Summary
5-Aza-2ʹ-deoxycytidine (5-azaCdR) inhibits growth of breast cancer cells but increases their metastatic potential. Combining methylated DNA binding protein 2 depletion and 5-azaCdR has a synergistic antitumor effect enhancing tumor growth, while inhibiting invasiveness, pointing to a potential new epigenetic therapeutic for breast cancer.
Abstract
5-Aza-2ʹ-deoxycytidine (5-azaCdR) not only inhibits growth of non-invasive breast cancer cells but also increases their invasiveness through induction of pro-metastatic genes. Methylated DNA binding protein 2 (MBD2) is involved in silencing methylated tumor suppressor genes as well as activation of pro-metastatic genes. In this study, we show that a combination of MBD2 depletion and DNA methyltransferases (DNMT) inhibition in breast cancer cells results in a combined effect in vitro and in vivo, enhancing tumor growth arrest on one hand, while inhibiting invasiveness triggered by 5-azaCdR on the other hand. The combined treatment of MBD2 depletion and 5-azaCdR suppresses and augments distinct gene networks that are induced by DNMT inhibition alone. These data point to a potential new approach in targeting the DNA methylation machinery by combination of MBD2 and DNMT inhibitors.
Introduction
DNA methylation is a chemical modification of DNA involved in gene expression programming. One of the hallmarks of cancer is aberrant DNA methylation (1). Overexpression of DNA methyltransferases (DNMT1) as well as deregulation of the proper cell cycle-coordinated expression of DNMT1 causes cellular transformation (2). On the other hand, knock out of dnmt1 protects mice from colorectal cancer (3). Taken together, these data support the idea that inhibition of DNMT1 should be a reasonable strategy for anticancer therapeutics. The anticancer effects of DNMT1 inhibition were demonstrated both pharmacologically using antisense oligonucleotide inhibitors (4) and genetically using dnmt1−/− mice (3). The main mechanism of action of DNMT1 inhibitors was believed to be inhibition of DNA methylation and activation of tumor suppressor genes that were silenced by DNA methylation (5). The first DNA methylation inhibitor 5-azacytidine (AC; Vidaza) (6) was approved by the Food and Drug Administration for treatment of myelodysplastic syndromes (7). Vidaza is considered a new and promising approach to cancer therapy.
Although the focus in the field has been on the role of hypermethylation of tumor suppressor genes, screens for hypomethylated genes in different cancers revealed several promoters of pro-metastatic genes that were characteristically unmethylated in different types of cancer (8–11). A large number of promoters of genes that are members of networks involved in cancer growth and metastasis are demethylated and induced in liver cancer (12). Indeed, AC has been known for three decades to induce metastasis and invasive phenotypes in animal models and cell culture (13–15). Notwithstanding the critical clinical implications of such observations, particularly with the expanding clinical use of AC, this has oddly received very little attention. As AC and other DNMT inhibitors are emerging as novel and significant drugs in cancer therapy, this poses the challenge of how to take full advantage of the clinical benefits of DNMT inhibitors as inducers of silenced tumor suppressor genes, while avoiding the potential critical adverse side effects resulting from activation of pro-metastatic genes.
DNA methylation in promoters is believed to silence gene expression through attracting ‘readers’ of DNA methylation methylated DNA binding proteins (MBD) that in turn recruit chromatin-silencing chromatin modifying complexes (16). MBD2 binds methylated DNA and was shown to silence methylated genes (17). Therefore, inhibition of MBD2, a ‘reader’ of DNA methylation, should result in similar consequences for gene expression as inhibition of DNA methylation. Indeed, a recent study has shown that MBD2 depletion adds to the activation of several tumor suppressor genes that are induced by 5-aza-2ʹ-deoxycytidine (5-azaCdR) in breast cancer cell lines (18).
MBD2 is involved on the other hand also in activation of gene expression and thus has been proposed to have a bimodal mechanism of action (19). MBD2 could activate certain promoters through interaction with cAMP response element-binding protein transcriptional coactivator complexes (20) or through interaction with histone acetyltransferases that is mediated by the protein TACC3 (21). MBD2 has been suggested to be involved in demethylation of DNA (22), but this activity has been disputed by others (23,24). MBD2 was later shown to be associated with the conserved non-coding sequence 1, which is required for demethylation of TH2 cytokine genes, suggesting a role in DNA demethylation of cytokine genes during maturation of CD4+T cells (25). Overexpression of MBD2 in liver cells triggers demethylation and induction of U-Plasminogen Activator (uPA) (12). More recently, MBD2 was shown to be required for demethylation and transcriptional activation of FOXP3 regulatory regions and differentiation of T regulatory cells; this role of MBD2 in demethylation was proposed to be mediated through interaction with tet methylcytosine dioxygenase 2 (26).
This bimodal mode of action of MBD2 was recently confirmed in genome wide studies with exogenous expressed MBD2; MBD2 was shown to interact with both methylated inactive regions of the genome as well as active unmethylated promoters (27). We have recently shown that MBD2 has a bimodal mode of action on genes in HePG2 liver cancer cells and that interaction of MBD2 with transcription factors CCAAT/enhancer-binding protein α is associated with gene activation and demethylation (28). We have shown previously that MBD2 was required for expression of the pro-metastatic genes uPA and MMP2 in several invasive cancer cell lines including breast, prostate and liver cancers (11,12,29). MBD2 depletion by antisense oligonucleotides resulted in silencing of these genes and inhibition of invasiveness and metastasis of breast, prostate and liver cancer cell lines (11,12,30). Therefore, we tested here the possibility that a combination of 5-azaCdR and MBD2 depletion would have both an antagonistic and additive effect on gene expression that will result in a combined anticancer growth through silencing of tumor suppressor genes and antimetastasis effect, whereby MBD2 depletion would block the induction of pro-metastatic genes by 5-azaCdR, while maintaining and even enhancing the growth suppression activity.
Materials and methods
Cell culture, transfection treatments, cell invasion, growth and apoptotic assays
Human non-invasive breast cancer cell lines MCF-7 and ZR-75-1 were purchased from American Type Culture Collection. MCF-7 cells were cultured in minimum Eagle’s medium with 10 μg/ml of insulin (Invitrogen). ZR-75-1 cells were cultured in RPMI1640 (Invitrogen). Both media were supplemented with 10% fetal bovine serum, 2mM glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin. For 5-azaCdR (Sigma) treatment, cells were grown in a regular culture medium in the presence of different concentrations of 5-azaCdR (0.05–5 µM). 5-AzaCdR was replenished every 48h. Short interference RNA (siRNA) toward Mbd2 (siMBD2) and scrambled siRNA control (siCon; Dharmacon; sequence ordered: siMbd2 5ʹ GAAAGAUGAUGCCUAGUAA 3ʹ, siMbd2 sequence 2 5ʹ AAGAGGAUGGAUUGCCCGGCC 3ʹ and siCon 5ʹ GCCUUGGCAGCCUAGGCGA 3ʹ) were transfected at a final concentration of 70 nM using lipofectin (Invitrogen) as carrier. Cells were plated at a density of 3 × 105 in a 10 cm culture dish 24 h before transfection. On the day of transfection, 15 µl of lipofectin was incubated in 100 µl of reduced serum media Opti-MEM® (Invitrogen) for 45 min at room temperature. SiRNA was mixed with 60 µl of Opti-MEM® to reach a final concentration in the transfection culture medium of 70 (siMbd2) or 90 nM (siMbd2 sequence 2). The siRNA/Opti-MEM® mixture was then added to the lipofectin/Opti-MEM® mixture and incubated for 15 min at room temperature. About 4 ml of Opti-MEM® were then added to the siRNA mixture and it was transferred onto the culture dishes that contained cells that were twice pre-washed with phosphate-buffered saline. After 4 h of incubation at 37°C, 4 ml of Opti-Mem/siRNA mixture was replaced with fresh media containing fetal bovine serum albumin (10%) and other supplements as described. For 5-azaCdR treatments, the drug was added to the fresh media after the 4 h incubation period of the first siRNA transfection. SiRNA transfections were repeated 48 h and 96 h following initiation of treatment. Fresh 5-azaCdR was added to the media after each siRNA transfection and in control treatments every 48 h to maintain 5-azaCdR concentrations. Six days after initiation of treatment, which was found in preliminary experiments in MCF-7 cells to be required for maximal 5-azaCdR effects on cell growth and global DNA methylation, the following outcomes were measured. The invasive capacity in vitro was measured using Boyden chamber Invasion assay (Chemicon) following the manufacturer’s protocol. An equal number of treated and control viable cells (1.5 × 105; as determined by trypan blue exclusion) were plated onto each Matrigel Boyden Chamber. After incubation for 48 h in the invasion chambers, the invaded cells at the bottom of membrane were stained and counted under light microscope with ×400 magnification. Five randomly selected fields were counted and averaged. The number of live cells was determined using trypan blue exclusion assays. Non-viable cells that were stained blue were excluded. To determine anchorage-independent growth, a measure of transformation in vitro, 3 × 103 viable treated cells (as determined by trypan blue exclusion) were plated in triplicates onto six-well dishes containing 4 ml of complete medium with 0.33% BD Bacto™ agar solution at 37°C. The cells were fed with 2 ml of media every 2 days and the total number of colonies (>10 cells) that formed on soft agar was counted under a light microscope after 2 weeks of plating. For apoptotic assays, 2 × 103 treated cells were plated in 24-well culture dish. The number of apoptotic cells was determined 24 h after plating using the TUNEL Universal Apoptosis Detection Kit (biotin-labeled POD; GenScript) following manufacturers protocol.
The siRNA conditions used here were first optimized by testing different concentrations of Mbd2 siRNA; 70 and 100 nM of SiMbd2 and different transfection conditions: (i) 4 hours with SiMbd2 followed by refreshment with original medium supplemented with 5-azaCdR (SiMbd2 70 nM-4 h + 5-azaCdR); (ii) SiMbd2 (100 nM-4 h + 5-azaCdR) or without 5-azaCdR (SiMbd2 70 nM-4h), SiMbd2 (100nM-4 h) and (iii) SiMBD2–lipofectin complex in Opti-MEM with 5-azaCdR overnight (SiMbd2 70 nM-o/n), SiMBD2 (100 nM-o/n) or without 5-azaCdR (SiMbd2 70 nM-o/n + 5-azaCdR), SiMbd2 (100 nM-o/n + 5-azaCdR) followed by replenishing with the original medium supplemented or not with fresh 5-azaCdR. The most significant reduction was obtained with 70 nM SiMbd2 and 4 h of incubation.
Trypan blue cell viability assay
About 2 × 104 cells, treated with 5-azaCdR and/or shRNAs for 6 days were plated in six-well plates in triplicate. At specific time points (24, 72 and 120h) after treatment, the cells were trypsinized and stained with trypan blue. Viable cells were counted under a light microscope. An equal number of viable cells was subjected to the different tests of cell growth, anchorage independent growth on soft agar and cell invasiveness in a Boyden chamber (no 5-azaC or siRNA were added during these assays). All experiments were performed in triplicate.
ShRNA treatment
To suppress Mbd2 gene in non-invasive ZR-75-1 cancer cell line, we used lentivirus-mediated human pGIPZ shRNA plasmids (3ʹ ATTAC TAGGATGATTTGTG 5ʹ) and control pGIPZ-scrambled shRNA (#RHS4346 non-silencing GIPZ lentiviral shRNAmir control; contains no homology to known mammalian genes; Open Biosystems). Lentiviruses were assembled using the following three vectors: green fluorescent protein expression pGIPZ transfer vector—includes the insert (Open Biosystems); pMD2.G (VSV-G envelope expressing plasmid); PAX (packaging plasmid). The day before transfection, 106 HEK293T cells were plated in a 10cm dish (20–30% confluence). Next day, 5 µg of each vectors were transfected using FuGene HD transfection reagent (Roche) according to the manufacturer’s protocol. Cells were incubated for 48h followed by collection of the medium containing the virus filtered and used to infect the target cells. Selection with the 1mg/ml puromycin (Sigma) was started after 48 h of postinfection.
Flow cytometry
To measure cell cycle kinetics, cells were fixed by adding 70% of ice-cold ethanol at the end of treatment (6 days). Fixed cells were washed with phosphate-buffered saline and then treated with 1U of DNase-free RNase and stained with 0.5 mg of propidium iodide overnight. Fluorescence-activated cell sorting analysis was performed on a Calibur machine. Results were further analyzed using the FlowJo Software.
RNA extraction and quantitative real-time PCR
RNA from cell lines and mice tumor cells was extracted using TRIZOL reagent as described previously (31). RNA was extracted from three tumors derived from three different mice xenografts per treatment group (except for the Combined 5-azaC-siMBD2 treatment because only a single tumor developed in this treatment group). The outer layer and the core of the tumors were excluded to eliminate confounding normal tissue. Reverse transcription was performed using 3 µg of RNA and 20 U avian myeloblastosis virus reverse transcriptase [reverse transcriptase (Roche)], as recommended by the manufacturer. Two microliter of complementary DNA was used in a 20 µl reaction with SYBR green mix, 0.5 µM forward and reverse primers (Supplementary Table S1, available at Carcinogenesis Online, for primers sequences). Quantitative PCR amplification was performed in Roche LC480 LightCycler using the following conditions: denaturation at 95°C for 10min; amplification at 95°C for 10 s; annealing temperature for 10 s; extension at 72°C for 10 s, cycle 45; and final extension at 72°C for 10min.
Western blot analysis
About 25µg of protein extracts were loaded and resolved on a 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis with standard molecular size markers, which were used to determine the molecular size of the visualized bands. After transferring to nitrocellulose membrane, non-specific binding was blocked with 5% milk in Tris-buffered saline. After blocking, the membrane was incubated in a polyclonal MBD2 antibody generated in our laboratory (12) in Tris-buffered saline for 1 h followed by anti-rabbit immunoglobulin G (IgG; Sigma) and an enhanced chemiluminescence detection kit (Amersham Pharmacia Biotech). Nucleolin was used as loading control and was detected using a specific antibody (sc8031; Santa Cruz) in Tris-buffered saline followed by anti-mouse IgG (Jackson ImmunoResearch Labs) in Tris-buffered saline.
Nearest neighbor analysis
Global genomic DNA methylation was determined by nearest neighbor analysis, measuring methylation at the CG dinucleotide as described previously (32). Global methylation levels are represented as percentage (methylcytosine)/(cytosine + methylcytosine).
Expression microarrays
For transcriptome analysis, 1 µg of MCF-7 RNA from the same treatments used for MeDIP was prepared and subjected to microarray expression analysis using Affymetrix Human Genome U133_Plus 2.0 (array hybridization was performed at the Génome Québec Innovation Centre, Montréal, Canada). To compare dose effects of 5-azaCdR, cells treated with either 0.3 or 5 µM of 5-azaCdR were used for the arrays.
Data analysis of messenger RNA expression arrays
Biological replicates were normalized using the RMA method. Differentially expressed genes were chosen to be those with >1.5-fold increase or <0.5-fold decrease in each sample compared with untreated control and false discovery rate (FDR) was set at 0.2 (t-test, P < 0.05, FDR < 0.2). Over- and under-expressed gene sets were then analyzed for enrichment of different cellular processes, biological pathways and functional classes using Gene Ontology (33), L2L Microarray Analysis Tool (34), KEGG (35) and Ingenuity™ Pathway Analysis software (Ingenuity Systems, Redwood City, CA).
Quantitative chromatin immunoprecipitation
Three million treated MCF-7 cells were fixed with 1% formaldehyde for 10 min at 37°C in the presence of protease inhibitor. Fixed cells were then lysed and subjected to sonication. Each sample was divided into input, bound and IgG negative control. Antibody-bound and IgG fractions were incubated with protein G agarose and 10 µg of respective antibody (MBD2, Santa Cruz sc-9397, Upstate AB3467) overnight at 4°C. The next day, the unbound fraction was removed, and the beads were subjected to a series of salt washes. The bound fractions were then eluted and the antibodies were degraded by protease K treatment. Bound DNA was then obtained after phenol/chloroform precipitation. Chromatin immunoprecipitation DNA samples were used as template for quantitative PCR reactions. About 20 ng of DNA was used as starting material in all conditions. Level of antibody binding was expressed as (bound-IgG)/input. Primer sequences are provided in Supplementary Table S1, available at Carcinogenesis Online.
Quantification of DNA methylation by pyrosequencing
Three microgram of DNA samples were sodium bisulfite-treated as described previously (12). Treated DNA was subjected to PCR amplification using biotinylated primers. Twenty-five microliters of PCR products were used to perform pyrosequencing in PyroMark™ Q24 (Biotage). Data were analyzed by the PyroMark™ Q24 software. Primers used are provided in Supplementary Table S1, available at Carcinogenesis Online.
Determining tumor growth rates in vivo
For xenograft tumor growth studies, 4–6-weeks-old female Balb/c nude mice (NCI Research Resources, Frederick, MD) were inserted with estrogen pellets (Innovative Research, Sarasota, FL) subcutaneously in the back of the neck. About 70 nM of siMbd2 and/or 5 µM of 5-azaCdR was used to treat MCF-7 cells for a period of 6 days before injection (see ‘Cell culture, transfection treatments’ for treatment details). An equal number of viable and control MCF-7 cells (1.5 × 106) were counted under light microscopy using trypan blue staining and were inoculated into the mammary fat pad in 20% Matrigel. Tumor volume was measured at weekly intervals starting at week 3 using the formula: tumor volume = (length × width2)/2. At the end of the experiment, mice were killed and tumors were dissected and frozen in −80°C for further analyses. All the experimental animal protocols were in accordance with the McGill University Animal Care Committee guidelines.
Statistics
Student’s t-tests were performed to determine statistical significance of the quantitative PCR and pyrosequencing results. Individual treatments are compared with untreated controls. In addition, we compared DNA methylation and expression values for 5-azaCdR + siCon and 5-azaCdR + SiMbd2. Significance threshold is set at P-value < 0.05 and indicated by asterisks (*).
Results
Combined 5-azaCdR and MBD2 depletion has a joint anticancer and antimetastatic effect on breast cancer cells
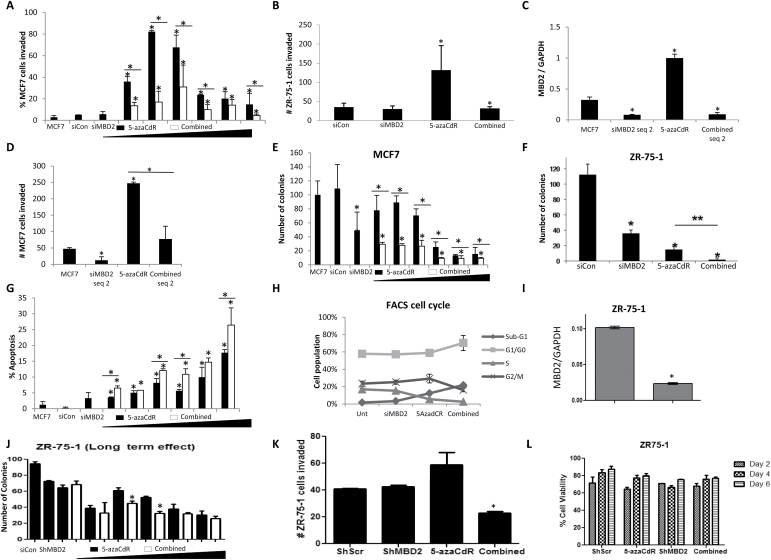
5-AzaCdR induces invasiveness in MCF-7 cells at a concentration as low as 50 nM (Figure 1A). Depletion of MBD2 by siRNA concurrently with 5-azaCdR treatment (combined) blocks invasiveness induced by 5-azaCdR treatment alone (Figure 1A). These effects of 5-azaCdR, Mbd2 depletion and their combination treatments are also seen in another non-invasive human breast cancer cell line ZR-75-1 (Figure 1B) as well as when an independent siMbd2 sequence is used to deplete MBD2, suggesting that the effect is not an idiosyncrasy of a particular cell line or siMbd2 sequence (Figure 1C and D). MBD2 depletion on the other hand enhances the reduction in anchorage-independent growth in MCF-7 (Figure 1E) and ZR-75-1 (Figure 1F) cells, increased apoptosis (Figure 1G) and alterations in cell cycle kinetics leading to increased fraction of cells in G1/G0 and reduction in S phase (Figure 1H) triggered by 5-azaCdR as has been reported previously (18).
Fig. 1.
(A–H) MBD2 depletion reduces 5-azaCdR-induced invasion, while maintaining anchorage-independent growth suppression. MCF-7 cells were treated with either 70nM siCon, 70nM siRNA targeting MBD2 (siMBD2) and an increasing dose of 5-azaCdR (0.05, 0.1, 0.3, 0.5, 1 and 5µM), (which was replenished every 2 days) in the presence of 70nM siCon (5-azaCdR; black bars) or siMBD2 (Combined; white bars) for 6 days. Untreated controls were also included. (A) Boyden chamber invasion assays as described in Supplementary Materials and Methods, available at Carcinogenesis Online. (B) ZR-75-1 cells were treated with 70nM siMbd2 and 5 µM of 5-azaCdR, alone or in combination, for 6 days. Treated cells were subjected to Boyden chamber invasion assays. (C and D) An independent sequence of siRNA against MBD2 (siMBD2 seq 2) was used to treat MCF-7 cells (90nM, 6 days). MBD2 expression (C) and cell invasion (D) were measured as previously described. (E) Soft agar anchorage-independent growth assay: 3×103 treated MCF-7 cells were re-suspended into a single cell suspension and were allowed to grow in 0.33% BD Bacto™ agar at 37°C. Colonies were counted after 2 weeks. (F) ZR-75-1 cells were treated with 70nM siMbd2 and 5 µM 5-azaCdR, alone or in combination, for 6 days. Treated cells were subjected to soft agar assay. (G) TUNEL assays: 2×103 treated MCF-7 cells were labeled with biotinylated dTTPs, which incorporated into the 3ʹ-OH of the cleaved DNA. The brown apoptotic cells were counted and the percentage of apoptotic cells was determined. All values are presented as mean ± standard error of the mean (SEM) of triplicates per dose (*P < 0.05, two-tailed Student’s t-test). (H) Cell cycle analysis was performed by flow cytometry 6 days after initiation of treatment of MCF-7 cells. Graph represents percentage of cells in sub-G1, G1/G0, S and G2M for each condition. SEM is calculated from triplicate experiments. Values are presented as means ± SEM of triplicate wells at each dose (*P < 0.05, two-tailed Student’s t-test in comparison with control or as otherwise indicated by a line). (I) Quantitative reverse transcriptase–PCR determination of Mbd2 messenger RNA levels in lenti-ShMbd2 and lenti-ShScr- stable ZR-75-1 breast cancer cell lines normalized to GAPDH. (J) Anchorage-independent growth of lenti-ShMbd2 and lenti-ShScr- stable ZR-75-1 breast cancer cell lines with and without treatment with 5 μM 5-azaCdR for 6 days. (K) Invasiveness as measured by a Boyden chamber invasion assay of lenti-ShMbd2 and lenti-ShScr- stable ZR-75-1 breast cancer cell lines with and without treatment with 5 μM 5-azaCdR for 6 days. (L) Control (ShScr) and MBD2 knockdown (ShMbd2) ZR-75-1 breast cancer cell were plated in six-well plates in triplicate and were treated with 5 μM 5-azaCdR for 6 days every second day. At specific time points (days 2, 4 and 6), cells were trypsinized and stained with trypan blue. Viable cells were counted under a light microscope. SEM is calculated from triplicate experiments.
Further, to study the long-term effect of MBD2 reduction, we performed lentivirus-mediated MBD2 knockdown in the ZR-75-1 cell line and identified transfectants with the highest stable knockdown of MBD2. We confirmed downregulation of Mbd2 messenger RNA in ZR-75-1 cells with quantitative reverse transcriptase–PCR (Figure 1I). MBD2 depletion enhances the reduction in anchorage-independent growth (Figure 1J) and blocks invasiveness induced by 5-azaCdR treatment (Figure 1K). These effects, however, are less significant than what was observed with transient siMbd2 depletion (Figure 1B and F). This difference can be explained by the higher reduction in MBD2 levels (>90%) in siMbd2 knockdown cells (Figure 2B) compared with >75% reduction in stable ZR-75-1 shMbd2 knockdown cells (Figure 1I). The lower reduction of MBD2 in stable knockdown might result from selection against low MBD2 expressing cells in long-term cultures.
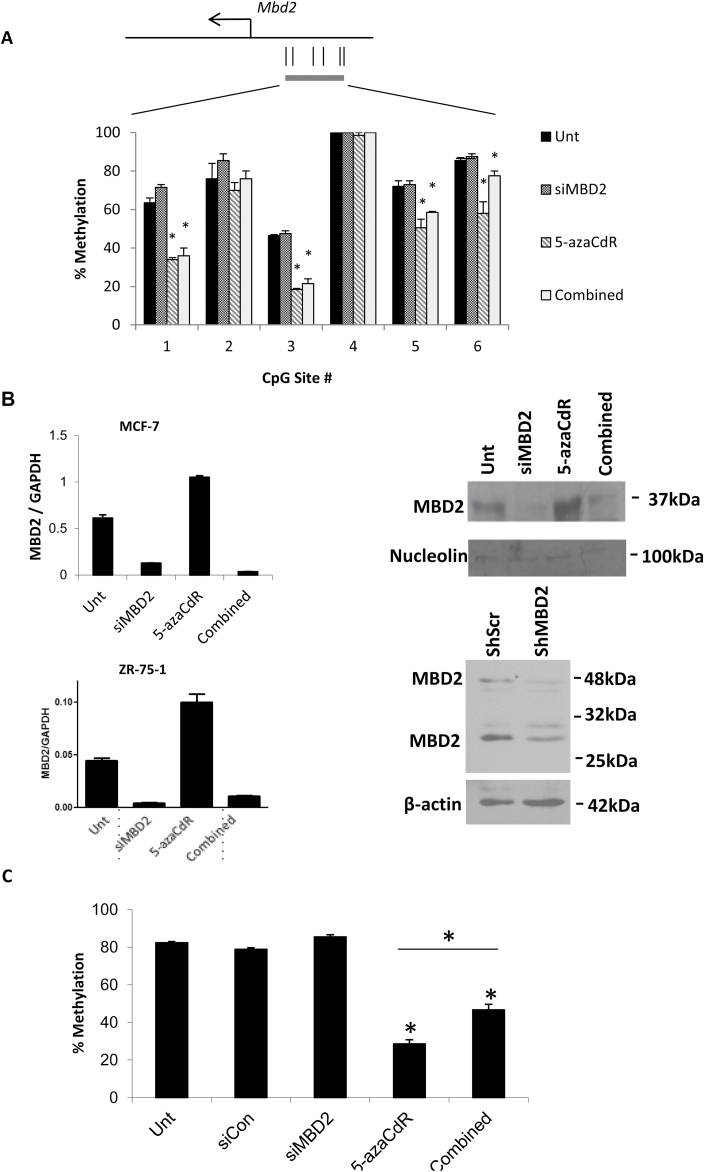
Fig. 2.
Analysis of MBD2 expression, promoter methylation, global methylation and transcriptional changes induced by 5-azaCdR, Mbd2 siRNA and combination treatments. (A) DNA was extracted from MCF-7 cells treated with 70nM of siMbd2 and/or 5 µM of 5-azaCdR. Purified DNA was subjected to bisulfite conversion and pyrosequencing of the MBD2 5ʹ region. (B) Quantitative PCR (MCF-7 and ZR-75-1) and western blot analysis (MCF-7) show efficient knockdown of MBD2 in the siMBD2-treated cells and the Combination of 5-azaCdR and siMBD2 treatment. The position of the proteins as determined using molecular weight markers is indicated. (C) Measurement of overall genome DNA methylation at the dinucleotide sequence CpG by nearest neighbor analysis. Percentage methylation is presented as mean ± SEM of triplicate experiments (*P < 0.05, two-tailed Student’s t-test).
We conclude that MBD2 depletion inhibits the increased invasiveness triggered by 5-azaCdR without compromising the growth inhibitory activity of 5-azaCdR.
5-AzaCdR demethylates the Mbd2 promoter in MCF-7 cells as determined by pyrosequencing (Figure 2A) and induces its expression at both messenger RNA and protein levels (Figure 2B). 5-AzaCdR also induces the expression of Mbd2 gene in another non-invasive breast cancer cell line, ZR-75-1 (Figure 2B). These data support the hypothesis that MBD2 is potentially a downstream effector of 5-azaCdR. MBD2 depletion partially inhibits global demethylation effects of 5-azaCdR (Figure 2C) consistent with the idea that MBD2 depletion limits 5-azaCdR effects on global DNA methylation.
MBD2 depletion activates gene networks involved in apoptosis and cell growth and suppresses metastasis and invasion networks that are induced by 5-azaCdR
To understand the mechanisms involved in these effects of MBD2 depletion and 5-azaCdR treatment on the invasive and transformed phenotypes, we examined the effects of these treatments (5-azaCdR, siMbd2 and siMbd2 + 5-azaCdR) on the state of the transcriptome using Affymetrix human expression arrays at the same time point when control and treated cells were tested for either invasion or anchorage-independent growth. Thus, we mapped the state of the transcriptome at the same time that the biological effects of the treatment were measured to delineate the changes in transcription that might explain these biological differences. The summary of the array data could be found in Supplementary Table S2, available at Carcinogenesis Online.
Examination of the transcriptional changes triggered by knockdown of MBD2 alone is consistent with the previously documented bimodal mode of action of MBD2. Depletion of MBD2 resulted in both induction (201 genes) and suppression (108) of gene expression (Supplementary Figure S1, available at Carcinogenesis Online). Importantly for cancer therapy, although MBD2 depletion affects gene expression in both ways, the functional gene pathways that are either activated or silenced by MBD2 depletion fall into distinct functional gene networks and canonical pathways suggesting higher level organization of MBD2 roles in breast cancer cells (Supplementary Table S4, available at Carcinogenesis Online). 5-AzaCdR, which targets all DNMTs in the cell, has a broader impact on gene transcription. A total of 1945 genes are induced and 1080 are repressed (Supplementary Figure S1, available at Carcinogenesis Online). The observation that 5-azaCdR treatment results also in gene silencing should not be surprising because this drug is a ubiquitous DNMT inhibitor and might cause changes in DNA methylation that are positively correlated with gene expression such as demethylation of gene bodies (36). In addition, activation of negative regulatory genes by demethylation might result in indirect suppression of other genes. Interestingly, the combination of 5-azaCdR and MBD2 depletion results in a different list of changes in gene expression than each treatment on its own; importantly, hundreds of genes that are affected by the combination are not anticipated by either treatment on its own suggesting a true synergistic response of the combination creating a new transcription landscape that could not be anticipated by either treatment on its own (Supplementary Figure S1, available at Carcinogenesis Online).
Sixty-five genes that were induced by 5-azaCdR were silenced by combined MBD2 depletion, whereas 50 genes that were induced by 5-azaCdR were further induced by MBD2 depletion (Supplementary Figure S1 and Table S2, available at Carcinogenesis Online). We reasoned that some of the genes that are induced by 5-azaCdR and repressed by combined depletion of MBD2 would be involved with the antagonistic effects of MBD2 depletion on 5-azaCdR-triggered invasiveness (Figure 1A). As has been observed previously, 5-azaCdR induces several genes known to be involved in metastasis explaining the increased invasiveness of 5-azaCdR-treated breast cancer cells (Supplementary Table S3, available at Carcinogenesis Online) (37). Interestingly, MBD2 depletion inhibits 5-azaCdR-triggered induction of pro-metastatic genes (Supplementary Table S3, available at Carcinogenesis Online; t-tests, P < 0.05 and Wilcoxon rank-sum test FDR < 0.25) but does not affect the induction by 5-azaCdR of several tumor suppressing genes (Supplementary Table S3, available at Carcinogenesis Online; t-tests, P < 0.05 and Wilcoxon rank-sum test FDR < 0.25). The genes that are induced by 5-azaCdR and are repressed by a combination of 5-azacdR and siMbd2 fall into different gene networks and Gene Ontology terms than the group of genes that are induced by 5-azaCdR alone and remain upregulated after combining siMBD2 and 5-azaCdR (Supplementary Tables S5 and S6, available at Carcinogenesis Online). The gene networks induced by 5-azaCdR and repressed by siMbd2 include invasion of cells, invasion of tumor cell lines and metastasis of breast cancer (Fisher’s exact tests, P < 10–5), regulation of cell–cell adhesion as well as epithelial-to-mesenchymal transition (EMT). EMT refers to the process by which epithelial cells lose polarity and develop characteristics of mesenchymal cells, which often, but not always, have increased translocation ability (38,39). Although EMT has been shown to increase invasion in vitro in many systems, the involvement of EMT in cancer progression has been debated due to lack of evidence of EMTs in clinical tumors (40). Several studies showed, however, that EMT is indeed present in vivo at the ‘invasive front’ of the tumors (41–43). Not all EMT genes are identical in all cases, but the expression of several genes, termed EMT markers, are always altered. Particularly in breast cancers, alterations in the expressions of EMT markers are significantly correlated with poor prognosis (40). Gene pathways induced by 5-azaCdR and remain induced when MBD2 is depleted include cell death and apoptosis as well as inhibition of cell invasion pathways (Fisher’s exact tests P < 10–5) explaining the concurrent additive effect of MBD2 depletion on apoptosis (Figure 1C) and its inhibition of invasiveness (Figure 1A, B, and K). The impact of 5-azaCdR on the transcriptome and reversal of some of the effects by MBD2 depletion are seen not only in the micromolar (µM) range of concentration used in our experiments but also in the nanomolar (nM) range (Supplementary Table S2, available at Carcinogenesis Online).
Analysis of the set of genes that are induced by 5-azaCdR and silenced by concurrent MBD2 depletion using the L2L Microarray Analysis expression database tool reveals enrichment of gene sets that are highly expressed in invasive breast cancer (Supplementary Table S7, available at Carcinogenesis Online), high-grade tumors and poor outcome patients (Supplementary Table S7, available at Carcinogenesis Online). This supports the hypothesis that 5-azaCdR induces genes that are involved in high-grade tumors and that MBD2 depletion inhibits these genes. In summary, MBD2 depletion has a bimodal effect on gene expression in response to 5-azaCdR. Bimodal effects of MBD2 on gene expression were demonstrated before and our data are consistent with previous reports that MBD2 silences tumor suppressor genes (18) and activates pro-metastatic genes (11,12,30).
Quantification of the impact of MBD2 depletion and 5-azaCdR treatment on the expression of specific pro-metastatic, tumor suppressor and apoptosis inhibitory genes
We validated the transcriptome analysis obtained from MCF-7 cells by quantitative PCR in both non-invasive breast cancer cell lines, MCF-7 and ZR-75-1. The results presented in Figure 3 show that although 5-azaCdR induces genes involved in EMT (Figure 3A) and metastasis (Figure 3B), these are suppressed when MBD2 is concurrently depleted. In contrast, the induction of several genes involved in tumor suppression by 5-azaCdR (Figure 3C) is either enhanced by MBD2 depletion (CST6), induced exclusively (INHA) or unaffected (MAEL; Figure 3C). SiMbd2 treatment inhibits 5-azaCdR-induced genes that are involved in inhibition of apoptosis XIAP (44) and BIRC3 (only in MCF-7 cells, Birc3 expression in ZR-75-1 was below detection level; Figure 3D), which can explain why siMbd2 treatment enhances the apoptotic activity of 5-azaCdR (Figure 1D). BORIS, a testes-specific paralog of CTCF (45), an early marker of tumorigenesis and epigenetic regulator implicated in induction of cancer testes-specific genes (46), is induced by 5-azaCdR as has been demonstrated previously (47). This induction is suppressed by MBD2 depletion (Figure 3D). Similar effects on expression were obtained with a different siMbd2 sequence (Figure 3E) suggesting that these results are not an idiosyncrasy of siRNA sequence.
Fig. 3.
Quantitative PCR validations of expression arrays in MCF-7 and ZR-75-1 cell lines. Columns in each of the graphs are presented in the following order: control, siMBD2, 5 µM 5-azaCdR and Combined. (A) EMT markers, (B) pro-metastatic genes and (C) tumor suppressor genes. (D) Important biological functions. GAPDH was used as a reference gene in all cases. Values presented as mean ± SEM of triplicate experiments (*P < 0.05, two-tailed Student’s t-test). (E) siMbd2 seq 2 depletion (90nM): Expression profiles of uPA, MMP2 and MAEL were measured by quantitative PCR. (F) Correlation between Mbd2 normalized expression levels in ZR-75-1 in the different individual control and treatment samples and relative expression of Vimentin, FABP7, HAS3, SPARC and uPA (Axe X) in the same samples as determined by linear regression. GAPDH was used as a reference gene for normalization.
Figure 3F demonstrates that expression of pro-metastatic genes (uPA and FABP7) and EMT markers (SAPRC, Vimentin and HAS3) in different treated cell samples are highly correlated with MBD2 expression levels in the same samples.
MBD2 binds the promoters of several pro-metastatic genes and its depletion partially inhibits 5-azaCdR-triggered promoter demethylation
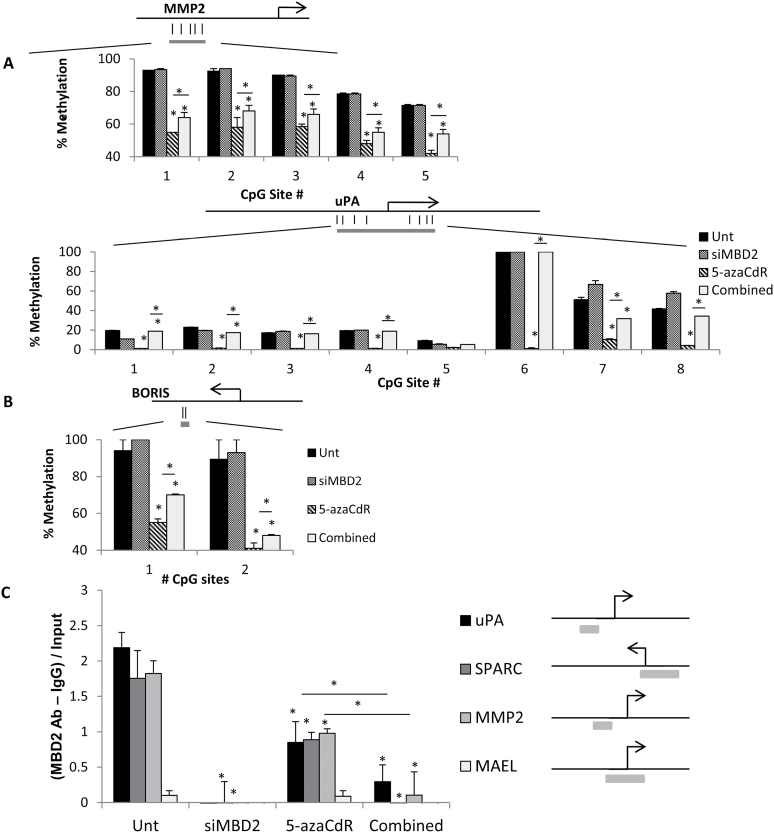
The mechanisms involved in MBD2 effects on silencing tumor suppressor genes in breast cancer cells were previously extensively described (18,48–52). We focused here on the mechanisms involved in activation of pro-metastatic genes by 5-azaCdR and their silencing by depletion of MBD2. 5-AzaCdR direct mechanism of action involves DNA demethylation (53). We, therefore, examined the state of methylation of MMP2, uPA and BORIS that were induced by 5-azaCdR and were repressed by concurrent MBD2 depletion. The analysis presented in Figure 5A and 5B shows that several CGs were demethylated in response to 5-azaCdR as expected and were partially protected from demethylation when MBD2 was depleted. MBD2 was shown previously to be required for maintaining several pro-metastatic genes hypomethylated (29,54). For example, CpG sites 1, 2, 3, 4, 6, 7 and 8 in the uPA promoter are completely demethylated by 5-azaCdR and their levels of methylation is similar to control untreated cells following concurrent MBD2 depletion. In sites 7 and 8, MBD2 depletion alone results in increased methylation above the levels of methylation in untreated control cells.
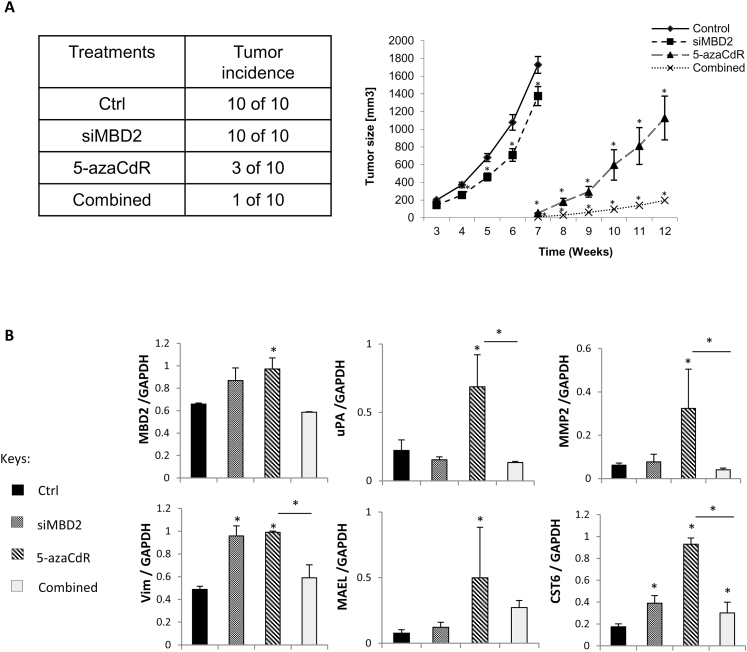
Fig. 5.
Combination of transient 5-azaCdR treatment and MBD2 depletion effectively reduces xenograft tumor volume in nude mice. (A) Control (Ctrl) and MCF-7 cells treated with 70nM siMBD2 and/or 5 µM 5-azaCdR were injected into the mammary fat pad of female Balb/c nude mice as described in ‘Materials and Methods’. Tumor volume was monitored starting from postinjection week 3 until the mice were killed. (B) Quantitative PCR analysis of expression of pro-metastatic genes in xenograft tumors. RNA was extracted from the xenograft tumors from three mice in each treatment group (except in the Combined treatment where there was only one incidence of tumor). Expression of MBD2, pro-metastatic genes (uPA, MMP2 and vimentin) and tumor suppressor genes (MAEL and CST6) were measured by quantitative reverse transcriptase–PCR. GAPDH gene was used as reference in all cases. Results represent the mean ± SEM of at least four biological replicates from each group. Values that are significantly different from control are represented by asterisks (P < 0.05).
To understand whether the pro-metastatic genes we investigated were indeed MBD2 targets, we performed chromatin immunoprecipitation assays using antibody against MBD2. Figure 4C shows that MBD2 is bound to three of the promoters of pro-metastatic genes induced by 5-azaCdR and silenced by MBD2 depletion in the untreated state, whereas MBD2 binds poorly to MAEL, a tumor suppressor that is not affected by MBD2 depletion in MCF-7 cells (Figure 3C). Following 5-azaCdR treatment, the promoters are still bound with MBD2, albeit at a lower level, which could be explained by the reduced methylation of the promoters. Depletion of MBD2 results in disappearance of MBD2 from the promoter (Figure 4C). This suggests that MBD2 is in direct interaction with the promoters of the pro-metastatic genes or the chromatin associated with these promoters and that MAEL, which is not affected by MBD2 depletion, is not a target for MBD2 binding. Nevertheless, MBD2 occupancy of the promoter per se is insufficient to cause demethylation or activation of these genes as evidenced by the fact that MBD2 is bound to the methylated uninduced promoter (Figure 4C). Other factors must be involved. Interestingly, similarly Wang et al. (26) have recently shown that MBD2, which is required for activation and demethylation of the foxp3 gene in T regulatory cells, is nevertheless bound to the inactive methylated foxp3 regulatory region in T effector cells (26), suggesting that factors in addition to MBD2 are required to cause demethylation and activation of its targets. Nevertheless, in both cases in T regulatory cells in Wang et al. (26) and in our MCF-7 cells (Figure 4A and C; Figure 3B), elimination of MBD2 results in partial reversal of demethylation and in gene activation suggesting that MBD2 is necessary but not sufficient for activation of its targets.
Fig. 4.
State of methylation and MBD2 binding of target genes in response to 5-azaCdR treatment alone or combined MBD2 depletion and 5-azaCdR. DNA was extracted from MCF-7 cells treated with 70nm of siMbd2 and/or 5 µM of 5-azaCdR and was subjected to (A) site-specific methylation analysis by pyrosequencing of MMP2 and uPA promoter regions. (B) Pyrosequencing of BORIS differentially methylated region. A schematic diagram is shown for each sequenced region (gray bar) with CpG sites indicated as straight lines. Percentage methylation is presented as means ± SEM of triplicate experiments (*P < 0.05, twi-tailed Student’s t-test). (C) Chromatin immunoprecipitation assay with anti-MBD2 antibody 2 in MCF-7- and 5-azaCdR-treated cells. Knockdown of MBD2 released MBD2 binding. Data represented as (bound fraction – IgG background) divided by total input for each condition (*P < 0.05, two-tailed Student’s t-test). The schematic diagrams show the quantitative PCR region amplified for each gene.
Long-term impact of MBD2 depletion and 5-azaCdR treatment in vitro on cancer growth and invasiveness in vivo
Epigenetic treatments are believed to reprogram cells and this reprogramming is maintained long after the initial trigger is gone. MCF-7 cells were therefore treated transiently in vitro as described in Materials and Methods and equal number of living cells were then injected into mammary fat pads of Balb/c nude mice in vivo. Pre-treating MCF-7 cells with either 5-azaCdR or siMbd2 had a significant effect on tumor growth in vivo, whereas a combination of MBD2 depletion and 5-azaCdR dramatically blocked the ability of MCF-7 cells to form tumors in mice in vivo (Figure 5A). Transient treatment of cancer cells with 5-azaCdR resulted in a stable lasting change in vivo in transcription of pro-metastatic genes uPA and MMP2 that was stably inhibited by combination of transient 5-azaCdR treatment and transient MBD2 depletion in vitro (Figure 5B).
Discussion
AC (Vidaza) and 5-azaCdR are the first representatives of DNA methylation inhibitors class of anticancer agents that are in clinical use (7). However, early animal data that have surprisingly received little attention showed that 5-azaCdR could also trigger cancer metastasis (13,14). This potential risk has obviously important clinical and mechanistic implications. We show here that 5-azaCdR indiscriminately causes widespread demethylation and has conflicting modes of action such as inducing apoptotic and antiapoptotic genes as well as pro-metastatic genes (Figure 3A and D and Supplementary Table S3, available at Carcinogenesis Online). The clinical relevance of this effect in our study is underscored by the fact that several of the genes that are induced by 5-azaCdR in non-invasive breast cancer cell lines MCF-7 and ZR-75-1 are known to be upregulated in high-grade breast tumors and poor outcome patients (Supplementary Table S7, available at Carcinogenesis Online). The molecular effects reported here cannot be dismissed as non-specific off-target activity of the drug (55) because the changes in gene expression reported here are observed with nanomolar concentrations of 5-azaCdR at concentration where very limited apoptosis is observed (Figure 1G; Supplementary Table S2, available at Carcinogenesis Online), are related to its in bona fide mechanism of action, DNA demethylation and have exquisite effects on gene networks associated with invasiveness and EMT transition (56). Our study focused mainly on promoter methylation. However, recent data suggest that gene body methylation might have an opposite effect on gene expression, which might confound the overall impact of 5-azaCdR on the transcriptome (36). The role of gene body methylation in regulating gene expression is still unclear, but future studies will need to address this question to fully understand the effects of 5-azaCdR on the transcriptome.
The well-documented pro-apoptotic effect of 5-azaCdR (57) is enhanced by combined MBD2 depletion. The increased apoptosis observed with combined (siMbd2 with 5-azaCdR) treatment relative to 5-azaCdR on its own is possibly the result of silencing of apoptotic inhibitory genes that are induced by 5-azaCdR and tamper the apoptotic effect of 5-azaC (Xiap and Birc3; Figure 3D). Thus, not only does MBD2 depletion not inhibit the pro-apoptotic effect of 5-azaCdR, it enhances this effect.
The idea pursued here was to combine DNMT inhibitors with inhibitors of a different epigenetic protein. MBD2 was chosen as a target because previous studies from our laboratory indicated that it was required for activation pro-metastatic genes in breast cancer (11), prostate cancer (29) and liver cancer cell lines (12), but it was also shown to silence tumor suppressor genes and cancer growth (48,50). Thus, we anticipated that MBD2 depletion would enhance the tumor suppressor silencing effects of 5-azaCdR as has been published previously (18), while antagonizing its pro-metastatic effects at the same time. Our results presented here are consistent with this hypothesis; MBD2 depletion antagonizes the pro-metastatic effects of 5-azaCdR (Figure 1A, B, D, and K), while supporting its anticancer growth and pro-apoptotic effects (Figures 1E–H and 5).
Although our data presented here shows that depletion of MBD2 could result in changes in transcription and DNA methylation (Figures 3 and 4; Supplementary Table S3 and Figure S1, available at Carcinogenesis Online), they obviously do not necessarily imply that MBD2 per se has either methylase or demethylase activities. The observation that MBD2 depletion has antithetical effects on different promoters is consistent with a significant number of previously published data that show that MBD2 could in certain contexts suppress gene activity of DNA methylated genes (17,19,20) and in other contexts promote demethylation (26), bind unmethylated promoters (27) and associate with active transcription complexes (20,21,28). Our data presented in this study show that in breast cancer cells, this property of MBD2 results in potentially highly favorable consequences for a pharmacological combination of MBD2 depletion and 5-azaCdR treatment because the gene activation and gene silencing activities of MBD2 target different gene networks with different roles in cancer.
It is interesting to note that mbd2 deficiency protects Apc+/− mice from developing intestinal tumors (58) and this is believed to be partially mediated through inhibition of the Wnt pathway, which is highly implicated in invasiveness and metastasis (59). This is consistent with the strong effects of MBD2 depletion on suppression of metastasis described here.
Here, we show that distinct and selective functional pathways are activated (apoptosis, cell growth, inhibition of metastasis) and silenced (cell movement, invasion and metastasis) in response to MBD2 depletion in 5-azaCdR-treated MCF-7 cells (Supplementary Tables S3, S5 and S6, available at Carcinogenesis Online). The simple explanation for the selectivity of MBD2 action is differential binding to different genes as shown in Figure 4C. Selective binding of factors to specific positions in DNA is a common feature of most DNA binding proteins and involves most probably sequence as well as chromatin context, but the precise mechanism is not fully clear. The mechanisms involved in specific targeting of MBD2 to repressive or activating sites in the genome are of particular interest and were addressed in previous investigations. Interaction of MBD2 with particular transcription regulatory complex was shown previously to be required for transcriptional activation (20,21,28) and we have recently shown colocalization of the transcription factor enhancer-binding protein α and MBD2 on promoters that are activated and maintained demethylated by MBD2 in liver cancer cells (28). Future studies beyond the scope of this study are needed to identify the transcription factors that confer upon MBD2 its selectivity in breast cancer cells.
Our data point to the possibility of a combined 5-azaCdR siMbd22 depletion in breast cancer therapy to synergize growth suppression, while blocking the conversion of non-invasive breast cancer cells into highly invasive cancers. It should be noted, however, that in contrast with non-invasive breast cancer cell lines, which become invasive with 5-azaCdR treatment as demonstrated here, previous studies have shown that 5-azaCdR treatment in invasive cell lines demethylates and re-expresses metastasis suppressor genes such as RECK and EPHB6 (60,61) and hence suppresses invasion in metastatic cell lines. We also obtained similar results when we treated the metastatic breast cancer cell line MDA-MB-231 with 5-azaCdR and observed a decrease in invasion (62). In these cells, pro-metastatic genes such as uPA and MMP2 are demethylated prior to 5-azaCdR (11,12,30) and thus are not further activated by 5-azaCdR. But even in a situation where invasive breast cancer cells are present in the tumor, co-depletion of MBD2 is warranted. We have demonstrated previously that MBD2 depletion in invasive prostate liver and breast cancer cell lines blocks their invasiveness in vitro and metastasis in vivo (11,12,30). Thus, although the data suggest that 5-azaCdR would not increase invasiveness of already invasive cells, tumors are characterized by their heterogeneous nature (63), and are often composed of mixtures of populations of non-invasive and invasive cells. Therefore, blocking the effects of 5-azaCdR treatment on non-invasive cancer cells within tumors by MBD2 depletion should prevent the possibility of non-invasive cells converting to highly invasive cancers with 5-azaCdR, while being highly effective in blocking metastasis of the invasive cancer cells population in the tumor. Future preclinical studies in vivo that are beyond the scope of this study are required to establish the clinical plausibility of the approach that is proposed here.
We demonstrate here the strong combined antigrowth and antimetastatic effect that could be accomplished by combining DNA methylation modulators acting at different levels of the DNA methylation regulatory network. Comprehensive transcriptome analyses reveal that the combination of DNMT inhibition and MBD2 depletion results in induction and suppression of hundreds of genes that are not touched by either agent on its own (Supplementary Figures S1 and Table S2, available at Carcinogenesis Online). The breadth of the effects of the combination versus single treatment illustrates that consequences of drug combinations are poorly predicted from the mechanism of action of each drug on its own and constitute a new entity that is qualitatively different from the sum of the effects of single treatments. If this applies to other combinations of epigenetic therapeutics, this approach could broaden and diversify the therapeutic potential of epigenetic agents. This observation has important general implications for predicting the toxic adverse effects and clinical benefits of drug combinations. Although additive combinations of histone deacetylase inhibitors and DNA methylation inhibitors were tested before (64), combinations of agents that target two different components of the DNA methylation machinery where one agent antagonizes the adverse effects of the other agent were not tested previously. We demonstrate here that a combination of epigenetic modulators creates a joint anticancer and antimetastatic activity through opposite effects on different cellular pathways that either promote or suppress cancer growth and metastasis. Our study could serve as a model for this new approach to epigenetic modulation in cancer.
Epigenetic reprogramming results in stable long-term changes in gene expression. Epigenetic drugs that reprogram DNA methylation are expected to produce long-lasting changes in gene expression. In accordance with this prediction, our results show that the effects of 5-azaCdR on induction of pro-metastatic gene expression are maintained in tumor xenografts in vivo in the absence of further treatment (Figure 5B). Similarly, the effects of concurrent MBD2 depletion on inhibition of expression of genes induced by 5-azaCdR are maintained in tumor grafts in vivo in the absence of further MBD2 depletion. Interestingly, although the transient knockdown of Mbd2 is not maintained in the tumor several months later as expected and Mbd2 expression returns to normal in the combined treatment as expected, nevertheless the induced expression of Mbd2 that is seen with 5-azaCdR alone is gone. Levels of Mbd2 in the combined treatment are similar to the control and lower than the 5-azaCdR-treated tumors. This supports the hypothesis that the transient treatment with siMbd2 resulted in reprogramming even of Mbd2 expression in 5-azaCdR-treated tumors and blocked its induction with 5-azaCdR. These results have important implications on the therapeutic use of 5-azaCdR and the long-term risk of induction of pro-metastatic genes as well as on the therapeutic potential of combining MBD2 depletion with 5-azaCdR treatment.
Supplementary material
Supplementary Tables S1–S7 and Figure S1 can be found at http://carcin.oxfordjournals.org/
Funding
Canadian Institute for Health Research (CIHR) (MOP-42411 to M.S.); GlaxoSmithKline CIHR professorship (to M.S.); Israel Cancer Research Foundation (to D.C.).
Conflict of Interest Statement: None declared.
Supplementary Material
Glossary
Abbreviations:
- AC
5-azacytidine
- 5-azaCdR
5-aza-2ʹ-deoxycytidine
- DNMT
DNA methyltransferases
- EMT
epithelial-to-mesenchymal transition
- FDR
false discovery rate
- IgG
immunoglobulin G
- MBD2
methylated DNA binding protein 2
- siCon
scrambled siRNA control
- SEM
standard error of the mean
- siRNA
short interference RNA.
References
- 1. Baylin S.B., et al. (2001). Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum. Mol. Genet., 10, 687–692 [DOI] [PubMed] [Google Scholar]
- 2. Detich N., et al. (2001). A conserved 3’-untranslated element mediates growth regulation of DNA methyltransferase 1 and inhibits its transforming activity. J. Biol. Chem., 276, 24881–24890 [DOI] [PubMed] [Google Scholar]
- 3. Laird P.W., et al. (1995). Suppression of intestinal neoplasia by DNA hypomethylation. Cell, 81, 197–205 [DOI] [PubMed] [Google Scholar]
- 4. Ramchandani S., et al. (1997). Inhibition of tumorigenesis by a cytosine-DNA, methyltransferase, antisense oligodeoxynucleotide. Proc. Natl Acad. Sci. USA, 94, 684–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Merlo A., et al. (1995). 5’ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat. Med., 1, 686–692 [DOI] [PubMed] [Google Scholar]
- 6. Jones P.A., et al. (1980). Cellular differentiation, cytidine analogs and DNA methylation. Cell, 20, 85–93 [DOI] [PubMed] [Google Scholar]
- 7. Kuendgen A., et al. (2008). Current status of epigenetic treatment in myelodysplastic syndromes. Ann. Hematol., 87, 601–611 [DOI] [PubMed] [Google Scholar]
- 8. Sato N., et al. (2003). Identification of maspin and S100P as novel hypomethylation targets in pancreatic cancer using global gene expression profiling. Oncogene, 23, 1531–1538 [DOI] [PubMed] [Google Scholar]
- 9. Rauch T.A., et al. (2008). High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc. Natl Acad. Sci. USA, 105, 252–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shteper P.J., et al. (2003). Role of promoter methylation in regulation of the mammalian heparanase gene. Oncogene, 22, 7737–7749 [DOI] [PubMed] [Google Scholar]
- 11. Pakneshan P., et al. (2004). Reversal of the hypomethylation status of urokinase (uPA) promoter blocks breast cancer growth and metastasis. J. Biol. Chem., 279, 31735–31744 [DOI] [PubMed] [Google Scholar]
- 12. Stefanska B., et al. (2011). Definition of the landscape of promoter DNA hypomethylation in liver cancer. Cancer Res., 71, 5891–5903 [DOI] [PubMed] [Google Scholar]
- 13. Olsson L., et al. (1984). Induction of the metastatic phenotype in a mouse tumor model by 5-azacytidine, and characterization of an antigen associated with metastatic activity. Proc. Natl Acad. Sci. USA, 81, 3389–3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Habets G.G., et al. (1990). Induction of invasive and metastatic potential in mouse T-lymphoma cells (BW5147) by treatment with 5-azacytidine. Clin. Exp. Metastasis, 8, 567–577 [DOI] [PubMed] [Google Scholar]
- 15. Ateeq B., et al. (2008). Pharmacological inhibition of DNA methylation induces proinvasive and prometastatic genes in vitro and in vivo . Neoplasia, 10, 266–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nan X., et al. (1997). MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell, 88, 471–481 [DOI] [PubMed] [Google Scholar]
- 17. Ng H.H., et al. (1999). MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat. Genet., 23, 58–61 [DOI] [PubMed] [Google Scholar]
- 18. Mian O.Y., et al. (2011). Methyl-binding domain protein 2-dependent proliferation and survival of breast cancer cells. Mol. Cancer Res., 9, 1152–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Detich N., et al. (2002). Promoter-specific activation and demethylation by MBD2/demethylase. J. Biol. Chem., 277, 35791–35794 [DOI] [PubMed] [Google Scholar]
- 20. Fujita H., et al. (2003). Antithetic effects of MBD2a on gene regulation. Mol. Cell. Biol., 23, 2645–2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Angrisano T., et al. (2006). TACC3 mediates the association of MBD2 with histone acetyltransferases and relieves transcriptional repression of methylated promoters. Nucleic Acids Res., 34, 364–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bhattacharya S.K., et al. (1999). A mammalian protein with specific demethylase activity for mCpG DNA. Nature, 397, 579–583 [DOI] [PubMed] [Google Scholar]
- 23. Ng H.H., et al. (1999). MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat. Genet., 23, 58–61 [DOI] [PubMed] [Google Scholar]
- 24. Hendrich B., et al. (2001). Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev., 15, 710–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aoki K., et al. (2009). Regulation of DNA demethylation during maturation of CD4+ naive T cells by the conserved noncoding sequence 1. J. Immunol., 182, 7698–7707 [DOI] [PubMed] [Google Scholar]
- 26. Wang L., et al. (2013). Mbd2 promotes Foxp3 demethylation and T-regulatory cell function. Mol Cell Biol, 33, 4106–4115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Baubec T., et al. (2013). Methylation-dependent and -independent genomic targeting principles of the MBD protein family. Cell, 153, 480–492 [DOI] [PubMed] [Google Scholar]
- 28. Stefanska B., et al. (2013). Transcription onset of genes critical in liver carcinogenesis is epigenetically regulated by methylated DNA-binding protein MBD2. Carcinogenesis, 34, 2738–2749 [DOI] [PubMed] [Google Scholar]
- 29. Shukeir N., et al. (2006). Alteration of the methylation status of tumor-promoting genes decreases prostate cancer cell invasiveness and tumorigenesis in vitro and in vivo . Cancer Res., 66, 9202–9210 [DOI] [PubMed] [Google Scholar]
- 30. Nemr A.E., et al. (2006). Distribution and sources of polycyclic aromatic hydrocarbons in surface sediments of the Suez Gulf. Environ Monit Assess, 111, 333–358 [Google Scholar]
- 31. Rio D.C., et al. (2010). Enrichment of poly(A)+ mRNA using immobilized oligo(dT). Cold Spring Harb. Protoc., 2010, pdb.prot5454. [DOI] [PubMed] [Google Scholar]
- 32. Ramsahoye B.H., et al. (2000). Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl Acad. Sci. USA, 97, 5237–5242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ashburner M., et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet., 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Newman J.C., et al. (2005). L2L: a simple tool for discovering the hidden significance in microarray expression data. Genome Biol., 6, R81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kanehisa M., et al. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res., 28, 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hellman A., et al. (2007). Gene body-specific methylation on the active X chromosome. Science, 315, 1141–1143 [DOI] [PubMed] [Google Scholar]
- 37. Chik F., et al. (2011). Effects of specific DNMT gene depletion on cancer cell transformation and breast cancer cell invasion; toward selective DNMT inhibitors. Carcinogenesis, 32, 224–232 [DOI] [PubMed] [Google Scholar]
- 38. Thompson E.W., et al. (2005). Carcinoma invasion and metastasis: a role for epithelial-mesenchymal transition? Cancer Res., 65, 5991–5; discussion 5995 [DOI] [PubMed] [Google Scholar]
- 39. Wicki A., et al. (2006). Tumor invasion in the absence of epithelial-mesenchymal transition: podoplanin-mediated remodeling of the actin cytoskeleton. Cancer Cell, 9, 261–272 [DOI] [PubMed] [Google Scholar]
- 40. Thiery J.P., et al. (2009). Epithelial-mesenchymal transitions in development and disease. Cell, 139, 871–890 [DOI] [PubMed] [Google Scholar]
- 41. Prall F. (2007). Tumour budding in colorectal carcinoma. Histopathology, 50, 151–162 [DOI] [PubMed] [Google Scholar]
- 42. Wyckoff J.B., et al. (2007). Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res., 67, 2649–2656 [DOI] [PubMed] [Google Scholar]
- 43. Brabletz T., et al. (2001). Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc. Natl Acad. Sci. USA, 98, 10356–10361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rajcan-Separovic E., et al. (1996). Assignment of human inhibitor of apoptosis protein (IAP) genes xiap, hiap-1, and hiap-2 to chromosomes Xq25 and 11q22-q23 by fluorescence in situ hybridization. Genomics, 37, 404–406 [DOI] [PubMed] [Google Scholar]
- 45. Pugacheva E.M., et al. (2010). The structural complexity of the human BORIS gene in gametogenesis and cancer. PLoS One, 5, e13872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Klenova E.M., et al. (2002). The novel BORIS + CTCF gene family is uniquely involved in the epigenetics of normal biology and cancer. Semin. Cancer Biol., 12, 399–414 [DOI] [PubMed] [Google Scholar]
- 47. Vatolin S., et al. (2005). Conditional expression of the CTCF-paralogous transcriptional factor BORIS in normal cells results in demethylation and derepression of MAGE-A1 and reactivation of other cancer-testis genes. Cancer Res., 65, 7751–7762 [DOI] [PubMed] [Google Scholar]
- 48. Magdinier F., et al. (2001). Selective association of the methyl-CpG binding protein MBD2 with the silent p14/p16 locus in human neoplasia. Proc. Natl Acad. Sci. USA, 98, 4990–4995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stirzaker C., et al. (2004). Transcriptional gene silencing promotes DNA hypermethylation through a sequential change in chromatin modifications in cancer cells. Cancer Res., 64, 3871–3877 [DOI] [PubMed] [Google Scholar]
- 50. Zhu D., et al. (2011). Overexpression of MBD2 in glioblastoma maintains epigenetic silencing and inhibits the antiangiogenic function of the tumor suppressor gene BAI1. Cancer Res., 71, 5859–5870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Singal R., et al. (2001). Cytosine methylation represses glutathione S-transferase P1 (GSTP1) gene expression in human prostate cancer cells. Cancer Res., 61, 4820–4826 [PubMed] [Google Scholar]
- 52. Lin X., et al. (2003). Methyl-CpG-binding domain protein-2 mediates transcriptional repression associated with hypermethylated GSTP1 CpG islands in MCF-7 breast cancer cells. Cancer Res., 63, 498–504 [PubMed] [Google Scholar]
- 53. Jones P.A., et al. (1983). Inhibition of DNA methylation by 5-azacytidine. Recent Results Cancer Res., 84, 202–211 [DOI] [PubMed] [Google Scholar]
- 54. Pakneshan P., et al. (2004). Demethylation of urokinase promoter as a prognostic marker in patients with breast carcinoma. Clin. Cancer Res., 10, 3035–3041 [DOI] [PubMed] [Google Scholar]
- 55. Karimi M., et al. (2006). Using LUMA: a Luminometric-based assay for global DNA-methylation. Epigenetics, 1, 45–48 [DOI] [PubMed] [Google Scholar]
- 56. Kokkinos M.I., et al. (2007). Vimentin and epithelial-mesenchymal transition in human breast cancer–observations in vitro and in vivo . Cells. Tissues. Organs, 185, 191–203 [DOI] [PubMed] [Google Scholar]
- 57. Hackanson B., et al. (2014). Decitabine. Recent Results Cancer Res., 201, 269–297 [DOI] [PubMed] [Google Scholar]
- 58. Sansom O.J., et al. (2003). Deficiency of Mbd2 suppresses intestinal tumorigenesis. Nat. Genet., 34, 145–147 [DOI] [PubMed] [Google Scholar]
- 59. Phesse T.J., et al. (2008). Deficiency of Mbd2 attenuates Wnt signaling. Mol. Cell. Biol., 28, 6094–6103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cho C.Y., et al. (2007). Epigenetic inactivation of the metastasis suppressor RECK enhances invasion of human colon cancer cells. J. Cell. Physiol., 213, 65–69 [DOI] [PubMed] [Google Scholar]
- 61. Yu J., et al. (2010). The EPHB6 receptor tyrosine kinase is a metastasis suppressor that is frequently silenced by promoter DNA hypermethylation in non-small cell lung cancer. Clin. Cancer Res., 16, 2275–2283 [DOI] [PubMed] [Google Scholar]
- 62. Chik F., et al. (2014). Synergistic anti-breast cancer effect of a combined treatment with the methyl donor S-adenosyl methionine and the DNA methylation inhibitor 5-aza-2’-deoxycytidine. Carcinogenesis, 35, 138–144 [DOI] [PubMed] [Google Scholar]
- 63. Badve S., et al. (2012). Breast-cancer stem cells-beyond semantics. Lancet. Oncol., 13, e43–e48 [DOI] [PubMed] [Google Scholar]
- 64. Cameron E.E., et al. (1999). Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat. Genet., 21, 103–107 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.