Abstract
For longitudinal data, the modeling of a correlation matrix R can be a difficult statistical task due to both the positive definite and the unit diagonal constraints. Because the number of parameters increases quadratically in the dimension, it is often useful to consider a sparse parameterization. We introduce a pair of prior distributions on the set of correlation matrices for longitudinal data through the partial autocorrelations (PACs), each of which vary independently over [−1,1]. The first prior shrinks each of the PACs toward zero with increasingly aggressive shrinkage in lag. The second prior (a selection prior) is a mixture of a zero point mass and a continuous component for each PAC, allowing for a sparse representation. The structure implied under our priors is readily interpretable for time-ordered responses because each zero PAC implies a conditional independence relationship in the distribution of the data. Selection priors on the PACs provide a computationally attractive alternative to selection on the elements of R or R−1 for ordered data. These priors allow for data-dependent shrinkage/selection under an intuitive parameterization in an unconstrained setting. The proposed priors are compared to standard methods through a simulation study and a multivariate probit data example. Supplemental materials for this article (appendix, data, and R code) are available online.
Keywords: Bayesian methods, Correlation matrix, Longitudinal data, Multivariate probit, Partial autocorrelation, Selection priors, Shrinkage
1. Introduction
Determining the structure of an unknown J × J covariance matrix Σ is a long standing statistical challenge, including in settings with longitudinal data. A key difficulty in dealing with the covariance matrix is the positive definiteness constraint. This is because the set of values for a particular element σij that yield a positive definite Σ depends on the choice of the remaining elements of Σ. Additionally, because the number of parameters in Σ is quadratic in the dimension J, methods to find a parsimonious (lower-dimensional) structure can be beneficial.
One of the earliest attempts in this direction is the idea of covariance selection (Dempster, 1972). By setting some of the off-diagonal elements of the concentration matrix Ω = Σ−1 to zero, a more parsimonious choice for the covariance matrix of the random vector Y is achieved. A zero in the (i, j)-th position of Ω implies zero correlation (and further, independence under multivariate normality) between Yi and Yj, conditional on the remaining components of Y. This property, along with its relation to graphical model theory (e.g., Lauritzen, 1996), has led to the use of covariance selection as a standard part of analysis in multivariate problems (Wong et al., 2003; Yuan and Lin, 2007; Rothman et al., 2008). However, one should be cautious when using such selection methods as not all produce positive definite estimators. For instance, thresholding the sample covariance (concentration) matrix will not generally be positive definite, and adjustments are needed (Bickel and Levina, 2008).
Model specification for Σ may depend on a correlation structure through the so-called separation strategy (Barnard et al., 2000). The separation strategy involves reparameterizing Σ by Σ = SRS, with S a diagonal matrix containing the marginal standard deviations of Y and R the correlation matrix. Let
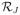 denote the set of valid correlation matrices, that is, the collection of J × J positive definite matrices with unit diagonal. Separation can also be performed on the concentration matrix, Ω = TCT so that T is diagonal and C ∈
denote the set of valid correlation matrices, that is, the collection of J × J positive definite matrices with unit diagonal. Separation can also be performed on the concentration matrix, Ω = TCT so that T is diagonal and C ∈
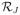 . The diagonal elements of T give the partial standard deviations, while the elements cij of C are the (full) partial correlations. The covariance selection problem is equivalent to choosing elements of the partial correlation matrix C to be null. Several authors have constructed priors to estimate Σ by allowing C to be a sparse matrix (Wong et al., 2003; Carter et al., 2011).
. The diagonal elements of T give the partial standard deviations, while the elements cij of C are the (full) partial correlations. The covariance selection problem is equivalent to choosing elements of the partial correlation matrix C to be null. Several authors have constructed priors to estimate Σ by allowing C to be a sparse matrix (Wong et al., 2003; Carter et al., 2011).
In many cases the full partial correlation matrix may not be convenient to use. In cases where the covariance matrix is fixed to be a correlation matrix such as the multivariate probit case, the elements of the concentration matrix T and C are constrained to maintain a unit diagonal for Σ (Pitt et al., 2006). Additionally, interpretation of parameters in the partial correlation matrix can be challenging, particularly for longitudinal settings as the partial correlations are defined conditional on future values. For example, c12 gives the correlation between Y1 and Y2 conditional on the future measurements Y3, …, YJ. An additional issue with Bayesian methods that promote sparsity in C is calculating the volume of the space of correlation matrices with a fixed zero pattern; see Section 4.2 for details.
In addition to the role R plays in the separation strategy, in some data models the covariance matrix is constrained to be a correlation matrix for identifiability. This is the case for the multivariate probit model (Chib and Greenberg, 1998), Gaussian copula regression (Pitt et al., 2006), certain latent variables models (e.g. Daniels and Normand, 2006), among others. Thus, it is necessary to develop methods specific for estimating and/or modeling a correlation matrix.
We consider this problem of correlation matrix estimation in a Bayesian context where we are concerned with choices of an appropriate prior distribution p(R) on
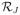 . Commonly used priors include a uniform prior over
. Commonly used priors include a uniform prior over
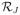 (Barnard et al., 2000) and Jeffrey’s prior p(R) ∝ |R|−(J+1)/2. In these cases the sampling steps for R can sometimes benefit from parameter expansion techniques (Liu, 2001; Zhang et al., 2006; Liu and Daniels, 2006). Liechty et al. (2004) develop a correlation matrix prior by specifying each element ρij of R as an independent normal subject to R ∈
(Barnard et al., 2000) and Jeffrey’s prior p(R) ∝ |R|−(J+1)/2. In these cases the sampling steps for R can sometimes benefit from parameter expansion techniques (Liu, 2001; Zhang et al., 2006; Liu and Daniels, 2006). Liechty et al. (2004) develop a correlation matrix prior by specifying each element ρij of R as an independent normal subject to R ∈
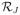 . Pitt et al. (2006) extend the covariance selection prior (Wong et al., 2003) to the correlation matrix case by fixing the elements of T to be constrained by C so that T is the diagonal matrix such that R = (TCT)−1 has unit diagonal.
. Pitt et al. (2006) extend the covariance selection prior (Wong et al., 2003) to the correlation matrix case by fixing the elements of T to be constrained by C so that T is the diagonal matrix such that R = (TCT)−1 has unit diagonal.
The difficulty of jointly dealing with the positive definite and unit diagonal constraints of a correlation matrix has led some researchers to consider priors for R based on the partial autocorrelations (PACs) in settings where the data are ordered. PACs suggest a practical alternative by avoiding the complication of the positive definite constraint, while providing easily interpretable parameters (Joe, 2006). Kurowicka and Cooke (2003, 2006) frame the PAC idea in terms of a vine graphical model. Daniels and Pourahmadi (2009) construct a flexible prior on R through independent shifted beta priors on the PACs. Wang and Daniels (2013a) construct underlying regressions for the PACs, as well as a triangular prior which shifts the prior weight to a more intuitive choice in the case of longitudinal data. Instead of setting partial correlations from C to zero to incorporate sparsity, our goal is to encourage parsimony through the PACs. As the PACs are unconstrained, selection does not lead to the computational issues associated with finding the normalizing constant for a sparse C. We introduce and compare priors for both selection and shrinkage of the PACs that extends previous work on sensible default choices (Daniels and Pourahmadi, 2009).
The layout of this article is as follows. In the next section we will review the relevant details of the partial autocorrelation parameterization. Section 3 proposes a prior for R induced by shrinkage priors on the PACs. Section 4 introduces the selection prior for the PACs. Simulation results showing the performance of the priors appear in Section 5. In Section 6 the proposed PAC priors are applied to a data set from a smoking cessation clinical trial. Section 7 concludes the article with a brief discussion.
2. Partial autocorrelations
For a general random vector Y = (Y1,…, YJ)′ the partial autocorrelation between Yi and Yj (i < j) is the correlation between the two given the intervening variables (Yi+1, …, Yj−1). We denote this PAC by πij, and let Π be the upper-triangular matrix with elements πij. Because the PACs are formed by conditioning on the intermediate components, there is a clear dependence on the ordering of the components of Y. In many applications such as longitudinal data modeling, there is a natural time ordering to the components. With an established ordering of the elements of Y, we refer to the lag between Yi and Yj as the time-distance j − i between the two.
We now describe the relationship between R and Π. For the lag-1 components (j − i = 1) πij = ρij since there are no components between Yi and Yj. The higher lag components are calculated from the formula (Anderson, 1984, Section 2.5),
| (1) |
where , and R3(i, j) is the sub-correlation matrix of R corresponding to the variables (Yi+1,…, Yj−1). The scalars rl (l = 1, 2) are . Equivalent to (1), we may define the partial autocorrelation in terms of the distribution of the (mean zero) variable Y. Let Ỹ = (Yi+1,…, Yj−1)′ be the vector (possibly empty or scalar) of the intermediate responses, and and be the linear least squares predictors of Yi and Yj given Ỹ, respectively. Then, , and it is reasonable to consider πij to define the correlation between Yi and Yj after correcting for Ỹ.
Examination of formula (1) shows that the operation from R to Π is invertible. By inverting the previous operations recursively over increasing lag j − i, one obtains the correlation matrix from the PACs by ρi,i+1 = πi,i+1 and
for j − i > 1. As the relationship between R and Π is one-to-one, the Jacobian for the transformation from R to Π can be computed easily. The determinant of the Jacobian is given by
| (2) |
(Joe, 2006, Theorem 4). Notationally, we let R(Π) denote correlation matrix corresponding to the PACs Π. Similarly, Π(R) represents the set of PACs corresponding to correlation matrix R. When it is clear from context, we continue to use only the matrix R or Π and not the functional notation.
The key advantage in using PACs is that parameters are unconstrained (Joe, 2006). For the correlation matrix R, the subset of values in (−1, 1) that ρij can take satisfying the positive definite constraint is determined by the configuration of the other elements of R. For a geometric interpretation of this phenomenon, see Rousseeuw and Molenberghs (1994). For the PACs, each πij can take any value in (−1, 1), regardless of the choice of the remaining π’s. This is especially important in the selection context, as setting certain elements of R (or the partial correlation matrix C) to zero can greatly restrict the sets of values that yield a positive definite matrix for other elements in R (C).
Define SBeta(α, β) to be the beta distribution shifted to the support (−1, 1), i.e., the density proportional to (1 + y)α−1(1 − y)β−1 for y ∈ (−1, 1). Daniels and Pourahmadi (2009) use the PACs to form a prior on R by letting each πij come from this shifted beta distribution where the two shape parameters depend on the lag j − i, with the special case where each πij ~ SBeta(1, 1). We call this the flat-PAC (or flat-Π) prior since it specifies a uniform distribution for each of the PACs. Wang and Daniels (2013a) advise using a triangular prior with SBeta(2,1) which (weakly) encourages positive values for the PACs.
The result in (2) shows that we can write the flat prior of Barnard et al. (2000) in terms of a prior on the PACs. We call the prior pfR(R) ∝ I(R ∈
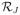 ) the flat-R prior since it is uniform over the space
) the flat-R prior since it is uniform over the space
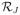 . Hence, the flat-R is equal to pfR(Π) ∝ |J(Π)|−1, which has a contribution from πij of
. Note that pfR(Π) is the product of independent SBeta(αij, βij) distributions for each πij, where αij = βij = 1+[J −1−(j −i)]/2. This provides an unconstrained representation of the flat-R prior.
. Hence, the flat-R is equal to pfR(Π) ∝ |J(Π)|−1, which has a contribution from πij of
. Note that pfR(Π) is the product of independent SBeta(αij, βij) distributions for each πij, where αij = βij = 1+[J −1−(j −i)]/2. This provides an unconstrained representation of the flat-R prior.
In longitudinal/ordered data contexts, we expect the PACs to be negligible for elements that have large lags. We exploit this concept via two types of priors. First, we introduce priors that shrink PACs toward zero with the aggressiveness of the shrinkage depending on the lag. Next, we propose, in the spirit of Wong et al. (2003), a selection prior that will stochastically choose PACs to be set to zero.
3. Partial autocorrelation shrinkage priors
3.1. Specification of the shrinkage prior
Using the PAC framework, we form priors that will shrink the PAC πij toward zero. It has long been known that shrinkage estimators can produce greatly improved estimation (James and Stein, 1961). As previously noted, πij = 0 implies that Yi and Yj are uncorrelated given the intervening variables (Yi+1, …, Yj−1). In the case where Y has a multivariate normal distribution, this implies independence between Yi and Yj, given (Yi+1, …, Yj−1). We anticipate that variables farther apart in time (and conditional on more intermediate variables) are more likely to be uncorrelated, so we will more aggressively shrink πij for larger values of the lag j − i.
We let each πij ~ SBeta(αij, βij) independently. As we wish to shrink toward zero, we want E{πij} = 0, so we fix αij = βij. It is easily shown that
which we denote by ξij. We recover the SBeta shape parameters by . Hence, the distribution of πij is determined by its variance ξij. Rather than specifying these J(J − 1)/2 different variances, we parameterize them through
| (3) |
where ε0 ∈ (0, 1) and γ > 0. Clearly, ξij is decreasing in lag so that higher lag terms will generally be closer to zero. We let the positive γ parameter determine the rate that ξij decreases in lag.
To fully specify the Bayesian set-up, we must introduce prior distributions on the two parameters, ε0 and γ. To specify these hyperpriors, we use a uniform (or possibly a more general beta) for ε0 and a gamma distribution for γ. We require γ > 0, so ξij = ε0|j − i|−γ remains an decreasing function of lag. In the simulations and data analysis of Sections 5 and 6, we use γ ~ Gamma(5,5), so that γ has a prior mean of 1 and prior variance of 1/5. We use a moderately informative prior to keep γ from dominating the role of ε0 in ξij = ε0|j − i|−γ. A large value of γ will force all ξij of lag greater than one to be approximately zero, regardless of the value of ε0.
3.2. Sampling under the shrinkage prior
The utility of our prior depends on our ability to incorporate it into a Markov chain Monte Carlo (MCMC) scheme. For simplicity we assume that the data consists of Y1, …, YN, where each Yi is a J-dimensional normal vector with mean zero and covariance R, which is a correlation matrix so as to mimic the computations for the multivariate probit case. Let
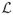 (Π|Y) denote the likelihood function for the data, parameterized by the PACs, Π.
(Π|Y) denote the likelihood function for the data, parameterized by the PACs, Π.
The MCMC chain we propose involves sequentially updating each of the J(J − 1)=2 PACs, followed by updating the hyperparameters determining the variance of the SBeta distributions. To sample a particular πij, we must draw the new value from the distribution proportional to
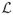 (πij, Π(−ij)|Y) pij(πij), where pij(πij) is the SBeta(αij, βij) density and Π(−ij) represents the set of PACs except πij. Due to the subtle role of πij in the likelihood piece, there is no simple conjugate sampling step. In order to sample from
(πij, Π(−ij)|Y) pij(πij), where pij(πij) is the SBeta(αij, βij) density and Π(−ij) represents the set of PACs except πij. Due to the subtle role of πij in the likelihood piece, there is no simple conjugate sampling step. In order to sample from
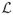 (πij, Π(−ij)|Y) pij(πij), we introduce an auxiliary variable Uij (Damien et al., 1999; Neal, 2003), and note that we can rewrite the conditional distribution as
(πij, Π(−ij)|Y) pij(πij), we introduce an auxiliary variable Uij (Damien et al., 1999; Neal, 2003), and note that we can rewrite the conditional distribution as
| (4) |
suggesting a method to sample πij in two steps. First, sample Uij uniformly over the interval [0,
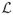 (πij, Π(−ij)|Y)pij(πij)], using the current value of πij. We then draw the new πij uniformly from the slice set
(πij, Π(−ij)|Y)pij(πij)], using the current value of πij. We then draw the new πij uniformly from the slice set
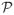 = {π : uij <
= {π : uij <
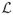 (π, Π(−ij)|Y)pij(π)}. Because this set lies within the compact set [−1, 1],
(π, Π(−ij)|Y)pij(π)}. Because this set lies within the compact set [−1, 1],
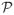 could be calculated numerically to within a prespecified level of accuracy, but this is not generally necessary due to the “stepping out” algorithm of Neal (2003).
could be calculated numerically to within a prespecified level of accuracy, but this is not generally necessary due to the “stepping out” algorithm of Neal (2003).
The variance parameters, ε0 and γ, are not conjugate so sampling new values in the MCMC chain requires a non-standard step. We also update them using the auxiliary variable technique.
4. Partial autocorrelation selection priors
4.1. Specification of the selection prior
Having developed a prior that shrinks the partial autocorrelations toward zero, we now consider prior distributions that give positive probability to the event that the PAC πij is equal to zero. Again, this zero implies that Yi and Yj are uncorrelated given the intervening variables (Yi+1, …, Yj−1) with independence under multivariate normality. The selection priors are formed by independently specifying the prior for each πij as the mixture distribution,
| (5) |
where δ0 represents a degenerate distribution with point mass at zero. In the shrinkage prior we parameterize the shifted beta parameters αij, βij to depend on lag, but here we generally let α = αij and β = βij and incorporate structure through the modeling choices on εij. While there is flexibility to make any choice of these shifted beta parameters α, β, we recommend as default choices either a uniform distribution on [−1, 1] through α = β = 1 (Daniels and Pourahmadi, 2009) or the triangular prior of Wang and Daniels (2013a) by α = 2, β = 1; alternatively, independent hyperpriors for α, β could be specified.
The value of εij gives the probability that πij will be non-zero, i.e. will be drawn from the continuous component in the mixture distribution. Hence, we have the probability that Yi and Yj are uncorrelated, given the interceding variables, is 1 − εij. As the values of the ε’s decrease, the selection prior places more weight on the point-mass δ0 component of the distribution (5), yielding more sparse choices for Π. As with our parameterizations of the variance ξij in Section 3.1, we make a structural choice of the form of εij so that this probability depends on the lag-value. We let
| (6) |
similar to our choice of ξij in the shrinkage prior.
This choice (6) specifies the continuous component probability to be an polynomial function of the lag. Because εij is decreasing as the lag j − i increases, P(πij = 0) increases. Conceptually, this means that we anticipate that variables farther apart in time (and conditional on more intermediate variables) are more likely to be uncorrelated. As with the shrinkage prior, we choose hyperpriors of ε0 ~ Unif(0, 1) and γ ~ Gamma(5,5).
4.2. Normalizing constant for priors on R
One of the key improvements of our selection prior over other sparse priors for R is the simplicity of the normalizing constant, as mentioned in the introduction. Previous covariance priors with a sparse C (Wong et al., 2003; Pitt et al., 2006; Carter et al., 2011) place a flat prior on the non-zero components cij for a given pattern of zeros. However, the needed normalizing constant requires finding the volume of the subspace of
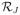 corresponding to the pattern of zeros in C. This turns out to be a quite difficult task and provides much of the challenge in the work of the three previously cited papers.
corresponding to the pattern of zeros in C. This turns out to be a quite difficult task and provides much of the challenge in the work of the three previously cited papers.
We are able to avoid this issue by specifying our selection prior in terms of the unrestricted PAC parameterization. As the value of any of the πij’s does not effect the support of the remaining PACs, the volume of [−1, 1]J(J−1)/2 corresponding to any configuration of Π with J0 (≤ J(J − 1)/2) non-zero elements is 2J0, the volume of a J0-dimensional hypercube. Because this constant does not depend on which elements are non-zero, we need not explicitly deal with it in the MCMC algorithm to be introduced in the next subsection. Further, we are able the exploit structure in the order of the PACs in selection (i.e. higher lag terms are more likely to be null), whereas in Pitt et al. (2006), the probability that cij is zero is chosen to minimize the effort required to find the normalizing constant.
An additional benefit of performing selection on the partial autocorrelation as opposed to the partial correlations C is that the zero patterns hold under marginalizations of the beginning and/or ending time points. For instance, if we marginalize out the Jth time point, the corresponding matrix of PACs is the original Π after removing the last row and column. However, any zero elements in C will not be preserved because corr(Y1, Y2|Y3, …, YJ) = 0 does not generally imply that corr(Y1, Y2|Y3, …, YJ−1) = 0.
4.3. Sampling under the selection prior
Sampling with the selection prior proceeds similarly to the shrinkage prior scheme with the main difference being the introduction of the point mass in (5). As before we sequentially update each of the PACs, by drawing the new value from the distribution proportional to
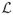 (πij, Π(−ij)|Y) pij(πij), where pij(πij) gives the density corresponding the prior distribution in (5) (with respect to the appropriate mixture dominating measure). We cannot use the slice sampling step according to (4) but must write the distribution as
(πij, Π(−ij)|Y) pij(πij), where pij(πij) gives the density corresponding the prior distribution in (5) (with respect to the appropriate mixture dominating measure). We cannot use the slice sampling step according to (4) but must write the distribution as
| (7) |
For the selection prior, we sample Uij uniformly over the interval from zero to
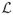 (πij, Π(−ij)|Y), using the current value of πij, and then draw πij from pij(·), restricted to the slice set
(πij, Π(−ij)|Y), using the current value of πij, and then draw πij from pij(·), restricted to the slice set
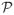 = {π: uij <
= {π: uij <
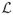 (π, Π(−ij)|Y)}.
(π, Π(−ij)|Y)}.
To sample from pij(·) restricted to
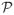 , let F(x) = P(πij ≤ x) denote the (cumulative) distribution function for the prior (5) of πij. Note that F (x) is available in closed form when the SBeta distribution is uniform or triangular. We then draw a random variable Z uniformly over the set F(
, let F(x) = P(πij ≤ x) denote the (cumulative) distribution function for the prior (5) of πij. Note that F (x) is available in closed form when the SBeta distribution is uniform or triangular. We then draw a random variable Z uniformly over the set F(
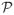 ) ⊂ [0, 1], and the updated value of πij is F−1(Z) = inf{π : F(π) ≥ Z}. This is simply a version of the probability integral transform. It is relatively straight-forward to verify that sampling according to (7) instead of (4) using the “stepping out” algorithm of Neal (2003) leaves the stationary distribution invariant.
) ⊂ [0, 1], and the updated value of πij is F−1(Z) = inf{π : F(π) ≥ Z}. This is simply a version of the probability integral transform. It is relatively straight-forward to verify that sampling according to (7) instead of (4) using the “stepping out” algorithm of Neal (2003) leaves the stationary distribution invariant.
The similarity between the sampling steps for the shrinkage and selection priors is notable. Consider the situation when the parameter of concern is the vector of regression coefficients for a linear regression model. With a shrinkage prior these regression coefficients may be drawn simultaneously. But when using a selection prior, each coefficient must be sampled one at a time, and each step requires finding the posterior probability it should be set to zero. For linear models the computational effort required for selection is often much greater than under shrinkage.
In the PAC context, this is not the case. We cannot update the PACs in blocks under the shrinkage prior, so there is no computational benefit relative to selection. Because we sample from the probability integral transform restricted to
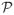 , there is also no need to compute the posterior probability that the parameter is selected. Hence, the computational effort for the shrinkage and selection is roughly equivalent. Finally, with the exception of the minor step of updating the hyperparameters, the non-sparse flat-Π and triangular priors also require a similar level of computational time as the selection and shrinkage priors.
, there is also no need to compute the posterior probability that the parameter is selected. Hence, the computational effort for the shrinkage and selection is roughly equivalent. Finally, with the exception of the minor step of updating the hyperparameters, the non-sparse flat-Π and triangular priors also require a similar level of computational time as the selection and shrinkage priors.
To sample the parameters ε0 and γ defining the mixing proportions εij, we introduce the set of dummy variable ζij = I(πij ≠ 0), which have the property that P(ζij = 1) = εij. The sampling distributions of ε0 and γ depend on Π only through the set of indicator variables ζij. As with the variance parameters of the shrinkage priors, we incorporate a pair of slice sampling steps to update the hyperparameters.
5. Simulations
To better understand the behavior of our proposed priors, we conducted a simulation study to assess the (frequentist) risk of their posterior estimators. We consider four choices A–D for the true covariance matrix in the case of six-dimensional (J = 6) data. RA will have an autoregressive (AR) structure with . The corresponding ΠA has values of 0.7 for the lag-1 terms and zero for the others, a sparse parameterization. For the second correlation matrix RB we choose the identity matrix so that all of PACs are zero in this case. The ΠC has a structure that decays to zero. For the lag-1 terms , and for the remaining terms, , j − i > 1. Neither ΠC nor RC have zero elements, but decrease quickly in lag j − i. Finally, we consider a correlation matrix that comes from a sparse ΠD,
where the upper-triangular elements correspond to ΠD and the lower-triangular elements depict the marginal correlations from RD. Note that while ΠD is somewhat sparse, RD has only non-zero elements.
For each of these four choices of the true dependence structure and for sample sizes of N = 20, 50, and 200, we simulate 100 datasets. For each dataset a posterior sample for Π (and hence, R) is obtained by running an MCMC chain for 5000 iterations, after a burn-in of 1000. We use every tenth iteration for inference, giving a sample of 500 values for each dataset. We consider the performance of both the selection and shrinkage priors on Π. For the selection prior, we perform analyses with SBeta(1, 1) (i.e., Unif(−1; 1)) and SBeta(2, 1) (triangular prior) for the continuous component of the mixture distributions (5). In both the selection and shrinkage priors, the hyperpriors are ε0 ~ Unif(0, 1) and γ ~ Gamma(5,5). The estimators from the shrinkage and selection priors are compared with the estimators resulting from the flat-R, flat-PAC, and triangular priors. Finally, we consider a naive shrinkage prior where γ is fixed at zero in (3). Here, all PACs are equally shrunk with variance ξij = ε0 independently of the lag.
We consider two loss functions in comparing the performance of the seven prior choices: L1(R̂, R) = tr(R̂ R−1) − log |R̂ R−1| − p and L2(Π̂, Π) = Σi<j(π̂ij − πij)2. The first loss function is the standard covariance log-likelihood loss (Yang and Berger, 1994), whose Bayes estimator is E{R−1}−1. Because this quantity generally does not have a unit diagonal, we use R̂1= S E{R−1}−1 S, where S = [diag(E{R−1})]1/2 is the diagonal matrix that guarantees R̂1 is a correlation matrix. The Bayes estimator for L2 is R̂2 = R (E{Π}), the correlation matrix corresponding to the posterior mean of Π.
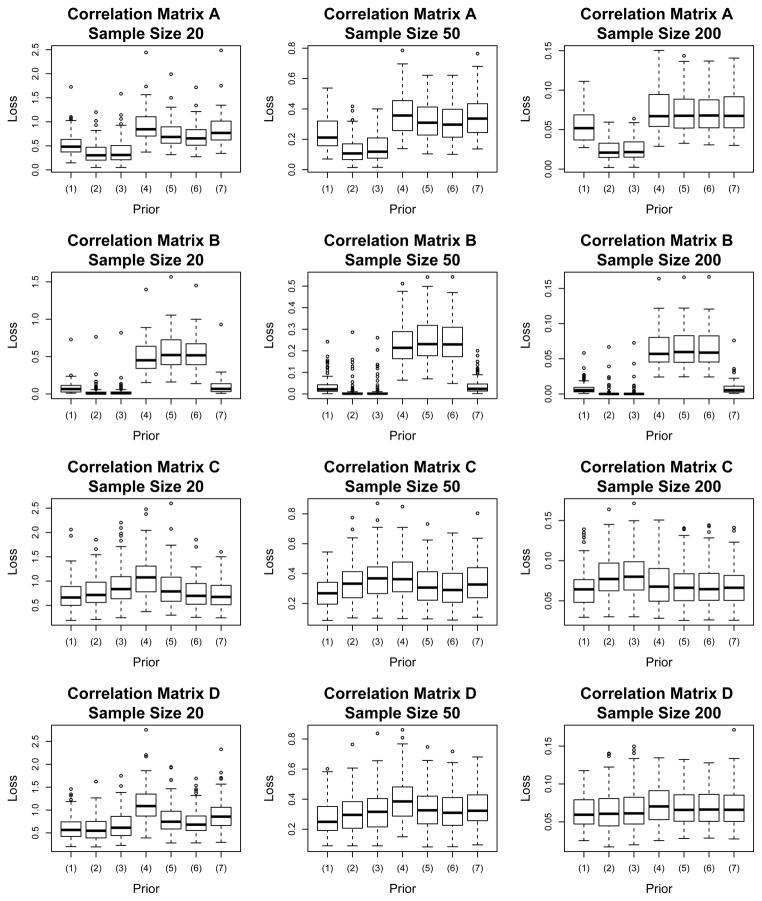
We estimate the frequentist risk for Rk, k ∈ {A, B, C, D}, by averaging the loss over the 100 datasets. Table 1 contains the estimated risk by loss function, prior choice, sample size, and true correlation matrix. When evaluating the risk for loss function l, we are using the estimator R̂l for l = 1, 2. Figure 1 contains the box plots of the observed losses for L1 with R̂1. Plots using loss function 2 look similar and have been excluded for brevity. The Monte Carlo standard errors for the risk estimates are contained in the online supplementary materials.
Table 1.
Risk estimates for simulation study with dimension J = 6. Correlation matrices: A autoregressive structure; B independence; C non-zero decaying; D sparse. Loss functions: L1(R̂, R) = tr(R̂ R−1) − log |R̂ R−1| − p; L2(Π̂, Π) = Σi<j(π̂ij − πij)2.
| R | N | Loss Fcn | Risk Estimates by Prior | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Shrinkage | Selection (2,1) | Selection (1,1) | flat-R | flat-Π | Triangular | Naive Shrink | |||
| A | 20 | 1 | 0.54 | 0.36 | 0.40 | 0.91 | 0.75 | 0.70 | 0.84 |
| A | 50 | 1 | 0.24 | 0.13 | 0.15 | 0.37 | 0.32 | 0.32 | 0.35 |
| A | 200 | 1 | 0.055 | 0.025 | 0.025 | 0.074 | 0.072 | 0.071 | 0.073 |
| B | 20 | 1 | 0.087 | 0.031 | 0.029 | 0.49 | 0.56 | 0.54 | 0.100 |
| B | 50 | 1 | 0.038 | 0.015 | 0.014 | 0.231 | 0.250 | 0.249 | 0.039 |
| B | 200 | 1 | 0.008 | 0.002 | 0.002 | 0.064 | 0.065 | 0.065 | 0.009 |
| C | 20 | 1 | 0.73 | 0.79 | 0.93 | 1.10 | 0.87 | 0.76 | 0.73 |
| C | 50 | 1 | 0.28 | 0.34 | 0.37 | 0.38 | 0.33 | 0.31 | 0.35 |
| C | 200 | 1 | 0.066 | 0.082 | 0.084 | 0.073 | 0.070 | 0.070 | 0.069 |
| D | 20 | 1 | 0.62 | 0.61 | 0.68 | 1.13 | 0.83 | 0.75 | 0.90 |
| D | 50 | 1 | 0.28 | 0.30 | 0.32 | 0.40 | 0.34 | 0.33 | 0.35 |
| D | 200 | 1 | 0.063 | 0.064 | 0.066 | 0.073 | 0.071 | 0.070 | 0.071 |
|
| |||||||||
| A | 20 | 2 | 0.26 | 0.13 | 0.14 | 0.52 | 0.46 | 0.44 | 0.48 |
| A | 50 | 2 | 0.13 | 0.039 | 0.045 | 0.23 | 0.21 | 0.20 | 0.22 |
| A | 200 | 2 | 0.035 | 0.0057 | 0.0059 | 0.052 | 0.051 | 0.051 | 0.051 |
| B | 20 | 2 | 0.070 | 0.019 | 0.017 | 0.41 | 0.47 | 0.45 | 0.077 |
| B | 50 | 2 | 0.034 | 0.012 | 0.011 | 0.210 | 0.227 | 0.227 | 0.035 |
| B | 200 | 2 | 0.008 | 0.002 | 0.002 | 0.063 | 0.064 | 0.063 | 0.008 |
| C | 20 | 2 | 0.39 | 0.44 | 0.51 | 0.65 | 0.52 | 0.44 | 0.42 |
| C | 50 | 2 | 0.15 | 0.20 | 0.22 | 0.22 | 0.19 | 0.18 | 0.20 |
| C | 200 | 2 | 0.040 | 0.055 | 0.057 | 0.044 | 0.044 | 0.043 | 0.043 |
| D | 20 | 2 | 0.30 | 0.29 | 0.32 | 0.56 | 0.47 | 0.42 | 0.47 |
| D | 50 | 2 | 0.15 | 0.17 | 0.18 | 0.22 | 0.20 | 0.19 | 0.20 |
| D | 200 | 2 | 0.037 | 0.037 | 0.039 | 0.044 | 0.043 | 0.043 | 0.044 |
Figure 1.
Box plots of the observed loss using L1 (R̂1, R) for the J = 6 cases. The prior distributions compared are (1) shrinkage, (2) selection (2,1), (3) selection (1,1), (4) flat-R, (5) flat-Π, (6) triangular, and (7) naive shrinkage.
It is immediately clear that the shrinkage and selection priors dominate the two flat priors for correlation matrices A and B. These are the matrices that have the most sparsity. From the box plots we see the losses for the middle 50% of datasets for the selection priors fall completely below the middle 50% for the four competitors. For RA we see risk reductions between 29 and 60% for the sparse estimators over the estimators from the flat priors with N = 20; for N = 200 the improvements range from 24 to 66%. In the independence case, the estimators from the shrinkage and selection priors outperform the flat estimators by margins between 82 and 97%. While our focus is mainly on the comparison of the sparse priors to the others, we note that generally the triangular and flat-Π choices are best among the four competitors, with the naive shrinkage prior performing quite well for RB.
For ΠC all of the seven prior choices perform comparably. From Figure 1 we see that the middle 50% of the losses fall in the same range for each of the sample sizes. For all sample sizes the shrinkage prior is (slightly) favored, and for N = 20 the estimated risk for flat-R is visibly worse than the others. Recall that is decreasing in lag but is not equal to zero. In fact, the smallest element which may not be close enough to zero to be effectively zeroed out, explaining why the selection priors are less effective for ΠC than in the other scenarios.
When we consider estimating the sparse correlation matrix ΠD, the shrinkage and selection priors outperform the four other priors. From Table 1 we see that for loss function 1 and the N = 20 sample size the estimated risk decreases by 45 (25), 46 (27) and 40 (18) percent for the estimates from the shrinkage, selection (2,1), and selection (1,1) priors over the flat-R (flat-Π) priors. This is quite a substantial drop for the small sample size. For the other sample sizes we still observed a clear decrease over the flat priors. For N = 50 there is a drop of 30 (19), 24 (11), and 19 (5) percent for the sparse priors over the flat priors, and with N = 200 a decrease of 13 (10), 12 (9), and 10 (6) percent.
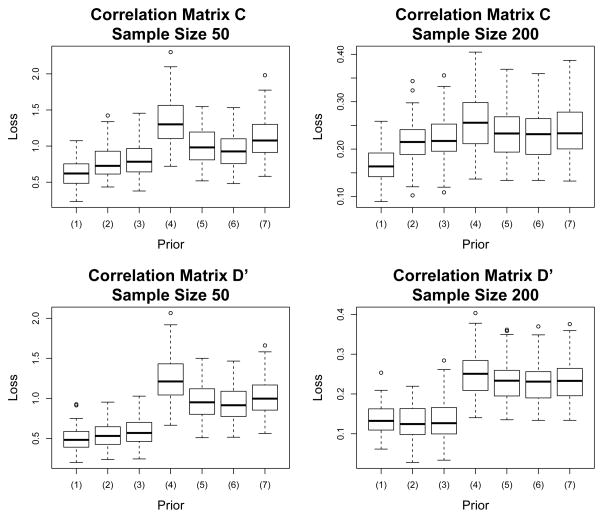
To investigate how our priors behave as J increases, we repeat the analysis using the non-sparse decaying RC and a sparse RD′ with the dimension of the matrix increased to J = 10. Again, for the lag-1 terms and for all j − i > 1, and we expand the previous RD to the 10 × 10 RD′ shown in Table 2. As before the above diagonal elements are from ΠD′ and the below diagonal elements from the corresponding RD′. ΠD′ is very sparse, while RD′ has no zero elements. We consider sample sizes of 50 and 200. Risk estimates and box plots for this simulation are displayed in Table 3 and Figure 2.
Table 2.
10 × 10 PAC matrix ΠD′ shown above the diagonal and its respective correlation matrix RD′ shown below the diagonal.
|
|
Table 3.
Risk estimates for simulation study with dimension J = 10. Correlation matrices: C nonzero decaying; D′ sparse. Loss functions: L1(R̂, R) = tr(R̂ R−1) − log |R̂ R−1| − p; L2(Π̂, Π) = Σi<j(π̂ij − πij)2.
| R | N | Loss Fcn | Risk Estimates by Prior | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Shrinkage | Selection (2,1) | Selection (1,1) | flat-R | flat-Π | Triangular | Naive Shrink | |||
| C | 50 | 1 | 0.63 | 0.77 | 0.83 | 1.33 | 1.00 | 0.94 | 1.12 |
| C | 200 | 1 | 0.165 | 0.215 | 0.221 | 0.254 | 0.232 | 0.229 | 0.238 |
| D′ | 50 | 1 | 0.49 | 0.54 | 0.59 | 1.26 | 0.97 | 0.93 | 1.03 |
| D′ | 200 | 1 | 0.134 | 0.130 | 0.133 | 0.250 | 0.230 | 0.227 | 0.235 |
|
| |||||||||
| C | 50 | 2 | 0.38 | 0.48 | 0.51 | 0.88 | 0.72 | 0.67 | 0.75 |
| C | 200 | 2 | 0.110 | 0.155 | 0.159 | 0.184 | 0.174 | 0.171 | 0.174 |
| D′ | 50 | 2 | 0.27 | 0.31 | 0.34 | 0.77 | 0.69 | 0.66 | 0.69 |
| D′ | 200 | 2 | 0.085 | 0.080 | 0.082 | 0.184 | 0.178 | 0.175 | 0.178 |
Figure 2.
Box plots of the observed loss using L1(R̂1,R) for J = 10. The prior distributions compared are (1) shrinkage, (2) selection (2,1), (3) selection (1,1), (4) flat-R, (5) flat-Π, (6) triangular, and (7) naive shrinkage.
From both Table 3 and Figure 2 it is clear that estimation of the correlation matrix is greatly improved under the sparse priors. In the simulations of both dimensions we find that the estimators from the triangular selection prior tend to be slightly better than the selection prior with SBeta(1,1). With the sparse correlation matrix RD′ the risk under the sparse priors are about half of the risk of the flat prior under both sample sizes. Recall that ΠC is not sparse but has elements which decay exponentially. Because many of the large lag components are very small, the selection priors provide stability by explicitly zeroing many of these out. For the larger sample size, the flat priors do comparatively better although still worse than the sparse priors.
We have demonstrated that the sparse priors yield improved estimation of the correlation matrix in a variety of data situations. In order to investigate the performance in the standard situation where the true dependence structure is unknown, we apply the sparsity and shrinkage priors to a data set obtained from a smoking cessation clinical trail.
6. Data analysis
The first Commit to Quit (CTQ I) study (Marcus et al., 1999) was a clinical trial designed to encourage women to stop smoking. As weight gain is often a viewed as a factor decreasing the effectiveness of smoking cessation programs, a treatment involving an exercise regimen is utilized to try to increase the quit rate. The control group received an educational intervention of equal time. The study ran for twelve weeks, and patients were encouraged to quit smoking at week 5. As the study required a significant time commitment (three exercise/educational sessions a week), there is substantial missingness due to study dropout. As in previous analyses of this data (Daniels and Hogan, 2008), we assume this missingness is ignorable.
For patient i = 1,…, N (N = 281), we denote the vector of quit statuses by Qi = (Qi1,…, QiJ)′. We only consider the responses after patients are asked to quit, weeks 5 through 12 (J = 8). Here Qit = 1 indicates a success (not smoking) for patient i at time t (1 ≤ t ≤ J, corresponding to week t + 4), Qit = −1 for a failure (smoking during the week), and Qit = 0 if the observation is missing. Following the usual conventions of the multivariate probit regression model (Chib and Greenberg, 1998), we let Yi be the J-dimensional vector of latent variables corresponding to Qi. Thus, Qit = 1 implies that Yit ≥ 0, and Qit = −1 gives Yit < 0. When Qit = 0, the sign of Yit represents the (unobserved) quit status for the week.
We assume the latent variables follow a multivariate normal distribution Yi ~ NJ (μi, R) for i = 1,…, N, where μi = Xiβ, Xi is a J × q matrix of covariates and β a q-vector of regression coefficients. As the scale of Y is unidentified, the covariance matrix of Y is constrained to be a correlation matrix R. We consider two choices of Xi: ‘time-varying’ which specifies a different μit for each time within each treatment group (q = 2J) and ‘time-constant’ which gives the same value of μit across all times within treatment group (q = 2).
With the time-constant and time-varying choices of the mean structure, we consider the following priors for R: shrinkage, selection, flat-R, flat-Π, triangular, naive shrinkage, and an autoregressive (AR) prior. The AR prior assumes an AR(1) structure for R, that is, ρij = ρ|j−i| and πi,i+1 = ρ and πij = 0 if |j − i| > 1. We assume a Unif(−1, 1) distribution for ρ. As in the risk simulation, we consider the selection prior with both SBeta(1, 1) and with SBeta(2, 1) for the continuous component. The remaining prior distributions to be specified are ε0 ~ Unif(0, 1), γ ~ Gamma(5, 5), and the prior on the regression coefficients β is flat.
To analyze the data we run an MCMC chain for 12,000 iterations after a burn-in of 3000. There are three sets of parameters to sample in the MCMC chain: the regression coefficients, the correlation matrix, and the latent variables. The conditional for β given Y and R is multivariate normal. Sampling the correlation matrix evolves as discussed in Sections 3.2 and 4.3 using the residuals Yi − μi. The latent variables Yi, which are constrained by Qi, are sampled according to the strategy of Liu et al. (2009, Proposition 1). With the shrinkage prior the autocorrelation of all PACs was less than 0.1 within 20 iterations. With the higher lag terms of the selection prior, the autocorrelation does not decrease as quickly due to discrete component of the distribution (πij may be equal to zero for many iterations), but the lag 1 and 2 terms also have autocorrelations less than 0.1 in 20 iterations. Based on these autocorrelation values, we retain every tenth iteration of each chain to use for inference. Trace plots and other graphical diagnostics further confirm good mixing of the chain.
To compare the specification based on our prior choices, we make use of the deviance information criterion (DIC; Spiegelhalter et al., 2002). The DIC statistic can be viewed similarly to the Bayesian or Akaike information criterion, but the DIC does not require the user to “count” the number of model parameters. This is key for Bayesian models that utilize shrinkage and/or sparsity priors as it is not clear whether or how one should count a parameter that has been set to or shrunk toward zero. To that end, let
| (8) |
be the deviance or twice the negative log-likelihood with the parameters β̂ and R̂. Here β̂ is the posterior mean, and for the correlation estimate R̂, we use the first of the estimators we considered in Section 5, R̂ = S E{R−1}−1 S with S = [diag(E{R−1})]1/2. The complexity of the model is measured by the term pD, sometimes called the effective number of parameters. This pD is calculated as
| (9) |
where the expectation is over the posterior distribution of the parameters (β, R). The DIC model comparison statistics is DIC = Dev + 2pD, the sum of terms measuring model fit and complexity. Smaller values of DIC are preferred.
As Wang and Daniels (2011) point out, the DIC should be calculated using the observed data, which in this case is the quit status responses Qi not the latent variables Yi. Hence the log-likelihood for Qi at parameters (β, R) is equal to
| (10) |
where φ(·|μ, Σ) is the J-dimensional multivariate normal density with mean μ and covariance matrix Σ. The integral in (10) is not tractable but can be estimated using importance sampling (Robert and Casella, 2004, Section 3.3). See the appendix in the online supplementary materials for details about estimating the DIC. The model fit (Dev), complexity (pD), and comparison (DIC) statistics are in Table 4; DIC statistics were estimated with a standard error of approximately 0.5.
Table 4.
Model comparison statistics for the CTQ data.
| Mean Structure | Correlation Prior | Dev | pD | DIC |
|---|---|---|---|---|
| Time-constant | Shrinkage | 1031 | 14 | 1060 |
| Time-constant | Selection (2,1) | 1042 | 12 | 1066 |
| Time-constant | Selection (1,1) | 1044 | 12 | 1068 |
| Time-constant | Triangular | 1029 | 20 | 1068 |
| Time-constant | flat-Π | 1029 | 20 | 1069 |
| Time-constant | Naive shrinkage | 1033 | 20 | 1074 |
| Time-constant | AR | 1071 | 3 | 1078 |
| Time-constant | flat-R | 1043 | 21 | 1086 |
|
| ||||
| Time-varying | Shrinkage | 1022 | 25 | 1071 |
| Time-varying | Triangular | 1017 | 30 | 1077 |
| Time-varying | Selection (2,1) | 1033 | 22 | 1077 |
| Time-varying | Selection (1,1) | 1036 | 22 | 1080 |
| Time-varying | flat-Π | 1019 | 30 | 1080 |
| Time-varying | Naive shrinkage | 1023 | 31 | 1085 |
| Time-varying | AR | 1068 | 13 | 1093 |
| Time-varying | flat-R | 1034 | 31 | 1097 |
We see that the models that use a mean structure that depends only on treatment and not time t tend to have lower DIC values. The time-varying models are penalized in the pD term for having to estimate the additional 14 regression coefficients. Of the correlation priors the flat-R and AR priors perform much worse than the shrinkage, selection, triangular, and flat-PAC priors with the same mean structure. Additionally, the selection prior that uses the triangular form for SBeta (α = 2, β = 1) tend to have a smaller DIC than the SBeta(1,1) priors. From Table 4 we determine the prior choice that best balances model fit with parsimony is clearly the model with time-constant mean structure and the shrinkage prior on the correlation matrix prior.
Using this best fitting model, the posterior mean of β is (−0.504, −0.295) implying that the marginal probability (95% credible interval) of not smoking during a given study week is Φ(−0.504) = 0.307 (0.24, 0.37) for the control group and Φ(−0.295) = 0.384 (0.32, 0.45) for the exercise group, where Φ(·) is the distribution function of the standard normal distribution. The test of the hypothesis that the control treatment is as effective as the exercise treatment (i.e., H0 : β1 ≥ β2) has a posterior probability of 0.06, providing some evidence to the claim that exercise improves cessation results.
We now examine in more detail the effect the shrinkage prior has on modeling the correlation matrix. The posterior means (95% credible interval) of the shrinkage parameters are ε̂0 = 0.406 (0.25, 0.60) and γ̂ = 2.44 (1.6, 3.4). With a value of γ greater than 1, the variance of πij is decaying to zero fairly rapidly. The posterior mean of Π is
with the lower diagonal values giving the elements of R̂. We see that the PACs are far from zero in only the first two lags and the remaining π’s are close to zero. This is because these partial autocorrelations have been shrunk almost to zero in most iterations.
7. Discussion
In this paper we have introduced two new priors for correlation matrices, a shrinkage prior and a selection prior. These priors choose a sparse parameterization of the correlation matrix through the set of PACs. In the selection context, by stochastically selecting the elements of Π to zero out, our model finds interpretable independence relationships for normal data and avoids the need for complex model selection of the dependence structure. A key improvement of the selection prior over existing methods for sparse correlation matrices is that our approach avoids the complex normalizing constants seen in previous work. Additionally, in settings with time-ordered data, the partial autocorrelations are more interpretable than the full partial correlations, as they do not involve conditioning on future values.
While the examples we have considered here involve situations where the covariance matrix was constrained (as in the data example) or known (as in the simulations) to be a correlation matrix, the extension to arbitrary Σ is simple. Returning to the separation strategy Σ = SRS (Barnard et al., 2000), a prior for Σ can be formed by placing independent priors on S and R, i.e. p(Σ) = p(R)p(S). Using one of the proposed priors for p(R), sensible choices of p(S) include an independent inverse gamma for each of the or a flat prior on {S = diag(σ11, …, σJJ) : σjj > 0}. This leads to a prior on Σ with sparse PACs.
The simulations and data we have considered here deal with Y of low or moderate dimension. We provide a few comments regarding scaling of our approach for data with larger J. As we believe that PACs of larger lag play a progressively smaller role in describing the (temporal) dependence, it may be reasonable to specify a maximum allowable lag for non-zero PACs. That is, we choose some k such that πij = 0 for all j − i > k and sample πij (j − i ≤ k) from either our shrinkage or selection prior. Banding the Π matrix is related to the idea of banding the covariance matrix (Bickel and Levina, 2008), concentration matrix (Rothman et al., 2008), or the Cholesky decomposition of Σ−1 (Rothman et al., 2010). Banding Π has also been studied by Wang and Daniels (2013b). In addition to reducing the number of parameters that must be sampled, other matrix computations will be faster by using properties of banded matrices.
Related to this, modifications to the shrinkage prior may be needed for larger dimension J. Recall that the variance of πij is ξij = ε0|j − i|−γ. For large lags, this can be very close to zero leading to numerical instability; recall the parameters of the SBeta distribution are inversely related to ξij through . Replacing (3) with ξij = ε0 min{|j − i|, k}−γ or ξij = ε0 + ε1|j −i|−γ to bound the variances away from zero or banding Π after the first k lags provide two possibilities to avoid such numerical issues.
Further, we have parametrized the variance component and the selection probability in similar ways in our two sparse priors. The quantity is of the form ε0|j − i|−γ for both ξij in (3) and εij in (6), but other parameterizations are possible. We have considered some simulations (not included) allowing the variance/selection probability to be unique for lag, i.e. εij = ε|j−i|. A prior needs to be specified for each of these J − 1 ε’s, ideally decreasing in lag. Alternatively, one could use ε0/|j − i|, which can be viewed as a special case where the prior on γ is degenerate at 1. In our experience results were not very sensitive to the choice of the parameterization, and posterior estimates of Π and R were similar.
In addition, we have focused our discussion on the correlation estimation problem in the context of analysis with multivariate normal data. We note that these priors are additionally applicable in the context of estimating a constrained scale matrix for the multivariate Student t-distribution. Consider the random variable Y ~ tJ (μ, R, ν). That is, Y follows a J-dimensional t-distribution with location (mean) vector μ, scale matrix R (constrained to be a correlation matrix), and ν degrees of freedom (either fixed or random). Using the gamma-mixture-of-normals technique (Albert and Chib, 1993), we rewrite the distribution of Y to be Y|τ ~ NJ (μ, τ−1R) and τ ~ Gamma(ν/2, ν/2). Sampling for R as part of an MCMC chain follows as in Sections 3.2 and 4.3 using as the data. However, one should note that a zero PAC πij implies that Yi and Yj are uncorrelated given Yi+1, …, Yj−1, but this is not equivalent to conditional independence as in the normal case.
Supplementary Material
Acknowledgments
This research was partially supported by NIH CA-85295.
Contributor Information
J. T. Gaskins, Email: jeremy.gaskins@louisville.edu.
M. J. Daniels, Email: mjdaniels@austin.utexas.edu.
B. H. Marcus, Email: bmarcus@ucsd.edu.
References
- Albert JH, Chib S. Bayesian analysis of binary and polychotomous response data. Journal of the American Statistical Association. 1993;88(422):669–679. [Google Scholar]
- Anderson T. An Introduction to Multivariate Statistical Analysis. 2. Wiley; 1984. [Google Scholar]
- Barnard J, McCulloch R, Meng XL. Modeling covariance matrices in terms of standard deviations and correlations, with application to shrinkage. Statistica Sinica. 2000;10(4):1281–1311. [Google Scholar]
- Bickel P, Levina E. Regularized estimation of large covariance matrices. The Annals of Statistics. 2008;36(1):199–227. [Google Scholar]
- Carter CK, Wong F, Kohn R. Constructing priors based on model size for non-decoposable Gaussian graphical models: A simulation based approach. Journal of Multivariate Analysis. 2011;102:871–883. [Google Scholar]
- Chib S, Greenberg E. Analysis of multivariate probit models. Biometrika. 1998;85(2):347–361. [Google Scholar]
- Damien P, Wakefield J, Walker S. Gibbs sampling for Bayesian non-conjugate and hierarchical models by using auxiliary variables. Journal of the Royal Statistical Society, Series B: Statistical Methodology. 1999;61:331–344. [Google Scholar]
- Daniels MJ, Hogan JW. Missing Data in Longitudinal Studies: Strategies for Bayesian Modeling and Sensitivity Analysis. Chapman & Hall; 2008. [Google Scholar]
- Daniels MJ, Normand SL. Longitudinal profiling of health care units based on mixed multivariate patient outcomes. Biostatistics. 2006;7:1–15. doi: 10.1093/biostatistics/kxi036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels MJ, Pourahmadi M. Modeling covariance matrices via partial autocorrelations. Journal of Multivariate Analysis. 2009;100(10):2352– 2363. doi: 10.1016/j.jmva.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempster AP. Covariance selection. Biometrics. 1972;28:157–75. [Google Scholar]
- James W, Stein C. Estimation with quadratic loss. Proceedings of the Fourth Berkeley Symposium on Mathematical Statistics and Probability; Univesity of California Press; 1961. pp. 311–319. [Google Scholar]
- Joe H. Generating random correlation matrices based on partial correlations. Journal of Multivariate Analysis. 2006;97:2177–2189. [Google Scholar]
- Kurowicka D, Cooke R. A parameterization of positive definite matrices in terms of partial correlation vines. Linear Algebra and its Applications. 2003;372:225–251. [Google Scholar]
- Kurowicka D, Cooke R. Completion problem with partial correlation vines. Linear Algebra and its Applications. 2006;418:188–200. [Google Scholar]
- Lauritzen SL. Graphical Models. Clarendon Press; 1996. [Google Scholar]
- Liechty J, Liechty M, Muller P. Bayesian correlation estimation. Biometrika. 2004;91:1–14. [Google Scholar]
- Liu C. Comment on “The art of data augmentation” by D. A. van Dyk and X.-L. Meng. Journal of Computational and Graphical Statistics. 2001;10(1):75–81. [Google Scholar]
- Liu X, Daniels MJ. A new algorithm for simulating a correlation matrix based on parameter expansion and re-parameterization. Journal of Computational and Graphical Statistics. 2006;15:897–914. [Google Scholar]
- Liu X, Daniels MJ, Marcus B. Joint models for the association of longitudinal binary and continuous processes with application to a smoking cessation trial. Journal of the American Statistical Association. 2009;104(486):429–438. doi: 10.1198/016214508000000904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus B, Albrecht A, King T, Parisi A, Pinto B, Roberts M, Niaura R, Abrams D. The efficacy of exercise as an aid for smoking cessation in women: A randomized controlled trial. Archives of Internal Medicine. 1999;159:1229–1234. doi: 10.1001/archinte.159.11.1229. [DOI] [PubMed] [Google Scholar]
- Neal RM. Slice sampling. The Annals of Statistics. 2003;31(3):705–767. [Google Scholar]
- Pitt M, Chan D, Kohn R. Efficient Bayesian inference for Gaussian copula regression models. Biometrika. 2006;93:537–554. [Google Scholar]
- Robert CP, Casella G. Monte Carlo Statistical Methods. 2 Springer-Verlag; 2004. [Google Scholar]
- Rothman AJ, Bickel PJ, Levina E, Zhu J. Sparse permutation invariant covariance estimation. Electronic Journal of Statistics. 2008;2:494–515. [Google Scholar]
- Rothman AJ, Levina E, Zhu J. A new approach to Cholesky-based covariance regularization in high dimensions. Biometrika. 2010;97(3):539–550. [Google Scholar]
- Rousseeuw PJ, Molenberghs G. The shape of correlation matrices. The American Statistician. 1994;48(4):276–279. [Google Scholar]
- Spiegelhalter DJ, Best NG, Carlin BP, van der Linde A. Bayesian measures of model complexity and fit (with discussion) Journal of the Royal Statistical Society, Series B. 2002;64(4):583–639. [Google Scholar]
- Wang C, Daniels MJ. A note on MAR, identifying restrictions, model comparison, and sensitivity analysis in pattern mixture models with and without covariates for incomplete data (with correction) Biometrics. 2011;67(3):810–818. doi: 10.1111/j.1541-0420.2011.01565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Daniels MJ. Bayesian modeling of the dependence in longitudinal data via partial autocorrelations and marginal variances. Journal of Multivariate Analysis. 2013a;116:130–140. doi: 10.1016/j.jmva.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Daniels MJ. Technical report. University of Florida; Gainesville, Florida: 2013b. Estimating large correlation matrices by banding the partial autocorrelation matrix. [Google Scholar]
- Wong F, Carter CK, Kohn R. Efficient estimation of covariance selection models. Biometrika. 2003;90(4):809–830. [Google Scholar]
- Yang R, Berger JO. Estimation of a covariance matrix using the reference prior. The Annals of Statistics. 1994;22:1195–1211. [Google Scholar]
- Yuan M, Lin Y. Model selection and estimation in the Gaussian graphical model. Biometrika. 2007;94(1):19–35. [Google Scholar]
- Zhang X, Boscardin WJ, Belin TR. Sampling correlation matrices in Bayesian models with correlated latent variables. Journal of Computational and Graphical Statistics. 2006;15(4):880–896. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.