Abstract
Epigenetic mechanisms such as DNA methylation (DNAm) are essential for regulation of gene expression. DNAm is dynamic, influenced by both environmental and genetic factors. Epigenetic drift is the divergence of the epigenome as a function of age due to stochastic changes in methylation. Here we show that epigenetic drift may be constrained at many CpGs across the human genome by DNA sequence variation and by lifetime environmental exposures. We estimate repeatability of DNAm at 234,811 autosomal CpGs in whole blood using longitudinal data (2–3 repeated measurements) on 478 older people from two Scottish birth cohorts—the Lothian Birth Cohorts of 1921 and 1936. Median age was 79 yr and 70 yr, and the follow-up period was ∼10 yr and ∼6 yr, respectively. We compare this to methylation heritability estimated in the Brisbane Systems Genomics Study, a cross-sectional study of 117 families (offspring median age 13 yr; parent median age 46 yr). CpG repeatability in older people was highly correlated (0.68) with heritability estimated in younger people. Highly heritable sites had strong underlying cis-genetic effects. Thirty-seven and 1687 autosomal CpGs were associated with smoking and sex, respectively. Both sets were strongly enriched for high repeatability. Sex-associated CpGs were also strongly enriched for high heritability. Our results show that a large number of CpGs across the genome, as a result of environmental and/or genetic constraints, have stable DNAm variation over the human lifetime. Moreover, at a number of CpGs, most variation in the population is due to genetic factors, despite some sites being highly modifiable by the environment.
Epigenetics refers to the chemical modifications of DNA that regulate gene expression without altering the underlying DNA sequence. DNA methylation (DNAm), the covalent addition of a methyl group to a cytosine nucleotide primarily in the context of a CpG dinucleotide, is among the best-studied epigenetic mechanisms and plays a crucial role in mammalian development (Li et al. 1992; Tate et al. 1996), X-chromosome inactivation (Panning and Jaenisch 1998), imprinting (Li et al. 1993), cell-lineage specification (Bröske et al. 2009; Trowbridge et al. 2009; Challen et al. 2012), and maintaining genome stability (Lengauer et al. 1997; Bellacosa et al. 1999). In mammalian development, shortly after fertilization, DNAm marks are largely erased. Genome-wide and tissue-specific de novo methylation takes place following blastocyst formation. Primordial germ cells also undergo demethylation, followed by de novo methylation during gametogenesis and establishment of sex-dependent imprints. In contrast with sequence variants, DNAm is dynamic and is influenced by many factors, including lifestyle, environment, and genetics (Jirtle and Skinner 2007; Mathers et al. 2010; Alegría-Torres et al. 2011).
Two forms of inheritance of epigenetic marks have been demonstrated: genetic inheritance and epigenetic inheritance. The latter is independent of DNA sequence and may occur through incomplete erasure of epigenetic marks during epigenetic reprogramming at the gamete and zygote stages. The genetic inheritance of an epigenetic state occurs when an individual’s DNA sequence affects the epigenetic state. A large number of common single nucleotide polymorphisms (SNPs) have been shown to affect methylation at CpG sites that are typically in close proximity to the SNP, i.e., located in cis (Kerkel et al. 2008; Gibbs et al. 2010; Bell et al. 2011; Grundberg et al. 2012), with some SNPs explaining up to 90% of the variation in methylation at the associated CpG site (Gibbs et al. 2010). These genetic variants are referred to as DNA methylation quantitative trait loci (mQTL), and methylation at the associated CpG sites is highly heritable (McRae et al. 2014). Recent estimates of mean genome-wide DNAm heritability (proportion of variance explained by additive genetic factors) are 12%–18% in blood (Bell et al. 2012; Grundberg et al. 2013; McRae et al. 2014), but locus-specific levels have shown to be variable. Mean heritability across tissues vary, with estimates of 5% in placenta and 7% in human umbilical vein endothelial cells (HUVECs) (Gordon et al. 2012).
Age-related changes in DNAm are well documented, and a large number of age-associated methylation sites have been identified across the genome (Bocklandt et al. 2011; Hannum et al. 2013; Horvath 2013). The epigenome diverges with age, even within monozygotic twins, with older twins having less similar epigenetic profiles than younger twins (Fraga et al. 2005). The differences in the epigenome of genetically identical individuals have been attributed to differences in external factors, such as changes in lifestyle and environment, but also to internal stochastic factors, such as small defects in transmitting epigenetic information through successive cell divisions or maintaining it in differentiated cells, a process termed “epigenetic drift” (Fraga et al. 2005). Epigenetic differences in genetically identical twins may be key to understanding disease discordance in monozygotic twins.
Understanding what factors contribute to DNAm variation is important not only for understanding the role of DNAm in complex disease traits, but also for informing future epigenetic studies that take a cross-sectional approach. There have been relatively few longitudinal studies on DNAm, and these have focused either on a handful of selected loci (Talens et al. 2010; Wong et al. 2010) or only on loci whose DNAm changes with age (Madrigano et al. 2012; Florath et al. 2013), and others have used only global measures of methylation (Bjornsson et al. 2008). Therefore, it is still not fully known how stable DNAm levels across the genome are over time and to what extent these are influenced by heritable versus environmental factors.
When longitudinal data are available, one measure of stability over time is the repeatability (Rm) of methylation levels. Rm is the correlation between repeated measures of the same individual (Falconer and Mackay 1996) and is calculated as the ratio of the between-individual variance over the total variance. Variance between individuals can be partitioned into variance due to genetic factors and variance due to permanent differences in the environment between individuals. The genetic variance can be further partitioned into additive and nonadditive (dominant and epistatic effects) genetic variance, and the proportion of phenotypic variance due to additive genetic factors is the (narrow sense) heritability (h2). Since heritability is one of the determining factors of repeatability, h2 estimates should not exceed Rm estimates in the same sample, and Rm therefore sets an upper bound for h2 (see Methods). By comparing the Rm and h2, we can assess the extent to which variation between individuals is due to additive genetic factors.
To assess the stability of DNAm across the genome over time, we estimated methylation Rm at 234,811 autosomal CpG probes in whole blood using longitudinal data on 478 older, initially healthy, Scottish individuals from two birth cohorts—the Lothian Birth Cohorts of 1921 (LBC1921) and 1936 (LBC1936) (Deary et al. 2004, 2007, 2012). Participants had two or three repeated measures of DNAm over a period of ∼10 yr in LBC1921 and ∼6 yr in LBC1936. Median ages at baseline (wave 1) were 79 yr and 70 yr, in LBC1921 and LBC1936, respectively. To assess the extent to which additive genetic factors contribute to DNAm variation, we compared the Rm for each CpG site to their h2 estimated in the Brisbane Systems Genomics Study (BSGS). The BSGS is a cross-sectional study of 117 families comprising adolescent monozygotic (MZ) and dizygotic (DZ) twins, their siblings and their parents, with an age range of 9–75 yr (offspring median age 13 yr; parent median age 46 yr) (Powell et al. 2012).
Results
Data
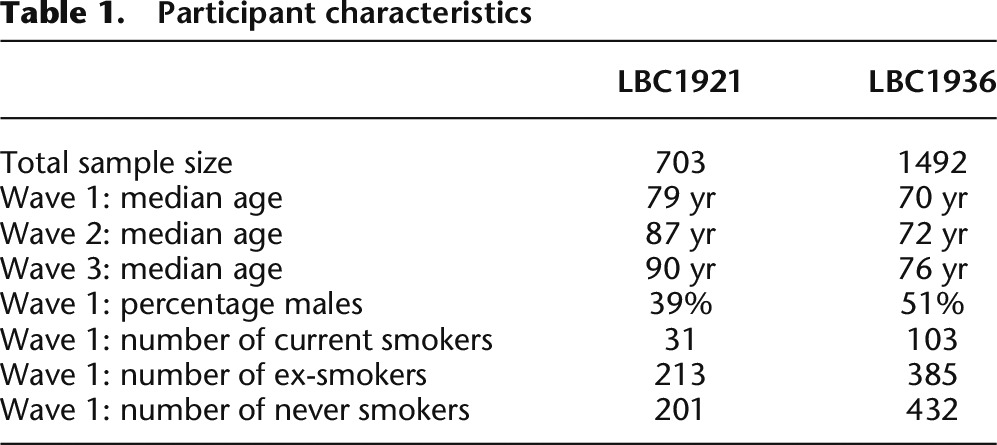
In LBC, DNAm was measured in 2460 whole blood samples (from 1520 individuals) at 485,512 CpG sites across the genome using the Infinium HumanMethylation450 array. After quality control (QC) analysis and probe filtering (see Methods), 2195 samples (from 1366 individuals) and 249,741 methylation probes remained. Only 478 of the 1366 individuals in the QC'ed data had repeated measures of DNAm (186 individuals with two measures and 292 individuals with three measures), and these were used for estimating repeatability. At the three waves of sample collection, LBC1921 participants had median age 79 (n = 446), 87 (n = 175), and 90 (n = 82) yr, and LBC1936 participants had median age 70 (n = 920), 72 (n = 299), and 76 (n = 273) yr (Table 1). In BSGS, DNAm was measured in 614 blood lymphocyte samples using the Infinium HumanMethylation450 array. All 614 samples passed QC with DNAm data available on 261,450 autosomal CpG sites.
Table 1.
Participant characteristics
Beta value heteroscedasticity
Methylation levels are summarized as beta values ranging between 0 and 1, where a value close to zero indicates that all copies of the CpG site in the sample were completely unmethylated (no methylated molecules were measured), and a value close to one indicates that every copy of the site was methylated. Six percent of CpG sites (14,930 of 249,741) were either unmethylated (beta ≤ 0.1) across all samples or completely methylated (beta values ≥ 0.9) across all samples. The beta value has significant heteroscedasticity, and the standard deviation of the beta value in these very low and very high beta ranges is greatly compressed (Supplemental Fig. S1; Du et al. 2010). Any variation in methylation signal at these high and low ranges is likely to be measurement noise. Since Rm is a function of both within-individual and between-individual variance, individual consistency will only be evident in the repeatability if there is between-individual variation in the sample (Supplemental Fig. S2). Therefore, despite these CpGs having extremely stable methylation in the entire sample, their estimates of repeatability were very low (Supplemental Fig. S3A). This was also the case for heritability estimates (Supplemental Fig. S3B). Based on assessments of the relationship between mean beta values, beta variance, and Rm, we excluded these 14,930 probes. Rm estimates for 234,811 probes were therefore considered in further analysis.
Genome-wide repeatability
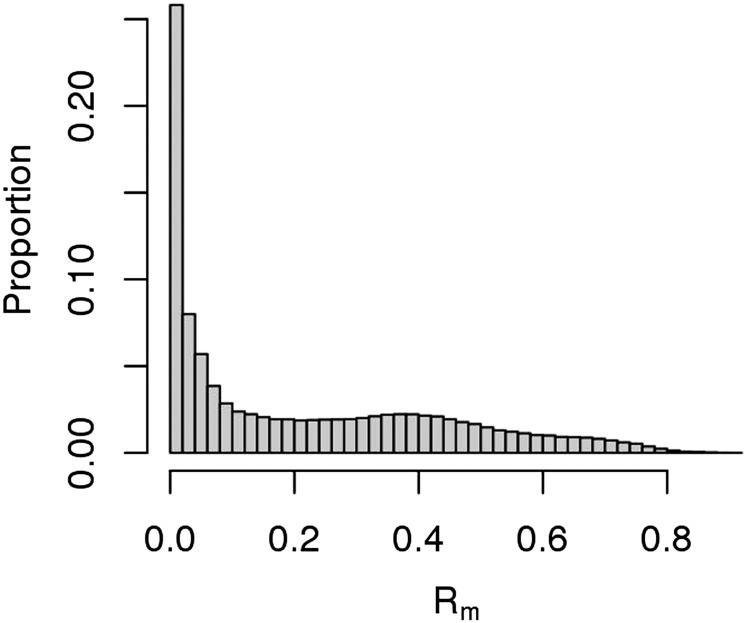
The distribution of Rm for the 234,811 autosomal CpG sites is shown in Figure 1. We found that 13% (31,273 probes) had Rm ≥ 0.5. To show that the CpG sites with higher Rm were not observed simply by chance, we used permutation to derive the null distribution. The empirical distribution of Rm values was not consistent with the null hypothesis of chance associations (Supplemental Fig. S4). For comparison, we also looked at the distribution of Rm for the 234,811 CpG sites in seven pairs of technical replicates available in the BSGS data (no replicate data was available in the LBCs). The Rm distribution was shifted to the right, with the majority of probes having high Rm values (60% with Rm ≥ 0.5) (Supplemental Fig. S5). However, around 25,000 probes had repeatability of zero even in technical replicates. These may represent poor-performing probes. A much larger number of replicate samples would be needed to accurately estimate the true proportion of repeatable probes and identify those that warrant removal from further analyses. With a larger number of replicates, the standard error in the Rm estimation will decrease, and it is likely the Rm distribution would shift further to the right, with most probes either having repeatability close to one or close to zero, providing greater power to distinguish the poor-performing probes.
Figure 1.
Distribution of Rm for 234,811 autosomal CpG sites in the Lothian Birth Cohorts (LBC).
Correlation between repeatability and heritability
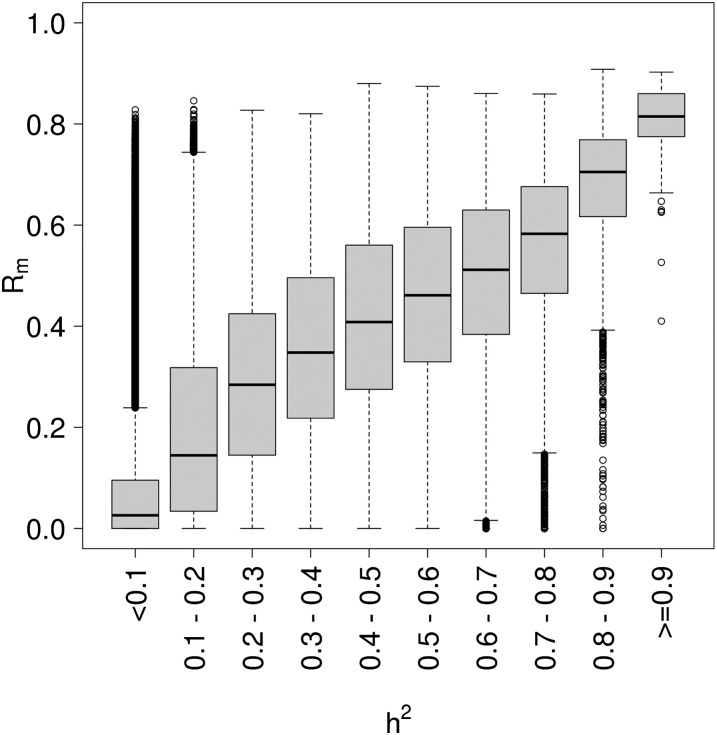
We then correlated probe Rm to probe h2, which had been estimated in the younger BSGS family cohort (McRae et al. 2014). All 234,811 probes were present in the QC'ed BSGS data set. We observed a high correlation of 0.68 (P-value < 1.0 × 10−16) between Rm and h2 (Fig. 2). Given the relationship of both Rm and h2 with mean beta values (Supplemental Fig. S3) in the two separate data sets, one might expect the correlation between probe Rm and h2 to be confounded by the mean beta values of the CpG probes. However, we found a strong positive correlation between Rm and h2 in different bins of mean beta values (Supplemental Fig. S7), suggesting that the observed correlation between Rm and h2 is not confounded by mean beta values.
Figure 2.
Correlation between Rm and h2. The distribution of Rm estimated in the LBC cohorts by bins of h2 estimated in the BSGS cohort. Correlation between the two measures was 0.68.
Both genetic and/or environmental factors may be responsible for high DNAm repeatability. Probes with high h2 generally had high Rm, consistent with a genetic constraint. To demonstrate this, we used the sum of ranks to select 10 CpG sites with the highest Rm and h2 and looked for associations with SNPs in the BSGS data. For all 10 CpG sites, we identified significant associations between methylation levels and SNPs (association P-value < 8.3 × 10−9, representing significance at the 5% level after Bonferroni correction for 6,005,138 SNP association tests performed on the BSGS genotype data). For nine of 10 CpG sites, the associated SNP was located <150 kb from the CpG site; and for seven of these, the SNP was located within 1 kb. The effect sizes of these single cis-acting SNPs were very large, explaining 50%–85% of the variation in methylation at the CpG site (Supplemental Table S1). For one of these associations, the SNP was located on a different chromosome to the CpG site but still explained 30% of the variation in methylation at this site. Using the sum of ranks, we also selected 10 probes that had highest Rm and lowest h2 (ranking probes by decreasing Rm and increasing h2), and none had significant associations with any SNPs, suggesting that the high repeatability may be due to environmental effects.
Repeatability and heritability of smoking-associated methylation sites
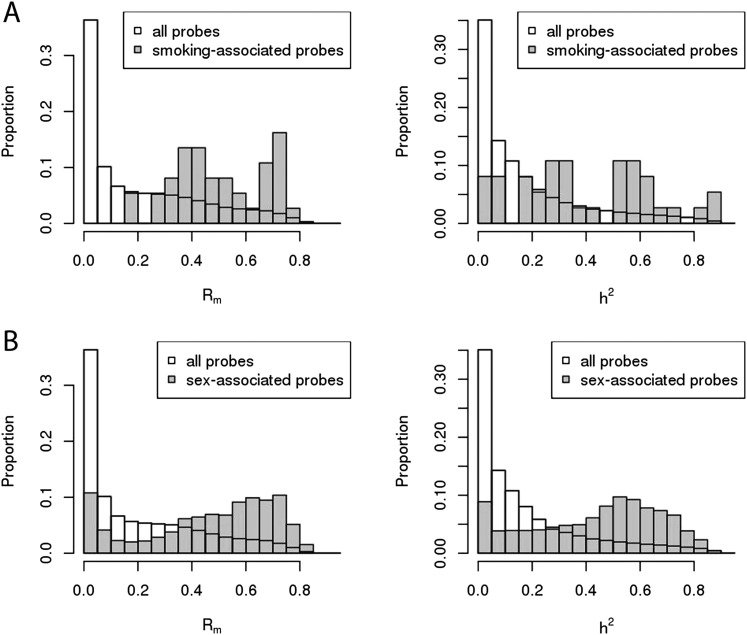
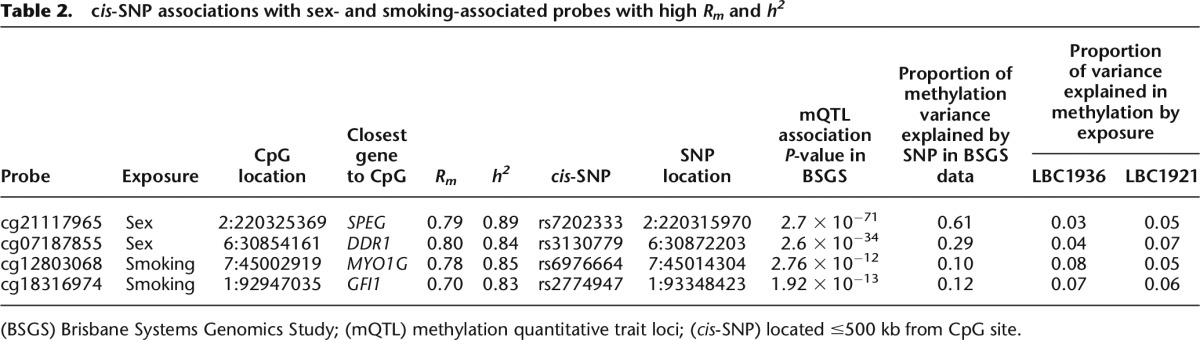
We hypothesized that CpG sites with high Rm but low h2 may be under strong influence of environmental factors. To test this, we assessed the distribution of Rm and h2 of CpG sites whose methylation levels were associated with an environmental factor—smoking. Using LBC1936 as a discovery sample (731 never smokers versus 139 current smokers), we identified 305 significantly associated autosomal CpG sites (at P-value < 2.13 × 10−7, representing significance at the 5% level after Bonferroni correction for 234,811 tests). Using LBC1921 as a replication sample (333 never-smokers versus 39 current smokers), 37 sites in or near 27 genes were validated (P-value < 0.00016; Bonferroni correction for 305 tests). Of the identified genes, 16 had previously been identified to contain CpGs that were differentially methylated in response to smoking status (Supplemental Table S2; Joubert et al. 2012; Shenker et al. 2013; Zeilinger et al. 2013). Looking at the distribution of Rm and h2, we observed a strong enrichment for probes with high Rm (Fig. 3). Although some of these CpG sites also had high h2, the enrichment for high heritability probes was not as substantial (Fig. 3). The discordance in the distribution of Rm and h2 for smoking-associated CpG sites was more obvious when looking at the larger subset of the 305 CpG sites that were identified in the LBC1936 discovery sample. For this set of 305 probes, a much larger proportion had high Rm compared to those with high h2 (Supplemental Fig. S8). The correlation between Rm and h2 for the 37 smoking-associated probes was 0.68 (P-value = 3.0 × 10−6), suggesting that even for sites where methylation is influenced by environment, additive genetic factors have a strong influence. We found significant SNP associations for 12 of the 37 smoking probes (32%), 10 of which were with cis-acting SNPs (located within 500 kb of CpG site), one was with a SNP on the same chromosome but >500 kb away, and one with a SNP on a different chromosome. A lookup of two independent smoking-associated probes with the highest Rm and h2 identified strong underlying cis-SNP effects, each explaining ∼10% of the methylation variation at these CpG sites, similar to that explained by smoking status itself (Table 2). The most significant association with smoking status in our data was with a previously reported CpG probe (cg05575921) in the AHRR gene. This probe had high Rm of 0.74 but relatively low h2 of 0.24, with no significant SNP associations found in the BSGS data.
Figure 3.
Distribution of Rm and h2 for smoking- and sex-associated CpG sites. (A) 37 smoking-associated autosomal probes. (B) 1687 sex-associated autosomal probes.
Table 2.
cis-SNP associations with sex- and smoking-associated probes with high Rm and h2
Repeatability and heritability of sex-associated methylation sites
We also identified and assessed the distribution of Rm and h2 of CpG sites that were differentially methylated between males and females. Sex can be considered as a genetic factor, but also as an unmodifiable lifelong “exposure,” and differences between males and females would be confounded by any gender-related differences in environment or lifestyle. Using LBC1936 as a discovery sample (732 females and 760 males) and modeling an individual as a random effect, 3810 autosomal CpGs were significantly associated with sex (P-value < 2.13 × 10−7; Bonferroni correction for 234,811 tests). Of these, 1687 were replicated (P-value < 1.30 × 10−5; Bonferroni correction for 3810 tests) in the LBC1921 data (409 females and 294 males). These were located in or near 1157 genes (Supplemental Table S3). Sex-associated CpG probes were significantly enriched for probes with high Rm and h2 (Fig. 3; Supplemental Fig. S8). It is important to note that Rm and h2 were calculated after adjusting for sex; therefore, the high Rm and h2 are not being driven simply by mean differences between the two groups. The correlation between Rm and h2 for these CpG sites was 0.76 (P-value < 1.0 × 10−16). This suggests that methylation at these sites is strongly influenced by genetic factors. Of the 1687 probes, 561 (33%) had significant underlying SNP associations, of which 519 (93%) were located within 500 kb from the CpG site. A lookup of two sex-associated probes with high Rm and h2 identified strong underlying cis-SNP effects explaining 29% and 61% of the variation in methylation (Table 2), which was much higher than the proportion of variance explained by sex (3%–7%). One of these CpG sites (cg07187855) is located within a regulatory region upstream of the discoidin domain receptor tyrosine kinase 1 (DDR1) gene, which based on mice studies, plays an essential role in mammary gland development (Vogel et al. 2001).
Discussion
Although observations of changes in global (or average) methylation over time have been previously reported (Fraga et al. 2005; Bjornsson et al. 2008) using population-based, longitudinal methylation analyses, we have identified a large number of autosomal CpG sites across the genome whose methylation levels are stable over time. It has been suggested by studies on global methylation that epigenetic drift with age is a genome-wide phenomenon that may occur due to the accumulation of small defects in the transmission of epigenetic marks through successive cell divisions (Fraga et al. 2005). We identify genetic and environmental factors that appear to be responsible for DNAm stability over time at a number of CpG sites. Such factors may therefore be constraining drift at these CpG sites.
The strong correlation between repeatability estimated in older individuals to heritability estimated in younger individuals suggests, first, that additive genetic factors are the main contributors to methylation variation at a large proportion of the measured CpG sites; and second, that CpG sites under strong genetic influence appear to be stable almost over the human life course. Heritability estimates for any given CpG site can change, and can be age-, tissue-, and population-specific. Methylation at the same CpG site measured over an individual’s lifetime can have different genetic and environmental effects influencing it so that the variances become a function of age. However, despite the differences between the LBC and BSGS participant characteristics, especially the age of the participants, we find that CpG sites under strong genetic influence have stable methylation even later on in life. Heritability can be thought of as the repeatability within related individuals. For example, if stability is solely due to genetic factors, then the repeatability within a person over time is the same as the correlation between monozygotic twins (and this correlation is the heritability). Since h2 is partly derived from the correlation between parents and offspring, high h2 in BSGS data indicates that methylation levels are stable within the age range of the BSGS samples (9–75 yr). If the same probes have high Rm in LBC, this indicates that methylation is also stable from 70–90 yr. Therefore, for probes with both high h2 and Rm, methylation repeatability is high throughout the age ranges in BSGS and LBC (9–90 yr).
The stability of DNA methylation over time is an important factor to consider when drawing conclusions about causality in epidemiological studies, especially in prospective cohorts, where only a single biological sample has been collected but phenotypic information on participants is collected at subsequent phases. For CpG sites where environment/lifestyle plays the major role, the length or continuity of exposure may be key. For example, for cg05575921, the most strongly associated probe with smoking status, we observed that the mean methylation level at baseline in ex-smokers (unadjusted beta mean = 0.83; SD = 0.087) was significantly lower than those who had never smoked (unadjusted beta mean = 0.88; SD = 0.05) (P < 1 × 10−16), but also significantly higher than current smokers (unadjusted beta mean = 0.61; SD = 0.11) (P < 1 × 10−16). Information on when ex-smokers gave up smoking would allow us to determine the longevity of the effects of smoking on methylation, which would be important in determining the long-term exposure risk in diseases where smoking is a known risk factor. CpG sites influenced by smoking were relatively stable over time. However, although some CpGs had high heritability, overall the genetic contribution was not as strong as that observed for the sex-associated CpG sites. This was evident in the higher correlation between Rm and h2 for the sex-associated CpGs compared to that for the smoking-associated CpGs. CpG sites with differential methylation between males and females will be of interest in the context of diseases that are more prevalent in one sex compared to the other. It remains to be seen whether the stability and heritability of DNAm translates to gene expression levels. A comparison between methylation QTLs and expression QTLs would be needed to determine if common genetic variation affects gene expression via changes in DNAm.
There are a number of factors to consider when interpreting the results from these analyses: First, repeatability is a well-established measure for assessing reproducibility (Sokal and Rohlf 1995), but it is only evident if there is between-individual variation in the population. It is therefore not a suitable method for estimating reproducibility in extremely stable measures where there is little between-individual variation, and is likely to underestimate the true number of repeatable CpGs. Our analyses on replicate samples did identify a number of probes with repeatability of zero. These may be probes that are truly stable over time or probes that due to technical reasons do not perform well. Under the null distribution of the test statistic for repeatability, the expected proportion of probes with zero repeatability is 50%. Using this, we can estimate a lower limit of the true proportion of probes that have a nonzero repeatability as 2*(0.5 – P), where P is the proportion of observed probes with Rm = 0. In the LBC data, P = 0.16, thus we conclude that at least 68% of probes have nonzero repeatability over the time course studied. Similarly, from our analysis of seven replicates (P = 0.1), the upper-bound of the proportion of probes that have zero repeatability through either showing no variation in DNA methylation levels or through technical issues is 20%. This indicates that although our estimated proportion of repeatable results in LBC is downwardly biased by the presence of probes with technical issues or having no variation in DNA methylation, it is not markedly biased.
Second, CpGs with mean beta values from 0.4–0.6 showed the highest Rm and h2 values (Supplemental Fig. S3). This may partly reflect the heteroscedasticity of the beta value and the reliance of Rm estimation on the presence of between-individual variation. However, CpGs with strong underlying SNP effects, especially if these are common SNPs, have high between-individual variation with mean methylation around 0.5 (individuals in the three genotype groups tend to have beta values around 0, 0.5, and 1). Therefore, high Rm and h2 for probes with mean beta values in the medium methylation range (0.4–0.6) is likely to reflect underlying mQTLs.
Third, although the Infinium HumanMethylation450 array comprehensively evaluates promoter regions and CpG islands as well as other potentially relevant intergenic regions, such as regulatory regions (Slieker et al. 2013), it only interrogates a small subset of the ∼28 million CpG sites in the human genome. Therefore, our results may not be representative of all CpG sites across the entire human genome. Finally, whole blood, from which the DNA samples were extracted, consists of a heterogenous cell population, and it has been shown that these functionally distinct populations have unique DNA methylation profiles (Reinius et al. 2012) which could confound analyses. However, we found a very high correlation (0.98) between Rm estimated from data adjusted and unadjusted for cell count.
In summary, we show that genetic and environmental factors may constrain epigenetic drift. We also show that methylation at CpG sites measured in blood has a number of the same characteristics as other “complex traits.” First, variation in methylation at any given CpG site can be influenced by both genetic and environmental factors; second, heritable variation can be mapped by associations with SNPs; and third, robust and replicable associations with environmental factors can be found. Therefore, methylation levels at a single CpG site can be treated as a complex trait.
Methods
Subjects
The Lothian Birth Cohort 1921 (LBC1921): This cohort is the basis of a longitudinal study of aging (Deary et al. 2004, 2012). All participants were born in 1921, and most completed a cognitive ability test (Moray House Test No. 12) at about age 11 yr in the Scottish Mental Survey 1932 (SMS1932) (Scottish Council for Research in Education 1933). The LBC1921 study attempted to follow up individuals who might have completed the SMS1932 and resided at about age 79 yr in the Lothian region (Edinburgh and its surrounding areas) of Scotland; 550 people (n = 234; 43% men) were successfully traced and participated in the study from age 79 yr. To date, there have been four additional follow-up waves at average ages of 83, 87, 90, and 92 yr. The cohort has been deeply phenotyped during the later-life waves, including blood biomarkers, white blood cell counts, cognitive testing, and psychosocial, lifestyle, and health measures. DNA methylation was measured in subjects at an average age of 79 yr (n = 515), 87 yr (n = 181), and 90 yr (n = 87), and data from these waves were used for analyses.
Lothian Birth Cohort 1936 (LBC1936): All participants were born in 1936 (Deary et al. 2007, 2012). Most had taken part in the Scottish Mental Survey 1947 at mean age 11 yr as part of national testing of almost all children born in 1936 who attended Scottish schools on June 4, 1947 (Scottish Council for Research in Education 1949). The cognitive test administered was the same Moray House Test No. 12 used in the SMS1932. A total of 1091 participants (n = 548; 50% men) who were mostly living in the Lothian area of Scotland were recontacted in later life. Extensive phenotyping, including white blood cell counts, has also been carried out in this study, with data collection waves at three time points. DNA methylation was measured at mean age 70 yr (n = 1005), 73 yr (n = 336), and 76 yr (n = 332), and data from these waves were used for analyses.
Brisbane Systems Genetics Study (BSGS): Participants were originally recruited into an ongoing study of the genetic and environmental factors influencing cognition and pigmented nevi. DNA methylation was measured on 614 individuals from 117 families of European descent. Families consist of adolescent monozygotic (MZ; n = 67 pairs) and dizygotic (DZ; n = 111 pairs) twins, their siblings (n = 119), and their parents (n = 139). Children have a mean age of 14 yr (range 9–23) and parents 47 yr (range 33–75) (Powell et al. 2012).
Ethics
Following informed consent, venesected whole blood was collected for DNA extraction in both LBC1921 and LBC1936. Ethics permission for the LBC1921 was obtained from the Lothian Research Ethics Committee (Wave 1: LREC/1998/4/183). Ethics permission for the LBC1936 was obtained from the Multi-Center Research Ethics Committee for Scotland (Wave 1: MREC/01/0/56), the Lothian Research Ethics Committee (Wave 1: LREC/2003/2/29). Written informed consent was obtained from all subjects. The BSGS study was approved by the Queensland Institute for Medical Research Human Research Ethics Committee. All participants gave informed written consent.
DNA methylation
LBC1921 and LBC1936
DNA was extracted from whole blood samples in LBC1921 at MRC Technology, Western General Hospital, Edinburgh (LBC1921), and the Wellcome Trust Clinical Research Facility (WTCRF), Western General Hospital, Edinburgh (LBC1936), using standard methods. Methylation typing of 485,512 probes was performed at the WTCRF. Bisulphite converted DNA samples were hybridized to the Infinium HumanMethylation450 array using the Infinium HD Methylation protocol and Tecan robotics (Illumina). Raw intensity data were background-corrected and normalized using internal controls, and methylation beta values were generated using the R minfi package (Aryee et al. 2014). Quality control (QC) analysis was performed to remove probes with a low (<95%) detection rate at P < 0.01. Manual inspection of the array control probe signals was used to identify and remove low quality samples (e.g., samples with inadequate hybridization, bisulfite conversion, nucleotide extension, or staining signal). The Illumina recommended threshold was used to eliminate samples with a low call rate (samples with <450,000 probes detected at P < 0.01). Samples whose predicted sex, based on XY probes, did not match reported sex were excluded. Probes on the X and Y chromosomes were also removed. After these QC steps, 450,726 autosomal probes remained. Beta values were corrected for effects of sample plate, BeadChip, position on BeadChip, hybridization date, white blood cell counts (basophils, eosinophils, monocytes, lymphocytes, neutrophils), and sex using a generalized linear model with a logistic link function. Residuals from this model were used in further analyses.
BSGS
DNA was extracted from peripheral blood lymphocytes by the salt precipitation method (Miller et al. 1988) from samples that were time matched to a sample collection of PAXgene tubes for gene expression studies in the Brisbane Systems Genetics Study. Bisulphite converted DNA samples were hybridized to the 12 sample Infinium HumanMethylation450 array using the Infinium HD Methylation protocol and Tecan robotics (Illumina). The proportion of probes with detection P-value < 0.01 was examined to confirm strong binding of the sample to the array. The probability of a probe within a sample either being called as missing or with a detection P-value < 0.001 was estimated from the average rate across all probes and samples. A threshold for probes showing significant deviation from random missingness (or excess poor binding) was determined by testing against a binomial distribution for the number of samples at the 0.05 significance level with a Bonferroni correction for the number of probes. Any probe with more than 11 individuals with missing data or more than five individuals with detection P-values > 0.001 were removed. Probes on the X and Y chromosomes were also removed. After QC, 614 samples and 458,836 autosomal probes remained. Raw intensity values were background corrected and normalized to internal controls using the Genome Studio software and beta values generated. Percentage of monocytes, B cells, NK cells, CD4+ T cells, CD8+ T cells, and granulocytes were predicted using a published prediction algorithm (Houseman et al. 2012). Beta values were corrected for array, sex, age, age2, sex × age, sex × age2, and predicted white blood cell counts using a generalized linear model with a logistic link function, and residuals used in subsequent analyses. BSGS methylation data are available from the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE56105.
Genotyping and methylation quantitative trait loci (mQTL) analysis
All BSGS individuals were genotyped on the Illumina Human610-Quad BeadChip. Full details of genotyping procedures is given elsewhere (Medland et al. 2009). Standard QC filters were applied, leaving 528,509 SNPs. The remaining genotyped SNPs were phased using HAPI-UR (Williams et al. 2012) and imputed using 1000 Genomes Phase I Version 3 (The 1000 Genomes Project Consortium 2010) with IMPUTE2 (Howie et al. 2011, 2012). Raw imputed SNPs were filtered to remove any SNPs with low imputation quality as defined by an R2 < 0.8. Subsequent quality control removed SNPs with minor allele frequency (MAF) < 0.05, those with Hardy–Weinberg equilibrium (HWE) P < 1 × 10−6, and a missing rate >10%. After filtering, 6,005,138 SNPs remained for further analysis. Association of methylation data (residuals after adjusting for covariates) with imputed genotype probabilities was performed using Merlin (Abecasis et al. 2002).
Repeatability
The repeatability measure (Rm) for each methylation probe was calculated as the ratio of the between-individual variance over the total variance (Vtotal). These quantities can be extracted from a random effects linear regression with individual as random effect
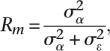 |
where 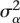 and
and 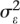 are the between-subject and residual (within-subject) variances, respectively; and Vtotal =
are the between-subject and residual (within-subject) variances, respectively; and Vtotal = 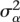 +
+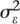 . The lmer() function in the lme4 R CRAN package (Bates et al. 2014) was used to perform the random effects linear regression.
. The lmer() function in the lme4 R CRAN package (Bates et al. 2014) was used to perform the random effects linear regression.
Since the variance between individuals can be partitioned into variance due to genetic factors (VG) and variance due to permanent differences in environment between individuals (VEp), we can define Rm as
The genetic variance can be further partitioned into additive and nonadditive (dominant and epistatic effects) genetic variance, so that Rm can be defined as
The proportion of phenotypic variance due to additive genetic factors (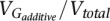 ) is the (narrow sense) heritability (h2). Based on this equation, we can see that h2 estimates should not exceed Rm estimates in the same sample, and Rm therefore sets an upper bound for h2. By comparing the Rm and h2, we can assess the extent to which variation between individuals is due to additive genetic factors.
) is the (narrow sense) heritability (h2). Based on this equation, we can see that h2 estimates should not exceed Rm estimates in the same sample, and Rm therefore sets an upper bound for h2. By comparing the Rm and h2, we can assess the extent to which variation between individuals is due to additive genetic factors.
Probes that contained SNPs or had been identified previously as potentially cross-hybridizing (Price et al. 2013) showed some inflation in Rm and h2 (Supplemental Fig. S9). Therefore, to avoid potential biases due to effects of SNPs on array binding, we removed all probes that encompass SNPs annotated in the 1000 Genomes Project data (The 1000 Genomes Project Consortium 2010) and high-quality SNPs in dbSNP (Price et al. 2013). Probes where all samples had beta ≤ 0.1 or all samples had beta ≥ 0.9 were excluded from analysis, as the lack of between-individual variance means Rm or h2 cannot be accurately estimated for these probes. The null distribution for Rm was derived using data permutation.
Heritability
The heritability for each probe was estimated in BSGS by partitioning its variance into additive genetic (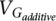 ) and environmental (VEp) components by fitting a linear mixed model of the form
) and environmental (VEp) components by fitting a linear mixed model of the form
where y is the vector of the covariate-adjusted methylation level; μ is the mean value; a is the additive genetic effects; and e is the unique environmental effects (residuals). The model was fitted using QTDT (Abecasis et al. 2000), which uses maximum likelihood to estimate the variance components. Probes that contained SNPs or had been identified previously as potentially cross-hybridizing (Price et al. 2013) were excluded from analysis.
Association analysis
To identify probes associated with smoking status and sex, data from all waves was analyzed using a linear mixed model, where individual was modeled as a random effect. LBC1936 samples were used for discovery and LBC1921 as a replication sample. Association with smoking (never smokers versus current smokers) was performed on residuals corrected for sample plate, BeadChip, position on BeadChip, hybridization date, sex, cell count, and wave. For sex-association analysis, residuals uncorrected for sex were used, and smoking status (current/ex/never) was added as a covariate in the model. Smoking information was available at all waves for 693 of the 1366 (51%) individuals; and for the vast majority of these individuals, smoking status did not change across the different waves. Therefore, baseline smoking status was used for all samples in the association analysis. In a mixed model analysis, the proportion of variance explained by fixed effects (marginal R2 value) for each probe can be extracted using the r.squaredGLMM() command in the MuMIn R package (Barton 2014). For the sex-association analysis, since both sex and smoking were fitted as fixed effects, to get the proportion of variance explained by sex alone, the marginal R2 had to be extracted from a linear mixed model that included only sex but not smoking. Probes significant at the 5% level after Bonferroni correction in the LBC1936 discovery sample (P < 2.13 × 10−7 for 234,811 tests) were validated in the LBC1921 replication sample.
Data access
LBC data have been submitted to the European Genome-phenome Archive (EGA; https://www.ebi.ac.uk/ega/home) under accession number EGAS00001000910.
Acknowledgments
We thank the cohort participants and team members who contributed to these studies. Phenotype collection in the Lothian Birth Cohort 1921 was supported by the UK’s Biotechnology and Biological Sciences Research Council (BBSRC), The Royal Society, and The Chief Scientist Office of the Scottish Government. Phenotype collection in the Lothian Birth Cohort 1936 was supported by Age UK (The Disconnected Mind project). Methylation typing was supported by Centre for Cognitive Ageing and Cognitive Epidemiology (Pilot Fund award), Age UK, The Wellcome Trust Institutional Strategic Support Fund, The University of Edinburgh, and The University of Queensland. R.E.M., I.J.D., S.E.H., S.R.C., J.M.S., and P.M.V. are members of the University of Edinburgh Centre for Cognitive Ageing and Cognitive Epidemiology (CCACE). CCACE is supported by funding from the BBSRC, the Economic and Social Research Council (ESRC), the Medical Research Council (MRC), and the University of Edinburgh as part of the cross-council Lifelong Health and Wellbeing Initiative (MR/K026992/1). Research reported in this publication was supported by National Health and Medical Research Council (NHMRC) project grants 613608, APP496667, APP1010374, and APP1046880; NHMRC Fellowships to G.W.M., P.M.V., and N.R.W. (613602); and Australia Research Council (ARC) Future Fellowship to N.R.W. (FT0991360). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NHMRC or ARC.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.176933.114.
References
- The 1000 Genomes Project Consortium . 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis GR, Cardon LR, Cookson WO. 2000. A general test of association for quantitative traits in nuclear families. Am J Hum Genet 66: 279–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. 2002. Merlin-rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30: 97–101 [DOI] [PubMed] [Google Scholar]
- Alegría-Torres JA, Baccarelli A, Bollati V. 2011. Epigenetics and lifestyle. Epigenomics 3: 267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, Irizarry RA. 2014. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30: 1363–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton K. 2014. MiMIn: Multi-model inference. http://cran.r-project.org/web/packages/MuMIn/index.html
- Bates D, Maechler M, Bolker B, Walker S. 2014. lme4: Linear mixed-effects models using Eigen and S4. http://cran.r-project.org/web/packages/lme4/index.html
- Bell JT, Pai AA, Pickrell JK, Gaffney DJ, Pique-Regi R, Degner JF, Gilad Y, Pritchard JK. 2011. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol 12: R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JT, Tsai PC, Yang TP, Pidsley R, Nisbet J, Glass D, Mangino M, Zhai G, Zhang F, Valdes A, et al. 2012. Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS Genet 8: e1002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellacosa A, Cicchillitti L, Schepis F, Riccio A, Yeung AT, Matsumoto Y, Golemis EA, Genuardi M, Neri G. 1999. MED1, a novel human methyl-CpG-binding endonuclease, interacts with DNA mismatch repair protein MLH1. Proc Natl Acad Sci 96: 3969–3974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H, Yu W, Rongione MA, Ekström TJ, Harris TB, et al. 2008. Intra-individual change over time in DNA methylation with familial clustering. JAMA 299: 2877–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocklandt S, Lin W, Sehl ME, Sánchez FJ, Sinsheimer JS, Horvath S, Vilain E. 2011. Epigenetic predictor of age. PLoS ONE 6: e14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröske AM, Vockentanz L, Kharazi S, Huska MR, Mancini E, Scheller M, Kuhl C, Enns A, Prinz M, Jaenisch R, et al. 2009. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet 41: 1207–1215 [DOI] [PubMed] [Google Scholar]
- Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, Bock C, Vasanthakumar A, Gu H, Xi Y, et al. 2012. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet 44: 23–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deary IJ, Whiteman MC, Starr JM, Whalley LJ, Fox HC. 2004. The impact of childhood intelligence on later life: following up the Scottish mental surveys of 1932 and 1947. J Pers Soc Psychol 86: 130–147 [DOI] [PubMed] [Google Scholar]
- Deary IJ, Gow AJ, Taylor MD, Corley J, Brett C, Wilson V, Campbell H, Whalley LJ, Visscher PM, Porteous DJ, et al. 2007. The Lothian Birth Cohort 1936: a study to examine influences on cognitive ageing from age 11 to age 70 and beyond. BMC Geriatr 7: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deary IJ, Gow AJ, Pattie A, Starr JM. 2012. Cohort profile: the Lothian Birth Cohorts of 1921 and 1936. Int J Epidemiol 41: 1576–1584 [DOI] [PubMed] [Google Scholar]
- Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, Lin SM. 2010. Comparison of β-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics 11: 587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer DS, Mackay TFC. 1996. Variance. In Introduction to quantitative genetics, 4th ed., pp 122–144. Benjamin Cummings, San Francisco, CA
- Florath I, Butterbach K, Müller H, Bewerunge-Hudler M, Brenner H. 2013. Cross-sectional and longitudinal changes in DNA methylation with age: an epigenome-wide analysis revealing over 60 novel age-associated CpG sites. Hum Mol Genet 23: 1186–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suñer D, Cigudosa JC, Urioste M, Benitez J, et al. 2005. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci 102: 10604–10609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs JR, van der Brug MP, Hernandez DG, Traynor BJ, Nalls MA, Lai SL, Arepalli S, Dillman A, Rafferty IP, Troncoso J, et al. 2010. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet 6: e1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon L, Joo JE, Powell JE, Ollikainen M, Novakovic B, Li X, Andronikos R, Cruickshank MN, Conneely KN, Smith AK, et al. 2012. Neonatal DNA methylation profile in human twins is specified by a complex interplay between intrauterine environmental and genetic factors, subject to tissue-specific influence. Genome Res 22: 1395–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundberg E, Small KS, Hedman ÅK, Nica AC, Buil A, Keildson S, Bell JT, Yang TP, Meduri E, Barrett A, et al. 2012. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat Genet 44: 1084–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundberg E, Meduri E, Sandling JK, Hedman AK, Keildson S, Buil A, Busche S, Yuan W, Nisbet J, Sekowska M, et al. 2013. Global analysis of DNA methylation variation in adipose tissue from twins reveals links to disease-associated variants in distal regulatory elements. Am J Hum Genet 93: 876–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan JB, Gao Y, et al. 2013. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell 49: 359–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S. 2013. DNA methylation age of human tissues and cell types. Genome Biol 14: R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT. 2012. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Marchini J, Stephens M. 2011. Genotype imputation with thousands of genomes. G3 (Bethesda) 1: 457–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. 2012. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet 44: 955–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirtle RL, Skinner MK. 2007. Environmental epigenomics and disease susceptibility. Nat Rev Genet 8: 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubert BR, Håberg SE, Nilsen RM, Wang X, Vollset SE, Murphy SK, Huang Z, Hoyo C, Midttun Ø, Cupul-Uicab LA, et al. 2012. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ Health Perspect 120: 1425–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkel K, Spadola A, Yuan E, Kosek J, Jiang L, Hod E, Li K, Murty VV, Schupf N, Vilain E, et al. 2008. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat Genet 40: 904–908 [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. 1997. DNA methylation and genetic instability in colorectal cancer cells. Proc Natl Acad Sci 94: 2545–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69: 915–926 [DOI] [PubMed] [Google Scholar]
- Li E, Beard C, Jaenisch R. 1993. Role for DNA methylation in genomic imprinting. Nature 366: 362–365 [DOI] [PubMed] [Google Scholar]
- Madrigano J, Baccarelli A, Mittleman MA, Sparrow D, Vokonas PS, Tarantini L, Schwartz J. 2012. Aging and epigenetics: longitudinal changes in gene-specific DNA methylation. Epigenetics 7: 63–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathers JC, Strathdee G, Relton CL. 2010. Induction of epigenetic alterations by dietary and other environmental factors. Adv Genet 71: 3–39 [DOI] [PubMed] [Google Scholar]
- McRae AF, Powell JE, Henders AK, Bowdler L, Hemani G, Shah S, Painter JN, Martin NG, Visscher PM, Montgomery GW. 2014. Contribution of genetics variation to transgenerational inheritance of DNA methylation. Genome Biol 15: R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medland SE, Nyholt DR, Painter JN, McEvoy BP, McRae AF, Zhu G, Gordon SD, Ferreira MAR, Wright MJ, Henders AK, et al. 2009. Common variants in the trichohyalin gene are associated with straight hair in Europeans. Am J Hum Genet 85: 750–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF. 1988. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16: 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panning B, Jaenisch R. 1998. RNA and the epigenetic regulation of X chromosome inactivation. Cell 93: 305–308 [DOI] [PubMed] [Google Scholar]
- Powell JE, Henders AK, McRae AF, Caracella A, Smith S, Wright MJ, Whitfield JB, Dermitzakis ET, Martin NG, Visscher PM, et al. 2012. The Brisbane Systems Genetics Study: genetical genomics meets complex trait genetics. PLoS ONE 7: e35430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price ME, Cotton AM, Lam LL, Farré P, Emberly E, Brown CJ, Robinson WP, Kobor MS. 2013. Additional annotation enhances potential for biologically-relevant analysis of the Illumina Infinium HumanMethylation450 BeadChip array. Epigenetics Chromatin 6: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlén SE, Greco D, Söderhäll C, Scheynius A, Kere J. 2012. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS ONE 7: e41361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scottish Council for Research in Education . 1933. The intelligence of Scottish children: a national survey of an age-group. University of London Press, London
- Scottish Council for Research in Education . 1949. The trend of Scottish intelligence. University of London Press, London
- Shenker NS, Polidoro S, van Veldhoven K, Sacerdote C, Ricceri F, Birrell MA, Belvisi MG, Brown R, Vineis P, Flanagan JM. 2013. Epigenome-wide association study in the European Prospective Investigation into Cancer and Nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Hum Mol Genet 22: 843–851 [DOI] [PubMed] [Google Scholar]
- Slieker RC, Bos SD, Goeman JJ, Bovée JV, Talens RP, van der Breggen R, Suchiman HED, Lameijer EW, Putter H, van den Akker EB, et al. 2013. Identification and systematic annotation of tissue-specific differentially methylated regions using the Illumina 450k array. Epigenetics Chromatin 6: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal R, Rohlf F. 1995. Biometry: the principles and practice of statistics in biological research, 3rd ed. W.H. Freeman and Company, New York [Google Scholar]
- Talens RP, Boomsma DI, Tobi EW, Kremer D, Jukema JW, Willemsen G, Putter H, Slagboom PE, Heijmans BT. 2010. Variation, patterns, and temporal stability of DNA methylation: considerations for epigenetic epidemiology. FASEB J 24: 3135–3144 [DOI] [PubMed] [Google Scholar]
- Tate P, Skarnes W, Bird A. 1996. The methyl-CpG binding protein MeCP2 is essential for embryonic development in the mouse. Nat Genet 12: 205–208 [DOI] [PubMed] [Google Scholar]
- Trowbridge JJ, Snow JW, Kim J, Orkin SH. 2009. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell 5: 442–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel WF, Aszodi A, Alves F, Pawson T. 2001. Discoidin domain receptor 1 tyrosine kinase has an essential role in mammary gland development. Mol Cell Biol 21: 2906–2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AL, Patterson N, Glessner J, Hakonarson H, Reich D. 2012. Phasing of many thousands of genotyped samples. Am J Hum Genet 91: 238–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CCY, Caspi A, Williams B, Craig IW, Houts R, Ambler A, Moffitt TE, Mill J. 2010. A longitudinal study of epigenetic variation in twins. Epigenetics 5: 516–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeilinger S, Kühnel B, Klopp N, Baurecht H, Kleinschmidt A, Gieger C, Weidinger S, Lattka E, Adamski J, Peters A, et al. 2013. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS ONE 8: e63812. [DOI] [PMC free article] [PubMed] [Google Scholar]