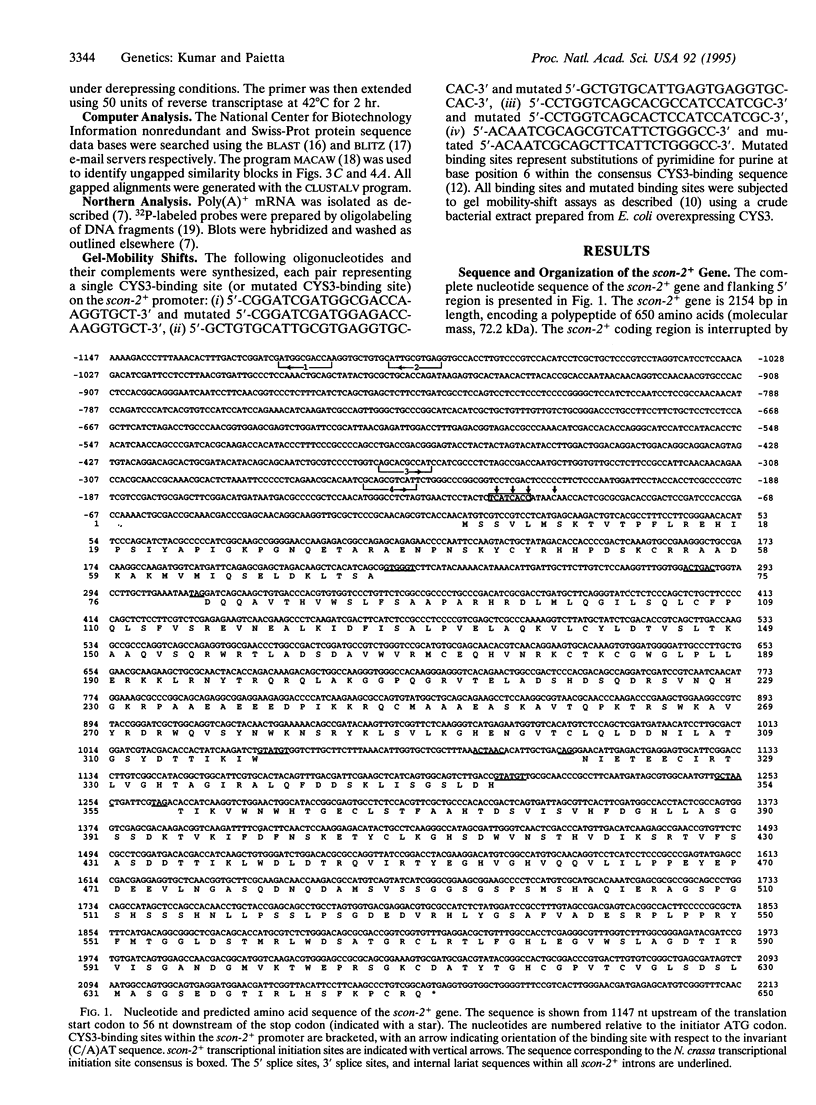
Abstract
The sulfur regulatory system of Neurospora crassa is composed of a set of structural genes involved in sulfur catabolism controlled by a genetically defined set of trans-acting regulatory genes. These sulfur regulatory genes include cys-3+, which encodes a basic region-leucine zipper transcriptional activator, and the negative regulatory gene scon-2+. We report here that the scon-2+ gene encodes a polypeptide of 650 amino acids belonging to the expanding beta-transducin family of eukaryotic regulatory proteins. Specifically, SCON2 protein contains six repeated G beta-homologous domains spanning the C-terminal half of the protein. SCON2 represents the initial filamentous fungal protein identified in the beta-transducin group. Additionally, SCON2 exhibits a specific amino-terminal domain that potentially defines another subfamily of beta-transducin homologs. Expression of the scon-2+ gene has been examined using RNA hybridization and gel mobility-shift analysis. The dependence of scon-2+ expression on CYS3 function and the binding of CYS3 to the scon-2+ promoter indicate the presence of an important control loop within the N. crassa sulfur regulatory circuit involving CYS3 activation of scon-2+ expression. On the basis of the presence of beta-transducin repeats, the crucial role of SCON2 in the signal-response pathway triggered by sulfur limitation may be mediated by protein-protein interactions.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Burton E. G., Metzenberg R. L. Novel mutation causing derepression of several enzymes of sulfur metabolism in Neurospora crassa. J Bacteriol. 1972 Jan;109(1):140–151. doi: 10.1128/jb.109.1.140-151.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X. W., Matsui M., Wei N., Wagner D., Chu A. M., Feldmann K. A., Quail P. H. COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell. 1992 Nov 27;71(5):791–801. doi: 10.1016/0092-8674(92)90555-q. [DOI] [PubMed] [Google Scholar]
- Duronio R. J., Gordon J. I., Boguski M. S. Comparative analysis of the beta transducin family with identification of several new members including PWP1, a nonessential gene of Saccharomyces cerevisiae that is divergently transcribed from NMT1. Proteins. 1992 May;13(1):41–56. doi: 10.1002/prot.340130105. [DOI] [PubMed] [Google Scholar]
- Dynlacht B. D., Weinzierl R. O., Admon A., Tjian R. The dTAFII80 subunit of Drosophila TFIID contains beta-transducin repeats. Nature. 1993 May 13;363(6425):176–179. doi: 10.1038/363176a0. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fong H. K., Hurley J. B., Hopkins R. S., Miake-Lye R., Johnson M. S., Doolittle R. F., Simon M. I. Repetitive segmental structure of the transducin beta subunit: homology with the CDC4 gene and identification of related mRNAs. Proc Natl Acad Sci U S A. 1986 Apr;83(7):2162–2166. doi: 10.1073/pnas.83.7.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. H., Paietta J. V., Mannix D. G., Marzluf G. A. cys-3, the positive-acting sulfur regulatory gene of Neurospora crassa, encodes a protein with a putative leucine zipper DNA-binding element. Mol Cell Biol. 1989 Mar;9(3):1120–1127. doi: 10.1128/mcb.9.3.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley D. A., Preiss A., Artavanis-Tsakonas S. A deduced gene product from the Drosophila neurogenic locus, enhancer of split, shows homology to mammalian G-protein beta subunit. Cell. 1988 Dec 2;55(5):785–795. doi: 10.1016/0092-8674(88)90134-1. [DOI] [PubMed] [Google Scholar]
- Icho T., Wickner R. B. The MAK11 protein is essential for cell growth and replication of M double-stranded RNA and is apparently a membrane-associated protein. J Biol Chem. 1988 Jan 25;263(3):1467–1475. [PubMed] [Google Scholar]
- Keleher C. A., Redd M. J., Schultz J., Carlson M., Johnson A. D. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992 Feb 21;68(4):709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- Legerton T. L., Yanofsky C. Cloning and characterization of the multifunctional his-3 gene of Neurospora crassa. Gene. 1985;39(2-3):129–140. doi: 10.1016/0378-1119(85)90306-3. [DOI] [PubMed] [Google Scholar]
- Marzluf G. A. Regulation of sulfur and nitrogen metabolism in filamentous fungi. Annu Rev Microbiol. 1993;47:31–55. doi: 10.1146/annurev.mi.47.100193.000335. [DOI] [PubMed] [Google Scholar]
- Metzenberg R. L. Implications of some genetic control mechanisms in Neurospora. Microbiol Rev. 1979 Sep;43(3):361–383. doi: 10.1128/mr.43.3.361-383.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neer E. J., Schmidt C. J., Nambudripad R., Smith T. F. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994 Sep 22;371(6495):297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- Paietta J. V., Akins R. A., Lambowitz A. M., Marzluf G. A. Molecular cloning and characterization of the cys-3 regulatory gene of Neurospora crassa. Mol Cell Biol. 1987 Jul;7(7):2506–2511. doi: 10.1128/mcb.7.7.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paietta J. V. Molecular cloning and analysis of the scon-2 negative regulatory gene of Neurospora crassa. Mol Cell Biol. 1990 Oct;10(10):5207–5214. doi: 10.1128/mcb.10.10.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paietta J. V. Molecular cloning and regulatory analysis of the arylsulfatase structural gene of Neurospora crassa. Mol Cell Biol. 1989 Sep;9(9):3630–3637. doi: 10.1128/mcb.9.9.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paietta J. V. Production of the CYS3 regulator, a bZIP DNA-binding protein, is sufficient to induce sulfur gene expression in Neurospora crassa. Mol Cell Biol. 1992 Apr;12(4):1568–1577. doi: 10.1128/mcb.12.4.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner O., Carrozzo R., Shen Y., Wehnert M., Faustinella F., Dobyns W. B., Caskey C. T., Ledbetter D. H. Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature. 1993 Aug 19;364(6439):717–721. doi: 10.1038/364717a0. [DOI] [PubMed] [Google Scholar]
- Rice C. M., Fuchs R., Higgins D. G., Stoehr P. J., Cameron G. N. The EMBL data library. Nucleic Acids Res. 1993 Jul 1;21(13):2967–2971. doi: 10.1093/nar/21.13.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D., Chen C. H., Caldwell J., Jamieson L., Orr E., Mochly-Rosen D. Cloning of an intracellular receptor for protein kinase C: a homolog of the beta subunit of G proteins. Proc Natl Acad Sci U S A. 1994 Feb 1;91(3):839–843. doi: 10.1073/pnas.91.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler G. D., Altschul S. F., Lipman D. J. A workbench for multiple alignment construction and analysis. Proteins. 1991;9(3):180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- Sethi N., Monteagudo M. C., Koshland D., Hogan E., Burke D. J. The CDC20 gene product of Saccharomyces cerevisiae, a beta-transducin homolog, is required for a subset of microtubule-dependent cellular processes. Mol Cell Biol. 1991 Nov;11(11):5592–5602. doi: 10.1128/mcb.11.11.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood P. W., Tsang S. V., Osley M. A. Characterization of HIR1 and HIR2, two genes required for regulation of histone gene transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Jan;13(1):28–38. doi: 10.1128/mcb.13.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spevak W., Keiper B. D., Stratowa C., Castañn M. J. Saccharomyces cerevisiae cdc15 mutants arrested at a late stage in anaphase are rescued by Xenopus cDNAs encoding N-ras or a protein with beta-transducin repeats. Mol Cell Biol. 1993 Aug;13(8):4953–4966. doi: 10.1128/mcb.13.8.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams F. E., Trumbly R. J. Characterization of TUP1, a mediator of glucose repression in Saccharomyces cerevisiae. Mol Cell Biol. 1990 Dec;10(12):6500–6511. doi: 10.1128/mcb.10.12.6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Voorn L., Ploegh H. L. The WD-40 repeat. FEBS Lett. 1992 Jul 28;307(2):131–134. doi: 10.1016/0014-5793(92)80751-2. [DOI] [PubMed] [Google Scholar]