Abstract
Until recently, acquired resistance to cytostatics had mostly been attributed to biochemical mechanisms such as decreased intake and/or increased efflux of therapeutics, enhanced DNA repair, and altered activity or deregulation of target proteins. Although these mechanisms have been widely investigated, little is known about membrane barriers responsible for the chemical imperviousness of cell compartments and cellular segregation in cytostatic-treated tumors. In highly heterogeneous cross-resistant and radiorefractory cell populations selected by exposure to anticancer agents, we found a number of atypical recurrent cell types in (1) tumor cell cultures of different embryonic origins, (2) mouse xenografts, and (3) paraffin sections from patient tumors. Alongside morphologic peculiarities, these populations presented cancer stem cell markers, aberrant signaling pathways, and a set of deregulated miRNAs known to confer both stem-cell phenotypes and highly aggressive tumor behavior. The first type, named spiral cells, is marked by a spiral arrangement of nuclei. The second type, monastery cells, is characterized by prominent walls inside which daughter cells can be seen maturing amid a rich mitochondrial environment. The third type, called pregnant cells, is a giant cell with a syncytium-like morphology, a main nucleus, and many endoreplicative functional progeny cells. A rare fourth cell type identified in leukemia was christened shepherd cells, as it was always associated with clusters of smaller cells. Furthermore, a portion of resistant tumor cells displayed nuclear encapsulation via mitochondrial aggregation in the nuclear perimeter in response to cytostatic insults, probably conferring imperviousness to drugs and long periods of dormancy until nuclear eclosion takes place. This phenomenon was correlated with an increase in both intracellular and intercellular mitochondrial traffic as well as with the uptake of free extracellular mitochondria. All these cellular disorders could, in fact, be found in untreated tumor cells but were more pronounced in resistant entities, suggesting a natural mechanism of cell survival triggered by chemical injury, or a primitive strategy to ensure stemming, self-renewal, and differentiation under adverse conditions, a fact that may play a significant role in chemotherapy outcomes.
Background
Acquired multidrug resistance is defined as the refractiveness of tumors to multiple xenobiotics and can be conferred by noncellular and cellular mechanisms, which appear to be evolutionary strategies involved in the detoxification of organisms to ensure survival. Noncellular mechanisms include poor vascularization of tumors as well as the colonization of niches, resulting in limited accessibility of drugs and hypoxic environments that promote tumor growth via stemness. Cellular mechanisms encompass nonclassical biochemical processes such as DNA repair, altered activity or overexpression of target proteins, and cellular detoxification systems, for example, glutathione. Classical biochemical processes include the efflux of xenobiotics by energy-dependent proteins such as ABC-type transporters counteracting the buildup of therapeutic intracellular concentrations. Since many organ systems require a high expression of such transport proteins in order to maintain physiological integrity, the administration of classical or tailored multiple drug resistance (MDR)-modulators to overcome multidrug resistance often results in therapy failure due to fatal systemic toxicity (Tannock, 2001; Donnenberg and Donnenberg, 2005; Lu and Shervington, 2008; Nakai et al., 2009; Sabisz and Skladanowski, 2009; Baguley, 2010; Gillet and Gottesman, 2010; Bao et al., 2012, 2014; Chau and Figg, 2012; Cheng et al., 2012; De Palma and Nucera, 2012; Hu et al., 2012; Cao et al., 2013; Chekhonin et al., 2013; Prigione et al., 2014).
Current cancer therapies may also be rendered inefficient by the selection or induction of cancer stem cells (CSCs) and the associated cellular heterogeneity (Hansen et al., 1997; Erenpreisa et al., 2008; Bartkowiak et al., 2009; McCord et al., 2009; Díaz-Carballo et al., 2010; Mo and Zhang, 2012; Mascre et al., 2012). The still evolving term CSC is based on both in vivo and in vitro tumor-initiating processes. It is assumed that CSCs derive from normal stem cells which undergo successive re-programming steps in response to physical, biological, or chemical stress, resulting in the generation of diverse cell phenotypes with a hierarchical structure (Walton et al., 2004; Spees et al., 2006; Lobo et al., 2007; Harris et al., 2008; Bartkowiak et al., 2009; Nakai et al., 2009; Sabisz and Skladanowski, 2009; Díaz-Carballo et al., 2010; Marques et al., 2010; Kitambi, 2011; Economopoulou et al., 2012; Mascre et al., 2012; Martin-Padura et al., 2012; Shyh-Chang et al., 2013; Zheng et al., 2013). While the presence of heterogeneity in tumors regardless of the therapy status is a well-documented fact, it should be emphasized that it can be increased by therapeutics. The administration of anticancer agents leads to the selection and clonal expansion of morphologically atypical malignant stem cell-like cells that then boost cellular heterogeneity and disseminate to produce new tumors. Common phenomena associated with cytostatic and irradiation-induced heterogeneity are endoreplication (Plasmodia) and cell–cell fusions (Syncytia), both of which give rise to multinucleated entities (Merlin et al., 2000; Makarovskiy et al., 2002; Díaz-Carballo et al., 2008a; Weihua et al., 2011). Some of these multinucleated cells will suffer a mitotic catastrophe and consequently have short lifespans, while others survive under persistent cell division, which promotes tumor resistance, regrowth, and tumor progression. Such polyploidization events as a result of DNA damage can be reversible when TP53 is absent or dysfunctional. Moreover, they are associated with reversible senescence, generating mitotically cycling survivors that provide clonogenicity in vitro and rapid malignant growth in vivo (Merlin et al., 2000; Erenpreisa and Cragg, 2007, 2013; Díaz-Carballo et al., 2008a; Erenpreisa et al., 2008; Weihua et al., 2011). To explain the events in cell survivors after genotoxic exposure, a theory called neosis has emerged in recent years. Neosis is considered as occurring in multinucleated postsenescent cells and as being characterized by karyokinesis via nuclear budding and asymmetric cytokinesis, producing aneuploid mononuclear cells with extended lifespans and transient stem cell features. It is believed that polyploid mother cells die after these events (Rajaraman et al., 2006; Erenpreisa and Cragg, 2013). Consequently, genotoxic resistance seems to be closely associated with reversible senescence, polyploidization, and stemness. Certain events observed during the evolution of cell heterogeneity from multinucleated polyploid cells appear to be linked to the oncogerminative theory of tumor formation suggested by Vinnitsky, who elegantly proposed that the malignant transformation of somatic cells is based on the activation of embryogenic programs that confer phenotypical features of germ cells, the ability to multiply via parthenogenesis, and the formation of oncospheroids containing three different cell types: oncogerminative (stem), oncotrophoblastic, and oncosomatic entities. Oncospheroids pass through two phases: vascularization (in vivo) and the development of metastatic secondary cells (Vinnitsky, 1993). This process can be regarded as an analogy to the embryonic migration and further differentiation of germ cells in a strictly coordinated balance of senescence/apoptosis and cell survival. In fact, it has been proposed that the role of the oncogene mos, a key driver of meiosis in animals, could be crucial in spawning oocyte features in somatic cells, as observed in DNA-damaged cells without functional TP53 (Erenpreisa and Cragg, 2007, 2013).
Hence, polyploid cells resulting from anticancer treatments and their proliferative potential may play an important role in tumor persistence and metastasis formation (Erenpreisa and Cragg, 2007, 2013). Unfortunately, the post-therapy recurrence of multinucleated atypical cells and its clinical implications has been underestimated by the scientific community. The biological heterogeneity of the resulting cells changes the landscape of the molecular biology and physiology of the whole tumor. In this regard, Molina et al. as well as our own group recently established a link between endopolyploidy and CSCs. Molina has characterized, from the point of view of the CSC phenotype, a kind of highly invasive polyploid doughnut-like glioblastoma cells but did not establish a relationship with therapy resistance (Molina et al., 2010).
In order to isolate CSCs, they have to be unequivocally identified. However, the presence of membrane proteins and specific stem cell markers is not a reliable distinguishing feature between CSCs and non-CSCs so that, in order to enrich CSCs, a different approach based on the exclusion of a “side population” (SP) by Hoechst 33258 staining has been established. It is believed that SP fractions are sorted out by the most straightforward mechanism, that is, the efflux of xenobiotics via ABC transporters, a common feature of both normal stem cells and CSCs. Contrary to that, recent reports revealed that CSC phenotyping should not solely be based on SP exclusion (Patrawala et al., 2005; Molina et al., 2010; Broadley et al., 2011; Richard et al., 2013). Nevertheless, it remains questionable whether this method is appropriate for the isolation of multinucleated cells, as they are larger and therefore more fragile than cells of the main population.
A virgin aspect in oncology is the role of mitochondria in acquired resistance. Mitochondria are highly dynamic organelles that move inside the cell along microtubules in an anterograde way driven by the kinesin-1 motor (KHC, Kif5b) and a dynein-driven retrograde mechanism. Moreover, mitochondria can shuttle between cells, suggesting an exchange of genetic and protein material (Spees et al., 2006; Pasquier et al., 2013). Of particular relevance is the transfer of mitochondria from endothelial to cancer cells and its role in chemoresistance, a process that is not yet fully understood. An important link between mitochondria and multinucleated cells can be found in the work of Chan et al. in which they analyzed different populations of mitochondria for mitofusin (MFN1/2) expression in placental trophoblasts. Mitofusins are nuclear encoded proteins involved in mitochondrial fusion–fission processes. Fusion is likely to safeguard mitochondrial functioning by mixing contents, whereas fission ensures a uniform distribution along cytoskeletal axes. Placental syncytiotrophoblasts hold a mitochondria population which is particularly rich in mitofusin-2 (MFN2), a fact that is strongly correlated to the genesis of these giant multinucleated cell entities and the outcome of embryonic development. It is worthy to note that placental trophoblasts show stemness in their lineage development (Koch et al., 2012). Furthermore, during fertilization and embryogenesis, the mitochondria adopt a perinuclear arrangement, creating a spatial proximity to the nucleus that makes the coordinated cross-talk between both organelles more efficient. This suggests that energy-requiring processes such as the differentiation of pluripotent cells and CSCs depend on the specific spatial arrangement of mitochondria to supply the energy necessary to sustain specialized functions in the different tissues (Chan et al., 2006, 2006a, 2006b, 2007; Spees et al., 2006; Xu et al., 2013).
Besides the physiological distribution of mitochondria around the nucleus (subsarcolemmal mitochondria), the formation of net-like perinuclear structures consisting of elongated mitochondria has recently been reported to be induced by mitochondrial fusion inhibitors that are capable of decreasing the efficiency of Yamanaka's three-factor cocktail (OCT4, KLF4, and SOX2) by more than 95% in fibroblast reprogramming to pluripotency (Vazquez-Martin et al., 2012).
Another aspect that remains poorly understood with regard to CSCs is the post-therapy nuclear fate of multinucleated cells and the destiny of tumor nuclei in general. Differences in nuclear staining observed by Lison in the 1950s probably provide an important hint about differences of cell populations in their nuclear status and could be related to the sub-population identified by Hoechst 33258 (Lison, 1955).
Materials and Methods
Cell cultures and determination of IC50 values
Cells employed in this study were obtained from the cell and tumor bank of the University Duisburg Essen, Medical School. Wild-type (WT) and induced cancer stem cells (iCSCs) were cultured in DMEM medium (Invitrogen) containing 10% heat-inactivated fetal calf serum and 15 μg/mL Ciprobay (Bayer AG). Cells stably expressing green fluorescent protein (GFP) were previously established in our lab using the vectors TurboGFP (BioCat GmbH) that were transfected using the TurboFect Trasfection Reagent (Thermo Scientific, Inc.) as per standard protocol.
IC50 values were determined using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) proliferation assay as previously described, and reported as the mean of three independent experiments. Briefly, cells in exponential growth phase were harvested, washed with medium, and seeded in 96-well plates at appropriate densities according to their growth kinetics. After a conditioning period of 24 h, cells were exposed to increasing concentrations of cytostatics for 72 h. The cultures were then incubated with MTT (Sigma-Aldrich) and dissolved in phosphate-buffered saline (PBS) at a final concentration of 1 mg/mL for 4 h. Supernatants were aspirated, and the purple formazan crystals were dissolved in 100 μL of solubilization solution (10% SDS in DMSO; Sigma-Aldrich). The absorbance was measured in a microtiter plate reader (Infinite F200 Tecan) at 570 nm (Díaz-Carballo et al., 2014).
Time-lapse videography, microscopic analysis, and cell structure measurements
For the documentation of cell behavior by direct light microscopy, we used a TE2000 Nikon and a DMI Leica inverted contrasting microscope coupled with a PeCom incubator (5% CO2, 37°C). Immunostained sections were examined on a Nikon Eclipse 50i microscope and a DS-Fi1c camera. Images were processed with the NIS-Elements BR software (Nikon Vision). Area and diameter measurements of cell structures were also carried out by applying the tool Annotations and Measurements for manual measurement enclosed in the same software.
Induction of CSCs from tumor cell lines by exposure to etoposide
Induction of CSCs from tumor cell lines was performed by the selection of subpopulations using etoposide as previously described (Díaz-Carballo et al., 2010, 2014). Briefly, IC50 values for cytostatics were determined by MTT assay. Exponentially growing cells were then exposed to 2×IC50 for 24 h. For recovery, cells were washed and incubated with drug-free culture medium until new colonies had formed. This procedure was repeated several times, each time doubling the original IC50 until 64×IC50 was reached. The surviving cells were subjected to a resistance selection by incubation with increasing concentrations of the respective drugs (16× to 512×IC50) for 24 h. Cells that proliferated at higher drug concentrations (128×) within 1 week were considered chemotherapy refractory. Resistant colonies were then expanded in the continuous presence of cytostatics and used for molecular-biological analysis, in particular for studying the expression of CSC features. The resistance factor was determined by MTT proliferation assay and reported as the IC50 iCSCs/IC50 parental ratio.
Induction of mitochondrial accumulation in the perinuclear compartments using etoposide
Near-confluent U87 human glioblastoma cells were stressed with 1–10 μg/mL etoposide for at least 30 days. After this time, nuclear encapsulation could be directly observed by light microscopy as a dense mass of mitochondria in the perinuclear compartment. Cells could be cultured under these conditions for long periods if the medium was replaced from time to time. With slight differences, metaplastic phases could be observed in cell culture for long periods. Mitochondrial accumulation was visualized by incubating the cells with MTT at a final concentration of 1 mg/mL.
Isolation of mitochondrial and nuclear fractions and labeling of mitochondria with GFP
Mitochondria and nuclei were isolated according to standard protocols that permitted the recovery of functionally intact organelles (Franko et al., 2013). U87 mitochondria-providing cells were grown to 90% confluence. The medium was discarded, and cultures were rinsed twice in cold PBS. Then, PBS was completely removed and 1.4 mL prechilled lysis buffer (0.01 M Tris-HCl pH 7.8, 0.001 M EGTA, 0.250 M sucrose, and 1% bovine serum albumin [BSA]) was added while keeping the flasks on ice for a few minutes. Cells were completely detached using a scraper, transferred to a prechilled Dounce homogenizator, and incubated on ice for another 15 min. Afterward, they were homogenized on ice by applying 40 strokes. Cell homogenates were transferred to 2 mL Eppendorf tubes and centrifuged for 5 min at 2500 g, 4°C (nuclear fraction). The upper phase was transferred to 2-mL tubes; the pellet was resupended in 700 μL lysis buffer, transferred to the douncer, homogenized, and spun down by applying the same conditions. The upper phase containing the mitochondrial fraction was centrifuged for 5 min at 5000 g, 4°C and the resulting pellet was maintained on ice. Supernatants were transferred to new 2-mL tubes and examined by light microscopy for nuclear contamination. The mitochondrial fraction was spun down for 15 min at 10,000 g, 4°C. The mitochondrial pellet was washed once in 700 μL lysis buffer and again centrifuged at 10,000 g for 15 min. A small portion of the mitochondrial and nuclear fractions was lysated in radioimmunoprecipitation assay (RIPA) buffer, and the protein content was determined by Pierce BCA Protein Assay Kit (Thermo Scientific, Inc.). In order to define the organelle content, the protein concentration was adjusted to 10 mg/mL. Physiological experiments were performed immediately after organelle isolation. Protein extracts were portioned and stored at −80°C for later western blot (WB) analysis. For cell experiments, mitochondrial fractions were resuspended in DMEM medium and added to the cultures. Mitochondrial pellets were then resuspended in 1.6 mL DMEM medium in 5-mL tubes and kept at 37°C. Transfection mixes were previously prepared by employing 160 μL serum-free DMEM, 1.6 μg pTurboGFP-C vector (Evrogen), and 3.2 μL TurboFect Reagent (Thermo Scientific, Inc.). These mixes were incubated for 20 min at room temperature and afterward, they were gently added to 1.6 mL mitochondrial fractions. The resulting mixes were then incubated for 2 h at 37°C under culture conditions. After this step, the mitochondria were spun down as described earlier. Pellets containing organelles were resuspended in 20 μL DNAse buffer (10 mM Tris-HCl pH 7.6, 2.5 mM NaCl2, and 0.5 mM CaCl2). DNAse I (2 U) was added to the suspensions and incubated in a water bath for 10 min at 37°C. Thereafter, 1 mL DMEM was added and the samples were placed on ice. Mitochondria were centrifuged as described, and the pellets were resuspended in DMEM medium for culture purposes.
Mitochondrial labeling with MitoTracker
To track exogenous mitochondria and their uptake by the cells, organelles extracted from U87 cells were labeled with 0.01 M green or red MitoTracker (Life Technologies) in serum-free medium at 37°C for 60 min.
Hoechst 33258 dye uptake
U87 cells were incubated with 0.05 μg/mL Hoechst 33258 fluorescent dye, and the cellular incorporation of this fluorescent tracer was continuously monitored at 465 nm for 6 h at 37°C in an Infinite F200 reader (Tecan).
Ionizing irradiation of cell cultures
Radioresistance was analyzed as previously reported (Banaz-Yasar et al., 2012). In brief, cells were seeded in 96-well microtiter plates at a density of 250 to 1000 cells per well. After 24 h, cultures were irradiated at room temperature using a Pantak x-ray machine (Pantak) operated at 310 kV, 10 mA with a 2 mm AI filter (effective photon energy ∼90 kV), at a distance of 75 cm and a dose rate of 2.7 Gy/min ultrasoft x-rays for approximately a final dose of 10 and 20 Gy, respectively. Dosimetry was performed with a Victoreen dosimeter that was used to calibrate an in-field ionizing monitor. The culture plates were returned to the incubator immediately after irradiation; after 72 h, cell proliferation was determined by MTT assay.
Tumor xenograft studies
All animal experiments were carried out in accordance with the German Animal Welfare Act (approval number: AZ 87-51.04.2010.A210). Eight- to 10 week-old pathogen-free female NMRI nu/nu immunoincompetent mice were housed in semi-sterile cages with a regimen of 12 h of light and darkness, respectively, with water and food supply ad libitum. 1×106 tumor cells suspended in 100 μL PBS were injected in the upper flanks. Once the tumors had reached a volume of 1 cm3, the mice were anaesthetized with isofluoran (Actavis) and killed by cervical dislocation. Tumors were extracted, embedded in 4% formalin for 24 h, and then rinsed in 70% ethanol for immunohistochemical (IHC) staining.
Generation of polyclonal antibodies against resistant whole tumor cell populations
All animal experiments were done according to the German ethical guidelines for animal care (approval number: G1141/10 and AZ 1295/12). Polyclonal antibodies were raised by an injection of resistant cells into Wistar rats (♀, circa 250 g, circa 8 weeks old). 30×106 cells were suspended in PBS, diluted 1:2 in GERBU LQ complete adjuvant (GERBU Biotechnik GmbH), and applied in a final volume of 250 μL four times every 2 weeks (1×intraperitoneally and 3×subcutaneously). Animals were then anaesthetized in an isofluoran (Actavis) atmosphere, bled by intracardiac puncture, and killed by cervical dislocation. Blood was maintained at 37°C for an hour to obtain the antisera, which were stored at −80°C until use.
Western and dotblot analysis
5×106 cells in exponential growth were washed with cold PBS and centrifuged; pellets were lysed in RIPA buffer [150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, and 50 mM Tris-HCl pH 7.4] in the presence of a proteinase inhibitor cocktail according to the manufacturer's instructions (Roche Diagnostics GmbH) for 30 min on ice and then centrifuged for 20 min at 14,000 g, 4°C. Supernatants (30 μg) were resolved by SDS-PAGE in a 4–12% gradient gel (Invitrogen) using Tris-glycine (0.025 M Tris-HCl, 0.192 M glycine pH 8.5) buffer, and transferred overnight to 0.2 μm nitrocellulose membrane (Pierce Protein; Thermo Scientific, Inc.). Blots were blocked with 5% BSA or nonfat milk taking into consideration the recommendations of the manufacturers of the primary and secondary antibodies. Primary antibodies were purchased as follows: CD44 v3, v10 from Abcam, CD133 from Biorbyt and Cell Signaling, and the rest from Cell Signaling. Conjugated secondary antibodies were obtained from Cell Signaling and Jackson ImmunoResearch Europe Ltd. Immunoblots were developed by Western Lightning® Plus-ECL (Perkin Elmer) using a ChemiDoc XRS+system with Image Lab Version 2.0.1 software (Biorad).
For dotblot analysis, 2.5 μg of cell lysate extracted in RIPA were transferred to 0.2 μm nitrocellulose paper. Dilution and visualization of the primary and secondary antibodies was carried out as described for WB. Spots were normalized by densitometric measurements of nitrocellulose-absorbed protein using standard fast green staining. The expression of the proteins analyzed was given as its relation to their respective WTs, which were reduced to zero.
Histochemical, IHC, and immunocytochemical staining
IHC and immunocytochemical (ICC) staining was performed according to standard protocols with some modifications (Klein et al., 2011; Muturi et al., 2013). Briefly, ICC cells were grown in glass dishes to appropriate densities, washed with 1× PBS, fixed with 4% formaldehyde in PBS for 20 min, rinsed twice with 1× PBS for 5 min, and blocked with 10% normal goat serum (AbD Serotec) at room temperature for 60 min. For IHC, tissue samples were fixed with 4% formaldehyde in PBS and embedded in paraffin. Paraffin tissue sections of 4 μm thickness were baked overnight at 60°C to firmly attach the sections to the slides. After baking, the sections were deparaffinized in two changes of xylene-substitute (Thermo Scientific) solution for 10–15 min and rehydrated in a series of graded ethanol solutions (100%, 100%, 95%, 70%, and 50%) for 3 min each. HE staining was performed using conventional techniques. For IHC, antigens were retrieved by heating the sections for 30 min in 10 mM sodium citrate buffer pH 9.0 at 95°C in a domestic vegetable steamer. The slides were washed twice in 1× PBS for 5 min and blocked for 60 min with 10% normal goat serum at room temperature. All primary antibodies were applied according to the manufacturers' recommendations. Sections were incubated overnight in PBS/0.05% Tween 20, 1.5% goat serum at final concentrations between 1 and 5 μg/mL. On the next day, the slides were washed thrice in PBST (PBS/0.05%Tween 20) for 5 min each and rinsed in 1× PBS for another 5 min. Conjugated secondary antibodies diluted in PBS/0.05% Tween 20/2.5% goat serum were incubated for 120 min at room temperature according to the manufacturers' recommendations. Next, the samples were stained for 15 min with 10 μg/mL propidium iodide or Hoechst 33258 diluted in PBS in order to visualize the nuclei. The slides were then washed thrice in PBST (PBS/0.05% Tween 20) for 5 min each and rinsed in 1× PBS for another 5 min. DAB (3,3′-diaminobenzidine; Dako) staining was done using standard IHC methods. Tissue specimens were mounted in Faramound Mounting medium (Dako) for visualization.
Transmission electron microscopy and three-dimensional reconstruction analysis
Tissue pieces (∼2 mm3) were fixed for 3 h in 2.5% 0.1 M glutaraldehyde-cacodylate buffer (cb) at room temperature, washed in cb 3×30 min, and embedded into a 1% osmiumtetroxide solution in cb, which was removed by washing 3×30 min with 30% ethanol/50% cb. Blocks were treated with 1% uranyl acetate in 70% ethanol in darkness for contrast enhancement and then incubated thrice for 20 min with 80%, 90%, 96%, and absolute ethanol, respectively. Samples were treated thrice with propylene oxide for 15 min followed by increasing EPON® concentrations (3:1, 1:1, and 1:3) for 60 min each and finally cast in pure EPON overnight at room temperature. Sample embedding was performed at 60°C for 2 days to enable polymerization. EPON blocks (1) were cut to 60 nm thick slices with a Reichert-Jung Ultracut® ultramicrotome for electron microscopy (EM). Sections were then mounted on 200 Mesh hexagonal cooper grids and treated with 1% aqueous uranyl acetate solution for 20 min at room temperature, followed by 5 min incubation with lead citrate (0.4% in water) for contrast enhancement. A Zeiss transmission electron microscope (EM 902A) was employed for observations applying 80 kV for magnification from 3000× to 140,000×. Digital images were taken with a MegaView II slow-scan CCD camera connected to a PC running ITEM® 5.0 software (Soft-imaging-systems) and documented. (2) were cut to 0.5 μm with an ultramicrotome for serial-section analysis. Fifty consecutive sections were mounted on glass slides and stained with 1% toluidine blue, azure II, methylene blue, and sodium tetraborate solution dissolved in distillated water, dried, mounted with xylene substitute, and covered with slips. Images were taken at 1000× magnification on a Leica DM 4000B light microscope equipped with a digital camera. Stacks of 50 serial images were taken of four representative areas, and aligned to the same position using Adobe Photoshop CS4. The regions of interest were finally processed with JASC Animation Shop 3.01 into animated clips.
MicroRNA screening
Total RNA isolated from 79HF6 WT and 79HF6 iCS cells was analyzed for integrity with a bioanalyzer (Agilent 2000). Two micrograms of RNA (RIN values between 8.7 and 9.8) were each mixed with 2.5 fmol of 1 of 18 RNA oligonucleotides reverse complementary to miRControl 3 probes and subsequently fluorescence labeled using a commercial kit (miRCuryTM LNA microRNA Array Power labeling kit; Exiqon) following the instructions of the manufacturer. The total RNA mix was hybridized in a dual-color approach to microarrays. Controls were labeled with Hy3, and experimental samples were labeled with Hy5. The fluorescence-labeled samples were hybridized overnight to miRXploreTM microarrays using the a-HybTM Hybridization Station (Miltenyi Biotec). Image capture and signal quantification of hybridized miRXploreTM microarrays was done with a laser scanner (Agilent Technologies) and ImaGene software Version 8.0 (BioDiscovery).
Mean signal and mean local background intensities were computed for each spot of the microarray images using the ImaGene® software (BioDiscovery). Low-quality spots were flagged and excluded from analysis. The unflagged spots were analyzed with the PIQOR™ Analyzer software performing background subtraction; variance stabilization was carried out by way of log2 scaling; and gene expression normalization was calculated with the quantile normalization method in Matlab®. Results were evaluated for statistical significance by applying Student's t-test, with a significance level α <0.05.
Hierarchical clustering was performed with the correlation metric and the average linkage method: the shorter the horizontal link that connects two branches, the closer the populations represented by the branches. Data postprocessing and creation of graphics was done with Matlab. Results were validated for fluctuations using standard reverse transcriptase-polymerase chain reaction (Banaz-Yasar et al., 2012; Grindberg et al., 2013; Han et al., 2013; Marthaler et al., 2013; Nieto-Estevez et al., 2013).
miRNA validation
miRNA microarray results were validated by employing the stem-loop quantitative real-time polymerase chain reaction technique published by Mhammadi-Yeganeh, which is a modified form of the technique from Life Technologies (Mohammadi-Yeganeh et al., 2013). Briefly, in a reverse transcription step, a so-called stem-loop primer binds with 6–8 bp of its 3′-end to the respective miRNA. The miRNA is reversely transcribed to cDNA and extended to the stem-loop primer sequence (∼60 bp). Specific cDNAs were amplified using a forward primer partially complementary to the miRNA to be analyzed, Taq polymerase, and a universal reverse primer, the sequence of which was integrated into the stem-loop primer. The design of forward primers contained a primer-specific 5′-tail (annealing temperature-dependent design element). The miRNA-specific sequence of the stem-loop primers was defined with the help of “By sequence” provided by the miRBase database (University of Manchester, United Kingdom). All sequences were checked for specificity by applying different combinations of software search sequences (Mature miRNAs, stem-loop sequences) and search methods (BLASTN, SSEARCH) as well as the maximum of E-value cutoffs. Finally, annealing temperatures and specificity of the forward primers were determined with Primer-BLAST (NCBI) using a sequence construct consisting of the forward primer sequence, a poly-A sequence, and the complementary sequence of the reverse primer.
Statistical analysis
Experiments were performed at least in triplicate and the data were given as means±standard error of means, unless stated otherwise. The comparison of medians between the groups was performed by Kruskal–Wallis one-way analysis of variance by ranks and a pairwise comparison using Dunn's test. In some cases, the Student's t-test with four degrees of freedom was applied. Statistical analyses were performed with Sigma Plot 12 (Systat Software, Inc.). Significance was accepted when p<0.05. EM, WB, ICC, IHC, and video microscopy studies were descriptive and, therefore, not analyzed statistically; the results shown are representative of at least three independent experiments.
Results
Biochemical features of etoposide-resistant cell populations
Resistance to cytostatics is associated with a string of closely related biological factors, as observable in vitro, animal models, and in oncologic patients. These factors, including resistance degrees, cross-resistance, radioresistance, and the formation of metastases, are those that lead to an uncontrolled clinical situation and, ultimately, death. As a consequence of cytostatic insults, tumors undergo profound changes with regard to cellular heterogeneity, tumorigenicity, metastasis formation, and the expression of markers specific for tumor initiating cells.
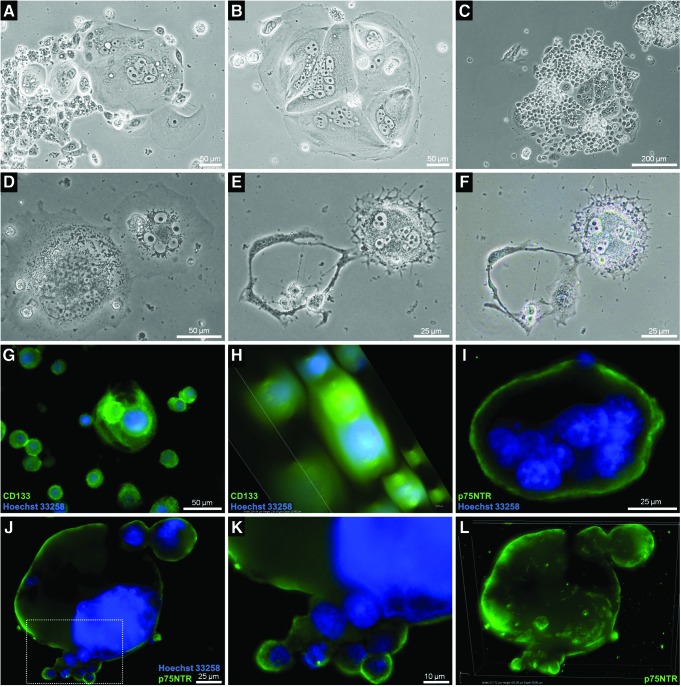
Induction of resistance via etoposide triggers expression of CSC phenotypes
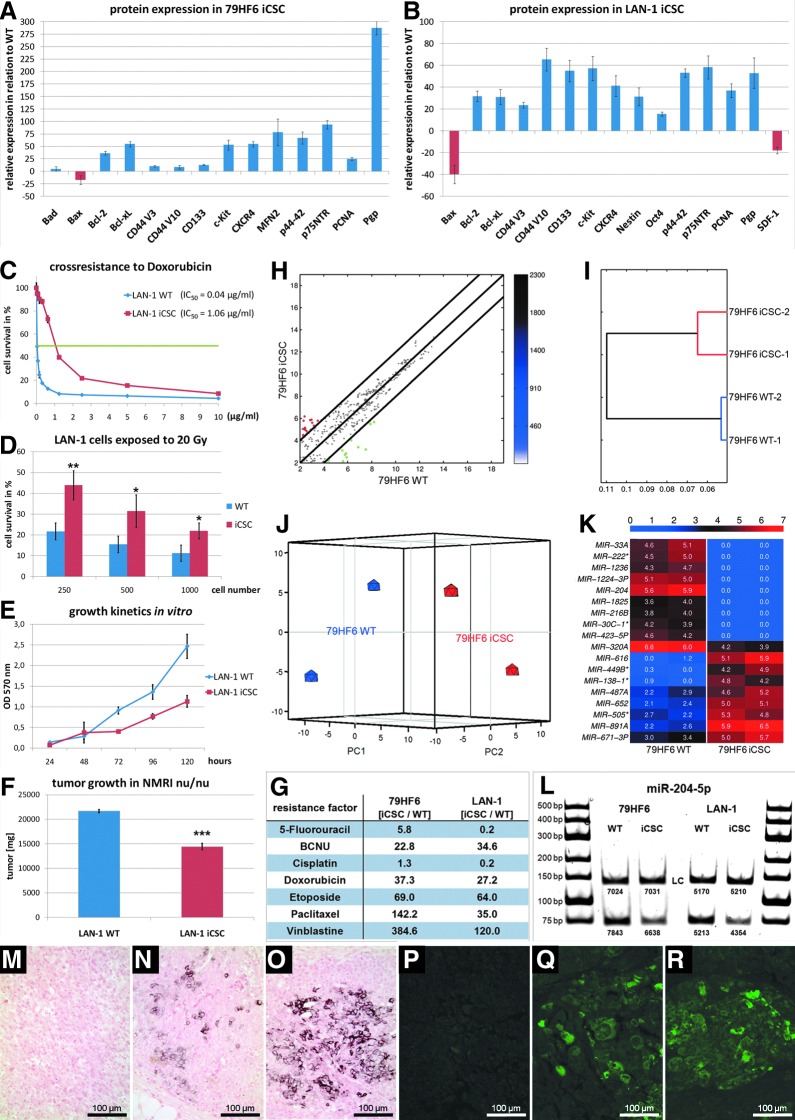
To induce chemotherapy resistance in vitro, several protocols mimicking clinical chemotherapy regimens have been designed to analyze the activity of new antitumoral agents as possible first- or second-line therapeutics. We noted that resistant tumor cells selected via exposure to cytostatics evolved CSC features which remained stable even after xenografting. We addressed this phenomenon (1) by analyzing whole resistant populations, and (2) by examining recurrent atypical cells. In particular, we focused on neuroblastoma and glioblastoma-derived resistant populations selected using etoposide (iCSCs). These subcultures did not show signs of apoptotic processes (Fig. 1A, B and Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/dna). Highly aggressive neuroblastoma and glioblastoma tumors are characterized by the expression of markers associated with poor prognosis and CSC markers such as p75NTR, CD44, CD133, c-kit, nestin, Oct-4, and the axis CXCR4/SDF1. All these proteins were found upregulated in iCSCs derived from human LAN-1 neuroblastoma and 79HF6 glioblastoma cells, among other tumors with the same histological origins. Interestingly, the CD44 variants v3 and v10 were found upregulated in comparison to their WT parental cells (Fig. 1A, B). Moreover, LAN-1 and 79HF6 iCSCs xenografted in NMRI nu/nu mice displayed large islands of cells expressing p75NTR and CD133 (Fig. 1M–R).
FIG. 1.
Principal biological characteristics of LAN-1 neuroblastoma and 79HF6 glioblastoma iCSCs after etoposide selection. (A, B) Global densitometric dotblot analysis. CSC markers CD44 v3/v10, p75NTR, c-kit, nestin, Oct-4, CD133, and p44/42 were the most prominent proteins found to be upregulated. Anti-apoptotic proteins such as Bcl-2 and Bcl-XL were also upregulated. Pro-apoptotic Bax was significantly downregulated. (C) Cross-resistance to doxorubicin as determined by MTT proliferation assay. (D) iCSCs were highly resistant to irradiation (20 Gy), as demonstrated in LAN-1-derived resistant populations (p-values for 250 cells, 0.009, for 500 cells, 0.035, and for 1000 cells, 0.027). (E, F) A characteristic trait in the very first weeks of resistance development is the slowdown of both in vitro and in vivo tumor growth (p≤0.001). (G) Resistant factors (RF) for some cytostatics analyzed in 79HF6 and LAN-1 iCSCs. (H–K) microRNA analysis revealing differences in microRNA profiles between 79HF6 WT and iCS cells. Results depicted as pairwise scatter plot (H), hierarchical clustering analysis (I), three-dimensional (3D) Principal Component Analysis (PCA) (J), and heat map (K) with 18 microRNAs deregulated to a highly significant degree. The validation of some miRNAs was consistent with the global analysis as shown for miR-204-5p (L). (M–R) paraffin sections labeled for p75NTR in LAN-1 (M–O, DAB) and for CD133 in 79HF6 iCSCs xenografted in nu/nu NMRI mice (Q–R, IF). CSC, cancer stem cell; iCSCs, induced CSCs; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; WT, wildtype. *: significant (p≤0.05%); **: very significant (p≤0.01%); ***: highly significant (p≤0.001%). Color images available online at www.liebertpub.com/dna
Cross-resistance, radiorefraction, and slowdown in tumor growth kinetics in iCSCs
Resistant tumors obtained via exposure to etoposide overexpressed the MDR1 transporter (Pgp) as reflected in Figure 1A and B. This explains both the resistance and cross-resistance phenomena observed for various structurally and/or functionally unrelated xenobiotics, for example, BCNU, anthracyclins, and alkaloids; while some cell systems remain relatively vulnerable to cytostatics such as cisplatin and 5FU (Fig. 1C, G). In addition, the vast majority of chemotherapy-refractory cells become highly resistant to single doses of radiation (Fig. 1D). In the early stages of resistance development, tumors undergo a remarkable slowdown of growth patterns both in vitro and in vivo (Fig. 1E, F). Interestingly, growth will accelerate again in the later stages even beyond the initial rate as the tumor becomes more resistant. This behavior is fully consistent with what is observed in clinical cancer therapy.
Glioblastoma 79HF6 WT and iCS cells display differences in microRNA fingerprints
The microRNA scatterplot revealed that 79HF6 WT and iCS cells had different microRNA profiles, with a Fisher's correlation coefficient between samples of R=0.971 (Fig. 1H). Hierarchical clustering and the three-dimensional (3D) Principal Component Analysis also indicated differences in microRNA profiles between samples and a tight correlation among the biological replicates (Fig. 1I, J). Applying log2 scaling, we found 323 differentially expressed microRNA transcripts, representing 9.16% of all the transcripts. After filtering out non-unique transcripts, 43 microRNAs showed statistically significant differences and 18 exhibited highly significant expression differences (see heat map in Fig. 1K). The validation of the expression of some microRNAs was congruent with the global fingerprint, of which miR-204-5p (associated with the degree of malignancy in glioblastoma) is shown as an example in Figure 1L.
Morphological aspects, cell hypertrophy, and atypical heterogeneity in response to cytostatic insult
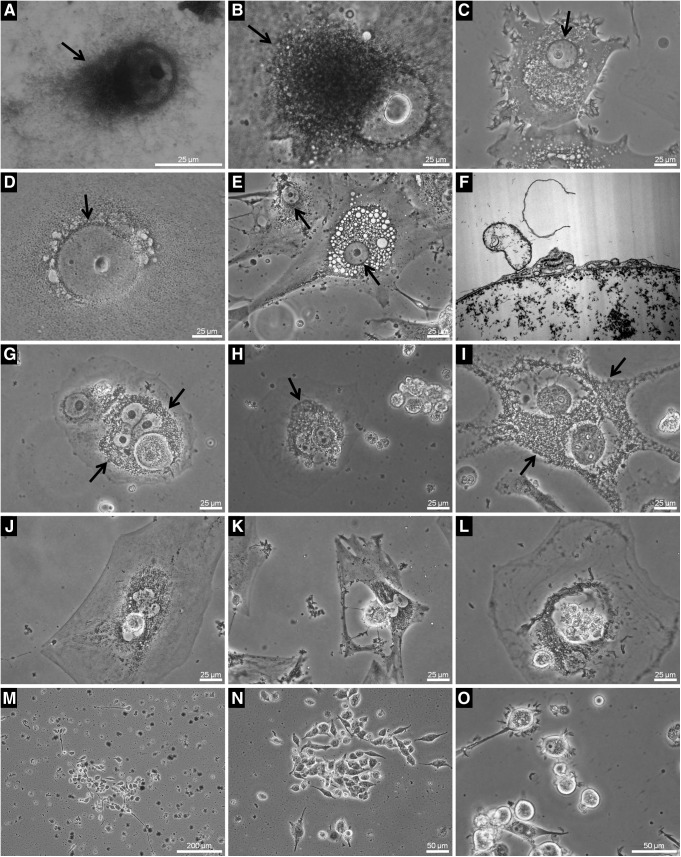
Cells in the process of acquiring resistance to cytostatics underwent a number of simultaneous morphological changes, among which hypertrophy and increased heterogeneity were the most prominent. These changes were observed in several tumor cell lines of different histological origins with similar patterns. It should be noted, however, that these events were also observed in WT cultures, albeit at very low rates (Fig. 2, Sequences I and II).
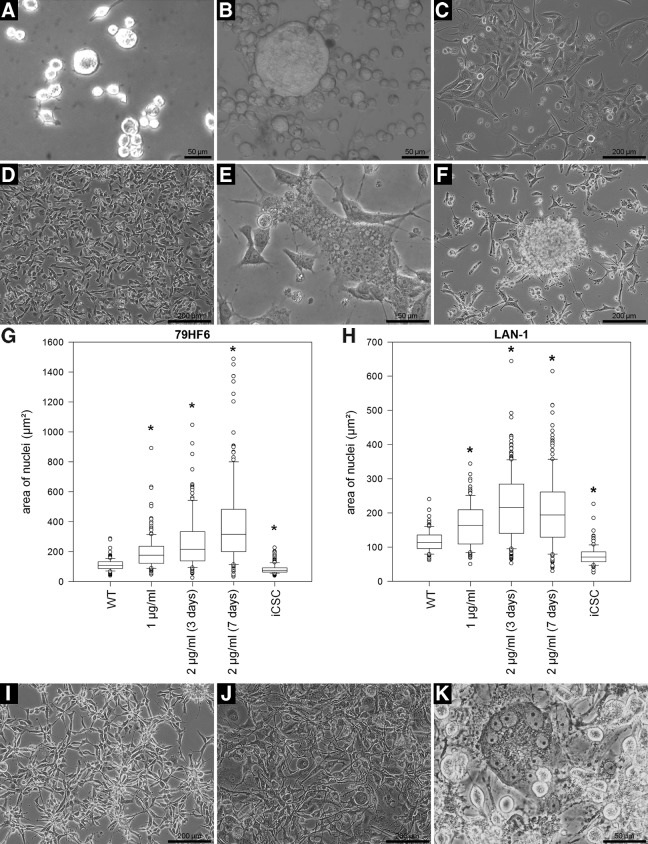
FIG. 2.
Cell heterogeneity, hypertrophy, and metaplastic events in resistant tumor cell lines. Sequence I: (A–C) 79HF6 human glioblastoma. (A, B) Cells trypsinized to highlight their contours; (A) untreated, (B) iCSCs depicting a giant cell. (C) Resistant cultures with normal morphology after several weeks of resistance induction. (D–F) LAN-1 human neuroblastoma. (D) WT cells, (E) syncytium-like entity, and (F) spherical colonies formed during resistance development. (G, H) Measurement of the nuclear area of 79HF6 and LAN-1 cells during resistance development (n>100, group differences by analysis of variance p≤0.001, pairwise differences in relation to WT by Dunn's Method, p≤0.05). Nuclear hypertrophy was a transitory event, with the cells and nuclei returning to their initial size once resistance was fully evolved. (I–K) U87 glioblastoma cells. (I) Untreated, (J, K) iCSCs. (J) Hypertrophic cells, and (K) spiral cells. Sequence II: (A–C) Y79 human retinoblastoma suspension cells. (A) Untreated, (B, C) etoposide-resistant; (B) hypertrophic cells, and (C) cells attached to a plastic culture flask; note the multinucleated cells. (D–F) Electron microscopic imaging of Jurkat ALL cells. (D) Untreated and (E, F) etoposide-resistant cells showing abnormal nuclear segmentation and hypertrophy. (G–I) Encapsulation events in K562 CML suspension cells during acquisition of resistance. Cells undergo transitory attachment to a plastic culture flask and start to encapsulate. (J–L) Jurkat-resistant cells with spherical colonies attached to the culture flask (J), hypertrophic (K) and spiral cells (SP) (L) showing a spiral arrangement of nuclei as visualized by hematoxylin staining. Note the syncytium-like entities (SL) and hypertrophic cells. Magnification: (D–F) 3000×. *: significant (p≤0.05%). Color images available online at www.liebertpub.com/dna
Hypertrophy and heterogeneity induced by exposure to etoposide
During in vitro resistance acquirement, the very first change we noticed was the hypertrophy of the nuclei, nucleoli, and cell volumes in certain cell populations (Fig. 2, Sequence I: B, E, G, H, J, K). These changes were transitory rather than permanent and took different courses depending on the cell type.
In suspension-growing leukemia and retinoblastoma cells, the hypertrophic period was followed by a metaplastic transition as the cells became temporarily attached to the plastic culture flasks but later returned to their original growth patterns (Fig. 2, Sequence II: B, C). This behavior was observed in both single-nucleus and syncytium-like cells (Fig. 2, Sequence II: C). Profound changes in the nuclear ultrastructure were observed in resistant Jurkat leukemia and other cell lines (Fig. 2, Sequence II: E, F). Another frequent event in response to drug insult was cell encapsulation (Fig. 2, Sequence II: G–I) and the emergence of spherical colonies with a morphology similar to what was obtained by hanging drops (Fig. 2, Sequence I: F and Sequence II: J). We observed that the cells, once attached to a surface, started forming colonies around the spherical colonies which spread rapidly.
The recurrence of atypical populations encompassing multinucleated cells was frequently observed throughout the process of resistance development (Fig. 2, Sequence I: B, E, K and Sequence II: E, F, K, L). Due to the large variety of atypical cells existing concurrently after drug exposure, we focused our efforts on the most remarkable and common entities, for example, as illustrated in Figure 2 (Sequence I: K and Sequence II: L and Supplementary Film S1), which showed a spiral arrangement of nuclei; in a first step, we subjected them to a morphological analysis and then addressed their possible stem-cell characteristics.
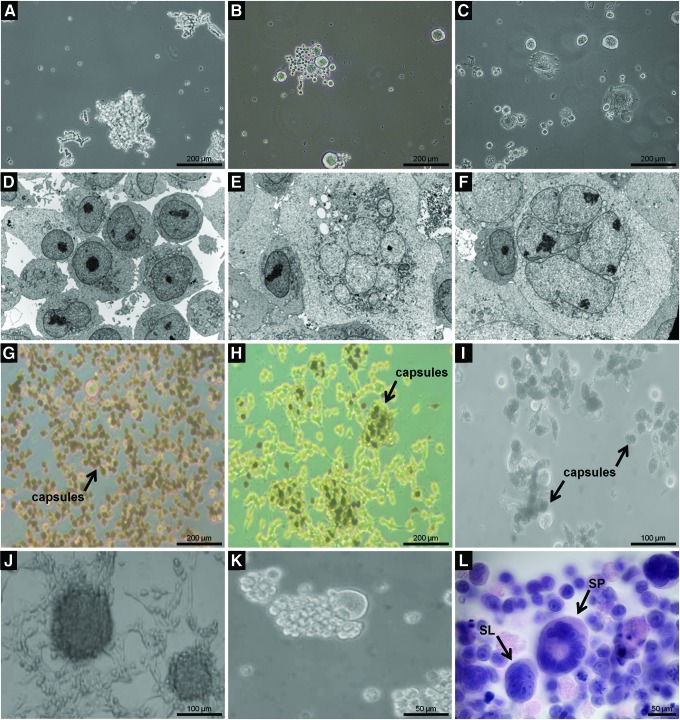
Spiral cells
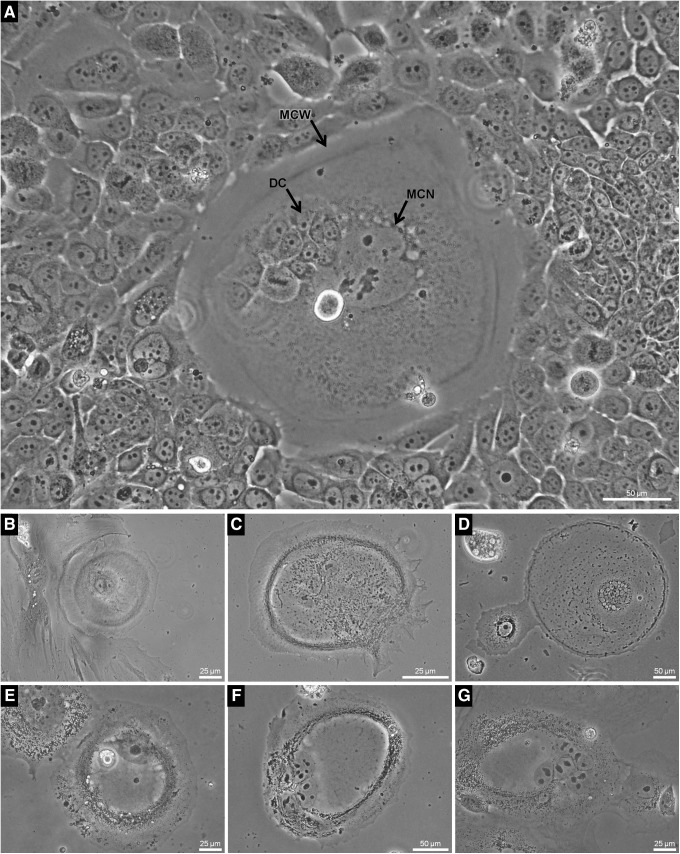
In accordance with the findings of Molina et al., in glioblastoma tumors, we observed a recurrent multinucleated cell type in several resistant cultures as well as in naïve cultures but even more frequently in chemotherapy-resistant tumors, with an approximate ratio of 1:10 (WT:R) (Fig. 2, Sequences I and II: K, L respectively and Fig. 3). The unique morphological feature of these oversized cells was the spiral arrangement of their nuclei in different planes (Fig. 3A–C) as shown by 3D reconstruction of serial sections (Fig. 3R–V and Supplementary Films S2, S3). Using polyclonal antibodies raised against glioblastoma/astrocytoma-derived iCSCs (GB, glioblastoma serum), we detected these cell entities in all analyzed systems (Fig. 3H–N). Interestingly, we noted not only an abnormal arrangement of mitochondria in the perinuclear compartments of these cells but also the expression of proliferating cell nuclear antigen (PCNA) (Fig. 3O, Q), suggesting a heightened proliferative readiness. Moreover, the cells expressed CSC phenotype markers such as CD44 and stem cell factor (SCF) (Fig. 3P, Q).
FIG. 3.
Spiral cells. This cell type was distinguished by a signature, spiral-like arrangement of nuclei and the presence of an internuclear space as revealed by 3D reconstruction in 79HF6 iCSCs (A–C, 40× and Supplementary Film S2). Cells of this type were identified in 79HF6 iCSC xenografts (D, 60×) and in glioblastoma patient paraffin sections (E, 60×). (F, G) Spiral cells in SKNAS iCSCs. Polyclonal antibodies raised against a pool of iCSCs derived from different glioblastoma cell lines clearly reveal the morphology of these cells (H, L–N). (O) Spiral cell in iCSCs isolated from SKNAS neuroblastoma cells. Nuclei and morphology were visualized using an antibody against lamin A and C (H–O). The spiral cells expressed CD44, PCNA, and SCF as shown in (P, Q). 3D reconstruction of nucleii (R–V and Supplementary Film S3) PCNA, proliferating cell nuclear antigen; SCF, stem cell factor. Color images available online at www.liebertpub.com/dna
Monastery cells
Another distinct cell type with plasmodia-like features, which we named monastery cells, was characterized by the formation of a conspicuous wall structure inside which cell replication took place. A striking feature was the accumulation of mitochondria around the daughter cells, suggesting a vital yet atypical role of these organelles in their biology (Fig. 4A). These cells were able to undergo asymmetrical cell division (Fig. 4D, E, G and Supplementary Film S4).
FIG. 4.
Monastery cells at different developmental stages. These cell entities were observed in different tumor cell populations that were acquiring resistance to etoposide; their chief characteristic was the presence of walls that defined their borders. During maturation of these cells, mitochondria appeared to play a key role in wall development, as these organelles assumed a centripetal layout helping to delimit borders. A monastery cell and progenitor cells at different stages of development (A, 5637 iCSC bladder carcinoma. Supplementary Film S4). (B, C) A549 iCSCs lung carcinoma. (D) MCF-7 iCSCs mamma carcinoma. (E–G) SKNAS iCSCs neuroblastoma. Cells undergoing asymmetric division (D–G). DC, daughter cells; MCN, monastery cell nucleus; MCW, monastery cell wall.
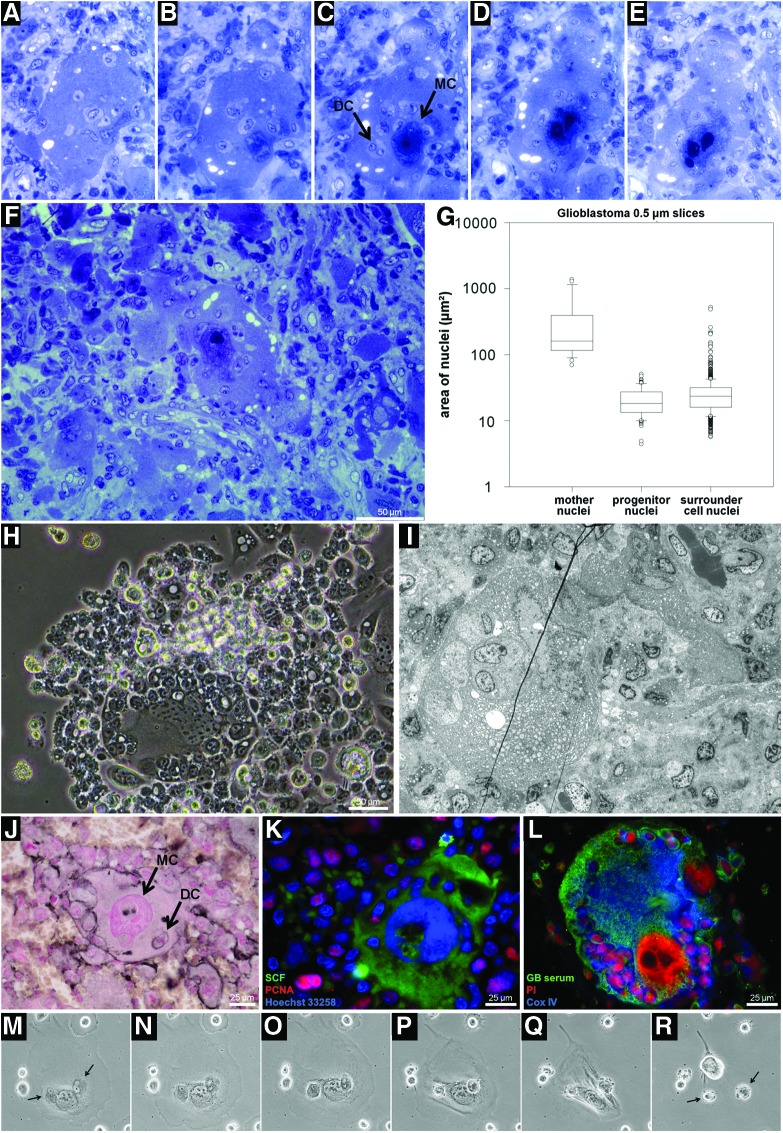
Pregnant cells
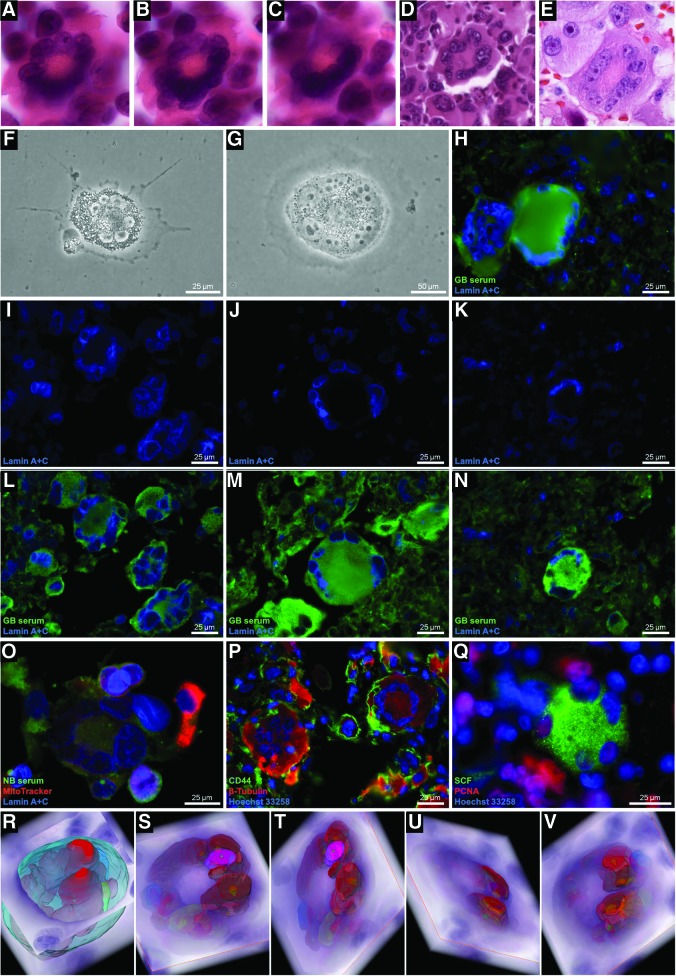
This cell type did not possess the walls described for monastery cells but shared other crucial characteristics such as a free main nucleus and numerous daughter cells with defined cell compartments detectable by both, vertical and horizontal 3D-reconstruction (Fig. 5A–E and Supplementary Films S5–10). Cells of this kind were found both in resistant cell cultures and in xenografted tumors, especially in glioblastoma multiforme and in astrocytomas where they were CD45 negative (ruling out potential leukocyte infiltrations. Supplementary Fig. S1). The cell progeny was seen embedded in the cytoplasm of the mother cells at different developmental stages, judging by their size. Moreover, they were similar in size to the cells that made up the tumor mass (Fig. 5F, G). Daughter cells contained cytoplasm as observed by differences in methylene blue staining and EM visualization (Fig. 5A–F, I).
FIG. 5.
Pregnant cells in human glioblastoma. (A–E) Patient paraffin serial sections (0.5 μm) showing the morphological structure of pregnant cells. A single big nucleus (ø 360 μm2) with well-defined nucleoli was a hallmark of these cells. Daughter cells (DC) maturing inside mother cells (A–F and Supplementary Films S5–6, S9–10, H–L). (H) U87 glioblastoma iCSC culture with a pregnant cell entity. Nuclei of DC localized inside their progenitors (n>100) had slightly smaller areas (ø 21 μm2) than the nuclei of cells forming the tumor surroundings (ø 27 μm2) (G, n>1000). DC were different in cytoplasmic content than their mother cells as observed by methylene blue staining and EM contrasting (A–F, I). (J) Membrane contours of daughter cells as visualized by CD44 labeling. These cells expressed high levels of SCF and PCNA (K). The morphology and distribution of this cell type was analyzed in patient paraffin sections using polyclonal antibodies against resistant glioblastoma cell populations (L). (M–R) Mother cells do not die after expulsing their progeny (M–R and Supplementary Film S11). Magnification: (A–E, M–R) 20×, (I) 300×. Arrows indicate MC for main cell and DC for daughter cells. EM, electron microscopy; MC, main cell. Color images available online at www.liebertpub.com/dna
Pregnant cells were differentially labeled using antibodies raised against different a pool of glioblastoma-derived iCS cell lines. The cells phenotypically expressed CS markers such as CD44 and SCF (Fig. 5J, K) and were characterized by large numbers of mitochondria in their cytoplasm and in the perimeter of their daughter cells (Fig. 5I, L). The cell membranes of the daughter cells could be delimited by CD44 labeling (Fig. 5J). In neosis, mother cells die after giving rise to daughter cells. Here, videographic observations clearly demonstrated that the fate of the mother cells does not necessarily end with the death (Fig. 5M–R and Supplementary Film S11).
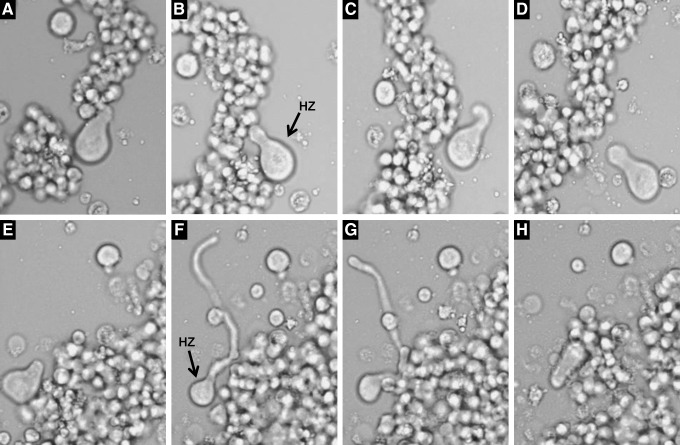
Hirtenzellen (shepherd cells)
Another phenomenon associated with resistance to cytostatics was the presence of pear-shaped cells in in vitro cultures of hematological malignancies. These cells appeared to be physically interacting with nearby colonies and were marked by a large single lobopodium many times the size of the cell body that occasionally showed great mobility (Fig. 6 and Supplementary Film S12). These extremely active cells were very elusive, which has precluded more in-depth studies until now.
FIG. 6.
Hirtenzellen (Shepherd cells) amid etoposide-resistant Jurkat cell populations. They were pear-shaped entities capable of rapidly growing a polarized, highly mobile lobopodium that interacted with nearby colonies. (A–H and Supplementary Film S12) Sequence selected from a 10-min clip at 40× magnification. HZ, Hirtenzellen.
Syncytium-like populations and syncytial division in resistant tumor cell cultures
Among highly heterogenic etoposide-resistant populations, we observed syncytium-like entities undergoing regular cell division (Fig. 7A–F), which has not been reported for syncytial systems until now. Moreover, in contrast to established theory that describes such cell systems as simple cell-membrane fusion products of adjoining cells or endoreplicative cell types without cytoplasmic membranes, cells living inside syncytium-like entities synthesized their own cytoplasmic membrane, identifiable by the detection of CD133 expression and 3D reconstruction techniques. These cells matured completely inside the capsules before the latter broke up (Fig. 7G, H). A typical endoreplicative (Plasmodium) entity among NB69 neuroblastoma cells expressing p75NTR is shown in Figure 7I. Moreover, p75NTR+ plasmodium-like cells were observed as undergoing asymmetrical divisions in which daughter cells dragged fragments of the parental cytoplasmic membranes for their own coating (Fig. 7J–L).
FIG. 7.
Syncytium-like cells among resistant populations. (A–F) Among highly heterogenic resistant colon carcinoma HCT8 iCSC (A–C), neuroblastoma NB69 iCSC populations (E, F), syncytium-like entities can be seen undergoing cell replication comparable to classical cell division, maintaining a syncytial-like unit. (D–F) Syncytium-like entity undergoing cell division, with one of the resulting cells breaking down into single cells that may form new colonies. (G, H) 79HF6 iCS cells inside progenitor cells with intact membrane as determined by CD133 labeling (H, 3D reconstruction). (I) Endoreplicative p75NTR+ entity. (J–L) Asymmetrical cell division in p75NTR+ NB69 iCSCs. (K) Magnification of marked area in (J). (L) 3D reconstruction. Color images available online at www.liebertpub.com/dna
Spatial differences in mitochondrial aggregation in iCSCs
We observed that U87 iCS cells displayed two different types of mitochondrial rearrangement in close spatial relation to the nuclei: there were cells with polarized tail-like structures (Fig. 8A, B, arrows), and others with a shell-like mitochondrial arrangement in the perinuclear spaces (Fig. 8C–E, arrows). It was found that, as a result of exposure to cytostatics, cells underwent a methuosis-like process, in which some nuclei appeared to have lost their associated cytoplasm in order to form new colonies. This was observed in both regular and syncytium or plasmodium-like entities. At least two different processes could be differentiated, as documented. In the simpler form, the nuclei shed their cytoplasm and formed new cell units probably from residual cytoplasmic material carried between the internal and external membranes (Fig. 8F, I). In the more complex variant (Fig. 8G–K), the nuclei appeared to become cocooned in a mitochondrial mass as described earlier, shed their cytoplasm, and remained encapsulated for a hibernation period (Fig. 8L), after which new colonies were formed (Fig. 8M, N).
FIG. 8.
Different patterns of mitochondrial accumulation in iCSCs. U87 iCSCs showing two different configurations of mitochondrial accumulation, a polarized tail-like structure (A, B), and a shell-like arrangement (C–E). Formation of methuosis-like process was observed as a preamble to cytoplasm shedding (C–E), with the nuclei entraining some cytoplasmic material in their intermembrane space (F). NB69 iCSCs showing perinuclear accumulation of mitochondria, shedding of cytoplasm, and nuclei encapsulated for a hibernation period (G–L). De novo colonies in NGP neuroblastoma (M, N) and U87 iCS glioblastoma cells (O). Magnification: (F) 3000×. Arrows indicate mitochondrial accumulation.
Overview of the principal features of cell entities used in this study
Table 1 reflects the main morphological and phenotypical characteristics of tumor cell entities analyzed in this work. In addition, a description of their recurrence in tumors was included.
Table 1.
Overview of Etoposide-Induced Types of Rare Cells, Pertinent Cell Lines, and Principal Morphological and Biochemical Characteristics
| Atypical cell type | Source cell line | Morphological hallmarks | Biological features | Molecular characteristics | Shown in |
|---|---|---|---|---|---|
| Spiral (doughnut-like or wreath-like) | Leukemia (Jurkat, K562) | Multinucleated | Detectable in cell cultures, xenografted tumors, and patient paraffin sections | CD44/PCNA/SCF (+) | Figure 2, Sequence I |
| Glioblastoma (U87, 79HF6) | Helical arrangement of nuclei | Detectable after xenotransplantation | TP53 (−) | Figure 2, Sequence II | |
| Retinoblastoma (Y79) | Accumulation of mitochondria in perinuclear compartment | More numerous in resistant systems | Figure 3 | ||
| Monastery | Bladder carcinoma (5637) | Multinucleated cells with striking wall structure similar to oocyte zona pellucida | Progeny is able to resume proliferative activity within the walls | CD44/SCF/CD133 (+) | Figure 4 |
| Lung carcinoma (A549) | Single main nucleus and several offspring cells within the wall | Marked mitochondrial accumulation | Bcl-2 and Bcl-xL (+) | ||
| Colon carcinoma (HCT8) | More numerous in resistant systems | TP53 (−) | |||
| Breast carcinomas (MCF-7) | |||||
| Pregnant | Glioblastoma (U87) | Multinucleated cells with single, giant nucleus and numerous daughter cells with clearly defined cytoplasmic compartments | Stable after xenotransplantation | CD44/SCF/CD133 (+) | Figure 5 |
| Colon carcinoma (HCT8) | Large amounts of mitochondria in perinuclear compartment and around daughter cells | Senescence markers missing | TP53/CD45 (−) | ||
| Neuroblastoma (SKNAS, LAN-1) | Parental cell survives after release of progeny | ||||
| Retinoblastoma (Y79) | More numerous in resistant systems | ||||
| Hirtenzellen (shepherd) | Leukemia (Jurkat) | Mononucleated | Strong physical interaction with adjacent colonies | N.A. | Figure 6 |
| Pear shaped | Considerable mobility and cell metamorphosis | ||||
| Highly mobile lobopodium |
N.A., not applicable; PCNA, proliferating cell nuclear antigen; SCF, stem cell factor.
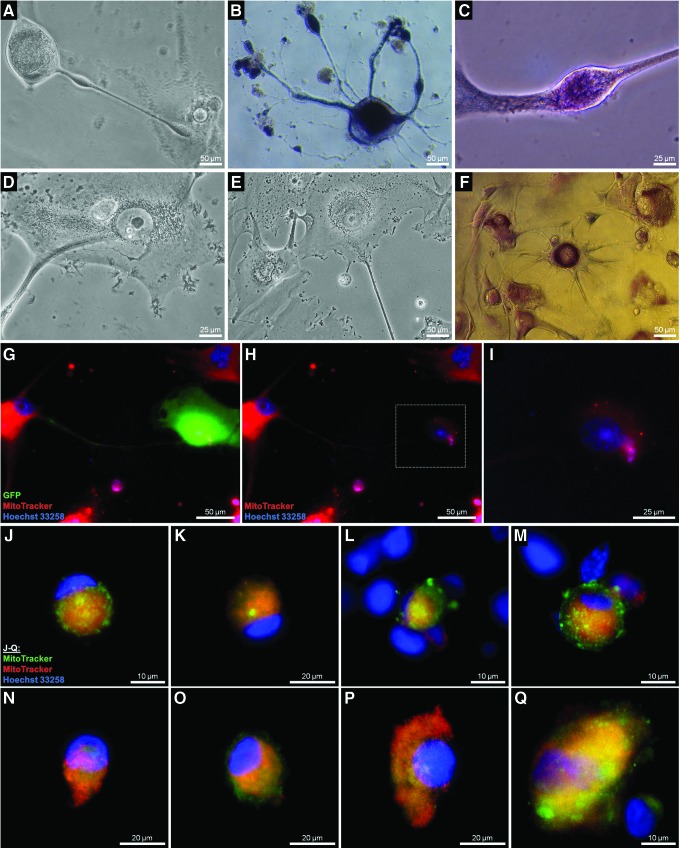
Intercellular exchange of mitochondria and nuclear uptake of exogenous mitochondria
We noticed that resistant cells exchanged mitochondria with other cells, as has been described for normal tissues. Some cells acted as donors by actively injecting or depositing mitochondria preferably in the nuclear perimeters of adjacent cells (Fig. 9A, D, E). In U87 and other iCSC cultures, these donor cells were marked by a massive perinuclear agglomeration of mitochondria (Fig. 9B, D–I) and developed a kind of varicosity that they used to inject the mitochondrial bolus (Fig. 9A–C). On the other hand, some resistant nuclei with different mitochondrial arrangements (shell-like, Fig. 9J–M, or tail-like, Fig. 9N–Q) acted as acceptors and incorporated exogenous mitochondria into their own perimeters, probably mediated by the expression of MFN2, a promoter of mitochondrial fusion that was found overexpressed in GB iCSCs (Fig. 1A). It was found that this process could only be harnessed by a minority of nuclei (Fig. 9L, M).
FIG. 9.
Mitochondrial exchange between adjacent cells and uptake of exogenous mitochondria by nuclei isolated from resistant U87 iCSCs. Donor cells depositing mitochondria in the nuclear perimeters of adjacent cells via a kind of varicosity (A–F). Mitochondrial injection into the perinuclear compartments of U87 GFP was documented by co-cultivation of U87 iCSCs labeled with red MitoTracker® and U87 GFP. Resistant cells transferred mitochondrial content to untreated cells (G–I): magnification of marked area in (H). (J–Q) Nuclei isolated from resistant U87 iCSCs previously labeled with red MitoTracker and exposed to exogenous mitochondria isolated from U87 iCSCs previously labeled with green MitoTracker. Incorporation of exogenous mitochondria led to the formation of a capsule inside which the nucleus continued its life cycle. GFP, green fluorescent protein. Color images available online at www.liebertpub.com/dna
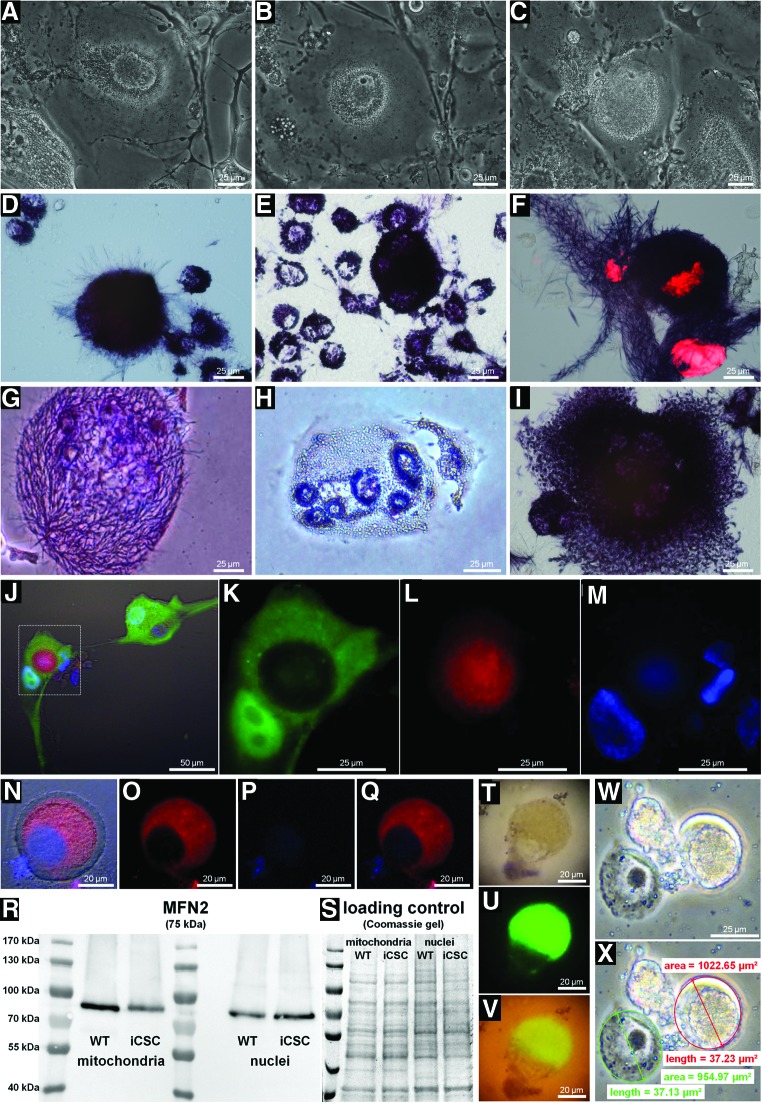
Mitochondrial enveloping of the nucleus
Remarkably, nuclear encapsulation via retrograde perinuclear accumulation of mitochondria in response to cytostatic insult was observed in cells with both single and multiple nuclei (Fig. 10A–I, N–Q, T–X). As mentioned, the mitochondrial enveloping of nuclei could be forced by resistant cells that preferentially injected mitochondrial boluses into untreated cells. This issue was addressed by co-cultivating GFP-U87 WT with resistant U87 iCSCs previously labeled with red MitoTracker (Fig. 10J–M). The process commenced with the earlier described recruiting of mitochondria to the perinuclear compartments until the nuclei were fully enveloped (Fig. 10A–M). In some cells, however, this process was incomplete (Fig. 10N–Q) and left a mitochondria-free annular zone. This suggests that the membrane in this annular zone may have a different biochemical composition for which mitochondria do not have tropism. We analyzed the mitochondrial populations by fractionating the perinuclear and cytoplasmic compartments and found that, despite possible contaminations, mitochondria attached to the nuclei of resistant cells expressed more MFN2 (Fig. 10R). Some cells with encapsulated nuclei remained dormant for prolonged periods of time before the nuclei were ejected along with some residual cytoplasm trapped in their intermembrane space (Fig. 10T–X), as measured by the size of capsules and free nuclei (Fig. 10X).
FIG. 10.
Nuclear encapsulation via mitochondrial perinuclear accumulation. Nuclear encapsulation took place in iCSCs with both single and multiple nuclei in response to drug exposure. (A–C) Different stages of nuclear encapsulation in U87 iCS single-nucleus cells. (D–I) Mitochondrial distribution in NB69 iCSCs with single and multiple nuclei as revealed by MTT incorporation (F: MTT and propidium iodide staining). (J–M) U87 iCSCs previously labeled with red MitoTracker depositing mitochondria to nuclear perimeters of GFP-labeled U87 WT cells (K–M) depict a magnification of the square of J. The mitochondria acted as a shield, interfering with visualization of the nuclei via Hoechst 33258 staining. (N–Q) Details of annular zones of the nuclei in some cells that were not invaded by mitochondria (red: MitoTracker, blue: Hoechst 33258). (R) MFN2 expression in mitochondria isolated from the perinuclear and cytoplasmic compartments of U87 iCSCs as analyzed by WB. Remarkably, the perinuclear mitochondria expressed greater quantities of this fusion protein. (S) Loading controls as visualized by Coomassie staining. (T–V) Eclosion of encapsulated nuclei of U87 iCSCs. Mitochondria were previously transfected with GFP. (W) Nucleus ejected from a U87 iCS cell, juxtaposed to empty capsule, (X) comparison of surface areas. MFN2, mitofusin-2. Color images available online at www.liebertpub.com/dna
Physiological role of mitochondrial encapsulation
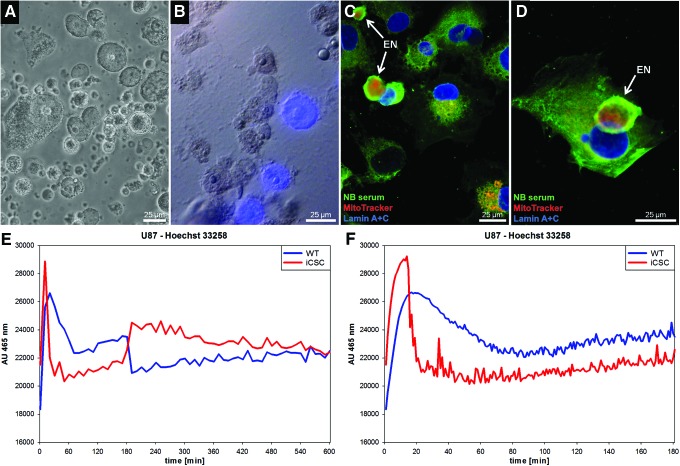
It has been known since the 1950s that, in normal tissues, there exist two kinds of nuclei which can be differentiated by their staining characteristics. We noted that in fixed preparations of resistant cell cultures stained with Hoechst 33258, some cells showed no nuclear staining although their nuclei were clearly visible by light microscopy (Fig. 11A–D). This was fully consistent with the finding that not all nuclei isolated from U87 iCSCs were susceptible to Hoechst 33258 staining (Fig. 11B). These phenomena can be explained by nuclear encapsulation via accumulation of mitochondria that shield the genetic material from the dye. In fact, the process is so efficient that nuclear dyes or antibodies which recognize nuclear membranes could not reach their target structures (Fig. 11C, D). We also have physiologically correlated this phenomenon with the differential kinetics in the uptake of Hoechst 33258 by resistant cells with nuclear encapsulation. In the first 20 min, the iCSCs were capable of eliminating Hoechst 33258 (Fig. 11E, F) probably via ABC-type transporters. Cell populations that had developed a nuclear defense via mitochondrial encapsulation could be effectively labeled with this nuclear dye only after >10 h of incubation, as concluded from their uptake kinetics (Fig. 11E).
FIG. 11.
Differential staining of nuclei and impermeability to nuclear dyes. (A) Unstained U87 iCSC nuclei. (B) U87 iCSC nuclei showing differences in permeability to Hoechst 33258 (blue). (C, D) SKNAS neuroblastoma iCSCs stained with MitoTracker (red) and labeled with anti-lamin A+C antibodies (EN, encapsulated nuclei). While most nuclei could be effectively labeled and stained, some appeared to be shielded. (E, F) Significant differences in Hoechst 33258 uptake kinetics between U87 WT and iCSC populations. (E) An iCSC subpopulation increased the uptake of the dye after ∼180 min. Both cell types showed full Hoechst 33258 staining after 10 h. F (close up of E): Elimination of the dye in the first 20 min probably by ABC-type transporters. NB serum, neuroblastoma antiserum. Color images available online at www.liebertpub.com/dna
Discussion
Since the advent of chemotherapy in cancer care, the development of new antineoplastic agents has been closely related to advancements in our understanding of the cellular and molecular biology of cancer cells, most notably of the altered elements that promote uncontrolled cell division. However, the curative effects of cytostatics were soon found to be all too easily foiled by a number of resistance mechanisms that the tumor cells can harness, which, in turn, has inspired many new efforts to identify modulators that may help overcome this acquired resistance. However, all past and present endeavors notwithstanding, the fact remains that several cytostatics routinely used in the clinic nowadays lead to the activation of ABC-type transporters, that is, components of the detoxification mechanisms naturally present in various organ systems. Targeting these transporters with modulators is, therefore, associated with a string of serious systemic side effects that are incompatible with life, meaning that the introduction of such modulators has not produced the desired clinical effect (Tannock, 2001; Lobo et al., 2007; Lu and Shervington, 2008; Stavrovskaya and Stromskaya, 2008; Nakai et al., 2009; Baguley, 2010; Cheng et al., 2012; Mo and Zhang, 2012; Nakanishi and Ross, 2012; Schumacher et al., 2012; Stacy et al., 2013).
The majority of studies on chemotherapy resistance have focused on elements such as ABC-type transporters, DNA repair mechanisms, or antioxidants such as glutathione, while factors underlying cellular heterogeneity and segregation have not been addressed in detail. As to the biology of CSCs, this aspect could become highly significant, as there is evidence that a number of cytostatics trigger CSC phenotypes as recently reported by some groups. Abductively, if anticancer agents induce CSC phenotypes and cellular heterogeneity, it is mandatory to analyze the biological role of these recurrent cell populations in tumor genesis and differentiation (Donnenberg and Donnenberg, 2005; Díaz-Carballo et al., 2010, 2014; Kitambi, 2011; Grimm et al., 2012).
Biochemical features of resistant tumor cell entities
Etoposide is a compound that finds widespread use as a part of polychemotherapy regimens directed against a spectrum of neoplasias, including neuroblastoma. Exposure to this cytostatic readily triggered all characteristics associated with chemoresistance such as cross-resistance to different antineoplastic agents, radioresistance, slowed cell growth both in vitro and in vivo, and the expression of ABC-type transporters. In almost all cell lines, it also forced the expression of resistant phenotypes within a very short time and proved to be more efficient in inducing MDR phenotypes than other cytostatics, for example, members of the anthracycline group (Díaz-Carballo et al., 2014). Furthermore, etoposide provoked the expression of tissue-specific stem cell markers, as globally analyzed in whole cell populations. In neuroblastoma and glioblastoma cell lines, acquired resistance was associated with the expression of p75NTR, CXCR4, c-Kit, nestin, Oct-4, CD44 variants 3 and 10, and CD133. Long-term studies have shown that the expression of these antigens, although cyclic, was clearly sustainable for long periods of time beyond in vitro CSC induction and persisted even after xenografting to immunoincompetent mice. Even more important was the global change in the physiology of these cells forced by etoposide toward acquirement of the CSC status. In addition, the analysis and validation of iCSC microRNAs isolated from 79HF6 glioblastoma cells revealed a subtle deregulation of different miRNAs such as miR204-5p known to contribute to tumor aggressiveness and metastatic behavior (Díaz-Carballo et al., 2008b; Bao et al., 2013; Qiu et al., 2013). We decided to take a descriptive rather than analytic approach to the miRNA results, as the understanding of the function of the studied representatives and this field in general is still evolving (Fig. 1).
In the course of the development of chemoresistance in vitro, the cell populations showed both qualitative and quantitative alterations, which were mostly cyclic: for example, nuclei underwent drastic morphological changes, the number of syncytium or plasmodium-like entities was increased, mitochondria showed pathological anterograde and retrograde migration patterns, cells were subject to methuosis-like processes, and neuronal morphology as well as spindle forms were observed in a large number of cell lines of different histological origins. In untreated tumors, the presence of a few oversized and mostly multinucleated cells was the rule. During drug exposure, however, these cells became more numerous and heterogeneous. They were also accompanied by populations with much smaller cell sizes than their parental cells and a variety of hypertrophic cells detectable by light microscopy. The latter showed a pathologically increased cell volume and nucleus/cytoplasm ratio but returned to normal after long periods in cultivation, as did the number of oversized or small cells (Figs. 2 and 3).
As widely described in the literature (Shahriyari and Komarova, 2013), resistant tumor cells undergo different forms of division, including asymmetrical ones, a common finding also seen in our resistant systems. In addition, we observed resistant syncytium-like entities undergoing a kind of cytokinesis, resulting in two similar daughter cells that either (1) proceeded to form new colonies or (2) died off (Fig. 7). This is in contrast to the accepted concept that syncytia emerge from cell–cell fusions rather than from cellular division. Moreover, the cells co-expressed stem cell markers such as SCF, p75NTR, and CD133, which led us to the idea that these entities are, in fact, CSCs (Walton et al., 2004; Harris et al., 2008; Wu et al., 2008; McCord et al., 2009; Jin et al., 2010; Han et al., 2013).
In addition to the deregulated cellular morphology and heterogeneity, we noted some changes in growth patterns, among which the formation of spherical colonies, slowed growth, and metaplastic behavior were the most prominent (Fig. 2). The emergence of spherical colonies was observed in both solid and hematological malignancies and was comparable to colonies obtained by hanging drop cultures. During the metaplastic process, a peculiar temporary form of attachment of suspension cultures to culture flask surfaces was noted. The growth kinetics of etoposide-iCSCs differed depending on tumor type and level of resistance. Cells grew much slower or remained dormant for several months until they returned to their initial division rates.
Spiral cells
One of the most strikingly amplified cell types among the resistant populations we generated, the spiral cells, was marked by a number of nuclei arranged in a helical pattern that left an empty central space, as shown by 3D reconstruction techniques. These cells were found among tumor cell lines of different embryonic origins and proved to be etoposide-inducible, as they already existed in naïve tumors but were more abundant in resistant tumors both in vitro and in vivo. Another striking feature was the accumulation of mitochondria around their nuclei, which was also noted in resistant single-nucleus nonspiral cells. In H79F6, a highly malignant glioblastoma cell line, spiral cells survived even after xenografting to immunoincompetent mice. We noticed the co-expression of stem cell markers such as SCF, CD44, and CD133 by these cells (in cultures and xenografts) and by morphologically similar cells in patient paraffin sections. Undoubtedly, cells of this kind are identical to those identified as CSCs by Molina et al. (2010), who studied their metastatic behavior, however without linking them to chemotherapy resistance. Moreover, cells with spirally arranged nuclei were already described as “doughnut” or “wreath” cells present among anaplastic large-cell lymphoma cells. Since we have deeply analyzed the morphological structure of their helical nuclei by 3D-reconstruction and found this cell type in a different set of tumor entities, that is, in (1) chemotherapy-resistant leukemia, colon carcinoma, neuroblastoma, glioblastoma, and astrocytoma cell lines, (2) in the respective xenografted tumors, and in (3) patient sections (Fig. 3D–Q), we termed them spiral cells (Fig. 3).
Pregnant and monastery cells
Another prominent cell type, the plasmodium-like pregnant cells, was ubiquitous both in resistant cell lines and in patient/xenograft material. These giant cells were characterized by the presence of many daughter cells inside their cytoplasm, which was delimited by a common external membrane. The fact that these cells were present both in cell cultures and in xenograft/patient tumors without being CD45 positive excludes the possibility of infiltrations into the in vivo or patient specimens by immune cells (Supplementary Fig. S1). We also noted that, in contrast to classical syncytia which are a product of cell–cell fusion, the daughter cells were capable of dividing in an intramural manner, retaining their own cytoplasmic membranes or entraining membrane material from the parental cell during eclosion. Another key feature was the presence of a main giant nucleus, which too is incongruous with the syncytial concept. This morphologic peculiarity was shared by the monastery cells, another atypical variety possibly representing a pregnant cell subtype with the distinguishing feature of a sturdy perimeter wall. Daughter cells possessed a well-developed cytoplasmic membrane as detected by labeling for markers such as CD44 and CD133. Spatial studies such as 3D reconstructions revealed that the daughter cells living inside those entities are ejected from the mother cells by two principal virus-like mechanisms, a lytic and lysogenic form. The lytic form was associated with membrane disruption and outright disintegration of the mother cell; in the lysogenic form, daughter cells dragged with them a part of the parental membranes. Interestingly, these cells expressed anti-apoptotic proteins such as Bcl-2 and Bcl-xL rather than pro-apoptotic ones such as Bax or Bad and harbored large accumulations of mitochondria in both their cytoplasmic and perinuclear compartments (Supplementary Fig. S2). These findings suggest that major apoptotic events are unlikely to occur in these cell populations, opening a front for studying the physiological role of mitochondria in these highly resistant cells (Figs. 4 and 5).
Hirtenzellen or shepherd cells
In the pool of heterogeneous populations, a very rare cell type first detected in Jurkat [an acute lymphoid leukemia (ALL) cell line] was the Hirtenzellen (shepherd cells). These elusive, pear-shaped cells were capable of sprouting a single, highly mobile lobopodium that interacted with nearby colonies. These impressively vigorous movements and deformations were the hallmark of extremely active cells capable of rapid cytoskeletal rearrangement, suggesting a very high energy turnover. Unfortunately, the scarcity of these cells made them difficult to characterize (Fig. 6).
Relationship between etoposide-induced atypical cell types and current theories of cancer formation
Polyploidization can be considered a reversible event caused by DNA-damaging insults, especially where TP53 is absent or dysfunctional, and it is strictly associated with reversible senescence (Erenpreisa and Cragg, 2007, 2013). Since polyploid cells were already detected in naïve tumors but were found to be much more numerous in chemoresistant ones, we have to consider that their number was increased simply by exposure to genotoxic agents. In cell cultures and tumor material, we noted different recurrent multinucleated TP53 negative cell types that we categorized according to their morphological characteristics (Supplementary Fig. S3). This enables us to determine their role in tumor formation and chemotherapy resistance in a better way in order to develop new specific therapies.
Undoubtedly, studying the biochemical mechanisms underlying the replication and segregation of multinucleated cell populations in a tumor is a very difficult task. In recent years, a number of illustrative descriptions were provided by Erenpreisa, who summarized many aspects of this problem by integrating observations made by herself and others (Erenpreisa and Cragg, 2007, 2013; Erenpreisa et al., 2008).
It is evident that certain events reported in this work concerning the generation of cell heterogeneity through polyploid cells appear to be linked to the oncogerminative theory of tumor formation proposed by Vinnitsky. The author suggests that malignant transformation of somatic cells is based on activated embryogenic programs which are evolutionary conserved that govern the expression of germ cell phenotypes. Particularly attractive, if theoretical, is the relationship he established between embryonic development and the functionality/heterogeneity (oncogerminative, oncotrophoblastic, and oncosomatic entities) of cancer cells. Some of our findings, in particular those related to pregnant and monastery cells, are consistent with his theory (Vinnitsky, 1993).
Rajaraman has introduced the neosis theory that tries to explain the replication mechanism of multinucleated cells. Neosis is thought to occur in postsenescent multinucleated cells, and is characterized by karyokinesis via nuclear budding followed by asymmetric cytokinesis. This process gives rise to aneuploid mononuclear progenies with extended lifespans and transient stem cell features, while the polyploid mother cells die (Rajaraman et al., 2006; Erenpreisa and Cragg, 2007, 2013). Although attractive, the neosis concept does not provide a satisfactory explanation of three major aspects of our observations: our cell populations show no senescence as determined by the low expression of beta galactosidase, the mother cells do not die, and the stem cell phenotype persists even when xenografted, suggesting it is stable rather than transient.
General considerations with regard to etoposide-induced rare cell types
Atypical cells as described in this work were, in fact, seen in naïve cell cultures too, although they were not nearly as numerous; there is no doubt that exposure to etoposide played a pivotal role in their numerical increase. We hypothesize that etoposide triggers a mechanism that drives some cells into initiating endoreplicative events rather than simply fusing membranes with neighboring cells. Only a few cells showed this behavior, indicating that they possessed an individual genetic reprogramming capacity. Interestingly, events like deregulated cellular morphology and heterogeneity were not associated with etoposide exposure in normal cells isolated from heart (Supplementary Fig. S4). In the case of pregnant cells, the existence of a big main nucleus and the parallel intracytoplasmic development of daughter cells points to a new cell type. Daughter cells possessed a cell membrane and emerged from the mother cell by disrupting its membrane or by wrapping themselves into it. These are highly relevant findings if we look at viral life cycles and keep in mind that some cancer entities have a viral etiology, such as glioblastomas and neuroblastomas (Flægstad et al., 1999; Dziurzynski et al., 2012). This leads us to hypothesize that exposure of CSCs to etoposide may activate a set of integrated functional viral genes that govern transformation and growth. Thus, molecular signatures for such processes should be studied in the future.
An aspect in our morphologic studies was the specific identification of these cell entities with antisera raised against whole resistant populations. Using polyclonal antibodies, we were able to detect these cells in glioblastoma patient material and in paraffin sections obtained from xenografted resistant cells in immunoincompetent mice. Moreover, using the same antibodies, we found that cell lysates from resistant tumors showed different protein expression patterns than lysates from naïve material.
The role of mitochondria in chemotherapy-refractory cell lines and tumors
A first clue to an atypical role of mitochondria in resistant populations was the pathological fusion and exchange of these organelles between cells via different routes. In one form, needle-like projections inject the mitochondrial bolus preferentially into the perinuclear space of adjacent cells. Another form is the ability of resistant cells to incorporate free mitochondria exogenously added to the culture. Even more impressive was the finding that isolated nuclei with or without perinuclear mitochondria were able to recruit exogenous mitochondria to their perinuclear spaces (Figs. 8 and 9). Etoposide-induced or selected pregnant cells and syncytium-like entities showed enormous agglomerations of mitochondria around their nuclei and/or daughter cells; while MFN2, a protein involved in mitochondrial fusion, was principally localized on mitochondria in the perinuclear compartment of resistant cells (Fig. 10). Moreover, our EM studies suggest that nuclei may “archive” organelles in their intermembrane space to ensure the continuity of cellular metabolism (Fig. 8). Combining the earlier findings, we hypothesize that some nuclei, in response to cytostatic therapy, can develop a mitochondrial protective capsule which spatially separates them from the cytoplasm and prevents drugs from penetrating. Nuclei can hibernate inside this structure for long periods of time, possibly in order to raise more sophisticated resistance mechanisms. The nucleus then abandons the vehicle to resume activity and form new colonies (Figs. 2 and 10). The biochemical constitution of these capsules may, therefore, be a product of mitochondrial degradation around the nuclei. Moreover, their minute size may help cell-free nuclei travel into regions of the body that are impervious to regular cells, with significant implications for metastasis formation. Interestingly, some encapsulated nuclei retained a mitochondria-free region, indicating that the mitochondria have no tropism for this annular zone probably due to its biochemical composition. Nuclei with a complete mitochondrial envelope were completely impermeable to dyes such as Hoechst 33258 or antibodies that recognize nuclear associated proteins such as lamin C (Fig. 11). This is consistent with a forgotten observation dating back to the 1950s that indicates the existence of heterogeneous nuclei according to differential histochemical staining (Lison, 1955). Thus, we conclude that the perinuclear accumulation of mitochondria may explain why resistant CSCs show divergent kinetics in the uptake of nuclear dyes such as Hoechst 33258. Highly resistant cells are capable of eliminating dyes within the first 20 min probably via ABC transports. Moreover, there is a population that remains impermeable to this dye at the same time and is only detectable after many hours of dye exposure, as deduced from IF and Hoechst 33258 kinetics.
In order to propose a reconstruction of the mitochondrial accumulation process around nuclei, it is necessary to take into consideration the existence of two different nuclear morphologies as observed in etoposide-treated U87 cells. One type is marked by a comet-like tail of mitochondria adjunct to the endoplasmatic reticulum, while others accumulate these organelles uniformly in the perimeter of the nucleus. After a process similar to methuosis, which is probably caused by fused giant mitochondria, the nucleus starts an encapsulation process that ends in its separation from the rest of the cytoplasmic environment (Fig. 8). It appears that nuclei with comet-like tails have better chances of creating a capsule, hibernating inside, and afterward emerging to form de novo colonies. Hypothetically, this type of nuclei could signify a new form of cell migration, because a nucleus-sized cell may be better equipped to travel by the blood stream and settle at its final destination, where it actively incorporates other organelles or factors necessary for further development, such as mitochondria.
All these inducible biological events resistance, heterogeneity, new cell entities expressing CSCs-markers, and nuclear encapsulation as a defensive response to xenobiotics were also observed with other antineoplastic agents we have studied. Although cytostatics may prolong the life of oncologic patients, they could select or trigger the differentiation of CSCs into more aggressive tumor entities, as has been widely observed in clinical oncology. Thus, new therapeutics that target structures unique to cancer stem cells are urgently needed (Díaz-Carballo et al., 2014).
Authors' Contribution
David Díaz-Carballo: Principal investigator. Study conception, design, and planning. Biological experiments, data analysis, and interpretation. Article conception and preparation. Holger Jastrow: Morphological studies on CSCs, EM, and article revision. Ali Haydar Acikelli: Biological studies, data acquisition. Sebastian Gustmann: Biological studies, IHC, ICC, data acquisition, and statistics. Jacqueline Klein: Biological studies, data acquisition. Ulrike Dembinski: Biological studies, data acquisition. Walter Bardenheuer: Biological studies, article revision, and corrections. Sascha Malak: Article revision and corrections. Philipp Dammann: Animal experiments. Marcos J. Araúzo-Bravo: microRNA analysis and statistics. Beate Schultheis Article revision and corrections. Constanze Aldinger: Biological studies, data acquisition. Dirk Strumberg: Article revision and corrections.
Supplementary Material
Acknowledgment
This investigation was supported by grants afforded by the Mildred Scheel Foundation (106993, 108608 Bonn, Germany).
Disclosure Statement
The authors declare that there are no conflicts of interest.
References
- Baguley B.C. (2010). Multiple drug resistance mechanisms in cancer. Mol Biotechnol 46,308–316 [DOI] [PubMed] [Google Scholar]
- Banaz-Yasar F., Gedik N., Karahan S., Diaz-Carballo D., Bongartz B.M., and Ergun S. (2012). LINE-1 retrotransposition events regulate gene expression after X-ray irradiation. DNA Cell Biol 31,1458–1467 [DOI] [PubMed] [Google Scholar]
- Bao B., Ali S., Ahmad A., Azmi A.S., Li Y., Banerjee S., Kong D., Sethi S., Aboukameel A., Padhye S.B., and Sarkar F.H. (2012). Hypoxia-induced aggressiveness of pancreatic cancer cells is due to increased expression of VEGF, IL-6 and miR-21, which can be attenuated by CDF treatment. PLoS One 7,e50165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao B., Azmi A.S., Li Y., Ahmad A., Ali S., Banerjee S., Kong D., and Sarkar F.H. (2014). Targeting CSCs in tumor microenvironment: the potential role of ROS-Associated miRNAs in tumor aggressiveness. Curr Stem Cell Res Ther 9,22–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W., Wang H.H., Tian F.J., He X.Y., Qiu M.T., Wang J.Y., Zhang H.J., Wang L.H., and Wan X.P. (2013). A TrkB-STAT3-miR-204-5p regulatory circuitry controls proliferation and invasion of endometrial carcinoma cells. Mol Cancer 12,155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkowiak D., Stempfhuber M., Wiegel T., and Bottke D. (2009). Radiation- and chemoinduced multidrug resistance in colon carcinoma cells. Strahlenther Onkol 185,815–820 [DOI] [PubMed] [Google Scholar]
- Broadley K.W., Hunn M.K., Farrand K.J., Price K.M., Grasso C., Miller R.J., Hermans I.F., and McConnell M.J. (2011). Side population is not necessary or sufficient for a cancer stem cell phenotype in glioblastoma multiforme. Stem Cells (Dayton, Ohio) 29,452–461 [DOI] [PubMed] [Google Scholar]
- Cao Y., Eble J.M., Moon E., Yuan H., Weitzel D.H., Landon C.D., Yu-Chih Nien C., Hanna G., Rich J.N., Provenzale J.M., and Dewhirst M.W. (2013). Tumor cells upregulate normoxic HIF-1alpha in response to doxorubicin. Cancer Res 73,6230–6242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan D., Frank S., and Rojo M. (2006). Mitochondrial dynamics in cell life and death. Cell Death Differ 13,680–684 [DOI] [PubMed] [Google Scholar]
- Chan D.C. (2006a). Mitochondria: dynamic organelles in disease, aging, and development. Cell 125,1241–1252 [DOI] [PubMed] [Google Scholar]
- Chan D.C. (2006b). Mitochondrial fusion and fission in mammals. Annu Rev Cell Dev Biol 22,79–99 [DOI] [PubMed] [Google Scholar]
- Chan D.C. (2007). Mitochondrial dynamics in disease. N Engl J Med 356,1707–1709 [DOI] [PubMed] [Google Scholar]
- Chau C.H., and Figg W.D. (2012). Angiogenesis inhibitors increase tumor stem cells. Cancer Biol Ther 13,586–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekhonin V.P., Shein S.A., Korchagina A.A., and Gurina O.I. (2013). VEGF in tumor progression and targeted therapy. Curr Cancer Drug Targets 13,423–443 [DOI] [PubMed] [Google Scholar]
- Cheng C., Liu Z.G., Zhang H., Xie J.D., Chen X.G., Zhao X.Q., Wang F., Liang Y.J., Chen L.K., Singh S., et al. (2012). Enhancing chemosensitivity in ABCB1- and ABCG2-overexpressing cells and cancer stem-like cells by an aurora kinase inhibitor CCT129202. Mol Pharm 9,1971–1982 [DOI] [PubMed] [Google Scholar]
- De Palma M., and Nucera S. (2012). Circulating endothelial progenitors and tumor resistance to vascular-targeting therapies. Cancer Discov 2,395–397 [DOI] [PubMed] [Google Scholar]
- Díaz-Carballo D., Acikelli A.H., Bardenheuer W., Gustmann S., Malak S., Stoll R., Kedziorski T., Nazif M.A., Jastrow H., Wennemuth G., et al. (2014). Identification of compounds that selectively target highly chemotherapy refractory neuroblastoma cancer stem cells. Int J Clin Pharmacol Ther 2014June6. PMID: [DOI] [PubMed] [Google Scholar]
- Díaz-Carballo D., Acikelli A.H., Gustmann S., Bardenheuer W., and Strumberg D. (2010). Acquired resistance to cytostatics triggers cancer stem-cell-like phenotype in different tumor entities. J Stem Cells Regen Med 6,146–147 [PubMed] [Google Scholar]
- Díaz-Carballo D., Malak S., Bardenheuer W., Freistuehler M., and Peter Reusch H. (2008a). The contribution of plukenetione A to the anti-tumoral activity of Cuban propolis. Bioorg Med Chem 16,9635–9643 [DOI] [PubMed] [Google Scholar]
- Díaz-Carballo D., Malak S., Bardenheuer W., Freistuehler M., and Reusch H.P. (2008b). Cytotoxic activity of nemorosone in neuroblastoma cells. J Cell Mol Med 12,2598–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnenberg V.S., and Donnenberg A.D. (2005). Multiple drug resistance in cancer revisited: the cancer stem cell hypothesis. J Clin Pharmacol 45,872–877 [DOI] [PubMed] [Google Scholar]
- Dziurzynski K., Chang S.M., Heimberger A.B., Kalejta R.F., McGregor Dallas S.R., Smit M., Soroceanu L., and Cobbs C.S. (2012). Consensus on the role of human cytomegalovirus in glioblastoma. Neuro Oncol 14,246–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economopoulou P., Kaklamani V.G., and Siziopikou K. (2012). The role of cancer stem cells in breast cancer initiation and progression: potential cancer stem cell-directed therapies. Oncologist 17,1394–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erenpreisa J., and Cragg M.S. (2007). Cancer: a matter of life cycle? Cell Biol Int 31,1507–1510 [DOI] [PubMed] [Google Scholar]
- Erenpreisa J., and Cragg M.S. (2013). Three steps to the immortality of cancer cells: senescence, polyploidy and self-renewal. Cancer Cell Int 13,92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erenpreisa J., Ivanov A., Wheatley S.P., Kosmacek E.A., Ianzini F., Anisimov A.P., Mackey M., Davis P.J., Plakhins G., and Illidge T.M. (2008). Endopolyploidy in irradiated p53-deficient tumour cell lines: persistence of cell division activity in giant cells expressing Aurora-B kinase. Cell Biol Int 32,1044–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flægstad T., Andresen P.A., Johnsen J.I., Asomani S.K., Jørgensen G.-E., Vignarajan S., Kjuul A., Kogner P., and Traavik T. (1999). A possible contributory role of BK virus infection in neuroblastoma development. Cancer Res 59,1160–1163 [PubMed] [Google Scholar]
- Franko A., Baris O.R., Bergschneider E., von Toerne C., Hauck S.M., Aichler M., Walch A.K., Wurst W., Wiesner R.J., Johnston I.C., and de Angelis M.H. (2013). Efficient isolation of pure and functional mitochondria from mouse tissues using automated tissue disruption and enrichment with anti-TOM22 magnetic beads. PLoS One 8,e82392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet J.P., and Gottesman M.M. (2010). Mechanisms of multidrug resistance in cancer. Methods Mol Biol (Clifton, NJ) 596,47–76 [DOI] [PubMed] [Google Scholar]
- Grimm M., Krimmel M., Polligkeit J., Alexander D., Munz A., Kluba S., Keutel C., Hoffmann J., Reinert S., and Hoefert S. (2012). ABCB5 expression and cancer stem cell hypothesis in oral squamous cell carcinoma. Eur J Cancer 48,3186–3197 [DOI] [PubMed] [Google Scholar]
- Grindberg R.V., Yee-Greenbaum J.L., McConnell M.J., Novotny M., O'Shaughnessy A.L., Lambert G.M., Arauzo-Bravo M.J., Lee J., Fishman M., Robbins G.E., et al. (2013). RNA-sequencing from single nuclei. Proc Natl Acad Sci U S A 110,19802–19807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D.W., Tapia N., Arauzo-Bravo M.J., Lim K.T., Kim K.P., Ko K., Lee H.T., and Scholer H.R. (2013). Sox2 level is a determinant of cellular reprogramming potential. PLoS One 8,e67594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen O., Overgaard J., Hansen H.S., Overgaard M., Hoyer M., Jorgensen K.E., Bastholt L., and Berthelsen A. (1997). Importance of overall treatment time for the outcome of radiotherapy of advanced head and neck carcinoma: dependency on tumor differentiation. Radiother Oncol 43,47–51 [DOI] [PubMed] [Google Scholar]
- Harris M.A., Yang H., Low B.E., Mukherjee J., Guha A., Bronson R.T., Shultz L.D., Israel M.A., and Yun K. (2008). Cancer stem cells are enriched in the side population cells in a mouse model of glioma. Cancer Res 68,10051–10059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y.L., Jahangiri A., De Lay M., and Aghi M.K. (2012). Hypoxia-induced tumor cell autophagy mediates resistance to anti-angiogenic therapy. Autophagy 8,979–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F., Zhao L., Guo Y.J., Zhao W.J., Zhang H., Wang H.T., Shao T., Zhang S.L., Wei Y.J., Feng J., et al. (2010). Influence of Etoposide on anti-apoptotic and multidrug resistance-associated protein genes in CD133 positive U251 glioblastoma stem-like cells. Brain Res 1336,103–111 [DOI] [PubMed] [Google Scholar]
- Kitambi S.S., and Chandrasekar G. (2011). Stem cells: a model for screening, discovery and development of drugs. Stem Cells Cloning 27,51–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein D., Weisshardt P., Kleff V., Jastrow H., Jakob H.G., and Ergun S. (2011). Vascular wall-resident CD44+multipotent stem cells give rise to pericytes and smooth muscle cells and contribute to new vessel maturation. PLoS One 6,e20540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch Y., van Furden B., Kaiser S., Klein D., Kibschull M., Schorle H., Carpinteiro A., Gellhaus A., and Winterhager E. (2012). Connexin 31 (GJB3) deficiency in mouse trophoblast stem cells alters giant cell differentiation and leads to loss of oxygen sensing. Biol Reprod 87,37. [DOI] [PubMed] [Google Scholar]
- Lison L. (1955). Staining differences in cell nuclei. Q J Microsc Sci 96,227–237 [Google Scholar]
- Lobo N.A., Shimono Y., Qian D., and Clarke M.F. (2007). The biology of cancer stem cells. Annu Rev Cell Dev Biol 23,675–699 [DOI] [PubMed] [Google Scholar]
- Lu C., and Shervington A. (2008). Chemoresistance in gliomas. Mol Cell Biochem 312,71–80 [DOI] [PubMed] [Google Scholar]
- Makarovskiy A.N., Siryaporn E., Hixson D.C., and Akerley W. (2002). Survival of docetaxel-resistant prostate cancer cells in vitro depends on phenotype alterations and continuity of drug exposure. Cell Mol Life Sci 59,1198–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques D.S., Sandrini J.Z., Boyle R.T., Marins L.F., and Trindade G.S. (2010). Relationships between multidrug resistance (MDR) and stem cell markers in human chronic myeloid leukemia cell lines. Leuk Res 34,757–762 [DOI] [PubMed] [Google Scholar]
- Marthaler A.G., Tiemann U., Arauzo-Bravo M.J., Wu G., Zaehres H., Hyun J.K., Han D.W., Scholer H.R., and Tapia N. (2013). Reprogramming to pluripotency through a somatic stem cell intermediate. PLoS One 8,e85138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Padura I., Marighetti P., Agliano A., Colombo F., Larzabal L., Redrado M., Bleau A.M., Prior C., Bertolini F., and Calvo A. (2012). Residual dormant cancer stem-cell foci are responsible for tumor relapse after antiangiogenic metronomic therapy in hepatocellular carcinoma xenografts. Lab Invest 92,952–966 [DOI] [PubMed] [Google Scholar]
- Mascre G., Dekoninck S., Drogat B., Youssef K.K., Brohee S., Sotiropoulou P.A., Simons B.D., and Blanpain C. (2012). Distinct contribution of stem and progenitor cells to epidermal maintenance. Nature 13,257–262 [DOI] [PubMed] [Google Scholar]
- McCord A.M., Jamal M., Williams E.S., Camphausen K., and Tofilon P.J. (2009). CD133+ glioblastoma stem-like cells are radiosensitive with a defective DNA damage response compared with established cell lines. Clin Cancer Res 15,5145–5153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlin J.L., Bour-Dill C., Marchal S., Bastien L., and Gramain M.P. (2000). Resistance to paclitaxel induces time-delayed multinucleation and DNA fragmentation into large fragments in MCF-7 human breast adenocarcinoma cells. Anticancer Drugs 11,295–302 [DOI] [PubMed] [Google Scholar]
- Mo W., and Zhang J.T. (2012). Human ABCG2: structure, function, and its role in multidrug resistance. Int J Biochem Mol Biol 3,1–27 [PMC free article] [PubMed] [Google Scholar]
- Mohammadi-Yeganeh S., Paryan M., Mirab Samiee S., Soleimani M., Arefian E., Azadmanesh K., Mostafavi E., Mahdian R., and Karimipoor M. (2013). Development of a robust, low cost stem-loop real-time quantification PCR technique for miRNA expression analysis. Mol Biol Rep 40,3665–3674 [DOI] [PubMed] [Google Scholar]
- Molina J.R., Hayashi Y., Stephens C., and Georgescu M.-M. (2010). Invasive glioblastoma cells acquire stemmness and increased Akt activation. Neoplasia 12,453–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muturi H.T., Dreesen J.D., Nilewski E., Jastrow H., Giebel B., Ergun S., and Singer B.B. (2013). Tumor and endothelial cell-derived microvesicles carry distinct CEACAMs and influence T-cell behavior. PLoS One 8,e74654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai E., Park K., Yawata T., Chihara T., Kumazawa A., Nakabayashi H., and Shimizu K. (2009). Enhanced MDR1 expression and chemoresistance of cancer stem cells derived from glioblastoma. Cancer Invest 27,901–908 [DOI] [PubMed] [Google Scholar]
- Nakanishi T., and Ross D.D. (2012). Breast cancer resistance protein (BCRP/ABCG2): its role in multidrug resistance and regulation of its gene expression. Chin J Cancer 31,73–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Estevez V., Pignatelli J., Arauzo-Bravo M.J., Hurtado-Chong A., and Vicario-Abejon C. (2013). A global transcriptome analysis reveals molecular hallmarks of neural stem cell death, survival, and differentiation in response to partial FGF-2 and EGF deprivation. PLoS One 8,e53594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquier J., Guerrouahen B.S., Al Thawadi H., Ghiabi P., Maleki M., Abu-Kaoud N., Jacob A., Mirshahi M., Glas L., Rafii S., and Le foll F. (2013). Preferential transfer of mitochondria from endothelial to cancer cells through tunneling nanotubes modulates chemoresistance. J Transl Med 11,94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrawala L., Calhoun T., Schneider-Broussard R., Zhou J., Claypool K., and Tang D.G. (2005). Side population is enriched in tumorigenic, stem-like cancer cells, whereas ABCG2+ and ABCG2- cancer cells are similarly tumorigenic. Cancer Res 65,6207–6219 [DOI] [PubMed] [Google Scholar]
- Prigione A., Rohwer N., Hoffman S., Mlody B., Drews K., Bukowiecki R., Blumlein K., Wanker E.E., Ralser M., Cramer T., and Adjaye J. (2014). HIF1alpha modulates cell fate reprogramming through early glycolytic shift and up-regulation of PDK1-3 and PKM2. Stem Cells (Dayton, Ohio) 32,364–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y.H., Wei Y.P., Shen N.J., Wang Z.C., Kan T., Yu W.L., Yi B., and Zhang Y.J. (2013). miR-204 inhibits epithelial to mesenchymal transition by targeting slug in intrahepatic cholangiocarcinoma cells. Cell Physiol Biochem 32,1331–1341 [DOI] [PubMed] [Google Scholar]
- Rajaraman R., Guernsey D.L., Rajaraman M.M., and Rajaraman S.R. (2006). Stem cells, senescence, neosis and self-renewal in cancer. Cancer Cell Int 6,25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard V., Nair M.G., Santhosh Kumar T.R., and Pillai M.R. (2013). Side population cells as prototype of chemoresistant, tumor-initiating cells. Biomed Res Int 2013,517237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabisz M., and Skladanowski A. (2009). Cancer stem cells and escape from drug-induced premature senescence in human lung tumor cells: implications for drug resistance and in vitro drug screening models. Cell Cycle (Georgetown, TX) 8,3208–3217 [DOI] [PubMed] [Google Scholar]
- Schumacher U., Nehmann N., Adam E., Mukthar D., Slotki I.N., Horny H.P., Flens M.J., Schlegelberger B., and Steinemann D. (2012). MDR-1-overexpression in HT 29 colon cancer cells grown in SCID mice. Acta Histochem 114,594–602 [DOI] [PubMed] [Google Scholar]
- Shahriyari L., and Komarova N.L. (2013). Symmetric vs. asymmetric stem cell divisions: an adaptation against cancer? PLoS One 8,e76195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N., Daley G.Q., and Cantley L.C. (2013). Stem cell metabolism in tissue development and aging. Development (Cambridge, England) 140,2535–2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spees J.L., Olson S.D., Whitney M.J., and Prockop D.J. (2006). Mitochondrial transfer between cells can rescue aerobic respiration. Proc Natl Acad Sci U S A 103,1283–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacy A.E., Jansson P.J., and Richardson D.R. (2013). Molecular pharmacology of ABCG2 and its role in chemoresistance. Mol Pharmacol 84,655–669 [DOI] [PubMed] [Google Scholar]
- Stavrovskaya A.A., and Stromskaya T.P. (2008). Transport proteins of the ABC family and multidrug resistance of tumor cells. Biochemistry 73,592–604 [DOI] [PubMed] [Google Scholar]
- Tannock I.F. (2001). Tumor physiology and drug resistance. Cancer Metastasis Rev 20,123–132 [DOI] [PubMed] [Google Scholar]
- Vazquez-Martin A., Cufi S., Corominas-Faja B., Oliveras-Ferraros C., Vellon L., and Menendez J.A. (2012). Mitochondrial fusion by pharmacological manipulation impedes somatic cell reprogramming to pluripotency: new insight into the role of mitophagy in cell stemness. Aging 4,393–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinnitsky V.B. (1993). Oncogerminative hypothesis of tumor formation. Med Hypotheses 40,19–27 [DOI] [PubMed] [Google Scholar]
- Walton J.D., Kattan D.R., Thomas S.K., Spengler B.A., Guo H.-F., Biedler J.L., Cheung N.-K.V., and Ross R.A. (2004). Characteristics of stem cells from human neuroblastoma cell lines and in tumors. Neoplasia 6,838–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihua Z., Lin Q., Ramoth A.J., Fan D., and Fidler I.J. (2011). Formation of solid tumors by a single multinucleated cancer cell. Cancer 117,4092–4099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Oh S., Wiesner S.M., Ericson K., Chen L., Hall W.A., Champoux P.E., Low W.C., and Ohlfest J.R. (2008). Persistence of CD133+ cells in human and mouse glioma cell lines: detailed characterization of GL261 glioma cells with cancer stem cell-like properties. Stem Cells Dev 17,173–184 [DOI] [PubMed] [Google Scholar]
- Xu X., Duan S., Yi F., Ocampo A., Liu G.H., and Izpisua Belmonte J.C. (2013). Mitochondrial regulation in pluripotent stem cells. Cell Metab 18,325–332 [DOI] [PubMed] [Google Scholar]
- Zheng X., Naiditch J., Czurylo M., Jie C., Lautz T., Clark S., Jafari N., Qiu Y., Chu F., and Madonna M.B. (2013). Differential effect of long-term drug selection with doxorubicin and vorinostat on neuroblastoma cells with cancer stem cell characteristics. Cell Death Dis 4,e740. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.