger that is pivotal for substrate binding (Rychlik et al., 2010), indicating that the mutant allele most probably encodes a nonfunctional protein. To confirm this hypothesis, we analyzed an available putative knockout line (GABI-139H04, nominated hereafter trd1-2) harboring a T-DNA insertion in intron 3 (Figure 2A). The trd1-2 mutation was crossed in wee1-1 plants and tested for HU hypersensitivity. Root growth analysis showed that the trd1-2 mutation partially rescued the HU hypersensitivity of WEE1KO mutants (Figures 1A to 1C). Similarly, the root cell death phenotype was suppressed in the double mutant, confirming that it is a lack of RNase H2 subunit A activity that rescues the replication defect of the WEE1 checkpoint mutant.
Figure 2.
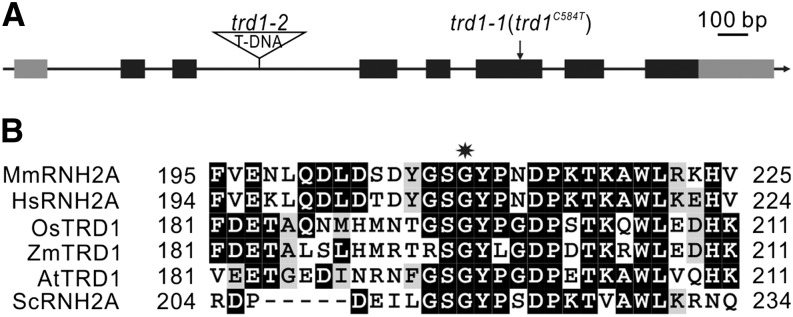
TRD1 Encodes the Catalytic Subunit of the RNase H2 Complex.
(A) Intron-exon organization of TRD1. Black and gray boxes represent exons and untranslated regions, respectively. The position of the mutated base pair (trd1-1) and T-DNA insertion site (trd1-2) are indicated.
(B) Sequence alignment of the RNase H2 subunit A from different species highlighting the conserved position of the Gly residue (indicated by a star). Mm, Mus musculus; Hs, Homo sapiens; Os, Oryza sativa; Zm, Zea mays; At, Arabidopsis thaliana; Sc, Saccharomyces cerevisiae.
The Plant RNase H2 Complex Holds Three Subunits
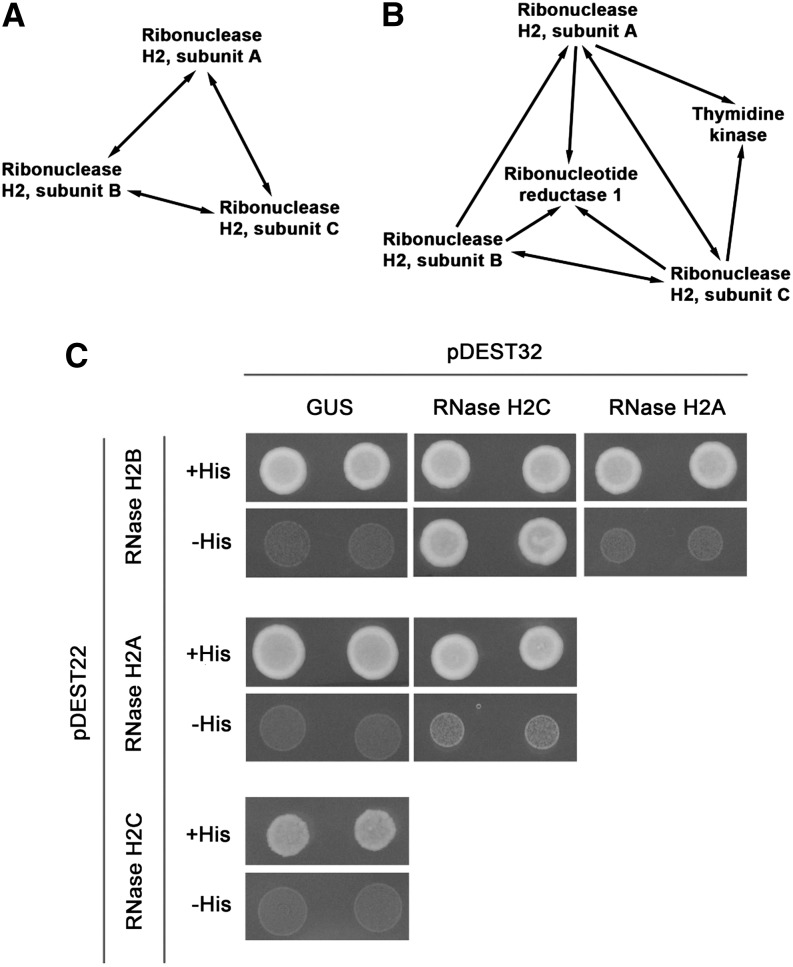
TRD1-interacting proteins were screened for by tandem affinity purification (TAP) using cell cultures (Supplemental Data Set 1). Under control conditions the RNase H2 subunit A pulled down proteins homologous to the regulatory B and C subunits of the RNase H2 complex from other species (Figures 3A; Supplemental Data Set 2), revealing that the Arabidopsis RNase H2 complex comprises three subunits. The interaction between the three RNase H2 subunits was confirmed by reverse TAP experiments using the subunit B or subunit C as bait. Intriguingly, when performing TAP in the presence of HU, the thymidine kinase and the large subunit of ribonucleotide reductase 1 also were isolated (Figure 3B; Supplemental Data Set 2). Through yeast two-hybrid interaction assays, a direct connection could be detected only between the RNase H2 B and C subunits (Figure 3C), suggesting that most interactions between the different subunits occur through cooperative binding.
Figure 3.
The Arabidopsis RNase H2 Complex Comprises Three Subunits.
(A) and (B) Protein-protein interaction between different RNase H2 subunits as identified by tandem affinity purification from cell cultures cultivated in the absence (A) or presence of 10 mM HU for 24 h (B). Arrows point from bait to prey and correspond to interactions that were confirmed in a repeat experiment (Supplemental Data Set 2).
(C) Yeast two-hybrid interactions between the different subunits of RNase H2. The GUS protein was used as negative control.
To check whether plants deficient in RNase H2 subunit B and C could revert the wee1-1 mutant phenotype, T-DNA insertion lines of both genes were identified. Similar to the trd1 mutants, absence of the RNase H2 subunit B or C rescued the HU hypersensitivity of the WEE1KO line (Supplemental Figure 1).
RNase H2 Subunit A Mutants Display Constitutive High Recombination Rates
To better understand how the lack of RNase H2 activity overcomes the replication defects of WEE1KO plants, an RNA sequencing experiment was conducted, comparing the transcriptome of wee1-1 and wee1-1 trd1-2 mutant root meristems grown in the absence and presence of HU. Only 25 genes were differentially regulated between wee1-1 and wee1-1 trd1-2 under control conditions (Supplemental Data Set 3), indicating that the trd1 mutation induces only a limited transcriptional response. Moreover, all genes within a genomic region spanning At2g38120 to At2g38290 showed a uniform 2-fold induction, indicating a genome duplication, confirmed by whole-genome sequencing (Supplemental Figure 2). The transcriptional increase of the genes within this locus likely correlated with the increased gene copy number, rather than with RNase H2 deficiency, as no transcriptional induction was observed in the independent wee1-1 trd1-1 mutant or in a trd1-2 mutant in which the duplicated region was segregated from the mutation (Supplemental Figure 2). Among the seven remaining genes being transcriptionally upregulated, five appeared in a coexpression cluster (Supplemental Figure 3). The same genes were strongly activated in both genotypes after treatment for 24 h with HU (Supplemental Figure 4) and were differentially expressed between control plants and trd1-2 mutants (Supplemental Figure 3). To pinpoint the underlying process that triggered the transcriptional response of these genes, we collected a list of genes being coexpressed with at least three of five genes present within the coexpression cluster (Supplemental Data Set 4), revealing an enrichment for DNA repair-related processes (Supplemental Data Set 5). In the presence of HU, none of the genes being differentially expressed between wee1-1 and wee1-1 trd1-2 displayed coexpression clusters or Gene Ontology enrichment (Supplemental Data Set 6).
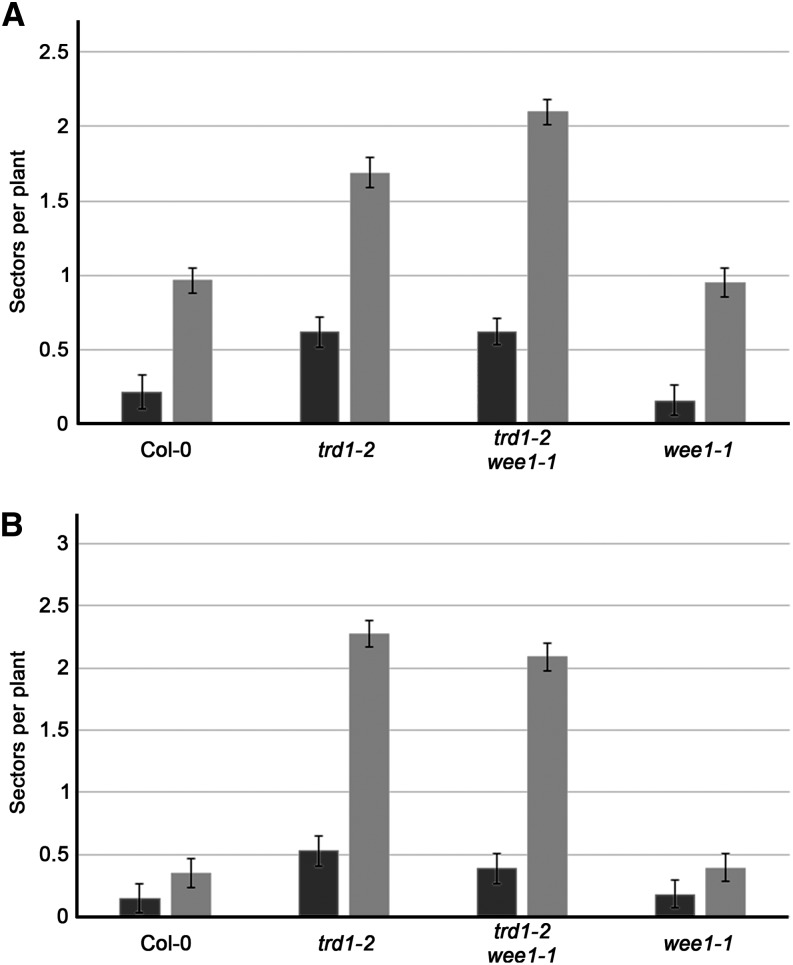
As DNA repair commonly occurs through homologous recombination (HR), we introduced two different β-glucuronidase (GUS) recombination substrates (651 and IC9C) into the trd1-2 mutant background. These recombination substrates contain an inactive GUS gene whose activity can be restored by either intra- and interchromosomal recombination in the 651 line or by interchromosomal recombination in IC9C (Swoboda et al., 1994; Molinier et al., 2004). Recombination frequencies can be deduced from the number of blue sectors after histochemical staining. When comparing plants grown with and without HU, an increase in recombination could be observed with both reporter lines in all genotypes tested, illustrating that HU treatment triggers HR (Figure 4). Remarkably, HR frequencies of trd1-2 and trd1-2 wee1-1 were already significantly increased (P value < 0.01) in comparison with the wild type and wee1-1 in the absence of HU (2- to 4-fold in 651 line and 2- to 6-fold in IC9C line) (Figure 4), indicating that loss of RNase H2 enhances HR in Arabidopsis. Adding HU to the medium stimulated HR but did not alter the overall trend among the lines, demonstrating an independence of the increased HR levels in the RNase H2 knockout from the HU treatment.
Figure 4.
RNase H2 Deficiency Triggers Increased HR.
Recombination frequencies of untreated (black bars) and HU-treated (0.75 mM; gray bars) control (Col-0), trd1-2, trd1-2 wee1-1, and wee1-1 seedlings using the 651 (A) or IC9C (B) reporters. Data represent mean number of GUS sectors ± se (n = 4, minimum 50 plants per repeat).
Resolving Holliday junctions that arise during HR requires the MUS81 endonuclease (Mannuss et al., 2010). To test the impact of the increased HR in RNase H2-deficient plants, the mus81-1 mutation was introduced into the trd1-2 mutant. While the trd1-2 and mus81-1 mutants were phenotypically indistinguishable from control plants, double mutants showed a severe root and shoot growth inhibition phenotype (Supplemental Figure 5), demonstrating that HR resolution is essential for plant survival of RNase H2-deficient plants.
RNase H2 Subunit A Mutants Accumulate γ-H2AX Foci
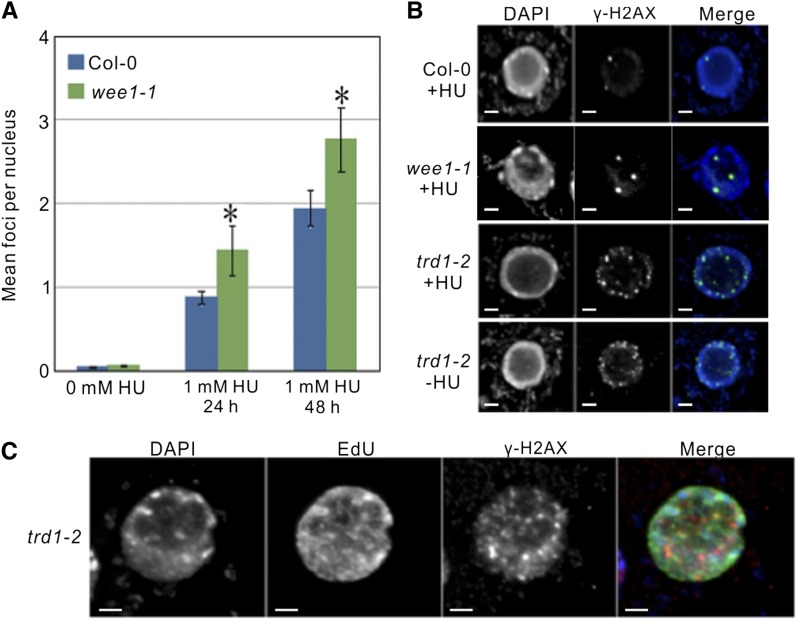
HR is induced upon the occurrence of DSBs or replication fork destabilization that can be detected through immunodetection of γ-H2AX foci, representing a phosphorylated form of the histone variant H2AX (Kinner et al., 2008; Sirbu et al., 2011). To detect such genomic problems in the RNase H2-defective background, we performed in situ immunostaining experiments in wild-type, wee1-1, trd1-2, and wee1-1 trd1-2 Arabidopsis root tip nuclei, treated or untreated with HU, using a γ-H2AX antibody. As expected, no γ-H2AX foci were detected in mitotic root tip nuclei of untreated wild-type plants and wee1-1 mutants. By contrast, upon HU treatment, nuclei of both genotypes displayed γ-H2AX foci (Figures 5A and 5B). Furthermore, the number of γ-H2AX foci per nucleus was significantly higher in the WEE1KO background (Figure 5A). Surprisingly, trd1-2 mutants showed a high number of γ-H2AX foci regardless of being HU treated or not (Figure 5B). Thus, in agreement with the transcriptomic analysis and the observed higher HR rates, plants lacking a functional RNase H2 complex accumulate chromosomal instability that can be visualized as γ-H2AX foci.
Figure 5.
Lack of RNase H2 Activity Triggers the Accumulation of γ-H2AX Foci.
(A) Average number of γ-H2AX foci per nucleus of wild-type (Col-0) and wee1-1 mutants, untreated or treated with HU. Data represent mean ± sd (n = 100, *P value < 0.05, two-sided Student’s t test).
(B) Detection of γ-H2AX foci in wild-type (Col-0), wee1-1, trd1-2, and wee1-1 trd1-2 root tip cells untreated (−HU) or treated with 1 mM HU (+HU). Bar = 2 μm.
(C) γ-H2AX immunofluorescence in S- or early G2-phase nuclei of trd1-2 mutants (being Edu positive). Bar = 2 μm.
To analyze whether the DSBs in the RNase H2 mutant could be the result of replication stress, we screened for a specific enrichment of γ-H2AX foci in replicating nuclei (Figure 5C). Root tips of trd1-2 mutants were incubated for 1 h with the thymidine analog ethynyl deoxyuridine (EdU), labeling ∼20% of all nuclei, representing the S- and early G2-phase nuclei that underwent replication during the EdU treatment. A total of 53% of all nuclei showed γ-H2AX foci and among these 86% scored Edu positive, illustrating a significant enrichment for replicating nuclei. These data clearly suggest that replication defects lay at the basis of the DSBs occurring in RNase H2-deficient plants.
Increased HR Does Not Rescue WEE1KO HU Hypersensitivity
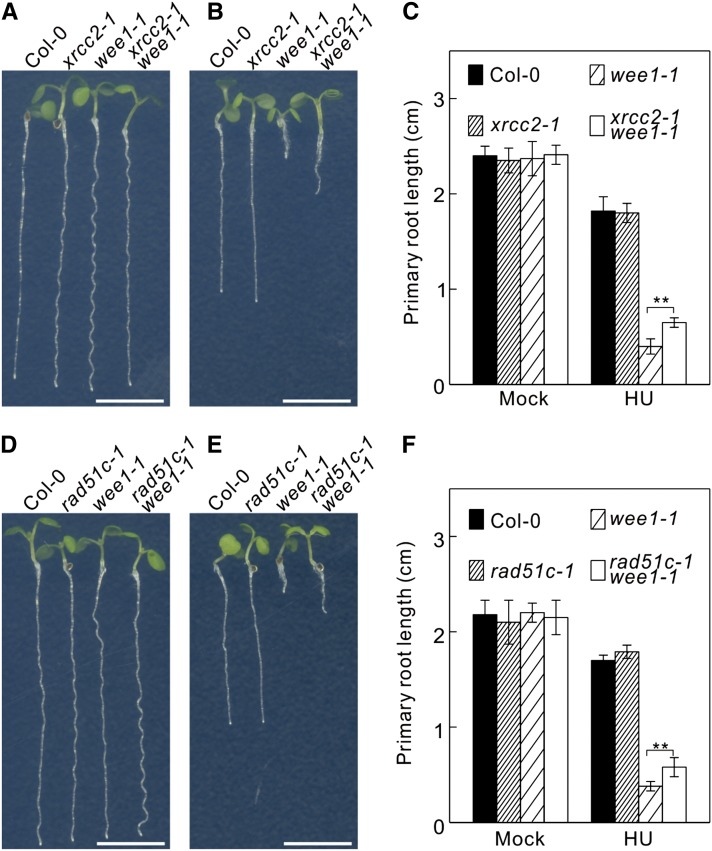
The observed increase of HR in RNase H2 mutant plants might account for the suppression of the WEE1 mutant phenotype in the presence of HU. To test this hypothesis, we suppressed HR in the double trd1-2 wee1-1 mutant by introducing a mutant XRCC2 allele, encoding an inactive RAD51 paralogous gene that causes hypersensitivity to the DNA cross-linking drug MMC (Bleuyard et al., 2005). Surprisingly, the xrcc2-1 mutation did not remove the HU resistance phenotype of the trd1-2 wee1-1 double mutants, but instead enhanced it, conferring root growth on HU-supplemented medium that was equal to that of control plants (Supplemental Figure 6). Similarly, xrcc2-1 partially rescued the HU hypersensitive phenotype of the single wee1-1 mutants (Figures 6A to 6C). Similar data were obtained using a mutation in the paralogous RAD51C gene (Figures 6D to 6F). Thus, HR contributes to the HU sensitivity of the wee1-1 lines, rather than rescuing it.
Figure 6.
Mutations in XRCC2 and RAD51C Partially Rescue WEE1KO HU Hypersensitivity.
(A) and (B) Root growth of 7-d-old wild-type (Col-0) and xrcc2-1, wee1-1, and xrcc2-1 wee1-1 mutant plants grown on control medium (A) or medium supplemented with 0.75 mM HU (B). Bar = 0.5 cm.
(C) Quantification of the root length of plants shown in (A) and (B). Data represent mean ± sd (n > 10, **P value < 0.01, two-sided Student’s t test).
(D) and (E) Root growth of 7-d-old wild-type (Col-0) and rad51c-1, wee1-1, and rad51c-1 wee1-1 mutant plants grown on control medium (D) or medium supplemented with 0.75 mM HU (E). Bar = 0.5 cm.
(F) Quantification of the root length of plants shown in (D) and (E). Data represent mean ± sd (n > 10, **P value < 0.01, two-sided Student’s t test).
[See online article for color version of this figure.]
RNase H2 Deficient Plants Incorporate rNMPs into Their Genome
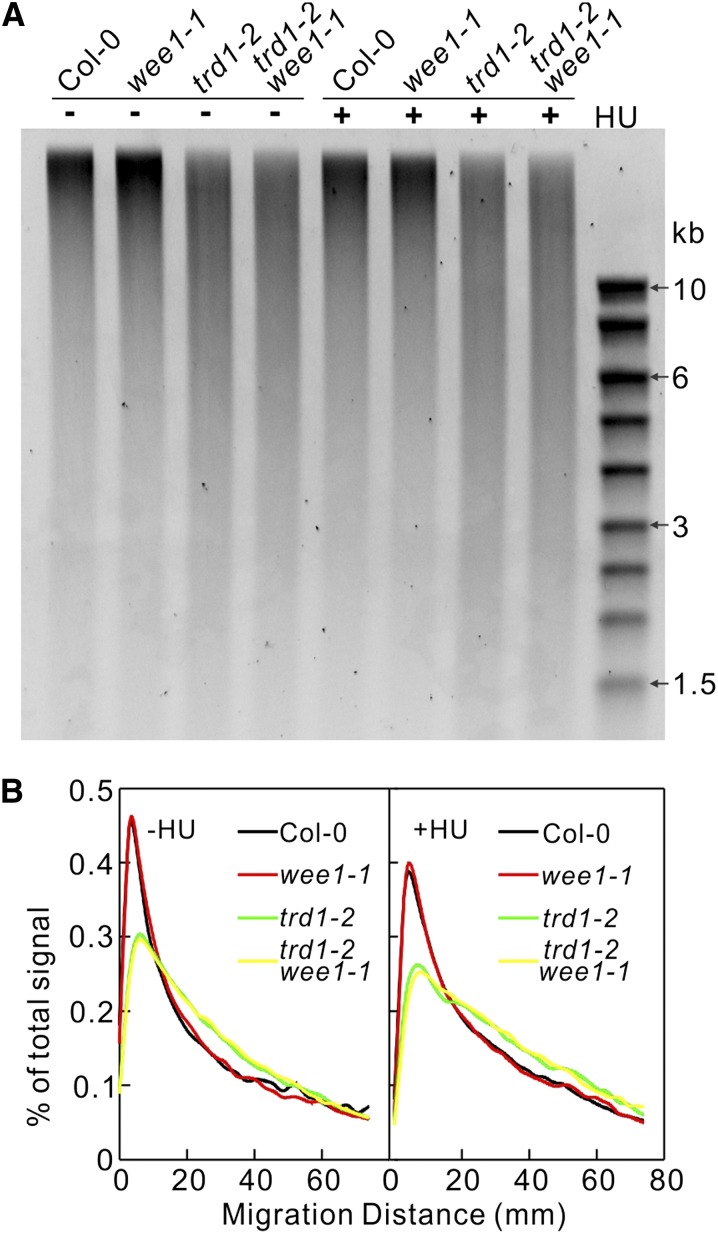
Yeast and mouse genomes defective in RNase H2 activity incorporate rNMPs (Nick McElhinny et al., 2010a; Miyabe et al., 2011; Reijns et al., 2012). Because rNMPs possess a reactive 2’OH on the sugar part, their incorporation makes the DNA backbone more susceptible to strand cleavage by alkali. Therefore, incubating genomic DNA in KOH and subsequently monitoring the resulting fragmentation by alkaline agarose gel electrophoresis can detect the presence of ribonucleotides in DNA. We analyzed the level of rNMP incorporation in the genomic DNA of wild-type, wee1-1, trd1-2, and trd1-2 wee1-1 plants grown under control conditions or in the presence of HU. Under both conditions, the genomic DNA samples isolated from trd1-2 and trd1-2 wee1-1 mutants were more sensitive to alkaline hydrolysis than DNA samples isolated from the wild type and wee1-1, as observed by the accumulation of short DNA fragments (Figure 7). No strong effect of HU application on the DNA fragmentation was observed, indicating that the absence of RNase H2 activity is the main cause of DNA instability. Similar as observed for trd1-2, DNA fragmentation indicative of rNMP incorporation was detected for DNA isolated from RNase H2 subunit B or subunit C mutant plants (Supplemental Figure 7).
Figure 7.
RNase H2-Deficient Plants Accumulate rNMPs in DNA.
(A) Alkaline cleavage products of genomic DNA extracted from 7-d-old wild-type (Col-0), wee1-1, trd1-2, and trd1-2 wee1-1 seedlings grown under control conditions or in the presence of 0.75 mM HU.
(B) Densitometry plot of lanes in (A).
Lack of RNase H2 Activity Results in an Increased Mutation Rate
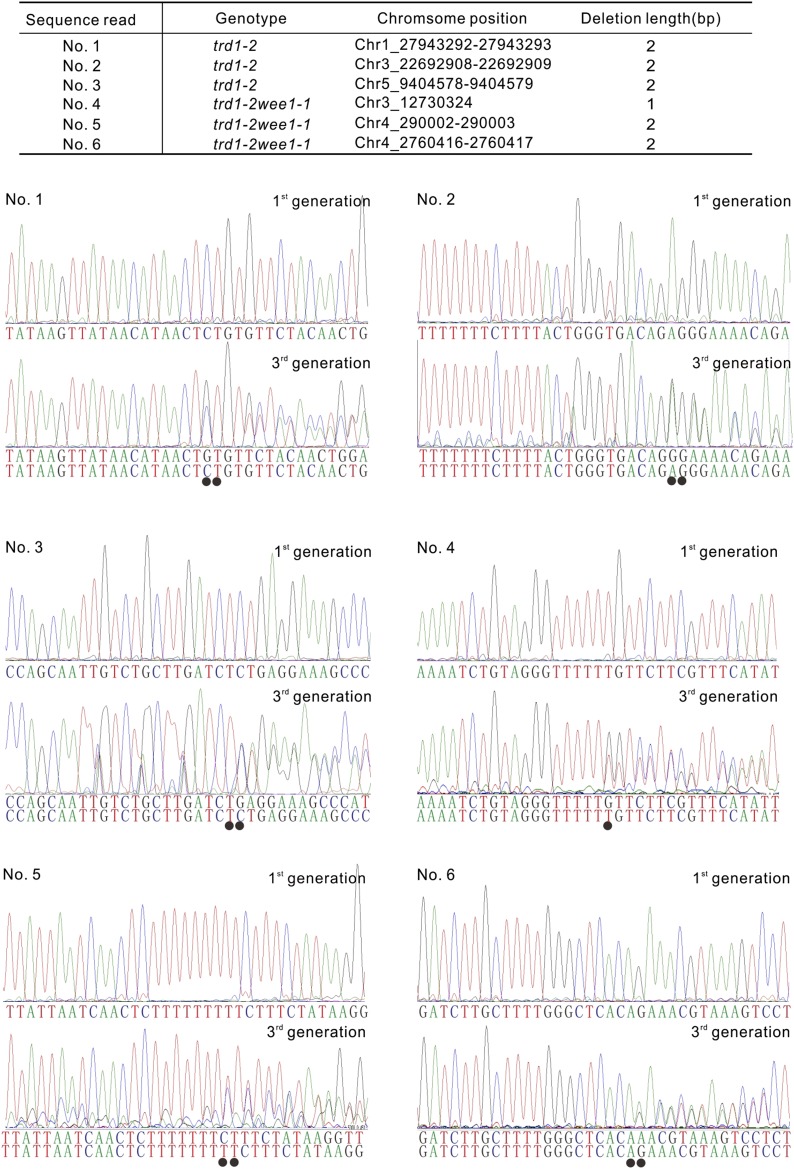
Root growth analysis of the RNase H2-deficient plants suggested that these plants appear to suffer no growth penalties for substituting dNTPs for rNTPs. However, short-term growth analyses do not exclude genomic defects over the long term. Therefore, we sequenced and compared the genome of a single wild-type, wee1-1, trd1-2, and trd1-2 wee1-1 plants, grown side-by-side over three generations. The sequence reads were mapped to the Arabidopsis reference genome (TAIR10) and scored for base pair changes and short base pair deletions. The number of base substitutions was not significantly different among the different genotypes (Table 1). By contrast, whereas both wild-type and wee1-1 plants showed no base pair deletions, trd1-2 and trd1-2 wee1-1 displayed three and two 2-bp deletions, respectively. Additionally, the trd1-2 mutant showed a 3-bp deletion (Table 1). Sequencing of the mutant loci over different generations confirmed the presence of the small deletions and illustrated that all but the 3-bp deletion were generated within less than two generations (Figure 8).
Table 1. Number of Base Substitutions and Deletions in Control (Col-0), wee1-1, trd1-2, and trd1-2 wee1-1 Plants Compared to the Reference Genome (TAIR10).
| Genotype | Base Substitution | 1 bp Δ | 2 bp Δ | 3 bp Δ |
|---|---|---|---|---|
| Col-0 | 9 | 0 | 0 | 0 |
| wee1-1 | 6 | 0 | 0 | 0 |
| trd1-2 | 8 | 0 | 3 | 1 |
| trd1-2 wee1-1 | 9 | 1 | 2 | 0 |
Figure 8.
Lack of RNase H2 Activity Causes Small Base Pair Deletions.
Sequencing reads of mutant loci in first versus third generation plants. Deleted base pairs (indicated by dots) result in dual sequence reads.
A parallel genome sequencing experiment was conducted on plants grown in the constant presence of HU. Again, plants lacking RNase H2 activity (trd1-2 and trd1-2 wee1-1) did accumulate small base pair deletions that were demonstrated by direct sequencing to be absent in the grandparent lines (Supplemental Data Set 7 and Supplemental Figure 8). By contrast, no deletions were found in the RNase H2 proficient plants (wild type and wee1-1). Compared with the control-grown plants, the HU treatment appeared to have no effect on the number of deletions. Indeed, linear regression analysis showed that only the presence of the trd1-2 mutation correlated with the total number of deletions (estimated coefficient of 2.36, P value < 0.001). Thus, absence of RNase H2 appeared to trigger genome instability in the form of small deletions.
DISCUSSION
The Arabidopsis WEE1 gene is essential for growth adaptation under replication stress, both from exogenous or endogenous origin (De Schutter et al., 2007; Takahashi et al., 2008). Here, we show that deficiency in RNase H2 activity reverts the replication stress hypersensitivity of WEE1KO mutants and delimits the cell death phenotype that is caused by premature vascular cell differentiation. WEE1-deficient plants display a smaller root meristem upon replication stress, which is due to a reduction in the number of proliferating cells, resulting in a short root phenotype. We previously demonstrated that a prolonged S-phase underlies this growth phenotype (Cools et al., 2011). Absence of RNase H2A reverts the short root phenotype by restoring meristem size, implying that mutation of the RNase H2 gene restores the replication rate of the WEE1 knockouts.
The TAP experiment revealed that the RNase H2 complex of Arabidopsis comprises three subunits that are homologs of the mammalian and yeast catalytic and regulatory subunits, confirming their importance during evolution. An intriguing observation in the TAP experiment is the pull-down of thymidine kinase and the large subunit of ribonucleotide reductase 1 (RNR1) in the presence of HU. Thymidine kinase supplies an important precursor, deoxythymidine monophosphate, for nucleic acid biosynthesis. RNR catalyzes the formation of dNTPs from rNTPs. As HU limits the available sources of dNTPs for DNA synthesis, it is possible that the association of thymidine kinase and RNR with the RNase H2 complex represents a mechanism to obtain a local enrichment of dNTPs to be used to replace the excised rNTPs.
In contrast to mouse and yeast, knockout of RNase H2 in plants does not result in a slow growth phenotype (Nick McElhinny et al., 2010a; Hiller et al., 2012; Reijns et al., 2012) that has been attributed to the activation of a DNA damage checkpoint, as visualized by the accumulation of γ-H2AX foci. Such foci are also observed in the Arabidopsis RNase H2 mutant plants. The lack of growth arrest in Arabidopsis indicates that the plant DNA damage checkpoints might be less robust or repair more efficiently. Alternatively, the impact of rNMP incorporation in non-plant species might be higher, which would correspond with the limited effect of RNase H2 deficiency on the Arabidopsis transcriptome, whereas the equivalent knockout in yeast results in changes in expression of hundreds of genes (Arana et al., 2012). The few genes being activated in the trd1 mutant appear in a coexpression cluster functionally linked to DNA repair. Correspondingly, an increased HR rate was observed in the trd1-2 background. These results are consistent with observations in yeast in which lack of RNase H2 activity increases the recombination frequency (Ii et al., 2011). Given the accumulation of γ-H2AX foci in S-phase nuclei, single-strand breaks encountered by the replication fork could be converted into DSBs during DNA synthesis. The phenotype of the trd1-2 mus81-1 double mutant supports such a need for increased HR to deal with rNMP incorporation. MUS81 is a conserved endonuclease involved in the resolution of Holliday-like DNA junctions (Mannuss et al., 2010), and the synthetic lethality of trd1 with mus81 indicates that MUS81 is required to resolve the resulting replication intermediates arising in the absence of RNase H2.
The increased HR rate caused by absence of RNase H2 activity cannot account for the observed rescue of the wee1-1 phenotype. Contrary, triple mutant trd1-2 wee1-1 xrcc2-1 plants displayed a stronger recovery phenotype than that seen in trd1-2 wee1-1 under replication stress (Supplemental Figure 6). Similarly, wee1-1 xrcc2-1 and wee1-1 rad51c-1 double mutants tolerated HU better than the single wee1-1 mutant. Possibly, unwinding of double-stranded DNA at a stalled replication fork in the absence of replication results in artificial HR substrates that trigger inaccurate recombinational repair, similar as postulated for plants deficient in DNA polymerase δ activity (Schuermann et al., 2009). Therefore, it is tempting to speculate that amelioration of the wee1-1 and trd1-2 wee1-1 phenotype in the absence of XRCC2 or RAD51C might be due to a decrease in the level of toxic genome rearrangements induced by HR. This suggests that one of the roles of WEE1 upon replication stress is to coordinate replication fork unwinding with replication fork progression. In yeast and mammals, this is accomplished by reducing the activity of the S-phase CDKs through action of the ATR-CHK1 pathway that inhibits CDC25 (Cook, 2009; Willis and Rhind, 2009; Zegerman and Diffley, 2009). Since in plants the CDC25 phosphatase is not present and its antagonist WEE1 has a prominent role in the S-phase (Boudolf et al., 2006; Cools et al., 2011), it is very likely that in plants the inhibition of CDKs during a compromised S-phase is executed by an activation of WEE1. The resulting decrease in CDK activity might help in adjusting the replication pace to the depletion of dNTPs triggered by the treatment with HU. The inability to control the activity of the replication forks probably results in the generation of long strands of ssDNA, as DNA replication halts but unwinding continues, eventually resulting in inappropriate HR substrates (Lopes et al., 2001; Sogo et al., 2002; Branzei and Foiani, 2010). These data are substantiated by the observed increase in the number of γ-H2AX foci in WEE1KO plants in the presence of HU.
If the increased HR rate of the trd1-2 mutants does not account for the rescue of the HU sensitivity of the wee1-1 plants, what does? One possible explanation might be that in wild-type plants, RNase H2-dependent repair, triggered by the incorporation of rNMPs in DNA, might generate some type of DNA structure that induces a WEE1-dependent checkpoint. In this scenario in the absence of RNase H2, these aberrant DNA structures might not occur, reducing the need for the WEE1 kinase. However, this hypothesis appears unlikely, given that wild-type and RNase H2-deficient plants do not display a differential level of WEE1 expression. Moreover, both in the absence and presence HU, no difference in meristem size between wild-type and trd1-2 mutant plants is observed (Supplemental Figure 9), indicating the lack of an RNase H2-dependent cell cycle checkpoint. Rather, we postulate that the ability to tolerate substitution of dNTPs with rNTPs in the RNase H2-deficient mutant facilitates fork progression at a normal pace in WEE1KO plants, thereby limiting the above-mentioned ssDNA that would be prone to produce toxic structures through recombination. Alternatively, unwounded ssDNA on the lagging strand might be stabilized by the continuous presence of RNA primers of Okazaki fragments or hybrids between transcripts and DNA. Incorporating rNMP into the DNA, however, results in a fragile genome, as the reactive 2’-hydroxyl on the ribose ring sensitizes the DNA backbone to cleavage. In budding yeast, rNMP incorporation in the absence of RNase H2 causes the accumulation of 2- to 5-bp deletions within short tandem repeats, which is largely dependent on topoisomerase 1 (TOP1) activity (Nick McElhinny et al., 2010a; Kim et al., 2011; Williams et al., 2013). In general, TOP1 mediates the removal of replication- and transcription-associated supercoils (Wang, 2002). In addition to this function, TOP1 acts as an endonuclease at RNA-DNA junctions (Sekiguchi and Shuman, 1997). Ligation of the generated ends within short directed repeats might cause misalignment of the complementary strands, resulting in the loss of one repeat unit following replication (Williams and Kunkel, 2014). Similarly, we found that over less than three generations, plants lacking a functional RNase H2 complex accumulated slippage mutations, mostly in simple base pair repeats, whereas the number of base substitutions was not affected. The number of small deletions did not increase when plants were grown in the continuous presence of HU, suggesting that the number of rNMPs incorporated is mainly determined by the absence of RNase H2 activity, supported by the observation that the DNA fragmentation pattern was not more pronounced in the HU-treated samples (Figure 7). Interestingly, a large genome duplication was found in the trd1-2 mutant. Although at this stage it cannot be excluded that this duplication might originate from an event independent of the absence of RNase H2 activity, large chromosomal rearrangements have also been observed in RNase H2-defective mouse embryonic fibroblasts (Reijns et al., 2012). These rearrangements have been speculated to result from DNA breaks due to unrepaired rNMPs in DNA (Williams and Kunkel, 2014). Thus, although RNase H2 deficiency allows overcoming a shortage in dNTPs, it increases genome instability and would be expected to affect fitness of the organism over multiple generations.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana plants were grown in vitro vertically under long-day conditions (16 h light/8 h darkness) at 21°C on half-strength Murashige and Skoog medium (2.151 g/L) (Duchefa), 10 g/L sucrose, and 0.5 g/L MES, pH 5.7, adjusted with 1 M KOH and 10 g/L agar. For drug treatments, the HU concentration used was 0.75 mM for direct germination and 1 mM for transfer experiments. The wee1-1, rad51c-1, xrcc2-1, and mus81-1 lines have been described previously (Bleuyard et al., 2005; De Schutter et al., 2007; Hartung et al., 2007). The trd1-2 allele (GABI-139H04) was obtained from the GABI-Kat T-DNA mutant collection (Li et al., 2003), whereas the rnh2b-1 (SAIL_609_A02) and rnh2c-1 (SALK_043851) alleles were obtained from the Salk Institute T-DNA Express database. Genotyping primers are listed in Supplemental Data Set 9.
Ethyl Methanesulfonate Mutagenesis and Mapping
WEE1KO mutant seeds were soaked for 12 h in 0.25% (v/v) ethyl methanesulfonate (Sigma-Aldrich) and then washed two times for 15 min with 0.1 M sodium thiosulfate (Sigma-Aldrich) and two times for 15 min with water, dried, and sown in 200 pools of each 250 seeds. After self-fertilization of the M0, the M1 seeds were grown, and the M2 seeds were collected from individual M1 plants. The M2 plants were screened for restoration of root growth on vertical plates containing HU.
Leaf samples of wee1-1 trd1-1 plants were used for nuclear DNA extraction according to Schneeberger et al. (2009). The leaves were bulked prior to DNA extraction. Illumina True-Seq libraries were generated from extracted DNA according to the manufacturer’s protocol and sequenced on an Illumina HiSeq2000 50-bp single read run. The SHORE pipeline (Ossowski et al., 2008) was used for the alignment to the reference genome (Col-0; TAIR8). Base counts per position and single nucleotide polymorphisms are also determined based on the alignments. The candidate zone is narrowed based on the relative allele frequencies of the two parents (Col-0 and Landsberg erecta) by SHOREmap (Schneeberger et al., 2009).
Confocal Microscopy and Root Growth Measurements
To visualize meristems, root tips were stained for 2 min in a 10 μM propidium iodide solution (Sigma-Aldrich) and were analyzed with either an LSM 510 or LSM 5 exciter confocal microscope (Zeiss). For root growth measurements, the position of the root tips was marked daily on the plate and length of the roots was measured and analyzed by ImageJ.
Tandem Affinity Purification and Liquid Chromatography-Tandem Mass Spectrometry Analysis
Cloning of transgenes encoding tag fusions under control of the constitutive cauliflower mosaic virus 35S promoter and transformation of Arabidopsis cell suspension cultures were performed as previously described (Van Leene et al., 2007). Tandem affinity purification of protein complexes was done using the GS tag (Van Leene et al., 2008) followed by a downscaled purification protocol and liquid chromatography-tandem mass spectrometry analysis on LTQ Orbitrap Velos (Thermo Fisher Scientific) as described by Cuéllar Pérez et al. (2014). A list of nonspecific background proteins was assembled by combining our previous background list (Van Leene et al., 2010) with background proteins from control GS purifications on mock, GFP-GS, and GUS-GS cell culture extracts identified with LTQ Orbitrap Velos. To obtain the final list of interactors, these background proteins were subtracted from the list of identified proteins.
Yeast Two-Hybrid Analysis
Plasmids encoding the bait (pDEST32) and prey (pDEST22) were transformed into the yeast strain PJ69-4α (MATα; trp1-901, leu2-3,112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2::GAL1-HIS3, GAL2-ADE2, met2GAL7-lacZ) and PJ69-4a (MATa; trp1-901, leu2-3,112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2TGAL1-HIS3, GAL2-ADE2, met2TGAL7-lacZ) by the LiAc method (Gietz et al., 1992). Transformed yeast cells were selected on synthetic dextrose plates without Leu (pDEST 32) or without Trp (pDEST22). Interactions between proteins were assayed by the mating method (Bendixen et al., 1994).
RNA Sequencing
The WEE1 knockout (wee1-1) and the RNH2A/WEE1 double knockout (trd1-2 wee1-1) seeds were germinated on control medium on a nylon mesh and transferred 5 d after germination to control medium or medium supplemented with 2 mM HU. Each sample had three independent biological repeats. Twenty-four hours after the transfer, 200 root tips (<2 to 3 mm) were collected and frozen in liquid nitrogen. RNA was extracted from root tissue with the RNeasy Plant Kit (Qiagen). Illumina True-Seq libraries were generated from cDNA according to the manufacturer’s protocol and sequenced on a HiSeq2000. Quality of the reads (Phred quality score) was calculated by FASTQC from Babraham Bioinformatics. After filtering and trimming using a FASTX toolkit (by Assaf Gordon at Cold Spring Harbor Laboratory), reads were aligned to the Arabidopsis genome (TAIR10) using Genomic Short-read Nucleotide Alignment Program (Wu and Nacu, 2010). Tables of counts were produced using the Python software htseq-counts. Afterwards, empirical analysis of gene expression data was calculated and normalized in R environment using edgeR from Bioconductor.
Quantitative RT-PCR
RNA was extracted from root tip tissue with the RNeasy plant kit (Qiagen), and cDNA was prepared from 500 ng of total RNA with the iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer’s instructions. Quantitative RT-PCR was performed with LightCycler 480 SYBR Green I Master (Roche) in a final volume of 5 μL and 0.2 mM primer concentration and analyzed with a LightCycler 480 (Roche). For each reaction, three technical repeats and two to three biological repeats were done. The primer sequences were 5′-CTCTCGTTCCAGAGCTCGCAAAA-3′ and 5′-AAGAACACGCATCCTACGCATCC-3′ for EMB2386 (AT1G02780), 5′-CCGTACCGGGAAAGATAACGAAGA-3′ and 5′-CACTTGAGCCACTTGGTTAGATGC-3′ for AT2G38120, 5′-GGTTTGCCTTGAAGGTTCTTCACC-3′ and 5′-CCATGACCTGAAGGATGAGGATGA-3′ for AT2G38180, 5′-CTTCTCTTCCAGATGCCACTCCTT-3′ and 5′-AAGCTCTCCCTTCAGATGGCGTAT-3′ for AT2G38280, 5′-TGTTTAAATCGCTGCCCAACCTGG-3′ and 5′-CAAAATGCGGGTGGAAGAAGTCATC-3′ for AT5G60250, 5′-TGTACCCCCACGAAGCTCCTAA-3′ and 5′-TGCAGCTGCTTCATGGTTCAGAG-3′ for AT2G18600, 5′-TCCCAAAGGCGGTAAGGCAAAC-3′ and 5′-GTCCGATCACCGCGAGGTATTT-3′ for NAC103 (AT5G64060), 5′-TCCCTCGATTCGCTTCTGGATG-3′ and 5′-AGTCAACCCCGCAACAACGAGA-3′ for TRFL10 (AT5G03780), 5′-ATGGCGTTCTGCTCCTCTGC-3′ and 5′-GGTGCTGTTTTCCCCACACC-3′ for PARP2 (AT4G02390), 5′-TCTCTTTGCAGGATGGGACAAGC-3′ and 5′-AGACTGAGCCGCCTGATTGTTTG-3′ for PAC1 (AT3G22110), and 5′-GACTTTCAAGCGCAGGAATGGTG-3′ and 5′-CCTTGTCCTTGGGGCAACACTTT-3′ for RPS26C (AT3G56340). EMB2386, PAC1, and RPS26C were used as reference genes. Statistical analysis was executed with the Statistical Analysis Software (SAS Enterprise Guide 5.1; SAS Institute) using the mixed model procedure, and P values were Bonferroni adjusted for multiple measurements.
Homologous Recombination Assay
Two recombination substrates, 651 (Swoboda et al., 1994) and IC9C (Molinier et al., 2004), were crossed to trd1-2 wee1, trd1-2, wee1-1, and Col-0. For the HR recombination assay, 50 seeds of each line were germinated on half-strength Murashige and Skoog medium with or without 0.75 mM HU. The restoration of the reporter gene was visualized by histochemical GUS staining according to the standard protocol (Jefferson et al., 1987). HR events of individual plants were assessed visually using a binocular microscope. The HR assays were repeated three times, and the mean values were calculated. Statistical analysis was executed with the Statistical Analysis Software (SAS Enterprise Guide 5.1) using the mixed model procedure, and P values were Bonferroni adjusted for multiple measurements.
Detection of Alkali-Sensitive Sites in Genomic DNA
Genomic DNA was isolated using a DNeasy plant kit (Qiagen). Either KOH or KCl was added to genomic DNA to a final concentration of 0.3 M in 40 μL volumes and incubated at 55°C for 2 h. Following treatment, 6× alkaline loading buffer (300 mM KOH, 6 mM EDTA, 18% Ficoll [Type 400], 0.15% bromocresol green, and 0.25% xylene cyanol) was added to KOH-treated samples. Neutral loading buffer (30% glycerol in TE buffer, 0.25% bromophenol blue, and 0.25% xylene cyanol) was added to KCl-treated samples. Electrophoresis of alkaline-treated samples was performed using a 50 mM NaOH, 1 mM EDTA, and 1% agarose alkaline gel with 50 mM NaOH, 1 mM EDTA electrophoresis buffer (Sambrook et al., 2006). Electrophoresis of KCl-treated samples was performed using a 1% agarose gel and TBE buffer. Electrophoresis of the samples was at 1 V/cm for 18 h. Alkaline gels were neutralized by soaking in 1 M Tris HCl, pH 8.0, and 1.5 M NaCl for 1 h and then stained with SYBR Gold (Invitrogen). Signal intensity per lane was measured with ImageJ and normalized to the total signal intensity per lane. Data were smoothed using a LOESS (Local Regression) algorithm.
Immunostaining Using γ-H2AX Antibodies
Slide preparation, immunostaining, quantification of γ-H2AX foci of mitotic nuclei, and EdU staining were performed as previously described (Amiard et al., 2010).
Whole-Genome Sequencing
Col-0, wee1-1, trd1-2, and trd1-2 wee1-1 plants were grown side-by-side using the Araponics system (http://www.araponics.com) for three generations, either in the absence or continuous presence of 0.75 mM HU. Medium was renewed once a week. We sequenced two genomes for each genotype: one from a plant grown under control conditions and one from a plant grown in the presence of HU (eight genomes in total). Genomic DNA was extracted using a DNeasy plant kit (Qiagen). Paired-end Illumina True-Seq libraries were generated from extracted DNA according to the manufacturer’s protocol and sequenced in multiplexes of four on a HiSeq2000 leading to an average coverage between 43 and 47× per sample. Each genome was sequenced to nearly the same genome-wide sequencing coverage (Supplemental Data Set 10). To estimate the sequencing error rate in each sample, we followed the approach described by Ossowski et al. (2008). Briefly, we used sites covered by at least 10 and <70 reads, in which the consensus base was the same in all eight genomes sequenced, and no more than 20% of reads call a discordant base in any of the sequenced genomes. We subsampled five times 1% of the sites in each genome and estimated error frequencies. We found that error rates were very similar across replicates, indicating no positional bias. Estimated error frequencies were also very similar across genomes. The average error frequency was 0.08% and ranged from 0.076 to 0.086% (Supplemental Data Set 10). The sequencing errors identified showed the same base pair spectrum in all sequenced genomes (paired Mann-Whitney U Test, P value > 0.05). Moreover, there was no indication that single base pair deletions have a higher probability to be erroneously called in any of the genomes, also not when comparing the classes of genomes with TRD1 and without (Mann-Whitney U Test, P value > 0.05). Therefore, the detection of an increased number of mutations in one genome is most likely not due to an increased sequencing error rate for this genome.
To identify newly induced single base pair substitutions, we followed the consensus approach described by Ossowski et al. (2008). Base changes were called if one of the sequenced genomes differed from all others (control genomes). We excluded sites suspected to be noise sources, like repetitive regions. Each site had to be covered by at least five reads in every genome. No control genome was allowed to contain erroneous reads, defined as a subset of reads that report a base different from the majority. If more than one base was reported for one site, the variant base had to be supported by 20% of the reads but at least three reads. Between 88.1 and 88.2 million sites matched those quality criteria in each sequenced genome reflecting a similar sequencing quality. The dependency of the number of genomic deletions on the different mutations (wee1-1 and trd1-2) and treatments (−HU and +HU) was statistically analyzed by applying linear regression on log-transformed count data.
Accession Numbers
RNA sequencing data have been submitted to ArrayExpress (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-2590. Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: WEE1 (At1g02970), TRD1/RNH2A (At2g25100), RNH2B (At4g20325), RNH2C (At2g39440), RAD51C (At2g45280), XRCC2 (At5g64520), MUS81 (At4g30870), MmRNH2A (NP_081463), HsRNH2A (NP_006388), OsTRD1 (NP_001065783), ZmTRD1 (NP_001152248), and ScRNH2A (NP_014327).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Mutations in the Regulatory Subunits of the RNase H2 Complex Partially Rescue the HU Hypersensitivity Phenotype of WEE1KO Plants.
Supplemental Figure 2. The trd1-2 Mutant Holds a Large DNA Duplication.
Supplemental Figure 3. Absence of RNase H2 Activates a DNA Repair Coexpression Cluster.
Supplemental Figure 4. Transcriptional Induction of trd1-2 Differentially Expressed Genes by HU.
Supplemental Figure 5. The trd1-2 Mutant Is Synthetically Lethal in a mus81 Mutant Background.
Supplemental Figure 6. Simultaneous Knockout of XRCC2 and TRD1 Rescues the HU Hypersensitivity Phenotype of WEE1KO Plants Completely.
Supplemental Figure 7. RNase H2 Mutant Plants Accumulate rNMPs in DNA.
Supplemental Figure 8. Confirmation of Small Base Pair Deletions in RNase H2-Deficient Plants Grown in the Presence of HU.
Supplemental Figure 9. Number of Meristematic Cortex Cells in Wild-Type (Col-0) and trd1-2 Mutant Roots.
Supplemental Data Set 1. Protein Identification Details Obtained with the LTQ Orbitrap Velos (Thermo Fisher Scientific) and Mascot Distiller Software (Version 2.4.1; Matrix Science) Combined with the Mascot Search Engine (Version 2.3; Matrix Science) and Database TAIR10.
Supplemental Data Set 2. Proteins Identified in TAP Experiments with as Baits Ribonuclease H2, Subunits A, B, and C, Either without and with HU Treatment.
Supplemental Data Set 3. Differentially Expressed Genes in trd1-2 wee1-1 versus wee1-1 Root Tips under Control Conditions.
Supplemental Data Set 4. Genes Coexpressed with at Least Three Genes Induced in trd1-2 Mutants.
Supplemental Data Set 5. GO Enrichment of Coexpressed Genes.
Supplemental Data Set 6. Differentially Expressed Genes in trd1-2 wee1-1 versus wee1-1 Root Tips upon HU Treatment.
Supplemental Data Set 7. Number of Base Substitutions and Deletions in HU-Treated Control (Col-0), wee1-1, trd1-2, and trd1-2 wee1-1 Plants.
Supplemental Data Set 8. Statistical Analysis of the Variables Linked to Genome Deletions.
Supplemental Data Set 9. List of Primers Used for Genotyping.
Supplemental Data Set 10. Sequencing Summary.
Supplementary Material
Acknowledgments
We thank Annick Bleys for help in preparing the article. This work was supported by grants from the Research Foundation Flanders (G.0C72.14N) and the Interuniversity Attraction Poles Programme (IUAP P7/29 “MARS”), initiated by the Belgian Science Policy Office. T.C. is a Postdoctoral Fellow of the Research Foundation-Flanders.
AUTHOR CONTRIBUTIONS
P.K., Z.H., G.D.J., K.S., C.I.W., and L.D.V. conceived and designed the research. P.K., Z.H., T.C., S.A., N.D.W., and E.-M.W. performed the experiments. P.K., Z.H., T.C., S.A., E.-M.W., G.D.J., K.G., K.S., C.I.W., and L.D.V. analyzed the data and wrote the article. All authors read, revised, and approved the article.
Glossary
- ssDNA
single-stranded DNA
- DSB
double-strand break
- CDK
cyclin-dependent kinase
- HU
hydroxyurea
- RNR
ribonucleotide reductase
- rNTP
ribonucleotide
- dNTP
deoxyribonucleotide
- rNMP
ribonucleoside monophosphate
- Col-0
Columbia-0
- TAP
tandem affinity purification
- HR
homologous recombination
- EdU
ethynyl deoxyuridine
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Amiard, S., Charbonnel, C., Allain, E., Depeiges, A., White, C.I., Gallego, M.E. (2010). Distinct roles of the ATR kinase and the Mre11-Rad50-Nbs1 complex in the maintenance of chromosomal stability in Arabidopsis. Plant Cell 22: 3020–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arana, M.E., Kerns, R.T., Wharey, L., Gerrish, K.E., Bushel, P.R., Kunkel, T.A. (2012). Transcriptional responses to loss of RNase H2 in Saccharomyces cerevisiae. DNA Repair (Amst.) 11: 933–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendixen, C., Gangloff, S., Rothstein, R. (1994). A yeast mating-selection scheme for detection of protein-protein interactions. Nucleic Acids Res. 22: 1778–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleuyard, J.Y., Gallego, M.E., Savigny, F., White, C.I. (2005). Differing requirements for the Arabidopsis Rad51 paralogs in meiosis and DNA repair. Plant J. 41: 533–545 [DOI] [PubMed] [Google Scholar]
- Boudolf, V., Inzé, D., De Veylder, L. (2006). What if higher plants lack a CDC25 phosphatase? Trends Plant Sci. 11: 474–479 [DOI] [PubMed] [Google Scholar]
- Branzei, D., Foiani, M. (2010). Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 11: 208–219 [DOI] [PubMed] [Google Scholar]
- Brown, J.A., Suo, Z. (2011). Unlocking the sugar “steric gate” of DNA polymerases. Biochemistry 50: 1135–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubeck, D., Reijns, M.A., Graham, S.C., Astell, K.R., Jones, E.Y., Jackson, A.P. (2011). PCNA directs type 2 RNase H activity on DNA replication and repair substrates. Nucleic Acids Res. 39: 3652–3666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuéllar Pérez, A., Nagels Durand, A., Vanden Bossche, R., De Clercq, R., Persiau, G., Van Wees, S.C.M., Pieterse, C.M.J., Gevaert, K., De Jaeger, G., Goossens, A., Pauwels, L. (2014). The non-JAZ TIFY protein TIFY8 from Arabidopsis thaliana is a transcriptional repressor. PLoS ONE 9: e84891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerritelli, S.M., Crouch, R.J. (2009). Ribonuclease H: the enzymes in eukaryotes. FEBS J. 276: 1494–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia, A., Elledge, S.J. (2010). The DNA damage response: making it safe to play with knives. Mol. Cell 40: 179–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen, A.R., Zhang, S., Burgers, P.M., Lee, M.Y., Kunkel, T.A. (2013). Ribonucleotide incorporation, proofreading and bypass by human DNA polymerase δ. DNA Repair (Amst.) 12: 121–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, J.G. (2009). Replication licensing and the DNA damage checkpoint. Front Biosci (Landmark Ed) 14: 5013–5030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cools, T., Iantcheva, A., Weimer, A.K., Boens, S., Takahashi, N., Maes, S., Van den Daele, H., Van Isterdael, G., Schnittger, A., De Veylder, L. (2011). The Arabidopsis thaliana checkpoint kinase WEE1 protects against premature vascular differentiation during replication stress. Plant Cell 23: 1435–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan, K., Tissier, A., Britt, A. (2004). ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell 16: 1091–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan, K.M., Robertson, C.E., Foreman, J., Doerner, P., Britt, A.B. (2006). ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J. 48: 947–961 [DOI] [PubMed] [Google Scholar]
- De Schutter, K., Joubès, J., Cools, T., Verkest, A., Corellou, F., Babiychuk, E., Van Der Schueren, E., Beeckman, T., Kushnir, S., Inzé, D., De Veylder, L. (2007). Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. Plant Cell 19: 211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraro, P., Franzolin, E., Pontarin, G., Reichard, P., Bianchi, V. (2010). Quantitation of cellular deoxynucleoside triphosphates. Nucleic Acids Res. 38: e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, V., Bruchet, H., Camescasse, D., Granier, F., Bouchez, D., Tissier, A. (2003). AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell 15: 119–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, D., St Jean, A., Woods, R.A., Schiestl, R.H. (1992). Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, N., Hernould, M., Delmas, F., Gévaudant, F., Duffe, P., Causse, M., Mouras, A., Chevalier, C. (2004). Molecular characterization of a WEE1 gene homologue in tomato (Lycopersicon esculentum Mill.). Plant Mol. Biol. 56: 849–861 [DOI] [PubMed] [Google Scholar]
- Harper, J.W., Elledge, S.J. (2007). The DNA damage response: ten years after. Mol. Cell 28: 739–745 [DOI] [PubMed] [Google Scholar]
- Hartung, F., Suer, S., Puchta, H. (2007). Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 18836–18841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller, B., Achleitner, M., Glage, S., Naumann, R., Behrendt, R., Roers, A. (2012). Mammalian RNase H2 removes ribonucleotides from DNA to maintain genome integrity. J. Exp. Med. 209: 1419–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ii, M., Ii, T., Mironova, L.I., Brill, S.J. (2011). Epistasis analysis between homologous recombination genes in Saccharomyces cerevisiae identifies multiple repair pathways for Sgs1, Mus81-Mms4 and RNase H2. Mutat. Res. 714: 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., Bevan, M.W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, N., Huang, S.N., Williams, J.S., Li, Y.C., Clark, A.B., Cho, J.E., Kunkel, T.A., Pommier, Y., Jinks-Robertson, S. (2011). Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science 332: 1561–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinner, A., Wu, W., Staudt, C., Iliakis, G. (2008). Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 36: 5678–5694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Rosso, M.G., Strizhov, N., Viehoever, P., Weisshaar, B. (2003). GABI-Kat SimpleSearch: a flanking sequence tag (FST) database for the identification of T-DNA insertion mutants in Arabidopsis thaliana. Bioinformatics 19: 1441–1442 [DOI] [PubMed] [Google Scholar]
- Lopes, M., Cotta-Ramusino, C., Pellicioli, A., Liberi, G., Plevani, P., Muzi-Falconi, M., Newlon, C.S., Foiani, M. (2001). The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412: 557–561 [DOI] [PubMed] [Google Scholar]
- Mannuss, A., Dukowic-Schulze, S., Suer, S., Hartung, F., Pacher, M., Puchta, H. (2010). RAD5A, RECQ4A, and MUS81 have specific functions in homologous recombination and define different pathways of DNA repair in Arabidopsis thaliana. Plant Cell 22: 3318–3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyabe, I., Kunkel, T.A., Carr, A.M. (2011). The major roles of DNA polymerases epsilon and delta at the eukaryotic replication fork are evolutionarily conserved. PLoS Genet. 7: e1002407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinier, J., Ries, G., Bonhoeffer, S., Hohn, B. (2004). Interchromatid and interhomolog recombination in Arabidopsis thaliana. Plant Cell 16: 342–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny, S.A., Kumar, D., Clark, A.B., Watt, D.L., Watts, B.E., Lundström, E.B., Johansson, E., Chabes, A., Kunkel, T.A. (2010a). Genome instability due to ribonucleotide incorporation into DNA. Nat. Chem. Biol. 6: 774–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny, S.A., Watts, B.E., Kumar, D., Watt, D.L., Lundström, E.B., Burgers, P.M.J., Johansson, E., Chabes, A., Kunkel, T.A. (2010b). Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. USA 107: 4949–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski, S., Schneeberger, K., Clark, R.M., Lanz, C., Warthmann, N., Weigel, D. (2008). Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 18: 2024–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns, M.A.M., et al. (2012). Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell 149: 1008–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricaud, L., Proux, C., Renou, J.P., Pichon, O., Fochesato, S., Ortet, P., Montané, M.H. (2007). ATM-mediated transcriptional and developmental responses to gamma-rays in Arabidopsis. PLoS ONE 2: e430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rychlik, M.P., Chon, H., Cerritelli, S.M., Klimek, P., Crouch, R.J., Nowotny, M. (2010). Crystal structures of RNase H2 in complex with nucleic acid reveal the mechanism of RNA-DNA junction recognition and cleavage. Mol. Cell 40: 658–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydberg, B., Game, J. (2002). Excision of misincorporated ribonucleotides in DNA by RNase H (type 2) and FEN-1 in cell-free extracts. Proc. Natl. Acad. Sci. USA 99: 16654–16659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Russell, D.W., and Sambrook, J. (2006). The Condensed Protocols from Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press). [Google Scholar]
- Schneeberger, K., Ossowski, S., Lanz, C., Juul, T., Petersen, A.H., Nielsen, K.L., Jørgensen, J.E., Weigel, D., Andersen, S.U. (2009). SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat. Methods 6: 550–551 [DOI] [PubMed] [Google Scholar]
- Schuermann, D., Fritsch, O., Lucht, J.M., Hohn, B. (2009). Replication stress leads to genome instabilities in Arabidopsis DNA polymerase delta mutants. Plant Cell 21: 2700–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi, J., Shuman, S. (1997). Site-specific ribonuclease activity of eukaryotic DNA topoisomerase I. Mol. Cell 1: 89–97 [DOI] [PubMed] [Google Scholar]
- Sirbu, B.M., Couch, F.B., Feigerle, J.T., Bhaskara, S., Hiebert, S.W., Cortez, D. (2011). Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 25: 1320–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo, J.M., Lopes, M., Foiani, M. (2002). Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297: 599–602 [DOI] [PubMed] [Google Scholar]
- Sorrell, D.A., Marchbank, A., McMahon, K., Dickinson, J.R., Rogers, H.J., Francis, D. (2002). A WEE1 homologue from Arabidopsis thaliana. Planta 215: 518–522 [DOI] [PubMed] [Google Scholar]
- Sparks, J.L., Chon, H., Cerritelli, S.M., Kunkel, T.A., Johansson, E., Crouch, R.J., Burgers, P.M. (2012). RNase H2-initiated ribonucleotide excision repair. Mol. Cell 47: 980–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, H., Hausen, P. (1969). Enzyme from calf thymus degrading the RNA moiety of DNA-RNA Hybrids: effect on DNA-dependent RNA polymerase. Science 166: 393–395 [DOI] [PubMed] [Google Scholar]
- Sun, Y., Dilkes, B.P., Zhang, C., Dante, R.A., Carneiro, N.P., Lowe, K.S., Jung, R., Gordon-Kamm, W.J., Larkins, B.A. (1999). Characterization of maize (Zea mays L.) Wee1 and its activity in developing endosperm. Proc. Natl. Acad. Sci. USA 96: 4180–4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swoboda, P., Gal, S., Hohn, B., Puchta, H. (1994). Intrachromosomal homologous recombination in whole plants. EMBO J. 13: 484–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, N., Lammens, T., Boudolf, V., Maes, S., Yoshizumi, T., De Jaeger, G., Witters, E., Inzé, D., De Veylder, L. (2008). The DNA replication checkpoint aids survival of plants deficient in the novel replisome factor ETG1. EMBO J. 27: 1840–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene, J., et al. (2007). A tandem affinity purification-based technology platform to study the cell cycle interactome in Arabidopsis thaliana. Mol. Cell. Proteomics 6: 1226–1238 [DOI] [PubMed] [Google Scholar]
- Van Leene, J., Witters, E., Inzé, D., De Jaeger, G. (2008). Boosting tandem affinity purification of plant protein complexes. Trends Plant Sci. 13: 517–520 [DOI] [PubMed] [Google Scholar]
- Van Leene, J., et al. (2010). Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol. Syst. Biol. 6: 397–, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J.C. (2002). Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 3: 430–440 [DOI] [PubMed] [Google Scholar]
- Williams, J.S., Kunkel, T.A. (2014). Ribonucleotides in DNA: origins, repair and consequences. DNA Repair (Amst.) 19: 27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, J.S., Clausen, A.R., Nick McElhinny, S.A., Watts, B.E., Johansson, E., Kunkel, T.A. (2012). Proofreading of ribonucleotides inserted into DNA by yeast DNA polymerase ɛ. DNA Repair (Amst.) 11: 649–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, J.S., Smith, D.J., Marjavaara, L., Lujan, S.A., Chabes, A., Kunkel, T.A. (2013). Topoisomerase 1-mediated removal of ribonucleotides from nascent leading-strand DNA. Mol. Cell 49: 1010–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis, N., Rhind, N. (2009). Regulation of DNA replication by the S-phase DNA damage checkpoint. Cell Div. 4: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, T.D., Nacu, S. (2010). Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26: 873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman, P., Diffley, J.F.X. (2009). DNA replication as a target of the DNA damage checkpoint. DNA Repair (Amst.) 8: 1077–1088 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.