The accommodation of thousands of nitrogen-fixing rhizobia in legume root nodules requires changes in functionality of the vacuoles of infected cells. Suppression of HOPS tethering complex genes and tonoplast aquaporin retargeting to symbiosomes is part of the underlying mechanism. Defunctionalization of host vacuoles contributes to the expansion and maintenance of intracellular rhizobia.
Abstract
In legume–rhizobia symbioses, the bacteria in infected cells are enclosed in a plant membrane, forming organelle-like compartments called symbiosomes. Symbiosomes remain as individual units and avoid fusion with lytic vacuoles of host cells. We observed changes in the vacuole volume of infected cells and thus hypothesized that microsymbionts may cause modifications in vacuole formation or function. To examine this, we quantified the volumes and surface areas of plant cells, vacuoles, and symbiosomes in root nodules of Medicago truncatula and analyzed the expression and localization of VPS11 and VPS39, members of the HOPS vacuole-tethering complex. During the maturation of symbiosomes to become N2-fixing organelles, a developmental switch occurs and changes in vacuole features are induced. For example, we found that expression of VPS11 and VPS39 in infected cells is suppressed and host cell vacuoles contract, permitting the expansion of symbiosomes. Trafficking of tonoplast-targeted proteins in infected symbiotic cells is also altered, as shown by retargeting of the aquaporin TIP1g from the tonoplast membrane to the symbiosome membrane. This retargeting appears to be essential for the maturation of symbiosomes. We propose that these alterations in the function of the vacuole are key events in the adaptation of the plant cell to host intracellular symbiotic bacteria.
INTRODUCTION
Legumes can establish symbioses with the N2-fixing bacteria that are collectively named rhizobia. The symbiosis leads to the formation of a new organ, the root nodule. Unique in higher plants, the nodule cells contain thousands of bacteria, which are kept in individual membrane compartments provided by the host. The membrane-bound bacterial units are called symbiosomes and show structural similarities to microbes housed in mammalian pathogenic vacuoles (Brumell and Scidmore, 2007; Isberg et al., 2009; von Bargen et al., 2009). However, unlike mammals, legumes have specialized cells that promote intracellular bacteria accommodation, whereas in mammalian tissues such cells do not exist.
In nitrogen-fixing infected cells, symbiosomes do not fuse with the lytic vacuole and remain as individual units within the cytosol. The mechanisms that inhibit this fusion and subsequently enhance lytic clearance in senescing infected cells are unknown. To clarify the mechanisms of symbiotic cell adaptation to intracellular bacteria, we first quantified cell, vacuole, and microsymbiont surface–volume dynamics during nodule development. This showed that vacuole modification plays a crucial role in symbiotic cell progression. We hypothesized that the maintenance of symbiosomes requires a major adjustment of the vacuole formation pathway and tonoplast-targeted trafficking. Therefore, we characterized the vacuoles of host cells during intracellular bacterial accommodation.
We selected for our studies the model legume Medicago truncatula. M. truncatula nodules have a persistent meristem; as a result, the nodule is composed of zones representing subsequent stages of development. The apical part of the nodule consists of the meristem and the infection zone. At this site, bacteria are released from infection threads into the host cell cytoplasm. Upon release, bacteria are surrounded by a host cell–derived membrane to form symbiosomes. The release requires a specific exocytotic pathway (Ivanov et al., 2012), and the symbiosomes continue to share some properties of the plasma membrane during their lifespan (Catalano et al., 2007). After release, rhizobia grow, divide, and gradually colonize the entire host cell.
Next, mature infected cells form in the so-called fixation zone. In these cells, the rhizobial enzyme nitrogenase is induced, allowing the bacteria to reduce atmospheric nitrogen to ammonia, and the bacterial differentiation process is terminated (Vasse et al., 1990; Maagd et al., 1994; Farkas et al., 2014). At later stages of maturation, the symbiosome membrane acquires tonoplast and late endosomal identity markers (Behnia and Munro, 2005), including the small GTPase Rab7 and vacuolar SNAREs (Limpens et al., 2009). Symbiosomes have some vacuolar properties, but they do not fuse with the vacuole in nitrogen-fixing infected cells. To test our hypothesis that the pathway of vacuole formation in infected cells is impaired, we examined the expression and localization of proteins belonging to the tethering complex HOPS (for homotypic fusion and vacuole protein sorting complex). HOPS is the key regulator involved in formation of the vacuole (Nickerson et al., 2009; Balderhaar and Ungermann, 2013). In yeast, the HOPS complex consists of six vacuolar sorting proteins (VPS): VPS11, VPS16, VPS18, VPS33, VPS39, and VPS41. The HOPS complex ensures specificity during the fusion of membranes with the vacuole (Balderhaar and Ungermann, 2013). In plants, HOPS proteins also function in vacuole formation and localize to the tonoplast and prevacuolar compartments. A null mutation of VPS16 causes embryonic lethality in Arabidopsis thaliana (Rojo et al., 2001, 2003).
To test whether default tonoplast-targeted trafficking is compromised in infected cells, we investigated the localization of the vacuolar aquaporin TIP1g. Aquaporins are membrane proteins that facilitate the transport of small molecules such as water, glycerol, and ammonia (Kaldenhoff and Fischer, 2006; Chaumont and Tyerman, 2014). Higher plant aquaporins are subdivided into five main subfamilies: the plasma membrane intrinsic proteins, the tonoplast intrinsic proteins (TIPs), the Nodulin26-like intrinsic proteins, the small basic intrinsic proteins, and the X intrinsic proteins (Forrest and Bhave, 2007; Maurel et al., 2009; Wudick et al., 2009; Hwang et al., 2010).
The transition from infection to nitrogen fixation requires a developmental switch that controls the adaptation of host cells to the accommodation of nitrogen-fixing rhizobia. At this transition, rhizobial nif genes are expressed and the HOPS complex is turned off, a process strictly correlated with the collapse and defunctionalization of vacuoles in the infected cells. As a consequence, the tonoplast aquaporin TIP1g is retargeted toward symbiosomes. This retargeting is in line with the loss of fusion specificity of tonoplast-targeted vesicles due to temporary suppression of HOPS. Retargeting appears to be essential for the functional maturation of symbiosomes and the maintenance of turgor pressure in the infected cells. Our study shows that a major adjustment in vacuole formation and tonoplast targeting occurs during the development of the infected cells, and we discuss how this contributes to the mechanisms by which symbiosomes are maintained in infected cells of root nodules.
RESULTS
Host Cell Architecture during the Development of Infected Cells
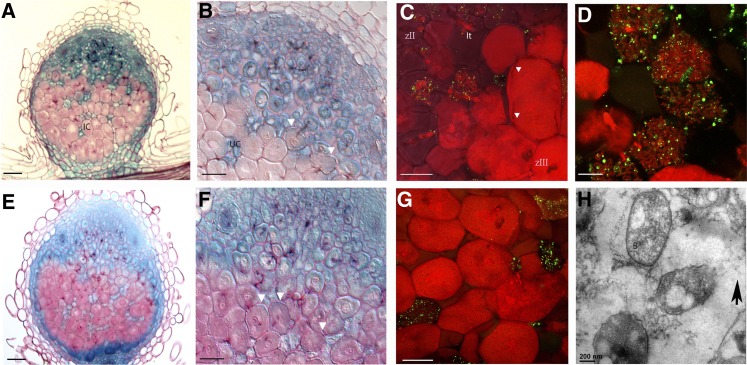
To determine the adaptation of infected cells to accommodate intracellular rhizobia, we quantified the volume and surface area of cells, symbiosomes, and vacuoles. For this, we used confocal microscopy and 3D reconstruction in combination with Imaris 7.5 software to image transgenic roots expressing green fluorescent protein (GFP)–tagged SYP22 (vacuolar SNARE), which specifically labels the vacuoles (Limpens et al., 2009). Nodules were stained with propidium iodide (PI) to contrast the cell wall, nuclei, and symbiosomes. Z-stacks obtained by confocal microscopy were used for 3D reconstruction and quantification of the volumes occupied by vacuoles and symbiosomes. Infected and noninfected cells were analyzed in subsequent developmental stages. An overview of nodule zones and the developmental stages of symbiosomes is provided as a cartoon in Figure 1A and as light microscopy images in Figures 1B to 1E. Infected and noninfected cells were analyzed at four stages of development: (1) young (distal) cells of the infection zone (Figures 1A to 1C and 1F); (2) the most proximal cell layer of the infection zone; (3) the (adjacent) first cell layer of the fixation zone (interzone 2/3 according to Vasse et al., 1990) (Figures 1A, 1B, 1D, and 1G); and (4) more mature infected cells of the fixation zone (Figures 1A, 1B, 1E, and 1H). For 3D reconstruction, Z-stacks covering whole cells were obtained for at least eight infected/noninfected cells at each selected developmental stage (Figures 1F to 1H). Figure 1I shows an example of 3D reconstructions of infected cells.
Figure 1.
Nodule Zonation, Stages of Symbiosome Development, and 3D Reconstructed Cells of M. truncatula Nodules.
(A) Scheme of the nodule zones: M, meristem; zII, infection zone; zIII, zone of nitrogen fixation. The stages of symbiotic cells in development are shown next to each other (1 to 4). Zones described here correspond to the light microscopy image of the nodule
(B) The arrow points to the zone of transition between the infection and fixation zones. The vacuoles of infected cells in this zone are highlighted by red. Here, the arrows point to the cells in consequent stages of development given in high-magnification images ([C] to [E]) and as examples of 3D reconstruction ([F] to [H]).
(C) and (F) Cells from the zone of bacteria release.
(D) and (G) The transition zone (interzone 2/3). It is recognized by the appearance of starch grains.
(E) and (H) Mature infected cells. Flaccid vacuoles of a mature symbiotic cell have a “scalloped” surface, whereas vacuoles of an uninfected cell are roundish and turgid.
(I) An example of Imaris rendering based on a confocal Z-stack of a GFP-SYP22–expressing nodule. Frames show the reconstructed images of vacuoles from the neighboring cells. Samples were contrasted by PI (red).
IC, infected cell; It, infection thread; s, symbiosomes; SG, starch grains; UC, uninfected cell; Uv, vacuole of uninfected cell; v, vacuole. Bar in (B) = 100 µm; bar in (C) = 25 µm; bars in (D) and (E) = 75 µm; bar in (H) = 12.5 µm; bar in (F) = 5 µm; bar in (G) = 10 µm; bar in (I) = 50 µm.
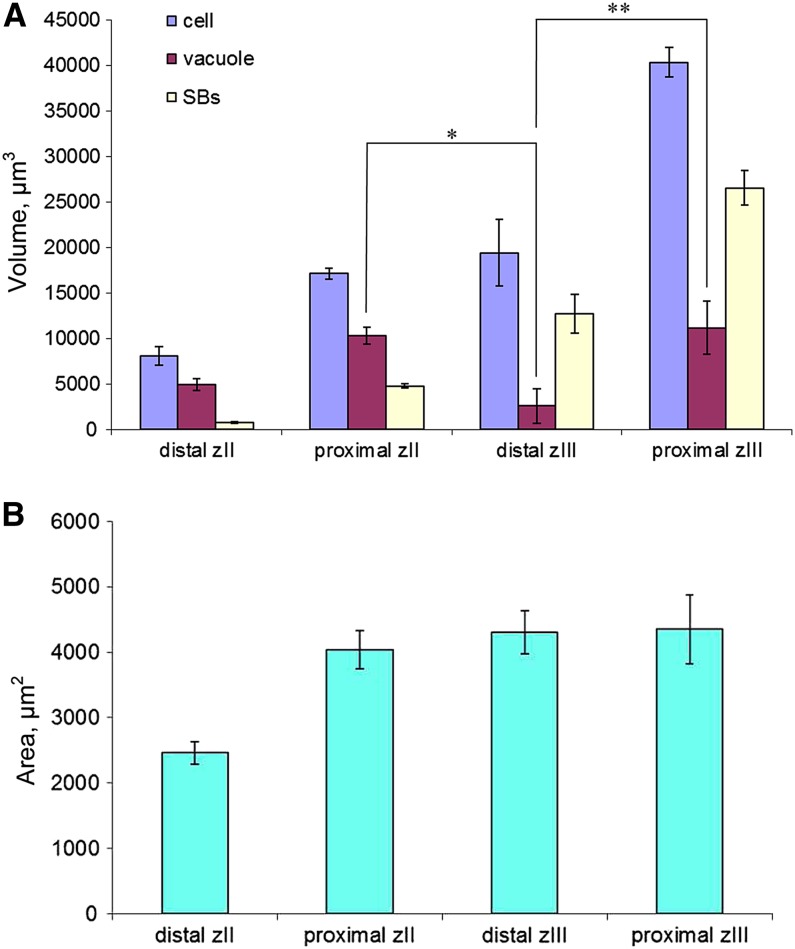
Young infected cells of the infection zone are relatively small, compared with mature cells, and ∼65% of their cell volume is occupied by vacuoles. The vacuole size is comparable with that of noninfected cells (Figures 1C, 1F, and 2A). In the most proximal (oldest) layer of the infection zone, cells have increased in size, with ∼60% of the cell volume occupied by the vacuole and 30% by the symbiosomes. In the adjacent cell layer, the first cell layer of the nitrogen-fixing zone, the vacuoles of infected cells collapse as their volume decreases (Figures 1D, 1G, and 2A). The infected cells have only slightly increased in size in this transition layer. By contrast, the absolute volume of the vacuoles was reduced 4-fold and the absolute volume of symbiosomes increased 2.5-fold to occupy ∼65% of the cell volume (Figure 2A). Hence, the absolute volume of the vacuoles in this zone decreased and the total volume of microsymbiont increased.
Figure 2.
Quantification of Cell, Vacuole, and Symbiosome Volume and Vacuole Surface Area from Imaris 3D Reconstructed Z-Stacks of Infected Cells.
(A) Dynamics of vacuole, symbiosome (SBs), and cell volume. *P < 0.001 and **P < 0.05.
(B) Dynamics of vacuole surface area.
Error bars indicate sd calculated from the data sets
[See online article for color version of this figure.]
In the cell layers after the termination of microsymbiont expansion, infected cells have doubled in size. The vacuoles are again clearly present; hence, the vacuole volume returns to the preinfection level. However, vacuoles remain flaccid in the mature infected cells of the fixation zone (Figures 1E and 1H). The change in vacuole volume suggests that during the transition from infection to fixation zone the vacuoles are maintained and only their volume has been reduced (Figures 1D and 1E). To determine whether this is the case, we quantified the surface of the vacuoles using Imaris 7.5 software (Reisen et al., 2005). This showed that the surface of the tonoplast indeed remained similar, whereas the volume of vacuoles decreased 4-fold (Figure 2B). This experiment was repeated twice, and both experiments showed a similar change in the vacuole volume in infected cells. The total vacuole volume in mature infected cells is ∼30% of total cell volume, whereas for plant cells it is normally ∼80 to 90% (Reisen et al., 2005). This change in vacuole morphology prompted us to determine whether other vacuolar properties are also modified.
Vacuoles of Infected Cells Are Not Acidic
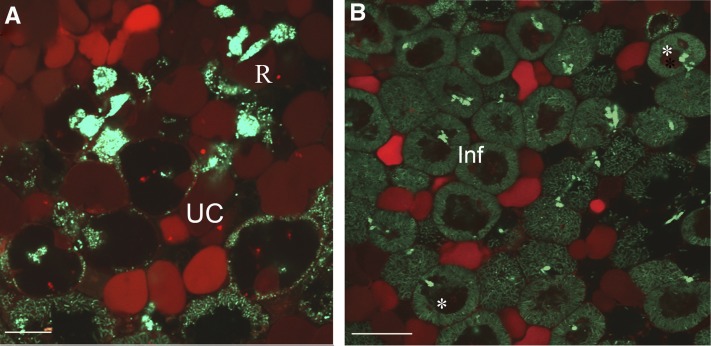
Functional lytic vacuoles have a characteristic acidic pH. To determine the pH in infected cells, we stained them with Neutral Red (NR), a supravital stain and pH marker. In its unprotonated form, NR penetrates the plasma membrane and tonoplast and accumulates in acidic compartments, resulting in red staining of these compartments. However, the detection of NR staining in root nodules by light microscopy (Fedorova and Brown, 2007; Sujkowska et al., 2011) has disadvantages, due to the multilayer structure of nodule sections. Therefore, hand sections of the nodules were analyzed by confocal microscopy, which permits the observation of a single cell layer and shows the cell zones with different pH. NR accumulated in the vacuoles of noninfected cells but not in vacuoles of infected cells (Figures 3A and 3B). To verify this, we stained two different sets of nodules (n = 10 to 12 nodules per experiment), obtaining the same result for each. Our results indicate that the pH of infected cell vacuoles is not acidic. We suggest that this nonacidic vacuolar pH is already established in cells where bacteria are released from the infection thread and is maintained throughout the development of infected cells, whereas vacuoles of neighboring uninfected cells remain acidic. Vacuoles in infected cells often contained some rhizobia. In nodules stained with NR, the bacteria had red fluorescence, most likely due to the dye sticking to dead bacteria (see Discussion). This strongly suggests that NR enters the vacuoles of infected cells but does not change color because the pH of these vacuoles is not acidic. The difference in pH status between noninfected and infected cells was further verified by using the ratiometric probe LysoSensor Yellow/Blue DND-160 (Han and Burgess, 2010). In its acidic form, this dye fluoresces brightly yellow and in neutral pH the fluorescence becomes blue. The nodules were hand-sectioned, immediately loaded with LysoSensor Yellow/Blue, and observed. Vacuoles of noninfected cells emitted fluorescence corresponding to an acidic pH 5 to 5.5 according to the calibration (Supplemental Figures 1A and 1B). By contrast, vacuoles of infected cells emitted fluorescence corresponding to neutral pH (Supplemental Figures 1A and 1B). This analysis was repeated twice with 10 to 12 nodules each time. Twenty-five infected and 25 uninfected cells from zone III of wild-type nodules were analyzed for each experiment.
Figure 3.
NR Staining to Determine Vacuolar pH in Nodule Cells.
(A) Infection zone.
(B) Fixation zone.
Confocal images display the red color of the acidotrophic dye showing acid compartments–vacuoles. Symbiosomes were counterstained by Sytox Green. inf, infected cells; R, bacteria release; UC uninfected cells. Asterisks indicate dead bacteria stained by NR inside the vacuole lumen. Bar in (A) = 20 µm; bar in (B) = 50 µm.
The HOPS Complex Is Temporarily Repressed in Young Nodule Infected Cells of the Fixation Zone
The HOPS complex is essential for the tethering and fusion of vesicles to vacuoles, a process that ensures the specificity of fusion (Balderhaar and Ungermann, 2013). To determine the role of the HOPS complex in symbiotic cell development, two M. truncatula genes encoding subunits of the HOPS complex, VPS11 and VPS39, were analyzed.
The promoter regions (2.5 kb upstream of the translation start) of VPS11 and VPS39 were fused to β-glucuronidase (GUS) and introduced into M. truncatula roots by Agrobacterium rhizogenes–mediated transformation. Analysis of GUS staining in transgenic root nodules expressing ProVPS11:GUS (Figures 4A and 4B) or ProVPS39:GUS (Figures 4E and 4F) showed that these promoters were active in the meristem, in cells of the peripheral tissues, and in the infection zone. Surprisingly, the promoters were not expressed in the infected cells at the transition of the infection to the fixation zone (interzone 2/3 according to Vasse et al., 1990) but were expressed in the neighboring noninfected cells (Figures 4A and 4E).
Figure 4.
Expression of ProVPS11:GUS and ProVPS39:GUS in Young (14-DAI) Nodules and the Localization of GFP-VPS11 and GFP-VPS39 in Mature and Senescent Infected Cells Undergoing Lysis.
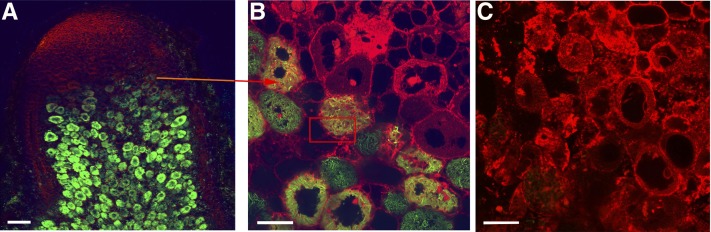
(A) and (B) GUS staining shows the expression of VPS11 in all cells of the apical part of nodules and only in uninfected cells in the zone of nitrogen fixation.
(C) and (G) GFP-VPS labels young infected cells during bacteria release and only noninfected cells in the fixation zone.
(D) and (H) VPS11 appears on symbiosomes in senescent infected cells where symbiosomes are scattered in a disintegrated cytoplasm, a bacteria in a vacuole-like structure formed by the fusion of senescent symbiosomes. VPS signal is marked by 15-nm gold particles in (H).
(E) and (F) GUS staining shows the expression of VPS39 in all cells of the apical part of nodules and only in uninfected cells in the zone of nitrogen fixation.
Membranes are contrasted by staining with FM4-64 in (C), (D), and (G). B, bacteroid; IC, infected cell; It, infection thread; UC, uninfected cell; zII, infection zone; zIII, fixation zone. Arrowheads indicate starch grains of infected cells in the first cell layer of the fixation zone. Bars in (A) and (D) = 75 µm; bars in (B) and (E) = 25 µm; bars in (C), (D), and (G) = 5 µm; bar in (H) = 200 nm.
To check whether the repression of the HOPS complex is specific to the legume–rhizobia symbiosis, we inoculated the transgenic roots expressing ProVPS11:GUS or ProVPS39:GUS with Glomus intraradices, fungi that form arbuscular mycorrhizal symbiosis in the roots. VPS11 and VPS39 promoters were active in the inner root cortical cells where the fungus forms the arbuscules (Supplemental Figures 2A and 2B). The vacuoles in the cells, containing arbuscules, also retained their acidic pH (Supplemental Figure 2C). Hence, we conclude that the suppression of HOPS does not occur in the mycorrhizal interaction.
The repression of VPS genes is rather unusual, as the absence of HOPS is generally lethal (Rojo et al., 2001, 2003). Therefore, we analyzed the distribution of VPS11 and VPS39 proteins in nodules. We used nodules formed on transgenic roots expressing ProVPS11:GFP-VPS11 and ProVPS39:GFP-VPS39 harvested 14 and 35 d after inoculation (DAI). As VPS proteins are not abundant in the cells, the analysis required enhancement of the GFP signal with anti-GFP antiserum. Hand sections of transgenic nodules were analyzed by confocal microscopy. At 14 DAI, nodules showed GFP-VPS signal in endosomes and the tonoplast of cells in the zone where bacteria are released. However, the labeling was not detected in infected cells of the fixation zone, while neighboring uninfected cells displayed the signal. VPS proteins were absent or their level was drastically reduced in all infected cells of the fixation zone (Figures 4C and 4G). This suggests that the maintenance of symbiosomes in the fixation zone correlates with the temporary repression of the HOPS tethering complex in infected cells.
In senescing nodule cells, lytic compartments form de novo (Dupont et al., 2012). This likely requires a HOPS complex. Confocal and immunogold labeling electron microscopy (EM) of senescing nodules at 35 DAI showed that the GFP-VPS proteins indeed accumulate in infected cells undergoing lysis (Figures 4D and 4H). Reappearance of the VPS proteins in infected cells during senescence implies that the fusion of symbiosomes and the formation of lytic vacuole–like structures, typical for nodule senescence, require the presence of a functional HOPS tethering complex.
To verify the correct localization of GFP-VPS proteins, we examined their colocalization with VTI11, a member of the vacuolar SNAREpin complex, in ProVPS11:GFP-VPS11 and ProVPS39:GFP-VPS39 transgenic roots and nodules. VTI11 is involved in membrane fusion to the tonoplast and interaction with the HOPS complex (Balderhaar and Ungermann, 2013). Using an M. truncatula–specific anti-VTI11 antibody (Limpens et al., 2009), we showed that VTI11 was generally colocalized with GFP-tagged VPS proteins (Supplemental Figures 3A and 3B), confirming that GFP-VPS–tagged proteins maintain their correct localization in ProVPS11:GFP-Vps11 and ProVPS39:GFP-VPS39 transgenic roots and nodules.
To validate the localization of VPS11 on the tonoplast, we used a construct where GFP-VPS11 was expressed under the control of the Ubiquitin3 promoter (ProUBQ3). The strong expression under the control of the UBQ3 promoter permitted clear visualization of GFP-VPS11 on the tonoplast membrane (Supplemental Figure 3C).
Functional Analysis of VPS Proteins
RNA interference (RNAi) was used to analyze the function of VPS. Initially, the cauliflower mosaic virus 35S promoter (Pro35S) was used to direct expression of the constructs, but transgenic roots expressing either Pro35S:RNAi-VPS39 or Pro35S:RNAi-VPS11 were not obtained. This suggested that the VPS proteins are essential for root growth and development, confirming previous studies (Rojo et al., 2001). Therefore, we used the promoter of Nodulin E12 (ProE12), which is active only in nodule primordia and in the youngest part of the infection zone of the nodule (Vijn et al., 1995), to express the RNAi constructs. The level of silencing, as measured by quantitative RT-PCR (qRT-PCR) is shown in Supplemental Figures 4A and 4B.
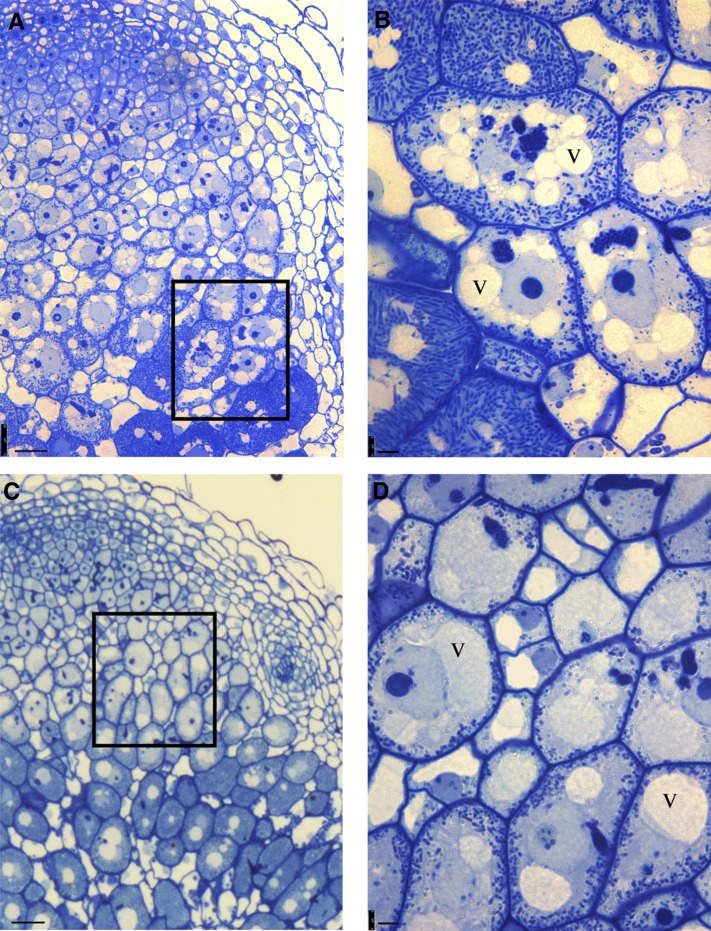
Nodules from ProE12:RNAi-VPS11 and ProE12:RNAi-VPS39 transgenic roots showed a higher number of small nonfused vacuoles (8.12 ± 3.8 across three sections from individual ProE12:RNAi-VPS39 nodules; Figures 5A and 5B) relative to empty vector controls (2.40 ± 1.21 [P < 0.005]; Figures 5C and 5D). In two separate experiments, where a total of 20 random nodules were examined, the vacuole phenotype was detected in nine nodules. The significant increase of nonfused vacuole numbers supports the hypothesis that the identified VPS genes encode a part of a HOPS tethering complex that controls homotypic and heterotypic fusion of vacuoles. This raised the question of whether vesicles normally targeted to the tonoplast are mistargeted in infected cells or, alternatively, the specificity of their fusion is lost. To test this, we selected and used a tonoplast aquaporin as a marker for vesicle targeting.
Figure 5.
Silencing of VPS39 Disturbs the Formation of the Central Vacuole in the Infected Cell.
(A) In the zone of infection, infected cells have numerous small vacuoles.
(B) Magnification of (A).
(C) and (D) Empty vector control.
v, vacuole. Bars in (A) and (C) = 25 µm; bars in (B) and (D) = 5 µm.
[See online article for color version of this figure.]
MtTIP1g as a Membrane Marker for Vacuolar Protein Targeting in Infected Cells
TIPs have been used as vacuolar markers in a variety of plant species (Gattolin et al., 2009). M. truncatula TIPs were identified based on homology with Arabidopsis TIPs. Seven homologs were found in the available genomic and cDNA sequences. As shown in Supplemental Figure 5A, all of them cluster with Arabidopsis TIPs in a phylogenetic analysis (Johanson et al., 2001; Tamura et al., 2011).
The predicted amino acid sequences of M. truncatula TIPs were aligned and compared with aquaporins that previously had been functionally analyzed. The M. truncatula TIPs share with these aquaporins six α-helical transmembrane domains and five interhelical loops (predicted by TMHMM; http://www.cbs.dtu.dk/services/TMHMM/) and two highly conserved NPA (for Asn-Pro-Ala) motifs (Forrest and Bhave, 2007) (Supplemental Figure 5B). To select an M. truncatula TIP for the study, we first determined the expression of TIP genes in nodules (14 DAI) compared with roots using qRT-PCR analysis (Supplemental Figure 6). Most examined TIP genes had low expression in nodules, with the exception of TIP1g, which displayed a high level of expression in nodules. ClustalW alignments against other TIP aquaporins suggest that TIP1g is most likely a tonoplast intrinsic protein. This conclusion was supported by the functional characterization of TIP1g in Xenopus laevis oocytes.
Water permeability by TIP1g was measured using an X. laevis oocyte heterologous expression system. We confirmed the localization of TIP1g to the oocyte plasma membrane by injecting the oocytes with a yellow fluorescent protein (YFP)–tagged version of TIP1g (Supplemental Figure 7). Water permeability was measured by incubating oocytes in a 5-fold-diluted ND96 solution (osmolarity of 47 mosmol/kg). Oocyte volume change was derived from images captured at 3-s intervals for 2 min using a dissecting microscope. The value of the water permeability coefficient for TIP1g-expressing oocytes was 0.00413 ± 0.00125 cm/s, while water-injected and AQP1-expressing oocytes exhibited water permeability coefficients of 0.00096 ± 0.00093 and 0.00878 ± 0.0023 cm/s, respectively. On average, 10 oocytes were tested for each trial (Supplemental Figure 8). These results suggest that TIP1g is a functional water transporter. Whole-oocyte current measurements revealed that TIP1g does not transport malate or ammonia (Supplemental Figures 9A and 9B).
To determine whether TIP1g is targeted to the tonoplast, we analyzed its subcellular localization in young nodule cells. TIP1g was fused to the N terminus of GFP and expressed under the control of either the ProUBQ3 or Leghemoglobin (ProLB) promoter. Confocal microscopy of ProUBQ3:GFP-TIP1g–expressing nodules showed that it is located on the tonoplast (Figure 6A). Under the control of the ProLB promoter, GFP-TIP1g signal was found on the tonoplast and symbiosomes of infected nodule cells (Figure 6B). Collectively, these data show that TIP1g can be used as a marker to study the targeting of tonoplast-residing proteins as well as those localized to the symbiosome membrane.
Figure 6.
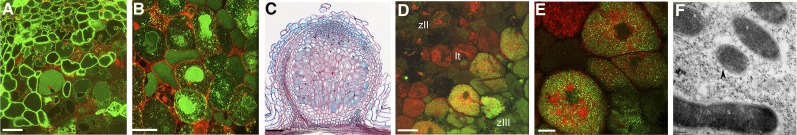
Localization of TIP1g.
(A) Confocal image of ProUBQ3:GFP-TIP1g–expressing nodules. GFP-TIP1g labels the tonoplast of the apical nodule cells.
(B) Confocal image of ProLB:GFP-TIP1g–expressing nodules. GFP-TIP1g labels the tonoplast and symbiosome membrane in infected cells of the fixation zone.
(C) TIP1g promoter activity. Histochemical GUS staining is observed throughout the developing and fixing nodule zones on young 14-DAI nodules.
(D) Confocal image of the apical part of a ProTIP1g:GFP-TIP1g–expressing nodule. GFP-TIP1g labels the symbiosome membrane from the first cell layer of the fixation zone.
(E) Magnification of the first cell layer of the fixation zone.
(F) EM immunogold labeling with anti-GFP antibody. Signal (arrowhead) is present over the symbiosome membrane. The immunogold labeling has been quantified from five different frames, each containing 6 to 8 symbiosomes (37 symbiosomes in total), and 85% of the symbiosomes showed the labeling.
Inf, infected cell; It, infection thread; zII, infection zone; zIII, fixation zone. Bars in (A), (B), and (D) = 20 µm; bar in (C) = 75 µm; bar in (E) = 5 µm; bar in (F) = 200 nm.
TIP1g Is Retargeted to the Symbiosome Membrane
To examine the targeting of TIP1g in infected cells of M. truncatula nodules, a new construct was made, with the GFP-TIP1g fusion driven by the TIP1g promoter (ProTIP1g). ProTIP1g:GUS expression analysis showed that this promoter is active in all zones of the nodule, including the fixation zone (Figure 6C). Anti-GFP antibodies were used to enhance the GFP-TIP1g signal. Confocal microscopy of transgenic nodules expressing ProTIP1g:GFP-TIP1g showed that TIP1g is located in the tonoplast of infected and noninfected cells in the infection zone, although the level of signal was not high (Figure 6D). The signal of GFP-TIP1g was abundant in infected cells of the fixation zone (Figures 6D and 6E). In these cells, it appeared that TIP1g is localized on the symbiosome membrane, indicating that this protein might be retargeted toward the symbiosomes from its default route to the tonoplast. EM and immunogold labeling of ProTIP1g:GFP-TIP1g–expressing nodules confirmed that GFP-TIP1g is present on the symbiosome membrane (Figure 6F). According to the estimation from five EM frames, 80% of the symbiosomes showed GFP labeling.
Silencing of TIP1g Affects Symbiosome Development
Since TIP1g is targeted to the symbiosome, we investigated whether this is essential for symbiosome maturation. RNAi constructs under the control of either the Pro35S or ProLB promoter were introduced into M. truncatula hairy roots. These promoters are active in different zones of the nodule. Pro35S is active in the meristem and in the proximal infection zone, and ProLB is most active in the fixation zone, which contains mature, N2-fixing symbiosomes, although expression already begins in the distal part of the infection zone (Yang et al., 1991).
To check whether the partial silencing of TIP1g affects nitrogen fixation in symbiosomes, transgenic roots were inoculated with a Sinorhizobium meliloti line expressing GFP under the control of the nifH promoter (nifH:GFP). In nodules that are not fixing nitrogen, the rhizobial nifH gene is not induced, allowing discrimination between Fix+ and Fix− nodules on ProLB:RNAi-TIP1g transgenic roots by analysis of GFP fluorescence in infected cells (Supplemental Figure 10). The Fix+ and Fix− nodules (14 DAI) from transgenic ProLB:RNAi-TIP1g roots were harvested separately and analyzed by confocal and light microscopy. In control nodules, the expression of nifH:GFP was clearly detectable (Figure 7A) at the transition between the zone of infection and the zone of nitrogen fixation (interzone 2/3) (Vasse et al., 1990). The first cell layer of the fixation zone can be distinguished by the appearance of starch grains on the periphery of the infection cells (Figure 7B). Previously, this zone was named interzone 2/3, as it was presumed that symbiosomes of these cell layers were not able to fix nitrogen (Vasse et al., 1990). However, as nifH is specifically induced at this transition, this shows that, functionally, it belongs to the fixation zone (Figures 7A and 7B). Fix+ ProLB:RNAi-TIP1g transgenic nodules showed an identical pattern to the control nodules. However, in Fix− transgenic nodules, the zone of nitrogen fixation did not develop, nifH:GFP was not detectable, and the zone of transition, which contains undeveloped symbiosomes, expanded to four to six cell layers (Figure 7C). The Fix+ and Fix− transgenic nodules were harvested separately, and the expression level of TIP1g was estimated by qRT-PCR (Supplemental Figure 11). In Fix− nodules, TIP1g expression was 5% of that in control nodules, while Fix+ nodules have expression similar to that in control nodules (Supplemental Figure 11).
Figure 7.
nifH Expression Is Not Induced in ProLB:RNAi-TIP1g Transgenic Nodules.
(A) and (B) nifH expression is switched on in the first cell layer of the fixation zone in control nodules. (B) is a magnification of (A); the rectangle highlights starch grains in the infected cell of the first cell layer of the fixation zone.
(C) In ProLB:RNAi-TIP1g–expressing nodules, the zone of nitrogen fixation is not developed, such that the zone of transition, which contains small undeveloped symbiosomes, is expanded to four to six cell layers.
Hand sections of M. truncatula nodules formed by S. meliloti (nifH:GFP) were counterstained by FM4-64. zII, infection zone; zIII, fixation zone. Bar in (A) = 100 µm; bars in (B) and (C) = 20 µm.
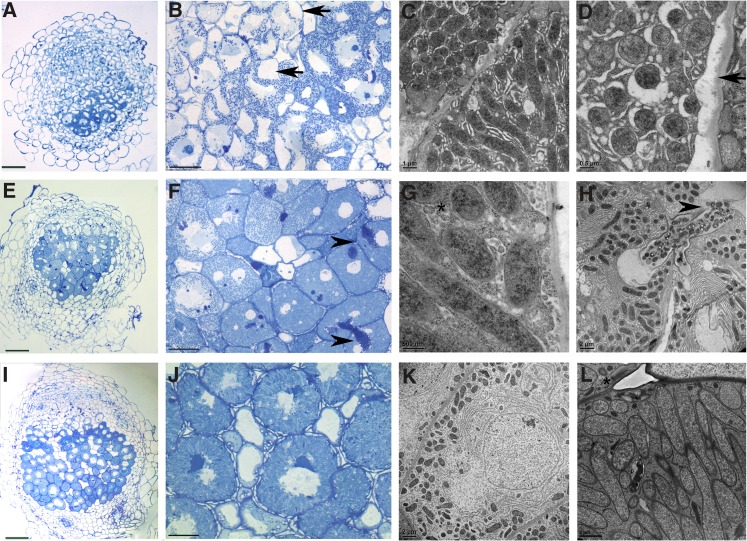
RNAi-TIP1g transgenic nodules (14 DAI) were analyzed by light microscopy. No effect on nodule development was observed in Fix+ ProLB:RNAi-TIP1g and Pro35S:RNAi-TIP1g transgenic root nodules. However, each Fix− nodule (12 nodules analyzed) from ProLB:RNAi-TIP1g–expressing roots displayed an interruption in the development of symbiosomes: maturation of symbiosomes from elongation stage 3 to nitrogen-fixing stage 4 (classification of Vasse et al., 1990) did not occur (Figures 8A, 8B, 8E, and 8F). This suggests that TIP1g is required specifically for symbiosome maturation between stages 3 and 4. In some nodules, small vacuoles in the nitrogen fixation zone were observed (Figure 8F). Sections of the nodules also displayed more pronounced defects, including the distortion of plant cell turgor and the detachment of the plasma membrane from the cell wall of symbiotic cells. These differences in phenotype may reflect the level of silencing of TIP1g (Figure 8B).
Figure 8.
TIP1g Is Required for Symbiosome Development and Infected Cell Turgidity.
(A), (B), (E), and (F) Light microscopy of ProLB:RNAi-TIP1g 14-DAI nodules. Symbiosome development in transgenic nodules did not proceed further than stage 3. Detachment of the plasma membrane from the cell wall in infected cells is indicated by arrows. Secondary release of bacteria in infected cells is indicated by arrowheads.
(C), (D), (G), and (H) Electron microscopy of ProLB:RNAi-TIP1g 14-DAI nodules. Symbiosomes reached only developmental stage 2 or 3, and cells showed extreme ER proliferation (C). Detachment of plasma membrane formed a gap between the plasma membrane and the cell wall (D). The asterisk indicates autophagic bodies containing small parts of the cytoplasm (G). Secondary release of rhizobia into already populated cells is indicated in (H).
(I) and (J) Light microscopy of empty vector control 14-DAI nodules.
(K) and (L) EM of empty vector control 14-DAI nodules. A young infected cell populated by symbiosomes in stage 2 to 3 is shown in (K), and mature radially aligned symbiosomes are shown in (L). The asterisk indicates starch grains.
Bars in (A), (E), and (I) = 75 µm; bars in (B), (F), and (J) = 25 µm; bar in (C) = 1 µm; bars in (D) and (G) = 0.5 µm; bars in (H), (K), and (L) = 2 µm.
[See online article for color version of this figure.]
EM analysis of the effects of TIP1g silencing on nodule ultrastructure showed that symbiosomes from Fix− ProLB:RNAi-TIP1g transgenic root nodules reached only stage 2 or 3 in their maturation (Figures 8C and 8D), even in nodules where plasma membrane–cell wall detachments were not observed by light microscopy (Figures 8G and 8H). In most symbiotic cells, the proliferation of endoplasmic reticulum (ER) occurred and the ER lumen widened. Structures similar to autophagosomes were present in symbiotic cells (Figure 8G). These autophagosomes were situated peripherally in the host cell cytoplasm, forming a vacuole-like structure adjacent to the cell wall. ER membranes encased symbiosomes and parts of host cell cytoplasm. We consider that in these cells, the ER is involved in the formation of membranes to isolate the autophagic bodies, as was shown recently for animal cell omegasomes (Li et al., 2012). Some infected cells displayed an electron-dense cytoplasm that indicates premature senescence. In some cells, “secondary” bacterial release occurred, with rhizobia entering already populated cells, a process that may be facilitated by low turgor pressure and gaps between the cell wall and the plasma membrane. Bacteria released into these cells have to be considered saprophytic and not symbiotic, as the cell was already undergoing lytic clearance (Figure 8H). Since symbiosomes in the RNAi nodules did not reach functional maturity as in control nodules (Figures 8I to 8L), they were not able to fix atmospheric nitrogen and the symbiosis appears to be terminated prematurely.
DISCUSSION
We report here that host cell architecture and vacuole formation are modified in nodule infected cells. In root nodules in the transition from the zone of infection to the zone of nitrogen fixation, several vacuolar changes occur. The host cell vacuole volume is reduced by 75%, and this is accompanied by tonoplast folding. The HOPS complex in infected cells is temporarily repressed, and the integral tonoplast aquaporin TIP1g accumulates at relatively high levels and is retargeted to the symbiosomes. The collapse of the vacuoles in this transition may permit the expansion of bacteria in the host cell cytoplasm at the expense of vacuole volume.
The cell volume change related to the modulation of the vacuole volume is not uncommon for plant cells. This process is best studied in guard cells. Stomatal opening is partly caused by vacuolar convolution (Zouhar and Rojo, 2009; Bak et al., 2013). The conditions that induce stomatal closing result in fragmentation of the central vacuole and loss of guard cell turgor. Loss of vacuole volume and tonoplast folding have also been described in cells exposed to hyperosmotic stress (Reisen et al., 2005; Brett and Merz, 2008). Hence, cell architecture remodeling, which is mediated by the change in volume, may be a common reaction during plant development. Vermeer et al. (2014) showed that volume loss in endodermal cells, and vacuole remodeling via fragmentation, take place during lateral root formation in Arabidopsis. They considered this to be a coordinated response reflecting the mechanical stresses at neighboring cell layers, which might affect tissue and organ patterning.
The collapse of the vacuole in the transition zone of M. truncatula root nodules is most likely due to the combination of suppression of the HOPS complex, high water demand from proliferating bacteria, and a functionally compromised nonacidic vacuole. It may also reflect the reaction of nodule cells to mechanical stress due to rhizobial expansion. We assume that the pathway of vacuole formation in infected nodule cells is modified and traffic to the tonoplast is impaired. This is confirmed by the accumulation of TIP1g and its retargeting to the symbiosome membrane in the transition between the infection and fixation zones. It is interesting that the increase of TIP protein content and the retargeting of TIP aquaporin to other endomembranes from the tonoplast were previously reported as a result of osmotic stress in leaves of Mesembryanthemum crystallinum (Vera-Estrella et al., 2004). In that case, it was shown to be a developmental adaptation by which cells are preprogrammed for such a response. By analogy, symbiotic cells may also have evolved such reprogramming mechanisms.
To clarify the role of the HOPS tethering complex during the development of symbiosis, we studied VPS11 and VPS39, which encode putative HOPS subunits. The expression of both VPS11 and VPS39 was temporarily switched off at the transition of the infection to the fixation zone and the proteins were not detected in infected cells of the fixation zone, while neighboring uninfected cells display the signal. The mechanism for this quite specific block of expression remains to be elucidated, but the analysis of the expression of VPS11 and VPS39 in arbuscular mycorrhizal symbiosis shows that this block is nodule specific. Functional silencing of VPS11 and VPS39 in the infection zone results in infected cells that have an increased number of small nonfused vacuoles instead of a large central vacuole. Furthermore, both VPS proteins are localized on endosomes and tonoplast membranes. We considered that the localization of VPS11 and VPS39 is similar to the localization reported for VPS proteins in yeast and Arabidopsis (Rojo et al., 2003; Balderhaar and Ungermann, 2013). We did not find GFP-tagged VPS proteins on symbiosomes during the infection stage, despite the presence of the protein in the cell. The explanation is that the identity of young symbiosomes is similar to that of the plasma membrane (Ivanov et al., 2012), and the vacuolar identity is acquired at later stages (Limpens et al., 2009). In the zone of fixation, VPS proteins are not detected. However, during the termination of symbiosis and the senescence of infected cells, VPS proteins accumulate. In these cells, symbiosomes lose the ability to fix nitrogen and are transformed into lytic compartments that fuse and form vacuole-like units (Van de Velde et al., 2006; Dupont et al., 2012). These results show that the vacuolar fusion machinery controls symbiosome lysis.
The retargeting of TIP1g indicates a loss of specificity of tonoplast-targeted vesicle fusion, probably due to the absence of the HOPS complex. The TIP1g RNAi experiments indicate that this retargeting is essential for symbiosome expansion and maturation to the nitrogen-fixing stage, as partial silencing of TIP1g causes a block in symbiosome maturation. The presence of TIP1g on symbiosomes might have a direct function in symbiosome maturation, for example by enhancing the availability of water. A similar block in symbiosome development was observed after silencing of the small GTPase Rab7, a nonintegral tonoplast protein involved in membrane fusion (Limpens et al., 2009). This underlines the importance of retargeting of tonoplast proteins to symbiosomes.
In TIP1g RNAi experiments, the nodules with high levels of silencing (95%) are Fix−. In these nodules, the symbiosis is not functional, and the symbiosomes do not mature to reach the nitrogen-fixing stage. In the majority of infected cells of TIP1g:RNAi, nodules in which nifH expression is blocked, a massive burst of autophagy occurs. Macroautophagy involves the formation of autophagosomes, double-membrane structures that sequester part of the cytoplasm or organelles (Bassham et al., 2006; Avila-Ospina et al., 2014). The mechanisms for the formation of the autophagosome isolation membrane are not completely understood, but its formation always begins with the ER surrounding the part of the cytoplasm that later appears inside the autophagosome. Recently, Uemura et al. (2014) suggested the mechanisms for transformation of the ER to autophagic isolation membranes. In TIP1g:RNAi–expressing infected cells, we have observed the formation of autophagic bodies, and massive proliferation of ER, surrounding the symbiosomes and part of the cytoplasm. Therefore, it is quite possible that formation of the isolation membrane of autophagic bodies from the ER in the nodules is similar to the process described for animal cells (Li et al., 2012; Uemura et al., 2014).
The spatial and temporal patterns of autophagic body formation in TIP1g:RNAi nodules are quite different from macroautophagy events in wild-type nodules. In wild-type nodules, the formation of autophagic bodies occurs in young infected cells and in senescing cells, but not in the efficient nitrogen-fixing zone. In these cells, some parts of the cytoplasm or small freshly released symbiosomes, being in close proximity with the vacuole, become engulfed in the vacuole lumen (Fedorova and Brown, 2007). In some cases, such as nodules formed on the defective in nitrogen fixation mutant, where symbiosomes do not mature, autophagic bodies become quite numerous (Wang et al., 2010), but they do not specifically entrap symbiosomes.
In wild-type root nodules, the quick expansion of the microsymbiont helps to maintain the tight contact of the plasma membrane with the cell wall even after the collapse of the vacuole. However, in TIP1g:RNAi–infected cells when symbiosome growth and maturation was blocked, the plasma membrane contact with the cell wall was impaired. It is possible that this may cause the starvation of these cells. The arrest of symbiosome development and their consequent lysis might also be partly due to the starvation.
In other plants, for example in Arabidopsis, single and multiple knockouts of TIP aquaporins are not lethal (Wudick et al., 2009). The severe phenotype, which we observed in TIP1g:RNAi nodules, is most likely due to the precarious situation of symbiotic cells, which lack a functional vacuole but at the same time are “burdened” by thousands of symbiosomes. This phenotype suggests that infected cells do not have a robust mechanism to maintain contact between the plasma membrane and the cell wall and thus may be easily subjected to disruption of water transport from the apoplast to the infected cell.
The presence of tonoplast or late endosome proteins on symbiosomes raises the question of how they can fuse to this membrane in the absence of a HOPS tethering complex. We showed that VPS11, which encodes the core subunit of the HOPS complex, is temporarily repressed in the first cell layer of the fixation zone. This subunit is shared with another tethering complex, CORVET (Balderhaar and Ungermann, 2013). Therefore, it is unlikely that CORVET can replace the HOPS complex in the symbiotic cells of the fixation zone. How the fusion of vesicles is regulated in mature symbiosomes in the zone of fixation remains to be solved.
The nonacidic pH of the vacuole in the symbiotic cells reduces its lytic properties and may affect endocytotic trafficking (Dettmer et al., 2006). The alteration in endocytosis probably suppresses the fusion of symbiosomes with themselves as well as with other endosomes and young vacuoles. This is similar to the suppression of fusion of bacteria-containing vacuoles with lysosomes in mammalian cells, as this also involves the deacidification of phagosomes as well as lysosomes (Huynh and Grinstein, 2007; von Bargen et al., 2009). A further consequence of an increased vacuolar pH is that it may lead to impaired malate uptake by the vacuole, as the inward transport is dependent on the acidic pH of the vacuolar lumen (Hurth et al., 2005; Etienne et al., 2013). Malate is a primary carbon source for symbiosomes (White et al., 2007); thus, vacuole deacidification may boost the availability of malate for symbiosomes by negatively affecting the transport of malate into the vacuole. The mechanism by which the vacuolar pH is increased remains to be determined, but it seems that a V-ATPase, regulating vacuolar pH, could be involved (Xu et al., 2010; Schnitzer et al., 2011; Tarsio et al., 2011).
In conclusion, we report here that infected cell adjustment to accommodate rhizobia involves the suppression and defunctionalization of its vacuole and retargeting of some tonoplast proteins to symbiosomes. The temporary suppression of the HOPS tethering complex is most likely part of the mechanism that facilitates symbiosome expansion and maintenance as nitrogen-fixing organelles.
METHODS
Plant Materials, Transformation, and Inoculation
Agrobacterium rhizogenes–based root transformation of Medicago truncatula cv Jemalong A17 was performed according to Limpens et al. (2005). Roots were inoculated by Sinorhizobium meliloti 2011 and with S. meliloti expressing nifH:GFP. M. truncatula plants that were inoculated with Glomus intraradices were cocultivated with Allium schoenoprasum nurse plants in a sand/hydrobead mixture saturated with Hoagland medium according to Ivanov et al. (2012).
Cloning
VPS11 and VPS39 open reading frames and their 2.5-kb 5′ regulatory sequence were amplified via PCR from 7-DAI nodule cDNA and genomic DNA, respectively, using Phusion high-fidelity polymerase (Finnzymes). Primers are listed in Supplemental Table 1.
Coding sequences of VPS11 and VPS39 were directionally cloned with SmaI-KpnI and KpnI-BamHI, respectively, into a modified pENTR vector (pENTR2) containing a multiple cloning site. Entry clones for VPS11 and VPS39 promoters were generated by TOPO cloning (Invitrogen). The Gateway cloning system (Invitrogen) was used to create genetic constructs for RNAi, promoter-GUS, and GFP fusion. pENTR clones of VPS11 and VPS39 were recombined into the following destination vectors using LR Clonase (Invitrogen): pE12-pK7WGF2-R (containing the M. truncatula ENOD12 promoter) (Limpens et al., 2009), pKGW-GGRR, and UBQ3-pK7WGF2-R, creating N-terminal GFP-X fusions. GFP-VPS11 and GFP-VPS39 translational fusions driven by their own promoters were generated by multiple cloning of gene and promoter sequences into pKGW-MGW.
The TIP1g open reading frame and its 2-kb 5′ regulatory sequence were amplified via PCR from 7-DAI nodule cDNA and genomic DNA, respectively, using Phusion high-fidelity polymerase (Finnzymes). TIP1g pENTR clones were recombined into the following destination vectors using LR Clonase (Invitrogen): pLBpK7WGF2-R (containing the Pisum sativum LB promoter) (Limpens et al., 2009), pKGWGGRR, UBQ3-pK7WGF2-R, and LB-pK7WGF2-R, creating N-terminal GFP-X fusions. The GFP-TIP1g translational fusion driven by its own promoter was generated by multiple cloning of gene and promoter sequences into pKGW-MGW. Primers are listed in Supplemental Table 1.
GUS Staining, Sectioning, and Light Microscopy
Transgenic roots and nodules were collected and washed twice in 0.1 M sodium phosphate buffer, pH 7.2, incubated in GUS buffer under vacuum at room temperature for 30 min to allow the buffer to replace oxygen in the tissue, incubated at 37°C for 2 h or overnight to enable the enzymatic reaction, and embedded for sectioning following the Technovit 7100 protocol (Technovit). Sections were mounted on microscope slides, counterstained with ruthenium red, and analyzed using Nikon Optiphot-2 and Leica DM 5500 Flu microscopes.
The selection of transgenic roots and nodules was performed on a Leica MZFLIII fluorescence macroscope equipped with filter cubes for the detection of GFP, YFP, red fluorescent protein, excitation/emission EGFP (excitation, 470/40 D; 495 emission, 525/50), EYFP (excitation, 510/20 D; 530 emission, 560/40), and DsRED (excitation, 545/30 D; 570 emission, 620/60).
Confocal Microscopy
Confocal imaging of GFP-fused proteins was done on transgenic hand-sectioned nodules by using a Zeiss LSM 5 Pascal confocal laser-scanning microscope (Carl Zeiss) and the Zeiss Meta LSM 510 microscope. For enhancement of GFP-VPS11, GFP-VPS39, and GFP-TIP1g signal, driven by their own promoters, we used polyclonal rabbit anti-GFP antibody (Molecular Probes) (1:100 dilution) and secondary anti-rabbit Alexa 488 antibody (Molecular Probes) (1:200 dilution); normal goat serum or 3% BSA was used for blocking. Sections were counterstained with FM4-64 (30 μg/mL) or PI. In cases where the constructs were created using strong promoters like ubiquitin, the localization was observed without enhancement. For immunolocalization of VTI11-specific M. truncatula, anti-VTI11 antibodies developed in rabbit (diluted 1:100) were used (Limpens et al., 2009), and as a secondary antibody, an anti-rabbit CY3 antibody (Molecular Probes) (diluted 1:200) was used. To enhance the GFP signal in this experiment, anti-GFP antibodies developed in mouse (1:50) were used, followed by secondary anti-mouse Alexa 488 (1:200). A mix of 0.5% skim-milk powder with 2% BSA was used as a blocking solution. The protocol for NR staining was adapted from Dubrovsky et al. (2006). Confocal microscopy settings with the window for NR excitation/emission (564/595 to 615 nm) were used. Bacteroids were contrasted by staining with SYTO 16 (green).
3D Reconstruction and Volume-Surface Measurements
3D reconstruction of Z-stacks obtained by confocal microscopy of root nodules expressing GFP-SYP22 driven by the ProUBQ3 or ProLB promoter or wild-type root nodules counterstained by Sytox Green (Molecular Probes) was used for quantitative estimation of the volume and surface area of the vacuole, bacteroids, and cell. Bacteroids and nuclei were counterstained either by PI (red fluorescence) or Sytox Green. To obtain 3D reconstructions, confocal image stacks (50 images with 0.5 μm of Z-step) were imported to Imaris 7.5 (Bitplane). After baseline subtraction, a subregion with a cell of interest was defined. The isosurface module was used to reconstitute the 3D pictures. Volume and surface area measurements were performed using the Imaris MeasurementPro module.
Sample Preparation for Light Microscopy and EM, and EM Immunodetection
The protocol for tissue processing was described previously (Limpens et al., 2009). Semithin sections (0.6 μm) for light microscopy and thin sections (60 nm) for EM of transgenic nodules were cut using a Leica Ultracut microtome. Nickel grids with the sections were blocked in normal goat serum with 1% milk or 2% BSA in PBS and incubated with the primary antibody at the dilutions given above. Goat anti-rabbit antibody coupled with 10-nm gold (BioCell) (1:50 dilution) was used as secondary antibody. Sections were examined using a JEOL JEM 2100 transmission electron microscope equipped with a Gatan US4000 4K×4K camera.
Expression in Xenopus laevis Oocytes
The cDNA of TIP1g was cloned into the expression vector pGEMHE using the restriction enzymes BamHI and XbaI. To linearize the plasmid, pGEMHE-TIP1g was digested with NheI. Complementary RNA (cRNA) was transcribed using 1 μg of linearized DNA with the mMESSAGE mMACHINE kit (Ambion). X. laevis oocytes were surgically removed and defolliculated, then injected with 30 ng of cRNA or water, using a microinjector (Drummond Nanoject II automatic nanoliter injector; Drummond Scientific). After injection, oocytes were incubated in ND96 for 48 h at 18°C. To measure water permeability, oocytes were transferred to a 5-fold-diluted solution of ND96 (osmolarity of 47 mosmol/kg). The volume change in the oocyte was derived from images captured at 3-s intervals for 2 min using a dissecting microscope with IC Capture 2.0 software (The Imaging Source) as AVI format video files. ImageJ software (http://rsbweb.nih.gov/ij/) was used to calculate the change in the total area of the oocytes captured in the AVI video file. The rate of oocyte swelling was plotted as V/V0 versus time, where V is a volume at a certain time point and V0 is the initial volume. Water permeability coefficient values for oocytes injected with cRNA encoding TIP1g, AQP1, or water were determined as described by Fetter et al. (2004). The significance of results was analyzed by Tukey’s multiple comparison test.
qRT-PCR Analysis
Total RNA was extracted from roots and different zones of 14-DAI root nodules using the E.Z.N.A. Plant RNA Mini Kit (Omega Bio-Tek) and transcribed into cDNA using the iScript cDNA synthesis kit (Bio-Rad). Real-time PCR was set up in a 20-μL reaction system using iQ SYBR Green Supermix (Bio-Rad). Gene-specific primers were designed with Primer-3-Plus software (Untergasser et al., 2007). Gene expression profiles were normalized against the transcription level of the reference gene UBQ10. Primers are listed in Supplemental Table 1.
Accession Numbers
Sequence data from this article can be found in the Phytozome and GenBank websites and databases under the following accession numbers: VPS11, Medtr1g019500; VPS39, Medtr5g020140; and TIP1g, Medtr4g063090.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. LysoSensor Yellow/Blue Staining to Determine Vacuolar pH in Nodule Cells.
Supplemental Figure 2. The Expression of ProVPS11:GUS and ProVPS39:GUS in Transgenic Roots Inoculated by G. intraradices.
Supplemental Figure 3. Colocalization of Endosome/Vacuole Molecular Marker VTI11 with VPS Proteins.
Supplemental Figure 4. The Level of Gene Silencing of VPS11 and VPS39 by RNAi.
Supplemental Figure 5. Transporters from M. truncatula and Arabidopsis.
Supplemental Figure 6. Expression of M. truncatula TIP Genes.
Supplemental Figure 7. YFP-TIP1g Localizes on the Plasma Membrane of Injected Oocytes.
Supplemental Figure 8. Water-Channel Activity of TIP1g.
Supplemental Figure 9. TIP1g Does Not Transport Ammonia and Malate.
Supplemental Figure 10. The Induction of the Rhizobial nifH Gene, Detectable Due to GFP Fluorescence, Permits Discrimination between Fix+ and Fix− Nodules on ProLB:RNAi-TIP1g Transgenic Roots.
Supplemental Figure 11. The Level of TIP1g Gene Silencing in Fix+ and Fix− Nodules.
Supplemental Table 1. Primers Used for Cloning and PCR Analysis of VPS11, VPS39, and Tonoplast Aquaporins.
Supplemental Methods 1. Calibration of LysoSensor Yellow/Blue, Targeting of TIP1g in X. laevis Oocytes, NH3 Flux Experiment, and C14 Malate Flux.
Supplemental Data Set 1. Text File of Alignment Used for Phylogenetic Analysis Shown in Supplemental Figure 5.
Supplementary Material
Acknowledgments
We thank T.W.J. Gadella (University of Amsterdam) for help with the analysis of Imaris 3D reconstructed images. We thank P. Smith (University of Sydney) for help in editing the article. We thank our colleagues Norbert de Ruijter for assistance with confocal imaging, Rene Geurts for helpful discussions, and Jan Hontelez for providing the line of S. meliloti expressing nifH:GFP. A.G. received a Ph.D. fellowship from the EPS School of Biological Sciences (Wageningen University).
AUTHOR CONTRIBUTIONS
A.G. and E.E.F. designed and performed the experiments and participated in the writing and editing of the article. B.N.K. and D.G. performed the analysis of MtTIP1g aquaporin water transport capacity and participated in editing of the article. S.D.T. and Z.W. participated in the work with X. laevis oocytes and performed the analysis of malate and ammonia transport. T.B. participated in designing the experiments and writing and editing of the article.
Glossary
- PI
propidium iodide
- NR
Neutral Red
- DAI
days after inoculation
- EM
electron microscopy
- RNAi
RNA interference
- qRT-PCR
quantitative RT-PCR
- ER
endoplasmic reticulum
- cRNA
complementary RNA
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Avila-Ospina, L., Moison, M., Yoshimoto, K., Masclaux-Daubresse, C. (2014). Autophagy, plant senescence, and nutrient recycling. J. Exp. Bot. 65: 3799–3811 [DOI] [PubMed] [Google Scholar]
- Bak, G., Lee, E.-J., Lee, Y., Kato, M., Segami, S., Sze, H., Maeshima, M., Hwang, J.-U., Lee, Y. (2013). Rapid structural changes and acidification of guard cell vacuoles during stomatal closure require phosphatidylinositol 3,5-bisphosphate. Plant Cell 25: 2202–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balderhaar, H.J., Ungermann, C. (2013). CORVET and HOPS tethering complexes—Coordinators of endosome and lysosome fusion. J. Cell Sci. 126: 1307–1316 [DOI] [PubMed] [Google Scholar]
- Bassham, D.C., Laporte, M., Marty, F., Moriyasu, Y., Ohsumi, Y., Olsen, L.J., Yoshimoto, K. (2006). Autophagy in development and stress responses of plants. Autophagy 2: 2–11 [DOI] [PubMed] [Google Scholar]
- Behnia, R., Munro, S. (2005). Organelle identity and the signposts for membrane traffic. Nature 438: 597–604 [DOI] [PubMed] [Google Scholar]
- Brett, C.L., Merz, A.J. (2008). Osmotic regulation of Rab-mediated organelle docking. Curr. Biol. 18: 1072–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumell, J.H.Scidmore, M.A. (2007). Manipulation of rab GTPase function by intracellular bacterial pathogens. Microbiol. Mol. Biol. Rev. 71: 636–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalano, C.M., Czymmek, K.J., Gann, J.G., Sherrier, D.J. (2007). Medicago truncatula syntaxin SYP132 defines the symbiosome membrane and infection droplet membrane in root nodules. Planta 225: 541–550 [DOI] [PubMed] [Google Scholar]
- Chaumont, F., Tyerman, S.D. (2014). Aquaporins: Highly regulated channels controlling plant water relations. Plant Physiol. 164: 1600–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer, J., Hong-Hermesdorf, A., Stierhof, Y.-D., Schumacher, K. (2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky, J.G., Guttenberger, M., Saralegui, A., Napsucialy-Mendivil, S., Voigt, B., Baluska, F., Menzel, D. (2006). Neutral red as a probe for confocal laser scanning microscopy studies of plant roots. Ann. Bot. (Lond.) 97: 1127–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont, L., Alloing, G., Pierre, O., Msehli, S.E., Hopkins, J., and Hérouart, D.P.F. (2012). The legume root nodule: From symbiotic nitrogen fixation to senescence. In Senescence, T. Nagata, ed (InTech), pp. 137–168. [Google Scholar]
- Etienne, A., Génard, M., Lobit, P., Mbeguié-A-Mbéguié, D., Bugaud, C. (2013). What controls fleshy fruit acidity? A review of malate and citrate accumulation in fruit cells. J. Exp. Bot. 64: 1451–1469 [DOI] [PubMed] [Google Scholar]
- Farkas, A., Maróti, G., Durgő, H., Györgypál, Z., Lima, R.M., Medzihradszky, K.F., Kereszt, A., Mergaert, P., Kondorosi, É. (2014). Medicago truncatula symbiotic peptide NCR247 contributes to bacteroid differentiation through multiple mechanisms. Proc. Natl. Acad. Sci. USA 111: 5183–5188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova, E.E., Brown, S. (2007). Cytochemistry of proteolytic activity and pH status of vacuoles in Medicago truncatula root nodules. Russ. J. Plant Physiol. 54: 25–31 [Google Scholar]
- Fetter, K., Van Wilder, V., Moshelion, M., Chaumont, F. (2004). Interactions between plasma membrane aquaporins modulate their water channel activity. Plant Cell 16: 215–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest, K.L., Bhave, M. (2007). Major intrinsic proteins (MIPs) in plants: A complex gene family with major impacts on plant phenotype. Funct. Integr. Genomics 7: 263–289 [DOI] [PubMed] [Google Scholar]
- Gattolin, S., Sorieul, M., Hunter, P.R., Khonsari, R.H., Frigerio, L. (2009). In vivo imaging of the tonoplast intrinsic protein family in Arabidopsis roots. BMC Plant Biol. 9: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, J., Burgess, K. (2010). Fluorescent indicators for intracellular pH. Chem. Rev. 110: 2709–2728 [DOI] [PubMed] [Google Scholar]
- Hurth, M.A., Suh, S.J., Kretzschmar, T., Geis, T., Bregante, M., Gambale, F., Martinoia, E., Neuhaus, H.E. (2005). Impaired pH homeostasis in Arabidopsis lacking the vacuolar dicarboxylate transporter and analysis of carboxylic acid transport across the tonoplast. Plant Physiol. 137: 901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh, K.K., Grinstein, S. (2007). Regulation of vacuolar pH and its modulation by some microbial species. Microbiol. Mol. Biol. Rev. 71: 452–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, J.H., Ellingson, S.R., Roberts, D.M. (2010). Ammonia permeability of the soybean nodulin 26 channel. FEBS Lett. 584: 4339–4343 [DOI] [PubMed] [Google Scholar]
- Isberg, R.R., O’Connor, T.J., Heidtman, M. (2009). The Legionella pneumophila replication vacuole: Making a cosy niche inside host cells. Nat. Rev. Microbiol. 7: 13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov, S., Fedorova, E.E., Limpens, E., De Mita, S., Genre, A., Bonfante, P., Bisseling, T. (2012). Rhizobium-legume symbiosis shares an exocytotic pathway required for arbuscule formation. Proc. Natl. Acad. Sci. USA 109: 8316–8321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson, U., Karlsson, M., Johansson, I., Gustavsson, S., Sjövall, S., Fraysse, L., Weig, A.R., Kjellbom, P. (2001). The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiol. 126: 1358–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaldenhoff, R., Fischer, M. (2006). Aquaporins in plants. Acta Physiol. (Oxf.) 187: 169–176 [DOI] [PubMed] [Google Scholar]
- Li, L., et al. . (2012). The invasion of tobacco mosaic virus RNA induces endoplasmic reticulum stress-related autophagy in HeLa cells. Biosci. Rep. 32: 171–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens, E., Ivanov, S., van Esse, W., Voets, G., Fedorova, E., Bisseling, T. (2009). Medicago N2-fixing symbiosomes acquire the endocytic identity marker Rab7 but delay the acquisition of vacuolar identity. Plant Cell 21: 2811–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens, E., Mirabella, R., Fedorova, E., Franken, C., Franssen, H., Bisseling, T., Geurts, R. (2005). Formation of organelle-like N2-fixing symbiosomes in legume root nodules is controlled by DMI2. Proc. Natl. Acad. Sci. USA 102: 10375–10380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maagd, R.A.d., Yang, W.-C., Roo, G.-d., Mulders, I.H.M., Roest, H.P., Spaink, H.P., Bisseling, T., Lugtenberg, B.J.J. (1994). Down-regulation of expression of the Rhizobium leguminosarum outer membrane protein gene ropA occurs abruptly in interzone II–III of pea nodules and can be uncoupled from nif gene activation. Mol. Plant Microbe Interact. 7: 279–281 [Google Scholar]
- Maurel, C., Santoni, V., Luu, D.-T., Wudick, M.M., Verdoucq, L. (2009). The cellular dynamics of plant aquaporin expression and functions. Curr. Opin. Plant Biol. 12: 690–698 [DOI] [PubMed] [Google Scholar]
- Nickerson, D.P., Brett, C.L., Merz, A.J. (2009). Vps-C complexes: Gatekeepers of endolysosomal traffic. Curr. Opin. Cell Biol. 21: 543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisen, D., Marty, F., Leborgne-Castel, N. (2005). New insights into the tonoplast architecture of plant vacuoles and vacuolar dynamics during osmotic stress. BMC Plant Biol. 5: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo, E., Gillmor, C.S., Kovaleva, V., Somerville, C.R., Raikhel, N.V. (2001). VACUOLELESS1 is an essential gene required for vacuole formation and morphogenesis in Arabidopsis. Dev. Cell 1: 303–310 [DOI] [PubMed] [Google Scholar]
- Rojo, E., Zouhar, J., Kovaleva, V., Hong, S., Raikhel, N.V. (2003). The AtC-VPS protein complex is localized to the tonoplast and the prevacuolar compartment in Arabidopsis. Mol. Biol. Cell 14: 361–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnitzer, D., Seidel, T., Sander, T., Golldack, D., Dietz, K.-J. (2011). The cellular energization state affects peripheral stalk stability of plant vacuolar H+-ATPase and impairs vacuolar acidification. Plant Cell Physiol. 52: 946–956 [DOI] [PubMed] [Google Scholar]
- Sujkowska, M., Górska-Czekaj, M., Bederska, M., Borucki, W. (2011). Vacuolar organization in the nodule parenchyma is important for the functioning of pea root nodules. Symbiosis 54: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., Kumar, S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarsio, M., Zheng, H., Smardon, A.M., Martínez-Muñoz, G.A., Kane, P.M. (2011). Consequences of loss of Vph1 protein-containing vacuolar ATPases (V-ATPases) for overall cellular pH homeostasis. J. Biol. Chem. 286: 28089–28096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura, T., Yamamoto, M., Kametaka, A., Sou, Y.S., Yabashi, A., Yamada, A., Annoh, H., Kametaka, S., Komatsu, M., Waguri, S. (2014). A cluster of thin tubular structures mediates transformation of the endoplasmic reticulum to autophagic isolation membrane. Mol. Cell. Biol. 34: 1695–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser, A., Nijveen, H., Rao, X., Bisseling, T., Geurts, R., Leunissen, J.A.M. (2007). Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35: W71–W74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Velde, W., Guerra, J.C.P., De Keyser, A., De Rycke, R., Rombauts, S., Maunoury, N., Mergaert, P., Kondorosi, E., Holsters, M., Goormachtig, S. (2006). Aging in legume symbiosis: A molecular view on nodule senescence in Medicago truncatula. Plant Physiol. 141: 711–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasse, J., de Billy, F., Camut, S., Truchet, G. (1990). Correlation between ultrastructural differentiation of bacteroids nitrogen fixation in alfalfa nodules. J. Bacteriol. 172: 4295–4306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera-Estrella, R., Barkla, B.J., Bohnert, H.J., Pantoja, O. (2004). Novel regulation of aquaporins during osmotic stress. Plant Physiol. 135: 2318–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeer, J.E.M., von Wangenheim, D., Barberon, M., Lee, Y., Stelzer, E.H.K., Maizel, A., Geldner, N. (2014). A spatial accommodation by neighboring cells is required for organ initiation in Arabidopsis. Science 343: 178–183 [DOI] [PubMed] [Google Scholar]
- Vijn, I., Christiansen, H., Lauridsen, P., Kardailsky, I., Quandt, H.-J., Broer, I., Drenth, J., Ostergaard Jensen, E., van Kammen, A., Bisseling, T. (1995). A 200 bp region of the pea ENOD12 promoter is sufficient for nodule-specific and nod factor induced expression. Plant Mol. Biol. 28: 1103–1110 [DOI] [PubMed] [Google Scholar]
- von Bargen, K., Polidori, M., Becken, U., Huth, G., Prescott, J.F., Haas, A. (2009). Rhodococcus equi virulence-associated protein A is required for diversion of phagosome biogenesis but not for cytotoxicity. Infect. Immun. 77: 5676–5681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D., Griffitts, J., Starker, C., Fedorova, E., Limpens, E., Ivanov, S., Bisseling, T., Long, S. (2010). A nodule-specific protein secretory pathway required for nitrogen-fixing symbiosis. Science 327: 1126–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, J., Prell, J., James, E.K., Poole, P. (2007). Nutrient sharing between symbionts. Plant Physiol. 144: 604–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wudick, M.M., Luu, D.-T., Maurel, C. (2009). A look inside: Localization patterns and functions of intracellular plant aquaporins. New Phytol. 184: 289–302 [DOI] [PubMed] [Google Scholar]
- Xu, L., Shen, X., Bryan, A., Banga, S., Swanson, M.S., Luo, Z.Q. (2010). Inhibition of host vacuolar H+-ATPase activity by a Legionella pneumophila effector. PLoS Pathog. 6: e1000822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, W.C., Horvath, B., Hontelez, J., VanKammen, A., Bisseling, T. (1991). In situ localization of Rhizobium mRNAs in pea root nodules: nifA and nifH localization. Mol. Plant Microbe Interact. 4: 464–468 [Google Scholar]
- Zouhar, J., Rojo, E. (2009). Plant vacuoles: Where did they come from and where are they heading? Curr. Opin. Plant Biol. 12: 677–684 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.