Abstract
Post-translational modifications (PTMs) of histones constitute a major chromatin indexing mechanism, and their proper characterization is of highest biological importance. So far, PTM-specific antibodies have been the standard reagent for studying histone PTMs despite caveats such as lot-to-lot variability of specificity and binding affinity. Herein, we successfully employed naturally occurring and engineered histone modification interacting domains for detection and identification of histone PTMs and ChIP-like enrichment of different types of chromatin. Our results demonstrate that histone interacting domains are robust and highly specific reagents that can replace or complement histone modification antibodies. These domains can be produced recombinantly in Escherichia coli at low cost and constant quality. Protein design of reading domains allows for generation of novel specificities, addition of affinity tags, and preparation of PTM binding pocket variants as matching negative controls, which is not possible with antibodies.
The unstructured N-terminal tails of histones protrude from the core nucleosome and harbor complex patterns of post-translational modifications (PTMs) (Kouzarides 2007; Margueron and Reinberg 2010; Bannister and Kouzarides 2011; Tan et al. 2011). These PTMs regulate a multitude of chromatin-templated transactions, play a central role in development, and are implicated in many diseases, such as cancer (Suva et al. 2013). Therefore, understanding the role of histone marks in chromatin-dependent processes is of paramount importance. So far, techniques based on the specific binding of antibodies to modified histone proteins have been the only method available for genome-wide analyses of histone modifications with locus-specific resolution. The central role of antibodies for the characterization of histone PTMs in chromatin research makes the quality and reliability of these reagents a very important scientific issue. In general, antibodies are very powerful and important reagents in biomolecular research, but the validation of commercial antibodies is not always sufficiently rigorous (Bordeaux et al. 2010). This is particularly important in the chromatin field, where specific recognition and discrimination of subtle epitopes defined only by the presence of distinct PTMs is required. Moreover, several important modifications occur in very similar amino acid sequence motifs, like the methylation of H3K9 and H3K27, which are both placed in the context of an ARKS sequence. In addition, histone tails are hypermodified, as exemplified by the H3 tail, where the adjacent R8, K9, and S10 amino acid side chains are known to be methylated, acetylated, or phosphorylated. This implies that secondary modifications often occur on the peptide segment contacted by the antibody in the immediate vicinity of the target PTM and sometimes prevent the binding of antibodies in spite of the presence of the target modification, yielding false negative results. When undocumented, the cross-reactivity with related or unrelated marks and the combinatorial effect of neighboring marks compromise the application of antibodies, as illustrated in Figure 1. Additionally, different antibodies show distinct profiles of false positive and false negative signals, and even antibodies with the same catalog numbers regularly show lot-to-lot fluctuations of properties (also illustrated in Fig. 1). This variability is not unexpected for polyclonal antibodies, where new batches are produced by immunization of a new animal, but changes of purification procedure may cause variance of properties of monoclonal antibodies as well. Occasionally some lots of commercial antibodies even prefer to bind to secondary targets (see Fig. 1 and H3K36me3 antibodies documented in Bock et al. 2011a). This necessitates a detailed quality control and documentation of each antibody and each lot in order to give the user all relevant information for correct data interpretation, which is often not sufficiently provided. The urgency for better quality assessment and documentation of antibodies used in chromatin research has been widely recognized in the field (Bock et al. 2011a; Egelhofer et al. 2011; Fuchs et al. 2011; Nishikori et al. 2012; Peach et al. 2012; Hattori et al. 2013; Heubach et al. 2013), and the ENCODE Project Consortium has set up quality criteria for histone PTM antibodies (Egelhofer et al. 2011; Landt et al. 2012). According to these guidelines, antibodies must specifically detect modified histones in Western blots and fulfill one or more of the following secondary criteria: (1) specific binding to modified peptides in dot blot assays; (2) mass spectrometric detection of the modification in precipitated chromatin; (3) loss of signal upon knockdown of the corresponding histone modifying enzyme; (4) reproducibility of ChIP-seq; (5) similarity of ChIP-seq results of two different antibodies directed against the same modification; or (6) overlap of ChIP-seq peaks with expected genomic annotations.
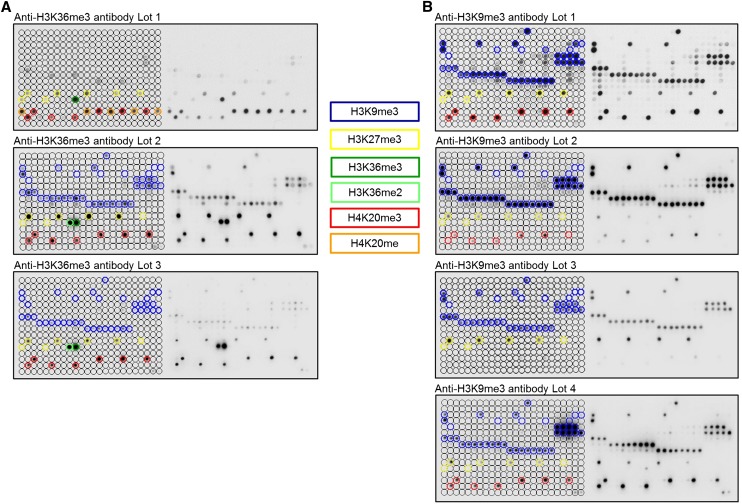
Figure 1.
Peptide array analyses showing lot-to-lot fluctuations, cross-reactivity, and effects of proximal marks on the binding of popular histone tail antibodies. Peptide spots are annotated on the left side of the glass slide. The color-coded boxes denote the presence of the designated modifications. (A) Anti-H3K36me3 antibodies showed significant cross-reactivity to H4K20me1, H4K20me3, H3K27me3, H3K9me3, and H3K36me2. All lots of the H3K36me3 antibody had a weak binding specificity, and they showed very different properties. (B) Anti-H3K9me3 antibodies displayed cross-reactivity to H3K27me3 (Lot 3 even prefers H3K27me3 peptides) and H4K20me3 (very overt in the case of Lot 1 and Lot 4). H3K9me3 peptide spots that were not bound by these antibodies generally also contained H3S10ph or H3T11ph, indicating that this secondary mark prevents binding. For a more detailed annotation of the modifications at each spot, refer to Supplemental File S2. Among the H3K9me3 antibodies, only Lot 2 showed a binding specificity of acceptable quality. Anti-H3K36me3 (Lot 1) and anti-H3K9me3 (Lot 1) data were taken from Bock et al. (2011a) and reprocessed. Additional examples of commercial H3K36me3 antibodies with insufficient specificity are shown in Bock et al. (2011a). Examples of the fundamental differences in the specificity profiles of different antibodies directed against H3K27me3 are given in Supplemental Figure S1.
To develop an alternative to antibodies for chromatin research, we assessed the applicative potential and utility of naturally occurring and engineered histone modification interacting domains (HMIDs). This approach has several distinct advantages over antibodies, such as the ease and cost-effectiveness of recombinant production of HMIDs in Escherichia coli, the amenability of HMIDs to protein engineering, and the possibility of producing them at constant quality, eliminating lot-to-lot variability. In support of this concept, affinity methods based on protein domains have been successfully employed for the enrichment of methylated or unmethylated CpG islands in the analysis of DNA methylation (Cross et al. 1994; Blackledge et al. 2012) or in proteome-wide analyses of non-histone lysine methylation (Liu et al. 2013; Moore et al. 2013). In this proof-of-principle study, we started by characterizing the specificity of several HMIDs and compared them with ENCODE-validated antibodies for the same PTM using peptide arrays. Specificities were further validated in Western blots by detecting histone tail PTMs using unmodified histones and histones specifically depleted with the target PTM as controls. Then, we investigated the applicative potential of HMIDs in ChIP-like experiments to enrich for chromatin with particular modifications, which represents one of the most important and commonly used applications of histone tail PTM antibodies.
Results
Specificity analysis of H3K9me3 binders on histone peptide arrays
Histone PTMs exert their biological effects mainly through the regulated binding of HMIDs, which are critical constituents of many chromatin-modifying complexes (Taverna et al. 2007; Patel and Wang 2013). HMIDs are often small domains with a stable fold that can be easily expressed in E. coli and purified with high yield by affinity chromatography (Supplemental Fig. 2A). Here, we aimed to investigate the applicative potential of HMIDs as an alternative to histone PTM-specific antibodies. Specific binding to modified peptide epitopes is a necessary prerequisite for histone PTM antibodies and HMIDs (Egelhofer et al. 2011; Landt et al. 2012). In order to obtain detailed information about the specificity of histone PTM antibodies and HMIDs, it is essential to test their binding to a large collection of potential substrate peptides with various combinations of PTMs. Such experiments can be ideally conducted using peptide arrays. We used the CelluSpots histone peptide arrays, which feature 384 peptides and 59 identified or hypothetical histone PTMs of all histone tails (Dhayalan et al. 2010, 2011; Zhang et al. 2010; Bock et al. 2011a,b). Each glass slide contains two copies of the same array to ensure reliability. As a first step toward comparing the properties of histone PTM antibodies and HMIDs, we conducted a direct side-by-side comparison of the CelluSpots binding patterns of the MPHOSPH8 (also known as MPP8) Chromo and ATRX ADD domains, which are well-characterized H3K9me3 binders (Bock et al. 2011a; Dhayalan et al. 2011; Li et al. 2011), and different batches of a widely used and ENCODE-validated anti-H3K9me3 antibody (Figs. 1, 2A). The H3K9me3 mark was selected because it is among the most widely studied histone PTMs.
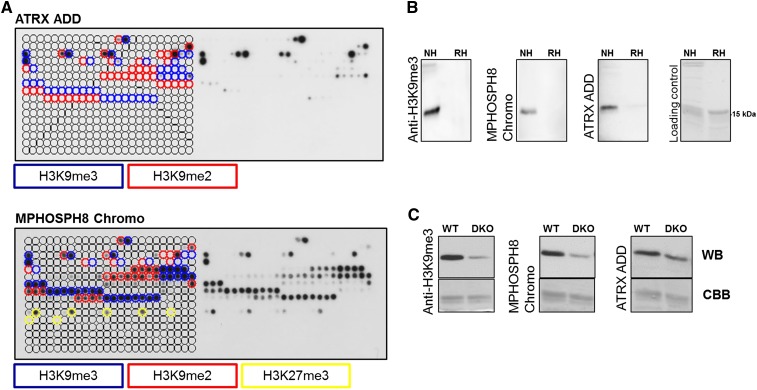
Figure 2.
Peptide array and Western blot specificity analyses of H3K9me3-specific reagents. (A) CelluSpots peptide array analyses of ATRX ADD and MPHOSPH8 Chromo. Peptide spots are annotated on the left side of the glass slide. The color-coded boxes denote the presence of the designated modifications. ATRX ADD did not bind to H3K9me2/3 peptides also containing H3K4me2/3, H310Sph, or H3T11ph. Binding of MPHOSPH8 Chromo to peptides containing H3K9me2/3 or H3K27me3 was inhibited by H3S10ph, H3T11ph, or H3S28ph. For a more detailed annotation of the modifications at each spot, refer to Supplemental File S2. For comparison with anti-H3K9me3 antibodies, refer to Figure 1. The MPHOSPH8 data set was taken from Bock et al. (2011b) and reprocessed. (B) Western blot analysis using ATRX ADD, MPHOSPH8 Chromo, and anti-H3K9me3 antibody (Lot 2) with native (NH) and recombinant histones (RH). The approximate position of the H3 protein at 15 kDa is indicated. (C) Western blot (WB) analysis using ATRX ADD, MPHOSPH8 Chromo, and the same anti-H3K9me3 antibody with nucleosomes isolated from wild-type and Suv39h1 and Suv39h2 double knockout (DKO) cells. The Coomassie brilliant blue (CBB)-stained gel after transfer is shown as a loading control.
The MPHOSPH8 Chromo domain binds specifically to histone H3 trimethylated on lysine 9 through the same conserved recognition mechanism as the Chromo domains of the HP1 protein family (including human CBX1) (Li et al. 2011). On the peptide array, MPHOSPH8 Chromo bound most prominently to H3K9me3 peptides and to a lesser extent to H3K9me2 and H3K27me3 peptides (Fig. 2A; Bock et al. 2011b). Akin to the Chromo domain of CBX1 (Supplemental Fig. S3A), peptide binding was inhibited by adjacent H3S10ph and H3T11ph, but not by H3R8citr. The observed weak binding to H3K27me3 is discordant with solution peptide binding studies, where binding to H3K27me3 was undetectable (Li et al. 2011). The ATRX ADD domain exhibited strong binding to peptides harboring H3K9me3 and weaker to peptides having H3K9me2 (Fig. 2A). The binding was abrogated by secondary modifications at the H3K4 position and H3S10ph or H3T11ph. These observations are in agreement with previous data where the dual readout of unmodified H3K4 and trimethylated H3K9 by the ATRX ADD domain was reported (Dhayalan et al. 2011; Eustermann et al. 2011; Iwase et al. 2011). The modification-dependent interaction of MPHOSPH8 Chromo and ATRX ADD with histone peptides was confirmed with methyllysine-binding pocket mutants, which all lost peptide binding (Supplemental Fig. S4A). The best lot of the Abcam ab8898 anti-H3K9me3 antibody (Lot 2) bound specifically to H3K9me3 modified peptides with a minute cross-reactivity to H3K27me3 and H4K20me3 (Fig. 1). Similarly to MPHOSPH8 Chromo, ATRX ADD and other commercial anti-H3K9me3 antibodies (Bock et al. 2011a) phosphorylation of S10 or T11 eliminated binding. We conclude that the specificity of MPHOSPH8 Chromo and ATRX ADD is comparable to good antibodies currently used in chromatin research, and the ENCODE criterion for specific binding to peptide epitopes is fulfilled by these HMIDs.
Western blot analyses of H3K9me3 binders
To further test the applicative potential of HMIDs as detection and affinity precipitation reagents, Western blot experiments were carried out in which an HMID was used instead of a primary antibody. This experiment allows investigation of the specificity of HMID binding, as well as cross-reactivity with other histone PTMs and nuclear proteins. MPHOSPH8 Chromo and ATRX ADD domains performed robustly as Western blot detection reagents by specifically binding to native histones prepared from human cell lines, but not to recombinant histones, which do not carry PTMs (Fig. 2B). In each case, the modification-dependent interaction was confirmed with methyllysine-binding pocket mutants, which lost binding (Supplemental Fig. S4B). To further verify the specificity of the interaction with H3K9me3, Western blot experiments were carried out with histones isolated from wild-type and Suv39h1/Suv39h2 double-knockout (DKO) immortalized MEF cells (Fig. 2C). With both MPHOSPH8 Chromo and ATRX ADD, the binding to DKO histones was reduced to a similar extent as observed with the corresponding antibody. Next, to address the minute cross-reactivity of MPHOSPH8 Chromo to H3K27me3 peptides on the peptide array, we performed Western blot experiments against cells treated with an EZH2 inhibitor (Fiskus et al. 2009), which leads to a strong reduction of global H3K27me3 (Fig. 5B, see below). No reduction of H3 interaction was observed (Supplemental Fig. S4C), which confirms that MPHOSPH8 Chromo does not bind H3 harboring H3K27me3 (Li et al. 2011). Based on the specificity and Western blot data, we conclude that MPHOSPH8 Chromo and ATRX ADD domains are as specific as antibodies and can be utilized as a powerful and specific readout tool in Western blot assays.
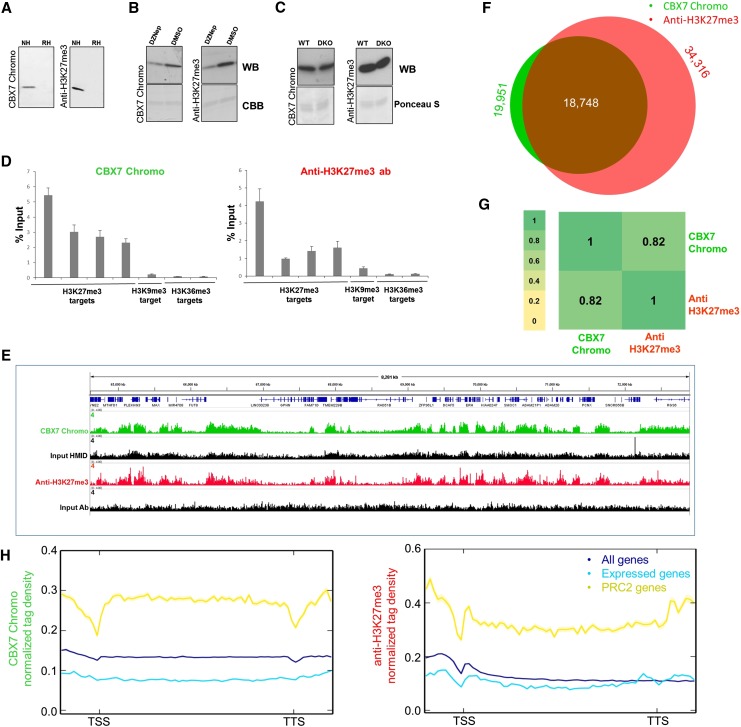
Figure 5.
Comparative analysis of results obtained with the CBX7 Chromo domain and anti-H3K27me3 antibody No. 1 Lot 1 (Supplemental Fig. S1). (A) Western blot analysis of CBX7 Chromo and anti-H3K27me3 antibody binding to native (NH) and recombinant histones (RH). (B) Western blot analysis of CBX7 Chromo and anti-H3K27me3 antibody binding to nucleosomes isolated from EZH2 inhibitor (DZNep)- or DMSO-treated HepG2 cells. (C) Western blot analysis of CBX7 Chromo and anti-H3K27me3 antibody binding to histones isolated from wild-type and Suv39h DKO immortalized MEFs. (D) Comparison of the CIDOP-qPCR and ChIP-qPCR results of the CBX7 Chromo domain and anti-H3K27me3 antibody analyzed with amplicons associated with H3K27me3, H3K9me3, and H3K36me3. All experiments were carried out in triplicates of biological duplicates. Error bars represent the standard errors of the mean. (E) Representative genome browser snapshot of CIDOP-seq and ChIP-seq data obtained with the CBX7 Chromo domain and anti-H3K27me3 antibody (taken from ENCODE). For more examples, refer to Supplemental Figures S8C and S9. (F) Venn diagram of the overlap of peak regions between CBX7 Chromo domain and anti-H3K27me3 antibody. (G) Spearman correlation coefficient heatmap of CIDOP-seq and ChIP-seq tags in 10-kb bins. (H) Metagene profiles of CBX7 Chromo and anti-H3K27me3 antibody ranked by association with all genes, all expressed genes, and PRC2 (EZH2)-associated genes. The shaded lines represent the standard errors of the mean.
Chromatin precipitation analyses using H3K9me3 binders
Next, we investigated if HMIDs have the ability to specifically precipitate discrete types of chromatin containing the target modifications in native ChIP-like assays, which represents one of the most important applications of histone PTM antibodies. We call these experiments chromatin interacting domain precipitation (CIDOP). The chromatin used in these experiments was prepared from HepG2 cells and consisted primarily of mononucleosomes (Supplemental Fig. S2B). CIDOP and ChIP were conducted with HMIDs and antibodies in parallel. Initially, the recovered DNA was analyzed by quantitative PCR of selected loci containing defined PTMs based on ENCODE ChIP-seq data of HepG2 cells (The ENCODE Project Consortium 2011). As shown in Figure 3A, the CIDOP-qPCR profiles of MPHOSPH8 Chromo and ATRX ADD are in very good agreement with the corresponding anti-H3K9me3 antibody (Lot 2), although the domains show a slightly higher signal for nontargets than the antibody. The precipitation of chromatin was modification-dependent, as indicated by experiments carried out with the methyllysine-binding pocket mutants, where no pull down was observed (Supplemental Fig. S4D).
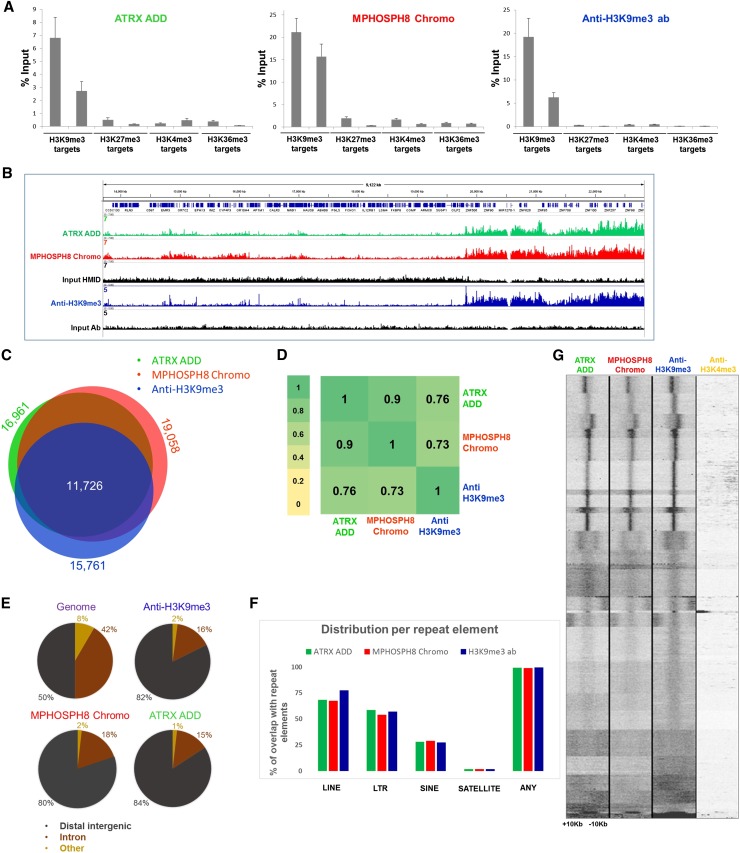
Figure 3.
CIDOP and ChIP of ATRX ADD, MPHOSPH8 Chromo, and anti-H3K9me3 antibody. (A) Comparison of CIDOP-qPCR and ChIP-qPCR signals with domains and anti-H3K9me3 antibody (Lot 2 from Fig. 1) using amplicons associated with H3K9me3, H3K27me3, H3K4me3, and H3K36me3. All experiments were carried out in triplicates of biological duplicates. Error bars represent the standard errors of the mean. (B) Representative genome browser snapshot comparing CIDOP-seq and ChIP-seq signals of domains and anti-H3K9me3 antibody (taken from ENCODE). For more examples, refer to Supplemental Figures S7A, S8A, and S8C. (C) Venn diagram of the overlap of peak regions between domains and anti-H3K9me3 antibody. The number of peaks in each area is given in Supplemental Table S3. (D) Spearman correlation coefficient heatmap of CIDOP-seq and ChIP-seq tags in 15-kb bins. (E) Distribution of CIDOP-seq and ChIP-seq peaks among different genomic elements. (F) Distribution of CIDOP-seq and ChIP-seq peaks among different repeat elements (cf. Supplemental Fig. S4E). (G) Clustering analysis of tag densities from MPHOSPH8 Chromo and ATRX ADD CIDOP-seq as well as anti-H3K9me3 and H3K4me3 antibody ChIP-seq data sets. Tags were collected within a 20-kb window, centered on the midpoints of anti-H3K9me3 antibody peaks, and sorted by k-means clustering (30 clusters). An analysis with 100 clusters is shown in Supplemental Figure S4F.
To extend the CIDOP-qPCR results to a genome-wide level, we performed chromatin interacting domain precipitation coupled with next-generation sequencing (CIDOP-seq). We compared our CIDOP-seq data with available ENCODE ChIP-seq data from HepG2 cells (Fig. 3B). It is worth pointing out that optimal standards for comparison and detection of broad marks such as H3K9me3, H3K27me3 and H3K36me3 are lacking or being developed (Landt et al. 2012). Identical data analysis procedures were used for CIDOP and ChIP experiments in our study. We compared peak regions obtained with ATRX ADD, MPHOSPH8 Chromo, and the anti-H3K9me3 antibody and observed a notable overlap (Fig. 3C), as well as high Spearman correlation coefficients of tags in 15-kb bins (Fig. 3D). The correlation of biological CIDOP-seq replicates was similar to typical ChIP-seq replicates (Supplemental Fig. S7A,B). As expected, the majority of peak regions were enriched in heterochromatin in all three experiments (Supplemental Fig. S4E), which reflects the known heterochromatic localization of H3K9me3 (Peters et al. 2001, 2002). Also, the distribution of peak regions among different genomic elements was very similar in all three experiments, with the majority of peak regions localizing in distal intergenic sites (Fig. 3E). We analyzed the distribution of peak regions in repeat elements, again yielding similar results (Fig. 3F). The strong similarity of ATRX ADD, MPHOSPH8 Chromo, and anti-H3K9me3 antibody binding profiles and the difference compared to H3K9me2 and H3K27me3 profiles (Supplemental Fig. S8A,C,D) indicates that HMIDs are as efficient in genome-wide chromatin studies as good antibodies.
Since the ATRX ADD domain simultaneously reads the absence of di- and trimethylation at H3K4 and the presence of H3K9me3, while MPHOSPH8 Chromo and the antibody only recognize H3K9me3, the coexistence of H3K4me2/3 and H3K9me3 on one H3 tail would result in loss of signal in ATRX ADD precipitated chromatin, while MPHOSPH8 Chromo and anti-H3K9me3 antibody should remain unchanged. As mentioned above, the ATRX ADD, MPHOSPH8 Chromo, and anti-H3K9me3 antibody binding profiles were very similar, indicating that such coexistence of H3K4me2/3 and H3K9me3 on one tail must be rare. To detect potential minor fractions of chromatin containing this double modification, we performed k-means clustering of the CIDOP-seq and ChIP-seq data, which allows for the detection of small subgroups of potentially biologically relevant loci. We did not observe prominent differences in the formation of clusters between ATRX ADD, MPHOSPH8 Chromo, and anti-H3K9me3, and the vast majority of clusters were depleted of H3K4me3 methylation (Fig. 3G), even when we extended the number of clusters to one hundred or aligned the reads around the TSS (Supplemental Data S4F,G). These results indicate that H3K9me3 and H3K4me2/3 do not or rarely coexist on the same histone tail, which is consistent with previous reports, where lack of correlation between H3K9me3 and H3K4me3 in global distribution profiles was observed (Barski et al. 2007).
Application of the DNMT3A PWWP domain as H3K36me3 binder
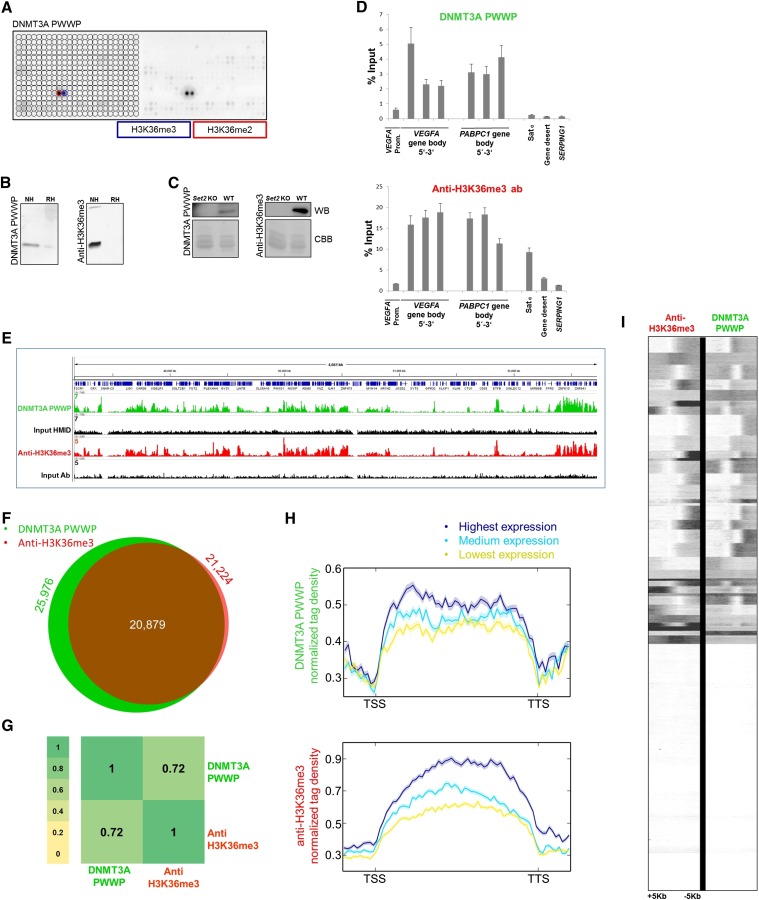
Since anti-H3K36me3 antibodies often showed weak specificity (see Fig. 1 and Bock et al. 2011a), novel tools for detection and enrichment of H3K36me3 marks are needed. For that reason, we decided to investigate the applicability of the DNMT3A PWWP domain, which has modest affinity to methylated H3K36 in vitro (Dhayalan et al. 2010; Bock et al. 2011a), as a reagent in chromatin research. On the peptide array, the PWWP domain of DNMT3A specifically bound to H3K36me2 and H3K36me3 peptides, and no other interaction was detected, while a popular anti-H3K36me3 antibody (Abcam, ab9050) displayed binding to H3K36me2/3, but also cross-reactivity to H3K27me3, H3K9me3, H4K20me, and H4K20me3 peptides (Figs. 1, 4A). The CelluSpots array contains only one H3K36me3 peptide spot, precluding an analysis of the potential effects of proximal marks. In Western blot experiments, the PWWP domain bound to native but not to recombinant histones, fulfilling the corresponding ENCODE quality criteria (Fig. 4B). Native histones isolated from wild-type and set2 knockout Saccharomyces cerevisiae cells were successfully used to validate the specificity of the interaction (Fig. 4C). However, with this experiment we were not able to address the cross-reactivity of the anti-H3K36me3 antibody with H3K27me3 and H4K20me3, due to the absence of these marks in S. cerevisiae. These promising results prompted us to conduct chromatin precipitation experiments with the PWWP domain as well. The precipitation of chromatin was modification-dependent, as indicated by experiments carried out with the PWWP methyllysine-binding pocket variant (Supplemental Fig. S5). CIDOP-qPCR and CIDOP-seq data resembled but were not fully identical to the corresponding ChIP-qPCR (H3K36me3 antibody Lot 2) and ChIP-seq data (ENCODE antibody) (Fig. 4D,E,G–I; Supplemental Fig. S10). The CIDOP-qPCR and ChIP-qPCR experiments (Fig. 4D) showed enrichment of signal in gene bodies as exemplified at the VEGFA and PABPC1 loci, although the individual intensities slightly differed. In addition, the recovery with PWWP was lower than with antibody, which may be related to the lower binding affinity of PWWP. Furthermore, in the anti-H3K36me3 antibody experiment, we observed enrichment of satellite alpha, which is likely due to the cross-reactivity of the antibody with H4K20me3. Next, we conducted CIDOP-seq and compared with ChIP-seq data obtained with H3K36me3 antibody and observed a clear overlap of peaks and high correlation coefficients of both data sets (Fig. 4F,G). The correlation between the PWWP domain and the anti-H3K36me3 antibody was similar to biological replicates of anti-H3K36me3 antibodies (Supplemental Fig. S7D). Genome browser snapshots verified the clear colocalization of signals (Fig. 4E; Supplemental Fig. S10), which was also observed in a metagene analysis and k-means clustering around transcription start sites (Fig. 4H,I). Both signals were correlated with the level of gene expression (Fig. 4H). As in the CIDOP-qPCR experiments, a slight difference in the distribution of intragenic signals between the domain and the antibody was observed, in which the domain tends to enrich signals in the 5′ end of genes, while the antibody signals are more enriched in the middle and 3′ part of genes. However, we want to raise caution by pointing out the insufficient specificity of the H3K36me3 antibodies (see Fig. 1 and Bock et al. 2011a). Therefore, some of the signals obtained by H3K36me3 ChIP experiments could result from an off-target binding of the anti-H3K36me3 antibody, as illustrated by the detection of satellite alpha signal.
Figure 4.
Comparative analysis of results obtained with the DNMT3A PWWP domain and anti-H3K36me3 antibody. (A) Peptide array analysis of the specificity of DNMT3A PWWP. For comparison with anti-H3K36me3 antibodies, refer to Figure 1. (B) Western blot analysis using DNMT3A PWWP and anti-H3K36me3 antibody (Lot 2 from Fig. 1) with native (NH) and recombinant histones (RH). (C) Western blot (WB) analysis using DNMT3A PWWP and anti-H3K36me3 antibody (Lot 3 from Fig. 1) with native histones isolated from wild-type and set2 KO S. cerevisiae. The Coomassie brilliant blue (CBB)-stained gel after transfer is shown as a loading control. (D) Comparison of CIDOP-qPCR and ChIP-qPCR results obtained with the DNMT3A PWWP domain and anti-H3K36me3 antibody (Lot 2) using amplicons associated with H3K27me3, H3K9me3, and the gene bodies of VEGFA and PABPC1. All experiments were carried out in triplicates of biological duplicates. Error bars represent the standard errors of the mean. (E) Representative genome browser snapshot comparing CIDOP-seq and ChIP-seq results obtained with the DNMT3A PWWP domain and anti-H3K36me3 antibody (ENCODE). For more examples, refer to Supplemental Figure S10. (F) Venn diagram of the overlap of peak regions between DNMT3A PWWP domain and anti-H3K36me3 antibody. (G) Spearman correlation coefficient heatmap of CIDOP-seq and ChIP-seq tags in 15-kb bins. (H) Metagene profiles of DNMT3A PWWP and anti-H3K36me3 antibody ranked by differential expression. The shaded lines represent the standard errors of the mean. Supplemental Figure S5D shows this image with the y-axis scale starting from 0. (I) Clustering analysis of tag densities from the DNMT3A PWWP and anti-H3K36me3 antibody CIDOP-seq and ChIP-seq data sets. Tags were collected within a 10-kb window around TSS and sorted by k-means clustering (50 clusters). The clusters with low DNMT3A PWWP and anti-H3K36me3 signals also lack H3K4me3 and likely are not expressed (not shown).
Application of the CBX7 Chromo domain as H3K27me3 binder
Finally, we investigated the applicability of the CBX7 Chromo domain as a potential reader of H3K27me3, because H3K27me3, like H3K9me3, is a widely studied chromatin PTM. CBX7 is a component of the Polycomb repressive complex 1 (PRC1) known to bind to H3K27me3. In spite of this, its Chromo domain was reported to bind indiscriminately to H3K27me3 and H3K9me3 on peptide arrays (Kaustov et al. 2011). Accordingly, we observed binding of CBX7 Chromo to both H3K27me3- and H3K9me3-containing peptides on the CelluSpots arrays, showing sensitivity to neighboring H3R26citr and H3S28ph (Supplemental Fig. S6). The H3K27me3 antibody (No.1 Lot 1) specifically bound to H3K27me3 peptides, and binding was abolished by adjacent H3S28ph marks (Supplemental Fig. S1). The CBX7 Chromo domain performed robustly in Western blot experiments (Fig. 5A). To determine its specificity in histone binding, we conducted experiments with histones isolated from wild-type and Suv39h DKO immortalized MEF cells and HepG2 cells treated with an EZH2 inhibitor and compared the results with anti-H3K9me3 and anti-H3K27me3 antibodies. As shown in Figure 5, B and C, CBX7 binding was lost after EZH2 inhibition, but it bound equally to histones isolated from wild-type and Suv39h DKO cells, indicating a specific readout of H3K27me3 by CBX7 Chromo. This observation was further validated with CIDOP-qPCR/CIDOP-seq and the corresponding ChIP-qPCR/ChIP-seq experiments, where the CBX7 Chromo domain displayed a clear mono-specific precipitation of H3K27me3-associated chromatin (Fig. 5D,E; Supplemental Figs. S8C,D, 9). A notable overlap of the peak regions of CBX7 Chromo and anti-H3K27me3 antibody was observed, but not with the anti-H3K9me3 antibody (Fig. 5F; Supplemental Fig. S8E). Furthermore, we observed a high Spearman correlation of the domain data with the anti-H3K27me3 antibody in 10-kb bins (Fig. 5G) as one would typically expect in ChIP-seq of biological replicates (Supplemental Fig. S7C), suggesting that the binding to H3K9me3 only occurs with peptides. It is likely that CBX7 Chromo requires a longer part of the histone tail for specific H3K27me3 recognition than the one present on the peptide arrays. The modification-dependent interaction of CBX7 Chromo with histones was verified with a methyllysine-binding pocket mutant (Supplemental Data S6). Finally, a metagene analysis of CBX7 Chromo and anti-H3K27me3 antibody as expected showed a higher enrichment of signal for PRC2-associated genes, compared to expressed genes in HepG2 or all annotated genes (Fig. 5H), again indicating the similarity between the two reagents.
Discussion
Chromatin is a highly dynamic and multifaceted structure embellished with more than 150 histone PTMs (Kouzarides 2007; Margueron and Reinberg 2010; Bannister and Kouzarides 2011; Tan et al. 2011). Chromatin precipitation studies with antibodies have been the workhorse of chromatin biology in deciphering the complex syntax of the histone modification language. However, these are indirect experiments, and the identity of the material captured in these studies fully depends on the reagent. As documented above, the properties and specificity reports of existing antibodies are imperfect, so that novel analytic tools are necessary. To this aim, we have developed a collection of native and engineered histone modification interaction domains (HMIDs), which strongly and specifically interact with H3 tails methylated at different lysine residues. The most promising domains were investigated with respect to the primary and secondary ENCODE criteria for antibody validation, such as specificity in Western blotting with native, recombinant, and PTM-depleted histones, binding to modified peptides, and ChIP-like experiments (Egelhofer et al. 2011; Landt et al. 2012). In all our experiments, the efficiency of HMIDs was directly compared side-by-side with popular commercial antibodies recognizing the same PTMs. Our results show that HMIDs fulfill all tested antibody quality criteria. Most importantly, the comparative analyses of HMIDs and ENCODE validated antibodies in CIDOP and ChIP experiments showed high correlation of signals, indicating the potential of using HMIDs alone or in concert with antibodies in genome-wide or locus-specific studies.
HMIDs bind strong enough to modified histone tails for applications in chromatin biology
One obvious difference between antibodies and HMIDs is in their binding affinity to modified histone tails. The KD values of HMIDs used here are in the high nanomolar to low micromolar range (MPHOSPH8 Chromo binding to H3K9me3 peptides: 120 nM [Bock et al. 2011a], 140 nM [Chang et al. 2011]; ATRX ADD-H3K9me3: 1.4 µM [Dhayalan et al. 2011]; DNMT3A PWWP-H3K36me3: 64 µM [Dhayalan et al. 2010]; CBX7-H3K27me3: 22 µM [Bernstein et al. 2006]). In contrast, typical antibody binding constants of commercial histone tail antibodies are in the low nanomolar to low micromolar range (for example, two papers reported KD values of 0.2, 4, 60, 83, 520, 820, 1000, 2200, and 2700 nM for several representative commercial antibodies [Nishikori et al. 2012; Hattori et al. 2013]). As indicated by these numbers, the ranges of affinities of antibodies and HMIDs overlap, but in general, antibodies bind more strongly. However, the specificity of a reagent is defined by the ratio of the specific and nonspecific interaction, so that the absolute binding affinity does not play a major role. The weaker equilibrium binding affinity of HMIDs may lead to lower fractional saturation of histone proteins with HMIDs as compared with antibodies, but this can be compensated by higher concentrations of these reagents in the experiment. Since all typical chromatin assays are heterophasic, the half-life of HMID or antibody complexes with peptides or histone proteins (as determined by the kinetic dissociation constant of the complex, koff) is another critical parameter. In this study we show experimentally that HMIDs also satisfy the applicative needs in this respect.
Strengths and pitfalls of first principle techniques for antibody and HMID specificity validation
There are two main hypothesis-free approaches to determine the binding specificity of histone tail interacting reagents—mass spectrometry and binding to modified peptides. Mass spectrometry is the only method allowing the study of the nature of the chromatin captured by a binding reagent directly. It is among the secondary criteria for quality control of antibodies suggested by the ENCODE Project Consortium. Currently, mass spectrometry is especially useful in the specificity analysis of antibodies for DNA interacting proteins, but less powerful for the analysis of histone PTM antibodies (Landt et al. 2012). The reason for this is that precipitation of one chromatin modification can simultaneously enrich associated secondary modifications on the same histone tail or on the same nucleosome. Since mass spectrometry cannot pinpoint the direct interaction of an antibody and a target modification, it is impossible to decide if coenrichment of a secondary modification is due to a cross-reactivity of the reagent or if the secondary mark is associated with the primary target. In addition, the identification of inhibitory secondary marks is difficult by mass spectrometry. However, future technological improvements may further increase the value of mass spectrometric analysis for histone PTM antibody and HMID quality assessment.
The second approach allowing for a directed determination of the specificity of histone tail interacting reagents is to investigate their binding to modified peptides. For a detailed analysis, modified histone tail peptide arrays are particularly powerful, since they contain a lot of histone tail peptides with different PTMs in different combinations. Hence, they allow for cross-reactivity analyses of histone binding reagents and detection of inhibitory effects of secondary modifications (Bock et al. 2011a). However, our data also illustrate some pitfalls of peptide-based specificity analysis of histone PTM binders, because even antibodies and HMIDs, which show some cross-reactivity with peptides, can still give clear-cut ChIP and CIDOP profiles. For example, the cross-reactivities of MPHOSPH8 Chromo toward H3K27me3 peptides and CBX7 Chromo toward H3K9me3 peptides were not detectable in chromatin experiments. Differences between peptide and histone binding of antibodies and HMIDs may be due to the fact that peptides are too short, which may lead to more promiscuous binding because HMIDs may form additional interactions to the histone outside of the peptide region. Moreover, peptides present artificial ends when compared to histone proteins, which, for example, may allow an H3K9me3 binder to interact with an H3K27me3 peptide, but not with the K27me3 modified histone H3 protein. However, modified peptides are one of the few ways to define the detailed specificity profile of an antibody or HMID by first principle. Hence peptide binding, in our view, will remain an important reference point when analyzing the specificity of these reagents.
HMIDs can be produced at constant quality with invariable properties
Collectively, our data show compelling evidence that HMIDs are powerful and versatile affinity and detection reagents that hold a genuine applicative potential. Like antibodies, these domains do show some cross-reactivity and dependence on secondary modifications. However, the recombinant expression and easy purification of HMIDs will allow for production of HMIDs at a constant quality, eliminating lot-to-lot variability. Therefore, detailed specificity documentation of an HMID, including data on cross-reactivity and the combinatorial effects of secondary modifications, once prepared, will be valid for all future lots of this reagent. The availability of detailed specificity documentation will allow for more reliable interpretation of gain and loss of signals in genome-wide and locus-specific chromatin studies. In addition, the unlimited availability of HMIDs with invariable properties will enable researchers to directly compare results obtained in different labs and reproduce data much more easily. Thereby, the application of HMIDs will improve the overall scientific quality and stringency in the field. This point is illustrated by the fact that we were unable to directly investigate the quality and cross-specificity of the particular antibody lots used by the ENCODE labs for ChIP-seq, because these lots were no longer available. Such problems would not occur with HMIDs, which can be produced with constant quality and invariable properties. Recently, recombinant antibodies have been described that also have the potential to solve the problem of lot-to-lot variability (Hattori et al. 2013), but they are much more difficult to generate than HMIDs. After submission of our work, another publication also raised concerns about histone PTM antibody quality and demonstrated the application of reading domains for the analysis of chromatin states (Su et al. 2014). However, the Su et al. (2014) work was not based on a systematic validation of HMIDs and side-by-side comparison with antibodies following ENCODE guidelines.
Potential limitations of the antibody- and HMID-based technologies
In spite of the fact that quality control measures will continue to evolve, along with novel domain-based technologies and more stringent antibody production conditions, the presence of hypermodified histone tails with similar amino acid sequences carrying chemically related PTMs might impose tight limits on the maximum specificity that can be obtained with these histone interacting reagents. Using HMIDs might alleviate this problem by providing detailed specificity documentation of each and every precipitation domain, such that researchers can decide if the properties of a certain domain will match its planned application.
Further perspectives of the HMID technology
In addition to providing a reliable and cost-effective experimental tool, HMIDs could open many novel research avenues due to their ease of manipulation and amenability to protein engineering. This allows for adding affinity tags for simplified detection, preparation of PTM binding pocket variants as ideal matching negative controls (as exemplified in this work), and design of novel specificities. For example, we show here that the di- and trimethyl pan-specific binding of the KDM4A double Tudor domain can be changed either to a trimethyl pan-specific or to an H4K20me2/3 monospecific readout (Supplemental Fig. S11). Along the same lines, the S10ph inhibition of H3K9me3 binders could be resolved by rationally designed or naturally occurring phosphorylation-insensitive H3K9me3 HMIDs (Rothbart et al. 2012). Additionally, there is an increased interest in the readout of complex histone PTM patterns. The differential readout of H3K9me3 in the context of modified or unmodified H3K4 tails by different HMIDs enabled us here to directly demonstrate that there is none or little coexistence of H3K9me3 and H3K4me2/3 on single H3 tails. In the future, it is even conceivable to design fusion domains with dual specificities for different marks, which in principle is not possible with antibodies. For instance, a combined readout of H3K9me3 and H4K20me3 may be used to probe the heterochromatic landscape and reveal the extent of codependency between these two marks. On the same note, the combined readout of H3K27me3 and H3K4me3 could assist the investigation of bivalent domains. We anticipate that HMID-based novel experimental tools will aid the community in answering important questions in chromatin biology.
Methods
Cloning, site-directed mutagenesis, expression, and purification
The sequences encoding the Chromo domain of human CBX7 (amino acids 7–61 of Swiss-Prot entry O95931), CBX1 (also known as HP1beta homolog) (17–76 of NP_001120700.1), MPHOSPH8 (also known as MPP8) (57–111 of NP_059990.2), the double Tudor domain of human KDM4A (also known as JMJD2A) (856–1047 NP_055478.2), the ADD domain of human ATRX (163–292 of NP_000480.2), and PWWP domain of murine DNMT3A (279–420 of NP_001258682) were amplified from cDNA and cloned as GST-fusion proteins into the pGEX-6p-2 vector (GE Healthcare). The sequences of the expression vectors are given in Supplemental File S3. All GST-tagged proteins were overexpressed at 18°C and purified essentially as described in Rathert et al. (2008), electrophoresed on 12% SDS-PAGE, and stained with colloidal Coomassie brilliant blue G-250. Mutations were introduced using the megaprimer method described in Jeltsch and Lanio (2002), and successful mutagenesis was confirmed by restriction analysis and Sanger DNA sequencing.
Cell culture
HEK293 cells were grown in Dulbecco’s modified Eagle’s medium with 5% fetal bovine serum at 37°C at 5% CO2 until they reached 90% confluence. HepG2 cells were grown in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum at 37°C and 5% CO2 until they reached 75% confluence. When necessary, the HepG2 cells were supplemented with 1 µM DZNep (Calbiochem, EMD Millipore) (Fiskus et al. 2009) for 72–96 h, collected, and processed. Immortalized MEFs were grown in DMEM supplemented with 10% FBS, L-glutamine, nonessential amino acids, sodium pyruvate, and 100 µM β-mercaptoethanol.
Binding of domains and antibodies on CelluSpots histone peptide arrays, Western blot, and peptide pull down
Experiments to determine the binding specificity and context dependence of the GST-fusion domains and antibodies were performed using the CelluSpots peptide array platform as described in Bock et al. (2011a,b). Popular antibodies (although not the same batches) were selected from the Antibody Validation Database (http://compbio.med.harvard.edu/antibodies). In this study, we used anti-H3K27me3 antibody (Active Motif, #39155), anti-H3K9me3 (Abcam, ab8898), anti-H3K36me3 (Abcam, ab9050), and H3K9me2 (Abcam, ab1220). The lots of the corresponding antibodies are specified in Supplemental Table S2. For Western blots, we applied the peptide array protocol. Native histones were isolated by acid extraction (Shechter et al. 2007) from HEK293 cells or S. cerevisiae grown in YPD medium overnight (after spheroplasting and extensive nuclei preparation), and recombinant histones H3 and H4 were purchased from New England Biolabs or purified from E. coli. Five micrograms of native histones and 2.5 µg of recombinant histones were loaded and electrophoresed on 16% SDS-PAGE and transferred onto nitrocellulose membranes. The amounts of GST-fused proteins (wild type and mutants) used in CelluSpots and Western blot analyses were the following: CBX7 Chromo (10–100 nM), CBX1 Chromo (10 nM), MPHOSPH8 Chromo (10–100 nM), KDM4A double Tudor (10 nM), ATRX ADD (10 nM) (for WB binding in 300 mM NaCl peptide array buffer), and DNMT3A PWWP (10–100 nM). The antibodies were incubated following the manufacturer’s recommendations. All experiments were performed in duplicates or triplicates. Pull-down experiments with biotinylated peptides were performed as described in Kim et al. (2006).
Native chromatin precipitation (CIDOP and ChIP) and quantitative PCR
Native nucleosomes were isolated from around 20 million HepG2 cells (which was sufficient for 5–15 CIDOP/ChIP experiments) by micrococcal nuclease digestion of nuclei as described (Brand et al. 2008) with minor modifications. In brief, following MNase digestion, the nuclei were centrifuged at 13,000g for 10 min, and the resulting supernatant which contained the soluble nucleosomal fraction (predominantly mononucleosomes) was collected and snap frozen. Then, native chromatin (7.5–60 µg based on DNA absorbance) was precleared for 1 h at 4°C in DP buffer (16.7 mM Tris-Cl, 167 mM NaCl, 1.1% Triton X-100, 1.2 mM EDTA, and protease inhibitors) and 20 µl glutathione sepharose 4B (GE Healthcare) or protein G magnetic Dynabeads (Invitrogen), the beads were removed, and the supernatant was incubated overnight with the domain (7.5–60 µg) or antibody of interest (following the manufacturer’s recommendations). On the next day, the domain- or antibody-chromatin complexes were immobilized on 20 µl glutathione sepharose 4B (GE Healthcare) or protein G magnetic Dynabeads (Invitrogen), respectively. The washing steps of MPHOSPH8 Chromo, CBX1 Chromo, CBX7 Chromo, and the antibodies were the following: 1× low salt buffer (20 mM Tris-Cl, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, and 2 mM EDTA), 1× high salt buffer (20 mM Tris-Cl, 500 mM NaCl, 1% Triton X-100, 0.1% SDS, and 2 mM EDTA), 1× LiCl buffer (10 mM Tris-Cl, 250 mM LiCl, 1% NP-40, 1% DOC, and 1 mM EDTA), and 2× TE buffer. The washing steps of DNMT3A PWWP and ATRX ADD were less stringent: 3× PB buffer (50 mM Tris-Cl, 200–400 mM NaCl, 1 mM EDTA, 0.5% NP-40, 2 mM DTT), 2× TE buffer. Bound chromatin was eluted in elution buffer (50 mM Tris-Cl, 50 mM NaCl, 1 mM EDTA, 1% SDS) for 45 min at room temperature with rotation. DNA was recovered using ChIP DNA purification columns (Active Motif). The quantitative PCR assays were performed on a CFX96 Touch or CFX96 Real-Time detection system (Bio-Rad) using SYBR fast qPCR mix (Kapa Biosystems) or SsoFast EvaGreen supermix (Bio-Rad). A standard curve was generated to calculate percent of precipitated DNA and test the efficiency of each primer set. The primer sequences can be found in Supplemental Table S1.
Deep sequencing and data analysis
Approximately 20–50 million 50- or 100-nt sequence reads obtained with Illumina’s HiSeq 2000/2500 were mapped to the human reference genome hg19 with Bowtie (Langmead et al. 2009) from the Chipster software tool (Kallio et al. 2011). Only uniquely mapped reads were retained and all duplicates were removed. SICER (Zang et al. 2009) was used for peak calling with a window size of 200, fragment size of 150, gap size of 1200, and statistical threshold value of 0.01 (FDR). The genome coverage files were obtained with BEDTools (Quinlan and Hall 2010) in Galaxy (Giardine et al. 2005; Blankenberg et al. 2010; Goecks et al. 2010), and the coverage was normalized by reads per million (RPM). The peak annotation was done with CEAS within the Cistrome platform (Shin et al. 2009; Liu et al. 2011). For overlapping pieces of peak regions, we used the common intersection tool in Galaxy and Cistrome. For peak overlap analyses of MPHOSPH8 Chromo and anti-H3K9me3 antibodies, only peaks with a twofold enrichment metric were selected to make the analysis more reliable. For analyses with ATRX ADD, peaks with 2.75-fold enrichment were used. EpiExplorer (Halachev et al. 2012) was used for annotation of repeats and chromatin segments (based on data from Ernst et al. 2011), and seqMINER (Ye et al. 2011) was used for k-means clustering and heatmap generation. To assess the overall pairwise similarity between domains and antibodies, we calculated Spearman’s rank correlation coefficient in window sizes of 10 or 15 kb with deepTools (Ramirez et al. 2014). The genome browser snapshots were taken with the Integrative Genomics Viewer (IGV).
The HepG2 cell line raw sequencing data for all ChIP-seq experiments (except for H3K9me2, which were carried out in our lab) were downloaded from ENCODE (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeBroadHistone/).
RNA-seq analysis was performed with the Tuxedo Suite package (Trapnell et al. 2012). The reads were mapped with TopHat using default settings, and the transcripts were assembled with Cufflinks. After assembly, all transcripts were ranked based on the FPKM (fragments per kilobase of exon per million fragments mapped) metric in three groups (highest expression FPKM > 50, medium expression FPKM 5–15, and lowest expression FPKM < 5). These transcripts were associated with genes and used as a reference for metagene analysis of DNMT3A PWWP and the anti-H3K36me3 antibody. In the case of CBX7 Chromo and the anti-H3K27me3 antibody, all UCSC genes, expressed genes from RNA-seq data, and genes associated with EZH2 peaks were used as a reference for metagene analysis. All genes were fitted in a 5-kb-long metagene with 1-kb regions upstream of and downstream from the TSS and TTS, respectively, and the composite profiles were obtained with deepTools.
RNA-seq data were downloaded from ENCODE (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeCaltechRnaSeq/) using wgEncodeCaltechRnaSeqHepg2R1x75dFastqRep1.fastq.gz.
Genes associated with EZH2 peaks were retrieved from ENCODE (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeAwgTfbsUniform/) using wgEncodeAwgTfbsBroadHepg2Ezh239875UniPk.narrowPeak.gz.
Data access
CIDOP-seq data for this study have been submitted to the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-2143. The HMID expression plasmids have been submitted to AddGene (www.addgene.org; deposit 71277) under plasmid ID numbers 59694–59699.
Competing interest statement
C.H. and P.L. are employees of Active Motif, Inc.
Acknowledgments
We thank Dr. Michaela Strotbek for help with the qPCR experiments. The S. cerevisiae wild-type and Set2 KO strains were a kind gift from Dr. Brian Strahl. The Suv39h DKO cells were kindly provided by Dr. Thomas Jenuwein. This work has been supported by the BMBF grant FKZ 0315886A.
Author contributions: G.K. contributed to all experiments, data analysis, and interpretation. I.K., R.T., S.K., and R.Z.J. contributed to the cloning and purification of domains, protein design, and specificity analysis. C.H., P.L., and R.R. were involved in the ChIP-seq and CIDOP-seq experiments and contributed to the data analysis. I.K. and P.L. contributed to the design of the study. G.K. and A.J. designed the study and prepared the manuscript draft. All authors read and approved the final manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.170985.113.
References
- Bannister AJ, Kouzarides T. 2011. Regulation of chromatin by histone modifications. Cell Res 21: 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD. 2006. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol 26: 2560–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge NP, Long HK, Zhou JC, Kriaucionis S, Patient R, Klose RJ. 2012. Bio-CAP: a versatile and highly sensitive technique to purify and characterise regions of non-methylated DNA. Nucleic Acids Res 40: e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. 2010. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol 89: 19.10.1–19.10.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock I, Dhayalan A, Kudithipudi S, Brandt O, Rathert P, Jeltsch A. 2011a. Detailed specificity analysis of antibodies binding to modified histone tails with peptide arrays. Epigenetics 6: 256–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock I, Kudithipudi S, Tamas R, Kungulovski G, Dhayalan A, Jeltsch A. 2011b. Application of Celluspots peptide arrays for the analysis of the binding specificity of epigenetic reading domains to modified histone tails. BMC Biochem 12: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordeaux J, Welsh A, Agarwal S, Killiam E, Baquero M, Hanna J, Anagnostou V, Rimm D. 2010. Antibody validation. Biotechniques 48: 197–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand M, Rampalli S, Chaturvedi CP, Dilworth FJ. 2008. Analysis of epigenetic modifications of chromatin at specific gene loci by native chromatin immunoprecipitation of nucleosomes isolated using hydroxyapatite chromatography. Nat Protoc 3: 398–409 [DOI] [PubMed] [Google Scholar]
- Chang Y, Horton JR, Bedford MT, Zhang X, Cheng X. 2011. Structural insights for MPP8 chromodomain interaction with histone H3 lysine 9: potential effect of phosphorylation on methyl-lysine binding. J Mol Biol 408: 807–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross SH, Charlton JA, Nan X, Bird AP. 1994. Purification of CpG islands using a methylated DNA binding column. Nat Genet 6: 236–244 [DOI] [PubMed] [Google Scholar]
- Dhayalan A, Rajavelu A, Rathert P, Tamas R, Jurkowska RZ, Ragozin S, Jeltsch A. 2010. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J Biol Chem 285: 26114–26120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhayalan A, Tamas R, Bock I, Tattermusch A, Dimitrova E, Kudithipudi S, Ragozin S, Jeltsch A. 2011. The ATRX-ADD domain binds to H3 tail peptides and reads the combined methylation state of K4 and K9. Hum Mol Genet 20: 2195–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelhofer TA, Minoda A, Klugman S, Lee K, Kolasinska-Zwierz P, Alekseyenko AA, Cheung MS, Day DS, Gadel S, Gorchakov AA, et al. 2011. An assessment of histone-modification antibody quality. Nat Struct Mol Biol 18: 91–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium . 2011. A user’s guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol 9: e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. 2011. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustermann S, Yang JC, Law MJ, Amos R, Chapman LM, Jelinska C, Garrick D, Clynes D, Gibbons RJ, Rhodes D, et al. 2011. Combinatorial readout of histone H3 modifications specifies localization of ATRX to heterochromatin. Nat Struct Mol Biol 18: 777–782 [DOI] [PubMed] [Google Scholar]
- Fiskus W, Wang Y, Sreekumar A, Buckley KM, Shi H, Jillella A, Ustun C, Rao R, Fernandez P, Chen J, et al. 2009. Combined epigenetic therapy with the histone methyltransferase EZH2 inhibitor 3-deazaneplanocin A and the histone deacetylase inhibitor panobinostat against human AML cells. Blood 114: 2733–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs SM, Krajewski K, Baker RW, Miller VL, Strahl BD. 2011. Influence of combinatorial histone modifications on antibody and effector protein recognition. Curr Biol 21: 53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, et al. 2005. Galaxy: a platform for interactive large-scale genome analysis. Genome Res 15: 1451–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J. 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11: R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halachev K, Bast H, Albrecht F, Lengauer T, Bock C. 2012. EpiExplorer: live exploration and global analysis of large epigenomic datasets. Genome Biol 13: R96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori T, Taft JM, Swist KM, Luo H, Witt H, Slattery M, Koide A, Ruthenburg AJ, Krajewski K, Strahl BD, et al. 2013. Recombinant antibodies to histone post-translational modifications. Nat Methods 10: 992–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heubach Y, Planatscher H, Sommersdorf C, Maisch D, Maier J, Joos TO, Templin MF, Poetz O. 2013. From spots to beads-PTM-peptide bead arrays for the characterization of anti-histone antibodies. Proteomics 13: 1010–1015 [DOI] [PubMed] [Google Scholar]
- Iwase S, Xiang B, Ghosh S, Ren T, Lewis PW, Cochrane JC, Allis CD, Picketts DJ, Patel DJ, Li H, et al. 2011. ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome. Nat Struct Mol Biol 18: 769–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeltsch A, Lanio T. 2002. Site-directed mutagenesis by polymerase chain reaction. Methods Mol Biol 182: 85–94 [DOI] [PubMed] [Google Scholar]
- Kallio MA, Tuimala JT, Hupponen T, Klemela P, Gentile M, Scheinin I, Koski M, Kaki J, Korpelainen EI. 2011. Chipster: user-friendly analysis software for microarray and other high-throughput data. BMC Genomics 12: 507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaustov L, Ouyang H, Amaya M, Lemak A, Nady N, Duan S, Wasney GA, Li Z, Vedadi M, Schapira M, et al. 2011. Recognition and specificity determinants of the human cbx chromodomains. J Biol Chem 286: 521–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, Zhang Y, Bedford MT. 2006. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep 7: 397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. 2007. Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, et al. 2012. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res 22: 1813–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li Z, Ruan J, Xu C, Tong Y, Pan PW, Tempel W, Crombet L, Min J, Zang J. 2011. Structural basis for specific binding of human MPP8 chromodomain to histone H3 methylated at lysine 9. PLoS ONE 6: e25104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Ortiz JA, Taing L, Meyer CA, Lee B, Zhang Y, Shin H, Wong SS, Ma J, Lei Y, et al. 2011. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol 12: R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Galka M, Mori E, Liu X, Lin YF, Wei R, Pittock P, Voss C, Dhami G, Li X, et al. 2013. A method for systematic mapping of protein lysine methylation identifies functions for HP1β in DNA damage response. Mol Cell 50: 723–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. 2010. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet 11: 285–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KE, Carlson SM, Camp ND, Cheung P, James RG, Chua KF, Wolf-Yadlin A, Gozani O. 2013. A general molecular affinity strategy for global detection and proteomic analysis of lysine methylation. Mol Cell 50: 444–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikori S, Hattori T, Fuchs SM, Yasui N, Wojcik J, Koide A, Strahl BD, Koide S. 2012. Broad ranges of affinity and specificity of anti-histone antibodies revealed by a quantitative peptide immunoprecipitation assay. J Mol Biol 424: 391–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel DJ, Wang Z. 2013. Readout of epigenetic modifications. Annu Rev Biochem 82: 81–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peach SE, Rudomin EL, Udeshi ND, Carr SA, Jaffe JD. 2012. Quantitative assessment of chromatin immunoprecipitation grade antibodies directed against histone modifications reveals patterns of co-occurring marks on histone protein molecules. Mol Cell Proteomics 11: 128–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, O’Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, et al. 2001. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107: 323–337 [DOI] [PubMed] [Google Scholar]
- Peters AH, Mermoud JE, O’Carroll D, Pagani M, Schweizer D, Brockdorff N, Jenuwein T. 2002. Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat Genet 30: 77–80 [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez F, Dundar F, Diehl S, Gruning BA, Manke T. 2014. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42: W187–W191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathert P, Dhayalan A, Murakami M, Zhang X, Tamas R, Jurkowska R, Komatsu Y, Shinkai Y, Cheng X, Jeltsch A. 2008. Protein lysine methyltransferase G9a acts on non-histone targets. Nat Chem Biol 4: 344–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, Barsyte-Lovejoy D, Martinez JY, Bedford MT, Fuchs SM, et al. 2012. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat Struct Mol Biol 19: 1155–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechter D, Dormann HL, Allis CD, Hake SB. 2007. Extraction, purification and analysis of histones. Nat Protoc 2: 1445–1457 [DOI] [PubMed] [Google Scholar]
- Shin H, Liu T, Manrai AK, Liu XS. 2009. CEAS: cis-regulatory element annotation system. Bioinformatics 25: 2605–2606 [DOI] [PubMed] [Google Scholar]
- Su Z, Boersma MD, Lee JH, Oliver SS, Liu S, Garcia BA, Denu JM. 2014. ChIP-less analysis of chromatin states. Epigenetics Chromatin 7: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suva ML, Riggi N, Bernstein BE. 2013. Epigenetic reprogramming in cancer. Science 339: 1567–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, et al. 2011. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146: 1016–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. 2007. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol 14: 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye T, Krebs AR, Choukrallah MA, Keime C, Plewniak F, Davidson I, Tora L. 2011. seqMINER: an integrated ChIP-seq data interpretation platform. Nucleic Acids Res 39: e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. 2009. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics 25: 1952–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jurkowska R, Soeroes S, Rajavelu A, Dhayalan A, Bock I, Rathert P, Brandt O, Reinhardt R, Fischle W, et al. 2010. Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Res 38: 4246–4253 [DOI] [PMC free article] [PubMed] [Google Scholar]