Significance
Developmental complexity often parallels expansion of transcription factor families, and highly similar proteins can play related but distinct roles in the development of cell lineages and cell types. Comparative sequence analyses can point to regions of proteins that may be involved in specialization, but complementary functional approaches are required to confirm these roles. Using the “natural laboratory” of stomatal development in which closely related basic helix–loop–helix (bHLH) transcription factors regulate distinct sequential processes, we assign functions to unique domains and identify functional divergences hidden within regions of sequence identity.
Keywords: stomata, bHLH transcription factors, Arabidopsis, evolution, SPEECHLESS
Abstract
Transcription factor duplication events and subsequent specialization can drive evolution by facilitating biological innovation and developmental complexity. Identification of sequences that confer distinct biochemical function in vivo is an important step in understanding how related factors could refine specific developmental processes over time. Functional analysis of the basic helix–loop–helix (bHLH) protein SPEECHLESS, one of three closely related transcription factors required for stomatal lineage progression in Arabidopsis thaliana, allowed a dissection of motifs associated with specific developmental outputs. Phosphorylated residues, shown previously to quantitatively affect activity, also allow a qualitative shift in function between division and cell fate-promoting activities. Our data also provide surprising evidence that, despite deep sequence conservation in DNA-binding domains, the functional requirement for these domains has diverged, with the three stomatal bHLHs exhibiting absolute, partial, or no requirements for DNA-binding residues for their in vivo activities. Using these data, we build a plausible model describing how the current unique and overlapping roles of these proteins might have evolved from a single ancestral protein.
Transcription factors regulate various processes integral to multicellular development, from framing the overall body plan to specifying the fates of individual cells. Paralogs within transcription factor families can exhibit distinct but related functions, and these unique functions can arise through alterations in expression patterns (via mutations in cis-regulatory elements) or through alterations of biochemical function via mutations in the gene’s coding sequence. The Drosophila homeobox gene paired, for example, is able to rescue the gooseberry larval cuticle phenotype when expressed under the gooseberry promoter, indicating divergent embryonic roles owing to cis-regulatory elements (1), but a later role of paired in male fertility cannot be rescued by ectopic expression of gooseberry, indicating that biochemical differences also exist between these two transcription factors (2). Investigating specific sequence alterations that functionally distinguish genes from one another is critical to understanding the evolution and diversification of biological processes.
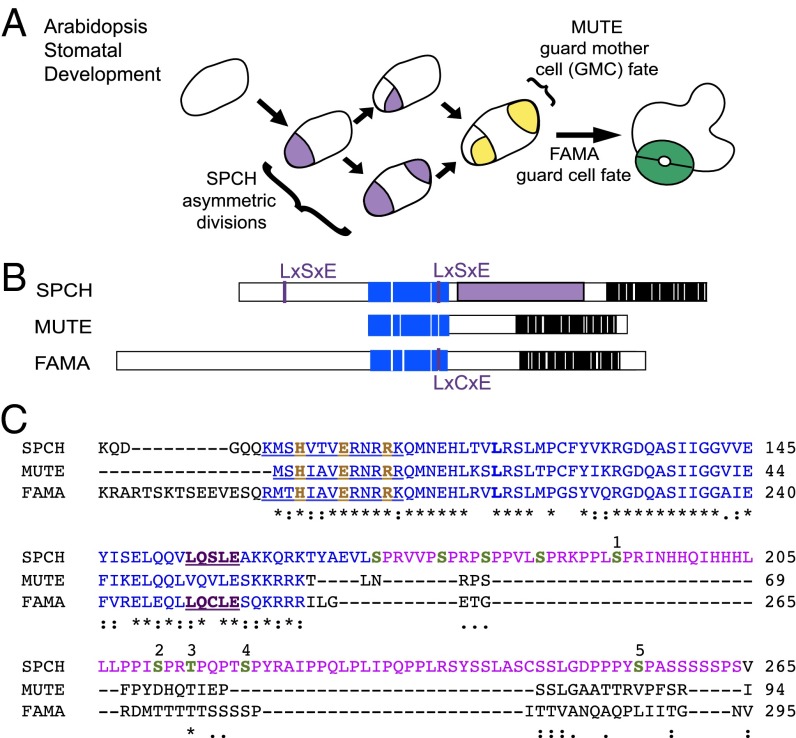
Stomatal development is a useful system for addressing how closely related transcription factors fulfill distinct functions. Stomata, pores in the plant epidermis that regulate gas exchange, develop via a series of divisions and cell fate transitions. Although stomatal development shows variation throughout the plant kingdom in terms of structure, precursors, and patterning (3), related basic-helix–loop–helix (bHLH) transcription factors are likely central to specifying cell fates in all stomata-producing plants (4, 5), and the evolutionary diversification of these bHLH proteins may be responsible for stomatal development variation (6). Among the ∼150 bHLH proteins in Arabidopsis [composing 26 subgroups (7)], five have established roles in stomatal development. Two members of bHLH subgroup IIIb, INDUCER OF COLD EXPRESSION (ICE1)/SCREAM (SCRM) and SCRM2, are expressed and function throughout stomatal development and exhibit functional redundancy (8), whereas three closely related subgroup Ia members, SPEECHLESS (SPCH), MUTE, and FAMA, have distinct functions regulating sequential steps of stomatal development. SPCH is required for the entry division into the stomatal lineage that generates a meristemoid, and MUTE promotes transition of the meristemoid to a guard mother cell (GMC). A GMC divides once more symmetrically, and the resulting daughter cells differentiate into guard cells (GCs) via FAMA activity (Fig. 1A) (9–11). Phenotypes resulting from overexpression of these three factors are the opposite of their loss-of-function phenotypes and are easily distinguishable from one another (9–11). SPCH, MUTE, and FAMA have distinct expression patterns, and promoter swaps between MUTE and FAMA and between SPCH and FAMA are consistent with the genes encoding proteins with distinct biochemical functions (9–11).
Fig. 1.
Closely related bHLHs SPCH, MUTE, and FAMA promote sequential transitions in Arabidopsis stomatal development. (A) Scheme of stomatal development. SPEECHLESS (SPCH) promotes asymmetric divisions that generate creating meristemoids (purple) and ground cells (white), which initiate and expand the lineage. Meristemoids differentiate into GMCs (yellow) via MUTE activity, and the GMCs divide and differentiate into GCs (green) via FAMA activity. (B) SPCH, MUTE, and FAMA protein structures with the bHLH domain in blue, conserved C-terminal residues in black, and SPCH’s MPKTD in purple. LxCxE and LxSxE are RBR-binding motifs. (C) Excerpt from ClustalW2 alignment of Arabidopsis SPCH, MUTE, and FAMA proteins from MUTE’s first residue through the SPCH MPKTD. The basic region of the bHLH domain (blue) is underlined, putative DNA-contacting residues are in orange, and the SPCH MPKTD is in purple. Putative phosphorylation sites are in green, numbered according to the notation in ref. 14. Asterisks indicate identical residues; colons, conserved substitutions; dots, semiconserved substitutions.
Sequence analysis of SPCH, MUTE, and FAMA reveals both common and unique elements that are conserved across species (Fig. 1 B and C). Along with other subgroup Ia members, these genes show high conservation in the extension following their bHLH domain and in their C-termini (6, 12, 13). Arabidopsis SPCH has a unique 93-aa domain known as the MAP KINASE TARGET DOMAIN (MPKTD) located between its bHLH domain and the conserved C-terminus in which multiple phosphorylatable residues are targets of MPK3 and MPK6 (14). The GSK3β-like kinase BRASSINOSTEROID-INSENSITIVE 2 (BIN2) targets overlapping MPKTD residues, as well as several sites in the N-terminus (15), indicating that multiple signaling pathways converge on posttranslational regulation of SPCH. FAMA also encodes a long N-terminal extension (conserved among orthologs, but distinct from SPCH), whereas MUTE lacks sequences N-terminal of its bHLH domain entirely (6, 9, 11). Sequence and functional analysis in the distantly related moss Physcomitrella patens revealed that the subgroup Ia member PpSMF1 partially complements both mute and fama, but not spch, when expressed under the respective Arabidopsis promoters (5), raising the questions of how MUTE and FAMA functions became subdivided and how the distinct asymmetric division-promoting function of SPCH arose.
The stomatal bHLHs regulate independent developmental transitions within a single plant, and provide an opportunity to examine transitions in protein specialization over evolutionary timescales. Here we conducted a detailed functional study of the Arabidopsis proteins anchored by analysis of SPCH. We examined both conserved (DNA-binding) and unique (phosphorylation site) motifs, and found that SPCH has some capacity to substitute for MUTE, but not for FAMA. We also found that neither SPCH nor MUTE requires canonical DNA-binding motifs for function, but FAMA does. Based on a recent sequence analysis of the family (6), these data are consistent with a model in which FAMA is representative of the ancestral state, from which SPCH and MUTE arose via duplication events and diminished the DNA-binding requirement. Subsequent divergence of SPCH and MUTE involved acquisition (or retention) of novel phosphorylation sites in SPCH.
Results
Removal of SPCH Phosphorylation Sites Increases SPCH’s Ability to Promote GMC Fate.
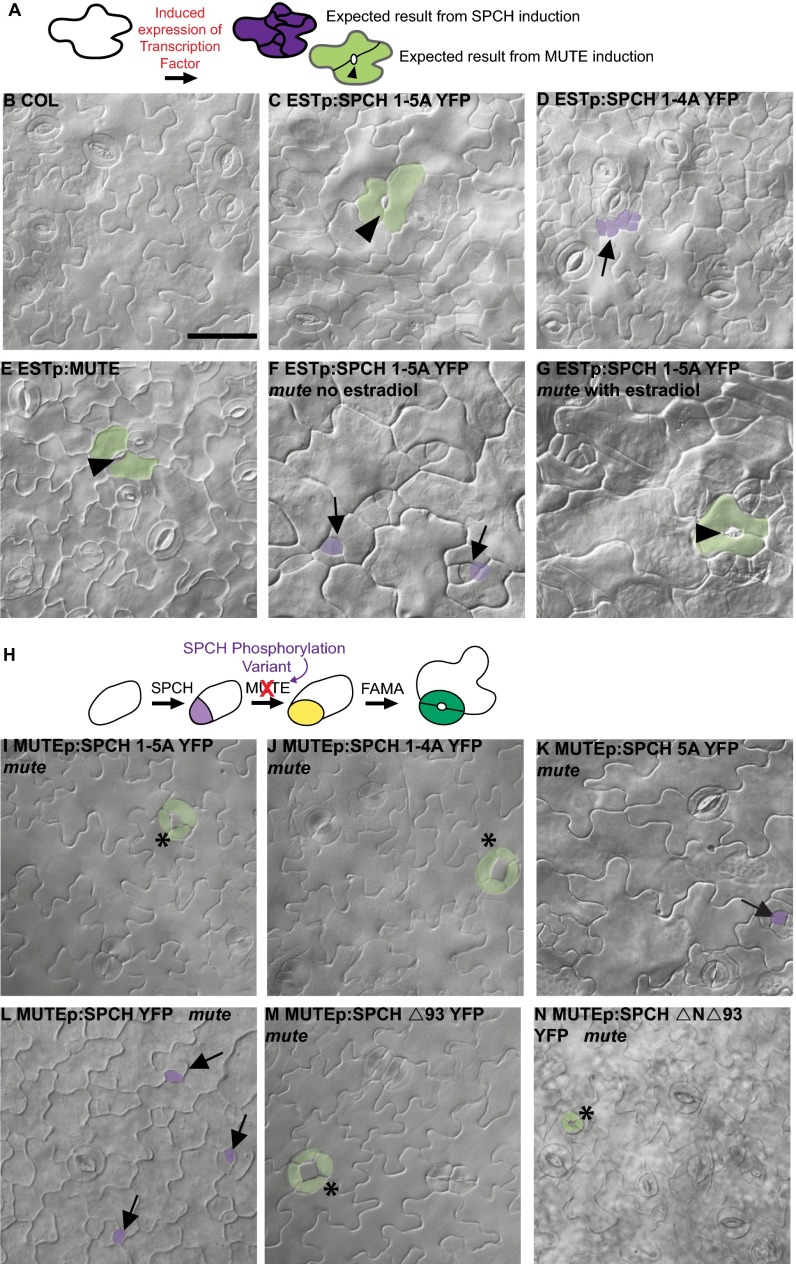
Previously, comparisons of the behaviors of SPCH variants in which normally phosphorylated serine/threonine residues were successively replaced by nonphosphorylatable alanines indicated that phosphorylation quantitatively inhibits SPCH activity, but also may qualitatively restrict SPCH behavior to promoting asymmetric divisions in early stomatal development (14, 15). Specifically, expression in spch of a SPCH variant in which five MAPK phosphorylation sites were eliminated (SPCH1-5A) promoted GC production but not asymmetric divisions, whereas a variant lacking four sites (SPCH1-4A) produced excessive asymmetric divisions (14). To clarify how the loss of phosphorylation sites alters SPCH’s developmental activity, we analyzed the phenotypic output of SPCH phosphorylation variants expressed under both ectopic and stomatal lineage promoters.
We used a broadly expressed estrogen-inducible promoter (Estp) (16) to overexpress SPCH variants in WT. When transferred to induction media at 5 d postgermination (dpg) and imaged 72 h later, SPCH1-5AYFP converted pavement cells into stomata-like complexes with pores formed between large lobed cells (Fig. 2C). In contrast, SPCH1-4AYFP generated supernumerary divisions in pavement cells (Fig. 2D). The conversion of pavement cells into stomatal complexes is an action normally associated with MUTE's activity in promoting GMC fate (Estp:MUTE lines; Fig. 2E) (11).
Fig. 2.
SPCH phosphorylation variants have increased ability to promote GMC fate. (A) Experimental setup for inducible transcription factor expression. SPCH activity promotes ectopic divisions, whereas MUTE promotes GMC (and subsequent stomatal) fate, including formation of stomatal pores (black arrowhead). (B–G) Differential interference contrast (DIC) images of 8-dpg abaxial cotyledons of indicated genotypes after 10 µM β-estradiol treatment (except F, noninduced control for G). (H) Experimental setup for replacement of MUTE with MUTE promoter-driven expression of SPCH variants. (I–N) DIC images of phenotypes in 8-dpg abaxial cotyledons of indicated genotypes. (Scale bar: 50 μm.) B–G and I–N are all at the same magnification. Arrowheads indicate stomatal pores; arrows, arrested meristemoid lineages; asterisks, aberrant divisions in GCs. Examples of cells with GC and meristemoid fate are shaded green and purple, respectively.
SPCH1-5AYFP could be promoting GMC fate directly (i.e., by substituting for MUTE) or indirectly by promoting the expression of endogenous MUTE. To distinguish between these possibilities, we induced SPCH1-5AYFP in a mute background. Pavement cells in induced Estp:SPCH1-5A YFP mute plants could indeed become GCs, suggesting that SPCH1-5A promotes GMC fate itself (Fig. 2 F and G).
To test whether only excessive levels of SPCH1-5A elicit this behavior, we generated MUTEp:SPCH1-5A YFP lines. Even when expressed with the weak and cell type-specific MUTE promoter (11), SPCH1-5A could rescue mute (Fig. 2I), consistent with it acquiring MUTE GMC-promoting activity rather than activating or overriding the requirement for endogenous MUTE. The phenotype generated by MUTEp:SPCH1-5A YFP in mute was not completely identical to rescue by MUTEp:MUTE-YFP, however (Table S1) (11, 17). Notably, MUTEp:SPCH1-5A YFP mute produced a novel phenotype of aberrant GC divisions (Fig. 2I). These extra divisions indicate that SPCH1-5A likely retains a division-promoting behavior characteristic of SPCH, and that this phosphovariant has SPCH-MUTE hybrid activity.
The ability of SPCH1-5A to rescue mute was surprising, given previous results indicating that stomatal bHLHs generally are not interchangeable; neither FAMA nor MUTE promotes asymmetric divisions when expressed under the SPCH promoter (10), and FAMA and MUTE are not able to compensate for each other when expressed under each others’ promoters (5). To address whether SPCH1-5A was hinting at functional interchangeability between SPCH and MUTE, we tested the ability of additional SPCH variants to promote GMC fate when expressed under the MUTE promoter in mute plants. We first tested SPCH1-4AYFP (14), the variant that promoted divisions rather than GMC fate when expressed ectopically (Fig. 2D). MUTEp:SPCH1-4A YFP was able to rescue stomatal development in mute seedlings (Fig. 2J). We next tested a SPCH phosphovariant missing a single phosphorylatable residue (MUTEp:SPCH5A YFP). Again, mute lethality was rescued (Table S1); most stomatal lineages arrested (Fig. 2K, arrows), but the ≥20 stomata per cotyledon were apparently sufficient for seedling growth. In contrast, WT SPCH exhibited a much more limited capacity to promote GMC fate. MUTEp:SPCH-YFP mute seedlings arrested, with most stomatal lineages exhibiting the mute phenotype of arrested meristemoids, although a few stomata per cotyledon developed (Fig. 2L). These results indicate that when placed in an appropriate spatiotemporal context, SPCH can promote GMC fate, albeit inefficiently. This GMC-promoting capacity is enhanced by the removal of MPKTD phosphorylation sites. The ability of SPCH variants to substitute for MUTE is unique to this pair, however, as demonstrated by the fact that similar SPCH variant swaps in a fama background (FAMAp:SPCH-YFP, FAMAp:SPCH2-4A YFP, FAMAp:SPCH1-4A YFP, and FAMAp:SPCH1-5A YFP) were unable to promote GC formation (Table S1).
In addition to its MPKTD, SPCH has phosphorylatable residues at its N-terminus that regulate activity (15) and could be important in distinguishing SPCH from MUTE, especially considering that MUTE lacks any extension N-terminal to its bHLH domain (Fig. 1 B and C). We were particularly interested in whether these N-terminal sites are responsible for the production of clusters of stomata or for GCs that undergo additional divisions (both evidence of retained SPCH-like division-promoting behavior) created when SPCH1-5A, SPCH1-4A, or SPCH5A was used to rescue mute (Fig. 2 I and J). We used the MUTE promoter to express SPCH variants lacking the entire MPKTD (SPCHΔ93) (14) or both the MPKTD and the N-terminal domain (SPCHΔNΔ93) (15). Constructs lacking these domains are able to rescue mute (Fig. 2 M and N and Table S1) but still exhibit divided GCs, indicating that differences in SPCH and MUTE behavior cannot be attributed entirely to the known phosphorylation domains of SPCH. Collectively, however, these alterations in SPCH behavior suggest that phosphorylation status is a defining characteristic in the functional distinction between SPCH and MUTE.
Another difference between SPCH and MUTE is the presence of two potential RETINOBLASTOMA RELATED (RBR)-binding motifs (LxSxE) (18) in SPCH (Fig. 1C). Excessive cell division phenotypes resembling those produced by SPCH hyperactivity result from depletion of Arabidopsis RBR (19), suggesting an alternative mechanism by which SPCH could specifically promote cell divisions; however, an SPCH variant lacking both LxSxE motifs (SPCHp:SPCHLGK) rescued spch and did not exhibit excessive cell divisions (Fig. S1 A and B).
SPCH Does Not Require DNA-Binding Residues for GMC Promotion.
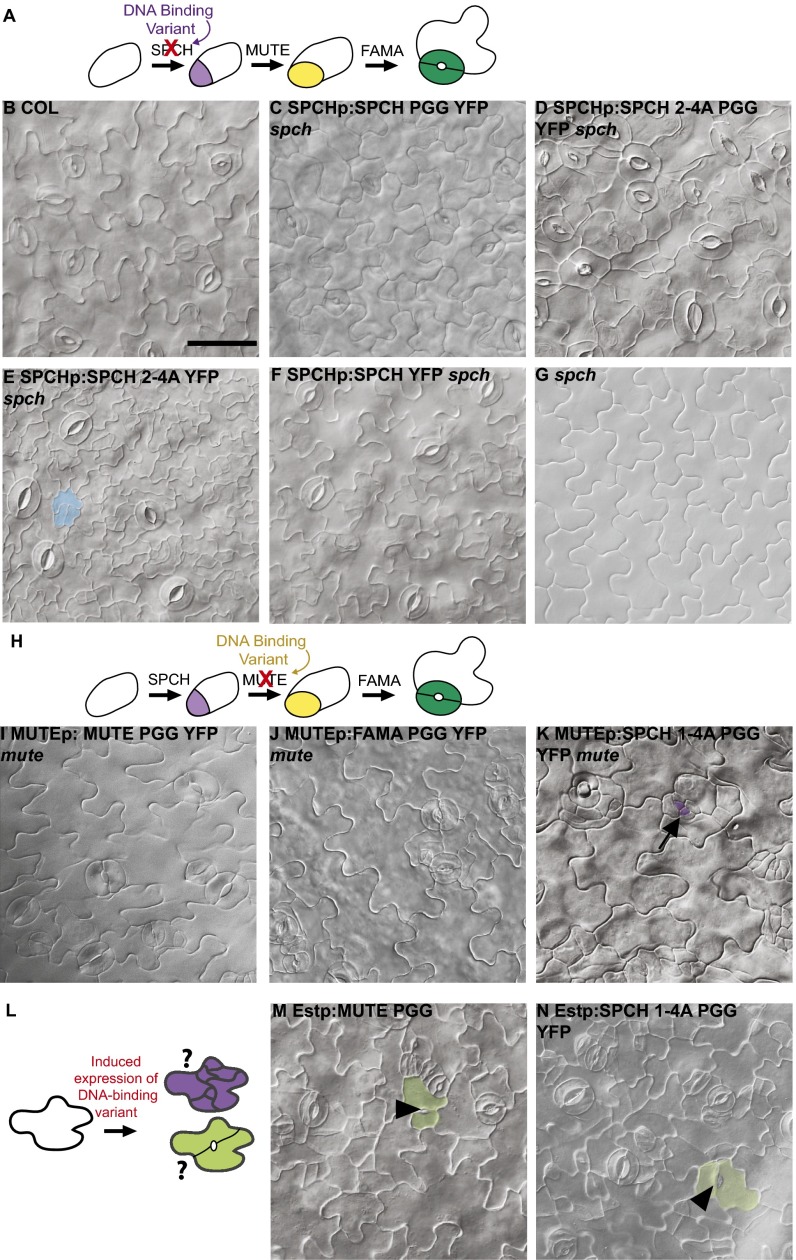
Because SPCH is a transcription factor, its ability to promote asymmetric division and cryptic ability to promote GMC fate are likely tied to its capacity to regulate gene expression. SPCH, MUTE, and FAMA belong to a subclass of plant bHLHs whose basic domain contains three characteristic amino acids—histidine (H), glutamic acid (E), and arginine (R), with conserved spacing—that have been shown in other proteins to make critical contacts with DNA and recognize G-box DNA elements (13). Substitution of these residues can render other bHLH proteins inactive and/or create dominant negative versions that sequester dimerization partners into nonfunctional complexes (20). Altering the HER residues in FAMA (FAMAPGG) renders the protein incapable of rescuing fama (9).
Given the previous data indicating that the DNA-binding residues provide specificity in some stomatal lineage functions, we mutated HER residues, creating SPCHp:SPCHPGGYFP and SPCHpro:SPCH2-4A PGGYFP, and tested whether these function normally as SPCH and/or if they gain MUTE-like activity. SPCHp:SPCHPGGYFP was able to rescue lethality and promote stomatal formation in spch (Fig. 3C). Expression of SPCHp:SPCH2-4A PGGYFP also was able to rescue spch lethality, and we observed both asymmetric divisions and stomatal production (Fig. 3D). In contrast to the SPCH2-4AYFP variant, however, SPCH2-4A PGGYFP did not lead to a striking overproliferation of small cells in spch, but instead exhibited aberrant pavement cells with GC morphologies and stomatal clusters (Fig. 3 D and E). Taken together, these results indicate that mutation of SPCH’s DNA-binding residues results in a shift of activity toward the MUTE-like activity of GMC specification.
Fig. 3.
SPCH and MUTE missing DNA-binding residues still promote divisions and GMC fate. (A) Experimental setup for replacement of SPCH with HER motif variants. (B–G) DIC images of 6-dpg abaxial cotyledons of indicated genotypes. (H) Experimental setup for replacement of MUTE with HER motif variants. (I–K) DIC images of 9-dpg abaxial cotyledons of indicated genotypes. (L) Experimental setup of inducible expression of DNA-binding variants. (M and N) DIC images of 8-dpg abaxial cotyledons of indicated genotypes. (Scale bar: 50 μm.) B–G, I–K, M, and N are at the same magnification. Black arrows indicate arrested lineages, and arrowheads denote stomatal pores. Shading indicates cells that have undergone ectopic divisions (blue) or that have GC (green) or meristemoid (purple) fate.
MUTE Does Not Require DNA-Binding Residues for GMC Fate, but FAMA Does So to Promote GC Fate.
The observation that both SPCHPGG and FAMAPGG (when expressed in WT) promote the production of ectopic two-celled stomata raises the possibility that MUTE normally promotes GMC identity without its presumed DNA-binding residues. We created MUTE promoter-driven versions of MUTE (as well as of SPCH and FAMA) missing the HER residues to test whether any was capable of promoting GMC fate in the absence of endogenous MUTE. Both MUTEp:MUTEPGGYFP and MUTEp:FAMAPGGYFP promoted stomatal development in mute, but with occasional stomatal clusters (Fig. 3 I and J). FAMAp:FAMAPGG YFP did not produce stomata in fama seedlings (Table S1), confirming previous reports (9) that mutating these residues precludes FAMA from specifying GC identity. These results indicate that DNA-binding is critical for FAMA GC-promoting activity and suggest the possibility that the posttranslational regulation of DNA-binding capacity of an ancestral FAMA-like protein could allow a single protein to mediate both GMC and GC fate transitions.
In contrast to the MUTE and FAMA variants, a form of SPCH with alternations to both the MPKTD and DNA-binding domain (SPCH1-4A, PGG) could not effectively promote GMC fate in mute (Fig. 3K). Although ESTp:SPCH1-4A PGGYFP converted pavement cells to stomata in WT (Fig. 3 M and N), the effects of MUTEp:SPCH1-4A PGGYFP are dependent on the presence of endogenous MUTE. In roughly one-half of MUTEp:SPCH1-4A PGGYFP mute seedlings, we could find between one and three stomata, but the majority of meristemoids arrested, and the stomatal lineage cells exhibited divisions in excess of those normally observed in mute plants (Fig. 3K, arrows and Table S1). Thus, versions of SPCH lacking several phosphorylation sites and DNA-binding residues no longer can effectively substitute for MUTE.
SPCH and MUTE Have Overlapping but Distinct Expression Patterns.
Despite the evidence indicating that features of the SPCH protein confer unique functions, there is considerable overlap in the GMC-promoting behavior of SPCH and MUTE. Thus, we next asked to what extent expressional differences distinguish SPCH and MUTE from each other. Although SPCH and MUTE control sequential steps of stomatal development and SPCH expression precedes MUTE expression (10, 11), it is not known if or to what extent SPCH persists in MUTE-expressing cells.
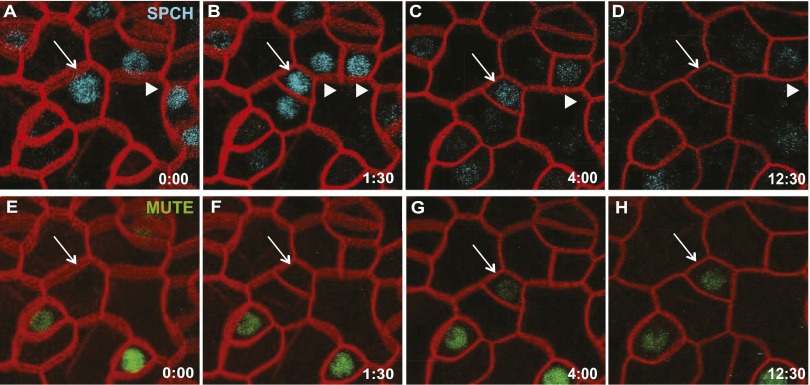
We developed a custom time-lapse microscopy chamber to monitor the expression of SPCHp:SPCH-CFP and MUTEp:MUTE-YFP simultaneously in the epidermis of true leaves of 5- to 6-d-old seedlings. Stomatal development is asynchronous, and we could follow multiple cells at each stage of stomatal lineage progression. We focused our analysis on the transitions of meristemoid cells as they were beginning to express MUTE-YFP. In this population, peak expression of SPCH-CFP expression clearly precedes the onset of MUTE-YFP expression (Fig. 4); however, in all six of the cells that we monitored, when MUTE expression became detectable, SPCH-CFP signal was still present (Fig. 4 C and G, arrows). The low levels of SPCH-CFP in strong MUTE-YFP–expressing cells indicate that the two proteins have distinct peak accumulation patterns, but that there is some potential for their coexistence. This coexistence appears to be sufficiently limited to functionally segregate SPCH and MUTE in Arabidopsis, because mute seedlings produce no stomata, but supplying SPCH in MUTE’s expression domain (MUTEp:SPCH mute) allows the production of a few stomata (Fig. 2L).
Fig. 4.
Time-lapse imaging of SPCH and MUTE reveals overlapping but distinct expression patterns. Confocal imaging of SPCHp:SPCH-CFP (A–D) and MUTEp:MUTE-YFP (E–H). Cell outlines (red) marked with ML1p:RCI2A-mCherry. White arrows follow a single cell through a SPCH-driven asymmetric division (A and E) until its conversion into a MUTE-expressing cell (D and H). Other cells lose SPCH expression without gaining MUTE expression in this time frame (arrowheads). Time is given in hours:minutes at the bottom right corner of each panel.
Discussion
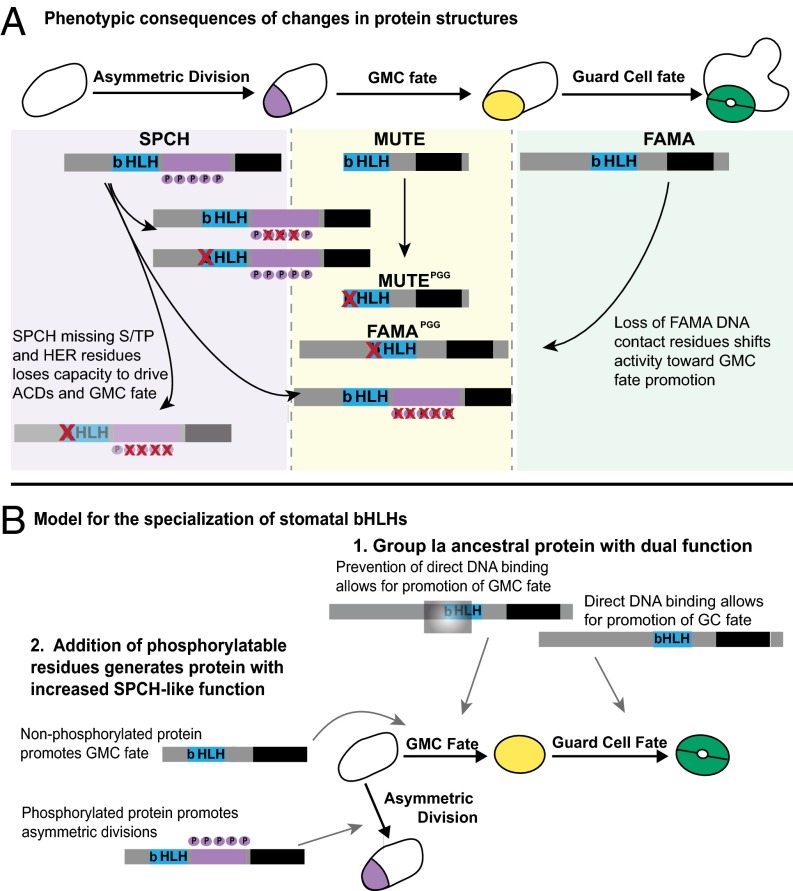
Arabidopsis SPCH, MUTE, and FAMA originated from a common ancestor, yet currently drive essential and discrete steps of stomatal development. In Arabidopsis, the development of paired GCs requires, at a minimum, the activities of MUTE and FAMA to first specify a GMC that will divide symmetrically and to then specify the GC identity of the resulting daughter cells. The addition of SPCH allows for flexible lineage expansion in response to intercellular signals and environmental inputs, but is not absolutely required for GC fate (10). Previous analysis indicates that the subgroup Ia genes in P. patens have MUTE/FAMA dual functionality in their ability to promote both GMC and GC fate when expressed in Arabidopsis (5). Our present results provide insight into motifs that may have been important for the functional diversification of the stomatal bHLHs (Fig. 5).
Fig. 5.
Summary of behavior-modifying alterations to stomatal bHLHs and a model for the evolution of specialized stomatal bHLH function. (A) Schematic of normal Arabidopsis stomatal progression with WT SPCH, MUTE, and FAMA positioned under the transition that they promote. P’s in purple circles indicate phosphorylation sites. The subgroup Ia-specific C-terminus is in black. Variants investigated in this study are illustrated, with red X’s marking elimination of specific residues. These modified proteins are positioned relative to the transitions that they promote. (B) A plausible evolutionary model supported by sequence comparisons (6) and experimental data from this study. 1, The ancestral subgroup Ia protein may promote both GMC and GC fates by existing in DNA-binding and non–DNA-binding states (here indicated by occlusion with a gray box); after gene duplication, these two functions later become associated with separate proteins (MUTE and FAMA). 2, After another gene duplication, the acquisition of phosphorylation sites changed the subgroup Ia ancestral activity by suppressing the GMC differentiation function and enabling a novel asymmetric division function.
Alterations in DNA binding may be important in distinguishing MUTE from FAMA. A MUTE variant lacking its DNA-binding residues is able to rescue mute (Fig. 3I), and a FAMA variant lacking its DNA-binding residues rescues mute (Fig. 3J) but not fama (9), indicating that the loss of FAMA’s DNA-binding activity confers the ability to promote GMC fate. These results suggest that for GC specification, FAMA must actively bind DNA, whereas for GMC specification, members of a larger transcription factor complex (in which MUTE is a part) may be the guiding factors for selecting targets. A protein could fulfill both MUTE and FAMA functions provided that it was capable of alternating between DNA-binding and non–DNA-binding states (Fig. 5B). In this case, posttranslational regulation via interacting partners may be an important means of controlling a protein that is required over multiple developmental transitions.
Our data also expand the roles for phosphorylation of SPCH. SPCH variants missing phosphorylation sites are able to promote GMC fate and rescue mute to nearly WT appearance; however, WT SPCH expressed under the same MUTE promoter cannot (Fig. 2 I–L). The phosphorylation sites may be important in modulating SPCH’s conformation or interacting partners. The ability of SPCH to promote GMC fate offers an explanation for the puzzling observation that the loss of the ERECTA family (ERf) LRR-RLKs can suppress mute (17). MPK3 and MPK6, downstream effectors of the ERf kinases, phosphorylate SPCH (14). Loss of ERf signaling would generate hypophosphorylated SPCH, resulting in increased GMC-promoting behaviors (Fig. 5A). Thus, phosphorylation of SPCH appears to have at least two regulatory functions; it down-regulates the protein, but also ensures that SPCH promotes asymmetric divisions rather than GMC identity. Because MUTEp:SPCH-YFP also weakly promotes GMC fate in mute, expressional differences likely contribute to the stage-specific action of these proteins (Fig. 4).
Although SPCH, MUTE, and FAMA have distinct functions, little is known about the larger transcriptional complexes through which they mediate cell fate transitions, except that all three appear to form obligate heterodimers with ICE1 and SCRM2 (8). ICE1 binds canonical E-box motifs (21), and the excessive stomatal production conferred by the ICE1-D mutation requires the conserved E residue of the protein’s DNA-binding region (8), indicating that ICE1 could select targets of SPCH, MUTE, and FAMA transcriptional complexes. Homologs of ICE1 are present in all extant plant species with complete genomes, indicating that the heterodimeric relationships between group Ia and group III bHLHs could be ancient (6). This partnership makes the contribution of specific residues to DNA-binding activity of SPCH, MUTE, and FAMA difficult to dissect, because in vitro binding assays become dominated by the activity of the group III partner. Recent improvements in ChIP of developmental regulators (22) make it possible to detect genome-scale binding activities, however. ChIP-seq revealed that SPCH binds to thousands of sites in the genome, many of them E-box containing, and SPCH RNA-seq revealed expression changes in the hundreds (22). Comparisons of similar ChIP-seq and RNA-seq profiles from MUTE and FAMA with those from SPCH variants would show how specific residues alter in vivo biochemical activity, and link gene expression changes with the different developmental outcomes.
FAMA was recently shown to recruit RBR through its LxCxE interaction motif (23). Because a version of SPCH missing its potential RBR-binding sites behaved indistinguishably from WT SPCH, FAMA is likely unique in its physical interaction with RBR. Interaction with RBR cannot be the sole defining feature of FAMA, however, given that elimination of RBR binding does not create a version of FAMA that behaves like SPCH or MUTE (Fig. S1C). Additional work on protein–protein interactions is needed to uncover the larger transcriptional complexes that contribute to specific functions and to identify how different motifs, such as SPCH phosphorylation, are involved in these interactions.
Collectively, our results identify motifs that functionally distinguish related transcription factors and point to a possible explanation for how multiple functions might have coexisted in ancestral proteins expressed across multiple developmental transitions (Fig. 5B). In a recent phylogenetic analysis of stomatal subgroup Ia bHLHs (6), no clear MUTE homologs were identified in gymnosperms, but there were SPCH homologs with truncated MPKTDs. Our demonstration of SPCH’s increased ability to promote GMC fate with the loss of even a single phosphorylation site (MUTEp:SPCH5AYFP compared with MUTEp:SPCH-YFP) is consistent with the possibility of gymnosperm proteins having dual functions. A hypothetical ancestral protein with SPCH/MUTE duality may have had phosphorylation sites that were lost after gene duplication (a model favored by some; ref 6). Alternatively, some or all of SPCH’s phosphorylation sites may have been acquired after duplication of a more MUTE-like ancestor, allowing for increased posttranslational regulation and, consequently, increased regulation of stomatal divisions in angiosperms. Overexpression of the P. patens subgroup Ia gene SMF1 in Arabidopsis recapitulated aspects of SPCH, MUTE, and FAMA overexpression, including the promotion of ectopic divisions, suggesting the presence of nascent SPCH-like activity even without the presence of an obvious MPKTD (5). Currently, Arabidopsis SPCH is a target of MAPK and brassinosteroid signaling, as well as an integration point for environmental information that allows for appropriate patterning and optimization of stomatal density in changing conditions. Interestingly, rice MUTE has potential MAPK phosphorylation sites and can promote divisions when expressed in Arabidopsis (4), indicating that rice MUTE may have dual SPCH/MUTE-like functions.
The evolutionary path that ultimately partitioned the specific SPCH, MUTE, and FAMA functions among individual proteins is likely complex, given the combination of cis-regulatory, coding sequence, and potential posttranslational regulation modifications that together generate their current distinct functions in Arabidopsis. Likewise, the Arabidopsis SPCH, MUTE, and FAMA paradigm described here might differ across the plant kingdom, and there may be several evolutionary routes to fine-tune stomatal development. As a complement to the acquisition of sequence data from diverse lineages, which can define novelties (like SPCH phosphorylation sites), functional studies such as this one in a single organism can reveal that conserved sequences (such as DNA-binding residues) nonetheless may diverge in activity. Thus, functional analysis of motifs in homologs is essential to further our understanding of how different sequence modifications can allow for alterations of biochemical function and diversification of a developmental process over evolutionary time.
Materials and Methods
Plant Materials.
Previously published mutants used in this study were spch-3 (SAIL36_B04) (10), mute-1 (11), and fama-1 (SALK_100073) (9), all in Columbia (Col) ecotype. Epidermal phenotypes were analyzed by clearing seedlings in a 7:1 ethanol:acetic acid solution and mounting in Hoyer’s solution (24). For estrogen inductions, 5-dpg seedlings were flooded with a solution of 10 μM β-estradiol (Sigma-Aldrich) in water for 6–8 h and scored for epidermal phenotypes 3 d later. For assaying rescue, T1s were genotyped for the mutation. If no homozygous mutant T1s were obtained, then progeny of heterozygotes were analyzed for the mutant phenotype. T2 seedling arrest was scored at 10–14 dpg, and deviation from 1/4 (no rescue) or 1/16 (rescue) of the mutant phenotype was determined using the χ2 test. For lines in which rescue was not observed, construct expression was verified by observation of YFP fluorescence. For time-lapse imaging, 5-dpg seedlings were transferred to a sterilized chamber (25) for imaging on a Leica SP5 confocal microscope. The chamber was perfused with one-quarter strength 0.75% (wt/vol) sucrose liquid MS media. Z-stacks through the epidermis were captured with Leica software every 30 min and processed with Fiji.
DNA Manipulations.
Constructs for this study were created through Gateway (Invitrogen) cloning using SPCH and MUTE variants in pENTR and pDONR lines in one of five backbones: (i) pMDC7 (16): ESTp:SPCH1-4A YFP, ESTp:SPCH 1–5A YFP, ESTp:MUTE, EStp:SPCH 1–4A PGG YFP, Estp:MUTE PGG; (ii) R4pGWB540 (26, 27): MUTEp:SPCH 1–4A YFP, MUTEp:SPCH 1–5A YFP, MUTEp:SPCHYFP, MUTEp:SPCH5AYFP, MUTEp:SPCHΔSPCH5AYFP ΜΥΤEp:SPCHΔNΔ93YFP, SPCHp:SPCH2-4A PGG YFP, SPCHp:SPCH PGG YFP, MUTEp:MUTE PGG YFP, MUTEp:FAMA PGG, SPCHp:SPCHLGKYFP; (iii) pHGC (28): SPCHp:SPCH-CFP; (iv) pHGY (28): MUTEp:MUTE YFP; or (v) R4pGWB501 (26, 27): MUTEp:SPCH 1–4A YFP and ML1p:mCherry RCI2A. Primer sequences are listed in Table S2.
Supplementary Material
Acknowledgments
We thank Drs. T. Nakagawa, L. Pilliterri, A. Roeder, C. MacAlister, C. Northover, D. Wengier, and S. Wenkel for reagents; E. Abrash for the time-lapse setup; C. Ballenger for technical help; and M. Rowe for comments on the manuscript. Funding was provided by the National Institutes of Health (Grant 1R01 GM086632). K.A.D. was supported by National Institutes of Health Grant 5T32GM007276 and by a National Science Foundation graduate research fellowship. D.C.B. is a Gordon and Betty Moore Foundation Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1411766111/-/DCSupplemental.
References
- 1.Li X, Noll M. Evolution of distinct developmental functions of three Drosophila genes by acquisition of different cis-regulatory regions. Nature. 1994;367(6458):83–87. doi: 10.1038/367083a0. [DOI] [PubMed] [Google Scholar]
- 2.Xue L, Li X, Noll M. Multiple protein functions of paired in Drosophila development and their conservation in the Gooseberry and Pax3 homologs. Development. 2001;128(3):395–405. doi: 10.1242/dev.128.3.395. [DOI] [PubMed] [Google Scholar]
- 3.Rudall PJ, Hilton J, Bateman RM. Several developmental and morphogenetic factors govern the evolution of stomatal patterning in land plants. New Phytol. 2013;200(3):598–614. doi: 10.1111/nph.12406. [DOI] [PubMed] [Google Scholar]
- 4.Liu T, Ohashi-Ito K, Bergmann DC. Orthologs of Arabidopsis thaliana stomatal bHLH genes and regulation of stomatal development in grasses. Development. 2009;136(13):2265–2276. doi: 10.1242/dev.032938. [DOI] [PubMed] [Google Scholar]
- 5.MacAlister CA, Bergmann DC. Sequence and function of basic helix-loop-helix proteins required for stomatal development in Arabidopsis are deeply conserved in land plants. Evol Dev. 2011;13(2):182–192. doi: 10.1111/j.1525-142X.2011.00468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ran JH, Shen TT, Liu WJ, Wang XQ. Evolution of the bHLH genes involved in stomatal development: Implications for the expansion of developmental complexity of stomata in land plants. PLoS ONE. 2013;8(11):e78997. doi: 10.1371/journal.pone.0078997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pires N, Dolan L. Origin and diversification of basic helix-loop-helix proteins in plants. Mol Biol Evol. 2010;27(4):862–874. doi: 10.1093/molbev/msp288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanaoka MM, et al. SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. Plant Cell. 2008;20(7):1775–1785. doi: 10.1105/tpc.108.060848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohashi-Ito K, Bergmann DC. Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell. 2006;18(10):2493–2505. doi: 10.1105/tpc.106.046136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacAlister CA, Ohashi-Ito K, Bergmann DC. Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature. 2007;445(7127):537–540. doi: 10.1038/nature05491. [DOI] [PubMed] [Google Scholar]
- 11.Pillitteri LJ, Sloan DB, Bogenschutz NL, Torii KU. Termination of asymmetric cell division and differentiation of stomata. Nature. 2007;445(7127):501–505. doi: 10.1038/nature05467. [DOI] [PubMed] [Google Scholar]
- 12.Heim MA, et al. The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol Biol Evol. 2003;20(5):735–747. doi: 10.1093/molbev/msg088. [DOI] [PubMed] [Google Scholar]
- 13.Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 2003;15(8):1749–1770. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lampard GR, Macalister CA, Bergmann DC. Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science. 2008;322(5904):1113–1116. doi: 10.1126/science.1162263. [DOI] [PubMed] [Google Scholar]
- 15.Gudesblat GE, et al. SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat Cell Biol. 2012;14(5):548–554. doi: 10.1038/ncb2471. [DOI] [PubMed] [Google Scholar]
- 16.Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133(2):462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pillitteri LJ, Bogenschutz NL, Torii KU. The bHLH protein, MUTE, controls differentiation of stomata and the hydathode pore in Arabidopsis. Plant Cell Physiol. 2008;49(6):934–943. doi: 10.1093/pcp/pcn067. [DOI] [PubMed] [Google Scholar]
- 18.Durfee T, et al. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7(4):555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 19.Borghi L, et al. Arabidopsis RETINOBLASTOMA-RELATED is required for stem cell maintenance, cell differentiation, and lateral organ production. Plant Cell. 2010;22(6):1792–1811. doi: 10.1105/tpc.110.074591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Massari ME, Murre C. Helix-loop-helix proteins: Regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20(2):429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chinnusamy V, et al. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003;17(8):1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lau OS, et al. Direct roles of SPEECHLESS in the specification of stomatal self-renewing cells. Science. 2014 doi: 10.1126/science.1256888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matos JL, et al. Irreversible fate commitment in the Arabidopsis stomatal lineage requires a FAMA and RETINOBLASTOMA-RELATED module. Science 2014 doi: 10.7554/eLife.03271. , in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu CM, Meinke DW. The titan mutants of Arabidopsis are disrupted in mitosis and cell cycle control during seed development. Plant J. 1998;16(1):21–31. doi: 10.1046/j.1365-313x.1998.00268.x. [DOI] [PubMed] [Google Scholar]
- 25.Robinson S, et al. Generation of spatial patterns through cell polarity switching. Science. 2011;333(6048):1436–1440. doi: 10.1126/science.1202185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakagawa T, et al. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng. 2007;104(1):34–41. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
- 27.Nakagawa T, et al. Development of R4 gateway binary vectors (R4pGWB) enabling high-throughput promoter swapping for plant research. Biosci Biotechnol Biochem. 2008;72(2):624–629. doi: 10.1271/bbb.70678. [DOI] [PubMed] [Google Scholar]
- 28.Kubo M, et al. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 2005;19(16):1855–1860. doi: 10.1101/gad.1331305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.