Abstract
Carotid atherosclerosis is a chronic inflammatory disease of the arterial wall. The present study aimed to identify changes in the gene expression and regulatory factors for atherosclerotic plaques of carotid atherosclerosis from an early to an advanced stage. The original data were downloaded from the NCBI GEO database under accession no. GSE28829. Differentially expressed genes (DEGs) were detected by the Robust Multiarray Average (RMA). The enriched Gene Ontology (GO) terms and pathways for DEGs using DAVID were subsequently identified. The transcriptional and microRNA (miRNA) regulatory network were constructed for the DEGs. Cis-regulatory signals were also investigated. More genes were activated in the advanced stage compared with the early stage. IGHG1 and SPP1 were upregulated, while MYBL1 and PLD were downregulated. The upregulated genes in the advanced stage were involved in atherosclerosis-related GO terms such as immune, vascular and cell movement homeostasis. The DEGs were significantly enriched in cell adhesion molecules (CAMs) and the focal adhesion pathway. MMP9 and CFL2 played key roles in the transcriptional regulatory network. Moreover, miR-328 was identified as one of the hubs in the miRNA regulatory network. The results may therefore be used to determine the mechanism involved in carotid atherosclerosis.
Keywords: carotid atherosclerosis, differentially expressed genes, transcription factors, miRNA
Introduction
Atherosclerosis is a chronic disease that remains asymptomatic for decades (1). It is caused by the formation of multiple plaques within the arteries. Attention has been focused on the ‘vulnerable plaque’ since the late 1990s onwards (2). Atherosclerosis can lead to ischemic heart disease, cerebrovascular accidents and peripheral vascular diseases (3). Carotid intima-media thickness (cIMT) level is known as a surrogate marker of atherosclerosis (4). A special type of carotid atherosclerosis with CagA-positive Helicobacter pylori (CagA+ HP) infection is common in China. An increased serum YKL-40 level suggests plaque instability and more severe clinical symptoms of carotid atherosclerosis with CagA+ HP infection (5). Inflammatory cytokines induced by VEGF, such as monocyte chemoattractant protein (MCP-1), have been previously shown to be involved in the pathogenesis and progression of carotid atherosclerosis (6).
Metabolic syndrome may be independently associated with the early stage but not the later and advanced stages of carotid atherosclerosis in community residents in China (7). An in vivo 3T MRI study was previously conducted to determine the effect of gender differences of high-risk carotid atherosclerotic plaque with <50% stenosis in asymptomatic patients (8). Evaluation of carotid atherosclerosis was therefore performed from the perspective of blood flow reflection (9). Results of a multivariate analysis revealed plaque number by ultrasonography (P=0.023), age (P=0.001), calcium-phosphate product (P=0.049) and serum albumin (P=0.009) as independent risk factors (10). Higher brachial-ankle pulse wave velocity was identified as a risk factor for carotid atherosclerosis in patients with end-stage renal disease (11). The relationship between levels of circulating intercellular cell-adhesion molecule-1 (cICAM-1) or P-selectin (cP-selectin) and the severity of carotid atherosclerosis was also examined (12). The findings of that study showed that cP-selectin did not increase until atherosclerosis was at an advanced stage (12).
Inactivation of the PDZK1 gene is known to promote the development of aortic root atherosclerosis in apolipoprotein E (apoE) KO mice fed with a high fat/high cholesterol diet (13). Additionally, SERPINA1 was found to be upregulated in atherosclerotic plaques (14). Angiogenesis, the process of new capillary formation from existing blood vessels, is dysregulated in many pathological disorders including atherosclerosis (15). In CD68+ cells from regressing plaque of atherosclerosis, Feig et al (16) observed that genes related to cell adhesion, such as cadherins and vinculin, were downregulated.
Transcription factors such as LDLR and TP53 are important in the development of atherosclerosis, as identified in a Malaysian study population (13). It has been previously reported that miRNAs are associated with atherosclerosis (17). However, the association between miRNAs and atherosclerosis has not yet been fully elucidated.
Immune-associated biological processes can affect atherosclerosis. Growth differentiation factor-15 deficiency inhibits the progression of atherosclerosis by controlling the interleukin-6-dependent inflammatory response to vascular injury (18). Vascular smooth muscle cell (VSMC) phenotypic modulation plays a key role in atherosclerosis. SMCs secrete cytokines and express cell adhesion molecules such as IL-8, IL-6 and VCAM-1 (19). Prenatal arsenic exposure alters gene expression in the adult liver to a proinflammatory state contributing to accelerated atherosclerosis by affecting pathways such as antigen processing and presentation (20). Vascular smooth muscle contraction is a main effect for artery (21). Morelloflavone, a biflavonoid and an active ingredient of the plant, has been shown to inhibit VSMC migration through its inhibition of multiple migration-related kinases such as focal adhesion kinase (22).
Therefore, in the present study, we initially detected differentially expressed genes (DEGs). Subsequently, we identified the enriched Gene Ontology (GO) terms and pathways for the DEGs. The transcriptional and miRNA regulatory network for the DEGs was also constructed. Cis-regulatory signals were also investigated.
Materials and methods
Data preprocessing
The original data were downloaded from the National Center for Biotechnology Gene Expression Omnibus (NCBI GEO) database for atherosclerotic plaques of carotid atherosclerosis under accession no. GSE28829 (23), including 13 chips at early stage (EAR, pathological, intimal thickening and intimal xanthoma) and 16 chips at advanced stage (ADV, thin or thick fibrous cap atheroma). The chip platform was GPL570, Affymetrix Human Genome U133 Plus 2.0 Array.
Background subtraction and quantile normalization was performed using Affymetrix Power Tools (APT) (http://www.affymetrix.com/) in the Robust Multiarray Average (RMA) algorithm (24). Genes with a low expression were filtered, ensuring that their plier was ≥100 in at least 2/3 of the samples (25).
Identifying differentially expressed genes
Criteria for a differential probe included: i) signal strength of one probe is >1.5-fold between the two groups; ii) t-test P-value of ≤0.01; and iii) plier ≥100 in at least 2/3 of the samples.
Functional enrichment of differential genes
Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was conducted with DAVID (26) for the differential genes between two groups. The threshold [P-value ≤0.05 and false discovery rate (FDR) ≤0.05] was calculated using Fisher’s Exact test.
Transcriptional regulatory network
The differential genes, especially atherosclerosis-related genes, may be regulated by different transcriptional factors and miRNAs in different stages. We initally extracted the sequences located at 1k bp upstream and 200 bp downstream of 5′UTR for the transcription start site, using JASPAR (27) (http://jaspar.genereg.net) (the score threshold was 0.95). The core transcriptional regulatory network was constructed for the upregulated genes, with a >4-fold change in the advanced stage and a >2-fold change in the early stage. Upregulated genes were regulated by transcriptional factors (TFs) in the early and advanced stages of carotid atherosclerosis. TFs act depending on cis-regulatory motif in 5′UTR of their target genes. In order to detect such significant enriched cis-regulatory signals, hypergeometric distribution test was implemented (28), with a P-value of ≤0.05.
miRNA regulatory network
The 1k bp 3′UTR sequence was extracted from the differentially expressed genes. Although the binding of miRNA and its targets in animals are not as conservative as in plant, while binding is relatively conservative in the seed region. RNAhybrid is based conservation in seed region, which makes it more suitable for animal miRNA target gene prediction. RNAhybrid (29) was subsequently used to identify miRNA binding sites (P-value ≤0.01).
Results
Differentially expressed genes
In the present study, the early stage sample was considered as the control group. We identified 889 differential probes (corresponding to 707 genes), of which 413 probes (322 genes) were downregulated, and 476 probes (385 genes) were upregulated in advanced stage samples.
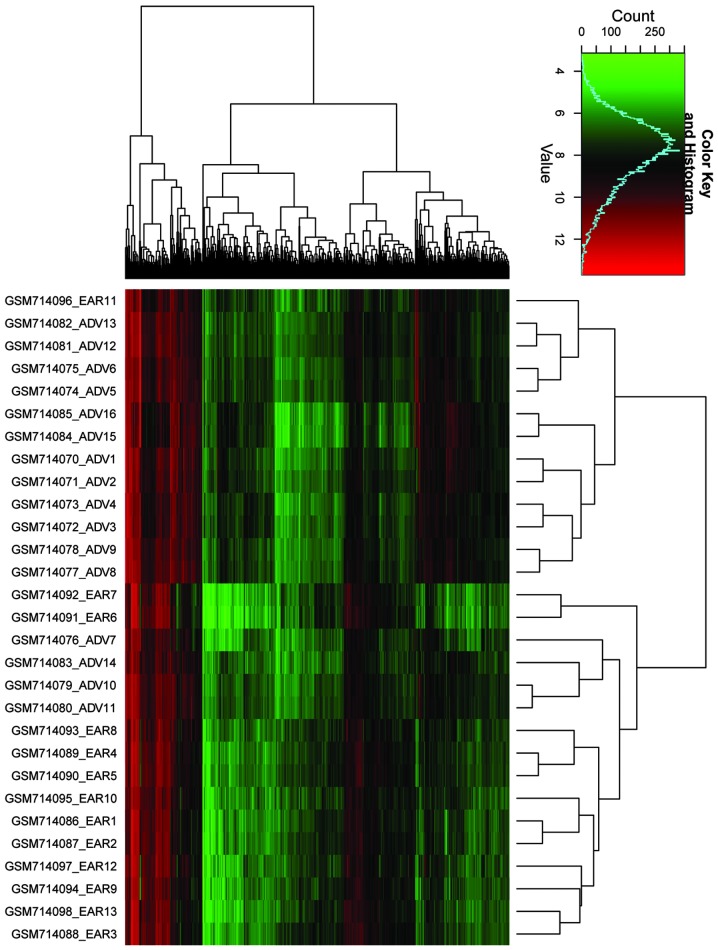
The 889 differential probes were clustered using R hclust based on RMA log2-transformed values (Fig. 1). Thirteen samples were clustered into one group, with only one early stage sample being mis-clustered in this group. The standard error of the mean distribution is shown in Fig. 2. Fig. 3 shows the up- and downregulation for the 889 differential probes (corresponding to 708 genes). As the absolute log fold change (|logFC|) becomes larger, the number of upregulated genes were reduced (Fig. 3A). However, the fold change increased as the |logFC| became larger (Fig. 3B). For example, when |logFC|≥1.5, the fold change was 0.58, whereas when |logFC|≥1, the fold change was 1. This finding suggested that a large number of genes were activated in the advanced stage.
Figure 1.
Hierarchical clustering analysis of 889 differentially expressed transcripts in all samples.
Figure 2.
The average expression value of 889 differentially expressed transcripts in each sample. Purple bars are the advanced samples and green bars are the early stage sample. Bar values are the standard error of the mean (SEM).
Figure 3.
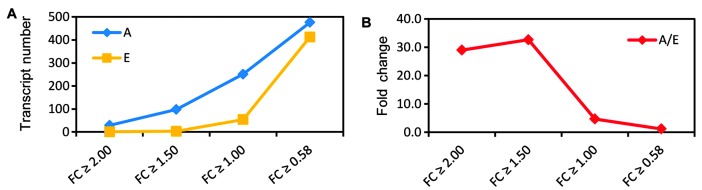
The distribution of 889 differentially expressed transcripts (DETs) in the early and advanced stages. (A) The number of transcripts with a different FC. Blue shows the advanced stage samples, while yellow shows the early stage samples. (B) The fold change of DETs in the advanced samples when compared with its expression in the early stage samples with a different FC. FC represents the log2 fold change.
Enriched functional terms of differential genes
The downregulated genes were enriched in biological processes including cytoskeleton organization, cell adhesion, muscle organ development, regulation of muscle contraction, regulation of cell growth, regulation of system process, regulation of metal ion transport, heart development, muscle contraction, muscle cell differentiation, negative regulation of cell growth, intracellular signaling cascade, myofibril assembly, cellular metal ion homeostasis, regulation of calcium ion transport, negative regulation of cell size and regulation of intracellular transport (Table I). The findings demonstrated that the biological processes do not correlate with atherosclerosis.
Table I.
Major biological processes enriched by upregulated genes in early stage.
| Term | Count | P-value | FDR |
|---|---|---|---|
| GO:0007010: Cytoskeleton organization | 25 | 2.12E-07 | 3.51E-04 |
| GO:0007155: Cell adhesion | 29 | 1.14E-05 | 1.88E-02 |
| GO:0007517: Muscle organ development | 14 | 4.57E-05 | 7.56E-02 |
| GO:0006937: Regulation of muscle contraction | 7 | 1.16E-03 | 1.90E+00 |
| GO:0001558: Regulation of cell growth | 11 | 1.38E-03 | 2.25E+00 |
| GO:0044057: Regulation of system process | 14 | 1.80E-03 | 2.93E+00 |
| GO:0010959: Regulation of metal ion transport | 7 | 2.13E-03 | 3.46E+00 |
| GO:0007507: Heart development | 11 | 2.92E-03 | 4.73E+00 |
P≤0.05 and false discovery rate (FDR) ≤0.05.
The upregulated genes enriched in biological processes associated with atherosclerosis were: i) immune-associated GO terms, including defense response, response to wounding, inflammatory response, immune effector process, leukocyte mediated immunity, antigen processing and presentation of peptide or polysaccharide antigen via the MHC class II, adaptive immune response, activation of immune response, lymphocyte mediated immunity, antigen processing and presentation, behavior, B cell-mediated immunity, antigen processing and presentation of exogenous peptide antigen, acute inflammatory response, chemotaxis, antigen processing and presentation of exogenous antigen; ii) vascular-related GO terms including blood vessel development, blood vessel morphogenesis, and angiogenesis; and iii) cell movement homeostasis including cell adhesion, regulation of cell motion, regulation of cell migration, regulation of locomotion, cation homeostasis and chemical homeostasis (Table II).
Table II.
Major biological processes enriched by upregulated genes in advanced stage.
| Term | Count | P-value | FDR |
|---|---|---|---|
| GO:0006955: Immune response | 86 | 4.85E-40 | 8.42E-37 |
| GO:0006952: Defense response | 65 | 1.40E-25 | 2.43E-22 |
| GO:0009611: Response to wounding | 57 | 1.10E-22 | 1.91E-19 |
| GO:0006954: Inflammatory response | 43 | 2.13E-20 | 3.69E-17 |
| GO:0006935: Chemotaxis | 27 | 2.20E-15 | 3.85E-12 |
| GO:0042330: Taxis | 27 | 2.20E-15 | 3.85E-12 |
| GO:0002252: Immune effector process | 21 | 2.08E-11 | 3.61E-08 |
| GO:0048584: Positive regulation of response to stimulus | 27 | 2.37E-11 | 4.11E-08 |
| GO:0007626: Locomotory behavior | 29 | 2.40E-11 | 4.16E-08 |
| GO:0002443: Leukocyte mediated immunity | 17 | 7.37E-11 | 1.28E-07 |
| GO:0002504: Antigen processing and presentation of peptide or polysaccharide antigen via MHC class II | 12 | 8.24E-11 | 1.43E-07 |
| GO:0002250: Adaptive immune response | 16 | 1.47E-10 | 2.55E-07 |
| GO:0002253: Activation of immune response | 17 | 2.97E-10 | 5.16E-07 |
| GO:0002449: Lymphocyte mediated immunity | 15 | 4.22E-10 | 7.33E-07 |
| GO:0019882: Antigen processing and presentation | 16 | 4.49E-10 | 7.79E-07 |
| GO:0007610: Behavior | 33 | 2.54E-08 | 4.41E-05 |
| GO:0019724: B cell-mediated immunity | 12 | 3.99E-08 | 6.92E-05 |
| GO:0002478: Antigen processing and presentation of exogenous peptide antigen | 7 | 5.36E-08 | 9.29E-05 |
| GO:0007155: Cell adhesion | 40 | 1.72E-07 | 2.98E-04 |
| GO:0001568: Blood vessel development | 22 | 1.75E-07 | 3.04E-04 |
| GO:0002526: Acute inflammatory response | 14 | 3.02E-07 | 5.23E-04 |
| GO:0019884: Antigen processing and presentation of exogenous antigen | 7 | 3.29E-07 | 5.70E-04 |
| GO:0001775: Cell activation | 23 | 6.13E-07 | 1.06E-03 |
| GO:0030334: Regulation of cell migration | 17 | 1.43E-06 | 2.48E-03 |
| GO:0048514: Blood vessel morphogenesis | 19 | 1.45E-06 | 2.51E-03 |
| GO:0051270: Regulation of cell motion | 18 | 1.83E-06 | 3.18E-03 |
| GO:0055066: Di-, tri-valent inorganic cation homeostasis | 20 | 2.13E-06 | 3.69E-03 |
| GO:0045321: Leukocyte activation | 20 | 2.56E-06 | 4.44E-03 |
| GO:0006956: Complement activation | 9 | 3.78E-06 | 6.56E-03 |
| GO:0002541: Activation of plasma proteins involved in acute inflammatory response | 9 | 4.55E-06 | 7.90E-03 |
| GO:0001525: Angiogenesis | 15 | 6.60E-06 | 1.14E-02 |
| GO:0042592: Homeostatic process | 38 | 6.64E-06 | 1.15E-02 |
| GO:0040012: Regulation of locomotion | 17 | 7.55E-06 | 1.31E-02 |
| GO:0009617: Response to bacterium | 17 | 8.07E-06 | 1.40E-02 |
| GO:0055080: Cation homeostasis | 21 | 8.17E-06 | 1.42E-02 |
| GO:0050865: Regulation of cell activation | 16 | 1.04E-05 | 1.80E-02 |
| GO:0048878: Chemical homeostasis | 29 | 1.48E-05 | 2.57E-02 |
| GO:0006874: Cellular calcium ion homeostasis | 16 | 1.77E-05 | 3.07E-02 |
P≤0.05 and false discovery rate (FDR) ≤0.05.
The enriched pathways of differential genes included systemic lupus erythematosus, antigen processing and presentation, complement and coagulation cascades, asthma, viral myocarditis, lysosome, intestinal immune network for IgA production (Table III).
Table III.
Enriched pathways of differentially expressed transcripts.
| Term | Count | P-value | FDR |
|---|---|---|---|
| hsa05322: Systemic lupus erythematosus | 21 | 3.65E-08 | 0.00004 |
| hsa04514: Cell adhesion molecules (CAMs) | 23 | 2.78E-07 | 0.00033 |
| hsa04612: Antigen processing and presentation | 18 | 3.26E-07 | 0.00039 |
| hsa04610: Complement and coagulation cascades | 16 | 7.32E-07 | 0.00087 |
| hsa05310: Asthma | 10 | 5.73E-06 | 0.00681 |
| hsa05416: Viral myocarditis | 15 | 5.92E-06 | 0.00704 |
| hsa04142: Lysosome | 19 | 1.13E-05 | 0.01347 |
| hsa04672: Intestinal immune network for IgA production | 12 | 1.66E-05 | 0.01978 |
P-value ≤0.05 and false discovery rate (FDR) ≤0.05.
Transcriptional regulatory network
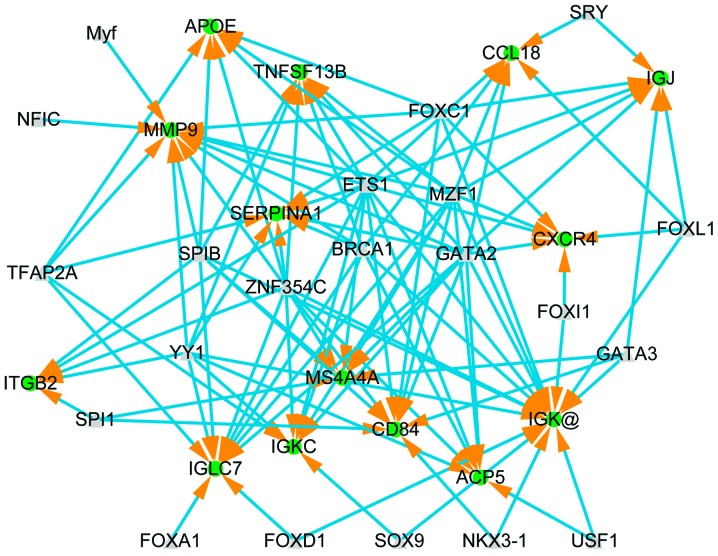
The core transcriptional regulatory network is shown in Fig. 4 for upregulated genes (RMA log2 transformed values >2). Fourteen upregulated genes are shown, targeted by 21 transcription factors, constructing 111 regulatory relationships. The hub genes are IGK (15 neighbors), MMP9 (11 neighbors) and IGLC7 (10 neighbors).
Figure 4.
The core transcriptional regulatory network of upregulated genes (RMA log2 transformed values >2). Green nodes are the target genes, and the other nodes are the transcription factors. Arrows show that transcription factors target the genes.
The core transcriptional regulatory network is shown in Fig. 5 for downregulated genes (RMA log2 transformed values <-1). Forty-five downregulated genes, targeted by 22 transcription factors, constructing 324 regulatory relationships. The hub genes are AW451999 (10 neighbors), CFL2 (10 neighbors) and PDZRN3 (10 neighbors).
Figure 5.
The core transcriptional regulatory network of downregulated genes (RMA log2 transformed values <-1). Green nodes are the target genes, and the other nodes are the transcription factors. Arrows show that transcription factors target the genes.
Significantly enriched cis-regulatory signals
Fig. 6A shows the 5′UTR of upregulated genes enriched in TF motif Myf (P-value 1.33E-02) and MZF1 (P-value 2.47E-02). By contrast, Fig. 6B shows the 5′UTR of downregulated genes enriched in TF motif FOXL1 (P-value, 5.95E-03) and NKX3-1 (P-value, 3.57E-02).
Figure 6.
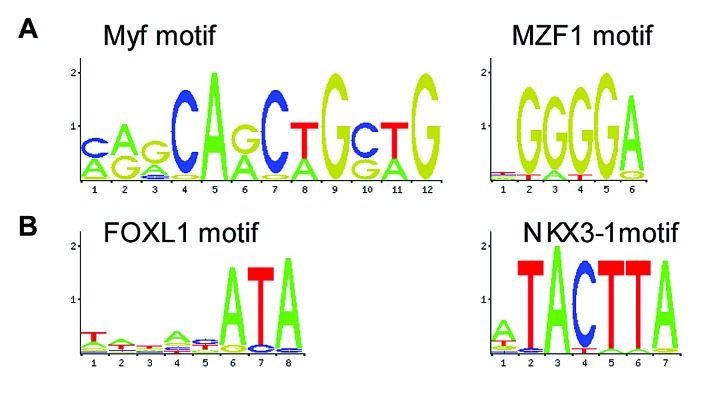
Significantly enriched TF motifs by DETs. Enriched motifs of (A) upregulated and (B) downregulated genes.
miRNA regulatory network
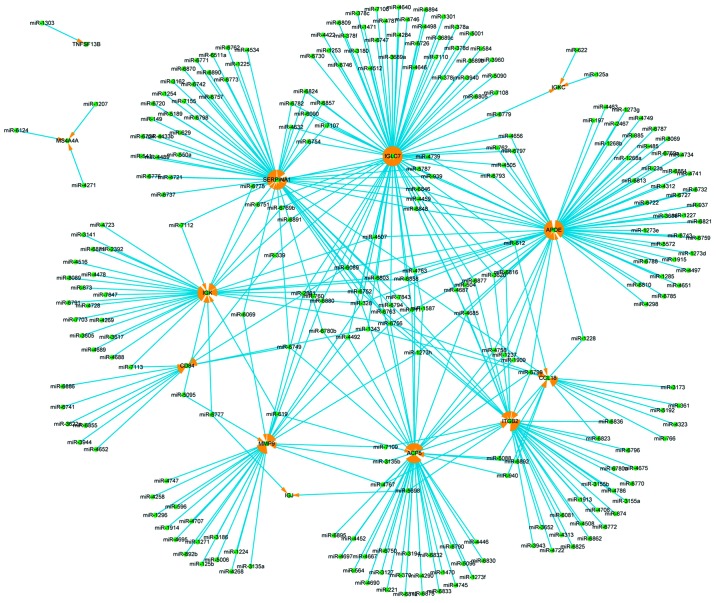
The core miRNA regulatory network is shown in Fig. 7 for the upregulated genes (RMA log2 transformed values >2). The genes identified by arrows are 13 upregulated genes, targeted by 262 miRNAs, comprising 372 regulatory relationships. The hub genes are IGLC7 (78 miRNAs target it), APOE (66 miRNAs) and SERPINA1 (53 miRNAs). The hub miRNAs are miR-6756 (8 target genes), miR-328 (6 genes) and miR-6803 (5 genes).
Figure 7.
The core miRNA regulatory network of upregulated genes (RMA log2 transformed values >2) in advanced stage samples.
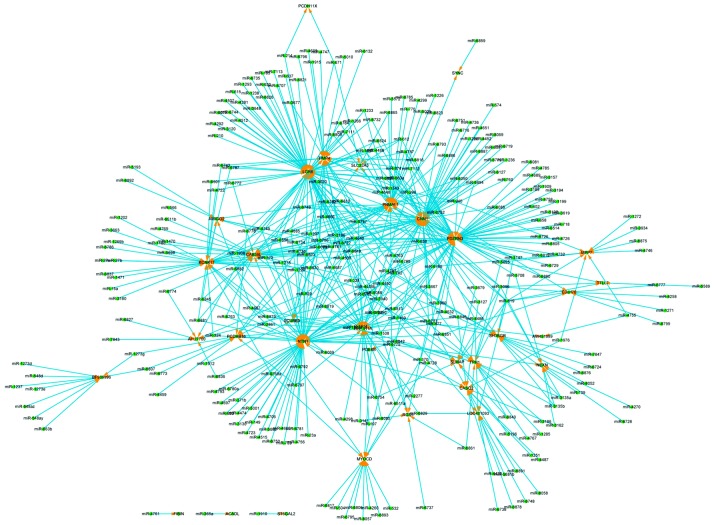
The core miRNA regulatory network is shown in Fig. 8 for the downregulated genes (RMA log2 transformed values >1). The genes identified by arrows are 33 downregulated genes, targeted by 295 miRNAs, comprising 561 regulatory relationships. The hub genes are LGR6 (71 miRNAs target it), NTN1 (61 miRNAs), CNN1 (59 miRNAs) and PDZRN3 (57 miRNAs). The hub miRNAs are miR-6756 (11 target genes), miR-619 (10 genes), miR-6089 (8 genes) and miR-6803 (8 genes).
Figure 8.
The core miRNA regulatory network of upregulated genes (RMA log2 transformed values >2) in the early stage samples.
Discussion
Carotid atherosclerosis is defined as the presence of atherosclerotic plaques in any of the carotid vessel segments (30). In the present study, we aimed to identify gene expression changes and regulatory factors for carotid atherosclerosis from an early to an advanced stage. More genes were activated in advanced stage compared with early stage. The upregulated genes in the advanced stage were involved in GO terms including immune, vascular and cell movement homeostasis. The differentially expressed genes (DEGs) were significantly enriched in cell adhesion molecules (CAMs) and focal adhesion. Genes such as MMP9 and CFL2 played key roles in the transcriptional regulatory network. Moreover, miR-328 was one of the hubs in the miRNA regulatory network.
A total of 889 transcripts were identified to be differentially expressed from early stage plaques of carotid atherosclerosis to advanced stage plaques. As shown in Fig. 3B, A/E increased when the fold change threshold was elevated. The majority of the DEGs were upregulated in the advanced stage, while they were inhibited in the early stage.
The DEGs activated in the advanced stage may correlate with plaques of carotid atherosclerosis and various types of cancer. A number of immune system-related cells were detected in human carotid atherosclerosis patients such as monocytes/macrophages, T cells and plasmacytoid dendritic cells (pDCs) (23). IGHG1 (Ig γ-1 chain C region) is one gene associated with ‘innate immune response’. IGHG1 expression has been reported to correlate with immune evasion mechanisms, which contribute to the proliferation of human pancreatic cancer (31). Furthermore, inhibiting IGHG1 expression by siRNA leads to cancer growth inhibition and apoptosis in prostate cancer (32). SPP1 (osteopontin) is involved as a cytokine in type I immunity to elevate the product of interferon-γ and interleukin-12. On the other hand, SPP1 reduces the expression of interleukin-10, and IFN-γ treatment, resulting in an increase of cytokines/cytokine receptors including CSF2, IL1R2 and SPP1 (33). In addition, smoking can increase SPP1 expression, and subsequently induce inflammation and emphysema (34). IGKC (Ig κ chain C region) participates in humoral immune response, which has been reported to play key roles in non-small cell lung cancer and breast cancer (35–37). The genes activated in early stage do not possess a similar function, including BTC, MYBL1 and PLD.
Immune-associated GO terms, vascular-related GO terms and cell movement homeostasis were identified to be associated with atherosclerosis. The IL-6-gp130 axis is a key regulator of inflammatory acute phase signaling in hepatocytes for the development of atherosclerosis (38). Blood flow is crucial for blood vessel development during embryogenesis and for the regulation of vessel diameter in adult life. It is also a key factor in atherosclerosis, which occurs mainly in regions of arteries that experience disturbances in fluid flow (39). Fibrinolytic balance and the potential contribution of PAI-1 to the regulation of cell migration are involved in the pathogenesis of the simple atherosclerotic lesions observed in the mouse (40).
Leukocyte transendothelial migration is one of the earliest events of immune inflammatory responses and may contribute to atherosclerosis (41). Immunologic arterial injury due to allograft rejection acting in synergy with hypercholesterolemia resulting from a dietary supplement of cholesterol can lead to rapidly developing atherosclerosis (42). Autoimmune thyroid disease has a causal relationship with atherosclerosis (even if mediated through traditional risk factors) (43). Focal adhesion plays key roles in VSMCs. Focal adhesion pathways may be expected to facilitate the formation of atherosclerotic plaques in ApoE-null mice (44). Systemic lupus erythematosus (SLE) is a systemic autoimmune disease that is characterized by autoantibody production and inflammatory disease involving multiple organs. Premature atherosclerosis is a common complication of SLE and results in substantial morbidity and mortality from cardiovascular disease (CVD) (45). Diabetes and atherosclerosis are associated with disorders of lipids and lipoproteins, notably high apolipoprotein B (apoB) and low apolipoprotein A1 (apoA1) are well established (46). Type I diabetes mellitus was also enriched by differential genes in this study. The passage of leukocytes across the endothelium and into arterial walls is a critical step in the development of atherosclerosis. It is consistent with our observation that DEGs were enriched in the pathway ‘leukocyte transendothelial migration’ (47).
The expression levels of matrix metallopeptidase 9 (MMP9) were assessed. MMP9 was potentially important in the development of atherosclerosis in a Malaysian study population (13). Cystic fibrosis transmembrane conductance regulator (CFTR) has a similar function to ABCA1. Schmitz and Buechler (48) identified ABCA1 as the major regulator of plasma high density lipoprotein (HDL) cholesterol. HDL metabolism is crucial in the prevention of the progression of atherosclerosis. CFL2 is an interactor of CFTR as reported by Wang and colleagues (49). An in vivo ApoE−/− mouse model was utilized to assess the effects of chronic moderate exposure to arsenic on plaque formation and composition in order to facilitate mechanistic investigations (50). Arsenic exposure increases oxidative stress, inflammation and atherosclerotic lesion formation in ApoE−/− mice (51). In human arterial tissue, SERPINA1 was upregulated (6.3-fold) in atherosclerotic plaques (14). In the present study, SERPINA1 was also upregulated in the advanced stage.
In this study, we also found that miR-328 may be crucial for atherosclerosis. miR-328 was linked to multiple upregulated genes in advanced stage samples and miR-328 has been found to be antiangiogenic (52). Anti-angiogenic perfluorocarbon nanoparticles has already used for diagnosis and treatment of atherosclerosis (53). Recently, in the ABCG2-positive cancer cells, miR-328 has been reported to regulate the expression of BCRP/ABCG2 (54). Additionally, miR-328 expression in plasma was significantly increased in atrial fibrillation (AF) patients (55).
Motifs enriched by upregulated genes in the early and advanced stages including Myf, MZF1, FOXL1 and NKX3-1, which were not investigated extensively were also investigated. These transcriptional factors may play pivotal roles in carotid atherosclerosis. In the present study, we identified gene expression changes and regulatory factors in carotid atherosclerosis. These results may facilitate in identifying the mechanism involved in carotid atherosclerosis.
References
- 1.Ross R. The pathogenesis of atherosclerosis: a perspective for the 1990s. Nature. 1993;362:801–809. doi: 10.1038/362801a0. [DOI] [PubMed] [Google Scholar]
- 2.Maseri A, Fuster V. Is there a vulnerable plaque? Circulation. 2003;107:2068–2071. doi: 10.1161/01.CIR.0000070585.48035.D1. [DOI] [PubMed] [Google Scholar]
- 3.Ross R. Atherosclerosis - an inflammatory disease. N Engl J Med. 1999;340:115–126. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 4.O’Leary DH, Polak JF, Kronmal RA, Manolio TA, Burke GL, Wolfson SK., Jr Carotid-artery intima and media thickness as a risk factor for myocardial infarction and stroke in older adults. Cardiovascular Health Study Collaborative Research Group. N Engl J Med. 1999;340:14–22. doi: 10.1056/NEJM199901073400103. [DOI] [PubMed] [Google Scholar]
- 5.Wu Y, Tao Z, Song C, et al. Overexpression of YKL-40 predicts plaque instability in carotid atherosclerosis with CagA-positive helicobacter pylori infection. PLoS One. 2013;8:e59996. doi: 10.1371/journal.pone.0059996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamada M, Kim S, Egashira K, et al. Molecular mechanism and role of endothelial monocyte chemoattractant protein-1 induction by vascular endothelial growth factor. Arterioscler Thromb Vasc Biol. 2003;23:1996–2001. doi: 10.1161/01.ATV.0000096208.80992.63. [DOI] [PubMed] [Google Scholar]
- 7.Leng XY, Chen XY, Chook P, et al. Association between metabolic syndrome and carotid atherosclerosis: a community-based study in Hong Kong. Metab Syndr Relat Disord. 2013;11:109–114. doi: 10.1089/met.2012.0099. [DOI] [PubMed] [Google Scholar]
- 8.Ota H, Reeves MJ, Zhu DC, et al. Sex differences of high-risk carotid atherosclerotic plaque with less than 50% stenosis in asymptomatic patients: an in vivo 3T MRI study. AJNR Am J Neuroradiol. 2013;34:1049–1055. doi: 10.3174/ajnr.A3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ino-Oka E, Sekino H, Kajikawa S, Satoh T, Inooka H. Evaluation of carotid atherosclerosis from the perspective of blood flow reflection. Clin Exp Hypertens. 2009;31:188–200. doi: 10.1080/10641960902822443. [DOI] [PubMed] [Google Scholar]
- 10.Maeda S, Sawayama Y, Furusyo N, Shigematsu M, Hayashi J. The association between fatal vascular events and risk factors for carotid atherosclerosis in patients on maintenance hemodialysis: plaque number of dialytic atherosclerosis study. Atherosclerosis. 2009;204:549–555. doi: 10.1016/j.atherosclerosis.2008.09.028. [DOI] [PubMed] [Google Scholar]
- 11.Munakata M, Sakuraba J, Tayama J, et al. Higher brachial-ankle pulse wave velocity is associated with more advanced carotid atherosclerosis in end-stage renal disease. Hypertens Res. 2005;28:9–14. doi: 10.1291/hypres.28.9. [DOI] [PubMed] [Google Scholar]
- 12.Hashimoto H, Kitagawa K, Kuwabara K, et al. Circulating adhesion molecules are correlated with ultrasonic assessment of carotid plaques. Clin Sci (Lond) 2003;104:521–527. doi: 10.1042/CS20020290. [DOI] [PubMed] [Google Scholar]
- 13.Blin J, Ahmad Z, Rampal LR, Mohtarrudin N, Tajudin AK, Adnan RS. Preliminary assessment of differential expression of candidate genes associated with atherosclerosis. Genes Genet Syst. 2013;88:199–209. doi: 10.1266/ggs.88.199. [DOI] [PubMed] [Google Scholar]
- 14.Inouye M, Ripatti S, Kettunen J, et al. Novel Loci for metabolic networks and multi-tissue expression studies reveal genes for atherosclerosis. PLoS Genet. 2012;8:e1002907. doi: 10.1371/journal.pgen.1002907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bateman HR, Liang Q, Fan D, Rodriguez V, Lessner SM. Sparstolonin B inhibits pro-angiogenic functions and blocks cell cycle progression in endothelial cells. PLoS One. 2013;8:e70500. doi: 10.1371/journal.pone.0070500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feig JE, Vengrenyuk Y, Reiser V, et al. Regression of atherosclerosis is characterized by broad changes in the plaque macrophage transcriptome. PLoS One. 2012;7:e39790. doi: 10.1371/journal.pone.0039790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang E, Wu Y. Dual effects of miR-155 on macrophages at different stages of atherosclerosis: LDL is the key? Med Hypotheses. 2014;83:74–78. doi: 10.1016/j.mehy.2014.04.004. 2014. [DOI] [PubMed] [Google Scholar]
- 18.Bonaterra GA, Zugel S, Thogersen J, et al. Growth differentiation factor-15 deficiency inhibits atherosclerosis progression by regulating interleukin-6-dependent inflammatory response to vascular injury. J Am Heart Assoc. 2012;1:e002550. doi: 10.1161/JAHA.112.002550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orr AW, Hastings NE, Blackman BR, Wamhoff BR. Complex regulation and function of the inflammatory smooth muscle cell phenotype in atherosclerosis. J Vasc Res. 2010;47:168–180. doi: 10.1159/000250095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.States JC, Singh AV, Knudsen TB, et al. Prenatal arsenic exposure alters gene expression in the adult liver to a proinflammatory state contributing to accelerated atherosclerosis. PLoS One. 2012;7:e38713. doi: 10.1371/journal.pone.0038713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bahls M, Bidwell CA, Hu J, et al. Gene expression differences during the heterogeneous progression of peripheral atherosclerosis in familial hypercholesterolemic swine. BMC Genomics. 2013;14:443. doi: 10.1186/1471-2164-14-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinkaew D, Hutadilok-Towatana N, Teng BB, Mahabusarakam W, Fujise K. Morelloflavone, a biflavonoid inhibitor of migration-related kinases, ameliorates atherosclerosis in mice. Am J Physiol Heart Circ Physiol. 2012;302:H451–H458. doi: 10.1152/ajpheart.00669.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doring Y, Manthey HD, Drechsler M, et al. Auto-antigenic protein-DNA complexes stimulate plasmacytoid dendritic cells to promote atherosclerosis. Circulation. 2012;125:1673–1683. doi: 10.1161/CIRCULATIONAHA.111.046755. [DOI] [PubMed] [Google Scholar]
- 24.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 25.Philip R. Semantic similarity in a taxonomy: an information-based measure and its application to problems of ambiguity in natural language. J Artif Intell Res. 1999;11:95–130. [Google Scholar]
- 26.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 27.Portales-Casamar E, Thongjuea S, Kwon AT, et al. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38:D105–D110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang K, Hu F, Xu K, et al. CASCADE_SCAN: mining signal transduction network from high-throughput data based on steepest descent method. BMC Bioinformatics. 2011;12:164. doi: 10.1186/1471-2105-12-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li LX, Zhao CC, Ren Y, et al. Prevalence and clinical characteristics of carotid atherosclerosis in newly diagnosed patients with ketosis-onset diabetes: a cross-sectional study. Cardiovasc Diabetol. 2013;12:18. doi: 10.1186/1475-2840-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X, Ni R, Chen J, et al. The presence of IGHG1 in human pancreatic carcinomas is associated with immune evasion mechanisms. Pancreas. 2011;40:753–761. doi: 10.1097/MPA.0b013e318213d51b. [DOI] [PubMed] [Google Scholar]
- 32.Pan B, Zheng S, Liu C, Xu Y. Suppression of IGHG1 gene expression by siRNA leads to growth inhibition and apoptosis induction in human prostate cancer cell. Mol Biol Rep. 2013;40:27–33. doi: 10.1007/s11033-012-1944-x. [DOI] [PubMed] [Google Scholar]
- 33.Kitaya K, Yasuo T, Yamaguchi T, Fushiki S, Honjo H. Genes regulated by interferon-γ in human uterine microvascular endothelial cells. Int J Mol Med. 2007;20:689–697. [PubMed] [Google Scholar]
- 34.Shan M, Yuan X, Song LZ, et al. Cigarette smoke induction of osteopontin (SPP1) mediates TH17 inflammation in human and experimental emphysema. Sci Transl Med. 2012;4:117ra9. doi: 10.1126/scitranslmed.3003041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lohr M, Edlund K, Botling J, et al. The prognostic relevance of tumour-infiltrating plasma cells and immunoglobulin kappa C indicates an important role of the humoral immune response in non-small cell lung cancer. Cancer Lett. 2013;333:222–228. doi: 10.1016/j.canlet.2013.01.036. [DOI] [PubMed] [Google Scholar]
- 36.Whiteside TL, Ferrone S. IGKC and prognosis in breast cancer - response to Schmidt. Clin Cancer Res. 2013;19:305. doi: 10.1158/1078-0432.CCR-12-3200. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt M, Micke P, Hengstler JG. IGKC and prognosis in breast cancer. Clin Cancer Res. 2013;19:304. doi: 10.1158/1078-0432.CCR-12-2818. [DOI] [PubMed] [Google Scholar]
- 38.Luchtefeld M, Preuss C, Ruhle F, et al. Gp130-dependent release of acute phase proteins is linked to the activation of innate immune signaling pathways. PLoS One. 2011;6:e19427. doi: 10.1371/journal.pone.0019427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hahn C, Schwartz MA. Mechanotransduction in vascular physiology and atherogenesis. Nat Rev Mol Cell Biol. 2009;10:53–62. doi: 10.1038/nrm2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sjoland H, Eitzman DT, Gordon D, Westrick R, Nabel EG, Ginsburg D. Atherosclerosis progression in LDL receptor-deficient and apolipoprotein E-deficient mice is independent of genetic alterations in plasminogen activator inhibitor-1. Arterioscler Thromb Vasc Biol. 2000;20:846–852. doi: 10.1161/01.atv.20.3.846. [DOI] [PubMed] [Google Scholar]
- 41.Languino LR, Duperray A, Joganic KJ, Fornaro M, Thornton GB, Altieri DC. Regulation of leukocyte-endothelium interaction and leukocyte transendothelial migration by intercellular adhesion molecule 1-fibrinogen recognition. Proc Natl Acad Sci USA. 1995;92:1505–1509. doi: 10.1073/pnas.92.5.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alonso DR, Starek PK, Minick CR. Studies on the pathogenesis of atheroarteriosclerosis induced in rabbit cardiac allografts by the synergy of graft rejection and hypercholesterolemia. Am J Pathol. 1977;87:415–442. [PMC free article] [PubMed] [Google Scholar]
- 43.McLeod DS. Autoimmune thyroid disease: a novel risk factor for atherosclerosis? Endocrine. 2013;44:8–10. doi: 10.1007/s12020-013-9952-8. [DOI] [PubMed] [Google Scholar]
- 44.Bu DX, Rai V, Shen X, et al. Activation of the ROCK1 branch of the transforming growth factor-β pathway contributes to RAGE-dependent acceleration of atherosclerosis in diabetic ApoE-null mice. Circ Res. 2010;106:1040–1051. doi: 10.1161/CIRCRESAHA.109.201103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richez C, Richards RJ, Duffau P, et al. The effect of mycophenolate mofetil on disease development in the gld.apoE−/− mouse model of accelerated atherosclerosis and systemic lupus erythematosus. PLoS One. 2013;8:e61042. doi: 10.1371/journal.pone.0061042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hashemi M, Saadat M, Behjati M, Kelishadi R. Comparison of serum apolipoprotein levels of diabetic children and healthy children with or without diabetic parents. Cholesterol. 2012;2012:490381. doi: 10.1155/2012/490381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samson T, van Buul JD, Kroon J, et al. The guanine-nucleotide exchange factor SGEF plays a crucial role in the formation of atherosclerosis. PLoS One. 2013;8:e55202. doi: 10.1371/journal.pone.0055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmitz G, Buechler C. ABCA1: regulation, trafficking and association with heteromeric proteins. Ann Med. 2002;34:334–347. doi: 10.1080/078538902320772098. [DOI] [PubMed] [Google Scholar]
- 49.Wang X, Venable J, LaPointe P, et al. Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell. 2006;127:803–815. doi: 10.1016/j.cell.2006.09.043. [DOI] [PubMed] [Google Scholar]
- 50.Lemaire M, Lemarie CA, Molina MF, Schiffrin EL, Lehoux S, Mann KK. Exposure to moderate arsenic concentrations increases atherosclerosis in ApoE−/− mouse model. Toxicol Sci. 2011;122:211–221. doi: 10.1093/toxsci/kfr097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Srivastava S, Vladykovskaya EN, Haberzettl P, Sithu SD, D’Souza SE, States JC. Arsenic exacerbates atherosclerotic lesion formation and inflammation in ApoE−/− mice. Toxicol Appl Pharmacol. 2009;241:90–100. doi: 10.1016/j.taap.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suarez Y, Sessa WC. MicroRNAs as novel regulators of angiogenesis. Circ Res. 2009;104:442–454. doi: 10.1161/CIRCRESAHA.108.191270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Caruthers SD, Cyrus T, Winter PM, Wickline SA, Lanza GM. Anti-angiogenic perfluorocarbon nanoparticles for diagnosis and treatment of atherosclerosis. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2009;1:311–323. doi: 10.1002/wnan.9. [DOI] [PubMed] [Google Scholar]
- 54.Li X, Pan YZ, Seigel GM, Hu ZH, Huang M, Yu AM. Breast cancer resistance protein BCRP/ABCG2 regulatory microRNAs (hsa-miR-328, -519c and -520h) and their differential expression in stem-like ABCG2+ cancer cells. Biochem Pharmacol. 2011;81:783–792. doi: 10.1016/j.bcp.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang X, Qiu CG, Huang ZW, Han ZY, Lu WJ, Chen XJ. The expression and clinical implication of plasma miR-328 in patients with atrial fibrillation. Zhonghua Xin Xue Guan Bing Za Zhi. 2013;41:126–129. (In Chinese) [PubMed] [Google Scholar]