Abstract
Although fibroblasts and multipotent stromal/stem cells, including adipose-derived stromal cells (ADSCs), have been extensively studied, they cannot be clearly distinguished from each other. We, therefore, investigated the cellular and molecular characteristics of ADSCs and fibroblasts. ADSCs and fibroblasts share several morphological similarities and surface markers, but were clearly found to be different types of cells. Contrary to previous reports, fibroblasts were not able to differentiate into adipocytes, osteoblasts, or chondrocytes. Polysome-bound mRNA profiling revealed that ∼1,547 genes were differentially expressed (DE) in the two cell types; the genes were related to cell adhesion, the extracellular matrix, differentiation, and proliferation. These findings were confirmed by functional analyses showing that ADSCs had a greater adhesion capacity than fibroblasts; the proliferation rate of fibroblasts was also higher than that of ADSCs. Importantly, 185 DE genes were integral to the plasma membrane and, thus, candidate markers for ADSC isolation and manipulation. We also observed that an established marker of fibroblasts and ADSCs, CD105, was overexpressed in ADSCs at both mRNA and protein levels. CD105 expression seemed to be related to differentiation capacity, at least for adipogenesis. This study shows that ADSCs and fibroblasts are distinct cell types. These findings should be taken into account when using these two cell types in basic and therapeutic studies.
Introduction
Multipotent stromal/stem cells (MSCs), including adipose-derived stromal cells (ADSCs), and fibroblasts not only share a similar morphology but also they both proliferate well and express many of the same cell surface markers. Fibroblasts usually express high levels of MSC markers and do not express hematopoietic markers [1–3]. Currently, the best approach to distinguishing between MSCs and fibroblasts is based on the analysis of their functional properties. MSCs self-renew and retain a multipotent differentiation capacity, whereas fibroblasts seem to display only limited, or no such, multipotent differentiation [2], although there is controversy in the literature over this issue. Indeed, it has been suggested that fibroblasts are able to differentiate [1,3–7] and could be used in cell therapy [8]. Recently, Blasi et al. [3] reported that fibroblasts may differentiate but lack anti-inflammatory and angiogenic capacity. Recent studies comparing MSCs and fibroblasts, however, indicate that fibroblasts have no differentiation capacity [2,9]. Thus, a more detailed and rigorous comparative analysis of ADSCs and fibroblasts is needed to document the identity of these cell types and to determine the multipotent differentiation capacity of fibroblasts, if any. Functional genomics studies using total mRNA and miRNA have established an MSC-specific molecular signature consisting of only 64 genes and 21 miRNAs: the expression of these genes is at least 10-fold higher and that of the miRNAs is 2-fold higher in MSCs than in fibroblasts [9].
Several studies have shown that MSCs are probably promiscuous transcribers [10–13] and, as previously stated, MSCs seem to be multi-differentiated cells at the molecular level, because they usually express markers and regulators of various differentiated cell lineages [14]. Most attempts to determine the mRNA profile of self-renewing cells have used total RNA for high throughput analyses [15,16]. Studies comparing mRNA and protein levels in eukaryotes indicate that, although transcript levels correlate with protein synthesis, the strength of the correlation is low, suggesting a high degree of post-transcriptional regulation (reviewed by Keene [17]). These various findings suggest that studies using the total population of transcripts do not necessarily represent the identity of these cells faithfully.
In this work, we show that fibroblasts do not differentiate or differentiate only very poorly. We conducted mRNA profiling analyses and identify ∼1,547 transcripts isolated from polysomal fractions (ie, associated with the translation machinery) that are differentially expressed (DE) between the two cell types. Thus, we describe 20 times more DE transcripts than previously reported. These transcripts are related to cell adhesion, the extracellular matrix, and differentiation. Functional assays confirmed our findings and clearly show that fibroblasts are a terminally differentiated cell type, but which are probably difficult to isolate with a high degree of purity. Thus, we demonstrate that dermal fibroblasts and ADSCs are functionally and molecularly different entities.
Materials and Methods
Cell culture
Tissue samples were obtained, and stem cells were isolated as previously described [10]. All samples were collected after informed consent had been obtained, in accordance with guidelines for research involving human subjects, and with the approval of the Ethics Committee of Fundação Oswaldo Cruz, Brazil (approval number 419/07). ADSCs were cultured in DMEM/F12 medium (Gibco Invitrogen), with 10% fetal calf serum (FCS; Gibco Invitrogen), 100 U/mL penicillin, and 100 μg/mL streptomycin (Sigma-Aldrich). The cell isolation protocols resulted in a population highly enriched (>95%) in adult MSCs, as defined by Dominici et al. [18]. Normal human adult dermal fibroblasts were obtained from the American Type Culture Collection (ATCC PCS-201-012) and were cultured in fibroblast growth basal medium, with 7.5 mM L-glutamine, 5 ng/mL rhFGF, 5 μg/mL recombinant human insulin, 1 μg/mL hydrocortisone, 50 μg/mL ascorbic acid, and 2% fetal bovine serum (all from ATCC). As required for the purposes of comparison, the different cell types were cultured in DMEM/F12 medium, with 10% FCS (Gibco Invitrogen), 100 U/mL penicillin, and 100 μg/mL streptomycin (Sigma-Aldrich). All cultures were maintained at 37°C, in a humidified atmosphere containing 5% CO2. The culture medium was changed every 3 or 4 days.
Phenotypic characterization by flow cytometry
Surface proteins on ADSC and fibroblasts were detected by cytofluorometry as previously described [10] with modifications. Three samples of each cell type, between the third and fifth passages, were first incubated with purified mouse IgG (used to block Fc receptors), and then incubated with anti-CD90-FITC, anti-CD105-PE, anti-CD73-APC, anti-CD45-FITC, anti-CD34-PE, anti-HLA-DR-APC, anti-CD31-FITC, anti-CD117-PE, anti-CD19-FITC or anti-CD200-APC mAb, or to the corresponding IgG matched negative controls. For the detection of intracellular keratins, cells were incubated for 30 min, after the fixation/permeabilization process, with unconjugated primary mouse-specific antibodies: anti-KRT18 or anti-KRT19. Cell samples were then stained with an anti-mouse secondary antibodie conjungate to Alexa Fluor 488. Cell fixation and permeabilization was performed with a commercial kit according to the manufacturer's instructions (Cytofix/Cytoperm Kit; BD Biosciences). The samples were subsequently analyzed using an FACSCanto II apparatus (Becton Dickinson). A cell gate excluding cell debris and nonviable cells was determined using forward and side scatter (SSC) parameters, and was confirmed in some experiments by propidium iodide staining and immediate analysis of unfixed cells. Analyses were done after recording at least 10,000 events for each sample. Results are expressed as percentages of fluorescence-positive cells (% cell+) and mean fluorescence intensity (MFI).
Differentiation into mesenchymal lineages
The capacity of ADSCs and fibroblasts to differentiate into adipocytes, osteoblasts, and chondroblasts was assessed.
For adipogenic differentiation, cultures were treated with hMSC Adipogenic Differentiation Bullet Kit (Lonza), in accordance with the manufacturer's instructions. Briefly, cells were grown in mesenchymal stem cell growth medium (MSCGM) until they reached confluence. At 100% confluence, three cycles of induction/maintenance were performed. Each cycle consisted of feeding cells with supplemented adipogenic induction medium (dexamethasone, indomethacin, human insulin, and IBMX) and culture for 3 days followed by 1–3 days of culture in supplemented adipogenic maintenance medium (human insulin). After completion of the three cycles, cells were cultured for an additional 7 days in adipogenic maintenance medium. Control cells were cultured only in adipogenesis maintenance medium. Oil Red O staining was used to visualize the accumulation of cytoplasmic triglycerides in the cells. Briefly, the cells were washed in phosphate buffered saline (PBS), fixed with 4% paraformaldehyde for 30 min, and incubated for 30 min with 0.5% Oil Red O solution (Sigma-Aldrich).
Osteogenic differentiation was induced with the hMSC Osteogenic Differentiation Bullet Kit (Lonza), in accordance with the manufacturer's instructions. Briefly, cells were grown in MSCGM medium until 60%–70% confluence. After that, the growth medium was replaced by supplemented osteogenic induction medium (dexamethasone, ascorbate, and β-glycerophosphate), which was changed twice a week for 21 days. Control cells were maintained in MSCGM medium. To determine the degree of osteogenic differentiation and calcium deposition, the culture was stained with Alizarin Red S (Sigma-Aldrich). Briefly, the cells were washed in PBS, fixed with 4% paraformaldehyde for 30 min, and incubated for 30 min with 2% Alizarin Red S solution, pH 4.2.
Micromass cultures and hMSC Chondrogenic Differentiation Bullet Kit (Lonza) were used to promote chondrogenic differentiation, in accordance with the manufacturer's instructions. Briefly, 2.5×105 cells were washed in incomplete chondrogenic induction medium (dexamethasone, ascorbate, ITS+supplement, sodium pyruvate, and proline) and finally centrifuged in complete chondrogenic induction medium [incomplete chondrogenic induction medium plus transforming growth factor-β3 (TGF-β3)] at 150 g for 5 min to form a pellet. Micromass cells were fed twice a week for 21 days. Chondrogenesis was visualized by toluidine blue staining. Briefly, the cells were washed in PBS, fixed with 10% formaldehyde for 1 h, dehydrated in serial ethanol dilutions, and embedded in paraffin blocks. Sections (4 μm thick) of the paraffin blocks were stained with toluidine blue solution (Sigma-Aldrich) for histological analysis to demonstrate the presence of intracellular matrix mucopolysaccharides.
The stained cells were examined and photographed under inverted Nikon Eclipse TE300 or Eclipse E600 microscopes.
Quantification of adipocyte differentiation by Nile Red staining
Cells were washed with PBS, fixed by incubation with 4% paraformaldehyde for 10 min, and washed again with PBS. They were then stained with a solution of Nile Red (Sigma-Aldrich), prepared immediately before use by diluting 1,000-fold a stock solution (1 mg/mL of Nile Red dissolved in dimethylsulfoxide) in PBS, for 30 min at 4°C. The cells were washed with PBS and stained with DAPI for 20 min, washed again with PBS, and photographed. Ten images of random fields were obtained at a magnification of two hundred with Nikon Eclipse TE300 fluorescence microscope. The area in pixels per nucleus was determined using ImageJ software (National Institutes of Health).
Sucrose density gradient separation and RNA purification
Polysomal fractions from hADSC and fibroblast cultures at 50% to 60% confluence were prepared according to [19]. In brief, cells were treated with 0.1 mg/mL cycloheximide (Sigma-Aldrich) for 10 min at 37°C, removed from the culture flasks with a cell scraper, and resuspended in 0.1 mg/mL cycloheximide in PBS. The suspension was centrifuged (2,000 g for 5 min), and the resulting pellet was washed twice with 0.1 mg/mL cycloheximide in PBS. The cells were lysed by incubation for 10 min on ice with polysome buffer (15 mM Tris-HCl pH 7.4, 1% Triton X-100, 15 mM MgCl2, 0.3 M NaCl, 0.1 μg/mL cycloheximide, and 1 mg/mL heparin), and the cell lysate was centrifuged at 12,000 g for 10 min at 4°C. The supernatant was carefully isolated, loaded onto 10% to 50% sucrose gradients, and centrifuged at 39,000 rpm (HIMAC CP80WX HITACHI) for 160 min at 4°C. The sucrose gradient was fractionated with the ISCO gradient fractionation system (ISCO Model 160 gradient former), connected to a UV detector to monitor the absorbance at 275 nm, and the polysome profile was recorded. The polysomal RNA fractions were extracted by a standard Trizol (Invitrogen) RNA isolation protocol.
cDNA library construction and RNA sequencing
Polysome-associated RNA samples were amplified using the Amino Allyl Message Amp II aRNA Amplification Kit (Ambion), to generate templates for SOLiD libraries. The cDNA libraries were prepared with the SOLiD Whole Transcriptome Analysis Kit (Applied Biosystems), and the purified products were evaluated with an Agilent Bioanalyzer (Agilent). Library molecules were subjected to clonal amplification according to the SOLiD Full-Scale Template Bead preparation protocol and sequenced with the SOLiD4 System (Applied Biosystems).
RNA sequencing data analysis
RNA sequencing (RNA-seq) data analysis was performed as previously described [19]. In brief, NGSQC [20] software was used for quality control analysis of sequencing data. Various quality indicators were explored visually for each sample (distribution of colors per sample/tile, genomic hit count per sample with different numbers of mismatches, sequencing read density, and mean quality values for each sample). All samples passed the quality control filters. Mapping and counting were performed with the R package Rsubread [21]. Hierarchical clustering of the samples (log of counts plus one) was performed to evaluate biological variability. Each sample was normalized to one million reads to account for library size. We also conducted a correspondence analysis (COA), involving a dimension reduction method, to the matrix of counts, to explore associations between variables. COA allows samples and genes to be visualized simultaneously, revealing associations between them: Genes, or samples, lying close to each other tend to behave similarly.
For the comparison of fibroblasts with ADSCs, we analyzed only those genes with counts of more than 1 per million. Genes DE between cell types were identified with the edge R bioconductor package [22]. This set of genes was used for gene ontology (GO) term analysis with the Gene Ontology enRIchment anaLysis and visuaLizAtion tool (Gorilla: http://cbl-gorilla.cs.technion.ac.il/), a web server that identifies enriched GO terms in long lists of genes. Gorilla results were then visualized using the REVIGO software (REViGO: http://revigo.irb.hr/), which summarizes long lists of GO terms with respective significance values by removing redundancy in terms.
Reverse transcription-polymerase chain reaction and western blotting
RT-PCR and quantitative polymerase chain reaction (qPCR) were performed as previously described [10]. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript was used as an internal control. Amplifications were performed with cells from different experiments, with technical triplicates. Student's t-test was used to assess the significance of differences between the cell populations. The gene ID, sequence of primers, number of cycles, amplicon size, and annealing temperature are indicated in Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/scd). We considered P values<0.05 statistically significant.
Western blotting (WB) was performed as previously described [11].
NanoLC-MS/MS analysis
Peptide mixtures from two samples (∼7×105 cells/sample) were separated by online RP nanoscale capillary LC (nanoLC) and analyzed by ESI MS/MS. The experiments were performed with an Ultra 1D Plus (Eksigent) system connected to the LTQ-Orbitrap XL ETD mass spectrometer equipped with a nano-electrospray ion source (Thermo Scientific). Peptides were chromatographically separated in a 15-cm fused silica emitter (75 μm inner diameter) packed in-house with reversed-phase ReproSil-Pur C18-AQ 3 μm resin (Dr. Maisch GmbH).
Peptide mixtures were injected onto the column at a flow rate of 250 nL/min and subsequently eluted at a flow rate of 250 nL/min from 5% to 40% ACN in 0.1% formic acid over 180 min. The mass spectrometer was operated in data-dependent mode to switch automatically between MS and MS/MS (MS2) acquisition. Survey full-scan MS spectra (at 350–1,650 m/z range) were acquired in the Orbitrap analyzer with resolution R=60,000 at m/z 400 (after accumulation to a target value of 1,000,000 in the linear ion trap). The ten most intense ions were sequentially isolated and fragmented in the linear ion trap using collision-induced dissociation at a target value of 10,000. Former target ions selected for MS/MS were dynamically excluded for 90 s. Total cycle time was ∼3 s. The general mass spectrometric conditions were spray voltage, 2.3 kV; no sheath and auxiliary gas flow; ion transfer tube temperature, 100°C; collision gas pressure, 1.3 mTorr; and normalized collision energy using wide-band activation mode 35% for MS2. Ion selection thresholds were 250 counts for MS2. An activation q=0.25 and activation time of 30 ms were applied for MS2 acquisitions. The “lock mass” option was enabled in all full scans to improve mass accuracy for precursor ions [23].
Proteomic data analysis
The MaxQuant platform (version 1.3.0.5) [24], which includes the algorithm Andromeda [25] for database searching, was used for Peaklist picking, and protein identification, quantification, and validation. Default parameters of the software were used for all analysis steps, unless stated otherwise. Proteins were searched against a “decoy database” prepared by reversing the sequence of each entry of the human protein sequence database [containing 87,061 protein sequences, downloaded in March 29, 2010 from IPI protein database (version 3.68)] and appending them to the forward sequences. This database was complemented with frequently observed contaminants (porcine trypsin, Achromobacter lyticus lysyl endopeptidase, and human keratins) and their reversed sequences. Search parameters specified an MS tolerance of 7 ppm, an MS/MS tolerance of 0.5 Da, and full trypsin specificity, allowing for approximately two missed cleavages. Carbamidomethylation of cysteine was set as a fixed modification, and oxidation of methionines and N-terminal acetylation (protein) were allowed as variable modifications.
For validation of the identifications, a minimum peptide length of six amino acids and two peptides per protein were required. In addition, a posterior error probability threshold of 0.01 was applied at both peptide and protein levels.
Bromodeoxyuridine proliferation assay
The proliferation assay was performed according to [26]. In brief, fibroblasts and ADSCs at 70% confluence were incubated with 100 μM bromodeoxyuridine (BrdU; Invitrogen) for 24 h. The cells were detached with trypsin and fixed by incubation with 100% ethanol for 30 min on ice. The samples were centrifuged, and the cell pellet was resuspended in 100 μL of distilled water, heated for 5 min at 95°C, and rapidly chilled in an ice-water bath. The cells were incubated with an Alexa Fluor 488-conjugated anti-BrdU antibody (Invitrogen) for 30 min at room temperature. An FACSCanto II flow cytometer (BD Bioscience) and Flow Jo software (Tree Star) were used for quantitative analyses of BrdU-labeled cells.
Cell adhesion assay
ADSCs and fibroblasts were tested for adhesion to 24-well culture plates for 20 and 40 min at 37°C as follows. The cells were first washed and then added to the wells at a concentration of 2.5×104 cells per well in 500 μL DMEM/F12 medium (Gibco Invitrogen) supplemented with 10% FCS. After the adhesion period (20 or 40 min), the plates were maintained for 5 min of incubation under shaking (100 rpm) and nonadherent cells were removed by washing thrice with ice-cold PBS. Adherent cells were fixed (100% ethanol for 5 min) and stained (2% toluidine blue for 15 min) for counting. Four wells for each cell type (three ADSC and three fibroblast samples) were analyzed and photographed for counting labeled cells with the ImageJ software (National Institutes of Health). Five representative fields from each well were used to calculate mean values.
ADSC and fibroblast cell sorting
Cytofluorometry was used for detection and separation of ADSC and fibroblast populations. A mixed sample of each cell type, between the fourth and fifth passages, was first incubated with anti-CD105-PE mAb, and subsequently sorted using an FACSaria II apparatus (Becton Dickinson). A cell gate discriminating between low and high CD105-PE fluorescence intensity was determined to separate cell populations. Doublets were excluded from the cell population using a sequential gating strategy relative to width versus height on SSC and forward scatter (FSC) dot plots. Finally, weak (CD151+) and bright (CD151++) CD105-positive cells were selected and sorted using the cell sorter's purity option at a rate of 2,500 events per second. Sorted populations were reanalyzed (5×103 events recorded) for purity and viability, and 4×105 cells of each population were culture expanded for imunophenotyping and cell differentiation assays.
Statistical analysis of functional assays
Statistical significance was determined by Student's t-test using GraphPad Prism software. Values of P<0.05 were considered significant. Data are expressed as means±standard deviation.
Results
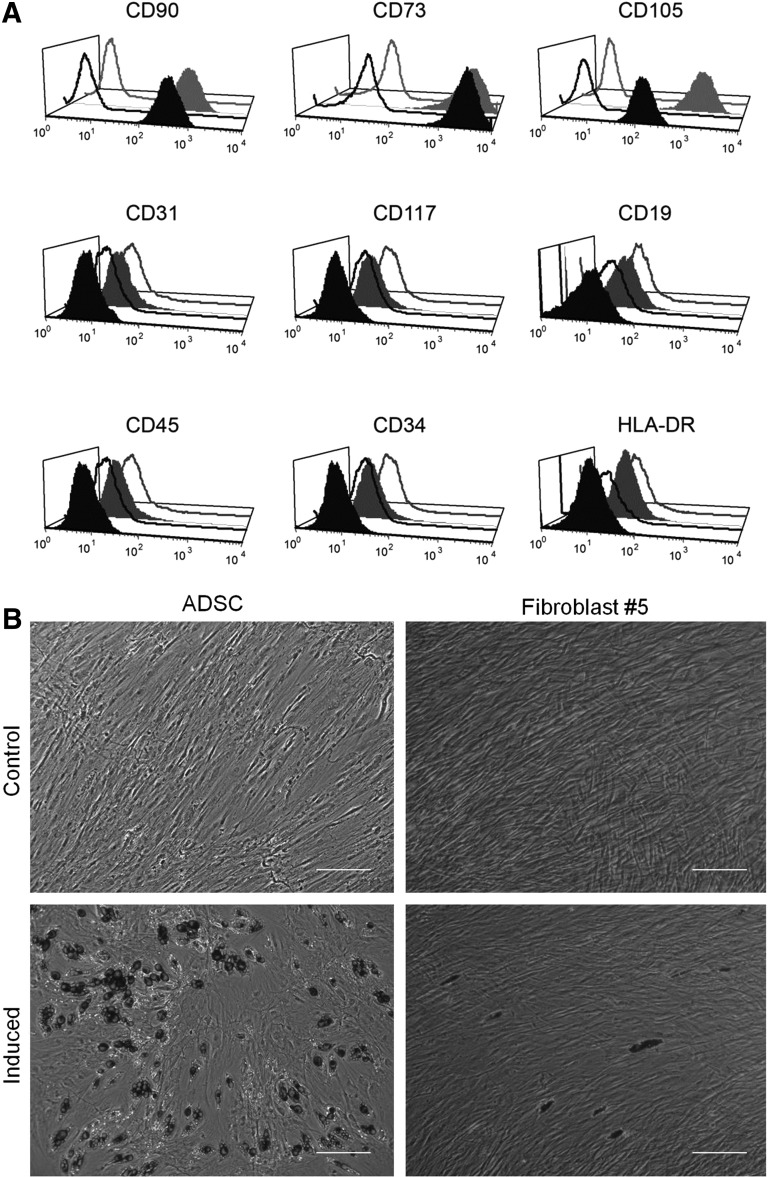
The expression of cell-surface antigens proposed by Dominici et al. [18] as a minimal set for stromal/stem cell characterization was evaluated by flow cytometry in three biological samples of ADSCs and three technical samples of dermal fibroblast at passage P5 (Fig. 1A). With one exception, the two types of cells displayed similar immunophenotypes for the markers analyzed. Cells were uniformly positive for the membrane glycoprotein CD90 and the surface enzyme ecto 5′ nucleotidase CD73. No detectable contamination by hematopoietic or endothelial cells was observed, as flow cytometry analysis was negative for CD19, CD31, CD34, CD45, CD117, and HLA-DR. More than 95% of the cells in both cell populations expressed the endoglin receptor CD105; however, the MFI was significantly higher for ADSCs than for fibroblasts (Fig. 1A).
FIG. 1.
Immune and functional phenotypes of ADSCs and fibroblasts. (A) Analysis of immune phenotypes by flow cytometry. ADSCs (gray lines) and fibroblasts (black lines) were labeled with antibodies against the indicated antigens, and analyzed by flow cytometry. Representative histograms are displayed. Isotype controls are shown as empty lines/areas, and the solid histograms indicate reactivity with the antibody. (B) Differentiation of ADSCs and fibroblasts. Both cell types at passage P5 were incubated for 21 days in the presence of specific agents inducing differentiation into adipocytes. Differentiation into the adipocyte lineage was demonstrated by staining with Oil Red O. Untreated control cultures without adipogenic differentiation stimuli are shown on the upper line of photographs. The scale bar indicates 200 μm. ADSCs, adipose-derived stromal cells.
Cells at passage P5 were compared for their multilineage differentiation plasticity by in vitro assays: specifically, differentiation to adipocytes, osteoblasts, and chondrocytes was studied. We used the presence of lipid-rich vacuoles stained with Oil Red O to analyze adipogenic induction on day 21. ADSCs presented large cells with cytoplasmic lipid-rich vacuoles; fibroblasts displayed very few differentiating cells, and they had smaller lipid droplets than those observed in ADSCs (Fig. 1B). Osteogenic differentiation was assessed by using Alizarin Red S to stain the mineralized extracellular matrix after 21 days of induction. Differentiation of ADSCs was easily detected. No differentiation was observed in the induced fibroblast cultures (Supplementary Fig. S1A). Similar results were obtained for the chondrogenic differentiation assays. ADSCs formed aggregates that became detached, floating in suspension in the culture. Sections of the aggregates stained with Toluidine Blue showed cuboidal cells and chondrocyte-like lacunae. Fibroblast differentiation was minimal or undetectable (Supplementary Fig. S1B). Untreated control cultures, which were growing in standard medium without adipogenic, osteogenic, or chondrogenic differentiation stimuli, did not exhibit spontaneous adipocyte (Fig. 1B and Supplementary Fig. S1) or osteoblast or chondroblast formation after 21 days of cultivation.
In the culture conditions used, the mean doubling time of fibroblasts is 18–24 h, and the cells were sub-cultured approximately every 3 days; thus, fibroblasts at passage P10–11 had undergone more than 30 population doublings. Consequently, fibroblasts at P10 are considered at a presenescence stage [27]. Since no evident signs of senescence were visible at P11, the adipogenic differentiation potential of fibroblasts at this passage was tested and compared with that of fibroblasts at P5. Surprisingly, there was a higher proportion of differentiated cells at P11 (Supplementary Fig. S2).
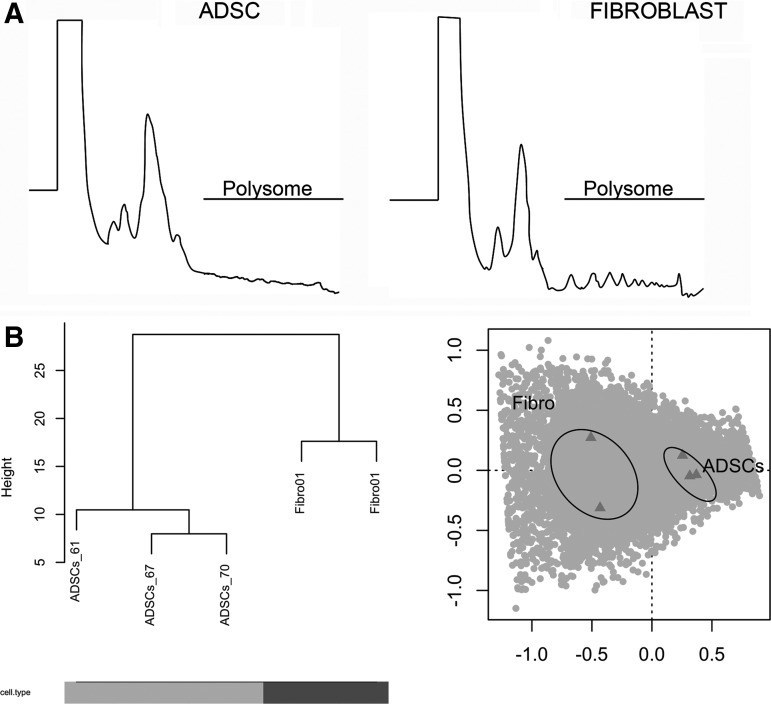
For a more detailed characterization of both cell lineages, gene expression patterns in each cell type were determined by studying mRNA associated with the translation machinery. The polysomes of ADSCs and fibroblasts were profiled by ultracentrifugation of cytoplasmic extracts onto sucrose density gradients (10%–50%) containing cycloheximide. Starting with extracts obtained from the same number of cells, the translation activity (measured as the height of polysome peaks) was lower in ADSCs than in fibroblasts (Fig. 2A). Polysome-associated mRNAs were isolated from the corresponding gradient fractions, and the mRNA fractions of ADSCs and fibroblasts were analyzed by RNA sequencing (RNA-Seq) using the SOLiD4 platform. The total number of reads obtained for each sample is shown in the Supplementary Data (Supplementary Table S2). The reads of all samples were mapped onto the reference genome (Hg19; NCBI Build 37.64), yielding a mean mapping percentage of ∼55%. To decrease ambiguity, the analyses were limited to reads mapping to a unique position in the genome. Hierarchical clustering and COA grouped the samples according to cell lineage (ADSCs and fibroblasts), rather donor, indicating that the cell-specific polysomal RNA populations are intrinsically more characteristic than donor particularities (Fig. 2B).
FIG. 2.
Polysome profiles and internal consistency of RNA sequencing data. (A) Polysome profile analysis of ADSCs (left) and fibroblasts (right) obtained by fractionation of cytoplasmic extracts in sucrose gradients 10%–50%. (B) Hierarchical clustering and correspondence analysis (COA) showing the internal consistency of the data. Left panel: dissimilarity-based (bottom-up) hierarchical clustering was performed on the log-transformed counts of genes for the various samples. Initially, each object is assigned to its own cluster and then the algorithm proceeds iteratively, at each stage joining the two most similar clusters, according to some distance measure (in this case, a complete linkage approach), continuing until there is one single cluster. The process is visualized as a dendrogram: the first branching event separates fibroblast (Fibro) from ADSC samples. The numbers of the samples correspond to the donors. The height axis represents the distance between each branching event. Cell type accounted for the largest proportions of the variance in both analyses, evidence of the consistency of the experiments. Right panel: COA with the samples. The x-axis represents the first component (that explaining the most variance 69.1%), and the y-axis represents the second component (representing 17.7% of the variance).
A total of 17,340 protein-coding transcripts were sequenced. Most transcripts (15,776) were detected in both ADSC and fibroblast polysomes, albeit with different levels of expression. Nevertheless, a substantial number of transcripts were exclusively found in only one or the other RNA population: 924 in fibroblast and 640 in ADSCs. Since polysomal-associated mRNA was used in this study, it was expected that most of the proteins identified in a proteomic data set of ADSCs would be represented in the mRNA population. To test whether this was the case, proteins from ADSCs from two samples were identified by LC-MS/MS and the results were compared with those the RNA-Seq data for the same ADSC donors. A total of 297 proteins were identified with at least two peptides in one of the ADSC samples (Supplementary Table S3) and, as expected, more than 85% of them were represented with at least five tags (after normalization) in each of the three biological samples analyzed by RNA-Seq. Moreover, of the 42 proteins (15%) not found in the RNA-Seq data, 10 were histone transcripts that lack poly-A tails and, thus, could not be detected by the RNA-Seq method used in this work.
The DE genes were identified by paired comparisons (fibroblasts vs. ADSCs) using the R package edgeR. We found 1,115 and 1,547 DE protein-coding genes in the polysomal fractions with false discovery rate (FDR) 0.0001 and <0.001, respectively (Supplementary Table S4). At these FDR values, most of the genes (>98%) showed logFC values >2 or <−2.
We confirmed the sequencing data at the mRNA level by qPCR of five genes: CAMK2N1, fibroblast specific; RPL10A, TMSB10, and ARPC2, equally expressed between the two cell types; SCRG1, ADSC specific. All five genes analyzed using the polysomal fractions confirmed our RNA-seq data (Supplementary Table S5, compare the third and fourth column). In addition, the nonpolysomal fraction was also used in qPCR with the same set of primers for calculating the translation efficiency (TE) of the five selected transcripts, that is, the ratio between the normalized qPCR data obtained from the polysomal versus the nonpolysomal fraction. The three genes equally expressed in both cell types showed a very similar TE for each transcript analyzed and the ratios between TEs of each cell type for each transcript (TE fibroblasts/TE ADSCs) were close to 1. The transcript of the gene CAMK2N1 that is over-represented in fibroblasts showed a TE 4.5 times higher in these cells than in ADSCs. Interestingly, the transcript over-represented in the polysomal fraction of ADSCs also showed a TE 4.8 times higher in fibroblasts than in ADSCs (Supplementary Table S5).
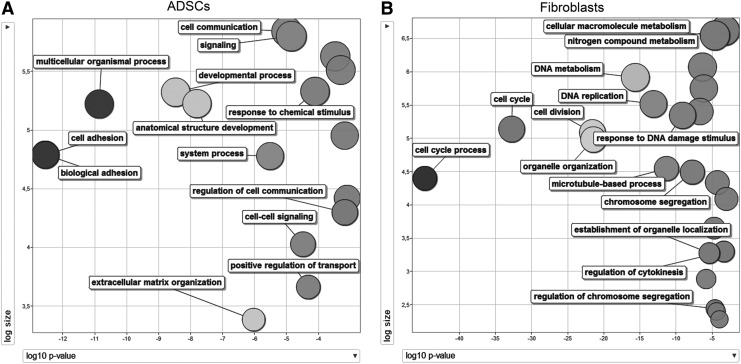
The DE genes in the polysomal fraction were analyzed in greater detail: several of the surface markers previously described to be present in both fibroblasts and ADSCs (for example, the activated leukocyte cell adhesion molecule CD166, the β1-integrin CD29, and the hyaluronate receptor CD44) were absent from the DE list (Supplementary Table S6), providing further validation of our results. To test our experimental approach as a tool to predict DE proteins between ADSCs and fibroblasts, we analyzed the expression of three proteins: one surface protein, CD200, upregulated in ADSCs and two cytoskeleton proteins, keratin (KRT) 18 upregulated in ADSCs and KRT 19 upregulated in fibroblasts. All three proteins confirmed our sequencing results by either western blot or flow cytometry (Supplementary Fig. S3). It is interesting to note that the same percentage of cells was positive for KRT18 in both types of cells; however, the MFI was twice higher in ADSCs than in fibroblasts. Two GO analyses were performed, one for the genes upregulated in fibroblasts and one for the genes upregulated in ADSC. A target gene list and a background list were considered for these analyses: the target list corresponds to DE genes with FDR<0.0001, and the background list included all 17,340 genes identified in this study. GOrilla identified 10,942 genes associated with a GO term. GOs with P<E-9 were extracted and used for REVIGO analysis and visualization. Sixteen over-represented GO terms were identified for ADSCs (P<1.9E-7), and most are related to processes such as cell adhesion, cell signaling, multicellular organism processes, and developmental processes (Fig. 3A). Thirty-seven terms were found to be over-represented in the analysis of the polysomal fraction of fibroblasts; these GO terms were similar to each other, and refer to cell cycle processes and proliferation (Fig. 3B).
FIG. 3.
GO analyses, one for the genes upregulated in ADSCs (A) and one for the genes upregulated in fibroblasts (B). REVIGO visualization of the GO analyses performed with GOrilla is shown. For these analyses, a target gene list and a background list were used: the target list is composed of genes represented with FDR<0.0001; the background list included all the 17,340 genes identified in this study. The log10 of the P value of each GO after REVIGO analyses is plotted on the x-axis, and the log10 of the size of GOs is plotted on the y-axis. FDR, false discovery rate; GO, gene ontology.
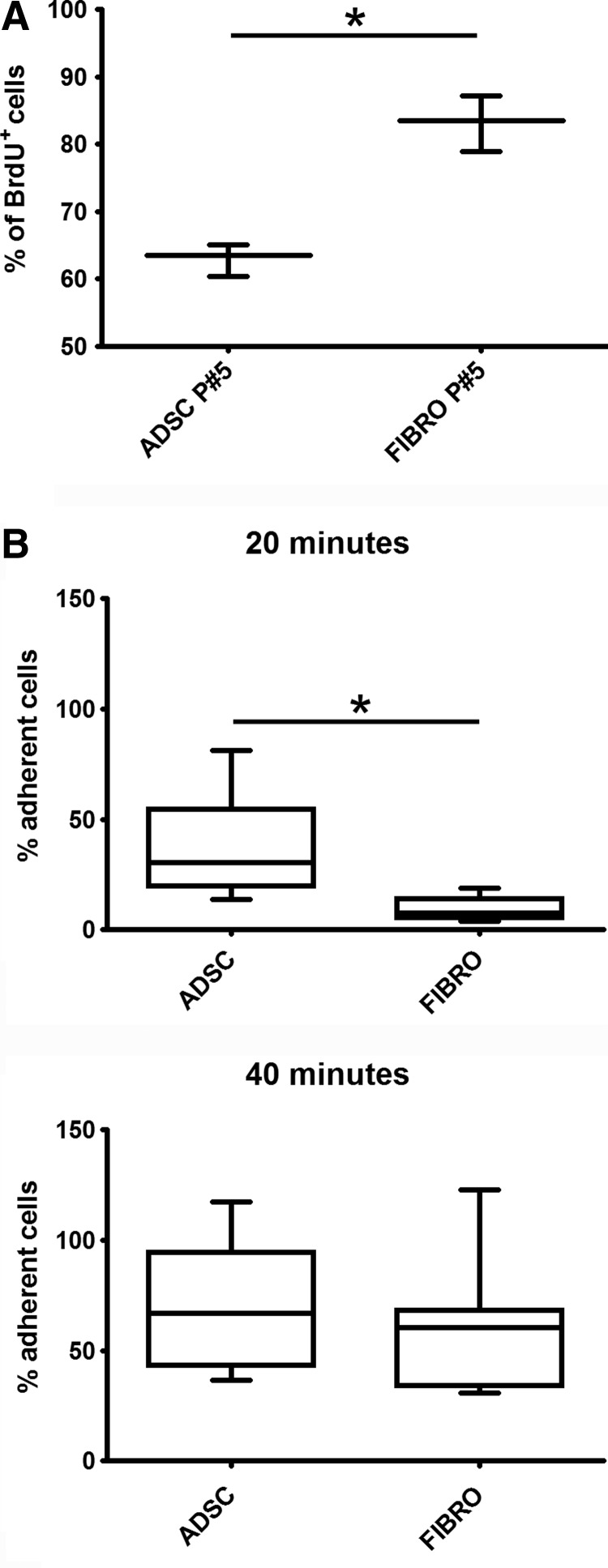
Three different approaches were used to confirm these findings. First, the RNA-Seq data was compared with the proteomic data published by Kim et al. [28]. All the transcripts represented with at least five normalized tags in each sample of ADSCs were compared with proteins in Kim's data with at least two spectra. Of the 188 proteins present in undifferentiated ADSCs, 165 (∼88%) were found to be DE by our RNA-Seq analysis. Second, differential expression was confirmed by reverse transcriptase-quantitative polymerase chain reaction with five selected transcripts (Supplementary Table S5) and by WB/cytometry of three proteins encoded by DE genes (see above and Supplementary Fig. S3). Lastly, some of the over-represented GO processes for each cell type were analyzed by functional assays. The proliferation of ADSCs was compared with that of dermal fibroblasts by studying BrdU incorporation at passage P5. Under exactly the same culture conditions (medium, temperature/CO2, confluence, and passage), fibroblasts showed significantly higher proliferation values than ADSCs: ∼20% more cells incorporated BrdU after 24 h of culture (Fig. 4A).In addition, the numbers of adherent cells after 20, 40, and 60 min of culture was determined by counting the number of nonadherent cells in the culture and subtraction from the total number. At 20 min, significantly more ADSCs than fibroblasts adhered to the plastic plates (Fig. 4B). This difference was maintained, but not statistically significant, after 40 min (Fig. 4B), and no difference was observed after 60 min of culture (data not shown). These results are in agreement with the results from the GO analysis for biological process terms obtained from RNA-Seq. Unfortunately, the number of proteins identified by the proteomic analysis was insufficient to obtain significant results in a GO analysis.
FIG. 4.
Comparison between ADSC and fibroblast proliferation and adhesion. (A) Proliferation was assessed by BrdU incorporation over 24 h. The mean numbers of BrdU-positive cells for three donors of ADSCs and for technical triplicates of fibroblasts. BrdU, bromodeoxyuridine. (B) Adhesion assay. Number of adherent cells after 20 and 40 min in culture was evaluated by counting the number of nonadherent cells in the culture and subtracting. *P<0.05.
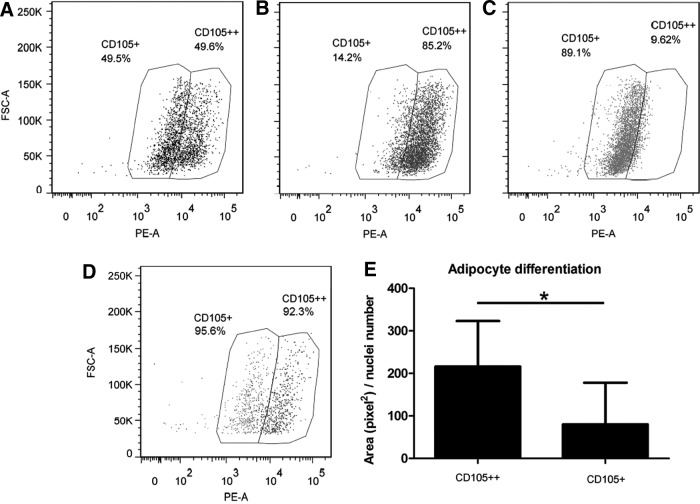
We analyzed the GO of cellular component terms: the terms associated with ADSCs concentrate mainly at the plasma membrane and extracellular location (Supplementary Fig. S4). For example, CD200 has a LogFC of 6.1 in ADSCs with an FDR=5.73E-14. In addition, the endoglin protein CD105, a well-defined surface marker for ADSCs and also for fibroblasts, is significantly overexpressed in ADSCs (LogFC=2.6). This result was confirmed by flow cytometry (Fig. 1): more than 95% of the cells of both lineages were positive for CD105; however, the level of expression was significantly higher in ADSCs than in fibroblasts (LogFC=3.7, Supplementary Table S7). We tested whether this difference would be sufficient to distinguish fibroblasts from ADSCs in a mixed population and whether it correlated with differentiation capacity. We, therefore, conducted a cell sorting assay with a mixed sample of ADSCs and fibroblasts. The mixed sample was stained with anti-CD105-PE mAb and, in an FSC versus FL2 dot plot, weakly and strongly CD105-positive cell populations were gated (Fig. 5A). There was an overlap between the CD105 expression by the two cell types impeding the definition of an unambiguous boundary between ADSC and fibroblast populations. We, therefore, established a cell gate dividing the total CD105-positive population into two equal halves (Fig. 5A). The percentages of CD105++ and CD105+ cells in each cell population before mixing are shown in Fig. 5B and C, respectively. A total of 4×105 cells of CD105++ and CD105+ cell populations were collected. By reference to the parental mixed cell population, their percentages were 92.3% and 95.6%, respectively, indicating sorting purity of more than 90% (Fig. 5D). We then subjected sorted cells to induction of adipocyte differentiation to evaluate whether these isolated populations, based on CD105 expression, display the same differentiation potential as ADSCs and fibroblasts. Sorted strongly and weakly CD105-positive populations showed, as expected, differentiation patterns resembling those exhibited by induced ADSCs and fibroblasts, respectively (Fig. 5E). The presence of rare cells with cytoplasmic lipids among induced CD105+ cells could be attributed to two phenomena: a small number of differentiating cells with small intracellular lipid droplets as observed among induced fibroblast; and the overlap between the two cell types as concerns CD105-positive events.
FIG. 5.
Isolation and differentiation of weakly and strongly CD105-positive cells from a mixed population of ADSCs and fibroblasts. (A) Representative FSC and FL2 plot of the 1:1 mixed sample of ADSCs and fibroblasts. CD105 expression by (B) ADSCs and (C) fibroblasts before sorting. (D) Representative overlay dot plot of sorted CD105+ and CD105++ cells. The percentages shown are according to the gating strategy defined in (A) and are referenced to the parental mixed cell population. (E) Sorted CD105+ and CCD105++ cells induced to differentiate into adipocytes. Columns represent the quantification of Nile Red stained area in pixel2 per nuclei number. The significance of differences between mean values was evaluated with Student's t-test. *P<0.05. FSC-A, forward scatter pulse area; PE-A, phycoerythrin area.
There were 183 genes, other than CD105 and CD200, corresponding to the “integral to plasma membrane” GO term. This means that there are more than a hundred putative cell surface markers that could be evaluated as tools or markers for ADSCs purification/manipulation (Supplementary Fig. S4 and Supplementary Table S8).
Discussion
We report data demonstrating that ADSCs and fibroblasts are different types of cells, despite sharing several morphological similarities and surface markers. They differ on the basis of hundreds of DE genes, as evaluated by the analyses of mRNAs associated with the translation machinery. We also show that, contradicting repeated previous suggestions [1,3–7], fibroblasts are not able to differentiate into adipocytes, osteoblasts, or condrocytes. We believe that the residual differentiation previously described was due to the fact that it is very difficult to obtain pure populations of dermal fibroblasts, as also suggested by Lennon et al. [29]. Thus, any differentiating “fibroblasts” observed at passage P11 may be the consequence of contaminating MSCs overgrowing the culture, as the fibroblasts themselves reach senescence, resulting in greater differentiation.
There is extensive post-transcriptional regulation in mammals and particularly in stem cells [30,31]; as a result, correlations between total mRNA and protein levels, although positive, are low, ranging from r=0.2 to 0.4 [19,32]. This is an important issue when analyzing mRNA levels in cells. Indeed, the RNAs associated with polysomes represent the cellular phenotype more faithfully than analyses of total mRNA; unfortunately, total mRNA is still widely used in most stem cell studies. Moreover, it has been observed in ADSCs differentiation assays that control cultures without differentiation inducers express several genes at the mRNA level, specific for diverse lineages such as cardiomyocytes, osteoblasts, adipocytes, and condrocytes [10,33]. In addition, promiscuous gene expression has been suggested or shown by others in MSCs, including ADSCs [13], and also in other stem cell populations [12,34,35]; these observations clearly indicate that total mRNA populations do not provide a good molecular representation of ADSCs.
It is well established that the TE of a particular mRNA could be quantified by measuring the ratio of polysomal and nonpolysomal abundance of that mRNA. The ratio will be high in case of over-representation and low in case of under-representation in a particular condition [36,37]. The mentioned correlation was observed for four out of five genes analyzed by qPCR: CAMK2N1, RPL10A, TMSB10, and ARPC2 (Supplementary Table S5). This was not true for the transcript of the SCRG1 gene, over-represented in ADSCs, as the TE was higher in fibroblasts than in ADSCs. However, the amount of transcript in the nonpolysomal fraction of ADSCs is 620 times higher than in fibroblasts (ADSCs=80.72; Fibroblasts=0.13), resulting in the over-representation observed in the polysomal fraction of ADSCs. It is important to point out that this work is comparing TEs obtained from two different kinds of cells and not from the same type of cells in two different conditions as has been the case in previous publications. Anyhow, this result is in accordance with recently published data showing the extent of post-transcriptional regulation in adult stem cells [19]. We could speculate, for example, that most of the SCRG1 mRNA present in the cell might be stored in ribonucleoprotein complexes present in the cytoplasm and a small portion access polysomes for translation.
To our knowledge, the proliferation rate of fibroblasts and ADSCs has not been considered at all when comparing these two types of cells. Here, we demonstrate that proliferation-related mRNA are better represented in the polysomal fraction of fibroblasts than ADSCs and, accordingly, under the same culture conditions at early passages (P3–5) the fibroblast proliferation rate is higher than that of ADSCs.
ADSCs adhered better than fibroblasts to a plastic surface during the first 20 min of our assay, in agreement with some of the GOs over-represented in ADSCs. Indeed, a subpopulation of MSCs derived from human bone marrow cells that exhibited enhanced adhesive and multipotent capacities has been recently identified by Bolontrade et al. [38]. It would be interesting to determine whether a subpopulation of this type also exists in ADSCs and whether it is responsible for the difference to fibroblasts in adherence.
Other functional genomics studies using MSCs under standard culture conditions, that is, without differentiation inducers, and considering total mRNA for transcriptome analyses [9,39] were only able to find a few dozen MSC-specific genes to establish a molecular signature for these cells. Conversely, the approach we used successfully identified several hundred DE genes, specific to each cell type, even under stringent criteria (FDR<0.0001). More importantly, 185 genes encoded products integral to the plasma membrane; these gene products are, therefore, candidate, easily accessible markers for ADSC isolation and manipulation. One of these genes was identified as CD200, previously detected in stromal/stem cells isolated from bone marrow, adipose tissue, and Wharton's jelly. It seems that this molecule and its receptor (CD200R, only present in myeloid-lineage cells) are responsible for two-way communication between MSCs and T-lymphocytes [40] and, probably, participate in mesenchymal stromal/stem cell immunosuppressant activity [41].
Previously, some markers were shown to distinguish MSCs from fibroblasts [40]. The expression level of CD166, CD106, integrin alpha 11, and insulin-like growth factor-2 was significantly higher in MSCs than in fibroblasts; while that of CD9, matrix metalloproteinase (MMP) 1, and MMP3 was significantly lower. CD146 was expressed only in MSCs. We had similar results for integrin alpha 11, MMP3, and CD146. The other few genes were absent in our list of DE genes (Supplementary Table S4). This might be explained, because these authors have used cells from a different source (bone marrow-derived mesenchymal stem cells) whereas we have used a very stringent set of conditions to select the DE genes; they have used total mRNA instead of mRNA associated to the translation machinery (polysomal fraction).
Halfon et al. [40] have also showed that some markers were regulated with passage: CD106, integrin alpha 11, and CD146 were downregulated in P6 of MSCs, and CD9 was upregulated; whereas other commonly used MSC markers, including CD105, did not change. This is something important to be taken into account when looking for new and solid MSC markers. We show that the established marker of fibroblasts and ADSCs, CD105, is overexpressed in ADSCs: this was demonstrated at the mRNA level by deep sequencing and at the protein level by flow cytometry. CD105 expression level seems to be related to differentiation capacity, at least for adipogenesis. A CD105++-enriched population differentiated thrice more than a CD105+-enriched population. Accordingly, it has been shown that downregulation of CD105 is associated with multilineage differentiation in human umbilical cord blood-derived mesenchymal stem cells [41]. CD105, also known as endoglin, is a type I integral membrane glycoprotein. It is abundant on proliferating cells and has been identified as an accessory receptor for TGF-β [42]. Nevertheless, it remains to be established whether CD105 is directly involved in multipotency.In conclusion, the work presented here demonstrates that ADSCs and fibroblasts are distinct cell types. Although they share various morphological and immunophenotypic similarities, there are vast differences between them in terms of gene expression and functionality/biological dynamics. These findings may be relevant for any applications of these two cell types in both basic and therapeutic studies.
Supplementary Material
Acknowledgments
This work was supported by grants from Ministério da Saúde and Conselho Nacional de Desenvolvimento Cientifico e Tecnológico—CNPq, FIOCRUZ-Pasteur Research Program, and Fundação Araucária. L.S. received fellowship from ANII (Agencia Nacional de Investigación e Innovación, Uruguay); S.G., J.Z., and B.D. from CNPq; P.S. from FIOCRUZ; and A.C. from Fundação Araucária.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Lorenz K, Sicker M, Schmelzer E, Rupf T, Salvetter J, Schulz-Siegmund M. and Bader A. (2008). Multilineage differentiation potential of human dermal skin-derived fibroblasts. Exp Dermatol 17:925–932 [DOI] [PubMed] [Google Scholar]
- 2.Alt E, Yan Y, Gehmert S, Song YH, Altman A, Gehmert S, Vykoukal D. and Bai X. (2011). Fibroblasts share mesenchymal phenotypes with stem cells, but lack their differentiation and colony-forming potential. Biol Cell 103:197–208 [DOI] [PubMed] [Google Scholar]
- 3.Blasi A, Martino C, Balducci L, Saldarelli M, Soleti A, Navone SE, Canzi L, Cristini S, Invernici G, Parati EA. and Alessandri G. (2011). Dermal fibroblasts display similar phenotypic and differentiation capacity to fat-derived mesenchymal stem cells, but differ in anti-inflammatory and angiogenic potential. Vasc Cell 3:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brohem CA, de Carvalho CM, Radoski CL, Santi FC, Baptista MC, Swinka BB, de AUC, de Araujo LR, Graf RM, Feferman IH. and Lorencini M. (2013). Comparison between fibroblasts and mesenchymal stem cells derived from dermal and adipose tissue. Int J Cosmet Sci 35:448–457 [DOI] [PubMed] [Google Scholar]
- 5.Lysy PA, Smets F, Sibille C, Najimi M. and Sokal EM. (2007). Human skin fibroblasts: From mesodermal to hepatocyte-like differentiation. Hepatology 46:1574–1585 [DOI] [PubMed] [Google Scholar]
- 6.Huang HI, Chen SK, Ling QD, Chien CC, Liu HT. and Chan SH. (2010). Multilineage differentiation potential of fibroblast-like stromal cells derived from human skin. Tissue Eng Part A 16:1491–1501 [DOI] [PubMed] [Google Scholar]
- 7.Haniffa MA, Wang XN, Holtick U, Rae M, Isaacs JD, Dickinson AM, Hilkens CM. and Collin MP. (2007). Adult human fibroblasts are potent immunoregulatory cells and functionally equivalent to mesenchymal stem cells. J Immunol 179:1595–1604 [DOI] [PubMed] [Google Scholar]
- 8.Osonoi M, Iwanuma O, Kikuchi A. and Abe S. (2011). Fibroblasts have plasticity and potential utility for cell therapy. Hum Cell 24:30–34 [DOI] [PubMed] [Google Scholar]
- 9.Bae S, Ahn JH, Park CW, Son HK, Kim KS, Lim NK, Jeon CJ. and Kim H. (2009). Gene and microRNA expression signatures of human mesenchymal stromal cells in comparison to fibroblasts. Cell Tissue Res 335:565–573 [DOI] [PubMed] [Google Scholar]
- 10.Rebelatto CK, Aguiar AM, Moretao MP, Senegaglia AC, Hansen P, Barchiki F, Oliveira J, Martins J, Kuligovski C, et al. (2008). Dissimilar differentiation of mesenchymal stem cells from bone marrow, umbilical cord blood, and adipose tissue. Exp Biol Med (Maywood) 233:901–913 [DOI] [PubMed] [Google Scholar]
- 11.Shigunov P, Sotelo-Silveira J, Kuligovski C, de Aguiar AM, Rebelatto CK, Moutinho JA, Brofman PS, Krieger MA, Goldenberg S, et al. (2012). PUMILIO-2 is involved in the positive regulation of cellular proliferation in human adipose-derived stem cells. Stem Cells Dev 21:217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tondreau T, Lagneaux L, Dejeneffe M, Massy M, Mortier C, Delforge A. and Bron D. (2004). Bone marrow-derived mesenchymal stem cells already express specific neural proteins before any differentiation. Differentiation 72:319–326 [DOI] [PubMed] [Google Scholar]
- 13.Zipori D. (2004). Mesenchymal stem cells: harnessing cell plasticity to tissue and organ repair. Blood Cells Mol Dis 33:211–215 [DOI] [PubMed] [Google Scholar]
- 14.Jiang Y, Vaessen B, Lenvik T, Blackstad M, Reyes M. and Verfaillie CM. (2002). Multipotent progenitor cells can be isolated from postnatal murine bone marrow, muscle, and brain. Exp Hematol 30:896–904 [DOI] [PubMed] [Google Scholar]
- 15.Menssen A, Haupl T, Sittinger M, Delorme B, Charbord P. and Ringe J. (2011). Differential gene expression profiling of human bone marrow-derived mesenchymal stem cells during adipogenic development. BMC Genomics 12:461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeong JA, Ko KM, Bae S, Jeon CJ, Koh GY. and Kim H. (2007). Genome-wide differential gene expression profiling of human bone marrow stromal cells. Stem Cells 25:994–1002 [DOI] [PubMed] [Google Scholar]
- 17.Keene JD. (2007). RNA regulons: coordination of post-transcriptional events. Nat Rev Genet 8:533–543 [DOI] [PubMed] [Google Scholar]
- 18.Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop D. and Horwitz E. (2006). Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 8:315–317 [DOI] [PubMed] [Google Scholar]
- 19.Spangenberg L, Shigunov P, Abud AP, Cofre AR, Stimamiglio MA, Kuligovski C, Zych J, Schittini AV, Costa AD, et al. (2013). Polysome profiling shows extensive posttranscriptional regulation during human adipocyte stem cell differentiation into adipocytes. Stem Cell Res 11:902–912 [DOI] [PubMed] [Google Scholar]
- 20.Dai M, Thompson RC, Maher C, Contreras-Galindo R, Kaplan MH, Markovitz DM, Omenn G. and Meng F. (2010). NGSQC: cross-platform quality analysis pipeline for deep sequencing data. BMC Genomics 11Suppl 4:S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao Y, Smyth GK. and Shi W. (2013). The subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acid Res 41:e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson MD, McCarthy DJ. and Smyth GK. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P. and Mann M. (2006). Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127:635–648 [DOI] [PubMed] [Google Scholar]
- 24.Cox J. and Mann M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372 [DOI] [PubMed] [Google Scholar]
- 25.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV. and Mann M. (2011). Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 10:1794–1805 [DOI] [PubMed] [Google Scholar]
- 26.Zych J, Stimamiglio MA, Senegaglia AC, Brofman PR, Dallagiovanna B, Goldenberg S. and Correa A. (2013). The epigenetic modifiers 5-aza-2′-deoxycytidine and trichostatin A influence adipocyte differentiation in human mesenchymal stem cells. Braz J Med Biol Res 46:405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makpol S, Zainuddin A, Chua KH, Yusof YA. and Ngah WZ. (2012). Gamma-tocotrienol modulation of senescence-associated gene expression prevents cellular aging in human diploid fibroblasts. Clinics (Sao Paulo) 67:135–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim J, Choi YS, Lim S, Yea K, Yoon JH, Jun DJ, Ha SH, Kim JW, Kim JH, et al. (2010). Comparative analysis of the secretory proteome of human adipose stromal vascular fraction cells during adipogenesis. Proteomics 10:394–405 [DOI] [PubMed] [Google Scholar]
- 29.Lennon DP, Haynesworth SE, Arm DM, Baber MA. and Caplan AI. (2000). Dilution of human mesenchymal stem cells with dermal fibroblasts and the effects on in vitro and in vivo osteochondrogenesis. Dev Dyn 219:50–62 [DOI] [PubMed] [Google Scholar]
- 30.Cheung TH. and Rando TA. (2013). Molecular regulation of stem cell quiescence. Nat Rev Mol Cell Biol 14:329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathieu J. and Ruohola-Baker H. (2013). Regulation of stem cell populations by microRNAs. Adv Exp Med Biol 786:329–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miranda HC, Herai RH, Thome CH, Gomes GG, Panepucci RA, Orellana MD, Covas DT, Muotri AR, Greene LJ. and Faca VM. (2012). A quantitative proteomic and transcriptomic comparison of human mesenchymal stem cells from bone marrow and umbilical cord vein. Proteomics 12:2607–2617 [DOI] [PubMed] [Google Scholar]
- 33.Rebelatto CK, Aguiar AM, Senegaglia AC, Aita CM, Hansen P, Barchiki F, Kuligovski C, Olandoski M, Moutinho JA, et al. (2009). Expression of cardiac function genes in adult stem cells is increased by treatment with nitric oxide agents. Biochem Biophys Res Commun 378:456–461 [DOI] [PubMed] [Google Scholar]
- 34.Miyamoto T, Iwasaki H, Reizis B, Ye M, Graf T, Weissman IL. and Akashi K. (2002). Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev Cell 3:137–147 [DOI] [PubMed] [Google Scholar]
- 35.Parekkadan B, Fletcher AL, Li M, Tjota MY, Bellemare-Pelletier A, Milwid JM, Lee JW, Yarmush ML. and Turley SJ. (2012). Aire controls mesenchymal stem cell-mediated suppression in chronic colitis. Mol Ther 20:178–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davidson LA, Wang N, Ivanov I, Goldsby J, Lupton JR. and Chapkin RS. (2009). Identification of actively translated mRNA transcripts in a rat model of early-stage colon carcinogenesis. Cancer Prev Res (Phila) 2:984–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas JD. and Johannes GJ. (2007). Identification of mRNAs that continue to associate with polysomes during hypoxia. RNA 13:1116–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bolontrade MF, Sganga L, Piaggio E, Viale DL, Sorrentino MA, Robinson A, Sevlever G, Garcia MG, Mazzolini G. and Podhajcer OL. (2012). A specific subpopulation of mesenchymal stromal cell carriers overrides melanoma resistance to an oncolytic adenovirus. Stem Cells Dev 21:2689–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jaager K, Islam S, Zajac P, Linnarsson S. and Neuman T. (2012). RNA-seq analysis reveals different dynamics of differentiation of human dermis- and adipose-derived stromal stem cells. PLoS One 7:e38833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Najar M, Raicevic G, Jebbawi F, De Bruyn C, Meuleman N, Bron D, Toungouz M. and Lagneaux L. (2012). Characterization and functionality of the CD200-CD200R system during mesenchymal stromal cell interactions with T-lymphocytes. Immunol Lett 146:50–56 [DOI] [PubMed] [Google Scholar]
- 41.Pietila M, Lehtonen S, Tuovinen E, Lahteenmaki K, Laitinen S, Leskela HV, Natynki A, Pesala J, Nordstrom K. and Lehenkari P. (2012). CD200 positive human mesenchymal stem cells suppress TNF-alpha secretion from CD200 receptor positive macrophage-like cells. PLoS One 7:e31671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheifetz S, Bellon T, Cales C, Vera S, Bernabeu C, Massague J. and Letarte M. (1992). Endoglin is a component of the transforming growth factor-beta receptor system in human endothelial cells. J Biol Chem 267:19027-19030 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.