Abstract
Recent advances in class II tetramer staining technology have allowed reliable direct ex vivo visualization of antigen-specific CD4 T cells. In order to define the frequency and phenotype of a prototype response to a nonpersistent pathogen, we have used such techniques to analyze influenza virus-specific memory CD4 T cells directly from blood. These responses are stably detectable ex vivo at low frequencies (range, 0.00012 to 0.0061% of CD4 T cells) and display a distinct “central memory” CD62L+ phenotype.
The development of major histocompatibility complex (MHC) class I tetramers has greatly facilitated the analysis of the frequency, function, and phenotype of antigen-specific CD8 cells (1). In contrast, difficulties in producing stable peptide-MHC class II complexes as well as the low frequency of antigen-specific CD4 cells in peripheral blood have contributed to a lag in the development of MHC class II tetramers for efficient detection of antigen-specific CD4 cells (11, 12). However, recent advances in class II tetramer technology have shown that human virus-specific memory CD4 cells can be detected directly ex vivo in peripheral blood (7). Such techniques can now be exploited to define differences in the phenotypes or frequencies of antigen-specific CD4 cells in different disease states for persistent infections, such as those caused by human immunodeficiency virus and hepatitis C virus, compared to nonpersistent viral infections such as those caused by influenza virus. Previous studies of virus-specific CD8 cells with the use of class I tetramers have indicated a broad spectrum of phenotypes associated with antigen-specific CD8 cells at various stages of different viral infections, including those caused by Epstein-Barr virus, cytomegalovirus, human immunodeficiency virus, and hepatitis C virus (2), although a comparative analysis of the ex vivo phenotypes of virus-specific CD4 cells has yet to be described in detail.
In order to analyze the frequency and phenotype of virus-specific memory CD4 cells directly ex vivo to a nonpersistent viral pathogen, we utilized both a previously described (5) and a commercially available (Beckman Coulter) DRB1*0101 class II tetramer bound to a peptide (amino acids [aa] 306 to 318) from the hemagglutinin (HA) protein of influenza virus. All results shown were obtained using the DR1-HA tetramer purchased from Beckman Coulter, with the exception of the results for the first three time points for subjects 1 and 4 shown in Fig. 1C, which were obtained using the DR1-HA tetramer constructed as previously described (5). Fresh peripheral blood mononuclear cells (PBMCs) were obtained from six DR1-positive healthy individuals. PBMCs were stained for 2 h at room temperature with phycoerythrin (PE)-conjugated DR1-HA tetramer, and allophycocyanin (APC)-conjugated anti-CD4 antibodies, peridinin chlorophyll protein (PerCP)-conjugated anti-CD14 and anti-CD19 antibodies (to exclude monocytes and B cells, respectively), Via-Probe (containing 7-amino-acinomycin D for dead cell exclusion), and fluorescein isothiocyanate (FITC)-conjugated antibodies for phenotypic analysis were added during the last 20 min of incubation. All antibodies were obtained from BD Biosciences PharMingen, except for anti-human FITC-conjugated CCR7 antibody, which was obtained from R & D Systems. Cells were subsequently washed and labeled with anti-PE microbeads (Miltenyi Biotec), and 10% of the cells were reserved for flow cytometric analysis to determine the total input number of cells, while the remaining 90% of anti-PE-labeled cells were applied to magnetic columns to enrich for PE-conjugated tetramer-positive cells and subsequently analyzed by flow cytometry. Cells were gated on CD4+ CD14− CD19− Via-Probe− cells in the live lymphocyte gate.
FIG. 1.
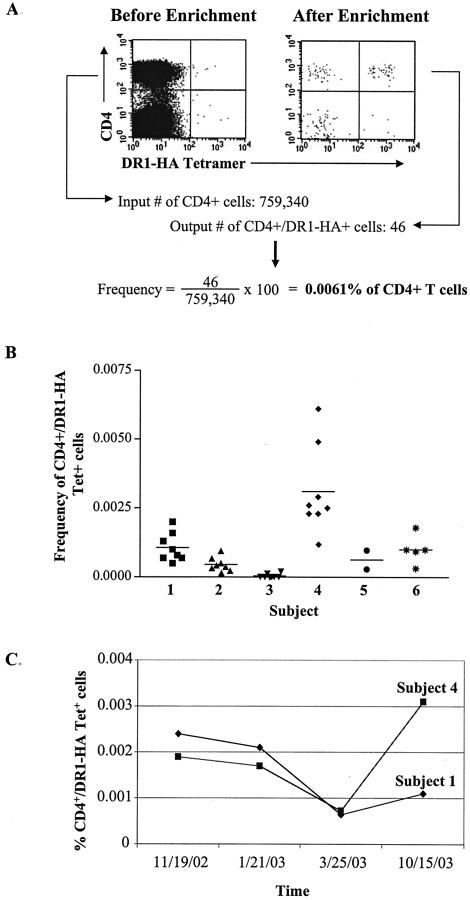
Frequencies of circulating DR1-HA tetramer-positive cells ex vivo. (A) Example of ex vivo DR1-HA tetramer staining of CD4 T cells before and after magnetic bead enrichment for PE-positive cells. PBMCs were stained with PE-conjugated DR1-HA tetramer, APC-conjugated anti-CD4 antibody, PerCP-conjugated anti-CD14 and -CD19 antibodies to exclude monocytes and B cells, respectively, and Via-Probe to exclude dead cells. Cells were incubated with anti-PE microbeads, and PE-positive cells were selected (enriched) on a magnetic column. Plots are gated on CD4+ CD14− CD19− Via-Probe− cells. The plot on the left shows DR1-HA tetramer staining of CD4+ cells before enrichment, and the plot on the right shows the same cell population after enrichment. The frequency of DR1-HA tetramer-positive cells after enrichment was calculated by dividing the number of CD4+ tetramer+ cells after enrichment by the input number of CD4 cells. In this example, the input number of CD4 cells was 759,340 and the output number of CD4+ DR1-HA+ cells was 46. (B) Reproducibility of the frequency estimations of DR1-HA tetramer-positive cells in six healthy DR1-positive individuals from a single time point. Between two and eight replicate tetramer staining assays were performed on a single PBMC collection from each subject. The frequency of tetramer-positive cells was analyzed as described above. The horizontal bar represents the mean tetramer-positive frequency for each individual. (C) Longitudinal analysis of DR1-HA tetramer-positive cells in two DR1-positive individuals. Four PBMC samples were obtained over a 12-month period and analyzed as described above. The mean frequency of DR1-HA tetramer-positive cells at each time point is plotted for subjects 1 and 4.
Figure 1 indicates the frequency of HA-specific CD4 cells after enrichment for DR1-HA tetramer-positive cells. Before enrichment, no distinct population of HA-specific CD4 cells could be detected in fresh PBMCs, although a population of CD4+ DR1-HA+ cells is clearly visible after enrichment with anti-PE microbeads (Fig. 1A). The frequencies of HA-specific CD4 cells were calculated by dividing the number of output CD4+ tetramer+ cells after enrichment by the input number of CD4 cells. Multiple replicates of the frequencies of HA-specific CD4 cells were performed in six DR1-positive individuals from a single blood draw. These data reveal that the frequencies of HA-specific CD4 cells are extremely low directly ex vivo, ranging from 0.00012 to 0.0061% of circulating CD4 cells (Fig. 1B). These frequencies are approximately 10-fold higher than those previously estimated based on CFSE labeling of PBMC-stimulated influenza virus peptides for 6 days in culture (6).
We next investigated the stability of HA-specific CD4 responses in two DR1-positive healthy individuals over time. Four PBMC samples were obtained over a 12-month period, and between two and eight replicate DR1-HA tetramer staining assays were performed for each individual. The mean frequencies of HA-specific CD4 cells in subject 1 ranged from 0.00064 to 0.0024% of CD4 cells and 0.00072 to 0.0031% of CD4 cells in subject 4 (Fig. 1C). These data indicate that HA-specific CD4 memory cells are persistently maintained at these low frequencies in peripheral blood in healthy individuals over time.
Having demonstrated that we can reliably and reproducibly detect circulating DR1-HA-specific CD4 cells in healthy individuals, we next examined the phenotype of these cells to further characterize a prototype human memory CD4 cell response to a nonpersistent viral pathogen. We examined phenotypic markers that have been reported to be associated with different memory or effector populations of antigen-specific T cells (2, 9, 15, 16, 21). DR1-HA+ CD4+ cells were found to be CCR7+ CD62L+ CD27+ CD28+ CD45RA− and CD25−; representative data from subjects 1 and 4 are shown in Fig. 2A. This phenotype is consistent with that of a “central memory” phenotype, as may be predicted in response to a nonpersistent antigen such as influenza virus that is cleared following each infection. Phenotypic analysis of DR1-HA tetramer-positive cells was performed in four DR1-positive individuals with sufficient frequencies of tetramer-positive cells directly ex vivo (subjects 1, 2, 4, and 6, Fig. 1B), and a very consistent phenotype indicative of central memory cells was found for HA-specific CD4 cells in all four subjects (Fig. 2B). In addition, the ability of HA-specific CD4 cells to produce gamma interferon (IFN-γ) and interleukin 2 (IL-2) was assessed in three subjects by incubating freshly isolated PBMCs with or without the HA peptide (aa 306 to 318; final concentration of 10 μg/ml) for 6 h at 37°C. HA-specific cells secreting IFN-γ and/or IL-2 were detected using the IL-2 (PE) and IFN-γ (APC) cell enrichment and detection kits according to the instructions of the manufacturer (Miltenyi Biotec) (4). The majority of HA-specific cells produced both IFN-γ and IL-2 and are present at frequencies similar to that detected with the DR1-HA tetramer. Representative data comparing DR1-HA tetramer staining to HA-specific IFN-γ and IL-2 production in subject 6 are shown in Fig. 2C.
FIG. 2.
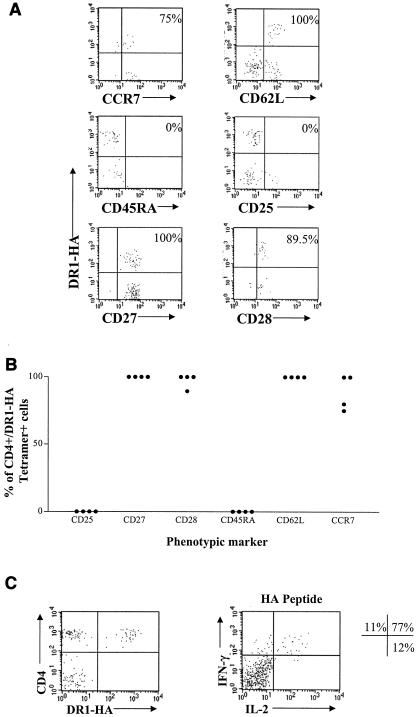
Ex vivo phenotypic and functional analysis of DR1-HA tetramer-positive cells. (A) Example of phenotypic analysis on DR1-HA tetramer-positive cells for six markers pertaining to memory status of CD4+ T cells. Fresh PBMCs were stained as described for Fig. 1A with the addition of an FITC-conjugated antibody for either CD25, CD27, CD28, CD45RA, CD62L, or CCR7. Plots are shown after enrichment for PE-positive cells and are gated on CD4+ CD14− CD19− Via-Probe− events as described for Fig. 1. The frequency shown indicates the percentage of CD4+ DR1-HA tetramer+ cells positive for each marker. These results were obtained from subjects 1 and 4. (B) Summary of phenotypic data for four DR1-positive individuals. Individual data points were obtained as described above. (C) Ex vivo functional analysis of HA-specific CD4 cells. The plot on the left indicates enrichment for DR1-HA tetramer-positive cells from subject 6; the frequency of tetramer-positive cells in this subject was 0.0018% of CD4 T cells. PBMCs from the same time point were stimulated with the DR1-HA peptide (aa 306 to 318) for 6 h at 37°C, followed by enrichment for IFN-γ- and IL-2-positive cells with the IL-2 (PE) and IFN-γ (APC) secretion assay cell enrichment and detection kits according to the instructions of the manufacturer (Miltenyi Biotec). The percentages shown indicate the relative proportions of CD4 cells producing IFN-γ and/or IL-2 following stimulation with the HA peptide; background levels of nonspecific cytokine production have been subtracted. The total frequency of cytokine-positive cells after subtraction of background in this subject was 0.0017%. The frequencies of IFN-γ single-positive, IFN-γ IL-2 double-positive, and IL-2 single-positive cells were 0.00018, 0.0013, and 0.00021%, respectively, of CD4 cells.
In this study we have used magnetic bead enrichment techniques to rapidly and reliably enrich for low-frequency class II tetramer-positive populations and have detected influenza virus-specific CD4 cells directly ex vivo with a frequency as low as 0.00012%. It should be noted that the frequencies reported here of circulating HA-specific CD4 cells represent a minimal ex vivo frequency, as some CD4 cells that bind the DR1-HA tetramer with low affinity may be lost during the enrichment procedure. Furthermore, frequencies of influenza virus-specific CD4 or CD8 T-cell responses may fluctuate over time due to reexposure to influenza virus antigens via reinfection or vaccination (6). The detection of HA-specific CD4 cells in this study with a frequency as low as 0.00012% represents a frequency 25-fold more sensitive than that in previous reports of the sensitivity of class II influenza virus tetramers, which were reported to be able to detect HA-specific CD4 cells in peripheral blood at a frequency of 0.003% (6). The phenotype of these cells as determined in three individuals ex vivo supports that of a central memory phenotype. These results are consistent with previous findings of markers expressed on HA-specific CD4 cells in a single subject analyzed after influenza virus vaccination, with the important exception of the expression of CD62L, where HA-specific CD4 cells from this individual were reported to be CD62L− (6). This may be due to the expansion of HA-specific CD4 cells with an effector phenotype following vaccination. This notion is supported by recent studies with mice, which indicated that CD4 effector cells downregulated CD62L during the peak of the immune response following infection with influenza virus (14). In the case of influenza virus-specific CD8 T cells, reversion to CD62L+ status can take several months following acute stimulation with influenza A virus (20). Despite repeated exposure to influenza virus naturally in humans, we found no evidence here of influenza virus-specific CD4 cells with a “late” or “effector” phenotype in the individuals analyzed. As previous studies have shown CD4 cells to play a vital role both in priming efficient antibody responses and in maintaining effective virus-specific memory CD8 responses (3, 8, 10, 13, 17-19, 22), further characterization of virus-specific CD4 cells ex vivo utilizing class II tetramers by sensitive techniques as described here will be important to begin to address potential mechanisms of immune failure in persistent uncontrolled viral infections.
Acknowledgments
This work was supported by grants from the Wellcome Trust and the European Union (QLK2-CT-2002-01329) to M.L. and P.K. and from the Royal Society to C.L.D.
REFERENCES
- 1.Altman, J. D., P. A. Moss, P. J. Goulder, D. H. Barouch, M. G. McHeyzer-Williams, J. I. Bell, A. J. McMichael, and M. M. Davis. 1996. Phenotypic analysis of antigen-specific T lymphocytes. Science 274:94-96. [DOI] [PubMed] [Google Scholar]
- 2.Appay, V., P. R. Dunbar, M. Callan, P. Klenerman, G. M. Gillespie, L. Papagno, G. S. Ogg, A. King, F. Lechner, C. A. Spina, S. Little, D. V. Havlir, D. D. Richman, N. Gruener, G. Pape, A. Waters, P. Easterbrook, M. Salio, V. Cerundolo, A. J. McMichael, and S. L. Rowland-Jones. 2002. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat. Med. 8:379-385. [DOI] [PubMed] [Google Scholar]
- 3.Battegay, M., D. Moskophidis, A. Rahemtulla, H. Hengartner, T. W. Mak, and R. M. Zinkernagel. 1994. Enhanced establishment of a virus carrier state in adult CD4+ T-cell-deficient mice. J. Virol. 68:4700-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brosterhus, H., S. Brings, H. Leyendeckers, R. A. Manz, S. Miltenyi, A. Radbruch, M. Assenmacher, and J. Schmitz. 1999. Enrichment and detection of live antigen-specific CD4+ and CD8+ T cells based on cytokine secretion. Eur. J. Immunol. 29:4053-4059. [DOI] [PubMed] [Google Scholar]
- 5.Cunliffe, S. L., J. R. Wyer, J. K. Sutton, M. Lucas, G. Harcourt, P. Klenerman, A. J. McMichael, and A. D. Kelleher. 2002. Optimization of peptide linker length in production of MHC class II/peptide tetrameric complexes increases yield and stability, and allows identification of antigen-specific CD4+ T cells in peripheral blood mononuclear cells. Eur. J. Immunol. 32:3366-3375. [DOI] [PubMed] [Google Scholar]
- 6.Danke, N. A., and W. W. Kwok. 2003. HLA class II-restricted CD4+ T cell responses directed against influenza viral antigens postinfluenza vaccination. J. Immunol. 171:3163-3169. [DOI] [PubMed] [Google Scholar]
- 7.Day, C. L., N. P. Seth, M. Lucas, H. Appel, L. Gauthier, G. M. Lauer, G. K. Robbins, Z. M. Szczepiorkowski, D. R. Casson, R. T. Chung, S. Bell, G. Harcourt, B. D. Walker, P. Klenerman, and K. W. Wucherpfennig. 2003. Ex vivo analysis of human memory CD4 T cells specific for hepatitis C virus using MHC class II tetramers. J. Clin. Investig. 112:831-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janssen, E. M., E. E. Lemmens, T. Wolfe, U. Christen, M. G. von Herrath, and S. P. Schoenberger. 2003. CD4+ T cells are required for secondary expansion and memory in CD8+ T lymphocytes. Nature 421:852-856. [DOI] [PubMed] [Google Scholar]
- 9.Lanzavecchia, A., and F. Sallusto. 2000. Dynamics of T lymphocyte responses: intermediates, effectors, and memory cells. Science 290:92-97. [DOI] [PubMed] [Google Scholar]
- 10.Matloubian, M., R. J. Concepcion, and R. Ahmed. 1994. CD4+ T cells are required to sustain CD8+ cytotoxic T-cell responses during chronic viral infection. J. Virol. 68:8056-8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McMichael, A. J., and A. Kelleher. 1999. The arrival of HLA class II tetramers. J. Clin. Investig. 104:1669-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Novak, E. J., A. W. Liu, G. T. Nepom, and W. W. Kwok. 1999. MHC class II tetramers identify peptide-specific human CD4+ T cells proliferating in response to influenza A antigen. J. Clin. Investig. 104:R63-R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Planz, O., S. Ehl, E. Furrer, E. Horvath, M. A. Brundler, H. Hengartner, and R. M. Zinkernagel. 1997. A critical role for neutralizing-antibody-producing B cells, CD4+ T cells, and interferons in persistent and acute infections of mice with lymphocytic choriomeningitis virus: implications for adoptive immunotherapy of virus carriers. Proc. Natl. Acad. Sci. USA 94:6874-6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roman, E., E. Miller, A. Harmsen, J. Wiley, U. H. Von Andrian, G. Huston, and S. L. Swain. 2002. CD4 effector T cell subsets in the response to influenza: heterogeneity, migration, and function. J. Exp. Med. 196:957-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sallusto, F., D. Lenig, R. Forster, M. Lipp, and A. Lanzavecchia. 1999. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401:708-712. [DOI] [PubMed] [Google Scholar]
- 16.Seder, R. A., and R. Ahmed. 2003. Similarities and differences in CD4+ and CD8+ effector and memory T cell generation. Nat. Immunol. 4:835-842. [DOI] [PubMed] [Google Scholar]
- 17.Shedlock, D. J., and H. Shen. 2003. Requirement for CD4 T cell help in generating functional CD8 T cell memory. Science 300:337-339. [DOI] [PubMed] [Google Scholar]
- 18.Sun, J. C., and M. J. Bevan. 2003. Defective CD8 T cell memory following acute infection without CD4 T cell help. Science 300:339-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomsen, A. R., J. Johansen, O. Marker, and J. P. Christensen. 1996. Exhaustion of CTL memory and recrudescence of viremia in lymphocytic choriomeningitis virus-infected MHC class II-deficient mice and B cell-deficient mice. J. Immunol. 157:3074-3080. [PubMed] [Google Scholar]
- 20.Tripp, R. A., S. Hou, and P. C. Doherty. 1995. Temporal loss of the activated l-selectin-low phenotype for virus-specific CD8+ memory T cells. J. Immunol. 154:5870-5875. [PubMed] [Google Scholar]
- 21.Wherry, E. J., V. Teichgraber, T. C. Becker, D. Masopust, S. M. Kaech, R. Antia, U. H. von Andrian, and R. Ahmed. 2003. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat. Immunol. 4:225-234. [DOI] [PubMed] [Google Scholar]
- 22.Zajac, A. J., J. N. Blattman, K. Murali-Krishna, D. J. Sourdive, M. Suresh, J. D. Altman, and R. Ahmed. 1998. Viral immune evasion due to persistence of activated T cells without effector function. J. Exp. Med. 188:2205-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]


