ABSTRACT
A defining characteristic of Chlamydia spp. is their developmental cycle characterized by outer membrane transformations of cysteine bonds among cysteine-rich outer membrane proteins. The reduction-oxidation states of host cell compartments were monitored during the developmental cycle using live fluorescence microscopy. Organelle redox states were studied using redox-sensitive green fluorescent protein (roGFP1) expressed in CF15 epithelial cells and targeted to the cytosol, mitochondria, and endoplasmic reticulum (ER). The redox properties of chlamydiae and the inclusion were monitored using roGFP expressed by Chlamydia trachomatis following transformation. Despite the large morphological changes associated with chlamydial infection, redox potentials of the cytosol (Ψcyto [average, −320 mV]), mitochondria (Ψmito [average, −345 mV]), and the ER (ΨER [average, −258 mV]) and their characteristic redox regulatory abilities remained unchanged until the cells died, at which point Ψcyto and Ψmito became more oxidized and ΨER became more reduced. The redox status of the chamydial cytoplasm was measured following transformation and expression of the roGFP biosensor in C. trachomatis throughout the developmental cycle. The periplasmic and outer membrane redox states were assessed by the level of cysteine cross-linking of cysteine-rich envelope proteins. In both cases, the chlamydiae were highly reduced early in the developmental cycle and became oxidized late in the developmental cycle. The production of a late-developmental-stage oxidoreductase/isomerase, DsbJ, may play a key role in the regulation of the oxidoreductive developmental-stage-specific process.
IMPORTANCE
Infectious Chlamydia organisms have highly oxidized and cysteine cross-linked membrane proteins that confer environmental stability when outside their host cells. Once these organisms infect a new host cell, the proteins become reduced and remain reduced during the active growth stage. These proteins become oxidized at the end of their growth cycle, wherein infectious organisms are produced and released to the environment. How chlamydiae mediate and regulate this key step in their pathogenesis is unknown. Using biosensors specifically targeted to different compartments within the infected host cell and for the chlamydial organisms themselves, the oxidoreductive states of these compartments were measured during the course of infection. We found that the host cell redox states are not changed by infection with C. trachomatis, whereas the state of the chlamydial organisms remains reduced during infection until the late developmental stages, wherein the organisms’ cytosol and periplasm become oxidized and they acquire environmental resistance and infectivity.
INTRODUCTION
Chlamydia spp. are obligate intracellular bacteria that are widely distributed in nature and as human pathogens impose a tremendous burden on global public health. Chlamydia trachomatis is the leading cause of sexually transmitted infection, responsible for an estimated 2.8 million new cases annually in the United States (1), and is also the etiologic agent of trachoma, a blinding eye disease that is of major concern in many developing countries (2). Chlamydiae undergo a distinctive biphasic developmental growth cycle, consisting of a condensed, osmotically resistant, extracellular form called the elementary body (EB), and an intracellular, osmotically sensitive and metabolically active form, called the reticulate body (RB). Infection begins with attachment of the Chlamydia EB to the host cell, followed by internalization of the bacterium into a vacuolar compartment, termed the inclusion, which evades endolysosomal fusion (3). Conversion from EB to RB occurs within this protective niche, and after 48 to 72 h of metabolic growth and replication of RB by binary fission, chlamydiae convert into EB and initiate their exit from the host cell by cellular lysis or extrusion of the inclusion vacuole (4).
The developmental conversion of chlamydiae from EB to RB, and subsequently from RB to EB, requires the reduction and oxidation of several cysteine-rich outer envelope proteins (5–8). One of these, the chlamydial major outer membrane protein (MOMP), is extensively disulfide cross-linked in EB and reduced in RB (9). Immediately after internalization into host cells, MOMP and other cysteine-rich proteins are reduced (6, 7, 10, 11). Consistent with these data, the porin function that has been described for MOMP also occurs only when the protein is reduced (5). The mechanism of reduction and oxidation of these outer membrane proteins is unknown; moreover, it has not been established whether the changes in redox states are an outcome of the host cell status or are defined selectively by chlamydiae.
Because the inclusion membrane is permeable to ions and small molecules (12), it is possible that redox changes that occur within the chlamydial inclusion will be echoed in the host cell cytoplasm and elicit effects on host cell function. Specific effects of intracellular chlamydial growth on this aspect of host cell physiology have not been determined. We investigated the production of oxidoreductant compounds following Chlamydia infection by measuring quantitative changes in the oxidoreductive status of the host cell cytosol, mitochondria, and ER by cellular compartment targeting of a redox-sensitive green fluorescent protein (GFP) biosensor.
Surprisingly, the steady-state redox potentials of these cellular compartments were unaffected by Chlamydia infection, suggesting that changes in redox potential necessary for modification of outer membrane proteins are confined to chlamydial organisms and within the inclusion vacuole. We tested this hypothesis by transformation and selection for stable expression of a redox-sensitive GFP biosensor within chlamydiae. Redox potentials were evaluated, and it was determined that the reduction and subsequent oxidation of chlamydial organisms required chlamydial protein synthesis and were developmental stage specific. It can be concluded that the alterations in reduction and oxidation necessary for the modified phenotypes of RB and EB are chlamydia directed and likely driven by rebalancing of oxidoreductant donors and acceptors.
RESULTS
Steady state and regulation of subcellular redox potential in Chlamydia-infected cells.
Chlamydiae undergo extensive disulfide-state modification of their outer membrane proteins during EB to RB conversion and subsequent RB to EB conversion, yet the biological basis of redox potential changes in the Chlamydia inclusion is unknown. We hypothesized that redox changes in the inclusion lumen, irrespective of their origin, would be mimicked by, or even a consequence of, redox changes in host cytoplasm or organelles. To directly test this question, we employed a redox-sensitive GFP mutant (roGFP1) that has been targeted to specific subcellular compartments (13). This GFP elicits ratiometric changes in fluorescent properties in response to changes in redox potential (14, 15). CF15 epithelial cells were transfected with one of three roGFP1 constructs designed to specifically target the host cytosol, mitochondria, or the endoplasmic reticulum (ER) (13). Cells were infected with C. trachomatis, and subcellular steady-state redox potentials were measured by fluorescence microscopy at 20, 44, and 68 h postinfection (hpi). Subcellular compartments were selectively targeted by their respective probes, and roGFP fluorescence was strong at all times prior to and after Chlamydia infection (Fig. 1).
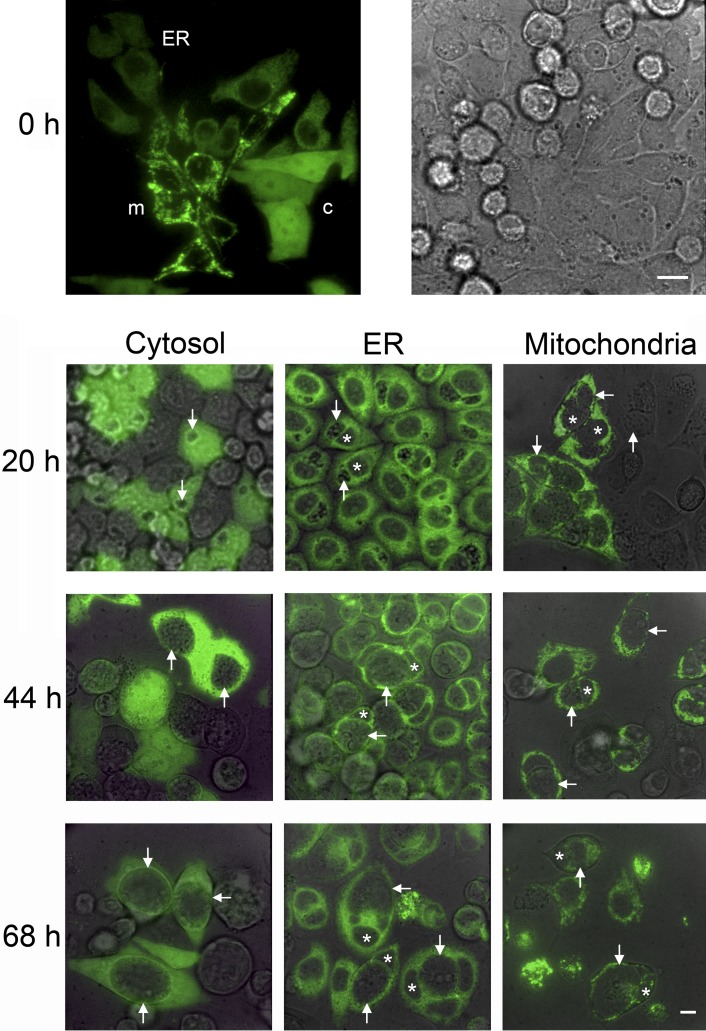
FIG 1 .
Organelle-specific localization of roGFP1 in CF15 cells infected with C. trachomatis. CF15 cells transiently expressing three different organelle-specific roGFPs were seeded together on one coverslip. Confocal microscopy (excitation, 488 nm; emission, 530 nm) indicates organelle-specific expression at 0 h (top panels). c, cytosol; ER, endoplasmic reticulum; m, mitochondria. The corresponding bright-field image is shown to the right. CF15 cells on separate coverslips expressing one type of roGFP1 were monitored at 20, 44, and 68 hpi. Arrows indicate chlamydial inclusions, and stars label the nucleus. The overlay of confocal and bright-field images shows a time-dependent increase in inclusion size, while the expression of roGFP1 remains restricted to its corresponding organelle. The scale bars represent 10 µm.
Steady-state redox potentials of the cytosol, mitochondria, and ER were determined and compared between uninfected CF15 cells and Chlamydia-infected cells at 20, 44, and 68 hpi. In uninfected cells, the cytosolic redox potential (Ψcyto) was −320 mV (Fig. 2A), the mitochondrial redox potential (Ψmito) was −345 mV (Fig. 2B), and the redox potential of the ER (ΨER) was −258 mV (Fig. 2C). Surprisingly, Chlamydia infection had no effect on steady-state redox for any of the subcellular compartments tested (Fig. 2A to C).
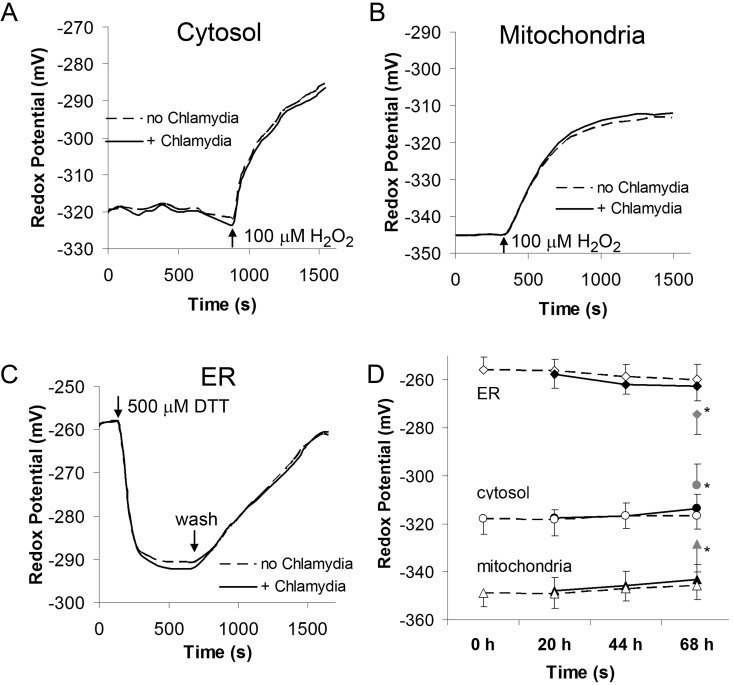
FIG 2 .
C. trachomatis does not affect organelle redox regulation in intact CF15 host cells. Redox responses of organelle-targeted roGFP1 in the absence or presence of infecting chlamydiae were tested by treating cells with 100 µM H2O2 (panel A for the cytosol and panel B for the mitochondria) or 500 µM DTT (panel C for the ER). Panel D shows steady-state redox potentials obtained for ER, cytosol, and mitochondria with (solid lines) and without (dashed lines) chlamydial infection over time. Chlamydial infection did not affect the organelle redox potentials of intact CF15 cells. Only CF15 cells at 68 hpi with a degenerating morphology showed significant redox changes. *, P < 0.05, based on Student’s t test. Raw data were background subtracted and calibrated into redox potentials. Data show averages and standard deviations based on >10 cells for each experiment.
Because steady-state redox potentials were unaffected following chlamydial infection, we next investigated whether differences in redox dynamics were elicited in response to intracellular Chlamydia growth and differentiation. Cells were transfected with appropriate roGFP constructs and treated with redox-active agents, and the dynamic responses of each compartment were measured by real-time fluorescence imaging. The reduced environments of the cytosol and mitochondria were oxidized with 100 µM H2O2 (Fig. 2A and B), and the effect of Chlamydia infection on the cytosolic and mitochondrial redox buffering capacity was determined. There was no apparent effect on oxidation times in either compartment due to Chlamydia infection at any stage of the developmental cycle (Fig. 2A and B and D). The ER was partially reduced by treating cells with 500 µM dithiothreitol (DTT), after which cells were rinsed and the ER was allowed to reoxidize. The occurrence of Chlamydia infection had no effect on ER redox regulatory behavior, and the observed recovery rates were in agreement with published findings (13). A summary of steady-state redox potentials and redox dynamics is shown in Fig. 2D. The only time at which differences in redox states between uninfected and Chlamydia-infected cells were observed occurred at 68 hpi—increased oxidation of the cytosol and mitochondria and slight reduction of the ER lumen. However, these data (indicated by asterisks in Fig. 2D) were limited to dead or dying host cells that exhibited a disparate morphology (not shown). These differences in redox potential are likely attributed to cellular death and lysis.
Redox dynamics of the chlamydial inclusion.
Given the paucity of redox changes in host cell compartments during chlamydial infection, we next tested redox dynamics of the chlamydial inclusion. This was addressed by expression of roGFP in transformed chlamydiae. A Chlamydia SW2 transformation vector (16) was constructed containing the roGFP gene placed under the control of the dnaK promoter from C. trachomatis (Fig. 3). Transformants of C. trachomatis with pdnaK-roGFP-SW2 were recovered after 3 passages in the presence of penicillin G. Chlamydia expressing roGFP displayed an apparently normal developmental cycle and inclusion size in HeLa cells (not shown), suggesting that there was little effect on chlamydial growth. The roGFP expression was detected by SDS-PAGE and showed the predicted protein size (Fig. 4A). The expression of roGFP was demonstrated by fluorescence within inclusions and was unambiguously detected as early as 12 hpi and until the end of the developmental cycle (Fig. 4B and C).
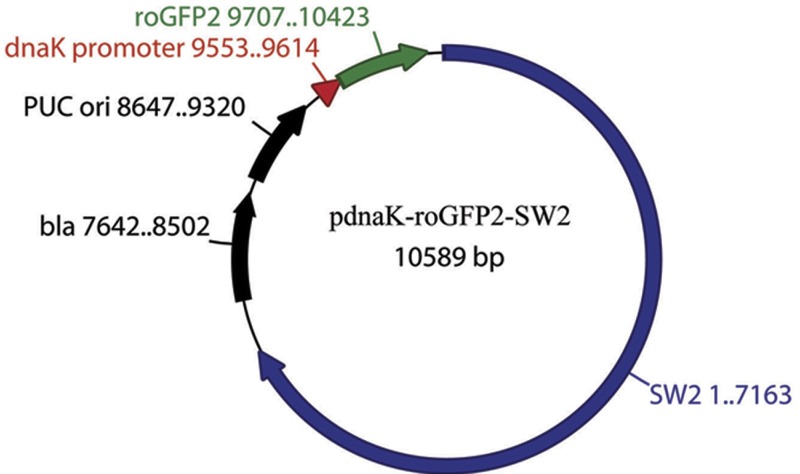
FIG 3 .
Map of the chlamydial roGFP expression vector. The roGFP gene (green) under the control of the chlamydial promoter dnaK (red) was inserted into the E. coli plasmid containing bla and ori fragments and then ligated with plasmid pSW2 from C. trachomatis SW2 (blue), as described in Materials and Methods.
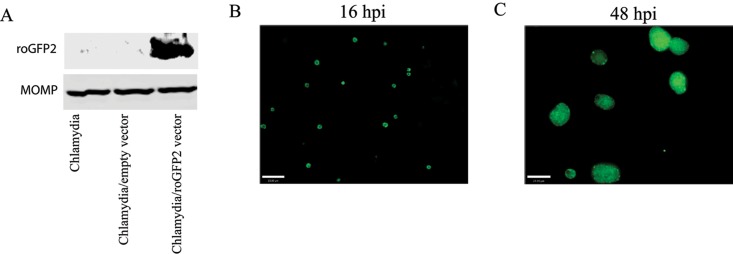
FIG 4 .
Expression of roGFP2 in C. trachomatis L2. HeLa cells were infected with the expression vector-transformed chlamydiae, and roGFP2 expression was detected by Western blotting and fluorescence microscopy. The cell lysates prepared from untransformed, or transformed chlamydia-infected cells were isolated by SDS-PAGE and then transferred to nitrocellulose membrane. roGFP2 expression was detected with an anti-GFP antibody, and the MOMP was used as a loading control (A). The roGFP2 fluorescence from chlamydiae within inclusions was also observed by fluorescence microscopy at 16 hpi and 48 hpi (B and C). Scale bars are 23 µm.
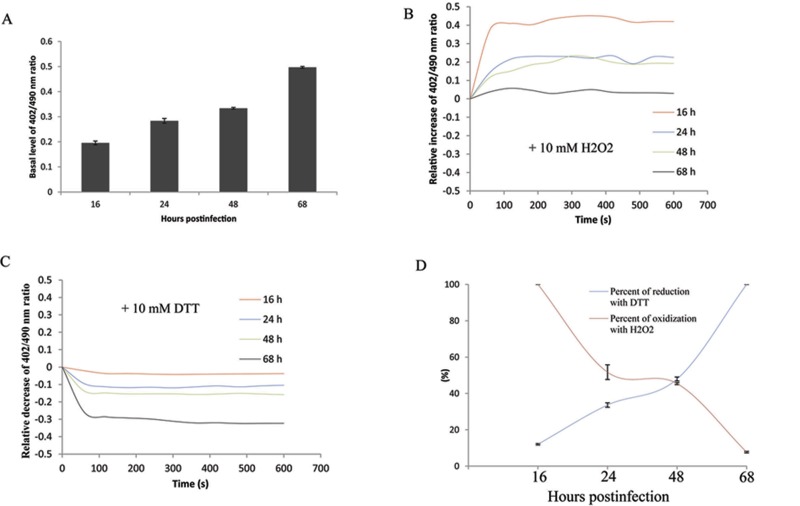
HeLa cells infected with chlamydiae expressing roGFP were challenged with oxidizing (H2O2) or reducing (DTT) agents at selected times during the infection, and the recovery response was measured. There was a significant decrease in the excitation ratio (402/490 nm) of roGFP observed in the presence of 10 mM DTT within 1 min. The response reached the maximum level of reduction at 5 to 6 min (Fig. 5C). Additionally, challenge of infected cells with H2O2 resulted in an increased 402/490-nm ratio, and the oxidation maximum was reached at 5 min (Fig. 5B). These data demonstrated the capability of roGFP to effectively monitor the redox status of chlamydial organisms within infected cells.
FIG 5 .
Response of roGFP2 expressed in chlamydiae upon exposure to redox agents. HeLa cells grown in 4-well chambered coverslips were infected with roGFP2-expressing chlamydiae and exposed to DTT (10 mM) or H2O2 (10 mM) in Ringer’s buffer at different times during the infection. The 402/490-nm ratio of roGFP2 at the basal level (A) and the relative change of the ratios under the different treatment conditions over time were calculated (B and C). The maximal percentages of reduction and oxidation in the presence of DTT or H2O2 at different times were also calculated (D) as described in Materials and Methods. Each experiment was repeated at least three times, and a representative result is shown.
Notably, it was shown that the basal level of the 402/490-nm ratio exhibited a significant increase during the time course from 16 to 68 hpi (Fig. 5A), consistent with increased oxidation during this process. Exposure of infected cells to 10 mM DTT resulted in a significant reduction of roGFP at late times compared to early times postinfection. The maximum reduction induced by DTT was achieved at 68 h (100%) (Fig. 5D), although a slight reduction induced by DTT was also observed at an early time. In concert with the reduction challenge data, treatment of infected cells with the oxidation agent, H2O2, dramatically elevated the oxidized level of roGFP at the early times postinfection, and maximal oxidation (100%) was reached at 16 hpi, while the inability of increasing the oxidization of roGFP was observed at 68 hpi (Fig. 5D). These data showed that chlamydiae were reduced at the early stage and switched to an oxidized state during the late stage of development.
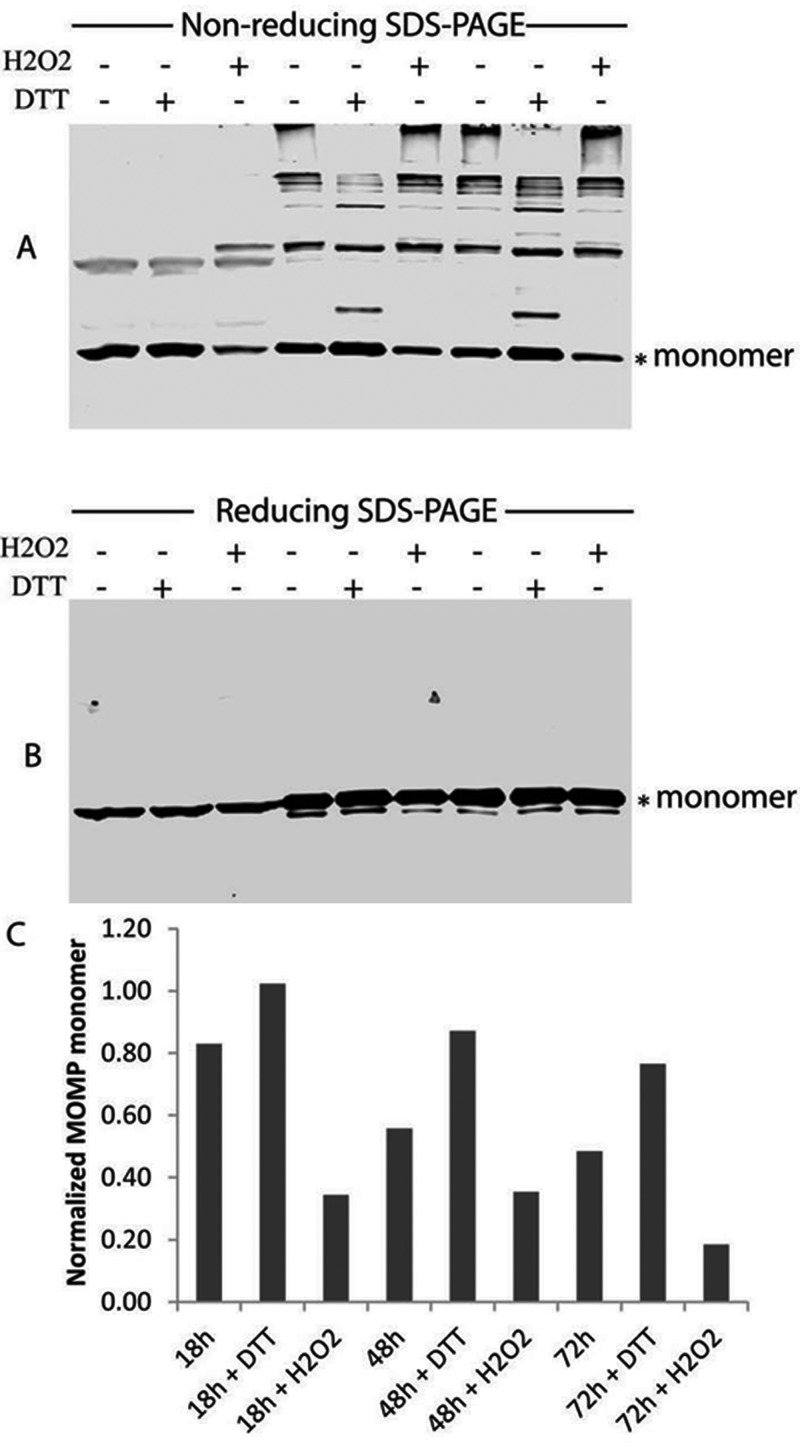
To support the redox state using roGFP expressed in the chlamydial cytoplasm, we used the multimeric status of the chlamydial MOMP and the cysteine-rich OmcB (17) proteins as biomarkers of the redox state of the outer envelope as previously shown by Hatch et al. (7). However, we additionally addressed their redox status by monitoring the cross-linked envelope proteins in the presence of different redox-active agents at different stages during the infection. Thus, the infected cells were treated under the same conditions used in the roGFP experiments. MOMP was predominantly present as a monomer in chlamydiae at early mid-growth-phase times (18 hpi), and reduction was not increased following treatment of infected cells with 10 mM DTT for 10 min. Treatment with 10 mM H2O2 at this time significantly increased the cross-linked MOMP and reduced the monomer form, showing that the chlamydial envelope proteins were highly reduced at 18 hpi. Exposure of infected cells to H2O2 significantly altered the redox status of chlamydial organisms from a reducing to an oxidizing status, resulting in a high degree of MOMP cross-linking (Fig. 6). MOMP in untreated cells was highly cross-linked at 48 and 68 hpi compared to 18 hpi (Fig. 6), indicating that MOMP was broadly oxidized at the late times postinfection. Treatment of infected cells with 10 mM DTT at these times significantly increased the monomer form of MOMP and reduced the multiple cross-linked proteins. A more pronounced reduction of cross-linked MOMP was observed at 48 hpi compared to 68 hpi, suggesting a highly oxidized environment for chlamydiae at 68 hpi. These data are consistent with the roGFP data.
FIG 6 .
Cross-linking of MOMP during the chlamydial developmental cycle. HeLa cells were infected with wild-type C. trachomatis L2 and lysed at the different times postinfection (see details in Materials and Methods). An equal amount of cell lysate containing chlamydial proteins was subjected to nonreducing (A) or reducing (B) SDS-PAGE, and the MOMP was detected by immunoblotting with an anti-MOMP antibody. The monomer form of MOMP under the nonreducing conditions was normalized with the reduced MOMP under the reducing condition (C). The monomer form of MOMP is indicated with asterisks (*). Where indicated, infected cells were treated with DTT (10 mM) or H2O2 (10 mM) for 10 min before being subjected to Western blotting.
DISCUSSION
A central defining phenotype for the genus Chlamydia is its classic developmental cycle, in which the osmotically resistant and metabolically inactive form of the organism, the EB, is required for infection of eukaryotic host cells. Upon entry, the EB begins a transformation to the osmotically sensitive, vegetative and metabolically active replicating RB form. The cycle is complete at approximately 48 to 72 hpi, when a large population of RB transition to the infectious EB form prior to their exit from the cell by lysis or extrusion (4). Chlamydiae are confined from the host cell cytoplasm throughout their developmental cycle in the inclusion. Redox-mediated disulfide modifications are an important and unique biological theme for Chlamydia as reduction of disulfides is essential for (i) initiation of entry of chlamydiae into their host cells (18), (ii) activation of the type III secretion system (19), and (iii) the EB-RB-EB developmental stage transitions. The EB-RB-EB transitions are characterized by the modification of several cysteine-rich envelope proteins that provide structural rigidity and environmental stability to the EB when oxidized and envelope plasticity for the RB in their reduced state (5, 6, 9, 20–22). The transformation from RB to EB is complex as two of the central cysteine-rich proteins are transcriptionally regulated and only expressed late in the developmental cycle and thus are present in EB and absent in RB (7). It is unambiguous that EB are dominated by cysteine-rich proteins whose disulfides are oxidized, resulting in an osmosis- and detergent-resistant envelope that is fully dissociable following chemical reduction.
We initially hypothesized that redox changes in the chlamydiae may be consequent to changes in the host cell, potentially even in discrete subcellular compartments because of differential redox buffering capacities among cellular organelles and because of the intersection of host cell exocytic pathways with the chlamydial inclusion (23, 24). However, an effect of chlamydial infection on redox-dependent fluorescence properties of roGFP in cytosol, mitochondria, or ER was not detected. Measurements of redox potentials and organelle-specific redox regulation obtained in this study match previously published values (13). The only time we observed changes in subcellular redox was after cells were dead and had released chlamydiae. Given the extensive interactions and modifications of the host cell machinery by chlamydiae (3), we consider these findings of a stably maintained and regulated redox status of the host cell during infection to be important as they rule out a model of generalized changes in cellular compartment redox during chlamydial development.
While the redox potentials of the host cell compartments were not perturbed by chlamydial infection, these measurements may not be applicable to the chlamydial inclusion vacuole status or, ultimately, the status of chlamydial organisms. We approached this challenge by employing an roGFP biosensor that could be stably expressed in chlamydial organisms following genetic transformation (16). Sustained expression of roGFP was monitored throughout the developmental cycle, thereby enabling determination of the chlamydial redox state during different developmental stages. The expression of roGFP was limited to the chlamydial cytoplasm; thus, these measurements were a direct determination of the basal redox state of the organism. Direct assessment of periplasmic redox state was obtained by determination of the level of cysteine-disulfide cross-linking among proteins in the outer envelope. We showed that chlamydial organisms (outer membrane, periplasm, and cytoplasm) were highly reduced at the early developmental stage and became oxidized at late stages of the developmental cycle.
There was no evidence that the expressed roGFP from chlamydiae was secreted into the inclusion lumen; thus, it is uncertain whether the inclusion lumen has the same redox conditions as the host cell cytoplasm during the developmental cycle. The process of chlamydial development and production of EB in the inclusion lumen is asynchronous; thus, this compartment contains a mixture of EB, RB, and intermediate forms simultaneously at later times postinfection. This suggests that the redox state of the inclusion lumen may become oxidized during late stages, and the remaining RB require reductase activity to remain reduced in this environment.
It is unclear how the chlamydiae modulate their redox states during the developmental cycle. Since the redox status of infected cells was not altered during the infection, we propose that chlamydiae manipulate this essential pathogenic process by regulating the intracellular redox status of individual chlamydial organisms at different developmental stages. The management of disulfide bonds in bacterial proteins is provided by two pathways that, for Gram-negative organisms, occur in the periplasm (25). The oxidative folding pathway forms disulfide bonds by oxidation of two cysteine residues. This pathway in Escherichia coli is mediated by DsbA and DsbB (25, 26). The periplasmic protein DsbA oxidizes its folded protein substrates and becomes reduced, wherein the inner membrane DsbB reoxidizes DsbA. DsbB is reduced by the production of reducing capacity generated in the cytoplasm as an outcome of metabolism. Chlamydiae have both DsbA and DsbB homologs (i.e., CT176 and CT177) (27). The measurement of increasing oxidative potential in the chlamydial cytoplasm coincident with EB development could simply reflect an outcome of shutting down their metabolic activity (e.g., glycolysis) necessary to maintain a reduced environment.
The second pathway is the disulfide isomerization pathway, in which the inner membrane DsbD protein reduces periplasmic substrate proteins DsbC and DsbG that then are activated to resolve incorrect disulfides in target proteins (25). Chlamydiae contain a relative of DsbD, but they do not contain homologs of DsbC or DsbG (28). The substrates for DsbD have been predicted to be unique (28), and chlamydiae encode two proteins with leader signal sequences and with predicted disulfide bond isomerase activity, CT780 and CT783 (27). The CT780 and CT783 homologs have been named DsbH and DsbJ, respectively, reflecting their unique character (29). The functional activity of DsbH was determined to be a reducing disulfide oxidoreductase with only weak isomerase activity and was thus proposed to be a periplasmic reductase (29). The function of DsbJ has not been tested. Unlike other oxidoreductases and thiol disulfide isomerases, DsbJ lacks a CXXC motif that is essential for function and resolution of mixed disulfides. Significantly, DsbJ is strongly regulated and highly expressed specifically late in the chlamydial developmental cycle (30), supporting a conclusion for an important role in regulating the redox state of chlamydial envelope proteins. Its periplasmic location, genomic linkage with DsbH reductase, late-stage expression, and lack of the canonical CXXC motif suggest that it functions as an inhibitor or competitor of DsbH, serving as a regulator of the oxidative state of the late developmental stage chlamydial organisms.
MATERIALS AND METHODS
Reagents.
Unless otherwise specified, all reagents and chemicals were obtained from Sigma (St. Louis, MO). The shuttle plasmid pGFP::SW2 was kindly provided by Ian N. Clarke, University of Southampton Medical School. The pRSETB-roGFP2 plasmid containing roGFP2 was purchased from the University of Oregon. The anti-GFP antibody was obtained from BD Biosciences. The anti-MOMP antibody was a stock from our laboratory and produced from a mouse. All of the enzymes used in the study were obtained from New England Biolabs. XL10-Gold ultracompetent cells were obtained from Agilent Technologies.
Cell culture and infections.
Chlamydia trachomatis LGV/434/Bu serovar L2 cells were grown and elementary bodies were purified as described previously (31). HeLa cells were cultured in RPMI. JME/CF15 nasal epithelial cells homozygous for the ΔF508 cystic fibrosis transmembrane conductance regulator (CFTR) (32), (termed “CF15” throughout the article) were cultured as previously described (13). Cells were plated on glass bottom culture dishes (MatTek, Ashland, MA) and typically infected with Chlamydia cells diluted in Hanks’s balanced salt solution (HBSS) at a multiplicity of infection (MOI) of 1 for 1.5 h at 25°C. Cells were washed with HBSS and incubated in normal growth medium for 6 h at 37°C. Cells were then cultured in medium containing 75 µg/ml vancomycin or 100 IU/ml penicillin for the times indicated in the text.
Solutions.
In experiments to measure cytosolic redox potentials and to monitor cellular localization of fluorophores, cells were incubated in Ringer’s solutions containing 145 mM NaCl, 1.2 mM MgSO4, 2 mM CaCl2, 2.4 mM K2HPO4, 0.6 mM KH2PO4, 10 mM HEPES, and 10 mM glucose (pH 7.4).
Redox potential measurements using roGFP1 and imaging microscopy.
Expression of roGFP1 in transiently transfected CF15 cells (either infected with Chlamydia or control) was analyzed on a Solamere spinning-disc confocal microscope with excitation at 488 nm. Cells were bathed in Ringer’s solution containing 145 mM NaCl, 1.2 mM MgSO4, 2 mM CaCl2, 2.4 mM K2HPO4, 0.6 mM KH2PO4, 10 mM HEPES, and 10 mM glucose (pH 7.4). Images were obtained using a 515-nm long pass emission filter and 40× objective. Differential inference contrast (DIC) images were also recorded to correlate cell morphology and roGFP1 fluorescence. Images were overlaid using Adobe Photoshop.
Measurements of cytosolic redox potentials in CF15 cells were performed as described recently (13). Briefly, CF15 cells grown on cover glasses were transiently transfected with plasmids coding for a redox-sensitve GFP mutant, roGFP1, using the Effectene transfection reagent according to the manufacturer’s protocol (Qiagen, Valencia, CA). roGFP1-expressing cells infected with Chlamydia were analyzed in Ringer’s solution at the times indicated in the text using a Nikon Diaphot microscope with a 40× Neofluar objective (1.4 NA). Ratiometric imaging was performed using a charge-coupled device (CCD) camera, filter wheel (Lambda-10, Sutter Instruments, Novato, CA), and Axon Instruments Imaging Workbench 4 (Axon Instruments, Foster City, CA) to collect emission (>510-nm) images during alternate excitation at 385 ±5 nm and 474 ±5 nm. Cells were bathed in Ringer’s solution, and roGFP1 ratios were recorded over time. Calibration of the roGFP1 ratios in terms of redox potentials was performed using a protocol that was described previously (13). At the end of each experiment, roGFP1 385/474 ratios were recorded during maximal oxidation by treatment with 10 mM H2O2 and then during maximal reduction by treatment with 10 mM DTT. Images were background subtracted, and the roGFP1 excitation ratios were normalized to the values measured using 10 mM DTTred as 0% oxidation and 10 mM H2O2 as 100% oxidation. The normalized excitation ratios were converted to redox potentials (mV) using an in situ calibration curve that was published previously (13).
Construction of pdnaK-roGFP2-SW2 vector.
The replication origin (ori) and β-lactamase (bla) genes from the pGFP::SW2 vector were amplified with the following primers: p1-F (5′ CGGGGATCCGGTTCTATAGTGTCACCTA 3′) and p1-R (5′ CAGCTGCTCGAGGCCGGTCTCCCTA 3′). The resulting PCR product was digested with the BamHI and XhoI enzymes and then purified by gel purification kit (Qiagen). The wild-type C. trachomatis dnaK promoter was synthesized by annealing the following primers: pdnaK-F, 5′(Phos)-TCGAGATTCTTGACCGGTGGAGACGGTTTTCTTATAATGACACCGACTTATGGAAAATAGAGGTTCCATGGGTGCTAGCGGTACCGTCGACG-3′; and pdnaK-R, 5′(Phos)-GATCCGTCGACGGTACCGCTAGCACCCATGGTACCTCTATTTTCCA TAAGTCGGTGTCATTATAAGAAAACCGTCTCCACCGGTCAAGAATC-3′. The annealed double-stranded dnaK promoter was ligated with purified ori-bla fragment and then transformed into the E. coli XL-10 strain; the resulting plasmid was named pdnaK. The roGFP2 gene was amplified from plasmid pRSETB-roGFP2 with the primers proGFP-F 5′ AGGGCTAGCATGAGTAAAGGAGAAGAAC 3′ and proGFP-R 5′ AGCGTCGACCTATTTGTATAGTTCATC 3′. The PCR product was digested with the NheI and SalI enzymes, inserted into the pdnaK plasmid digested with the same enzymes, and then transformed into the XL-10 strain. The resulting plasmid was named pdnaK-roGFP2. In order to construct the shuttle plasmid pdnaK-roGFP2-SW2, the pGFP::SW2 plasmid was digested with BamHI, and the chlamydial plasmid SW2 was purified from a DNA agarose gel. The plasmid pdanK-roGFP2 was also digested with BamHI and then dephosphorylated with calf intestinal alkaline phosphatase (CIP). The resulting product was ligated with digested SW2 fragment and then transformed into the XL-10 strain. The resulting shuttle vector was confirmed by PCR and sequencing. Transformation of C. trachomatis with pdnak-roGFP2-SW2 was performed as described previously (16).
Investigation of redox status of chlamydial organisms using fluorescence microscopy.
HeLa cells grown on the glass coverslips were infected with roGFP2-expressing C. trachomatis L2. At the different times (16, 24, 48, and 68 h) after infection, the cells were bathed in Ringer’s solution and treated with either 10 mM DTT or 10 mM H2O2 for 10 min. The roGFP fluorescence from chlamydial inclusions was excited sequentially at 402 nm and 490 nm, and images were acquired at an emission of 525 nm during the treatment on a Zeiss Axiovert 200 inverted microscope fitted with an Orca-R2 cooled digital CCD camera. The mean fluorescence intensity values from over 10 chlamydial inclusions were measured using the Volocity software. The ratio of excitation at 402 nm and 490 nm was calculated. Each image was corrected for background by subtracting the intensity of an adjacent cell-free region. The relative changes of the 402/490-nm ratio after addition of 10 mM DTT or H2O2 were also calculated by subtracting the basal level fluorescence ratio at different time points. Expression at the time points that exhibited a maximal increase under oxidized conditions was considered to represent full oxidation and set equal to 100%, and the minimal ratio measured under reduced conditions was considered to represent full reduction and set equal to 100%.
Measurement of cross-linked MOMP by immunoblotting.
HeLa cells were infected with C. trachomatis L2 at an MOI of 5 in a six-well plate for different time points (20, 48, and 72 h). At each time point, the infected cells were treated with 10 mM DTT or 10 mM H2O2 for 10 min and then lysed with phosphate-buffered saline (PBS) buffer containing 2% SDS and 50 mM N-ethylmaleimide (NEM). The lysates were sonicated with a sonicator (two times for 10 s each time) and incubated for 1 h at 37°C. The supernatant from cell lysates was collected by centrifugation at a speed of 14,000 × g for 10 min at 4°C and then equally divided into two portions, one of which was subjected to 12% SDS-PAGE under nonreducing conditions in the presence of 50 mM NEM in the sample buffer (0.25 M Tris, 8% SDS, 30% glycerol, 0.02% bromophenol blue [pH 6.8]), another part of samples was subjected to 12% SDS-PAGE under reducing conditions in the presence of 10% 2-mercaptoethanol (2ME) and heated at 100°C for 5 min before the samples were loaded. The proteins isolated by SDS-PAGE were transferred to nitrocellulose membranes (0.45-µm pore size; Bio-Rad), and MOMP was detected by anti-MOMP antibody using the LI-COR system according to the manufacturer’s instruction.
ACKNOWLEDGMENTS
We gratefully acknowledge Jeremy Thorner (Department of Molecular & Cell Biology, University of California, Berkeley) for generous support with LI-COR equipment in our experiments.
This research was supported by NIH award AI091851.
Footnotes
Citation Wang X, Schwarzer C, Hybiske K, Machen TE, Stephens RS. 2014. Developmental stage oxidoreductive states of Chlamydia and infected host cells. mBio 5(6):e01924-14. doi:10.1128/mBio.01924-14.
REFERENCES
- 1. Weinstock H, Berman S, Cates W. 2004. Sexually transmitted diseases among American youth: incidence and prevalence estimates, 2000. Perspec. Sex. Reprod. Health 36:6–10. 10.1363/3600604. [DOI] [PubMed] [Google Scholar]
- 2. Burton MJ, Mabey DC. 2009. The global burden of trachoma: a review. PLoS Negl. Trop. Dis. 3:e460. 10.1371/journal.pntd.0000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bastidas RJ, Elwell CA, Engel JN, Valdivia RH. 2013. Chlamydial intracellular survival strategies. Cold Spring Harb. Persp. Med. 3:a010256. 10.1101/cshperspect.a010256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hybiske K, Stephens RS. 2007. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc. Natl. Acad. Sci. U. S. A. 104:11430–11435. 10.1073/pnas.0703218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bavoil P, Ohlin A, Schachter J. 1984. Role of disulfide bonding in outer membrane structure and permeability in Chlamydia trachomatis. Infect. Immun. 44:479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hackstadt T, Todd WJ, Caldwell HD. 1985. Disulfide-mediated interactions of the chlamydial major outer membrane protein: role in the differentiation of chlamydiae? J. Bacteriol. 161:25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hatch TP, Miceli M, Sublett JE. 1986. Synthesis of disulfide-bonded outer membrane proteins during the developmental cycle of Chlamydia psittaci and Chlamydia trachomatis. J. Bacteriol. 165:379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Newhall WJ. 1987. Biosynthesis and disulfide cross-linking of outer membrane components during the growth cycle of Chlamydia trachomatis. Infect. Immun. 55:162–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Caldwell HD, Kromhout J, Schachter J. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 31:1161–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Plaunt MR, Hatch TP. 1988. Protein synthesis early in the developmental cycle of Chlamydia psittaci. Infect. Immun. 56:3021–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eissenberg LG, Wyrick PB, Davis CH, Rumpp JW. 1983. Chlamydia psittaci elementary body envelopes: ingestion and inhibition of phagolysosome fusion. Infect. Immun. 40:741–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grieshaber S, Swanson JA, Hackstadt T. 2002. Determination of the physical environment within the Chlamydia trachomatis inclusion using ion-selective ratiometric probes. Cell. Microbiol. 4:273–283. 10.1046/j.1462-5822.2002.00191.x. [DOI] [PubMed] [Google Scholar]
- 13. Schwarzer C, Illek B, Suh JH, Remington SJ, Fischer H, Machen TE. 2007. Organelle redox of CF and CFTR-corrected airway epithelia. Free Radic. Biol. Med. 43:300–316. 10.1016/j.freeradbiomed.2007.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hanson GT, Aggeler R, Oglesbee D, Cannon M, Capaldi RA, Tsien RY, Remington SJ. 2004. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J. Biol. Chem. 279:13044–13053. 10.1074/jbc.M312846200. [DOI] [PubMed] [Google Scholar]
- 15. Dooley CT, Dore TM, Hanson GT, Jackson WC, Remington SJ, Tsien RY. 2004. Imaging dynamic redox changes in mammalian cells with green fluorescent protein indicators. J. Biol. Chem. 279:22284–22293. 10.1074/jbc.M312847200. [DOI] [PubMed] [Google Scholar]
- 16. Wang Y, Kahane S, Cutcliffe LT, Skilton RJ, Lambden PR, Clarke IN. 2011. Development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS Pathog. 7:e1002258. 10.1371/journal.ppat.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Allen JE, Stephens RS. 1989. Identification by sequence analysis of two-site posttranslational processing of the cysteine-rich outer membrane protein 2 of Chlamydia trachomatis serovar L2. J. Bacteriol. 171:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Abromaitis S, Stephens RS. 2009. Attachment and entry of Chlamydia have distinct requirements for host protein disulfide isomerase. PLoS Pathog. 5:e1000357. 10.1371/journal.ppat.1000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Betts-Hampikian HJ, Fields KA. 2011. Disulfide bonding within components of the Chlamydia type III secretion apparatus correlates with development. J. Bacteriol. 193:6950–6959. 10.1128/JB.05163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Newhall WJ, Jones RB. 1983. Disulfide-linked oligomers of the major outer membrane protein of chlamydiae. J. Bacteriol. 154:998–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hatch TP. 1996. Disulfide cross-linked envelope proteins: the functional equivalent of peptidoglycan in chlamydiae? J. Bacteriol. 178:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hatch TP, Allan I, Pearce JH. 1984. Structural and polypeptide differences between envelopes of infective and reproductive life cycle forms of Chlamydia spp. J. Bacteriol. 157:13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moore ER, Fischer ER, Mead DJ, Hackstadt T. 2008. The chlamydial inclusion preferentially intercepts basolaterally directed sphingomyelin-containing exocytic vacuoles. Traffic 9:2130–2140. 10.1111/j.1600-0854.2008.00828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cocchiaro JL, Kumar Y, Fischer ER, Hackstadt T, Valdivia RH. 2008. Cytoplasmic lipid droplets are translocated into the lumen of the Chlamydia trachomatis parasitophorous vacuole. Proc. Natl. Acad. Sci. U. S. A. 105:9379–9384. 10.1073/pnas.0712241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shouldice SR, Heras B, Walden PM, Totsika M, Schembri MA, Martin JL. 2011. Structure and function of DsbA, a key bacterial oxidative folding catalyst. Antiox. Redox Signal. 14:1729–1760. 10.1089/ars.2010.3344. [DOI] [PubMed] [Google Scholar]
- 26. Dutton RJ, Boyd D, Berkmen M, Beckwith J. 2008. Bacterial species exhibit diversity in their mechanisms and capacity for protein disulfide bond formation. Proc. Natl. Acad. Sci. U. S. A. 105:11933–11938. 10.1073/pnas.0804621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, Koonin EV, Davis RW. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754–759. 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 28. Cho S-H, Parsonage D, Thurston C, Dutton RJ, Poole LB, Collet JF, Beckwith J. 2012. A new family of membrane electron transporters and its substrates, including a new cell envelope peroxiredoxin, reveal a broadened reductive capacity of the oxidative bacterial cell envelope. mBio 3(2):e00291-11. 10.1128/mBio.00291-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mac T-T, Von Hacht A, Hung K-C, Dutton RJ, Boyd D, Bardwell JC, Ulmer TS. 2008. Insight into disulfide bond catalysis in Chlamydia from the structure and function of DsbH, a novel oxidoreductase. J. Biol. Chem. 283:824–832. 10.1074/jbc.M707863200. [DOI] [PubMed] [Google Scholar]
- 30. Nicholson TL, Olinger L, Chong K, Schoolnik G, Stephens RS. 2003. Global stage-specific gene regulation during the developmental cycle of Chlamydia trachomatis. J. Bacteriol. 185:3179–3189. 10.1128/JB.185.10.3179-3189.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Koehler JE, Burgess RR, Thompson NE, Stephens RS. 1990. Chlamydia trachomatis RNA polymerase major sigma subunit. Sequence and structural comparison of conserved and unique regions with Escherichia coli sigma 70 and Bacillus subtilis sigma 43. J. Biol. Chem. 265:13206–13214. [PubMed] [Google Scholar]
- 32. Jefferson DM, Valentich JD, Marini FC, Grubman SA, Iannuzzi MC, Dorkin HL, Li M, Klinger KW, Welsh MJ. 1990. Expression of normal and cystic fibrosis phenotypes by continuous airway epithelial cell lines. Am. J. Physiol. 259:L496–L505. [DOI] [PubMed] [Google Scholar]