Background: Photosystem I is a large protein/pigment assembly required for photosynthesis.
Results: The PSA2 protein harbors a DnaJ-type zinc finger domain, is required for Photosystem I accumulation, and is found in a PsaG-containing complex in the thylakoid lumen.
Conclusion: PSA2 promotes Photosystem I biogenesis via interaction with a PsaG-containing complex in the thylakoid lumen.
Significance: These findings elucidate the biogenesis of the photosynthetic apparatus.
Keywords: Arabidopsis, Chaperone DnaJ (DnaJ), Chloroplast, Membrane Biogenesis, Photosynthesis, Protein-disulfide Isomerase, Maize, Thylakoid Lumen
Abstract
Photosystem I (PSI) is a large pigment-protein complex and one of the two photosystems that drive electron transfer in oxygenic photosynthesis. We identified a nuclear gene required specifically for the accumulation of PSI in a forward genetic analysis of chloroplast biogenesis in maize. This gene, designated psa2, belongs to the “GreenCut” gene set, a group of genes found in green algae and plants but not in non-photosynthetic organisms. Disruption of the psa2 ortholog in Arabidopsis likewise resulted in the specific loss of PSI proteins. PSA2 harbors a conserved domain found in DnaJ chaperones where it has been shown to form a zinc finger and to have protein-disulfide isomerase activity. Accordingly, PSA2 exhibited protein-disulfide reductase activity in vitro. PSA2 localized to the thylakoid lumen and was found in a ∼250-kDa complex harboring the peripheral PSI protein PsaG but lacking several core PSI subunits. PSA2 mRNA is coexpressed with mRNAs encoding various proteins involved in the biogenesis of the photosynthetic apparatus with peak expression preceding that of genes encoding structural components. PSA2 protein abundance was not decreased in the absence of PSI but was reduced in the absence of the PSI assembly factor Ycf3. These findings suggest that a complex harboring PSA2 and PsaG mediates thiol transactions in the thylakoid lumen that are important for the assembly of PSI.
Introduction
Photosystem I (PSI)2 is one of the two photosystems that drive oxygenic photosynthesis and is among the most complex macromolecular structures known in nature (1). In land plants, the PSI core complex consists of at least 16 different polypeptides (PsaA–P), three Fe-S clusters, and at least 173 chlorophyll molecules; this core associates with a peripheral light-harvesting complex consisting of four additional proteins (Lhcal–Lhca4) and numerous pigments (2). The biogenesis of PSI in chloroplasts is further complicated by the fact that the genes encoding its subunits are distributed between the nuclear and plastid genomes: PsaA, PsaB, PsaC, PsaI, and PsaJ are encoded in the chloroplast, whereas the remaining subunits are nucleus-encoded (3). In higher plants, PSI biogenesis is coupled to leaf development. This process necessitates the coordinated assembly of subunits and prosthetic groups at the thylakoid membrane where PSI is situated. Several factors have been described that are required for the assembly of PSI but that are not found in the mature complex; these include the chloroplast-encoded proteins Ycf3 and Ycf4 (4–6) and the nucleus-encoded proteins Y3IP1 (7), Pyg7 (8), and PPD1 (9,10). Additional proteins mediate the association of prosthetic groups with PSI subunits (11–14). Despite this progress, many aspects of PSI biogenesis remain poorly understood (3, 15).
To thoroughly characterize the nuclear gene complement required for the biogenesis of the photosynthetic apparatus, our groups are taking (i) a large scale forward genetic approach to identify nuclear genes required for photosynthesis in maize (16, 17) and (ii) a large scale reverse genetic analysis of genes that are conserved in photosynthetic organisms but absent from non-photosynthetic organisms, the so-called “GreenCut” gene set (18, 19). Here we describe how combining these two approaches revealed a function for a GreenCut protein that acts in the thylakoid lumen to promote the biogenesis of PSI.
EXPERIMENTAL PROCEDURES
Mutant Isolation and Plant Growth
The maize psa2 mutation is a recessive allele conditioning a pale green, non-photosynthetic phenotype that arose in maize lines harboring active Mu transposons. Heterozygotes were outcrossed to inbred lines, and the F1 progeny were then self-pollinated to recover ears segregating homozygous mutants. DNA was isolated from mutant individuals from each of four F2 ears and analyzed by Mu Illumina sequencing to map all of the Mu insertions in each plant (16). An insertion in GRMZM2G021687 was found in all four mutant individuals; subsequent gene-specific PCR showed this insertion to be absent from closely related +/+ ears. Maize seedlings used for phenotypic analysis were grown in a growth chamber in 16-h light (120 μmol of photons m−2 s−1), 8-h dark cycles. For exposure to high light conditions, illumination was increased to 800 μmol of photons m−2 s−1.
Three tDNA insertions in the Arabidopsis psa2 ortholog (AT2G34860) were obtained: At-psa2-1 (GABI_475C12 Col background) and At-psa2-2 (GABI_314G07 Col background) came from the GABI-Kat collection, and At-psa2-3 (SK32878 Col background) came from the Saskatoon collection. Plants were grown in a growth chamber under long day conditions (16-h light, 8-h dark) at 120 μmol of photons m−2 s−1. For growth on sucrose-containing synthetic medium, seeds were surface-sterilized and plated on Murashige and Skoog agar medium containing 0.8% sucrose.
Analysis of Chloroplast Gene Expression
The expression of all chloroplast genes in maize psa2 mutants was analyzed with a ribosome profiling method that provides a genome-wide and quantitative readout of chloroplast ribosome positions in vivo (20). The method uses high resolution microarrays to compare the positions and abundance of “ribosome footprints” in mutant and wild-type samples. The results report differences in both mRNA abundance and ribosome occupancy for every chloroplast gene.
Chloroplast Isolation and Fractionation
4-week-old Arabidopsis or 2-week-old maize plants were used for the preparation of chloroplasts and thylakoid membranes. 2 g of Arabidopsis leaf tissue or the leaves from three maize seedlings were collected either after 4 h of dark or after 3 h of illumination (120 μmol of photons m−2 s−1). Chloroplasts, thylakoid membranes, and subthylakoid fractions were isolated as described previously (21, 22). The lumenal fraction was recovered as the supernatant after treating isolated thylakoid membranes with 0.2% Triton X-100 for 2 min on ice followed by centrifugation at 140,000 × g for 10 min.
Antibodies
An antibody specific for PSA2 was produced by immunization of rabbits with a recombinant protein corresponding to amino acids 87–186 of Arabidopsis PSA2 (Agrisera AS132654). Antibodies against D1, D2, Lhcb1, Lhca1, Lhca2, and CP43 were obtained from Agrisera. Antibodies against CF1 were generated by immunization with the purified CF1 complex from spinach (23). PsaA- and PsaF-specific antibodies were kindly provided by Dr. Jean-David Rochaix (University of Geneva). PsaG- and PsaK-specific antibodies were kindly provided by Dr. Poul Erik Jensen (University of Copenhagen). Antibody to the Rubisco large subunit (RbcL) was a gift from Dr. Steve Rodermel (Iowa State University). Antibodies to PsaD and cytochrome f were described previously (24, 25). Antibody to PPD1 was generously provided by Johnna Roose and Terry Bricker (Louisiana State University).
Protein Analyses
Blue native polyacrylamide gel electrophoresis (BN-PAGE) of thylakoid membranes was performed as described (26). Prior to electrophoresis, thylakoid membranes were solubilized by incubation on ice (10 min) in 20% (w/v) glycerol, 25 mm BisTris-HCl, pH 7.0, 0.75% (w/v) n-dodecyl β-d-maltoside (DM) at a chlorophyll concentration of 2 mg/ml. For immunoblot assays, proteins were separated by SDS-PAGE in the presence of 6 m urea or by BN-PAGE, electrophoretically transferred to polyvinylidene difluoride (PVDF) membranes (Immobilon, Millipore), and then probed with the antibodies described above. Antibodies were detected with horseradish peroxidase-conjugated secondary antibody and the SuperSignal WestPico HRP detection kit (Thermo Scientific).
For immunoprecipitation, chloroplasts from 3-week-old Arabidopsis plants were resuspended in solubilization buffer (20 mm Tricine-NaOH, pH 7.8, 5 mm MgCl2, 100 mm sucrose, 10 mm DM) and incubated on ice for 15 min. Particulates were removed by centrifugation at 4 °C at 14,000 × g for 20 min. Pierce cross-link magnetic beads were coupled to PSA2 antibody according to the manufacturer's instructions (Pierce Crosslink Magnetic IP/Co-IP kit 88805). Immunoprecipitations were performed by incubating the antibody-coupled beads with 400 μl of solubilized chloroplasts at a protein concentration of 2 μg/μl for 60 min with slow rotation at room temperature. The beads were washed, and the bound proteins were eluted according to the manufacturer's instructions.
Chlorophyll Fluorescence Analyses
Plants were dark-adapted for 20 min before each experiment. Fluorescence parameters were measured using a JTS-10 light-emitting diode spectrometer (Bio-Logic Scientific Instruments) and calculated as follows: Fv/Fm = (Fm − F0)/Fm where Fv is the calculated variable fluorescence, Fm is the maximal fluorescence measured immediately after the saturating pulse, and F0 is the initial fluorescence of dark-adapted tissues. Data from different samples were normalized with JTS10 spectrophotometer software (Bio-Logic) using default parameters. The kinetics of PSI photooxidation were measured on detached leaves in absorbance detection mode. P700 was oxidized using 10-ms actinic flashes of far-red light-emitting diodes and a 705-nm interference filter. Baseline absorbances were obtained without actinic illumination and subtracted.
Assaying Protein-disulfide Reductase Activity
The ability of PSA2 to catalyze the reduction of disulfide bonds was tested with an assay for insulin precipitation that is often used as a proxy for protein-disulfide isomerase activity (27). Reactions contained 0.13 mm insulin (Sigma-Aldrich), 0.1 mm BSA, 0.1 m sodium phosphate, pH 7.0, 2.5 mm EDTA, and a 1 μm concentration of either PSA2 or RBF1. Reactions were initiated by adding dithiothreitol to a final concentration of 100 μm. Insulin precipitation was monitored by the increase in absorbance at 650 nm (27). RBF1, a ribosome-binding protein that contains no cysteines in the mature sequence (21), was used as a control.
RESULTS
The GreenCut Gene psa2 Is Required for the Accumulation of Photosystem I in Maize and Arabidopsis
The psa2 gene was identified during the systematic analysis of mutants in the Photosynthetic Mutant Library, a large collection of chloroplast-defective mutants that arose in maize lines with active Mu transposons (17). The homozygous mutants are pale green (Fig. 1A) and die after the development of three leaves as is typical of non-photosynthetic mutants in maize. These phenotypes were tentatively ascribed to a specific loss of PSI based on an initial immunoblot survey that probed the abundance of one core subunit of PSI, Rubisco, photosystem II (PSII), the cytochrome b6f complex, and the chloroplast ATP synthase. To identify the insertion responsible for these phenotypes, we used a deep sequencing method to sequence Mu-gene junctions in multiple mutant individuals (16). An insertion in GRMZM2G021687 cosegregated with the mutant phenotype and was an appealing candidate for the causal mutation for several reasons. (i) The product of this gene and its Arabidopsis ortholog (AT2G34860) have been detected in plastid proteome data sets (28). (ii) The ATTED-II database (29) showed coexpression of the Arabidopsis ortholog with several known assembly factors for photosynthetic complexes. (iii) The protein has a cysteine-rich domain that is characteristic of DnaJ chaperones (InterPro IPR001305) and found in several previously described proteins involved in the assembly or maintenance of the photosynthetic apparatus (30–33) (Fig. 2). A plastome-wide analysis of chloroplast gene expression by ribosome profiling indicated that all chloroplast genes encoding PSI subunits or assembly factors (psaA, psaB, psaC, psaJ, psaI, ycf3, and ycf4) are expressed at near normal levels in the mutant (supplemental Fig. 1). Based on these observations, we hypothesized that the product of GRMZM2G021687 promotes the assembly of PSI. On this basis, we assigned the name psa2 to GRMZM2G021687 to reflect its role in PSI function.
FIGURE 1.
Maize and Arabidopsis psa2 mutants. A, maize psa2-1 mutant. Plants were grown for 8 days in soil. The position of the Mu insertion is shown above with the nine-base pair target site duplication underlined. This insertion is 52 base pairs upstream of the start codon. B, Arabidopsis psa2 mutants. Triangles mark tDNA insertion sites, and the sequences flanking each insertion are shown below. The insertion in At-psa2-1 is 39 base pairs upstream of the start codon and is followed by a nine-base pair deletion. The insertion in At-psa2-2 is 544 base pairs downstream of the start codon and is followed by an eight-base pair deletion. The insertion in At-psa2-3 is 799 base pairs downstream of the start codon and is followed by a deletion of five base pairs. The deleted nucleotides are shown in bold, underlined italic font. Plants were grown at a photon flux density of 120 μmol m−2 s−1 (16-h light, 8-h dark) for 21 and 14 days (soil and sucrose, respectively). Scale bars, 1 cm.
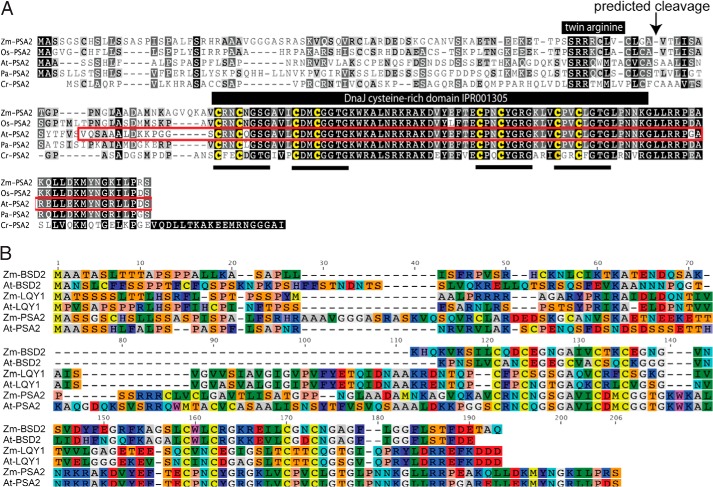
FIGURE 2.
Multiple sequence alignments of PSA2 orthologs and homologs. A, alignment of PSA2 orthologs. The alignment was generated with ClustalW. The DnaJ-like cysteine-rich domain predicted by InterPro (IPR001305) is marked with the four predicted zinc binding motifs underlined. The region used for antibody production is outlined in red. The pair of arginines characteristic of proteins that engage the TAT thylakoid import pathway are marked as is the predicted cleavage site that separates the targeting peptide from the mature protein. At-PSA2 corresponds to Arabidopsis thaliana gene model AT2G34860.1. Cr-PSA2 corresponds to Chlamydomonas reinhardtii gene model Cre11.g475850.t1.2. Os-PSA2 corresponds to Oryza sativa gene model Os08g36140.1. Pa-PSA2 corresponds to Populus alba gene model POPTR_0001s04720.1. Zm-PSA2 corresponds to Zea mays gene model GRMZM2G021687_P01. B, alignment of PSA2, LQY1, and BSD2 orthologs in maize and Arabidopsis. ZmBSD2 is GRMZM2G062788, AtBSD2 is AT3G47650, AtLQY1 is AT1G75690, and ZmLQY1 is GRMZM2G071996. Amino acids are colored according to the RasMol convention.
The psa2 ortholog in Arabidopsis, At-PSA2 (AT2G34860), is a member of the GreenCut gene set (18). PSA2 belongs to the subset of GreenCut genes that is found in photosynthetic eukaryotes but not in cyanobacteria. A multiple sequence alignment of orthologs in monocots, dicots, and Chlamydomonas (Fig. 2A) shows a high degree of sequence identity. As such, we anticipated that the function of this gene would be conserved among photosynthetic eukaryotes. To test this premise, we analyzed three insertion alleles of At-PSA2 (Fig. 1B). Plants homozygous for At-psa2-1 were pale green and grew very slowly when planted in soil. Plants homozygous for either At-psa2-2 or At-psa2-3 died shortly after germination on soil but grew to a small size on sucrose-containing growth medium (Fig. 1B).
Arabidopsis and Maize psa2 Mutants Specifically Lack the PSI Core Complex
Immunoblot analysis of thylakoid proteins (Fig. 3A) showed that the PSI subunits PsaA, PsaD, PsaF, PsaG, and PsaK were reduced in abundance in maize and Arabidopsis psa2 mutants, whereas subunits of the light-harvesting complexes (Lhcb1, Lhca1, and Lhca2), PSII (D1, D2, and CP43), the cytochrome b6f complex, and the chloroplast ATP synthase (CF1αβ) accumulated to normal levels. PsaA and PsaD were severely reduced in maize psa2 mutants even after 12 h of growth in the dark, whereas PsaG and PsaK accumulated to near normal levels under these conditions (Fig. 3A). Therefore, photooxidative damage was not the primary cause of the loss of the PSI core proteins PsaA and PsaD in psa2 mutants.
FIGURE 3.
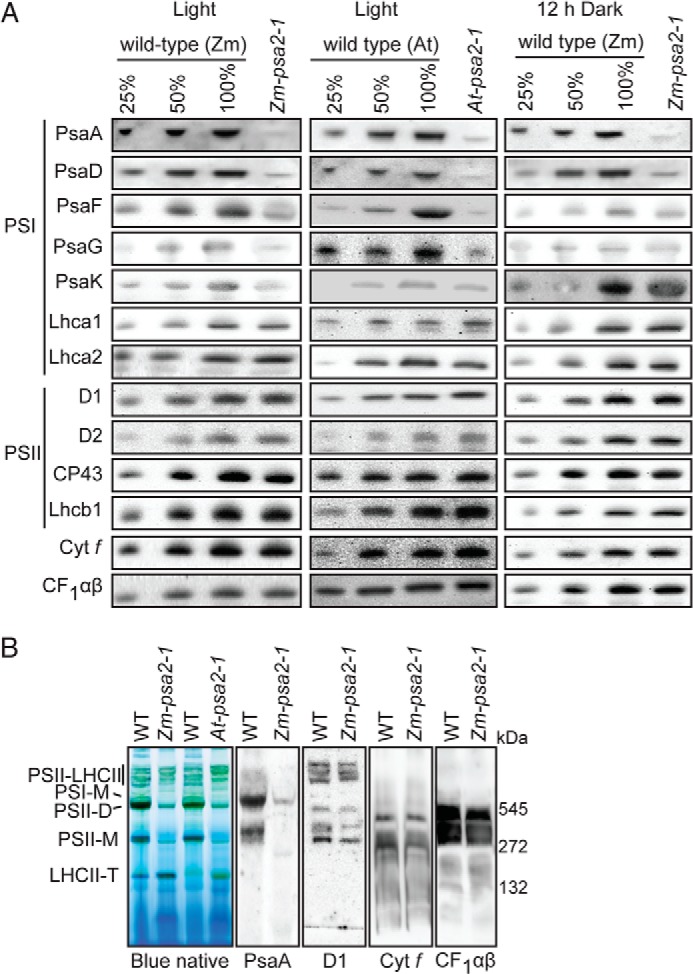
Specific loss of PSI core complex in maize and Arabidopsis psa2 mutants. A, immunoblot analyses of thylakoid proteins in maize (Zm-psa2-1) and Arabidopsis (At-psa2-1) mutants. A dilution series of material from wild-type siblings was included to aid in quantification. The panel to the right shows the results from maize seedlings that had been dark-adapted for 12 h. Antibodies were directed against PsaA, PsaF, PsaG, PsaK, and PsaD (PSI core subunits); Lhca1 and Lhca2 (PSI antenna proteins); D1, D2, and CP43 (PSII core subunits); Lhcb1 (PSII antenna protein); subunit of the cytochrome b6f complex (Cyt f), and CF1αβ (the α and β subunits of the membrane extrinsic portion of the ATP synthase complex). B, BN-PAGE analysis of thylakoid membrane complexes in maize and Arabidopsis psa2 mutants. Bands were assigned as the PSII-light-harvesting assemblies (PSII-LHCII), monomeric PSI (PSI-M), PSII dimer (PSII-D), PSII monomer (PSII-M), and trimeric light-harvesting complex II (LHCII-T) based on prior reports (26, 62, 63). Panels to the right show immunoblots of BN-PAGE-separated thylakoid proteins from wild-type and mutant maize seedlings.
Two additional assays support the view that psa2 mutants have a specific defect in PSI. First, BN-PAGE analysis of thylakoid membranes in maize and Arabidopsis psa2 mutants (Fig. 3B) revealed the loss of two major stained bands, at roughly 550 and 300 kDa. This pattern matches that reported for Arabidopsis ppd1 mutants, which specifically lack PSI due to a defect in PSI assembly (9). The upper band corresponds in position to monomeric PSI (9). The lower band comigrates with a small fraction of PsaA; although the band at this position is often described solely as PSII monomer, a PSI-related complex has been detected previously at this position (34). Immunoblots of BN-PAGE-resolved thylakoid membrane complexes were probed to detect subunits of PSII (D1), the cytochrome b6f complex, and the stromal portion of the thylakoid ATP synthase (CF1). The complexes harboring each of these proteins were not detectably altered in psa2 mutants (Fig. 3B).
Finally, we monitored several photosynthetic parameters by measuring chlorophyll fluorescence kinetics of intact leaves of psa2 mutants in both maize and Arabidopsis. Photooxidation of P700 was monitored in the presence of 3-(3,4-dichlorophenyl)-1,1-dimethylurea to block electron transfer from PSII. P700 was severely impaired in the mutants but less so in dark-adapted plants (Fig. 4A). To monitor PSII activity, the fluorescence rise between F0 and Fm was monitored (Fig. 4B). The Fv/Fm value was only slightly affected in dark-adapted mutant plants but showed a decrease under normal light conditions, indicating that PSII is light-sensitive in psa2 mutants. The decrease in PSII activity became more severe with increasing light intensity (Fig. 4C), suggesting that PSII is subject to increased photoinhibition in psa2 mutants. Increased photoinhibition has been observed previously in mutants with defects in PSI (9).
FIGURE 4.
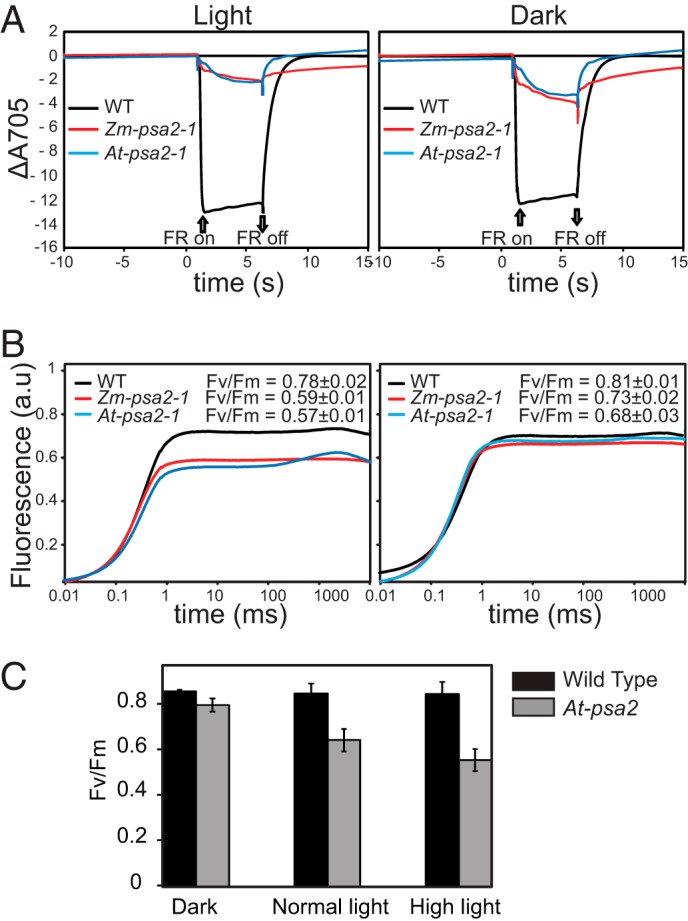
Spectroscopic characterization of PSI and PSII activity in psa2 mutants. A, PSI activity. The oxidation state of P700 was determined by measuring the absorbance at 705 nm after plants were illuminated with far-red light (720 nm). Plants of the indicated genotypes were monitored either during growth at normal photon flux density (120 μmol m−2 s−1) (left panel) or after 12 h in the dark (right panel). Leaves were soaked in 3-(3,4-dichlorophenyl)-1,1-dimethylurea for 10 min prior to the measurements. The plants were dark-adapted for 20 min and then preilluminated with far-red light for 2 min prior to the measurements (64). Wild-type (WT) maize and Arabidopsis performed very similarly in this assay (not shown); only the Arabidopsis data are shown. B, PSII activity. Time-resolved fluorescence kinetics of chlorophyll a from plants of the indicated genotypes grown at normal photon flux density (left panel) or dark-adapted for 12 h are shown. Leaves were soaked in 3-(3,4-dichlorophenyl)-1,1-dimethylurea for 10 min prior to the measurements. The leaves from the light-grown plants were dark-adapted for 20 min before the measurements. Wild-type (WT) maize and Arabidopsis performed very similarly in this assay (not shown). C, the ratio of variable to maximal chlorophyll fluorescence (Fv/Fm) measured in Arabidopsis plants adapted to dark (12 h), normal light (120 μmol of photons m−2 s−1), or high light (800 μmol of photons m−2 s−1 for 12 h). The plants were germinated and grown for 2 weeks under standard light conditions and then shifted to the indicated light conditions at the time of the normal dark to light transition. Readings were taken 12 h after the light shift. Error bars represent one S.D. (n = 8). FR, far-red; a.u., absorbance units.
These results demonstrate that the PSA2 orthologs in maize and Arabidopsis are required specifically for the accumulation and activity of PSI. It should be noted that AT2G34860 has been annotated as “embryo sac developmental arrest 3” due to a supposed role in embryo development (35). However, other than the seedling lethality that is an expected outcome of a defect in photosynthesis, we did not detect any defects in the development or inheritance of the mutants we analyzed.
PSA2 Localizes to the Thylakoid Lumen
We generated a polyclonal antibody to recombinant At-PSA2 lacking its predicted N-terminal targeting sequence. The antibody recognized a protein of the expected size (∼11 kDa) in leaf extracts from wild-type maize and Arabidopsis plants; this protein was reduced more than 4-fold in the maize and Arabidopsis psa2 mutants (Fig. 5A), providing strong evidence that it is PSA2. The residual accumulation of PSA2 in the maize mutant is not surprising given that Mu insertions in 5′-untranslated regions are typically hypomorphic alleles. PSA2 was below the limit of detection in the Arabidopsis At-psa2-1 mutant; however, the At-psa2-1 allele conditioned a less severe phenotype than do the At-psa2-2 and At-psa2-3 alleles (Fig. 1B), indicating that At-psa2-1 is not a null allele. PSA2 accumulated to near normal levels in non-allelic maize mutants lacking PSI, indicating that PSA2 accumulation is not coupled to that of PSI (Fig. 5B). Interestingly, however, PSA2 was absent in the Arabidopsis apo1 mutant (Fig. 5C), which lacks PSI due to a defect in the expression of the chloroplast gene encoding the PSI assembly factor Ycf3 (36, 37). This finding suggests a potential functional relationship between PSA2 and Ycf3 in PSI assembly.
FIGURE 5.
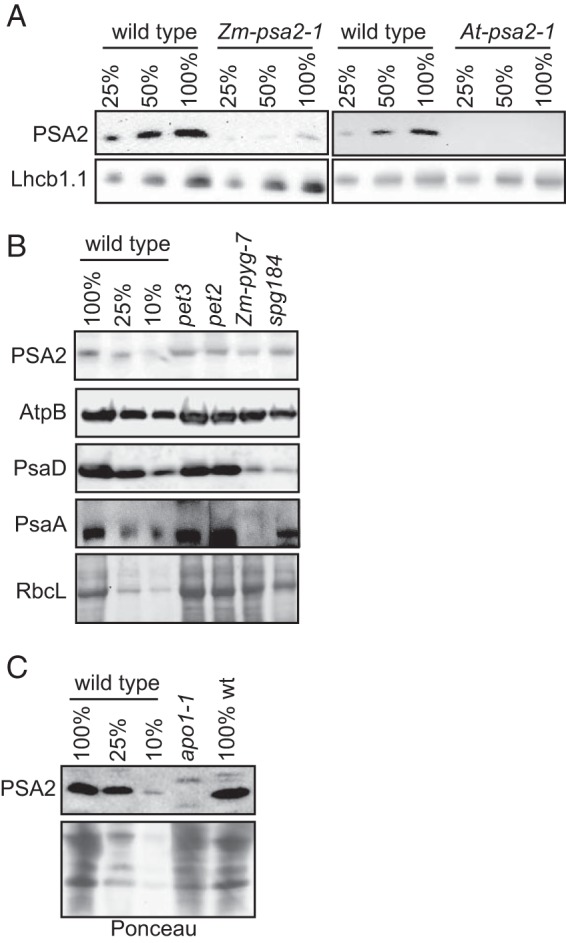
PSA2 abundance in mutants lacking PSI. A, characterization of a PSA2 antibody. Immunoblots of maize (left) or Arabidopsis (right) chloroplast extract were probed with a polyclonal antibody raised to At-PSA2. The panels below show the same blots probed to detect Lhcb1 as a loading control. B, PSA2 abundance in maize mutants lacking the cytochrome b6f complex or PSI. Total leaf extracts from maize seedlings of the indicated genotypes were analyzed by probing immunoblots with anti-PSA2 antibody. Zm-pyg7 mutants have an insertion in the maize ortholog of the Arabidopsis PSI biogenesis gene PYG7. spg184 has an insertion in a gene encoding a novel PSI biogenesis factor (R. Williams-Carrier and A. Barkan, manuscript in preparation). pet2 and pet3 mutants specifically lack the cytochrome b6f complex (25). Replicate blots were probed with antibodies to the PSI core subunits PsaD and PsaA or the AtpB subunit of the ATP synthase. A portion of the Ponceau S-stained blot used for the AtpB and PsaD probings is shown below. C, PSA2 fails to accumulate in an Arabidopsis mutant lacking the PSI assembly factor Ycf3. Total leaf extracts from Arabidopsis seedlings of the indicated genotypes were analyzed by probing immunoblots with anti-PSA2 antibody. A portion of the Ponceau S-stained blot is shown below to illustrate equal loading of the mutant and wild-type samples. apo1 mutants lack PSI due to a defect in the splicing of the chloroplast mRNA encoding the PSI assembly factor Ycf3 (37).
PSA2 was detected in two independent proteomic analyses of thylakoid membranes in Arabidopsis, the latter of which reported its enrichment specifically in non-appressed membranes (28, 38). We confirmed that PSA2 is localized to non-appressed membranes by probing immunoblots of leaf extracts, intact chloroplasts, and chloroplast subfractions with the PSA2 antibody (Fig. 6A). PSA2 lacks predicted transmembrane domains and had been proposed to localize to the thylakoid lumen based on a predicted lumenal targeting sequence (28). To test this possibility, soluble proteins in the thylakoid lumen were released by treating thylakoids with a low concentration of Triton X-100 (Fig. 6B). PSA2 and the soluble lumenal protein plastocyanin were both enriched in the supernatant, whereas an integral thylakoid protein (LHCB1) and a protein bound to the lumenal face (OEC33) remained with the membrane pellet (Fig. 6B). The lumenal targeting sequence of PSA2 includes a conserved “twin arginine” motif (Fig. 2), suggesting that it is translocated to the lumen via the cpTAT pathway (39). In support of this view, PSA2 failed to accumulate in maize hcf106 mutants, which lack a component of the TAT targeting machinery (Fig. 6C). Translocation via the TAT pathway is consistent with the fact that PSA2 harbors a predicted zinc finger, the assembly of which in the stroma would necessitate translocation across the thylakoid membrane in a folded state.
FIGURE 6.
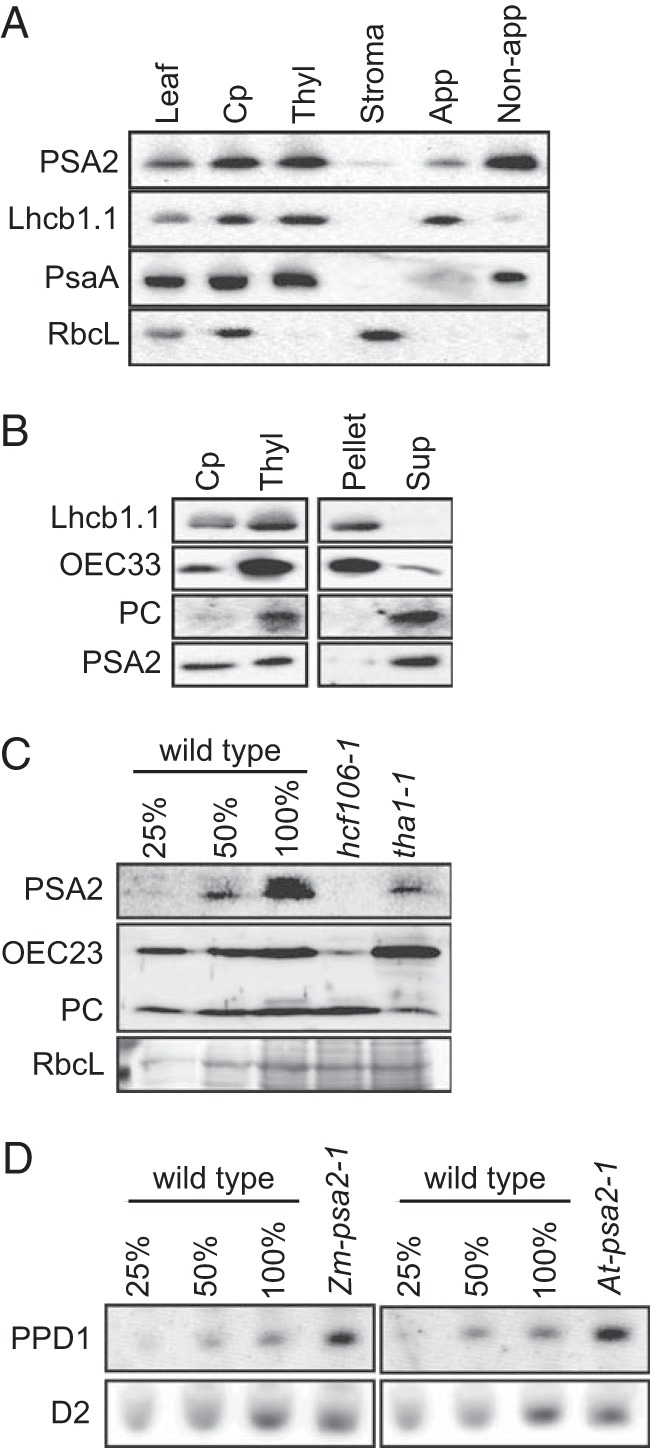
PSA2 is localized to the thylakoid lumen. A, intrachloroplast localization of PSA2. Proteins from Arabidopsis leaf, chloroplasts (Cp), thylakoid membranes (Thyl), stroma, appressed membranes (App), and non-appressed membranes (Non-app) were purified, and equivalent amounts of protein (15 μg/sample) were analyzed by immunoblotting using antibodies against the indicated proteins. Lhcb1, PsaA, and RbcL serve as markers for appressed thylakoids, non-appressed thylakoids, and stroma, respectively. B, separation of thylakoid membrane-bound and lumenal proteins. Proteins from Arabidopsis chloroplasts (Cp), thylakoids (Thyl), and the pellet and supernatant (Sup) recovered after gentle solubilization of thylakoid membranes were purified, and equivalent amounts of protein (15 μg/sample) were analyzed by immunoblotting using antibodies against the indicated proteins. OEC33, a subunit of the oxygen evolving complex, is bound to the lumenal face of the thylakoid membrane. Plastocyanin (PC) is a soluble protein in the thylakoid lumen. Lhcb1.1 is an integral thylakoid protein. C, immunoblot analysis of PSA2 in maize mutants with defects in the cpSec or cpTAT thylakoid targeting pathways. Total leaf extracts from maize seedlings of the indicated genotypes were analyzed by immunoblotting with the indicated antibodies. A portion of the Ponceau S-stained blot is shown below to illustrate the abundance of RbcL as a loading control. The hcf106 and tha1 mutants are defective in the cpTAT and cpSec thylakoid import pathways, respectively (65). D, immunoblot analysis of PPD1 in psa2 mutants. Total leaf extract from the indicated species and genotypes were probed with antibody to PPD1. A replicate blot probed with antibody to the photosystem II protein D2 serves as a loading control.
Another PSI biogenesis factor, PPD1, was shown previously to localize to the thylakoid lumen (9). Therefore, we considered the possibility that PSA2 mediates its effects via an effect on PPD1 abundance. However, PPD1 accumulated to elevated levels in psa2 mutants (Fig. 6D), suggesting the existence of a compensatory mechanism that increases PPD1 expression in the absence of PSA2 or PSI.
PSA2 Associates with a PSI-related Subcomplex That Includes PsaG
To determine whether PSA2 associates with mature PSI and/or with complexes containing subsets of PSI subunits, immunoblots of BN-PAGE-resolved, DM-solubilized thylakoid membranes from Arabidopsis (Fig. 7A) or maize (Fig. 7B) were probed with the PSA2 antibody. The antibody detected one major PSA2-containing complex in both species at roughly 250 kDa. PSA2 was not detected at the position of the stained band corresponding to mature PSI. Furthermore, immunoblot analyses showed that PSA2 did not comigrate with the core PSI subunits PsaA and PsaD. However, PSA2 did comigrate with PsaG, a peripheral subunit that links Lhc1 to the PSI core (2, 40). Immunoblot analyses of Arabidopsis extract that had been fractionated in two-dimensional gels (BN-PAGE followed by SDS-PAGE) corroborated these findings (Fig. 7C). Furthermore, the lumenal PSI assembly factor PPD1 also comigrated with PSA2 in these gels. These results show that PSA2 does not associate in a stable manner with the PSI complex and suggest that it may instead be found in a complex harboring PsaG and PPD1.
FIGURE 7.
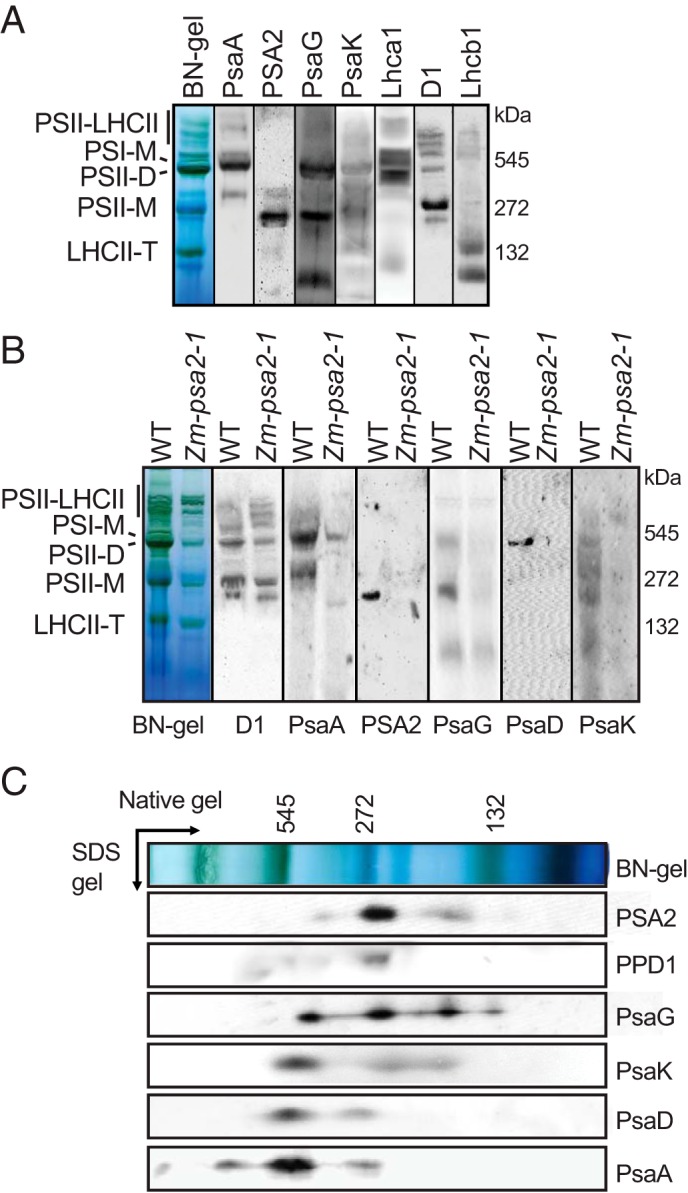
Analysis of multimeric complexes harboring PSA2. A, immunoblot analyses of BN-PAGE resolved thylakoid membrane protein complexes from Arabidopsis. Immunoblots were probed with antibodies to PSA2, subunits of PSI (PsaA, PsaG, and PsaK), Lhca1 (light-harvesting protein of PSI), D1 (core protein of PSII), and Lhcb1 (light-harvesting protein of PSII). B, immunoblot analyses of BN-PAGE-resolved thylakoid protein complexes from maize. Blots were probed with the indicated antibodies. The maize psa2 mutant was included as a control. C, immunoblot analysis of two-dimensionally resolved thylakoid membrane complexes in Arabidopsis. Thylakoid membranes were solubilized with DM and resolved by BN-PAGE (first dimension) followed by SDS-PAGE (second dimension). Replicate blots were probed with the indicated antibodies. PSII-LHCII, PSII-light-harvesting assemblies; PSI-M, monomeric PSI; PSII-D, PSII dimer; PSII-M, PSII monomer; LHCII-T, trimeric light-harvesting complex II.
To test whether PSA2, PsaG, and PPD1 are found in the same complex, the PSA2 antibody was used for coimmunoprecipitation assays with DM-solubilized thylakoid membranes from Arabidopsis (Fig. 8A). To reduce background, several assays were repeated with lysate that had been precleared by incubation with beads lacking antibody (Fig. 8B). The antibody to PSA2 coimmunoprecipitated PsaG but did not detectably coprecipitate PsaA, PsaK, or PPD1. Furthermore, PsaG was substantially depleted from the extract after immunoprecipitation, suggesting that a large fraction of PsaG is found in a complex together with PSA2. Taken together, the BN-PAGE and coimmunoprecipitation data provide strong evidence that PSA2 is found in a complex together with PsaG and that this complex lacks many other PSI components. Although PSA2 and PPD1 comigrated in native gels, the coimmunoprecipitation data indicate that they were not bound stably in the same complex.
FIGURE 8.
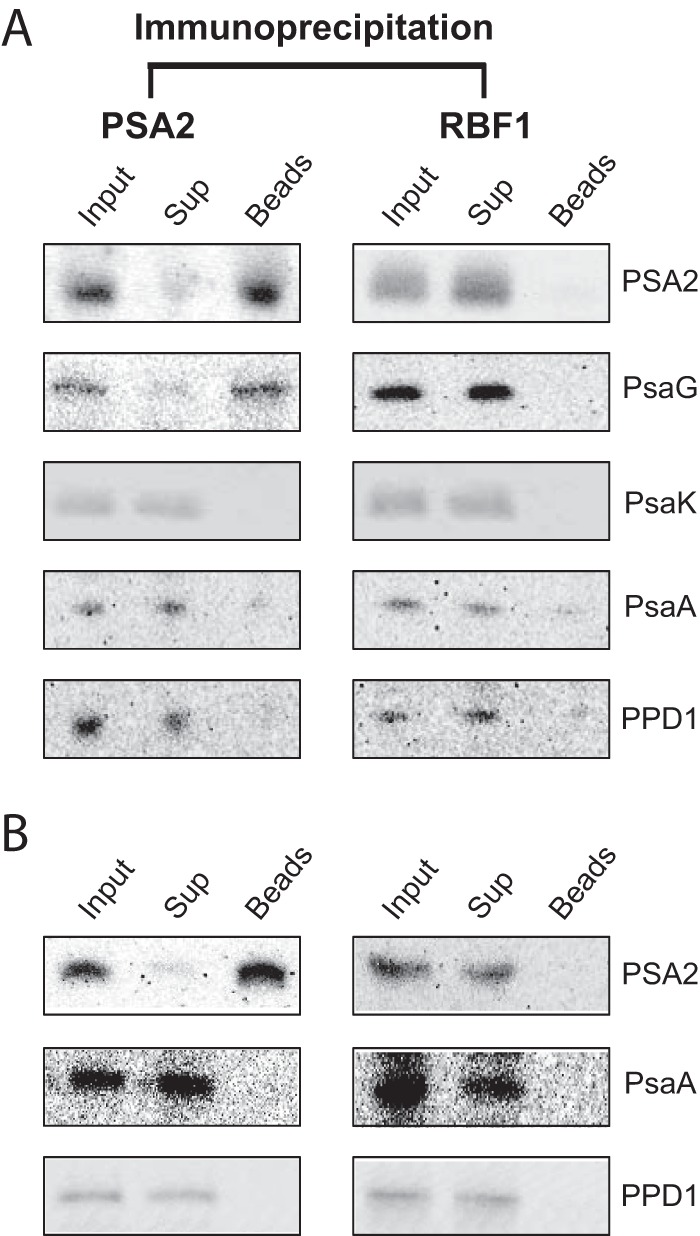
Analysis of proteins that coimmunoprecipitate with PSA2. DM-solubilized thylakoid membranes were subjected to immunoprecipitation with PSA2 antibody or with antibody to the stromal ribosome assembly factor RBF1. The immunoprecipitates were separated by SDS-PAGE, and specific proteins in the input, supernatant (Sup), and immunoprecipitate (Beads) were detected by probing immunoblots with the indicated antisera. A, assays performed without preclearing lysate by incubation with beads lacking antibody. B, assays performed with lysate that had been precleared with beads lacking antibody. The preclearing step reduced the background observed for PsaA and PPD1 in A.
PSA2 Exhibits Protein-disulfide Reductase Activity in Vitro
Mature PSA2 consists almost entirely of a domain annotated at InterPro as “heat shock protein DnaJ, cysteine-rich domain” (InterPro ID IPR001305) (Fig. 2A). This domain has been shown to exhibit protein-disulfide isomerase activity in the context of three different proteins: in DnaJ itself and in the thylakoid biogenesis/homeostasis factors CYO1/SCO2 and LQY1 (30, 32, 41). To test whether PSA2 can catalyze disulfide transactions, we assayed its ability to catalyze the reduction of disulfide bonds in insulin (Fig. 9), an activity that is often used as a proxy for protein-disulfide isomerase activity (27). Recombinant PSA2 stimulated insulin precipitation, whereas the ribosome biogenesis factor RBF1 had no effect (Fig. 9). These results support the view the PSA2 has protein-disulfide isomerase activity as do other proteins harboring the DnaJ-like cysteine-rich domain.
FIGURE 9.
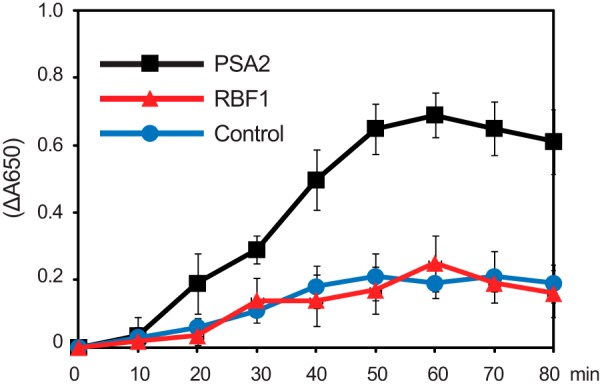
Protein-disulfide reductase activity of recombinant PSA2. The reduction of disulfide bonds in insulin was monitored by an increase in absorbance at 650 nm, which results from insulin precipitation. The reaction was initiated by the addition of dithiothreitol to a final concentration of 100 μm. The ribosome-binding protein RBF1 served as a negative control. All reactions contained BSA. Values represent the mean of three independent assays, each with three replicates. Error bars indicate one S.D.
DISCUSSION
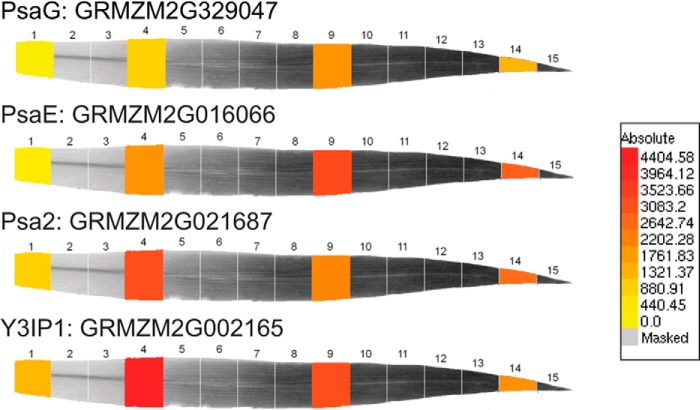
Results presented here show that PSA2, a highly conserved protein found specifically in photosynthetic eukaryotes, localizes to the thylakoid lumen and is required for the accumulation of PSI. We provide evidence that PSA2 is found in a complex harboring the PSI subunit PsaG and that it does not associate stably with mature PSI or influence chloroplast gene expression. Although the Arabidopsis PSA2 ortholog AT2G34860 was previously assigned the name embryo sac development arrest 3 (35), we did not detect any defect in the inheritance of the mutant alleles used in our study. Instead, our results strongly suggest that PSA2 promotes the assembly of PSI. A role in PSI assembly rather than (or in addition to) PSI homeostasis is supported by (i) the fact that PSA2 was required for PSI accumulation even after prolonged dark adaptation (Fig. 3A), (ii) the loss of PSA2 in a mutant lacking the PSI assembly factor Ycf3 (Fig. 5C), and (iii) the coexpression of PSA2 with proteins known to function in the assembly of the photosynthetic apparatus. Arabidopsis coexpression data can be viewed at the ATTED-II database (29), which reports seven genes that are directly connected with PSA2 in a coexpression network. Five of these genes are of known function; all are involved in chloroplast biogenesis and do not function directly in photosynthesis: CCB1, which is required for assembly of the cytochrome b6f complex (42); HCF136 and LPA3, which are involved in PSII assembly (43–45); At5g51110, which is orthologous to a Rubisco assembly factor in maize (46); and GC1, which is required for chloroplast division (47). Analogous coexpression databases are not available for maize. However, transcriptome data for maize seedling leaf segments representing a gradient of photosynthetic development (48) show that maize psa2 exhibits peak expression in a “chloroplast biogenesis” zone early in leaf development similar to known thylakoid biogenesis factors and distinct from genes encoding structural components of the photosynthetic apparatus, the expression of which peaks at a later stage (Fig. 10).
FIGURE 10.
Expression profile of psa2 during maize leaf development. Representative images for PSI biogenesis genes (psa2 and Zm-y3IP1) and structural genes (psaE and psaG) were derived from expression data in Li et al. (48). Known photosystem assembly factors typically exhibit peak expression in a chloroplast biogenesis zone relatively early in leaf development (leaf section 4), whereas proteins involved directly in photosynthesis and photosynthetic adaptation/homeostasis typically exhibit peak expression in mature chloroplasts (leaf sections 9 and 14) (A. Barkan, unpublished observations).
PSA2 is a small protein that consists almost entirely of a heat shock protein DnaJ, cysteine-rich domain (InterPro ID IPR001305) (Fig. 2A). Three previously characterized proteins in plants likewise consist of a stand-alone domain of this type, and all localize to chloroplasts: BSD2 (31), LQY1 (32), and CYO1/SCO2 (30, 33). BSD2 localizes to the stroma and mediates Rubisco assembly (31), whereas LQY1 and CYO1 are thylakoid membrane proteins that are involved in the assembly or maintenance of membrane-embedded photosynthetic complexes (30, 32, 33, 49). The cysteine-rich domains in both DnaJ and LQY1 have been shown to bind two zinc moieties and to form a zinc finger (32, 50, 51). The four zinc-coordinating motifs characteristic of this domain family (CXXCXGXG) are conserved in PSA2 orthologs (Fig. 2), suggesting that PSA2 likewise harbors a zinc finger. This domain in DnaJ has been demonstrated to have two biochemical activities. (i) It prevents protein aggregation (50, 51), and (ii) it has protein-disulfide isomerase activity (41). The chloroplast proteins CYO1/SCO2 and LQY1 also exhibit protein-disulfide isomerase activity in vitro (30, 32). That PSA2 likewise mediates thiol transactions is supported by our finding that recombinant PSA2 catalyzed the reduction of disulfide bonds in vitro. PSA2, like BSD2, LQY1, and CYO1/SCO2, lacks the other domains characteristic of DnaJ chaperones and is therefore unlikely to act as a cochaperone with Hsp70. It seems likely, therefore, that PSA2 mediates its effects on PSI biogenesis by preventing inappropriate protein-protein interactions and/or by influencing the formation of specific disulfide bonds in the thylakoid lumen. PSA2 may, for example, interact with lumen-exposed segments of PSI subunits that harbor cysteine residues. PsaG, the one PSI subunit we detected in the ∼250-kDa complex harboring PSA2, lacks cysteines. However, Lhca1 has two conserved cysteine residues on the lumenal side of the membrane, and these are in close proximity to PsaG in the PSI structure (52). Thus, a plausible hypothesis is that the PSA2-PsaG complex influences the formation of disulfide bonds involving these cysteines in Lhca1. It is important to note that the psa2 mutant phenotype is much more severe than is the phenotype resulting from the loss of PsaG (40, 53). This implies that the role of PSA2 goes beyond facilitating the assembly of PsaG into PSI. Examination of the complete composition of the PSA2 complex by mass spectrometry will be an important next step toward defining the precise functions of PSA2.
PSI is among the most complex membrane-embedded assemblies in nature, so it is to be expected that its biogenesis involves numerous accessory factors. Proteins that promote PSI assembly can be divided in two functional classes: (i) proteins involved in cofactor synthesis or attachment and (ii) proteins that assist the assembly of the complex by interacting with PSI subunits (3). Two chloroplast-encoded proteins (Ycf3 and Ycf4) and three nucleus-encoded proteins (Y3IP1, Pyg7/Ycf37, and PPD1) have been shown previously to fall into the latter category (4, 5, 7–9, 54). All of these proteins interact with subunits of the core PSI complex (3). By contrast, PSA2 is found in a multimeric complex that includes PsaG. PsaG is incorporated into PSI at a late stage of assembly and is replaced more frequently than are proteins in the PSI core (55, 56). The N and C termini of PsaG are exposed to the thylakoid lumen (57), providing a potential point of contact with PSA2. PsaG influences the dynamics of the interactions between plastocyanin and PSI (58) and has been proposed to undergo a conformational change in response to light quantity/quality that influences the rate of electron transfer to ferredoxin (40). Thus, in addition to a role for PSA2 in the buildup of PSI during chloroplast development, it is plausible that PSA2 acts in mature chloroplasts to regulate PSI activity by influencing the interactions of PsaG with the core complex. Our coimmunoprecipitation data (Fig. 8) indicate that a substantial fraction of PsaG is found in a PSA2-containing complex. Perhaps this represents a “storage” complex that is poised to donate PsaG to PSI.
Several observations suggest a functional link between PSA2 and the PSI assembly factor PPD1. First, PPD1, like PSA2, localizes to the thylakoid lumen (9). Second, the accumulation of PsaG is especially sensitive to the partial loss of PPD1 (10). Finally, PPD1 is found in a complex that comigrates with the PSA2-PsaG complex in native gels (Fig. 7C). Although our coimmunoprecipitation experiments did not detect an interaction between PPD1 and PSA2 (Fig. 8B), it remains possible that these two proteins function in the same biochemical processes or pathway.
The thylakoid lumen harbors a rich collection of proteins that function in the assembly, repair, and regulation of photosynthetic complexes (59). These include LQY1, the zinc finger of which is predicted to be lumenal (32), and PPD1, which promotes PSI assembly via interaction with lumenal loops of PsaA and PsaB (9). PSA2 and PPD1 are the only PSI assembly factors known to reside in the thylakoid lumen (3, 9). They are also the only known PSI assembly factors to be present in photosynthetic eukaryotes but not in cyanobacteria (3, 9). PSI in plants includes several subunits that are not found in cyanobacteria, including PsaG (60, 61). It will be interesting to explore whether the evolution of eukaryote-specific PSI assembly factors like PSA2 and PPD1 was driven by distinct lumenal environments in plants and cyanobacteria and/or by the need to incorporate novel subunits into the more elaborate PSI found in eukaryotes.
Supplementary Material
Acknowledgments
We are grateful to Susan Belcher (University of Oregon) for expert assistance with the genetic analysis of the maize psa2 mutant and to Reimo Zoschke (University of Oregon) for help with the ribosome profiling experiments. We thank Mark Arbing and Annie Shin (UCLA Department of Energy Protein Expression Technology Center) for assistance with the expression of recombinant PSA2.
This work was supported by United States Department of Energy Cooperative Agreement DE-FC02-02ER63421 (to David Eisenberg at UCLA), National Science Foundation Grant IOS-0922560 (to A. B.), and a postdoctoral fellowship from the WennerGren Foundation (to R. F.).

This article contains supplemental Fig. 1.
- PSI
- Photosystem I
- PSII
- Photosystem II
- DM
- n-dodecyl β-d-maltoside
- BN
- blue native
- Rubisco
- ribulose-bisphosphate carboxylase/oxygenase
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- (cp)TAT
- (chloroplast) twin arginine translocation
- At
- A. thaliana
- Zm
- Z. mays.
REFERENCES
- 1. Nelson N., Ben-Shem A. (2004) The complex architecture of oxygenic photosynthesis. Nat. Rev. Mol. Cell Biol. 5, 971–982 [DOI] [PubMed] [Google Scholar]
- 2. Amunts A., Drory O., Nelson N. (2007) The structure of a plant photosystem I supercomplex at 3.4 Å resolution. Nature 447, 58–63 [DOI] [PubMed] [Google Scholar]
- 3. Schöttler M. A., Albus C. A., Bock R. (2011) Photosystem I: its biogenesis and function in higher plants. J. Plant Physiol. 168, 1452–1461 [DOI] [PubMed] [Google Scholar]
- 4. Krech K., Ruf S., Masduki F. F., Thiele W., Bednarczyk D., Albus C. A., Tiller N., Hasse C., Schöttler M. A., Bock R. (2012) The plastid genome-encoded Ycf4 protein functions as a nonessential assembly factor for photosystem I in higher plants. Plant Physiol. 159, 579–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boudreau E., Takahashi Y., Lemieux C., Turmel M., Rochaix J. D. (1997) The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex. EMBO J. 16, 6095–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ruf S., Kössel H., Bock R. (1997) Targeted inactivation of a tobacco intron-containing open reading frame reveals a novel chloroplast-encoded photosystem I-related gene. J. Cell Biol. 139, 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Albus C. A., Ruf S., Schöttler M. A., Lein W., Kehr J., Bock R. (2010) Y3IP1, a nucleus-encoded thylakoid protein, cooperates with the plastid-encoded Ycf3 protein in photosystem I assembly of tobacco and Arabidopsis. Plant Cell 22, 2838–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stöckel J., Bennewitz S., Hein P., Oelmüller R. (2006) The evolutionarily conserved tetratrico peptide repeat protein pale yellow green7 is required for photosystem I accumulation in Arabidopsis and copurifies with the complex. Plant Physiol. 141, 870–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu J., Yang H., Lu Q., Wen X., Chen F., Peng L., Zhang L., Lu C. (2012) PsbP-domain protein1, a nuclear-encoded thylakoid lumenal protein, is essential for photosystem I assembly in Arabidopsis. Plant Cell 24, 4992–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Roose J. L., Frankel L. K., Bricker T. M. (2014) The PsbP domain protein 1 functions in the assembly of lumenal domains in photosystem I. J. Biol. Chem. 289, 23776–23785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lezhneva L., Amann K., Meurer J. (2004) The universally conserved HCF101 protein is involved in assembly of [4Fe-4S]-cluster-containing complexes in Arabidopsis thaliana chloroplasts. Plant J. 37, 174–185 [DOI] [PubMed] [Google Scholar]
- 12. Schwenkert S., Netz D. J., Frazzon J., Pierik A. J., Bill E., Gross J., Lill R., Meurer J. (2010) Chloroplast HCF101 is a scaffold protein for [4Fe-4S] cluster assembly. Biochem. J. 425, 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dall'Osto L., Piques M., Ronzani M., Molesini B., Alboresi A., Cazzaniga S., Bassi R. (2013) The Arabidopsis nox mutant lacking carotene hydroxylase activity reveals a critical role for xanthophylls in photosystem I biogenesis. Plant Cell 25, 591–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yabe T., Morimoto K., Kikuchi S., Nishio K., Terashima I., Nakai M. (2004) The Arabidopsis chloroplastic NifU-like protein CnfU, which can act as an iron-sulfur cluster scaffold protein, is required for biogenesis of ferredoxin and photosystem I. Plant Cell 16, 993–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chi W., Ma J., Zhang L. (2012) Regulatory factors for the assembly of thylakoid membrane protein complexes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 3420–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Williams-Carrier R., Stiffler N., Belcher S., Kroeger T., Stern D. B., Monde R. A., Coalter R., Barkan A. (2010) Use of Illumina sequencing to identify transposon insertions underlying mutant phenotypes in high-copy Mutator lines of maize. Plant J. 63, 167–177 [DOI] [PubMed] [Google Scholar]
- 17. Stern D. B., Hanson M. R., Barkan A. (2004) Genetics and genomics of chloroplast biogenesis: maize as a model system. Trends Plant Sci. 9, 293–301 [DOI] [PubMed] [Google Scholar]
- 18. Karpowicz S. J., Prochnik S. E., Grossman A. R., Merchant S. S. (2011) The GreenCut2 resource, a phylogenomically derived inventory of proteins specific to the plant lineage. J. Biol. Chem. 286, 21427–21439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grossman A. R., Karpowicz S. J., Heinnickel M., Dewez D., Hamel B., Dent R., Niyogi K. K., Johnson X., Alric J., Wollman F. A., Li H., Merchant S. S. (2010) Phylogenomic analysis of the Chlamydomonas genome unmasks proteins potentially involved in photosynthetic function and regulation. Photosynth. Res. 106, 3–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zoschke R., Watkins K. P., Barkan A. (2013) A rapid ribosome profiling method elucidates chloroplast ribosome behavior in vivo. Plant Cell 25, 2265–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fristedt R., Scharff L. B., Clarke C. A., Wang Q., Lin C., Merchant S. S., Bock R. (2014) RBF1, a plant homolog of the bacterial ribosome-binding factor RbfA, acts in processing of the chloroplast 16S ribosomal RNA. Plant Physiol. 164, 201–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fristedt R., Willig A., Granath P., Crèvecoeur M., Rochaix J. D., Vener A. V. (2009) Phosphorylation of photosystem II controls functional macroscopic folding of photosynthetic membranes in Arabidopsis. Plant Cell 21, 3950–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Selman-Reimer S., Merchant S., Selman B. R. (1981) Isolation, purification, and characterization of coupling factor 1 from Chlamydomonas reinhardi. Biochemistry 20, 5476–5482 [DOI] [PubMed] [Google Scholar]
- 24. Barkan A., Walker M., Nolasco M., Johnson D. (1994) A nuclear mutation in maize blocks the processing and translation of several chloroplast mRNAs and provides evidence for the differential translation of alternative mRNA forms. EMBO J. 13, 3170–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Voelker R., Barkan A. (1995) Nuclear genes required for post-translational steps in the biogenesis of the chloroplast cytochrome b6f complex in maize. Mol. Gen. Genet. 249, 507–514 [DOI] [PubMed] [Google Scholar]
- 26. Fristedt R., Vener A. V. (2011) High light induced disassembly of photosystem II supercomplexes in Arabidopsis requires STN7-dependent phosphorylation of CP29. PLoS One 6, e24565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lundström J., Holmgren A. (1990) Protein disulfide-isomerase is a substrate for thioredoxin reductase and has thioredoxin-like activity. J. Biol. Chem. 265, 9114–9120 [PubMed] [Google Scholar]
- 28. Friso G., Giacomelli L., Ytterberg A. J., Peltier J. B., Rudella A., Sun Q., Wijk K. J. (2004) In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. Plant Cell 16, 478–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Obayashi T., Hayashi S., Saeki M., Ohta H., Kinoshita K. (2009) ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res. 37, D987–D991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shimada H., Mochizuki M., Ogura K., Froehlich J. E., Osteryoung K. W., Shirano Y., Shibata D., Masuda S., Mori K., Takamiya K. (2007) Arabidopsis cotyledon-specific chloroplast biogenesis factor CYO1 is a protein disulfide isomerase. Plant Cell 19, 3157–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brutnell T. P., Sawers R. J., Mant A., Langdale J. A. (1999) BUNDLE SHEATH DEFECTIVE2, a novel protein required for post-translational regulation of the rbcL gene of maize. Plant Cell 11, 849–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lu Y., Hall D. A., Last R. L. (2011) A small zinc finger thylakoid protein plays a role in maintenance of photosystem II in Arabidopsis thaliana. Plant Cell 23, 1861–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tanz S. K., Kilian J., Johnsson C., Apel K., Small I., Harter K., Wanke D., Pogson B., Albrecht V. (2012) The SCO2 protein disulphide isomerase is required for thylakoid biogenesis and interacts with LHCB1 chlorophyll a/b binding proteins which affects chlorophyll biosynthesis in Arabidopsis seedlings. Plant J. 69, 743–754 [DOI] [PubMed] [Google Scholar]
- 34. Aro E. M., Suorsa M., Rokka A., Allahverdiyeva Y., Paakkarinen V., Saleem A., Battchikova N., Rintamäki E. (2005) Dynamics of photosystem II: a proteomic approach to thylakoid protein complexes. J. Exp. Bot. 56, 347–356 [DOI] [PubMed] [Google Scholar]
- 35. Pagnussat G. C., Yu H. J., Ngo Q. A., Rajani S., Mayalagu S., Johnson C. S., Capron A., Xie L. F., Ye D., Sundaresan V. (2005) Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132, 603–614 [DOI] [PubMed] [Google Scholar]
- 36. Amann K., Lezhneva L., Wanner G., Herrmann R. G., Meurer J. (2004) ACCUMULATION OF PHOTOSYSTEM ONE1, a member of a novel gene family, is required for accumulation of [4Fe-4S] cluster-containing chloroplast complexes and antenna proteins. Plant Cell 16, 3084–3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Watkins K. P., Rojas M., Friso G., van Wijk K. J., Meurer J., Barkan A. (2011) APO1 promotes the splicing of chloroplast group II introns and harbors a plant-specific zinc-dependent RNA binding domain. Plant Cell 23, 1082–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tomizioli M., Lazar C., Brugière S., Burger T., Salvi D., Gatto L., Moyet L., Breckels L. M., Hesse A. M., Lilley K. S., Seigneurin-Berny D., Finazzi G., Rolland N., Ferro M. (2014) Deciphering thylakoid sub-compartments using a mass spectrometry-based approach. Mol. Cell. Proteomics 13, 2147–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Celedon J. M., Cline K. (2013) Intra-plastid protein trafficking: how plant cells adapted prokaryotic mechanisms to the eukaryotic condition. Biochim. Biophys. Acta 1833, 341–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jensen P. E., Rosgaard L., Knoetzel J., Scheller H. V. (2002) Photosystem I activity is increased in the absence of the PSI-G subunit. J. Biol. Chem. 277, 2798–2803 [DOI] [PubMed] [Google Scholar]
- 41. de Crouy-Chanel A., Kohiyama M., Richarme G. (1995) A novel function of Escherichia coli chaperone DnaJ. Protein-disulfide isomerase. J. Biol. Chem. 270, 22669–22672 [DOI] [PubMed] [Google Scholar]
- 42. Lezhneva L., Kuras R., Ephritikhine G., de Vitry C. (2008) A novel pathway of cytochrome c biogenesis is involved in the assembly of the cytochrome b6f complex in Arabidopsis chloroplasts. J. Biol. Chem. 283, 24608–24616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cai W., Ma J., Chi W., Zou M., Guo J., Lu C., Zhang L. (2010) Cooperation of LPA3 and LPA2 is essential for photosystem II assembly in Arabidopsis. Plant Physiol. 154, 109–120 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44. Plücken H., Müller B., Grohmann D., Westhoff P., Eichacker L. A. (2002) The HCF136 protein is essential for assembly of the photosystem II reaction center in Arabidopsis thaliana. FEBS Lett. 532, 85–90 [DOI] [PubMed] [Google Scholar]
- 45. Meurer J., Plücken H., Kowallik K. V., Westhoff P. (1998) A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J. 17, 5286–5297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Feiz L., Williams-Carrier R., Belcher S., Montano M., Barkan A., Stern D. (2014) RAF2, a novel Rubisco biogenesis factor in maize. Plant J., in press [DOI] [PubMed] [Google Scholar]
- 47. Maple J., Fujiwara M. T., Kitahata N., Lawson T., Baker N. R., Yoshida S., Møller S. G. (2004) GIANT CHLOROPLAST 1 is essential for correct plastid division in Arabidopsis. Curr. Biol. 14, 776–781 [DOI] [PubMed] [Google Scholar]
- 48. Li P., Ponnala L., Gandotra N., Wang L., Si Y., Tausta S. L., Kebrom T. H., Provart N., Patel R., Myers C. R., Reidel E. J., Turgeon R., Liu P., Sun Q., Nelson T., Brutnell T. P. (2010) The developmental dynamics of the maize leaf transcriptome. Nat. Genet. 42, 1060–1067 [DOI] [PubMed] [Google Scholar]
- 49. Muranaka A., Watanabe S., Sakamoto A., Shimada H. (2012) Arabidopsis cotyledon chloroplast biogenesis factor CYO1 uses glutathione as an electron donor and interacts with PSI (A1 and A2) and PSII (CP43 and CP47) subunits. J. Plant Physiol. 169, 1212–1215 [DOI] [PubMed] [Google Scholar]
- 50. Szabo A., Korszun R., Hartl F. U., Flanagan J. (1996) A zinc finger-like domain of the molecular chaperone DnaJ is involved in binding to denatured protein substrates. EMBO J. 15, 408–417 [PMC free article] [PubMed] [Google Scholar]
- 51. Banecki B., Liberek K., Wall D., Wawrzynów A., Georgopoulos C., Bertoli E., Tanfani F., Zylicz M. (1996) Structure-function analysis of the zinc finger region of the DnaJ molecular chaperone. J. Biol. Chem. 271, 14840–14848 [DOI] [PubMed] [Google Scholar]
- 52. Amunts A., Toporik H., Borovikova A., Nelson N. (2010) Structure determination and improved model of plant photosystem I. J. Biol. Chem. 285, 3478–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Varotto C., Pesaresi P., Jahns P., Lessnick A., Tizzano M., Schiavon F., Salamini F., Leister D. (2002) Single and double knockouts of the genes for photosystem I subunits G, K, and H of Arabidopsis. Effects on photosystem I composition, photosynthetic electron flow, and state transitions. Plant Physiol. 129, 616–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Naver H., Boudreau E., Rochaix J. D. (2001) Functional studies of Ycf3: its role in assembly of photosystem I and interactions with some of its subunits. Plant Cell 13, 2731–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zygadlo A., Robinson C., Scheller H. V., Mant A., Jensen P. E. (2006) The properties of the positively charged loop region in PSI-G are essential for its “spontaneous” insertion into thylakoids and rapid assembly into the photosystem I complex. J. Biol. Chem. 281, 10548–10554 [DOI] [PubMed] [Google Scholar]
- 56. Ozawa S., Onishi T., Takahashi Y. (2010) Identification and characterization of an assembly intermediate subcomplex of photosystem I in the green alga Chlamydomonas reinhardtii. J. Biol. Chem. 285, 20072–20079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rosgaard L., Zygadlo A., Scheller H. V., Mant A., Jensen P. E. (2005) Insertion of the plant photosystem I subunit G into the thylakoid membrane. FEBS J. 272, 4002–4010 [DOI] [PubMed] [Google Scholar]
- 58. Zygadlo A., Jensen P. E., Leister D., Scheller H. V. (2005) Photosystem I lacking the PSI-G subunit has a higher affinity for plastocyanin and is sensitive to photodamage. Biochim. Biophys. Acta 1708, 154–163 [DOI] [PubMed] [Google Scholar]
- 59. Järvi S., Gollan P. J., Aro E. M. (2013) Understanding the roles of the thylakoid lumen in photosynthesis regulation. Front. Plant Sci. 4, 434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fromme P., Melkozernov A., Jordan P., Krauss N. (2003) Structure and function of photosystem I: interaction with its soluble electron carriers and external antenna systems. FEBS Lett. 555, 40–44 [DOI] [PubMed] [Google Scholar]
- 61. Kargul J., Nield J., Barber J. (2003) Three-dimensional reconstruction of a light-harvesting complex I-photosystem I (LHCI-PSI) supercomplex from the green alga Chlamydomonas reinhardtii. Insights into light harvesting for PSI. J. Biol. Chem. 278, 16135–16141 [DOI] [PubMed] [Google Scholar]
- 62. Järvi S., Suorsa M., Paakkarinen V., Aro E. M. (2011) Optimized native gel systems for separation of thylakoid protein complexes: novel super- and mega-complexes. Biochem. J. 439, 207–214 [DOI] [PubMed] [Google Scholar]
- 63. Caffarri S., Kouril R., Kereïche S., Boekema E. J., Croce R. (2009) Functional architecture of higher plant photosystem II supercomplexes. EMBO J. 28, 3052–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Joliot P., Joliot A. (2005) Quantification of cyclic and linear flows in plants. Proc. Natl. Acad. Sci. U.S.A. 102, 4913–4918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Voelker R., Barkan A. (1995) Two nuclear mutations disrupt distinct pathways for targeting proteins to the chloroplast thylakoid. EMBO J. 14, 3905–3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.