Abstract
Unfixed tissue specimens most frequently are stored for long term research uses at either −80° C or in vapor phase liquid nitrogen (VPLN). There is little information concerning the effects such long term storage on tissue RNA or protein available for extraction. Aliquots of 49 specimens were stored for 5–12 years at −80° C or in VPLN. Twelve additional paired specimens were stored for 1 year under identical conditions. RNA was isolated from all tissues and assessed for RNA yield, total RNA integrity and mRNA integrity. Protein stability was analyzed by surface-enhanced or matrix-assisted laser desorprion ionization time of flight mass spectrometry (SELDI-TOF-MS, MALDI-TOF-MS) and nano-liquid chromatography electrospray ionization tandem mass spectrometry (nLC-ESI-MS/MS). RNA yield and total RNA integrity showed significantly better results for −80° C storage compared to VPLN storage; the transcripts that were preferentially degraded during VPLN storage were these involved in antigen presentation and processing. No consistent differences were found in the SELDI-TOF-MS, MALDI-TOF-MS or nLC-ESI-MS/MS analyses of specimens stored for more than 8 years at −80° C compared to those stored in VPLN. Long term storage of human research tissues at −80° C provides at least the same quality of RNA and protein as storage in VPLN.
Keywords: assays, human tissues, proteomics, RNA, storage, temperatures
The current gold standard for preservation of RNA and protein analytes in tissue specimens is snap-freezing, with subsequent storage in either mechanical freezers at −80° C or in the vapor phase of liquid nitrogen (VPLN). Although RNA and protein can be isolated from tissue specimens processed for routine histologic analysis (formalin fixation and paraffin embedding), the processing can cause alterations in RNA and proteins; however, there is little difference in assay results when mRNA is analyzed by real-time, reverse transcriptase quantitative polymerase chain reaction technology (RT-Q-PCR) (Steg et al. 2006, 2007). Frozen samples of tissues, however, are preferred for some basic and translational studies, especially genome-wide sequencing experiments. Long term storage of samples sometimes is desirable, because the research value of human specimens, especially cancer specimens, increases over time if needed for longitudinal clinical and outcome data.
It is considered a theoretical advantage to store tissue below the glass transition phase of pure water, because aqueous based chemical reactions are thought to cease at the glass transition temperature (Tg). Such conditions are achieved in VPLN (−150° C) and by some mechanical freezers. Owing to logistics and expense, however, many investigators and biorepositories store samples in mechanical freezers at temperatures of −70 to −90° C. It is noteworthy that there is controversy about the exact Tg value of pure water; recent studies suggest that this is 165° K or −108° C, although it frequently is reported to be 136° K or −137° C (Giovambattista et al. 2004). Complicating this issue is the fact that cells are not filled with “pure” water, so the practical Tg values for the water in mammalian tissues are unknown. Nevertheless, enzymatic reactions, in general, are thought to continue at −80° C and cells do not remain viable when stored at −80° C.
To compare the relative effects of the two time-honored methods for storing frozen tissue on RNA and protein integrity, we performed a series of analyses to assess the content and quality of the RNA transcripts and proteins/peptides on a cohort of matched human tissue specimens stored for a number of years at both −80° C and in VPLN.
Materials and methods
Patients and specimens
Our study was approved by the Nationwide Children′s Hospital Institutional Review Board. The requirement for written informed consent from the participants was waived, because the study used de-identified specimens.
Aliquots of 49 paired human tissue specimens were stored at −80° C in mechanical freezers and matched aliquots were stored in VPLN. An additional 12 paired specimens were obtained from the member institutions of the Cooperative Human Tissue Network (CHTN) and were stored similarly in different types of freezers. The specimens were remnant solid tissues from surgeries that were available after clinical diagnoses had been made. Tumors, normal appearing tissues adjacent to trumors and other non-neoplasric tissue specimens were used (Table 1). The selection of specimens for the first cohort was based on the availability of matched frozen tissue stored in VPLN and at −80° C. The specimens obtained from the CHTN were procured prospectively for this analysis. All tissues were reviewed by pathologists to ensure that the matched specimens from both storage methods were the same histologically (see Specimen Processing section).
Table 1.
Tissue types used to assess RNA integrity after long term storage at −80° C and in VPLN
| Tissue | Storage period (years) |
Tissue | Storage period (years) |
|---|---|---|---|
| thyroid papillary carcinoma | 10 | choroid plexus carcinoma | 8 |
| lymphoma | 10 | neuroblastoma | 8 |
| benign neural tumor | 10 | colon-non neoplastic | 8 |
| dysgerminoma | 10 | lipoma | 8 |
| ganglioglioneuroma | 10 | hepatoblastoma | 8 |
| pilocytic astrocytoma | 10 | neuroblastoma | 8 |
| PNET | 10 | cellular fibroadenoma breast | 8 |
| embryonal rhabdomyosarcoma | 10 | neuroblastoma | 8 |
| ganglioglioneuroma | 10 | skull mass | 8 |
| spleen, normal | 10 | myxoid neoplasm-malignant | 8 |
| endodermal sinus tumor | 10 | Langerhans histiocytosis | 8 |
| pancreatic cystic/solid papillary neoplasm | 9 | neurofibroma | 8 |
| pilocytic astrocytoma | 9 | neuroblastoma | 8 |
| hepatoblastoma | 9 | paraganglioma | 7 |
| ganglioglioneuroma | 9 | ependymoma | 7 |
| neuroblastoma | 9 | kidney- non neoplastic | 7 |
| ganglioneuroma | 9 | Burkitt’s lymphoma | 7 |
| lymph node, follicular hyperplasia | 9 | meningioma | 6 |
| spleen, non-neoplastic | 9 | pilocytic astrocytoma | 6 |
| MPNST | 9 | ependymoma | 6 |
| alveolar rhabdomyosarcoma | 9 | papillary thyroid carcinoma | 6 |
| ganglioglioneuroma | 9 | Burkitt’s lymphoma | 6 |
| infantile myofibromatosis | 9 | osteosarcoma | 6 |
| neuroblastoma | 9 | ganglioglioma | 5 |
| pilocytic astrocytoma | 8 |
The tissue procured from the CHTN were obtained within 2 h of removal from the patient, split into identical aliquots, snap frozen or snap frozen in OCT, then shipped to a central facility on dry ice for storage at either −80° C or in VPLN for 1 year. All freezers used for this study were monitored electronically and manually and no failures in temperature maintenance occurred during the storage of the specimens.
Specimen processing for RNA isolation
Specimens not initially frozen in OCT were embedded by placing the frozen tissue in partially frozen OCT, covering with additional OCT, and freezing at −20° C immediately prior to analysis. All OCT embedded specimens were equilibrated at −20° C, histologic sections were cut using a cryostat, and the sections were stained with hematoxylin and eosin (H & E) for histologic assessment. Ten additional 10 µm frozen sections were obtained and placed on dry ice for isolation of either mRNA or protein.
For RNA isolation, the samples were incubated for 10 min in 1 ml Tri Reagent (Molecular Research Center, Inc., Cincinnati, OH) at 50° C and insoluble material was pelleted by centrifugation for 10 min at 3,000 × g. Nine hundred microliters of supernatant were used for RNA isolation according to the manufacturer′s recommendations. Each of the resulting RNA pellets was dissolved in 25 µl RNAse-free water. RNA was quantified using a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE).
Quantification of total RNA integrity
For microcapillary electrophoresis measurements of total RNA integrity, the Agilent 2100 Bioanalyzer was used in conjunction with the RNA 6000 Nano LabChip kits (Agilent, Waldbronn, Germany) following the manufacturer′s recommendations. Bioanalyzer electrophoresis report files were analyzed for RNA integrity number (RIN) and degradation factor as described earlier (Auer et al. 2003). For RIN analysis, 2100 Expert software version B.02.03.SI307 (Agilent) was used. RIN reports total RNA integrity on a scale of 1 to 10, where 1 represents complete degradation and 10 is the highest level of RNA integrity. A value of 1 (lowest integrity) was used for samples where no RIN values were available. For degradation factor analysis, results of individual samples were exported from the 2100 Expert software as CSV files and analyzed by Degradometer software version 1.4.2 (Auer et al. 2003). Degradation factor reports total RNA integrity on a scale of 1 to 100, where 1 is the highest level of RNA integrity and 100 is complete degradation. For samples where no degradation factors were available, 100 (lowest integrity) was used for further calculations.
Microarray expression profiling
Samples were used for expression profiling if both of the following criteria were met: 1) at least one of the paired samples showed RIN> 1, and 2) at least one of the paired samples showed a degradation factor < 100. From 37 paired samples that met these criteria, 25 ng of total RNA per sample were processed using isothermal SPIA Biotin System (NuGEN Technologies, Inc., San Carlos, CA) amplification; 2.2 µg cDNA resulting from the amplification were used for microarray hybridization (Affymetrix Human U133A2.0 GeneChips, Santa Clara, CA).The U133A2.0 microarray contains more than 22,000 probe sets to analyze 18,400 transcripts including 14,500 well-characterized genes. After 16 h hybridization at 45° C, washing and staining of microarrays was performed using a Fluidics Station 450 (Affymetrix); GeneChips were scanned in a GeneChip Scanner 3000 (Affymetrix). All steps of sample and microarray processing were performed according to manufacturer′s recommendations. CEL files were generated from DAT files using GCOS software (Affymetrix). Probe set expression estimates were calculated using RNA algorithm in Array Assist Lite software version 3.4 (Stratagene, Santa Clara, CA).
Characterization of mRNA integrity
To evaluate mRNA integrity of specimens stored under different conditions, ratios of signal intensities were calculated for probe sets that measure the 3′ and 5′ ends of two genes (GAPDH and ACTB). Because first strand cDNA synthesis uses oligo(dT), RNA degradation can cause reduced signal intensities at the 5′ end of transcripts and therefore can increase 3′:5′ ratios. To evaluate whether certain transcripts from specific genes showed consistent alterations due to storage conditions, significance analysis of microarrays (SAM) was used (Tusher et al. 2001).
Protein analysis by mass spectrometry
Frozen sections from matched pairs from eight cases were cut at 5 µm and mounted on glass microscope slides for histopathological analysis. These samples were selected from various tumor specimens in which aliquots of the same specimen were stored for ≥9 years at either −80° C or in VPLN. The frozen sections of both aliquots of a pair were reviewed by a pathologist to verify that each aliquot was equivalent by microscopic examination, i.e., those that contained approximately the same content and histology of the tumor. Ten micrometer sections then were cut from the equivalent paired samples and mounted for analysis by mass spectrometry. The analysis was blinded concerning which aliquot of each specimen was stored at one or the other temperature. When the 5 µm sections of both aliquots of a pair were equivalent by microscopic examination, e.g., contained approximately the same content of tumor, up to eight 10 µm frozen sections, depending upon the tumor in the respective frozen section, were scraped from the slide, avoiding the OCT, and were lysed with 100 µ1 of lysis buffer (20 mm Hepes, pH 8.0, 1% Tween 20). The supernatants were separated using a high speed microfuge at 10,000 × g for 10 min Eight pairs of samples of the specimens that had been stored ≥ 9 years were analyzed (Table 2).
Table 2.
Pairs of surgical specimens stored for at least 9 years and analyzed by mass spectrometry
| Case no. | Diagnosis | Condition | Storage period |
|---|---|---|---|
| 1A | Langerhans histiocytosis | −80° C | 10 years |
| 1B | Langerhans histiocytosis | liquid nitrogen | |
| 2A | ependymoma | −80° C | 9 years |
| 2B | ependymoma | liquid nitrogen | |
| 3A | spleen, non-neoplasia | −80° C | 11 years |
| 3B | spleen, non-neoplasia | liquid nitrogen | |
| 4A | lymphoma | −80° C | 12 years |
| 4B | lymphoma | liquid nitrogen | |
| 5A | hepatoblastoma | −80° C | 10 years |
| 5B | hepatoblastoma | liquid nitrogen | |
| 6A | lymph node, follicular hyperplasia | −80° C | 11 years |
| 6B | lymph node, follicular hyperplasia | liquid nitrogen | |
| 7A | MPNST | −80° C | 11 years |
| 7B | MPNST | liquid nitrogen | |
| 8A | PNET | −80° C | 12 years |
| 8B | PNET | liquid nitrogen |
The protein in each lysate supernatant was measured using the Pierce BCA protein assay and an equivalent amount of protein (10 µg) was loaded in triplicate with each of the aliquots loaded randomly on one of the eight sampling spots of an IMAC-3 copper activated metal chip (Bio-Rad, Hercules, CA). Each spot on the chip was analyzed by surfaced enhanced laser absorption/desorprion time of flight mass spectrometry (SELDI-TOF-MS, Protein Biosystem II; Bio-Rad). In general, the approach described earlier was used for copper activation and sample loading (Adam et al. 2002, Semmes et al. 2005, McLerran et al. 2008a,b, Grizzle et al. 2005a,b). Specifically, the blinded samples were processed using a Biomek 2000 (Beckman Coulter, Brea, CA) robotic sample preparation platform that diluted the samples and loaded them on the IMAC-3 copper activated surface. The robotic system also spotted sinapinic acid matrix on each sample. A control sample was loaded and analyzed in at least one of the eight wells of each metal chip; locations of cases and controls on chips were chosen randomly to minimize bias (Adam et al. 2002, Semmes et al. 2005, McLerran et al. 2008a,b, Grizzle et al. 2005a,b).
The metal IMAC-3 chips also were read on a MALDI-TOF-MS (Ultraflex III, Brüker Daltronics, Billerica, MA) using an adapter plate made specifically for the Brüker instrument. All samples were prepared according to the manufacturer′s instructions (see above). Our method of MALDI-TOF-MS analysis has been reported previously (Kojima et al. 2008). Specifically, 1600 shots were acquired automatically from 2000–100,000 m/z with high filtering set to on, deflector set to 1000 m/z, and detector gain set at 1769 V, using smart beam technology at a single (empirically determined) laser energy, while using a random walk command. Fuzzy acquisition was turned on to avoid summing poor spectra. Flexanalysis then was used to baseline subtract all spectra using a top hat approach followed by a batch text output processing script (written in house), thus allowing further processing to be carried out in MatLab (Mathworks, Natick, MA).
Specimens of lysate also were analyzed using nano-liquid chromatography electrospray ionization tandem mass spectrometry (nLC-ESI-MS/ MS) as described earlier (Wang et al. 2010). For these experiments, protein extracts were concentrated and exchanged using equal volumes of 50 mM ammonium bicarbonate three times using 3 K cutoff spin filters (EMD Millipore, Billerica, MA) and digested overnight with trypsin gold (Promega, Madison WI) according to the manufacturer′s recommendations. The resulting peptides were diluted to 100 ng/µ1 in 0.1% formic acid and 5 µl were subjected to nLC-ESI-MS/ MS analysis using a ThermoFisher Scientific, Inc., Waltham, MA LTQ-XL ion trap mass spectrometer equipped with a Thermo MicroAS autosampler and Thermo Surveyor HPLC pump, Nanospray source, and Xcalibur 1.4 instrument control (ThermoFisher Scientific, Inc.). Proteins were searched in species-specific subsets of the UniRef database (European Bioinformatics, Cambridge, UK). Tandem mass spectrometry data were converted to mzXML format using instrument-specific conversion software (Institute for Systems Biology, Seattle WA; Fred Hutchinson Cancer Center) and run separately through SEQUEST (ThermoFisher Scientific, Inc.) with a “no enzyme” setting so that non-trypsin cleavage sites were mapped, and also with MASCOT (Matrix Science Inc., Boston MA) using a “strict trypsin” setting so that only trypsin cleavage sites were mapped. ProteoIQ (Biolnquire, Athens, GA) was used to combine tandem mass spectrometry database search results to determine thresholds, which identify as many real proteins as possible while encountering a minimal number of false positive protein identifications. The numbers of unique peptides were calculated per sample based on no enzyme and strict trypsin database searches.
Statistical analysis
To evaluate the significance of differences between RNA and protein integrity from samples stored under the two storage conditions, a paired t-test was performed. Total RNA yield, RIN, Degradation Factor, and 3′/5′ ratios of GAPDH and ACTB were analyzed for significant differences. Transcripts with the highest susceptibility to storage-induced degradation were identified by paired analysis performed in SAM using a false discovery rate (FDR) <0.12 and running 100 permutations. The list of significant transcripts expressed differentially provided by SAM was used for EASE analysis (Hosack et al. 2003) to identify preferentially affected gene ontology categories. Results of EASE analysis are reported when the EASE score is less than 0.05 after Benjamini correction.
For the MALDI-TOF-MS analysis, the MatLab toolbox was used to align and peak-select mass spectra, thus producing a peak matrix file that then was applied to calculate mean intensities and coefficient of variance. For the LC-ESI-MS data, ProteoIQ (NuSep) was used to incorporate the two most common methods for statistical validation of large proteome datasets; false discovery rate (FDR) calculations combined with peptide and protein probability approaches were used (Keller et al. 2002, Nesvizhskii et al. 2003, Weatherly et al. 2005). The cutoff was selected at less than 1%.
Results
RNA
Microarray analysis was performed and mRNA integrity was assessed by 3′/5′ ratios of GAPDH and ACTB. Neither GAPDH nor ACTB showed significant integrity differences caused by different storage conditions (Table 3).
Table 3.
RNA yield and integrity of specimens after long term storage at −80° C and in VPLN
| −80° C | VPLN | ||||
|---|---|---|---|---|---|
| Mean | Standard deviation | Mean | Standard deviation | p value | |
| yield | 7100 ng | 6800 ng | 4400 ng | 5400 ng | 0.006 |
| RIN | 5.8 | 2.8 | 4.3 | 3.3 | 0.0002 |
| DF | 39 | 37 | 56 | 42 | 0.003 |
| 3′:5′ GAPDH | 2.6 | 6.2 | 2.2 | 5.8 | 0.9 |
| 3′:5′ ACTB | 52 | 49 | 54 | 55 | 0.5 |
Based on the assumption that certain transcripts could be especially susceptible to storage-induced RNA degradation, SAM was performed to identify transcripts with significant differences between paired samples. All 44 probe sets with the highest significant differences between the groups showed higher signals in the group of samples stored at −80° C compared to those stored in VPLN (Table 4). We conclude that certain transcripts are especially susceptible to storage-induced degradation, which causes signal loss in the VPLN stored samples. Gene Ontology analysis of the 44 transcripts identified by SAM showed significant over-representation of specific biological themes (Table 5). This means that it is very unlikely that the differences between the two storage conditions could have been caused by random variation between samples.
Table 4.
Probe sets with significantly lower signals in specimens stored in VPLN compared to −80° C according to SAM analysis
| Probe set ID | Gene name |
|---|---|
| 202087_s_at | cathepsin L |
| 209581_at | HRAS-like suppressor 3 |
| 211911_x_at | major histocompatibility complex, class I, B |
| 201272_at | aldo-keto reductase family 1, member B1 (aldose reductase) |
| 209612_s_at | alcohol dehydrogenase IB (class I), beta polypeptide |
| 201649_at | ubiquitin-conjugating enzyme E2L 6 |
| 208729_x_at | major histocompatibility complex, class I, B |
| 209059_s_at | endothelial differentiation-related factor 1 |
| 211991_s_at | major histocompatibility complex, class II, DP alpha 1 |
| 202675_at | succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| 214864_s_at | glyoxylate reductase/hydroxypyruvate reductase |
| 200725_x_at | ribosomal protein L10 |
| 210972_x_at | T cell receptor alpha locus /// T cell receptor alpha constant |
| 206559_x_at | eukaryotic translation elongation factor 1 alpha 1 |
| 209036_s_at | malate dehydrogenase 2, NAD (mitochondrial) |
| 211529_x_at | HLA-G histocompatibility antigen, class I, G |
| 211799_x_at | major histocompatibility complex, class I, C |
| 209613_s_at | alcohol dehydrogenase IB (class I), beta polypeptide |
| 208870_x_at | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| 202201_at | biliverdin reductase B (flavin reductase (NADPH)) |
| 201231_s_at | enolase 1, (alpha) |
| 213366_x_at | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| 202746_at | integral membrane protein 2A |
| 200737_at | phosphoglycerate kinase 1 |
| 210460_s_at | proteasome (prosome, macropain) 26S subunit, non-ATPase, 4 |
| 212085_at | solute carrier family 25, member 6 |
| 201931_at | electron-transfer-flavoprotein, alpha polypeptide (glutaric aciduria II) |
| 200820_at | proteasome (prosome, macropain) 26S subunit, non-ATPase, 8 |
| 204599_s_at | mitochondrial ribosomal protein L28 |
| 209244_s_at | kinesin family member 1C |
| 217933_s_at | leucine aminopeptidase 3 |
| 202474_s_at | host cell factor C1 (VP16-accessory protein) |
| 204806_x_at | major histocompatibility complex, class I, F |
| 218893_at | isochorismatase domain containing 2 |
| 200663_at | CD63 antigen (melanoma 1 antigen) |
| 217972_at | coiled-coil-helix-coiled-coil-helix domain containing 3 |
| 218232_at | complement component 1, q subcomponent, alpha polypeptide |
| 202343_x_at | cytochrome c oxidase subunit Vb |
| 214836_x_at | HRV Fab N8-VL |
| 217408_at | mitochondrial ribosomal protein S18B |
| 215313_x_at | major histocompatibility complex, class I, A |
| 210547_x_at | islet cell autoantigen 1, 69 kDa |
| 213160_at | dedicator of cytokinesis 2 |
| 200752_s_at | calpain 1, (mu/l) large subunit |
Table 5.
Gene ontology categories over-represented in the 44 probe sets with significantly lower signals in specimens stored at VPLN
| System | Gene category | EASE score | Benjamini |
|---|---|---|---|
| GO Biological process | antigen presentation | 2×10−8 | 3×10−6 |
| antigen processing | 2×10−8 | 3×10−6 | |
| antigen presentation\, endogenous antigen | 4×10−8 | 4×10−6 | |
| antigen processing\, endogenous antigen via MHC class I | 6×10−8 | 5×10−6 |
Owing to the unexpected finding that samples stored at −80° C showed greater RNA integrity, we investigated whether removing the samples from storage boxes could be a confounding factor that affects RNA integrity. When samples from a storage box were removed, the entire box was stored for several minutes on dry ice and this could expose the samples to a temporary temperature increase. The biorepository′s database was used to determine the number of times that a storage box was moved from VPLN to dry ice because of routine specimen retrieval during the period of sample storage. Neither the number of accessions of a box nor the number of accessions of aliquots from the same sample was related to the loss of the ten most degradation-sensitive transcripts identified by SAM (data not shown).
Protein
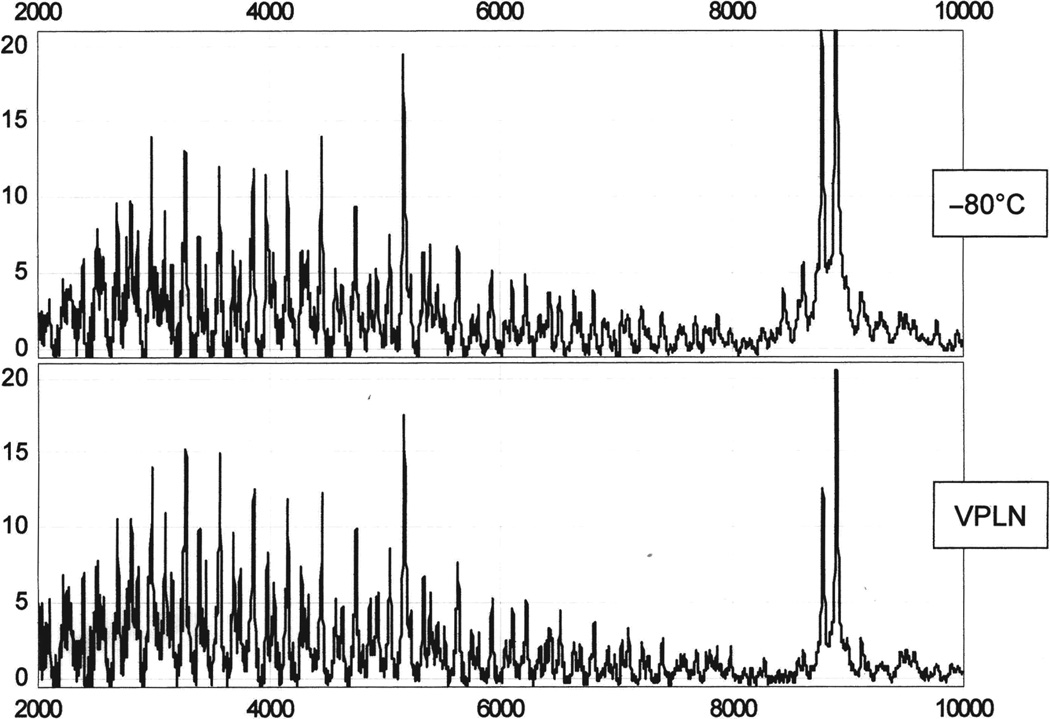
The peaks generated from the various aliquots analyzed by SELDI-TOF-MS, which was utilized to assess whole “non-enzyme digested” low molecular weight proteins and peptides, were similar to the equivalent triplicate measurements of each aliquot (Fig. 1). In some cases, the visual analysis of a blinded aliquot stored at −80° C yielded a higher signal to noise ratio for specific features, while this observation was reversed for other features depending on the type of tissue specimen stored in VPLN. We could not identify consistently which storage condition gave the best results for analyzing these samples; however, the spectra obtained from both groups were qualitatively similar and reproducible despite the different conditions and times of storage.
Fig. 1.
Spectra from a pair of lysed tissues from separate aliquots of the same human specimen stored under different conditions and for different times at −80° C or in VPLN. The spectra are similar.
Based on the qualitative results from SELDI-TOF-MS analysis, we initiated a more extensive analysis on the same samples using a high-end MALDI-TOF-MS instrument and a Brüker adapter plate specifically made to fit the SELDI-MS probes. All spectra were compared across each matched pair with 102 consistent features identified with a signal to noise ratio >4 (n = 4). Similar results were obtained; features within the −80° C group had a coefficient of variation (CV%) = 0.92, and the VPLN group had a CV% = 0.86. Therefore, the two groups were consistent overall. Specifically, comparing the spectra of matched pairs, the majority of peaks were similar within each aliquot of the matched pairs. Some peaks were observed consistently at one storage temperature (e.g., −80° C), but not at the other temperature (e.g., VPLN) and vice versa (i.e., absent at −80° C, but present at VPLN). Thus, an optimal temperature for storage could not be determined using either of these approaches (data not shown).
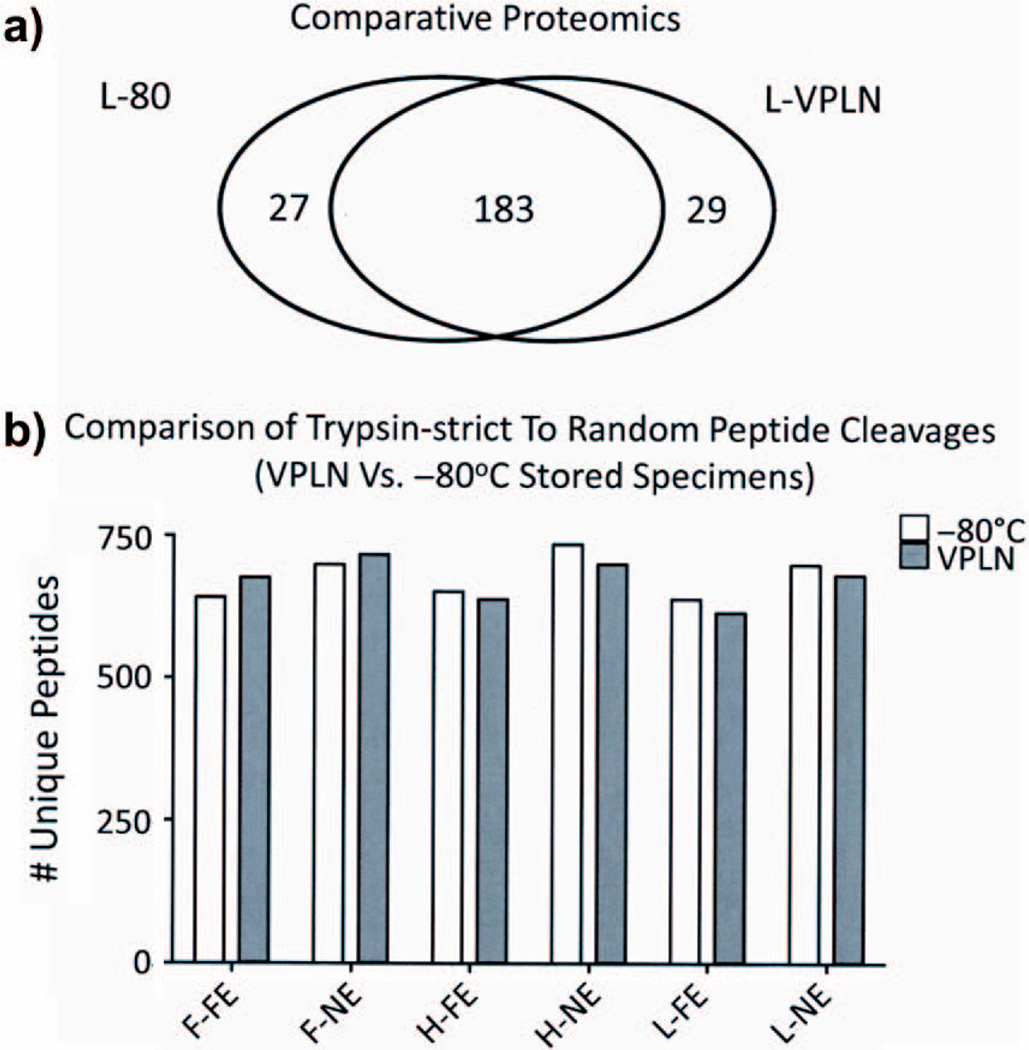
Finally, using LC-ESI-MS/MS analyses, we compared the protein identification numbers (ID’s) of matched pairs and a majority (183 of 239; 77%) was observed consistently in all specimens. The total number of proteins and the number of protein ID′s that were independent of each storage group also were similar (total: independent, 210:27 for −80° C and 211:29 for VPLN) (Fig. 2a). All tandem MS/MS spectra generated by the nLC-ESI-MS analysis were matched at the peptide level against all potential “no enzyme” cleavage sites to assess differences that likely are due to either continued endogenous enzymatic degradation or shearing effects caused by the storage method. This value is estimated by assigning MS/MS spectra that match peptide sequences with “strict trypsin” cleavage sites found only at the C-terminal residue of lysine and/or arginine vs. “no enzyme” cleavage sites found at any C- or N- terminal amino acid. While there was a slight increase in all runs using the no enzyme vs. the strict trypsin approach (702 vs. 655, an approximately 7% lower value than 702), there were no consistent differences between the two storage groups (Fig. 2b). Examples of the most abundant proteins identified by mass spectrometry are listed in Table 6.
Fig. 2.
a) Number of unique peptide ID’s generated by LCMS are shown within a strict Venn diagram for the two storage groups (−80° C and VPLN) derived from lymphoma (L) specimens as an example. b) Number of unique peptide identification numbers (ID′s) are shown in the figure for each sample for all groups. L, lymphoma; H, hepatoblastoma; F, follicular hyperplasia. Non-neoplastic lymph node comparing full enzyme (FE) and no enzyme (NE) searches in SEQUEST. In this way, we can estimate the number of shearing sites to be expected from freezing thawing or from a less optimal freezing approach.
Table 6.
Twenty-six most abundant proteins evaluated in the LC-ESI-MS-MS study of −80° C. VPLN storage for three specimens
| UniRef100 ID |
Sequence name | Protein weight (kDa) |
No. unique peptides |
H/VPLN | H/ −80C | %CV | L/VPLN | L/−80C | %CV | S/VPLN | S/ −80C | %CV |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B4DMF5 | glutamate dehydrogenase 1 | 56.55 | 10 | 28.4 | 34.2 | 0.00 | 2.5 | 0.0 | 0.00 | 0.0 | 0.0 | 0.00 |
| B3KQT2 | Protein disulfide-isomerase A3 |
54.85 | 15 | 22.2 | 26.8 | 18.71 | 10.1 | 7.0 | 35.87 | 8.3 | 7.8 | 6.71 |
| B3KML9 | tubulin beta-2C chain | 44.56 | 10 | 8.7 | 9.8 | 12.06 | 11.3 | 12.0 | 5.92 | 8.3 | 11.7 | 33.52 |
| P10809 | 60 kDa heat shock protein | 61.00 | 20 | 50.7 | 61.0 | 18.47 | 18.9 | 12.0 | 44.46 | 2.1 | 2.6 | 21.84 |
| P11021 | 78 kDa glucose-regulated protein |
72.27 | 25 | 64.3 | 63.4 | 1.32 | 18.9 | 19.0 | 0.74 | 10.4 | 22.0 | 71.85 |
| P06733 | alpha-enolase | 47.12 | 25 | 33.4 | 40.2 | 18.70 | 44.0 | 42.0 | 4.65 | 14.6 | 16.9 | 14.65 |
| B3VL17 | beta globin | 11.24 | 11 | 106.3 | 80.5 | 27.61 | 30.2 | 45.0 | 39.46 | 150.7 | 132.2 | 13.06 |
| A8K4K6 | disulfide isomerase family A, member 4 |
72.80 | 12 | 22.2 | 8.5 | 0.00 | 0.0 | 1.0 | 0.00 | 0.0 | 0.0 | 0.00 |
| P14625 | endoplasmin | 92.39 | 25 | 34.6 | 45.1 | 26.39 | 15.1 | 14.0 | 7.49 | 12.5 | 13.0 | 3.85 |
| P04406 | glyceraldehyde-3- phosphate dehydrogenase |
36.01 | 13 | 9.9 | 23.2 | 80.34 | 31.4 | 42.0 | 28.79 | 32.2 | 25.9 | 21.63 |
| P11142 | heat shock cognate 71 kDa protein |
70.84 | 20 | 38.3 | 42.7 | 10.79 | 377 | 31.0 | 19.53 | 20.8 | 31.1 | 39.80 |
| P07900 | heat shock protein HSP 90-alpha |
84.59 | 15 | 40.8 | 28.1 | 36.99 | 16.3 | 22.0 | 29.53 | 13.5 | 15.6 | 14.10 |
| P08238 | heat shock protein HSP 90-beta |
83.19 | 16 | 48.2 | 39.0 | 21.03 | 25.1 | 26.0 | 3.36 | 9.4 | 13.0 | 32.26 |
| P69905 | hemoglobin alpha | 15.23 | 10 | 70.4 | 48.8 | 36.32 | 11.3 | 30.0 | 90.49 | 139.3 | 105.0 | 28.06 |
| Q3LR79 | hemoglobin beta | 11.46 | 10 | 60.6 | 54.9 | 9.82 | 176 | 31.0 | 55.14 | 69.6 | 51.9 | 29.27 |
| P02042 | hemoglobin subunit delta | 16.03 | 12 | 64.3 | 46.3 | 32.39 | 10.1 | 20.0 | 66.13 | 110.2 | 106.3 | 3.58 |
| P68871 | LVV-hemorphin-7 | 15.97 | 16 | 116.2 | 93.9 | 21.19 | 32.7 | 55.0 | 50.88 | 189.2 | 159.5 | 17.04 |
| Q06830 | peroxiredoxin-1 | 22.08 | 13 | 2.5 | 8.5 | 110.26 | 16.3 | 11.0 | 39.06 | 9.4 | 9.1 | 3.04 |
| P00558 | phosphoglycerate kinase 1 | 44.57 | 11 | 11.1 | 73 | 41.21 | 13.8 | 14.0 | 1.22 | 5.2 | 13.0 | 85.46 |
| P13796 | plastin-2 | 70.23 | 11 | 0.0 | 0.0 | 0.00 | 18.9 | 15.0 | 22.80 | 13.5 | 14.3 | 5.40 |
| P14618 | pyruvate kinase isozymes M1/M2 |
57.88 | 16 | 1.2 | 3.7 | 98.78 | 23.9 | 24.0 | 0.46 | 11.4 | 2.6 | 126.11 |
| Q9Y490 | talin-1 | 269.58 | 13 | 0.0 | 0.0 | 0.00 | 1.3 | 3.0 | 81.69 | 16.6 | 14.3 | 15.34 |
| P67936 | tropomyosin alpha-4 chain | 28.49 | 13 | 14.8 | 13.4 | 10.06 | 6.3 | 20.0 | 104.30 | 19.8 | 16.9 | 15.85 |
| P23381 | tryptophanyl-tRNA synthetase |
53.11 | 11 | 0.0 | 0.0 | 0.00 | 0.0 | 15.0 | 200.00 | 73 | 13.0 | 56.13 |
| P07437 | tubulin beta chain | 49.62 | 17 | 16.1 | 20.7 | 25.39 | 15.1 | 24.0 | 45.59 | 22.9 | 18.2 | 23.01 |
| P08670 | vimentin | 53.60 | 24 | 44.5 | 378 | 16.24 | 41.5 | 48.0 | 14.55 | 39.5 | 50.6 | 24.56 |
| sum = 909.4 | 8378 | 25.93 | 470.2 | 583.0 | 38.16 | 936.6 | 866.0 | 27.16 | ||||
| avg. % rsd |
avg. % rsd |
avg. % rsd |
H, hepatoblastoma; LP, lymphoma; S, non-neoplastic spleen; rsd, relative standard deviation.
Discussion
Storage at −80° C and in VPLN are the two most common methods for long term storage of fresh-frozen tissue samples. Little information is available about the relative benefits of these long term storage methods for analysis of RNA or proteins, except that storage near VPLN temperatures is required to maintain long term viability of cells.
Some molecules may be affected by freezing, others by thawing and others by both. Similarly, the duration of long term storage under either condition probably results in molecular changes as indicated by the RIN scores we found for specimens whose average RINs were less than expected for specimens stored for a short time. The freezing and thawing procedures for the two storage methods were equivalent overall. The effects of storage time on RIN were based on paired samples with each tissue specimen of a pair stored for the same time. Thus, the most common difference between each of the paired samples and the differences we observed in RNA and proteins we attribute to storage temperature.
We found that for 49 specimens of various types of tissues and cancers, storage at −80° C maintained at least the same RNA integrity as VPLN storage. For RNA yield and total RNA integrity, −80° C storage provided significantly better results than VPLN storage. For 37 specimens analyzed by expression profiling, mRNA integrity measured by 3′:5′ ratios of GAPDH and ACTB showed no significant differences; by contrast, SAM showed that a number of transcripts seemed to be especially susceptible to degradation in samples stored by VPLN. These transcripts were significantly lower in samples stored in VPLN compared to storage at −80° C. Transcripts that are highly sensitive to VPLN storage frequently are involved in antigen presentation and processing, which suggests that these transcripts may be useful for assessing storage-induced RNA degradation.
With the development of real-time qualitative PCR (RT-Q-PCR), the effects of degradation of RNA may be minimized if short amplicons (≤ 100 bp) are used. Using this technology, gene expression even in paraffin blocks can be analyzed reliably (Steg et al. 2006, 2007). Use of RT-Q-PCR also reduces some concern about long term storage of tissues below −80° C.
We reported earlier that multiple freeze-thaw cycles cause degradation of proteins in samples of serum. We demonstrated also that storage at −80° C for 18 months does not cause significant degradation of many proteins; however, storage of serum samples at −20° C for more than 6 months does cause significant protein degradation (Grizzle et al. 2005a,b). We have shown here that in the absence of thawing, there were no appreciable differences in protein integrity between the two storage methods. While these approaches to mass spectrometry do not evaluate proteins and peptides that are present at low concentrations, for most of the proteins we evaluated, there were no differences between the two storage temperatures; however, it should be emphasized that the proteins analyzed by SEL-DI-MS, MALDI-MS, and LC-ESI-MS/MS represent only a sample of some of the more abundant proteins of the proteome. Our study was limited to global low resolution studies in which the amounts of protein were small and the resources available for analysis were limited; therefore, specific proteins that were not observed may be more or less susceptible to storage conditions.
The effects of storage temperature on human tissue have not been studied previously to the extent that we present here and our results are informative with regard to the lack of major changes in the most abundant proteins. There is no reason to expect that changes during storage of more abundant proteins do not mirror those of less abundant proteins. Together with the RNA studies, our results suggest that more extensive, high resolution studies are warranted. It is important that most unique proteins identified by mass spectrometry in each member of the paired samples were the same. A relatively few unique proteins were not detected in one of the storage conditions compared to the other storage condition (Fig. 1). Neither method of storage was clearly better than the other with regard to preserving specific proteins (Table 6).
Our results are surprising in view of our current understanding and assumptions regarding temperature and biospecimen integrity. Theoretically, samples stored in VPLN should be below the glass transition temperature of water and no enzymatic or hydrolytic breakdown of RNA or protein should occur. We currently have no hypothesis about why VPLN storage does not conserve RNA integrity as well as −80° C storage. Some of the specimens stored in VPLN were removed from the freezer more frequently for distribution of specimens to researchers and this could have affected RNA integrity adversely if the specimens were warmed above the glass transition phase for short periods of time. If this hypothesis were true, however, one would expect that samples that were moved more frequently would show a greater degree of RNA degradation, but we found no such correlation in our cohort. There is controversy concerning the exact Tg of pure water (Giovambattista et al. 2004) and the Tg of the water phase of mammalian tissues is unknown; however, enzymatic reactions are thought to continue at −80° C.
We conclude that the long term storage of fresh-frozen human tissue specimens in mechanical −80°C freezers preserves at least the same RNA and protein quality as specimens stored in VPLN.
Acknowledgements
Drs. Ramirez and Grizzle are co-senior authors on this manuscript. Supported by all the individual grants of the Cooperative Human Tissue Network (CHTN) and the University of Alabama at Birmingham Mass Spectrometry/Proteomics (MSP) and Tissue Procurement Shared Facilities of the University of Alabama at Birmingham Comprehensive Cancer Center.
Footnotes
Dedication
This manuscript is dedicated to Dr. Steven J. Qualman, who is deceased. Without Steve′s vision and leadership, this manuscript would not have been possible. David L. Newsom, who is deceased, also contributed to this manuscript.
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for its contents.
References
- Adam BL, Qu Y, Davis JW, Ward MD, Clements MA, Cazares LH, Semmes OJ, Schellhammer PF, Yasui Y, Feng Z, Wright GL., Jr Serum protein fingerprinting coupled with a pattern-matching algorithm distinguishes prostate cancer from benign prostate hyperplasia and healthy men. Cancer Res. 2002;62:3609–3614. [PubMed] [Google Scholar]
- Auer H, Lyianarachchi S, Newsom D, Klisovic MI, Marcucci G, Kornacker K. Chipping away at the chip bias: RNA degradation in microarray analysis. Nat. Genet. 2003;35:292–293. doi: 10.1038/ng1203-292. [DOI] [PubMed] [Google Scholar]
- Giovambattista N, Angell CA, Sciortino F, Stanley HE. Glass-transition temperature of water: a simulation study. Am. Phys. Soc. 2004;93 doi: 10.1103/PhysRevLett.93.047801. 047801-1-047801-4. [DOI] [PubMed] [Google Scholar]
- Grizzle WE, Semmes OJ, Bigbee W, Zhu L, Malik G, Oelschlager DK, Marine B, Marine U. The need for the review and understanding of SELDI/MALDI mass spectroscopy data prior to analysis. Cancer Informat. 2005a;1:86–97. [PMC free article] [PubMed] [Google Scholar]
- Grizzle WE, Semmes OJ, Bigbee WL, Malik G, Miller E, Manne B, Oelschalger DK, Zhu L, Manne U. Use of mass spectrographic methods to identify disease processes. In: Patrinos G, Ansorg W, editors. Molecular Diagnostics, Chapter 17. Waltham, MA: Academic Press/ Elsevier; 2005b. pp. 211–222. [Google Scholar]
- Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. Epub 2003 Sep 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Kojima K, Asmellash S, Klug CA, Grizzle WE, Mobley JA, Christein JD. Applying proteomic-based biomarker tools for the accurate diagnosis of pancreatic cancer. J Gastrointest. Surg. 2008 Jul; doi: 10.1007/s11605-008-0632-6. [DOI] [PubMed] [Google Scholar]
- McLerran D, Grizzle WE, Feng Z, Thompson IM, Bigbee WL, Cazares LH, Chan DW, Dahlgren J, Diaz J, Kagan J, Lin DW, Malik G, Oelschlager D, Partin A, Randolph TW, Sokoll L, Srivastava S, Srivastava S, Thornquist M, Troyer D, Wright GL, Zhang Z, Zhu L, Semmes OJ. SELDI-TOF MS whole serum proteomic profiling with IMAC surface does not reliably detect prostate cancer. Clin. Chem. 2008a;54:53–60. doi: 10.1373/clinchem.2007.091496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLerran D, Grizzle WE, Feng Z, Bigbee WL, Banez LL, Cazares LH, Chan DW, Diaz J, Izbicka E, Kagan J, Malehorn DE, Malik G, Oelschlager D, Partin A, Randolph T, Rosenzweig N, Srivastava S, Srivastava S, Thompson IM, Thornquist M, Troyer D, Yasui Y, Zhang Z, Zhu L, Semmes OJ. Analytical validation of serum proteomic profiling for diagnosis of prostate cancer: Sources of sample bias. Clin. Chem. 2008b;54:44–52. doi: 10.1373/clinchem.2007.091470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Semmes OJ, Feng Z, Adam B-L, Banez LL, Bigbee WL, Campos D, Cazares LH, Chan DW, Grizzle WE, Izbicka E, Kagan J, Malik G, McLerran D, Moul JW, Partin A, Prasanna P, Rosenzweig J, Sokoll LJ, Srivastava S, Srivastava S, Thompson I, Welsh MJ, White N, Winget M, Yasui Y, Zhang Z, Zhu L. Evaluation of serum protein profiling by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry for the detection of prostate cancer: I. Assessment of platform reproducibility. Clin. Chem. 2005;51:102–112. doi: 10.1373/clinchem.2004.038950. [DOI] [PubMed] [Google Scholar]
- Steg A, Wang W, Blanquicett C, Grunda JM, Eltoum IA, Wang K, Buchsbaum DJ, Vickers SM, Russo S, Diasio RB, Frost AR, Grizzle WE, Johnson MR. Multiple gene expression analyses in paraffin-embedded tissues by Taqman low density array: application to Hedgehog and Wnt pathway analysis in ovarian endometrioid adenocarcinoma. J. Mol. Diagn. 2006;8:76–83. doi: 10.2353/jmoldx.2006.040402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steg A, Vickers SM, Eloubeidi M, Wang W, Eltoum IA, Grizzle WE, Saif MW, Lobuglio AF, Frost AR, Johnson MR. Hedgehog pathway expression in heterogeneous pancreatic adenocarcinoma: implications for the molecular analysis of clinically available biopsies. Diagn. Mol. Pathol. 2007;16:229–237. doi: 10.1097/PDM.0b013e31811edc7e. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Clark ME, Crossman DK, Kojima K, Messinger JD, Mobley JA, Curcio CA. Abundant lipid and protein components of drusen. PLoS ONE. 2010;5:el0329. doi: 10.1371/journal.pone.0010329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherly DB, Atwood JA, III, Minning TA, Cavola C, Tarleton RL, Orlando R. A heuristic method for assigning a false-discovery rate for protein identifications from Mascot database search results. Mol. Cell. Proteom. 2005;4:762–772. doi: 10.1074/mcp.M400215-MCP200. [DOI] [PubMed] [Google Scholar]