Abstract
Cilia are hair-like organelles extending from the cell surface with important sensory and motility functions. Ciliary defects can result in a wide range of human diseases known as ciliopathies. However, the molecular mechanisms controlling ciliogenesis remain poorly defined. Here we show that cylindromatosis (CYLD), a tumor suppressor protein harboring deubiquitinase activity, plays a critical role in the assembly of both primary and motile cilia in multiple organs. CYLD knockout mice exhibit polydactyly and various ciliary defects, such as failure in basal body anchorage and disorganization of basal bodies and axenomes. The ciliary function of CYLD is partially attributed to its deconjugation of the polyubiquitin chain from centrosomal protein of 70 kDa (Cep70), a requirement for Cep70 to interact with γ-tubulin and localize at the centrosome. In addition, CYLD-mediated inhibition of histone deacetylase 6 (HDAC6), which promotes tubulin acetylation, constitutes another mechanism for the ciliary function of CYLD. Small-molecule inhibitors of HDAC6 could partially rescue the ciliary defects in CYLD knockout mice. These findings highlight the importance of protein ubiquitination in the modulation of ciliogenesis, identify CYLD as a crucial regulator of this process, and suggest the involvement of CYLD deficiency in ciliopathies.
Keywords: CYLD, ciliogenesis, centrosome, ubiquitination, acetylation
Introduction
Cilia are present in most eukaryotes, ranging from unicellular flagellates to humans, and are traditionally classified into two groups, primary and motile cilia. The primary cilium is an immotile, solitary structure that emanates from the surfaces of most mammalian cell types and can sense various extracellular signals1,2. By contrast, motile cilia are typically found in large numbers on differentiated epithelial cells, such as those in the trachea, brain ventricle, and oviduct; ciliary motility is critical for the flow of mucus and cerebrospinal fluid and the movement of eggs3. Flagella present on sperm and unicellular eukaryotes represent a special type of motile cilia, but they are longer and exist in fewer numbers on the cell surface. Consistent with the diverse sensory and motility functions of cilia, defects in ciliary assembly and function have been implicated in a variety of human diseases, which are collectively known as ciliopathies4,5.
A microtubule-based apparatus called the axoneme serves as the scaffold of cilia. The axoneme of a typical motile cilium has a pair of central microtubules surrounded by nine outer microtubule doublets (termed a 9+2 axoneme), whereas the axoneme of a primary cilium usually contains the outer doublets in the absence of the central pair (termed a 9+0 axoneme). Ciliary assembly begins with the nucleation of axonemal microtubules from the basal body, which consists of nine microtubule triplets and is derived from the mother centriole and anchored to the plasma membrane6. The cilium then grows due to the elongation of axonemal microtubules and the transport of protein complexes along the axoneme, a process known as intraflagellar transport (IFT)7. As cilia possess a wide spectrum of physiological functions and their defects result in ciliopathies4,5, the assembly of cilia must undergo exquisite regulation. The past decade has witnessed great progress in understanding the composition and structural features of cilia8; however, the molecular details for the regulation of ciliogenesis remain largely unknown.
Cylindromatosis (CYLD) is a tumor suppressor protein harboring deubiquitinase activity that primarily removes lysine 63-linked polyubiquitin chains from a number of signaling components9. CYLD also contains cytoskeleton-associated protein glycine-rich (CAP-Gly) domains that mediate its interaction with microtubules and histone deacetylase 6 (HDAC6)10,11,12. Several cilium-related symptoms, such as male infertility, osteoporosis, and impaired lung maturation, have been reported in CYLD knockout mice or in mice carrying truncated CYLD13,14,15. In addition, we found that a significant proportion of CYLD knockout mice had extra digits on the preaxial side of hind feet (Supplementary information, Figure S1), a condition frequently associated with ciliary abnormalities. These findings prompted us to hypothesize that CYLD might be a critical regulator of ciliogenesis.
Results
CYLD knockout mice have ciliary defects in multiple organs
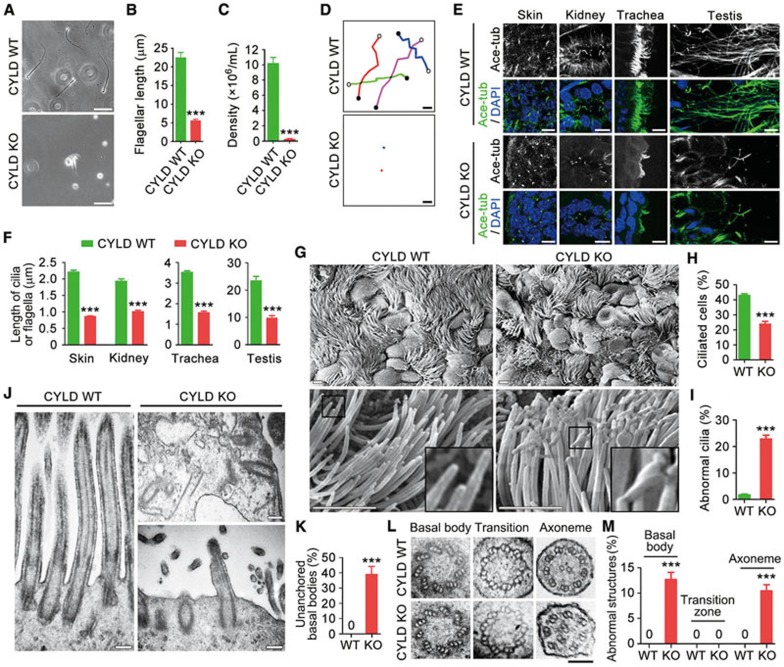
To test the potential role of CYLD in ciliogenesis, we first examined the sperm of CYLD wild-type and knockout mice. We found that the flagella were much shorter or even absent in CYLD knockout sperm (Figure 1A and 1B). In addition, consistent with the previous finding15, we found that CYLD deficiency dramatically impaired sperm production; the density of sperm in CYLD knockout mice was much lower than that in wild-type mice (Figure 1C). Examination of sperm swimming trajectories with time-lapse microscopy revealed that CYLD was critically required for sperm motility (Figure 1D). We then analyzed primary cilia in mouse skin and kidney tissues, motile cilia in the trachea, and flagella in the testis by immunostaining acetylated α-tubulin, a well-characterized ciliary marker16. We found that the length of cilia/flagella was greatly decreased in these tissues in CYLD knockout mice, as compared to wild-type mice (Figure 1E and 1F).
Figure 1.
CYLD knockout mice have ciliary defects in multiple organs. (A-D) Images (A), flagellar length (B, n = 60), density (C, n = 12), and swimming trajectories (D) of sperm isolated from CYLD wild-type (WT) and knockout (KO) mice. Experiments were performed 3 times. Scale bar, 10 μm. (E, F) Immunofluorescence images (E) and length (F, n = 120) of cilia/flagella in mouse tissues, stained with anti-acetylated α-tubulin (ace-tubulin) antibody and DAPI. Experiments were performed 3 times. Scale bar, 5 μm. (G-I) Scanning electron microscopy images of cilia (G), percentage of ciliated cells (H, n = 120), and percentage of abnormal cilia (I, n = 300) in the mouse tracheal epithelium. Experiments were performed 3 times. Scale bar, 2.5 μm. (J) Transmission electron microscopy images of the longitudinal sections of cilia in the tracheal epithelium. Scale bar, 200 nm. (K) Quantification of basal bodies that fail to anchor to the plasma membrane. n = 200. Experiments were performed 3 times. (L) Transmission electron microscopy images of the cross sections of cilia in the tracheal epithelium. Scale bar, 100 nm. (M) Quantification of cilia with abnormal basal bodies (n = 200), transition zones (n = 30), or axonemes (n = 200). Experiments were performed 3 times. Student's t test for B, C, F, H, and I. Fisher's exact test for K and M. ***P < 0.001. Error bars indicate SEM.
Scanning electron microscopy was then performed to examine mouse tracheal surface epithelium, where ciliated cells are interspersed with non-ciliated goblet and Clara cells. The loss of CYLD significantly reduced the percentage of ciliated cells (Figure 1G and 1H). In addition, 22.8% of the tracheal epithelial cilia in CYLD knockout mice displayed abnormal morphology (e.g., winding at the distal tip) (Figure 1G and 1I).
To understand how CYLD deficiency affects ciliary ultrastructure, we examined the longitudinal sections of mouse tracheal epithelial cilia with transmission electron microscopy. In agreement with the immunofluorescence data, cilia were fewer and shorter in the tracheal epithelium of CYLD knockout mice (Figure 1J). Strikingly, 39.2% of the basal bodies failed to anchor to the plasma membrane in the absence of CYLD (Figure 1J and 1K). We also analyzed the cross sections of the tracheal epithelial cilia. Compared to the wild-type controls, a proportion of basal bodies and axonemes were severely disorganized in CYLD knockout cilia; 12.7% of the basal bodies lacked or had defects in one of the nine microtubule triplets (replaced by a doublet or quadruplet), and 10.5% of the axonemes displayed abnormal number and/or position of the outer microtubule doublets or the central microtubule pair (Figure 1L, 1M and Supplementary information, Figure S2).
The deubiquitinase and CAP-Gly domains of CYLD contribute to its role in ciliogenesis
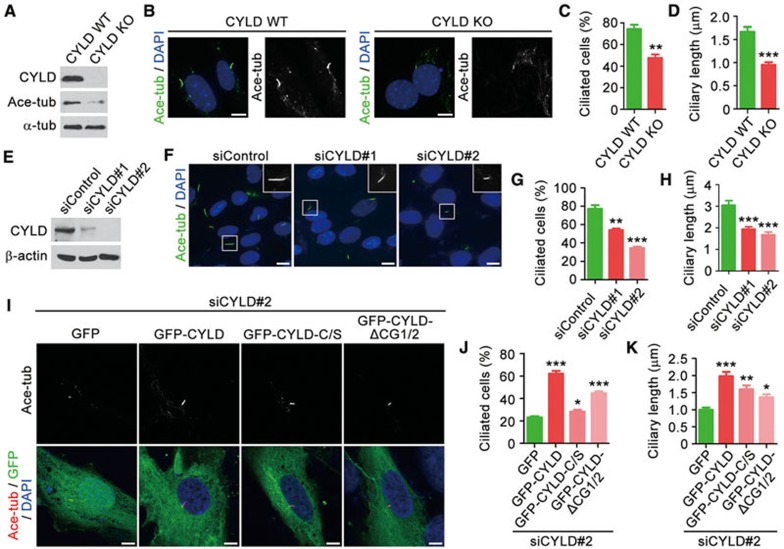
To investigate whether CYLD is required for ciliogenesis in vitro, CYLD wild-type and knockout mouse embryonic fibroblasts (MEFs) were serum-starved for 48 h. Immunoblotting revealed that the loss of CYLD significantly reduced the level of tubulin acetylation in MEFs (Figure 2A). Following serum starvation, primary cilia were evident on 74.4% of wild-type MEFs; by contrast, only 47.5% of CYLD knockout MEFs had primary cilia (Figure 2B and 2C). In addition, the average length of cilia was remarkably decreased in CYLD knockout MEFs, as compared to wild-type MEFs (Figure 2B and 2D). The decrease in the percentage of ciliated cells and in ciliary length was also observed in RPE-1 retinal pigment epithelial cells that were transfected with CYLD siRNAs (Figure 2E-2H). Taken together, the above results reveal a critical role for CYLD in the control of ciliogenesis both in mice and in vitro.
Figure 2.
The deubiquitinase and CAP-Gly domains of CYLD contribute to its role in ciliogenesis. (A) Immunoblots for CYLD, ace-tubulin, and α-tubulin expression in CYLD wild-type and knockout MEFs serum-starved for 48 h. (B-D) Immunofluorescence images (B), percentage of ciliated cells (C, n = 100), and ciliary length (D, n = 60) of MEFs serum-starved for 48 h and stained with anti-ace-tubulin antibody and DAPI. Experiments were performed 3 times. Scale bar, 5 μm. (E) Immunoblots for CYLD and β-actin expression in control and CYLD siRNA-treated RPE-1 cells. (F-H) Immunofluorescence images (F), percentage of ciliated cells (G, n = 200), and ciliary length (H, n = 80) of RPE-1 cells transfected with control or CYLD siRNAs, followed by serum starvation for 48 h and staining with anti-ace-tubulin antibody and DAPI. Experiments were performed 4 times. Scale bar, 5 μm. (I-K) Immunofluorescence images (I), percentage of ciliated cells (J, n = 50), and ciliary length (K, n = 40) of RPE-1 cells transfected with CYLD siRNA and GFP, GFP-CYLD, GFP-CYLD-C/S, or GFP-CYLD-ΔCG1/2, followed by serum starvation for 48 h and staining with anti-ace-tubulin antibody and DAPI. C/S, mutation of cysteine 601 to serine; ΔCG1/2, without the two amino-terminal CAP-Gly domains. Experiments were performed 4 times. Scale bar, 5 μm. Student's t test for all graphs. *P < 0.05, **P < 0.01, ***P< 0.001. Error bars indicate SEM.
To determine the molecular mechanisms of how CYLD regulates ciliogenesis, RPE-1 cells were transfected with CYLD siRNA and plasmids that express wild-type or mutant CYLD. Two different CYLD mutants were used: the C/S mutant, in which cysteine 601 was mutated to serine to disrupt the deubiquitinase activity17, and the ΔCG1/2 mutant, which lacked the two amino-terminal CAP-Gly domains. We found that wild-type CYLD significantly rescued CYLD siRNA-induced ciliogenesis defects; by contrast, only partial rescue effects were seen for the C/S and ΔCG1/2 mutants (Figure 2I-2K). Thus, both the deubiquitinase domain and the two amino-terminal CAP-Gly domains of CYLD contribute to its role in ciliogenesis.
Depletion of CYLD impairs the localization of Cep70 at the centrosome
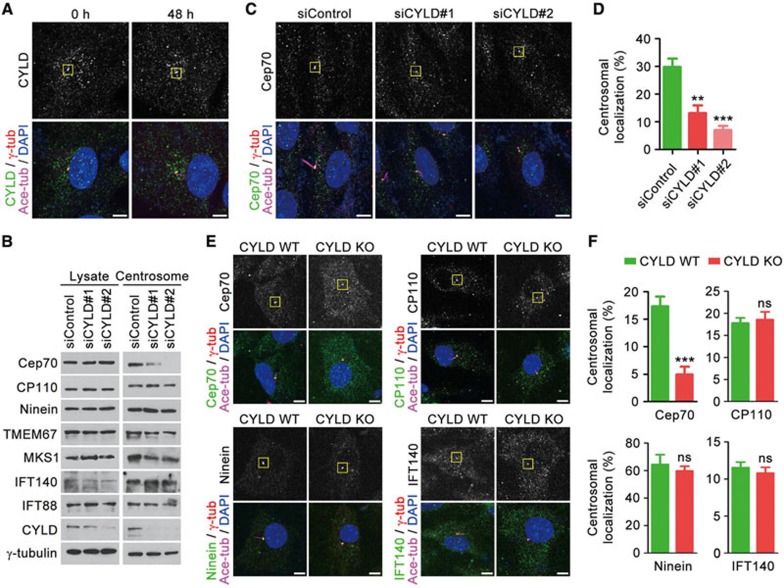
We then assessed the subcellular localization of CYLD with immunofluorescence microscopy. A subset of CYLD signal was detected at the centrosome in both serum-starved and non-starved RPE-1 cells (Figure 3A). To identify proteins mediating the ciliary function of CYLD, we analyzed the effect of CYLD siRNAs on the expression of several key regulators of ciliogenesis8. Immunoblotting revealed that depletion of CYLD did not obviously affect the level of CP110 (centrosomal protein of 110 kDa), ninein, TMEM67 (transmembrane protein 67), MKS1 (Meckel syndrome 1), IFT140, IFT88, or γ-tubulin in the total cell lysate or in the centrosomal preparation (Figure 3B). However, the level of Cep70 (centrosomal protein of 70 kDa), a recently identified regulator of ciliogenesis18,19,20, at the centrosome was significantly decreased upon the depletion of CYLD, although the level of Cep70 in the total cell lysate was not affected (Figure 3B). Immunofluorescence microscopy further confirmed that CYLD was important for Cep70 localization at the centrosome in RPE-1 cells (Figure 3C and 3D). The centrosomal localization of Cep70 was also dramatically reduced in CYLD knockout MEFs, as compared to wild-type MEFs, whereas no significant difference was found for CP110, ninein, or IFT140 (Figure 3E and 3F).
Figure 3.
Depletion of CYLD impairs the localization of Cep70 at the centrosome. (A) Immunofluorescence images of RPE-1 cells serum-starved for 0 or 48 h, followed by staining with anti-CYLD, anti-ace-tubulin, and anti-γ-tubulin antibodies and DAPI. The boxed regions indicate the centrosomal localization of CYLD. Scale bar, 5 μm. (B) Immunoblots for the expression of selected proteins involved in ciliogenesis, in the total lysate, or centrosomal preparation of cells transfected with control and CYLD siRNAs followed by serum starvation for 48 h. (C) Immunostaining with anti-Cep70, anti-ace-tubulin, and anti-γ-tubulin antibodies and DAPI in control and CYLD siRNA-treated RPE-1 cells. The boxed regions indicate the centrosomal localization of Cep70. Scale bar, 5 μm. (D) Quantification of the relative centrosomal localization of Cep70 in cells treated as in C. n = 40. Experiments were performed 4 times. (E) Immunostaining with anti-Cep70 (anti-CP110, anti-ninein, or anti-IFT140), anti-ace-tubulin, and anti-γ-tubulin antibodies and DAPI in CYLD wild-type and knockout MEFs. The boxed regions indicate the centrosomal localization of Cep70, CP110, ninein, and IFT140. Scale bar, 5 μm. (F) Quantification of the relative centrosomal localization of Cep70, CP110, ninein, and IFT140 in cells treated as in E. n = 20. Experiments were performed 4 times. Student's t test for all graphs. **P < 0.01, ***P < 0.001; ns, not significant. Error bars indicate SEM.
CYLD-mediated deubiquitination of Cep70 promotes its interaction with γ-tubulin and localization at the centrosome
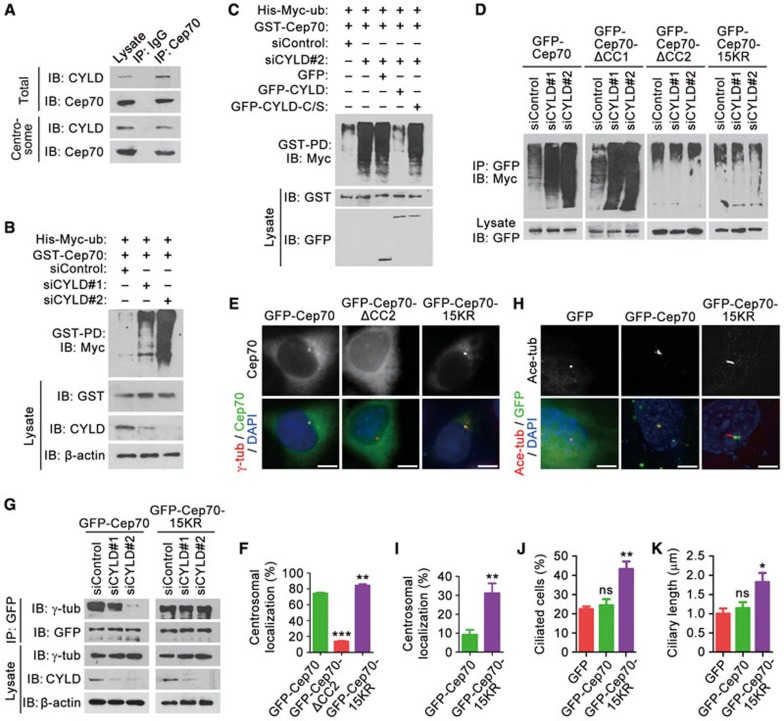
We next asked how CYLD regulates the centrosomal localization of Cep70. Immunoprecipitation by using either the total cell lysate or the centrosomal preparation revealed an interaction between endogenous CYLD and Cep70 in RPE-1 cells (Figure 4A). The interaction between them was observed throughout the cell cycle and was more evident in S phase as compared to G1, G2, and M phases (Supplementary information, Figure S3A). By using purified His-CYLD and MBP-Cep70, we further found a direct interaction between these two proteins (Supplementary information, Figure S3B). Since the deubiquitinase activity of CYLD is required for ciliogenesis, we asked whether CYLD alters the level of Cep70 ubiquitination. By GST pull-down and immunoblotting, we found that depletion of CYLD remarkably enhanced Cep70 ubiquitination (Figure 4B). CYLD siRNA-induced increase of Cep70 ubiquitination was rescued by wild-type CYLD, but not by CYLD-C/S (Figure 4C). Thus, CYLD is able to deubiquitinate Cep70.
Figure 4.
CYLD-mediated deubiquitination of Cep70 promotes its interaction with γ-tubulin and localization at the centrosome. (A) Immunoprecipitation (IP) and immunoblotting (IB) showing the interaction between endogenous CYLD and Cep70 in RPE-1 cells, by using either the total cell lysate or the centrosomal preparation. (B) Examination of Cep70 ubiquitination by GST pull-down (PD) and immunoblotting in cells transfected with control or CYLD siRNAs, together with His-Myc-ubiquitin and GST-Cep70. (C) Immunoblots showing that transfection of GFP-CYLD, but not its C/S mutant, rescues the level of Cep70 ubiquitination in CYLD-depleted cells. (D) Examination of Cep70 ubiquitination in cells transfected with control or CYLD siRNAs, together with His-Myc-ubiquitin and GFP-Cep70 or its mutants. ΔCC1, without the first coiled-coil domain. ΔCC2, without the second coiled-coil domain. 15KR, mutation of all the 15 lysines in the CC2 domain to arginines. (E) Immunofluorescence images of cells transfected with GFP-Cep70 or its mutants, followed by staining with anti-γ-tubulin antibody and DAPI. Scale bar, 5 μm. (F) Quantification of the relative centrosomal localization of Cep70 or its mutants in cells treated as in E. n= 30. Experiments were performed 3 times. (G) Immunoprecipitation and immunoblotting showing the interaction of γ-tubulin with GFP-Cep70 and its 15KR mutant in RPE-1 cells transfected with control or CYLD siRNAs. (H) Immunofluorescence images of RPE-1 cells transfected with CYLD siRNA and GFP, GFP-Cep70, or GFP-Cep70-15KR and serum-starved for 48 h, followed by staining with anti-ace-tubulin antibody and DAPI. Scale bar, 5 μm. (I-K) Relative centrosomal localization of GFP-Cep70 and its 15KR mutant (I, n = 30), percentage of ciliated cells (J, n = 50), and ciliary length (K, n = 30) in cells treated as in H. Experiments were performed 3 times. Student's t test for all graphs. *P < 0.05, **P < 0.01, ***P < 0.001; ns, not significant. Error bars indicate SEM.
We found previously that Cep70 interacts, through its two coiled-coil domains, with γ-tubulin to localize at the centrosome20. It is also known that CYLD can modulate protein-protein interactions by removing polyubiquitin chains from its substrates9. Therefore, we investigated the possibility that CYLD might deconjugate the polyubiquitin chain of Cep70 from its coiled-coil domains to regulate its interaction with γ-tubulin and its localization at the centrosome. We found that CYLD siRNA-induced elevation of Cep70 ubiquitination was abolished by deletion of its second coiled-coil domain (ΔCC2), but not by deletion of its first coiled-coil domain (ΔCC1) (Figure 4D). In addition, mutation of all the 15 lysines in the second coiled-coil domain of Cep70 to arginines (15KR) also abrogated the effect of CYLD siRNA on Cep70 ubiquitination (Figure 4D). These data indicate that CYLD mainly deubiquitinates Cep70 at its second coiled-coil domain.
Consistent with the previous finding20, immunofluorescence microscopy revealed that the centrosomal localization of Cep70 was greatly dependent on its second coiled-coil domain (Figure 4E and 4F). Importantly, the 15KR mutation significantly promoted Cep70 localization at the centrosome (Figure 4E and 4F), suggesting that the deubiquitination of Cep70 stimulates its centrosomal localization. By immunoprecipitation, we further found that depletion of CYLD decreased the interaction of γ-tubulin with wild-type Cep70, but not with the 15KR mutant (Figure 4G). These results suggest that CYLD-mediated deubiquitination of Cep70 enhances its interaction with γ-tubulin, hence promoting its centrosomal localization.
To corroborate the above findings in the context of ciliogenesis, RPE-1 cells were transfected with CYLD siRNA and Cep70 or its 15KR mutant, and then serum-starved to allow ciliary assembly. The 15KR mutant showed dramatically increased centrosomal localization, as compared to wild-type Cep70 (Figure 4H and 4I). Moreover, the 15KR mutant, but not wild-type Cep70, significantly rescued CYLD siRNA-induced ciliogenesis defects, including the percentage of ciliated cells and the length of cilia (Figure 4H, 4J, and 4K). Thus, CYLD-mediated deubiquitination of Cep70, by stimulating Cep70 localization at the centrosome, contributes to the function of CYLD in ciliogenesis.
HDAC6 is involved in the role of CYLD in regulating ciliary length
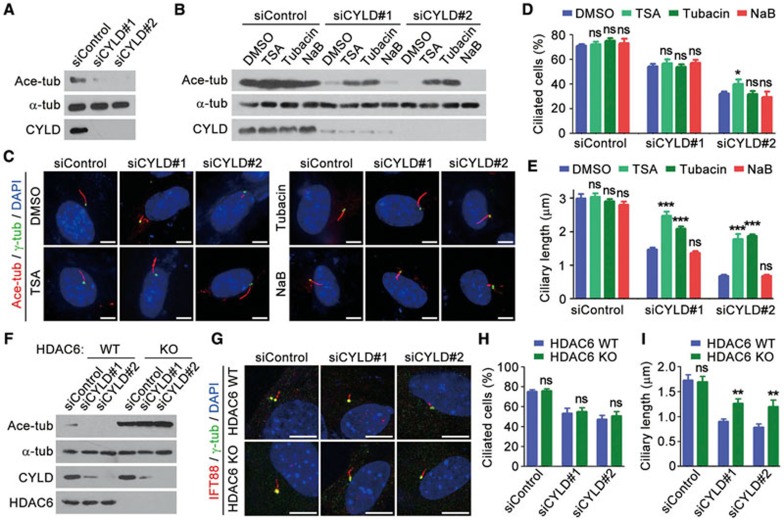
We next investigated how the two amino-terminal CAP-Gly domains of CYLD mediate its action in ciliogenesis. These CAP-Gly domains are important for CYLD to interact with HDAC611, and for CYLD to inhibit HDAC6 activity toward tubulin deacetylation (Supplementary information, Figure S4). We therefore investigated whether CYLD promotes ciliogenesis by inhibiting the tubulin deacetylase activity of HDAC6. Immunoblotting revealed that knockdown of CYLD significantly reduced tubulin acetylation in RPE-1 cells (Figure 5A), consistent with the result obtained in MEFs (Figure 2A). CYLD siRNA-induced decrease of tubulin acetylation was largely rescued by trichostatin A (TSA), an inhibitor of class I, IIa, and IIb HDACs including HDAC6, and by tubacin, a selective inhibitor of HDAC6, but not by sodium butyrate, an inhibitor of class I and IIa HDACs, which do not include HDAC6 (Figure 5B). Strikingly, inhibition of HDAC6 activity by TSA or tubacin could significantly rescue CYLD siRNA-induced decrease of ciliary length in RPE-1 cells, but did not have obvious effect on the percentage of ciliated cells (Figure 5C-5E).
Figure 5.
HDAC6 is involved in the role of CYLD in regulating ciliary length. (A) Immunoblots showing the levels of ace-tubulin, α-tubulin, and CYLD in RPE-1 cells transfected with control or CYLD siRNAs. (B) Immunoblots showing that trichostatin A (TSA) and tubacin, but not sodium butyrate (NaB), rescue CYLD siRNA-induced decrease of tubulin acetylation. (C-E) Immunofluorescence images (C), percentage of ciliated cells (D, n = 100), and ciliary length (E, n = 50) of RPE-1 cells transfected with control or CYLD siRNAs, treated with the indicated compounds, serum-starved for 48 h, and stained with anti-ace-tubulin and anti-γ-tubulin antibodies and DAPI. Experiments were performed 4 times. Scale bar, 2 μm. (F) Immunoblots for the expression of ace-tubulin, α-tubulin, CYLD, and HDAC6 in HDAC6 wild-type and knockout MEFs transfected with control or CYLD siRNAs. (G-I) Immunofluorescence images (G), percentage of ciliated cells (H, n = 100), and ciliary length (I, n= 50) of HDAC6 wild-type and knockout MEFs transfected with control or CYLD siRNAs, serum-starved for 48 h, and stained with anti-IFT88 and anti-γ-tubulin antibodies and DAPI. Experiments were performed 4 times. Scale bar, 5 μm. Student's t test for all graphs. *P < 0.05, **P < 0.01, ***P < 0.001; ns, not significant. Error bars indicate SEM.
To provide more insights into the role of HDAC6 in CYLD-mediated ciliogenesis, we assessed the effects of CYLD siRNAs using HDAC6 wild-type and knockout MEFs. Immunoblotting showed that depletion of CYLD decreased tubulin acetylation in HDAC6 wild-type MEFs, but not in the HDAC6-knockout MEFs (Figure 5F). This finding suggests that the effect of CYLD on tubulin acetylation is primarily due to its inhibition of HDAC6 activity, rather than its binding to microtubules via the CAP-Gly domains. We further found that CYLD siRNAs reduced the percentage of ciliated cells and ciliary length in both HDAC6 wild-type and knockout MEFs; notably, HDAC6-knockout MEFs had much longer cilia than wild-type MEFs upon treatment with CYLD siRNAs (Figure 5G-5I). Together, these results demonstrate that inhibition of HDAC6 activity contributes to the function of CYLD in regulating ciliogenesis.
HDAC6 inhibitors partially rescue the ciliary length defects in CYLD-knockout mice
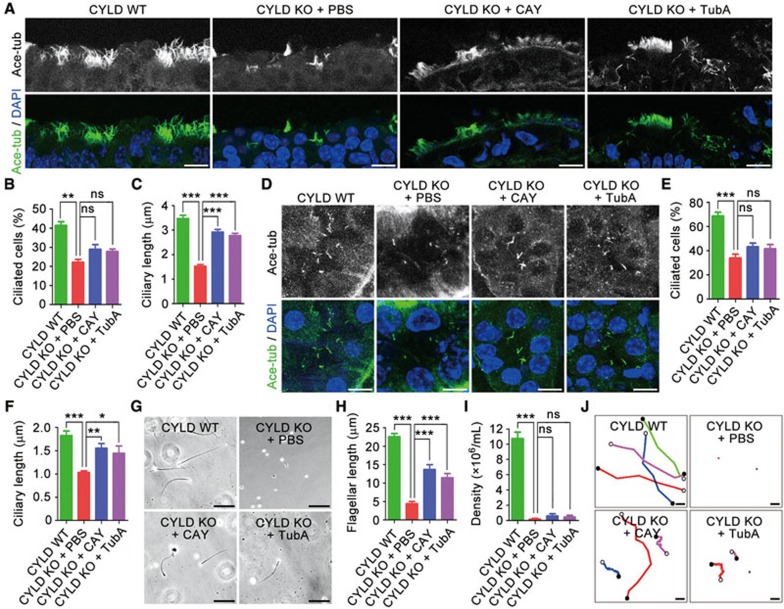
We next explored the potential of targeting HDAC6 to rescue the ciliogenesis defects in CYLD knockout mice. Mice were treated intraperitoneally and daily for 7 days with CAY10603 (CAY) or tubastatin A (TubA), two specific inhibitors of HDAC621,22. Analysis of motile cilia in the tracheal surface epithelium and primary cilia in the renal tubular epithelium revealed that these HDAC6 inhibitors partially rescued the ciliary length defect of CYLD knockout mice, but they did not have obvious effect on the percentage of ciliated cells (Figure 6A-6F). CAY and TubA did not have obvious effect on sperm production in CYLD knockout mice; however, the flagellar length and motility of sperm in CYLD knockout mice were increased by these HDAC6 inhibitors (Figure 6G-6J). Thus, inhibition of HDAC6 activity could partially restore ciliogenesis defects in CYLD-deficient mice.
Figure 6.
HDAC6 inhibitors partially rescue the ciliary length defects in CYLD knockout mice. (A-C) Immunofluorescence images of cilia (A), percentage of ciliated cells (B, n = 100), and ciliary length (C, n = 150) in the tracheal epithelial cells of 6-week-old CYLD wild-type mice untreated or CYLD knockout mice treated intraperitoneally and daily with CAY10603 (CAY), tubastatin A (TubA), or vehicle (PBS) for 7 days and stained with anti-ace-tubulin antibody and DAPI. Experiments were performed 3 times. Scale bar, 5 μm. (D-F) Immunofluorescence images of cilia (D), percentage of ciliated cells (E, n = 100), and ciliary length (F, n = 60) in the renal tubular epithelial cells of 6-week-old mice treated as in A-C. Experiments were performed 3 times. Scale bar, 5 μm. (G-J) Images (G), flagellar length (H, n = 30), density (I, n = 12), and swimming trajectories (J) of sperm isolated from 6-week-old mice treated as in A-C. Experiments were performed 3 times. Scale bar, 10 μm. Student's t test for all graphs. *P < 0.05, **P < 0.01, ***P < 0.001; ns, not significant. Error bars indicate SEM.
Discussion
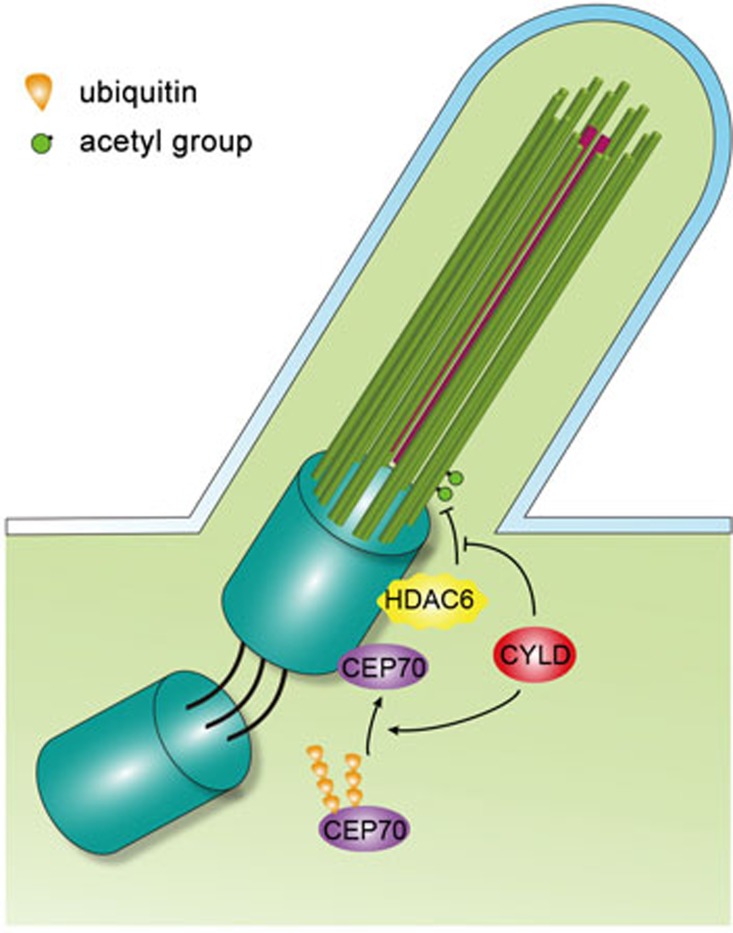
This work identifies an important role for CYLD in the control of ciliogenesis through deconjugating the polyubiquitin chain from Cep70 and inhibiting HDAC6 activity (Figure 7). The Chlamydomonas homolog of Cep70, CRC70, has been shown to localize preferentially at immature centrioles and promote centriole assembly by recruiting spindle assembly abnormal protein 6 (SAS-6) and Bld10p18, two cartwheel proteins required for the formation of the microtubule triplets of centrioles and basal bodies23,24. It will be interesting to investigate in the future whether CYLD regulates basal body organization by acting on the Cep70/SAS-6/Bld10p axis. In Chlamydomonas, a subset of CRC70 has also been detected at the transition fibers of the mother centriole/basal body18, sheet-like structures that are projected from the mother centriole/basal body and connect the microtubule triplets to the plasma membrane. It is tempting to speculate that CYLD may promote basal body anchorage to the plasma membrane via a Cep70/transition fiber-related mechanism.
Figure 7.
Molecular model for CYLD function in ciliogenesis.
Growth of the cilium requires the elongation of axonemal microtubules and protein transport along the axoneme, and defects in axoneme integrity can result in abnormal ciliary length. In this scenario, it is easy to understand that both reduced ciliary length and disorganized axonemes are found in CYLD knockout mice. Interestingly, shortened cilia have also been observed in zebrafish embryos injected with Cep70 morpholinos19, and Cep70 has been shown to stimulate microtubule assembly25,26. In addition, HDAC6 has been implicated in the regulation of ciliary disassembly/shortening by modulating the acetylation of axonemal microtubules27,28. Therefore, the role of CYLD in ciliary length control is likely to be mediated by the coordinated effect of Cep70 and HDAC6 on axonemal microtubules. However, given the involvement of CYLD in multiple signaling pathways and the complex nature of ciliogenesis, it would not be surprising if a mechanism independent of Cep70 and HDAC6 were uncovered in the future.
It is worthy of note that HDAC6 wild-type and knockout MEFs do not show obvious difference in the percentage of ciliated cells or in ciliary length. Similarly, small-molecule inhibition of HDAC6 activity does not significantly affect the percentage of ciliated cells or ciliary length. These results are consistent with the findings that HDAC6 knockout mice are viable and fertile and develop normally, although they show hyperacetylated tubulin in many tissues29,30. It is possible that some redundancy exists in the regulation of ciliogenesis by tubulin deacetylases. Another interesting phenomenon is that depletion of HDAC6 expression or inhibition of HDAC6 activity is able to partially rescue CYLD siRNA-induced decrease of ciliary length, but does not have obvious effect on the percentage of ciliated cells. It is possible that the elongation of axonemal microtubules, as compared to their nucleation, is more sensitive to the modulation of tubulin acetylation by HDAC6. In addition, given that HDAC6 has a variety of substrates, mechanisms independent of tubulin acetylation may also contribute to the complex action of HDAC6 in mediating the role of CYLD in ciliogenesis.
Consistent with the critical function of CYLD in ciliogenesis, CYLD-deficient mice exhibit several cilium-related disorders13,14,15, together with polydactyly, as revealed in this study. The polydactylous symptom may result from the impairment of the Sonic hedgehog (Shh) pathway, a cilium-dependent pathway important for vertebrate limb development31, since Shh-induced nuclear localization and transcriptional activity of the Gli family of proteins are decreased in CYLD siRNA-treated RPE-1 cells (Supplementary information, Figure S5). To evaluate the clinical value of CYLD, it would be important to analyze whether patients with ciliopathies have mutation or abnormal expression of this protein. In addition, given that cilia are frequently lost in tumor cells32 and that CYLD is mutated in various malignancies33, whether the ciliary function of CYLD contributes to its tumor-suppressing activity merits further investigation.
During the review of this manuscript, Eguether et al. demonstrated by using transgenic mice carrying CYLD with a truncated deubiquitinase domain (CYLDΔ932), that CYLD was important for the formation of motile cilia in the mouse trachea34. In the present study, we show that CYLD knockout mice exhibit polydactyly and have defects in the formation of primary cilia in skin and kidney tissues and flagella in the testis, in addition to motile ciliary defects in the trachea. While both studies demonstrate that CYLD regulates the docking of basal bodies to the apical plasma membrane, our study reveals that CYLD is also required for proper organization of basal bodies and axenomes. In addition, while the study of Eguether et al. is focused on how the centrosomal localization of CYLD is controlled (i.e., by interaction with centrosome-associated protein 350), our study explores downstream events underlying the role of CYLD in ciliogenesis (i.e., deubiquitination of Cep70 and inactivation of HDAC6). Together, these findings highlight an important function for CYLD-mediated protein deubiquitination in the regulation of ciliogenesis.
Materials and Methods
Antibodies, chemicals, siRNAs and plasmids
Antibodies against CYLD, ninein, Gli1 (Santa Cruz Biotechnology), TMEM67, CP110, MKS1, IFT140, IFT88 (Proteintech), acetylated α-tubulin, β-actin, Myc, HA, GST (Sigma-Aldrich), α-tubulin, γ-tubulin, Gli2 (Abcam), GFP (Roche), and HDAC6 (Millipore) were purchased from the indicated sources. Cep70 antibody was generated as described previously20. Horseradish peroxidase-conjugated secondary antibodies were from Amersham Biosciences, and fluorescein-, rhodamine-, or Cy5-conjugated secondary antibodies were from Jackson ImmunoResearch Laboratories. TSA, tubacin, sodium butyrate, CAY10603, and tubastatin A were from Santa Cruz Biotechnology, and DAPI was from Sigma-Aldrich. The sequences for control and CYLD siRNAs were described previously12. Mammalian expression plasmids for GFP-CYLD, GFP-Cep70, GST-Cep70, HA-HDAC6, and His-Myc-ubiquitin were described previously12,20,35, and various mutants were generated by PCR and site-directed mutagenesis. MBP-Cep70 was purified from bacteria expressing the pMALp2T-Cep70 plasmid as described previously20, with amylose resin (New England Biolabs). His-CYLD was purified from insect cells using the Bac-to-Bac baculovirus expression system (Life Technologies), with nickel resin (Bio-Rad). The Gli-responsive firefly luciferase reporter plasmid pGliBS-Luc and the amino-terminal Shh (ShhN)-expressing plasmid pRK5-ShhN were from Steven Y Cheng (Nanjing Medical University, China), and the thymidine kinase promoter driven-Renilla luciferase reporter plasmid pRL-TK was from Promega.
Mice
CYLD knockout mice (in C57BL6/DBA mixed genetic background) were generated and genotyped as described previously36. CYLD heterozygous mice were intercrossed to generate CYLD wild-type and knockout littermates. Animal experiments were performed in accordance with protocols approved by the Animal Care and Use Committee of Nankai University.
Cell culture and transfection
RPE-1 cells were obtained from the American Type Culture Collection and grown in the DMEM/F12 medium supplemented with 10% fetal bovine serum. CYLD wild-type and knockout MEFs were isolated from E13.5 mouse embryos. HDAC6 wild-type and knockout MEFs were kindly provided by Dr Tso-Pang Yao (Duke University, USA)30. All MEFs were cultured in DMEM medium supplemented with 10% fetal bovine serum. To induce ciliary formation, RPE-1 cells were cultured in serum-free medium for 48 h. Cells were synchronized at G1, S, G2, or M phase as described previously12. Plasmids were transfected to cells using TurboFect (Thermo Scientific), and siRNAs were transfected using Lipofectamine RNAiMAX (Invitrogen).
Immunofluorescence microscopy
Mouse tissues were fixed in 4% (wt/vol) paraformaldehyde/PBS, embedded in Tissue-Tek OCT (Sakura), and snap-frozen in liquid nitrogen. Sections were stained with primary antibody, fluorescein-conjugated secondary antibody, and then with DAPI. Sections were subsequently mounted onto slides and examined with a TCS SP5 confocal microscope (Leica). Cells grown on glass coverslips were fixed with 4% (wt/vol) paraformaldehyde/PBS for 30 min and permeabilized in 0.5% (vol/vol) Triton X-100/PBS for 20 min. Cells were blocked and incubated with primary antibodies and then rhodamine-, fluorescein-, or Cy5-conjugated secondary antibodies followed by staining with DAPI. Coverslips were then mounted onto slides and examined as above. The length of cilia and the percentage of ciliated cells were measured with ImageJ (National Institutes of Health, USA), and only cilia with distinguishable ends were included for the measurement of average cilia length. The fluorescence intensity of proteins at specific regions was quantified as described previously20. To quantify the relative centrosomal localization of Cep70, we measured the intensity of centrosomal Cep70 and the intensity of total cellular Cep70. The relative centrosomal localization of Cep70 was then calculated as the intensity of centrosomal Cep70 divided by the intensity of total cellular Cep70. The relative centrosomal localization of CP110, ninein, and IFT140 was also measured by this method.
Electron microscopy
For scanning electron microscopy, mouse trachea was isolated and prefixed with 2.5% (vol/vol) glutaraldehyde in 0.1 M sodium cacodylate at 4 °C for 5 min. The trachea was then cut open along the longitudinal axis, unfolded, stapled to a plastic sheet, and immersed in the above fixation buffer at 4 °C overnight. Samples were post-fixed in 1% (wt/vol) osmium tetroxide for 2 h, dehydrated in ethanol, and dried. The samples were then gold-coated by the sputter technique and examined by a QUANTA 200 scanning electron microscope at an accelerating voltage of 15 kV. For transmission electron microscopy, 2-mm pieces of the trachea were fixed as described above and stained with 2% (wt/vol) uranyl acetate. After dehydration and embedding in epoxy resin, 50-nm sections were cut and stained with uranyl acetate and lead citrate. Samples were examined by a Hitachi H-7650B scanning electron microscope at 80 kV.
Immunoblotting, immunoprecipitation, and GST or MBP pull-down
Proteins were resolved by SDS-PAGE and transferred onto polyvinylidene difluoride membranes (Millipore). The membranes were blocked and incubated with primary antibodies and then with horseradish peroxidase-conjugated secondary antibodies. Specific proteins were visualized with enhanced chemiluminescence detection reagent (Thermo Fisher Scientific). For immunoprecipitation and GST or MBP pull-down, cell lysates or purified proteins were incubated with antibody-coated agarose beads, glutathione-conjugated Sepharose 4B beads, or amylose-conjugated agarose beads at 4 °C for 2 h. The beads were washed and boiled in the SDS loading buffer, and the proteins were detected by immunoblotting.
Sperm analysis
The cauda epididymis was isolated from mice, immersed in normal saline, and cut into pieces. Samples were incubated at 37 °C for 15 min to allow sperm to escape from the epididymis. Sperm motility was examined by a Zeiss Observer Z1 microscope equipped with a live-cell imaging system and recorded with 50 frames per second. The morphology and swimming trajectory of sperm were analyzed with ImageJ.
Luciferase reporter assay
Cells were transfected with pGliBS-Luc and pRL-TK for 24 h and subsequently serum-starved for 48 h. Cells were then cultured for 12 h in the ShhN-conditioned medium, which were generated by transfection of 293T cells with pRK5-ShhN. The firefly luciferase activity was then measured and normalized to the Renilla luciferase activity.
Statistics
Analysis of statistical significance was performed by the Student's t-test and the Fisher's exact test in Microsoft Excel.
Acknowledgments
We thank Xueliang Zhu for discussion, Shiyi Guo, Di An, Ruming Liu, Long Miao, and Chuanmao Zhang for technical assistance, and Tso-Pang Yao and Steven Y Cheng for reagents. This work was supported by grants from the National Basic Research Program of China (2012CB945002) and the National Natural Science Foundation of China (31130015, 31271437, and 31371382).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Polydactyly in CYLD knockout mice.
Transmission electron microscopy images of the cross sections of axonemes in the tracheal epithelial cilia of CYLD knockout mice.
Examination of the interaction between CYLD and Cep70 in different phases of the cell cycle and in vitro.
CYLD interacts with HDAC6 and inhibits its activity toward tubulin deacetylation.
CYLD regulates the Shh signaling.
References
- Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley ES, Nachury MV. The perennial organelle: assembly and disassembly of the primary cilium. J Cell Sci. 2010;123:511–518. doi: 10.1242/jcs.061093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Marshall WF. Ciliogenesis: building the cell's antenna. Nat Rev Mol Cell Biol. 2011;12:222–234. doi: 10.1038/nrm3085. [DOI] [PubMed] [Google Scholar]
- Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- Oh EC, Katsanis N. Cilia in vertebrate development and disease. Development. 2012;139:443–448. doi: 10.1242/dev.050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Dynlacht BD. Regulating the transition from centriole to basal body. J Cell Biol. 2011;193:435–444. doi: 10.1083/jcb.201101005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L, Scholey JM. Intraflagellar transport at a glance. J Cell Sci. 2009;122:889–892. doi: 10.1242/jcs.023861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Sun Z. Expanding horizons: ciliary proteins reach beyond cilia. Annu Rev Genet. 2013;47:353–376. doi: 10.1146/annurev-genet-111212-133243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SC. CYLD: a tumor suppressor deubiquitinase regulating NF-kappaB activation and diverse biological processes. Cell Death Differ. 2010;17:25–34. doi: 10.1038/cdd.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Huo L, Sun X, et al. The tumor suppressor CYLD regulates microtubule dynamics and plays a role in cell migration. J Biol Chem. 2008;283:8802–8809. doi: 10.1074/jbc.M708470200. [DOI] [PubMed] [Google Scholar]
- Wickstrom SA, Masoumi KC, Khochbin S, Fassler R, Massoumi R. CYLD negatively regulates cell-cycle progression by inactivating HDAC6 and increasing the levels of acetylated tubulin. EMBO J. 2010;29:131–144. doi: 10.1038/emboj.2009.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Liu M, Li D, et al. CYLD regulates spindle orientation by stabilizing astral microtubules and promoting dishevelled-NuMA-dynein/dynactin complex formation. Proc Natl Acad Sci USA. 2014;111:2158–2163. doi: 10.1073/pnas.1319341111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Chang M, Paul EM, et al. Deubiquitinating enzyme CYLD negatively regulates RANK signaling and osteoclastogenesis in mice. J Clin Invest. 2008;118:1858–1866. doi: 10.1172/JCI34257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompouki E, Tsagaratou A, Kosmidis SK, et al. Truncation of the catalytic domain of the cylindromatosis tumor suppressor impairs lung maturation. Neoplasia. 2009;11:469–476. doi: 10.1593/neo.81424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright A, Reiley WW, Chang M, et al. Regulation of early wave of germ cell apoptosis and spermatogenesis by deubiquitinating enzyme CYLD. Dev Cell. 2007;13:705–716. doi: 10.1016/j.devcel.2007.09.007. [DOI] [PubMed] [Google Scholar]
- L'Hernault SW, Rosenbaum JL. Chlamydomonas alpha-tubulin is posttranslationally modified by acetylation on the epsilon-amino group of a lysine. Biochemistry. 1985;24:473–478. doi: 10.1021/bi00323a034. [DOI] [PubMed] [Google Scholar]
- Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature. 2003;424:793–796. doi: 10.1038/nature01803. [DOI] [PubMed] [Google Scholar]
- Shiratsuchi G, Kamiya R, Hirono M. Scaffolding function of the Chlamydomonas procentriole protein CRC70, a member of the conserved Cep70 family. J Cell Sci. 2011;124:2964–2975. doi: 10.1242/jcs.084715. [DOI] [PubMed] [Google Scholar]
- Wilkinson CJ, Carl M, Harris WA. Cep70 and Cep131 contribute to ciliogenesis in zebrafish embryos. BMC Cell Biol. 2009;10:17. doi: 10.1186/1471-2121-10-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Sun X, Liu M, Li D, Aneja R, Zhou J. CEP70 protein interacts with gamma-tubulin to localize at the centrosome and is critical for mitotic spindle assembly. J Biol Chem. 2011;286:33401–33408. doi: 10.1074/jbc.M111.252262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler KV, Kalin J, Brochier C, Vistoli G, Langley B, Kozikowski AP. Rational design and simple chemistry yield a superior, neuroprotective HDAC6 inhibitor, tubastatin A. J Am Chem Soc. 2010;132:10842–10846. doi: 10.1021/ja102758v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozikowski AP, Tapadar S, Luchini DN, Kim KH, Billadeau DD. Use of the nitrile oxide cycloaddition (NOC) reaction for molecular probe generation: a new class of enzyme selective histone deacetylase inhibitors (HDACIs) showing picomolar activity at HDAC6. J Med Chem. 2008;51:4370–4373. doi: 10.1021/jm8002894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraki M, Nakazawa Y, Kamiya R, Hirono M. Bld10p constitutes the cartwheel-spoke tip and stabilizes the 9-fold symmetry of the centriole. Curr Biol. 2007;17:1778–1783. doi: 10.1016/j.cub.2007.09.021. [DOI] [PubMed] [Google Scholar]
- Nakazawa Y, Hiraki M, Kamiya R, Hirono M. SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Curr Biol. 2007;17:2169–2174. doi: 10.1016/j.cub.2007.11.046. [DOI] [PubMed] [Google Scholar]
- Shi X, Wang J, Yang Y, Ren Y, Zhou J, Li D. Cep70 promotes microtubule assembly in vitro by increasing microtubule elongation. Acta Biochim Biophys Sin (Shanghai) 2012;44:450–454. doi: 10.1093/abbs/gms017. [DOI] [PubMed] [Google Scholar]
- Shi X, Liu M, Li D, Wang J, Aneja R, Zhou J. Cep70 contributes to angiogenesis by modulating microtubule rearrangement and stimulating cell polarization and migration. Cell Cycle. 2012;11:1554–1563. doi: 10.4161/cc.19954. [DOI] [PubMed] [Google Scholar]
- Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell. 2007;129:1351–1363. doi: 10.1016/j.cell.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez de Diego A, Alonso Guerrero A, Martinez AC, van Wely KH. Dido3-dependent HDAC6 targeting controls cilium size. Nat Commun. 2014;5:3500. doi: 10.1038/ncomms4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kwon S, Yamaguchi T, et al. Mice lacking histone deacetylase 6 have hyperacetylated tubulin but are viable and develop normally. Mol Cell Biol. 2008;28:1688–1701. doi: 10.1128/MCB.01154-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao YS, Hubbert CC, Lu J, Lee YS, Lee JY, Yao TP. Histone deacetylase 6 regulates growth factor-induced actin remodeling and endocytosis. Mol Cell Biol. 2007;27:8637–8647. doi: 10.1128/MCB.00393-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa YI, Lin C, Chuang PT. Hedgehog signaling from the primary cilium to the nucleus: an emerging picture of ciliary localization, trafficking and transduction. Curr Opin Genet Dev. 2013;23:429–437. doi: 10.1016/j.gde.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans DA, Voest EE, Giles RH. All along the watchtower: is the cilium a tumor suppressor organelle. Biochim Biophys Acta. 2008;1786:114–125. doi: 10.1016/j.bbcan.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Massoumi R. CYLD: a deubiquitination enzyme with multiple roles in cancer. Future Oncol. 2011;7:285–297. doi: 10.2217/fon.10.187. [DOI] [PubMed] [Google Scholar]
- Eguether T, Ermolaeva MA, Zhao Y, et al. The deubiquitinating enzyme CYLD controls apical docking of basal bodies in ciliated epithelial cells. Nat Commun. 2014;5:4585. doi: 10.1038/ncomms5585. [DOI] [PubMed] [Google Scholar]
- Li D, Sun X, Zhang L, et al. Histone deacetylase 6 and cytoplasmic linker protein 170 function together to regulate the motility of pancreatic cancer cells. Protein Cell. 2014;5:214–223. doi: 10.1007/s13238-013-0010-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiley WW, Zhang M, Jin W, et al. Regulation of T cell development by the deubiquitinating enzyme CYLD. Nat Immunol. 2006;7:411–417. doi: 10.1038/ni1315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Polydactyly in CYLD knockout mice.
Transmission electron microscopy images of the cross sections of axonemes in the tracheal epithelial cilia of CYLD knockout mice.
Examination of the interaction between CYLD and Cep70 in different phases of the cell cycle and in vitro.
CYLD interacts with HDAC6 and inhibits its activity toward tubulin deacetylation.
CYLD regulates the Shh signaling.