Abstract
Latency-associated nuclear antigen 1 (LANA1) of Kaposi's sarcoma-associated herpesvirus (KSHV) is implicated in the persistence of the viral genome during latent infection. It has been suggested that LANA1 tethers the viral genome to the host chromosome and also participates actively in DNA replication from the terminal repeat of KSHV. Here we show by mutational analysis that the mitotic chromosome-binding activity of LANA1 is tightly coupled to its replication activity. Thus, KSHV appears to have evolved a unique tactic for its stable maintenance.
Kaposi's sarcoma-associated herpesvirus (KSHV) is the etiological agent of Kaposi's sarcoma and some lymphoproliferative diseases (9, 11, 47); it is closely related to Epstein-Barr virus (EBV) and herpesvirus saimiri. The circularized viral genome is maintained as an extrachromosomal element in latently infected cells, whereas the linearized form of the viral genome is packaged into infectious virus particles during lytic infection (10, 16). Latency-associated nuclear antigen 1 (LANA1) is a viral protein expressed during latent infection (22, 33, 51, 55). As a transcriptional modulator, LANA1 interacts with several cellular transcription factors and influences the activity of viral and cellular promoters (3, 20, 23, 25, 30, 35, 36, 40, 41, 50, 54). When tethered to promoters via a heterologous DNA-binding domain, LANA1 acts as a transcriptional repressor, possibly by interacting with the mSin3 complex and heterochromatin protein 1 (36, 43, 57). As a replication factor, LANA1 colocalizes with the viral genome on the host chromosome and has been shown to be responsible for the stable maintenance of plasmids containing the KSHV terminal repeat (TR); it may therefore link the viral genome to a host chromosome, retaining the viral genome in the nucleus during mitosis and permitting equipartition to the progeny (5, 14). The chromosome-binding activity of LANA1 has been mapped to its N-terminal 22 amino acids and has been designated the chromosome-binding sequence (CBS) (49). Its C terminus binds to sequences within the TRs located at both ends of the KSHV genome and represses TR-dependent transcription (6, 15, 24, 42). As expected from the chromosome-tethering model, the LANA1 CBS is necessary for long-term replication of a KSHV TR-containing plasmid (59). In addition, we and others have shown that LANA1 is required for the transient replication of KSHV TR-containing plasmids, indicating that it may play an essential role not only in plasmid maintenance but also in DNA replication from the KSHV TR (26, 28, 42). Using a panel of LANA1 deletion mutants, we found that the CBS is necessary and sufficient for the C-terminal DNA-binding domain to support the replication of a KSHV TR-containing plasmid, suggesting that LANA1 must bind to the chromosome to fulfill its replication function (C. Lim, T. Seo, J. Jung, and J. Choe, submitted for publication). To examine this possibility further, we have generated several LANA1 derivatives with point mutations in their CBS and characterized their activities.
We reasoned that, because of its functional importance, the LANA1 CBS may be evolutionarily conserved among herpesviruses. Multiple sequence alignment of herpesvirus LANA1 homologues disclosed a few conserved residues within the CBS, which is usually followed by basic nuclear localization signal (NLS) (Fig. 1A). We generated point mutations in these conserved residues by PCR with primers containing mutated sequence. To exclude errors during the amplification of the internal repeat sequences of LANA1, we first made cDNAs which encompassed 1 to 1,020 nucleotides of LANA1 sequence with intended mutations by PCR. Then, the region corresponding to nucleotides 1 to 820 of the wild-type LANA1 sequence cloned in pFLAG-CMV2 was replaced with the same region carrying point mutations, taking advantage of the internal BamHI site in LANA1 cDNA. Point mutations in pFLAG-CMV2 LANA1 derivatives were confirmed by sequencing. To check the expression level of the LANA1 point mutants, 293T cells were transfected with a vector expressing FLAG-tagged LANA1 wild-type and point mutant derivatives. After 36 h, FLAG-tagged protein was immunoprecipitated and immunoblotted with anti-FLAG monoclonal antibody as described previously (41). As shown in Fig. 1B, the FLAG-tagged LANA1 point mutants were expressed at a level comparable to that of wild-type LANA1.
FIG. 1.
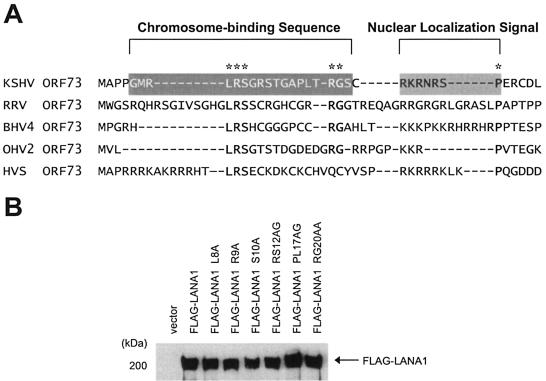
Construction of LANA1 point mutants. (A) Multiple sequence alignment of herpesvirus LANA1 homologues. The N-terminal amino acid sequences are aligned. The CBS and NLS of KSHV LANA1 are shown. Conserved residues are indicated by asterisks. RRV, rhesus rhadinovirus; BHV4, bovine herpesvirus 4; OHV2, ovine herpesvirus 2; HVS, herpesvirus saimiri; ORF, open reading frame. (B) Expression of FLAG-tagged LANA1 point mutants in transiently transfected 293T cells.
We next examined the subcellular localization of the LANA1 point mutants by an immunofluorescence assay (43). To visualize the interphase nuclei and mitotic chromosomes, we cotransfected the FLAG-tagged expression vectors with an expression vector for histone 2B protein fused to N-terminal green fluorescent protein (GFP). In a previous report (34), GFP-fused histone was shown to behave like its natural counterparts and was used to study chromosome dynamics in living cells. As described previously (5, 14, 49), wild-type LANA1 is localized to interphase nuclei, where it forms a heterogeneous pattern (Fig. 2A), and associates tightly with mitotic chromosomes (Fig. 2B). Similar results were obtained using H3-GFP and H4-GFP (data not shown). When we examined the LANA1 point mutants, we found that they were localized exclusively to the nucleus in interphase cells. An exception was the RG20AA mutant, which was present in both cytoplasm and the nucleus (Fig. 2C). In mitotic cells, however, point mutations between the 8th and 13th amino acid of LANA1 completely abolished association with mitotic chromosomes (Fig. 2D). These data indicate that residues 8 to 13 of the LANA1 CBS are critical for mitotic chromosome binding. The mutation of RG to AA may impair the NLS function of LANA1, and the mutant may have access to the nucleus only after the nuclear membrane breaks down during mitosis.
FIG. 2.
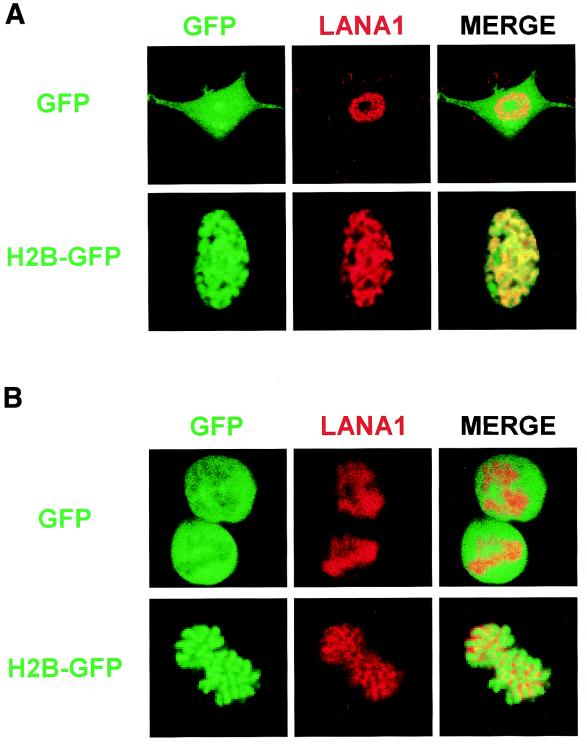
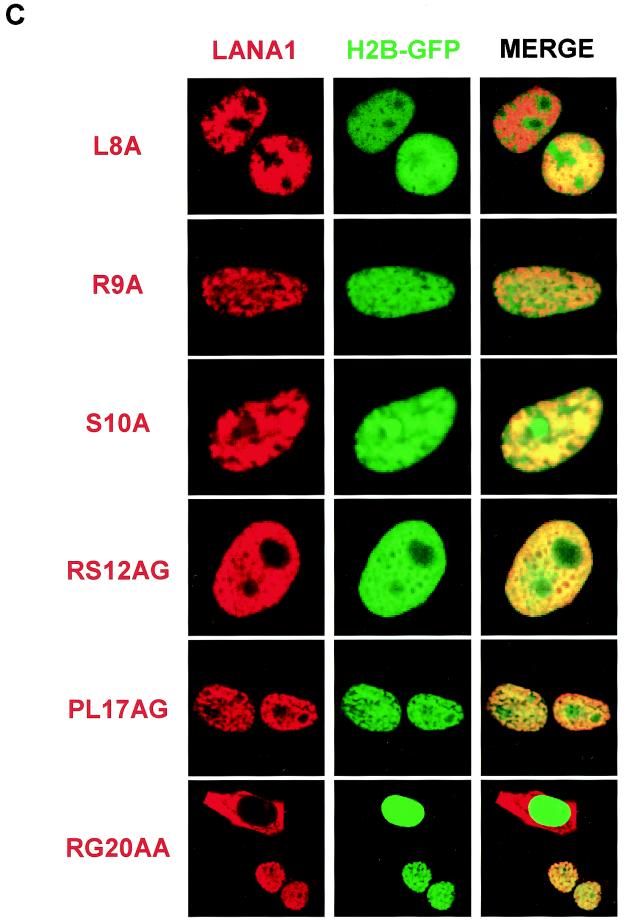
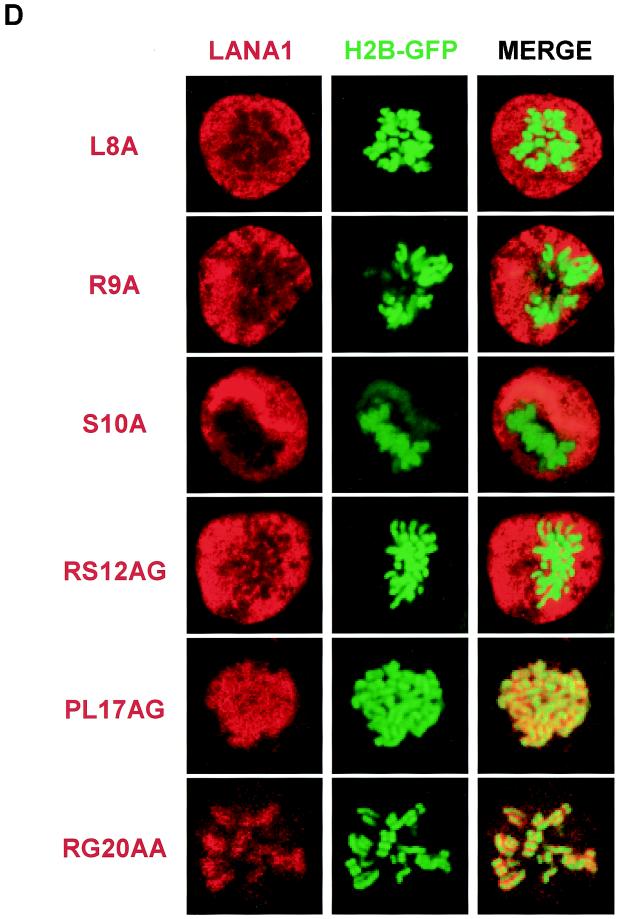
Subcellular localization and mitotic chromosome association of LANA1 point mutants. 293T cells grown on coverglasses were transfected with expression vectors for FLAG-tagged wild-type or point mutants of LANA1 and GFP-fused histone. FLAG-tagged proteins were detected with anti-FLAG monoclonal antibody and secondary rhodamine-conjugated anti-mouse antibody. (A and B) Interphase localization (A) and mitotic chromosome association (B) of FLAG-tagged wild-type LANA1 were examined using GFP-fused histone 2B protein under a confocal laser microscope. (C and D) Interphase localization (C) and mitotic chromosome association (D) of FLAG-tagged LANA1 point mutants were similarly investigated.
Due to the limited resolution of our immunofluorescence assay, it was not clear whether each LANA1 point mutant associated with the chromosome and/or occupied different subnuclear locations in interphase cells. To examine this, we fractionated total proteins from 293T cells transiently expressing each FLAG-tagged LANA1 point mutant and analyzed their distributions within cells (43). The cells were harvested 36 h after transfection and lysed in cytoskeleton (CSK) buffer (10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) [pH 6.8], 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 1 mM dithiothreitol, 0.1 mM ATP, 0.2 mM phenylmethylsulfonyl fluoride, 1 mM Na3VO4, and 10 mM NaF) containing 0.5% Triton X-100. The lysate was centrifuged at low speed, and the supernatant was clarified by high-speed centrifugation (fraction S). The pellet from the low-speed centrifugation was washed once in CSK buffer, subjected to extensive digestion with DNase I, and again centrifuged at low speed. The supernatant and pellet corresponded to the DNase I-extractable fraction (fraction C) and DNase I-resistant fraction (fraction M), respectively. As shown in Fig. 3, the majority of wild-type LANA1 appeared in the DNase I-resistant fraction (fraction M). In contrast, LANA1 point mutants defective for mitotic chromosome binding were found exclusively in the Triton X-100-extractable fraction (fraction S). The PL17AG and RG20AA mutants, which associate with mitotic chromosomes, were distributed between the Triton X-100-extractable fraction and the DNase I-resistant fraction. Interestingly, transiently expressed FLAG-tagged EBNA-1, a functional analog of KSHV LANA1 in the latent replication of EBV, was exclusively found in the Triton X-100-extractable fraction. We also used antibodies to examine the partitioning of endogenous proteins during fractionation to confirm that other cellular proteins showed a specific distribution and that each of the FLAG-tagged LANA1 point mutants was fractionated in an equivalent manner. Under our experimental conditions, p53 and histone proteins were exclusively found in fraction M, while β-actin was equivalently distributed to all fractions (data not shown). These data indicate that LANA1 point mutants defective for mitotic chromosome binding also fail to cofractionate with histone proteins in transfected cells. Since cells from unsynchronized populations are largely engaged in interphase, this suggests that they have different subnuclear compartments during not only mitosis but also interphase, at the time when DNA synthesis occurs.
FIG. 3.

Biochemical fractionation of LANA1 point mutants. 293T cells in 60-mm dishes were transfected with 4 μg of expression vectors encoding FLAG-tagged wild-type LANA1 or point mutant derivatives and harvested 36 h after transfection. Total proteins were fractionated as described in the text. LANA1 was detected by immunoblotting with rabbit polyclonal anti-LANA1 serum. Distribution of transiently expressed FLAG-tagged EBNA-1 was similarly revealed by immunoblotting with anti-FLAG monoclonal antibody. WT, wild type; fraction S, Triton X-100-extractable fraction; fraction C, DNase I-extractable fraction; fraction M, DNase I-resistant fraction.
LANA1 represses transcription from the KSHV TR by binding to sequences within the TR via its C-terminal DNA-binding domain (6, 15, 24, 42). From the organization of the functional domains of LANA1, it appeared likely that point mutations within the LANA1 CBS would not alter the DNA-binding and transcriptional repression activities of the C terminus of LANA1. To test this expectation, we performed a transient-reporter assay using p4TR-luc to examine the transcriptional repression activity of the LANA1 point mutants. As shown in Fig. 4A, all the point mutants, like wild-type LANA1, inhibited TR-dependent transcription, although the efficiency of repression varied. The mutants defective in chromosome binding inhibited the transcriptional activity of TR more strongly than wild type. In contrast, the RG20AA mutant, possibly because of its different subcellular localization during interphase, repressed TR-dependent transcription less efficiently. In a parallel experiment, we also examined the effect of LANA1 point mutants on the transcription from pGL2-basic, a backbone reporter of p4TR-luc (data not shown). Consistent with previous reports (23, 42, 54), LANA1 activated TR-independent transcription up to twofold. However, point mutants defective for mitotic chromosome binding failed to activate the transcription from pGL2-basic, partly explaining their enhanced transcriptional repression activities on TR-dependent transcription.
FIG. 4.
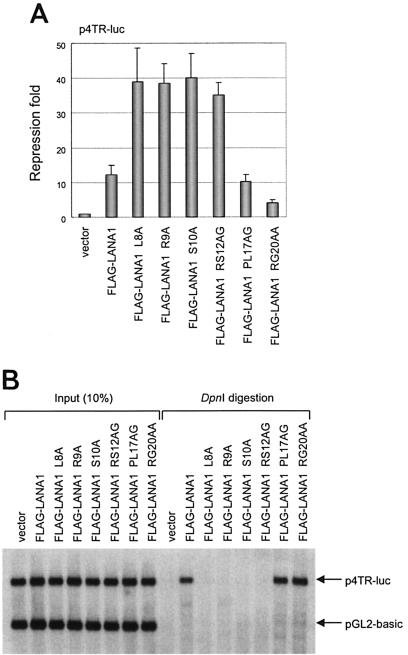
Transcription and replication activities of LANA1 point mutants. (A) 293T cells in 60-mm dishes were cotransfected with 1 μg of p4TR-luc and 4 μg of blank or FLAG-tagged LANA1 expression vector encoding wild type or point mutants. After 36 h, cells were harvested and luciferase assays were performed (42). The relative activation was calculated by normalizing to the luciferase activity obtained with the blank vector. The results are averages of three independent experiments, with standard deviations depicted by the error bars. (B) 293T cells in 100-mm dishes were transiently transfected with 2 μg of p4TR-luc containing four copies of KSHV TR, 2 μg of pGL2-basic as a nonreplicating internal control, and 8 μg of expression vector for FLAG-tagged LANA1 or its point mutants. Low-molecular-weight DNA was purified 96 h after transfection and subjected to digestion with Alw44I alone (left) or Alw44I/DpnI (right). Digested DNA was separated on an 0.8% agarose gel, transferred to a nylon membrane, and detected by Southern blotting with a luciferase-specific probe. Input, 10% of the DNA used in the Alw44I/DpnI digestion.
In order to examine the ability of the LANA1 point mutants to promote the replication of a KSHV TR-containing plasmid in a transient-replication assay, we cotransfected 293T cells with p4TR-luc containing four TR sequences of the KSHV genome and pGL2-basic, together with expression vectors for the wild-type or point mutants of LANA1. The cells were split 36 h after transfection to remove free DNA and harvested 96 h after transfection. Low-molecular-weight DNA was purified from the transfected cells as described previously (42). Using the methylation-sensitive restriction enzyme DpnI, which does not cut the plasmid DNA replicated in mammalian cells, we estimated the extent of replication of the TR-containing plasmid in the presence of the wild-type or point mutant LANA1. We included nonreplicating pGL2-basic as an internal control to monitor the efficiency of transfection and of plasmid recovery, as well as the completeness of the digestion of unreplicated DNA by DpnI. As shown in Fig. 4B, mutants of LANA1 defective in binding to mitotic chromosomes did not support the replication of the TR-containing plasmid. Longer exposure could hardly reveal any DpnI-resistant plasmid in the presence of chromosome-binding-defective mutants, making it difficult to quantitatively analyze the decrease in the relative level of replication (data not shown).
In order to survive in latently infected cells, DNA viruses with extrachromosomal genomes must replicate their DNA and ensure segregation of the daughter molecules to the two progeny cells. In the case of papillomavirus a viral helicase, E1 (67), in concert with a viral auxiliary protein, E2, unwinds the viral origin of replication and initiates DNA synthesis (13, 17, 18, 62). However, long-term maintenance of a viral oriP-containing plasmid requires an additional viral cis-acting element designated a minichromosome maintenance element (MME) (48). The MME is located within the long control region, which is distinct from the replication origin and contains multiple high-affinity E2-binding sites. Accumulating data suggest that chromatin-bound E2 tethers the viral genome to the host chromosome via the MME, thereby ensuring equipartition of the replicated viral genomes at mitosis (8, 31, 39, 61, 63). In the case of EBV, two cis elements constitute the origin of replication during latent replication (44, 46, 53, 68). These are members of a family of repeat (FR) and dyad symmetry sequences separated by ∼1 kb within the EBV genome. Both sequences contain binding sites for EBNA-1 (2, 4, 52), the viral trans-acting element required for the latent replication of EBV (69). In contrast to papillomavirus E1, EBNA-1 does not have any enzymatic activity for DNA replication, but instead it recruits a prereplicative complex to the viral oriP (12, 19, 56). DNA synthesis initiates at or near low-affinity EBNA-1-binding sites within dyad symmetry sequences (21), and these latter are necessary and sufficient for transient DNA replication mediated by EBNA-1 (27, 38, 44, 53). However, the stable maintenance of a plasmid containing the EBV oriP requires, in addition, multiple high-affinity EBNA-1-binding sites within FR sequences (1, 38, 46, 64). As in the case of papillomavirus E2, EBNA-1 associates with host chromosomes (45), and it has been suggested that the action of the FR in retaining the plasmid involves the viral genome hitchhiking on the host chromosome via EBNA-1.
It seems likely that KSHV has evolved a similar chromosome-tethering mechanism for its persistence in host cells (5, 14, 49, 59) and that LANA1 is responsible not only for the stable maintenance but also for the replication of KSHV TR-containing plasmids (26, 28, 42). In a transient-replication assay using a panel of LANA1 deletion mutants, the minimal domain of LANA1 able to support the replication of a TR-containing plasmid was identified as LANA1 Δ23-950, which contains the N-terminal CBS in addition to the C-terminal DNA-binding and dimerization domain (Lim et al., submitted). The requirement for the N-terminal CBS for replication activity was unexpected, since (i) the N-terminal CBS was thought to function mainly in the equal segregation and nuclear retention of the viral genome and (ii) the C terminus of LANA1, which can dimerize, bind TR sequences, and interact with the components of prereplicative complex (42), seemed likely to be sufficient for the replication activity of LANA1. In the present work we have generated several point mutations in the N-terminal CBS and characterized their activity in order to confirm that the replication activity of LANA1 requires its ability to bind to the chromosome. We found that point mutants, which were unable to associate with mitotic chromosomes, also failed to exclusively cofractionate with histone proteins in an unsynchronized population and facilitate the transient replication of a KSHV TR-containing plasmid, supporting the idea that chromosome binding of LANA1 is a prerequisite for its replication activity. It is noteworthy that all LANA1 point mutations used in this study did not abolish the nuclear localization in interphase cells (with some variation in the RG20AA mutant), the transcriptional repression activity of TR-dependent transcription, and the association with SAP30, which was previously shown to interact with the N terminus of LANA1 (36) (data not shown). Therefore, it seems unlikely that point mutations in the LANA1 CBS globally affect its structure or stability. The following evidence also argues against the possibility that the failure of the mutants defective in chromosome binding to support the replication of a KSHV TR-containing plasmid results from their inability to retain the KSHV TR-containing plasmid in the nucleus when nuclear membranes are reassembled at the end of mitosis. First, the LANA1 point mutations have an all-or-nothing effect on plasmid replication, whereas if the replication activity were intact but only the chromosome tethering and nuclear retention activities were abolished, the replication activities of those mutants should have been reduced rather than completely abolished. Second, trans-acting elements of papillomavirus and EBV lacking chromosome-binding and/or plasmid maintenance activity retain replication activity comparable to wild type in transient-replication assays (29, 39, 60, 65, 66), although there are some reports that mitotic chromosome binding of EBV EBNA-1 is also important for its replication activity (32, 58). Furthermore, viral cis-acting elements responsible for plasmid retention that probably act by tethering themselves to chromosomes are not required for replication activity in those systems (27, 38, 44, 48, 53). These data imply that defects in segregation and nuclear retention should not affect the replication of viral oriP-containing plasmids in transient-replication assays.
Instead we favor the idea that the N-terminal CBS of LANA1 targets TR-containing plasmids to the host chromosomes by interacting with chromatin-bound cellular factors. These could be replication factors specifically required for initiating replication from the KSHV TR or auxiliary factors facilitating the stable assembly of a prereplicative complex on the viral origin of DNA replication. MeCP-2 and H1 could be proteins targeted by the LANA1 CBS (14, 37, 59), although their functional relevance in the latent DNA replication of KSHV remains to be further investigated. While it cannot be excluded that the possibility that different cellular proteins are individually involved in DNA replication and the chromosome association activity of LANA1, our data suggest that the replication activity of LANA1 is tightly coupled to its chromosome association, and the replication of the latent KSHV genome may be intimately tied up with its equal segregation and nuclear retention. Such a mechanism for ensuring the persistence of the KSHV genome in latently infected cells would be unique among DNA viruses. In papillomavirus and EBV, replication of the viral genome and its partitioning are independent events mediated by different viral cis elements, and multiple high-affinity binding sites for the viral trans-acting element are required for stable maintenance of viral oriP-containing plasmids. In contrast, the fact that a single unit of KSHV TR is sufficient for plasmid persistence (6), but does not have multiple high-affinity binding sites for LANA1, may explain the peculiar mechanism of replication of latent KSHV.
While this report was under review, Barbera et al. (7) reported their point mutational analysis in chromosome association, DNA replication, and plasmid maintenance activity of LANA1. They sequentially replaced residues within the LANA1 CBS with alanine and obtained data consistent with our study.
Acknowledgments
We thank P. R. Cook and H. Kimura for providing us with histone-GFP vectors.
This work was supported in part by grants from the National Research Laboratory Program of the Korea Institute of Science and Technology Evaluation and Planning and from the Molecular and Cellular BioDiscovery Research Program of the Ministry of Science and Technology, Korea.
REFERENCES
- 1.Aiyar, A., C. Tyree, and B. Sugden. 1998. The plasmid replicon of EBV consists of multiple cis-acting elements that facilitate DNA synthesis by the cell and a viral maintenance element. EMBO J. 17:6394-6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambinder, R. F., W. A. Shah, D. R. Rawlins, G. S. Hayward, and S. D. Hayward. 1990. Definition of the sequence requirements for binding of the EBNA-1 protein to its palindromic target sites in Epstein-Barr virus DNA. J. Virol. 64:2369-2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.An, J., A. K. Lichtenstein, G. Brent, and M. B. Rettig. 2002. The Kaposi sarcoma-associated herpesvirus (KSHV) induces cellular interleukin 6 expression: role of the KSHV latency-associated nuclear antigen and the AP1 response element. Blood 99:649-654. [DOI] [PubMed] [Google Scholar]
- 4.Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, et al. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207-211. [DOI] [PubMed] [Google Scholar]
- 5.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 6.Ballestas, M. E., and K. M. Kaye. 2001. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 75:3250-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbera, A. J., M. E. Ballestas, and K. M. Kaye. 2004. The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 N terminus is essential for chromosome association, DNA replication, and episome persistence. J. Virol. 78:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bastien, N., and A. A. McBride. 2000. Interaction of the papillomavirus E2 protein with mitotic chromosomes. Virology 270:124-134. [DOI] [PubMed] [Google Scholar]
- 9.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 10.Cesarman, E., P. S. Moore, P. H. Rao, G. Inghirami, D. M. Knowles, and Y. Chang. 1995. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 86:2708-2714. [PubMed] [Google Scholar]
- 11.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 12.Chaudhuri, B., H. Xu, I. Todorov, A. Dutta, and J. L. Yates. 2001. Human DNA replication initiation factors, ORC and MCM, associate with oriP of Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 98:10085-10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiang, C. M., M. Ustav, A. Stenlund, T. F. Ho, T. R. Broker, and L. T. Chow. 1992. Viral E1 and E2 proteins support replication of homologous and heterologous papillomaviral origins. Proc. Natl. Acad. Sci. USA 89:5799-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cotter, M. A., II, and E. S. Robertson. 1999. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 264:254-264. [DOI] [PubMed] [Google Scholar]
- 15.Cotter, M. A., II, C. Subramanian, and E. S. Robertson. 2001. The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen binds to specific sequences at the left end of the viral genome through its carboxy-terminus. Virology 291:241-259. [DOI] [PubMed] [Google Scholar]
- 16.Decker, L. L., P. Shankar, G. Khan, R. B. Freeman, B. J. Dezube, J. Lieberman, and D. A. Thorley-Lawson. 1996. The Kaposi sarcoma-associated herpesvirus (KSHV) is present as an intact latent genome in KS tissue but replicates in the peripheral blood mononuclear cells of KS patients. J. Exp. Med. 184:283-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Del Vecchio, A. M., H. Romanczuk, P. M. Howley, and C. C. Baker. 1992. Transient replication of human papillomavirus DNAs. J. Virol. 66:5949-5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Demeret, C., M. Le Moal, M. Yaniv, and F. Thierry. 1995. Control of HPV 18 DNA replication by cellular and viral transcription factors. Nucleic Acids Res. 23:4777-4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dhar, S. K., K. Yoshida, Y. Machida, P. Khaira, B. Chaudhuri, J. A. Wohlschlegel, M. Leffak, J. Yates, and A. Dutta. 2001. Replication from oriP of Epstein-Barr virus requires human ORC and is inhibited by geminin. Cell 106:287-296. [DOI] [PubMed] [Google Scholar]
- 20.Friborg, J., Jr., W. Kong, M. O. Hottiger, and G. J. Nabel. 1999. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402:889-894. [DOI] [PubMed] [Google Scholar]
- 21.Gahn, T. A., and C. L. Schildkraut. 1989. The Epstein-Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell 58:527-535. [DOI] [PubMed] [Google Scholar]
- 22.Gao, S. J., L. Kingsley, M. Li, W. Zheng, C. Parravicini, J. Ziegler, R. Newton, C. R. Rinaldo, A. Saah, J. Phair, R. Detels, Y. Chang, and P. S. Moore. 1996. KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi's sarcoma. Nat. Med. 2:925-928. [DOI] [PubMed] [Google Scholar]
- 23.Garber, A. C., M. A. Shu, J. Hu, and R. Renne. 2001. DNA binding and modulation of gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:7882-7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garber, A. C., J. Hu, and R. Renne. 2002. Latency-associated nuclear antigen (LANA) cooperatively binds to two sites within the terminal repeat, and both sites contribute to the ability of LANA to suppress transcription and to facilitate DNA replication. J. Biol. Chem. 277:27401-27411. [DOI] [PubMed] [Google Scholar]
- 25.Groves, A. K., M. A. Cotter, C. Subramanian, and E. S. Robertson. 2001. The latency-associated nuclear antigen encoded by Kaposi's sarcoma-associated herpesvirus activates two major essential Epstein-Barr virus latent promoters. J. Virol. 75:9446-9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grundhoff, A., and D. Ganem. 2003. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus permits replication of terminal repeat-containing plasmids. J. Virol. 77:2779-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrison, S., K. Fisenne, and J. Hearing. 1994. Sequence requirements of the Epstein-Barr virus latent origin of DNA replication. J. Virol. 68:1913-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu, J., A. C. Garber, and R. Renne. 2002. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus supports latent DNA replication in dividing cells. J. Virol. 76:11677-11687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hung, S. C., M. S. Kang, and E. Kieff. 2001. Maintenance of Epstein-Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proc. Natl. Acad. Sci. USA 98:1865-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hyun, T. S., C. Subramanian, M. A. Cotter, 2nd, R. A. Thomas, and E. S. Robertson. 2001. Latency-associated nuclear antigen encoded by Kaposi's sarcoma-associated herpesvirus interacts with Tat and activates the long terminal repeat of human immunodeficiency virus type 1 in human cells. J. Virol. 75:8761-8771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ilves, I., S. Kivi, and M. Ustav. 1999. Long-term episomal maintenance of bovine papillomavirus type 1 plasmids is determined by attachment to host chromosomes, which is mediated by the viral E2 protein and its binding sites. J. Virol. 73:4404-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanda, T., M. Otter, and G. M. Wahl. 2001. Coupling of mitotic chromosome tethering and replication competence in Epstein-Barr virus-based plasmids. Mol. Cell. Biol. 21:3576-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kedes, D. H., E. Operskalski, M. Busch, R. Kohn, J. Flood, and D. Ganem. 1996. The seroepidemiology of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat. Med. 2:918-924. [DOI] [PubMed] [Google Scholar]
- 34.Kimura, H., and P. R. Cook. 2001. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 153:1341-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knight, J. S., M. A. Cotter II, and E. S. Robertson. 2001. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus transactivates the telomerase reverse transcriptase promoter. J. Biol. Chem. 276:22971-22978. [DOI] [PubMed] [Google Scholar]
- 36.Krithivas, A., D. B. Young, G. Liao, D. Greene, and S. D. Hayward. 2000. Human herpesvirus 8 LANA interacts with proteins of the mSin3 corepressor complex and negatively regulates Epstein-Barr virus gene expression in dually infected PEL cells. J. Virol. 74:9637-9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krithivas, A., M. Fujimuro, M. Weidner, D. B. Young, and S. D. Hayward. 2002. Protein interactions targeting the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus to cell chromosomes. J. Virol. 76:11596-11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krysan, P. J., S. B. Haase, and M. P. Calos. 1989. Isolation of human sequences that replicate autonomously in human cells. Mol. Cell. Biol. 9:1026-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lehman, C. W., and M. R. Botchan. 1998. Segregation of viral plasmids depends on tethering to chromosomes and is regulated by phosphorylation. Proc. Natl. Acad. Sci. USA 95:4338-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim, C., H. Sohn, Y. Gwack, and J. Choe. 2000. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) binds ATF4/CREB2 and inhibits its transcriptional activation activity. J. Gen. Virol. 81:2645-2652. [DOI] [PubMed] [Google Scholar]
- 41.Lim, C., Y. Gwack, S. Hwang, S. Kim, and J. Choe. 2001. The transcriptional activity of cAMP response element-binding protein-binding protein is modulated by the latency associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Biol. Chem. 276:31016-31022. [DOI] [PubMed] [Google Scholar]
- 42.Lim, C., H. Sohn, D. Lee, Y. Gwack, and J. Choe. 2002. Functional dissection of latency-associated nuclear antigen 1 of Kaposi's sarcoma-associated herpesvirus involved in latent DNA replication and transcription of terminal repeats of the viral genome. J. Virol. 76:10320-10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lim, C., D. Lee, T. Seo, C. Choi, and J. Choe. 2003. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus functionally interacts with heterochromatin protein 1. J. Biol. Chem. 278:7397-7405. [DOI] [PubMed] [Google Scholar]
- 44.Lupton, S., and A. J. Levine. 1985. Mapping genetic elements of Epstein-Barr virus that facilitate extrachromosomal persistence of Epstein-Barr virus-derived plasmids in human cells. Mol. Cell. Biol. 5:2533-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marechal, V., A. Dehee, R. Chikhi-Brachet, T. Piolot, M. Coppey-Moisan, and J. C. Nicolas. 1999. Mapping EBNA-1 domains involved in binding to metaphase chromosomes. J. Virol. 73:4385-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Middleton, T., and B. Sugden. 1994. Retention of plasmid DNA in mammalian cells is enhanced by binding of the Epstein-Barr virus replication protein EBNA1. J. Virol. 68:4067-4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moore, P. S., and Y. Chang. 1995. Detection of herpesvirus-like DNA sequences in Kaposi's sarcoma in patients with and without HIV infection. N. Engl. J. Med. 332:1181-1185. [DOI] [PubMed] [Google Scholar]
- 48.Piirsoo, M., E. Ustav, T. Mandel, A. Stenlund, and M. Ustav. 1996. cis and trans requirements for stable episomal maintenance of the BPV-1 replicator. EMBO J. 15:1-11. [PMC free article] [PubMed] [Google Scholar]
- 49.Piolot, T., M. Tramier, M. Coppey, J. C. Nicolas, and V. Marechal. 2001. Close but distinct regions of human herpesvirus 8 latency-associated nuclear antigen 1 are responsible for nuclear targeting and binding to human mitotic chromosomes. J. Virol. 75:3948-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Radkov, S. A., P. Kellam, and C. Boshoff. 2000. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat. Med. 6:1121-1127. [DOI] [PubMed] [Google Scholar]
- 51.Rainbow, L., G. M. Platt, G. R. Simpson, R. Sarid, S. J. Gao, H. Stoiber, C. S. Herrington, P. S. Moore, and T. F. Schulz. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol. 71:5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rawlins, D. R., G. Milman, S. D. Hayward, and G. S. Hayward. 1985. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell 42:859-868. [DOI] [PubMed] [Google Scholar]
- 53.Reisman, D., J. Yates, and B. Sugden. 1985. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol. Cell. Biol. 5:1822-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Renne, R., C. Barry, D. Dittmer, N. Compitello, P. O. Brown, and D. Ganem. 2001. Modulation of cellular and viral gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:458-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schepers, A., M. Ritzi, K. Bousset, E. Kremmer, J. L. Yates, J. Harwood, J. F. Diffley, and W. Hammerschmidt. 2001. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus. EMBO J. 20:4588-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwam, D. R., R. L. Luciano, S. S. Mahajan, L. Wong, and A. C. Wilson. 2000. Carboxy terminus of human herpesvirus 8 latency-associated nuclear antigen mediates dimerization, transcriptional repression, and targeting to nuclear bodies. J. Virol. 74:8532-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sears, J., J. Kolman, G. M. Wahl, and A. Aiyar. 2003. Metaphase chromosome tethering is necessary for the DNA synthesis and maintenance of oriP plasmids but is insufficient for transcription activation by Epstein-Barr nuclear antigen 1. J. Virol. 77:11767-11780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shinohara, H., M. Fukushi, M. Higuchi, M. Oie, O. Hoshi, T. Ushiki, J. Hayashi, and M. Fujii. 2002. Chromosome binding site of latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus is essential for persistent episome maintenance and is functionally replaced by histone H1. J. Virol. 76:12917-12924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shire, K., D. F. Ceccarelli, T. M. Avolio-Hunter, and L. Frappier. 1999. EBP2, a human protein that interacts with sequences of the Epstein-Barr virus nuclear antigen 1 important for plasmid maintenance. J. Virol. 73:2587-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Skiadopoulos, M. H., and A. A. McBride. 1998. Bovine papillomavirus type 1 genomes and the E2 transactivator protein are closely associated with mitotic chromatin. J. Virol. 72:2079-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ustav, M., and A. Stenlund. 1991. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 10:449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Voitenleitner, C., and M. Botchan. 2002. E1 protein of bovine papillomavirus type 1 interferes with E2 protein-mediated tethering of the viral DNA to mitotic chromosomes. J. Virol. 76:3440-3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wade-Martins, R., J. Frampton, and M. R. James. 1999. Long-term stability of large insert genomic DNA episomal shuttle vectors in human cells. Nucleic Acids Res. 27:1674-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu, H., D. F. Ceccarelli, and L. Frappier. 2000. The DNA segregation mechanism of Epstein-Barr virus nuclear antigen 1. EMBO Rep. 1:140-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu, H., P. Kapoor, and L. Frappier. 2002. Separation of the DNA replication, segregation, and transcriptional activation functions of Epstein-Barr nuclear antigen 1. J. Virol. 76:2480-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang, L., I. Mohr, E. Fouts, D. A. Lim, M. Nohaile, and M. Botchan. 1993. The E1 protein of bovine papilloma virus 1 is an ATP-dependent DNA helicase. Proc. Natl. Acad. Sci. USA 90:5086-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yates, J., N. Warren, D. Reisman, and B. Sugden. 1984. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc. Natl. Acad. Sci. USA 81:3806-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yates, J. L., N. Warren, and B. Sugden. 1985. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature 313:812-815. [DOI] [PubMed] [Google Scholar]


