Abstract
The β-galactoside-binding protein galectin-9 is critical in regulating the immune response, but the mechanism by which it functions remains unclear. We have demonstrated that galectin-9 is highly expressed by induced regulatory T cells (iTreg) and was crucial for the generation and function of iTreg cells but not natural regulatory T (nTreg) cells. Galectin-9 expression within iTreg cells was driven by the transcription factor Smad3, forming a feed-forward loop, which further promoted Foxp3 expression. Galectin-9 increased iTreg cell stability and function by directly binding to its receptor CD44, which formed a complex with transforming growth factor-β (TGF-β) receptor I (TGF-βRI), and activated Smad3. Galectin-9 signaling was further found to regulate iTreg cell induction by dominantly acting through the CNS1 region of Foxp3 locus. Our data suggest that exogenous galectin-9, in addition to being an effector molecule for Treg cells, acts synergistically with TGF-β to enforce iTreg cell differentiation and maintenance.
Introduction
The role of Foxp3+ regulatory T (Treg) cells in immune tolerance and homeostasis has been extensively studied. Treg cells comprise a specific T cell lineage, which can suppress effector T cell responses during infection, inflammation and autoimmunity (Josefowicz et al., 2012a; Sakaguchi et al., 2010). As a master transcription factor of Treg cells, Foxp3 plays a critical role in their development and regulates a wide spectrum of Treg cell functions (Sakaguchi et al., 2010; Zheng and Rudensky, 2007). At least two different types of Foxp3+ Treg cells have been defined. Natural Treg (nTreg) cells develop in the thymus and recognize self-antigen with intermediate affinity, leading to their differentiation towards regulatory cells. In contrast, adaptive or induced Treg (iTreg) cells can differentiate from naïve T cells in the periphery and are especially important in regulating immune responses and autoimmunity in the gut (Bluestone and Abbas, 2003; Josefowicz et al., 2012b). Interestingly, both of these Treg cell subsets express Foxp3. Previous studies have shown that the expression of Foxp3 mainly depends on transforming growth factor-β receptor (TGF-βR) and interleukin 2 receptor (IL-2R) signaling (Fontenot et al., 2005; Kim et al., 2005; Ouyang et al., 2010). While TGF-β is critical for induction of Foxp3 expression, IL-2 supports the growth of iTreg cells. Loss of either TGF-β or IL-2 signaling results in a defect in Treg cell generation. TGF-β signaling largely activates Smad proteins, transcription factors known to promote the induction of a number of molecules required for Treg cell generation including Foxp3 (Ruan et al., 2009; Tone et al., 2008). The activation of TGF-βR directly triggers the phosphorylation and nuclear translocation of receptor-regulated Smad proteins, which subsequently mediate their binding to the Foxp3 locus, leading to the transactivation of Foxp3 expression (Lagna et al., 1996; Liu et al., 1997; Macias-Silva et al., 1996; Massague, 1998). Moreover, besides the transcriptional regulation of Foxp3, activation of Smad3 induces appropriate histone modifications, which result in the promotion and stabilization of the Foxp3 polypeptide (Ohkura et al., 2012; Toker and Huehn, 2011). A recent study demonstrates a critical role of non-coding DNA elements in the Foxp3 locus for the Treg cell lineage commitment, and epigenome-dependent regulation of these regions largely determines the function and stability of natural or induced Treg cells (Zheng et al., 2010).
Galectin-9 (encoded by Lgals9) is a member of the tandem-repeat group of galectins with multiple biological functions such as chemoattraction, cell aggregation and apoptosis (Alam et al., 2011; Rabinovich and Toscano, 2009). It is localized on the cell membrane, in the cytoplasm and nucleus (Hirashima et al., 2004). In vivo treatment with galectin-9 leads to the suppression of pro-inflammatory cytokines and an increase of Treg cells (Arikawa et al., 2009). Moreover, recent studies also indicate that administration of exogenous galectin-9 can regulate Th17 and Treg cell development (Kared et al., 2013; Oomizu et al., 2012). However, the precise molecular mechanism by which galectin-9 regulates Foxp3+ Treg cell differentiation is still largely unknown.
We and others have previously identified galectin-9 as a ligand for Tim-3, a T helper-1 (Th1) cell-specific type 1 membrane protein that can induce cell death in Th1 cells thereby downregulating effector Th1 cell responses (Zhu et al., 2005). Besides Tim-3, CD44 is another known cell surface molecule that can potentially interact with galectin-9 (Bollyky et al., 2009; Bourguignon et al., 2002; Liu et al., 2009; Tanikawa et al., 2010). CD44 is a highly glycosylated cell adhesion molecule, which binds not only to galectin-9 but also to hyaluronic acid (HA). It has been reported that galectin-9 can bind to CD44 and regulate leukocyte migration during allergic lung inflammation via modulation of CD44-HA interactions (Katoh et al., 2007; Nagahara et al., 2008).
In this study, by utilizing galectin-9 deficient mice, we have demonstrated that the genetic loss of galectin-9 leads to a reduction in Foxp3 expression and suppressor function of iTreg cells both in vitro and in vivo. We also have shown that galectin-9 synergizes with TGF-β to promote Foxp3 expression by interacting with a CD44-TGF-βRI complex. This interaction promoted not only the expression but also the stability of Foxp3, leading to enhanced suppressive function in iTreg cells. Moreover, galectin-9 signaling regulated iTreg generation via the CNS1 region of the Foxp3 genomic locus. Our results reveal a positive feed-forward loop employed by iTreg cells, which involves galectin-9-driven upregulation of Foxp3, which in turn strengthens the iTreg cell phenotype.
Results
Galectin-9 deficiency reduces iTreg but not nTreg cells
It has been previously reported that administration of galectin-9 promotes Treg cell numbers during the development of collagen-induced arthritis (CIA) (Arikawa et al., 2009; Seki et al., 2008), suggesting that galectin-9 might play an important role in Treg cell development in vivo. By employing galectin-9 deficient (Lgals9−/−) mice, we first found normal T cell development in thymus, spleen and lymph nodes (LN) in these mice (data not shown).
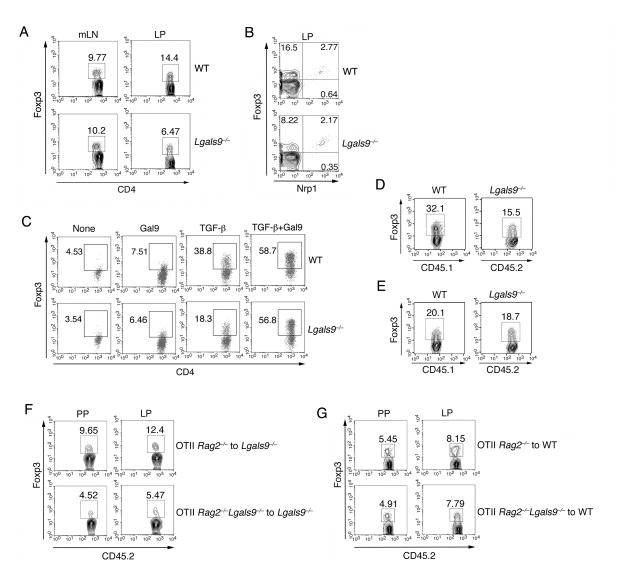
Under either steady state or inflammatory conditions, iTreg cells have been described to be generated and accumulate mainly in the gut (Coombes et al., 2007; Siddiqui and Powrie, 2008; Sun et al., 2007). Although we found a similar number of Foxp3+ Treg cells in the mesenteric LN (mLN), the cells isolated from the lamina propria (LP) of Lgals9−/− mice showed a lower frequency of Treg cells compared to WT littermates (Figure 1A and S1A). Neuropilin 1 (Nrp1) has been recently described as a surface marker to distinguish nTreg cells from iTreg cells (Weiss et al., 2012; Yadav et al., 2012). We thus analyzed the nTreg and iTreg population in the small bowel and found a lower frequency of Nrp1loFoxp3+ iTreg cells in LP of galectin-9 deficient mice, whereas there was no difference in Nrp1hiFoxp3+ nTreg cells between WT and Lgals9−/− mice (Figure 1B and S1B). These data suggest a defect in iTreg differentiation in the absence of galectin-9 in vivo.
Figure 1. Galectin-9 promotes Foxp3 expression during iTreg cell differentiation.
(A) The percentage of Foxp3+ Treg cells in mesenteric LN (mLN) and lamina propria (LP) was determined by flow cytometry; (B) Flow cytometry analysis of CD4+ T cells from the LP of WT or Lgals9−/− mice; (C) Activated naïve CD4+ T cells were stimulated with TGF-β and/or recombinant galectin-9. The frequency of Foxp3+ cells was determined by flow cytometry; CD45.1+ WT and CD45.2+ Lgals9−/− naïve T cells were cultured (D) separately or (E) together in the presence of TGF-β. The frequency of Foxp3+ cells was then determined by flow cytometry; OT-II Rag2−/− and OT-II Rag2−/−Lgals9−/− CD45.2+CD4+ T cells were transferred into congenic CD45.1 (F) Lgals9−/− or (G) WT recipient mice, followed by administration of OVA in the drinking water for 5 days. CD45.2+Foxp3+ iTreg cells from Peyer’s patches (PP) and LP of Lgals9−/− recipient mice were determined by flow cytometry. Data are representative of three independent experiments with n≥4 mice each group.
During in vitro differentiation, Lgals9−/− T cells displayed impaired Foxp3 expression (Figure 1C). Addition of exogenous galectin-9 alone only induced a modest increase in Foxp3 expression in activated naïve T cells, whereas we observed a substantial synergistic effect of galectin-9 with TGF-β in inducing Foxp3 expression. Furthermore, the exogenous addition of galectin-9 with TGF-β restored Foxp3 expression in Lgals9−/− iTreg cells (Figure 1C). We also discovered that galectin-9 promoted iTreg cells differentiation in a carbohydrate-dependent manner in that additional lactose, which breaks the β-galactoside bond of galectin-9, inhibited iTreg cell generation by TGF-β and galectin-9 (Figure S1C). Furthermore, this galectin-9 dependent effect was specific for iTreg cell differentiation, as we could not detect any defects of differentiation within other T cell subsets (data not shown). Overall, these results suggest that galectin-9 facilitates the expression of Foxp3 during iTreg cell differentiation both in vivo and in vitro.
Galectin-9 extrinsically regulates the generation of iTreg cells
We next asked the question whether such production of galectin-9 could function as an intrinsic or extrinsic mechanism for iTreg cell differentiation. We polarized CD45.1+ WT and CD45.2+ Lgals9−/− T cells in the presence of TGF-β either separately or together. The difference of Foxp3 expression between WT and Lgals9−/− T cells was diminished when the two types of cells were cultured together (Figure 1D and 1E). A similar phenotype was also observed in vivo. Ten weeks after reconstitution of congenic CD45.1 Lgals9−/− host mice with either WT or galectin-9-deficient (CD45.2) bone marrow (BM), we observed reduced Treg cells in LP with Lgals9−/− BM when compared to the WT BM chimeras (Figure S1D and S1E). However, when WT mice were used as hosts, where galectin-9 is present exogenously, Treg cells in different organs was found to be comparable (Figure S1F and S1G). To better understand whether the lack of galectin-9 could specifically impair iTreg cell generation in vivo, a model of ovalbumin (OVA)-induced generation of iTreg cells was employed (Coombes et al., 2007). We generated Lgals9−/− OVA specific T cells by crossing Lgals9−/− mice with Rag2−/− OT-II mice. OVA-specific T cells purified from these mice were transferred into congenic CD45.1 Lgals9−/− recipient mice. In response to the administration of OVA as a specific oral antigen, the frequency of Foxp3+ iTreg cells in the CD45.2+ donor cell compartment isolated from Peyer’s patches (PP) and LP was lower in the absence of galectin-9 compared to the frequency observed after WT cell transfer (Figure 1F and S1H). However, this difference was blunted when we transferred the cells into WT recipient mice (Figure 1G and S1I). Taken together, these data suggest that galectin-9 specifically promotes the generation of iTreg cells both in vitro and in vivo via a cell-extrinsic mechanism.
Smad3 regulates galectin-9 during iTreg cell differentiation
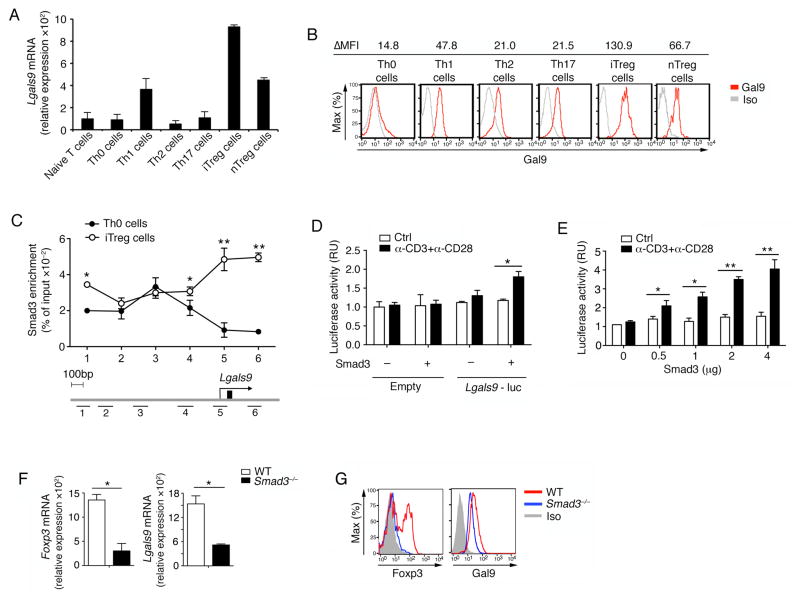
Galectin-9 is a ubiquitously expressed protein and has been found to be upregulated upon activation in various immune cells (Liu and Rabinovich, 2010; Vasta, 2009). We found that CD4+ T cells expressed higher galectin-9 (Figure S2A). Next we determined Lgals9 expression by q-PCR more specifically in different CD4+ T cell subsets. We noticed that Lgals9 mRNA and protein expression were both higher in iTreg cells than the other T cell subsets, including nTreg cells (Figure 2A and 2B). We further found that galectin-9 was expressed as early as 12 hours after activation in the presence of TGF-β, and its mRNA and protein expression were sustained high throughout iTreg cell differentiation (Figure S2B and S2C). These data thus demonstrate that galectin-9 is highly expressed during TGF-β-driven iTreg cell differentiation.
Figure 2. Smad3 regulates galectin-9 during iTreg cell differentiation.
(A) Naïve CD4+CD44loCD62L+CD25− T cells from WT mice were differentiated into different T cell subsets. Quantitative real-time PCR analysis of Lgals9 mRNA in different subsets is presented relative to the expression of GAPDH mRNA; (B) Protein expression of galectin-9 was determined in different T cell subsets by flow cytometry; (C) The binding of Smad3 to the Lgals9 promoter in Th0 and iTreg cells was assayed by ChIP-PCR. Six horizontal bars represent the locations of Smad3 binding sites on Lgals9 locus detected by real-time PCR; (D–E) Luciferase assay using a Lgals9 promoter-driven reporter in EL4 LAF cells transfected with a control or Smad3-expressing vector under the indicated conditions. Promoter activity was determined 24 h after the addition of stimuli; (F) mRNA and (G) protein expression levels of Foxp3 and galectin-9 within WT and Smad3−/− iTreg cells; Data are representative of three independent experiments with n ≥ 4 mice each group (A–B, F–G) or are pooled from three independent experiments (C–E). *P< 0.05 and **P< 0.01 (Student’s t-test, error bars, SD).
The upregulation of galectin-9 in iTreg cells suggests that there might be a positive feed-forward loop activated by TGF-β and galectin-9 to enhance the iTreg cell differentiation. It has been previously reported that Smad3 is critical for optimal induction of Foxp3 in response to TGF-β (Takimoto et al., 2010; Xu et al., 2010). We therefore addressed whether the activation of Smad3 directly results in the transactivation of Lgals9. Indeed, the chromatin immunoprecipitation (ChIP) assay showed that Smad3 interacted with multiple binding sites in the Lgals9 promoter in iTreg cells but not in Th0 cells (Figure 2C). To determine whether Smad3 transactivates galectin-9 expression, we expressed an Lgals9 promoter luciferase reporter construct in EL4 LAF cells, an EL4 subclone which maintains T cell activation properties (Tone et al., 2008; Xu et al., 2010). In support of our hypothesis, Smad3 transactivated the Lgals9 promoter with simultaneous T cell receptor (TCR) signaling in a dose-dependent manner (Figure 2D and 2E). Retroviral overexpression of Smad3 in WT naïve T cells under Th0 and iTreg conditions increased Foxp3 and galectin-9 mRNA as well as protein (Figure S2D and S2E). Consistently, Smad3−/− iTreg cells exhibited reduced expression of both Foxp3 and galectin-9 (Figure 2F and 2G). These data provide functional evidence that Smad3 transcriptionally regulates the expression of Lgals9 during iTreg cell differentiation, generating a positive autocrine loop to further enhance Foxp3 expression.
Lgals9−/− iTreg cells have reduced suppressive activity
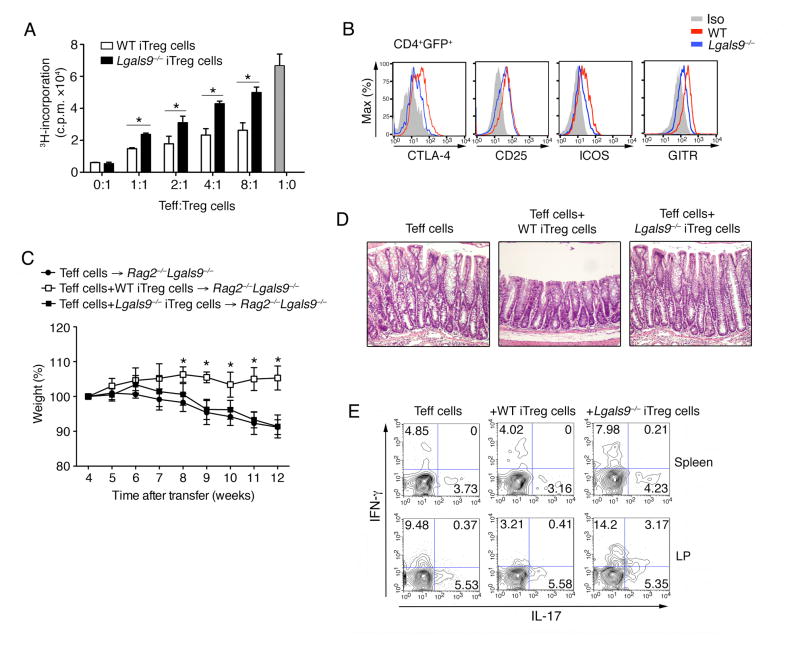
Using in vitro and in vivo approaches, multiple studies have shown that iTreg cells are important to maintain immune homeostasis and regulate effector T cells during immune responses (Fontenot et al., 2003; Williams and Rudensky, 2007). To assess the functionality of iTreg cells in the absence of galectin-9, we carried out in vitro suppression assays. To this end galectin-9-deficient mice were crossed to the Foxp3-GFP reporter background to obtain naïve CD4+ T cells and exclude GFP+ nTreg cells from the analysis. Naïve T cells (CD4+CD62L+CD44−GFP−) from the WT and Lgals9−/− mice were differentiated in vitro into iTreg cells for 3 days, sorted for GFP+ expression and then tested in functional assays (Figure S3A). Galectin-9-deficient Foxp3+ iTreg cells exhibited lower suppressive capacity, in that they were less efficient in suppressing the proliferative response of WT T effector cells (Figure 3A). The expression of a series of co-stimulatory molecules expressed on iTreg cells were reduced on Lgals9−/− iTreg cells (Figure 3B). The production of IL-10 in iTreg cells, a well-characterized anti-inflammatory effector cytokine produced by iTregs (Chaudhry et al., 2011; Rubtsovet al., 2008), was also found to be reduced in the absence of galectin-9 (Figure S3B and S3C). The reduction in regulatory function of Lgals9−/− iTreg cells was further studied in a T cell co-transfer colitis disease model. WT CD4+CD45RBhi cells were transferred into Rag2−/−Lgals9−/− mice to eliminate the exogenous galectin-9, together with either WT or Lgals9−/− in vitro differentiated Foxp3+ (GFP) iTreg cells at a ratio of 5:1. Consistent with previous study (Toms et al., 2008), WT iTreg cells efficiently suppressed the development of intestinal inflammation as measured by weight loss, whereas galectin-9-deficient iTreg cells didn’t protect the mice from the development of a wasting disease (Figure 3C, 3D and S3D). In fact Lgals9−/− iTreg cells failed to suppress Th1 but not Th17 cell responses in the gut during colitis (Figure 3E). These data suggest that galectin-9 promotes iTreg cell suppressive function and potentially prevents them from developing into effector T cells.
Figure 3. Lgals9−/− iTreg cells have reduced suppressive activity.
(A) Proliferation of WT naïve CD4+ T cells in the presence of anti-CD3 and anti-CD28 and GFP+ (Foxp3+) iTreg cells from Foxp3GFP and Foxp3GFPLgals9−/− mice. Data shown are presented as mean [3H]-thymidine incorporation; (B) The expression of indicated co-stimulatory molecules on Foxp3GFP and Foxp3GFPLgals9−/− CD4+GFP+ iTreg cells was determined by flow cytometry; (C) Body weight of Rag2−/−Lgals9−/− mice transferred with WT CD4+CD25−CD45RBhi T cells with or without WT or Lgals9−/− Foxp3+ (GFP) iTreg cells; (D) Hematoxylin and eosin staining of colon samples from the different groups as in (C) (original magnification, ×20); (E) Flow cytometry of IL-17 and IFN-γ secretion by CD4+ T cells isolated in spleen and LP from indicated groups as in (C) 10 weeks after colitis induction. The data are representative of three independent experiments with n ≥ 4 mice each group.* P< 0.05 (Student’s t-test, error bars, SD).
Galectin-9 enhances iTreg cell stability
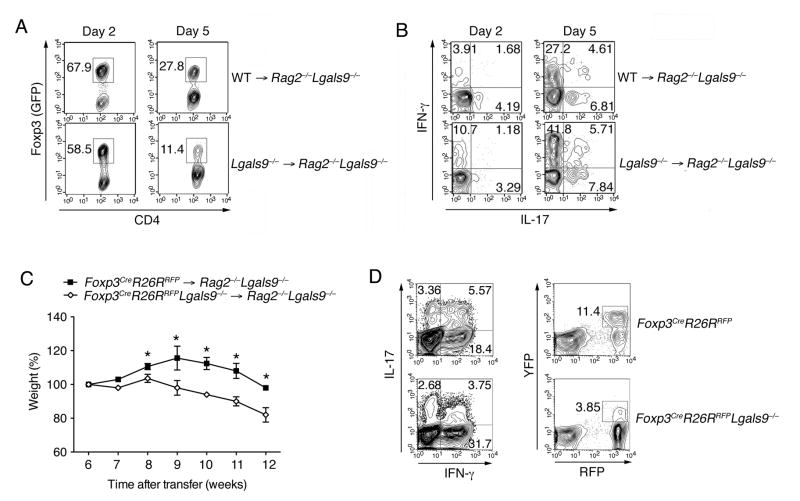
To address the role of galectin-9 in mediating the stability of iTreg cells, we transferred purified in vitro differentiated GFP+ iTreg cells from WT or Lgals9−/− mice into Rag2−/−Lgals9−/− mice. We analyzed the presence of Foxp3+ (GFP) cells 2 and 5 days after transfer, and found that there was a lower frequency of Foxp3+ cells in recipients of Lgals9−/− donor cells (Figure 4A). We further found that Lgals9−/− iTreg cells preferentially converted into IFN-γ but not IL-17-producing cells when compared to WT iTreg (Figure 4B). These results implicate that the loss of galectin-9 could impair iTreg cell stability and that these cells preferentially acquire a Th1 cell phenotype in vivo. To further assess the correlation of galectin-9 and iTreg cell fate, we took advantage of a Foxp3 fate-mapping (FM) mouse by crossing the Foxp3Cre (Foxp3-YFP-Cre) mice (Zheng et al., 2009) with Rosa26RFP (Rosa26 genetically targeted of loxP-stop-loxP-tdTomato) mice. In this mouse, YFP expression is synchronized with Foxp3 expression, and Cre activity is monitored by RFP from the Rosa26 promoter. Thus, this allowed us to visualize cells that have initiated the Foxp3 program (RFP) regardless of their present production of this protein. We then transferred CD4+CD45RBhiYFP−RFP− cells from WT or Lgals9−/− FM mice into Rag2−/−Lgals9−/− mice to induce colitis. Lgals9−/− FM cells exhibited enhanced development of intestinal inflammation (Figure 4C and S4A). A stronger Th1 cell response and a reduced frequency of Treg (CD4+RFP+ and CD4+RFP+YFP+) cells were observed in the gut within the Lgals9−/− FM mice 10 weeks after transfer (Figure 4D and S4B). More importantly, there was a dramatic loss of YFP+ T cells in the Lgals9−/− FM mice, indicating the iTreg cells could not maintain Foxp3 expression during intestinal inflammation in the absence of galectin-9 (Figure S4C and S4D).
Figure 4. Galectin-9 enhances iTreg cell stability.
(A) Purified GFP+ iTreg from Foxp3GFP and Foxp3GFPLgals9−/− mice were i.v. transferred into Rag2−/−Lgals9−/− recipient mice. 2 and 5 days after transfer, frequency of GFP+ cells were determined by flow cytometry; (B) IL-17 and IFN-γ production in the GFP− fraction in (A) were determined by flow cytometry; (C) Body weight of Rag2−/−Lgals9−/− mice transferred i.p. with CD4+CD25−CD45RBhiRFP−YFP− T cells from WT or Lgals9−/− FM mice; (D) Flow cytometry of IL-17 and IFN-γ, YFP and RFP by CD4+ T cells isolated in LP from indicated groups as in (C) 10 weeks after colitis induction. The data are representative of three independent experiments with n ≥ 5 mice each group. *P < 0.05, **P< 0.01 (Student’s t-test, error bars, SD).
CD44 mediates galectin-9 signaling during iTreg cell differentiation
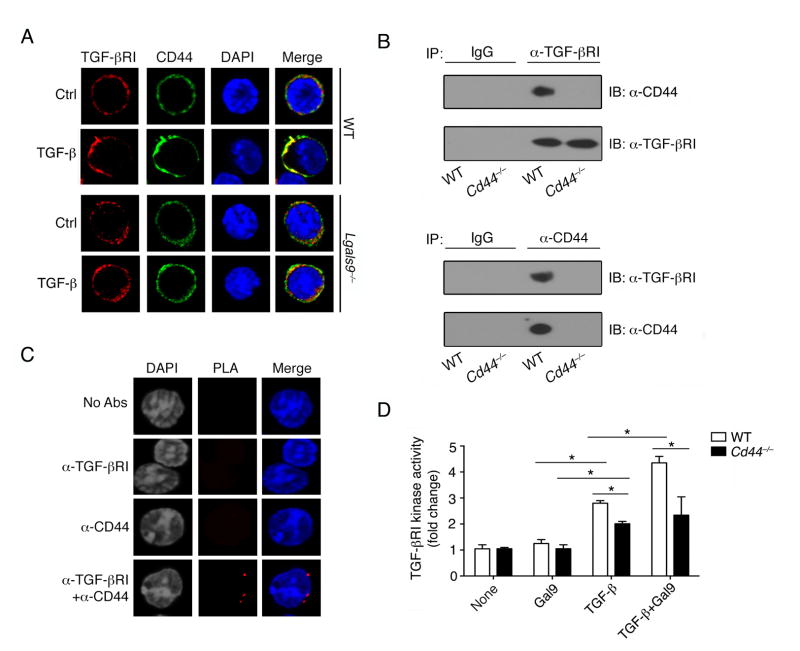
Tim-3 and CD44 have been both described as the receptors for galectin-9 on various cell types (Bourguignon et al., 2002; Chou et al., 2009; Katoh et al., 2007; Tanikawa et al., 2010; Wang et al., 2009; Zhu et al., 2005). We first found that Tim-3 was expressed on Th1 but not iTreg cells. In contrast, CD44 was observed to be highly expressed on both Th1 and iTreg cells in vitro (Figure S5A and S5B). We also showed here the protein-protein interaction between galectin-9 and its receptor CD44 on the differentiated iTreg cells (Figure S5C). It has been shown that in breast cancer cells, CD44 forms a complex with TGF-β receptor I (TGF -βRI), and in this setting the CD44 ligand HA can function as a co-signal enhancing the activity of TGF-βRI (Bourguignon et al., 2002). After 3 days culture with TGF-β, we found increased recruitment and co-clustering of TGF-βRI and CD44 on WT but not Lgals9−/− iTreg cells (Figure 5A). In WT iTregs, CD44 and TGF-βRI interacted with each other to form a complex (Figure 5B). In situ proximity ligation assay (PLA) confirmed the interaction of TGF-βRI and CD44 as evidenced by the presence of multiple associated dots appearing on the iTreg cell membrane (Figure 5C). As a consequence of such association, kinase activity of TGF-βRI in WT T cells was induced by TGF-β stimulation, which was further promoted by synergistic stimulation with galectin-9 while such activity was significantly reduced in Cd44−/− iTregs cells (Figure 5D). These data suggest that CD44 functions as one of the galectin-9 receptors on iTreg cells, transducing the signal from galectin-9 via a heterodimer complex of CD44-TGF-βRI.
Figure 5. CD44 mediates galectin-9 signaling during iTreg differentiation.
(A) Confocal images of WT or Lgals9−/− Th0 and iTreg cells stained with anti-TGF-βRI and anti-CD44 antibodies; (B) Immunoprecipitation (with control IgG, anti-TGF-βRI or anti-CD44) of lysates of WT and Cd44−/− iTreg cells, followed by immunoblot analysis with the indicated antibodies; (C) TGF-βRI and CD44 association was visualized in iTreg cells with an in situ proximity ligation assay. Punctate staining (red) indicates a TGF-βRI-CD44 interaction as detected by the assay; (D) Anti-CD3 and anti-CD28 activated WT and Cd44−/− naïve CD4+ T cells were stimulated with TGF-β or recombinant galectin-9 and TGF-βRI kinase activity was determined. Data are representative of two independent experiments (A–C) or are pooled from three independent experiments (D) with n≥4 mice each group.* P< 0.05 (Student’s t-test, error bars, SD).
The CNS1 region is crucial for mediating the galectin-9 signal within iTreg but not nTreg cells
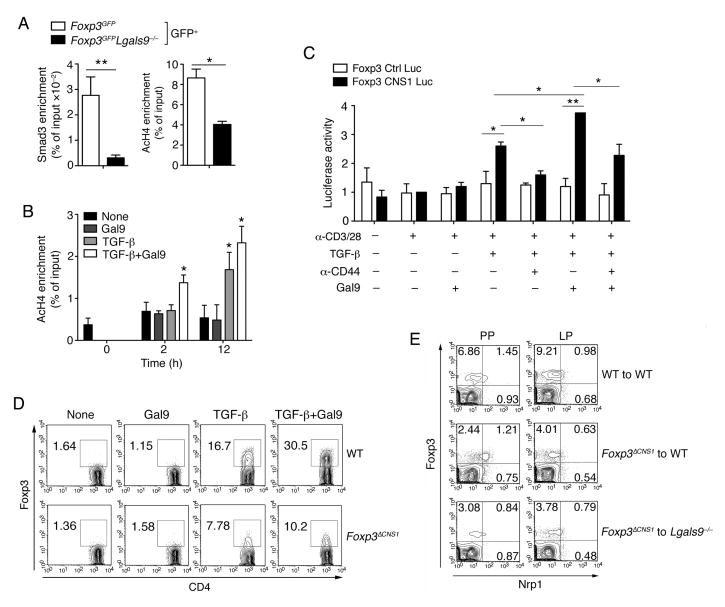
It has been described that Smad3 functions as a direct downstream target of TGF-βRI signaling (Takimoto et al., 2010; Xu et al., 2010). Immunoblot showed that phosphorylation of Smad3 was reduced in galectin-9-deficient T cells upon TGF-β stimulation compared with WT T cells (Figure S6A). It has been reported that the expression and stability of Foxp3 is dependent on a core promoter and several evolutionarily conserved noncoding sequences (CNS) at the Foxp3 locus (Zheng et al., 2010). The Smad3 binding site is harbored in the CNS1 region and has been shown to be critical for Foxp3 induction in iTreg cells (Tone et al., 2008). ChIP assays showed that Smad3 interaction with the CNS1 region is reduced in Lgals9−/− iTreg cells (Figure S6B). An accessible CNS1 region is associated with acetylated histones and considered as an enhancer for Foxp3 induction. ChIP analysis of acetylated histone H4 (AcH4) showed lower AcH4 modifications at the CNS1 site in Lgals9−/− iTreg cells, correlating with the Foxp3 expression (Figure S6C and 6A). Nevertheless, histone modification is rather a dynamic process than a poised status. We therefore analyzed Foxp3 mRNA at different time points under various iTreg induction conditions. We found that Foxp3 expression started to elevate under TGF-β stimulation at 12 h. Such induction was further enhanced with additional galectin-9 (Figure S6D). However, enhanced acetylated histone H4 was already detected under stimulation of TGF-β with additional galectin-9 but not other conditions as early as 2 h, while Foxp3 mRNA didn’t increase at this time (Figure 6B, Figure S6D). These results indicate that during the initial phase of iTreg cell differentiation, histone modification could be accelerated by the presence of galectin-9 signal, increasing the accessibility of the Foxp3 locus.
Figure 6. CNS1 region is essential for galectin-9 signaling within iTreg but not nTreg cells.
(A) The binding of Smad3 and AcH4 to the Foxp3 CNS1 region from sorted GFP+T cells of Figure S6C from Foxp3GFP and Foxp3GFPLgals9−/− iTreg cells was determined by ChIP-PCR; (B) The binding of AcH4 to the Foxp3 CNS1 region from activated naïve T cells as in Figure S6D was determined by ChIP-PCR; (C) EL4 LAF cells were transfected with a Foxp3 promoter construct with or without the CNS1 region and stimulated with anti-CD3 and anti-CD28 antibodies in the presence of the indicated stimulations. Luciferase activity was measured 48 hours later. Data are representative of two independent experiments; (D) Anti-CD3 and anti-CD28 activated naïve CD4+ T cells from WT or Foxp3ΔCNS1 mice were stimulated with different combinations of TGF-β and recombinant galectin-9 and the frequency of Foxp3+ cells was determined by flow cytometry; (E) Chimeric mice were generated by transferring WT or Foxp3ΔCNS1 BM into WT or Lgals9−/− host mice. 10 weeks after reconstitution, the frequency of Foxp3+Nrp1lo iTreg cells or Foxp3+Nrp1hi nTreg cells in PP and LP was determined by flow cytometry. Data are pooled from three independent experiments (A–C) or are representative of two independent experiments (D–E) with n≥4 mice each group. *P < 0.05 and **P < 0.01 (Student’s t-test, error bars, SD).
To determine the galectin-9 effect on Foxp3 promoter and/or enhancer activity, EL4 LAF cells were transfected with a Foxp3 luciferase reporter constructs with or without the CNS1 enhancer region under different stimuli. The luciferase activity was higher in the presence of exogenous galectin-9 than TGF-β alone and this enhancement was blocked by an anti-CD44 antibody (Figure 6C). The synergistic effect of galectin-9 with TGF-β on promoting Foxp3 expression via CNS1 enhancer was also found to be dose dependent (Figure S6E). Thereby, galectin-9 enhances the binding affinity of Smad3 to the Foxp3 CNS1 region and facilitates the permissiveness of the Foxp3 enhancer, leading to the upregulation of Foxp3 expression.
The CNS1 region has been reported to be critical in iTreg cell generation. CNS1-deficient mice (Foxp3ΔCNS1) exhibit comparable nTreg cells, while the number of TGF-β dependent iTreg cells is affected (Josefowicz et al., 2012b; Zheng et al., 2010). We observed reduced Foxp3 expression in the Foxp3ΔCNS1 Treg cells following in vitro differentiation with TGF-β. Addition of galectin-9 enhanced the expression of Foxp3 in WT but not CNS1-deficient T cells (Figure 6D). We further analyzed the role of galectin-9 in the generation of iTreg and nTreg cells in vivo by reconstituting congenic CD45.1 WT or Lgals9−/− host mice with WT or Foxp3ΔCNS1 (CD45.2) BM. Ten weeks after reconstitution, we observed similar thymic nTreg cells in mice received WT or Foxp3ΔCNS1 cells (Figure S6F and S6G). Analysis of the Treg cell population in PP and LP showed that the lack of the CNS1 region led to reduced development of iTreg cells in both WT and Lgals9−/− hosts (Figure 6E and S6H). More importantly, the presence of galectin-9 had no effect on the frequency of nTreg cells regardless of the presence of CNS1 (Figure 6E). Both in vitro and in vivo analysis of CNS1-deficient T cells confirmed that galectin-9 regulates the development of iTreg cells by engaging the CNS1 enhancer region, which exclusively regulates the development of iTreg but not nTreg cells.
Galectin-9-CD44 interaction regulates iTreg differentiation and function
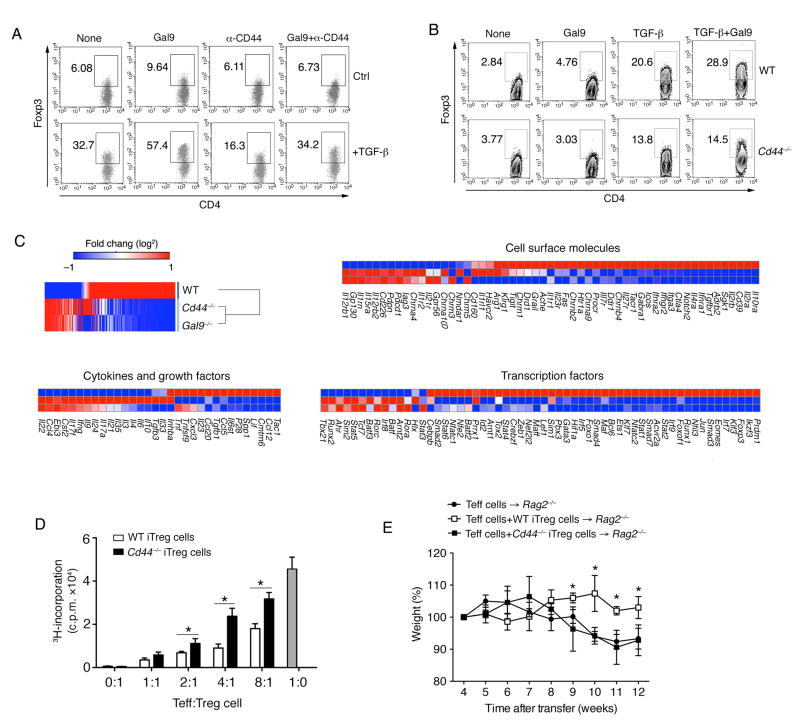
To illustrate whether the galectin-9 and CD44 interaction affects the differentiation of iTreg cells, we treated T cells with an anti-CD44 blocking antibody during in vitro iTreg cell differentiation. The synergistic effect of galectin-9 and TGF-β in inducing Foxp3 expression was abrogated by anti-CD44 antibody treatment (Figure 7A). Moreover, by utilizing CD44-deficient mice, we observed Foxp3 expression during iTreg cell differentiation was attenuated (Figure 7B). Similar to Lgals9−/− iTreg cells, the expression of different co-stimulatory molecules on Cd44−/− iTreg cells was also reduced (Figure S7A). Lastly nanostring multiplex expression analysis of Lgals9−/− iTreg cells revealed an overlap in the gene expression with Cd44−/− iTreg cells, and their expression pattern was different from that of WT iTreg cells (Figure 7C). In line with this observation, Cd44−/− iTreg cells displayed impaired suppressive capacity in vitro (Figure 7D), and failed to suppress disease in a T cell-induced colitismodel (Figure 7E, S7B and S7C). Together these genomic and functional data strongly support the relatedness of the galectin-9 and CD44 signaling pathways in inducing iTreg cells.
Figure 7. Galectin-9-CD44 interaction regulates iTreg differentiation and function.
(A) Activated WT naïve CD4+ T cells were stimulated with combinations of anti-CD44 antibody and recombinant galectin-9 with or without TGF-β for 3 days. The frequency of Foxp3+ cells was determined by flow cytometry; (B) Anti-CD3 and anit-CD28 activated naïve CD4+ T cells from WT or Cd44−/− mice were stimulated with different combinations of TGF-β and recombinant galectin-9 and the frequency of Foxp3+ cells was determined by flow cytometry; (C) Heat map displaying nanostring data, fold change of selected gene subsets in the experimental settings: Cd44−/− versus Lgals9−/− versus WT iTreg cells; (D) Proliferation of naïve CD4+ T cells in the presence of anti-CD3 and anti-CD28 and GFP+ (Foxp3+) iTreg cells from WT or Cd44−/− mice. Data shown are presented as mean [3H]-thymidine incorporation (cpm ± SEM, performed in triplicate); (E) Body weight of Rag2−/− mice transferred with WT CD4+CD25−CD45RBhi T cells with or without WT or Cd44−/− Foxp3+ (GFP) iTreg cells. Data are representative of three independent experiments with n ≥ 5 mice each group. *P < 0.05 (Student’s t-test, error bars, SD).
Discussion
Galectins are conserved glycan-binding proteins, which play various critical roles in both innate and adaptive immunity. Galectin-9 is a tandem repeat-type galectin, expressed ubiquitously in different tissues and cells in both humans and mice (Li and Turka, 2010; Zhu et al., 2005). Galectin-9 functions in both effector and regulatory phases of the immune response (Anderson et al., 2007; Arikawa et al., 2010; Su et al., 2011; Wang et al., 2009; Zhu et al., 2005). During inflammation, the production of galectin-9 in vivo generates a microenvironment that limits effector T cell responses. We and others have demonstrated that galectin-9 negatively regulates Th1 cell responses by binding to Tim-3, resulting in the induction of T cell apoptosis and exhaustion (Chou et al., 2009; Zhu et al., 2005). The binding of galectin-9 to Tim-3 on T cells induces intracellular Ca2+ flux and activates caspase 1 or apoptosis signaling pathways (Bi et al., 2008; Lu et al., 2007; Zhu et al., 2005). Further evidence has suggested that exogenous galectin-9 also suppresses the differentiation and function of Th17 cells (Seki et al., 2008). Our work presented here shows that instead of crosslinking Tim-3, galectin-9 contributes to the amplification of TGF-β signaling by clustering the CD44 and TGF-βRI, to re-enforce Foxp3 expression and stability, and is essential for iTreg-mediated suppressor function. Thus, galectin-9 signaling regulates the reciprocal balance of effector and regulatory T cells by not only inducing T cell death or exhaustion in previously differentiated effector T cells, but also promoting the generation of Foxp3+ regulatory T cells. Moreover, galectin-9 is induced by IFN-γ in inflamed tissue during Th1 cell responses. Galectin-9 in turn induces apoptosis of Th1 cells via Tim-3 and thus can control inflammation (Pender et al., 1992; Zhu et al., 2005). IFN-γ-deficient mice exhibit exaggerated autoimmunity, which is possibly caused by disruption of this regulatory feedback loop (Ferber et al., 1996; Wang et al., 2006). We have shown here that galectin-9-deficient iTreg cells are not as effective as WT iTreg cells in suppressing effector T cell responses in vitro and in vivo, suggesting that galectin-9 potentially acts as an effector molecule that delivers the immunosuppressive effects of Treg cells. This effect might not be limited to only CD4+ T cells, but also influences other immune cells, such as restraining the CD8+ T cell response during viral infection (Sehrawat et al., 2010). Therefore, galectin-9 could serve as part of a signaling loop in effector T cells and Treg cells, balancing the pro- and anti-inflammatory cell population and preventing protracted inflammation.
TGF-β signaling through the heterodimeric TGF-βRI-RII leads to the activation of the Smad pathway, which is essential for the induction of Foxp3 and development of Treg cells (Derynck and Zhang, 2003). Unexpectedly, the Foxp3 promoter region contains few TGF-β responsive elements and therefore has only limited influence on the induction of Foxp3 upon TGF-β stimulation. Instead, the intronic enhancer I (CNS1) region, which contains both NFAT and Smad3 binding sites, was determined to be the more dominant regulatory site and largely controls the expression of Foxp3 following TGF-β signaling (Tone et al., 2008; Xu et al., 2010). It has been described that CNS1 deficient mice exhibit dysregulated immune response on the mucosal surface, due to the iTreg cell deficiency while Th1 and Th17 cells are not altered (Josefowicz et al., 2012b; Katoh et al., 2007). Although these phenotypes are similar to the Lgals9−/− mice, we didn’t observe spontaneous inflammation in the intestines of the Lgals9−/− mice. This might be because CNS1 deficient mice have almost a complete loss of iTreg cell s while galectin-9 acts only an amplification factor of iTregs in the presence of TGF-β. We found that the loss of galectin-9 causes impaired Smad3 binding on the CNS1 region. Reporter assays further showed that the CNS1 region was crucial for the response to galectin-9 during iTreg cell differentiation. A recent study provides unexpected data indicating that even though Smad3 binding to CNS1 is critical for Foxp3 initiation in vitro, the specific deletion of the Smad3-binding site from the CNS1 region in vivo has no impact on the development of nTreg cells but has profound effects on the generation of iTreg cells in the gut (Schlenner et al., 2012). Our results with CNS1 null mice suggest that galectin-9 signaling acts dominantly through the CNS1 region of the Foxp3 locus, therefore specifically affecting the generation of iTreg, but not nTreg cells.
In vivo conversion of Treg cells into effector T cells has been described in a setting of inflammation during a mouse diabetes model (Zhou et al., 2009). In vitro generation of iTreg cells under TGF-β stimulation has been employed in various studies. However, their suppressive capability has been debated. The unstable Foxp3 expression in the absence of continued TGF-β signaling is one of the many reasons why iTreg cells can change their phenotype during an immune response (Floess et al., 2007). nTreg cells, on the other hand, have been shown to sustain Foxp3 expression and suppressive function in vivo even in the face of inflammation (Rubtsov et al., 2010). Here we observed impact of galectin-9 deficiency on Foxp3 maintenance specifically in iTreg cells, supporting the notion that an important function of galectin-9 is to stabilize iTreg cells. The expression of Foxp3 alone is not sufficient to promote the stability and suppressive capacity of the Treg cell lineage. It has been shown that the epigenome-dependent programming is critical for determining the sustainability and fate of Treg cells (Floess et al., 2007; Ohkura et al., 2012). We have shown that there is a reduction of histone acetylation in the Foxp3 locus in Lgals9−/− Foxp3+ iTreg cells. Galectin-9 signaling was further proved to be essential for initiating histone modification at the CNS1 site during iTreg cells differentiation. These data therefore provide evidence that galectin-9 promotes iTreg cell differentiation via increasing the accessibility of the enhancer to Foxp3.
Tim-3 has been reported as a well-characterized receptor for galectin-9 (Jayaraman et al., 2010; Reddy et al., 2011; Zhu et al., 2005). The Tim-3-galectin-9 interaction induces signals that can mediate Th1 cell apoptosis and inhibition of cytotoxic T lymphocytes (CTL), leading to the attenuation of autoimmune diseases and prolongation of allograft survival (Sehrawat et al., 2010; Xu et al., 2010; Zhu et al., 2005). Even though it has been reported that the Tim-3-galectin-9 interaction promotes the suppressive activity of Treg cells and induces a higher frequency of Treg cells during allogeneic skin grafts (Rabinovich and Toscano, 2009; Wang et al., 2009), we found no Tim-3 expression on iTreg cells in vitro. Another known receptor for galectin-9, CD44, is highly upregulated on activated T cells (Katoh et al., 2007; Tanikawa et al., 2010). Interestingly, its expression correlates with the upregulation of Foxp3 and the suppressive function of Treg cells (Liu et al., 2009). CD44 was reported to deliver a co-stimulatory signal to Treg cells upon the activation of TCR (Bollyky et al., 2009). Our data suggests that such a costimulatory signal may be mediated by galectin-9-CD44 interaction. Moreover, the fact that Lgals9−/− T cells show a specific defect in iTreg cell differentiation regardless of the ubiquitous expression of CD44 in activated T cells, suggests that galectin-9 plays an indispensable role in TGF-β signaling by bridging CD44 and TGF-βRI, hence promoting iTreg cell differentiation. Moreover, we observed a striking relatedness between the Lgals9−/− and Cd44−/− iTreg cells at the level of cell phenotype, genetic signatures and functionality. These data therefore implicate the direct interaction between galectin-9 and CD44 upon TGF-β stimulation in regulating the development of iTreg cells.
In summary, we show here that galectin-9 promotes Foxp3 expression and stability upon TGF-β stimulation in iTreg cells. Furthermore, our data demonstrate that during iTreg cell differentiation, a CD44-TGF-βRI complex is responsible for transducing the galectin-9 signal, which in turn increases Smad3-driven Foxp3 transactivation. Therefore, galectin-9 appears to inhibit autoimmunity and induce tolerance by multiple mechanisms, including inhibition of effector T cells by interacting with Tim-3 and inducing iTreg cells via CD44-TGF-βRI. Considering the critical role of iTreg cells in autoimmunity and tissue inflammation, the identification of a galectin-9-CD44 interaction in iTreg development and function may offer potential new targets for inhibiting pathogenic immune responses and promoting peripheral tolerance.
Material and Methods
Animals
C57BL/6 (B6), Cd44−/−, Rosa26RFP, Rag2−/− and Rag2−/− OT-II mice were purchased from Jackson Laboratory; Foxp3GFP, Foxp3Cre, Lgals9-Tg, Lgals9−/−, Smad3−/− and Foxp3ΔCNS1 mice have been described previously (Bettelli et al., 2006; Datto et al., 1999; Seki et al., 2007; Tsuboi et al., 2007; Zheng et al., 2009; Zheng et al., 2010). All experiments were carried out in accordance with guidelines prescribed by the Institutional Animal Care and Use Committee (IACUC) at Harvard Medical School.
T cell-induced colitis
Naïve (CD44loCD62L+GFP−) CD4+ T cells were purified from WT or Lgals9−/− Foxp3gfp mice, as described above, and were then stimulated with plate-bound anti-CD3 and anti-CD28 and 5 ng/ml TGF-β1 for 3 days. CD4+CD25−CD45RBhi naive T cells (7 × 105) were injected intraperitoneally (i.p.) with or without in vitro differentiated and re-sorted WT or Lgals9−/− iTreg CD4+Foxp3+ (1.5 × 105) into age- and sex-matched Rag2−/− mice and weight loss was monitored.
Statistical analysis
Statistical analysis was performed using Prism software (Graph Pad software, La Jolla, CA, USA). P values<0.05 were considered significant.
Supplementary Material
Figure S1, related to Figure 1. (A) Quantification of the frequency of Foxp3+ Treg cells within indicated organs as in Figure 1A; (B) Quantification of frequency of Foxp3+Nrp1lo iTreg cells or Foxp3+Nrp1hi nTreg cells as in Figure 1B; (C) Activated WT naïve CD4+ T cells were stimulated with combinations of TGF-β and recombinant galectin-9 with or without β-lactose (100mM) for 3 days. The frequency of Foxp3+ cells was determined by flow cytometry; Chimeric mice were generated by transferring WT or Lgals9−/− BM into (D–E) Lgals9−/− or (F–G) WT host mice. 10 weeks after reconstitution, the percentage of CD4+Foxp3+ Treg cells in mLN and LP was determined by flow cytometry and quantification; (H–I) Quantification of the frequency of CD45.2+CD4+Foxp3+ iTreg cells in PP and LP of recipient mice as in Figure 1F and Figure 1G. Data are representative of three independent experiments with n ≥ 4 mice each group. *P < 0.05 (Student’s t-test, error bars, SD).
Figure S2, related to Figure 2. (A) Protein expression level of galectin-9 was determined in CD4+, CD8+ T cells, CD11b+ macrophages, and CD19+ B cells by flow cytometry; (B) Quantitative real-time PCR analysis of the time course of the expression of Lgals9 mRNA in iTreg cells; (C) Time course of galectin-9 protein expression in iTreg cells was determined by flow cytometry; (D) mRNA and (E) protein expression levels of Foxp3 and galectin-9 in naive CD4+ T cells transduced with retrovirus expressing control vector (MIG) or Smad3-expressing vector and differentiated into Th0 or iTreg cells; Data are representative of two independent experiments with n ≥ 5 mice each group. *P < 0.05 (Student’s t-test, error bars, SD).
Figure S3, related to Figure 3. (A) Naïve Foxp3GFP or Foxp3GFPLgals9−/− T cells were differentiated into iTreg cells with TGF-β. The frequency of GFP (Foxp3+) cells was determined by flow cytometry; (B) Intracellular staining of IL-10 production by differentiated WT and Lgals9−/− CD4+Foxp3+ iTreg cells was determined by flow cytometry; (C) WT and Lgals9−/− iTreg cells were stimulated for 24 h with PMA and ionomycin, and IL-10 production in the supernatant was determined by ELISA; (D) Left: Colon lengths of Rag2−/−Lgals9−/− mice which had received the indicated cells for transfer as in Figure 3C, measured from the colocecal junction to the anal verge; Right: Quantification of pathological changes in the colon of mice as in Figure 3C; The data are representative of three independent experiments (A, B, D) or are pooled of three independent experiments (C) with n ≥ 4 mice each group. *P < 0.05, **P < 0.01 (Student’s t-test, error bars, SD).
Figure S4, related to Figure 4. (A) Left: Colon lengths of Rag2−/−Lgals9−/− mice which had received the indicated cells for transfer as in Figure 4C, measured from the colocecal junction to the anal verge; Right: Quantification of pathological changes in the colon of mice as in Figure 4C 10 weeks after colitis induction; (B) Quantification of the frequency of RFP+ among CD4+ T cells in the LP of WT and Lgals9−/− fate mapping mice; YFP expression within CD4+RFP+ T cells isolated from LP of indicated mice as in Figure 4C was determined by (C) flow cytometry and (D) quantification. Data are representative of two independent experiments with n ≥ 5 mice each group. *P < 0.05 (Student’s t-test, error bars, SD).
Figure S5, related to Figure 5. Flow cytometry of protein expression levels of (A) Tim3 and (B) CD44 on indicated T cell subsets; (C) Immunoprecipitation (with control IgG, anti-galectin-9 or anti-CD44) of lysates of WT and Cd44−/− or Lgals9−/− iTreg cells, followed by immunoblot analysis with the indicated antibodies. Data are representative of three independent experiments with n ≥ 3 mice each group.
Figure S6, related to Figure 6. (A) The level of phosphorylated Smad3 was assessed in WT and Lgals9−/− iTreg cells by immunoblot; (B) The binding of Smad3 to the Foxp3 CNS1 region in WT and Lgals9−/− iTreg cells was determined by ChIP-PCR; (C) Foxp3 expression frequency and mean fluorescence intensity (MFI) from Foxp3GFP and Foxp3GFPLgals9−/− iTreg cells; (D) Activated naïve CD4+ T cells were stimulated with TGF-β and/or recombinant galectin-9 for 2, 12 and 24 hours. Foxp3 mRNA expression was assessed by quantitative real-time PCR analysis; (E) EL4 LAF cells were transfected with a Foxp3 promoter reporter construct containing the CNS1 enhancer and stimulated with anti-CD3 and anit-CD28 antibodies and TGF-β in the presence of increasing concentrations of galectin-9. Luciferase activity was measured 48 hours later; Chimeric mice were generated by transferring WT or Foxp3ΔCNS1 BM into WT or Lgals9−/− host mice. 10 weeks after reconstitution, the frequency of Foxp3+ Treg cells in thymus was determined by (F) flow cytometry and (G) quantification; (H) Quantification of the frequency of Foxp3+Nrp1lo iTreg cells or Foxp3+Nrp1hi nTreg cells in PP and LP as in Figure 6E. Data are representative of three independent experiments (A, C–D, F–H) or are pooled of three independent experiments (B, E) with n ≥ 4 mice each group. *P < 0.05, **P < 0.01 (Student’s t-test, error bars, SD).
Figure S7, related to Figure 7. (A) The expression of indicated co-stimulatory molecules on Foxp3GFP or Foxp3GFPCd44−/− CD4+GFP+ iTreg cells was determined by flow cytometry; (B) Left: Colon lengths of Rag2−/− mice which had received the indicated cells for transfer as in Figure 7E, measured from the colocecal junction to the anal verge; Right: Quantification of pathological changes in the colon of mice as in Figure 7E; (c) Hematoxylin and eosin staining of colon samples from the different groups as in Figure 7E 10 weeks after colitis induction (original magnification, ×20). The data are representative of three independent experiments with n ≥ 5 mice each group. *P < 0.05, (Student’s t-test, error bars, SD).
Acknowledgments
We thank Deneen Kozoriz for cell sorting. Dr. Alexander Rudensky provided the CNS1-deficient mice. C.W. was supported by a Postdoctoral Fellowship from the National Multiple Sclerosis Society (FG2046). T.T. was supported by the Austrian Science Fund (FWF, J 3091-B12). S.X. was supported by the US National Institutes of Health (K01DK090105). F.O.F. was supported by Sao Paulo Research Foundation FAPESP (2012/14924-7). A.C.A was supported by the American Cancer Society (RSG-11-057-01-LIB) and the Slomo and Cindy Silvian Foundation. V.K.K. was supported by the US National Institutes of Health (P01AI073748, P01AI056299, P01AI039671 and R01NS045937).
Footnotes
Authors’ contribution
C.W. performed experiments and wrote the manuscript. T.T., S.X., R.F., C.W., C.H., C.Z. and A.C.A. performed experiments. M.H. provided Lgals9−/− mice. V.K.K. supervised the study and edited the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alam S, Li H, Margariti A, Martin D, Zampetaki A, Habi O, Cockerill G, Hu Y, Xu Q, Zeng L. Galectin-9 protein expression in endothelial cells is positively regulated by histone deacetylase 3. The Journal of biological chemistry. 2011;286:44211–44217. doi: 10.1074/jbc.M111.242289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson AC, Anderson DE, Bregoli L, Hastings WD, Kassam N, Lei C, Chandwaskar R, Karman J, Su EW, Hirashima M, et al. Promotion of tissue inflammation by the immune receptor Tim-3 expressed on innate immune cells. Science. 2007;318:1141–1143. doi: 10.1126/science.1148536. [DOI] [PubMed] [Google Scholar]
- Arikawa T, Saita N, Oomizu S, Ueno M, Matsukawa A, Katoh S, Kojima K, Nagahara K, Miyake M, Yamauchi A, et al. Galectin-9 expands immunosuppressive macrophages to ameliorate T-cell-mediated lung inflammation. European journal of immunology. 2010;40:548–558. doi: 10.1002/eji.200939886. [DOI] [PubMed] [Google Scholar]
- Arikawa T, Watanabe K, Seki M, Matsukawa A, Oomizu S, Sakata KM, Sakata A, Ueno M, Saita N, Niki T, et al. Galectin-9 ameliorates immune complex-induced arthritis by regulating Fc gamma R expression on macrophages. Clinical immunology. 2009;133:382–392. doi: 10.1016/j.clim.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, Kuchroo VK. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- Bi S, Earl LA, Jacobs L, Baum LG. Structural features of galectin-9 and galectin-1 that determine distinct T cell death pathways. The Journal of biological chemistry. 2008;283:12248–12258. doi: 10.1074/jbc.M800523200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluestone JA, Abbas AK. Natural versus adaptive regulatory T cells. Nature reviews Immunology. 2003;3:253–257. doi: 10.1038/nri1032. [DOI] [PubMed] [Google Scholar]
- Bollyky PL, Falk BA, Long SA, Preisinger A, Braun KR, Wu RP, Evanko SP, Buckner JH, Wight TN, Nepom GT. CD44 costimulation promotes FoxP3+ regulatory T cell persistence and function via production of IL-2, IL-10, and TGF-beta. Journal of immunology. 2009;183:2232–2241. doi: 10.4049/jimmunol.0900191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourguignon LY, Singleton PA, Zhu H, Zhou B. Hyaluronan promotes signaling interaction between CD44 and the transforming growth factor beta receptor I in metastatic breast tumor cells. The Journal of biological chemistry. 2002;277:39703–39712. doi: 10.1074/jbc.M204320200. [DOI] [PubMed] [Google Scholar]
- Chaudhry A, Samstein RM, Treuting P, Liang Y, Pils MC, Heinrich JM, Jack RS, Wunderlich FT, Bruning JC, Muller W, Rudensky AY. Interleukin-10 signaling in regulatory T cells is required for suppression of Th17 cell-mediated inflammation. Immunity. 2011;34:566–578. doi: 10.1016/j.immuni.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou FC, Shieh SJ, Sytwu HK. Attenuation of Th1 response through galectin-9 and T-cell Ig mucin 3 interaction inhibits autoimmune diabetes in NOD mice. European journal of immunology. 2009;39:2403–2411. doi: 10.1002/eji.200839177. [DOI] [PubMed] [Google Scholar]
- Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, Belkaid Y, Powrie F. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. The Journal of experimental medicine. 2007;204:1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datto MB, Frederick JP, Pan L, Borton AJ, Zhuang Y, Wang XF. Targeted disruption of Smad3 reveals an essential role in transforming growth factor beta-mediated signal transduction. Molecular and cellular biology. 1999;19:2495–2504. doi: 10.1128/mcb.19.4.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- Ferber IA, Brocke S, Taylor-Edwards C, Ridgway W, Dinisco C, Steinman L, Dalton D, Fathman CG. Mice with a disrupted IFN-gamma gene are susceptible to the induction of experimental autoimmune encephalomyelitis (EAE) Journal of immunology. 1996;156:5–7. [PubMed] [Google Scholar]
- Floess S, Freyer J, Siewert C, Baron U, Olek S, Polansky J, Schlawe K, Chang HD, Bopp T, Schmitt E, et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS biology. 2007;5:e38. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nature immunology. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- Fontenot JD, Rasmussen JP, Gavin MA, Rudensky AY. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nature immunology. 2005;6:1142–1151. doi: 10.1038/ni1263. [DOI] [PubMed] [Google Scholar]
- Hirashima M, Kashio Y, Nishi N, Yamauchi A, Imaizumi TA, Kageshita T, Saita N, Nakamura T. Galectin-9 in physiological and pathological conditions. Glycoconjugate journal. 2004;19:593–600. doi: 10.1023/B:GLYC.0000014090.63206.2f. [DOI] [PubMed] [Google Scholar]
- Jayaraman P, Sada-Ovalle I, Beladi S, Anderson AC, Dardalhon V, Hotta C, Kuchroo VK, Behar SM. Tim3 binding to galectin-9 stimulates antimicrobial immunity. The Journal of experimental medicine. 2010;207:2343–2354. doi: 10.1084/jem.20100687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annual review of immunology. 2012a;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefowicz SZ, Niec RE, Kim HY, Treuting P, Chinen T, Zheng Y, Umetsu DT, Rudensky AY. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 2012b;482:395–399. doi: 10.1038/nature10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kared H, Fabre T, Bedard N, Bruneau J, Shoukry NH. Galectin-9 and IL-21 mediate cross-regulation between Th17 and Treg cells during acute hepatitis C. PLoS pathogens. 2013;9:e1003422. doi: 10.1371/journal.ppat.1003422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh S, Ishii N, Nobumoto A, Takeshita K, Dai SY, Shinonaga R, Niki T, Nishi N, Tominaga A, Yamauchi A, Hirashima M. Galectin-9 inhibits CD44-hyaluronan interaction and suppresses a murine model of allergic asthma. American journal of respiratory and critical care medicine. 2007;176:27–35. doi: 10.1164/rccm.200608-1243OC. [DOI] [PubMed] [Google Scholar]
- Kim HP, Kim BG, Letterio J, Leonard WJ. Smad-dependent cooperative regulation of interleukin 2 receptor alpha chain gene expression by T cell receptor and transforming growth factor-beta. The Journal of biological chemistry. 2005;280:34042–34047. doi: 10.1074/jbc.M505833200. [DOI] [PubMed] [Google Scholar]
- Lagna G, Hata A, Hemmati-Brivanlou A, Massague J. Partnership between DPC4 and SMAD proteins in TGF-beta signalling pathways. Nature. 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- Li XC, Turka LA. An update on regulatory T cells in transplant tolerance and rejection. Nature reviews Nephrology. 2010;6:577–583. doi: 10.1038/nrneph.2010.101. [DOI] [PubMed] [Google Scholar]
- Liu F, Pouponnot C, Massague J. Dual role of the Smad4/DPC4 tumor suppressor in TGFbeta-inducible transcriptional complexes. Genes & development. 1997;11:3157–3167. doi: 10.1101/gad.11.23.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu FT, Rabinovich GA. Galectins: regulators of acute and chronic inflammation. Annals of the New York Academy of Sciences. 2010;1183:158–182. doi: 10.1111/j.1749-6632.2009.05131.x. [DOI] [PubMed] [Google Scholar]
- Liu T, Soong L, Liu G, Konig R, Chopra AK. CD44 expression positively correlates with Foxp3 expression and suppressive function of CD4+ Treg cells. Biology direct. 2009;4:40. doi: 10.1186/1745-6150-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu LH, Nakagawa R, Kashio Y, Ito A, Shoji H, Nishi N, Hirashima M, Yamauchi A, Nakamura T. Characterization of galectin-9-induced death of Jurkat T cells. Journal of biochemistry. 2007;141:157–172. doi: 10.1093/jb/mvm019. [DOI] [PubMed] [Google Scholar]
- Macias-Silva M, Abdollah S, Hoodless PA, Pirone R, Attisano L, Wrana JL. MADR2 is a substrate of the TGFbeta receptor and its phosphorylation is required for nuclear accumulation and signaling. Cell. 1996;87:1215–1224. doi: 10.1016/s0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- Massague J. TGF-beta signal transduction. Annual review of biochemistry. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- Nagahara K, Arikawa T, Oomizu S, Kontani K, Nobumoto A, Tateno H, Watanabe K, Niki T, Katoh S, Miyake M, et al. Galectin-9 increases Tim-3+ dendritic cells and CD8+ T cells and enhances antitumor immunity via galectin-9-Tim-3 interactions. Journal of immunology. 2008;181:7660–7669. doi: 10.4049/jimmunol.181.11.7660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura N, Hamaguchi M, Morikawa H, Sugimura K, Tanaka A, Ito Y, Osaki M, Tanaka Y, Yamashita R, Nakano N, et al. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity. 2012;37:785–799. doi: 10.1016/j.immuni.2012.09.010. [DOI] [PubMed] [Google Scholar]
- Oomizu S, Arikawa T, Niki T, Kadowaki T, Ueno M, Nishi N, Yamauchi A, Hattori T, Masaki T, Hirashima M. Cell surface galectin-9 expressing Th cells regulate Th17 and Foxp3+ Treg development by galectin-9 secretion. PloS one. 2012;7:e48574. doi: 10.1371/journal.pone.0048574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang W, Beckett O, Ma Q, Li MO. Transforming growth factor-beta signaling curbs thymic negative selection promoting regulatory T cell development. Immunity. 2010;32:642–653. doi: 10.1016/j.immuni.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pender MP, McCombe PA, Yoong G, Nguyen KB. Apoptosis of alpha beta T lymphocytes in the nervous system in experimental autoimmune encephalomyelitis: its possible implications for recovery and acquired tolerance. Journal of autoimmunity. 1992;5:401–410. doi: 10.1016/0896-8411(92)90001-7. [DOI] [PubMed] [Google Scholar]
- Rabinovich GA, Toscano MA. Turning ‘sweet’ on immunity: galectin-glycan interactions in immune tolerance and inflammation. Nature reviews Immunology. 2009;9:338–352. doi: 10.1038/nri2536. [DOI] [PubMed] [Google Scholar]
- Reddy PB, Sehrawat S, Suryawanshi A, Rajasagi NK, Mulik S, Hirashima M, Rouse BT. Influence of galectin-9/Tim-3 interaction on herpes simplex virus-1 latency. Journal of immunology. 2011;187:5745–5755. doi: 10.4049/jimmunol.1102105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Q, Kameswaran V, Tone Y, Li L, Liou HC, Greene MI, Tone M, Chen YH. Development of Foxp3(+) regulatory t cells is driven by the c-Rel enhanceosome. Immunity. 2009;31:932–940. doi: 10.1016/j.immuni.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsov YP, Niec RE, Josefowicz S, Li L, Darce J, Mathis D, Benoist C, Rudensky AY. Stability of the regulatory T cell lineage in vivo. Science. 2010;329:1667–1671. doi: 10.1126/science.1191996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsov YP, Rasmussen JP, Chi EY, Fontenot J, Castelli L, Ye X, Treuting P, Siewe L, Roers A, Henderson WR, Jr, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- Sakaguchi S, Miyara M, Costantino CM, Hafler DA. FOXP3+ regulatory T cells in the human immune system. Nature reviews Immunology. 2010;10:490–500. doi: 10.1038/nri2785. [DOI] [PubMed] [Google Scholar]
- Schlenner SM, Weigmann B, Ruan Q, Chen Y, von Boehmer H. Smad3 binding to the foxp3 enhancer is dispensable for the development of regulatory T cells with the exception of the gut. The Journal of experimental medicine. 2012;209:1529–1535. doi: 10.1084/jem.20112646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehrawat S, Reddy PB, Rajasagi N, Suryawanshi A, Hirashima M, Rouse BT. Galectin-9/TIM-3 interaction regulates virus-specific primary and memory CD8 T cell response. PLoS pathogens. 2010;6:e1000882. doi: 10.1371/journal.ppat.1000882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Oomizu S, Sakata KM, Sakata A, Arikawa T, Watanabe K, Ito K, Takeshita K, Niki T, Saita N, et al. Galectin-9 suppresses the generation of Th17, promotes the induction of regulatory T cells, and regulates experimental autoimmune arthritis. Clinical immunology. 2008;127:78–88. doi: 10.1016/j.clim.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Seki M, Sakata KM, Oomizu S, Arikawa T, Sakata A, Ueno M, Nobumoto A, Niki T, Saita N, Ito K, et al. Beneficial effect of galectin 9 on rheumatoid arthritis by induction of apoptosis of synovial fibroblasts. Arthritis and rheumatism. 2007;56:3968–3976. doi: 10.1002/art.23076. [DOI] [PubMed] [Google Scholar]
- Siddiqui KR, Powrie F. CD103+ GALT DCs promote Foxp3+ regulatory T cells. Mucosal immunology. 2008;1(Suppl 1):S34–38. doi: 10.1038/mi.2008.43. [DOI] [PubMed] [Google Scholar]
- Su EW, Bi S, Kane LP. Galectin-9 regulates T helper cell function independently of Tim-3. Glycobiology. 2011;21:1258–1265. doi: 10.1093/glycob/cwq214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun CM, Hall JA, Blank RB, Bouladoux N, Oukka M, Mora JR, Belkaid Y. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. The Journal of experimental medicine. 2007;204:1775–1785. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takimoto T, Wakabayashi Y, Sekiya T, Inoue N, Morita R, Ichiyama K, Takahashi R, Asakawa M, Muto G, Mori T, et al. Smad2 and Smad3 are redundantly essential for the TGF-beta-mediated regulation of regulatory T plasticity and Th1 development. Journal of immunology. 2010;185:842–855. doi: 10.4049/jimmunol.0904100. [DOI] [PubMed] [Google Scholar]
- Tanikawa R, Tanikawa T, Hirashima M, Yamauchi A, Tanaka Y. Galectin-9 induces osteoblast differentiation through the CD44/Smad signaling pathway. Biochemical and biophysical research communications. 2010;394:317–322. doi: 10.1016/j.bbrc.2010.02.175. [DOI] [PubMed] [Google Scholar]
- Toker A, Huehn J. To be or not to be a Treg cell: lineage decisions controlled by epigenetic mechanisms. Science signaling. 2011;4:pe4. doi: 10.1126/scisignal.2001783. [DOI] [PubMed] [Google Scholar]
- Toms C, Jessup H, Thompson C, Baban D, Davies K, Powrie F. Gpr83 expression is not required for the maintenance of intestinal immune homeostasis and regulation of T-cell-dependent colitis. Immunology. 2008;125:302–312. doi: 10.1111/j.1365-2567.2008.02857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tone Y, Furuuchi K, Kojima Y, Tykocinski ML, Greene MI, Tone M. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nature immunology. 2008;9:194–202. doi: 10.1038/ni1549. [DOI] [PubMed] [Google Scholar]
- Tsuboi Y, Abe H, Nakagawa R, Oomizu S, Watanabe K, Nishi N, Nakamura T, Yamauchi A, Hirashima M. Galectin-9 protects mice from the Shwartzman reaction by attracting prostaglandin E2-producing polymorphonuclear leukocytes. Clinical immunology. 2007;124:221–233. doi: 10.1016/j.clim.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Vasta GR. Roles of galectins in infection. Nature reviews Microbiology. 2009;7:424–438. doi: 10.1038/nrmicro2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Wan L, Zhang C, Zheng X, Li J, Chen ZK. Tim-3-Galectin-9 pathway involves the suppression induced by CD4+CD25+ regulatory T cells. Immunobiology. 2009;214:342–349. doi: 10.1016/j.imbio.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Wang Z, Hong J, Sun W, Xu G, Li N, Chen X, Liu A, Xu L, Sun B, Zhang JZ. Role of IFN-gamma in induction of Foxp3 and conversion of CD4+ CD25- T cells to CD4+ Tregs. The Journal of clinical investigation. 2006;116:2434–2441. doi: 10.1172/JCI25826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss JM, Bilate AM, Gobert M, Ding Y, Curotto de Lafaille MA, Parkhurst CN, Xiong H, Dolpady J, Frey AB, Ruocco MG, et al. Neuropilin 1 is expressed on thymus-derived natural regulatory T cells, but not mucosa-generated induced Foxp3+ T reg cells. The Journal of experimental medicine. 2012;209:1723–1742. S1721. doi: 10.1084/jem.20120914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LM, Rudensky AY. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nature immunology. 2007;8:277–284. doi: 10.1038/ni1437. [DOI] [PubMed] [Google Scholar]
- Xu L, Kitani A, Stuelten C, McGrady G, Fuss I, Strober W. Positive and negative transcriptional regulation of the Foxp3 gene is mediated by access and binding of the Smad3 protein to enhancer I. Immunity. 2010;33:313–325. doi: 10.1016/j.immuni.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav M, Louvet C, Davini D, Gardner JM, Martinez-Llordella M, Bailey-Bucktrout S, Anthony BA, Sverdrup FM, Head R, Kuster DJ, et al. Neuropilin-1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo. The Journal of experimental medicine. 2012;209:1713–1722. S1711–1719. doi: 10.1084/jem.20120822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Chaudhry A, Kas A, deRoos P, Kim JM, Chu TT, Corcoran L, Treuting P, Klein U, Rudensky AY. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458:351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463:808–812. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Rudensky AY. Foxp3 in control of the regulatory T cell lineage. Nature immunology. 2007;8:457–462. doi: 10.1038/ni1455. [DOI] [PubMed] [Google Scholar]
- Zhou X, Bailey-Bucktrout SL, Jeker LT, Penaranda C, Martinez-Llordella M, Ashby M, Nakayama M, Rosenthal W, Bluestone JA. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nature immunology. 2009;10:1000–1007. doi: 10.1038/ni.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Anderson AC, Schubart A, Xiong H, Imitola J, Khoury SJ, Zheng XX, Strom TB, Kuchroo VK. The Tim-3 ligand galectin-9 negatively regulates T helper type 1 immunity. Nature immunology. 2005;6:1245–1252. doi: 10.1038/ni1271. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1, related to Figure 1. (A) Quantification of the frequency of Foxp3+ Treg cells within indicated organs as in Figure 1A; (B) Quantification of frequency of Foxp3+Nrp1lo iTreg cells or Foxp3+Nrp1hi nTreg cells as in Figure 1B; (C) Activated WT naïve CD4+ T cells were stimulated with combinations of TGF-β and recombinant galectin-9 with or without β-lactose (100mM) for 3 days. The frequency of Foxp3+ cells was determined by flow cytometry; Chimeric mice were generated by transferring WT or Lgals9−/− BM into (D–E) Lgals9−/− or (F–G) WT host mice. 10 weeks after reconstitution, the percentage of CD4+Foxp3+ Treg cells in mLN and LP was determined by flow cytometry and quantification; (H–I) Quantification of the frequency of CD45.2+CD4+Foxp3+ iTreg cells in PP and LP of recipient mice as in Figure 1F and Figure 1G. Data are representative of three independent experiments with n ≥ 4 mice each group. *P < 0.05 (Student’s t-test, error bars, SD).
Figure S2, related to Figure 2. (A) Protein expression level of galectin-9 was determined in CD4+, CD8+ T cells, CD11b+ macrophages, and CD19+ B cells by flow cytometry; (B) Quantitative real-time PCR analysis of the time course of the expression of Lgals9 mRNA in iTreg cells; (C) Time course of galectin-9 protein expression in iTreg cells was determined by flow cytometry; (D) mRNA and (E) protein expression levels of Foxp3 and galectin-9 in naive CD4+ T cells transduced with retrovirus expressing control vector (MIG) or Smad3-expressing vector and differentiated into Th0 or iTreg cells; Data are representative of two independent experiments with n ≥ 5 mice each group. *P < 0.05 (Student’s t-test, error bars, SD).
Figure S3, related to Figure 3. (A) Naïve Foxp3GFP or Foxp3GFPLgals9−/− T cells were differentiated into iTreg cells with TGF-β. The frequency of GFP (Foxp3+) cells was determined by flow cytometry; (B) Intracellular staining of IL-10 production by differentiated WT and Lgals9−/− CD4+Foxp3+ iTreg cells was determined by flow cytometry; (C) WT and Lgals9−/− iTreg cells were stimulated for 24 h with PMA and ionomycin, and IL-10 production in the supernatant was determined by ELISA; (D) Left: Colon lengths of Rag2−/−Lgals9−/− mice which had received the indicated cells for transfer as in Figure 3C, measured from the colocecal junction to the anal verge; Right: Quantification of pathological changes in the colon of mice as in Figure 3C; The data are representative of three independent experiments (A, B, D) or are pooled of three independent experiments (C) with n ≥ 4 mice each group. *P < 0.05, **P < 0.01 (Student’s t-test, error bars, SD).
Figure S4, related to Figure 4. (A) Left: Colon lengths of Rag2−/−Lgals9−/− mice which had received the indicated cells for transfer as in Figure 4C, measured from the colocecal junction to the anal verge; Right: Quantification of pathological changes in the colon of mice as in Figure 4C 10 weeks after colitis induction; (B) Quantification of the frequency of RFP+ among CD4+ T cells in the LP of WT and Lgals9−/− fate mapping mice; YFP expression within CD4+RFP+ T cells isolated from LP of indicated mice as in Figure 4C was determined by (C) flow cytometry and (D) quantification. Data are representative of two independent experiments with n ≥ 5 mice each group. *P < 0.05 (Student’s t-test, error bars, SD).
Figure S5, related to Figure 5. Flow cytometry of protein expression levels of (A) Tim3 and (B) CD44 on indicated T cell subsets; (C) Immunoprecipitation (with control IgG, anti-galectin-9 or anti-CD44) of lysates of WT and Cd44−/− or Lgals9−/− iTreg cells, followed by immunoblot analysis with the indicated antibodies. Data are representative of three independent experiments with n ≥ 3 mice each group.
Figure S6, related to Figure 6. (A) The level of phosphorylated Smad3 was assessed in WT and Lgals9−/− iTreg cells by immunoblot; (B) The binding of Smad3 to the Foxp3 CNS1 region in WT and Lgals9−/− iTreg cells was determined by ChIP-PCR; (C) Foxp3 expression frequency and mean fluorescence intensity (MFI) from Foxp3GFP and Foxp3GFPLgals9−/− iTreg cells; (D) Activated naïve CD4+ T cells were stimulated with TGF-β and/or recombinant galectin-9 for 2, 12 and 24 hours. Foxp3 mRNA expression was assessed by quantitative real-time PCR analysis; (E) EL4 LAF cells were transfected with a Foxp3 promoter reporter construct containing the CNS1 enhancer and stimulated with anti-CD3 and anit-CD28 antibodies and TGF-β in the presence of increasing concentrations of galectin-9. Luciferase activity was measured 48 hours later; Chimeric mice were generated by transferring WT or Foxp3ΔCNS1 BM into WT or Lgals9−/− host mice. 10 weeks after reconstitution, the frequency of Foxp3+ Treg cells in thymus was determined by (F) flow cytometry and (G) quantification; (H) Quantification of the frequency of Foxp3+Nrp1lo iTreg cells or Foxp3+Nrp1hi nTreg cells in PP and LP as in Figure 6E. Data are representative of three independent experiments (A, C–D, F–H) or are pooled of three independent experiments (B, E) with n ≥ 4 mice each group. *P < 0.05, **P < 0.01 (Student’s t-test, error bars, SD).
Figure S7, related to Figure 7. (A) The expression of indicated co-stimulatory molecules on Foxp3GFP or Foxp3GFPCd44−/− CD4+GFP+ iTreg cells was determined by flow cytometry; (B) Left: Colon lengths of Rag2−/− mice which had received the indicated cells for transfer as in Figure 7E, measured from the colocecal junction to the anal verge; Right: Quantification of pathological changes in the colon of mice as in Figure 7E; (c) Hematoxylin and eosin staining of colon samples from the different groups as in Figure 7E 10 weeks after colitis induction (original magnification, ×20). The data are representative of three independent experiments with n ≥ 5 mice each group. *P < 0.05, (Student’s t-test, error bars, SD).