Significance
Germ-line mutation in the tumor suppressor TP53 causes Li–Fraumeni syndrome (LFS), a complex predisposition to multiple cancers. Types of cancers and ages at diagnosis vary among subjects and families, with apparent genetic anticipation: i.e., earlier cancer onset with successive generations. It has been proposed that anticipation is caused by accumulation of copy-number variations (CNV) in a context of TP53 haploinsufficiency. Using genome/exome sequencing, we found no evidence of increased rates of CNVs in two successive generations of TP53 mutation carriers and in successive generations of Trp53-deficient mice. We propose a stochastic model called “genetic regression” to explain apparent anticipation in LFS, caused by segregation of rare SNP and de novo mutations rather than by cumulative DNA damage.
Keywords: Li–Fraumeni syndrome, whole genome sequencing, genetic anticipation, p53 mutation, copy number variation
Abstract
The Li–Fraumeni syndrome (LFS) and its variant form (LFL) is a familial predisposition to multiple forms of childhood, adolescent, and adult cancers associated with germ-line mutation in the TP53 tumor suppressor gene. Individual disparities in tumor patterns are compounded by acceleration of cancer onset with successive generations. It has been suggested that this apparent anticipation pattern may result from germ-line genomic instability in TP53 mutation carriers, causing increased DNA copy-number variations (CNVs) with successive generations. To address the genetic basis of phenotypic disparities of LFS/LFL, we performed whole-genome sequencing (WGS) of 13 subjects from two generations of an LFS kindred. Neither de novo CNV nor significant difference in total CNV was detected in relation with successive generations or with age at cancer onset. These observations were consistent with an experimental mouse model system showing that trp53 deficiency in the germ line of father or mother did not increase CNV occurrence in the offspring. On the other hand, individual records on 1,771 TP53 mutation carriers from 294 pedigrees were compiled to assess genetic anticipation patterns (International Agency for Research on Cancer TP53 database). No strictly defined anticipation pattern was observed. Rather, in multigeneration families, cancer onset was delayed in older compared with recent generations. These observations support an alternative model for apparent anticipation in which rare variants from noncarrier parents may attenuate constitutive resistance to tumorigenesis in the offspring of TP53 mutation carriers with late cancer onset.
Li–Fraumeni syndrome (LFS) [Online Mendelian Inheritance in Man (OMIM) 151623], as well as its variant form, Li–Fraumeni-like (LFL), is one of the most disparate forms of familial cancer. This autosomal dominant disorder is characterized by clustering of early-onset cancers of the central nervous system, soft tissue sarcoma, osteosarcoma, and premenopausal breast cancer and by increased risk of a wide spectrum of common cancers in midlife (1). The most characteristic tumor phenotypes are rhabdomyosarcoma, adrenal cortical carcinoma, and choroid plexus carcinoma in early childhood, defining an LFS-specific cancer triad.
Patients from families with LFS/LFL traits often carry germ-line TP53 mutations (2). Currently, no other recurrent germ-line alteration has been associated with this disease pattern. TP53 is a tumor suppressor gene encoding the protein p53, exerting integrated antiproliferative effects through control of cell cycle, apoptosis, senescence, DNA repair, differentiation, and basal energy metabolism (3). Partial or complete loss of function may lead to decreased genetic stability in cells exposed to DNA damage and deficient capacity of stem/progenitor cells to enter differentiation and senescence, thus predisposing carriers of germ-line TP53 mutations to a defined spectrum of cancers.
Anticipation is commonly observed in multigenerational LFS/LFL pedigrees. This phenomenon takes two forms. First, the average age at cancer diagnosis seems to decrease from one generation to the other. Second, the type of cancer diagnosis changes from one generation to the other, with a switch from less LFS-specific adult cancers to more LFS-specific, early-childhood cancers (4). However, evaluating these effects is prone to multiple biases in ascertaining cancer over several generations. Moreover, observing anticipation requires that pedigrees originate from subjects who have not developed (or have survived) childhood cancer. Therefore, the exact nature of anticipation in LFS remains to be clarified.
Individual and familial variability in LFS/LFL has led to the hypothesis that, aside from differences in germ-line TP53 mutations (5), genetic or epigenetic traits may act as modifiers in determining age of disease onset. A single nucleotide polymorphism (SNP), SNP309 (rs2279744) in MDM2 (5), a negative regulator of TP53, a nonsynonymous SNP in exon 4 of TP53 (rs1042522) (6), and a 16-bp duplication polymorphism in intron 3 (rs17878362) (7) have all been associated with an earlier age of first cancer onset in LFS/LFL families. Other genetic mechanisms that have been suggested include accumulation of copy-number variations (CNVs) with successive generations, a hallmark of germ-line genome instability, and progressive telomere shortening (8). The latter mechanisms have also been documented in pedigrees with anticipation features: namely, in breast cancer susceptibility syndromes (BRCA1 carriers) (9) and in Lynch syndrome (10).
In this study, we used data on germ-line TP53 mutation compiled in the International Agency for Research on Cancer (IARC) TP53 mutation database to evaluate anticipation in pedigrees with germ-line TP53 mutations. Next, we have investigated the genetic basis of phenotypic heterogeneity by sequencing the entire genome of 13 members of an LFS kindred showing evidence of acceleration in age at cancer onset and increased severity of the syndrome over two generations. Our results suggest that anticipation is not driven by the accumulation of CNV but may result from Mendelian segregation of rare susceptibility/resistance variants affecting the penetrance of germ-line TP53 mutations. To our knowledge, this is the first report of using whole-genome sequencing (WGS) to assess genetic heterogeneity in a kindred with LFS.
Results
Anticipation Patterns in Germ-Line TP53 Mutation Carriers.
Table 1 compares the mean age at first cancer onset in different generations for the entire dataset of families with germ-line TP53 mutations documented in the R16 version of the IARC TP53 mutation database. For any two successive generations, and independently of the number of generations in the families, the mean age at first cancer onset tended to be lower in the generation of offspring than in the generation of parents. This observation recapitulates on the entire dataset of 269 families the observations made on many individual pedigrees showing decrease in age at onset and/or increasing severity of symptoms in successive generations. Statistical analysis showed that the acceleration in disease onset was significant over three generations in families with four generations (generation 1–3, from 51.8 ± 15.4 y in generation 1 to 33.5 ± 12.5 y in generation 3, P < 0.001) and in families with three generations (from 45.7 ± 16.2 y in generation 1 to 20.1 ± 15.1 y in generation 3, P < 0.032). The same trend was seen between generation 1 and generation 2 in families with two generations, but this difference was not statistically significant. It should be noted that pooled data for generation 4 in families with four generations show a very low age at cancer onset (10.0 ± 8.0 y). However, these data were excluded from the analysis because the median age in this generation was 11.5 y (average 10.8 ± 7.8), precluding the evaluation of age at diagnosis for adult forms of LFS/LFL, which represent over 25% of all diagnoses in LFS/LFL. On the other hand, Table 1 shows no significant difference in age at cancer onset when comparing equivalent generations between families with one, two, three, and four generations. Thus, age at cancer onset in families with a single documented generation is similar to the second generation of families with two generations and of the third generation of families with three generations. The same phenomenon is observed for any generation, independently of family structure. For example, the age at first cancer onset is 33.5 ± 12.5 y in the third generation of families with four documented generations, 36.9 ± 16.8 y in the second generation of families with three documented generations, and 35.9 ± 14.5 y in the first generation of families with two documented generations (differences are nonsignificant). This observation suggests that apparent anticipation is caused by delayed occurrence of cancer in the first generations of pedigrees with three or four documented generations, rather than by acceleration of cancer onset in successive generations.
Table 1.
Mean age at first cancer onset in different generations of LFS families with 1–4 documented generations
| Records of families with N generations with available data | |||||
| Generation | 431, n = 4 | 797, n = 3 | 570, n = 2 | 169, n = 1 | P (t test) |
| 1 | 51.8 ± 15.4 | — | — | — | — |
| 2 | 45.6 ± 15.3 | 45.7 ± 16.2 | — | — | P(4-3) = n.s. |
| 3 | 33.5 ± 12.5 | 36.9 ± 16.8 | 35.9 ± 14.5 | — | P(4-2) = n.s. |
| 4 | (10.0 ± 8.0) | 20.1 ± 15.1 | 22.5 ± 18.9 | 21.2 ± 23.9 | P(3-1) = n.s. |
| P (t test) | P(1-3) < 0.001 | P(1-3) < 0.032 | P(1-2) = n.s. | — | — |
P value for significance of difference in mean age of cancer onset between the first and last generation of multigeneration families are listed in the bottom row, and between comparable generations across different generation families are listed in the rightmost column. The mean age for generation 4 is in parenthesis because the median age of this generation is 11.5 years, so is excluded from analysis. n.s., nonsignificant.
The same observation is supported by Table 2, which compares the proportion of childhood cancers of the “LFS-specific” triad (rhabdomyosarcoma, adrenal cortical carcinoma, and choroid plexus carcinoma) at each generation in families with one to four documented generations. These early cancers can be considered as the most severe manifestation of the syndrome. Whereas there is a tendency for an increased proportion of these cancers in more recent generations of families with two, three, and four documented generations, the proportion of these cancers is similar in equivalent generations independently of family structure. Thus, these childhood cancers represent 2.9%, 2.8%, and 4.9% of all diagnoses in, respectively, generation 3 of families with four generations, generation 2 of families with three generations, and generation 1 of families with two generations. In the more recent generation documented, the proportion of “triad” cancers is of 24.0%, 17.4%, 8%, and 17.0% in families with four generations, three generations, two generations, or one documented generation, respectively. Further analyses according to sex did not reveal different patterns between males and females, with respect to age at cancer onset or type of cancer. The same analyses were repeated independently for subsets of data from Northern America and from Western Europe, with no differences between the two datasets. Furthermore, the proportion of patients with two or more cancer diagnoses, another possible hallmark of disease severity, was lower in recent generations of multigeneration families than in single-generation families (Fig. S1). This proportion was 57.1% in single-generation families, 23% in generation 2 of families with two generations, 19% in generation 3 of families with three generations, and 10.3% in generation 4 of families with four generations.
Table 2.
Frequency of childhood cancer (rhabdomyosarcoma, adrenal cortical carcinoma, choroid plexus carcinoma) in each generation of LFS families with 1–4 documented generations
| Records of families with N generations with available data | ||||
| Generation | 22, n = 4 | 59, n = 3 | 27, n = 2 | 24, n = 1 |
| 1 | 0 | — | — | — |
| 2 | 0 | 0 | — | — |
| 3 | 2.9 | 2.8 | 4.9 | — |
| 4 | 24.0 | 17.4 | 8.0 | 17.0 |
Finally, we examined whether the accrual with age of childhood cancer (until age 20) was accelerated in recent generations of families with multiple generations, compared with single-generation families. This analysis did not reveal any tendency for childhood cancer to occur at an earlier age in recent generations of multigeneration families. Overall, this analysis suggests that, in multigeneration families, cancer onset is delayed in the older generations compared with single-generation families and that there is no strictly defined anticipation effect associated with germ-line TP53 mutation carriage.
Whole-Genome Sequencing of an LFS Kindred.
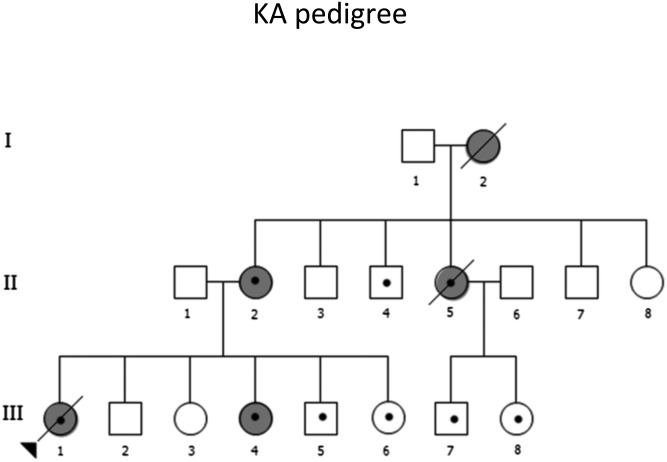
The above observations are consistent with the hypothesis that TP53 mutation carriers who develop cancer only at a late age may have a form of genetic resistance to early cancer, which may be attenuated by Mendelian segregation in successive generations. Alternatively, spouses who are not carriers may introduce into the carrier lineages susceptibility alleles that suppress the resistance observed in the founders of large pedigrees. These hypotheses are distinct from, but not incompatible with, the notion that, in some pedigrees, acceleration of cancer onset might be caused by increased genetic instability, accumulation of CNV, and progressive telomere shortening. To address this question in a global manner, we undertook the complete genome sequencing of 13 members of an LFS pedigree (of Malay origin) in which a deleterious germ-line TP53 mutation had been previously identified (6-bp insertion, residues 334 and 336) (Fig. 1) (11). This family showed a clear tendency for accelerated cancer onset over three successive generations, with a probable (unconfirmed) carrier grandmother who developed breast cancer at age 38, a carrier mother who developed bilateral breast cancer at age 26–27, and a sister who developed osteosarcoma at age 26, and carrier children who developed rhabdomyosarcoma at age 8 mo and adrenal cortical carcinoma at age 6 mo, whereas two other siblings and two cousins are carriers who have yet to develop a cancer (current ages 6–14).
Fig. 1.
Pedigree of LFS kindred KA. I:2 and II:2 had breast cancer at ages 38 y and 26 y, respectively. II:5 developed osteosarcoma at age 26 y. III:1 presented at 8 mo with embryonal rhabdomyosarcoma of the trunk. III:4 developed adrenocortical carcinoma (ACC) at 6 mo. There was no reported cancer history for II:1 and his parents and siblings. All members with validated mutation in TP53 are denoted with a dot. The family members II:1, II:2, III:1 to III:6, II:4, II:5, II:8, and III:7 and III:8 were selected for WGS, and II:1, II:2, III:2, III:4, and III:6 were also selected for aCGH.
The mean mapped depth of whole genomes ranged from 24 to 32, and average read length was 46 bp. Between 68% and 75% of the genome was covered with at least 20 reads and 90% of the genome was covered with at least 10 reads.
Because it has been suggested that TP53 haplotypes may exert a modifier effect on age at cancer onset (12), we phased TP53 genotypes into haplotypes for this family (Table S1) and examined their distribution in relation with age at cancer onset. Using single nucleotide polymorphisms (SNPs) identified in TP53 and its 10-kb surrounding regions, we selected 20 SNPs that are of high confidence and informative for phasing the genotypes into maternal haplotypes M1 and M2 of subjects II:2 (mother) (M1 carrying the TP53 mutation) and paternal haplotypes P1 and P2 of II:1 (father). Haplotypes M1 and P1 are identical haplotypes based on the 20 SNPs and are distinguished only by the presence of the mutation on M1.
The two carrier children with early cancer inherited different paternal TP53 haplotypes (III:1, haplotype P1; and III:4, haplotype P2). Similarly, the two carrier siblings without early cancer also inherited different paternal haplotypes (III:5, haplotype P1; and III:6, haplotype P2). This observation excludes the possibility that the paternal haplotype alone could explain the difference in cancer status between carrier siblings. Similarly, neither the MDM2-SNP309 G allele nor the rare (duplicated) allele of the TP53 PIN3 polymorphism was found in this LFS kindred, excluding these two polymorphisms as contributors to differences in age at cancer onset. Independently of WGS, we used The TeloTAGGG telomere length assay kit (Roche Diagnostics) to evaluate changes in telomere length between successive generations and among siblings within the same generation. Results did not reveal any significant difference in the size of average telomere restriction fragments (TRFs) among members of this LFS kindred (Fig. S2).
Analysis of CNV Distribution.
WGS data were extensively analyzed for CNV exceeding 10 kb in size. Results (Table 3) show that CNV composition did not show significant variation among family members, either within the same generation or between successive generations. In particular, our results show no significant difference in CNV composition among the four carrier siblings (III:1, III:4, III:5, and III:6) or between them and the two noncarrier siblings (III:2 and III:3) despite their differences in TP53 mutation carriage and in cancer status. For children III:1 to III:6, de novo CNVs were investigated by comparing their individual sequencing data with those of their parents (II:1 and II:2) (Table S2). Any CNV from a child that overlapped with a CNV from either parent was considered as inherited and otherwise as occurring de novo. The vast majority of CNV was inherited. At most one de novo CNV was found in any child, consistent with a low false-positive rate for CNV detection and independent of TP53 mutation carrier and cancer status. The average total number of CNVs of the six siblings (123 ± 8) was similar to the average of their parents (124), consistent with inheritance with no bias in selection of CNV numbers. Furthermore, the uncle (II:4) and two cousins (III:7 and III:8), all of whom are carriers, had higher total CNV numbers although they did not develop early cancer. The results were identical when restricting the analysis to either deletions or duplications, as well as to CNV greater than 100 kb or 500 kb. Thus, we conclude that there is no germ-line instability in the form of CNV larger than 10 kb in this LFS kindred and that total CNV accrual does not occur in relation with acceleration of cancer onset.
Table 3.
Number of CNV for LFS kindred KA from WGS
| CNV number | Subjects | ||||||||||||
| II:1 | II:2 | II:4 | II:5 | II:8 | III:1 | III:2 | III:3 | III:4 | III:5 | III:6 | III:7 | III:8 | |
| L > 10 kb | 134 | 114 | 157 | 144 | 137 | 123 | 118 | 114 | 137 | 125 | 122 | 132 | 151 |
| L > 100 kb | 46 | 43 | 40 | 37 | 38 | 47 | 46 | 38 | 40 | 41 | 39 | 45 | 45 |
| L > 500 kb | 3 | 6 | 5 | 4 | 5 | 5 | 5 | 6 | 3 | 8 | 7 | 5 | 3 |
The number of CNV with size L called using read-depth coverage and allele frequencies.
Rare Variants Cosegregating with Early Cancer Onset.
WGS analysis among family members identified rare single nucleotide variants (SNVs) from the father (noncarrier TP53) that were transmitted to the two TP53 mutation carrier children with early cancer but not to the two TP53 mutation carrier children who have not yet developed cancer. SNV found in II:1, III:1, and III:4 (but not found in III:5 and III:6) were identified and are listed in Table S3. The whole-genome list of rare SNV was narrowed down to 11 candidate genes by selecting only exonic or splicing variants in conserved regions and by removing nonsynonymous variants and common variants found in dbSNP, 1000 Genome, and 100 Southeast Asian Malay whole-genome databases (13). Additionally, we detected one nonsynonymous de novo SNV in the child with adrenocortical carcinoma (ACC) (III:4), which was not present in any other member of the family. Of these SNVs, nine missense mutations and one nonsense mutation were further confirmed by conventional Sanger sequencing (Table S3). Such rare SNV may be considered as candidate-modifier genes that may modulate age at cancer onset in subjects who carry a TP53 mutation in the germ line.
Whole-Exome Sequencing in an LFL Kindred.
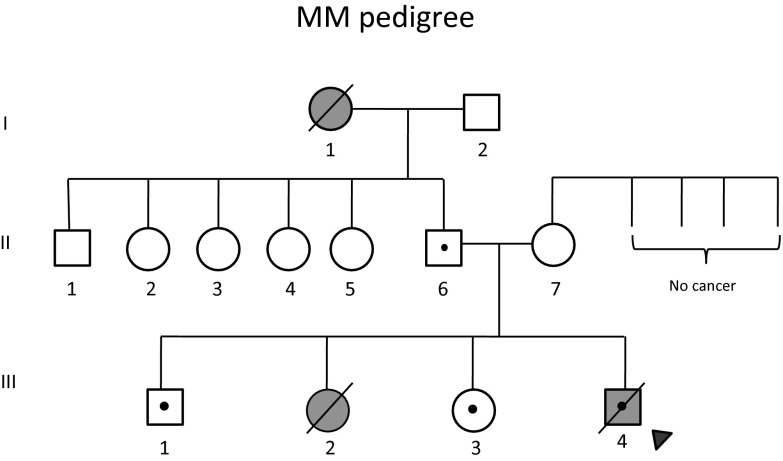
De novo mutation analysis was repeated in another independent LFS family with five members using whole-exome sequencing (WES). This family (Fig. 2) harbored a germ-line TP53 mutation at codon 245 (p.G245S), a classical “hotspot” TP53 mutation in LFS/LFL. The average sequencing read depth on target for WES was 60 and the fraction of target covered with at least 20 reads was 90%.
Fig. 2.
Pedigree of LFL kindred MM. Proband (III:4) was diagnosed with rhabdomyosarcoma at the age of 29 mo. III:2 had succumbed to a brain tumor at age of 32 mo. I:1 died at 55 y of age from breast cancer. The remaining family members are cancer-free. All members with validated mutation in TP53 are denoted with a dot. The family members II:6, II:7, III:1, III:3, and III:4 were selected for WES and aCGH.
De novo SNVs were identified in the offspring. The expected number of de novo SNV mutations in an offspring was one per exome. We identified one candidate de novo mutation in the proband (III:4) in Dync2h1:NM_001080463:exon35:c.G5518A:p.V1840I and one in his brother (III:1) in Fgf21:NM_0191113:exon2:c.C263T:p.P88L. No de novo mutation was found in the exome of his sister (III:3). There is no evidence that the affected child had a higher germ-line point mutation rate than unaffected siblings, but the numbers were too small for statistical significance.
CNV Detection Using Array Comparative Genomic Hybridization.
We performed array comparative genomic hybridization (aCGH) on five members of the KA family (II:1, II:2, III:2, III:4, and III:6) to validate our CNV findings from WGS. The numbers of CNVs detected from WGS and aCGH, respectively, were in strong agreement [II:1 (134 vs. 131), II:2 (114 vs. 118), III:2 (118 vs. 126), III:4 (137 vs. 133), and III:6 (122 vs. 121)] and, again, did not show any relation with TP53 status or age of first cancer (Dataset S1). Of note, III:6 had three large amplifications (4 Mb in 8q24.3, 11p15.5-p15.4, and 19p13.3-p13.11) and III:5 had one large amplification (11p15.5-p15.4) and one large deletion (9p13.1-p11.2). All large amplifications and the deletion were determined to be somatic based on the ratio of probe signals, showing that the gains or loss occur in a fraction of the total cell population.
We also performed aCGH on five members of the MM family and found no significant difference in the number of CNVs between carriers, noncarriers, and cancer status: II:6 (116), II:7 (117), III:1 (117), III:3 (99), and III:4 (126) (Dataset S2).
Rates of de Novo CNV in trp53 Knockout Versus WT Mice.
We used an experimental mouse model to further evaluate whether constitutive p53 deficiency in the germ line may increase the rate of de novo CNV in the offspring. C57BL/6 mice with defined trp53 status (either +/+, +/−, or −/−) were crossed with 129SvSL trp53+/+ mice, and 129SvSL trp53−/− mice were crossed with C57BL/6 trp53+/+ mice (Dataset S3). CNVs were analyzed in DNA isolated from whole F1 embryos at 17.5 d and compared with liver DNA from their parents. One out of 36 embryos from trp53+/+ parents was detected to have a potential de novo CNV (Dataset S3). No de novo CNV was detected in 84 embryos with a p53-deficient parent. These observations suggest that, in this mouse model, partial or complete deficiency of p53 function in the germ line does not increase the rate of CNV formation in the offspring.
Discussion
A decrease in age at first cancer onset is frequently observed with successive generations in individual LFS/LFL families (4). In the present study, we developed an assessment of this apparent anticipation effect using a curated dataset of 269 pedigrees of TP53 germ-line mutation carriers, including 1,771 cancer cases. We confirmed the statistical reality of decrease in age at first cancer onset in multigeneration pedigrees. However, our observations did not fit with a classical model of anticipation, which predicts that cancer would develop at a late and relatively constant age in the first generation of any pedigree, with a decrease in age of onset in successive generation. In fact, our results showed the opposite effect: compared with pedigrees with only one or two generations with documented cancer, pedigrees with three or four generations showed a delayed age at first cancer onset in the older generations of TP53 mutation carriers. This observation may be affected by a strong ascertainment bias due to underdiagnosis or underreporting of early cancer onset in older generations. A simple explanation for this phenomenon is that the founder of such pedigrees may carry, in addition to germ-line TP53 mutation, rare independent genetic modifiers that attenuate the risk of early cancer, allowing cancer-free survival until postreproduction age. Mendelian segregation may dilute this resistance trait during successive generations, leading to a progressive decrease in age at first cancer onset, as well as to a shift from adult to childhood cancer forms. Alternatively, acceleration of cancer onset may be caused by the introduction of susceptibility alleles from the noncarrier parent in the germ line of their TP53 mutation carrier progeny. We suggest the term of “genetic regression” to define this pattern of cancer inheritance, distinguishing it from strictly defined “genetic anticipation.” This term underlines the analogy between this apparent cancer acceleration pattern and the mathematical concept of regression to the mean.
It has been suggested that, in multigeneration pedigrees, a form of genetic instability may develop over successive generations, leading to the accumulation of alterations that accelerate cancer onset. In agreement with this hypothesis, Shlien et al. (8) have documented an accumulation of CNVs between generations within the same pedigree with anticipation traits, as well as increased CNV in subjects with early cancer compared with cancer-free subjects within the same generation of a pedigree.
In this study, we used the power of WGS to explore the extent of genetic and genomic variations among 13 members of two successive generations of an LFS kindred displaying anticipation and heterogeneous tumor phenotypes over three generations. This kindred was selected for its extremely disparate patterns of age at cancer onset. Although two of three TP53 mutation carriers in generation II developed cancer in their mid-twenties, in generation III, two carrier subjects developed cancer in the first year of life whereas two carrier siblings and two carrier cousins have yet to develop cancer at ages 6–14. Furthermore, this family is also characterized by the absence of any of the polymorphisms that have been so far associated with a possible effect on age at cancer onset in TP53 mutation carriers. Haplotyping of the TP53 locus and flanking areas ruled out that the differences in age at cancer onset might be associated with inheritance of a specific TP53 haplotype of paternal origin, modulating the effect of the mutant, maternal haplotype. Analysis of the size of average telomere restriction fragments did not reveal any difference among members of the family, either in relation with position in the pedigree, tumor phenotype, and carriage of germ-line TP53 mutation or age at cancer onset.
Systematic analysis of whole-genome CNV > 10 kb did not detect any significant difference among the 13 subjects. These results are at variance with previously reported data that increased submicroscopic copy-number alterations may correlate with earlier age at first cancer onset. This discrepancy may indicate that different mechanisms may contribute to phenotypic heterogeneity in different families and populations. However, it may also have a technical basis because microarray-based methods previously used for detecting CNV had low resolution compared with WGS, with detection of a mean of 3 and 12 CNVs per genome for control and TP53 mutation carriers, respectively (8). In contrast, WGS allowed the detection of about 130 CNVs per genome. Moreover, variation in total CNV is high in the population, and sampling errors may result in a case-control comparison of unrelated people showing spurious difference. Our comparison among siblings eliminates this potential confounder. Another intriguing possibility is that some of the CNVs detected in previous reports may be somatic instead of germ line, and blood of LFS carriers, especially those having had cancer, has more detectable somatic CNVs (14). Absence of significant differences in CNV accrual between carriers with and without early cancer was confirmed by aCGH of five members, including TP53 mutation carriers over two generations in a second, independent kindred with LFS/LFL tumor patterns. Furthermore, no evidence for increase of de novo SNV was detected for proband with early cancer compared with the sibling (carrier) without early cancer.
We propose that, rather than by CNV accrual, the peculiar and atypical anticipation pattern observed in LFS may be explained by the modifier effects of a wide range of rare variants that influence the penetrance in germ-line TP53 mutation carriers. According to this model, only a small number of de novo TP53 mutation carriers may have appropriate tolerant genetic backgrounds that would confer them long enough cancer-free survival to have children and become founders for LFS families. In subsequent generations, their offspring would inherit half of this tolerant genome whereas the genes inherited from the other parent may contain a number of variants that makes it less tolerant than the founder’s genome. Thus, these children may develop cancer at earlier ages, and this process may be cumulative over generations. This model is consistent with data from studies in Trp53-null mice. A study of over 100 breeding Trp53 knockout mice over 15 generations has not revealed evidence for increased accumulation of CNV and of germ-line genetic instability. In mice deficient for Trp53, age at onset and type of cancer is remarkably dependent upon genetic background (15) as well as of the presence of germ-line mutations in other genes such as BRCA1 (16) or RB1 (17–19). We anticipate that the variants involved in modulating heterogeneity may be complex and may differ from one family to the other. Individual cancer risk may therefore be determined by a complex interplay between the inherited mutant TP53 allele and the segregation of independent susceptibility and resistance alleles inherited from each parent. The new etiological model we propose for phenotypic heterogeneity and anticipation in LFS predisposition syndrome predicts many more childhood cancers from individuals with de novo germ-line TP53 mutations than are currently reported. As TP53 mutation testing and screening becomes more common, this model will be tested. Genetic regression may provide a paradigm for heterogeneity and anticipation in other autosomal dominant diseases with variable age of onset where manifestation of disease at prereproductive age will result in negative reproductive fitness.
Materials and Methods
This study was approved by the institutional ethics committee at the University of Malaya Medical Center, and written informed consent was obtained from all patients. Germ-line TP53 mutation and anticipation data are from the IARC TP53 database version R16, and their analyses are described in SI Materials and Methods. WGS, WES, aCGH, and their analyses on the KA and MM families are described in SI Materials and Methods. The Trp53 knockout and WT mice and their CNV analyses are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We acknowledge the contributions of Drs. Teo Soo Hwang, Teo Yik Ying, and Wong Lai Ping. This work was funded by a High-Impact Research Grant from the Ministry of Education, Malaysia (High Impact Research-Ministry of Higher Education Grant Scheme). C.S.C. is grateful for support from the Rutgers Cancer Institute of New Jersey. M.I.A. is grateful for support from National Institute of Oncogenomics (CNPq/MCT/FAPESP).
Footnotes
The authors declare no conflict of interest.
Data deposition: The mice data reported in this paper has been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo/ (accession no. GSE61660).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1417322111/-/DCSupplemental.
References
- 1.Mai PL, et al. Li-Fraumeni syndrome: Report of a clinical research workshop and creation of a research consortium. Cancer Genet. 2012;205(10):479–487. doi: 10.1016/j.cancergen.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malkin D, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250(4985):1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 3.Scoumanne A, Chen X. Protein methylation: A new mechanism of p53 tumor suppressor regulation. Histol Histopathol. 2008;23(9):1143–1149. doi: 10.14670/hh-23.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trkova M, Hladikova M, Kasal P, Goetz P, Sedlacek Z. Is there anticipation in the age at onset of cancer in families with Li-Fraumeni syndrome? J Hum Genet. 2002;47(8):381–386. doi: 10.1007/s100380200055. [DOI] [PubMed] [Google Scholar]
- 5.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: Origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010;2(1):a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bougeard G, et al. Impact of the MDM2 SNP309 and p53 Arg72Pro polymorphism on age of tumour onset in Li-Fraumeni syndrome. J Med Genet. 2006;43(6):531–533. doi: 10.1136/jmg.2005.037952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marcel V, et al. TP53 PIN3 and MDM2 SNP309 polymorphisms as genetic modifiers in the Li-Fraumeni syndrome: Impact on age at first diagnosis. J Med Genet. 2009;46(11):766–772. doi: 10.1136/jmg.2009.066704. [DOI] [PubMed] [Google Scholar]
- 8.Shlien A, et al. Excessive genomic DNA copy number variation in the Li-Fraumeni cancer predisposition syndrome. Proc Natl Acad Sci USA. 2008;105(32):11264–11269. doi: 10.1073/pnas.0802970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinez-Delgado B, et al. Genetic anticipation is associated with telomere shortening in hereditary breast cancer. PLoS Genet. 2011;7(7):e1002182. doi: 10.1371/journal.pgen.1002182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bozzao C, Lastella P, Stella A. Anticipation in lynch syndrome: Where we are where we go. Curr Genomics. 2011;12(7):451–465. doi: 10.2174/138920211797904070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ariffin H, Martel-Planche G, Daud SS, Ibrahim K, Hainaut P. Li-Fraumeni syndrome in a Malaysian kindred. Cancer Genet Cytogenet. 2008;186(1):49–53. doi: 10.1016/j.cancergencyto.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 12.Sagne C, et al. Age at cancer onset in germline TP53 mutation carriers: association with polymorphisms in predicted G-quadruplex structures. Carcinogenesis. 2014;35(4):807–815. doi: 10.1093/carcin/bgt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong LP, et al. Deep whole-genome sequencing of 100 southeast Asian Malays. Am J Hum Genet. 2013;92(1):52–66. doi: 10.1016/j.ajhg.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laurie CC, et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat Genet. 2012;44(6):642–650. doi: 10.1038/ng.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donehower LA, et al. Effects of genetic background on tumorigenesis in p53-deficient mice. Mol Carcinog. 1995;14(1):16–22. doi: 10.1002/mc.2940140105. [DOI] [PubMed] [Google Scholar]
- 16.Liu X, et al. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc Natl Acad Sci USA. 2007;104(29):12111–12116. doi: 10.1073/pnas.0702969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harvey M, Vogel H, Lee EY, Bradley A, Donehower LA. Mice deficient in both p53 and Rb develop tumors primarily of endocrine origin. Cancer Res. 1995;55(5):1146–1151. [PubMed] [Google Scholar]
- 18.Williams BO, et al. Cooperative tumorigenic effects of germline mutations in Rb and p53. Nat Genet. 1994;7(4):480–484. doi: 10.1038/ng0894-480. [DOI] [PubMed] [Google Scholar]
- 19.Donehower LA. Insights into wild-type and mutant p53 functions provided by genetically engineered mice. Hum Mutat. 2014;35(6):715–727. doi: 10.1002/humu.22507. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.