Abstract
Adipose tissue inflammation increases with obesity, but adipocyte vs. immune cell contributions are unclear. In the present study, transcriptome analyses were performed on highly-purified subcutaneous adipocytes from lean and obese women, and differentially expressed genes/pathways were determined in both adipocyte and stromal vascular fraction (SVF) samples. Adipocyte but not SVF expression of NOD-like receptor pathway genes, including NLRP3 and PYCARD, which regulate caspase-1-mediated IL-1β secretion, correlated with adiposity phenotypes and adipocyte class II major histocompatibility complex (MHCII) gene expression, but only MHCII remained after adjusting for age and body mass index. IFNγ stimulated adipocyte MHCII, NLRP3 and caspase-1 expression, while adipocyte MHCII-mediated CD4+ T cell activation, an important factor in adipose inflammation, induced IFNγ-dependent adipocyte IL-1β secretion. These results uncover a dialogue regulated by interactions among T cell IFNγ and adipocyte MHCII and NLRP3 inflammasome activity that appears to initiate and escalate adipose tissue inflammation during obesity.
Keywords: Obesity, adipocyte, transcriptome analysis, network modeling, NOD-like receptor pathway, inflammasome
1. Introduction
Obesity is characterized by adipose tissue expansion due to adipocyte hypertrophy and a dramatic accumulation of immune cells (Han and Levings, 2013; Olefsky and Glass, 2010). These pro-inflammatory adipose changes profoundly impact multiple organs to promote insulin resistance, nonalcoholic steatohepatitis, atherosclerosis, and pancreatic beta-cell loss, which leads to type 2 diabetes (Shoelson et al., 2006). However, little is known about which adipose cell types regulate specific inflammatory pathways. Adipocytes are highly responsive to changes in caloric load, and are thus strong candidates to regulate adipose immune cell changes. We have reported that adipocyte MHCII expression increases early during high fat diet (HFD) administration and can activate adipose resident CD4+ T cells to adopt a pro-inflammatory phenotype (Deng et al., 2013b). Adipocytes also express innate immunity proteins, including toll-like receptors (TLRs) and NOD-like receptors (NLRs), and activation of adipocyte TLR4 signaling by saturated free fatty acids (FFAs) stimulates pro-inflammatory cytokine production, and adipocyte insulin resistance (Song et al., 2006). Thus, adipocytes can contribute to adipose inflammation via both adaptive and innate pathways during obesity.
Previous studies of obese vs. lean whole adipose tissue samples, did not directly address adipocyte-dependent contributions to adipose inflammation. We therefore analyzed highly-purified adipocytes of lean and obese postmenopausal women to identify pathways differentially regulated in obesity and correlations of candidate genes with obesity-related phenotypes. Our data indicate that multiple NLR genes increase in adipocyte but not SVF samples of obese women, and suggest that adipocyte NLRP3 inflammasome activity is a major contributor to adipose inflammation and dysfunction during obesity.
2. Materials and Methods
2.1 Study subjects
Subcutaneous adipose was obtained from 44 women (34–78 years-of-age, BMI 16.3–48.5 kg/m2, 35 Caucasian, 6 Hispanic, and 3 African-American) undergoing elective abdominal surgery at the Houston Methodist Hospital (Table 1). Exclusion criteria included fever or infection, renal or neoplastic disease, organ transplantation, steroid use, AIDS, >10% weight change within the previous 3 months, type-1 diabetes, hemochromatosis, or lipodystrophy. Men were excluded to avoid hormonal differences that can affect inflammatory processes. Subjects were assessed for systolic and diastolic blood pressure (SBP and DBP), body mass index (BMI), waist circumference, waist-hip-ratio (WHR), body adiposity index (BAI,[(hip circumference)/(height)1.5]-18) (Bergman et al., 2011). Pre-surgery plasma samples were analyzed for fasting glucose (FPG), insulin (FPI; EZHI-14K; Millipore, Billerica MA), triglycerides (TG), total cholesterol (Chol), high- and low-density lipoprotein cholesterol (HDL and LDL), high-sensitivity C-reactive protein (hsCRP), aspartate and alanine aminotransferase (AST and ALT), and TNFα, IL-6, CCL2 and leptin concentrations (MADPK-71K, Millipore). Subcutaneous adipose was biopsied near the umbilicus, immediately transferred to ice-cold saline and processed to isolate adipocytes and stromal vascular fraction (SVF) samples (Halleux et al., 1999). This study was approved by the Institutional Review Board of the Houston Methodist Hospital. All subjects gave written informed consent prior to participation.
Table 1.
Phenotypic parameters of the study populations.
| Total population (N=44) Mean ± SE |
N | Population | Microarray | ||||
|---|---|---|---|---|---|---|---|
| Lean (N=13) Mean ± SE |
Obese (N=27) Mean ± SE |
Lean (N=7) Mean ± SE |
Obese (N=7) Mean ± SE |
||||
| Age (years) | 57.6 ± 1.5 | 44 | 54.3 ± 2.4 | 59.1 ± 1.8 | 57.7 ± 2.78 | 60.7 ± 2.53 | |
| Obesity | BMI (kg/m2) | 31.7 ± 1.3 | 44 | 22.0 ± 0.7 | 36.8 ± 1.2a | 20.8 ± 1.0 | 32.8 ± 0.9ce |
| BAI (cm/m1.5) | 38.0 ± 1.3 | 43 | 28.7 ± 0.9 | 43.3 ± 1.2a | 26.6 ± 1.2 | 40.2 ± 1.0cf | |
| WHR | 0.87 ± 0.01 | 43 | 0.82 ± 0.02 | 0.89 ± 0.02a | 0.83 ± 0.03 | 0.88 ± 0.03 | |
| Waist (cm) | 99.8 ± 3.0 | 43 | 77.3 ± 2.4 | 112 ± 2.6a | 76 ± 3.5 | 104 ± 4.1d | |
| MS factors | FPG (mmol/L) | 5.15 ± 0.23 | 44 | 4.48 ± 0.19 | 5.40 ± 0.32a | 4.15 ± 0.15 | 4.72 ± 0.41 |
| TG (mmol/L) | 1.43 ± 0.12 | 43 | 1.08 ± 0.16 | 1.50 ± 0.16 b | 1.02 ± 0.22 | 1.21 ± 0.17 | |
| HDL (mmol/L) | 1.41 ± 0.05 | 43 | 1.51 ± 0.11 | 1.40 ± 0.06 | 1.57 ± 0.18 | 1.43 ± 0.08 | |
| SBP (mmHg) | 130 ± 3.1 | 44 | 120 ± 6.1 | 135 ± 3.7a | 122 ± 8.6 | 132 ± 6.9 | |
| DBP (mmHg) | 70.2 ± 2.4 | 44 | 67.5 ± 3.7 | 72.7 ± 3.1 | 65.1 ± 5.5 | 69.7 ± 5.6 | |
| plasma factors | FPI (pmol/L) | 72.1 ± 9.7 | 38 | 50.4 ± 15.1 | 84.7 ± 12.9b | 44.0 ± 22.1 | 78.7 ± 17.1 |
| Chol (mmol/L) | 5.08 ± 0.15 | 43 | 4.6 ± 0.2 | 5.2 ± 0.2b | 4.6 ± 0.4 | 5.2 ± 0.4 | |
| LDL (mmol/L) | 2.87 ± 0.13 | 43 | 2.5 ± 0.2 | 3.0 ± 0.2a | 2.4 ± 0.2 | 3.0 ± 0.4 | |
| AST µkat/L) | 0.58 ± 0.06 | 44 | 0.58 ± 0.08 | 0.52 ± 0.04 | 0.66 ± 0.13 | 0.47 ± 0.04e | |
| ALT (µkat/L) | 0.48 ± 0.10 | 44 | 0.36 ± 0.05 | 0.42 ± 0.05 | 0.32 ± 0.06 | 0.26 ± 0.05 | |
| hsCRP (nmol/L) | 48.5 ± 7.4 | 37 | 16.9 ± 5.2 | 63.3 ± 9.9a | 17.2 ± 6.2 | 54.7 ± 18.5d | |
| HOMA-IR | 2.5 ± 0.4 | 38 | 1.5 ± 0.4 | 3.0 ± 0.5a | 1.2 ± 0.6 | 2.4 ± 0.5 | |
(ap<0.05 or bp<0.1 lean vs. obese in the total population, cp<0.05 or dp<0.1 lean vs. obese in the microarray population, and ep<0.05 or fp<0.1 in obese microarray vs. obese subjects in the total population.
See Materials and Methods for abbreviations.
2.2 RNA analyses
Adipocytes and SVF were isolated, and adipocyte samples were leukocyte-depleted to achieve <0.1% leukocyte contamination as previously described (Deng et al., 2013b). Adipocyte RNA samples isolated from consecutive post-menopausal lean (BMI <25 kg/m2) and obese (BMI 30–40 kg/m2) women (Table 1) that met RNA integrity criteria were analyzed by MOgene LLC (St. Louis, MO) using Agilent Human Whole Genome 4x44K arrays (Santa Clara, CA). Results were processed with the R-2.13.2 platform and the Bioconductor Agi4x44Preprocess package under default settings and deposited in the NCBI Gene Expression Omnibus (GEO) database under accession number GSE44000. All qualified probes were consolidated using the gene-set enrichment analysis (GSEA) platform CollapseDataset tool with default settings (Subramanian et al., 2005). Genes showing significant differential expression were identified using SAM (Tusher et al., 2001). RT-PCR analyses used reagents from Applied Biosystems (Grand Island, NY), with gene expression normalized to PPIA expression.
2.3 Network analysis
GSEAs were performed using GSEA v2.07 with 186 KEGG human pathways (Subramanian et al., 2005). Fisher's exact test and statistical corrections for multiple comparisons (Fisher, 1922) were calculated in MATLAB to identify pathways enriched with significant genes. Pathway overlap was analyzed with the Cytoscape 2.8.2 network visualization platform with Enrichment Map plugin v1.2 (Merico et al., 2010).
2.4 Capase-1 activity
Caspase-1 activity in protein lysates of total human adipose tissue and purified mouse adipocytes was measured using a fluorometric assay kit (BioVision, San Francisco, CA).
2.5 Mouse Studies
Male C57BL/6J mice (Jackson Laboratory, Bar Harbor, ME) were randomly assigned to chow (8904, Harlan Teklad) or high-fat diet (HFD: 60% kcal fat; D12492, Research Diets, New Brunswick, NJ) for 12 weeks then sacrificed for adipocyte and SVF isolation from epididymal fat (Deng et al., 2013b). Mouse SVF and adipocyte samples were generated using pooled adipose tissue from 3–4 chow-fed mice or 2 obese mice per sample. All animal procedures were conducted in a specific pathogen–free facility at the Houston Methodist Research Institute in accordance with institutional animal care and use committee guidelines.
2.6 Cell culture studies
Post-confluent 3T3-L1 cells were induced to differentiate in DMEM with 10% FBS, 1 µM dexamethasone, 0.5 mM IBMX, and 10 µg/ml insulin for 2 days, then maintained in DMEM/10% FBS/10 µg/ml insulin for 8 days. Cells were then incubated with PBS, 5ng/ml TGFβ or TNFα, 200 µM BSA-conjugated-palmitate, 2 ng/ml IFNγ, 10 ng/ml IL-1β or IL-6, or 100 ng/ml lipopolysaccharide (LPS); increasing doses (0–1000 pg) of IFNγ; or 2 ng/ml IFNγ plus 100 ng/ml LPS for 24 hrs prior to RNA isolation. TGFβ, IFNγ, TNFα, IL-1β and IL-6 were obtained from R&D Systems (Minneapolis, MN). Palmitate and LPS were purchased from Sigma-Aldrich (St. Louis, MO). Primary mouse adipocytes were purified from epididymal fat pads of HFD-fed C57BL/6J mice (Deng et al., 2013b), mixed with ice-cold Matrigel (BD Biosciences; San Jose, CA) using a 1:5 adipocytes to gel ratio that was seeded into chilled 96-well plates (40µl/well) for 5 min on ice then congealed 10 min at 37 C. CD4+ T cells of ovalubmin-reactive B6.Cg-Tg(TcraTcrb)425Cbn/J mice (Jackson Laboratory) were added to primary adipocyte cultures at 5×105 cells/well in 150ul RPMI-1640/10% FBS/β-mercaptoethanol with or without 500ug/ml ovalbumin (Sigma-Aldrich) or 2ng/ml IFNγ (R&D Systems), and supernatant IL-1β and IFNγ concentrations were analyzed by ELISA (BD Biosciences) after 3 days culture.
2.7 Statistical analyses
For correlation analyses, continuously-scaled variables were skew-zero transformed to remove scale and range effects by converting each variable’s percentile scores into inverse cumulative standard normal variates. Pearson partial correlation analysis was then run on the skew-zero variables with adjustment for age, BMI or age+BMI. Stata Version 11 was used for skew-zero transforms and Statistica Version 10 for partial correlation.
3. Results
3.1 The NOD-like receptor (NLR) pathway is upregulated in adipocytes from obese subjects
Subcutaneous adipose biopsies from women undergoing elective abdominal surgery (Table S1) were processed as previously described to isolate purified adipocytes (Deng et al., 2013b). Macrophage- (Emr1) and T-cell-specific (Cd3d) mRNAs were substantially depleted in purified adipocyte fractions (Fig. S1), consistent with previous results of <0.1% leukocyte contamination (Deng et al., 2013b), while SVF samples were depleted of adipocyte-specific (Adipoq) mRNA. Adipocyte RNA samples isolated from the first 7 consecutive lean (BM1<25 kg/m2) and obese (BMI >30 and <40 kg/m2) postmenopausal subjects were subjected to microarray analyses. Subjects whose samples were chosen for microarray analysis were similar to their corresponding groups in the total population (Table 1). No differences were apparent among the two lean groups, but the BMI, BAI and waist values were greater in the total vs. microarray obese populations, due to the more restrictive BMI criteria used to define the latter group. There was also a difference in ALT among the two obese groups, although not between lean vs. obese subjects in either group. The lean vs. obese groups of the total population demonstrated differences in parameters associated with obesity (BMA, BAI, WHR, waist) metabolic syndrome (FPG, TG, SBP), plasma factors (FPI, Chol, LDL, hsCRP) and insulin resistance. Microarray subjects demonstrated corresponding trends for change in all these parameters, but likely due to the smaller sample size these reached significance only for BMI, BAI, and hsCRP.
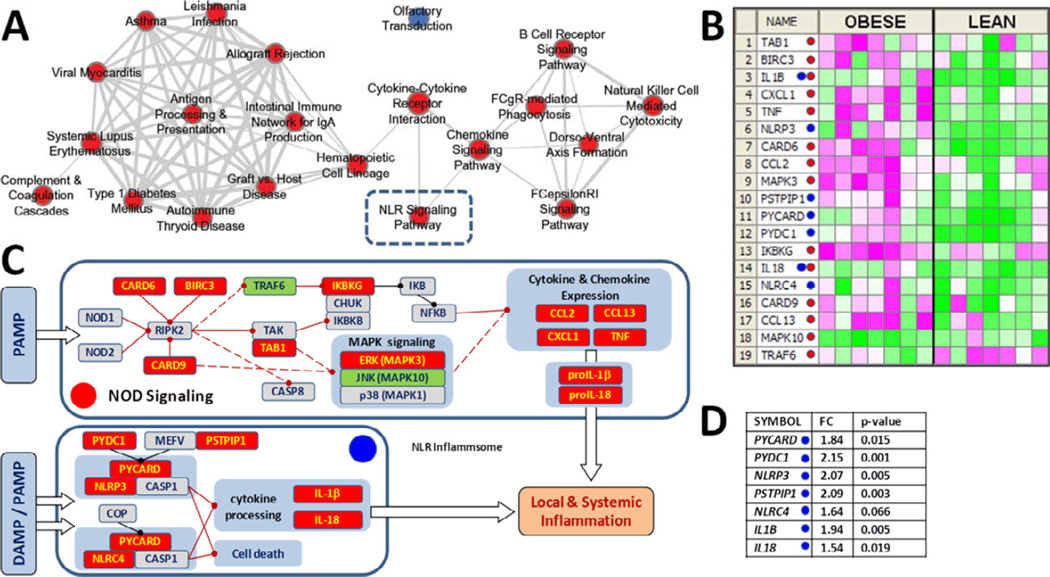
The microarray analysis detected 15 177 genes, of which 2 309 significantly differed (>1.5-fold absolute change, p-value <0.05) with obesity. GSEA of KEGG pathways using a nominal enrichment score (NES) p-value <0.01 and false discovery rate (FDR) q-value <0.01 identified 27 pathways enriched with genes up-regulated in obesity and 2 pathways enriched with down-regulated genes. Fisher’s exact test found that 21 of the 27 pathways were enriched with genes demonstrating significant changes between lean vs. obese groups (Benjamini-adjusted NES p<0.1 and FDR q<0.05). Shared genes connected 20 of these 21 pathways (Fig. 1A), segregating into two highly-interactive clusters.
Figure 1. Adipocyte NLR signaling pathway expression increases with obesity.
A) Network analysis of KEGG pathways enriched in SQ adipocytes of obese vs. lean women indicating pathways enriched with up- (red) or down-regulated (blue) genes. Line thickness depicts gene overlap (minimum 30% overlap with the smaller pathway). B) Heat map of up-(red) and down-regulated (green) NOD-signaling- and NLR-inflammasome-related genes (red and blue dots, respectively). C) Schematic of the NLR pathway. Red lines indicate direct (solid) and indirect (dashed) activation, while black lines denote inhibitory regulations (adapted from KEGG map). D) NLR inflammasome genes differentially-regulated in array data of obese vs. lean subjects.
Gene sets in the smaller cluster shared only one gene, the Erk MAPK family member MAPK3, while those in the larger cluster shared 5 genes of the class II major histocompatibility complex (HLA-DRA and HLA-DRB1, -3, -4, and -5), which we have reported is a key instigator of adipose inflammation during caloric excess (Deng et al., 2013b). The KEGG NOD-like receptor signaling pathway linked two nodes: cytokine-cytokine receptor interaction, which linked the two clusters, and the chemokine signaling pathway node. The toll-like receptor signaling pathway, which can regulate NOD-like receptor (NLR) signaling (Becker and O'Neill, 2007), missed the cut-off for enriched KEGG pathways (FDR q=0.025).
Multiple NOD signaling genes (15/57) were differentially regulated in adipocytes of obese subjects (>1.5-fold change, p<0.05), while four more (BIRC3, CARD9, TAB1 and NLRC4) revealed similar fold-changes at p<0.1 (Fig. 1B–C, red dots). Further, 7 of 18 components of the NLR inflammasome pathway increased (p<0.01) with obesity (Fig. 1C–D, blue dots), including 2 of 3 genes required for NLRP3 inflammasome activity (NLRP3 and PYCARD), as well as the inflammasome substrates IL-1β and IL-18.
3.2 Adipocyte, but not SVF, inflammasome expression increases with obesity
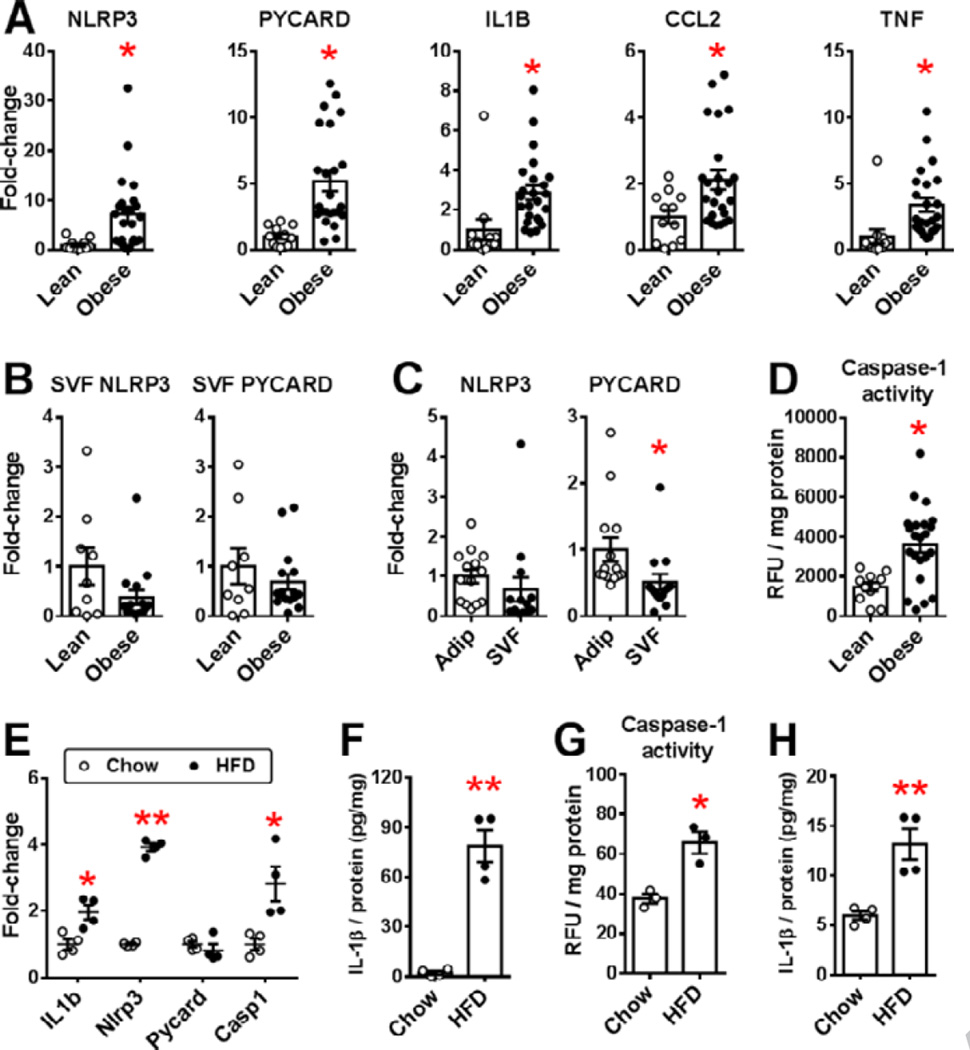
Adipocytes express all three components required for NLRP3 inflammasome activity (NLRP3, PYCARD and CASP1), and adipocyte NLRP3 and PYCARD expression significantly increased in obese versus lean subjects, as did other TLR-regulated genes (IL1B, CCL2 and TNF), validating our array results (Fig. 2A). However, SVF NLRP3 and PYCARD expression did not increase with obesity (Fig. 2B), and was similar in matched adipocyte and SVF samples of obese subjects (Fig. 2C). Caspase-1 activity increased in obese versus lean adipose tissue samples (Fig. 2D), but was not measured in human adipocyte and SVF fractions due to insufficient sample.
Figure 2. NLR expression in adipose tissue of lean and obese humans and mice.
NLR-related gene expression in human SQ A) adipocytes, B) SVF C) paired adipocyte (Adip) and SVF samples of obese subjects and D) adipose tissue caspase-1 activity. (Means±SE, *p<0.05, A: 12 lean/24 obese; B: 9 lean/16 obese; C: 14 paired samples; D: 11 lean/22 obese). Expression of E) NLRP3 family mRNA F) IL-1β protein (pg/mg lysate), G) caspase-1 activity (relative fluorescence units (RFU) /mg protein lysate) and H) cell culture IL-1β secretion (pg secreted/mg adipocyte lysate) in purified epididymal adipocytes of chow-fed lean and HFD-fed obese C57BL/6 male mice. (Mean±SE, N=4 (E,F,H) or N=3 (G) pooled samples/group, using 3–4 chow-fed mice or 2 HFD-fed mice for each pooled sample, *p<0.05 or **p<0.01 vs. chow-fed group).
Purified epididymal adipocytes of obese versus lean C57BL/6 mice revealed Nlrp3 and Il1B increases similar to those in human adipocytes, and increased Casp1 expression, but no increase in Pycard mRNA level (Fig. 2E). Adipocyte IL-1β protein levels markedly increased with obesity, as did caspase-1 activity, corresponding with increased caspase-1-dependent IL-1β secretion (Fig. 2F–H). Taken together, these data suggest that adipocytes substantially contribute to increased NLRP3 inflammasome activity in adipose tissue of obese humans and mice.
3.3 NLRP3 and PYCARD correlate with adiposity, inflammation and adipocyte MHCII
NLRP3 and PYCARD expression strongly correlated in human adipocytes, and both correlated with age, all measures of adiposity (BMI, WHR, BAI), SBP, FPI, hsCRP and leptin, and adipocyte expression of select adipokine (LEP, CCL2, TNF, IL1B), anti-oxidant (NQO1) and MHCII (CIITA and HLA-DPB) genes (Table 2). However, only PYCARD expression significantly correlated with HOMA-IR. Adipose caspase-1 activity correlated with NLRP3 and PYCARD, adiposity (BMI, BAI, but not WHR), plasma LDL, IL-6 and hsCRP, but not age or any other factors correlated with adipocyte NLRP3 and PYCARD expression. Adjusting for age and BMI, however, abrogated all correlations except NLRP3 with PYCARD, and their correlations with adipocyte MHCII expression (Table 2).
Table 2.
| Non-adjusted | BMI+Age-adjusted | ||||
|---|---|---|---|---|---|
| SA.NLRP3 | SA.PYCARD | SA.NLRP3 | SA.PYCARD | N | |
| Age | 0.41a | 0.39a | -- | -- | 40 |
| BMI | 0.44a | 0.64a | -- | -- | 40 |
| BAI | 0.36a | 0.59a | −0.08 | 0.08 | 39 |
| WHR | 0.38a | 0.44a | 0.15 | 0.19 | 39 |
| FPG | 0.13 | 0.17 | 0.01 | 0.00 | 40 |
| TG | 0.23 | 0.23 | 0.08 | 0.03 | 39 |
| HDL | −0.11 | −0.08 | −0.06 | 0.00 | 39 |
| SBP | 0.39a | 0.37a | 0.14 | 0.03 | 40 |
| DBP | 0.09 | 0.03 | 0.08 | −0.08 | 40 |
| FPI | 0.35a | 0.51a | 0.13 | 0.29 | 35 |
| Cholesterol | 0.28 | 0.22 | 0.21 | 0.06 | 39 |
| LDL | 0.23 | 0.21 | 0.16 | 0.05 | 39 |
| TNFA | 0.15 | 0.26 | −0.13 | 0.00 | 35 |
| Leptin | 0.47a | 0.64a | 0.20 | 0.33 | 35 |
| IL-6 | 0.05 | 0.10 | −0.19 | −0.24 | 35 |
| MCP-1 | 0.14 | −0.05 | 0.23 | 0.04 | 35 |
| hsCRP | 0.38a | 0.36a | 0.17 | −0.03 | 35 |
| HOMA-IR | 0.31 | 0.50a | 0.08 | 0.26 | 35 |
| Caspase-1 activity | 0.39a | 0.38a | 0.33 | 0.24 | 35 |
| SA.NLRP3 | 1.00 | 0.83a | 1.00 | 0.74a | 40 |
| SA.PYCARD | 0.83a | 1.00 | 0.74a | 1.00 | 40 |
| SA.SOD2 | 0.09 | 0.07 | 0.10 | 0.10 | 40 |
| SA.NQO1 | 0.39a | 0.56a | 0.11 | 0.24 | 40 |
| SA.TNFA | 0.57a | 0.66a | 0.22 | 0.16 | 40 |
| SA.CCL2 | 0.39a | 0.48a | 0.15 | 0.19 | 40 |
| SA.LEP | 0.47a | 0.63a | 0.20 | 0.30 | 40 |
| SA.IL1B | 0.45a | 0.56a | 0.07 | 0.06 | 40 |
| SA.CIITA | 0.68a | 0.80a | 0.51a | 0.70a | 40 |
| SA.HLA-DPB | 0.59a | 0.74a | 0.36a | 0.59a | 40 |
Non-adjusted and BMI-adjusted Pearson correlation coefficients of SQ adipocyte (SA) NLRP3 and PYCARD expression, indicating significant correlation (ap<0.5).
The number of paired values available for each correlation is shown, with smaller numbers indicating missing values.
See Materials and Methods for abbreviations.
3.4 SVF NLRP3 and PYCARD expression does not correlate with adiposity
In sharp contrast, SVF NLRP3 and PYCARD expression correlated with each other, but not with age, adiposity or metabolic syndrome factors, or adipose caspase-1 activity (Table 3). Macrophages invade adipose tissue during weight gain and polarize to a pro-inflammatory M1 versus an anti-inflammatory M2 phenotype (Han and Levings, 2013; Olefsky and Glass, 2010). SVF NLRP3 and PYCARD expression strongly correlated with the macrophage marker CD68, and trended (p<0.1) to negatively correlate with CD68-normalized expression of the M2 marker ARG1 (ARG1/CD68), but did not positively correlate with the M1 marker ITGAX (ITGAX/CD68). NLRP3 and PYCARD strongly correlated with SVF IL1B, but not other pro-inflammatory cytokines, including IFNG, suggesting that SVF NLRP3 inflammasome expression was not stimulated by these cytokines, which are known to increase in adipose tissue with weight gain.
Table 3.
| SS.NLRP3 | SS.PYCARD | N | |
|---|---|---|---|
| Age | 0.14 | 0.01 | 23 |
| BMI | −0.11 | 0.13 | 23 |
| BAI | −0.13 | 0.14 | 22 |
| WHR | −0.13 | −0.15 | 22 |
| FPG | −0.09 | 0.03 | 23 |
| TG | −0.18 | −0.27 | 22 |
| HDL | 0.16 | 0.19 | 22 |
| SBP | 0.15 | 0.01 | 23 |
| DBP | 0.16 | 0.21 | 23 |
| FPI | −0.09 | 0.15 | 20 |
| Cholesterol | 0.18 | 0.06 | 22 |
| LDL | 0.12 | 0.05 | 22 |
| TNFα | −0.09 | −0.08 | 21 |
| Leptin | −0.09 | 0.00 | 21 |
| IL-6 | −0.42a | −0.34 | 21 |
| MCP-1 | 0.32 | 0.24 | 21 |
| hsCRP | −0.02 | −0.09 | 20 |
| HOMA-IR | −0.18 | 0.07 | 20 |
| Caspase-1 activity | −0.05 | 0.10 | 20 |
| SS.Nlrp3 | 1.00 | 0.85a | 23 |
| SS.Pycard | 0.85a | 1.00 | 23 |
| SS.CD68 | 0.75a | 0.69a | 23 |
| SS.CD11/CD68 | 0.26 | 0.28 | 18 |
| SS.ARG1/CD68 | −0.46b | −0.41b | 18 |
| SS.TNFA | 0.14 | 0.28 | 15 |
| SS.CCL2 | −0.10 | −0.18 | 15 |
| SS.IFNg | 0.14 | 0.29 | 23 |
| SS.IL1B | 0.63a | 0.77a | 23 |
Pearson correlation coefficients of SQ SVF (SS) NLRP3 and PYCARD expression, indicating significant correlation (ap<0.5) and trend (bp<0.1) towards correlation.
The number of paired values available for each correlation is shown, with smaller numbers indicating missing values.
3.5 Adipocyte NLRP3 and PYCARD expression is regulated by IFNγ
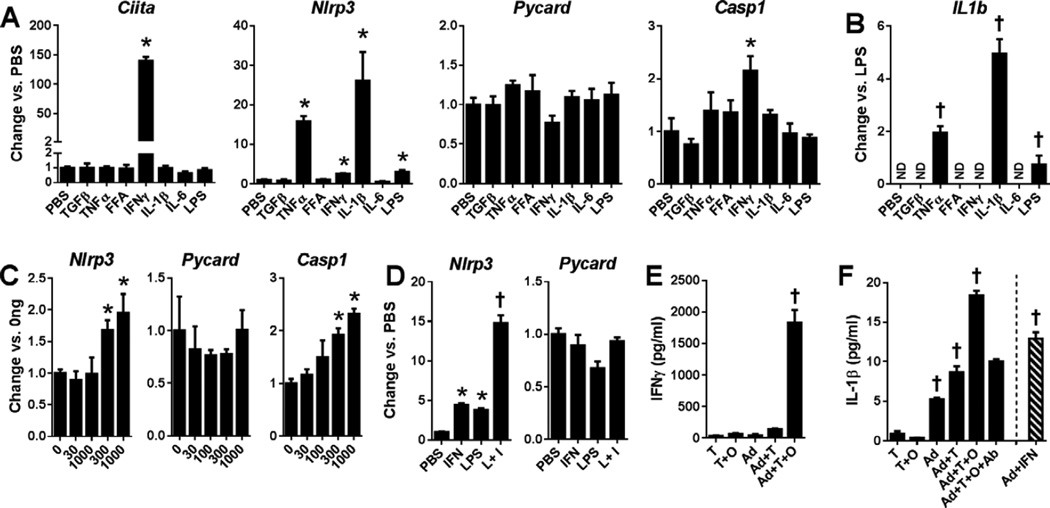
MHCII expression in adipocytes is primarily regulated by IFNγ (Deng et al., 2013b). Thus, strong correlations among adipocyte MHCII and inflammasome expression in human adipocytes may reflect common regulation by IFNγ primarily produced by adipose-resident T cells. Mouse 3T3-L1 adipocytes were incubated with pro-inflammatory adipokines, FFA and LPS to analyze the effect of stimuli present during obesity. Similar to adipocyte expression in obese vs. lean mice, IFNγ increased Nlrp3 and Casp1 expression, but not Pycard (Fig. 3A), while also increasing Ciita, the master regulator of the MHCII pathway. The TLR4 ligand LPS stimulated Nlrp3, as did TNFα and IL-1β, while the saturated FFA palmitate, which can directly and indirectly activate TLR4 signaling (Pal et al., 2012), had no effect (Fig. 3A). IL-1β expression was not induced by IFNγ but, like Nlrp3, was stimulated by IL-1β, TNFα and LPS (Fig. 3B). Nlrp3 and Casp1 upregulation by IFNγ was dose-dependent and Nlrp3 expression was synergistically increased upon exposure to both IFNγ and LPS (Fig 3C–D).
Figure 3. Regulation of NRL-related gene expression in adipocytes.
RT-PCR analysis of A) Ciita and NLRP3 inflammasome and B) IL1β gene expression in 3T3-L1 adipocytes after 24hr induction with the indicated stimuli, C) 0–1000 pg/ml IFNγ or D) PBS, IFNγ, LPS or LPS+IFNγ (L+I). E) IFNγ and F) IL-1β concentrations in primary mouse adipocyte (Ad) and T cell culture supernatants 3 days post-induction ± ovalbumin (O) and/or IFNγ blocking antibody (Ab). (Mean±SE, N=2–4/group, *p<0.05 vs. PBS by 2-tailed t-test, †p<0.05 vs. all others by ANOVA).
Adipocytes of HFD-fed mice promoted antigen-dependent CD4+ T cell IFNγ secretion and adipocyte IL-1β production; IFNγ-neutralizing antibodies fully inhibited IL-1β secretion, and IFNγ directly induced adipocyte IL-1β expression (Fig. 3E–F). Correspondingly, we found that NLRP3 and PYCARD in human adipocytes expression correlated or trended towards correlation with SVF IFNG expression, as did adipocyte MHCII gene expression and adipose tissue caspase-1 activity (Table S1). Strong NLRP3 and PYCARD correlations with MHCII genes in human adipocyte samples thus likely results from shared regulation by IFNγ. SVF IFNG expression, similar to adipocyte NLRP3 and PYCARD expression, also correlated or trended to correlate with age and measures of adiposity, insulin resistance and adipose tissue inflammation (Table S2), but did not correlate with SVF MHCII, NLRP3 or PYCARD.
4. Discussion
Secretion of pro-inflammatory factors by adipocytes and immune cells in adipose tissue profoundly alters the adipose tissue microenvironment in obesity, leading to defects in adipocyte differentiation, insulin action and lipid storage, and enhanced lipolysis. Resulting FFA increases promote ectopic lipid accumulation and metabolic derangements, including systemic insulin resistance, mitochondrial dysfunction, and oxidative stress in multiple tissues (Cusi, 2009). Both adipocytes and immune cells are implicated to induce adipose inflammation in obesity, but few studies have analyzed isolated adipocytes. Our microarray analysis of adipocytes of lean and obese post-menopausal women with no differences in metabolic syndrome components, but with clinically different systemic inflammation (hsCRP), identified 20 pathways increased in adipocytes of obese subjects, including an intriguing association of the NLR signaling pathway with pathways involved in cytokine signaling and MHCII function.
NLRP3 inflammasome activation requires the recruitment of PYCARD, which must exit the nucleus and combine with pro-caspase-1 to stimulate autocatalytic caspase-1 activation (Bryan et al., 2009). Adipose tissue caspase-1 expression increases with obesity (Stienstra et al., 2010), while NLRP3-, PYCARD- or caspase-1-deficient mice are resistant to HFD-induced obesity, have smaller adipocytes and reveal reduced accumulation of adipose tissue macrophages (Stienstra et al., 2011). NLRP3-deficient mice also demonstrate less adipose tissue caspase-1 activity, less insulin resistance and fewer adipose CD4+ and CD8+ T cells upon HFD challenge (Vandanmagsar et al., 2011). Recent work indicates that adipose caspase-1 activation localizes to hypertrophic and degenerating adipocytes surrounded by immune cells (Giordano et al., 2013). NRLP3 inflammasome activity inhibits adipocyte differentiation, while caspase-1-deficient adipocytes exhibit greater insulin sensitivity, differentiation and fat oxidation capacity than wild-type adipocytes (Stienstra et al., 2010). Mouse studies thus indicate that adipose NLRP3 inflammasome activity strongly impacts metabolic changes associated with obesity, but not whether adipocytes or macrophages are the primary source of the increased adipose inflammasome activity in obesity.
Equivalent studies are not available in humans, but in a cohort of obese men with type-2 diabetes weight loss correlated with decreased adipose expression of NLRP3 and IL-1β, but not PYCARD, with a positive correlation between adipose IL-1β and plasma glucose reductions (Vandanmagsar et al., 2011). Adipose tissue IL-1β, but not NLRP3, expression decreased in a second weight loss study (Moschen et al., 2011). A third study reported increased adipose expression of caspase-1 but not NLRP3, IL-1β or IL-18 in obese vs. lean subjects, where insulin sensitivity correlated with caspase-1 and IL-18 and tended to correlate with NLRP3 (Goossens et al.). Intriguingly, this study also found NLRP3, IL-1β and caspase-1 correlations with multiple T cell pro-inflammatory markers.
A microarray analysis of human subcutaneous and visceral adipose tissue performed by Klimcakova et al found that immune response genes were the most differentially-enriched among five detected functional groups upon comparison of obese women with metabolic syndrome vs. lean women, (Klimcakova et al., 2011). This included genes involved in TLR, TNF and NF-κB signaling, apoptosis and antigen processing and presentation, whose expression positively correlated with adiposity and plasma insulin and inversely correlated with glucose disposal rate. Although not discussed by the authors, their data also revealed an increase in 14/57 NLR signaling pathway genes, including PYCARD and IL1B, but no increase in NLRP3 or NLRC4. Similar results were not found by Elbein et al who analyzed subcutaneous adipose tissue from insulin-resistant and -sensitive men and women and detected 50 differentially expressed genes (≥1.5-fold and p<0.1), none of which were found in our analyses (Elbein et al., 2011). Sex hormone effects may, however, increase inflammatory gene variation in mixed gender populations to obscure differences between insulin-resistant and -sensitive groups. One early microarray study found expression differences in male vs. female subcutaneous adipose tissue, supporting this possibility (Linder et al., 2004).
There are few microarray studies of isolated human adipocytes. One study of subcutaneous adipocytes from obese vs. non-obese Pima Indians prior to GSEA software development found that inflammation/immune response genes were highly represented in the population of genes up-regulated in obesity, but did not identify any inflammasome-related genes. (Lee et al., 2005) Two later microarray studies found increased expression of amyloid precursor protein (Lee et al., 2008) and inhibin B (Sjoholm et al., 2006) in adipocytes of obese vs. lean subjects, but did not perform gene pathway analyses.
NLRP3 expression and inflammasome activation are triggered by hyperglycemia, saturated FFAs, ceramide, reactive oxygen species and urate, calcium and cholesterol crystals (Latz, 2010; Vandanmagsar et al., 2011), many of which increase with obesity (Deng et al., 2013a). Adipocyte NLRP3 and PYCARD expression did not correlate with plasma glucose, triglycerides or cholesterol in our study, but associated with adipocyte NQO1 expression, which correlates with BMI and adipocyte diameter and decreases with weight loss, consistent with attenuation of obesity-induced oxidative lipid and protein damage by weight loss (Palming et al., 2007). SVF IFNγ expression correlated with NLRP and PYCARD expression in adipocytes but not in SVF. Macrophages are a primary component of SVF during obesity (Weisberg et al., 2003). Some studies indicate that IFNγ can suppress macrophage NLRP3 inflammasome expression and activity (Guarda et al., 2011; Mishra et al., 2013), although this regulation appears to partially depend upon cell differentiation and/or polarization status. Thus, adipocytes and adipose tissue macrophages can differentially respond to IFNγ, to produce an adipocyte-dominated NRLP3 inflammasome response in obesity.
Adipocyte NLRP3 and PYCARD expression, however, most closely correlated with adipocyte MHCII expression, which is regulated by pro-inflammatory CD4+ TH1 cell IFNγ secretion during obesity (Deng et al., 2013b). IFNγ and TLR4 activation synergistically increased NLRP3 expression, although IL-1β revealed the strongest effect, suggesting that obesity-associated increases in IFNγ and TLR4 signaling may induce NLRP3 inflammasome activity that is subsequently maintained by an autocrine feedback mechanism (Fig. S2). Further, the NLRP3 effector cytokines IL-1β and IL-18 stimulate IFNγ production by TH1 and CD8+ T cells (Ben-Sasson et al., 2011; Iwai et al., 2008; Nakanishi et al., 2001), both of which accumulate in adipose tissue during obesity (Winer et al., 2009; Yang et al., 2010), suggesting that adipocyte inflammasome activity may contribute to the activation state of adipose resident T-cells.
We propose that IFNγ secretion from activated adipose-resident T cells induces adipocyte NLRP3 inflammasome expression to promote IL-1β and IL-18 secretion that further stimulates T cell IFNγ production, creating a spiraling pro-inflammatory cascade (Fig. S2). This putative adipocyte-T cell interaction may represent a new therapeutic target for the treatment of obesity-induced insulin resistance and other complications resulting from adipose inflammation. Further investigation of adipocyte NLR pathway changes in both men and women will be important to elucidate the impact of this novel adipocyte-T cell dialogue on obesity-associated metabolic and inflammatory phenotypes. NLRP3 inflammasome activity directly regulates IL-1β secretion (Sutterwala et al., 2006), which is implicated in beta-cell dysfunction and systemic inflammation in type 2 diabetes (Cavelti-Weder et al., 2012; Larsen et al., 2007) as well as unstable angina (Ridker et al., 2011). Attenuating adipose NLRP3 inflammasome activity by targeting pro-inflammatory pathways that regulate its increased expression in obesity may be useful to mitigate adipose tissue inflammation and its complications.
Supplementary Material
Highlights.
• NLRP3 inflammasome expression increases in adipocytes but not SVF during obesity. • After adjusting for BMI, inflammasome genes correlate only with adipocyte MHCII. • IFNγ regulates both adipocyte MHCII and NLRP3 expression. • CD4 T cell activation via adipocyte MHCII induces IFNγ-dependent NLRP3 expression. • This interaction appears likely to impact adipose and systemic inflammation.
Acknowledgements
This work was supported by a generous grant from the MacDonald Foundation and an NIH grant R24DK087723 to WAH; and JS Dunn Research Foundation, TT & WF Chao Foundation and NIH grants R01CA121225 and R01AG028928 to STCW.
Abbreviations
- SVF
stromal vascular fraction
- MHCII
class II major histocompatibility complex
- TLR
toll-like receptor
- NLR
NOD-like receptor
- FFA
free fatty acid
- SBP and DBP
systolic and diastolic blood pressure
- BMI
body mass index
- WHR
waist-hip-ratio
- BAI
body adiposity index
- FPG and FPI
fasting plasma glucose and insulin
- HDL and LDL
high- and low-density lipoprotein cholesterol
- hsCRP
high-sensitivity C-reactive protein
- AST and ALT
aspartate and alanine aminotransferase
- HFD
high-fat diet
- LPS
lipopolysaccharide
- GSEA
gene-set enrichment analysis
- NES
nominal enrichment score
- FDR
false discovery rate
- NLR
NOD-like receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURE STATEMENT: The authors have nothing to disclose.
Conflict of Interest
The authors disclose no conflict of interest.
References
- Becker CE, O'Neill LA. Inflammasomes in inflammatory disorders: the role of TLRs and their interactions with NLRs. Seminars in immunopathology. 2007;29:239–248. doi: 10.1007/s00281-007-0081-4. [DOI] [PubMed] [Google Scholar]
- Ben-Sasson SZ, Caucheteux S, Crank M, Hu-Li J, Paul WE. IL-1 acts on T cells to enhance the magnitude of in vivo immune responses. Cytokine. 2011;56:122–125. doi: 10.1016/j.cyto.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman RN, Stefanovski D, Buchanan TA, Sumner AE, Reynolds JC, Sebring NG, Xiang AH, Watanabe RM. A better index of body adiposity. Obesity (Silver Spring, Md. 2011;19:1083–1089. doi: 10.1038/oby.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan NB, Dorfleutner A, Rojanasakul Y, Stehlik C. Activation of inflammasomes requires intracellular redistribution of the apoptotic speck-like protein containing a caspase recruitment domain. J Immunol. 2009;182:3173–3182. doi: 10.4049/jimmunol.0802367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavelti-Weder C, Babians-Brunner A, Keller C, Stahel MA, Kurz-Levin M, Zayed H, Solinger AM, Mandrup-Poulsen T, Dinarello CA, Donath MY. Effects of gevokizumab on glycemia and inflammatory markers in type 2 diabetes. Diabetes care. 2012;35:1654–1662. doi: 10.2337/dc11-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusi K. Role of insulin resistance and lipotoxicity in non-alcoholic steatohepatitis. Clinics in liver disease. 2009;13:545–563. doi: 10.1016/j.cld.2009.07.009. [DOI] [PubMed] [Google Scholar]
- Deng T, Cui J, Lyon CJ, Zhang N, Wang HY, Wang RF, Hsueh WA. Inflammasomes and Obesity. In: Dannenberg AJ, Berger NA, editors. Obesity, Inflammation and Cancer. New York: Springer; 2013a. pp. 25–60. [Google Scholar]
- Deng T, Lyon CJ, Minze LJ, Lin J, Zou J, Liu JZ, Ren Y, Yin Z, Hamilton DJ, Reardon PR, et al. Class II major histocompatibility complex plays an essential role in obesity-induced adipose inflammation. Cell metabolism. 2013b;17:411–422. doi: 10.1016/j.cmet.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbein SC, Kern PA, Rasouli N, Yao-Borengasser A, Sharma NK, Das SK. Global gene expression profiles of subcutaneous adipose and muscle from glucose-tolerant, insulin-sensitive, and insulin-resistant individuals matched for BMI. Diabetes. 2011;60:1019–1029. doi: 10.2337/db10-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. On the interpretation of χ2 from contingency tables, and the calculation of P. Journal of the Royal Statistical Society. 1922;85:87–94. [Google Scholar]
- Giordano A, Murano I, Mondini E, Perugini J, Smorlesi A, Severi I, Barazzoni R, Scherer PE, Cinti S. Obese adipocytes show ultrastructural features of stressed cells and die of pyroptosis. Journal of lipid research. 2013;54:2423–2436. doi: 10.1194/jlr.M038638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens GH, Blaak EE, Theunissen R, Duijvestijn AM, Clement K, Tervaert JW, Thewissen MM. Expression of NLRP3 inflammasome and T cell population markers in adipose tissue are associated with insulin resistance and impaired glucose metabolism in humans. Molecular immunology. 2012;50:142–149. doi: 10.1016/j.molimm.2012.01.005. [DOI] [PubMed] [Google Scholar]
- Guarda G, Braun M, Staehli F, Tardivel A, Mattmann C, Forster I, Farlik M, Decker T, Du Pasquier RA, Romero P, et al. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity. 2011;34:213–223. doi: 10.1016/j.immuni.2011.02.006. [DOI] [PubMed] [Google Scholar]
- Halleux CM, Declerck PJ, Tran SL, Detry R, Brichard SM. Hormonal control of plasminogen activator inhibitor-1 gene expression and production in human adipose tissue: stimulation by glucocorticoids and inhibition by catecholamines. J Clin Endocrinol Metab. 1999;84:4097–4105. doi: 10.1210/jcem.84.11.6127. [DOI] [PubMed] [Google Scholar]
- Han JM, Levings MK. Immune regulation in obesity-associated adipose inflammation. J Immunol. 2013;191:527–532. doi: 10.4049/jimmunol.1301035. [DOI] [PubMed] [Google Scholar]
- Iwai Y, Hemmi H, Mizenina O, Kuroda S, Suda K, Steinman RM. An IFN-gamma-IL-18 signaling loop accelerates memory CD8+ T cell proliferation. PloS one. 2008;3:e2404. doi: 10.1371/journal.pone.0002404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimcakova E, Roussel B, Marquez-Quinones A, Kovacova Z, Kovacikova M, Combes M, Siklova-Vitkova M, Hejnova J, Sramkova P, Bouloumie A, et al. Worsening of obesity and metabolic status yields similar molecular adaptations in human subcutaneous and visceral adipose tissue: decreased metabolism and increased immune response. J Clin Endocrinol Metab. 2011;96:E73–E82. doi: 10.1210/jc.2010-1575. [DOI] [PubMed] [Google Scholar]
- Larsen CM, Faulenbach M, Vaag A, Volund A, Ehses JA, Seifert B, Mandrup-Poulsen T, Donath MY. lnterleukin-1-receptor antagonist in type 2 diabetes mellitus. The New England journal of medicine. 2007;356:1517–1526. doi: 10.1056/NEJMoa065213. [DOI] [PubMed] [Google Scholar]
- Latz E. The inflammasomes: mechanisms of activation and function. Current opinion in immunology. 2010;22:28–33. doi: 10.1016/j.coi.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Nair S, Rousseau E, Allison DB, Page GP, Tataranni PA, Bogardus C, Permana PA. Microarray profiling of isolated abdominal subcutaneous adipocytes from obese vs non-obese Pima Indians: increased expression of inflammation-related genes. Diabetologia. 2005;48:1776–1783. doi: 10.1007/s00125-005-1867-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Tharp WG, Maple RL, Nair S, Permana PA, Pratley RE. Amyloid precursor protein expression is upregulated in adipocytes in obesity. Obesity (Silver Spring, Md. 2008;16:1493–1500. doi: 10.1038/oby.2008.267. [DOI] [PubMed] [Google Scholar]
- Linder K, Arner P, Flores-Morales A, Tollet-Egnell P, Norstedt G. Differentially expressed genes in visceral or subcutaneous adipose tissue of obese men and women. Journal of lipid research. 2004;45:148–154. doi: 10.1194/jlr.M300256-JLR200. [DOI] [PubMed] [Google Scholar]
- Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PloS one. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra BB, Rathinam VA, Martens GW, Martinot AJ, Kornfeld H, Fitzgerald KA, Sassetti CM. Nitric oxide controls the immunopathology of tuberculosis by inhibiting NLRP3 inflammasome-dependent processing of IL-1beta. Nature immunology. 2013;14:52–60. doi: 10.1038/ni.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschen AR, Molnar C, Enrich B, Geiger S, Ebenbichler CF, Tilg H. Adipose and liver expression of interleukin (IL)-1 family members in morbid obesity and effects of weight loss. Molecular medicine (Cambridge, Mass. 2011;17:840–845. doi: 10.2119/molmed.2010.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K, Yoshimoto T, Tsutsui H, Okamura H. lnterleukin-18 regulates both Th1 and Th2 responses. Annual review of immunology. 2001;19:423–474. doi: 10.1146/annurev.immunol.19.1.423. [DOI] [PubMed] [Google Scholar]
- Olefsky JM, Glass CK. Macrophages, inflammation, and insulin resistance. Annual review of physiology. 2010;72:219–246. doi: 10.1146/annurev-physiol-021909-135846. [DOI] [PubMed] [Google Scholar]
- Pal D, Dasgupta S, Kundu R, Maitra S, Das G, Mukhopadhyay S, Ray S, Majumdar SS, Bhattacharya S. Fetuin-A acts as an endogenous ligand of TLR4 to promote lipid-induced insulin resistance. Nature medicine. 2012;18 doi: 10.1038/nm.2851. [DOI] [PubMed] [Google Scholar]
- Palming J, Sjoholm K, Jernas M, Lystig TC, Gummesson A, Romeo S, Lonn L, Lonn M, Carlsson B, Carlsson LM. The expression of NAD(P)H:quinone oxidoreductase 1 is high in human adipose tissue, reduced by weight loss, and correlates with adiposity, insulin sensitivity, and markers of liver dysfunction. J Clin Endocrinol Metab. 2007;92:2346–2352. doi: 10.1210/jc.2006-2476. [DOI] [PubMed] [Google Scholar]
- Ridker PM, Thuren T, Zalewski A, Libby P. Interleukin-1beta inhibition and the prevention of recurrent cardiovascular events: rationale and design of the Canakinumab Anti-inflammatory Thrombosis Outcomes Study (CANTOS) American heart journal. 2011;162:597–605. doi: 10.1016/j.ahj.2011.06.012. [DOI] [PubMed] [Google Scholar]
- Shoelson SE, Lee J, Goldfine AB. Inflammation and insulin resistance. The Journal of clinical investigation. 2006;116:1793–1801. doi: 10.1172/JCI29069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoholm K, Palming J, Lystig TC, Jennische E, Woodruff TK, Carlsson B, Carlsson LM. The expression of inhibin beta B is high in human adipocytes, reduced by weight loss, and correlates to factors implicated in metabolic disease. Biochemical and biophysical research communications. 2006;344:1308–1314. doi: 10.1016/j.bbrc.2006.04.030. [DOI] [PubMed] [Google Scholar]
- Song MJ, Kim KH, Yoon JM, Kim JB. Activation of Toll-like receptor 4 is associated with insulin resistance in adipocytes. Biochemical and biophysical research communications. 2006;346:739–745. doi: 10.1016/j.bbrc.2006.05.170. [DOI] [PubMed] [Google Scholar]
- Stienstra R, Joosten LA, Koenen T, van Tits B, van Diepen JA, van den Berg SA, Rensen PC, Voshol PJ, Fantuzzi G, Hijmans A, et al. The inflammasome-mediated caspase-1 activation controls adipocyte differentiation and insulin sensitivity. Cell metabolism. 2010;12:593–605. doi: 10.1016/j.cmet.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stienstra R, van Diepen JA, Tack CJ, Zaki MH, van de Veerdonk FL, Perera D, Neale GA, Hooiveld GJ, Hijmans A, Vroegrijk I, et al. Inflammasome is a central player in the induction of obesity and insulin resistance. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:15324–15329. doi: 10.1073/pnas.1100255108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterwala FS, Ogura Y, Szczepanik M, Lara-Tejero M, Lichtenberger GS, Grant EP, Bertin J, Coyle AJ, Galan JE, Askenase PW, et al. Critical role for NALP3/CIAS1/Cryopyrin in innate and adaptive immunity through its regulation of caspase-1. Immunity. 2006;24:317–327. doi: 10.1016/j.immuni.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandanmagsar B, Youm YH, Ravussin A, Galgani JE, Stadler K, Mynatt RL, Ravussin E, Stephens JM, Dixit VD. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nature medicine. 2011;17:179–188. doi: 10.1038/nm.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr Obesity is associated with macrophage accumulation in adipose tissue. The Journal of clinical investigation. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winer S, Chan Y, Paltser G, Truong D, Tsui H, Bahrami J, Dorfman R, Wang Y, Zielenski J, Mastronardi F, et al. Normalization of obesity-associated insulin resistance through immunotherapy. Nature medicine. 2009;15:921–929. doi: 10.1038/nm.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Youm YH, Vandanmagsar B, Ravussin A, Gimble JM, Greenway F, Stephens JM, Mynatt RL, Dixit VD. Obesity increases the production of proinflammatory mediators from adipose tissue T cells and compromises TCR repertoire diversity: implications for systemic inflammation and insulin resistance. J Immunol. 2010;185:1836–1845. doi: 10.4049/jimmunol.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.