Abstract
Glycoproteins derived from most retroviruses and from several families of enveloped viruses can form infectious pseudotypes with murine leukemia virus (MLV) and lentiviral core particles, like the MLV envelope glycoproteins (Env) that are incorporated on either virus type. However, coexpression of a given glycoprotein with heterologous core proteins does not always give rise to highly infectious viral particles, and restrictions on pseudotype formation have been reported. To understand the mechanisms that control the recruitment of viral surface glycoproteins on lentiviral and retroviral cores, we exploited the fact that the feline endogenous retrovirus RD114 glycoprotein does not efficiently pseudotype lentiviral cores derived from simian immunodeficiency virus, whereas it is readily incorporated onto MLV particles. Our results indicate that recruitment of glycoproteins by the MLV and lentiviral core proteins occurs in intracellular compartments and not at the cell surface. We found that Env and core protein colocalization in intracytoplasmic vesicles is required for pseudotype formation. By investigating MLV/RD114 Env chimeras, we show that signals in the cytoplasmic tail of either glycoprotein differentially influenced their intracellular localization; that of MLV allows endosomal localization and hence recruitment by both lentiviral and MLV cores. Furthermore, we found that upon membrane binding, MLV core proteins could relocalize Env glycoproteins in late endosomes and allow their incorporation on viral particles. Thus, intracellular colocalization, as well as interactions between Env and core proteins, may influence the recruitment of the glycoprotein onto viral particles and generate infectious pseudotyped viruses.
Enveloped viruses consist of nucleocapsids that are surrounded by a host-derived membrane envelope onto which are placed the viral surface glycoproteins. While the nucleocapsid contains the determinants that specify replication of the viral genome, the primary role of the envelope glycoproteins is to allow the entry of the virion into new cells through binding to cell surface receptors and subsequent triggering of fusion between the viral and cell membranes. The two types of viral structural determinants are expressed in distinct locations, in the cytosol for the nucleocapsid proteins and in the secretory pathway for the envelope glycoproteins (59). For retroviruses, it is not clear how the Gag and Env proteins, which form their nucleocapsid and envelope proteins, respectively, as well as the viral genomic RNAs, reach common sites to assemble, bud, and form infectious virions. In particular, the mechanisms that allow the recruitment of the viral glycoproteins onto retroviral cores are poorly understood.
So far, we know of at least two mechanisms that lead to the incorporation of viral or cellular glycoproteins onto viral particles. In the active model of Env glycoprotein incorporation (59), the assembly of viral particles depends either on direct interaction between the cytoplasmic tail of the pseudotyping glycoprotein and components of the virion core or on indirect interactions via a cellular factor. There is ample evidence in the literature to support the critical role of such interactions in viral assembly (reviewed in references 18 and 59), at least for lentiviruses (14, 33, 64, 67, 68). The model of passive Env incorporation, however, implies nonspecific interactions between the pseudotyping glycoprotein and the viral core, as long as the cytoplasmic tail of Env is not sterically incompatible with viral assembly or virion morphology (59) and as long as there is a sufficient abundance of Env at the site of virus budding. This model is substantiated by the formation of retroviral pseudotypes with heterologous Env and their colocalization in raft cell surface microdomains (10, 44). Surface proteins derived from most retroviruses and from many families of enveloped viruses allow pseudotype formation with retroviral particles. Nonretroviral glycoproteins include those derived from vesiculoviruses, lyssaviruses, arenaviruses, hepadnaviridae, flaviviruses, paramyxoviruses, orthomyxoviruses, filoviruses, and alphaviruses (reviewed in reference 53). However, even within the family Retroviridae, coexpression of a given glycoprotein with heterologous viral core proteins does not always give rise to highly infectious viral particles (12, 27, 30, 52, 54, 58, 62).
In previous studies, it was found that the feline endogenous virus RD114 glycoprotein did not allow efficient pseudotype formation with lentiviral cores derived from simian immunodeficiency virus (SIV) (52), whereas it was readily incorporated on murine leukemia virus (MLV) particles (13, 61). The RD114 glycoprotein is typical of those of beta retroviruses (42, 49), with which it shares the same cell surface receptor, RDR (48, 60), and bears significant homology in the transmembrane (TM) Env subunit, in particular. Thus, the RD114 glycoprotein differs significantly from that of MLV, which readily pseudotypes both MLV and lentiviruses (34). Here, we investigated the determinants carried by either MLV or RD114 glycoproteins that are responsible for this differential behavior. Our results support the notions that (i) recruitment of glycoproteins onto the viral core occurs in intracellular compartments and not at the cell surface, as proposed previously; (ii) intracellular colocalization between Gag and Env is required for pseudotype formation; and (iii) Gag-Env interactions may occur, directly or indirectly, and influence the manner in which pseudotyped viral particles are assembled intracellularly and released into the cell supernatant. By investigating chimeras between the MLV and RD114 Env glycoproteins, we found that signals harbored by the cytoplasmic tail of either glyco-protein differentially influenced their intracellular localizations; that of MLV allowed endosomal localization and hence recruitment by both lentiviral and MLV Gag proteins. Furthermore, we found that upon membrane binding, Gag proteins could relocalize Env glycoproteins harboring homologous cytoplasmic tails in late endosomal vesicles. Thus, intracellular colocalization and/or Gag-Env interactions influence the assembly of the glycoprotein on viral cores and lead to infectious pseudotyped particles.
MATERIALS AND METHODS
Cells.
The 293T human embryo kidney cell line (ATCC CRL-1573), the COS-7 African green monkey fibroblast kidney cell line (ATCC CRL-1651), and the TE671 human rhabdomyosarcoma cell line (ATCC CRL-8805) were grown in Dulbecco's modified Eagle's medium Invitrogen, Cergy Pontoise, France) supplemented with 10% fetal bovine serum and antibiotics.
Viral glycoprotein expression constructs.
phCMV-G, encoding the vesicular stomatitis virus (VSV) G protein (71), was used as an expression vector backbone to subclone the glycoproteins derived from the feline endogenous virus RD114 (GenBank accession no. X87829) (13) and from the 4070A strain of amphotropic MLV (MLV-A) (41). The phCMV-RD114 expression vector, expressing the RD114 virus envelope glycoprotein (RD114 glycoprotein), was further modified to generate a series of mutants that harbor modifications in the RD114 glycoprotein transmembrane domain and/or cytoplasmic tail. All subsequent constructs were generated by PCR-mediated and oligonucleotide site-directed mutagenesis (details and sequences are available upon request) and cloned in the phCMV-RD114 plasmid.
Production of virions.
The production of a pseudotyped SIV-based vector was decribed in a previous report (52). Briefly, 293T cells plated in 10-cm-diameter plates were transfected by 8.1 μg of the pR4SA green fluorescent protein (GFP) vector construct (31), 8.1 μg of the pSIV-3+ packaging construct (35), and 2.7 μg of the construct expressing the viral glycoprotein. Pseudotyped MLV-derived vectors were generated in a similar manner by DNA transfection of the pTG5349 MLV packaging construct (35), the pTG13077 MLV GFP vector construct (35), and the glycoprotein-expressing construct (2.7 μg). The expression construct encoding a myristoylation-defective MLV Gag mutant (the G2A mutation) (50) was reported elsewhere (3). A calcium-phosphate transfection protocol (Clontech, St. Quentim en Yvelines, France) was used according to the manufacturer's instructions. The medium (8 ml/plate) was replaced 16 h after transfection, and the supernatant was harvested 24 h later and filtered through 0.45-μm-pore-size membranes.
Infection assays.
To determine the infectious-virus titers of the different pseudotyped vectors, serial dilutions of vector preparations were added to TE671 target cells in the presence of 8 μg of Polybrene/ml for 4 h at 37°C. The medium was then replaced with normal culture medium for 72 h at 37°C. The transduction efficiency, determined as the percentage of GFP-positive cells, was then measured by fluorescence-activated cell sorter analysis, as previously described (52).
Antibodies.
Anti-RD114 glycoprotein (ViroMed Biosafety Labs), a goat antiserum raised against the RD114 gp70 envelope surface protein (SU), was used at 1/5,000 dilution for Western blotting and 1/4,000 dilution for immunofluorescence (IF) studies. Anti-SIV capsid protein (CA) (National Institutes of Health AIDS Research and Reference Reagent Program) is a mouse monoclonal antibody (2F12) raised against the SIVmac251 p27 CA, and it was used at 1/500 dilution for Western blotting. A mouse antiserum against SIV MAp17 (National Institutes of Health AIDS Research and Reference Reagent Program) was used at 1/1,000 dilution to visualize SIV Gag in IF studies. Anti-MLV CA (ViroMed Biosafety Labs) is a goat antiserum raised against the Rausher leukemia virus p30 CA, and it was used at 1/10,000 dilution for Western blotting. A rabbit antiserum against MLV CAp30 (a kind gift of A. Rein) was used at 1/10,000 dilution to identify MLV Gag in IF studies. Anti-CD25 (R&D Systems Europe, Abingdon, United Kingdom) is a goat antiserum, and it was used at 1/500 dilution for Western blotting. The Lamp-1 mouse monoclonal antibody was used to stain the lysosomes.
Expression of viral glycoproteins and virus incorporation.
Forty hours posttransfection, virion producer cells were chilled on ice, washed twice with cold phosphate-buffered saline++ (PBS++) (PBS, pH 8.0, supplemented with 0.7 mM CaCl2 and 0.25 mM MgSO4) and incubated with 0.5 mg of sulfo-NHS-LC-LC-biotin (Pierce, Rockford, Ill.)/ml for 30 min at 4°C. Biotinylation was stopped by incubating the cells with 1 M glycine in PBS++ for 5 min at 4°C. The cells were then washed with PBS-0.1 M glycine, lysed with MacDougal buffer (20 mM Tris-HCl, pH 8.0, 120 mM NaCl, 200 μM EGTA, 0.2 μM NaF, 0.2% sodium deoxycholate, 0.5% Nonidet P-40) containing a protease inhibitor cocktail (Complete mini; Roche Diagnostic, Meylon, France) and 0.1 M glycine, and centrifuged at 13,000 × g for 30 min; 80% of the cell lysates were incubated overnight at 4°C with streptavidin-Sepharose beads (Pierce). The beads were then washed with MacDougal glycine buffer, resuspended in a denaturing buffer (1% β-mercaptoethanol, 0.5% sodium dodecyl sulfate [SDS]), and boiled for 5 min. Purified virus samples were obtained by ultracentrifugation of viral supernatants through a 1.5-ml 20% sucrose cushion in a Beckman SW41 rotor (25,000 rpm; 2.5 h; 4°C) and suspended in PBS. All samples were mixed 5:1 (vol/vol) in a loading buffer (375 mM Tris-HCl [pH 6.8] containing 6% SDS, 30% β-mercaptoethanol, 10% glycerol, and 0.06% bromophenol blue), boiled for 5 min, and then run on SDS-13% polyacrylamide gel electrophoresis. PNGaseF (New England Biolabs, Hitchin, United Kingdom) digestions of protein samples were performed according to the manufacturer's procedures. Western blotting was performed using standard procedures. An ECL+ system (Amersham Biosciences) was used for visualization. Membrane scanning was performed with the Storm 860 device, and signal densities were calculated with the ImageQuant program (Molecular Dynamics Inc.).
IF and confocal microscopy imaging.
Fugene 6-transfected 293T or COS-7 cells were grown on 35-mm-diameter coverglass dishes coated with d-lysine (Mattek Corporation, Ashland, Mass.) or on uncoated 14-mm-diameter glass coverslips, respectively; 40 h posttransfection, IF staining was performed at room temperature. The cells were washed with PBS, fixed with 3% paraformaldehyde in PBS for 15 min, quenched with 50 mM NH4Cl, and permeabilized by 0.2% Triton X-100 for 5 min. The fixed cells were incubated for 1 h with the primary antibody in 1% bovine serum albumin-PBS, washed, and stained for 1 h with the corresponding fluorescent Alexa-conjugated secondary antibody (at 0.5 μg/ml) in 1% bovine serum albumin-PBS. The cells were then washed several times with PBS and mounted on microscope slides with the antifading agent Prolong (Molecular Probes) prior to image acquisition. Images were acquired with an LSM 510 confocal microscope equipped with an Axiovert 100 M microscope (Carl Zeiss Inc., Thornwood, N.Y.) and a 40× 1.3-numerical-aperture Apocromat objective. Alexa 488 was excited with an argon laser line at 488 nm, and emissions were collected with a band pass filter (BP505-550). Alexa 546 was excited, independently of Alexa 488, with an HeNe laser line at 543 nm, and emissions were collected with a long-pass filter (LP560). The secondary antibodies Alexa 488 anti-rabbit, Alexa 488 anti-goat, Alexa 488 anti-mouse, and Alexa 546 anti-goat antisera were purchased from Molecular Probes, Inc. Expression vectors encoding specific endosomal marker proteins, cellubrevine-GFP and TiVamp-GFP (2) (kind gifts from T. Galli), were cotransfected with Env and Gag vectors.
RESULTS
Modifications of the RD114 glycoprotein cytoplasmic tail modulate pseudotyping of lentiviral or MLV particles.
The glycoproteins of both amphotropic MLV and RD114 can efficiently pseudotype MLV core particles (13, 61). Since the MLV glycoprotein, but not RD114 Env, efficiently pseudotypes lentiviral particles derived from SIV (35, 52), we assumed that the former should contain elements that control Env incorporation and/or the infectivity of lentiviral particles. Thus, to define determinants that restrict the capacity of the RD114 glycoprotein to pseudotype lentiviral particles, we generated a panel of RD114 glycoprotein mutants into which subregions derived from its ectodomain, transmembrane domain, and/or cytoplasmic tail were replaced by their MLV counterparts (Fig. 1). The RD/MTRMLV chimera carried the MLV transmembrane domain in addition to the MLV cytoplasmic tail. Mutant RD/TRMLV harbored the MLV cytoplasmic tail only. The RD-Rless glycoprotein was a truncated version of the RD114 glycoprotein and was generated by inserting a stop codon at a position corresponding to a putative cleavage site of its cytoplasmic tail (see below). The RD/EMLV chimera contained the MLV ectodomain and transmembrane domain.
FIG. 1.
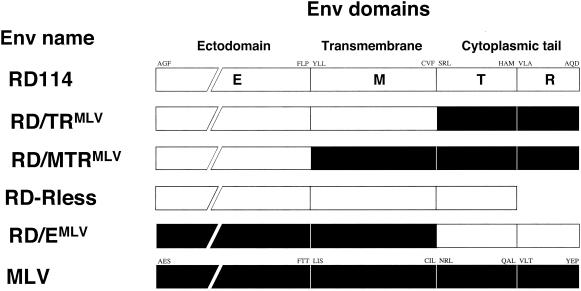
Schematic representation of RD114/MLV Env chimeras showing the domain organization of parental Env and chimeras. The open and solid boxes represent domains derived from RD114 (RD) and amphotropic MLV glycoproteins, respectively. The cytoplasmic region consists of the cytoplasmic tails (T) and the p2R peptides (R). E, ectodomain; M, transmembrane domain. The first and last amino acids of each domain are indicated.
The capacity of the RD114 glycoprotein chimeras to pseudotype either SIV or MLV particles was determined in comparison to wild-type RD114 glycoprotein. Viral particles were generated by transient expression in 293T human cells, as well as in COS-7 simian cells, transfected with constructs encoding the viral components, core proteins (Gag), Env glycoproteins, and a replication-defective viral genome harboring the GFP marker gene. Amphotropic MLV and VSV G glycoproteins, which efficiently pseudotype MLV and lentiviral particles (Fig. 2), were used as controls. Results obtained from either cell type (COS-7 or 293T cells) were similar, yet the overall production of infectious particles from COS-7 cells was lower than that from 293T cells (data not shown). The infectivities of viruses generated in the presence of the different glycoproteins were assessed using TE671 cells, which express receptors for all three glycoproteins, as target cells (Fig. 2). Consistent with previous results (52), the unmodified RD114 glycoprotein poorly pseudotyped lentiviral particles, resulting in infectious-virus titers of 3 × 104 infectious units (i.u.)/ml, on average. Lentiviral particles generated with the RD/EMLV chimera, harboring the same cytoplasmic tail, did not achieve titers higher than those obtained with the wild-type RD114 glycoprotein. Titers of viruses generated with the hyperfusogenic mutant RD-Rless were low, yet the strong cytopathic effect exerted by the mutant might have impaired the formation of viral particles. Indeed, strong cytopathic effects were noticed for the RD-Rless mutant (data not shown), suggesting that the carboxy-terminal part of the cytoplasmic tail negatively controls cell-cell fusion, as for other gamma- and beta-retrovirus glycoproteins (11, 46, 51). Importantly, the RD/TRMLV chimeric glycoprotein efficiently pseudotyped the lentiviral particles. This resulted in infectious-virus titers of up to 106 i.u./ml, i.e., 25-fold higher than those obtained with the wild-type RD114 glycoproteins and in the same range as infectious-virus titers obtained with lentiviruses pseudotyped with the amphotropic MLV glycoprotein (Fig. 2). Interestingly, MLV particles generated with all RD114 Env-derived chimeras except RD-Rless had infectious-virus titers higher than 106 i.u./ml, similar to those of viruses pseudotyped with wild-type RD114 glycoprotein (Fig. 2). Since the cytoplasmic tail was the minimal domain modified in these RD114 glycoprotein chimeras that led to increased infectivity of pseudotyped lentiviruses, these data suggested that the cytoplasmic domain of MLV Env, rather than its ecto- and transmembrane domains, harbored a component(s) that facilitated pseudotype formation with lentiviral particles. Subsequent investigations aimed at determining the role of the MLV cytoplasmic tail in pseudotyping of the lentiviral core were therefore achieved with the wild-type RD114 and RD/TRMLV glycoproteins only.
FIG. 2.
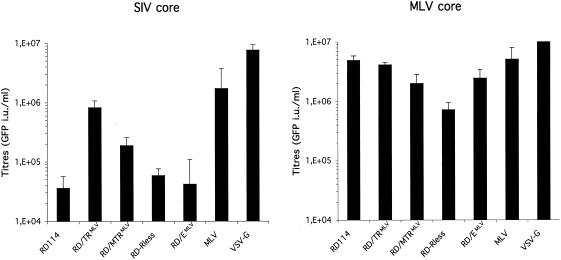
Results of infection assays. TE671 target cells were infected using diluted supernatants harvested from 293T cells transfected with GFP marker-gene-expressing SIV or MLV vectors and with the indicated viral glycoproteins and core components. The transduction efficiency, expressed as the percentage of GFP-positive cells, was measured by fluorescence-activated cell sorter analysis 72 h postinfection and was used to determine the number of infectious viral particles present in the producer cell supernatants. The infectious-virus titers were then expressed as the number of GFP infectious units per milliliter of viral supernatant. The values are the means plus standard deviations of up to six independent experiments. Similar results were obtained with COS-7 cells as the producer cells (not shown).
RD114 glycoprotein maturation, trafficking, and virus incorporation are influenced by the nature of the cytoplasmic tail.
We examined the expression of the viral proteins, i.e., glycoproteins and core components, both by quantitative Western blot analysis from total cell lysates, cell surface, and virions (Fig. 3 and 4) and by IF microscopy (Fig. 5 and 6). Western blot detection of RD114 glycoproteins resulted in three different bands (Fig. 3A): (i) an uncleaved precursor protein; (ii) a mature SU product at ca. 76 kDa, processed from the precursor protein by the subtilisin-like endoprotease furin (24), that could be readily distinguished after the removal of N-linked carbohydrates by PNGaseF treatment (Fig. 3) or on gels of greater resolution (data not shown); and (iii) a TM protein at ca. 18 kDa that was further processed in virions to a 16-kDa protein following cleavage by both the lentiviral and MLV proteases (data not shown). As summarized in Fig. 4, important variations in the proportions of these different forms were detected (i) in the different cell compartments analyzed, i.e., in total cell extracts, at the cell surface, and in the virions purified on high-density sucrose cushions; (ii) when comparing the expression of the wild-type RD114 glycoprotein to that of the RD/TRMLV chimera harboring the MLV cytoplasmic tail; and (iii) when coexpressing lentiviral Gag, MLV Gag, or assembly defective cores.
FIG. 3.
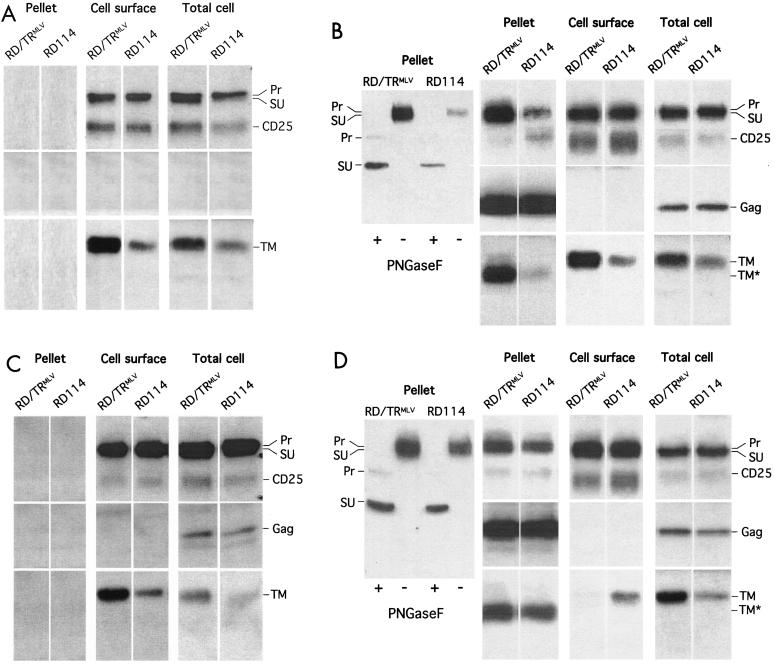
Characterization of glycoproteins and pseudotypes. Detection of precursor (Pr), surface (SU), and two transmembrane forms (TM and TM*, deleted from the p2R carboxy-terminal peptide) of the RD114 and RD/TRMLV glycoproteins and of SIV or MLV core proteins (Gag) was performed by Western blotting in crude cell lysates (Total cell), cell surface biotinylated proteins (Cell surface), and viral particles purified by ultracentrifugation of the cell supernatants on sucrose cushions (Pellet). As a control, the CD25 interleukin-2 receptor gamma chain that is not expressed in the producer cells was coexpressed with the viral proteins. The viral glycoproteins were expressed in 293T cells individually (A) or in the presence of Gag proteins derived from SIV (B) or MLV (D) or of a myristoylation-defective MLV G2A-Gag mutant (C). In separate experiments, the samples were digested with PNGaseF before being immunoblotted with anti-SU antibodies to readily distinguish SU from precursor proteins and to facilitate the relative quantification of the different Env forms (Pr, SU, and TM) as displayed in Fig. 4. Thus, upon deglycosylation by PNGaseF, the SU and Pr proteins appeared as 60- and 40-kDa bands (e.g., viral particles in B and C, insets). In this transient expression system, the budding of MLV particles was ∼5-fold more efficient than that of lentiviral particles, as evidenced both by different ratios between intracellular and virion-associated Gag proteins (data not shown) and by the difference between the infectious-virus titers of the two virus types pseudotyped with VSV G. Thus, to facilitate the comparison of the incorporations of the different glycoproteins on either type of virus core, equivalent amounts of vector particles were loaded on gels, using capsid proteins as the standard. The processing of the RD114 and RD/TRMLV TM in TM* was blocked when the producer cells were treated with saquinavir, an inhibitor of the retroviral proteases (not shown). Similar results were obtained when COS-7 cells were used as the producer cells (not shown).
FIG. 4.
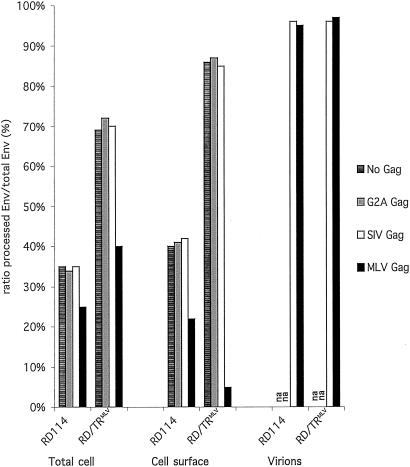
Quantification of mature glycoproteins. Upon expression with the indicated core components (Gag) in 293T cells, the SU, TM, and precursor forms of the RD114 and RD/TRMLV glycoproteins were quantified by Western blot analysis performed in total cell lysates, cell surface, and purified viral particles (Fig. 3). Scanning of Western blot membranes was performed with the Storm 860 device, and signal densities were calculated with the ImageQuant program. Transfection efficiencies were normalized using the CD25 interleukin-2 receptor gamma-chain marker. The ratios of the mature (processed) glycoproteins to total glycoproteins are provided. na, not applicable.
FIG. 5.
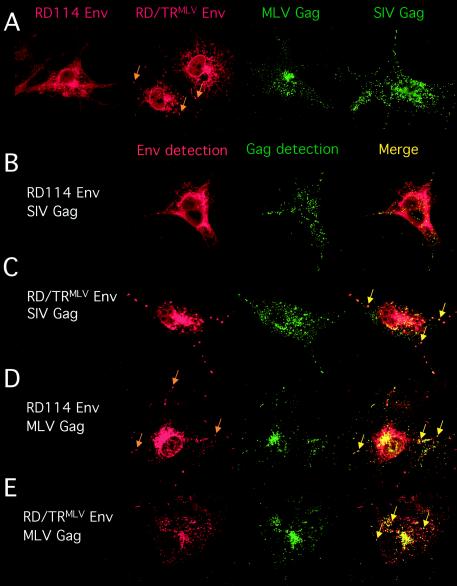
Detection of viral glycoproteins and core proteins by confocal microscopy. (A) COS-7 cells expressing either the RD114 and RD/TRMLV glycoproteins (Env) or the MLV and SIV core proteins (Gag), as indicated, were fixed, permeabilized, and stained using RD114 SU antibodies, MLV capsid protein antibodies, or SIV matrix protein antibodies. The transfected cells were grown on glass coverslips, fixed in paraformaldehyde, stained at room temperature, and imaged by confocal microscopy. The arrows indicate intracytoplasmic vesicles in which the RD/TRMLV glycoprotein localized. Similar staining patterns were observed with 293T cells (not shown). Expression of myristoylation-defective MLV Gag proteins (G2A Gag mutant) resulted in diffuse staining throughout the cytoplasm and an absence of punctate staining (not shown). (B to E) Colocalization of Env and Gag viral components. COS-7 cells coexpressing the RD114 (B and D) or RD/TRMLV (C and E) glycoprotein and the MLV (D and E) or SIV (B and C) core protein, as indicated, were fixed, permeabilized, and stained using RD114 SU antibodies (B to E), SIV matrix protein antibodies (B and C), or MLV capsid protein antibodies (D and E). Env and Gag coexpression are shown in the red (Env detection) and green (Gag detection) channels, respectively. The arrows in the red channel show intracytoplasmic vesicles in which the RD114 glycoprotein localized upon expression of MLV Gag protein (D). Colocalization of both viral components was assessed by confocal microscopy analysis (Merge). The arrows indicate intracytoplasmic vesicles in which the Env and Gag protein colocalized. Similar colocalization patterns were detected in 293T cells (not shown). Neither Env/Gag colocalization nor redistribution of Env localization was detected upon expression of myristoylation-defective MLV Gag proteins (data not shown).
FIG. 6.
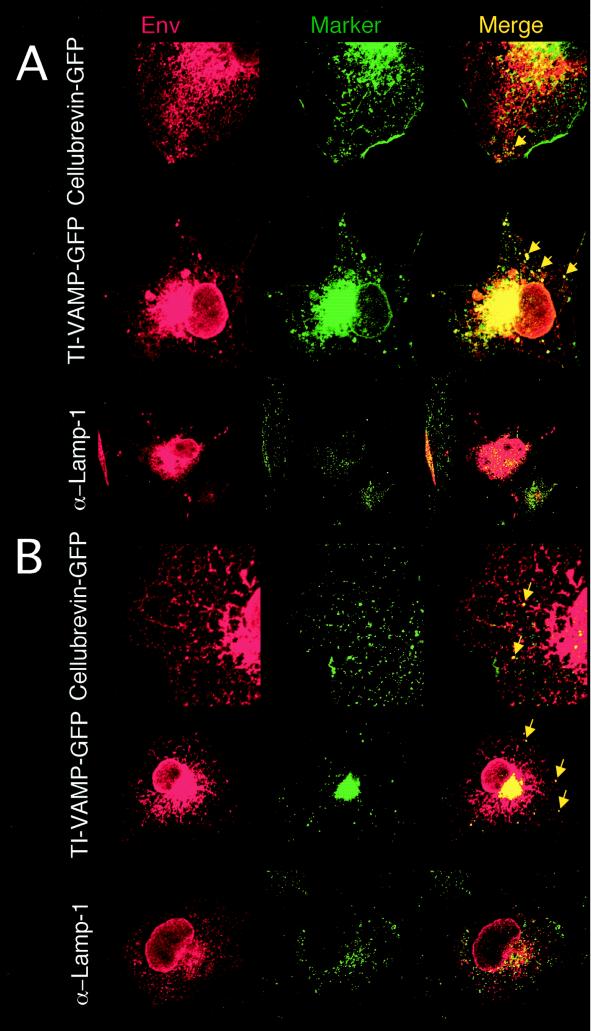
Colocalization of Env and cellular markers of the endosomal pathway. COS-7 cells coexpressing MLV Gag proteins and RD/TRMLV (A) or RD114 (B) glycoproteins were imaged by confocal microscopy analysis. Localization of the Env glycoproteins detected in the red channel (Env) in marked intracellular vesicles shown in the green channel (Marker) was assessed upon coexpression of vectors encoding the GFP-tagged cellular markers cellubrevin (Cellubrevin-GFP) and TI-VAMP (TI-VAMP-GFP), which localize in recycling and late endosomes, respectively, or upon costaining with Lamp-1 antibodies (α-Lamp-1) for detection of lysosomes. Colocalization of both Env and cellular markers is shown (Merge). The arrows indicate localization of Env in recycling or late endosomes.
In the absence of viral core proteins, while the overall glycoprotein expressions were identical for RD114 and RD/TRMLV, as detected in total cell lysates (Fig. 3A), ∼35% of the RD114 glycoprotein was detected as mature SU and TM forms compared to 70% for the RD/TRMLV chimera. This was in good agreement with the two- to threefold-increased amounts of TM proteins detected for RD/TRMLV relative to the wild type (Fig. 3A). The two glycoproteins exhibited comparable densities at the cell surface (Fig. 3A), indicating that they were transported and expressed at the plasma membrane. However, significant amounts of protein precursors were detected at the cell surface (Fig. 4), indicating incomplete cleavage in the trans-Golgi network (TGN). The more efficient maturation of the RD/TRMLV glycoprotein versus the wild type detected in cell lysates was readily apparent at the cell surface. This resulted in mature forms of the RD/TRMLV glycoprotein that were ∼6-fold more abundant at the cell surface than the mature forms of the wild-type glycoprotein (Fig. 3A). This pointed out the influence of the cytoplasmic tail on the processing efficiency of the viral glycoprotein. The difference may be explained by a modulation of the conformation of the ectodomain and thus of the accessibility of the SU/TM cleavage site or by differential trafficking of either glycoprotein to cell compartments where processing occurs, i.e., the TGN and the endosomes (63). The latter hypothesis was supported by the fact that the RD/TRMLV glycoprotein had a faster maturation kinetic than the parental RD114 glycoprotein, as evidenced in cell lysates by the decreased half-life of its precursor (2 versus 3 h) and the earlier detection of mature products by pulse-chase experiments (data not shown).
The level of viral glycoprotein cell surface expression is influenced not only by the rate of biosynthesis and the efficiency of transport through the secretory pathway but also by the rate at which the glycoproteins are removed from the cell surface and/or traffic to intracellular compartments. Thus, since furins are located in the TGN at steady state but traffic to the cell surface and recycle to the TGN via endocytosis (63), the discrepancy of maturation between the RD114 and RD/TRMLV glycoproteins suggests that their cytoplasmic tails influence their localization and/or dynamic cycling. Compared to its RD114 counterpart, and owing to different signals harbored by its cytoplasmic tail, the RD/TRMLV glycoprotein might cycle faster or accumulate in furin-rich compartments. Thus, to directly detect the influence of either cytoplasmic tail on Env trafficking, we analyzed their localization in producer cells by IF microscopy. As shown in Fig. 5A, the wild-type RD114 envelope was detected throughout the cytoplasm and was particularly abundant in the perinuclear region, reminiscent of an endoplasmic reticulum and Golgi area localization. In contrast, the RD/TRMLV glycoprotein displayed a marked vesicular pattern within the cytoplasm (Fig. 5A), in addition to being detected around the nucleus, both in the endoplasmic reticulum and in the Golgi apparatus. Localization of the RD/TRMLV glycoprotein in endosomes, as well as in the median and trans-Golgi apparatus, was further demonstrated using specific markers (Fig. 6A) (see below). Consistent with the more efficient maturation of the RD/TRMLV glycoprotein (Fig. 3A and 4), these observations suggested that while the majority of the RD114 glycoprotein localized in the endoplasmic reticulum at steady state, the RD/TRMLV glycoprotein was more efficiently trafficked in the endosomal pathway (Fig. 5A).
Coexpression of lentiviral core proteins with the glycoproteins did not change these patterns. Indeed, no modifications in the processing, cell surface expression (Fig. 3B), and intracellular localization (Fig. 5B and C) of the RD114 and RD/TRMLV glycoproteins were detected, whether lentiviral core proteins were coexpressed or not. Interestingly, despite the presence of precursors of the RD114 and RD/TRMLV glycoproteins at the cell surface, only the mature form of either glycoprotein was detected on the lentiviral particles purified from the producer cell supernatants (Fig. 3B). Consistent with the greater amounts of mature forms detected for the RD/TRMLV glycoprotein than for the wild-type glycoprotein, the former was ∼10-fold more abundant on the lentiviral particles (Fig. 3B). This strong difference in glycoprotein density on the virions was consistent with the >20-fold-increased infectious-virus titers (Fig. 2). The fact that lentiviral core particles incorporate only mature glycoproteins may be due to better compatibility of mature Env cytoplasmic tails with Gag proteins or, more likely, to a preferential localization of mature glycoprotein forms in an intracellular compartment that allows recruitment by core proteins. Differences in the signals harbored by the MLV and RD114 cytoplasmic tails may thus influence the trafficking of the glycoproteins to this intracellular compartment and hence their recruitment by lentiviral Gag proteins.
RD114 glycoprotein maturation, trafficking, and virus incorporation are influenced by the type of viral core.
Contrasting results were obtained when the RD114 and RD/TRMLV glycoproteins were coexpressed with MLV Gag proteins rather than with lentiviral core proteins. Indeed, MLV Gag proteins negatively interfered with the formation of mature products of either glycoprotein. As detected in total cell lysates (Fig. 3C and 4), the RD/TRMLV glycoprotein coexpressed with MLV Gag proteins exhibited a twofold-decreased ratio of mature Env to total glycoproteins: 40% of mature forms versus ca. 70% of mature Env, which was detected upon coexpression with lentiviral core proteins or in the absence of viral core proteins. These results were corroborated by a delayed processing of RD/TRMLV glycoprotein precursor when Gag proteins were coexpressed, as detected by pulse-chase labeling experiments (data not shown). While a significant, though limited, influence of MLV Gag on the maturation of the wild-type RD114 glycoprotein was detected, the cell surface densities of mature products of the RD/TRMLV glycoprotein were strongly reduced in the presence of MLV core proteins (Fig. 3C and 4) compared to that detected in the presence of lentiviral core proteins (Fig. 3B) or in the absence of viral core proteins (Fig. 3A). This resulted in cell surface densities of mature forms ∼10-fold lower than those detected for the wild-type RD114 Env. Such an effect was particularly marked for MLV Gag proteins coexpressed with RD/TRMLV glycoproteins harboring a homologous cytoplasmic tail, suggesting that specific interactions between these two types of viral components might influence their intracellular trafficking.
Importantly, the influence of MLV Gag proteins on the processing of either glycoprotein was triggered upon membrane binding of Gag. Indeed coexpression of either glycoprotein with assembly-defective MLV Gag proteins, harboring the G2A mutation that prevents Gag myristoylation (50), and thus its membrane binding, did not influence their maturation and resulted in ratios identical to those detected in the absence of core proteins or in the presence of lentiviral Gag proteins (Fig. 3D and 4). These results indicated that upon binding to membranes and/or upon subsequent events (e.g., Gag multimerization), MLV Gag proteins direct Env targeting in the appropriate compartment that allows interactions with the cytoplasmic tail of either glycoprotein and hence influence their dynamic trafficking.
Similar to lentiviral core particles, MLV particles incorporated only mature forms of either glycoprotein (Fig. 3C and 4). However, in contrast to lentiviral particles, MLV particles incorporated similar amounts of mature RD114 or RD/TRMLV glycoproteins (Fig. 3C), resulting in similar infectious-virus titers (Fig. 2). This indicated that lentiviruses and MLV recruit and/or assemble glycoproteins by different mechanisms. The similar viral densities of the glycoproteins were in great contrast to the much lower cell surface abundance of the mature RD/TRMLV forms relative to wild-type RD114. This indicated their enrichment in a particular intracellular compartment where mature forms of the Env glycoproteins are present in comparable amounts and are recruited by MLV cores.
RD/TRMLV and MLV Gag-induced localization of RD114 Env in late endosomes.
To gain further insights into Env intracellular trafficking, we compared the localization of the RD114 or RD/TRMLV glycoprotein in the presence of lentiviral or MLV core protein. MLV and lentiviral Gag proteins expressed alone showed punctate staining patterns dispersed in the cytoplasm and beneath the plasma membrane, with a marked preference of MLV Gag proteins for internal vesicles over lentiviral Gag proteins (Fig. 5A), as previously described (38, 56). Almost no colocalization was observed between lentiviral core proteins and wild-type RD114 glycoproteins at the level of the Gag-containing vesicles (Fig. 5B). In contrast, coexpression of lentiviral core proteins with the RD/TRMLV glycoprotein resulted in frequent colocalization of Env and Gag proteins in vesicles (Fig. 5C). Altogether, these results could explain the fact that the weak incorporation of RD114 glycoproteins on lentiviral particles (Fig. 3B) and the low resulting infectivity of the virions (Fig. 2) are likely due to poor intracellular colocalization of the two viral components. In agreement with this hypothesis, strong colocalization of RD/TRMLV glycoprotein and MLV Gag proteins was detected in vesicles (Fig. 5E).
Interestingly, the intracellular localization of the wild-type RD114 glycoprotein was markedly modified upon coexpression with MLV core proteins. Although the glycoprotein retained a significant diffuse staining pattern, similar to the pattern when it was expressed alone (Fig. 5A) or when it was coexpressed with lentiviral core proteins (Fig. 5B), the coexpression of MLV Gag proteins induced vesicular localization of the RD114 glycoprotein (Fig. 5D). Strong colocalization of RD114 and MLV core proteins was detected in these vesicles (Fig. 5D). This modification of RD114 Env cellular localization was induced by membrane binding of Gag proteins, since it was not observed upon coexpression of RD114 Env with the G2A myristoylation-defective Gag proteins that displayed a diffuse staining pattern throughout the cytosol (data not shown).
To identify the nature of these vesicles, we tested different markers of the endosomal and lysosomal pathway. We observed that the vesicular pattern of RD/TRMLV Env colocalized partially with recycling endosomes, rarely with the lysosome marker Lamp-1, and strongly with late endosomes and multivesicular bodies (MVBs) (Fig. 6A). Finally, we tested these markers to gain insight into the vesicular pattern of the RD114 Env induced by the expression of MLV Gag proteins. In that case, the cytoplasmic vesicles colocalized mainly with the late-endosome-MVB marker and rarely with the lysosome marker (Fig. 6B). These results indicated that the presence of MLV Gag proteins in cells expressing RD114 glycoproteins modified its trafficking and localized it in an endosomal pathway. Furthermore, they strongly suggested specific interactions between MLV Gag proteins and the RD114 cytoplasmic tail, the nature of which remains to be elucidated.
DISCUSSION
In this study, we have investigated the parameters that influence the assembly of a viral surface glycoprotein onto core particles from two different retroviruses. We examined several combinations of Gag and Env proteins: (i) an illegitimate combination between a lentiviral core and the glycoprotein from an endogenous gammaretrovirus, (ii) a homologous combination in which the glycoprotein cytoplasmic tail and the viral core are derived from the same gamma retrovirus, and (iii) an intermediate combination in which Gag and Env proteins were derived from two heterologous gamma retroviruses.
Our results are consistent with the notion that recruitment of the glycoprotein by the core proteins does not occur at the cell surface, as previously thought, but occurs intracellularly in the endosomal pathway. Indeed, although significant levels of furin-processed mature glycoproteins, as well as uncleaved glycoproteins, were detected at the cell surface, only the processed forms were incorporated onto the viral particles. This indicates, first, that recruitment of the glycoprotein occurs in intracellular compartments that are furin rich, among which the TGN, recycling, and late endosomes are candidates (63), and second, that cell surface expression of both processed and unprocessed Env may be only the consequence of abundant glycoprotein synthesis, particularly in transfected cells (22). Trafficking of the viral glycoproteins to early and late endosomes can occur by retrieval from the cell surface via endocytosis or directly from the Golgi apparatus by anterograde or retrograde transport (5, 8, 9, 39, 69). The punctate staining of Env that we observed by confocal microscopy imaging indicates the presence of Env in such vesicles, in particular, in late endosomes and MVBs (Fig. 5 and 6). It is likely that these organelles represent the sites where Gag recruits Env. Indeed, confocal microscopy analysis revealed that there are different localizations for the RD/TRMLV and RD114 Env glycoproteins (Fig. 5A). Only the glycoproteins that colocalized with Gag in these intracellular vesicles were efficiently incorporated on virions (Fig. 5B to E). Furthermore, in the homologous Env/Gag combination (RD/TRMLV Env-MLV Gag), for which the possibility of interactions between the Env cytoplasmic tail and Gag are most favorable, cell surface expression of the mature viral glycoprotein was almost completely abolished in the presence of Gag (Fig. 3). This cell surface depletion correlated well with optimal intracellular Gag-Env colocalization and with strong Env incorporation onto viral particles. Finally, in the intermediate situation, combining Env and Gag from heterologous gamma retroviruses (RD114 Env/MLV Gag), for which less optimal Env/Gag interactions occur, we could also detect Gag-induced intracellular relocalization of the Env glycoprotein (Fig. 5), mainly to late endosomes and MVBs (Fig. 6), in correlation with their virus incorporation (Fig. 3).
Altogether, our results seem to contradict the early proposal that preformed or nascent core particles pick up the glycoproteins in the course of budding from the plasma membrane (59). This pathway was supported by electron microscopy data (reviewed in references 18 and 20), as well as by the colocalization of the two types of viral components in glycolipid-enriched membrane domains on the cell surface (37, 40). It was also suggested that human immunodeficiency virus (HIV) Gag-Env interactions may result in disruption of the mechanisms that lead to down-regulation of Env cell surface expression (17), leading to optimal cell surface expression of the glycoproteins required to assemble fully infectious viruses (23). Our results with gamma-retroviral Env seem to contradict this notion, as we detected a strong reduction of cell surface expression of mature Env in the presence of the homologous Gag proteins (Fig. 3C). Furthermore, one implication of this previous model (17) is that optimal densities of glycoproteins should be found at the cell surface so as to favor Env incorporation, which may appear to contradict the fact that most retroviral glycoproteins exhibit fusogenic and cytopathic properties that need to be circumvented. Recent reports have in fact shed light on the mechanisms that limit Env cytotoxicity and that are achieved both by down-regulating the expression of glycoproteins from the cell surface and by reducing their intrinsic fusogenicity. Indeed the cytoplasmic tails of beta- and gamma-retroviral glycoproteins harbor a carboxy-terminal determinant, p2R, that inhibits their intrinsic fusogenicity until it is cleaved off by the viral protease during or shortly after virus budding (11, 46, 51). Similar to these other retroviral glycoproteins, the RD114 glycoprotein cytoplasmic tail contains a p2R peptide that inhibits cell-cell fusion, as well as virus-cell fusogenicity (V. Sandrin and F.-L. Cosset, unpublished data) and that can be removed by the MLV and lentiviral proteases after budding (Fig. 3B and C). Additionally, signals that control Env endocytosis and removal from the cell surface are found in the cytoplasmic tails of several retroviral Env glycoproteins (see below), as well as that of the RD114 glycoprotein (Sandrin and Cosset, unpublished). Thus, such dual control strongly suppresses cell-cell fusion, as demonstrated by cytoplasmic-tail mutants (1, 11, 25, 26, 46, 51, 70) and by the discovery of highly cell-cell-fusogenic endogenous retroviral glycoproteins that lack such determinants (6, 7).
Several motifs in the cytoplasmic tails of retroviral Env glycoproteins have been shown to regulate their intracellular trafficking via their interactions with intracellular effectors (1, 5, 16, 22, 28, 57). A tyrosine motif is implicated in the basolateral sorting of Env in polarized cells (28) and in the endocytosis of Env from the plasma membrane via the recruitment of the cellular clathrin-adaptor complex AP-2 (5, 9, 39). Another cellular partner, AP-1, acts in retrograde-anterograde transport of Env between early endosomes and the TGN via interaction with dileucine motifs present in the cytoplasmic tails of several cellular and retroviral glycoproteins (5, 69). Recently, a diaromatic motif located in the cytoplasmic tail of HIV Env was shown to interact with TIP47, a cellular protein that drives its retrograde transport from endosomes to the TGN (8). Finally, an acidic motif in the cytoplasmic tail of the human cytomegalovirus gB surface glycoprotein was also found to be required for its intracellular trafficking (15). Upon phosphorylation of the gB cytoplasmic tail by casein kinase 2, PACS-1 acts as a connector protein, linking acidic cluster motifs to the AP-1 adaptor complex and the subsequent retrieval of gB from endosomal compartments to the TGN. All these signals allow the recycling of the viral glycoproteins between the TGN and the cell surface via the endosomal pathway. They may not only lead to depletion of Env cell surface expression but also provide the meeting point(s) of Env and Gag components in order to dictate the sites of assembly and release of viral particles without compromising cell viability.
In previous reports, Env/Gag coexpression was shown to alter the intracellular localization of the Gag proteins and to modify the site of budding from polarized epithelial cells and neurons (4, 29, 55, 66). Env expression relocalized MLV Gag from lysosomes to late endosomes (4) and mediated the export of Macon-Pfizer monkey virus Gag from the pericentriolar assembly site (55). However, these studies did not investigate Gag-induced modification of Env intracellular trafficking. Reciprocally, our results show that MLV Gag relocated the RD114 glycoproteins in endosomes, where they colocalized. Soon after synthesis, Gag proteins bind to membranes at the cell surface, as well as in endosomal vesicles (4, 36, 38, 43, 56, 65). Comparison of the expression of wild-type versus G2A MLV Gag indicates that this event and/or a subsequent event, such as Gag multimerization, is critical to direct Env trafficking (Fig. 3) and for accumulation in endosomes (Fig. 5 and 6). However, the precise mechanism is not yet clear, as direct or indirect interactions between gamma-retrovirus Env and Gag proteins have not been reported. A possibility is that competition between the cellular factors implicated in the Env trafficking and the viral core could take place and hijack the trafficking of Gag or Env (4, 55). Although at steady state RD114 Env is predominantly localized in the endoplasmic reticulum, the evidence shows that it is also expressed at the cell surface (Fig. 3A) and is most likely recycled through the endosomal pathway. In contrast to lentiviral Gag, interactions of MLV Gag with RD114 Env are favorable (yet less intense than with RD/TRMLV Env) and likely redistribute RD114 Env equilibrium by pumping it out of a recycling pathway. Such Env/Gag interactions may accelerate the trafficking of the viral glycoproteins to endosomes, for example, by directly stimulating their endocytosis from the cell surface or their retrograde transport from the TGN to the endosomes, or they may inhibit the recycling of the glycoproteins from endosomes to the Golgi by interfering with adaptor complexes that mediate Env trafficking. We favor the last possibility, first, because Env/Gag interactions at the level of the plasma membrane would alter the cell surface localization of both mature and unprocessed Env forms, which was not detected for the latter (Fig. 3), and second, because direct Env/Gag interactions may sterically impair the association of the Env cytoplasmic tail with host adaptor complexes responsible for endocytosis, which would result in augmented cell surface expression.
Altogether, these results indicate that Env/Gag interactions modify the trafficking and intracellular localization of each protein's components in order to facilitate virus assembly. This is in agreement with reports by others investigating retroviral budding. Indeed, it is now becoming increasingly clear that the cell surface cannot be the only place where retroviral cores assemble or where Env and Gag meet. Proline-rich domains, called late domains, have been identified in the Gag proteins of a number of retroviruses, as well as in rhabdoviruses and filoviruses, as playing a critical role in viral egress (reviewed in reference 19). These motifs interact with cellular proteins implicated in the vacuolar-protein-sorting pathway, and recent studies have shown the ability of retroviruses, as well as lentiviruses, to exploit the vacuolar-protein-sorting machinery for viral assembly (reviewed in reference 45). In the case of HIV, by binding Tsg101, as well as other factors that recruit endosomal sorting complexes required for transport (ESCRT), Gag hijacks a cellular pathway for the formation of virion particles (32, 45, 65). Most recently, a number of groups have reported retrovirus budding into the vacuolar lumen of MVBs in both primary cells and standard tissue culture cell lines, such as the 293T cells used in this work (4, 21, 36, 38, 43, 47, 56, 65). The intraluminal vesicles of MVBs can then be released from the cells as exosomes by fusion of the MVBs with the plasma membrane, providing a preexisting cellular pathway for assembly and egress of the viral particles. Thus, our results are compatible with these recent findings, as they show that Gag and Env colocalization in the endocytic pathway, and more particularly in late endosomes, including MVBs (Fig. 6), correlates with the level of Env incorporation onto virions and with infectivity.
Acknowledgments
We thank Andrea Cimarelli for stimulating discussions and for critical reading of the manuscript. We thank Edouard Bertrand and Thierry Galli for the gift of TI-VAMP-GFP and cellubrevin-GFP vectors, Mariam Andrawiss and Mary Collins for the myristoylation-defective G2A mutant MLV Gag expression vector, Alan Rein for MLV Gag antibodies, Pierre Boulanger for the gift of Lamp-1 antibodies, and Fabienne Simian-Lermé and Claire Lionnet of the PLATIM platform for technical assistance in confocal microscopy.
This work was supported by the Agence Nationale pour la Recherche contre le SIDA (ANRS); the European Community (contracts QLK3-CT-1999-00386 and QLK3-CT-1999-00859); the Association Française contre les Myopathies (AFM), Vaincre La Mucoviscidose (VLM), Région Rhône-Alpes, Centre National de la Recherche Scientifique (CNRS); and the Institut National de la Santé et de la Recherche Médicale (INSERM). D.M. is supported by the Fondation pour la Recherche Médicale (FRM). V.S. is supported by VLM.
REFERENCES
- 1.Aguilar, H. C., W. F. Anderson, and P. M. Cannon. 2003. Cytoplasmic tail of Moloney murine leukemia virus envelope protein influences the conformation of the extracellular domain: implications for mechanism of action of the R peptide. J. Virol. 77:1281-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alberts, P., and T. Galli. 2003. The cell outgrowth secretory endosome (COSE): a specialized compartment involved in neuronal morphogenesis. Biol. Cell 95:419-424. [DOI] [PubMed] [Google Scholar]
- 3.Andrawiss, M., Y. Takeuchi, L. Hewlett, and M. Collins. 2003. Murine leukemia virus particle assembly quantitated by fluorescence microscopy: role of Gag-Gag interactions and membrane association. J. Virol. 77:11651-11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basyuk, E., T. Galli, M. Mougel, J. M. Blanchard, M. Sitbon, and E. Bertrand. 2003. Retroviral genomic RNAs are transported to the plasma membrane by endosomal vesicles. Dev. Cell 5:161-174. [DOI] [PubMed] [Google Scholar]
- 5.Berlioz-Torrent, C., B. L. Shacklett, L. Erdtmann, L. Delamarre, I. Bouchaert, P. Sonigo, M. C. Dokhelar, and R. Benarous. 1999. Interactions of the cytoplasmic domains of human and simian retroviral transmembrane proteins with components of the clathrin adaptor complexes modulate intracellular and cell surface expression of envelope glycoproteins. J. Virol. 73:1350-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaise, S., N. deParseval, L. Benit, and T. Heidmann. 2003. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc. Natl. Acad. Sci. USA 100:13013-13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blond, J.-L., D. Lavillette, V. Cheynet, O. Bouton, G. Oriol, S. Chapel-Fernandes, B. Mandrand, F. Mallet, and F.-L. Cosset. 2000. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J. Virol. 74:3321-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blot, G., K. Janvier, S. LePanse, R. Benarous, and C. Berlioz-Torrent. 2003. Targeting of the human immunodeficiency virus type 1 envelope to the trans-Golgi network through binding to TIP47 is required for env incorporation into virions and infectivity. J. Virol. 77:6931-6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boge, M., S. Wyss, J. S. Bonifacino, and M. Thali. 1998. A membrane-proximal tyrosine-based signal mediates internalization of the HIV-1 envelope glycoprotein via interaction with the AP-2 clathrin adaptor. J. Biol. Chem. 273:15773-15778. [DOI] [PubMed] [Google Scholar]
- 10.Briggs, J. A., T. Wilk, and S. D. Fuller. 2003. Do lipid rafts mediate virus assembly and pseudotyping? J. Gen. Virol. 84:757-768. [DOI] [PubMed] [Google Scholar]
- 11.Brody, B. A., S. S. Rhee, and E. Hunter. 1994. Postassembly cleavage of a retroviral glycoprotein cytoplasmic domain removes a necessary incorporation signal and activates fusion activity. J. Virol. 68:4620-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christodoulopoulos, I., and P. Cannon. 2001. Sequences in the cytoplasmic tail of the gibbon ape leukemia virus envelope protein that prevent its incorporation into lentivirus vectors. J. Virol. 75:4129-4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cosset, F.-L., Y. Takeuchi, J. Battini, R. Weiss, and M. Collins. 1995. High titer packaging cells producing recombinant retroviruses resistant to human serum. J. Virol. 69:7430-7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cosson, P. 1996. Direct interaction between the envelope and matrix proteins of HIV-1. EMBO J. 15:5783-5788. [PMC free article] [PubMed] [Google Scholar]
- 15.Crump, C. M., C. H. Hung, L. Thomas, L. Wan, and G. Thomas. 2003. Role of PACS-1 in trafficking of human cytomegalovirus glycoprotein B and virus production. J. Virol. 77:11105-11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delamarre, L., C. Pique, A. R. Rosenberg, V. Blot, M. P. Grange, I. L. Blanc, and M. C. Dokhelar. 1999. The Y-S-L-I tyrosine-based motif in the cytoplasmic domain of the human T-cell leukemia virus type 1 envelope is essential for cell-to-cell transmission. J. Virol. 73:9659-9663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egan, M., L. Carruth, J. Rowell, X. Yu, and R. Siliciano. 1996. Human immunodeficiency virus type 1 envelope protein endocytosis mediated by a highly conserved intrinsic internalization signal in the cytoplasmic domain of gp41 is suppressed in the presence of the Pr55gag precursor protein. J. Virol. 70:6547-6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freed, E. O. 1998. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 19.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garoff, H., R. Hewson, and D. J. Opstelten. 1998. Virus maturation by budding. Microbiol. Mol. Biol. Rev. 62:1171-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gould, S. J., A. M. Booth, and J. E. Hildreth. 2003. The Trojan exosome hypothesis. Proc. Natl. Acad. Sci. USA 100:10592-10597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grange, M., V. Blot, L. Delamarre, I. Bouchaert, A. Rocca, A. Dautry-Varsat, and M. Dokhelar. 2000. Identification of two intracellular mechanisms leading to reduced expression of oncoretrovirus envelope glycoproteins at the cell surface. J. Virol. 74:11734-11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hermida-Matsumoto, L., and M. Resh. 2000. Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imaging. J. Virol. 74:8670-8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunter, E. 1997. Viral entry and receptors, p. 71-120. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 25.Januszeski, M. M., P. M. Cannon, D. Chen, Y. Rozenberg, and W. F. Anderson. 1997. Functional analysis of the cytoplasmic tail of Moloney murine leukemia virus envelope protein. J. Virol. 71:3613-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, M., C. Yang, and R. W. Compans. 2001. Mutations in the cytoplasmic tail of murine leukemia virus envelope protein suppress fusion inhibition by R peptide. J. Virol. 75:2337-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindemann, D., M. Bock, M. Schweizer, and A. Rethwilm. 1997. Efficient pseudotyping of murine leukemia virus particles with chimeric human foamy virus envelope proteins. J. Virol. 71:4815-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lodge, R., L. Delamarre, J. P. Lalonde, J. Alvarado, D. A. Sanders, M. C. Dokhelar, E. A. Cohen, and G. Lemay. 1997. Two distinct oncornaviruses harbor an intracytoplasmic tyrosine-based basolateral targeting signal in their viral envelope glycoprotein. J. Virol. 71:5696-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lodge, R., H. Gottlinger, D. Gabuzda, E. Cohen, and G. Lemay. 1994. The intracytoplasmic domain of gp41 mediates polarized budding of human immunodeficiency virus type 1 in MDCK cells. J. Virol. 68:4857-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mammano, F., F. Salvatori, S. Indraccolo, A. de Rossi, L. Chieco-Bianchi, and H. G. Göttlinger. 1997. Truncation of the human immunodeficiency virus type 1 envelope glycoprotein allows efficient pseudotyping of Moloney murine leukemia virus particles and gene transfer into CD4+ cells. J. Virol. 71:3341-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mangeot, P.-E., D. Nègre, B. Dubois, A. J. Winter, P. Leissner, M. Methali, D. Kaiserlian, F.-L. Cosset, and J.-L. Darlix. 2000. Development of minimal lentiviral vectors derived from simian immunodeficiency virus (SIVmac251) and their use for gene transfer in human dendritic cells. J. Virol. 74:8307-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin-Serrano, J., A. Yaravoy, D. Perez-Caballero, and P. D. Bieniasz. 2003. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl. Acad. Sci. USA 100:12414-12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murakami, T., and E. O. Freed. 2000. Genetic evidence for an interaction between human immunodeficiency virus type 1 matrix and alpha-helix 2 of the gp41 cytoplasmic tail. J. Virol. 74:3548-3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nègre, D., G. Duisit, P.-E. Mangeot, P. Moullier, J.-L. Darlix, and F.-L. Cosset. 2002. Lentiviral vectors derived from simian immunodeficiency virus (SIV). Curr. Top. Microbiol. Immunol. 261:53-74. [DOI] [PubMed] [Google Scholar]
- 35.Nègre, D., P. Mangeot, G. Duisit, S. Blanchard, P. Vidalain, P. Leissner, A. Winter, C. Rabourdin-Combe, M. Mehtali, P. Moullier, J.-L. Darlix, and F.-L. Cosset. 2000. Characterization of novel safe lentiviral vectors derived from simian immunodeficiency virus (SIVmac251) that efficiently transduce mature human dendritic cells. Gene Ther. 7:1613-1623. [DOI] [PubMed] [Google Scholar]
- 36.Nguyen, D. G., A. Booth, S. J. Gould, and J. E. Hildreth. 2003. Evidence that HIV budding in primary macrophages occurs through the exosome release pathway. J. Biol. Chem. 278:52347-52354. [DOI] [PubMed] [Google Scholar]
- 37.Nguyen, D. H., and J. E. Hildreth. 2000. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 74:3264-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nydegger, S., M. Foti, A. Derdowski, P. Spearman, and M. Thali. 2003. HIV-1 egress is gated through late endosomal membranes. Traffic 4:902-910. [DOI] [PubMed] [Google Scholar]
- 39.Ohno, H., R. C. Aguilar, M. C. Fournier, S. Hennecke, P. Cosson, and J. S. Bonifacino. 1997. Interaction of endocytic signals from the HIV-1 envelope glycoprotein complex with members of the adaptor medium chain family. Virology 238:305-315. [DOI] [PubMed] [Google Scholar]
- 40.Ono, A., and E. O. Freed. 2001. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. USA 98:13925-13930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ott, D., R. Friedrich, and A. Rein. 1990. Sequence analysis of amphotropic and 10A1 murine leukemia virus: close relationship to mink cell focus forming viruses. J. Virol. 64:757-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Overbaugh, J., A. D. Miller, and M. V. Eiden. 2001. Receptors and entry cofactors for retroviruses include single and multiple transmembrane-spanning proteins as well as newly described glycophosphatidylinositol-anchored and secreted proteins. Microbiol. Mol. Biol. Rev. 65:371-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pelchen-Matthews, A., B. Kramer, and M. Marsh. 2003. Infectious HIV-1 assembles in late endosomes in primary macrophages. J. Cell Biol. 162:443-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pickl, W. F., F. X. Pimentel-Muinos, and B. Seed. 2001. Lipid rafts and pseudotyping. J. Virol. 75:7175-7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pornillos, O., J. E. Garrus, and W. I. Sundquist. 2002. Mechanisms of enveloped RNA virus budding. Trends Cell Biol. 12:569-579. [DOI] [PubMed] [Google Scholar]
- 46.Ragheb, J. A., and W. F. Anderson. 1994. pH-independent murine leukemia virus ecotropic envelope-mediated cell fusion: implications for the role of the R peptide and p12E TM in viral entry. J. Virol. 68:3220-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raposo, G., M. Moore, D. Innes, R. Leijendekker, A. Leigh-Brown, P. Benaroch, and H. Geuze. 2002. Human macrophages accumulate HIV-1 particles in MHC II compartments. Traffic 3:718-729. [DOI] [PubMed] [Google Scholar]
- 48.Rasko, J. E., J. L. Battini, R. J. Gottschalk, I. Mazo, and A. D. Miller. 1999. The RD114/simian type D retrovirus receptor is a neutral amino acid transporter. Proc. Natl. Acad. Sci. USA 96:2129-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reeves, R. H., and S. J. O'Brien. 1984. Molecular genetic characterization of the RD114 gene family of endogenous feline retroviral sequences. J. Virol. 52:164-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rein, A., M. R. McClure, N. R. Rice, R. B. Luftig, and A. M. Schultz. 1986. Myristylation site in Pr65gag is essential for virus particle formation by Moloney murine leukemia virus. Proc. Natl. Acad. Sci. USA 83:7246-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rein, A., J. Mirro, J. G. Haynes, S. M. Ernst, and K. Nagashima. 1994. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus env protein. J. Virol. 68:1773-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sandrin, V., B. Boson, P. Salmon, W. Gay, D. Nègre, R. LeGrand, D. Trono, and F.-L. Cosset. 2002. Lentiviral vectors pseudotyped with a modified RD114 envelope glycoprotein show increased stability in sera and augmented transduction of primary lymphocytes and CD34+ cells derived from human and non-human primates. Blood 100:823-832. [DOI] [PubMed] [Google Scholar]
- 53.Sandrin, V., S. J. Russell, and F.-L. Cosset. 2003. Targeting retroviral and lentiviral vectors. Curr. Top. Microbiol. Immunol. 281:137-178. [DOI] [PubMed]
- 54.Schnierle, B. S., J. Stitz, V. Bosch, F. Nocken, H. Merget-Millitzer, M. Engelstadter, R. Kurth, B. Groner, and K. Cichutek. 1997. Pseudotyping of murine leukemia virus with the envelope glycoproteins of HIV generates a retroviral vector with specificity of infection for CD4-expressing cells. Proc. Natl. Acad. Sci. USA 94:8640-8645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sfakianos, J. N., and E. Hunter. 2003. M-PMV capsid transport is mediated by Env/Gag interactions at the pericentriolar recycling endosome. Traffic 4:671-680. [DOI] [PubMed] [Google Scholar]
- 56.Sherer, N. M., M. J. Lehmann, L. F. Jimenez-Soto, A. Ingmundson, S. M. Horner, G. Cicchetti, P. G. Allen, M. Pypaert, J. M. Cunningham, and W. Mothes. 2003. Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic 4:785-801. [DOI] [PubMed] [Google Scholar]
- 57.Song, C., S. R. Dubay, and E. Hunter. 2003. A tyrosine motif in the cytoplasmic domain of Mason-Pfizer monkey virus is essential for the incorporation of glycoprotein into virions. J. Virol. 77:5192-5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stitz, J., C. Buchholz, M. Engelstadter, W. Uckert, U. Bloemer, I. Schmitt, and K. Cichutek. 2000. Lentiviral vectors pseudotyped with envelope glycoproteins derived from gibbon ape leukemia virus and murine leukemia virus 10A1. Virology 273:16-20. [DOI] [PubMed] [Google Scholar]
- 59.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, New York, N.Y. [PubMed]
- 60.Tailor, C. S., A. Nouri, Y. Zhao, Y. Takeuchi, and D. Kabat. 1999. A sodium-dependent neutral-amino-acid transporter mediates infections of feline and baboon endogenous retroviruses and simian type D retroviruses. J. Virol. 73:4470-4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takeuchi, Y., F. L. Cosset, P. J. Lachmann, H. Okada, R. A. Weiss, and M. K. L. Collins. 1994. Type C retrovirus inactivation by human complement is determined by both the viral genome and producer cell. J. Virol. 68:8001-8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takeuchi, Y., G. Simpson, R. Vile, R. Weiss, and M. Collins. 1992. Retroviral pseudotypes produced by rescue of Moloney murine leukemia virus vector by C-type, but not D-type, retroviruses. Virology 186:792-794. [DOI] [PubMed] [Google Scholar]
- 63.Thomas, G. 2002. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell Biol. 3:753-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vincent, M. J., L. R. Melsen, A. S. Martin, and R. W. Compans. 1999. Intracellular interaction of simian immunodeficiency virus Gag and Env proteins. J. Virol. 73:8138-8144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.vonSchwedler, U. K., M. Stuchell, B. Muller, D. M. Ward, H. Y. Chung, E. Morita, H. E. Wang, T. Davis, G. P. He, D. M. Cimbora, A. Scott, H. G. Krausslich, J. Kaplan, S. G. Morham, and W. I. Sundquist. 2003. The protein network of HIV budding. Cell 114:701-713. [DOI] [PubMed] [Google Scholar]
- 66.Weclewicz, K., M. Ekstrom, K. Kristensson, and H. Garoff. 1998. Specific interactions between retrovirus Env and Gag proteins in rat neurons. J. Virol. 72:2832-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.West, J. T., S. K. Weldon, S. Wyss, X. Lin, Q. Yu, M. Thali, and E. Hunter. 2002. Mutation of the dominant endocytosis motif in human immunodeficiency virus type 1 gp41 can complement matrix mutations without increasing Env incorporation. J. Virol. 76:3338-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wyma, D. J., A. Kotov, and C. Aiken. 2000. Evidence for a stable interaction of gp41 with Pr55(Gag) in immature human immunodeficiency virus type 1 particles. J. Virol. 74:9381-9387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wyss, S., C. Berlioz-Torrent, M. Boge, G. Blot, S. Honing, R. Benarous, and M. Thali. 2001. The highly conserved C-terminal dileucine motif in the cytosolic domain of the human immunodeficiency virus type 1 envelope glycoprotein is critical for its association with the AP-1 clathrin adaptor. J. Virol. 75:2982-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang, C., and R. W. Compans. 1997. Analysis of the murine leukemia virus R peptide: delineation of the molecular determinants which are important for its fusion inhibition activity. J. Virol. 71:8490-8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yee, J. K., T. Friedmann, and J. C. Burns. 1994. Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol. 43:99-112. [DOI] [PubMed] [Google Scholar]


