Summary
Beyond their contribution to basic metabolism, the major cellular organelles, in particular mitochondria, can determine whether cells respond to stress in an adaptive or suicidal manner. Thus, mitochondria can continuously adapt their shape to changing bioenergetic demands as they are subjected to quality control by autophagy, or they can undergo a lethal permeabilization process that initiates apoptosis. Along similar lines, multiple proteins involved in metabolic circuitries including oxidative phosphorylation and transport of metabolites across membranes may participate in the regulated or catastrophic dismantling of organelles. Many factors that were initially characterized as cell death regulators are now known to physically or functionally interact with metabolic enzymes. Thus, several metabolic cues regulate the propensity of cells to activate self-destructive programs, in part by acting on nutrient sensors. This suggests the existence of “metabolic checkpoints” that dictate cell fate in response to metabolic fluctuations. Here, we discuss recent insights into the intersection between metabolism and cell death regulation that have major implications for the comprehension and manipulation of unwarranted cell loss.
Keywords: apoptosis, ATP synthasome, BCL-2, caspases, cyclophilin D, regulated necrosis
Introduction
All living things metabolize nutrients as a source of energy and building blocks for their components to survive, and ultimately all living things die. When we apply these simple concepts to cells, the notion that the biochemistries of metabolism and cell death intimately interact is obvious. In this review, we take a largely mammal-centric view of the interplay between metabolic and cell death pathways.
The survival of animal cells critically relies on an energy source (be it extrinsic or, if necessary, provided by the autophagic, lysosomal or proteasomal degradation of cellular components). In the absence of such a source, cells lose the control of plasma membrane channels and are doomed to passive necrosis (which can also occur upon excessive physical damage to the plasma membrane). This said, our discussion focuses on those forms of cellular demise that require an active engagement of the cell. In this regard, it is useful to consider that such a participation can be of two types: (i) cellular “suicide”, in which cells engage molecular circuitries that have (presumably) evolved to precipitate their demise (i.e., apoptosis and some forms of regulated necrosis); and (ii) “subversion”, in which a cellular process drives cell death when disrupted, although (at least presumably) it did not evolve as a cell death mechanism (1). The latter is akin to a sabotage: dislodging a railway tie can destroy a train only if is moving. This dichotomy can help in understanding how there can be so many forms of cell death (Supplemental Discussion), although from some perspectives (e.g., the design of anticancer therapies, the control of pathological cell death) such a distinction may not be of overwhelming concern.
Apoptosis as well as regulated necrosis are accompanied by a sudden loss of metabolic functions that, once a threshold of deterioration has been trespassed, seal the cell’s irreversible fate (2). Beyond the cell death-associated loss of essential bioenergetic functions, metabolism and its derangement have a major impact on the avoidance, initiation, and execution of cell death at multiple levels. Here, we consider how metabolic signals are sensed and transduced to impact on active cell death pathways.
Metabolic checkpoints in cell fate: signals, sensors, transducers, and effectors
Nutrient and oxygen supply, activation (reflecting extracellular signals or intracellular events, such as oncogenic mutations), as well as the differentiation state of a cell influence the metabolic pathways engaged for the production of energy and biomaterials. In response to changes in these conditions, cells either adapt or die. Extending upon a classical convention referring to the cell cycle and DNA damage response (3), we propose the existence of metabolic checkpoints that dictate the consequences of such alterations on cell fate. Metabolic checkpoints can be defined as molecular mechanisms that regulate cellular functions in response to metabolic fluctuations, and comprise four components: signals, sensors, transducers, and effectors (4). In our discussion of the metabolic control of cell death, we consider these in terms of either the signal that promotes downstream events (perhaps through different sensors) or the sensor that coordinates one or more signals. Although this nomenclature is admittedly arbitrary, we suggest that the checkpoints we propose are useful starting blocks to probe how different metabolic processes feed into the cell fate decision, engaging processes that promote active death (Fig. 1).
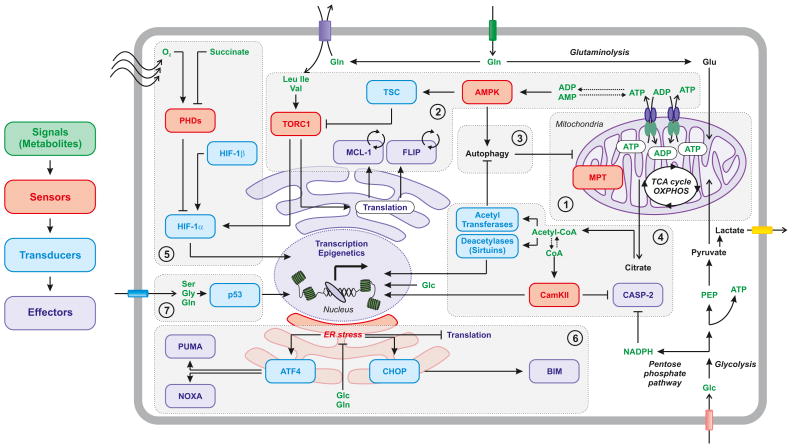
Figure 1. Metabolic checkpoints in cell death regulation.
Several metabolic checkpoints are in place to convert metabolic perturbations (signals), which are detected by specific systems (sensors), into vital or lethal stimuli that are dispatched to components of the cell death-regulatory machinery (effectors) through one or more signaling nodes (transducers). These include (but are not limited to): the mitochondrial checkpoint, in part impinging on the so-called mitochondrial permeability transition (MPT) (1); the AMPK-TORC1 checkpoint, which is based on the very short half-life of anti-apoptotic proteins such as FLIPL and MCL-1 (2); the autophagy checkpoint, which is extensively interconnected with other checkpoints (3); the acetyl-CoA/CoA checkpoint, which control cell death through both transcriptional and post-translational mechanisms (4); the HIF-1 checkpoint, integrating signals about oxygen availability and tricarboxylic acid (TCA) cycle proficiency (5); the endoplasmic reticulum (ER) stress checkpoint, which operates by altering the abundance of multiple BH3-only proteins (6); as well as the p53 checkpoint, detecting the availability of non-essential amino acids and converting it into an adaptive or lethal response (7). Glc, glucose; MPT, mitochondrial permeability transition; OXPHOS, oxidative phosphorylation; PEP, phosphoenolpyruvate.
Major metabolic signals that arise as a consequence of changes in nutrient availability or intracellular metabolic pathways include the adenosine triphosphate/adenosine diphosphate (ATP/ADP) ratio, acetyl-coenzyme A (acetyl-CoA)/CoA ratio, the ratios of oxidized and reduced nicotinamide adenine dinucleotide (NAD+/NADH) and NAD phosphate (NADP+/NADPH), as well as the amounts of lipid products, glycosylated proteins, and reactive oxygen species (ROS). For illustrative purposes, we distinguish these signals from second messengers, such as cAMP, phosphoinositides, and ion (including Ca2+) fluxes. However, the frontier between metabolism and signaling may be less defined than previously thought (5). Specific sensors directly interact with these metabolic cues to initiate downstream events, thereby impacting on signal transducers, including those involved in cell death regulation. Of note, for a sensor to be considered so, it must possess a Km for the signal that allows it to function in physiological (or pathophysiological) conditions. Our consideration of sensors within metabolic checkpoints attempts to take this concept into account, but at least in some cases this has not been formally determined. We discuss specific examples below.
The mitochondrial checkpoints: MOMP, MPT, and mitochondrial dynamics
Mitochondria are central to the control of cell life and death, and are fundamentally involved in metabolism as they are responsible for energy production through the tricarboxylic acid (TCA) cycle and oxidative phosphorylation (fueled by glycolysis, glutaminolysis, β oxidation, and other sources), as well as for the synthesis of lipids, pyrimidines, heme moieties, some amino acids, and other biomolecules. Moreover, mitochondria are the major intracellular source of ROS. As such, they are under extensive metabolic control, as is their biogenesis and removal.
Mitochondria control cell fate in four fundamental ways: (i) through mitochondrial outer membrane permeabilization (MOMP), leading to apoptosis; (ii) through the mitochondrial permeability transition (MPT), leading to regulated necrosis; (iii) by providing an energy supply; and (iv) by participating in the synthesis of several products, including lipid precursors, iron-sulfur clusters, and nucleotides (Fig. 2). Cells that have been depleted of mitochondria through an artificial widespread wave of mitophagy are resistant to apoptosis (6). However, despite assertions that a non-apoptotic form of cell death, necroptosis (Supplemental Discussion), is executed by mitochondrial alterations, cells lacking the vast majority of their mitochondria remain sensitive to this form of cellular demise (6). In contrast, mitochondria can precipitate other forms of necrosis via the MPT.
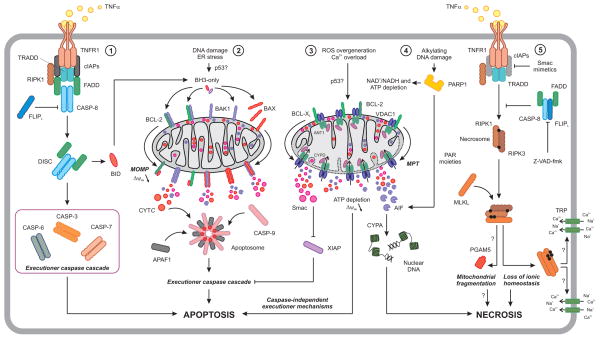
Figure 2. Major signal transduction cascades leading to active cell death.
The ligation of death receptor such as TNFR1 generally results in the assembly of a supramolecular complex at the inner leaflet of the plasma membrane known as a death-inducing signaling complex (DISC), which is under the control of FLIPL. By promoting the activation of CASP-8, the DISC directly ignites the caspase cascade that is responsible for apoptotic cell death (1). Similarly, the executioner caspase cascade can be set off upon mitochondrial outer membrane permeabilization (MOMP) (2) or the so-called mitochondrial permeability transition (MPT) (3), owing to the release into the cytosol of caspase activators as well as to the engagement of caspase-independent mechanisms. Death receptor-elicited apoptosis and MOMP are not entirely disconnected, as in some cells CASP-8 cleaves BID to generate a MOMP-promoting factor. In some settings, MPT can trigger necrotic forms of cell death, at least in part owing to the release of AIF, a mitochondrial protein with latent endonucleolytic activity that is also involved in the PARP1-dependent lethal cascade elicited by alkylating DNA damage (4). Of note, the ligation of TNFR1 in the presence of Z-VAD-fmk (a pan caspase inhibitor), alone or combined with a Smac mimetic, drives a specific instance of regulated necrosis known as necroptosis (5). Necroptosis, which is inhibited by a supramolecular complex involving FADD, CASP-8 and FLIPL, obligatorily relies on the formation of MLKL oligomers that relocalize to the plasma membrane and disrupt ionic homeostasis. CYPA, cyclophilin A (official name: peptidylprolyl isomerase A); CYTC, holocytochrome c; ER, endoplasmic reticulum; ROS, reactive oxygen species; TRADD, TNFRSF1A-associated via death domain; TRP, transient receptor potential cation channel; XIAP, X-linked inhibitor of apoptosis.
Mitochondria are the only cellular source of holocytochrome c, which is normally sequestered in the mitochondrial intermembrane space where it operates as an essential electron shuttle of the respiratory chain. In response to some lethal stimuli, however, mitochondria can undergo MOMP, a process that is executed and regulated by Bcl-2 family proteins (Fig. 2). Upon MOMP, holocytochrome c and other mitochondrial proteins are released into the cytosol, where they cooperate with cytosolic factors to promote the activation of caspases. This function of holocytochrome c is regulated by its redox state. Oxidized holocytochrome c does not activate caspases (7), and some cells can survive MOMP owing to an enhanced rate of glycolysis (7, 8). The survival of cells undergoing MOMP is facilitated by the enforced expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), enabling the regeneration of the mitochondrial pool from a small pool of organelles that resisted permeabilization (8, 9). The demise of cells in which MOMP does not (or cannot) activate caspases occurs by energetic catastrophe (10), or upon the release of mitochondrial factors that trigger caspase-independent cell death mechanisms, such as apoptosis-inducing factor (AIF) (11).
The continuous fission and fusion of the mitochondrial network (generally referred to as “mitochondrial dynamics”) is crucial for the biogenesis of these organelles as well as for the segregation and autophagic removal of dysfunctional mitochondria, thereby playing a key role in the control of intracellular homeostasis (12, 13) Fission relies on the small GTPase dynamin 1-like (DNM1L, best known as DRP1), which operates by interacting with mitochondrial fission factor (MFF) on the OMM to constrict both mitochondrial membranes and eventually divide them (14). Fusion requires the interaction of mitofusin 1 (MFN1) and MFN2, two OMM proteins, with optic atrophy 1 (OPA1), which is found at the inner mitochondrial membrane (IMM). Fusion (but not fission) of the IMM depends on the transmembrane potential (Δψm) generated by the respiratory chain. Thus, Δψm dissipation favors mitochondrial fragmentation (15). Mitochondrial dynamics is highly sensitive to metabolic cues. In line with this notion, mitochondria form a hyperfused network characterized by an increased amount of IMM cristae in conditions of low nutrient availability. This is presumed to increase the efficiency of the respiratory chain while restricting the autophagic removal of mitochondria. Conversely, mitochondria generally fragment in cells exposed to excess nutrients, a situation that presumably decreases the bioenergetic efficacy of the organelles and favors the removal of lipids and other potentially toxic molecules (16).
Mitochondrial dynamics reportedly influences the propensity of the organelles to undergo MOMP, but the precise mechanisms remain partially understood. MOMP has been suggested to preferentially occur at fission sites (17), which are also points of contact with the endoplasmic reticulum (ER) (18). Studies in cells (19) and isolated mitochondria (20) revealed that at least one of the pro-apoptotic Bcl-2 family members BAX and BAK1 is necessary for mitochondrial fusion through interaction with MFN1 and MFN2, a function that is lost upon the induction of MOMP (21). Along similar lines, the anti-apoptotic protein BCL-XL appears to regulate mitochondrial fusion, either by interacting with MFN2 (22) or by sequestering the active forms of BAX and BAK1 (23). Conversely, silencing or inhibiting DRP1 dampens BAX- or BAK1-dependent MOMP (17, 24, 25), but this effect may be unrelated to the role of DRP1 in the regulation of mitochondrial dynamics. It has indeed been suggested that DRP1 may favor MOMP by increasing the tension of the OMM (25). Thus, alterations in nutrient availability or other metabolic conditions affecting mitochondrial dynamics may influence the propensity of cells to undergo MOMP-driven apoptosis. Conversely, shifts in the mutual interactions between pro- and anti-apoptotic Bcl-2 family members as induced by other metabolic checkpoints (see below) may impact mitochondrial dynamics, hence affecting their bioenergetic efficacy. Intriguingly, the ability of BAX to control mitochondrial dynamics has been proposed to lower the threshold for MPT, hence affecting regulated variants of necrosis (26). This possibility, however, remains to be explored in detail.
The ATP synthasome is a multiprotein complex that produce ATP and export it from mitochondria in exchange of ADP and inorganic phosphate (27). The ATP synthasome comprises the F1FO-ATP synthase and two mitochondrial solute carrier (SLC) family members: adenine nucleotide translocase (ANT), which exists in three isoforms (official names: SLC25A4, SLC25A5 and SLC25A6, best known as ANT1, ANT2 and ANT3, respectively) and exchanges ADP for ATP, and the inorganic phosphate carrier (PIC, official name: SLC25A3), which mediates the mitochondrial uptake of phosphate. Beyond their function in ATP synthesis, the components of the ATP synthasome, individually or together, may regulate cell death as driven by the MPT, a sudden increase in the permeability of the IMM to solutes <1.5 kD. This occurs in response to increased intracellular Ca2+ concentrations or oxidative stress, and results in Δψm dissipation, uncoupling of the respiratory chain, influx of H2O and ions, osmotic swelling of the mitochondrial matrix, and consequent mechanical breakdown of the OMM (28).
The concentration of Ca2+ ions required for induction of the MPT is increased by ~2-fold upon addition of cyclosporine A (CsA) or knockout of the gene coding for cyclophilin D (CYPD, official name: peptidyl-prolyl isomerase F, PPIF), a CsA-inhibitable enzyme of the mitochondrial matrix. CYPD can be immunoprecipitated with ANT in cells responding to MPT inducers, and this interaction can be blocked by CsA (28). Therefore, CYPD appears to bind the ATP synthasome to form the so-called permeability transition pore complex (PTPC), the supramolecular entity that is responsible for the MPT (Fig. 2). In stringent conditions of co-immunoprecipitation, CYPD interacts with the oligomycin sensitivity-conferring protein (OSCP), but not with other subunits of the F1FO-ATP synthase lateral stalk (29). Among various proteins that have been suggested to compose the PTPC, including ANT and voltage-dependent anion channel (VDAC, an OMM-embedded protein), so far only CYPD has been ascribed with a critical role in MPT (30) Partial depletion of PIC also fails to affect MPT. Conversely, the knockdown of either OSCP or the c subunit of FO alters the propensity of mitochondria to undergo MPT in vitro and in cellula (29, 31). These observations suggest a major role for the F1FO-ATP synthase in MPT.
ANT also interacts with proteins that are not involved in the ATP synthasome, including VDAC (Fig. 3). In turn, VDAC (which constitutes a preferential channel for the export of mitochondrial ATP across the OMM) binds cytosolic hexokinase II (HKII). This interaction provides HKII (which catalyzes the initial, ATP-consuming step of glycolysis) with a privileged access to mitochondrial ATP. Moreover, the binding of HKII to VDAC reduces the propensity of cells to undergo lethal MPT (32). It is an open conundrum whether this effect is direct or rather involves fluctuations in the local gradients of ATP, ADP, inorganic phosphate and protons, all of which regulate MPT (28).
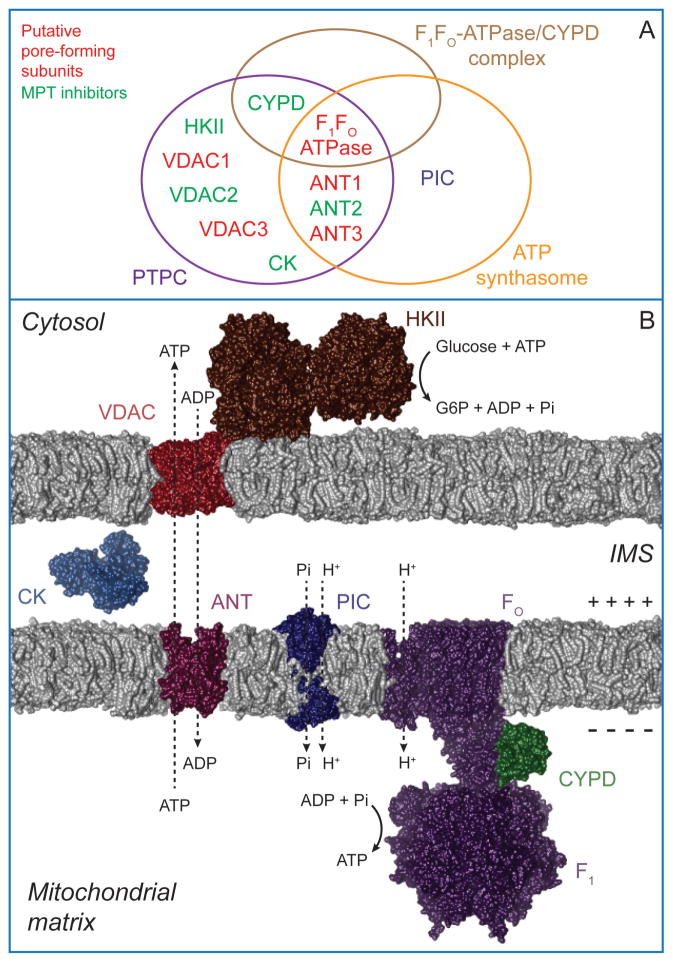
Figure 3. The ATP synthasome and its interacting partners.
A,B. The term ‘ATP synthasome’ refers to the supramolecular complex composed of the F1FO-ATP synthase, ANT and PIC. The F1FO-ATP synthase harnesses the electrochemical gradient generated by the respiratory chain across the inner mitochondrial membrane (IMM) to catalyze the synthesis of ATP, whereas PIC and ANT ensure the availability of inorganic phosphate and ADP, respectively. The ATP synthasome interacts with CYPD, a protein of the mitochondrial matrix, and the VDAC1-HKII complex, which is assembled at the outer mitochondrial membrane (OMM). On one hand, these interactions allow ATP molecules produced by the F1FO-ATP synthase to be channeled to the mitochondrial surface and support the HKII-mediated conversion of glucose into glucose-6-phosphate (G6P). On the other hand, they place the ATP synthasome in a privileged position to contribute to (or regulate) the so-called ‘permeability transition pore complex’ (PTPC), the supramolecular entity that mediates the mitochondrial permeability transition (MPT). CK, creatine kinase (official name: creatine kinase, mitochondrial 1, ubiquitous) IMS, intermembrane space. Molecular graphics and analyses were performed with the UCSF Chimera package. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311).
Rapid openings (‘flickering’) of the PTPC may avoid a potentially toxic accumulation of Ca2+ ions in the mitochondrial matrix, which may explain why the myocardium of Ppif−/− mice ages with an accelerated kinetics (33). Conversely, widespread (affecting most mitochondria within a cell) and irreversible MPT has been linked to necrosis. Indeed, several organs from Ppif−/− mice are relatively protected against ischemia-reperfusion damage, which is mostly necrotic. Such a phenotype is even more pronounced when Ppif−/− mice are crossed with mice lacking the pro-necroptotic kinase receptor-interacting protein kinase 3 (RIPK3) or treated with the anti-necroptotic agent necrostatin-1 (34), supporting the notion that CYPD and RIPK3 induce necrosis via mutually independent signaling pathways (6).
The mitochondrial checkpoint also influences the propensity of cells to die as it regulates the activation of caspase-1 by the NLR family, pyrin domain containing 3 (NLRP3) inflammasome (35). A number of clinically relevant stimuli, including monosodium urate crystals (associated with gout) (36), calcium pyrophosphage dihydrate crystals (associated with pseudogout) (36), and cholesterol crystals (associated with atherosclerosis) (37) activate the NLRP3 inflammasome through a mechanism that relies on mitochondrial ROS and VDAC1 (38). This may reflect the role of VDAC1 in mitochondrial metabolism. Of note, the activation of other inflammasomes, such as those orchestrated by absent in melanoma 2 (AIM2) and NLR family, CARD domain containing 4 (NLRC4), is independent of VDAC1, but may require VDAC2 (38). How the NLRP3 inflammasome is activated remains unknown. In some cases, this indeed occurs independently of ROS (39), and may rely on the translocation of cardiolipin (CL), a mitochondrion-restricted lipid, from the IMM to the OMM (40). Thus, alterations in mitochondrial functions, including ROS output, can have direct effects on the activation of the inflammasome, and hence on inflammation and cell death.
The AMPK-TORC1 checkpoint
The target of rapamycin complex 1 (TORC1) is a major regulatory node that controls cell growth, proliferation, and autophagy through the kinase activity of mammalian target of rapamycin (mTOR). TORC1 is activated on the surface of lysosomes when amino acids, especially leucine, are available. Several signal transducers can inhibit mTOR, including glycogen synthase kinase 3 (GSK3), which operates only when AKT1 is inactive (e.g., upon growth factor withdrawal), and 5′ AMP-activated protein kinase (AMPK), which responds to declining ATP levels (owing to the adenylate kinase-dependent conversion of ADP into AMP) by phosphorylating (and thus activating) the mTOR-inhibitory protein tuberous sclerosis 2 (TSC2) (41), as well as by phosphorylating (and thus inactivating) regulatory associated protein of MTOR, complex 1 (RPTOR), a component of TORC1 (42). AMPK is activated even by mild perturbations of ATP synthesis, acting as a primary sensor of mitochondrial stress in concert with other mitochondrial checkpoints.
Besides regulating TORC1, AMPK controls a variety of metabolic processes. In particular, AMPK inhibits the synthesis of fatty acids, sterols, and ribosomal RNA, while promoting catabolic processes such as glycolysis and fatty acid oxidation (43). In conditions of limited amino acid or growth factor availability, the activity of TORC1 gets reduced independently of AMPK (44). Conversely, the inhibition of glycolysis by glucose deprivation, 2-deoxyglucose (45) or caloric restriction (46) promotes the apoptotic demise of many cell types upon the AMPK-induced inhibition of TORC1. In both settings, TORC1 inhibition causes the arrest of several anabolic processes, including protein translation. As a result, the abundance of the short-lived anti-apoptotic Bcl-2 family member MCL-1 decreases, sensitizing the cell to mitochondrial apoptosis (46). In line with this notion, an unbiased approach identified MCL-1 as a target for the chemotherapy-enhancing effects of the TORC1 inhibitor rapamycin (47)
As glucose uptake by plasma membrane transporters relies on growth factor receptor signaling, the withdrawal of growth factors also promotes apoptosis (48). This effect reflects the activation of GSK3, which phosphorylates and hence further destabilizes MCL-1. In line with this notion, cells expressing a MCL-1 variant that cannot be phosphorylated by GSK3 or exposed to GSK3 inhibitors resist growth factor withdrawal-induced cell death (49–52). Therefore, this metabolic checkpoint controls cell fate at least in part by influencing the abundance of MCL-1. Of note, in cells lacking the pro-apoptotic BCL-2 proteins BAX and BAK1, the apoptotic response to growth factor withdrawal is substantially delayed and viability is preserved owing to the activation of the autophagic machinery (discussed below) (48).
Another short-lived protein is CASP8 and FADD-like apoptosis regulator (CFLAR, best known as cFLIP), a caspase-like protein that limits the pro-apoptotic activity of caspase-8 (as initiated by death receptors), and cooperates with caspase-8 to inhibit a form of regulated necrosis called necroptosis (which manifests upon death receptor ligation in the presence of caspase inhibitors) (53). Glucose deprivation sensitizes cells to death receptor-induced apoptosis (45, 54), a phenomenon that may involve alterations in the abundance of both cFLIP and MCL-1. Indeed, cells lacking BAX and BAK1 (which are expected to be resistant to declining amounts of MCL-1) can succumb to caspase-8-dependent apoptosis in response to glucose deprivation (55), perhaps as a consequence of alterations in the amounts of cFLIP. Moreover, although glucose deprivation impacts on the expression of c-FLIP, this may be independent of AMPK, at least in some cases (56). Thus, other metabolic checkpoints (as outlined below) are likely to influence cFLIP expression.
The overexpression of MYC, which occurs in some cancers, can cause an imbalance in ATP/ADP ratios and hence promote AMPK activation. In line with this notion, the increased sensitivity of MYC-expressing cells to the pro-apoptotic activity of a number of agents reportedly relies on this kinase (57). These effects of MYC overexpression, however, were ascribed to the phosphorylation-dependent stabilization of a cytosolic pool of the tumor suppressor p53, resulting in the activation of BAK1 (57). Conversely, the roles of TORC1 and MCL-1 (which inhibits BAK1) in this setting were not examined.
Given the profound effects of the AMPK-TORC1 checkpoint on protein translation and the abundance of MCL-1, many metabolic conditions that result in TORC1 inhibition are expected to sensitize cells to death. These include amino acid deprivation, which – among many other effects (discussed below) – inhibits the catalytic activity of TORC1 in an AMPK-independent manner. Finally, AMPK also promotes, while TORC1 inhibits, autophagy, as discussed in the next section.
The autophagy checkpoint
Macroautophagy (herein referred to as autophagy) is a major mechanism for the maintenance of intracellular homeostasis that can be dramatically upregulated by nutrient deprivation and organellar damage (12, 58). Thus, autophagy is controlled by several metabolic cues, including the ATP/ADP ratio (by the AMPK checkpoint, see above) and the availability of acetyl-CoA, which impact on the enzymatic activity of several acetyltransferases, notably E1A binding protein p300 (EP300) (59). The ability of autophagy to inhibit or promote cell death is an area of intense investigation (60, 61). Interestingly, permeabilized mitochondria are rapidly sequestered by autophagic vacuoles and degraded by mitophagy, a process that favors the maintenance of intracellular homeostasis (12, 13). Moreover, part of the mitochondrial network elongates and is hence spared from autophagic degradation in the course of adaptive responses to stress (62). These observations indicate that the mitochondrial and autophagy checkpoints are intimately interconnected.
Autophagy limits the cytotoxicity of tumor necrosis factor (ligand) superfamily, member 10 (TNFSF10, best known as TRAIL) in cells that either do not (Type I) or do (Type II) require the mitochondrial machinery to succumb to death receptor-induced apoptosis. In contrast, FAS ligand (FASLG, also known as CD95L) promotes apoptosis upon the autophagic degradation of protein tyrosine phosphatase, non-receptor type 13 (PTPN13, also known as FAP-1), which results in an increased expression of FAS (also known as CD95) on the cell surface (63). Autophagy can also promote cell death by degrading the antioxidant enzyme catalase, resulting in increased ROS production (64).
Type II cell death (which should not be confounded with Type II cells, see above) has been defined as a cell death subroutine accompanied by autophagy, although whether or not the autophagic machinery truly contributes to (rather than accompanies) cell death cannot be determined by morphological parameters alone (65). Type II death often manifests with large vacuoles that may be enlarged lysosomes or other structures. A form of non-apoptotic cell death that has recently been dubbed “autosis” is promoted by starvation (and therefore is likely to involve the AMPK-TORC1 checkpoint), hypoxia, ischemic insults, and autophagy-promoting peptides (66). Because autosis critically relies on several factors that are essential for autophagy, it may qualify as bona fide autophagic cell death (2). Similarly, HRAS induces the demise of several cell types through a pathway that manifests with Type II morphological features, and depends on essential autophagic mediators as well as on the BH3-only protein phorbol-12-myristate-13-acetate-induced protein 1 (PMAIP1, best known as NOXA) (67). This cell death subroutine can be inhibited by anti-apoptotic BCL-2 proteins, most likely as they sequester the critical autophagic factor Beclin-1 (BECN1). NOXA as well as other BH3-only proteins promote autophagy by releasing Beclin 1 from inhibitory interactions with BCL-2 and its homologue BCL-XL (67, 68). It is therefore possible that the complex interactions between Bcl-2 family members and components of the autophagic machinery control not only apoptosis but also autophagic cell death.
How autophagy may promote cell death is unclear. Cells lacking BAX and BAK1 (which are unable to undergo MOMP) survive growth factor deprivation for long periods of time in an autophagy-dependent manner. These cells manifest increasing degrees of atrophy and eventually succumb without displaying overt features of Type II cell death (48). It is possible, but remains to be formally demonstrated, that (at least in some settings) autophagy promotes cell death by favoring lysosomal membrane permeabilization. Lysosome-disrupting agents can actually provoke a non-apoptotic form of cell death (69), leaving this possibility open.
The acetyl-CoA/CoA checkpoint
The acetyl CoA/CoA ratio is dynamically regulated in response to metabolic functions. In mammalian cells, acetyl-CoA is mainly generated by glycolysis, and a large fraction of the intracellular acetyl-CoA pool is sequestered within mitochondria. In this setting, the extramitochondrial demand for acetyl-CoA is satisfied by a cytosolic variant of citrate lyase, which generates acetyl-CoA from citrate (an intermediate of the TCA cycle actively exported by mitochondria). Alternatively, acetyl-CoA can be produced from acetate by acetyl-CoA synthetase. A nuclear isoform of citrate lyase is required for the acetylation of histones and other proteins (70). Similarly, the deacetylating activity of sirtuins, which affects hundreds of proteins, is under a strict metabolic control.
From our perspective, acetylation and deacetylation regulate cell death by virtue of specific effectors. During the synthesis of fatty acids, acetyl-CoA is consumed and CoA is produced. CoA acts directly on Ca2+/calmodulin-dependent protein kinase II (CamKII) to reduce its requirement for Ca2+ ions, thus effectively activating it (71). One target of the enzymatic activity of CamKII is caspase-2, which upon phosphorylation is sequestered and kept inactive by 14-3-3ζ (72). As the phosphorylation of caspase-2 requires NADPH, a product of the pentose phosphate pathway (PPP), caspase-2 is released from 14-3-3ζ when the flow through the PPP is limited, resulting in the sensitization of cells to various stress signals (72). Sirtuin 1 (SIRT1) is one of the enzymes that deacetylate 14-3-3ζ, thus counteracting this effect (73). Declines in the abundance of acetyl-CoA also result in the inhibition of the acetyltransferase EP300, hence promoting autophagy (59).
An unbiased screen in Drosophila cells identified the human homolog of Ard1, which functions in irreversible N-terminal acetylation, as an essential regulator of caspase activation (74). N-terminal acetylation can promote protein degradation by the N-end rule pathway (75). Intriguingly, the anti-apoptotic protein BCL-XL appears to inhibit the production of acetyl-CoA (and hence limit N-acetylation) in a BAX- and BAK1-independent, but citrate- and acetate-sensitive manner (76). Remarkably, the anti-apoptotic effects of BCL-XL are partially inhibited by citrate and acetate, suggesting that N-acetylation also regulates apoptosis.
The HIF-1 checkpoint
The α chain of the transcription factor HIF-1 is regulated by specific proline hydroxylases (PHDs), which can target it to ubiquitin-dependent degradation. PHDs function to split molecular oxygen (O2) into two atoms, one of which hydroxylates its target (either of two highly conserved HIF-1α proline residues) and the other oxidizes α-ketoglutarate (α-KG) into succinate. Hypoxia or high amounts of succinate inhibit the activity of PHDs, whereas increased concentrations of α-KG stimulate it (77). When the functions of PHDs are compromised, HIF-1α accumulates and forms transcriptionally active heterodimers with HIF-1β.
Prolonged hypoxia (78, 79) as well as an increased availability of succinate (which occurs, for example, in individuals bearing inactivating loss-of-function mutations in succinate dehydrogenase or fumarate hydratase) (77) promotes a HIF-1-dependent state of resistance to apoptosis. This effect reflects the ability of HIF-1 to drive the expression of MCL-1 (80) BCL-XL (81), as well as many enzymes involved in glycolysis, including HKII (82). An increased glycolytic flux exerts cytoprotective effects, especially in hypoxic conditions, as it expands the pool of antioxidants generated by the pentose phosphate pathway (5). Finally, HIF-1 stimulates mitophagy and inhibits mitochondrial biogenesis (83, 84). This may impact on cell survival directly (by limiting the mitochondrial involvement in apoptotic and non-apoptotic cell death, see below) or indirectly, upon engagement of other checkpoints.
The ER stress checkpoint
The accumulation of unfolded proteins in the ER triggers a stress response that inhibits protein synthesis and activates several transcription factors (85). The ER stress response can originate from alterations in protein synthesis (e.g., the course of viral infection), degradation (e.g., when lysosomal proteases are defective), or folding (e.g., when the availability of chaperones is limited or in the presence of Ca2+, ROS, or increased temperatures). From our perspective, the ER stress response represents a checkpoint that promotes cell survival or cell death in a context-dependent manner.
Because protein glycosylation is critical to the normal processing of proteins in the ER and Golgi apparatus, defects in this process provoke ER stress responses. Protein glycosylation requires nucleotide sugars and hexosamines, which are synthesized from glucose (or other sugars) and various amino acids, including glutamine. Thus, glucose and amino acid deprivation can trigger ER stress. Along similar lines, an ER stress response can be elicited by hypoxia, because oxygen is required for the formation of disulfide bonds as well as for oligosaccharide modifications. Another source of ER stress is represented by dysregulated TORC1 activity (as caused by the absence of TSC2). In these conditions, cell survival is highly dependent on unsaturated lipids that sustain the expansion of the ER during the stress response (86).
In the course of ER stress, the eukaryotic translation initiation factor 2α (eIF2α) kinase 3 (EIF2AK3, best known as PERK) phosphorylates eIF2α, restricting the translation of a majority of proteins while stimulating the synthesis of activating transcription factor 4 (ATF4) and DNA-damage-inducible transcript 3 (DDIT3, best known as CHOP), which are necessary and sufficient for the death of cells exposed to tunicamycin, an inhibitor of protein glycosylation (87). Along similar lines, the cytotoxic response to glutamine deprivation of neuroblastoma cells overexpressing the MYC family proto-oncogene MYCN depends on ATF4 (88). ER stress as induced by ROS overgeneration also relies, at least in part, on the PERK-mediated induction of CHOP. Nonetheless, the cytotoxic activity of thapsigargin, which depletes ER Ca2+ stores, appears to be enhanced in PERK-deficient cells (89). These observations indicate that PERK may regulate cell death in opposing ways, depending on the initiating stimulus and perhaps other cell-intrinsic or cell-extrinsic variables.
It is likely that the ER stress machinery promotes cell survival by limiting protein synthesis, favoring the production of chaperones, enhancing protein degradation in the ER, and stimulating the expression of amino acid transporters (which support glycosylation), globally inhibiting the accumulation of additional unfolded proteins in this compartment. When this is insufficient to reestablish homeostasis, the cells die, a process that is exacerbated when eIF2α cannot be inhibited by PERK (87). The ER stress appears to promote apoptosis by inducing changes in the abundance of Bcl-2 proteins. ATF4 transactivates genes encoding the BH3-only proteins BCL2-binding component 3 (BBC3, best known as PUMA) and NOXA (88), whereas CHOP stimulates the expression of BCL2-like 11 (BCL2L11, best known as BIM) (90). BIM is further stabilized by the ER stress-dependent activation of protein phosphatase 2A (PP2A), resulting in the inhibition of anti-apoptotic BCL-2 proteins coupled to the activation of their pro-apoptotic counterparts and MOMP. Another, poorly understood pro-apoptotic BCL-2 protein, BOK, which structurally resembles BAX and BAK1, may be important in the response of some cells to tunicamycin (91). The molecular mechanisms whereby BOK is activated by ER stress remain to be elucidated. Of note, the accumulation of unfolded proteins within the ER can also cause the production of ROS by luminal NADPH oxidases that are activated by local protein disulfide isomerases and PPP-derived NADPH (87). In this setting, ATF4 and CHOP can further exacerbate ROS production and hence contribute to cell death.
While ER stress-induced apoptosis appears to proceed predominantly through the mitochondrial pathway, a number of studies suggest that caspases other than caspase-9 and caspase-3 are linked to cell death as induced by ER stress. These include caspase-2 (perhaps acting upstream of the mitochondrial pathway) (92), caspase-4 (93), and caspase-12 (94). However, caspase-12 is expressed in only a subset of humans (95) and murine Casp12−/− cells (96) as well as human cells lacking caspase-12 or caspase-4 (15975932) are fully susceptible to ER stress-induced apoptosis (97). The ER protein B-cell receptor-associated protein 31 (BCAP31) can interact with pro-caspase-8 during the ER stress response, and apparently activate it in a FADD-independent manner (98). In line with this notion, cells lacking the BAP31-interacting molecule cell death-inducing p53 target 1 (CDIP1) are resistant to the ER stress-induced activation of caspase-8 and consequent cell death (99). How these pathways contribute to the metabolic control of ER stress-induced cell death has not been fully resolved.
p53 and the non-essential amino acid checkpoint
The availability of amino acids in the extracellular milieu influences not only the activity of TORC1, but also protein glycosylation, as discussed above. In addition, several non-essential amino acids can promote cell survival (or death) by alternative mechanisms. Rapidly dividing and transformed cells can actually be “addicted” to these amino acids, which include (at least in some circumstances) glutamine, serine, glycine, arginine and asparagine (5).
Serine and glycine are interconvertible and serve as precursors for single carbon folate metabolism. Strikingly, the overexpression of several enzymes that participate in serine-glycine metabolism promotes oncogenic transformation, an effect that depends on their catalytic activity (100, 101) In this scenario, protein kinase Cζ (PKCζ), which represses the activity of many of these enzymes, acts as a tumor suppressor (102). Malignant cells as well as activated T lymphocytes consume large amounts of glutamine, representing a major source of carbon for the TCA cycle as well as for the synthesis of hexosamines, lipids and polyamines (103). Glutamine is also important for uptake of other amino acids, as its concentration gradient drives specific antiporters expressed at the cell surface.
When cells are deprived of serine or glycine, the enzymes of the serine-glycine synthetic pathway accumulate and p53 is temporarily stabilized, resulting in the transactivation of multiple target genes, including cyclin-dependent kinase inhibitor 1A (CDKN1A), coding for the cell cycle inhibitor p21Cip1 (104). Glutamine deprivation appears to trigger a similar signaling pathway (105). Cells in which p53 functions are compromised (such as many cancer cells) succumb to serine-glycine or glutamine deprivation (104, 105), suggesting that p53 actually suppresses (rather than activates) cell death in these settings. Whether the engagement of p53 by amino acid deprivation might also promote cell death upon the transactivation of pro-apoptotic p53 targets such as PUMA and NOXA is currently unexplored.
Some neoplasms lack the enzymes that generate arginine and asparagine, i.e., argininosuccinate synthetase and asparagine synthetase, respectively, thereby depending on external supplies for survival (5). The successful use of bacterial L-asparaginase (which degrades extracellular asparagine) in patients with acute lymphoblastic leukemia indicates that such a metabolic liability can be harnessed for therapeutic purposes. However, it seems that the cytotoxic effect of L-asparaginase stems (at least in part) from its ability to catabolize not only asparagine but also glutamine, hence activating the AMPK-TORC1 checkpoint (106).
Besides regulating cell cycle progression and cell death, p53 controls the expression of several proteins that are directly or indirectly involved in metabolism (107). These p53 targets do not include enzymes of the serine-glycine transformation pathway, but encompass TP53-induced glycolysis and apoptosis regulator (TIGAR, official name: C12orf5), which diverts the glycolytic flux toward the PPP (hence promoting nucleotide synthesis and the scavenging of ROS by the glutathione system) (108); glutaminase 2, which converts glutamine into glutamate, providing NH3+ for amino acid synthesis (109); SCO2, which regulates mitochondrial respiration (110), signal transducers implicated in the AMPK-TORC1 checkpoint, such as the β1 subunit of AMPK and tuberous sclerosis 2 (TSC2) (111); as well as several components of the autophagic machinery (112). Thus, p53 sustains cell survival in response to amino acid deprivation, as it does in conditions of limited DNA damage. Conversely, p53 may promote cell death when the effects of amino acid deprivation are severe, although this possibility has not yet been explored.
Several mechanisms may account for the impact of non-essential amino acids on p53 stabilization. In response to glutamine deprivation and the ER stress-dependent production of ROS, PP2A dephosphorylates (and hence inactivates) an E3 ubiquitin ligase that normally inhibits p53 (105). Importantly, amino acid deprivation also causes a disruption in ribosome biogenesis, mainly due to the inhibition of rRNA synthesis coupled to the ongoing production of ribosomal proteins. Accumulating rRNA-free ribosomal proteins promote the stabilization of p53 by interfering with its interaction with MDM2 (113). Thus, when the availability of amino acids is limited, the p53 checkpoint may be transiently activated to promote survival and permanently engaged to stimulate cell death (if homeostasis cannot be re-established). The p53 system is highly interconnected with various other metabolic checkpoints, including those orchestrated around the MPT and autophagy (114–116). Moreover, p53 appears to participate in the regulation of organismal homeostasis in a non-cell-autonomous manner, by both immunological (117) and metabolic circuitries (118). In particular, p53 has been proposed to control the synthesis of the adenosine A2b receptor (ADORA2B), hence regulating the cellular sensitivity to the stress-related extracellular accumulation of adenosine (118). Thus, the p53 checkpoint may be sensitive to metabolic fluctuations of both the intracellular and extracellular microenvironments.
Lipid synthesis as a metabolic checkpoint
Lipoapoptosis, i.e., the apoptotic death of cells exposed to lipids, is thought to provide a major etiological contribution to multiple diseases that develop when long-chain fatty acids (LCFAs) accumulate in non-adipose tissues, including non-alcoholic fatty liver disease, diabetes mellitus, and cardiovascular disorders. LCFAs such as palmitate can induce a lethal ER stress response (see above) (119), whose execution may rely on caspase-2 (120). Intriguingly, the ribonuclease endoplasmic reticulum to nucleus signaling 1 (ERN1, best known as IRE1α), which is usually activated upon ER stress, cleaves microRNA precursors that normally inhibit the translation of caspase-2 (121), corroborating the existence of a link between LCFA-induced ER stress and caspase-2 activation. Additionally, palmitate activates potentially lethal kinases such as c-JUN N-terminal kinase 1 (JNK1) and GSK3 (122).
The accumulation of cholesterol and its oxidation products also can have cytotoxic consequences. The cardiac damage inflicted by ischemia-reperfusion damage is accompanied by an increase in cholesterol uptake by mitochondria and the generation of auto-oxidized oxysterols, a phenomenon that can be avoided by ligands of translocator protein (18kDa) (TSPO), an OMM protein that interacts with VDAC1 and has been suggested to regulate MPT (123).
Cardiolipin (CL) is responsible for the positive curvature of the IMM and the proper function of several IMM enzymes. The CL molecules present at contact sites between the IMM and OMM reportedly facilitate the insertion into the latter of several Bcl-2 family members (124). In addition, CL usually adsorbs a large fraction of holocytochrome c to the IMM. Thus, besides facilitating MPT (see above) (125), the cell death-associated oxidation of polyunsaturated CL species favors the release of holocytochrome c from mitochondria upon MOMP (126). Recently, OMM-exposed CL has been shown to constitute a signal for the autophagic removal of permeabilized mitochondria (127), hence playing a major role in the maintenance of intracellular homeostasis.
Sphingolipid metabolism also has a prominent role in the regulation of cell death. Ceramide is generated from sphingosine or sphingomyelin by the enzymatic activity of ceramide synthase or various sphingomyelinases, respectively (128). Ceramide can trigger distinct apoptotic pathways depending on the subcellular compartment in which it accumulates. In response to ionizing radiation, acidic spingomyelinase is activated, catalyzing the generation of ceramide on the plasma membrane of endothelial intestinal cells, where it coalesces to form microdomains that transmit apoptotic signals (129). Ceramide that accumulates in mitochondria can trigger MOMP either by stimulating the insertion of BAX into the OMM (130) or by forming lipid pores that are under the control of proteins from the Bcl-2 family (131).
A physical and functional interaction between the ER and mitochondria regulates MOMP as it impacts on sphingolipid metabolism. In this setting, two products of the hydrolysis of sphingomyelin (which is mediated by ER and OMM enzymes), namely, sphingosine-1-phosphate and trans-2-hexadecenal, promote the pore-forming activity of BAK1 and BAX, respectively, whereas no role for ceramide other than as a precursor was identified (132). In line with this notion, the inhibition of the sphingomyelin pathway at any point upstream of the synthesis of trans-2-hexadecenal efficiently preserved cell survival in response to stress. Although it is not known how the ER and mitochondria precisely interact to regulate sphingolipid metabolism, it is interesting to note that palmitate stimulates sphingomyelin hydrolysis, implying that some of its cytotoxic effects may stem from direct MOMP stimulation.
Additional contributions of lipid metabolism to apoptosis are likely. The mitochondrial enzyme 3-oxoacyl-ACP synthase (OXSM) participates in stress-induced apoptosis in human cervical carcinoma cells (74). OXSM is required for the extension of long-chain fatty acids, and may have a role in the synthesis of lipoic acid, a potentially lipotoxic antioxidant. This said, how OXSM precisely contributes to apoptosis remains to be determined.
The PARP checkpoint
Poly (ADP-ribose) polymerases (PARPs) make up a large family of enzymes that catalyze the addition of poly (ADP-ribose) moieties (an NAD+-dependent reaction also known as ADP ribosylation) to multiple substrates, hence regulating a plethora of cellular processes, notably the repair of single and double-strand DNA breaks (133, 134). Thus, in the presence of extensive DNA damage, PARP1 hyperactivation may deplete the intracellular stores of NAD+, NADH, and ATP, resulting in a bioenergetic catastrophe often associated with necrotic cell death (135). Conversely, in the course of apoptosis, caspase-3 proteolytically inactivates PARP1, hence avoiding such a rapid drop in intracellular ATP concentrations leading to regulated necrosis. In line with this idea, cells that are unable to undergo MOMP because they lack BAX and BAK1 respond to an otherwise pro-apoptotic stimulus (alkylating DNA damage) by undergoing PARP1-dependent necrosis, in particular in conditions of intense proliferation (which are associated with an increase in glycolytic flux) (136). Conversely, quiescent cells (i.e., IL-3-dependent Bax−/−Bak1−/− cells deprived of growth factors or mouse embryonic fibroblasts at confluence), which manly obtain energy from oxidative phosphorylation, are resistant to such treatment (136). Thus, the metabolic status of the cell (notably the availability of NAD+, NADH and ATP) determines the subroutine by which cells die in response to (at least some) stimuli.
Hyperactivation of PARP1 may also promote non-apoptotic cell death by favoring the mitochondrial release of AIF (137), an NADH-consuming enzyme endowed with a latent endonuclease activity previously implicated in a MOMP- and calpain-dependent, but caspase-independent, form of apoptosis (11). Intriguingly, AIF appears to be released by mitochondria in response to PARP1 hyperactivation upon the binding of poly (ADP-ribose) moieties rather than as a result of calpain cleavage (138). How this would lead to the desorption of AIF from the IMM, in which it is stably inserted through a transmembrane domain, remain to be clarified. Irrespective of this unresolved issue, BCL-2 appears to suppress PARP1-dependent cell death by two mechanisms, namely, by inhibiting the mitochondrial release of AIF (11), and by directly blocking the enzymatic activity of PARP1 (139). Genetic ablation of Parp1 as well as a hypomorphic mutation in Aifm1 (which results in decreased AIF expression) exerts cytoprotective effects in several paradigms of acute cell loss in vivo, including various models of cerebral ischemia, supporting the potential pathophysiological relevance of this pathway, which has been named “parthanatos” (135).
Other metabolic checkpoints
Several metabolic enzymes may have roles in signaling that are controlled by the availability of their substrates. Although these have not been directly linked to cell death pathways, it is possible that such enzymes may represent metabolic checkpoints in cell fate decisions. This may account for the transforming activity of enzymes of the serine-glycine transformation pathway, although this has not been formally established. Two other examples involve glycolytic enzymes, namely, the M2 isoform pyruvate kinase, muscle (PKM2) and GAPDH.
PKM2 catalyzes the last step of glycolysis as it converts phosphoenolpyruvate into pyruvate. The glycolytic intermediate fructose-1,6-bisphosphate shifts the equilibrium between (enzymatically inactive) PKM2 dimers and (enzymatically active) PKM2 tetramers toward the latter. Conversely, by diverting the glycolytic flux toward the PPP, TIGAR (which operates as a fructose-2,6-bisphosphatase) favors PMK2 inactivation (5). The dimeric state of PKM2, which predominates over its tetrameric counterpart in many tumors, is also favored by direct interactions with oncogenic kinases (including RAF1 and SRC). Intriguingly, PKM2 physically interacts with, and thus modulates the transcriptional activity of, β catenin (140). In addition, dimeric PKM2 can operate as a protein kinase, thereby promoting the functions of signal transducer and activator of transcription 3 (STAT3), which control the synthesis of multiple anti-apoptotic proteins including BCL-2, BCL-XL and MCL-1 (141).
GAPDH converts glyceraldehyde-3-phosphate (G3P) into 1,3-bisphosphoglycerate. Under limited G3P availability, GAPDH binds specific mRNAs, such as that coding for interferon γ, and inhibits their translation (142). GAPDH also functions in a complex with other proteins to inhibit the translation of additional mRNAs in response to interferon γ (143). Moreover, a nuclear pool of GAPDH promotes transcriptional events through interactions with promyelocytic leukemia (PML) and OCT1 (144, 145). In one study, GADPH inhibited caspase-independent cell death by promoting the expression of the essential autophagic factor ATG12, thereby stimulating mitophagy (8). GADPH may accumulate at mitochondria in the course of apoptosis, thereby interacting with VDAC1 and promoting MPT (146). Except for the example cited above, however, the extent to which glycolysis and G3P contribute or modulate the extra-glycolytic functions of GAPDH is unknown. It remains possible that the metabolic state of the cell influences the ability of GAPDH to regulate cell death pathways. These and additional checkpoints based on metabolic enzymes may also function in non-canonical ways to control cell fate decisions in response to metabolic cues, although this remains to be formally demonstrated. Irrespective of such a possibility, the extra-glycolytic functions of PKM2 and GAPDH strongly corroborate the idea that metabolism and signaling are more intimately interconnected than previously thought.
Another possible metabolic checkpoint might involve Bcl-2 family members. These proteins have a number of functions that may impact on metabolism independently of their ability to control MOMP. For instance, BCL-2 and perhaps other members of the family physically interacts with the inositol 1,4,5-trisphosphate (IP3) receptor on the ER membrane, hence controlling reticular Ca2+ fluxes (147). The release of Ca2+ ions from reticular stores provides an important control on mitochondrial Ca2+ concentrations, which regulate a number of enzymes involved in the TCA cycle and in the electron transport chain (148). Pro-apoptotic Bcl-2 proteins also promote mitochondrial fusion (19) in a manner that is regulated by their anti-apoptotic counterparts (23, 149). This is relevant, as mitochondrial dynamics has an important role in preserving a metabolically efficient pool of mitochondria in response to stress (62). In addition, MCL-1 and BCL-XL appear to operate in the mitochondrial matrix or at the IMM, respectively, to promote respiratory functions and ATP production (150, 151). Finally, as mentioned above, (i) various members of the Bcl-2 protein family participate in the regulation of autophagy, mostly as they control the activity of BECN1, and (ii) BCL-XL appears to inhibit protein N-acetylation. Although such observations indicate that Bcl-2 proteins regulate various metabolic circuitries, the extent to which they do so is unclear. T lymphocytes lacking BAX and BAK1 exhibit activation defects that have been linked to a metabolic derangement (152), indicating that (at least in some circumstances) the metabolic activity of Bcl-2 proteins has a pathophysiological relevance.
If Bcl-2 proteins indeed represent an important hub for the regulation of cellular metabolism, we may ask how the detection of metabolic alterations feeds into this process. It is plausible that the mechanisms by which metabolic sensors impinge on the cell death-regulatory activity of Bcl-2 proteins also control the ability of this protein family to influence metabolic responses. As such, the interplay of metabolism and cell fate decisions may be “hard wired” through Bcl-2 proteins. It is an intriguing possibility that such interactions may have been ancestral, predating their role in cell death. In this context, it is interesting to note that the two Bcl-2 proteins identified so far in Drosophila probably do not regulate cell death, but rather exert metabolic functions (153).
Conclusions, implications and speculations
The relations between metabolism and cell death regulation are profound and intricate. On one hand, both apoptosis and necrosis can be initiated by metabolic perturbations (signals), mainly owing to the existence of refined detection systems (sensors) that are wired to the cell death machinery (effectors) through one or more signaling node(s) (transducers) (Fig. 1). Thus, small metabolites and metabolic by-products (e.g., ATP/ADP, NADH/NAD+, ROS), as well as various enzymes involved in metabolic circuitries (e.g., holocytochrome c, AIF, GAPDH) can influence cell fate decisions. On the other hand, several proteins originally characterized for their cell death-regulatory activity (e.g., Bcl-2 family members, p53) have a major metabolic functions (Suppl. Table 1). Thus, any distinction between metabolism and signaling has now decayed.
Two additional considerations can be made in this respect. First, as many factors that regulate or execute apoptosis and necrosis have roles unrelated to cell death, including roles in metabolism (154), these systems may have evolved to serve vital functions and only later may have acquired the ability to control cellular demise. Second, as many major regulators of cell death, including proteins and small metabolites, are also involved in steady-state metabolism, each of these molecules appears to operate in the context of a possible transition between physiological conditions, adaptive responses to stress (aimed at the re-establishment of homeostasis), and the activation of the cellular demise (when homeostasis cannot be restored). How the transition between the vital, adaptive, and lethal functions of each of these components is achieved remains unexplored. It is possible that the entire “network” built up at the interface between metabolism and cell death regulation may at some stage collapse as a result of an irreversible switch in the activity of one (or a few) of its components, provoking a domino effect. In this setting, multiple Bcl-2-like proteins may operate as central checkpoints that either interrupt (anti-apoptotic members) or amplify (pro-apoptotic members) the transition from homeostasis to death. Once these potential “firebreaks” have been trespassed, the fall is irreversible and cell death cannot be avoided.
How the various metabolic checkpoints are interconnected has just begun to emerge. It will be interesting to determine whether these systems are organized in a hierarchical manner and, if so, which are the most critical ones. Based on current knowledge, the checkpoints organized around MOMP, MPT, p53 and the Bcl-2 protein family stand out as prominent candidates for this title. A detailed understanding of the interplay between metabolism and cell death regulation is expected to drive the development of therapeutically relevant agents that either promote or inhibit cell death as they modulate metabolic circuitries (Suppl. Table 2). This knowledge will undoubtedly expand our armamentarium of anticancer agents as well as of drugs for avoiding (or at least minimizing) unwarranted cell death as it occurs the course of ischemic insults, neurodegeneration and various other clinical settings, including a large panel of genetic diseases in which specific metabolic alterations result in the pathological loss of post-mitotic cells (Suppl. Table 3).
Supplementary Material
Acknowledgments
We thank O. Kepp (INSERM, France) for critical help with figures and E. Gottlieb (Beatson Institute For Cancer Research, UK). GK is supported by the Ligue contre le Cancer (équipe labelisée); Agence National de la Recherche (ANR); Association pour la recherche sur le cancer (ARC); Cancéropôle Ile-de-France; AXA Chair for Longevity Research; Institut National du Cancer (INCa); Fondation Bettencourt-Schueller; Fondation de France; Fondation pour la Recherche Médicale (FRM); the European Commission (ArtForce); the European Research Council (ERC); the LabEx Immuno-Oncology; the SIRIC Stratified Oncology Cell DNA Repair and Tumor Immune Elimination (SOCRATE); the SIRIC Cancer Research and Personalized Medicine (CARPEM); and the Paris Alliance of Cancer Research Institutes (PACRI). DRG is supported by grants from the U.S. National Institutes of Health and the American Lebanese and Syrian Associated Charities.
References
- 1.Green DR, Victor B. Trends Cell Biol. 2012;22:555. doi: 10.1016/j.tcb.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galluzzi L, et al. Cell Death Differ. 2012;19:107. doi: 10.1038/cdd.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartwell LH, Weinert TA. Science. 1989;246:629. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 4.Wang R, Green DR. Nat Immunol. 2012;13:907. doi: 10.1038/ni.2386. [DOI] [PubMed] [Google Scholar]
- 5.Galluzzi L, Kepp O, Vander Heiden MG, Kroemer G. Nat Rev Drug Discov. 2013;12:829. doi: 10.1038/nrd4145. [DOI] [PubMed] [Google Scholar]
- 6.Tait SW, et al. Cell Rep. 2013 in press. [Google Scholar]
- 7.Vaughn AE, Deshmukh M. Nat Cell Biol. 2008;10:1477. doi: 10.1038/ncb1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colell A, et al. Cell. 2007;129:983. doi: 10.1016/j.cell.2007.03.045. [DOI] [PubMed] [Google Scholar]
- 9.Tait SW, et al. Dev Cell. 2010;18:802. doi: 10.1016/j.devcel.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lartigue L, et al. Mol Biol Cell. 2009;20:4871. doi: 10.1091/mbc.E09-07-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hangen E, et al. Trends Biochem Sci. 2010;35:278. doi: 10.1016/j.tibs.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 12.Green DR, Galluzzi L, Kroemer G. Science. 2011;333:1109. doi: 10.1126/science.1201940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green DR, Levine B. Cell. 2014;157:65. doi: 10.1016/j.cell.2014.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Bliek AM, Shen Q, Kawajiri S. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a011072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benard G, et al. J Cell Sci. 2007;120:838. doi: 10.1242/jcs.03381. [DOI] [PubMed] [Google Scholar]
- 16.Liesa M, Shirihai OS. Cell Metab. 2013;17:491. doi: 10.1016/j.cmet.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karbowski M, et al. J Cell Biol. 2002;159:931. doi: 10.1083/jcb.200209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedman JR, et al. Science. 2011;334:358. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karbowski M, et al. Nature. 2006;443:658. doi: 10.1038/nature05111. [DOI] [PubMed] [Google Scholar]
- 20.Hoppins S, et al. Mol Cell. 2011;41:150. doi: 10.1016/j.molcel.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karbowski M, et al. J Cell Biol. 2004;164:493. doi: 10.1083/jcb.200309082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheridan C, Delivani P, Cullen SP, Martin SJ. Mol Cell. 2008;31:570. doi: 10.1016/j.molcel.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 23.Llambi F, et al. Mol Cell. 2011;44:517. doi: 10.1016/j.molcel.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cassidy-Stone A, et al. Dev Cell. 2008;14:193. doi: 10.1016/j.devcel.2007.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montessuit S, et al. Cell. 2010;142:889. doi: 10.1016/j.cell.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whelan RS, et al. Proc Natl Acad Sci U S A. 2012;109:6566. doi: 10.1073/pnas.1201608109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C, et al. J Biol Chem. 2004;279:31761. doi: 10.1074/jbc.M401353200. [DOI] [PubMed] [Google Scholar]
- 28.Kroemer G, Galluzzi L, Brenner C. Physiol Rev. 2007;87:99. doi: 10.1152/physrev.00013.2006. [DOI] [PubMed] [Google Scholar]
- 29.Giorgio V, et al. Proc Natl Acad Sci U S A. 2013;110:5887. doi: 10.1073/pnas.1217823110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galluzzi L, Kroemer G. Nat Cell Biol. 2007;9:487. doi: 10.1038/ncb0507-487. [DOI] [PubMed] [Google Scholar]
- 31.Bonora M, et al. Cell Cycle. 2013;12:674. doi: 10.4161/cc.23599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pastorino JG, Hoek JB, Shulga N. Cancer Res. 2005;65:10545. doi: 10.1158/0008-5472.CAN-05-1925. [DOI] [PubMed] [Google Scholar]
- 33.Elrod JW, et al. J Clin Invest. 2010;120:3680. doi: 10.1172/JCI43171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Linkermann A, et al. Proc Natl Acad Sci U S A. 2013;110:12024. doi: 10.1073/pnas.1305538110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zitvogel L, Kepp O, Galluzzi L, Kroemer G. Nat Immunol. 2012;13:343. doi: 10.1038/ni.2224. [DOI] [PubMed] [Google Scholar]
- 36.Martinon F, et al. Nature. 2006;440:237. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 37.Duewell P, et al. Nature. 2010;464:1357. doi: 10.1038/nature08938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou R, Yazdi AS, Menu P, Tschopp J. Nature. 2011;469:221. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 39.Bauernfeind F, et al. J Immunol. 2011;187:613. doi: 10.4049/jimmunol.1100613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iyer SS, et al. Immunity. 2013;39:311. doi: 10.1016/j.immuni.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shackelford DB, Shaw RJ. Nat Rev Cancer. 2009;9:563. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gwinn DM, et al. Mol Cell. 2008;30:214. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Neill LA, Hardie DG. Nature. 2013;493:346. doi: 10.1038/nature11862. [DOI] [PubMed] [Google Scholar]
- 44.Inoki K, Kim J, Guan KL. Annu Rev Pharmacol Toxicol. 2012;52:381. doi: 10.1146/annurev-pharmtox-010611-134537. [DOI] [PubMed] [Google Scholar]
- 45.Pradelli LA, et al. Oncogene. 2010;29:1641. doi: 10.1038/onc.2009.448. [DOI] [PubMed] [Google Scholar]
- 46.Meynet O, et al. Blood. 2013;122:2402. doi: 10.1182/blood-2013-01-478651. [DOI] [PubMed] [Google Scholar]
- 47.Wei G, et al. Cancer Cell. 2006;10:331. doi: 10.1016/j.ccr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 48.Lum JJ, et al. Cell. 2005;120:237. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 49.Ding Q, et al. Mol Cell Biol. 2007;27:4006. doi: 10.1128/MCB.00620-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao Y, et al. Mol Cell Biol. 2007;27:4328. doi: 10.1128/MCB.00153-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maurer U, et al. Mol Cell. 2006;21:749. doi: 10.1016/j.molcel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 52.Ren H, et al. Mol Cancer. 2013;12:146. doi: 10.1186/1476-4598-12-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Green DR, et al. Mol Cell. 2011;44:9. doi: 10.1016/j.molcel.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Munoz-Pinedo C, et al. J Biol Chem. 2003;278:12759. doi: 10.1074/jbc.M212392200. [DOI] [PubMed] [Google Scholar]
- 55.Caro-Maldonado A, et al. Cell Death Differ. 2010;17:1335. doi: 10.1038/cdd.2010.21. [DOI] [PubMed] [Google Scholar]
- 56.Garcia-Garcia C, et al. Biochem Pharmacol. 2010;79:853. doi: 10.1016/j.bcp.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 57.Nieminen AI, et al. Proc Natl Acad Sci U S A. 2013;110:E1839. doi: 10.1073/pnas.1208530110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kroemer G, Marino G, Levine B. Mol Cell. 2010;40:280. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marino G, et al. Mol Cell. 2014 in press. [Google Scholar]
- 60.Gump JM, Thorburn A. Trends Cell Biol. 2011;21:387. doi: 10.1016/j.tcb.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marino G, Niso-Santano M, Baehrecke EH, Kroemer G. Nat Rev Mol Cell Biol. 2013 doi: 10.1038/nrm3735. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gomes LC, Di Benedetto G, Scorrano L. Nat Cell Biol. 2011;13:589. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gump JM, et al. Nat Cell Biol. 2014;16:47. doi: 10.1038/ncb2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu L, et al. Proc Natl Acad Sci U S A. 2006;103:4952. doi: 10.1073/pnas.0511288103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kroemer G, Levine B. Nat Rev Mol Cell Biol. 2008;9:1004. doi: 10.1038/nrm2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu Y, et al. Proc Natl Acad Sci U S A. 2013;110:20364. doi: 10.1073/pnas.1319661110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Elgendy M, Sheridan C, Brumatti G, Martin SJ. Mol Cell. 2011;42:23. doi: 10.1016/j.molcel.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 68.Maiuri MC, et al. EMBO J. 2007;26:2527. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boya P, Kroemer G. Oncogene. 2008;27:6434. doi: 10.1038/onc.2008.310. [DOI] [PubMed] [Google Scholar]
- 70.Wellen KE, et al. Science. 2009;324:1076. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McCoy F, et al. Mol Cell. 2013;52:325. doi: 10.1016/j.molcel.2013.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nutt LK, et al. Dev Cell. 2009;16:856. doi: 10.1016/j.devcel.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Andersen JL, et al. Mol Cell. 2011;43:834. doi: 10.1016/j.molcel.2011.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yi CH, et al. J Cell Biol. 2007;179:619. doi: 10.1083/jcb.200708090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hwang CS, Shemorry A, Varshavsky A. Science. 2010;327:973. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yi CH, et al. Cell. 2011;146:607. doi: 10.1016/j.cell.2011.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Selak MA, et al. Cancer Cell. 2005;7:77. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 78.Graeber TG, et al. Nature. 1996;379:88. doi: 10.1038/379088a0. [DOI] [PubMed] [Google Scholar]
- 79.Harris AL. Nat Rev Cancer. 2002;2:38. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 80.Piret JP, et al. J Biol Chem. 2005;280:9336. doi: 10.1074/jbc.M411858200. [DOI] [PubMed] [Google Scholar]
- 81.Chen N, et al. J Biol Chem. 2009;284:10004. doi: 10.1074/jbc.M805997200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Semenza GL. Nat Rev Cancer. 2003;3:721. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 83.Zhang H, et al. J Biol Chem. 2008;283:10892. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 84.Zhang H, et al. Cancer Cell. 2007;11:407. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 85.Wang S, Kaufman RJ. J Cell Biol. 2012;197:857. doi: 10.1083/jcb.201110131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Young RM, et al. Genes Dev. 2013;27:1115. doi: 10.1101/gad.198630.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Han J, et al. Nat Cell Biol. 2013;15:481. doi: 10.1038/ncb2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Qing G, et al. Cancer Cell. 2012;22:631. doi: 10.1016/j.ccr.2012.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Verfaillie T, et al. Cell Death Differ. 2012;19:1880. doi: 10.1038/cdd.2012.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Puthalakath H, et al. Cell. 2007;129:1337. doi: 10.1016/j.cell.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 91.Echeverry N, et al. Cell Death Differ. 2013;20:785. doi: 10.1038/cdd.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gu H, Chen X, Gao G, Dong H. Mol Cancer Ther. 2008;7:2298. doi: 10.1158/1535-7163.MCT-08-0186. [DOI] [PubMed] [Google Scholar]
- 93.Hitomi J, et al. J Cell Biol. 2004;165:347. doi: 10.1083/jcb.200310015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nakagawa T, et al. Nature. 2000;403:98. doi: 10.1038/47513. [DOI] [PubMed] [Google Scholar]
- 95.Saleh M, et al. Nature. 2004;429:75. doi: 10.1038/nature02451. [DOI] [PubMed] [Google Scholar]
- 96.Saleh M, et al. Nature. 2006;440:1064. doi: 10.1038/nature04656. [DOI] [PubMed] [Google Scholar]
- 97.Obeng EA, Boise LH. J Biol Chem. 2005;280:29578. doi: 10.1074/jbc.M502685200. [DOI] [PubMed] [Google Scholar]
- 98.Breckenridge DG, et al. Proc Natl Acad Sci U S A. 2002;99:4331. doi: 10.1073/pnas.072088099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Namba T, et al. Cell Rep. 2013;5:331. doi: 10.1016/j.celrep.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang WC, et al. Cell. 2012;148:259. doi: 10.1016/j.cell.2011.11.050. [DOI] [PubMed] [Google Scholar]
- 101.Possemato R, et al. Nature. 2011;476:346. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ma L, et al. Cell. 2013;152:599. doi: 10.1016/j.cell.2012.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang R, et al. Immunity. 2011;35:871. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Maddocks OD, et al. Nature. 2013;493:542. doi: 10.1038/nature11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Reid MA, et al. Mol Cell. 2013;50:200. doi: 10.1016/j.molcel.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 106.Willems L, et al. Blood. 2013;122:3521. doi: 10.1182/blood-2013-03-493163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vousden KH, Ryan KM. Nat Rev Cancer. 2009;9:691. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- 108.Bensaad K, et al. Cell. 2006;126:107. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 109.Hu W, et al. Proc Natl Acad Sci U S A. 2010;107:7455. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Matoba S, et al. Science. 2006;312:1650. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 111.Feng Z, et al. Cancer Res. 2007;67:3043. doi: 10.1158/0008-5472.CAN-06-4149. [DOI] [PubMed] [Google Scholar]
- 112.Kenzelmann Broz D, et al. Genes Dev. 2013;27:1016. doi: 10.1101/gad.212282.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Deisenroth C, Zhang Y. Oncogene. 2010;29:4253. doi: 10.1038/onc.2010.189. [DOI] [PubMed] [Google Scholar]
- 114.Tasdemir E, et al. Nat Cell Biol. 2008;10:676. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vaseva AV, et al. Cell. 2012;149:1536. doi: 10.1016/j.cell.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Green DR, Kroemer G. Nature. 2009;458:1127. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lujambio A, et al. Cell. 2013;153:449. doi: 10.1016/j.cell.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Long JS, et al. Mol Cell. 2013;50:394. doi: 10.1016/j.molcel.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 119.Ibrahim SH, Kohli R, Gores GJ. J Pediatr Gastroenterol Nutr. 2011;53:131. doi: 10.1097/MPG.0b013e31822578db. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Johnson ES, et al. J Biol Chem. 2013;288:14463. doi: 10.1074/jbc.M112.437210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Upton JP, et al. Science. 2012;338:818. doi: 10.1126/science.1226191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ibrahim SH, et al. J Hepatol. 2011;54:765. doi: 10.1016/j.jhep.2010.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Paradis S, et al. Cardiovasc Res. 2013;98:420. doi: 10.1093/cvr/cvt079. [DOI] [PubMed] [Google Scholar]
- 124.Katz C, et al. J Biol Chem. 2012;287:15016. doi: 10.1074/jbc.M111.328377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kagan VE, et al. Nat Chem Biol. 2005;1:223. doi: 10.1038/nchembio727. [DOI] [PubMed] [Google Scholar]
- 126.Ott M, Gogvadze V, Orrenius S, Zhivotovsky B. Apoptosis. 2007;12:913. doi: 10.1007/s10495-007-0756-2. [DOI] [PubMed] [Google Scholar]
- 127.Chu CT, et al. Nat Cell Biol. 2013;15:1197. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gault CR, Obeid LM, Hannun YA. Adv Exp Med Biol. 2010;688:1. doi: 10.1007/978-1-4419-6741-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rotolo J, et al. J Clin Invest. 2012;122:1786. doi: 10.1172/JCI59920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lee H, et al. PLoS One. 2011;6:e19783. doi: 10.1371/journal.pone.0019783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Perera MN, et al. Biochem J. 2012;445:81. doi: 10.1042/BJ20112103. [DOI] [PubMed] [Google Scholar]
- 132.Chipuk JE, et al. Cell. 2012;148:988. doi: 10.1016/j.cell.2012.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gibson BA, Kraus WL. Nat Rev Mol Cell Biol. 2012;13:411. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- 134.Michels J, et al. Oncogene. 2013 doi: 10.1038/onc.2012.623. [DOI] [PubMed] [Google Scholar]
- 135.Andrabi SA, et al. Nat Med. 2011;17:692. doi: 10.1038/nm.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zong WX, et al. Genes Dev. 2004;18:1272. doi: 10.1101/gad.1199904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang Y, et al. Sci Signal. 2011;4:ra20. doi: 10.1126/scisignal.2000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang Y, et al. J Neurochem. 2009;110:687. doi: 10.1111/j.1471-4159.2009.06167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Dutta C, et al. Cancer Res. 2012;72:4193. doi: 10.1158/0008-5472.CAN-11-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yang W, et al. Nature. 2011;480:118. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gao X, et al. Mol Cell. 2012;45:598. doi: 10.1016/j.molcel.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chang CH, et al. Cell. 2013;153:1239. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mukhopadhyay R, et al. Trends Biochem Sci. 2009;34:324. doi: 10.1016/j.tibs.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Colell A, Green DR, Ricci JE. Cell Death Differ. 2009;16:1573. doi: 10.1038/cdd.2009.137. [DOI] [PubMed] [Google Scholar]
- 145.Sirover MA. J Cell Biochem. 2012;113:2193. doi: 10.1002/jcb.24113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tarze A, et al. Oncogene. 2007;26:2606. doi: 10.1038/sj.onc.1210074. [DOI] [PubMed] [Google Scholar]
- 147.Rong Y, Distelhorst CW. Annu Rev Physiol. 2008;70:73. doi: 10.1146/annurev.physiol.70.021507.105852. [DOI] [PubMed] [Google Scholar]
- 148.Cardenas C, et al. Cell. 2010;142:270. doi: 10.1016/j.cell.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Delivani P, et al. Mol Cell. 2006;21:761. doi: 10.1016/j.molcel.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 150.Perciavalle RM, et al. Nat Cell Biol. 2012;14:575. doi: 10.1038/ncb2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chen YB, et al. J Cell Biol. 2011;195:263. doi: 10.1083/jcb.201108059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jones RG, et al. Immunity. 2007;27:268. doi: 10.1016/j.immuni.2007.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Monserrate JP, Chen MY, Brachmann CB. BMC Biol. 2012;10:63. doi: 10.1186/1741-7007-10-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Galluzzi L, et al. Cell Death Differ. 2008;15:1113. doi: 10.1038/cdd.2008.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.