Abstract
Riboswitches were discovered in 2002 in bacteria as RNA-based intracellular sensors of vitamin derivatives. During the last decade, naturally occurring RNA sensor elements have been found to bind a range of small metabolites and ions and to exert regulatory control of transcription, translation, splicing, and RNA stability. Extensive biochemical, structural, and genetic studies have established the basic principles underpinning riboswitch function in all three kingdoms of life with implications for developing antibiotics, designing new molecular sensors, and integrating riboswitches into synthetic circuits.
Introduction
For many years, bacteria and phages were the source of RNA-based paradigms of gene regulation, ranging from transcription attenuation and thermosensing to small regulatory RNAs (sRNAs). Even after seemingly exhaustive genetic and genomic explorations, bacteria continue to instigate some major advances in RNA biology. A case in point are the regulatory networks in which specific regions of mRNAs termed riboswitches directly sense cellular metabolites to modulate transcription or translation of mRNAs, which typically encode proteins involved in the biogenesis or transport of the metabolites. Following their first description 10 years ago (Mironov et al., 2002; Nahvi et al., 2002; Winkler et al., 2002), riboswitches have become recognized as important and widespread elements in the control of gene expression in numerous evolutionarily distant bacteria, with counterparts in archaea, plants, fungi, and algae.
Methods developed in the early 1990s for invitro selection and evolution of RNA sequences demonstrated that RNAs could be generated to bind a wide range of protein and small-molecule ligands (Ellington and Szostak, 1990; Robertson and Joyce, 1990; Tuerk and Gold, 1990). These “aptamers” bind their ligands with high selectivity and affinity, on par with proteins, while working with only four ribonucleotide building blocks as opposed to 20 amino acids. The discovery of riboswitches showed that organisms had capitalized on this ability of RNA and put it to good use.
Riboswitches are regions of mRNAs that contain specific evolutionarily conserved ligand-binding (sensor) domains along with a variable sequence, termed the expression platform, that enables regulation of the downstream coding sequences. The term “riboswitch” reflects the ability of these noncoding RNAs to function as genetic switches. When the metabolite abundance exceeds a threshold level, binding to the riboswitch sensor induces a conformational change in the expression platform, leading to modulation of downstream events (Figure 1). Switching RNA states as a way to control gene expression is shared by other mRNA-based regulators (attenuators) associated with metabolite sensors in a form of proteins (Gralla et al., 1974; Yanofsky, 1981) or tRNA (Grundy and Henkin, 1993). However, riboswitches are unique in their ability to directly bind diverse small molecules without intermediate molecules.
Figure 1. Diversity of Riboswitches and Mechanisms of Gene Control in Bacteria.
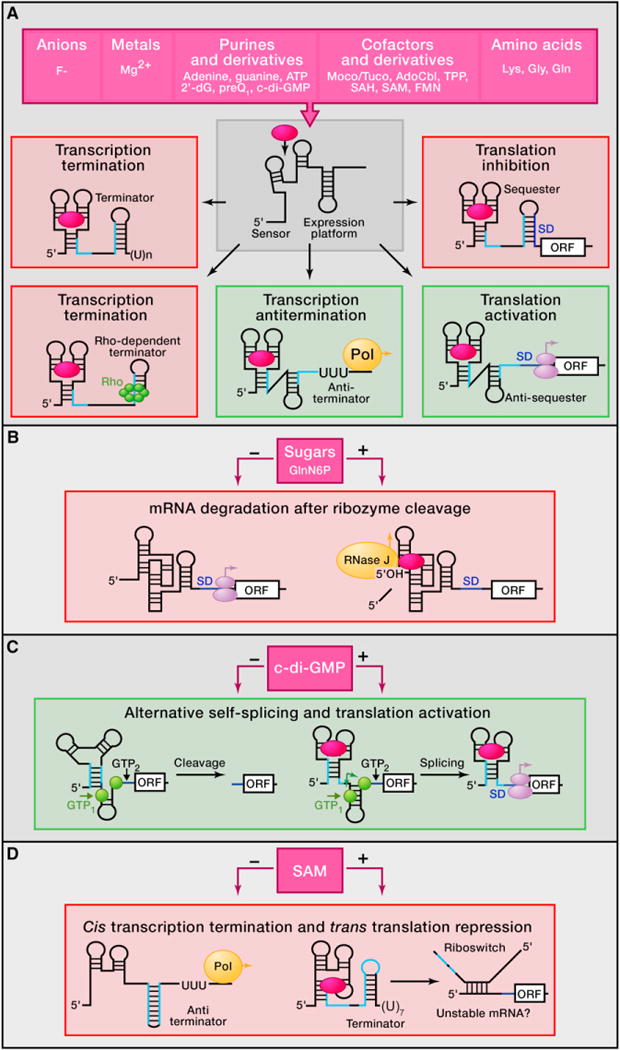
Mechanisms of modulation of gene expression are highly divergent in prokaryotes and involve control of transcription, translation, splicing, and mRNA stability.
(A) Various metabolites (violet, top) present in cells above threshold concentration can be directly sensed and specifically bound by sensor domains of riboswitches (white). Entrapment of a metabolite stabilizes the ligand-bound conformation of the riboswitch sensor and typically induces structural changes in the adjacent region (expression platform) that bears structural elements that stimulate (green) or repress (pink) gene expression. GlnN6P, glucosamine-6-phosphate; 2′-dG, 2′ -deoxyguanosine; preQ1, pre-queuosine-1; c-di-GMP, cyclic di-guanosyl-5′-monophosphate; Moco/Tuco, molybdenum and tungsten cofactors; SAH, S-adenosyl-L-homocysteine. Left: metabolite binding most often prevents formation of the antiterminator hairpin (complementary RNA regions in light blue) and promotes formation of the alternative Rho-independent termination hairpin (middle) or Rho binding site (bottom) that causes premature transcription termination. Center: in some cases, bound metabolites stabilize the antiterminator hairpin that allows RNA polymerase (Pol) to complete transcription of the gene (bottom). Right: expression of open reading frames (ORF) can be repressed by sequestration of the ribosome entry site (RBS or Shine-Dalgarno sequence, SD, dark blue) and blockage of translation initiation (middle). Metabolite binding to some riboswitches facilitates formation of the SD antisequester hairpin that opens up SD for ribosome binding and translation initiation (bottom).
(B) In gram-positive bacteria, bound GlcN6P induces cleavage by the glmS riboswitch-ribozyme in the 5′ UTR. The 5′-OH of the resulting fragment stimulates degradation of the message by RNase J.
(C) C. difficile exploits an allosteric ribozyme-riboswitch that combines self-splicing and translation activation. Left: in the absence of c-di-GMP, the intron uses GTP cleavage site 2 (black triangle, GTP2), thus yielding RNAs with truncated SDs that are not expressed. Right: binding of c-di-GMP to the riboswitch in the presence of GTP promotes cleavage at site 1 (green triangle, GTP1) and self-excision (green arrows) of the group I self-splicing intron using splicing sites shown in green circles. This self-cleavage brings together two halves of the SD, and the resulting mRNA is efficiently translated.
(D) In L. monocytogenes, the SreA and SreB riboswitches form antiterminator hairpins and allow normal transcription in the absence of SAM. Binding of SAM causes transcription termination. The resulting mRNA fragments base pair with the 5′ UTR of the mRNA and functions in trans as an antisense sRNA that destabilizes the target transcript, thus reducing protein production.
The discovery of riboswitches led to several questions. First, given the large assortment of metabolites in cells, how do riboswitches select their cognate ligands? Second, how is a metabolite binding signal communicated to the gene expression machineries? Riboswitches utilize different gene expression platforms and may follow different folding pathways to exert their function. Finally, how did riboswitches originate, and what is the relationship between riboswitches and other cellular regulatory mechanisms? These questions were the focus of numerous studies that resulted in the discovery of more than 20 classes of riboswitches distributed across many species, the determination of the X-ray structures of virtually all major validated riboswitch types, and the identification of the folding trajectories and ligand binding rules for several riboswitch classes. Here, we give a brief historical overview of riboswitches and highlight the diverse repertoire and structure/functional complexity of these ubiquitous natural RNA sensors.
Discovery of Riboswitches
Many bacterial species are able to either transport small organic molecules from the environment or synthesize them from simple precursors. Each process requires a distinct set of proteins, and bacteria often use feedback control by the end products of enzymatic pathways to repress synthesis of excess protein or to activate genes necessary for next biosynthetic steps. Cellular metabolites are typically sensed by proteins, which then interact with DNA or RNA to control the production of relevant enzymes. Therefore, when inhibition of vitamin B1-, B2-, and B12-biosynthetic genes by thiamine, riboflavin, and cobalamin, respectively, was elucidated, substantial efforts were undertaken to identify the relevant protein repressors (Miranda-Ríos et al., 2001; Nou and Kadner, 1998). Such hypothetical modulators, however, were not found. The negative results, nevertheless, pointed to a regulatory role for conserved mRNA sequences (“boxes”) in the regulation and suggested the intriguing possibility that mRNAs directly sense vitamin derivatives (Gelfand et al., 1999; Miranda-Ríos et al., 2001; Perkins and Pero, 2002; Stormo and Ji, 2001). Moreover, in vivo probing revealed alternative conformations of the Salmonella typhimurium cob mRNA leader region in the presence or absence of adenosylcobalamin (AdoCbl). However, attempts to directly test binding of cobalamin to RNA failed (Ravnum and Andersson, 2001). Similar indirect results emerged looking at the Escherichia coli btuB mRNA where addition of AdoCbl caused reverse transcriptase to pause at a site near the 3′ end of the mRNA leader during in vitro primer extension (Nou and Kadner, 2000), likely reflecting stabilization of the mRNA structure by metabolite binding.
Eventually, three vitamin derivatives, thiamine pyrophosphate (TPP) (Mironov et al., 2002; Winkler et al., 2002), flavin mononucleotide (FMN) (Mironov et al., 2002), and AdoCbl (Nahvi et al., 2002), were demonstrated to interact directly with their respective mRNAs to control the vitamin B1, B2, and B12 operons. These reports established that metabolite binding stabilizes the conformation of an evolutionarily conserved RNA sensor (natural aptamer) and induces folding of the nonconserved downstream RNA region (expression platform) to form a structure that affects transcription termination or translation initiation. Therefore, direct metabolite binding to RNA causes “riboswitching” between alternative mRNA conformations that alter gene expression.
Diversity of Riboswitches and Regulation Mechanisms
The first identifications of vitamin-specific riboswitches were followed by discoveries of many other riboswitch types. Currently, riboswitches are known to sense purines and their derivatives, protein coenzymes and related compounds, amino acids, and a phosphorylated sugar (Figure 1, top). Some riboswitches respond specifically to inorganic ligands, including metals (Mg2+ cations) (Cromie et al., 2006; Dann et al., 2007) that shield the negative charge of the sugar-phosphate backbone in RNA, and, most unexpectedly, to negatively charged fluoride anions (Baker et al., 2012).
Despite the plethora of riboswitch ligands, the regulatory activity of the vast majority of bacterial riboswitches is directed at either transcription or translation of metabolite transport and biosynthetic genes (Figure 1A). These regulatory activities are based on the ligand-dependent formation of mutually exclusive RNA conformations. In the case of transcription, the structures serve as Rho-independent terminator and antiterminator hairpins. In controlling translation, the structures sequester or release ribosome-binding sites (RBS) (Figure 1A). A recent study also demonstrated the control of Rho-dependent transcription termination by riboswitches (Hollands et al., 2012). This type of regulatory mechanism is likely to be widespread because a number of riboswitches appear to lack Rho-independent terminators and RBS-sequestering hairpins.
An unconventional mechanism is used by the glmS riboswitch-ribozyme that couples metabolite sensing with mRNA cleavage. This noncoding RNA is typically found in Gram-positive bacteria, where it interacts with glucosamine-6-phosphate (GlcN6P), which, after binding, cleaves the glmS mRNA within the riboswitch sequence (Figure 1B) (Winkler et al., 2004). RNase J then degrades the downstream cleavage product from the 5′-OH end, thus preventing translation of the mRNA (Collins et al., 2007). The glmS riboswitch-ribozyme defies the conventional view that a riboswitch recognizes a single compound. This riboswitch can sense an array of related chemical compounds and thereby functions to assess the overall metabolic state of a cell (Watson and Fedor, 2011).
Some riboswitches are capable of a broad regulatory function. The second messenger cyclic di-guanosyl-5′-monophosphate (c-di-GMP) triggers a wide range of physiological changes, and its respective riboswitches are positioned near genes involved in motility, virulence, and other processes (Sudarsan et al., 2008). Some c-di-GMP riboswitches are located adjacent to group I self-splicing introns (Lee et al., 2010). These RNA regulators elicit control by a complex cascade of events that requires collaboration of both RNA components: c-di-GMP binding to its aptamer induces folding changes that allow GTP to attack the intron’s 5′ splice site, resulting in self-excision of the intron, which brings two distantly located halves of the RBS together to produce a translatable mRNA (Figure 1C) (Chen et al., 2011). Such conjoined allosteric RNAs may constitute a two-input control system that senses c-di-GMP and GTP concentrations to trigger splicing events. This hypothesis awaits experimental validation.
Since the discovery of riboswitches, it has been speculated that these typical cis-regulatory elements can also function in trans as sRNAs. This appears to be true at least for the S-adenosylmethionine (SAM) riboswitches SreA and SreB of Listeria monocytogenes (Loh et al., 2009) (Figure 1D). After SAM-dependent transcription termination, these riboswitches pair with the 5′ UTR of the mRNA-encoding virulence regulator PrfA and downregulate its expression, mainly at the translational level.
In eukaryotes, the uncoupling of transcription from translation and the presence of introns necessitate different means to control gene expression. Instead of targeting translation and/or transcription, eukaryotic TPP riboswitches regulate genes via alternative splicing—a process in which introns are excised and exons rejoined in different combinations to yield alternatively spliced mRNAs. “Normal” splicing occurs when a segment within a riboswitch sensor, located in the intergenic region or 3′ UTR, base pairs with a complementary portion overlapping one of the splice sites in the absence of TPP (Figure 2). The resulting splicing product is translated into the full-length protein. If TPP is present in cells at the threshold concentration, it binds to the sensor and makes a previously obscured splice site accessible to the splicing machinery. Depending on the species, the alternatively spliced mRNAs contain internal stop codons that either cause translation of aberrant peptides (filamentous fungi) (Cheah et al., 2007) (Figure 2A) or premature translation termination (green algae) (Croft et al., 2007) (Figure 2B). In higher plants, alternative splicing produces species with long 3′ UTRs that destabilize the transcripts (Bocobza et al., 2007; Wachter et al., 2007) (Figure 2C).
Figure 2. TPP-Dependent Riboswitches in Eukaryotes.
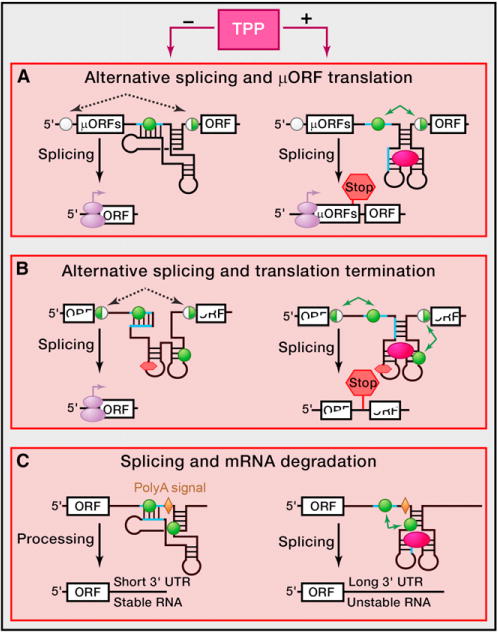
In some eukaryotes, TPP riboswitches modulate mRNA splicing by controlling accessibility to alternative splice sites. In the absence of TPP, the riboswitch is not folded, and complementary regions (light blue lines) of the riboswitch and the adjacent sequence interact with one another.
(A) In fungi, complementary base pairing results in preferential use of the distal set of splicing sites (black open circles) and elimination of the region between black dashed arrowed lines (left). The resulting mRNA is translated to yield full-length product. TPP binding to the riboswitch stabilizes the riboswitch fold, precludes the complementary base pairing, and opens different set(s) of splicing sites (green circles) that are otherwise sequestered. Splicing at the alternative splice sites removes sequences between the green arrowed lines. The resulting mRNAs retain micro ORFs, which preclude translation of the main ORF located downstream of the micro ORF (right).
(B) In algae, mRNA splicing in the absence of TPP eliminates a stop codon located within a riboswitch sequence. TPP binding promotes alternative splicing events that introduce a premature termination codon and disrupts translation of the ORF.
(C) In higher plants, sequestration of splice sites in the absence of TPP causes retention of the mRNA processing site (polyadenylation signal, yellow rhomb) and yields stable mRNA with a short 3′ UTR. TPP binding to the riboswitch sensor exposes the 5′ splice site, causing the removal of the fragment between the green arrows containing the polydenylation signal. The resulting mRNA contains long and less stable 3′ UTR, which compromises protein production.
Although validated eukaryotic riboswitches have been restricted to TPP-dependent systems, a recent study suggested the presence of adenosine-binding RNA aptamers in vertebrate genomes (Vu et al., 2012). A biological role of these RNAs remains to be elucidated. Some eukaryotic mRNAs are capable of responding to environmental signals via binary conformational changes analogous to riboswitches. For instance, an RNA switch in the 3′ UTR of human vascular endothelial growth factor-A (VEGF) mRNA integrates signals from interferon-γ and hypoxia to regulate VEGF translation in myeloid cells (Ray et al., 2009). However, the conformational interplay is metabolite independent and is dictated by stimulus-responsive binding of proteins.
Riboswitches do not always function as single regulatory units. Two sensor domains or entire riboswitches in so-called tandem riboswitches are occasionally located adjacent to one another. For example, many glycine riboswitches consist of two glycine sensors separated by a short linker (Mandal et al., 2004) and are capable of intricate tertiary interactions (Butler et al., 2011; Huang et al., 2010). Even though two sensor domains could possibly assist each other in folding and ligand binding (Kwon and Strobel, 2008; Mandal et al., 2004), the physiological purpose of such duplication remains to be elucidated (Kladwang et al., 2012; Sherman et al., 2012). The biological role of tandem riboswitches with different specificities is more apparent (Sudarsan et al., 2006). They modulate gene expression only when both cognate compounds are present in cells, thus operating as a two-input logic gate. The riboswitch-mediated control can be embedded in a multilayered configuration of regulation with an intricate and highly coordinated interplay of multiple regulatory strategies. For example, L. monocytogenes SAM riboswitches function only at a temperature permissive for infection, when the adjacent RNA thermosensor is unfolded (Loh et al., 2009). Another example involves ethanolamine utilization in Enteroccus faecalis, where an AdoCbl riboswitch cooperates with a protein regulator that affects stability of transcription terminators (Fox et al., 2009).
Riboswitch Architectures
The exceptional selectivity of riboswitches is entirely encoded in their conserved sensing domains. These recognition sites vary greatly in the size and complexity of their secondary and tertiary structures. High-resolution structures of metabolite-sensing domains have been determined for practically all major riboswitch classes in complex with validated ligands and for several riboswitch subclasses. Even though riboswitches adopt very different conformations with similarities only between closely related purine riboswitches (Batey et al., 2004; Pikovskaya et al., 2011; Serganov et al., 2004), most riboswitch structures involve multihelical junctions and pseudoknots (knot-shaped conformations typically formed through base pairing between a loop of an RNA stem-loop structure and an outside region), as was earlier described for ribozymes (Lilley and Eckstein, 2008). Therefore, we propose that the vast majority of riboswitches can be sorted into two types based on structural principles. The first type encompasses riboswitches folded into multihelical junctions (Figures 3A and 3B), and the second type adopts various pseudoknot folds (Figure 3C).
Figure 3. Structural Principles of Ligand Recognition by Riboswitches.

(A–C) Schematic representations of a “straight” junctional fold (A), observed in type Ia riboswitches; an “inverse” junctional fold (B) from type Ib riboswitches; and a pseudoknot fold (C), which is characteristic for type II riboswitches. Pink and blue shading depict ligand binding sites and long-distance tertiary interactions, respectively. The magenta segment in helix P1 designates regions capable of alternative base pairing. Representative structures in ribbon representation for TPP (Serganov et al., 2006; Thore et al., 2006), THF (Huang et al., 2011; Trausch et al., 2011), and fluoride (Ren et al., 2012) riboswitches are shown under the corresponding fold schematics. Ligands are in red surface representation. Mg2+ cations are depicted by green spheres. 3WJ, three-way junction; PK, pseudoknot.
(D) Recognition of TPP by the TPP riboswitch. PP, pyrophosphate; Thi, thiazole; and APy, aminopyrimidine moieties. Mg2+ cations are shown with coordination bonds depicted by green sticks. Water molecules are shown as red spheres.
“Junctional” riboswitches can be divided into two subtypes based on the location of a key junction with respect to the regulatory helix P1, which closes a metabolite-sensing domain and usually contains a segment involved in alternative pairing. In the first group (type Ia), a multihelical junction is positioned centrally and joins P1 with other stems, which typically form longdistance tertiary interactions stabilizing the overall conformation (Figure 3A), as observed in purine (Batey et al., 2004; Serganov et al., 2004) and TPP (Edwards and Ferré-D’Amaré, 2006; Serganov et al., 2006; Thore et al., 2006) riboswitches. One of the stems can be much longer than the others and must fold back toward the junction to make tertiary interactions, as in a lysine riboswitch (Garst et al., 2008; Serganov et al., 2008). Metabolite-binding pockets are formed within or adjacent to the junction so that ligand binding directly affects the stability of the junction and helix P1.
The second group (type Ib) is characterized by the “inverse” junctional architecture (Figure 3B), in which a key multihelical junction is positioned on the periphery, far from helix P1. A stem, radiating from the junction, folds back toward P1 and stabilizes P1 through long-range tertiary interactions. Metabolites bind RNA in the junctional region and/or in the vicinity of P1 and affect formation of P1 through stabilization of the global conformation and tertiary interactions. Typical type Ib RNAs are tetrahydrofolate (THF) (Huang et al., 2011; Trausch et al., 2011) and Mg2+ class II (Dann et al., 2007) riboswitches.
The type II group combines several metabolite-sensing RNAs, such as the SAM-II (Gilbert et al., 2008) and fluoride (Ren et al., 2012) riboswitches whose structures are entirely based on small pseudoknots. It should be noted that pseudoknots constitute integral and important parts of several junctional riboswitches and can participate in formation of metabolite-binding pockets, as in the glmS riboswitch-ribozyme (Cochrane et al., 2007; Klein and Ferré-D’Amaré, 2006) and in long-distance tertiary contacts, as in the SAM-I riboswitch (Montange and Batey, 2006).
It is becoming clear that the type of riboswitch architecture and the ligand structure are not correlated. Moreover, the three classes of SAM riboswitches adopt different junctional and pseudoknot folds (Gilbert et al., 2008; Lu et al., 2008; Montange and Batey, 2006), emphasizing the extraordinary ability of RNA to adopt different configurations that recognize the same ligand. Remarkably, many riboswitches contain recurrent structural motifs that are present in other natural and artificial RNAs (Jaeger et al., 2009; Montange and Batey, 2006; Serganov et al., 2009). Like other functional RNAs, riboswitches use these motifs as basic building blocks to adopt complex spatial conformations.
Metabolite Recognition
Riboswitches recognize chemically diverse ligands and do not possess a uniform metabolite recognition feature. Nonetheless, metabolite binding by riboswitches follows several common trends. The majority of riboswitches form tight binding pockets that possess excellent shape complementarity to parts of their cognate ligands (Figure 3D), with small ligands completely encapsulated in such pockets. The pockets are usually lined by conserved nucleotides and non-canonical base pairs that belong to a widened irregular helix or to converging stems. Most ligands, with only a few exceptions (Huang et al., 2011; Johnson et al., 2012; Peselis and Serganov, 2012; Trausch et al., 2011), rely on heteroatom functionalities to form specific hydrogen bonds and electrostatic interactions with the RNA. Often, specific hydrogen bonds involve ligand edges and conserved nonpaired nucleotides of RNA (for example, G40 in aminopurine sensor, Figure 3D). Planar groups of ligands are typically involved in stacking interactions and are sandwiched between purines (e.g., G42 and A43 in Figure 3D). Metal cations, Mg2+ or K+, can compensate for the negative charge of the ligand or its functional groups, including fluoride, carboxylate, and phosphate (PP in Figure 3D) moieties. Metal ions also mediate ligand-RNA interactions through direct and water-mediated coordination (Figure 3D). Observations of all these features in complexes between riboswitches and their cognate ligands have been supplemented by extensive information from X-ray structures of ligand-free riboswitches and riboswitches bound to near-cognate and noncognate ligands. These structures and related studies (Haller et al., 2011; Stoddard et al., 2010) have shown that riboswitches appear to bind cognate ligands through a combination of the “conformational selection” and induced-fit mechanisms. They discriminate against similar compounds primarily via steric hindrance and formation of specific interactions. Interestingly, riboswitches from the same class can be tuned to sense different concentrations of a single metabolite (Tomsic et al., 2008). They can also operate in thermodynamic and kinetic regimes—in other words, with and without equilibration between RNA and a cognate ligand (Lemay et al., 2011; Wickiser et al., 2005).
Origin of Riboswitches
The origin and evolution of riboswitches are among the most intriguing problems in the study of RNA. In vitro selection experiments revealed the relative ease with which RNA could be evolved to bind specific ligands, suggesting that it takes a relatively short time for natural selection to transform RNA sequences into metabolite-binding domains. Therefore, narrowly distributed riboswitches may have arisen late during evolution. Several such events could give rise to independent classes of riboswitches specific to the same compound, e.g., SAM (Corbino et al., 2005; Epshtein et al., 2003; Fuchs et al., 2006; McDaniel et al., 2003; Winkler et al., 2003). Alternatively, the presence of TPP riboswitches in all three kingdoms of life suggests an ancient origin of this riboswitch type. How early in evolution could riboswitches have appeared? According to the RNA world hypothesis (Gilbert, 1986), at some point, RNA evolved to act both as a carrier of genetic information and as a catalyst of chemical reactions. The catalytic capability of the glmS riboswitch-ribozyme and the ability of riboswitches to interact with “ancient” coenzymes, such as FMN, TPP, or SAM, which would have broadened the early repertoire of biochemical reactions, provide compelling reasons to suggest that riboswitch-like molecules were instrumental for the existence and evolution of the primordial RNA world (Breaker, 2006).
Future Challenges and Perspectives
Bioinformatic searches have identified many conserved mRNA elements that could potentially function as riboswitches but were missing their validated ligands (Weinberg et al., 2007, 2010). Some of these so-called “orphan” riboswitches are widespread in nature and may be associated with sensing of novel chemical cues (Breaker, 2011). For instance, recent studies suggested that the orphan ydaO motif is an ATP-sensing riboswitch (Watson and Fedor, 2012), whereas the yybP/ykoY motif is a pH sensor (Nechooshtan et al., 2009) that may also be involved in ATP sensing (Watson and Fedor, 2012). New riboswitches in bacteria and eukaryotes may not necessarily be highly conserved and could be present in genes expressed only under specific conditions in selected species, which would complicate their identification. One such example is the ATP-sensing virulence mRNA in Salmonella expressed under acidic pH (Lee and Groisman, 2012). Another hurdle in riboswitch validation is that the relationship between the structures of riboswitches and the nature of their cognate metabolites are not well understood, and growing evidence suggests that such interconnection may not exist. As a consequence, accumulated biochemical and structural information is not useful for predicting cognate metabolites for candidate riboswitches.
Despite technical limitations, we are beginning to understand the principles that confer specificity to metabolite binding by riboswitches as outlined above and have taken the first steps toward the manipulation of metabolite binding (Dixon et al., 2010) and the rational design of antibiotics that target riboswitches (Mulhbacher et al., 2010). Some riboswitches control vital metabolic and virulence genes in pathogenic species, and the identification of novel riboswitch-specific antibiotic scaffolds presents an attractive strategy for therapeutic intervention. Additionally, riboswitches are potential targets for the construction of artificial genetic circuits that could be controlled by nonnatural compounds (Dixon et al., 2012). Understanding the regulatory and structural principles exploited by natural riboswitches will help in the development of synthetic riboswitches that respond to a particular ligand (Suess et al., 2004; Verhounig et al., 2010) and the useful reprogramming of bacteria, for instance, to seek and destroy an unwanted herbicide (Sinha et al., 2010). Future studies will explore the full potential of riboswitches for medicinal and biotechnological applications and will reveal a whole spectrum of molecular mechanisms employed by natural RNA sensors.
Acknowledgments
This work was supported by the National Institutes of Health grant R01 GM58750 (E.N.) and by New York University Medical Center funds (A.S. and E.N.).
References
- Baker JL, Sudarsan N, Weinberg Z, Roth A, Stockbridge RB, Breaker RR. Widespread genetic switches and toxicity resistance proteins for fluoride. Science. 2012;335:233–235. doi: 10.1126/science.1215063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- Bocobza S, Adato A, Mandel T, Shapira M, Nudler E, Aharoni A. Riboswitch-dependent gene regulation and its evolution in the plant kingdom. Genes Dev. 2007;21:2874–2879. doi: 10.1101/gad.443907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker RR. Riboswitches and the RNA world. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006. pp. 89–108. [Google Scholar]
- Breaker RR. Prospects for riboswitch discovery and analysis. Mol Cell. 2011;43:867–879. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler EB, Xiong Y, Wang J, Strobel SA. Structural basis of cooperative ligand binding by the glycine riboswitch. Chem Biol. 2011;18:293–298. doi: 10.1016/j.chembiol.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- Chen AGY, Sudarsan N, Breaker RR. Mechanism for gene control by a natural allosteric group I ribozyme. RNA. 2011;17:1967–1972. doi: 10.1261/rna.2757311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane JC, Lipchock SV, Strobel SA. Structural investigation of the GlmS ribozyme bound to its catalytic cofactor. Chem Biol. 2007;14:97–105. doi: 10.1016/j.chembiol.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JA, Irnov I, Baker S, Winkler WC. Mechanism of mRNA destabilization by the glmS ribozyme. Genes Dev. 2007;21:3356–3368. doi: 10.1101/gad.1605307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbino KA, Barrick JE, Lim J, Welz R, Tucker BJ, Puskarz I, Mandal M, Rudnick ND, Breaker RR. Evidence for a second class of S-adenosylmethionine riboswitches and other regulatory RNA motifs in alpha-proteobacteria. Genome Biol. 2005;6:R70. doi: 10.1186/gb-2005-6-8-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft MT, Moulin M, Webb ME, Smith AG. Thiamine biosynthesis in algae is regulated by riboswitches. Proc Natl Acad Sci USA. 2007;104:20770–20775. doi: 10.1073/pnas.0705786105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie MJ, Shi Y, Latifi T, Groisman EA. An RNA sensor for intracellular Mg(2+) Cell. 2006;125:71–84. doi: 10.1016/j.cell.2006.01.043. [DOI] [PubMed] [Google Scholar]
- Dann CE, III, Wakeman CA, Sieling CL, Baker SC, Irnov I, Winkler WC. Structure and mechanism of a metal-sensing regulatory RNA. Cell. 2007;130:878–892. doi: 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- Dixon N, Duncan JN, Geerlings T, Dunstan MS, McCarthy JE, Leys D, Micklefield J. Reengineering orthogonally selective riboswitches. Proc Natl Acad Sci USA. 2010;107:2830–2835. doi: 10.1073/pnas.0911209107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon N, Robinson CJ, Geerlings T, Duncan JN, Drummond SP, Micklefield J. Orthogonal riboswitches for tuneable coexpression in bacteria. Angew Chem Int Ed Engl. 2012;51:3620–3624. doi: 10.1002/anie.201109106. [DOI] [PubMed] [Google Scholar]
- Edwards TE, Ferré-D’Amaré AR. Crystal structures of the thibox riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure. 2006;14:1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Epshtein V, Mironov AS, Nudler E. The riboswitch-mediated control of sulfur metabolism in bacteria. Proc Natl Acad Sci USA. 2003;100:5052–5056. doi: 10.1073/pnas.0531307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox KA, Ramesh A, Stearns JE, Bourgogne A, Reyes-Jara A, Winkler WC, Garsin DA. Multiple posttranscriptional regulatory mechanisms partner to control ethanolamine utilization in Enterococcus faecalis. Proc Natl Acad Sci USA. 2009;106:4435–4440. doi: 10.1073/pnas.0812194106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs RT, Grundy FJ, Henkin TM. The S(MK) box is a new SAM-binding RNA for translational regulation of SAM synthetase. Nat Struct Mol Biol. 2006;13:226–233. doi: 10.1038/nsmb1059. [DOI] [PubMed] [Google Scholar]
- Garst AD, Héroux A, Rambo RP, Batey RT. Crystal structure of the lysine riboswitch regulatory mRNA element. J Biol Chem. 2008;283:22347–22351. doi: 10.1074/jbc.C800120200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand MS, Mironov AA, Jomantas J, Kozlov YI, Perumov DA. A conserved RNA structure element involved in the regulation of bacterial riboflavin synthesis genes. Trends Genet. 1999;15:439–442. doi: 10.1016/s0168-9525(99)01856-9. [DOI] [PubMed] [Google Scholar]
- Gilbert W. Origin of life: the RNA world. Nature. 1986;319:618. [Google Scholar]
- Gilbert SD, Rambo RP, Van Tyne D, Batey RT. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nat Struct Mol Biol. 2008;15:177–182. doi: 10.1038/nsmb.1371. [DOI] [PubMed] [Google Scholar]
- Gralla J, Steitz JA, Crothers DM. Direct physical evidence for secondary structure in an isolated fragment of R17 bacteriophage mRNA. Nature. 1974;248:204–208. doi: 10.1038/248204a0. [DOI] [PubMed] [Google Scholar]
- Grundy FJ, Henkin TM. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell. 1993;74:475–482. doi: 10.1016/0092-8674(93)80049-k. [DOI] [PubMed] [Google Scholar]
- Haller A, Rieder U, Aigner M, Blanchard SC, Micura R. Conformational capture of the SAM-II riboswitch. Nat Chem Biol. 2011;7:393–400. doi: 10.1038/nchembio.562. [DOI] [PubMed] [Google Scholar]
- Hollands K, Proshkin S, Sklyarova S, Epshtein V, Mironov A, Nudler E, Groisman EA. Riboswitch controlof Rho-dependent transcription termination. Proc Natl Acad Sci USA. 2012;109:5376–5381. doi: 10.1073/pnas.1112211109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Serganov A, Patel DJ. Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch. Mol Cell. 2010;40:774–786. doi: 10.1016/j.molcel.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Ishibe-Murakami S, Patel DJ, Serganov A. Longrange pseudoknot interactions dictate the regulatory response in the tetrahydrofolate riboswitch. Proc Natl Acad Sci USA. 2011;108:14801–14806. doi: 10.1073/pnas.1111701108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger L, Verzemnieks EJ, Geary C. The UA_handle:a versatile submotif in stable RNA architectures. Nucleic Acids Res. 2009;37:215–230. doi: 10.1093/nar/gkn911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE, Jr, Reyes FE, Polaski JT, Batey RT. B12 cofactors directly stabilize an mRNA regulatory switch. Nature. 2012;492:133–137. doi: 10.1038/nature11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kladwang W, Chou FC, Das R. Automated RNA structure prediction uncovers a kink-turn linker in double glycine riboswitches. J Am Chem Soc. 2012;134:1404–1407. doi: 10.1021/ja2093508. [DOI] [PubMed] [Google Scholar]
- Klein DJ, Ferré-D’Amaré AR. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313:1752–1756. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- Kwon M, Strobel SA. Chemical basis of glycine riboswitch cooperativity. RNA. 2008;14:25–34. doi: 10.1261/rna.771608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EJ, Groisman EA. Control of a Salmonella virulence locus by an ATP-sensing leader messenger RNA. Nature. 2012;486:271–275. doi: 10.1038/nature11090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science. 2010;329:845–848. doi: 10.1126/science.1190713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay JF, Desnoyers G, Blouin S, Heppell B, Bastet L, St-Pierre P, Massé E, Lafontaine DA. Comparative study between transcriptionally- and translationally-acting adenine riboswitches reveals key differences in riboswitch regulatory mechanisms. PLoS Genet. 2011;7:e1001278. doi: 10.1371/journal.pgen.1001278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley DM, Eckstein F. Ribozymes and RNA catalysis: introduction and primer. In: Lilley DM, Eckstein F, editors. Ribozymes and RNA Catalysis. Cambridge: The Royal Society of Chemistry; 2008. pp. 1–8. [Google Scholar]
- Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, Repoila F, Buchrieser C, Cossart P, Johansson J. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell. 2009;139:770–779. doi: 10.1016/j.cell.2009.08.046. [DOI] [PubMed] [Google Scholar]
- Lu C, Smith AM, Fuchs RT, Ding F, Rajashankar K, Henkin TM, Ke A. Crystal structures of the SAM-III/S(MK) riboswitch reveal the SAM-dependent translation inhibition mechanism. Nat Struct Mol Biol. 2008;15:1076–1083. doi: 10.1038/nsmb.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- McDaniel BA, Grundy FJ, Artsimovitch I, Henkin TM. Transcription termination control of the S box system: direct measurement of S-adenosylmethionine by the leader RNA. Proc Natl Acad Sci USA. 2003;100:3083–3088. doi: 10.1073/pnas.0630422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Ríos J, Navarro M, Soberón M. A conserved RNA structure (thi box) is involved in regulation of thiamin biosynthetic gene expression in bacteria. Proc Natl Acad Sci USA. 2001;98:9736–9741. doi: 10.1073/pnas.161168098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- Montange RK, Batey RT. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441:1172–1175. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- Mulhbacher J, Brouillette E, Allard M, Fortier LC, Malouin F, Lafontaine DA. Novel riboswitch ligand analogs as selective inhibitors of guanine-related metabolic pathways. PLoS Pathog. 2010;6:e1000865. doi: 10.1371/journal.ppat.1000865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR. Genetic control by a metabolite binding mRNA. Chem Biol. 2002;9:1043–1049. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- Nechooshtan G, Elgrably-Weiss M, Sheaffer A, Westhof E, Altuvia S. A pH-responsive riboregulator. Genes Dev. 2009;23:2650–2662. doi: 10.1101/gad.552209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nou X, Kadner RJ. Coupled changes in translation and transcription during cobalamin-dependent regulation of btuB expression in Escherichia coli. J Bacteriol. 1998;180:6719–6728. doi: 10.1128/jb.180.24.6719-6728.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nou X, Kadner RJ. Adenosylcobalamin inhibits ribosome binding to btuB RNA. Proc Natl Acad Sci USA. 2000;97:7190–7195. doi: 10.1073/pnas.130013897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins JB, Pero J. Biosynthesis of riboflavin, biotin, folic acid, and cobalamin. In: Sonenshein AL, Hoch JA, Losick R, editors. Bacillus subtillis and Its Closest Relatives: From Genes to Cells. Washington: ASM Press; 2002. pp. 271–286. [Google Scholar]
- Peselis A, Serganov A. Structural insights into ligand binding and gene expression control by an adenosylcobalamin riboswitch. Nat Struct Mol Biol. 2012;19:1182–1184. doi: 10.1038/nsmb.2405. [DOI] [PubMed] [Google Scholar]
- Pikovskaya O, Polonskaia A, Patel DJ, Serganov A. Structural principles of nucleoside selectivity in a 2′-deoxyguanosine riboswitch. Nat Chem Biol. 2011;7:748–755. doi: 10.1038/nchembio.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravnum S, Andersson DI. An adenosyl-cobalamin (coenzyme-B12)-repressed translational enhancer in the cob mRNA of Salmonella typhimurium. Mol Microbiol. 2001;39:1585–1594. doi: 10.1046/j.1365-2958.2001.02346.x. [DOI] [PubMed] [Google Scholar]
- Ray PS, Jia J, Yao P, Majumder M, Hatzoglou M, Fox PL. A stress-responsive RNA switch regulates VEGFA expression. Nature. 2009;457:915–919. doi: 10.1038/nature07598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren A, Rajashankar KR, Patel DJ. Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch. Nature. 2012;486:85–89. doi: 10.1038/nature11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson DL, Joyce GF. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature. 1990;344:467–468. doi: 10.1038/344467a0. [DOI] [PubMed] [Google Scholar]
- Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Huang L, Patel DJ. Structural insights into amino acid binding and gene control by a lysine riboswitch. Nature. 2008;455:1263–1267. doi: 10.1038/nature07326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Huang L, Patel DJ. Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nature. 2009;458:233–237. doi: 10.1038/nature07642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman EM, Esquiaqui J, Elsayed G, Ye JD. An energetically beneficial leader-linker interaction abolishes ligand-binding cooperativity in glycine riboswitches. RNA. 2012;18:496–507. doi: 10.1261/rna.031286.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha J, Reyes SJ, Gallivan JP. Reprogramming bacteria to seek and destroy an herbicide. Nat Chem Biol. 2010;6:464–470. doi: 10.1038/nchembio.369. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Stoddard CD, Montange RK, Hennelly SP, Rambo RP, Sanbonmatsu KY, Batey RT. Free state conformational sampling of the SAM-I riboswitch aptamer domain. Structure. 2010;18:787–797. doi: 10.1016/j.str.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo GD, Ji Y. Do mRNAs act as direct sensors of small molecules to control their expression? Proc Natl Acad Sci USA. 2001;98:9465–9467. doi: 10.1073/pnas.181334498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Hammond MC, Block KF, Welz R, Barrick JE, Roth A, Breaker RR. Tandem riboswitch architectures exhibit complex gene control functions. Science. 2006;314:300–304. doi: 10.1126/science.1130716. [DOI] [PubMed] [Google Scholar]
- Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suess B, Fink B, Berens C, Stentz R, Hillen W. A theophylline responsive riboswitch based on helix slipping controls gene expression in vivo. Nucleic Acids Res. 2004;32:1610–1614. doi: 10.1093/nar/gkh321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thore S, Leibundgut M, Ban N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- Tomsic J, McDaniel BA, Grundy FJ, Henkin TM. Natural variability in S-adenosylmethionine (SAM)-dependent riboswitches: S-box elements in bacillus subtilis exhibit differential sensitivity to SAM In vivo and in vitro. J Bacteriol. 2008;190:823–833. doi: 10.1128/JB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trausch JJ, Ceres P, Reyes FE, Batey RT. The structure of a tetrahydrofolate-sensing riboswitch reveals two ligand binding sites in a single aptamer. Structure. 2011;19:1413–1423. doi: 10.1016/j.str.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Verhounig A, Karcher D, Bock R. Inducible gene expression from the plastid genome by a synthetic riboswitch. Proc Natl Acad Sci USA. 2010;107:6204–6209. doi: 10.1073/pnas.0914423107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu MMK, Jameson NE, Masuda SJ, Lin D, Larralde-Ridaura R, Lupták A. Convergent evolution of adenosine aptamers spanning bacterial, human, and random sequences revealed by structure-based bioinformatics and genomic SELEX. Chem Biol. 2012;19:1247–1254. doi: 10.1016/j.chembiol.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachter A, Tunc-Ozdemir M, Grove BC, Green PJ, Shintani DK, Breaker RR. Riboswitch control of gene expression in plants by splicing and alternative 3′ end processing of mRNAs. Plant Cell. 2007;19:3437–3450. doi: 10.1105/tpc.107.053645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson PY, Fedor MJ. The glmS riboswitch integrates signals from activating and inhibitory metabolites in vivo. Nat Struct Mol Biol. 2011;18:359–363. doi: 10.1038/nsmb.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson PY, Fedor MJ. The ydaO motif is an ATP-sensing riboswitch in Bacillus subtilis. Nat Chem Biol. 2012;8:963–965. doi: 10.1038/nchembio.1095. [DOI] [PubMed] [Google Scholar]
- Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, Gore J, Wang JX, Lee ER, Block KF, Sudarsan N, et al. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res. 2007;35:4809–4819. doi: 10.1093/nar/gkm487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Wang JX, Bogue J, Yang J, Corbino K, Moy RH, Breaker RR. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol. 2010;11:R31. doi: 10.1186/gb-2010-11-3-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat Struct Biol. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature. 1981;289:751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]


