Abstract
Processes leading to speciation in oceanic environments without obvious physical barriers remain poorly known. European and American eel (Anguilla anguilla and A. rostrata) spawn in partial sympatry in the Sargasso Sea. Larvae are advected by the Gulf Stream and other currents towards the European/North African and North American coasts, respectively. We analyzed 104 mitogenomes from the two species along with mitogenomes of other Anguilla and outgroup species. We estimated divergence time between the two species to identify major events involved in speciation. We also considered two previously stated hypotheses: one where the ancestral species was present in only one continent but was advected across the Atlantic by ocean current changes and another where population declines during Pleistocene glaciations led to increasing vicariance, facilitating speciation. Divergence time was estimated to ∼3.38 Mya, coinciding with the closure of the Panama Gateway that led to reinforcement of the Gulf Stream. This could have advected larvae towards European/North African coasts, in which case American eel would be expected to be the ancestral species. This scenario could, however, not be unequivocally confirmed by analyses of dN/dS, nucleotide diversity and effective population size estimates. Extended bayesian skyline plots showed fluctuations of effective population sizes and declines during glaciations, and thus also lending support to the importance of vicariance during speciation. There was evidence for positive selection at the ATP6 and possibly ND5 genes, indicating a role in speciation. The findings suggest an important role of ocean current changes in speciation of marine organisms.
Introduction
Understanding speciation is key in evolutionary biology. Yet, the processes involved encompass some of the most debated and controversial topics in the biological sciences (Mayr, 1963; Via, 2001; Coyne and Orr, 2004). The biology of the specific organisms and the properties of the environment that they inhabit are expected to have a major influence on speciation. Thus, understanding the intrinsic and extrinsic factors initiating speciation is crucial for understanding the speciation process itself.
Marine environments are characterized by few definitive geographical barriers to gene flow, and indeed intraspecific genetic differentiation is much lower in marine fishes as compared with freshwater fishes, normally ascribed to a combination of high gene flow and low genetic drift owing to high effective population sizes (Ward et al., 1994; Bernardi, 2013). Accordingly, it would also be expected that speciation-with-gene flow (Feder et al., 2012) and even sympatric speciation (Via, 2001) should be common in marine environments.
The two Atlantic species of the family of freshwater eels (Anguillidae), American and European eels (Anguilla anguilla and A. rostrata), spawn in the southern Sargasso Sea in a region characterized by distinct thermal fronts (the Subtropical Convergence Zone) (Tesch, 2003; van Ginneken and Maes, 2005). There is considerable overlap between their spawning regions, but with a clear dominance of American eels in the West and European eel in the East (McCleave et al., 1987b; Als et al., 2011). After hatching in the Sargasso Sea, the larvae are transported by the Gulf Stream and other currents towards the North American (American eel) and European and North African continents (European eel). Upon reaching the continental shelf, larvae metamorphose into the so-called glass eels and enter freshwater or coastal habitats. Adult eels complete their life cycle by returning to the Sargasso Sea to spawn and subsequently die. Despite widespread geographical distributions, molecular studies have demonstrated that both species are panmictic (Palm et al., 2009; Als et al., 2011; Côté et al., 2013).
European and American eels are morphologically almost indistinguishable, with differences in numbers of vertebrae being the most reliable diagnostic character (Tesch, 2003). Genetic differentiation at the nuclear level is also low, with reported FST-values at microsatellite loci of 0.09, 0.055 and 0.018 (Mank and Avise, 2003; Wirth and Bernatchez, 2003; Als et al., 2011) and 0.0685 for amplified fragment length polymorphism markers (Gagnaire et al., 2009). In sharp contrast, mitochondrial DNA (mtDNA) places the species as two reciprocally monophyletic lineages (Avise et al., 1986, 1988) that are sister species within Anguillidae (Tsukamoto and Aoyama, 1998; Lin et al., 2001; Tsukamoto et al., 2002; Minegishe et al., 2005; Teng et al., 2009). Furthermore, hybridization between the two species also occurs, with most hybrids being found in Iceland (Avise et al., 1990; Albert et al., 2006; Gagnaire et al., 2009).
The largely sympatric spawning regions but separate continental distribution of adult eels, combined with the distinct mitochondrial lineages, yet low nuclear differentiation and the occurrence of hybridization, strongly suggest a speciation-with-gene flow scenario for the origin of the two species. However, when this speciation occurred and which factors were involved remains unsolved. Avise et al. (1990) outlined two speciation scenarios based on the assumption that the two species diverged during the Pleistocene (ca. 2.6 Mya–12 Kya). The first ‘Vicariance' scenario assumed one single ancestral Northern distributed species. Cooling periods during the Pleistocene would have forced a southward retreat, as the Northern regions became uninhabitable, leading to disjunction of spawning areas and subsequently speciation. Although not explicitly stated by Avise et al. (1990), the decrease in available habitat might have been associated with population declines. Given the present distribution of larvae across the Sargasso Sea, where despite substantial overlap there are two different core areas for each species (McCleave et al., 1987b; Munk et al., 2010; Als et al., 2011), this could have resulted in numerically fewer eels from Europe spawning towards the West, and fewer eels from America spawning towards the East. The decreased overlap of spawning regions would thereby allow for allo- or parapatric speciation.
The second ‘Oceanic' scenario proposed that if the ancestral species initially inhabited a single continent, subsequent changes in the ocean currents during the Pleistocene could have advected some larvae to new habitats on the other continent. As these larvae would probably meet suitable spawning conditions in the region of the Sargasso Sea most proximate to their continent of distribution, and as strong disruptive selection might have been present for the duration of larval phase, assortative mating and finally speciation could have occurred in a parapatric or sympatric manner. Another event leading to a similar scenario, although not stated by Avise et al. (1990), is the formation of the Isthmus of Panama ca. 3.0–4.5 Mya, which disconnected the Pacific and Atlantic Oceans and reinforced the Gulf Stream (Haug and Tiedemann, 1998), the major current dispersing eel larvae towards the North American and European continents. If this scenario is correct, then it would be expected that the ancestral species was the American eel. Their larvae have a shorter dispersal distance from the Sargasso Sea to continental waters than the European eel, and therefore would be advected towards Europe by the reinforced Gulf Stream.
So far, several studies have attempted to estimate divergence time between the two Atlantic eel species and all yielded contradictory results. Avise et al. (1986) estimated the split to ca. 1.5 Mya based on restriction fragment length polymorphism analysis of total mtDNA and a standard substitution rate of 2% divergence per Myr. Subsequently, Tsukamoto and Aoyama (1998) used the mitochondrial cytochrome b gene to estimate divergence time to ca. 10.2 Mya. This was based on calibrating the phylogeny of eight Anguilla species, assuming that the ancestor of Atlantic eels used a Tethys Sea migration route into the present Atlantic region ca. 30 Mya. Finally, Minegishe et al. (2005) sequenced entire mitogenomes from 18 species of Anguillidae and estimated the divergence time using a mutation rate of 0.00073 sub per site per Myr (estimated for bony fishes from concatenated amino-acid sequences of nicotinamide adenine dinucleotide dehydrogenase subunit 2 and cytochrome b), resulting in a divergence estimate of 5.8 Mya for the Atlantic eels.
In this study, we sequenced mitogenomes from 53 European and 49 American eels. To test whether an ‘Oceanic' or ‘Vicariance' scenario is most compatible with speciation in Atlantic eels, we first estimated divergence time. We calibrated divergence time between Atlantic eels taking into account recent knowledge of outgroup relationships (Inoue et al., 2010; Johnson et al., 2012) and fossil information. This circumvents reliance on potentially misleading calibrations based on standard substitution rates (Pulquerio and Nichols, 2007) or a Tethys Sea dispersal scenario (Lin et al., 2001; Teng et al., 2009). In particular, (1) we tested if speciation occurred during the Pleistocene as suggested by Avise et al. (1990) or much earlier as suggested by other studies (Tsukamoto and Aoyama, 1998; Minegishe et al., 2005). (2) We assessed if divergence time coincides with major geological or climatic events that could explain the specific processes of speciation. (3) We determined the direction of nonsynonymous substitutions from the ancestral state and estimated dN/dS (rate of nonsynonymous substitutions compared to the rate of synonymous substitutions) to assess which of the two species are ancestral and derived. (4) We assessed the influence of glaciations on effective population size using a Bayesian skyline plot method (Heled and Drummond, 2008), thereby reconstructing the long-term demographic history of the two species. (5) We investigated the possibility of selection acting on the mitochondrial genome as an increasing number of studies have shown that positive selection may affect mtDNA variation (Ballard and Kreitman, 1995; Ballard and Whitlock, 2004). This may be the case in Atlantic eels, where Gagnaire et al. (2012) observed possible cytonuclear incompatibilities between American and European eels among genes involved in the ATP synthase complex. Hence, on one side, it is important to assess if positive selection has influenced mitogenome sequence divergence between European and American eel, possibly complicating estimates of divergence time and demographic history. On the other side, evidence for selection might indicate a role of the mitochondrial genome in the speciation process.
Materials and methods
Samples and locations
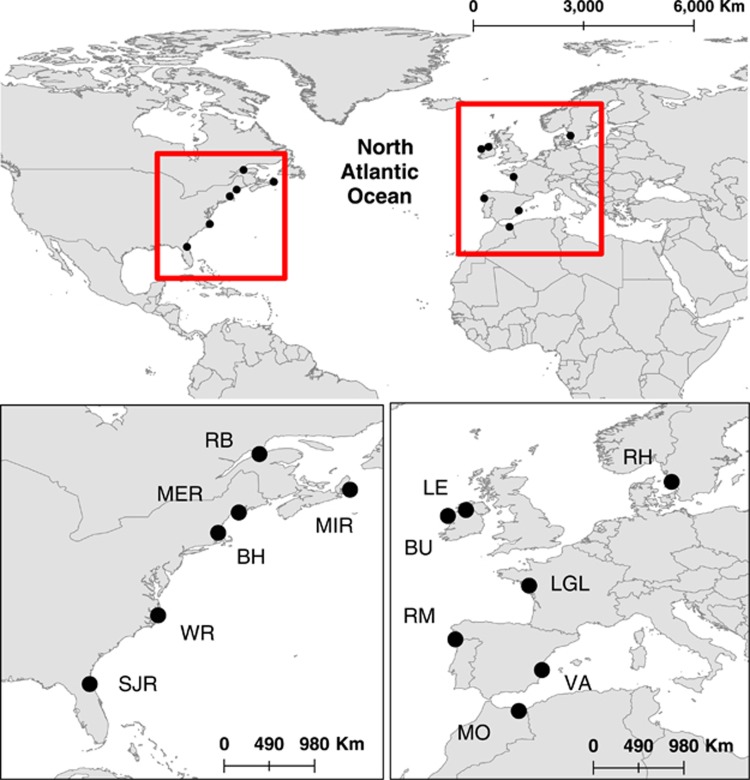
Fifty-three European and 49 American eels (sampled as glass eels and sub-adult yellow eels) were analyzed, representing 13 localities across their distributional ranges (Figure 1 and Table 1). Individuals were collected between 1999 and 2010 and have for some locations been used in a study based on amplified fragment length polymorphism variation (Albert et al., 2006). Mitogenome sequences from 21 Anguillid species, representing 18 species of freshwater eels (GenBank accession nos. NC_002707 and NC_006531–NC_006547; Minegishe et al., 2005) and three species of the closest related family of Anguillidae; Serrivomeridae (Johnson et al., 2012; GenBank accession nos. NC_013436 and NC_013627–NC_013628; Inoue et al., 2010), were included in the study to estimate divergence time and investigate selection. The sequences retrieved from GenBank included one European eel and one American eel, resulting in a total of 104 mitogenome sequences of Atlantic eels.
Figure 1.
Sampling locations of European and American eels. See Table 1 for population names and sample sizes.
Table 1. Information about the sampled locations, year of sampling, life stage and sample size.
| Sample (abbreviation) | Country | Year of sampling | Life stage | Sample size |
|---|---|---|---|---|
| Riviere Blanche (RB) | Canada | 2007 | Y/G | 21/5 |
| Mira River (MIR) | Canada | 2007 | G | 7 |
| Medomak River (MER) | USA | 1999 | Y | 5 |
| St. Johns River (SJR) | USA | 1999 | Y | 5 |
| Wye River (WR) | USA | 1999 | Y | 4 |
| Boston Harbour (BH) | USA | 1999 | G | 2 |
| Marylanda | USA | 1999 | Y | 1 |
| Ringhals (RH) | Sweden | 2008 | G | 15 |
| Lough Erne (LE) | Ireland | 2008 | G | 14 |
| Valencia (VA) | Spain | 2010 | Y | 11 |
| Burrishole (BU) | Ireland | 2010 | Y | 4 |
| Lac Grand Lieu (LGL) | France | 1999 | G | 3 |
| Rio Minho (RM) | Portugal | 1999 | G | 3 |
| Moulouya (MO) | Morocco | 1999 | G | 3 |
| Vilaineb | France | 2003 | Y | 1 |
Mitogenome sequencing
Tissue samples (fin clips, muscle or whole glass eels) were stored in 96% ethanol at −20 °C. DNA was extracted using standard phenol–chloroform extraction and kept frozen at −20 °C. Following Jacobsen et al. (2012), long-range PCRs were conducted, subdividing each mitogenome into two amplicons, each covering just over half of the mitogenome with small overlaps (for primer information and PCR conditions see Supplementary Table S1 and Supplementary Note S1). The concentrations of purified amplicons were measured using a Q-bit flourometer (Life Technologies, Carlsbad, CA, USA). Subsequently, the two amplicons from each DNA extract were pooled at equimolar concentration. The pooled PCR products were fragmented using a Bioruptor (Diagenode, Belgium, Europe) with eight cycles of 30 s of sonication, resulting in mean fragment sizes of ca. 200 bp. Individual samples were converted into Illumina index-tagged DNA libraries using the E6040S/L NEBnext Library Preparation Kit (New England BioLabs, Ipswich, MA, USA) with minor modifications. Subsequently, libraries were sequenced in pools of 12–46 per lane using either Illumina GA or HiSeq sequencing platforms and single-end chemistry to sequence 75 or 100 bases.
Mitogenome assembly and measurement of sequencing error
Mitogenome assembly was conducted using a suite of bioinformatic packages (see Supplementary Note S2 for details) to generate a file containing the highest quality reads from each position from each strand. Individual consensus sequences were generated adhering to a majority rule (>50%), at each genomic position with a minimum depth of coverage ≥5 (<5 calls an N). A given indel was considered a real event and included in the consensus sequence if at least 60% of the reads spanning that specific region contained the indel.
One technical replicate (same sample sequenced in two different lanes) was used to assess the degree of sequencing errors and refine the base calling process.
Neutrality test, nucleotide diversity and population structure
Number of haplotypes, nucleotide diversity (π), Tajimas D and Fu and Li's D and F (Tajima, 1989; Fu and Li, 1993) were calculated in DnaSP v5.1 (Librado and Rozas, 2009) for the whole mitogenome and separately for the control region for both species. All analyses were conducted without taking gaps/ambiguous bases into account as most of these were located in polymer regions and might represent sequencing errors. A neighbour-joining tree was constructed based on the whole mitogenome data set, using MEGA 5 (Tamura et al., 2011), assuming the TrN+G substitution model with 10 gamma categories as found using JMODELTEST version 2.11 (Posada, 2008), and 1000 replicates to calculate bootstrap support.
To test possible substructuring within species (although the species are assumed to be panmictic), two different tests were applied: (1) The genealogical sorting index (Cummings et al., 2008) using the other Atlantic species as outgroup. (2) Pairwise PhiST-values with their significances estimated in ARLEQUIN (version 3.5.2.1; Excoffier and Lischer, 2010) using the Tamura and Nei substitution model and 99 999 permutations.
Estimation of divergence time and substitution rate
Five mitogenome partitions were aligned and manually examined using GENEIOUS PRO 5.4.6 (BioMatters, 2012): the two concatenated rRNAs, the concatenated tRNAs and the concatenated 1st, 2nd and 3rd codon position for the 13 coding genes. The control region and non-coding parts varied in size, probably because of high mutation rate, and were omitted. Data sets comprised the 16 species of Anguillidae from Minegishe et al. (2005) (excluding Atlantic eels), three outgroup species from Serrivomeridae (Inoue et al., 2010; Johnson et al., 2012) and the two most divergent mitogenomes of each of the two Atlantic eel species, chosen using MEGA 5.1 (Tamura et al., 2011), and included to calculate the TMRCA (Time of Most Recent Common Ancestor) within each species for subsequent use in demographic analyses (see below). Mutational saturation was analysed using the method by Xia et al. (2003) implemented in DAMBE (Xia and Xie, 2001).
Estimates of divergence time were analysed using the Bayesian phylogenetic approach in BEAST (Bayesian evolutionary analysis by sampling trees) v1.7.4 (Drummond and Rambaut, 2007). Three different data sets were analysed: (1) all five partitions, (2) the two concatenated rRNAs and the concatenated tRNAs and (3) the 3rd base position for the 13 coding genes. The 3rd base position data set was chosen, as the rate of neutral mutation should be unaffected by selection in case of complete linkage as in mtDNA, and therefore may be used to estimate divergence in the presence of selection (Birky and Walsh, 1988). The YULE process was chosen as tree prior and a uniform prior for the TMRCA of all samples was set to 50–55 Myr ago (lower and upper bound) according to the oldest observed fossils of Anguillidae (Patterson, 1993). An uncorrelated log-normal clock was used to account for rate heterogeneity among branches as an initial test showed deviation from a constant clock. The final Markov chain Monte Carlo sample obtained was based on a run for 100 000 000 generations, with genealogies sampled every 10 000 generations with 10% discarded as burn-in.
As different demographic scenarios might affect divergence estimates (Ho et al., 2008), a second analysis was conducted in *BEAST (Heled and Drummond, 2010) to assess the robustness of the divergence estimate. It relies on two different tree priors: a speciation prior for between-species branch patterns and a coalescence prior for the within-species branch patterns. We used *BEAST for all 103 (one individual excluded, see Results) mitogenome sequences of Atlantic eels divided into six partitions: the two concatenated rRNAs; the concatenated tRNAs; the concatenated 1st, 2nd and 3rd codon position for the 13 coding genes and the control region. To calibrate a molecular clock, the mean rate of rRNAs estimated from the first analyses was used as a prior. A constant clock was chosen as a preliminary test in BEAST showed no significant rate heterogeneity among branches. Both the constant and linear piecewise change model of effective population size were used as coalescence priors to compare the robustness of the divergence estimates given differences in demographic history. The YULE prior was used as speciation prior. Only TMRCA was evaluated in the analyses.
All substitution models used were identified via the Akaike information criterion) with four gamma categories using JMODELTEST version 2.11 (Posada, 2008) (Supplementary Table S2) and partitions linked in all analyses reflecting clonal inheritance of mtDNA. Examination of convergence and effective sample size was conducted using TRACER v1.5 (Drummond and Rambaut, 2007). A second replicate was conducted for all analyses to check for convergence.
Demographic inference
Long-term trends in female effective population size (Nef) were analysed by extended Bayesian skyline plot (EBSP) analysis (Heled and Drummond, 2008) using BEAST (Drummond et al., 2005). EBSPs estimate the number of demographic changes directly from the data and thereby test for deviations from constant size. Each analysis was based on whole mitogenomes either divided into the same regions as in *BEAST or used as one whole locus. The two species were analyzed separately. Time was calibrated by estimates of mean TMRCA from the species analysis (using all regions) or mean TMRCA with associated 95% highest probability density (HPD) fitted to a log-normal distribution. Substitution model estimation, Markov chain Monte Carlo length and sampling and examination of convergence were performed as for the phylogenetic analysis (Supplementary Table S2). Constant clocks were chosen following rate heterogeneity tests. To unscale estimates of female effective population size, a female generation time of 8.73 years was assumed (Vollestad, 1992).
Test for selection
Mitochondrial genes are generally under strong purifying selection (Sun et al., 2011) and therefore unlikely to show overall dN/dS >1. Thus, the codon-based selection test FUBAR (Fast Unbiased Bayesian AppRoximation) (Murrell et al., 2013) was chosen to test for positive selection acting across Anguillidae. The test was conducted for all 13 mitochondrial genes using the 18 species of Anguillidae previously analysed in Minegishe et al. (2005). Compared with more classical random effects likelihood tests, FUBAR is less likely to produce false positives owing to assignment of codons experiencing different selection pressure into the same categories (Murrell et al., 2013). Using the same data set, all ancestral states of the 13 coding genes were reconstructed using the ASR (Ancestral State Reconstruction) software (Pond and Frost, 2005) (see Supplementary Note S3). The direction of nonsynonymous changes was inferred and average pairwise dN/dS was measured comparing the ancestral sequence to the European and American mitogenome samples using DnaSP v5.1.
To test for positive selection between the two Atlantic eel species, McDonald–Kreitman tests (Mcdonald and Kreitman, 1991), as implemented in DnaSP v5.1, were conducted for all 13 genes of European and American ancestry. The possibility of recombination acting among Atlantic eels was tested using the SBP software (Single Breakpoint reconstruction) (Pond et al., 2006).
FUBAR, ASR and SBP tests were conducted using the HYPHY package on the DataMonkey server (http://www.datamonkey.org/dataupload.php), using default parameters and an input topology similar to Teng et al. (2009).
Results
Sequencing and error estimates
A total of 102 mitogenomes were successfully sequenced (GenBank accession no. KJ564170–KJ564271). The average depth of coverage of uniquely mapped reads was >94.7% of the maximum possible (Supplementary Table S3). After refining the base calling method, the two technical replicates matched 100%, suggesting no significant impact of Illumina sequencing errors in the data set.
Neutrality test, nucleotide diversity and population structure within Atlantic eels
Ninety-six haplotypes were present among the 104 Atlantic eel mitogenome sequences (Table 2). Shared haplotypes were only present among American eel and represented a total of 11 individuals distributed across three different haplotypes. Of these haplotypes, several were shared across localities (Figure 2). American eel showed lower nucleotide diversity than European eel (0.00291 vs 0.00553 at the whole mitogenome level; Table 2).
Table 2. Nucleotide diversity estimates and neutrality tests.
| Species | Data | Haplotypes (h) | Nucleotide diversity (π) | Tajimas D | Fu and Li's D | Fu and Li's F |
|---|---|---|---|---|---|---|
| American eel (N=49)a | Mitogenome | 41 | 0.00291 | −1.94* | −4.24** | −4.03** |
| Control region | 37 | 0.00896 | −1.70 | −2.98* | −3.00* | |
| European eel (N=54) | Mitogenome | 54 | 0.00553 | −1.88* | −3.53** | −3.46** |
| Control region | 52 | 0.02283 | −1.42 | −2.26 | -2.3 |
Significance level: *P<0.05, **P<0.02
An individual with European ancestry was excluded from the analysis (see text).
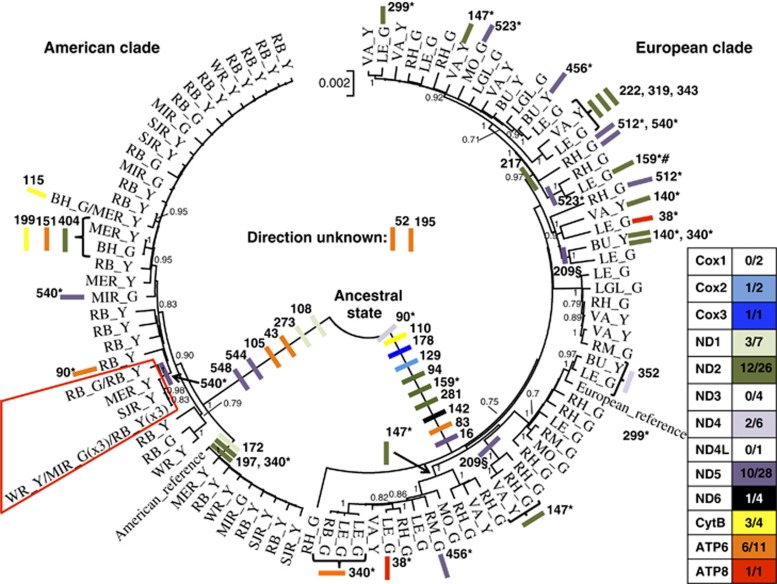
Figure 2.
Neighbour-joining tree of 96 mitogenome haplotypes. Bootstrap support >0.7 is shown above the branching points. The direction of nonsynonymous change is represented by coloured bars for all 13 genes along the respective branches and only given for mutations supported by at least two independent sequences. Numbers within gene boxes are the number of observed supported nonsynonymous changes compared with the total number of nonsynonymous changes. Amino-acid positions are shown next to the respective bar and a ‘*' denotes that the substitution is observed in sequences of independent ancestry. A ‘§' denotes that different amino-acid changes have occurred at this position and ‘#' denotes a back mutation. A ‘/' in the sequence name is used to separate between sampling location where haplotypes are shared between multiple samples. The red shape denotes the clade within the American lineage showing possible selection (see text). See Table 1 for sampling codes for individual sequences.
The neighbour-joining tree identified two reciprocally monophyletic clades corresponding to the two species. However, one glass eel sampled in Riviere Blanche (Québec, Canada) showed European ancestry (RBG10; Figure 2). As strong postzygotic selection possibly exists against hybrids of Atlantic eels (Pujolar et al., 2014), this sample was excluded from further analyses.
Neutrality tests showed excess of rare polymorphisms (negative values) (Table 2), compatible with either demographic expansion or selective sweeps. Neither genealogical sorting index nor PhiST showed evidence for genetic substructuring within each species (Supplementary Tables S4 and S5).
Estimation of divergence time and mutation rate
Mutation saturation tests did not reveal significant saturation in the five partitions. However, the 3rd base codons showed a considerable proportion of variable sites compared with the other data sets, with only 22% invariable sites (Supplementary Table S6).
Divergence time estimates obtained with BEAST were highly similar across different data sets, although the analysis of the 3rd base codon position supported a slightly older divergence time than the other results (Table 3). The most extensive data set encompassing all 13 coding genes, the RNAs and tRNAs suggested a divergence time for the Atlantic eels of 3.38 Myr (95% HPD 2.55–4.30) (for phylogeny see Supplementary Figure S1). In general, all three data sets supported divergence of American and European eels around 3–4.5 Mya. Analyses were also conducted using a Birth–Death prior. This resulted in highly similar results (Supplementary Table S7). The substitution rate of the concatenated rRNAs used in the *BEAST analysis was estimated to be 2.189 × 10−9 sub per site per year, whereas the substitution rate of the whole mitogenome was estimated to 5.463 × 10−9 sub per site per year (Supplementary Table S8). Estimates of TMRCA obtained with BEAST within each of the two Atlantic eel species showed overall deeper coalescence time in the European lineage (0.72 Myr, 95% HPD 0.48–0.99) compared with the American lineage (0.57 Myr, 95% HPD 0.36–0.78; Table 3), although with overlapping credible intervals. Analyses in *BEAST, using the substitution rate of the combined rRNAs, resulted in highly similar divergence time estimates compared with the more extensive species phylogeny (Table 3).
Table 3. Mean divergence time estimates (TMRCA) in million of years for different analyses and data partitions.
| Analysis | Groups | All data incl. control region | All data excl. control region | rRNAs+tRNAs | 3rd Base codon |
|---|---|---|---|---|---|
| BEAST analysis (N=23) | All freshwater eels | NA | 13.76 (11.14–16.50) | 14.92 (9.61–20.53) | 18.39 (12.74–24.47) |
| Atlantic eels | NA | 3.38 (2.55–4.30) | 3.32 (1.48–5.54) | 4.45 (2.58–6.79) | |
| American eela | NA | 0.57 (0.36–0.78) | 0.68 (0.18–1.29) | 0.67 (0.31–1.09) | |
| European eela | NA | 0.72 (0.48–0.99) | 0.99 (0.31–1.88) | 0.86 (0.41–1.38) | |
| *BEAST analysis (N=103) | Atlantic eelsb | 3.25 (2.62–3.91) | NA | NA | NA |
| Atlantic eelsc | 3.26 (2.63–3.92) | NA | NA | NA |
Abbreviations: BEAST, Bayesian evolutionary analysis by sampling trees; HPD, highest probability density; NA, no analysis was conducted for the partition; TMRCA, Time of Most Recent Common Ancestor.
95% HPD are given within parentheses.
TMRCA estimates for the two most divergent American and European samples.
YULE and piece-wise constant prior.
YULE and piece-wise linear prior.
Demographic inference
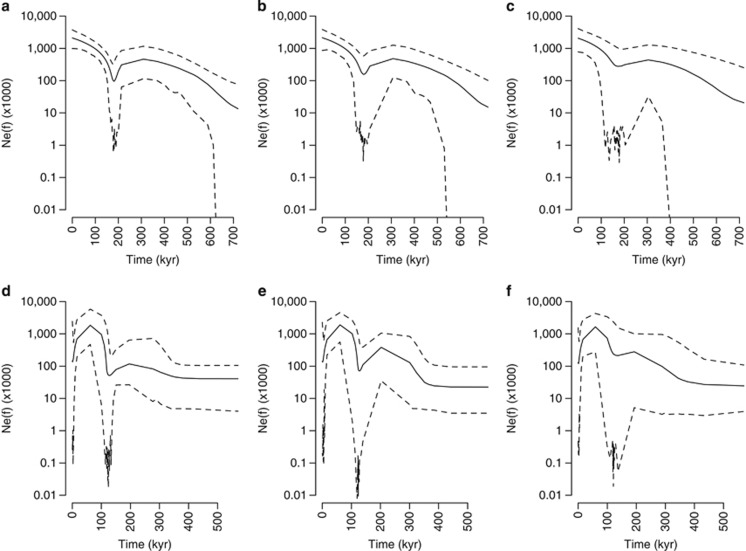
For both species, the EBSPs differed significantly from a model of constant population size with 3.3 (95% HPD 2.0–4.0) and 4.4 (95% HPD 3.0–6.0) estimated episodes of changes in female effective population size (Nef) for European and American eels, respectively (Figure 3). In general, long-term Nef showed the same trend in both species (Figure 3) and revealed a decline followed by expansion estimated to c. 180 Kya for European eel and c. 125 Kya for American eel with overlapping credible intervals. When incorporating the uncertainty of TMRCA from the species analysis, the declines became less pronounced compared with the analyses where the uncertainty of the TMRCA were estimated directly from the data (Figures 3c and f compared with Figures 3a, b and d, e). In contrast with the current population expansion inferred for European eel, a contemporary population decline was inferred for American eel, starting between 16–59 Kya. Accordingly, recent female effective population size was higher for European eel with an estimated size of ca. 2 066 000 (95% HPD 770 000–4 087 000) compared with ca. 130 000 (95% HPD 470–1 547 000) for American eel (as observed in Figures 3c and f).
Figure 3.
EBSPs of long-term trends in female effective population size (Nef). Solid lines represent median Nef-values, whereas dashed lines represent 95% HPD intervals. (a–c) European eel and (d–f) American eel. X-axes represent time and are scaled using estimates of TMRCA from the species phylogeny. (a, b and d, e) Time scaled by mean TMRCA using either the whole (a+d) or the partitioned mitogenome (b+e). (c and f) time scaled using the 95% HPD from the species analysis for the partitioned mitogenome.
Test for selection
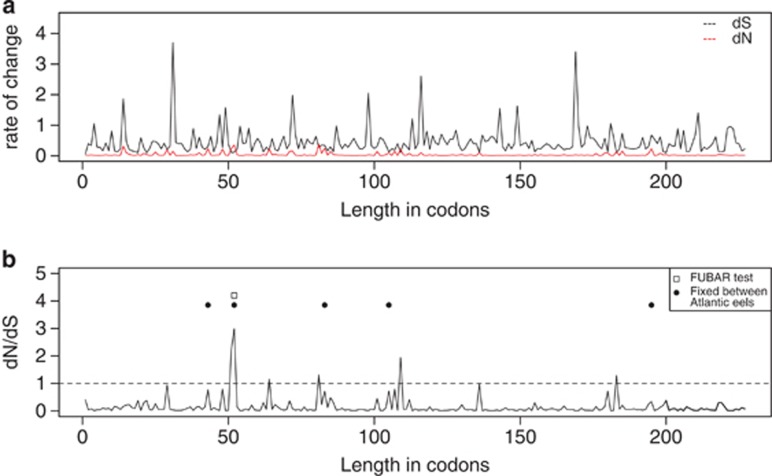
The FUBAR test suggested strong purifying selection acting on the mitochondrial genes (Supplementary Table S9). However, one site was found to be under positive selection (Bayes factor=30) (Figure 4). This site was detected in ATP6 codon position 52, one of five positions fixed between American and European eels previously observed by Gagnaire et al. (2012); (Figure 4 and Supplementary Table S9). McDonald–Kreitman tests also suggested positive selection acting within the ATP6 gene between the two species (Supplementary Table S10). No evidence for recombination was found.
Figure 4.
Results of the FUBAR test along the ATP6 gene. (a) Rate of synonymous (dS) and nonsynonymous change (dN), and (b) dN/dS. Position 52 that showed significantly elevated dN/dS is denoted by a square, whereas the five fixed nonsynonymous amino-acid changes between European and American eels are denoted by dots.
The analysis of the direction of change showed more fixed nonsynonymous changes along the European branch (Figure 2). In total, 10 mutations occurred in European direction, six in American direction, while for two mutations, both occurring in ATP6, the direction could not be assessed, as both species showed changes from the most likely ancestral state. Estimates of dN/dS were almost identical with 0.0338 for European and 0.0313 for American samples.
Although most replacements were found in terminal branches, one replacement of older ancestry was found in ND5 (position 540) in all descendants of a lineage comprising two of the three American haplotypes shared by two or more individuals (Figure 2), and moreover independently in one European and one American individual (Figure 2). This mutation changed the amino acid from valine (V) to isoleucine (I) (Supplementary Figure S2).
Discussion
Our results support previous findings by Avise et al. (1986, 1990) that first reported that European and Atlantic eel comprise two highly divergent mtDNA lineages, although with one North American individual showing European mitochondrial ancestry. They are furthermore congruent with recent studies showing that both species are panmictic (Palm et al., 2009; Als et al., 2011; Côté et al., 2013); (Supplementary Tables S4 and S5). However, the estimated species divergence time is different from previous studies, and points towards the closure of the Panama Gateway as the event initiating speciation. Moreover, there is evidence for positive selection at the mitogenome level. This raises the possibility that selection at mtDNA may have been involved in the speciation process, but also complicates interpretation of divergence time estimates and demographic history. We elaborate on these issues in the following.
Inferring speciation history from mtDNA
mtDNA has its own dynamics owing to its special properties of maternal inheritance, haploidy and, in most cases, absence of recombination. Parameters inferred for mtDNA may therefore not be representative for the organisms and their genomes as such (Ballard and Whitlock, 2004). One problem consists in an approximately fourfold lower effective population size for mtDNA compared with nuclear loci, thus rendering mtDNA more susceptible to drift (Birky et al., 1983). However, for Atlantic eels with expectedly very high effective population sizes in the thousands or higher (Côté et al., 2013; Pujolar et al., 2013), we do not expect fourfold lower effective population size at mtDNA as compared with nuclear genes to result in detectably higher drift. The phylogeny also does not indicate bias caused by mtDNA introgression between species. Nevertheless, large female effective population size would cause selection to have a particularly prominent role as compared with neutral processes. As recombination is generally absent in mtDNA, positive selection will affect the whole molecule by hitchhiking, thereby potentially affecting substitution rate and divergence time estimates. However, modelling and simulations suggest that complete linkage to either advantageous or deleterious mutations should not affect the substitution rate of selectively neutral mutations (Birky and Walsh, 1988). Given that FUBAR tests only suggested one positively selected site (Supplementary Table S9) and as estimates of divergence time estimated from 3rd base positions provided results comparable to estimates based on the full data set (Table 3), we tentatively conclude that our results have not been strongly affected by selection.
Comparison of divergence estimates and mutation rates
Divergence time (TMRCA) between European and American eel was estimated to be 3.38 Mya (95% HPD 2.55–4.30), using a data set comprising all mitochondrial regions (Table 3). The estimate appeared robust across analyses using smaller partitions of the data. Also, the separate *BEAST analyses conducted on the American and European eels (Table 3) supported a pre-Pleistocene divergence and furthermore showed little influence of demographic change, which otherwise have been shown to alter divergence time estimates considerably if not taken into account (Ho et al., 2008; Marino et al., 2011).
The fact that the 3rd codon position data resulted in slightly older divergence time estimates (Table 3) may reflect a faster mutation rate leading to slight saturation, supported by the lower percentage of invariable sites compared with the other regions (Supplementary Table S6). This may be a consequence of the deep fossil calibration used, which could lead to overestimation of divergence time. However, analysis based on the slowly evolving tRNAs and rRNA partitions gave almost identical estimates compared with the full data set analysis. The estimated substitution rate of 2.189 × 10−9 sub per site per year of the concatenated rRNAs is within the normal range of rRNA rates observed in other fish species of 0.9 × 10−10–6.0 × 10−9 sub per site per year (Alves-Gomes, 1999; Tringali et al., 1999; Domingues et al., 2005; Pujolar et al., 2012). Similarly, the substitution rate of the whole mitogenome (5.463 × 10−9 sub per site per year) is within the range of rates previously reported for salmonids of 5.0 × 10−9 sub per site per year, estimated from the restriction fragment length polymorphism analysis of mtDNA (Smith, 1992).
Our TMRCA estimate differs considerably from earlier studies. (Avise et al., 1986) estimated the split to 1.5 Mya (Tsukamoto and Aoyama, 1998) to 10.2 Mya and (Minegishe et al., 2005) to 5.8 Mya. However, both Avise et al. (1986) and Minegishe et al. (2005) relied on standard substitution rates, which are potentially misleading as substitution rates can vary considerably among species (Pulquerio and Nichols, 2007). Moreover, while Tsukamoto and Aoyama (1998) calibrated time directly based on their phylogeny, they relied on a Tethys Sea colonization route for the ancestors of the Atlantic eels. The studies based on the most extensive mtDNA data sets identify the closest relatives of Atlantic eels to be the Oceanian species A. dieffenbachii and A. australis (Supplementary Figure S1; Lin et al., 2001; Minegishe et al., 2005; Teng et al., 2009), supporting a scenario where the ancestor of Atlantic eels immigrated more recently through the Panama Gateway before the formation of the Isthmus (Lin et al., 2001; Teng et al., 2009). In total, we suggest that our estimate using the marine eels Serrivomer as outgroup species (Inoue et al., 2010; Johnson et al., 2012) combined with fossil calibration (Patterson, 1993) makes the best use of available data and circumvents the uncertainties of assuming either a Tethys Sea or Panama Gateway dispersal scenario as calibration point.
Divergence time and speciation mechanisms
The time of divergence of 3.38 Mya (95% HPD 2.55–4.30) estimated for Atlantic eels coincides with one of the most important geological events during the past ten million years, namely the formation of the Isthmus of Panama. The closure of the Panama Gateway was a gradual process occurring over millions of years but culminating 4.5–3.0 Mya (Haug and Tiedemann, 1998; Bartoli et al., 2005). Although the precise consequences are debated, it is generally assumed that this led to significant reinforcement of the Gulf Stream, which again may have contributed to the initiation of the Pleistocene cycles of glaciations by increasing evaporation and precipitation leading to increased snow cover in northern regions (Haug and Tiedemann, 1998; Bartoli et al., 2005). Even though the estimated divergence time of European and American eels of 3.38 Mya does not match with the assumption of speciation during the Pleistocene (2.6 Mya–12 Kya) suggested by Avise et al. (1986), a Panamanian speciation scenario, nevertheless, is in accordance with the ‘Oceanic' scenario suggested by Avise et al. (1990). If the ancestor of Atlantic eels dispersed into the Atlantic via the Panama Gateway (Lin et al., 2001; Teng et al., 2009), then an initial distribution in eastern North America would be expected and American eel should be the ancestral species. Reinforcement of the Gulf Stream after closure of the Panama Gateway would subsequently advect eel larvae from the Sargasso Sea to the European and North African coasts.
Two predictions can be made based on the scenario above: (1) If American eel is the ancestral species, then it would also be expected to be the most variable. (2) If European eel is the derived species, experiencing a novel selection regime, then it would be expected that the majority of nonsynonymous substitutions should represent the derived rather than ancestral state in this species. Unless rate differences exist between the two species, they should, however, be equally related to the ancestor at the neutrally evolving synonymous sites leading to a higher dN/dS in European eel.
This first prediction is not supported by this study as American eel showed lower nucleotide diversity than European eel (Table 2). In addition, coalescence time was in fact deeper within European than American eel. However, given a divergence time of 3.38 Myr (95% HPD 2.55–4.30) between the two species, it is likely that the lower nucleotide diversity and more shallow coalescence in American eel may be explained by the subsequent demographic history, as opposed to it being the derived species. This possibility finds support in the EBSP analyses, where American eels consistently showed lower female effective population size (Nef) as opposed to European eel (Figure 3). According to the second prediction, European eel showed more fixed nonsynonymous changes compared with American eel (Figure 2). However, dN/dS estimates were quite similar (0.0338 vs 0.0313) and the findings of more nonsynonymous changes in European eel may indicate an overall slightly faster substitution rate. Thus, to finally evaluate whether American eel is in fact the ancestral species, additional analyses at the nuclear level are required, preferably at a population genomics scale, for example, using RAD (Restriction-Associated DNA) sequencing (Baird et al., 2008; Hohenlohe et al., 2010) as has been previously applied to European eel (Pujolar et al., 2013).
The TMRCA of all freshwater eels was estimated to be around 14 Myr (11.14–16.50) ago (Table 3). This period coincides with the mid-Miocene, a period of particularly drastic global climate change causing major changes in sea level and deep ocean circulation patterns (Flower and Kennett, 1994). This is likely to have affected ancestral freshwater eels, possibly leading to reproductive isolation and speciation, thereby explaining the rapid radiation observed around the TMRCA of the freshwater eels (Minegishe et al., 2005; Teng et al., 2009).
Genetic variation and demographic history
The divergence time estimates provided limited support for speciation in the Pleistocene (2.6 Mya–12 Kya), although given the confidence intervals of the estimates, it cannot be fully refuted. However, the EBSP plots (Figure 3) support that declines occurred during glacial cycles. Hence, range contractions in the Sargasso Sea spawning region are likely to have occurred, as suggested in the ‘Vicariance' scenario, and could certainly have played a role in the later speciation process. This scenario moreover finds support in (1) subfossil data indicating the absence of European eel from Northern Europe during the last glacial maximum ca. 21–11 Kya (Kettle et al., 2008), thus suggesting drastic population decline; (2) microsatellite data suggesting historical declines in both species (Wirth and Bernatchez, 2003) and contemporary population size fluctuations in American eel coinciding with climatic cycles (Côté et al., 2013).
It is curious, although, that the declines in both species predate the Last Glacial Maximum 21–11 Kya where it would have been expected to be most severe (Kettle et al., 2008). Time dependency, where, for example, purifying selection causes substitution rates to decrease over long time spans (Ho et al., 2005) could potentially explain these results. Such an effect is supported by the observation of a high number of nonsynonymous changes in the terminal nodes (Figure 2). In fishes, intraspecific compared with interspecific rates have been estimated to be in the order of 3 to >10 times higher (Genner et al., 2007; Burridge et al., 2008; Jacobsen et al., 2012). Taking such differences in rates into account, it is possible that the decline and subsequent expansion correspond to the Last Glacial Maximum. Nef may have been overestimated for the same reason, although on the other side estimates in the hundreds of thousands or even millions seem possible given very high Ne estimates estimated from RAD sequencing data in European eel (132 000–1 320 000; Pujolar et al., 2013). Thus, the considerably lower estimates derived from microsatellite-based studies of 5–10 000 (Wirth and Bernatchez, 2003; Pujolar et al., 2011) may be a result of homoplasy in the fast evolving microsatellites causing an underestimate of historical Ne (Estoup et al., 2002), although it does not explain lower estimates of contemporary Ne obtained using linkage disequilibrium methods (Côté et al., 2013).
The recent decline suggested by EBSP in American eel is puzzling. Contrary to European eel where all haplotypes were unique, three haplotypes were represented by two or more individuals, amounting to a total of 11 individuals. This is likely to underlie the signal of recent decline, but the question is if the inferred decline is real. Nine of the eleven individuals were found in a single lineage characterized by a nonsynonymous substitution in the ND5 gene (position 540; Figure 2). This position is situated close to two fixed changes between American and European eels (positions 544 and 548; Figure 2) and is situated in a region of the ND5 gene, a ‘piston' arm, which previously has been suggested to be under positive selection in pacific salmon (Oncorhynchus sp.) (Supplementary Figure S2); (Garvin et al., 2011). However, similar mutations were observed in two individuals of independent ancestry (Figure 2) and the derived valine amino acid has almost identical physiochemical properties to the ancestral isoleucine. This does not support positive selection, although it cannot be disproved either. Knowledge on the exact function of the ‘piston' arm, the conformational changes inferred by the mutation and further analysis of American eel demography using nuclear markers are thus needed to support either scenario.
Selection on mitogenomes
Our study shows strong evidence for positive selection at ATP6 between the Atlantic eels as evidenced by both (1) McDonald–Kreitman tests conducted for the Atlantic eel samples (Supplementary Table S10) and (2) the codon-based selection test FUBAR searching for selection across the phylogeny of freshwater eels (Figure 4). The results are in agreement with Gagnaire et al. (2012), who also reported positive selection between American and European eels acting at ATP6. In our study, the single strongest candidate for selection was codon 52 that showed significantly elevated dN/dS in the FUBAR test and furthermore was fixed between European and American eels (Figure 4). As part of Complex V of the oxidative phosphorylation pathway, ATP6 is important for energy (ATP) production and replacement mutations have been shown to affect metabolic rate in humans (Jonckheere et al., 2012) and thermogenesis in white-toothed shrew (Fontanillas et al., 2005). However, also shortening in developmental time has been shown in Drosophila ATP6 mutants (Celotto et al., 2011) and the exact effect of mutations in ATP6 might be quite diverse. Both migratory distances and larval developmental time are highly variable between species of freshwater eels (Tesch, 2003). This is especially evident between Atlantic eels where European eel migrates 5000–6000 km to reach the Sargasso Sea compared with 2000–3000 km for American eel. Accordingly, results indicate a longer larval stage of ca. 2 years as opposed to ca. 7–9 months in American eels (Tesch, 2003), although this is still debated (Tesch, 2003; Zenimoto et al., 2011).
In total, selection may operate at ATP6 across freshwater eels in general and Atlantic eels in particular, rendering it possible that the evolution of ATP6, and thereby the entire mitogenome, is tightly linked to speciation. This is supported by the finding by Gagnaire et al. (2012) of potential cytonuclear incompatibility between the two Atlantic eel species. In the present study, we found evidence of nonsynonymous changes in ATP6 in both European and American direction. Similar results were achieved by Gagnaire et al. (2012), who furthermore found evidence for bi-directional change of the nuclear interactor atp5c1. As a consequence, we hypothesize that the reinforcement of the Gulf Stream following the closure of the Panama Gateway has led to strong differential selection at genes related to ATP synthase function. This would reflect strong selection at the phenotypic level on traits related to migratory endurance and optimal timing of larval development, caused by the different migration distances encountered by the two species.
Conclusions
Using an extensive set of mitogenome data from the two Atlantic freshwater eel species, our study revealed a divergence time of 3.38 Mya (95% HPD 2.54–4.29), which coincides with the closure of the Panama Gateway. Given the tight linkage between the life history of Atlantic eels, the frontal systems in the southern Sargasso Sea and major currents such as the Gulf Stream (McCleave et al., 1987a; Tesch, 2003; van Ginneken and Maes, 2005; Munk et al., 2010), this major geological event and its associated changes of oceanographic patterns provide a strong candidate for mechanisms initiating speciation. Under this scenario, European eel would be expected to be the derived species. However, this could not be conclusively validated in this study and resolving this issue must await further analysis at the nuclear genomic scale. The data also suggested that population declines have occurred during the past glaciations. Thus, vicariance associated with reduced overlap of spawning regions during periods of low population size could have further accelerated the speciation process. Finally, we identified regions in the mitochondrial genome under direct positive selection, which may have been part of the speciation process.
European and Atlantic eels exhibit some of the most peculiar life histories in the Animal Kingdom, with thousands of kilometres separating spawning and foraging regions. Yet, the mechanisms underlying their speciation could be representative also for other marine species. It is well established that pelagic marine fishes often spawn in frontal zones (Sabates et al., 2007; Munk et al., 2009), and ocean currents can have a strong influence on their population genetic structure (White et al., 2010). Major changes of ocean currents could therefore be an important factor to consider when testing hypotheses about speciation in marine organisms.
Data archiving
Sequence data have been submitted to GenBank: accession numbers: KJ564170–KJ564271.
Acknowledgments
We acknowledge the Danish Council for Independent Research, Natural Sciences (Grant 09-072120), Elisabeth og Knud Petersen's Foundation and the Danish National Research Foundation (Grant DNRF94) for funding. We thank Lillian Petersen, Marie-Louise Kampmann, Morten Rasmussen, Karen-Lise D Mensberg and the Danish National High-Throughput Sequencing Centre for technical assistance, Stefan Palm, Russell Poole and Håkan Wickström for providing glass eels, Rasmus Heller, Simon Ho, Pierre-Alexandre Gagnaire, C William Birky and Hans Siegismund for advice on data analysis, Virginia Settepani for assistance in preparing the map and two anonymous reviewers for valuable comments on the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Albert V, Jonsson B, Bernatchez L. Natural hybrids in Atlantic eels (Anguilla anguilla, A-rostrata): evidence for successful reproduction and fluctuating abundance in space and time. Mol Ecol. 2006;15:1903–1916. doi: 10.1111/j.1365-294X.2006.02917.x. [DOI] [PubMed] [Google Scholar]
- Als TD, Hansen MM, Maes GE, Castonguay M, Riemann L, Aarestrup K, et al. All roads lead to home: panmixia of European eel in the Sargasso Sea. Mol Ecol. 2011;20:1333–1346. doi: 10.1111/j.1365-294X.2011.05011.x. [DOI] [PubMed] [Google Scholar]
- Alves-Gomes JA. Systematic biology of gymnotiform and mormyriform electric fishes: phylogenetic relationships, molecular clocks and rates of evolution in the mitochondrial rRNA genes. J Exp Biol. 1999;202:1167–1183. doi: 10.1242/jeb.202.10.1167. [DOI] [PubMed] [Google Scholar]
- Avise JC, Ball RM, Arnold J. Current versus historical population sizes in vertebrate species with high gene flow—a comparison based on mitochondrial-DNA lineages and inbreeding theory for neutral mutations. Mol Biol Evol. 1988;5:331–344. doi: 10.1093/oxfordjournals.molbev.a040504. [DOI] [PubMed] [Google Scholar]
- Avise JC, Helfman GS, Saunders NC, Hales LS. Mitochondrial DNA differentiation in North Atlantic eels: population genetic consequences of an unusual life-history pattern. Proc Natl Acad Sci USA. 1986;83:4350–4354. doi: 10.1073/pnas.83.12.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avise JC, Nelson WS, Arnold J, Koehn RK, Williams GC, Thorsteinsson V. The evolutionary genetic status of Icelandic eels. Evolution. 1990;44:1254–1262. doi: 10.1111/j.1558-5646.1990.tb05229.x. [DOI] [PubMed] [Google Scholar]
- Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, et al. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE. 2008;3:e3376. doi: 10.1371/journal.pone.0003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard JWO, Kreitman M. Is mitochondrial-DNA a strictly neutral marker. Trends Ecol Evol. 1995;10:485–488. doi: 10.1016/s0169-5347(00)89195-8. [DOI] [PubMed] [Google Scholar]
- Ballard JWO, Whitlock MC. The incomplete natural history of mitochondria. Mol Ecol. 2004;13:729–744. doi: 10.1046/j.1365-294x.2003.02063.x. [DOI] [PubMed] [Google Scholar]
- Bartoli G, Sarnthein M, Weinelt M, Erlenkeuser H, Garbe-Schonberg D, Lea DW. Final closure of Panama and the onset of northern hemisphere glaciation. Earth Planet Sci Lett. 2005;237:33–44. [Google Scholar]
- Bernardi G. Speciation in fishes. Mol Ecol. 2013;22:5487–5502. doi: 10.1111/mec.12494. [DOI] [PubMed] [Google Scholar]
- BioMatters 2012. Geneious version 5.4.6. Downloaded at 21 July 2011. Available at: http://www.geneious.com
- Birky CW, Maruyama T, Fuerst P. An approach to population and evolutionary genetic theory for genes in mitochondria and chloroplasts, and some results. Genetics. 1983;103:513–527. doi: 10.1093/genetics/103.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birky CW, Walsh JB. Effects of linkage on rates of molecular evolution. Proc Natl Acad Sci USA. 1988;85:6414–6418. doi: 10.1073/pnas.85.17.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge CP, Craw D, Fletcher D, Waters JM. Geological dates and molecular rates: fish DNA sheds light on time dependency. Mol Biol Evol. 2008;25:624–633. doi: 10.1093/molbev/msm271. [DOI] [PubMed] [Google Scholar]
- Celotto AM, Chiu WK, Van Voorhies W, Palladino MJ. Modes of metabolic compensation during mitochondrial disease using the Drosophila model of ATP6 dysfunction. PLoS ONE. 2011;6:e25823. doi: 10.1371/journal.pone.0025823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. Speciation. Sinnauer Associates: Sunderland, MA, USA; 2004. [Google Scholar]
- Cummings MP, Neel MC, Shaw KL. A genealogical approach to quantifying lineage divergence. Evolution. 2008;62:2411–2422. doi: 10.1111/j.1558-5646.2008.00442.x. [DOI] [PubMed] [Google Scholar]
- Côté CL, Gagnaire PA, Bourret V, Verreault G, Castonguay M, Bernatchez L. Population genetics of the American eel (Anguilla rostrata): FST=0 and North Atlantic Oscillation effects on demographic fluctuations of a panmictic species. Mol Ecol. 2013;22:1763–1776. doi: 10.1111/mec.12142. [DOI] [PubMed] [Google Scholar]
- Domingues VS, Bucciarelli G, Almada VC, Bernardi G. Historical colonization and demography of the Mediterranean damselfish, Chromis chromis. Mol Ecol. 2005;14:4051–4063. doi: 10.1111/j.1365-294X.2005.02723.x. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A, Shapiro B, Pybus OG. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol. 2005;22:1185–1192. doi: 10.1093/molbev/msi103. [DOI] [PubMed] [Google Scholar]
- Estoup A, Jarne P, Cornuet JM. Homoplasy and mutation model at microsatellite loci and their consequences for population genetics analysis. Mol Ecol. 2002;11:1591–1604. doi: 10.1046/j.1365-294x.2002.01576.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Feder JL, Egan SP, Nosil P. The genomics of speciation-with-gene-flow. Trends Genet. 2012;28:342–350. doi: 10.1016/j.tig.2012.03.009. [DOI] [PubMed] [Google Scholar]
- Flower BP, Kennett JP. The middle Miocene climatic transition—East Antarctic ice-sheet development, deep-ocean circulation and global carbon cycling. Palaeogeogra Palaeoclimatol Palaeoecology. 1994;108:537–555. [Google Scholar]
- Fontanillas P, Depraz A, Giorgi MS, Perrin N. Nonshivering thermogenesis capacity associated to mitochondrial DNA haplotypes and gender in the greater white-toothed shrew, Crocidura russula. Mol Ecol. 2005;14:661–670. doi: 10.1111/j.1365-294X.2004.02414.x. [DOI] [PubMed] [Google Scholar]
- Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnaire PA, Albert V, Jonsson B, Bernatchez L. Natural selection influences AFLP intraspecific genetic variability and introgression patterns in Atlantic eels. Mol Ecol. 2009;18:1678–1691. doi: 10.1111/j.1365-294X.2009.04142.x. [DOI] [PubMed] [Google Scholar]
- Gagnaire PA, Normandeau E, Bernatchez L. Comparative genomics reveals adaptive protein evolution and a possible cytonuclear incompatibility between European and American eels. Mol Biol Evol. 2012;29:2909–2919. doi: 10.1093/molbev/mss076. [DOI] [PubMed] [Google Scholar]
- Garvin MR, Bielawski JP, Gharrett AJ. Positive Darwinian selection in the piston that powers proton pumps in complex i of the mitochondria of Pacific salmon. PLoS ONE. 2011;6:e24127. doi: 10.1371/journal.pone.0024127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genner MJ, Seehausen O, Lunt DH, Joyce DA, Shaw PW, Carvalho GR, et al. Age of cichlids: new dates for ancient lake fish radiations. Mol Biol Evol. 2007;24:1269–1282. doi: 10.1093/molbev/msm050. [DOI] [PubMed] [Google Scholar]
- Haug GH, Tiedemann R. Effect of the formation of the Isthmus of Panama on Atlantic Ocean thermohaline circulation. Nature. 1998;393:673–676. [Google Scholar]
- Heled J, Drummond AJ. Bayesian inference of population size history from multiple loci. BMC Evol Biol. 2008;8:289. doi: 10.1186/1471-2148-8-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heled J, Drummond AJ. Bayesian inference of species trees from multilocus data. Mol Biol Evol. 2010;27:570–580. doi: 10.1093/molbev/msp274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SYW, Larson G, Edwards CJ, Heupink TH, Lakin KE, Holland PWH, et al. Correlating Bayesian date estimates with climatic events and domestication using a bovine case study. Biol Lett. 2008;4:370–374. doi: 10.1098/rsbl.2008.0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SYW, Phillips MJ, Cooper A, Drummond AJ. Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol Biol Evol. 2005;22:1561–1568. doi: 10.1093/molbev/msi145. [DOI] [PubMed] [Google Scholar]
- Hohenlohe PA, Bassham S, Etter PD, Stiffler N, Johnson EA, Cresko WA. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genet. 2010;6:23. doi: 10.1371/journal.pgen.1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue JG, Miya M, Miller MJ, Sado T, Hanel R, Hatooka K, et al. Deep-ocean origin of the freshwater eels. Biol Lett. 2010;6:363–366. doi: 10.1098/rsbl.2009.0989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen MW, Hansen MM, Orlando L, Bekkevold D, Bernatchez L, Willerslev E, et al. Mitogenome sequencing reveals shallow evolutionary histories and recent divergence time between morphologically and ecologically distinct European whitefish (Coregonus spp.) Mol Ecol. 2012;21:2727–2742. doi: 10.1111/j.1365-294X.2012.05561.x. [DOI] [PubMed] [Google Scholar]
- Johnson GD, Ida H, Sakaue J, Sado T, Asahida T, Miya M. A 'living fossil' eel (Anguilliformes: Protanguillidae, fam. nov.) from an undersea cave in Palau. Philos R Soc Ser B. 2012;279:934–943. doi: 10.1098/rspb.2011.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonckheere AI, Smeitink JAM, Rodenburg RJT. Mitochondrial ATP synthase: architecture, function and pathology. J Inherit Metab Dis. 2012;35:211–225. doi: 10.1007/s10545-011-9382-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettle AJ, Heinrich D, Barrett JH, Benecke N, Locker A. Past distributions of the European freshwater eel from archaeological and palaeontological evidence. Quat Sci Rev. 2008;27:1309–1334. [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Lin YS, Poh YP, Tzeng CS. A phylogeny of freshwater eels inferred from mitochondrial genes. Mol Phylogenet Evol. 2001;20:252–261. doi: 10.1006/mpev.2001.0969. [DOI] [PubMed] [Google Scholar]
- Mank JE, Avise JC. Microsatellite variation and differentiation in North Atlantic eels. J Hered. 2003;94:310–314. doi: 10.1093/jhered/esg062. [DOI] [PubMed] [Google Scholar]
- Marino IAM, Pujolar JM, Zane L. Reconciling deep calibration and demographic history: Bayesian inference of post glacial colonization patterns in Carcinus aestuarii (Nardo, 1847) and C. maenas (Linnaeus, 1758) PLoS ONE. 2011;6:e28567. doi: 10.1371/journal.pone.0028567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr E. Animal Species and Evolution. Harvard University Press: Cambridge, MA, USA; 1963. [Google Scholar]
- McCleave JD, Kleckner RC, Castonguay M. Reproductive sympatry of American and European eels and implications for migration and taxonomy. Am Fish Soc Symp. 1987a;1:286–297. [Google Scholar]
- McCleave JD, Kleckner RC, Castonguay M. Reproductive sympatry of American and European eels and implications for migration and taxonomy. Am Fish Soc Symp. 1987b;1:286–297. [Google Scholar]
- Mcdonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Minegishe Y, Aoyama J, Inoue JG, Miya M, Nishida M, Tsukamoto K. Molecular phylogeny and evolution of the freshwater eels genus Anguilla based on the whole mitochondrial genome sequences. Mol Phylogenet Evol. 2005;34:134–146. doi: 10.1016/j.ympev.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Munk P, Fox CJ, Bolle LJ, van Damme CJG, Fossum P, Kraus G. Spawning of North Sea fishes linked to hydrographic features. Fish Oceanogr. 2009;18:458–469. [Google Scholar]
- Munk P, Hansen MM, Maes GE, Nielsen TG, Castonguay M, Riemann L, et al. Oceanic fronts in the Sargasso Sea control the early life and drift of Atlantic eels. Proc R Soc Ser B. 2010;277:3593–3599. doi: 10.1098/rspb.2010.0900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell B, Moola S, Mabona A, Weighill T, Sheward D, Pond SLK, et al. FUBAR: A Fast, Unconstrained Bayesian AppRoximation for Inferring Selection. Mol Biol Evol. 2013;30:1196–1205. doi: 10.1093/molbev/mst030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm S, Dannewitz J, Prestegaard T, Wickstrom H. Panmixia in European eel revisited: no genetic difference between maturing adults from southern and northern Europe. Heredity. 2009;103:82–89. doi: 10.1038/hdy.2009.51. [DOI] [PubMed] [Google Scholar]
- Patterson C. Osteichthyes: Teleostei. Chapman & Hall: London, UK; 1993. [Google Scholar]
- Pond SLK, Frost SDW. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- Pond SLK, Posada D, Gravenor MB, Woelk CH, Frost SDW. Automated phylogenetic detection of recombination using a genetic algorithm. Mol Biol Evol. 2006;23:1891–1901. doi: 10.1093/molbev/msl051. [DOI] [PubMed] [Google Scholar]
- Posada D. jModelTest: Phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Pujolar JM, Bevacqua D, Capoccioni F, Ciccotti E, De Leo GA, Zane L. No apparent genetic bottleneck in the demographically declining European eel using molecular genetics and forward-time simulations. Conserv Genet. 2011;12:813–825. [Google Scholar]
- Pujolar JM, Jacobsen MW, Als TD, Frydenberg J, Magnussen E, Jonsson B, et al. Assessing patterns of hybridization between North Atlantic eels using diagnostic single nucleotide polymorphisms. Heredity. 2014;112:627–637. doi: 10.1038/hdy.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujolar JM, Jacobsen MW, Frydenberg J, Als TD, Larsen PF, Maes GE, et al. A resource of genome-wide single-nucleotide polymorphisms generated by RAD tag sequencing in the critically endangered European eel. Mol Ecol Resour. 2013;13:706–714. doi: 10.1111/1755-0998.12117. [DOI] [PubMed] [Google Scholar]
- Pujolar JM, Zane L, Congiu L. Phylogenetic relationships and demographic histories of the Atherinidae in the Eastern Atlantic and Mediterranean Sea re-examined by Bayesian inference. Mol Phylogenet Evol. 2012;63:857–865. doi: 10.1016/j.ympev.2012.02.027. [DOI] [PubMed] [Google Scholar]
- Pulquerio MJF, Nichols RA. Dates from the molecular clock: how wrong can we be. Trends Ecol Evol. 2007;22:180–184. doi: 10.1016/j.tree.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Sabates A, Olivar MP, Salat J, Palomera I, Alemany F. Physical and biological processes controlling the distribution of fish larvae in the NW Mediterranean. Prog Oceanogr. 2007;74:355–376. [Google Scholar]
- Smith GR. Introgression in fishes—significance for Paleontology, Cladistics, and Evolutionary Rates. Syst Biol. 1992;41:41–57. [Google Scholar]
- Sun YB, Shen YY, Irwin DM, Zhang YP. Evaluating the roles of energetic functional constraints on Teleost mitochondrial-encoded protein evolution. Mol Biol Evol. 2011;28:39–44. doi: 10.1093/molbev/msq256. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng HY, Lin YS, Tzeng CS. A new Anguilla species and a reanalysis of the phylogeny of freshwater eels. Zool Stud. 2009;48:808–822. [Google Scholar]
- Tesch F. The Eel. Blackwell Science Lt: Oxford, UK; 2003. [Google Scholar]
- Tringali MD, Bert TM, Seyoum S, Bermingham E, Bartolacci D. Molecular phylogenetics and ecological diversification of the transisthmian fish genus Centropomus (Perciformes: Centropomidae) Mol Phylogenet Evol. 1999;13:193–207. doi: 10.1006/mpev.1999.0624. [DOI] [PubMed] [Google Scholar]
- Tsukamoto K, Aoyama J. Evolution of freshwater eels of the genus Anguilla: a probable scenario. Environ Biol Fish. 1998;52:139–148. [Google Scholar]
- Tsukamoto K, Aoyama J, Miller MJ. Migration, speciation, and the evolution of diadromy in anguillid eels. Can J Fish Aquat Sci. 2002;59:1989–1998. [Google Scholar]
- van Ginneken VJT, Maes GE. The european eel (Anguilla anguilla, Linnaeus), its lifecycle, evolution and reproduction: a literature review. Rev Fish Biol Fisher. 2005;15:367–398. [Google Scholar]
- Via S. Sympatric speciation in animals: the ugly duckling grows up. Trends Ecol Evol. 2001;16:381–390. doi: 10.1016/s0169-5347(01)02188-7. [DOI] [PubMed] [Google Scholar]
- Vollestad LA. Geographic-variation in age and length at metamorphosis of maturing European eel—environmental-effects and phenotypic plasticity. J Anim Ecol. 1992;61:41–48. [Google Scholar]
- Ward RD, Woodwark M, Skibinski DOF. A comparison of genetic diversity levels in marine, fresh-water, and anadromous fishes. J Fish Biol. 1994;44:213–232. [Google Scholar]
- White C, Selkoe KA, Watson J, Siegel DA, Zacherl DC, Toonen RJ. Ocean currents help explain population genetic structure. Philos R Soc Ser B. 2010;277:1685–1694. doi: 10.1098/rspb.2009.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth T, Bernatchez L. Decline of North Atlantic eels: a fatal synergy. Philos R Soc Ser B. 2003;270:681–688. doi: 10.1098/rspb.2002.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Xie Z. DAMBE: Software package for data analysis in molecular biology and evolution. J Hered. 2001;92:371–373. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- Xia XH, Xie Z, Salemi M, Chen L, Wang Y. An index of substitution saturation and its application. Mol Phylogenet Evol. 2003;26:1–7. doi: 10.1016/s1055-7903(02)00326-3. [DOI] [PubMed] [Google Scholar]
- Zenimoto K, Sasai Y, Sasaki H, Kimura S. Estimation of larval duration in Anguilla spp., based on cohort analysis, otolith microstructure, and Lagrangian simulations. Mar Ecol Prog Ser. 2011;438:219–228. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.