Abstract
The severe acute respiratory syndrome coronavirus (SARS-CoV) genome contains open reading frames (ORFs) that encode for several genes that are homologous to proteins found in all known coronaviruses. These are the replicase gene 1a/1b and the four structural proteins, nucleocapsid (N), spike (S), membrane (M), and envelope (E), and these proteins are expected to be essential for the replication of the virus. In addition, this genome also contains nine other potential ORFs varying in length from 39 to 274 amino acids. The largest among these is the first ORF of the second longest subgenomic RNA, and this protein (termed U274 in the present study) consists of 274 amino acids and contains three putative transmembrane domains. Using antibody specific for the C terminus of U274, we show U274 to be expressed in SARS-CoV-infected Vero E6 cells and, in addition to the full-length protein, two other processed forms were also detected. By indirect immunofluorescence, U274 was localized to the perinuclear region, as well as to the plasma membrane, in both transfected and infected cells. Using an N terminus myc-tagged U274, the topology of U274 and its expression on the cell surface were confirmed. Deletion of a cytoplasmic domain of U274, which contains Yxxφ and diacidic motifs, abolished its transport to the cell surface. In addition, U274 expressed on the cell surface can internalize antibodies from the culture medium into the cells. Coimmunoprecipitation experiments also showed that U274 could interact specifically with the M, E, and S structural proteins, as well as with U122, another protein that is unique to SARS-CoV.
The recent severe acute respiratory syndrome (SARS) epidemic, which affected over 30 countries, has profoundly disturbed social and economic activities regionally, as well as globally. A novel coronavirus was identified as the etiological agent of SARS (13, 14, 23, 36). Analysis of the nucleotide sequence of this novel SARS coronavirus (SARS-CoV) showed that the viral genome is nearly 30 kb in length and contains 14 potential open reading frames (ORFs) (31, 38, 52). Coronaviruses are positive-strand RNA viruses, and the virion consists of a nucleocapsid core surrounded by an envelope containing three membrane proteins—spike (S), membrane (M), and envelope (E)—that are common to all members of the genus (for reviews, see references 12, 24, 25, and 43). The RNA is packaged by the nucleocapsid (N) protein into a helical nucleocapsid (for a review, see reference 26). The S protein, which forms morphologically characteristic projections on the virion surface, mediates binding to host receptors and membrane fusion (for a review, see reference 9). The M protein is a triple-spanning integral membrane protein with a short ectodomain and a large carboxyl-terminal endodomain (for a review, see reference 39). More recently, the E protein was showed to play a major role in coronavirus assembly (2, 4, 10, 15, 27, 59, 60). Another structural protein, the hemagglutinin esterase glycoprotein (HE), is found in only a subset of coronaviruses, but its role in the virus life cycle has not been well established (for a review, see reference 7). The SARS-CoV genome does not appear to encode for a HE protein (38).
The genes for these structural proteins and the replicase 1a/1b gene, which is located at the 5′ end of the genome and constitutes two-thirds of it, are conserved among the subgroups of coronavirus so is their relative position in the genome (for reviews, see references 12, 25, 26, and 43). In addition, there are subgroup-specific accessory proteins that vary in size and position in the genome. These proteins are usually dispensable for viral replication, at least in the cell culture system, but may be important for virus-host interactions and thus contribute to viral fitness (for reviews, see references 8, 12, 24, and 28). For example, although the 7b gene of feline coronavirus is easily lost upon virus adaptation to cell culture, it is strictly maintained in naturally occurring strains, and its loss was correlated with reduced virulence (18, 58). A recent study also showed that some of the group-specific murine coronavirus genes are not essential for viral replication in cell culture, but their deletion, by reverse genetics, is attenuating in the natural host (11).
Initially, phylogenetic analysis of the SARS-CoV genome suggested that it does not belong to any of the preexisting coronavirus subgroups (31, 38). However, more extensive studies revealed that SARS-CoV is distantly related to the established group 2 coronaviruses (45). In addition to the replicase gene 1a/1b and the four structural proteins (S, E, M, and N), it contains another nine potential ORFs varying in length from 39 to 274 amino acids (aa) (31, 38, 52). Whether these ORFs are expressed or not and whether the expressed proteins are essential for viral replication awaits elucidation. The largest among these is the first ORF of the second-largest subgenomic RNA (sgRNA), which is 4.5 kbp in length. This sgRNA (termed sgRNA3) is found in high abundance in infected cells and contains a strong match to the transcription regulating consensus sequence close to and upstream of its first ORF (31, 38, 52). We have termed this protein U274 for unique protein of 274 aa since it does not show significant similarities to any known protein, although a region in its C terminus shows similarity to calcium-transporting adenosine triphosphatases (note that this ORF is named ORF3 in reference 31, X1 in reference 38, and ORF3a in references 45 and 52). Previously, we have analyzed 81 sera obtained from SARS patients in Singapore and found that most of the sera, particularly those from convalescent patients, contain antibodies against the U274 protein, suggesting that U274 could play an important role in the biogenesis of SARS-CoV (51).
In the present study, we used specific antibodies against U274 to show that it is indeed expressed in SARS-CoV-infected cells. Moreover, we further characterized its cellular localization and topology, as well as the effects of sorting signals in its C terminus cytoplasmic domain on its transport properties. The ability of U274 to interact with other SARS-CoV proteins was also determined.
MATERIALS AND METHODS
Materials.
All reagents used in the present study were purchased from Sigma (St. Louis, Mo.) unless otherwise stated. The 293T, HeLa, and Vero E6 cell lines were purchased from the American Type Culture Collection (Manassas, Va.) and cultured at 37°C in 5% CO2 in Dulbecco modified Eagle medium containing 1 g of glucose/liter, 2 mM l-glutamine, 1.5 g of sodium bicarbonate/liter, 0.1 mM nonessential amino acids, 0.1 mg of streptomycin/ml, 100 U of penicillin, and 5% fetal bovine serum (HyClone Laboratories, Inc., Logan, Utah).
Construction of plasmids.
For transient expression in mammalian cells, the vectors used were pXJ40HA and pXJ40myc for tagging proteins at the N terminus with a hemagglutinin (HA) or c-myc epitope, respectively, and pXJ40-3′HA for tagging proteins with three HA epitopes at the C terminus (GLAXO Group, Institute of Molecular and Cell Biology, Singapore [unpublished data]). For untagged U274, the pXJ40-3′HA vector was used, but a stop codon was inserted before the HA epitopes at the C terminus so that these epitopes would not be expressed.
The various viral genes used in the present study were obtained from SARS-CoV 2003VA2774, an isolate from a SARS patient in Singapore, as previously described (51). Briefly, RNA was extracted from SARS-CoV-infected Vero E6 cell culture supernatant, reverse transcribed, and used as a template in PCR with specific primers. Two mutated forms of U274 were used in the present study: U274mut1 and U274mut2. U274mut1 corresponds to 101 to 274 aa of U274, and this construct was obtained by PCR. U274mut2 corresponds to U274 with 160 to 173 aa (YNSVTDTIVVTEGD) deleted, and this construct was obtained by two rounds of PCR.
The sequences of all constructs used in the present study were confirmed by DNA sequencing performed at the core facility at the Institute of Molecular and Cell Biology by using the dideoxy method with the Taq DyeDeoxy terminator cycle sequencing kit and the automated DNA sequencer 373 from PE Applied Biosystems (Foster City, Calif.).
Raising polyclonal antibody against U274.
Glutathione S-transferase (GST)-U274(134-274) protein was expressed as previously described (51) and used to immunize a New Zealand White rabbit as previously described (47). This region corresponds to the C-terminal hydrophilic domain of U274. The final bleeding was carried out 10 days after the 10th injection, and the serum was used directly in the present study.
Transient transfection of mammalian cells and Western blot analysis.
Transient-transfection experiments were performed by using Lipofectamine reagent (Invitrogen, Carlsbad, Calif.) according to manufacturer's protocol. Typically, ∼106 cells were plated on 6-cm dish and allowed to attach overnight. Then, 1 to 3 μg of DNA was used per plate, and the cells were left for at least 16 h before the cells were washed with phosphate-buffered saline (PBS) and then lysed directly in Laemmli's sodium dodecyl sulfate (SDS) buffer and used for Western blot analysis. Approximately, ∼105 transfected cells were separated on SDS-polyacrylamide gel and transferred to nitrocellulose Hybond-C membrane (Amersham Pharmacia Biotech, Uppsala, Sweden). The membranes were blocked with 5% nonfat dry milk for 30 min, followed by overnight incubation at 4°C with the primary antibody. The membranes were then washed extensively with PBST (PBS containing 0.05% Tween 20), followed by incubation with an appropriate horseradish peroxidase-conjugated secondary antibody (Pierce, Rockford, Ill.) for 1 h at room temperature, washing, and detection by an enhanced chemiluminescence method (Pierce). The primary antibodies used were anti-HA polyclonal (Y-11), anti-myc polyclonal (A-14), and anti-myc monoclonal (9E10) antibodies (Santa Cruz Biotechnology, Santa Cruz, Calif.); anti-HA monoclonal antibody (12CA5; Roche Molecular Biochemicals, Indianapolis, Ind.); anti-actin antibody (AC-15; Sigma); anti-U274 rabbit polyclonal antibody (the present study); and anti-human transferrin receptor (TfR; BD Pharmingen, San Diego, Calif.).
When it was necessary to reprobe a membrane with another antibody, the membrane was stripped with stripping buffer (2% SDS, 100 mM β-mercaptoethanol, 62.5 mM Tris-HCl [pH 6.8]) for 30 min at 65°C and washed extensively with PBST before use.
Infection of Vero E6 cells with SARS-CoV.
SARS-CoV 2003VA2774, an isolate from a SARS patient in Singapore, was used to infect Vero E6 cells as previously described (33, 40, 51). Briefly, the virus stock used in the present study was from the third passage of the isolate, and the titer was 10−7 PFU ml−1. For the present study, monolayer of cells were infected at multiplicity of infection (MOI) of 1, and at approximately 8 h postinfection the cells showed 25% cytopathic effect (CPE). For another set of experiments, the cells were infected at an MOI of 0.1 and, at approximately 24 h postinfection, the cells showed 75% CPE. At these two time points (i.e., 25 and 75% CPE), the cells were washed with PBS and then lysed in 600 μl (for each 180-cm2 flask) of lysis buffer containing 50 mM Tris-HCl (pH 8), 150 mM NaCl, 0.5% NP-40 (BDH Laboratory Supplies), 0.5% sodium deoxycholate, and 0.005% SDS. A total of 20 μl of the lysate was used for Western blot analysis as described above.
For RNA extraction, infected cells were washed with PBS, and then total RNA was extracted with TRIzol reagent (Invitrogen) according to the manufacturer's protocol. The RNA was subjected to first strand cDNA synthesis with oligo(dT)20 primers and Superscript III RT (Invitrogen) according to the manufacturer's protocol. This product was used as a template for the first PCR. The first PCR (total volume of 25 μl) was carried out with the first set of primers (PCRfor1, ACCCAGGAAAAGCCAACCAACCTC [bp 2 to 25]; PCRrev1, AATGCCGTCACCTTCAGTAACGAC [bp 25776 to 25753]), and the second round of PCR was then performed with the second set of primers (PCRfor1 and PCRrev2, TTGTAGCGGTATCGTTGCTGTAGC [bp 25365 to 25342]) with 1 μl of the first PCR as a template in a total volume of 25 μl. These primers correspond to regions in the SARS-CoV genome (and the base pair numbering is given with reference to the 2003VA2774/SIN2774 isolate [GenBank accession no. AY28379]), which are close to the U274 encoding sequence. All PCRs were performed with High-Fidelity Taq Polymerase (Roche), and the PCR conditions were as follows: 95°C for 5 min, followed by 35 cycles of 95°C for 15 s, 60°C for 15 s, and 72°C for 30 s, with a final extension at 72°C for 10 min. Nested PCR products were cloned into the TOPO TA cloning vector, pCRII-TOPO (Invitrogen), and 15 independent clones were sequenced as described above.
For immunofluorescence, Vero E6 cells were infected with SARS-CoV at an MOI of 1 and grown on coverslips (Iwaki; Asahi Techno Glass, Tokyo, Japan) until the cells showed a CPE of ca. 25%. The medium was removed, and the coverslips were fixed in acetone for 20 to 30 min on ice. The acetone was then removed, and the coverslips were completely air dried and stored at −20°C until use. Before use, the coverslips were fixed again with 4% paraformaldehyde, permeabilized with Triton X-100, and incubated with antibodies as described below.
Comparison of U274 protein sequences of different SARS-CoV isolates.
Sequences of U274 were obtained from the different SARS-CoV isolates reported in the GenBank database and aligned by using MegAlign software (DNAStar, Inc., Madison, Wis.). The isolates used were TOR2 (AY274119), Urbani (AY278741), CUHK-W1 (AY278554), BJ01 (AY278488), HKU-39849 (AY278491), BJ02 (AY278487), BJ03 (AY278490), BJ04 (AY279354), GZ01 (AY278489), SIN2679 (AY283796), SIN2500 (AY283794), ZJ01 (AY297028), SIN2677 (AY283795), TW1 (AY29145), CUHK-Su10 (AY282752), SIN2748 (AY283797), SIN2774 (AY283798), Frankfurt1 (AY291315), SZ1 (AY304489), SZ3 (AY304486), SZ13 (AY304487), and SZ16 (AY304488).
Indirect immunofluorescence.
For transient transfection, cells grown on Permanox slide (Nalge Nunc International, Naperville, Ill.) were transfected as described above. The cells were then fixed with 4% paraformaldehyde for 10 min at room temperature and used directly or subjected to permeabilization with either digitonin (50 μg/ml; Sigma) or Triton X-100 (0.2%; BDH Laboratory Supplies, Poole, England) for 10 min at room temperature. Cells were blocked with PBS plus 1% bovine serum albumin (BSA) for 30 min and then incubated with the primary antibody (1:200) for 1.5 h, washed, and then incubated with fluorescein isothiocyanate (FITC)-conjugated or rhodamine-conjugated secondary antibody (1:200; Santa Cruz) for 1 h. All incubations and washes were performed at room temperature. Slides were mounted with FluorSave reagent (Oncogene Research Products, Cambridge, Mass.) and analyzed on a MRC1024 laser confocal microscope (Hercules, Calif.).
FACS analysis.
To determine the expression of proteins on the surface of live Vero E6 cells, fluorescence-activated cell sorting (FACS) analyses were performed as previously described (49, 50). Briefly, cells were treated with trypsin ca. 24 h after transfection, washed twice with PBS plus 1% BSA, and then incubated with an anti-myc monoclonal antibody (2.5 μg/ml; Santa Cruz Biotechnology) or an isotype-matched control (anti-Bcl-2, clone 100, immunoglobulin G1; Santa Cruz Biotechnology) for 4 h at 4°C. The cells were then washed three times with cold PBS plus 1% BSA and incubated with a goat FITC-conjugated anti-mouse F(ab′)2 antibody (15 μg/ml; Pierce) for 2 h at 4°C. Cells were washed as before and analyzed on a Becton Dickinson flow cytometer (San Jose, Calif.).
Internalization experiments.
To determine whether cell surface U274 can undergo endocytosis, internalization experiments were performed as previously described (49). Briefly, 5 × 105 cells (Vero E6 or HeLa) were plated on six-well plates and allowed to settle overnight. If required, transient transfection with pXJ40myc-U274, pXJ40myc-GST, or pXJ40-TfR (for expressing full-length human TfR protein; HWJ Laboratory, Institute of Molecular and Cell Biology, Singapore [unpublished data]) was performed as described above, and the cells were left for another 16 to 24 h. Then, 0.5 ml of a 2.5-μg/ml concentration of the appropriate antibody (anti-c-myc; Santa Cruz Biotechnology) or anti-human CD81 (Santa Cruz Biotechnology) or anti-human TfR (BD Pharmingen) monoclonal antibody was overlaid onto the cells for 3 h at 37°C. The plate was then cooled on ice for 10 min before any unbound antibody was removed with three washes of cold PBS plus 1% BSA. For experiments that involved the removal of surface-bound antibodies, the cells were further incubated with cold acid (0.2 M acetic acid [pH 2.0]-0.5 M NaCl) (17) for 10 min, followed by PBS washes. Finally, cells were harvested,d lysed in Laemmli SDS buffer (containing 10 mM dithiothreitol and 20 mM β-mercaptoethanol [BDH Laboratory Supplies]), and subjected to Western blot analysis.
Coimmunoprecipitation experiments.
293T cells were transiently transfected with pXJ40myc-GST or XJ40myc-U274 and the various HA-tagged constructs or pXJ3′-S(stop), which does not contain any tags. After ca. 16 to 24 h, the cells were harvested and used in coimmunoprecipitation experiments as previously described (48), except that the reaction buffer used here contained 10 mM HEPES (pH 7.2), 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, and 1% NP-40. Briefly, the lysate was incubated with an anti-myc polyclonal antibody (Santa Cruz Biotechnology) for 2 h at 4°C, followed by adsorption onto 10 μl of protein A-Sepharose beads (Roche). The beads were then washed three times with cold reaction buffer and subjected to Western blot analysis.
RESULTS
Polyclonal antibodies to the SARS-CoV unique protein U274.
The first protein encoded by the second-largest sgRNA (termed sgRNA3 as in reference 52) of the SARS-CoV consists of 274 aa and contains three putative transmembrane domains (Fig. 1A; this ORF is named ORF3 in reference 31, X1 in reference 38, and ORF3a in references 45 and 52), and we refer to it as U274 here. The C terminus hydrophilic domain (134 to 274 aa) of U274 was expressed as a GST fusion protein as previously described (51) and used to raise rabbit polyclonal antibodies. To determine the specificity of the antibody, U274, and three of the structural proteins, M, E, and N, were expressed as HA-tagged fusion proteins in mammalian cells. Another two unique SARS-CoV proteins with >100 aa, U154 (named ORF4 in reference 31, X2 in reference 38, and ORF3b in references 45 and 52) and U122 (named ORF8 in reference 31, X4 in reference 38, and ORF7a in references 45 and 52), were also tested. The expression levels of these proteins, which have the HA epitope fused either at the C terminus (U274-HA, U122-HA, M-HA, E-HA, and U154-HA) or the N terminus (HA-GST and HA-N), were showed in Fig. 1B (upper panel). The proteins that contain potential signal peptides are tagged at the C terminus to minimize any effects on their processing; otherwise, they are tagged at the N terminus. When the anti-U274 rabbit polyclonal antibody was used against these proteins, it recognizes U274 specifically in Western blot analysis, and there is no significant cross-reactivity to the other SARS-CoV proteins tested (Fig. 1B, lower panel). GST protein was also detected since a GST fusion protein was used for immunization.
FIG. 1.

Rabbit polyclonal antibody specific for U274, the first ORF in the second largest subgenomic RNA of SARS-CoV. (A) Schematic diagram showing the predicted topology of U274; (B) Western blot analysis of various HA-tagged SARS-CoV viral proteins expressed in mammalian cells and probed with an anti-HA polyclonal antibody (upper panel) or the anti-U274 rabbit polyclonal antibody (lower panel).
Expression of U274 in SARS-CoV-infected Vero E6 cells.
SARS-CoV 2003VA2774, an isolate from a SARS patient in Singapore, was used to infect Vero E6 cells as previously described (33, 40, 51). Infected cells showing 25 and 75% CPE, respectively, were harvested, lysed, and subjected to Western blot analysis with the anti-U274 antibody (Fig. 2A). Interestingly, three major protein bands (U274-a, -b, and -c) were observed in the infected cells that have 75% CPE (lane 5) but not in mock-infected cells (lane 3). Some weak signal was also observed in the infected cells that have shown 25% CPE (lane 4). The highest protein band (U274-a) at ∼35 kDa migrated at the same rate as U274 full-length protein expressed from a DNA construct (lane 1), suggesting that this is the full-length U274 expressed in infected cells. The two lower protein bands (U274-b and -c) could be processed forms of U274 containing the C terminus domain that is recognized by the antibody.
FIG. 2.
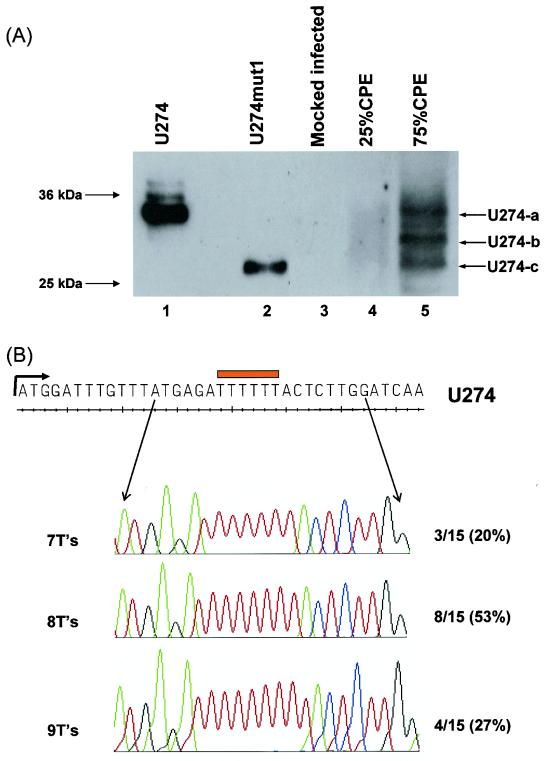
Expression and processing of U274 in SARS-CoV-infected Vero E6 cells. (A) Western blot analysis with anti-U274 rabbit polyclonal detected three protein bands in SARS-CoV-infected cells showing 25% (lane 4) and 75% (lane 5) CPE but not in mock-infected cells (lane 3). The highest (marked as U274-a) and lowest (marked as U274-c) molecular mass proteins in the infected cells migrated at the same rate as full-length U274 (lane 1) and U274mut1, which corresponds to the last 174 aa of U274 (lane 2), respectively. (B) Reverse transcription-PCR products obtained with RNA from infected cells were ligated into pCRII-TOPO vector and sequenced. Chromatographs showed that stretches of seven, eight, and nine T's were present in this region of sgRNA3, which corresponded to 16 to 21 bp downstream of the initiation codon (ATG) of U274. Analysis of the sequences of 18 different SARS-CoV isolates showed that all of them only have six T's in this region. The percentages of independent clones with the respectively number of T nucleotides are shown on the left.
An analysis of the RNA sequences of sgRNA3 isolated from infected Vero E6 cells showed that all of the clones contained insertions in a six-T stretch at 16 to 21 bp downstream of the ATG initiation codon of U274 (Fig. 2B). Of 15 independent clones, the percentages of clones containing seven, eight, and nine T's are 20, 53, and 27%, respectively. An insertion of three extra T nucleotides would result in an extra phenylalanine residue at eight residues from the N terminus of the protein, and the highest protein band observed in infected cells is likely to have been produced from the sgRNA3 containing nine T's in this region. The cellular localization of U274 is not affected by this additional residue (data not shown). On the other hand, an insertion of one or two T nucleotides in this region would result in premature stop codon at 20 or 18 residues from the N terminus of U274, respectively. However, these proteins would not be recognized by the U274 antibody, which was raised against the C terminus of U274. Several alternative initiation codons are found within the sgRNA3 sequences, but only two of them have the same reading frames as U274 and produce proteins that could be recognized by the U274 antibody. The first one translated into a protein of 270 aa, i.e., lacking the first 4 aa of U274, and would have a molecular size very similar to that of U274 (data not shown). The second one translated into a protein of 174 aa (termed U274mut1 in the present study), and this corresponded to the last 174 aa of U274 and would be recognized by the anti-U274 antibody. The U274mut1 protein expressed from a DNA construct migrated at the same rate as the lowest protein band (U274-c) observed in the infected cells (Fig. 2A, lanes 2 and 5), indicating that this truncated from of U274 can be expressed in the infected cells from sgRNA3, including those containing one or two T insertions. It is not clear how the second band (U274-b) observed in the infected cells arises, but it could be a processed form of full-length U274. In the present study, we refer to the U274 with six T's as full-length U274.
Anti-U274 rabbit polyclonal antibody was used in indirect immunofluorescence experiments to determine the localization of U274 in SARS-CoV-infected Vero E6 cells. As shown in Fig. 3A, clusters of cells expressing U274 can be detected in cells that have been infected with SARS-CoV, and no significant staining was observed in mock-infected cells (data not shown). Double labeling with a mouse polyclonal antibody against SARS-CoV S protein (S. Shen, unpublished data), a structural protein that is critical for viral assembly and packaging of coronavirus (for review, see reference 9), and anti-U274 antibody revealed that both proteins were expressed at high level in the same clusters of cells (Fig. 3A to C). In addition, the localization of U274 in infected cells (higher magnification, Fig. 3D) is similar to that observed in cells transfected with U274 (Fig. 3E).
FIG. 3.
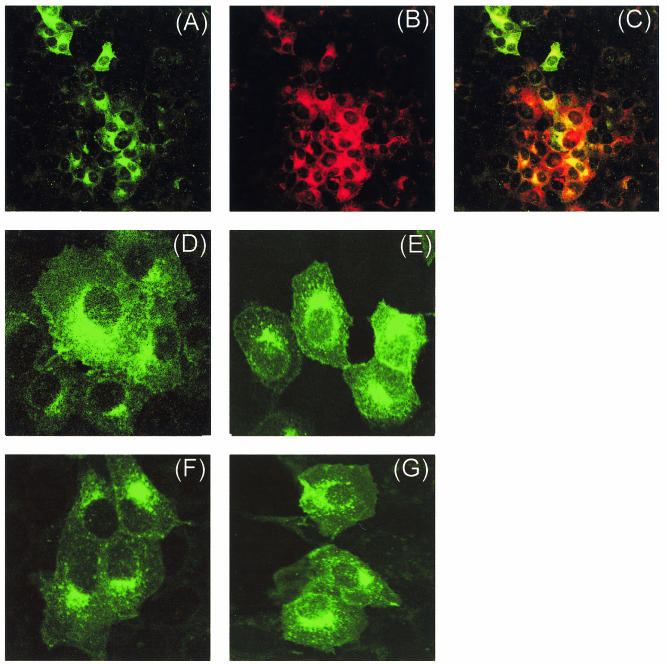
Cellular localization of U274 in infected and transfected Vero E6 cells as determined by indirect immunofluorescence. (A) SARS-CoV-infected cells stained with anti-U274 rabbit polyclonal antibody; (B) SARS-CoV-infected cells stained with anti-S mouse polyclonal antibody; (C) overlay of panels A and B showing cells that are expressing both S and U274 proteins (yellow); (D) SARS-CoV-infected cells (higher magnification); (E) Vero E6 cells transfected with untagged U274; (F) Vero E6 cells transfected with U274-HA; (G) Vero E6 cells transfected with myc-U274. Panels D to G are stained with anti-U274 rabbit polyclonal antibody.
Two fusion forms of U274 were also used in the present study. U274-HA refers to full-length U274 with three HA epitopes at the C terminus, and myc-U274 refers to full-length U274 with a c-myc epitope at the N terminus. As shown in Fig. 3F and G, the cellular localizations of these forms of U274 are similar to that of untagged U274 (Fig. 3E), indicating that these epitopes do not affect the processing of the protein.
Efficient transport of U274 to the cell surface.
Indirect immunofluorescence showed that U274 is localized to the perinuclear region, as well as to the plasma membrane of transfected and infected cells (see Fig. 3, where cells have been permeabilized with Triton X-100). To determine the topology of U274, Vero E6 cells were transfected with myc-U274 and analyzed with either an anti-myc monoclonal antibody, which recognizes the N terminus of the protein, or the anti-U274 rabbit polyclonal antibody, which recognizes the C terminus of the protein. In unpermeabilized cells, anti-myc antibody clearly stained the cell surface of transfected cells (Fig. 4A, lower panel, first column), whereas no signal was observed with anti-U274 rabbit antibody (Fig. 4A, upper panel, first column), suggesting that the N terminus of U274 is on the extracellular side of the plasma membrane and the C terminus is facing the cytoplasm (cf. Fig. 1A). When the cells were permeabilized with digitonin, which selectively permeabilized the plasma membrane (46), plasma membrane stainings were observed with both antibodies (Fig. 4A, upper and lower panels, second column). Similarly, in cells that were permeabilized with Triton X-100, both cell surface and intracellular stainings were observed with both antibodies (Fig. 4A, upper and lower panels, third column).
FIG. 4.

Detection of myc-U274 on the cell surface by indirect immunofluorescence and FACS analysis. (A) Vero E6 cells transiently transfected with myc-U274 were stained with either the anti-U274 rabbit polyclonal antibody (upper panel) that recognizes the C terminus of the protein or anti-myc monoclonal antibody (lower panel) that recognizes N terminus of the protein. Cells were fixed with 4% paraformaldehyde and used directly (no permeabilization) or permeabilized with digitonin or permeabilized with Triton X-100 before incubation with antibodies. (B) FACS analysis of live Vero E6 cells transfected with myc-U274, myc-U274mut2, myc-GST, or myc-N. Cells were incubated with a myc-monoclonal antibody (unfilled histogram) or an irrelevant antibody of the same isotype (filled histograms), followed by the addition of FITC-conjugated goat anti-mouse F(ab′)2 antibody. Surface expression of myc-U274 caused an increase in surface fluorescence upon staining with myc-monoclonal antibody. The expression levels of the proteins were determined by Western blot analysis (with anti-myc antibody).
FACS analysis was also performed to determine whether myc-U274 could be expressed on the surface of live cells. As shown in Fig. 4B (upper panel), an increase in surface fluorescence was observed when Vero E6 cells expressing myc-U274 were incubated with anti-myc monoclonal antibody (open histogram) compared to when these cells were incubated with an irrelevant antibody of the same isotype (solid histogram). Cells expressing myc-GST or myc-N did not show any increase in surface fluorescence (Fig. 4B, upper panel). In addition, a U274 mutant (U274mut2) that lacks sorting signals in the cytoplasmic domain (see Table 1 and Discussion) was also not transported to the cell surface, suggesting that this domain is essential for the transportation of U274 to the cell surface. Protein expression levels were determined by Western blot analysis (lower panel).
TABLE 1.
Comparison of Yxxφ and diacidic motifs found in the cytoplasmic domain of SARS-CoV U274 protein to those found in other known transmembrane proteinsa
| Protein | NCBI identifier (no.) | Yxxφ and (D/E)×(E/D) motif |
|---|---|---|
| U274 | Virus (37361915) | TM-38aa-YNSVTDTIVVTEGD-101aa |
| VSV-G | Virus (138311) | TM-18aa-YTDIEMNRLGK |
| VZV-GPI | Virus (138246) | TM-20aa-YAGLPVDDFEDSESTDTEEEF |
| TfR | Human (136378) | 19aa-YTRFSLARQVDGDNSHV-26aa-TM |
| LDLR (proximal) | Human (126073) | TM-17aa-YQKTTEDEVHICH-20aa |
| LDLR (distal) | Human (126073) | TM-34aa-YSYPSRQMVSLEDDVA |
| CI-M6PR | Mouse (1709091) | TM-26aa-YSKVSKEEETDENE-127aa |
| CD-M6PR | Mouse (127294) | TM-34aa-YRGVGDDGLGEESEERDDHLLPM |
| ASGPR (H1 subunit) | Human (126132) | MTKEYQDLQHLDNEES-24aa |
| CD3δ | Mouse (115986) | TM-21aa-YQPLRDREDTQ-14aa |
| CD3γ | Mouse (115994) | TM-21aa-YQPLKDREYDQ-12aa |
| LAP | Human (130727) | TM-8aa-YRHVADGEDHA |
The Yxxφ motif is underlined, and the acidic residues conforming to the diacidic [(D/E)×(E/D)] signal are in boldface. Sequences were obtained from National Center for Biotechnology Information (NCBI). Abbreviations: VSV-G, vesicular stomatitis virus glycoprotein; VZV-GPI, varicella-zoster virus glycoprotein; LDLR, low-density lipoprotein receptor; CI-M6PR, cation-independent mannose 6-phosphate receptor; CD-M6PR, cation-dependent mannose 6-phosphate receptor; ASGPR, asialoglycoprotein receptor. aa, amino acid(s).
U274 expressed on the cell surface can undergo endocytosis.
The C-terminal cytoplasmic domain of U274 contains a Yxxφ motif, which may be important for rapid internalization of the protein from the plasma membrane (for a review, see reference 56). As shown in Fig. 5 (lanes 3 and 4, upper panel), Vero E6 cells expressing myc-U274 were able to bind and internalize antibody in the culture medium into the cells since a high percentage of the antibodies were found to be internalized and cannot be washed away with the acid. There was no detectable unspecific binding of anti-myc antibody to Vero E6 cells transfected with myc-GST (upper panel, lanes 1 and 2). The experiment was repeated with anti-human CD81 and anti-human TfR antibodies to compare the internalization efficiency of U274 with these cell surface receptors. It has been shown that CD81 has poor internalization efficiency (37, 49), whereas TfR undergoes rapid internalization (for a review, see reference 56). As shown in Fig. 5 (upper panel, lanes 5 and 6), anti-CD81 antibodies bound to the surface of untransfected Vero E6 cells but was not internalized as an acid wash efficiently remove all of the surface-bound CD81 antibodies. Anti-human TfR antibody did not bind to Vero E6 cells (Fig. 5, upper panel, lanes 7 and 8), probably due to species difference for this gene. Therefore, the experiment was repeated by using Vero E6 cells transfected with a cDNA encoding for full-length human TfR (Fig. 5, upper panel, lanes 9 and 10), and the results showed that TfR is efficiently internalized as expected.
FIG. 5.
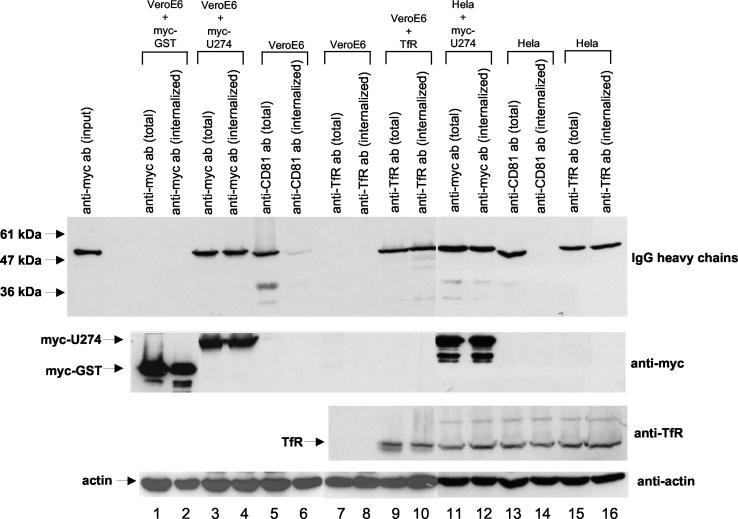
Internalization of antibodies by U274 and other cell surface receptors. Cells transfected with myc-GST (lanes 1 and 2 for Vero E6) or myc-U274 (lanes 3 and 4 for Vero E6 and lanes 11 and 12 for HeLa) were incubated with an anti-myc monoclonal antibody for 3 h at 37°C, followed by PBS washes or PBS-acid washes, and then the cells were harvested and lysed in Laemmli SDS buffer and subjected to Western blot analysis. The cell lysate for cells that were only washed with PBS would contain both surface-bound and internalized antibody (total). For cells that were subjected to an additional acid wash remove surface-bound antibody, the lysate would contain antibody that has been internalized (internalized). The heavy chains of the surface bound and internalized antibody (IgG heavy chains, upper panel) were detected by Western blotting. To compare the internalization efficiency of U274 with CD81 and TfR, the cells were overlaid with anti-human CD81 (lanes 5 and 6 for Vero E6 cells and lanes 13 and 14 for HeLa cells) or anti-human TfR (lanes 9 and 10 for Vero E6 cells transfected with a cDNA encoding for full-length human TfR and lanes 15 and 16 for HeLa cells) antibodies, and the amounts of total versus internalized antibodies were determined as described above. Expression levels of myc-GST or myc-U274 were determined by using anti-myc antibody (second panel). Expression levels of TfR were determined by using anti-TfR antibody (third panel). The level of endogenous actin protein was used to check for equal loading (lowest panel). This experiment was repeated three times, and similar results were obtained each time. A representative set of data is presented.
The same set of experiments was repeated with HeLa cells, which express high levels of endogenous CD81 and TfR. Again, HeLa cells transfected with myc-U274 were able to internalize anti-myc antibodies (Fig. 5, upper panel, lanes 11 and 12) just like endogenous TfR (Fig. 5, upper panel, lanes 15 and 16), whereas CD81 did not undergo efficient internalization (Fig. 5, upper panel, lanes 13 and 14). The expression levels of myc-GST and myc-U274 were determined by using an anti-myc polyclonal antibody (Fig. 5, second panel), and the expression levels of TfR were determined by using anti-TfR antibody. Anti-actin antibody (Fig. 5, lowest panel) was used to check for equal loading.
Interaction of U274 with other SARS-CoV proteins.
Coimmunoprecipitation experiments were performed to determine whether U274 could interact with the SARS-CoV structural proteins M, E, and N. As shown in Fig. 6A (upper panel), myc-U274 specifically coimmunoprecipitated E-HA (lane 6) and M-HA (lane 8), and neither of these showed any nonspecific binding to myc-GST (lanes 5 and 7). HA-N did not interact with myc-U274 or myc-GST (lanes 1 and 2). U274 also interacts strongly with U122 (lanes 3 and 4), another unique SARS-CoV protein that is expressed in infected cells (B. Fielding, unpublished data).
FIG. 6.
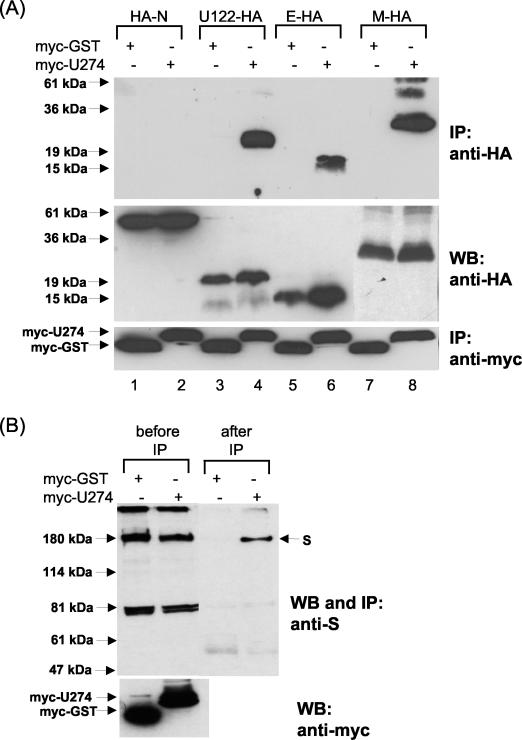
Interactions between U274 and other SARS-CoV proteins. (A) Cell lysates containing myc-GST or myc-U274 with either HA-N, U122-HA, E-HA, M-HA, or U274-HA proteins were immunoprecipitated with myc-polyclonal antibody and protein A-beads. The amounts of HA-tagged proteins that coimmunoprecipitated (IP) with myc-GST (lanes 1, 3, 5, and 7) or myc-U274 (lanes 2, 4, 6, and 8) were determined with an anti-HA monoclonal antibody (upper panel). Expression levels of the HA-tagged proteins were determined by subjecting aliquots of the lysates before immunoprecipitation to Western blot analysis (WB, middle panel). The first blot containing the immunocomplexes (i.e., upper panel) was stripped and reprobed with an anti-myc monoclonal antibody to determine the amount of myc-tagged proteins immunoprecipitated (lower panel). (B) Expression (before IP) and coimmunoprecipitation (after IP) of untagged S protein with myc-GST or myc-U274 were determined with an anti-S mouse polyclonal antibody (upper panel). Expression levels of myc-GST and myc-U274 in the lysates were determined by using anti-myc monoclonal antibody (before IP, lower panel).
In order to determine whether myc-U274 can interact with the S protein, the cells were transfected with myc-U274 and an untagged S construct, since the addition of HA tags to S resulted in poor expression (data not shown). After immunoprecipitation with anti-myc antibodies, the amount of S bound to myc-U274 or myc-GST was determined by using an anti-S mouse polyclonal antibody. As shown in Fig. 6B (upper panel), the S protein of ∼180-kDa interacts specifically with myc-U274 but not with myc-GST.
DISCUSSION
The sequence of the SARS-CoV reveals ORFs for the replicase gene 1a/1b and four structural proteins: S, M, E, and N (31, 38). These genes are expected to be essential for viral replication and are present in all coronaviruses (for reviews, see references 12, 24, 25, and 43). Besides these, there are numerous unique ORFs predicted from the SARS-CoV sequence (31, 38, 52). In the present study we analyzed the first ORF (termed U274 here) of sgRNA3 to determine whether it is expressed during viral infection and whether it serves a function in the viral replication cycle.
Using specific antibodies against U274 and S proteins, we showed by immunofluorescence that both of these proteins are expressed at high levels in Vero E6 cells that were infected by the SARS-CoV (Fig. 3). In infected cells, three forms of U274 could be detected by Western blot analysis: full-length U274 and two shorter products containing the same C terminus as the full-length protein (Fig. 2A). These shorter products may be have been processed from the full-length or could have translated from alternative initiation codons within the U274 encoding sequence. In addition, we also observed insertions in a six-T-nucleotide region found 16 to 21 bp downstream from the initiation codon of U274 (Fig. 2B). Analysis of the sequences of 18 different SARS-CoV isolates (obtained from GenBank database) revealed that none of them show insertion in this region, suggesting that this mutation may be specific to this particular laboratory strain of SARS-CoV. Alternatively, it may only be present at later passages since the third passage was used in the present study, whereas the previously reported sequence of this isolate contained only six T's and was determined at the first passage (SIN2774; GenBank accession no. AY283798 [40]). Alternatively, this may be due to errors during reverse transcription or PCR.
In both transfected and infected cells, U274 is localized to the perinuclear region as well as to the plasma membrane (Fig. 3). Immunofluorescence analysis on cells expressing U274 with a myc tag at the N terminus, by using either anti-myc monoclonal antibody or anti-U274 antibody that recognizes only the C terminus of U274, confirmed the topology of U274: its N terminus is facing the extracellular matrix, and its C terminus is facing the cytoplasm (Fig. 1A and 4). Interestingly, U274 is predicted to have a similar topology since M (31) and our data are consistent with the predicted topology but have not proven the presence of three transmembrane domains in U274. These results confirmed that U274 is a novel SARS-CoV protein that can be transported efficiently to the cell surface of infected cells. This would explain why anti-U274 antibodies were found in the majority of SARS patients (51).
Both S and HE proteins are transported to the cell surface in virus-infected cells and when expressed alone (20, 35, 57). Similarly, the E protein was observed in granular or punctuate cytoplasmic structures, as well as at the plasma membrane (15, 44, 60). On the other hand, when expressed alone, the M protein is found in the Golgi (for a review, see reference 39). However, when M is coexpressed with E, the two proteins form virus-like particles that are morphologically identical to spikeless virions and are released into the medium (2, 4, 10, 29, 59). For coronavirus, the viral envelope that packages the nucleocapsid is derived from the membrane of the intracellular budding site, which is the endoplasmic reticulum-Golgi intermediate compartment or cis-Golgi network (21, 22, 41, 55). After budding, virions are thought to move in vesicles through the secretory pathway and to exit the cells when these vesicles fuse with the plasma membrane (19, 53, 54). However, the exact mechanism(s) for viral release remains unknown, and it is not clear how the viral proteins (S, HE, or E) on the cell surface participate in this process (for reviews, see references 12, 24, 25, and 43).
Analysis of the C-terminal cytoplasmic domain of U274 revealed that it contains a Yxxφ (where x is any amino acid and φ is an amino acid with a bulky hydrophobic side chain) upstream of a ExD (diacidic) motif (see Table 1). The diacidic motif is required for efficient endoplasmic reticulum export (34), whereas the Yxxφ motif has been implicated in directing protein localization to various intracellular compartments (for reviews, see references 3, 30, and 56). Furthermore, most Yxxφ motifs are capable of mediating rapid internalization from the plasma membrane into the endosomes (for reviews, see references 3, 30, and 56). Interaction between the adaptor protein complex 2 (AP-2) with the Yxxφ motif present in the cytoplasmic domain of the internalizing protein concentrated the protein in the clathrin-coated vesicle, which then budded from the plasma membrane, resulting in internalization (for reviews, see references 3, 30, and 56). However, it appears that the Yxxφ motif can also bind other adaptor protein complexes, such as AP-1, -3, and -4, and the differential binding to the different adaptors will determine the pathway of a cargo protein containing a particular Yxxφ motif (for a review, see reference 3). Interestingly, the juxtaposition of these two different sorting motifs, i.e., the diacidic and Yxxφ motifs, have been found in the cytoplasmic tails of a number of plasma membrane proteins (for a review, see reference 1), suggesting that this may be important for transport of proteins to the plasma membrane. In the case of VSV-G, these domains overlapped and the efficient export of VSV-G from the endoplasmic reticulum requires both motifs (42). Indeed, deletion of these motifs from U274 (U274mut2) abolished the transport of U274 to the cell surface (Fig. 4B), suggesting that they are important for the exit of U274 from the endoplasmic reticulum and/or the subsequent transport to the plasma membrane. Further experiments demonstrated that U274 (with a myc tag at the N terminus) expressed on the cell surface was able to internalize anti-myc antibody from the culture medium into the cells, with a efficiency comparable to that of TfR (Fig. 5). The same results were obtained in both Vero E6 and HeLa cells, i.e., the internalization of U274 is not cell type specific (Fig. 5). Of note, an analysis of the sequences of 22 SARS-CoV isolates in the GenBank database, including the four (SZ1, SZ3, SZ13, and SZ16 [16]) that were recently isolated from market animals, showed that these two motifs of U274 are conserved in all but one. The only exception is GZ01 (GenBank accession no. AY278489), in which the ExD motif was AxD instead. It would be interesting to examine how the profile of U274 in this particular isolate differs from the others.
In conclusion, U274 is a novel SARS-CoV protein that can be transported efficiently to the cell surface of infected and transfected cells, and its cytoplasmic domain contained sorting signals that are important for this process. U274 expressed on the cell surface can also undergo endocytosis. It is intriguing to find that SARS-CoV has evolved to express a viral protein with endocytotic properties since endocytosis has been shown to play important roles in the replication of and adaptation to the host cell of a number of viruses (for a review, see reference 32). Taken together, with its ability to interact specifically with proteins M and E, which are two key players in the viral assembly of coronaviruses, as well as with the S protein (Fig. 6), it appears that U274 may play a role in viral assembly and/or release of virus from infected cells. Studies on the mouse hepatitis virus, a member of coronavirus group 2, have also shown that replicase gene products colocalized with structural proteins N and M at different times of infection, indicating that these structural and nonstructural viral proteins can form multiple complexes in infected cells (5, 6). Based on our observation that U274 interacts with U122, which is yet to be characterized, and with the structural proteins (M, E, and S), it is logical to hypothesize that these viral proteins may form multiple complexes in SARS-CoV-infected cells and that the formation of these complexes may be important for viral assembly. This hypothesis will be the subject of future studies. In addition, it will be important to determine the precise role of U274 in the biogenesis of SARS-CoV and to determine whether it has any effects on the host cells during an infection.
Acknowledgments
We thank Jian-Lin Fu, Puay-Yoke Tham, Choon-Tat Keng, and Kuo-Ming Lip for technical assistance and the In Vivo Mouse Unit, IMCB, for producing mouse anti-S antibodies.
This study was supported by grants from the Agency for Science, Technology, and Research (A*STAR) of Singapore.
REFERENCES
- 1.Bannykh, S. I., N. Nishimura, and W. E. Balch. 1998. Getting into the Golgi. Trends Cell Biol. 8:21-25. [DOI] [PubMed] [Google Scholar]
- 2.Baudoux, P., C. Carrat, L. Besnardeau, B. Charley, and H. Laude. 1998. Coronavirus pseudoparticles formed with recombinant M and E proteins induce alpha interferon synthesis by leukocytes. J. Virol. 72:8636-8643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonifacino, J. S., and L. M. Traub. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72:395-447. [DOI] [PubMed] [Google Scholar]
- 4.Bos, E. C., W. Luytjes, H. V. van der Meulen, H. K. Koerten, and W. J. Spaan. 1996. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology 218:52-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bost, A. G., R. H. Carnahan, X. T. Lu, and M. R. Denison. 2000. Four proteins processed from the replicase gene polyprotein of mouse hepatitis virus colocalize in the cell periphery and adjacent to sites of virion assembly. J. Virol. 74:3379-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bost, A. G., E. Prentice, and M. R. Denison. 2001. Mouse hepatitis virus replicase protein complexes are translocated to sites of M protein accumulation in the ERGIC at late times of infection. Virology 285:21-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brian, D. A., B. G. Hogue, and T. E. Kienzle. 1995. The coronavirus hemagglutinin esterase glycoprotein protein, p. 165-179. In S. G. Siddell (ed.), The Coronaviridae. Plenum Press, Inc., New York, N.Y.
- 8.Brown, T. D. K., and I. Brierly. 1995. The coronavirus nonstructural proteins, p. 191-217. In S. G. Siddell (ed.), The Coronaviridae. Plenum Press, Inc., New York, N.Y.
- 9.Cavanagh, D. 1995. The coronavirus surface glycoprotein protein, p. 73-113. In S. G. Siddell (ed.), The Coronaviridae. Plenum Press, Inc., New York, N.Y.
- 10.Corse, E., and C. E. Machamer. 2000. Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles. J. Virol. 74:4319-4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Haan, C. A. M., P. S. Masters, X. Shen, S. Weiss, and P. J. M. Rottier. 2002. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is atttenuating in the natural host. Virology 296:177-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Vries, A. A. F., M. C. Horzinek, P. J. M. Rottier, and R. J. de Groot. 1997. The genome organization of the Nidovirales: similarities and differences between arteri-, toro-, and coronaviruses. Semin. Virol. 8:33-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 14.Fouchier, R. A., T. Kuiken, M. Schutten, G. van Amerongen, G. J. van Doornum, B. G. van den Hoogen, M. Peiris, W. Lim, K. Stohr, and A. D. Osterhaus. 2003. Aetiology: Koch's postulates fulfilled for SARS virus. Nature 423:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Godet, M., R. L'Haridon, J. F. Vautherot, and H. Laude. 1992. TGEV coronavirus ORF4 encodes a membrane protein that is incorporated into virions. Virology 188:666-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan, Y., B. J. Zheng, Y. Q. He, X. L. Liu, Z. X. Zhuang, C. L. Cheung, S. W. Luo, P. H. Li, L. J. Zhang, Y. J. Guan, K. M. Butt, K. L. Wong, K. W. Chan, W. Lim, K. F. Shortridge, K. Y. Yuen, J. S. Peiris, and L. L. Poon. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276-278. [DOI] [PubMed] [Google Scholar]
- 17.Haigler, H. T., F. R. Maxfield, M. C. Willingham, and I. Pastan. 1980. Dansylcadaverine inhibits internalization of 125I-epidermal growth factor in BALB 3T3 cells. J. Biol. Chem. 255:1239-1241. [PubMed] [Google Scholar]
- 18.Herrewegh, A. A., H. Vennema, M. C. Horzinek, P. J. Rottier, and R. J. de Groot. 1995. The molecular genetics of feline coronaviruses: comparative sequence analysis of the ORF7a/7b transcription unit of different biotypes. Virology 212:622-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holmes, K. V., E. W. Doller, and L. S. Sturman. 1981. Tunicamycin resistant glycosylation of coronavirus glycoprotein: demonstration of a novel type of viral glycoprotein Virology 115:334-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kienzle, T. E., S. Abraham, B. G. Hogue, and D. A Brian. 1990. Structure and orientation of expressed bovine coronavirus hemagglutinin-esterase protein. J. Virol. 64:1834-1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klumperman, J., J. K. Locker, A. Meijer, M. C. Horzinek, H. J. Geuze, and P. J. Rottier. 1994. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 68:6523-6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krijnse-Locker, J., M. Ericsson, P. J. Rottier, and G. Griffiths. 1994. Characterization of the budding compartment of mouse hepatitis virus: evidence that transport from the RER to the Golgi complex requires only one vesicular transport step. J. Cell Biol. 124:55-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, L. J. Anderson, et al. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 24.Lai, M. M. C., and D. Cavanagh. 1997. The molecular biology of coronaviruses. Adv. Virus Res. 48:1-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai, M. M. C., and K. V. Holmes. 2001. Coronaviruses, p. 1163-1185. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 26.Laude, H., and P. S. Masters. 1995. The coronavirus nucleocapsid protein, p. 141-163. In S. G. Siddell (ed.), The Coronaviridae. Plenum Press, Inc., New York, N.Y.
- 27.Liu, D. X., and S. C. Inglis. 1991. Association of the infectious bronchitis virus 3c protein with the virion envelope. Virology 185:911-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luytjes, W. 1995. Coronavirus gene expression, p. 33-54. In S. G. Siddell (ed.), The Coronaviridae. Plenum Press, Inc., New York, N.Y.
- 29.Maeda, J., A. Maeda, and S. Makino. 1999. Release of coronavirus E protein in membrane vesicles from virus-infected cells and E protein-expressing cells. Virology 263:265-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marks, M. S., H. Ohno, T. Kirchhausen, and J. S. Bonifacino. 1997. Protein sorting by tyrosine-based signals: adapting to the Ys and wherefores. Trends Cell Biol. 7:124-128. [DOI] [PubMed] [Google Scholar]
- 31.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 32.Marsh, M., and A. Pelchen-Matthews. 2000. Endocytosis in viral replication. Traffic 1:525-532. [DOI] [PubMed] [Google Scholar]
- 33.Ng, M. L., S. H. Tan, E. E. See, E. E. Ooi, and A. E. Ling. 2003. Early events of SARS coronavirus infection in vero cells. J. Med. Virol. 71:323-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishimura, N., and W. E. Balch. 1997. A di-acidic signal required for selective export from the endoplasmic reticulum. Science 277:556-558. [DOI] [PubMed] [Google Scholar]
- 35.Parker, M. D., D. Yoo, G. J. Cox, and L. A. Babiuk. 1990. Primary structure of the S peplomer gene of bovine coronavirus and surface expression in insect cells. J. Gen. Virol. 71:263-270. [DOI] [PubMed] [Google Scholar]
- 36.Peiris, J. S., S. T. Lai, L. L. Poon, Y. Guan, L. Y. Yam, W. Lim, J. Nicholls, W. K. Yee, W. W. Yan, M. T. Cheung, V. C. Cheng, K. H. Chan, D. N. Tsang, R. W. Yung, T. K. Ng, K. Y. Yuen, et al. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petracca, R., F. Falugi, G. Galli, N. Norais, D. Rosa, S. Campagnoli, V. Burgio, E. Di Stasio, B. Giardina, M. Houghton, S. Abrignani, and G. Grandi. 2000. Structure-function analysis of hepatitis C virus envelope-CD81 binding. J. Virol. 74:4824-4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 39.Rottier, P. J. M. 1995. The coronavirus membrane glycoprotein, p. 115-139. In S. G. Siddell (ed.), The Coronaviridae. Plenum Press, Inc., New York, N.Y.
- 40.Ruan, Y. J., C. L. Wei, A. L. Ee, V. B. Vega, H. Thoreau, S. T. Su, J. M. Chia, P. Ng, K. P. Chiu, L. Lim, T. Zhang, C. K. Peng, E. O. Lin, N. M. Lee, S. L. Yee, L. F. Ng, R. E. Chee, L. W. Stanton, P. M. Long, and E. T. Liu. 2003. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet 361:1779-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salanueva, I. J., J. L. Carrascosa, and C. Risco. 1999. Structural maturation of the transmissible gastroenteritis coronavirus. J. Virol. 73:7952-7964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sevier, C. S., O. A. Weisz, M. Davis, and C. E. Machamer. 2000. Efficient export of the vesicular stomatitis virus G protein from the endoplasmic reticulum requires a signal in the cytoplasmic tail that includes both tyrosine-based and di-acidic motifs. Mol. Biol. Cell 11:13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siddell, S. G. 1995. The Coronaviridae. Plenum Press, Inc., New York, N.Y.
- 44.Smith, A. R., M. E. Boursnell, M. M. Binns, T. D. Brown, and S. C. Inglis. 1990. Identification of a new membrane-associated polypeptide specified by the coronavirus infectious bronchitis virus. J. Gen. Virol. 71:3-11. [DOI] [PubMed] [Google Scholar]
- 45.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stearns, M. E., and R. L. Ochs. 1982. A functional in vitro model for studies of intracellular motility in digitonin-permeabilized erythrophores. J. Cell Biol. 94:727-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan, B. H., J. Fu, R. J. Sugrue, E. H. Yap, Y. C. Chan, and Y. H. Tan. 1996. Recombinant dengue type 1 virus NS5 protein expressed in Escherichia coli exhibits RNA-dependent RNA polymerase activity. Virology 216:317-325. [DOI] [PubMed] [Google Scholar]
- 48.Tan, Y.-J., and A. E. Ting. 2000. Non-ionic detergent affects the conformation of a functionally active mutant of Bcl-X(L). Protein Eng. 13:887-892. [DOI] [PubMed] [Google Scholar]
- 49.Tan, Y.-J., S. P. Lim, P. Ng, P.-Y. Goh, S. G. Lim, Y. H. Tan, and W. Hong. 2003. CD81 engineered with endocytotic signals mediates HCV cell entry: implications for receptor usage by HCV in vivo. Virology 308:250-269. [DOI] [PubMed] [Google Scholar]
- 50.Tan, Y.-J., S. P. Lim, A. E. Ting, P.-Y. Goh, Y. H. Tan, S. G. Lim, and W. Hong. 2003. An anti-HIV-1 gp120 antibody expressed as an endocytotic transmembrane protein mediates internalization of HIV-1. Virology 315:80-92. [DOI] [PubMed] [Google Scholar]
- 51.Tan, Y. J., P.-Y. Goh, B. C. Fielding, S. Shen, C.-F. Chou, J.-L. Fu, H. N. Leong, Y. S. Leo, E. E. Ooi, A. E. Ling, S. G. Lim, and W. Hong. Profile of antibody responses against SARS-Coronavirus recombinant proteins and their potential use as diagnostic markers. Clin. Diagn. Lab. Immunol. 11:362-371. [DOI] [PMC free article] [PubMed]
- 52.Thiel, V., K. A. Ivanov, A. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weissbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84:2305-2315. [DOI] [PubMed] [Google Scholar]
- 53.Tooze, J., S. A. Tooze, and G. Warren. 1984. Replication of coronavirus MHV-A59 in sac− cells: determination of the first site of budding of progeny virions. Eur. J. Cell Biol. 33:281-293. [PubMed] [Google Scholar]
- 54.Tooze, J., S. A. Tooze, and S. D. Fuller. 1987. Sorting of progeny coronavirus from condensed secretory proteins at the exit from the trans-Golgi network of AtT20 cells. J. Cell Biol. 105:1215-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tooze, S. A., J. Tooze, and G. Warren. 1988. Site of addition of N-acetyl-galactosamine to the E1 glycoprotein of mouse hepatitis virus-A59. J. Cell Biol. 106:1475-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trowbridge, I. S., J. F. Collawn, and C. R. Hopkins. 1993. Signal-dependent membrane protein trafficking in the endocytic pathway. Annu. Rev. Cell Biol. 9:129-161. [DOI] [PubMed] [Google Scholar]
- 57.Vennema, H., L. Heijnen, A. Zijderveld, M. C. Horzinek, and W. J. Spaan. 1990. Intracellular transport of recombinant coronavirus spike proteins: implications for virus assembly. J. Virol. 64:339-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vennema, H., J. W. Rossen, J. Wesseling, M. C. Horzinek, and P. J. Rottier. 1992. Genomic organization and expression of the 3′ end of the canine and feline enteric coronaviruses. Virology 191:134-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vennema, H., G. J. Godeke, J. W. Rossen, W. F. Voorhout, M. C. Horzinek, D. J. Opstelten, and P. J. Rottier. 1996. Nucleocapsid-independent assembly of coronavirus-like particles by coexpression of viral envelope protein genes. EMBO J. 15:2020-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu, X., W. Bi, S. R. Weiss, and J. L. Leibowitz. 1994. Mouse hepatitis virus gene 5b protein is a new virion envelope protein. Virology 202:1018-1023. [DOI] [PubMed] [Google Scholar]


