Abstract
A role for microRNAs (miRs) in the physiologic regulation of sodium transport in the kidney has not been established. In this study, we investigated the potential of aldosterone to alter miR expression in mouse cortical collecting duct (mCCD) epithelial cells. Microarray studies demonstrated the regulation of miR expression by aldosterone in both cultured mCCD and isolated primary distal nephron principal cells. Aldosterone regulation of the most significantly downregulated miRs, mmu-miR-335–3p, mmu-miR-290–5p, and mmu-miR-1983 was confirmed by quantitative RT-PCR. Reducing the expression of these miRs separately or in combination increased epithelial sodium channel (ENaC)–mediated sodium transport in mCCD cells, without mineralocorticoid supplementation. Artificially increasing the expression of these miRs by transfection with plasmid precursors or miR mimic constructs blunted aldosterone stimulation of ENaC transport. Using a newly developed computational approach, termed ComiR, we predicted potential gene targets for the aldosterone-regulated miRs and confirmed ankyrin 3 (Ank3) as a novel aldosterone and miR-regulated protein. A dual-luciferase assay demonstrated direct binding of the miRs with the Ank3–3′ untranslated region. Overexpression of Ank3 increased and depletion of Ank3 decreased ENaC-mediated sodium transport in mCCD cells. These findings implicate miRs as intermediaries in aldosterone signaling in principal cells of the distal kidney nephron.
The epithelial sodium channel (ENaC) is a heterotrimeric sodium (Na+) selective ion channel composed of three homologous subunits: α, β, and γ.1,2 ENaC is prominently expressed in epithelial cells of kidneys, airways, salivary ducts, sweat ducts, and colon and constitutes the limiting step in the reabsorption of Na+ across epithelial cells.3–7 Because of its role in the regulation of Na+ homeostasis, ENaC has been associated with clinical defects of Na+ and water transport.8 In the kidney, ENaC is localized in principal cells of the distal nephron (distal convoluted tubule and collecting duct), where it participates in final Na+ uptake from the luminal urinary space. Conditional knockout of αENaC in mouse cortical collecting duct (mCCD) principal cells produced animals with a salt-wasting phenotype.9 In humans there is a direct link between defective ENaC physiology and disease in Liddle syndrome (hypertension) and pseudohypoaldosteronism (hypotension).5,10–16
ENaC is regulated by several intrinsic and external factors, the details of which have been reviewed previously.3,4,10,17,18 An essential ENaC regulator is the mineralocorticoid hormone aldosterone, which is central to Na+ homeostasis.19 The renin-angiotensin-aldosterone system (RAAS) is critical in the regulation of body fluid volume and therefore BP.20,21 Aldosterone binds to its mineralocorticoid receptor (MR) in the cytoplasm of target cells and translocates to the nucleus to interact with mineralocorticoid response elements. Aldosterone modulates the expression of many aldosterone-induced proteins, which ultimately leads to an increase in Na+ transport.22–24 In the distal nephron, aldosterone’s action is driven primarily through signaling of the serum and glucocorticoid-induced kinase (SGK1) and glucocorticoid-induced leucine zipper (GILZ) proteins.25,26 Both pathways converge on the E3 ligase Nedd4l. Phosphorylation of Nedd4l abrogates its interaction with PY motifs on ENaC intracellular C-termini and prevents the ligase’s action to ubiquitinate ENaC.27,28 Because ubiquitination acts as the internalization signal for ENaC, preventing the Nedd4l/ENaC interaction results in increased residence time for ENaC at the apical membrane and increased Na+ (and water) transport.10,27,29–31
Aldosterone exhibits differential regulation of targets in different epithelia, suggesting the possibility that mineralocorticoid response elements are differentially targeted or that there is an additional intermediate regulatory step in the MR signaling cascade.32,33 One such intermediate pathway may involve small noncoding microRNAs (miRs). The miRs are an extensive family of short (approximately 22 nucleotides), noncoding RNAs.34–39 They function as post-transcriptional repressors of gene expression by binding predominantly to the 3′-untranslated region (UTR) of mRNAs to interfere with protein production.40–42 The miRs differ significantly in their tissue distribution.43 Some miRs are enriched in the kidney, and a subset have greater expression in cortex than medulla.44 Because most miRs are transcribed by the same mechanism as protein coding genes,45,46 miR transcription is itself subject to regulation.47–49 However, a role for miRs in the physiologic regulation of Na+ transport has not been reported, and the ability of aldosterone to regulate miRs in the distal nephron is unknown.
Here we demonstrate that aldosterone alters the expression of miRs in CCD principal cells. This leads to a change in target gene expression to increase transepithelial Na+ transport. The regulation of miRs acts as an additional layer in the RAAS signal cascade that maintains Na+ homeostasis. Microarray analyses demonstrate that at least 20 miRs were significantly altered by aldosterone. Using our recently developed algorithm, ComiR,50,51 we predicted potential miR targets of the most significantly downregulated miRs and confirmed the regulation of ankyrin-G (Ank3). Ank3 expression was altered by aldosterone, and we demonstrate that this occurs through binding and regulation of the Ank3 3′-UTR by the miRs. Exogenous changes in Ank3 expression altered ENaC function and Na+ transport in mCCD cells. This study positions miRs as a new component in the RAAS signaling cascade that maintains Na+ transport in the kidney.
Results
Inhibition of miR Production Prevents Aldosterone Signaling
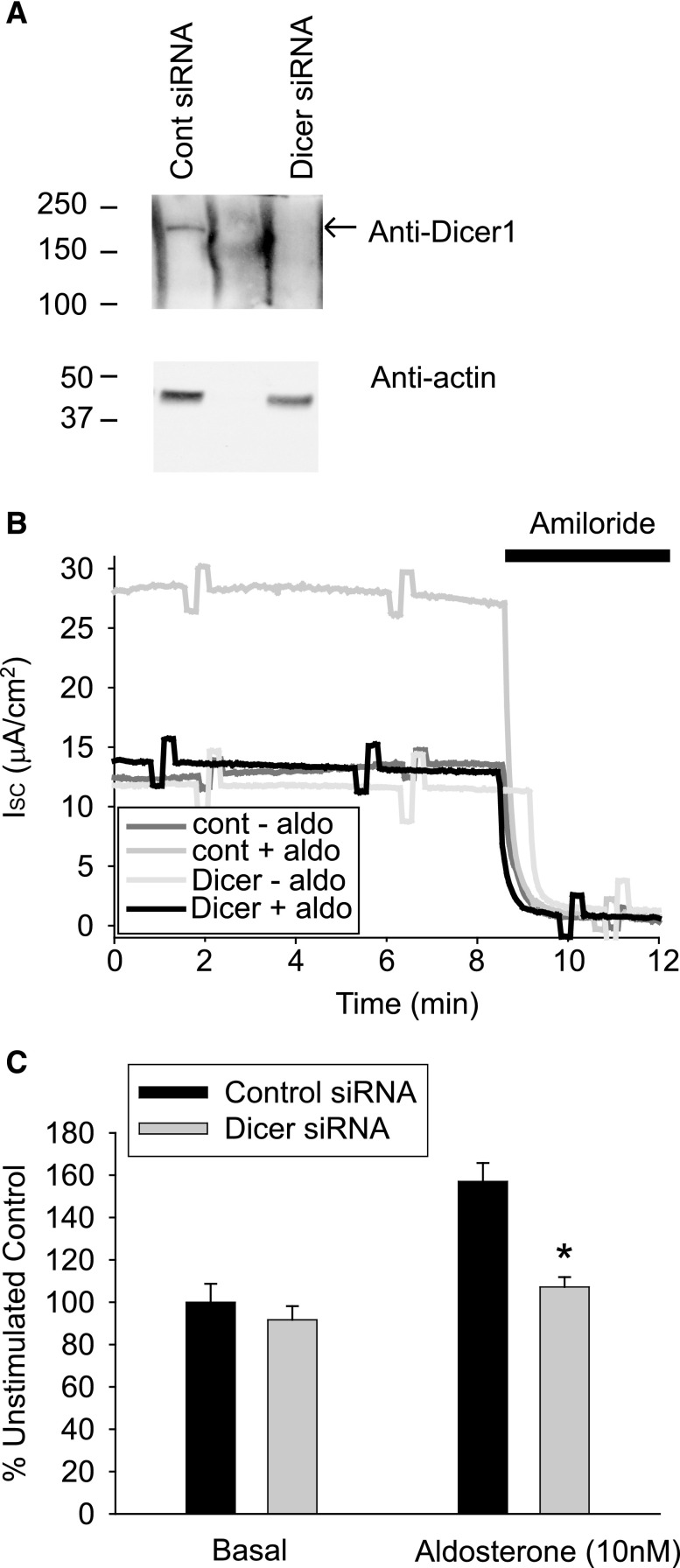
The expression of the miR processing enzyme Dicer1 was reduced in an mCCD cell line using small interfering RNA (siRNA) (Figure 1A). The average reduction in Dicer1 expression was 61.9%±14.8% (n=4). Dicer1 knockdown cells exhibited no significant change in basal levels of ENaC-mediated Na+ transport compared with scrambled siRNA transfected controls. However, when the cells were stimulated with aldosterone for 24 hours (10 nM), the Dicer1-depleted cells had a reduced response to aldosterone stimulation compared with control cells (Figure 1, B and C). This suggested that miR processing via the RNA-induced silencing complex was an essential component of the aldosterone signaling pathway.
Figure 1.
Dicer-1 depletion in mCCD cells reduces aldosterone stimulation of ENaC-mediated Na+ transport. (A) Western blot of whole-cell lysates collected from control siRNA and Dicer-1 knockdown mCCD cells grown on filter supports. (B) Representative short-circuit current (ISC) traces from mCCD cells mounted in modified Ussing chambers. Control and Dicer siRNA transfected cells stimulated with 10 nM aldosterone for 24 hours before ISC measurements. (C) Summarized data (mean±SEM) from similar experiments (n>12) to those presented in B. cont, control.
Aldosterone Alters miR Expression in mCCD Cells
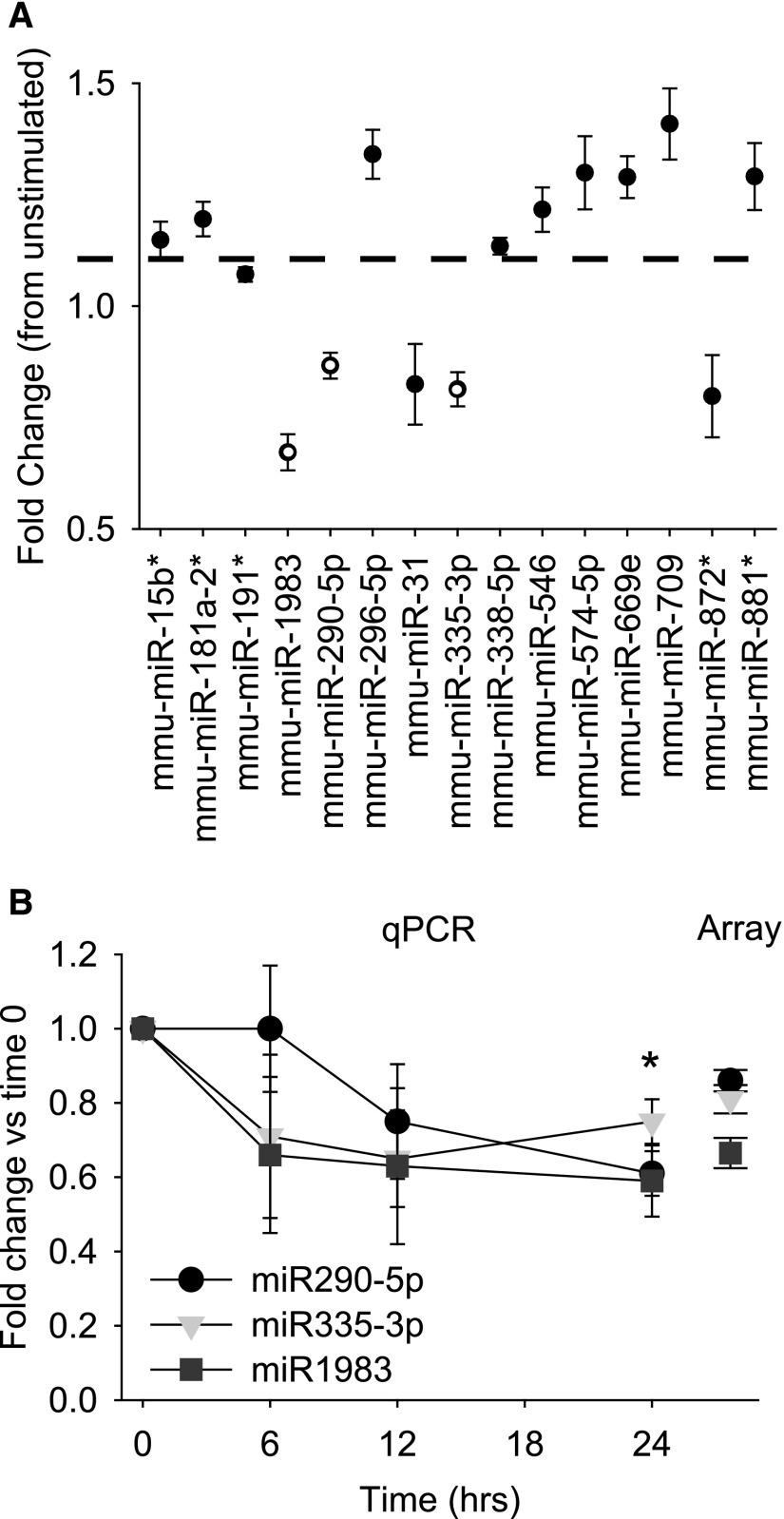
The miR expression from cultured mCCD cells, stimulated with aldosterone for 24 hours (50 nM, n=5) was determined for approximately 800 mouse miRs using microarray analysis. The complete results for control and aldosterone-treated mCCD cells are provided in the Supplemental Material (Supplemental Figure 1). We observed miR expression that was both increased and decreased following aldosterone stimulation (Figure 2A). Expression changes for the significantly downregulated miRs—mmu-miR-335–3p, mmu-miR-290–5p, and mmu-miR-1983—were confirmed by quantitative RT-PCR (qRT-PCR) (Figure 2B). Expression of these miRs decreased over a 24-hour time course following aldosterone stimulation (Figure 2B). Comparing microarray and qPCR data, we observed no significant differences in the fold change in expression at 24 hours (Figure 2B).
Figure 2.
Aldosterone stimulation alters miR expression in mCCD cells. (A) Summarized fold change in expression (mean±SEM) of listed miRs from microarray data (n=5) after 50 nM aldosterone stimulation (24 hours). All listed miRs had significantly altered expression levels (P<0.05) with the miRs in open circles the most significantly downregulated following aldosterone stimulation (P<0.01). (B) Fold change in expression of the significantly downregulated miRs (A) quantified by qRT-PCR for cells stimulated with 50 nM aldosterone for 6, 12, and 24 hours. Expression decreased (*P<0.05) for all three after 24 hours. Fold change did not significantly differ between PCR and array data at the 24-hour time point.
miRs Are Regulated by Aldosterone In Vivo
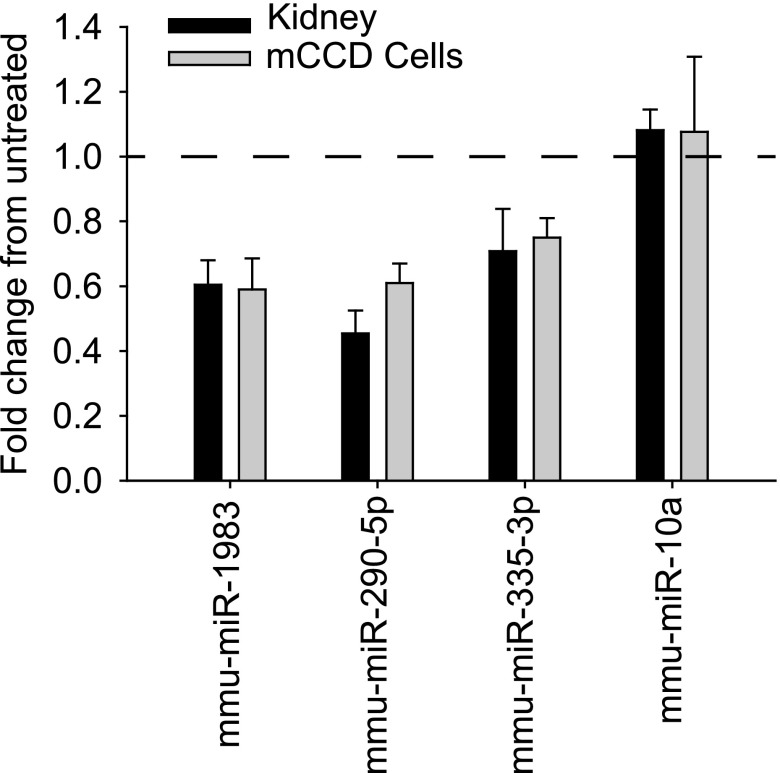
To verify that the miRs were expressed and regulated in CCD cells in vivo, we used a magnetic bead binding and purification procedure to selectively enrich CCD cells from the kidneys of mice maintained on standard and low-sodium diets for 7 days. Mice receiving low-sodium diets were housed in metabolic cages, and urine and plasma electrolyte concentrations were profiled to verify that they responded appropriately to the change in sodium diet, namely reduced Na+ output with lower urine osmolality due to aldosterone signaling (Table 1). Expression of the three miRs identified in the cultured mCCD cells was carried out from enriched distal nephron epithelial cells by qRT-PCR, and a reduction in expression was confirmed when mice were switched to low-sodium diets (Figure 3). As a control, the expression of highly expressed, CCD-localized miR, mmu-miR-10a was not altered by the low-sodium diet, in agreement with array and qPCR expression in vitro data from the aldosterone-treated mCCD cells.
Table 1.
Urine and plasma electrolyte values for mice receiving normal versus low-sodium diet (n=8)
| Variable | Normal Diet | Low-Sodium Diet |
|---|---|---|
| Body mass (g) | 20.0±0.6 | 19.7±0.4 |
| Water intake (μl/g per hr) | 7.0±1.3 | 6.0±0.5 |
| Urine output (μl/g per hr) | 1.5±0.3 | 1.8±0.2 |
| Serum Na+ (mM) | 153.2±0.7 | 151.5±1.0 |
| Serum K+ (mM) | 7.7±0.1 | 7.1±0.5 |
| Serum Cl− (mM) | 118.8±1.5 | 117.3±1.1 |
| Urinary osmolality (mOsm/kg H2O) | 1736.5±141.4 | 1312.2±135.8a |
| Urinary Na+ (mM) | 114.3±11.2 | 15.0±3.5a |
| Urinary K+ (mM) | 226.4±22.1 | 189.5±19.7a |
| Urinary Cl− (mM) | 160.9±15.0 | 50.9±5.9a |
Values are expressed as mean±SEM.
P<0.05.
Figure 3.
Expression of miRs is decreased in distal nephron epithelial cells isolated from mice receiving low-sodium diets. qRT-PCR miR expression in CCD cells isolated from kidneys of mice receiving low-sodium diets (black bars) or from cultured CCD cells stimulated with aldosterone (gray bars) was normalized to control expression. The fold change in miR expression did not significantly differ between in vivo and in vitro CCD cells. Expression of miR-1983, miR-335–3p, and miR-290–5p was significantly reduced from mice receiving normal diets or unstimulated mCCD cells. As a control, no difference in expression was observed for miR-10a after low-sodium diet or aldosterone stimulation (in agreement with the microarray data, see Supplemental Figure 1).
Altering miR Expression Changes ENaC Activity
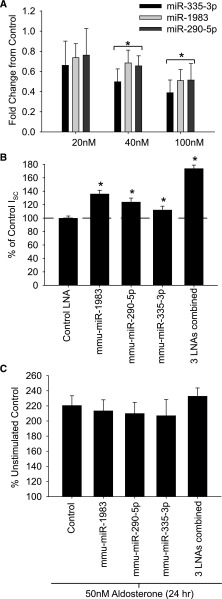
We exogenously altered the expression of the three miRs (mmu-miR-335–3p, mmu-miR-290–5p, and mmu-miR-1983) in the mCCD cell line to determine the effect on ENaC-mediated Na+ reabsorption. Expression of miRs was decreased by introducing interfering locked nucleic acids (LNAs) that target the three miRs either individually or in combination (Figure 4A). Reduction in miR expression was confirmed by qPCR, with no detectable change in off-target miRs (not shown). The amiloride-sensitive Na+ transport was assayed 72 hours after transfection. ENaC-mediated Na+ transport increased significantly for all downregulated miRs compared with control transfected LNAs, with the largest increase in Na+ current detected in cells in which expression of the three miRs was reduced together (Figure 4B). The increase in Na+ current never reached levels observed in aldosterone-stimulated control cells (see Figure 4C); however, these 3 miRs represent only a subset of the miRs altered in response to aldosterone.
Figure 4.
Reduction in miR expression increases ENaC-mediated Na+ transport. (A) A dose-dependent decrease in miR expression was observed in mCCD cells transfected with LNAs targeting each of the three miRs. qRT-PCR values were normalized to control LNA transfected cells. (B) Normalized ISC measurements from mCCD cells transfected with 50 nM LNA oligonucleotides, alone or in combination, expressed as a percentage of unstimulated control-transfected mCCDs (n=45). (C) Normalized ISC measurements from mCCD cells transfected as in B and stimulated with 50 nM aldosterone, expressed as a percentage of untreated, control transfected CCDs. Response did not significantly differ, and all cells had a significant increase (P<0.05) in ISC after aldosterone stimulation compared with unstimulated controls (n=9).
To determine the effect of aldosterone stimulation on these miR knockdown cells, mCCD cells were treated with 50 nM of aldosterone for 24 hours after LNA transfection (assayed at 72 hours). Aldosterone stimulation significantly increased Na+ currents in control LNA-treated cells, and in all cases of miR knockdown (alone or in combination) the ENaC-mediated current increased, but not to above the level of the controls (Figure 4C). These data suggest that aldosterone is likely signaling through a pathway that includes the miRs.
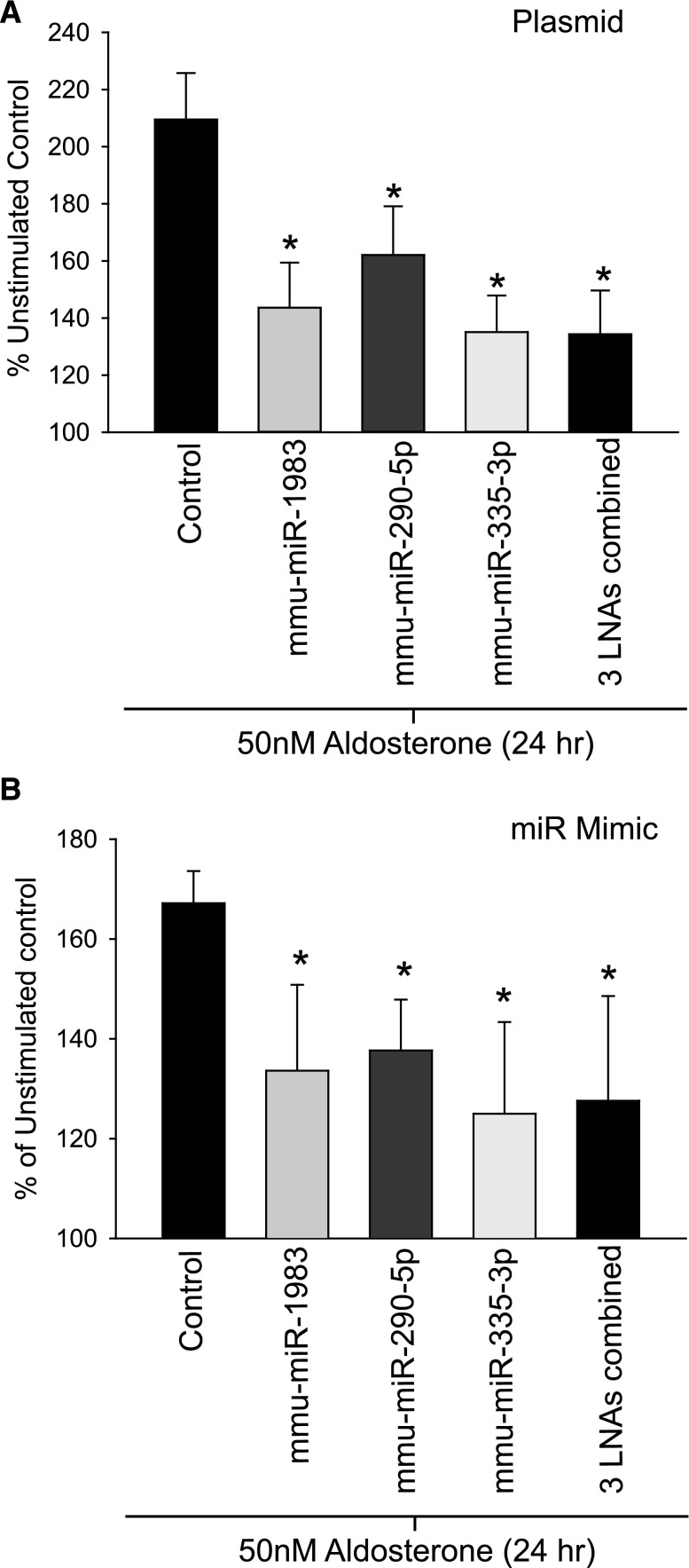
The three miR were next overexpressed alone or in combination using pre-miR plasmids or miR mimics (Figure 5). Cells were stimulated with 50 nM as above. For both pre-miR overexpression or miR mimics we observed a blunting of aldosterone stimulation (Figure 5A). The diminution of the aldosterone stimulation was most pronounced in cells in which all three miRs were overexpressed in combination (Figure 5, A and B). Again, this suggested that if aldosterone reduced miR expression to alter protein expression, overriding this regulation by exogenous overexpression of the miRs impaired aldosterone signaling.
Figure 5.
Exogenous increase in miR expression reduces aldosterone-stimulated ENaC transport. (A) Normalized ISC measurements from mCCD were transiently transfected with plasmids encoding pre-miR sequences, alone or in combination. Cells seeded onto filter supports were stimulated with aldosterone for 24 hours (50 nM), and ISC was normalized to unstimulated mCCD cells transfected with control plasmid. ISC response to aldosterone decreased significantly (*P<0.05) in cells overexpressing miR-1983, miR-290–5p, and miR-335–3p or the combination of the three miRs (n=30). (B) The same experiments as in A were carried out using miR mimics, and a significant reduction in aldosterone response was seen for all overexpressed miRs (n=10).
Target Prediction Identifies Ank3 as an miR Target
Our next goal was to find miR targets of these altered miRs. These miRs are not predicted to bind to the 3′-UTR of any ENaC subunits, or known ENaC regulators, Nedd4l, SGK1, or GILZ. Using target prediction sites, we found low consensus in predicted targets (see examples in Supplemental Figure 2A). We therefore used our newly developed algorithmic approach, named ComiR.51 The algorithm accounts for both the levels of miR expression, and the potential combinatorial effect of imperfect binding of multiple miRs into a given target UTR sequence, to get a more reliable prediction of target genes (see Coronnello et al.51 for details). Using the microarray data of the significantly downregulated miRs, a candidate list of targets was generated. Some of the predicted target mRNAs with a ComiR score>0.99 were screened by RT-PCR to test for a change in expression following aldosterone stimulation (Supplemental Figure 2B). We were able to identify new aldosterone-altered mRNAs, and validated five of them, including Ank3, with a ComiR score of 0.9998. By comparison, ENaC subunits were not predicted to be significant direct targets of the downregulated miRs, with scores for α,β,γ-ENaC of 0.3157, 0.2419, and 0.8252, respectively.
Ank3 Is Regulated by Aldosterone and miRs
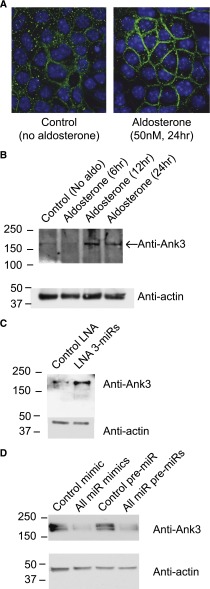
To verify that Ank3 is an aldosterone-induced protein, we performed both immunofluorescent labeling and Western blot analysis of Ank3 in mCCD cells after aldosterone stimulation. Whole-cell Ank3 expression was significantly increased by aldosterone, and we observed a closer membrane association of Ank3 after aldosterone stimulation (Figure 6A). Ank3 protein expression levels peaked at 12 hours after aldosterone stimulation (247%±55% of control) and remained elevated at 24 hours (209.3%±45% of control, n=3) (Figure 6B). To verify regulation of Ank3 by the miRs (mmu-miR-335–3p, mmu-miR-290–5p, and mmu-miR-1983), we reduced or increased their expression as described above. Whole-cell Ank3 expression was increased in the LNA miR knockdown cells by 265.0%±41.8% (n=3) compared with LNA control transfected cells (Figure 6C). Ank3 expression was reduced when the miRs were overexpressed using miR mimics (45.9%±15% of control, n=3) or pre-miR plasmids (56.9%±20.1% of control, n=3) demonstrating regulation of Ank3 by the miRs directly (Figure 6D).
Figure 6.
Ank3 is regulated by aldosterone and miRs. (A) Immunofluorescent images of mCCD cells cultured on filter supports and labeled for Ank3 (green) in cells without (left) and following aldosterone stimulation (right). Nuclei are counterstained in blue (DAPI); white bars=10 μm. (B) Western blots of Ank3 expression from whole cell lysates of mCCD cells cultured on filters and stimulated with aldosterone for 6, 12, and 24 hours. (C) Expression of the three miRs was reduced by LNA transfection (as in Figure 4) and cells seeded onto filters. A Western blot of Ank3 expression from whole cell lysates in control (unstimulated) and LNA transfected, miR reduced mCCD cells (no aldosterone) is presented. (D) The three miRs were overexpressed using miR mimics or plasmid pre-miR transfection. Expression of Ank3 from whole cell lysates is presented in the Western blot following miR overexpression or in control transfected mCCD cells. In both cases, miR overexpression reduced endogenous Ank3 expression.
Luciferase Assays Demonstrate miR Binding and Regulation of Ank3 3′-UTR
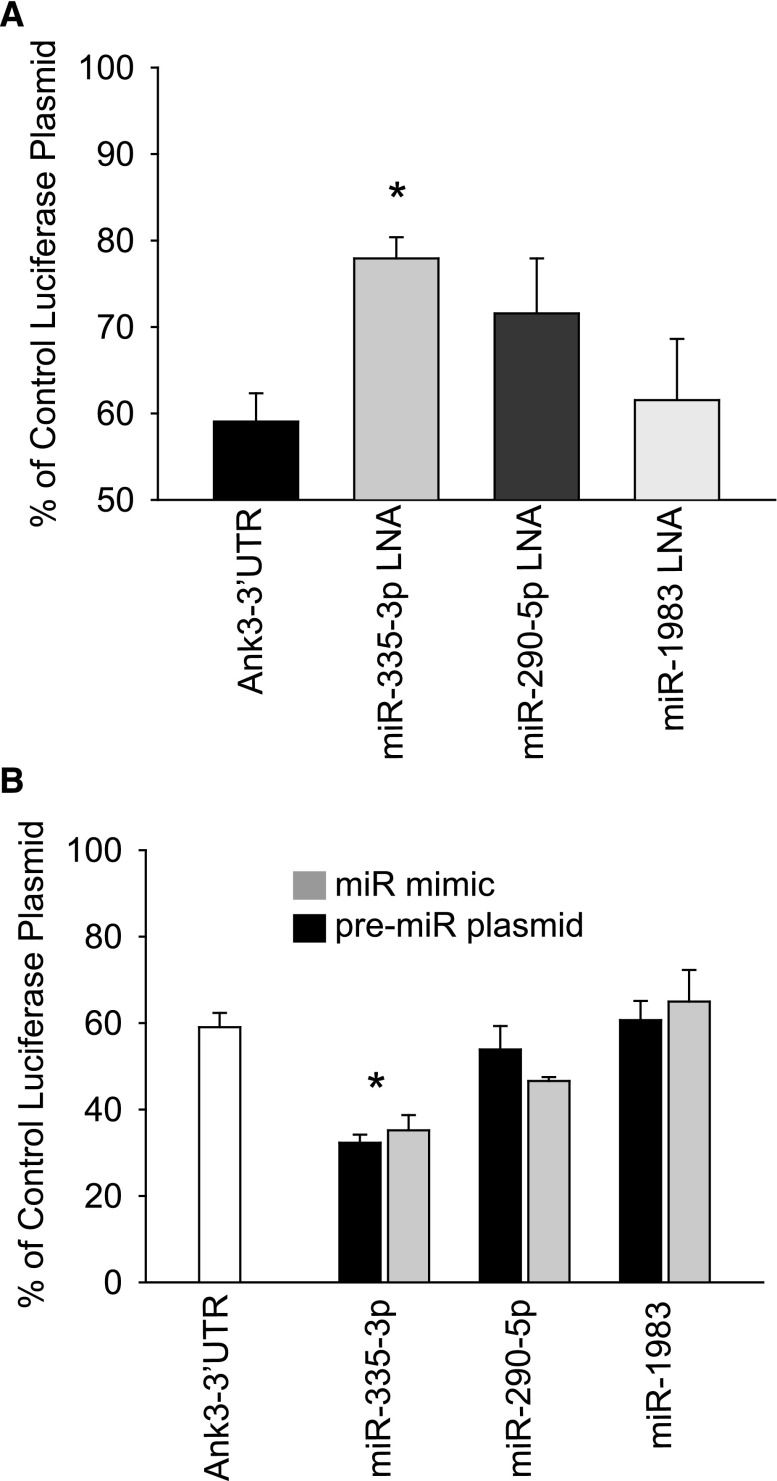
To determine the contribution of each miR to 3′-UTR binding and inhibition of Ank3 expression, we constructed dual luciferase reporter plasmids incorporating the Ank3 3′-UTR, which were then transfected into mCCD cells. The dual reporter plasmid allowed for the sequential detection of two luciferase signals to correct for variability in transfection efficiency. Data were normalized to a control reporter without the Ank3-UTR, which represented the maximum detectable signal obtained from the mCCD cells (set at 100%). MiR expression was altered by using LNA, pre-miR, or miR mimics transfected into mCCD cells as above (Figure 7). It should be noted that introduction of the Ank3-UTR reduced the normalized luciferase signal, indicating that binding of endogenous miRs was regulating Ank3-UTR expression in the mCCD cells even before we altered miR expression exogenously. Luciferase signal increased significantly when mmu-miR-335–3p expression was reduced (Figure 7A). In the converse experiment, overexpression of mmu-miR-335–3p using pre-miR plasmids or miR mimics further reduced the Ank3-UTR luciferase expression without affecting control luciferase expression (Figure 7B). Controls included nontargeting miRs and introduction of control LNA and miR mimics, which did not significantly alter the Ank3-UTR luciferase expression from baseline levels (not shown). These data confirm that Ank3 is regulated by miR binding to its 3′-UTR and that, of the three investigated miRs, mmu-miR-335–3p may be the predominant direct regulator of Ank3 expression through direct binding to its 3′-UTR.
Figure 7.
Luciferase reporter for Ank3 3′-UTR shows regulation by miRs. (A) Sequential luciferase signal was recorded from a dual luciferase reporter incorporating Ank3 3′-UTR transfected into mCCD cells, seeded onto filter supports, and normalized to empty vector controls (100%). The Ank3 3′-UTR signal alone cotransfected with control LNA was reduced compared with the empty vector. Expression of each miR was reduced by cotransfection of 50 nM LNA (as in Figure 4), and luciferase reporter expression increased significantly with reduction of miR-335–3p expression (n=9). (B) In the converse experiment to A, miR expression was increased using miR mimics and pre-miR plasmids (as in Figure 5), and a further, significant (P<0.05) reduction in normalized luciferase expression was observed in cells overexpressing miR-335–3p (n=9).
Altering Ank3 Expression Changes Sodium Transport through ENaC
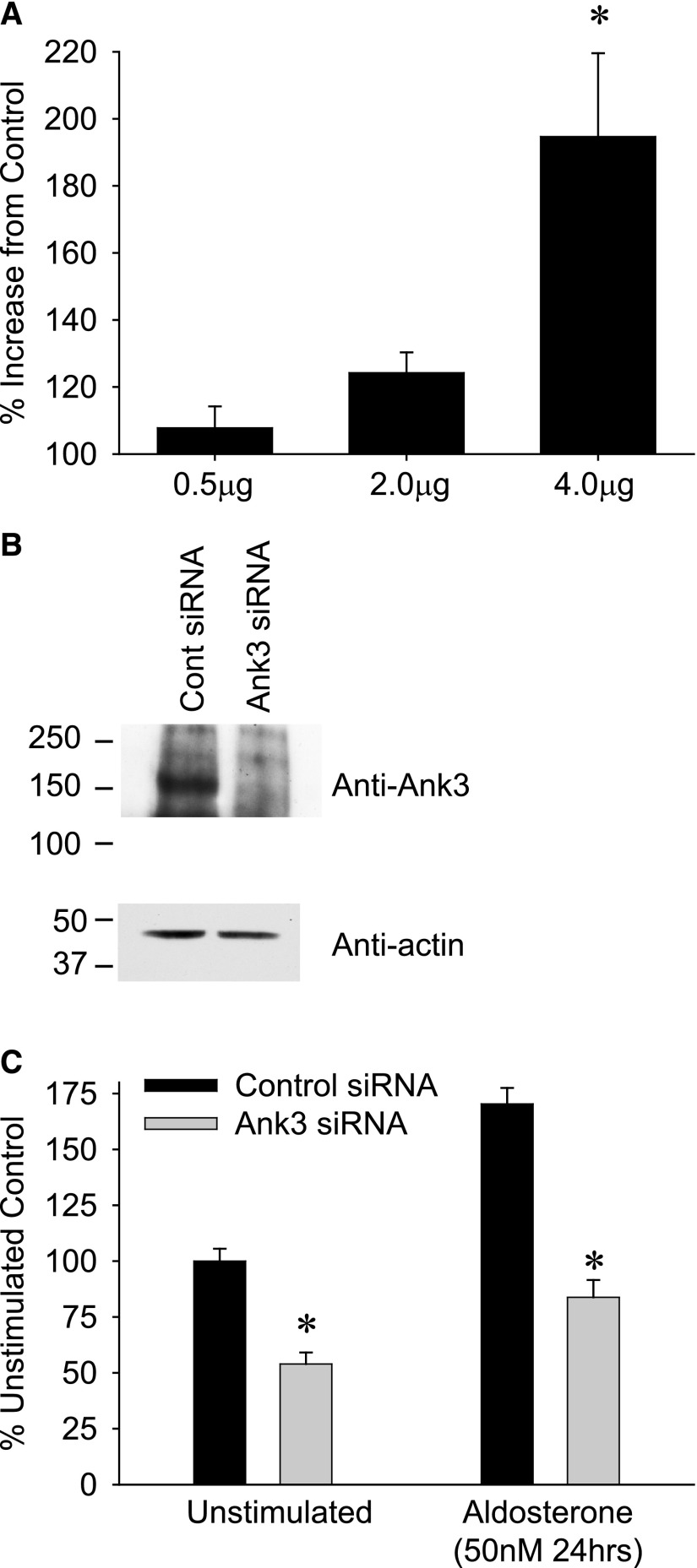
To confirm that Ank3 was directly involved in ENaC regulation, the expression of Ank3 was both increased and reduced in mCCD cells. Overexpression of exogenous human Ank3 by plasmid transfection caused a dose-dependent increase in ENaC-mediated sodium transport, in the absence of aldosterone stimulation (Figure 8A). By selectively reducing Ank3 expression using siRNA (44.8%±3% of control siRNA, n=5) (Figure 8B), both basal and aldosterone-stimulated ENaC currents were significantly decreased, thereby linking Ank3 expression to ENaC function (Figure 8C).
Figure 8.
Altering Ank3 expression changes ENaC-mediated Na+ transport. (A) mCCD cells were transfected with increasing amounts of human ANK3 plasmid and seeded onto filter supports; ISC was recorded after 72 hours. A significant increase in ENaC Na+ transport compared with control transfected cells was observed with 4 μg plasmid transfection (without aldosterone stimulation, n=5). (B) Ank3 expression was reduced in mCCD cells by siRNA transfection. Western blots of whole cell Ank3 expression in control siRNA and Ank3 targeting siRNA transfected cells are presented. (C) ISC recordings from cells transfected with control siRNA and Ank3-targeting siRNA (as in B) seeded onto filter supports and mounted into Ussing chambers. Both basal (unstimulated) and aldosterone stimulated ISC was significantly smaller (*P<0.05) in the Ank3 depleted cells compared with controls (n=12).
Discussion
Pathophysiologic roles of miRs have been described in many disease states, including heart disease, cancer, and hypertension.52–60 In the kidney, miRs have been linked to diabetic nephropathy, polycystic kidney disease, renal cancer, AKI, transplant rejection, and kidney development.59,61–66 Recently, several studies have focused on the role of miR-192, an miR known to be expressed in the kidney cortex, in the profibrotic progression of nephropathy.66,67 MiRs also appear to play a significant role in normal kidney physiology as conditional deletion of Dicer or Drosha in podocytes results in glomerular damage.61,65,68 Regulation of renal outer medullary potassium channels in kidney by miRs has been described,69 and they are implicated as potential regulators of RAAS70 and MR expression.71 It has been suggested that WNK1 may be regulated by miR-192, which in turn is regulated by aldosterone.72
The ability of miRs to act as intermediate targets in physiologic signaling networks has not been extensively explored, and there are no reports on the role of miRs in Na+ regulation in the kidney. By reducing the expression of Dicer-1 in an mCCD cell line, we demonstrate here that aldosterone signaling was compromised, and the typical increase in Na+ transport observed when these cells are stimulated was reduced. Signaling of all miRs would be compromised by Dicer knockdown, and it is likely that both down- and upregulated miRs participate in aldosterone response. At higher aldosterone doses or in cells stimulated by glucocorticoids, it was still possible to elicit a hormonal response (not shown). The miRs are therefore likely acting as modulators of aldosterone signaling, enhancing or repressing the expression of many targets in a complex network of regulation that maintains Na+ homeostasis.
The reduction in miR expression was observed at 6 hours after stimulation, and this fits with previous reports for aldosterone signaling. If miRs play a role in altering protein expression following aldosterone stimulation, the expression levels of these miRs would need to change before the ultimate change in protein expression is observed. To validate the in vivo regulation of miR by aldosterone signaling, we profiled the expression of miRs from isolated mouse CCD cells in animals placed on low-sodium diets, a well established maneuver to increase aldosterone release. When the expression levels of the three miRs were compared with mice on standard salt diets, a significant decrease in expression was observed for each miR, in line with the in vitro data. The relative change in expression of miRs was also similar between mCCD cultured cells and isolated ex vivo CCD cells, adding support for the utility of the cell line in these kinds of investigations.
The three significantly downregulated miRs that were chosen for further investigation do not necessarily represent the most abundantly expressed miRs in CCD, or those with the largest absolute change in expression, but rather were those that exhibited the most significant and consistent downregulation across biologic replicates. However, we focused exclusively on the downregulated miRs here, and by using the ComiR prediction algorithm, we could account for both the relative abundance and change in miR expression to make predictions of likely targets. A number of the target mRNA levels were shown to be increased by aldosterone, but the significance of these novel aldosterone-induced proteins in Na+ regulation has yet to be determined. While we chose to focus on one of the targets, Ank3, it is likely that a cohort of proteins will be targeted by the regulated miRs. It should be noted that the upregulated miRs may participate in the aldosterone signaling pathway, by repressing target protein expression. We have not explored targets that are downregulated by the increased miR expression, however these repressed proteins would form part of a network of interacting proteins regulated by miRs, and this will be the focus of future work.
Demonstrating miR signaling in a pathway that regulates Na+ transport provides a new role for miRs and aldosterone in the kidney. A dynamic interplay between external, hormonal factors and physiologic cellular responses should therefore consider the involvement of miRs as part of the signaling network. The diversity of miRs and large number of potential miR/target interactions will allow for fine tuning of hormonal responses and may account for the pleiotropic effects noted with aldosterone signaling. While not explored in this study, it would be of interest to determine whether other steroid hormones or regulators of sodium and water transport, such as long-term vasopressin stimulation, use miRs as part of their signaling cascade.
Concise Methods
Reagents and Antibodies
All reagents were obtained from Sigma-Aldrich (St. Louis, MO) unless otherwise stated. Antibodies used included anti-actin (Sigma-Aldrich), anti-Dicer1 (BioVision, Inc., San Francisco, CA), anti–ankyrin-G for Western blot (Invitrogen, Grand Island, NY), and anti–ankyrin-G for immunofluorescent staining (UC Davis/NIH NeuroMab Facility, clone N106/36). Primer oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA). A mouse Ank3 cDNA clone was obtained (BC021657, Clone ID:4188590; Thermo Fisher Scientific, Pittsburgh, PA) for use in overexpression studies and in construction of the 3′-UTR reporter assays (see below). The 3′-UTR was removed by restriction digest to obtain an expression construct lacking the UTR.
Metabolic Experiments
C57Bl/6 2-month-old mice (n=4) were fed in their home cage with standard diet for 7 days and were placed in metabolic cages for 24 hours to determine water and food consumption and urine and feces excretion and to collect urine. These mice were then switched to a low-salt diet (Sodium Deficient Diet; Harlan Laboratories Inc., Frederick, MD) for 7 days, and placed in metabolic cages for 24 hours to monitor the same metabolic measures. Kidneys and blood samples were collected from the animals. An equal number of animals were fed a low-salt diet first for 7 days, placed in metabolic cages for 24 hours, switched to standard diet for 7 days, and then placed in metabolic cages for 24 hours. Urine (n=8 for each group) and plasma (n=4 for each group) samples were sent to the Kansas State University Veterinary Diagnostic Laboratories, Clinical Pathology Laboratory (Manhattan, KS) to determine urine and plasma electrolytes and osmolality. All animals were housed in the vivarium at Rangos Research Center at Children’s Hospital of Pittsburgh of UPMC, Pittsburgh, Pennsylvania, and all animal experiments were carried out in accordance with the policies of the Institutional Animal Care and Use Committee at the Children’s Hospital of Pittsburgh.
Cell Culture
The mCCDc11 cells (kindly provided by B. Rossier and L. Schild, Université de Lausanne, Lausanne, Switzerland) were grown in flasks (passages 30–40) in defined (supplemented) medium at 37°C in 5% CO2 as described previously.73,74 The medium was changed every second day. For all electrophysiologic experiments, the mCCD cells were subcultured onto permeable filter supports (0.4 µm pore size, 0.33 cm2 or 4 cm2 surface area; Transwell, Corning, Lowell, MA). Cells were cultured in defined medium until a confluent transporting cell monolayer had developed. This was assessed by recording open circuit voltage and transepithelial resistance using an epithelial volt-ohmmeter and “chopstick” electrodes (EMD Millipore, Billerica, MA). Typically, 24 hours before use in any investigation, medium incubating filter-grown cells were replaced with a minimal medium (without drugs or hormones) that contained DMEM and Ham F12 only. In some experiments, cultures were subsequently stimulated with aldosterone for defined periods.
Immunofluorescence Imaging
CCD cells treated with or without aldosterone (50 nM, 24 hours) were fixed in a cold (4°C) paraformaldehyde buffer (4% in PBS at pH of 7.4). Following 30 minutes of fixation at 4°C, cells were washed in cold PBS with calcium and magnesium (+CM) three times, and permeabilized in PBS containing 0.01% Triton-X. Cells were incubated with primary anti–Ank3 antibody (1:50) for 12 hours in PBS with 10% skim milk at 4°C. Cells were washed three times in PBS+CM and incubated with a secondary antibody (1:1000, Alexa-488; Invitrogen) for 3 hours at 37°C. Following a final PBS+CM wash (three times), nuclei were counterstained using 10nM Hoechst 33342 (Trihydrochloride; Invitrogen). Cells were washed in PBS+CM (three times) and mounted onto slides using Fluoromount-G (Southern Biotech, Birmingham, AL) for imaging, as previously described.75,76 Images were captured using an Olympus IX81 fluorescent microscope (Olympus, Center Valley, PA) fitted with a DSU spinning disk and 300 W fluorescent light source, using a 60×, 1.4 numerical aperture oil objective. Single fluorescent images were captured using a Retiga cooled CCD camera (QImaging, Surrey, BC, Canada) at 1024×1024 resolution using MetaMorph (Molecular Devices Corp., Downingtown, PA). Linear adjustments of brightness and contrast were made offline in MetaMorph.
Short-Circuit Current Recordings
Inserts were mounted in modified Ussing chambers (P2300; Physiologic Instruments, San Diego, CA) and continuously short circuited with an automatic voltage clamp (VCC MC8; Physiologic Instruments) as described previously.77 The apical and basolateral chambers each contained 4 ml of Ringer solution (120 mM NaCl, 25 mM NaHCO3, 3.3 mM KH2PO4, 0.8 mM K2HPO4, 1.2 mM MgCl2, 1.2 mM CaCl2, and 10 mM glucose). Chambers were constantly gassed with a mixture of 95% O2, 5% CO2 at 37°C, which maintained the pH at 7.4 and established a circulating perfusion bath within the Ussing chamber. Simultaneous transepithelial resistance was recorded by applying a 2-mV pulse per minute via an automated pulse generator. Recordings were digitized and analyzed using PowerLab (AD Instruments, Colorado Springs, CO).
Transfections: RNA Interference, miRNA Overexpression, and Depletion
A number of DNA plasmids and RNA oligonucleotide constructs were transiently transfected into the mCCD cells using Lipofecamine 2000 (Invitrogen) according to the manufacturer’s instructions and as described previously.75,78 The target sequences for all siRNA constructs are listed in Supplemental Table 1. For miRNA overexpression, plasmids containing the stem-loop pre-miR sequences for mature miRs 335–3p, 1983 and 290–5p were obtained from GeneCopoeia (Rockville, MD) along with scrambled control clones. The plasmids contained an enhanced green fluorescent protein reporter and CMV promoter cassettes that allowed for detection of transfection efficiency and expression in mammalian cells. Double-stranded RNA miR mimics (miRIDIAN microRNA Mimics) were obtained from Thermo Fisher Scientific (Pittsburgh, PA) as an alternative to overexpress the miRs. To assay mimic delivery and transfection efficiency, nontargeting, fluorescently labeled control mimics were used (Thermo Fisher Scientific). To inhibit processing to mature miRs and reduce endogenous miR expression, LNA oligonucleotides targeting the three miRs along with nontargeting LNA controls were obtained from Exiqon, Inc. (Woburn, MA).
RNA Isolation and Microarray Analysis
RNA from cultured or primary CCD cells was isolated using the miRNeasy RNA isolation kit (Qiagen, Germantown, MD) according to the manufacturer’s protocol. The kit facilitated isolation of both miRNA and total RNA from each sample for use in qRT-PCR, RT-PCR, and microarray analysis. Total RNA (containing miRNAs) concentration and quality were evaluated for inclusion in subsequent in vitro transcription assays based on a spectrophotometric absorption ratio of 260/280 >1.8 (NanoDrop, Wilmington, DE) and an RIN (RNA integrity number) value of >5.0 via electrophoretic analysis (Agilent Bioanalyzer 2100; Agilent Technologies, Santa Clara, CA). Direct labeling of the miRNA using the Exiqon miRCURY LNA Hy3 Power Labeling Kit (Exiqon, Inc.) was performed on five paired expansions of mCCD cells cultured under standard conditions (n=5) or after 24 hours of 50 nM aldosterone treatment (n=5). Briefly, 1 μg of total RNA from each sample was incubated with Calf Intestinal Phosphatase (4 μl reaction volume) at 37°C for 30 minutes in an Eppendorf Thermostat Plus heat block (Eppendorf Inc., Hauppauge, NY). The samples were then denatured at 95°C for 5 minutes. Labeling Enzyme and Hy3 fluorescent label (Exiqon, Inc.) were added to each sample (12.5 μl final volume) for incubation at 16°C for 1 hour, followed by 15 minutes at 65°C. Hybridization buffer was added to each sample to a 400 μl volume, followed by denaturation at 95°C for 2 minutes before manual hybridization on Exiqon miRCURY LNA HSA, MMU, RNO spotted nucleotide arrays (Homo sapiens, Mus musculus, and Rattus norvegicus). For each sample, an Agilent gasket slide (1 microarray per slide format, part #G2534–60003; Agilent Technologies) was placed in an Agilent hybridization chamber and the entire sample was pipetted onto the gasket slide. The Exiqon array was then placed onto the gasket slide with the probe side facing down. The loaded hybridization chambers were clamped closed and placed into the hybridization oven for overnight incubation (18 hours at 56°C and 20 rpm).
The arrays were manually washed using CodeLink Parallel Processing Kits (Applied Microarrays Inc., Tempe, AZ) with Exiqon miRCURY LNA array Washing Buffer Kit (Exiqon, Inc.) according to the manufacturer’s specifications. The arrays were scanned on an Axon GenePix 4000B scanner (Molecular Devices Inc., Sunnyvale, CA) (settings: pixel size, 5 μm, 635 photomultiplier tube 600, 635 power 100, 532 photomultiplier tube 650, 532 power 100) and analyzed using GenePix Pro 6.0 software (Molecular Devices Inc.) with annotation of sequences from Sanger miRBase version 11.0. Local background subtraction was applied to each probe before averaging target replicates, and these values were compared with a threshold value determined from “negative” probes distributed throughout the array. Data from the 10 arrays were quantile normalized followed by testing for statistical differences using the paired t test function in the Significance Analysis of Microarrays software (SAM version 4.0).79
qRT-PCT
Primers and primer pairs used for all PCRs are listed in Supplemental Table 1. For qRT-PCR of miRNA, the nCode Express SYBR-Green miRNA with ROX qRT-PCR kit was used for reverse transcription and fist-strand DNA synthesis (Invitrogen). For all miRNA qPCRs, the miRNA-specific forward primers were paired to a universal reverse primer per the manufacturer’s protocol. Real-time PCR was carried out using an Applied Biosystems 7900HT Fast Real-Time PCR System (Applied Biosystems, Life Technologies, Grand Island, NY). Detected signals from miR amplifications were normalized to the relative expression of small nucleolar RNA (SNO-202 and SNO-135) with each reaction/sample run in triplicate. Negative controls included no template and no primer omissions. The standard qPCR protocol is provided in Supplemental Table 1. For qPCR of mRNA, primer pairs were used as listed (Supplemental Table 1), using the Express SYBR-Green with ROX qPCR kit (Invitrogen). Relative mRNA was normalized to the glyceraldehyde 3-phosphate dehydrogenase and/or actin message from each sample, and expression is presented as a fold change from control untreated samples (ΔΔCT).
Ex Vivo Kidney CCD Cell Isolation
Distal kidney nephron principal epithelial cells were isolated from a crude kidney tubules preparation using a lectin binding and magnetic bead isolation technique, similar to that described before with the following modifications80: briefly, before kidney harvest, streptavidin-conjugated magnetic beads (Dynabeads M-280; Invitrogen) were complexed to biotinylated Dolichos biflorus agglutinin (DBA; Vector Laboratories, Burlingame, CA) by incubation for 2 hours at room temperature with rotation. Beads were washed three times in in PBS with 0.1% BSA by magnetic separation and resuspended in PBS-BSA and stored at 4°C for use in epithelial cell isolation. Mice were euthanized, and kidneys were removed and immediately placed into HEPES/Ringer (HR) buffer at 4°C. Kidneys were crudely dissected into small pieces (approximately 3-mm cubes) by blunt dissection and incubated with agitation in digestion buffer (containing 0.2% collagenase and hyaluronidase) at 37°C for 45 minutes. Following addition of 0.001% DNase1, digestion proceeded for an additional 25 minutes at 37°C. At this stage the suspension contained separated cells and small tubule fragments. Large, undigested pieces were removed by passing through a cell strainer (100 μm; Thermo Fisher Scientific) and tubule fragments/cells were washed by centrifugation (75 g) and resuspended in HR buffer three times. The small tubule fragments were further digested to single cells by incubating in 0.25% trypsin-EDTA at 37°C for 5 minutes. Trypsin was neutralized by addition of equal volume of 5% FCS in PBS, and washed by centrifugation and resuspended in HR buffer as above. Cells isolated in the crude preparation were next labeled with DBA-conjugated beads by incubating beads/cells (30:1) for 30 minutes at room temperature with rotation. Finally, labeled cells linked to beads were isolated by magnetic isolation and resuspended in HR buffer (HR wash) five times. The isolated cells were immediately processed for RNA isolation; isolated RNA was used immediately or stored at −80°C until needed. Enrichment for distal nephron principal cells was confirmed by qRT-PCR using markers for proximal and distal nephron segments, normalized to whole kidney mRNA levels (see Supplemental Figure 3).
miRNA Target Prediction
The use of the miR target prediction algorithm combinatorial miRNA targeting (ComiR), along with detailed information on testing and development of the ComiR application, has been described in detail by us before.51 The algorithm integrates the expression of multiple miRs into the scores of four top target prediction tools using appropriate thermodynamic models. The scores of the four tools for each mRNA are then combined through a support vector machine with linear kernel trained on an established RNA-induced silencing complex immunoprecipitation dataset.51 The target prediction score of ComiR is the class probability value computed by using the trained support vector machine model. ComiR scores therefore range from 0 to 1, and higher scores correspond to higher probability of an mRNA being targeted by the particular set of miRNAs with given expression levels. A public web server has been made available for array analysis as described in this initial publication.
Dual Reporter Luciferase Plasmid
To demonstrate binding of miRs to the Ank3 3′UTR, an Ank3 image clone was obtained (BC021657, Clone ID:4188590; Thermo Fisher Scientific) and the 3′-UTR was isolated by digestion with SpeI and NheI. The digested fragment was gel extracted (Qiagen), and cloned into the NheI site of the pmirGLO Dual Luciferase miRNA Target Expression Reporter Vector (Promega, Madison, WI) according to the manufacturer’s protocol. A clone containing the insert in the correct orientation was isolated and the sequence confirmed (Genewiz, South Plainfield, NJ). This clone represents a stretch of approximately 850 bases distal to the stop codon in Ank3. Generated plasmids were transfected into mCCD cells (as above) and cells seeded onto filter supports. After 48 hours, luciferase activity was determined using the Dual-Glo Luciferase Assay System (Promega) according to the manufacturer’s protocol. Bioluminescence activity was recorded on a Synergy 1H plate reader (BioTek Instruments, Winooski, VT) using an integration time of 5 seconds and sensitivity of 200 for each sample.
Blot Quantification and Statistical Analyses
Densitometric quantification of protein band intensities was carried out in Adobe Photoshop CS5.1 (Adobe Systems, Inc., San Jose, CA), and values were expressed as a percentage of control signal, following background subtraction, and normalization to total protein expression (actin). Statistical analyses were performed using SigmaPlot software (Systat, San Jose, CA). All data are presented as mean±SEM. Data sets to be compared were tested for equal variance, and comparisons were performed using t tests or Mann–Whitney rank-sum tests. Groups were considered statistically significant different at P<0.05.
Disclosures
None.
Supplementary Material
Acknowledgments
We are grateful for the expert assistance of Xiaoning Liu, Maria A. Carananti, April Marrone, and Christin M. Sciulli. This study was supported financially by National Institutes of Health grants R00-DK078917 (M.B.B.), P30-DK079307 (pilot grant to M.B.B.), abd R01-DK047874 (J.P.J.); National Cancer Institute Cancer Center Support grants P30-CA47904 (W.A.L.), R01-LM009657 (P.V.B.), and R00-DK087922 (J.H.); and a Children’s Hospital of Pittsburgh of UPMC Research Advisory Committee startup grant (J.H.). C.C. was supported by Fondazione Ri.MED.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2013090931/-/DCSupplemental.
References
- 1.Garty H, Palmer LG: Epithelial sodium channels: Function, structure, and regulation. Physiol Rev 77: 359–396, 1997 [DOI] [PubMed] [Google Scholar]
- 2.Kellenberger S, Schild L: Epithelial sodium channel/degenerin family of ion channels: A variety of functions for a shared structure. Physiol Rev 82: 735–767, 2002 [DOI] [PubMed] [Google Scholar]
- 3.Bhalla V, Hallows KR: Mechanisms of ENaC regulation and clinical implications. J Am Soc Nephrol 19: 1845–1854, 2008 [DOI] [PubMed] [Google Scholar]
- 4.Butterworth MB: Regulation of the epithelial sodium channel (ENaC) by membrane trafficking. Biochim Biophys Acta 1802: 1166–1177, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butterworth MB, Edinger RS, Frizzell RA, Johnson JP: Regulation of the epithelial sodium channel by membrane trafficking. Am J Physiol Renal Physiol 296: F10–F24, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matalon S, Lazrak A, Jain L, Eaton DC: Invited review: Biophysical properties of sodium channels in lung alveolar epithelial cells. J Appl Physiol (1985) 93: 1852–1859, 2002 [DOI] [PubMed] [Google Scholar]
- 7.Rotin D, Kanelis V, Schild L: Trafficking and cell surface stability of ENaC. Am J Physiol Renal Physiol 281: F391–F399, 2001 [DOI] [PubMed] [Google Scholar]
- 8.Loffing J, Korbmacher C: Regulated sodium transport in the renal connecting tubule (CNT) via the epithelial sodium channel (ENaC). Pflugers Arch 458: 111–135, 2009 [DOI] [PubMed] [Google Scholar]
- 9.Christensen BM, Perrier R, Wang Q, Zuber AM, Maillard M, Mordasini D, Malsure S, Ronzaud C, Stehle JC, Rossier BC, Hummler E: Sodium and potassium balance depends on αENaC expression in connecting tubule. J Am Soc Nephrol 21: 1942–1951, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rotin D: Regulation of the epithelial sodium channel (ENaC) by accessory proteins. Curr Opin Nephrol Hypertens 9: 529–534, 2000 [DOI] [PubMed] [Google Scholar]
- 11.Gormley K, Dong Y, Sagnella GA: Regulation of the epithelial sodium channel by accessory proteins. Biochem J 371: 1–14, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hummler E: Epithelial sodium channel, salt intake, and hypertension. Curr Hypertens Rep 5: 11–18, 2003 [DOI] [PubMed] [Google Scholar]
- 13.Chang SS, Grunder S, Hanukoglu A, Rösler A, Mathew PM, Hanukoglu I, Schild L, Lu Y, Shimkets RA, Nelson-Williams C, Rossier BC, Lifton RP: Mutations in subunits of the epithelial sodium channel cause salt wasting with hyperkalaemic acidosis, pseudohypoaldosteronism type 1. Nat Genet 12: 248–253, 1996 [DOI] [PubMed] [Google Scholar]
- 14.Shimkets RA, Warnock DG, Bositis CM, Nelson-Williams C, Hansson JH, Schambelan M, Gill JR, Jr, Ulick S, Milora RV, Findling JW, et al. : Liddle’s syndrome: Heritable human hypertension caused by mutations in the beta subunit of the epithelial sodium channel. Cell 79: 407–414, 1994 [DOI] [PubMed] [Google Scholar]
- 15.Liddle GW, Bledsoe T, Coppage WS: A familial renal disorder simulating primary aldosteronism but with negligible aldosterone secretion. Trans Assoc Am Physicians 76: 199–213, 1963 [Google Scholar]
- 16.Abriel H, Loffing J, Rebhun JF, Pratt JH, Schild L, Horisberger JD, Rotin D, Staub O: Defective regulation of the epithelial Na+ channel by Nedd4 in Liddle’s syndrome. J Clin Invest 103: 667–673, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pochynyuk O, Tong Q, Staruschenko A, Ma HP, Stockand JD: Regulation of the epithelial Na+ channel (ENaC) by phosphatidylinositides. Am J Physiol Renal Physiol 290: F949–F957, 2006 [DOI] [PubMed] [Google Scholar]
- 18.Snyder PM: Minireview: Regulation of epithelial Na+ channel trafficking. Endocrinology 146: 5079–5085, 2005 [DOI] [PubMed] [Google Scholar]
- 19.Briet M, Schiffrin EL: Aldosterone: Effects on the kidney and cardiovascular system. Nat Rev Nephrol 6: 261–273, 2010 [DOI] [PubMed] [Google Scholar]
- 20.Hsueh WA, Wyne K: Renin-angiotensin-aldosterone system in diabetes and hypertension. J Clin Hypertens (Greenwich) 13: 224–237, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campese VM, Park J: The kidney and hypertension: Over 70 years of research. J Nephrol 19: 691–698, 2006 [PubMed] [Google Scholar]
- 22.Martel JA, Michael D, Fejes-Toth G, Naray-Fejes-Toth A: Melanophilin, a novel aldosterone-induced gene in mouse cortical collecting duct cells. Am J Physiol Renal Physiol 293: F904–F913, 2007 [DOI] [PubMed] [Google Scholar]
- 23.McCormick JA, Bhalla V, Pao AC, Pearce D: SGK1: A rapid aldosterone-induced regulator of renal sodium reabsorption. Physiology (Bethesda) 20: 134–139, 2005 [DOI] [PubMed] [Google Scholar]
- 24.Muller OG, Parnova RG, Centeno G, Rossier BC, Firsov D, Horisberger JD: Mineralocorticoid effects in the kidney: Correlation between alphaENaC, GILZ, and Sgk-1 mRNA expression and urinary excretion of Na+ and K+. J Am Soc Nephrol 14: 1107–1115, 2003 [DOI] [PubMed] [Google Scholar]
- 25.Bhalla V, Soundararajan R, Pao AC, Li H, Pearce D: Disinhibitory pathways for control of sodium transport: Regulation of ENaC by SGK1 and GILZ. Am J Physiol Renal Physiol 291: F714–F721, 2006 [DOI] [PubMed] [Google Scholar]
- 26.Soundararajan R, Zhang TT, Wang J, Vandewalle A, Pearce D: A novel role for glucocorticoid-induced leucine zipper protein in epithelial sodium channel-mediated sodium transport. J Biol Chem 280: 39970–39981, 2005 [DOI] [PubMed] [Google Scholar]
- 27.Rotin D, Staub O: Role of the ubiquitin system in regulating ion transport. Pflugers Archiv 461: 1–21, 2011. [DOI] [PubMed] [Google Scholar]
- 28.Zhou R, Patel SV, Snyder PM: Nedd4-2 catalyzes ubiquitination and degradation of cell surface ENaC. J Biol Chem 282: 20207–20212, 2007 [DOI] [PubMed] [Google Scholar]
- 29.Wiemuth D, Ke Y, Rohlfs M, McDonald FJ: Epithelial sodium channel (ENaC) is multi-ubiquitinated at the cell surface. Biochem J 405: 147–155, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Staub O, Verrey F: Impact of Nedd4 proteins and serum and glucocorticoid-induced kinases on epithelial Na+ transport in the distal nephron. J Am Soc Nephrol 16: 3167–3174, 2005 [DOI] [PubMed] [Google Scholar]
- 31.Debonneville C, Flores SY, Kamynina E, Plant PJ, Tauxe C, Thomas MA, Münster C, Chraïbi A, Pratt JH, Horisberger JD, Pearce D, Loffing J, Staub O: Phosphorylation of Nedd4-2 by Sgk1 regulates epithelial Na(+) channel cell surface expression. EMBO J 20: 7052–7059, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verrey F, Fakitsas P, Adam G, Staub O: Early transcriptional control of ENaC (de)ubiquitylation by aldosterone. Kidney Int 73: 691–696, 2008 [DOI] [PubMed] [Google Scholar]
- 33.Spindler B, Mastroberardino L, Custer M, Verrey F: Characterization of early aldosterone-induced RNAs identified in A6 kidney epithelia. Pflugers Arch 434: 323–331, 1997 [DOI] [PubMed] [Google Scholar]
- 34.Erson AE, Petty EM: MicroRNAs in development and disease. Clin Genet 74: 296–306, 2008 [DOI] [PubMed] [Google Scholar]
- 35.Flynt AS, Lai EC: Biological principles of microRNA-mediated regulation: Shared themes amid diversity. Nat Rev Genet 9: 831–842, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saal S, Harvey SJ: MicroRNAs and the kidney: Coming of age. Curr Opin Nephrol Hypertens 18: 317–323, 2009 [DOI] [PubMed] [Google Scholar]
- 37.Sontheimer EJ, Carthew RW: Silence from within: Endogenous siRNAs and miRNAs. Cell 122: 9–12, 2005 [DOI] [PubMed] [Google Scholar]
- 38.Bartel DP: MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Friedman RC, Farh KK, Burge CB, Bartel DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee Y, Jeon K, Lee JT, Kim S, Kim VN: MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J 21: 4663–4670, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diederichs S, Haber DA: Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell 131: 1097–1108, 2007 [DOI] [PubMed] [Google Scholar]
- 42.Jaskiewicz L, Filipowicz W: Role of Dicer in posttranscriptional RNA silencing. Curr Top Microbiol Immunol 320: 77–97, 2008 [DOI] [PubMed] [Google Scholar]
- 43.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foà R, Schliwka J, Fuchs U, Novosel A, Müller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T: A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tian Z, Greene AS, Pietrusz JL, Matus IR, Liang M: MicroRNA-target pairs in the rat kidney identified by microRNA microarray, proteomic, and bioinformatic analysis. Genome Res 18: 404–411, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guarnieri DJ, DiLeone RJ: MicroRNAs: A new class of gene regulators. Ann Med 40: 197–208, 2008 [DOI] [PubMed] [Google Scholar]
- 46.Kim VN, Han J, Siomi MC: Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139, 2009 [DOI] [PubMed] [Google Scholar]
- 47.Carthew RW, Sontheimer EJ: Origins and Mechanisms of miRNAs and siRNAs. Cell 136: 642–655, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim VN: MicroRNA biogenesis: Coordinated cropping and dicing. Nat Rev Mol Cell Biol 6: 376–385, 2005 [DOI] [PubMed] [Google Scholar]
- 49.Corcoran DL, Pandit KV, Gordon B, Bhattacharjee A, Kaminski N, Benos PV: Features of mammalian microRNA promoters emerge from polymerase II chromatin immunoprecipitation data. PLoS ONE 4: e5279, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coronnello C, Benos PV: ComiR: Combinatorial microRNA target prediction tool. Nucleic Acids Res 41[Web Server issue]: W159–164, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coronnello C, Hartmaier R, Arora A, Huleihel L, Pandit KV, Bais AS, Butterworth M, Kaminski N, Stormo GD, Oesterreich S, Benos PV: Novel modeling of combinatorial miRNA targeting identifies SNP with potential role in bone density. PLOS Comput Biol 8: e1002830, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuwabara Y, Ono K, Horie T, Nishi H, Nagao K, Kinoshita M, Watanabe S, Baba O, Kojima Y, Shizuta S, Imai M, Tamura T, Kita T, Kimura T: Increased microRNA-1 and microRNA-133a levels in serum of patients with cardiovascular disease indicate myocardial damage. Circ Cardiovasc Genet 4: 446–454, 2011 [DOI] [PubMed] [Google Scholar]
- 53.Rao PK, Toyama Y, Chiang HR, Gupta S, Bauer M, Medvid R, Reinhardt F, Liao R, Krieger M, Jaenisch R, Lodish HF, Blelloch R: Loss of cardiac microRNA-mediated regulation leads to dilated cardiomyopathy and heart failure. Circ Res 105: 585–594, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cao Q, Mani RS, Ateeq B, Dhanasekaran SM, Asangani IA, Prensner JR, Kim JH, Brenner JC, Jing X, Cao X, Wang R, Li Y, Dahiya A, Wang L, Pandhi M, Lonigro RJ, Wu YM, Tomlins SA, Palanisamy N, Qin Z, Yu J, Maher CA, Varambally S, Chinnaiyan AM: Coordinated regulation of polycomb group complexes through microRNAs in cancer. Cancer Cell 20: 187–199, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park JY, Helm J, Coppola D, Kim D, Malafa M, Kim SJ: MicroRNAs in pancreatic ductal adenocarcinoma. World J Gastroenterol 17: 817–827, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davis BN, Hata A: MicroRNA in cancer—The involvement of aberrant microRNA biogenesis regulatory pathways. Genes Cancer 1: 1100–1114, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Negrini M, Nicoloso MS, Calin GA: MicroRNAs and cancer—new paradigms in molecular oncology. Curr Opin Cell Biol 21: 470–479, 2009 [DOI] [PubMed] [Google Scholar]
- 58.Sotiropoulou G, Pampalakis G, Lianidou E, Mourelatos Z: Emerging roles of microRNAs as molecular switches in the integrated circuit of the cancer cell. RNA 15: 1443–1461, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lorenzen JM, Haller H, Thum T: MicroRNAs as mediators and therapeutic targets in chronic kidney disease. Nat Rev Nephrol 7: 286–294, 2011 [DOI] [PubMed] [Google Scholar]
- 60.Elton TS, Sansom SE, Martin MM: Cardiovascular disease, single nucleotide polymorphisms; and the renin angiotensin system: Is there a MicroRNA connection? IntJ Hypertens 2010: pii 281692, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhdanova O, Srivastava S, Di L, Li Z, Tchelebi L, Dworkin S, Johnstone DB, Zavadil J, Chong MM, Littman DR, Holzman LB, Barisoni L, Skolnik EY: The inducible deletion of Drosha and microRNAs in mature podocytes results in a collapsing glomerulopathy. Kidney Int 80: 719–730, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei Q, Bhatt K, He HZ, Mi QS, Haase VH, Dong Z: Targeted deletion of Dicer from proximal tubules protects against renal ischemia-reperfusion injury. J Am Soc Nephrol 21: 756–761, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krupa A, Jenkins R, Luo DD, Lewis A, Phillips A, Fraser D: Loss of MicroRNA-192 promotes fibrogenesis in diabetic nephropathy. J Am Soc Nephrol 21: 438–447, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kato M, Natarajan R: microRNA cascade in diabetic kidney disease: Big impact initiated by a small RNA. Cell Cycle 8: 3613–3614, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kato M, Arce L, Natarajan R: MicroRNAs and their role in progressive kidney diseases. Clin J Am Soc Nephrol 4: 1255–1266, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi JJ, Natarajan R: MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci U S A 104: 3432–3437, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chung AC, Huang XR, Meng X, Lan HY: miR-192 mediates TGF-beta/Smad3-driven renal fibrosis. J Am Soc Nephrol 21: 1317–1325, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harvey SJ, Jarad G, Cunningham J, Goldberg S, Schermer B, Harfe BD, McManus MT, Benzing T, Miner JH: Podocyte-specific deletion of dicer alters cytoskeletal dynamics and causes glomerular disease. J Am Soc Nephrol 19: 2150–2158, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin DH, Yue P, Pan C, Sun P, Wang WH: MicroRNA 802 stimulates ROMK channels by suppressing caveolin-1. J Am Soc Nephrol 22: 1087–1098, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nossent AY, Hansen JL, Doggen C, Quax PH, Sheikh SP, Rosendaal FR: SNPs in microRNA binding sites in 3′-UTRs of RAAS genes influence arterial blood pressure and risk of myocardial infarction. Am J Hypertens 24: 999–1006, 2011 [DOI] [PubMed] [Google Scholar]
- 71.Sõber S, Laan M, Annilo T: MicroRNAs miR-124 and miR-135a are potential regulators of the mineralocorticoid receptor gene (NR3C2) expression. Biochem Biophys Res Commun 391: 727–732, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Elvira-Matelot E, Zhou XO, Farman N, Beaurain G, Henrion-Caude A, Hadchouel J, Jeunemaitre X: Regulation of WNK1 expression by miR-192 and aldosterone. J Am Soc Nephrol 21: 1724–1731, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gaeggeler HP, Gonzalez-Rodriguez E, Jaeger NF, Loffing-Cueni D, Norregaard R, Loffing J, Horisberger JD, Rossier BC: Mineralocorticoid versus glucocorticoid receptor occupancy mediating aldosterone-stimulated sodium transport in a novel renal cell line. J Am Soc Nephrol 16: 878–891, 2005 [DOI] [PubMed] [Google Scholar]
- 74.Butterworth MB, Zhang L, Heidrich EM, Myerburg MM, Thibodeau PH: Activation of the epithelial sodium channel (ENaC) by the alkaline protease from Pseudomonas aeruginosa. J Biol Chem 287: 32556–32565, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Butterworth MB, Edinger RS, Silvis MR, Gallo LI, Liang X, Apodaca G, Frizzell RA, Johnson JP: Rab11b regulates the trafficking and recycling of the epithelial sodium channel (ENaC). Am J Physiol Renal Physiol 302: F581–F590, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Edinger RS, Bertrand CA, Rondandino C, Apodaca GA, Johnson JP, Butterworth MB: The epithelial sodium channel (ENaC) establishes a trafficking vesicle pool responsible for its regulation. PLoS ONE 7: e46593, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Myerburg MM, Harvey PR, Heidrich EM, Pilewski JM, Butterworth MB: Acute regulation of the epithelial sodium channel in airway epithelia by proteases and trafficking. Am J Respir Cell Mol Biol 43: 712–719, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Butterworth MB, Edinger RS, Ovaa H, Burg D, Johnson JP, Frizzell RA: The deubiquitinating enzyme UCH-L3 regulates the apical membrane recycling of the epithelial sodium channel. J Biol Chem 282: 37885–37893, 2007 [DOI] [PubMed] [Google Scholar]
- 79.Tusher VG, Tibshirani R, Chu G: Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98: 5116–5121, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grupp C, Troche I, Steffgen J, Langhans S, Cohen DI, Brandl L, Müller GA: Highly specific separation of heterogeneous cell populations by lectin-coated beads: Application for the isolation of inner medullary collecting duct cells. Exp Nephrol 6: 542–550, 1998 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.