Abstract
A search of the M1genome sequence, which includes 97% of the Myxococcus xanthus genes, identified 53 sequence homologs of σ54-dependent enhancer binding proteins (EBPs). A DNA microarray was constructed from the M1genome that includes those homologs and 318 other M. xanthus genes for comparison. To screen the developmental program with this array, an RNA extract from growing cells was compared with one prepared from developing cells at 12 h. Previous reporter studies had shown that M. xanthus has initiated development and has begun to express many developmentally regulated genes by 12 h. The comparison revealed substantial increases in the expression levels of 11 transcription factors that may respond to environmental stimuli. Six of the 53 EBP homologs were expressed at significantly higher levels at 12 h of development than during growth. Three were previously unknown genes, and they were inactivated to look for effects on fruiting body development. One knockout mutant produced fruiting bodies of abnormal shape that depended on the composition of the medium.
A program of fruiting body development is induced in Myxococcus xanthus by starving a culture at a high cell density on a solid surface. Induced M. xanthus cells glide to build dome-shaped, multicellular fruiting bodies of about 105 cells within 24 h. Inside the nascent fruiting body, the rod-shaped cells differentiate into dormant, spherical spores over the next few days (49). At the regulatory level, the developmental program is an ordered, temporal sequence of gene expression events that is coordinated by signaling between cells (28, 33, 36). A set of 36 developmentally regulated promoters has been identified by genomic insertions of the Tn5lac transposon (34). Each of these Tn5lac fusions reproducibly increases β-galactosidase expression at specific time points during the developmental program, ranging from the onset of starvation (time zero) to the beginning of sporulation (at 24 h). How developmental gene expression is temporally and spatially coordinated to build a fruiting body with spores inside remains a central question.
The transcriptional regulation of bacterial gene expression often employs regulatory proteins that act upon RNA polymerase. Sigma factors recruit RNA polymerase to specific promoters which then open for transcription. One sigma factor, σ54 encoded by rpoN, requires a special class of transcriptional activators called enhancer binding proteins (EBPs). EBPs increase the rates of DNA opening rather than increasing promoter occupancy (2). EBPs usually bind to regulatory DNA sequences upstream from the σ54 promoters, from which they next interact with the RNA polymerase already associated with σ54 (σ54-RNAP) at the promoter. Significantly, σ54-RNAP bound to a promoter is unable by itself to isomerize from the closed DNA complex to an open one and to initiate transcription. Conversion of the closed to an open complex depends on the interaction of the σ54-RNAP with a bound EBP and subsequent hydrolysis of ATP. An important consequence of this regulatory mechanism is that transcription of a gene can be turned off completely, a distinct advantage for developmental genes.
The single largest family of DNA binding regulatory proteins identified so far in M. xanthus are its EBPs. Thirty-seven genes predicted to encode EBPs have been identified, and 16 of these have been shown to be required for normal fruiting body development or motility (3, 8, 13, 14, 16, 17, 20, 32, 51, 56). Uniquely among bacteria, rpoN, the gene that encodes σ54, has been shown to be vital for both growth and development in M. xanthus (31). Several developmentally regulated genes in M. xanthus have been shown to have σ54-dependent promoters (12, 30, 44). The foregoing observations point to a regulon of σ54 promoters and their EBPs within the circuit that controls M. xanthus development and motivate this attempt to identify and characterize more of its constituents. For this purpose, we took advantage of a fourfold shotgun sequence coverage that includes 97% of the M. xanthus genome made available by the Monsanto Company. The sequence allowed us to identify homologs of the σ54-dependent EBPs and to construct a DNA microarray to monitor their expression. This microarray was designed to detect the RNA levels of several groups of genes in addition to the 53 putative EBP genes that were found. A total of 94 previously studied M. xanthus genes were included as controls to determine how well the microarray results correlate with previous data. The remaining PCR products, or amplicons, were chosen because they are putative DNA binding proteins, with most of them directly involved in the regulation of transcription. A total of 371 PCR products were spotted in duplicate on this array.
MATERIALS AND METHODS
Genome assembly.
The M. xanthus genomic DNA sequence was received as 1,273 nonoverlapping contigs totaling 8.9 Mbp of DNA sequence; it is estimated to include 97% of the total genome (48). To organize the sequence for analysis and annotation, the contigs were joined to each other roughly in order of size. The “M1genome” was assembled from the contig files in Artemis, a searchable sequence viewer (45) available from the Sanger Centre (http://www.sanger.ac.uk/Software/). The largest contig is 72,704 bp. Contigs were assembled into a linear sequence for the purpose of open reading frame (ORF) identification and annotation and for primer design. Because contig order, orientation, and intercontig spacing are unknown, the assembly was designed to preserve the integrity of each contig and to facilitate its translation to the completed genome sequence when it becomes available. To this end, contigs were separated by a special sequence, a 94-bp palindrome split by a 6-bp unique sequence. Each such intercontig sequence contains three components (listed from the outside of the palindrome to the inside): start codons in each reading frame, stop codons in each reading frame, and a unique identification sequence. The start and stop codons facilitate annotation: the stop codons isolate each contig and prevent ORFs from crossing contig boundaries, and the start codons allow ORFs that may have been truncated at their 5′ end to be identified. A sequence, or bar code, uniquely identifies and locates each intercontig sequence within the M1genome. The M. xanthus DNA sequence is available for BLAST search at http://tigrblast.tigr.org/ufmg/.
The M1genome was annotated with Glimmer 2.0 (TIGR) by using 215 previously published M. xanthus sequences as a training file. The following parameters constrained Glimmer: G+C proportion of 68.1% (48), minimum ORF size of 300 bp, minimum overlap length of 30 bp, minimum overlap of 10%, and threshold score of 90; use independent scores; ignore independent score on ORFs longer than 1,491 bp; and use first start codon. Glimmer found a total of 9,203 putative ORFs satisfying these constraints, but many of them overlapped. Overlaps were resolved by using several criteria: high-third codon position G+C bias, BLAST-P scores, and the degree of overlap. In M. xanthus the third codon position is occupied by a G or a C in more than 90% of all codons within ORFs (48). In addition, motifs were searched with specific Pfam (protein family database) sequences to facilitate the assignment of function to predicted ORFs; the assignments were recorded in an Artemis feature table. Application of these criteria verified 7,420 ORFs in the M1genome, each of which is identified by a four-digit number in the format Mx_iiii.
With a view toward their use in microarrays, the ORFs were chosen conservatively and always included the region that was common to a set of overlapping ORF calls. The rationale behind conservatism is that it is easier to add features to a microarray than it is to remove them. The M1genome data has less than 0.5% sequence errors, estimated by comparison with published M. xanthus gene sequences, always assuming the latter to be correct. Many short contigs contain only one ORF. Due to high gene density, the end of any contig is likely to be only part of an ORF.
Microarray construction.
Primers for the PCR amplification of ORF fragments to be spotted on the array were designed by Microarray Architect (C. Cummings, I. Yoon, and D. Relman, unpublished data), a program that combines features of BLAST and Primer3 to optimize the choice of amplicons and primers for a cDNA array. Microarray Architect first performs a BLAST search of each ORF against the entire genomic sequence to determine a unique segment of the ORF and then chooses primers for that segment by using Primer3. Microarray Architect thereby aims to minimize cross-hybridization between sequences on the microarray. For the M1genome, the optimum amplicon size was set at 275 bp, and the optimum primer size was set at 19 bp. Primers were received from Illumina of San Diego in aqueous solution premixed in pairs at a concentration of 50 μM. Each primer pair was diluted to 5 μM, combined 1:10 (vol/vol) with a PCR Master Mix to a final volume of 50 μl (1× PCR buffer, 2.5 mM MgCl2, 1.3 M betaine, 27 mg of dimethyl sulfoxide per ml, a 10 mM concentration of each deoxynucleoside triphosphate, 50 ng of genomic DNA per μl, 1.5 U of Taq DNA polymerase), incubated at 95°C for 5 min, and then cycled 35 times through the following conditions: 95°C for 1 min, 55°C for 1 min, 72°C for 3 min, with a final extension cycle of 72°C for 10 min. Reaction products were ethanol precipitated and redissolved prior to electrophoresis. The efficiency of the PCR was tested by agarose gel electrophoresis, and products were stored at −20°C in 50 μl of 3× SSC (1× SSC is 0.15M NaCl plus 0.015 M sodium citrate).
Microarray experiments.
To test developmental regulation, M. xanthus strain DK1622 was grown vegetatively at 32°C in CTT broth (34) with aeration. The culture was harvested in mid-exponential phase, sedimented, and resuspended in MC7 (10 mM morpholinepropanesulfonic acid [pH 7.0], 1 mM CaCl2) starvation media (34). One half of the culture was sedimented at 10,000 × g, frozen, and set aside as the growth sample. The remaining 4 ml of Klett 1000 cells was added to 28 ml of sterile MC7 medium and allowed to develop in a 15-cm petri dish. After 12 h at 32°C, the cells were harvested from the bottom of the plate with a Costar cell scraper, sedimented at 10,000 × g, and frozen as the development sample.
(i) Definition of a developmentally regulated gene.
The phrase “developmentally regulated gene” is frequently used in the text. It refers to a gene whose expression changes in the course of fruiting body development and sporulation. The direct inducer of the gene must be present in the developing cell, but the phrase does not indicate where that inducer came from. In the laboratory, fruiting body development is induced by starvation via the stringent response; the phrase includes starvation-induction.
(ii) Isolation of total RNA.
Frozen cell pellets were resuspended in 300 μl of solution I (0.3 M sucrose, 0.01 M sodium acetate [pH 4.5]), transferred to an Eppendorf tube containing 300 μl of solution II (2% sodium dodecyl sulfate [SDS], 0.01 M sodium acetate [pH 4.5]) and mixed well. Samples were incubated at 65°C for 1 to 2 min; 400 μl of phenol was then added, and the samples were thoroughly mixed and incubated at 65°C for 3 min. The samples were quick-frozen in liquid N2 and then centrifuged at 16,100 × g for 5 min. The aqueous layer was transferred to another tube already containing 600 μl of hot phenol; after thorough mixing, the samples were incubated at 65°C for 3 min, snap-frozen in liquid N2, and centrifuged as before. The aqueous layer was removed to a new tube containing 600 μl of a 1:1 mix of phenol and chloroform, mixed, and centrifuged as before. The aqueous layer was transferred to another tube containing 600 μl of chloroform, shaken thoroughly, and centrifuged as before. The aqueous layer was transferred to another tube containing 40 μl of 3 M sodium acetate, pH 4.5, and 900 μl of 96% ethanol and incubated at −20°C for 30 min, followed by centrifugation at 16,100 × g for 20 min at 4°C. The supernatant was discarded, and the pellet was washed with 200 μl of ice-cold 70% ethanol and centrifuged at 16,100 × g for 5 min. After removal of the supernatant, the pellet was dried in a SpeedVac and then suspended in 50 μl of water.
Each RNA sample was treated with 4 μl of RNase-free DNase I (2 units/μl; Ambion) after the addition of 10 μl of 10× DNase I buffer and 36 μl of water and then incubated for 30 min at 37°C. The RNA was then purified by use of a QIAGEN RNeasy Mini Kit. The concentration of RNA was measured by the absorbance at 260 nm, where an absorbance of 1 unit at 260 nm corresponds to 40 μg of RNA per ml.
(iii) Labeling of cDNA and hybridization to DNA arrays.
Five micrograms of pdN6 primer (Amersham Pharmacia) was mixed with 25 μg of RNA in 14.5 μl of water on ice. After 10 min of heating at 70°C and 10 min on ice, a transcription mix was added, consisting of 5 μl of water, 3 μl of 0.1 M dithiothreitol, 3 μl of 10× reverse transcription buffer, 0.6 μl of 50× aminoallyl-dUTP-deoxynucleoside triphosphate mix (25 mM concentrations of dATP, dCTP, and dGTP and 10 mM dTTP; Amersham Pharmacia) at a ratio of 3:2, 15 mM aminoallyl-dUTP (Sigma-Aldrich), and 3 μl of Stratascript reverse transcriptase (50 units/μl; Stratagene). These samples were incubated at 42°C for 2 h, and then the RNA was hydrolyzed by the addition of 10 μl of 1 M NaOH and 10 μl of 0.5 M EDTA, followed by incubation at 65°C for 15 min. The cDNA was purified on a Zymo spin column and eluted from the column with 8 μl of 50 mM sodium bicarbonate at pH 9.0. The cDNA was coupled to either Cy3 or Cy5 (Amersham Pharmacia) by the addition of 2 μl of dye dissolved in 10 μl of dimethyl sulfoxide and incubated for 1 h in the dark. The labeled cDNA was purified on a Zymo spin column, after the probes were eluted with 12 μl of water. The Cy3- and Cy5-labeled cDNAs were combined into a final volume of 22.5 μl, and 4.5 μl of 20× SSC (46), 2.25 μl of yeast tRNA (10 μg/μl), 0.75 μl of 1 M HEPES (pH 7.0), and 0.75 μl of 10% SDS were added. After heating to 100°C for 2 min, the labeled PCR product was centrifuged and incubated for 5 to 10 min at room temperature. ArrayIt SMP3 pins (TeleChem International) were used to print the PCR products onto poly-l-lysine-coated glass slides.
Relative levels of mRNA were determined by a parallel two-color hybridization to spotted DNA microarrays that contained a duplicate set of 371 ORFs from the M. xanthus genome on glass slides. Samples were hybridized for 12 to 14 h at 63°C and then washed in solution I (340 ml of water, 10 ml of 20× SSC, 1 ml of 10% SDS), followed by washing in solution II (350 ml of water plus 1 ml of 20× SSC), and finally dried by centrifugation (72 × g for 5 min). Triplicate hybridizations were performed by using RNA from three different batches of 12-h developing cells.
(iv)Data analysis.
Dried arrays were scanned in a GenePix 4000B microarray scanner controlled by the GenePix 4.0 software package (Axon Instruments, Union City, Calif.). A photo of a typical array is shown as Fig. 1. Intensities were normalized to the mean intensity of all spots. Compiled Cy3 and Cy5 intensities as well as their corresponding background signals were imported into a Microsoft Excel spreadsheet. Background signals were subtracted, and ratios were calculated. The calculated ratios and their standard deviations were averaged over three independent experiments.
FIG. 1.
A typical sector of an array after hybridization.
Construction of EBP mutant strains.
The M. xanthus wild-type strain DK1622 was used as the starting point for the construction of strains with integrated plasmids pLOJ3320 and pLOJ3098. To this end, internal fragments of ORFs Mx_3320 and Mx_3098 were amplified by PCR with the primer pair 3320fwd1 (5′-ACGTGCCGCTCGTGGTGCAGG-3′) and 3320rev1 (5′-GCAACGCGTCCACCGCGTCC-3′) and the pair 3098fwd1 (5′-CCTTCGTGGCGGTGAACTGC-3′) and 3098rev1 (5′-GATGCTCGACGTGGCCAGGC-3′), respectively. The resulting PCR products were cloned into plasmid pCR2.1-TOPO (Invitrogen), resulting in plasmids pLOJ3320 and pLOJ3098, respectively. The plasmids were purified from Escherichia coli TOP10 (Invitrogen) and introduced into M. xanthus wild-type strain DK1622 by electroporation as described previously (14). Because the plasmids used in this work cannot replicate in M. xanthus, drug-resistant electroporants resulted from a single-crossover homologous recombination event that incorporated the plasmid into the chromosome, thereby creating gene disruption mutants. Each structure was verified by PCR by using plasmid-specific primers TOPOfwd1 (5′-TTGGTACCGAGCTCGGATCC-3′) and TOPOrev1 (5′-CCTCTAGATGCATGCTGAGC-3′) together with the gene-specific primer pair 3320fwd2 (5′-GCGCATCTTCCGCGCGCTCG-3′) and 3320rev2 (5′-GGCCTCGCGGATGAACTCCCG-3′) and the pair 3098fwd2 (5′-TGGGGACGGGGGATGCGAGC-3′) and 3098rev2 (5′-TCGGCGGACTGGAAACAGGC-3′).
Mutant characterization.
Strains were tested for motility by growing them in CTT broth to a density of 5 × 108 cells/ml; they were then harvested and resuspended in TPM buffer (10 mM Tris-HCl [42], 1 mM KH2PO4, 8 mM MgSO4) to a calculated density of 5 × 109 cells/ml. A 10-μl cell suspension was spotted on 0.5% CTT plates containing either 0.5 or 1.5% agar. The swarm expansion and swarming edge morphology were inspected after 24 h. Aggregation was monitored on CF agar [0.15% Casitone, 10 mM Tris-HCl (10), 10 mM KPO4, 8 mM MgSO4, 1.5 mM (NH4)2SO4, 0.1% NaPyruvate, 0.2% NaCitrate, 1.5% agar] and on TPM agar as described previously (14, 47). Briefly, cells were grown to a density of 5 × 108 cells/ml in CTT, harvested, and resuspended in TPM medium to a calculated density of 5 × 109 cells/ml. Aliquots (25 μl) of concentrated cells were spotted on CF agar or TPM agar and incubated at 32°C. For development in submerged culture, cells were treated as described for development on agar except that the cells were concentrated in MC7 buffer. Aliquots (25 μl) of concentrated cells were mixed with 375 μl of MC7 buffer and transferred to a microtiter well with a diameter of 15 mm and incubated at 32°C. Aggregation was monitored visually by using a Nikon Eclipse E800 microscope equipped with a Spot RT camera (Diagnostic Instruments). For the calculation of fruiting body roundness, the area and perimeter of individual fruiting bodies were measured from microscope images by using the ImageJ 1.30 software (developed at the U.S. National Institutes of Health and available at http://rsb.info.nih.gov/ij/). Roundness is calculated as 4π (area/perimeter2). A circle has a roundness of 1.0.
The efficiency of sporulation was determined as the number of sonication- and heat-resistant CFU, as described previously (14).
RESULTS AND DISCUSSION
Assembly and annotation of the M1genome.
To prepare for the microarray experiment, the contig sequences as received from Monsanto were placed in linear order, as specified in Materials and Methods. As one test of the quality of the annotation of the M1genome, the overall average gene density was calculated and was determined to be 0.83 genes/kbp. That density was compared with the gene density of a fully sequenced 36.6-kbp fragment of the M. xanthus genome covering the dsp/dif locus (GenBank accession no. AF449411), reported to have a total of 29 genes and a gene density of 0.79 genes/kbp (38). An examination of four other sequenced microbial genomes revealed gene densities between 0.87 genes/kbp for Streptomyces avermitilis and 0.97 genes/kbp for Bacillus subtilis. In view of a (slight) inverse correlation between genome size and gene density and given the more than 9-Mbp size of the M1genome, its gene density seems to fit with published bacterial sequence data.
To assess the accuracy of ORF assignments in the M1genome, the genome was used to select amplicons for an array of all M. xanthus genes. A list of 294 previously published M. xanthus genes was then compared to the M1genome list, and 98% of these were found among the selected amplicons, which is consistent with the estimate that the M1genome includes 97% of all M. xanthus sequence. The M1genome is available as a set of Artemis files at http://cmgm.stanford.edu/devbio/kaiserlab/.
Identification of EBPs.
ORFs encoding possible EBPs in the M1genome were identified by using the presence of the “Sigma54_activat” Pfam domain (accession number PF00158). This domain is involved in the ATP-dependent interaction with the σ54-holoenzyme that is common to all EBPs (2). A total of 53 predicted proteins in the M1genome were found to contain this domain with an E value less than or equal to 10−21. A TBlastN search (i.e., a protein query against a translated nucleotide database) using the σ54 activator domain from E. coli NtrC (amino acids 140 to 369) identified the same 53 proteins. All previously recognized EBP genes were among these 53, except for Mxa_249 (14).
Microarray experiment.
By 12 h poststarvation, M. xanthus will have evaluated its capacity for protein synthesis, synthesized ppGpp, and will have assessed the local cell concentration with A-signaling (21). It will have begun the C-signaling that initiates behavioral changes in motility and builds the fruiting body (27, 28). Early movements align cells, and by 12 h cells are streaming (25). Moreover, Horiuchi et al. have shown that by 12 h the synthesis rates of more than 150 proteins are down-regulated and more than 100 proteins are up-regulated with reference to growing cells (24). A microarray was constructed to investigate the expression of potential EBP genes by comparing total RNA samples from vegetatively growing cells with those from bacteria that had been starved for 12 h. The 12-h time point is expected to provide a representative sample of developmental gene regulation.
Table 1 lists all the differentially expressed genes found with an increase in expression of greater than 2.5-fold between 12-h developing cells and growing cells. Given the observed standard deviations shown for the genes in Table 1, a 2.5-fold change in the level of expression is statistically significant.
TABLE 1.
Genes with high developmental expression
| M1genome identifier | Gene | Increase in gene expression (n-fold) | SD | Descriptiona |
|---|---|---|---|---|
| Mx_2385 | cglB | 6.8 | 1.7 | A-motility gene |
| Mx_1813 | cheA3 | 6.8 | 2.1 | Histidine kinase (CheA family) |
| Ω4469 | 5.7 | 2.2 | B- and D-signal reporter | |
| Mx_1080 | Ω4442 | 5.4 | 1.8 | Developmental reporter |
| Mx_6506 | Ω4404 | 5.3 | 1.5 | Developmental reporter |
| Mx_5723 | 5.2 | 2.4 | Transcriptional regulator (GntR family) | |
| Mx_2284 | dsgA | 5.0 | 1.5 | D signal |
| Mx_5260 | asgE | 4.9 | 1.1 | A signal |
| Mx_4601 | asgA | 4.9 | 2.5 | A signal |
| Mx_1652 | 4.6 | 1.4 | Methylesterase (CheB family) | |
| Mx_3098 | 4.6 | 1.8 | σ54 EBP | |
| Mx_5366 | devS | 4.5 | 1.3 | C-signal-dependent gene |
| Mx_7294 | 4.1 | 1.7 | Transcriptional regulator (GerE family) | |
| Mx_4935 | espB | 4.1 | 1.2 | Timing of sporulation |
| Mx_1930 | sasR | 4.0 | 1.6 | σ54 EBP |
| Mx_5217 | 3.9 | 1.6 | Transcriptional regulator (LysR family) | |
| Mx_3899 | Ω4521 | 3.9 | 1.9 | A-signal reporter |
| Mx_6494 | 3.9 | 2.1 | Transcriptional regulator (TetR family) | |
| Mx_1409 | dnaC | 3.6 | 2.4 | Replication initiation |
| Mx_4934 | espA | 3.6 | 1.4 | Timing of sporulation |
| Mx_6588 | difB | 3.6 | 1.3 | S motility (Fibrils) |
| Mx_4072 | asgD | 3.5 | 2.2 | A signal |
| Mx_1227 | vegA | 3.4 | 1.3 | No homology |
| Mx_4330 | relA | 3.4 | 1.2 | ppGpp production |
| devR | 3.4 | 0.9 | C-signal-dependent gene | |
| Mx_5256 | sdeK | 3.4 | 1.0 | C-signal-dependent gene |
| Mx_2420 | 3.3 | 1.8 | ABC transporter | |
| Mx_3013 | pilR | 3.3 | 2.1 | σ54 EBP |
| Mx_5206 | 3.2 | 0.8 | Transcriptional regulator (Arg_repressor family) | |
| Mx_4548 | mbhA | 3.1 | 0.9 | Bacterial hemagglutinin |
| Mx_2765 | 3.1 | 1.2 | Transcriptional regulator (TetR family) | |
| Mx_6087 | Ω4403 | 3.1 | 0.9 | C-signal reporter |
| Mx_4756 | 3.1 | 0.9 | σ54 EBP | |
| Mx_5869 | 3.1 | 1.8 | Putative histidine kinase | |
| Mx_7214 | 3.0 | 1.1 | Transcriptional regulator (MarR family) | |
| Mx_3320 | 3.0 | 1.6 | σ54 EBP | |
| Mx_5367 | devT | 3.0 | 1.0 | C-signal-dependent gene |
| Mx_4918 | 2.9 | 1.8 | σ70 ECF | |
| Mx_1846 | 2.9 | 1.1 | Transcriptional regulator (ArsR family) | |
| Mx_2076 | 2.9 | 0.7 | Transcriptional regulator (trans_reg_C family) | |
| Mx_3132 | 2.9 | 0.9 | Histidine kinase (CheY family) | |
| Mx_6587.3 | difC | 2.9 | 0.5 | S motility (Fibrils) |
| Mx_1969 | pilP | 2.8 | 0.8 | S motility (Type IV pili) |
| Mx_1086 | spdR | 2.7 | 0.9 | σ54 EBP |
| Mx_2396 | 2.7 | 0.8 | Transcriptional regulator (Fis family) | |
| Mx_2391 | csgA | 2.6 | 0.7 | C signal |
| Mx_4590 | 2.6 | 0.9 | Polysaccharide synthesis | |
| Mx_5303 | dnaA | 2.6 | 0.7 | Replication initiation |
| Mx_2691 | 2.5 | 0.9 | Transcriptional regulator (AraC family) |
Function or annotation.
EBPs.
As described in the introduction, EBPs bind specific DNA enhancer sequences and hydrolyze ATP to allow σ54-holoenzyme to open the promoter (18, 54). One might expect that some of the 53 EBP genes on the array would be needed during growth and some during development. Table 1 shows that, after 12 h of starvation, six EBP genes have increased their expression by 2.5-fold or more over their growth levels. These include three previously characterized genes, sasR (17), spdR (20), and pilR (56). The sasR and spdR genes have been implicated in the A- and B-signaling processes, respectively, whereas the pilR gene regulates the expression of the pilin gene pilA that is important for cell movement during aggregation. No other developmental transcription data have been published for these genes, but Western blots of the SpdR protein indicate that it is present at about the same level during development as in growth (20). Some genes are expected to be down-regulated by development, but none met the criterion of having a change in the level of expression of ≥2.5-fold, given the standard deviations in Table 1. Additional experiments will be required to find expected down-regulated genes.
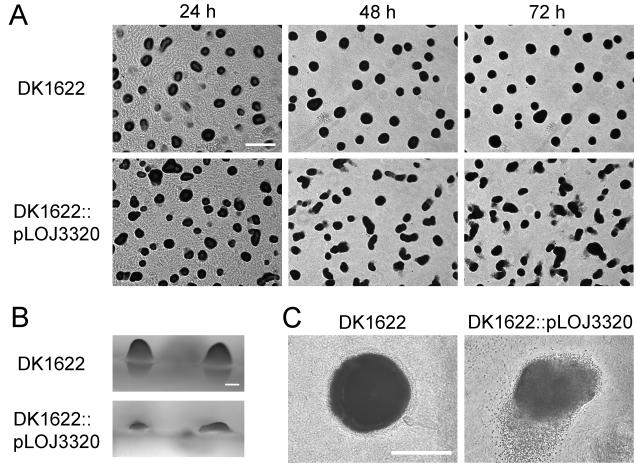
The transcription of three potentially new EBPs was observed to increase in the microarray experiment: Mx_3320, Mx_3098, and Mx_4756. We have disrupted two of these genes, and A. Garza has disrupted the third to see how they affect fruiting body development. Strains with a disruption of Mx_3320 are defective in mounding; that is, the mounds are less round than in strain DK1622 and often have a tail of lower density protruding from one end, as shown in Fig. 2. The mutant mounds had a roundness, which is defined in Materials and Methods, of 0.44 ± 0.19 (number of mounds, 305) compared with a roundness of 0.71 ± 0.08 (number of mounds, 236) for the mounds of strain DK1622. Also, the mutant fails to elevate a mound (Fig. 2B). In addition, the mutant strain sporulates about 12 h prematurely and in wrong positions. Spores are found not only within the aggregate, as for the wild type, but also around its periphery, often clustering as a tail at one end of the aggregate (Fig. 2C). At 48 h, the mutant presented 0.5 spores per sample whereas DK1622 did not; at 72 h, the mutant presented more than a 17-fold higher number of spores than strain DK1622. Significantly, these aggregation and sporulation abnormalities depend on the nutrient content of the medium used to support fruiting body development. Abnormalities were consistently observed on clone-fruiting (CF) medium which contains a low level of nutrients (19); defects were not observed on TPM medium or submerged in MC7 buffer, both of which are devoid of nutrient. More specifically, CF medium contains 0.15% Casitone, 1.5 mM (NH4)2SO4, 0.1% Na pyruvate, and 0.2% Na citrate. To test whether EBP Mx_3320 might respond to particular constituents of CF medium, single constituents were removed from CF medium or they were added to TPM medium. The effects on aggregation and sporulation are shown in Table 2. These results confirm a medium effect and suggest that it involves the source of nitrogen because the mutant develops normally on TPM agarose but abnormally on TPM agarose plus (NH4)Cl or (NH4)2SO4. The Mx_3320 disruptions exhibited normal motility during growth on both 0.4 and 1.5% agar plates, showing that the aggregation defect is not secondary to defective A or S motility. Disruption of Mx_3320 by plasmid integration could, in theory, have a polar effect on any downstream genes. But there is no downstream gene. The adjacent genes, Mx_3319 and Mx_3321, are both transcriptionally divergent from Mx_3320. Moreover, the three ORFs are close to each other, and there is little space between them for a cryptic gene. Thus, it is unlikely that the developmental defect is due to a polar effect.
FIG. 2.
Aggregation phenotypes of DK1622 (wild type) and DK1622::pLOJ3320 (Mx_3320 gene disruption mutant) on CF agar. (A) Top view of culture developing for the indicated time. Bar, 0.5 mm. (B) Side view of culture after 72 h. Bar, 0.1 mm. (C) Top view of culture after 72 h. Bar, 0.1 mm.
TABLE 2.
Fruiting body development of an Mx_3320-deficient mutant
| Medium | Aggregationa | Sporulationb |
|---|---|---|
| TPM agar | + | + |
| CF agar | − | d |
| CF agar lacking casitone | − | d |
| CF agar lacking (NH4)2SO4 | − | d |
| CF agar lacking pyruvate | − | d |
| CF agar lacking citrate | − | d |
| TPM agar + (NH4)Cl | − | d |
| TPM agar + (NH4)2SO4 | − | d |
| TPM agarose | − | d |
+, fruiting bodies similar to those of DK1622 in Fig. 1; −, aggregates of abnormal shape, similar to those of DK1622::pLOJ3320.
+, sporulation frequency and time like DK 1622 (see text); d, abnormal spore number or distribution (see text).
We observed no developmental phenotype after the disruption of Mx_3098 on either CF or TPM medium. Mx_4756 was disrupted by Anthony Garza at Washington State University, and his related experiments are published (3). He observed no phenotype of the disruption on TPM medium (A. Garza, personal communication).
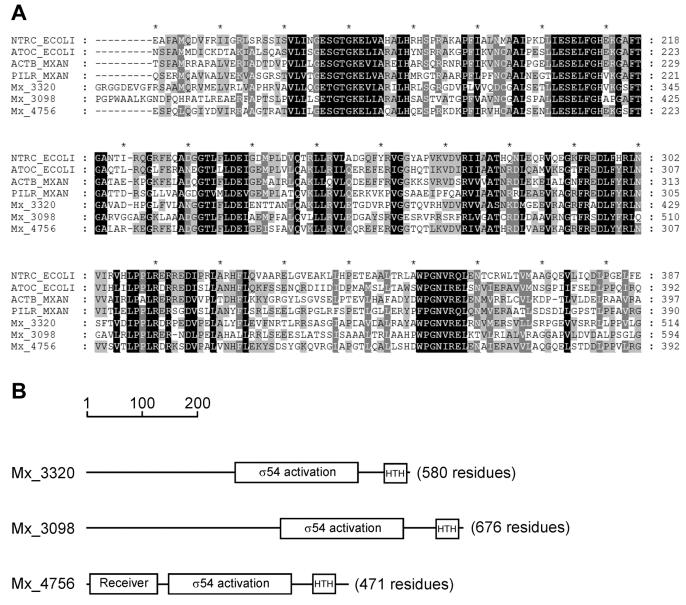
That Mx_3320, Mx_3098, and Mx_4756 encode EBPs is shown by the presence in each of a σ54 interaction domain (Pfam number 00158) (http://www.sanger.ac.uk/Software/Pfam/). Figure 3A displays an alignment of that domain with four established members of the EBP family, NtrC (41) and AtoC (4) from E. coli and the M. xanthus proteins ActB (13) and PilR (56). The alignment shows that the Mx_3320, Mx_3098, and Mx_4756 proteins have all the elements that are conserved among the established EBPs. In addition to a σ54 interaction domain, many EBPs have a C-terminal DNA binding domain thought to allow the protein to bind its specific DNA enhancer sequence. All three predicted proteins have a helix-turn-helix DNA binding domain (PF02954) located near their C termini (Fig. 3B). Some EBPs also have an N terminus that receives sensory input to regulate the activity of the protein (50). Only Mx_4756 had a recognizable signaling domain at its N terminus, a two-component response regulator receiver (PF00072) (Fig. 3B), suggesting that it may be modulated by sensor-kinase-mediated phosphorylation (43). Examples of EBPs with a domain architecture similar to that of Mx_4756 include NtrC and a regulatory protein of acetoacetate metabolism, AtoC from E. coli (50). Over the entire length of the protein, Mx_4756 is 44 and 38% identical to E. coli AtoC and NtrC, respectively. The N-terminal part of the predicted Mx_3098 protein (residues 65 to 213) is homologous, with 29% identity and 40% similarity, to the acetoin catabolism regulatory protein AcoR from Ralstonia eutrophus (35). Mx_3320 appears to be a sequence pioneer, because a BLAST search in January 2004 using the 250 N-terminal amino acids found no significant hits in the database.
FIG. 3.
Structure of the putative σ54 EBPs Mx_3320, Mx_3098, and Mx_4756. (A) Amino acid sequence alignment of the central portion of Mx_3320, Mx_3098, and Mx_4756 with the four documented σ54-EBPs E. coli NtrC (GenBank accession no. P06713), E. coli AtoC (GenBank accession no. Q06065), M. xanthus ActB (GenBank accession no. AAK38650), and M. xanthus PilR (GenBank accession no. S70529). Light gray bands indicate 60 to 79% similarity, dark gray bands indicate 80 to 99% similarity, and black indicates identity. The sequences were aligned by Clustal W (7) by using default settings; the alignment is presented by Genedoc (http://www.psc.edu/biomed/genedoc). (B) Domain architecture of Mx_3320, Mx_3098, and Mx_4756. The domains represented as boxes shown to scale were predicted by Pfam. Matches to known domains are indicated. σ54 activation, σ54 interaction domain (PF00158); Receiver, response regulator receiver domain (PF00072); HTH, helix-turn-helix domain (PF02954).
Motility proteins.
M. xanthus has two polar engines, called A and S, that propel its gliding motility (23). It moves to seek food for growth, and it moves to build its fruiting bodies. To assess the importance of movement during fruiting body development compared to growth, several components of both engines were included on the array. CglB is an outer membrane lipoprotein component of the A engine (22, 55). cglB expression is increased almost sevenfold during development (Table 1).
S motility is produced by the retraction of the polar type IV pili projecting from the leading end of the cell (29, 40, 53). Fibrils consisting of polysaccharide and protein provide the distal anchor sites for pili when they retract and are among the S-motility genes (11). Four S-motility genes were expressed more highly at 12 h of development: difB, difC, pilP, and pilR (Table 1). The dif mutants are fibril negative and pilus positive (57), while pilR (56) and pilP (E. Nudleman, unpublished data) mutants are pilus negative. Considering that developing cells are starving, these expression increases suggest that both A and S motility are given an even higher priority during development than they have during growth.
Cell-cell signaling and signal-dependent genes.
Progress from one developmental stage to the next is coordinated in M. xanthus by cell-to-cell signals. The A and C signals are extracellular, have been chemically identified, and are needed during the first 12 h of development. The bsg and dsg mutants can be rescued by codevelopment with wild-type cells (19), although no signal molecules have been identified for these two. The B-signal-dependent gene Ω4469 (33), the A-signal response gene sasR (17), and an A- and B-signal suppressor gene spdR (20) showed increased expression at 12 h of development (Table 1). The level of expression of the dsg gene, which encodes translation initiation factor 3 (IF3), was found to be increased by fivefold (Table 1). This increase suggests that M. xanthus may require more IF3 for the synthesis of developmental proteins than it does for growth that may be related to the stringent response. The dsg mutation, which is missense in a conserved region of IF3, allows normal growth but slows aggregation and delays sporulation (5, 6). It is as if there is more than enough IF3 for growth but a deficiency for development. The D-signal-dependent genes Ω4469 (which is also B dependent) and mbhA also increased their expression at 12 h (Table 1).
Of the A-signal genes which have been shown to be activated within a couple of hours of starvation (37), asgA, asgD, asgE, and sdeK showed increased expression on the microarray (Table 1). A-signal production depends on ppGpp and stringent control (21). This may explain why transcription of the relA gene is increased more than threefold at 12 h (Table 1). Expression of the structural gene for the C signal, csgA, also increased (Table 1), in accord with the measurements of C-signal biological activity, which increases during the course of development (15, 16, 39). The C-signal-dependent reporter Ω4403, whose lacZ expression increases at 12 h, is also seen to have increased transcription (Table 1). Three members of the dev operon, devT, devR, and devS, whose expression is C-signal dependent, are spatially localized to the fruiting body (26); these genes showed an increased level of expression at 12 h (Table 1). The dev operon appears to trigger sporulation (1, 52). The espAB operon, which affects the timing of sporulation (9), significantly increased expression at 12 h (Table 1). The espA gene may encode a histidine kinase (9). Another histidine kinase homolog that showed increased transcription is cheA3 (Table 1). CheA3 is thought to catalyze phosphorylation of the σ54 EBP, CrdA (32).
To conclude, by comparing RNA from rapidly growing cells with 12-h developing cells, increases of 2.5-fold or more in the expression levels were revealed in 11 transcription factors among the 371 genes deposited on the array. Among the 53 EBP homologs on the array, 6 showed increased expression levels at 12 h, suggesting that they play important roles in fruiting body development. Half of the six are new identifications, and they illustrate the power of microarrays for gene discovery in M. xanthus at this juncture. All three new genes were knocked out, and one (Mx_3320) seems to respond to the level of nutrient available. Although no phenotype was found for the knockouts of Mx_3098 or Mx_4756 on standard media, tests on a wider range of media seem in order, considering that the defect of Mx_3320 was only observed on other media and that M. xanthus has a need to manage its resources carefully to synthesize protein even as it starves.
Overall, the six EBPs whose expression increases point to a significant network of genes regulated by σ54 activator proteins that receive various sensory inputs. More work is needed to define those inputs. The identification of new EBP genes in the microarray experiment is validated by the three previously recognized σ54 activator proteins, SasR, SpdR, and PilR, whose expression was observed to increase in the same experiment. Also, the finding of expression increases in known cell-cell signaling genes and signal-responsive genes further validates the method of identification. Together, these validations for transcription factors add significance to the increased expression levels of A- and S-motility genes. Since there is extensive movement during growth, these significant increases suggest that cell movement in M. xanthus development is comparable in importance to changes in gene expression.
Acknowledgments
This work was supported by Public Health Service grant GM23441 to D.K. and by postdoctoral fellowship GM20356 to R.D.W., both from the National Institute of General Medical Sciences.
We thank the members of the Myxococcus Microarray Consortium for their evaluation of the M1genome.
REFERENCES
- 1.Boysen, A., E. Ellehauge, B. Julien, and L. Søgaard-Andersen. 2002. The DevT protein stimulates synthesis of FruA, a signal transduction protein required for fruiting body morphogenesis in Myxococcus xanthus. J. Bacteriol. 184:1540-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buck, M., M. T. Gallegos, D. J. Studholme, Y. Guo, and J. D. Gralla. 2000. The bacterial enhancer-dependent σ54 (σN) transcription factor. J. Bacteriol. 182:4129-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caberoy, N. B., R. D. Welch, J. S. Jakobsen, S. C. Slater, and A. G. Garza. 2003. Global mutational analysis of NtrC-like activators in Myxococcus xanthus: identifying activator mutants defective for motility and fruiting body development. J. Bacteriol. 185:6083-6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Canellakis, E., A. Paterakis, S. Huang, C. Panagiotidis, and D. Kyriakidis. 1993. Identification, cloning, and nucleotide sequencing of the ornithine decarboxylase antizyme gene of Escherichia coli. Proc. Natl. Acad. Sci. USA 90:7129-7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng, Y., and D. Kaiser. 1989. dsg, a gene required for cell-cell interaction early in Myxococcus development. J. Bacteriol. 171:3719-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, Y., and D. Kaiser. 1989. dsg, a gene required for Myxococcus development, is necessary for cell viability. J. Bacteriol. 171:3727-3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. Gibson, D. Higgins, and J. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho, K., A. Treuner-Lange, K. A. O'Connor, and D. R. Zusman. 2000. Developmental aggregation of Myxococcus xanthus requires frgA, an frz-related gene. J. Bacteriol. 182:6614-6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho, K., and D. R. Zusman. 1999. Sporulation timing in Myxococcus xanthus is controlled by the espAB locus. Mol. Microbiol. 34:714-725. [DOI] [PubMed] [Google Scholar]
- 10.Dufour, A., R. B. Furness, and C. Hughes. 1998. Novel genes that upregulate the Proteus mirabilis flhDC master operon controlling flagellar biogenesis and swarming. Mol. Microbiol. 29:741-751. [DOI] [PubMed] [Google Scholar]
- 11.Dworkin, M. 1999. Fibrils as extracellular appendages of bacteria: their role in contact-mediated cell-cell interactions in Myxococcus xanthus. BioEssays 21:590-595. [DOI] [PubMed] [Google Scholar]
- 12.Garza, A. G., J. S. Pollack, B. Z. Harris, A. Lee, I. M. Keseler, E. F. Licking, and M. Singer. 1998. SdeK is required for early fruiting body development in Myxococcus xanthus. J. Bacteriol. 180:4628-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorski, L., T. Gronewold, and D. Kaiser. 2000. A σ54 activator protein necessary for spore differentiation within the fruiting body of Myxococcus xanthus. J. Bacteriol. 182:2438-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorski, L., and D. Kaiser. 1998. Targeted mutagenesis of σ54 activator proteins in Myxococcus xanthus. J. Bacteriol. 180:5896-5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gronewold, T. M. A., and D. Kaiser. 2002. act operon control of developmental gene expression in Myxococcus xanthus. J. Bacteriol. 184:1172-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gronewold, T. M. A., and D. Kaiser. 2001. The act operon controls the level and time of C-signal production for M. xanthus development. Mol. Microbiol. 40:744-756. [DOI] [PubMed] [Google Scholar]
- 17.Guo, D., Y. Wu, and H. B. Kaplan. 2000. Identification and characterization of genes required for early Myxococcus xanthus developmental gene expression. J. Bacteriol. 182:4564-4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo, Y., C. M. Lew, and J. D. Gralla. 2000. Promoter opening by σ54 and σ70 RNA polymerases: σ factor-directed alterations in the mechanism and tightness of control. Genes Dev. 14:2242-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagen, D. C., A. P. Bretscher, and D. Kaiser. 1978. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev. Biol. 64:284-296. [DOI] [PubMed] [Google Scholar]
- 20.Hager, E., H. Tse, and R. E. Gill. 2001. Identification and characterization of spdR mutations that bypass the BsgA protease-dependent regulation of developmental gene expression in Myxococcus xanthus. Mol. Microbiol. 39:765-780. [DOI] [PubMed] [Google Scholar]
- 21.Harris, B. Z., D. Kaiser, and M. Singer. 1998. The guanosine nucleotide (p)ppGpp initiates development and A-factor production in Myxococcus xanthus. Genes Dev. 12:1022-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hodgkin, J., and D. Kaiser. 1979. Genetics of gliding motility in M. xanthus (Myxobacterales): genes controlling movement of single cells. Mol. Gen. Genet. 171:167-176. [Google Scholar]
- 23.Hodgkin, J., and D. Kaiser. 1979. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): two gene systems control movement. Mol. Gen. Genet. 171:177-191. [Google Scholar]
- 24.Horiuchi, T., M. Taoka, T. Isobe, T. Komano, and S. Inouye. 2002. Role of fruA and csgA genes in gene expression during development of Myxococcus xanthus: analysis by two-dimensional gel electrophoresis. J. Biol. Chem. 277:26753-26760. [DOI] [PubMed] [Google Scholar]
- 25.Jelsbak, L., and L. Sogaard-Andersen. 2002. Pattern formation by a cell surface-associated morphogen in M. xanthus. Proc. Natl. Acad. Sci. USA 99:2032-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Julien, B., A. D. Kaiser, and A. Garcia. 2000. Spatial control of cell differentiation in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 97:9098-9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaiser, D. 2003. Coupling cell movement to multicellular development in myxobacteria. Nat. Rev. Microbiol. 1:45-54. [DOI] [PubMed] [Google Scholar]
- 28.Kaiser, D. 2004. Signaling in Myxobacteria. Annu. Rev. Microbiol. 58:75-98. [DOI] [PubMed] [Google Scholar]
- 29.Kaiser, D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 76:5952-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keseler, I. M., and D. Kaiser. 1995. An early A-signal-dependent gene in Myxococcus xanthus has a σ54-like promoter. J. Bacteriol. 177:4638-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keseler, I. M., and D. Kaiser. 1997. σ54, a vital protein for Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 94:1979-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirby, J. R., and D. R. Zusman. 2003. Chemosensory regulation of developmental gene expression in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 100:2008-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kroos, L., and D. Kaiser. 1987. Expression of many developmentally regulated genes in Myxococcus depends on a sequence of cell interactions. Genes Dev. 1:840-854. [DOI] [PubMed] [Google Scholar]
- 34.Kroos, L., A. Kuspa, and D. Kaiser. 1986. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev. Biol. 117:252-266. [DOI] [PubMed] [Google Scholar]
- 35.Kruger, N., and A. Steinbuchel. 1992. Identification of acoR, a regulatory gene for the expression of genes essential for acetoin catabolism in Alcaligenes eutrophus H16. J. Bacteriol. 174:4391-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuspa, A., and D. Kaiser. 1989. Genes required for developmental signaling in Myxococcus xanthus: three asg loci. J. Bacteriol. 171:2762-2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuspa, A., L. Kroos, and D. Kaiser. 1986. Intercellular signaling is required for developmental gene expression in Myxococcus xanthus. Dev. Biol. 117:267-276. [DOI] [PubMed] [Google Scholar]
- 38.Lancero, H., J. E. Brofft, J. Downard, B. W. Birren, C. Nusbaum, J. Naylor, W. Shi, and L. J. Shimkets. 2002. Mapping of Myxococcus xanthus social motility dsp mutations to the dif genes. J. Bacteriol. 184:1462-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, S., B. U. Lee, and L. Shimkets. 1992. csgA expression entrains Myxococcus xanthus development. Genes Dev. 6:401-410. [DOI] [PubMed] [Google Scholar]
- 40.Merz, A. J., M. So, and M. P. Sheetz. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98-102. [DOI] [PubMed] [Google Scholar]
- 41.Miranda-Rios, J., R. Sanchez-Pescador, M. Urdea, and A. A. Covarrubias. 1987. The complete sequence of the glnALG operon of Escherichia coli K12. Nucleic Acids Res. 15:2757-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mueller, C., and M. Dworkin. 1991. Effects of glucosamine on lysis, glycerol formation, and sporulation in Myxococcus xanthus. J. Bacteriol. 173:7164-7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parkinson, J. S., and E. C. Kofoid. 1992. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 26:71-112. [DOI] [PubMed] [Google Scholar]
- 44.Romeo, J. M., and D. R. Zusman. 1991. Transcription of the myxobacterial hemagglutinin gene is mediated by a σ54-like promoter and a cis-acting upstream regulatory region of DNA. J. Bacteriol. 173:2969-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M.-A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Shimkets, L., and D. Kaiser. 1982. Induction of coordinated movement of Myxococcus xanthus cells. J. Bacteriol. 152:451-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimkets, L. J. 1993. The myxobacterial genome, p. 85-107. In M. Dworkin and D. Kaiser (ed.), Myxobacteria II. American Society for Microbiology, Washington, D.C.
- 49.Shimkets, L. J. 1990. Social and developmental biology of the Myxobacteria. Microbiol. Rev. 54:473-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Studholme, D. J., and R. Dixon. 2003. Domain architectures of σ54-dependent transcriptional activators. J. Bacteriol. 185:1757-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun, H., and W. Shi. 2001. Analyses of mrp genes during Myxococcus xanthus development. J. Bacteriol. 183:6733-6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thöny-Meyer, L., and D. Kaiser. 1993. devRS, an autoregulated and essential genetic locus for fruiting body development in Myxococcus xanthus. J. Bacteriol. 175:7450-7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wall, D., and D. Kaiser. 1999. Type IV pili and cell motility. Mol. Microbiol. 32:1-10. [DOI] [PubMed] [Google Scholar]
- 54.Weiss, D. S., J. Batut, K. E. Klose, J. Keener, and S. Kustu. 1991. The phosphorylated form of the enhancer-binding protein NTRC has an ATPase activity that is essential for activation of transcription. Cell 67:155-167. [DOI] [PubMed] [Google Scholar]
- 55.Wolgemuth, C., E. Hoiczyk, D. Kaiser, and G. Oster. 2002. How myxobacteria glide. Curr. Biol. 12:1-20. [DOI] [PubMed] [Google Scholar]
- 56.Wu, S. S., and D. Kaiser. 1997. Regulation of expression of the pilA gene in Myxococcus xanthus. J. Bacteriol. 179:7748-7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang, Z., X. Ma, L. Tong, H. B. Kaplan, L. J. Shimkets, and W. Shi. 2000. The Myxococcus xanthus dif genes are required for the biogenesis of cell surface fibrils essential for social gliding motility. J. Bacteriol. 182:5793-5798. [DOI] [PMC free article] [PubMed] [Google Scholar]