Abstract
Homologous recombination (HR) maintains genomic integrity against DNA replication stress and deleterious lesions, such as double-strand breaks (DSBs). Rad51 recombinase is critical for HR events that mediate the exchange of genetic information between parental chromosomes in eukaryotes. Additionally, Rad51 and HR accessory factors may facilitate replication fork progression by preventing replication fork collapse and repair DSBs that spontaneously arise during the normal cell cycle. In this study, we demonstrated a novel role for Rad51 during the cell cycle in mouse embryonic stem cells (mESCs). In mESCs, Rad51 was constitutively expressed throughout the cell cycle, and the formation of Rad51 foci increased as the cells entered S phase. Suppression of Rad51 expression caused cells to accumulate at G2/M phase and activated the DNA damage checkpoint, but it did not affect the self-renewal or differentiation capacity of mESCs. Even though Rad51 suppression significantly inhibited the proliferation rate of mESCs, Rad51 suppression did not affect the replication fork progression and speed, indicating that Rad51 repaired DNA damage and promoted DNA replication in S phase through an independent mechanism. In conclusion, Rad51 may contribute to G2/M transition in mESCs, while preserving genomic integrity in global organization of DNA replication fork.
Introduction
Embryonic stem cells (ESCs) are derived from the inner cell mass of the early stage embryo [1]. They can remain in a pluripotent state indefinitely under optimal culture conditions [2]. During the process of asymmetric cell division and self-renewal to establish a cellular continuum, stem cells undergo chronological aging caused by the accumulation of damaged or aberrant molecules. Aberrant chromosomes are observed in up to 50% of human ESCs in long-term culture [3,4]. Aging and the accumulation of mutations in stem cells can change the fate or cellular function of stem cell progeny. To avoid the accumulation of mutations and to prevent their transmission to subsequent generations, ESCs have developed robust systems to maintain genomic stability, including DNA repair machineries. In addition to active DNA damage repair mechanisms, faithful DNA replication is essential for maintaining genomic integrity in the normal cell cycle. In asynchronous, exponentially growing cells, up to 60% of mouse ESCs (mESCs) were in S phase, compared with 20% of mouse embryonic fibroblasts (MEFs) [5–8]. Obstacles on the DNA template, caused by exogenous or endogenous factors, such as ultraviolet light, reactive oxygen species, nutrient deficiency, and deregulation of replication activity, frequently impede replication fork progression, which can result in replication fork collapse and the formation of replication-dependent DNA double-strand breaks (DSBs) [9,10]. Many redundant pathways preserve the integrity of the replication fork and thereby prevent the lethal effects caused by complete dissociation of the replication machinery on stalled or collapsed replication forks.
Homologous recombination (HR) is the predominant mechanism for the repair of DSBs and recovery of stalled DNA replication. HR is a high-fidelity form of repair because the mechanism uses a sister chromatid template containing homologous sequences to repair lesions [11]. HR predominantly occurs in the late S and G2 phases of the cell cycle, when sister chromatids are more readily available as repair templates. Competition between HR and nonhomologous end joining (NHEJ) in DSB repair or at a stalled replication fork is specifically caused by template usage in S/G2 phase [12]. Failed DSB repair or inaccurate DNA repair causes chromosomal rearrangement, chromosome loss, or carcinogenesis [13,14]. In mESCs, DSBs are predominantly repaired through the high-fidelity HR pathway, which occurs throughout the cell cycle [6,15]. The essential role of HR in mESCs is supported by the fact that basal levels of proteins involved in HR are higher in mESCs than in fibroblasts. The protein levels correlate with HR repair activity, which is two- to fourfold higher in mESCs than in MEFs [16]. In addition, knockout of genes involved in HR leads to early embryonic lethality in mice [17,18].
Rad51, the eukaryotic ortholog of RecA in Escherichia coli, is a key player in the HR pathway. Rad51 has an essential role in homology recognition and strand exchange between two homologous templates during mitotic DSB repair and meiotic recombination [19]. The Mre11-Rad50-Nbs1 complex resects initial DSBs to generate 3′ single-stranded DNA (ssDNA) tails that invade the duplex template DNA. Replication protein A (RPA) initially binds to 3′ ssDNA overhangs to produce stable RPA-coated ssDNA [12,20]; Rad51 cofactors then dissociate the RPA-ssDNA filaments. A loading factor, BRCA2, helps Rad51 bind efficiently to ssDNA [21,22]. Rad51 plays a role in replication fork progression, which is critical for maintaining the structural integrity of chromosomes and ensuring cell proliferation in vertebrates [18,23,24]. Rad51 mediates two distinct pathways that suppress replication fork disruption. One pathway promotes replication restart when a replication fork encounters DNA damage or reduced nucleotide pools [25]. The other pathway uses HR to repair DSBs that occur after exposure to some genotoxins or at broken replication forks. To promote HR, Rad51 forms a filament on the 3′ ssDNA, which then invades and anneals to a homologous template provided by replicating sister chromatids or homologous chromatids [12]. ATPase activity of Rad51 is critical for stabilizing the catalytically active nucleoprotein filament [26]. Rad51 mutants defective for either ATP binding or ATP hydrolysis are unable to restart stalled replication forks and repair DSBs in human ESCs [27]. Recently, it was reported that Rad51 plays a direct role in replication fork progression by preventing the accumulation of ssDNA gaps at replication forks, which occurs independent of HR activity [28].
In this study, we reveal a novel function of Rad51 in the cell cycle progression of mESCs. Unlike differentiated cells, mESCs constitutively express Rad51 protein throughout the cell cycle. Suppression of Rad51 led to the activation of the DNA damage checkpoint and the accumulation of cells at G2/M phase. Rad51 siRNA treatment did not slow the replication fork progression time and speed, even though it significantly inhibited cell proliferation. Based on these results, we conclude that Rad51 regulates HR in mESCs to overcome single-strand breaks, possibly caused by the rapid replication of mESCs, after the completion of DNA replication at S phase.
Materials and Methods
Cell culture
The murine embryonic stem cell line J1 (Cat. No. SCR-1010) derived from a male agouti 129S4/SvJae embryo was obtained from ATCC. J1 mESCs were grown in Dulbecco's modified Eagle's medium (DMEM) plus GlutaMax1 (Cat. No. 10569; Gibco) supplemented with 10% horse serum (Cat. No. 16050-122; Gibco), 2 mM l-glutamine (Gibco), 10 mM HEPES (Gibco), 0.1 mM minimal essential medium–nonessential amino acids (MEM-NEAA; Gibco), 0.1 mM β-mercaptoethanol (Gibco), 100 U/mL penicillin, 100 μg/mL streptomycin (penicillin/streptomycin, Cat. No. 15140; Gibco), and 1,000 U/mL mouse ESGRO leukemia inhibitory factor (LIF; Cat. No. ESG 1107; Millipore) at 37°C in a 5% CO2 atmosphere. Before each experiment, mESCs were plated on culture plates briefly coated with 0.1% gelatin and without a feeder layer. MEFs generated from the blastocyst embryo of a CF1 pregnant female mouse on embryonic day 12.5 were used between passages 3 and 5 in all experiments. MEFs were cultured in DMEM (Cat. No. 11995; Gibco) supplemented with 10% fetal bovine serum (Cat. No. 16000-044; Gibco) and penicillin/streptomycin (Cat. No. 15140; Gibco). To maintain mESCs in a low-serum environment, cells growing in 10% serum were transferred and serially adapted to serum concentrations of 5%, 2.5%, and 1% for 4 days.
Cell synchronization and fluorescence-activated cell sorting analysis
For G1/S phase synchronization, cells were treated with thymidine (Sigma) at a final concentration of 2 mM for 16 h, washed twice with prewarmed phosphate-buffered saline (PBS), and then grown in fresh medium. After a 6-h release, thymidine was added again. Cells were incubated with thymidine for another 16 h, washed, and released from the thymidine block with the addition of fresh medium. For synchronous release from G2-phase arrest, cells were treated with 5 μM diphenyleneiodonium chloride (DPI; Sigma) for 16 h, washed, and released with the addition of fresh medium. Cells were collected at the indicated times for fluorescence-activated cell sorting (FACS), immunoblot, and immunofluorescence analyses. For FACS analysis, the collected cells were immediately fixed in 70% ethanol and stained with propidium iodide (PI; Sigma) for 30 min at room temperature in the dark. The distribution of cell cycle phases was quantified with flow cytometry analysis software (FlowJo; Tree Star, Inc.).
RNA isolation and quantitative PCR
Total RNA was isolated using TRIzol (Invitrogen), and 2 μg of total RNA was reverse transcribed using Superscript II Reverse Transcriptase (Invitrogen) and oligo (dT) primers (Invitrogen). Quantitative PCR was performed with a QuantiTect SYBR Green PCR kit (Qiagen) and the Bio-Rad CFX96 Real-Time System. Primer sequences are listed in Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/scd).
BrdU FACS analysis
Cells were pulsed with BrdU (Sigma) to a final concentration of 10 μM for 20 min before harvesting. The harvested cells were washed with PBS and then fixed in 70% ethanol at 4°C for 2 h. The fixed cells were incubated with denaturation buffer (2 N HCl and 0.5% Triton X-100) for 30 min. After washes with PBS, cells were recovered with neutralization buffer (0.1 M Na2B4O7·10H2O, pH 8.5) for 30 min and incubated with BrdU antibody in PBS containing 1% bovine serum albumin (BSA) and 0.5% Tween-20 for 1 h. Fluorescein isothiocyanate (FITC)–conjugated secondary antibody was added to BrdU-treated samples. After 1 h, cells were washed with PBS, stained with PI (including RNase A), and then analyzed using an FACSCalibur flow cytometer (Becton Dickinson).
RNAi
For small interfering RNA (siRNA) knockdown of mouse Rad51 (siRad51), an siGENOME SMARTpool was used (Cat. No. M-062730-01-0005; Dharmacon). The siRNA pool contained a mixture of four targeting nucleotides with the following sequences: 5′-CAUCAUCGCUCAUGCGUCA-3′, 5′-UGUCAUACGUUGGCUGUUA-3′, 5′-GGUAAUCACCAACCAGGUA-3′, and 5′-GAGAUCAUACAGAUAACUA-3′. siRNA was purchased from Dharmacon, and cells were transfected using DharmaFECT 1 (Dharmacon) and Lipofectamine 2000 (Invitrogen) according to the manufacturers' instructions. A nonspecific siRNA (siNS) was used as a negative control (ON-TARGETplus Nontargeting pool; Dharmacon). Cells were incubated for 48 h and collected for experiments (cell cycle analysis, immunoblot analysis, and DNA labeling).
Embryoid body formation and in vitro differentiation
mESCs were trypsinized to achieve a single-cell suspension and cultured in the absence of LIF. The medium was changed every 2 days and embryoid body formation was checked after 4 days from siRad51 transfection. In vitro differentiation of mESCs was induced by removing LIF and adding 0.2 μM all-trans retinoic acid (RA) in culture medium.
Extract preparation, antibodies, and immunoblotting
Samples were washed twice with PBS and lysed in cell lysis buffer (Cat. No. 9803; Cell Signaling Technology) containing a protease inhibitor cocktail (Calbiochem) or 1 mM PMSF. Protein samples (20–50 μg) were resolved by 10% SDS-PAGE. Antibodies against the following proteins were used: Rad51 (Cat. No. sc-8349), Cyclin B1 (Cat. No. sc-752), Cyclin A (Cat. No. sc-751), CDK1 (Cat. No. sc-54), Chk1 (Cat. No. sc-8408), Oct3/4 (Cat. No. sc-5279), STAT3 (Cat. No. sc-482), Sox2 (Cat. No. sc-20088), p53 (Cat. No. sc-6243), and β-actin (Cat. No. sc-47778) from Santa Cruz Biotechnology; Phospho-Histone H3Ser10 (Cat. No. 06-570) from Millipore; Phospho-Chk1Ser317 (Cat. No. 2344), Phospho-STAT3 (Cat. No. 9131), and α-tubulin (Cat. No. 2144) from Cell Signaling; and Rad51 (Cat. No. ab63801) and γH2AX (Cat. No. ab22551) from Abcam. Proteins were transferred to polyvinylidene difluoride (PVDF) membranes (Millipore) and blocked for 1 h with 5% skim milk in tris-buffered saline (TBS) containing 0.1% Tween-20. The membrane was incubated with primary antibodies overnight at 4°C, washed with TBS containing 0.1% Tween-20 three times for 10 min each, and then incubated with horseradish-peroxidase-conjugated secondary antibodies for 1 h at room temperature. Immunoreactivity was detected with a WEST-ZOL immunoblot detection system (Cat. No. 16024; iNtRON Biotechnology). The relative amount of each protein was quantified using Quantity One software (Bio-Rad).
Immunofluorescence
Cells attached to poly-L-lysine-coated coverslips were fixed in 1% paraformaldehyde for 15 min after and then permeabilized with 0.1% Triton X-100 in PBS for 15 min. Samples were washed three times with PBS between each step. Cells were blocked with 3% BSA in PBST (PBS+0.1% Tween-20) for 30 min and then immunostained with the following primary antibodies, diluted with 3% BSA in PBST for 1 h: Rad51 (Cat. No. sc-8349), RPA (Cat. No. sc-28709), Ki67 (Cat. No. sc-7846), Cyclin B1 (Cat. No. sc-752), and Geminin (Cat. No. sc-53923) from Santa Cruz Biotechnology; Phospho-Chk1Ser317 (Cat. No. 2344) from Cell Signaling; Rad51 (Cat. No. ab63801) and γH2AX (Cat. No. ab22551) from Abcam; and ORC2 (Cat. No. NA73) and Cdt1 (Cat. No. 07-1383) from Millipore. The cells were washed with PBST three times, incubated with fluorescence-conjugated secondary antibodies (Cy3, FITC, and Alexa 488) for additional 40 min, and then mounted on glass slides with a DAPI-containing mounting solution. Samples were visualized with an Olympus BX51 fluorescence microscope equipped with a DAPI filter and fluorescent channels. Digital images were obtained with Image Pro-Express software. Images of Rad51 and ORC2 foci in Fig. 3 were acquired with a confocal microscope (Leica TCS SP5 II) and processed with Leica Application Suite Advanced Fluorescence (LAS AF) software (Leica Microsystems). Nuclear foci of IdU and CldU were quantified with GraphPad Prism Software.
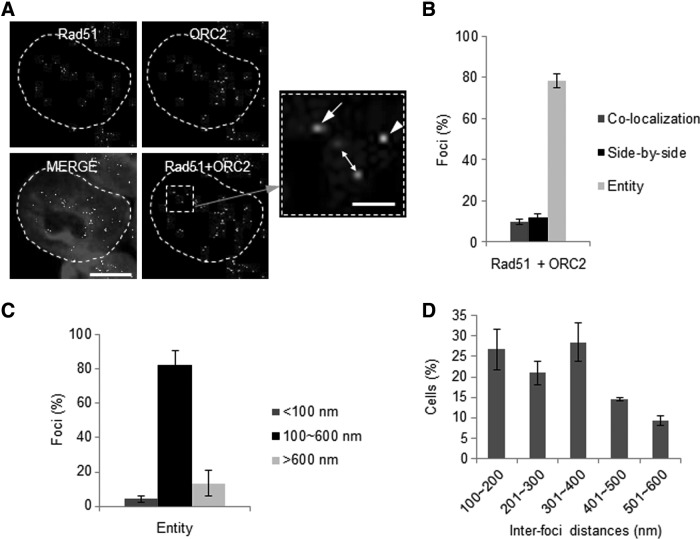
FIG. 3.
Analysis of Rad51 foci in DNA replication sites. (A) Representative images of confocal microscopy showing the formation of Rad51 and ORC2 (marker for DNA replication initiation) nuclear foci. The interfoci distances between Rad51 and ORC2 in each cell were measured using Leica Application Suite Advanced Fluorescence software. The scale bar indicates 10 μm. Foci inside the dotted square box were magnified (arrowhead: colocalization; arrow: side-by-side; full duplex arrow: entity). The scale bar in the magnified image is 1 μm. (B) Based on the pattern of nuclear foci formation, cells were divided into three groups and quantified as in (A). (C) Cell populations existing as an independent entity were subdivided into three groups based on the estimated replication factory size [50]. (D) Rad51 foci inside the replication factory. The interfoci distances of entity foci were categorized and quantified every 100 nm. Error bars indicate mean±SD.
Dynamic analysis of replication foci using thymidine analog
mESCs were synchronized at G1/S phase by double-thymine-block method after siRNA transfection and then released for 2 h. Cells were pulse labeled with 100 μM IdU (Cat. No. 347580; BD Biosciences) for 30 min. After 30-min chase time, cells were pulse labeled with 250 μM CldU (Cat. No. ab6326; Abcam) for 30 min. And then, cells were washed with PBS and fixed with cold 70% ethanol. For antibody staining, cells were incubated with primary antibodies for IdU (mouse anti-BrdU, diluted for 1:20; Cat. No. 347580; BD Biosciences) and CldU (rat anti-BrdU, diluted for 1:250; Cat. No. ab6326; Abcam) for 2 h. And then, the samples were incubated with sheep anti-mouse Cy3 (Cat. No. C2181; Sigma) for IdU or goat anti-rat Alexa Fluor488 (Cat. No. A11006; Invitrogen) for CldU for 1 h. Images were acquired with a confocal microscope (Leica TCS SP5 II) and processed with LAS AF software. Colocalization of IdU and CldU foci was quantified with GraphPad Prism Software.
DNA fiber assay
J1 cells were synchronized with a double-thymidine-block method after siRNA treatment. Cells were then pulse labeled with 100 μM IdU for 20 min, washed with prewarmed medium, and pulsed with 250 μM CldU for 20 min. At the end of the CldU pulse, cells were harvested and resuspended at a final concentration of 8×105 cells/mL in PBS. Cell lysis, DNA spreading, and immunofluorescence staining were performed as described previously [29]. In brief, the cell suspension (2.5 μL) was mixed with 7.5 μL of cell lysis buffer (50 mM EDTA and 0.5% SDS in 200 mM Tris-HCl, pH 7.5). IdU was detected using mouse anti-BrdU antibody (diluted for 1:25; Cat. No. 347580; BD Biosciences), and CldU was detected using rat anti-BrdU antibody (diluted for 1:50; Cat. No. ab6326; Abcam). Slides were incubated with primary antibodies diluted in 5% BSA for 2 h and then with sheep anti-mouse Cy3 (Cat. No. C2181; Sigma) and goat anti-rat Alexa Fluor488 (Cat. No. A11006; Invitrogen) secondary fluorescent antibodies diluted 1:250 in 5% BSA for 1 h. Fiber images were acquired using a Leica TCS SP5 II confocal microscope and analyzed with Image J software (NCI/NIH).
Flow cytometric analysis of annexin V
For apoptosis assays, the annexin V assay was performed using an ApoScan Annexin V-FITC apoptosis detection kit (Cat. No. LS-02-100; BioBud). The cells were stained with a combination of annexin V-FITC and PI and analyzed by flow cytometry. The distributions of live cells (annexin V−, PI−), early apoptotic cells (annexin V+, PI−), and late apoptotic cells (annexin V+, PI+) were analyzed with FlowJo software (Tree Star, Inc.). Both early and late apoptotic cells were classified as apoptotic cells.
Results
Rad51 is expressed throughout the cell cycle in mESCs
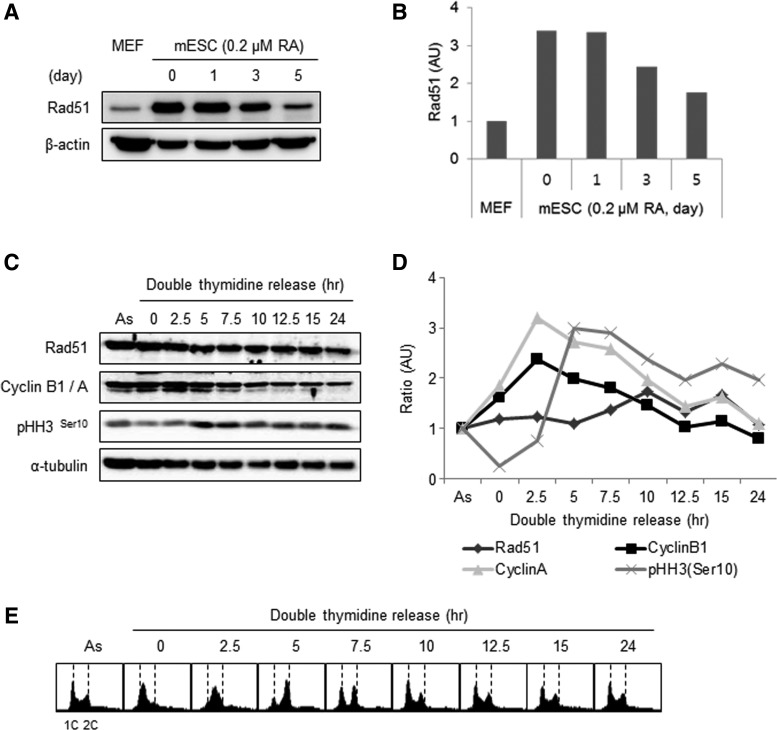
Compared with differentiated somatic cells, mESCs express higher levels of Rad51 proteins [6,8,16]. To observe the expression pattern of Rad51 during mESC differentiation, cells were treated with 0.2 μM RA to induce direct spontaneous differentiation of pluripotent stem cells. The level of Rad51 protein was ∼3.5-fold higher in mESCs than in MEFs, and it gradually decreased as mESCs differentiated (Fig. 1A, B). In differentiated cells, the level of Rad51 protein increases during S to G2 phases, but it is relatively low in thymidine- and nocodazole-arrested cells, as well as in asynchronous cells [30,31]. To understand the Rad51 expression pattern in mESCs, cells were arrested at G1/S phase with a double-thymidine block and released synchronously into the cell cycle (Fig. 1C, E). Unlike differentiated cells, mESCs maintained a steady level of Rad51 protein during the cell cycle (Fig. 1C, D). Cell cycle protein expression and FACS profiles indicated that cells entered mitosis and then returned to an asynchronous state in 5 and 10 h, respectively, after release from the double-thymidine block (Fig. 1E). The constitutive expression of Rad51 in mESCs during the cell cycle was confirmed by cell cycle arrest and release with DPI, which blocks cell cycle progression at G2 phase by downregulating cyclin B1 ([32]; Supplementary Fig. S1). Based on these results, we conclude that Rad51 protein is expressed at a high level throughout the cell cycle in mESCs.
FIG. 1.
Expression of Rad51 throughout the cell cycle in mouse embryonic stem cells (mESCs). (A) Rad51 protein levels in mESCs and mouse embryonic fibroblasts (MEFs). mESCs were spontaneously differentiated by removing leukemia inhibitory factor (LIF) and adding retinoic acid (0.2 μM). (B) Quantification of Rad51 expression in mESCs and MEFs. (C) mESCs were synchronized with a double thymidine and then released from G1/S phase. The cells were collected at 2.5-h intervals, as indicated. Cyclin B1/A and phospho-histone H3Ser10 were used as markers for cell cycle progression. α-Tubulin was used as a loading control. (D) The level of each protein was quantified. Relative ratio of each protein band over the band of α–tubulin was described in each time point. The numerical value of each sample at indicated time point was normalized by the value of asynchronous cells (As). (E) Cell cycle profile was assessed by fluorescence-activated cell sorting (FACS) analysis.
Rad51 specifically localizes on chromosomes during S phase in mESCs
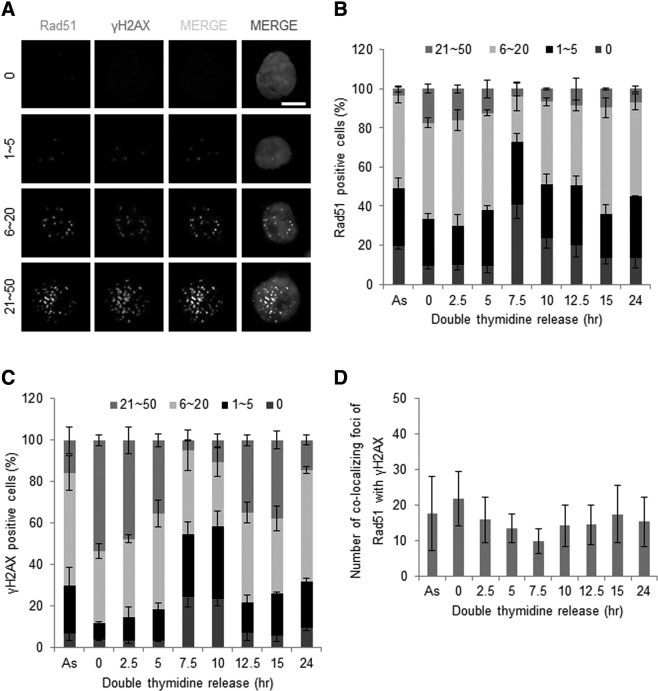
To understand the function of Rad51 of mESCs during cell cycle progression, we examined the stage-specific cellular localization of Rad51 by immunofluorescence. When cells were costained for Rad51 and RPA, which binds to ssDNA to prevent rewinding of the DNA double helix after unwinding by helicase [33], most RPA colocalized with Rad51 foci ([34]; Supplementary Fig. S2A). This result indicates that Rad51 stably binds to ssDNA in mESCs. Unchallenged mESCs and MEFs contain approximately five Rad51 chromosomal foci per nucleus [35]. In our analysis, we grouped Rad51-foci-positive cells into four categories, as shown in Fig. 2A. In the asynchronous state, ∼5% of Rad51-positive cells of total mESCs contained more than 20 Rad51 foci (Fig. 2B). To further examine the pattern of Rad51 foci formation during cell cycle progression, mESCs were synchronized with a double-thymidine-block method and then released as in Fig. 1E. Interestingly, unlike Rad51 protein levels, Rad51 foci frequency oscillated during cell cycle in mESCs, and numerous foci formed at S phase. Approximately, more than 70% of Rad51-foci-positive cells in S phase had more than five Rad51 foci; among these cells, 20% contained more than 20 Rad51 foci (Fig. 2B and Supplementary Fig. S2B).
FIG. 2.
Rad51 foci formation during the cell cycle in synchronized mESCs. (A) mESCs were synchronized with a double thymidine and then released from G1/S phase as in Fig. 1. Rad51 and γH2AX foci in mESCs were immunostained and visualized with fluorescence microscopy (×1000). Cells displaying fluorescent signals were categorized according to the number of foci per nucleus. The scale bar indicates 10 μm. The number of cells possessing Rad51 (B) and γH2AX (C) foci at each cell cycle phase was quantified. Three independent experiments were performed and, at least, 200 cells were counted for each experiment. (D) The colocalization pattern for the number of Rad51 and γH2AX foci at the indicated time points after release from the double-thymidine block. Error bars indicate mean±standard deviation (SD).
The phosphorylated form of H2AX, denoted γH2AX, is commonly used to detect DNA DSB sites. To determine whether the frequency of Rad51 foci was related to the frequency of DSB sites, we analyzed the number of γH2AX foci in γH2AX-foci-positive mESCs. Similar to our results with Rad51, ∼90% of γH2AX-positive mESCs in S phase had more than five γH2AX foci, indicating that γH2AX foci accumulated from early S phase and decreased as the cell cycle progressed to mitosis (Fig. 2C and Supplementary Fig. S2B). The pattern of Rad51 and γH2AX foci colocalization resembled the pattern of Rad51 and γH2AX foci fluctuation during cell cycle progression in mESCs ([36]; Fig. 2D). These results imply that functional Rad51 and γH2AX foci formed mainly during the DNA replication process in mESCs. In MEFs, by contrast, the percentage of Rad51-positive MEFs containing more than five Rad51 foci and the percentage of γH2AX-positive MEFs containing more than five γH2AX foci did not exceed 40% and 20%, respectively (data not shown).
The increase in Rad51 foci during early S phase suggested that Rad51 might be recruited to replication fork area, including the origin of replication initiation. For DNA replication, a prereplicative complex (pre-RC) composed of origin recognition complex (ORC), CDC6, and CDT1 must assemble at the replication origin [37]. To uncover the relevancy of Rad51 localization with DNA replication, ORC2 as a key component of ORC was used as a marker of the replication origin. As shown in Fig. 3A and B, most Rad51 foci did not colocalize with ORC2 foci, implying that Rad51 is not particularly recruited to the replication fork area in undamaged mESCs. Approximately 80% of Rad51 and ORC2 existed as separate foci (entities) with some distance between them (Fig. 3B). In general, the components of the replication factory, including cell cycle regulators, are localized within an area 0.1–1 μm in diameter [38–40]. Thus, we next analyzed the distance between Rad51 and ORC2 foci (described in Supplementary Fig. S4). For most entities, the interfoci distance between Rad51 and ORC2 was <600 nm (Fig. 3C). Rad51 foci were randomly distributed within 100–600 nm of ORC2 (Fig. 3D). These results suggest that the majority of Rad51 localizes near the replication origin, but not exactly at replication origin.
Rad51 knockdown causes a proliferation defect in mESCs
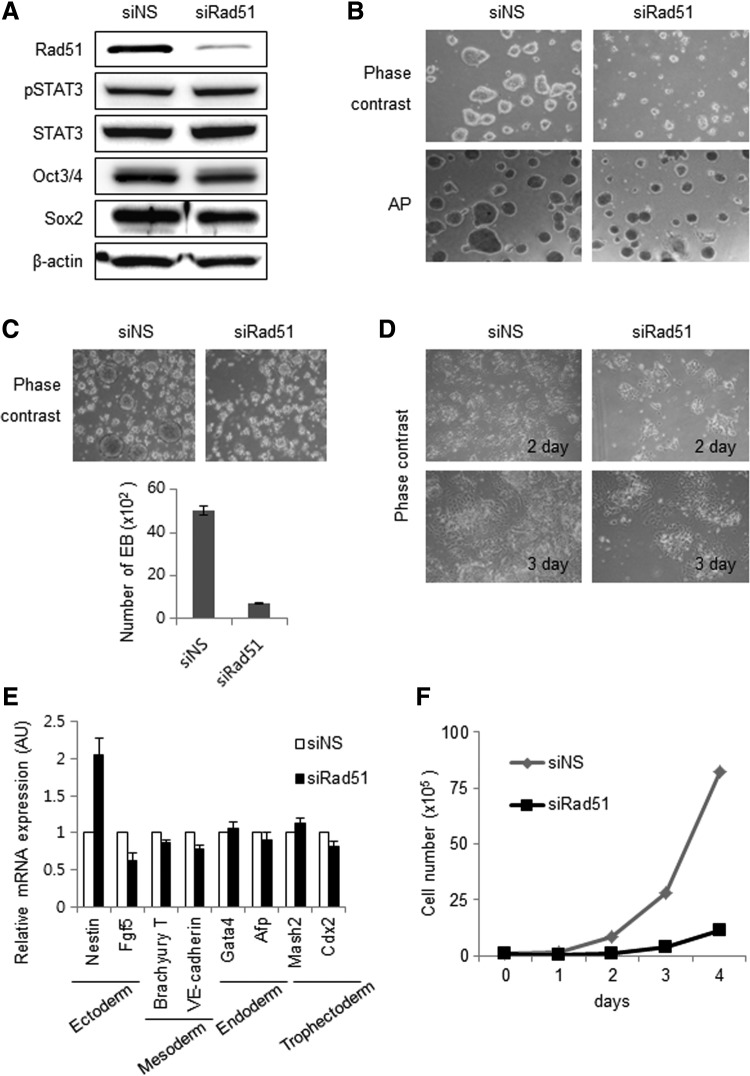
To assess whether Rad51 regulates stem cell characteristics, such as self-renewal and pluripotency, we studied the effect of Rad51 depletion on mESCs using siRNA against Rad51 (siRad51). The expression of self-renewal factors, such as Oct3/4, Sox2, and pSTAT3, was not affected by Rad51 depletion (Fig. 4A), nor was alkaline phosphatase activity, a widely used stemness marker (Fig. 4B). The number of embryonic bodies was significantly reduced by Rad51 suppression, but RA-mediated spontaneous differentiation was not affected by siRad51 treatment (Fig. 4C, D).
FIG. 4.
Rad51 knockdown affects the proliferation rate, but not the differentiation of mESCs. (A) The expression of pSTAT3, STAT3, Oct3/4, and Sox2 proteins in mESCs transfected with siNS or siRad51 was detected by immunoblot analysis. β-Actin was used as a loading control. (B) mESCs transfected with siNS or siRad51 were stained to measure alkaline phosphatase (AP) activity. (C) The effect of Rad51 on embryoid body (EB) formation. After transfection with siNS or siRad51, mESCs were incubated in EB media for 72 h, and the degree of EB formation was quantified (bottom). (D) The effect of Rad51 on differentiation of mESCs. mESCs were transfected with siNS or siRad51 and then spontaneously differentiated by the removal of LIF and addition of 0.2 μM of retinoic acid. (E) Quantitative RT-PCR analysis of expression of lineage-specific marker genes in spontaneously differentiated mESCs after siNS or siRad51 transfection. Most of lineage markers, except Nestin, in siRad51-treated cells showed similar expression level with those of control cells transfected with nonspecific siRNA. (F) Effect of Rad51 on proliferation of mESCs. mESCs were transfected with siNS or siRad51, and the viable cells were counted at the indicated times.
The expression of most of lineage markers in siRad51-treated cells was not significantly different from those of control cells, suggesting that Rad51 depletion does not significantly affect the expression of self-renewal genes or the differentiation capacity of mESCs (Fig. 4E). Since the expression of Nestin was increased more than twofold in Rad51-suppressed mESCs, we do not rule out the possibility that subpopulation of Rad51-depleted mESCs underwent spontaneous differentiation with a bias toward a neuroepithelial precursor fate.
The colony sizes of siRad51-treated mESCs were significantly smaller than those of control cells (Fig. 4B). This observation raised the possibility that depletion of Rad51 affected the proliferation rate of mESCs. Consistently, cell-counting analysis revealed that Rad51 suppression significantly delayed the proliferation of mESCs (Fig. 4F). We confirmed that the lower cell number and smaller colony size after Rad51 depletion were not caused by apoptosis (Supplementary Fig. S5). In addition, to check the relationship between cell proliferation and cellular senescence, we measured the level of Ki67 protein, a marker for senescence. Ki67 protein is highly expressed in actively dividing cells, but is absent from resting cells [41]. Ki67 staining analysis revealed that the proliferation defect caused by Rad51 suppression was not attributable to cellular senescence of mESCs (Supplementary Fig. S6).
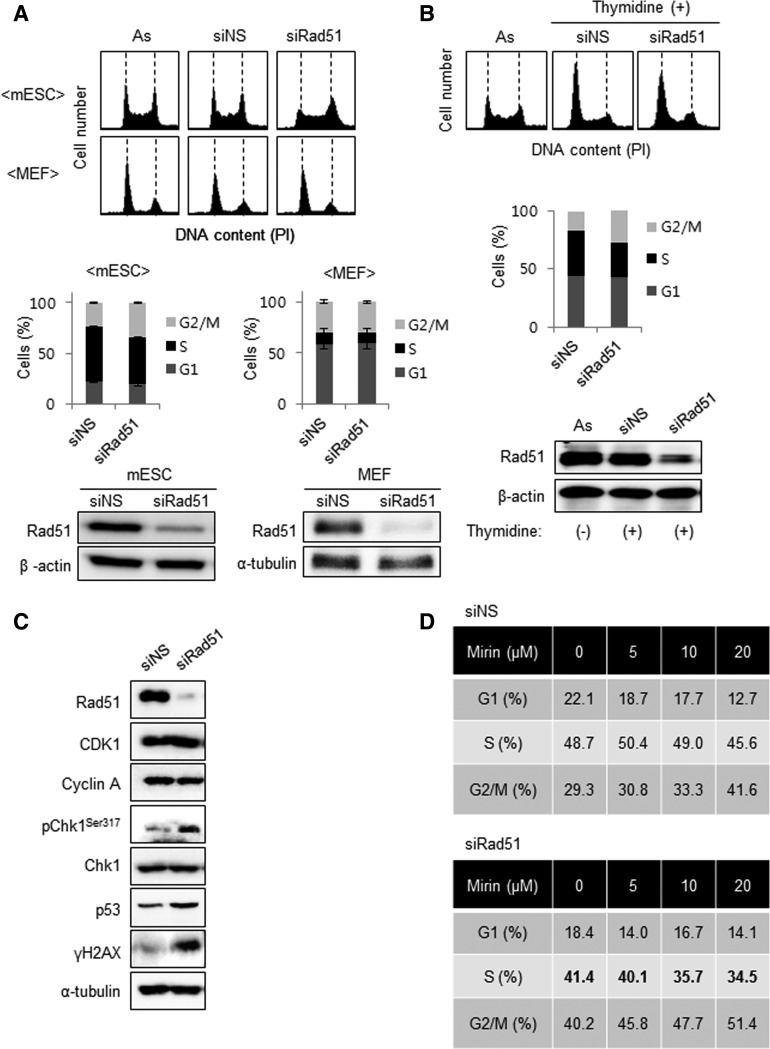
Proliferation delay in Rad51 knockdown is due to activation of the DNA damage checkpoint
To investigate the mechanism underlying the effects of Rad51 activity on cell proliferation in mESCs, Rad51 was depleted using siRNA-knockdown method. Then, cell cycle progression and checkpoint activation were analyzed. When cells were treated with siRad51, the number of mESCs, but not of MEFs, in G2/M phase increased by ∼10% (Fig. 5A). Immunofluorescence assay also showed that the level of cell cycle markers reflects the cell cycle profiles analyzed by FACS on Rad51-knockdown mESCs (Supplementary Fig. S7). We hypothesized that these G2/M-phase-accumulated mESCs from siRad51 would not be externally influenced by G1/S phase synchronization. A double-thymidine-block method used in Fig. 1E would not affect the cells that accumulated at G2/M phase. When the cell cycle of mESCs was experimentally induced at G1/S phase using thymidine, the number of cells in G2/M phase was determined. As we predicted, the increased populations of Rad51-depleted mESCs in G2/M phase were still remained at G2/M phase even in the presence of thymidine (Fig. 5B). These results suggest that Rad51 performs an essential role in normal cell cycle progression in mESCs.
FIG. 5.
Accumulation of Rad51-knockdown mESCs in G2/M and its effect on DNA damage checkpoint activation. (A) Analysis of cell cycle profiles after Rad51 knockdown in mESCs and MEFs. After siRNA transfection for 48 h, cells were harvested and stained with PI for FACS analysis as described in “Materials and Methods” (As; asynchronous cells). Data were quantified using FlowJo software (bottom). Error bars indicate mean±SD. (B) mESCs transfected with siNS or siRad51 were attempted to synchronize at G1/S phase using double thymidine. The cell cycle profiles of the cells were then analyzed using FACS Calibur. The population of each cell cycle phase was quantified with FlowJo software (middle). The level of Rad51 was determined by immunoblot analysis (bottom). (C) Activation of the DNA damage checkpoint by depletion of Rad51. Cells were harvested 48 h after siRNA transfection, and markers involved in cell cycle control and DNA damage checkpoint signaling were detected by immunoblot analysis. (D) The relative proportion of cells at indicated cell cycle phase after Rad51 siRNA transfection in the presence of mirin.
We next examined whether DNA damage checkpoint activation is the main cause of G2/M accumulation among siRad51-treated mESCs. As expected, the phosphorylation of Chk1, as well as H2AX in company with the total amount of p53, was upregulated in siRad51-treated mESCs (Fig. 5C). Immunofluorescence analysis confirmed that Chk1 was phosphorylated and it showed that the number of phospho-Chk1 foci increased approximately threefold when Rad51 expression was suppressed (Supplementary Fig. S8).
Rad51 is known to protect nascent single-strand DNA at replication forks from Mre11-dependent degradation and promotes continuous DNA synthesis [28]. Therefore, we asked whether the accumulation of siRad51-treated mESCs in G2/M phase results from Mre11-dependent degradation of DNA in S phase. To answer this question, we analyzed the effect of mirin, which disrupts the nuclease activity of Mre11 [42], on the cell cycle progression of siRad51-treated cells. Contrary to our expectation, mirin treatment amplified the effect of siRad51, further increasing the G2/M population in a dose-dependent manner (Fig. 5D). This result suggests that the role of Rad51 in unperturbed mESCs is not related to the protection of DNA from the nuclease activity of Mre11.
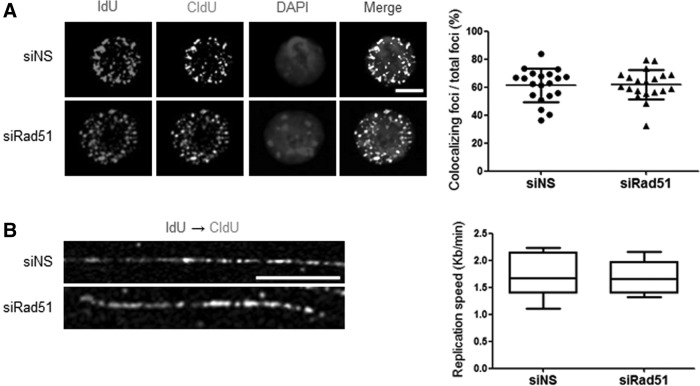
Rad51 depletion does not disturb DNA replication in mESCs
As shown earlier, Rad51 was constitutively expressed throughout the cell cycle (Fig. 1C–E and Supplementary Fig. S1) in mESCs, but Rad51 foci were formed specifically at S phase (Fig. 2 and Supplementary Fig. S2). These results implied that Rad51 proteins localize on chromosomes as multiple foci during DNA synthesis. To examine the effect of Rad51 depletion on replication in S phase, we analyzed the progression of the replication fork. Cells were sequentially treated with the thymidine analogs IdU and CldU to observe incorporated DNA fibers after transfecting siRad51 or nonspecific siRNA (siNS) in mESCs synchronized by a double-thymidine block. The labeling foci of IdU and CldU were stained and the degree of overlapping signals between IdU and CldU replication foci was then quantified. As reported previously, overlap of the two markers (IdU and CldU) indicates that DNA replication fork progression has stalled [43]. Unexpectedly, the extent of IdU-CldU foci colocalization was not significantly different between control and Rad51-knockdown mESCs (Fig. 6A). We further analyzed the effects of Rad51 suppression on replication fork progression with the DNA fiber assay (see Materials and Methods section). As shown in Fig. 6B, the mean replication speed was ∼1.71 kb/min in both control- and Rad51-knockdown cells, which indicates that the amount of Rad51 protein in the cells did not affect the replication speed.
FIG. 6.
S-phase progression after the depletion of Rad51 protein. (A) mESCs transfected with siNS or siRad51 were incubated with 2 mM thymidine for 16 h and then released into fresh media. Two hours later, the thymidine analogs IdU (50 μM) and CldU (200 μM) were successively incorporated into the DNA replication sites. Subsequently, the colocalization of IdU and CldU replication foci was stained with corresponding antibodies and the degree of colocalizing foci areas was quantified in each isolated cell using Image J software (right). (B) mESCs transfected with siNS or siRad51 were successively pulse labeled with IdU and CldU to final concentrations of 100 and 250 μM, respectively, for 20 min. DNA fibers were immunostained with antibodies specific for IdU and CldU. The replication speed was quantified from the mean fork extension rate (kb/min) during sequential pulse labels with IdU (1st label, 20 min) and CldU (2nd label, 20 min) in mESCs.
To examine whether Rad51 is required for DNA replication and S-phase progression, BrdU was incorporated into siRad51-transfected cells for 20 min, and the cells were analyzed with FACS. There was no obvious difference in the S-phase population both in control- and Rad51-siRNA mESCs (data not shown). These results show that Rad51 knockdown does not severely inhibit DNA replication.
Finally, we examined the S-phase progression of Rad51-depleted cells using a double-thymidine block and release to synchronize cells at G1/S phase. Cells were treated with thymidine and siRNA as described in Supplementary Fig. S9A. BrdU was incorporated for 20 min before harvest, and the cells were harvested at 2.5-h intervals after release. Cell cycle index of BrdU-stained cells showed that Rad51 protein levels did not affect cell cycle progression through S phase (Supplementary Fig. S9B). Quantification of BrdU-positive cells revealed that cell cycle progression was not significantly delayed by siRad51 (Supplementary Fig. S9C). These findings provide an expected conclusion that Rad51 activity does not affect DNA replication during S phase in mESCs.
Discussion
ESCs possess robust machineries to preserve their self-renewal capacity, sustain pluripotency, and maintain a stable genome. In recent studies, 50–70% of asynchronous mESCs were in S phase, compared with ∼30% of asynchronous MEFs. To respond to genomic instability within short cell cycles, ESCs utilize high-fidelity regulatory repair machineries to combat DNA damage. In ESCs, HR is the main pathway for maintaining genomic integrity, repairing DSBs, and reactivating stalled DNA replication forks when sister chromatids are available. In the replication dynamics of unchallenged mammalian somatic cells, Rad51 may promote continuous replication in an HR-independent manner to protect nascent ssDNA formed at replication forks from Mre11-dependent degradation or in an HR-dependent manner to ensure replication fork progression [28,44].
In this study, we demonstrated that Rad51 plays a novel role in the cell cycle progression of unperturbed mESCs and established a link between its activity in HR and checkpoint activation in the G2/M phase. Our results suggested that activation of the G2/M checkpoint by depletion of Rad51 was not related to the rate of replication fork progression, even though a deficiency in Rad51 HR activity caused cells to accumulate in G2/M phase (Figs. 5 and 6). In contrast, HR abnormalities in differentiated cell lines reduce the replication speed and increase the density of replication forks [44]. This strongly suggests that the HR activity of Rad51 is uncoupled from replication fork progression in S phase in mESCs. It also implies that Rad51 activity in mESCs is not restricted to the restart of collapsed replication forks [25]; rather, it might extend to the repair of replication defects. Because HR activity in mESCs is not related to replication fork progression, the replication speed in mESCs is similar to that in MEFs [45]. The localization of Rad51 within 100–600 nm of ORC2 indicates that the majority of Rad51 foci formed near the replication origin. This further supports that Rad51 foci formation is not restricted to the regions of replication origin, and thus Rad51 may play a role at postreplication stages. ORC2, the main component of the pre-RC, mainly functions during the assembly of the ORC to recruit proteins needed for replication initiation. Therefore, our results also suggest that genomic DNA in the region of the replication origin is prone to DSBs in mESCs.
A critical question is what underlies the differences in the mechanisms of Rad51 activity during the replication of unperturbed DNA in mESCs and differentiated cells. One possibility is that the high rate of proliferation in mESCs increases the risk of accumulating harmful DSBs during the replication of genomic DNA, thus necessitating the HR function of Rad51. Recent reports showed that expression of a dominant-negative Rad51 mutant in mESCs increased the level of spontaneous chromatid breaks, which further supports that DNA DSBs occur during S phase in mESCs [27]. We also speculate that the unique cell cycle pattern of mESCs underlies the constant expression of high levels of Rad51 throughout the cell cycle (Fig. 1C, D). Given that it takes ∼12 h for mESCs to complete one round of cell cycle [46], expeditious HR activity to repair DSBs that occur spontaneously during replication is essential. When the serum concentration was gradually decreased from 10% to 1%, Rad51 protein levels and the proliferation rate of mESCs decreased in proportion to the serum concentration, which indicates that Rad51 expression is positively related to the proliferation rate of mESCs (Supplementary Fig. S10). The molecular mechanism by which mESCs maintain high levels of Rad51 is another important question for investigation. The results of quantitative RT-PCR analysis suggest that constant transcription of Rad51 attributes to the high level of Rad51 in mESCs (data not shown). The Rad51 promoter is positively regulated by a STAT5-dependent pathway and negatively regulated by p53 [47,48]. It will be important to determine whether mESCs have a characteristic mechanism for the constitutive activation of Rad51 transcription.
Although Rad51 was expressed throughout the cell cycle, the number of Rad51 foci oscillated depending on the cell cycle phase, and Rad51 appeared to dissociate from chromosomes during mitosis (Fig. 2B and Supplementary Fig. S3). This pattern of Rad51 localization at mitosis was previously described, regardless of cell types or species [49]. In mitosis, Rad51 protein has strongly shown to localize in cytoplasm distinct from chromosomes, as diffuse, in human primary fibroblasts and MEFs [49]. We also observed Rad51 signals in the nucleus immediately after cytokinesis (data not shown). These results suggest that the association of Rad51 with chromosomes is strictly regulated during the cell cycle to prevent unexpected HR activity during mitosis.
Of note, the self-renewal and differentiation capacities of mESCs were not significantly affected by Rad51 expression (Fig. 4). However, we do not rule out the possibility that genomic DNA related to self-renewal or pluripotency of mESCs sustains DNA DSBs during replication. Our study provides insights into the mechanism by which mESCs respond to replication stress (collapsed replication forks, endogenous DNA damage, etc.) to maintain genome stability. Human ESCs (hESCs) also express higher levels of Rad51 than differentiated somatic cells (data not shown). Whether the function of Rad51 in cell cycle progression is conserved between mESCs and hESCs remains to be determined.
Supplementary Material
Acknowledgments
This research was supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Korean government (MEST) (no. 2012M3A9C6050367). This research was also supported by the Priority Research Centers Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2012-0006679). This work was also carried out with the support of “Cooperative Research Program for Agriculture Science & Technology Development (PJ010033)” Rural Development Administration, Republic of Korea.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Rossant J. (2001). Stem cells from the Mammalian blastocyst. Stem Cells 19:477–482 [DOI] [PubMed] [Google Scholar]
- 2.Andrews PW. (2002). From teratocarcinomas to embryonic stem cells. Philos Trans R Soc Lond B Biol Sci 357:405–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker DE, Harrison NJ, Maltby E, Smith K, Moore HD, Shaw PJ, Heath PR, Holden H. and Andrews PW. (2007). Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat Biotechnol 25:207–215 [DOI] [PubMed] [Google Scholar]
- 4.Maitra A, Arking DE, Shivapurkar N, Ikeda M, Stastny V, Kassauei K, Sui G, Cutler DJ, Liu Y, et al. (2005). Genomic alterations in cultured human embryonic stem cells. Nat Genet 37:1099–1103 [DOI] [PubMed] [Google Scholar]
- 5.Savatier P, Lapillonne H, Jirmanova L, Vitelli L. and Samarut J. (2002). Analysis of the cell cycle in mouse embryonic stem cells. Methods Mol Biol 185:27–33 [DOI] [PubMed] [Google Scholar]
- 6.Serrano L, Liang L, Chang Y, Deng L, Maulion C, Nguyen S. and Tischfield JA. (2011). Homologous recombination conserves DNA sequence integrity throughout the cell cycle in embryonic stem cells. Stem Cells Dev 20:363–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tichy ED. (2011). Mechanisms maintaining genomic integrity in embryonic stem cells and induced pluripotent stem cells. Exp Biol Med (Maywood) 236:987–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tichy ED, Pillai R, Deng L, Tischfield JA, Hexley P, Babcock GF. and Stambrook PJ. (2012). The abundance of Rad51 protein in mouse embryonic stem cells is regulated at multiple levels. Stem Cell Res 9:124–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Branzei D. and Foiani M. (2010). Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol 11:208–219 [DOI] [PubMed] [Google Scholar]
- 10.Burhans WC. and Weinberger M. (2007). DNA replication stress, genome instability and aging. Nucleic Acids Res 35:7545–7556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paques F. and Haber JE. (1999). Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 63:349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.San Filippo J, Sung P. and Klein H. (2008). Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 77:229–257 [DOI] [PubMed] [Google Scholar]
- 13.Blow JJ. and Gillespie PJ. (2008). Replication licensing and cancer—a fatal entanglement?. Nat Rev Cancer 8:799–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sieber OM, Heinimann K. and Tomlinson IP. (2003). Genomic instability—the engine of tumorigenesis?. Nat Rev Cancer 3:701–708 [DOI] [PubMed] [Google Scholar]
- 15.Adams BR, Golding SE, Rao RR. and Valerie K. (2010). Dynamic dependence on ATR and ATM for double-strand break repair in human embryonic stem cells and neural descendants. PLoS One 5:e10001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tichy ED, Pillai R, Deng L, Liang L, Tischfield J, Schwemberger SJ, Babcock GF. and Stambrook PJ. (2010). Mouse embryonic stem cells, but not somatic cells, predominantly use homologous recombination to repair double-strand DNA breaks. Stem Cells Dev 19:1699–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim DS. and Hasty P. (1996). A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol Cell Biol 16:7133–7143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, Sekiguchi M, Matsushiro A, Yoshimura Y. and Morita T. (1996). Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc Natl Acad Sci U S A 93:6236–6240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.West SC. (2003). Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol 4:435–445 [DOI] [PubMed] [Google Scholar]
- 20.Krejci L, Altmannova V, Spirek M. and Zhao X. (2012). Homologous recombination and its regulation. Nucleic Acids Res 40:5795–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holloman WK. (2011). Unraveling the mechanism of BRCA2 in homologous recombination. Nat Struct Mol Biol 18:748–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J, Doty T, Gibson B. and Heyer WD. (2010). Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol 17:1260–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carr AM, Paek AL. and Weinert T. (2011). DNA replication: failures and inverted fusions. Semin Cell Dev Biol 22:866–874 [DOI] [PubMed] [Google Scholar]
- 24.Sonoda E, Sasaki MS, Buerstedde JM, Bezzubova O, Shinohara A, Ogawa H, Takata M, Yamaguchi-Iwai Y. and Takeda S. (1998). Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J 17:598–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petermann E, Orta ML, Issaeva N, Schultz N. and Helleday T. (2010). Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell 37:492–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chi P, Van Komen S, Sehorn MG, Sigurdsson S. and Sung P. (2006). Roles of ATP binding and ATP hydrolysis in human Rad51 recombinase function. DNA Repair (Amst) 5:381–391 [DOI] [PubMed] [Google Scholar]
- 27.Kim TM, Ko JH, Hu L, Kim SA, Bishop AJ, Vijg J, Montagna C. and Hasty P. (2012). RAD51 mutants cause replication defects and chromosomal instability. Mol Cell Biol 32:3663–3680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hashimoto Y, Ray Chaudhuri A, Lopes M. and Costanzo V. (2010). Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat Struct Mol Biol 17:1305–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwab RAV. and Niedzwiedz W. (2011). Visualization of DNA replication in the vertebrate model system DT40 using the DNA fiber technique. J Vis Exp 56:e3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Badie S, Escandell JM, Bouwman P, Carlos AR, Thanasoula M, Gallardo MM, Suram A, Jaco I, Benitez J, et al. (2010). BRCA2 acts as a RAD51 loader to facilitate telomere replication and capping. Nat Struct Mol Biol 17:1461–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yata K, Lloyd J, Maslen S, Bleuyard JY, Skehel M, Smerdon SJ. and Esashi F. (2012). Plk1 and CK2 act in concert to regulate Rad51 during DNA double strand break repair. Mol Cell 45:371–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scaife RM. (2004). G2 cell cycle arrest, down-regulation of cyclin B, and induction of mitotic catastrophe by the flavoprotein inhibitor diphenyleneiodonium. Mol Cancer Ther 3:1229–1237 [PubMed] [Google Scholar]
- 33.Wold MS. (1997). Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem 66:61–92 [DOI] [PubMed] [Google Scholar]
- 34.Buisson R, Dion-Cote AM, Coulombe Y, Launay H, Cai H, Stasiak AZ, Stasiak A, Xia B. and Masson JY. (2010). Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat Struct Mol Biol 17:1247–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sioftanos G, Ismail A, Fohse L, Shanley S, Worku M. and Short SC. (2010). BRCA1 and BRCA2 heterozygosity in embryonic stem cells reduces radiation-induced Rad51 focus formation but is not associated with radiosensitivity. Int J Radiat Biol 86:1095–1105 [DOI] [PubMed] [Google Scholar]
- 36.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M. and Bonner WM. (2000). A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol 10:886–895 [DOI] [PubMed] [Google Scholar]
- 37.Bell SP. and Dutta A. (2002). DNA replication in eukaryotic cells. Annu Rev Biochem 71:333–374 [DOI] [PubMed] [Google Scholar]
- 38.Leonhardt H, Page AW, Weier HU. and Bestor TH. (1992). A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 71:865–873 [DOI] [PubMed] [Google Scholar]
- 39.Murti KG, He DC, Brinkley BR, Scott R. and Lee SH. (1996). Dynamics of human replication protein A subunit distribution and partitioning in the cell cycle. Exp Cell Res 223:279–289 [DOI] [PubMed] [Google Scholar]
- 40.Leonhardt H, Rahn HP, Weinzierl P, Sporbert A, Cremer T, Zink D. and Cardoso MC. (2000). Dynamics of DNA replication factories in living cells. J Cell Biol 149:271–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scholzen T. and Gerdes J. (2000). The Ki-67 protein: from the known and the unknown. J Cell Physiol 182:311–322 [DOI] [PubMed] [Google Scholar]
- 42.Dupre A, Boyer-Chatenet L, Sattler RM, Modi AP, Lee JH, Nicolette ML, Kopelovich L, Jasin M, Baer R, Paull TT. and Gautier J. (2008). A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nat Chem Biol 4:119–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bugler B, Schmitt E, Aressy B. and Ducommun B. (2010). Unscheduled expression of CDC25B in S-phase leads to replicative stress and DNA damage. Mol Cancer 9:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Daboussi F, Courbet S, Benhamou S, Kannouche P, Zdzienicka MZ, Debatisse M. and Lopez BS. (2008). A homologous recombination defect affects replication-fork progression in mammalian cells. J Cell Sci 121:162–166 [DOI] [PubMed] [Google Scholar]
- 45.Vannier JB, Sandhu S, Petalcorin MI, Wu X, Nabi Z, Ding H. and Boulton SJ. (2013). RTEL1 is a replisome-associated helicase that promotes telomere and genome-wide replication. Science 342:239–242 [DOI] [PubMed] [Google Scholar]
- 46.Nagy A, Gertsenstein M, Vintersten K. and Behringer R. (2003). Manipulating the Mouse Embryo: A Laboratory Manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 47.Pauklin S, Kristjuhan A, Maimets T. and Jaks V. (2005). ARF and ATM/ATR cooperate in p53-mediated apoptosis upon oncogenic stress. Biochem Biophys Res Commun 334:386–394 [DOI] [PubMed] [Google Scholar]
- 48.Slupianek A, Schmutte C, Tombline G, Nieborowska-Skorska M, Hoser G, Nowicki MO, Pierce AJ, Fishel R. and Skorski T. (2001). BCR/ABL regulates mammalian RecA homologs, resulting in drug resistance. Mol Cell 8:795–806 [DOI] [PubMed] [Google Scholar]
- 49.Cappelli E, Townsend S, Griffin C. and Thacker J. (2011). Homologous recombination proteins are associated with centrosomes and are required for mitotic stability. Exp Cell Res 317:1203–1213 [DOI] [PubMed] [Google Scholar]
- 50.Koberna K, Ligasova A, Malinsky J, Pliss A, Siegel AJ, Cvackova Z, Fidlerova H, Masata M, Fialova M, Raska I. and Berezney R. (2005). Electron microscopy of DNA replication in 3-D: evidence for similar-sized replication foci throughout S-phase. J Cell Biochem 94:126–138 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.