Abstract
Podocytes are kidney cells with specialized morphology that is required for glomerular filtration. Diseases, such as diabetes, or drug exposure that causes disruption of the podocyte foot process morphology results in kidney pathophysiology. Proteomic analysis of glomeruli isolated from rats with puromycin-induced kidney disease and control rats indicated that protein kinase A (PKA), which is activated by adenosine 3′,5′-monophosphate (cAMP), is a key regulator of podocyte morphology and function. In podocytes, cAMP signaling activates cAMP response element–binding protein (CREB) to enhance expression of the gene encoding a differentiation marker, synaptopodin, a protein that associates with actin and promotes its bundling. We constructed and experimentally verified a β-adrenergic receptor–driven network with multiple feedback and feedforward motifs that controls CREB activity. To determine how the motifs interacted to regulate gene expression, we mapped multicompartment dynamical models, including information about protein subcellular localization, onto the network topology using Petri net formalisms. These computational analyses indicated that the juxtaposition of multiple feedback and feedforward motifs enabled the prolonged CREB activation necessary for synaptopodin expression and actin bundling. Drug-induced modulation of these motifs in diseased rats led to recovery of normal morphology and physiological function in vivo. Thus, analysis of regulatory motifs using network dynamics can provide insights into pathophysiology that enable predictions for drug intervention strategies to treat kidney disease.
INTRODUCTION
Kidney podocytes are terminally differentiated epithelial cells with a specialized morphology of interdigitating foot processes that form the slit diaphragm, which constitutes a critical layer of the glomerular filtration barrier (1). Some diseases, exposure to certain toxins, or clinically used drugs disrupt this morphology through a process called effacement of foot processes, which results in breach of the filtration barrier and increased urinary protein excretion. Increased protein in the urine, called proteinuria, is an early clinical indicator of kidney injury (2). Little is known about the molecular mechanisms that produce the loss of the differentiated state and how it can be reversed. Understanding how control of cellular regulation relates to podocyte morphology and consequently to physiological function will enable the development of treatment strategies for kidney disease.
A well-established and widely used animal model for kidney disease is puromycin-induced nephropathy, which is associated with foot process effacement and proteinuria (3). We performed proteomic analysis of glomeruli from control rats and rats with puromycin-induced nephropathy to assess disease-induced differences in the proteome. Through network analysis of the proteomic data and kinases that could target the altered proteins, we identified protein kinases associated with the differentiated state. In particular, we found that the adenosine 3′,5′-monophosphate (cAMP) to protein kinase A (PKA) network had a central role in defining the biochemical state associated with normal podocyte morphology and function in vivo.
We wanted to understand how the network topology and network motifs, such as feedback and feedforward loops, could contribute to the establishment and maintenance of the differentiated and morphologically distinct state of the functional podocyte. In many cellular contexts, prolonged activation of signaling proteins and transcription factors arises from bistability resulting from positive feedback loops (FBLs) (4–6) or from the presence of feedforward motifs (FFMs) (7). Positive FBLs can act as switches to enable physiological processes as diverse as long-term depression in neurons (8), reentry into the cell cycle in frog oocytes (9), and hunger control in mice (10). Because podocyte differentiation and expression of the differentiation marker synaptopodin require prolonged cAMP response element–binding protein (CREB) activation (11), we hypothesized that a network of interconnected feedback and feedforward loops centered on PKA and ending in CREB activation could define the biochemical state associated with foot process morphology and normal physiological function. Here, we describe how construction of a network of interconnected feedback and FFMs can define biochemical states associated with normal morphology and function, and how this information can be used to treat pathological processes.
RESULTS
Proteomic analysis of differentiated and injured podocytes
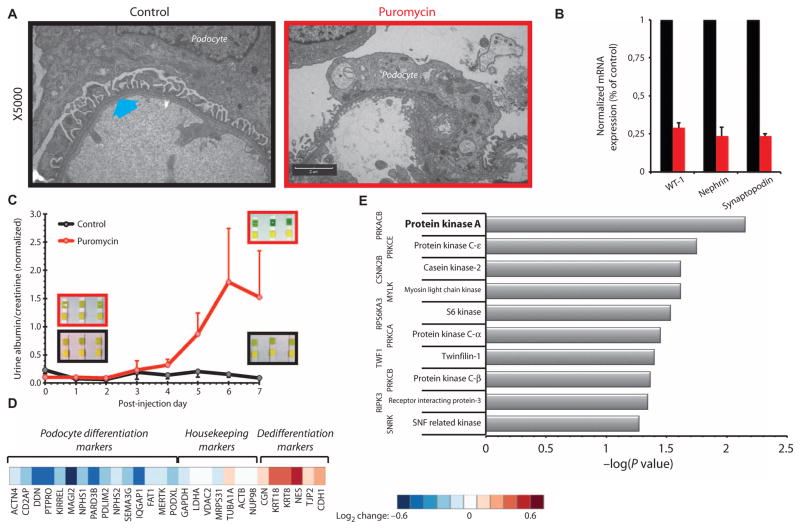
Treatment of rats with puromycin resulted in loss of differentiated podocyte morphology and effacement of foot processes (Fig. 1A). Isolated glomeruli had decreased expression of the differentiation markers encoding WT-1 and synaptopodin, as well as slit diaphragm marker nephrin (Fig. 1B). Consistent with loss of glomerular barrier function, rats treated with puromycin exhibited proteinuria, detected as increased albumin in the urine (Fig. 1C). Shotgun proteomics with amine-specific isobaric tags (12) identified more than 2800 unique proteins, 170 of which exhibited differential abundance in the glomeruli of puromycin-injected animals (91 down and 79 up; z test, 0.05) (table S1). Within these proteins, known podocyte differentiation markers were significantly reduced (down-regulated), whereas dedifferentiation markers, such as epithelial tight junction proteins, were increased (up-regulated) (Fig. 1D). Furthermore, according to the Mouse Genome Informatics (MGI) Pathways (13), the 91 down-regulated components were significantly enriched for mouse phenotype terms “abnormal urine homeostasis” (MP9643), “abnormal kidney morphology” (MP2135), and “abnormal kidney physiology” (MP2136), in agreement with loss of specialized podocyte morphology (fig. S1).
Fig. 1. Proteomic analysis indicates that PKA is important for differentiated podocyte morphology in vivo.
(A) Transmission electron microscopy images of glomerular podocytes from control [phosphate-buffered saline (PBS)–injected] and puromycin-induced nephropathy rats (×5000). Interdigitating foot processes in the control images are marked with a cyan arrow and are absent from the electron micrographs of puromycin-injected animals. Scale bar, 2 μm. (B) Glyceraldehyde-3-phosphate dehydrogenase (GAPDH)–normalized expression of podocyte differentiation markers WT-1, nephrin, and synaptopodin in control (black) and puromycin-injected (red) rats, as quantified by reverse transcription polymerase chain reaction [RT-PCR; all values are means ± SEM; P < 0.05, one-way analysis of variance (ANOVA); n = 3 rats]. (C) Loss of podocyte morphology is associated with functional impairment as quantified by the significant increase of protein in the urine (P < 0.05, two-way ANOVA; n = 3 rats). Insets: Urinalysis results with Chemstrip indicate clinical proteinuria when top strips turn from yellow to deep green. (D) Heatmap of changes in the abundance of known podocyte-associated markers as determined by proteomics. (E) Negative log of P values for the top 10 highest ranked upstream protein kinases computationally identified by X2K using differentially expressed proteins as input nodes.
Identification of network that may promote the podocyte differentiation
We used the 91 down-regulated proteins as seed nodes and the kinase enrichment analysis (KEA) (14) module of the Expression2Kinases (X2K) suite (15) to connect these proteins to potentially relevant protein kinases. In this analysis, PKA ranked as the highest protein kinase in one experiment (Fig. 1E) and the second highest in an independent second experiment (fig. S2). Construction of a protein-protein interaction network, using the 91 down-regulated proteins as seed nodes and adding one intermediate on the basis of known interactions (16), produced a highly connected network of 212 proteins (fig. S3). KEA also identified PKA as a significantly enriched protein kinase for this protein-protein interaction network. The proteomic data confirmed that the abundance of PKA subunits was not changed (table S2), indicating that the regulation of PKA was mediated by changes in its catalytic activity, not its abundance. Together, all of these analyses indicate that the cAMP-PKA signaling could be important for podocyte differentiation and function, which is in agreement with the importance of the cAMP pathway for differentiation of podocytes (17). Synaptopodin, a CREB-regulated gene product, is an actin-binding protein associated with differentiated podocytes in vivo (11, 18), and expression of synaptopodin is a marker of differentiation of podocytes in culture (19). We hypothesized that a cAMP network could be part of the regulatory mechanisms required for the differentiated state of podocytes and constructed a signaling network from the β-adrenergic receptor, which signals through cAMP and PKA, to CREB to understand how PKA might contribute to the differentiated state.
The amount and duration of transcription factor activities are widely used as markers of differentiation in many cell types. We hypothesized that upon β-adrenergic receptor stimulation, the duration of CREB activity could be an indicator of the cell state, with longer durations of CREB activity indicating differentiation and shorter times indicating loss of the differentiated state. The cAMP pathway can stimulate CREB activity through at least two mechanisms. First, cAMP binds to the regulatory subunits and releases the catalytic subunits of PKA to translocate to the nucleus, where they phosphorylate CREB at Ser133 (20). Second, cAMP can bind and activate EPAC, a guanine nucleotide–binding protein exchange factor (GEF), which ultimately stimulates the mitogen-activated protein kinase (MAPK) cascade to stimulate the MAPK substrate mitogen- and stress-activated kinase-1 (MSK-1). MSK-1 can also phosphorylate CREB at Ser133 (21). Thus, the activation of PKA and MAPK by cAMP forms a set of nested FFMs that converge on CREB, and the activity through either or both of these subpathways could promote podocyte differentiation.
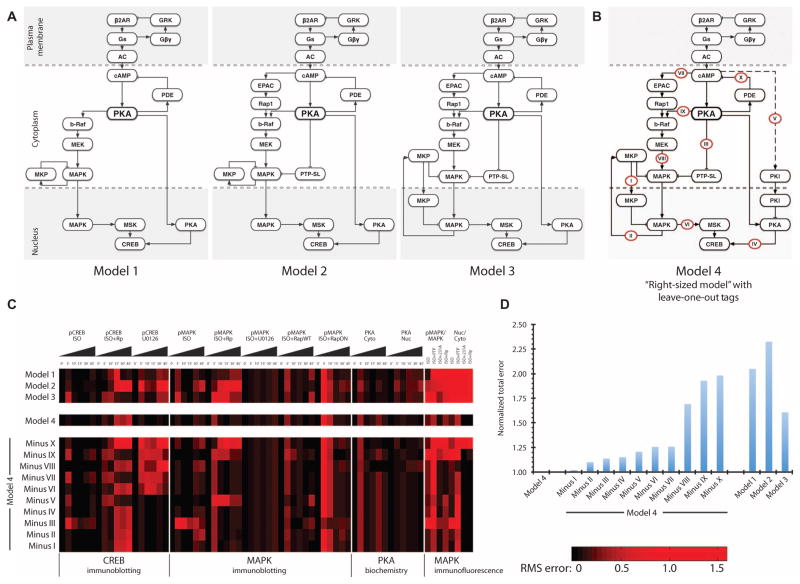
Network models with varying amounts of detail can be developed to determine the mechanistic relationship between β-adrenergic receptor activation and the duration of CREB activation. To identify the minimum number of components and interactions necessary to describe the appropriate amplitude and duration of CREB activation that we experimentally observed in podocytes, we used an iterative model-building process. We parameterized the model as a multicompartment system of ordinary differential equations (ODEs) using either previously published values for the parameters or values constrained by experimental data (table S3). Once these kinetic parameters were set, we did not change them during the simulations. Individual components were progressively added to the network (Fig. 2A, models 1 to 3) and compared against constraints presented by the experimental measurements (fig. S4). We started with the assumption of PKA as the primary regulator of CREB activity (Fig. 2A, model 1), which failed to account for the transient CREB activity during PKA or MEK (mitogen-activated or extracellular signal–regulated protein kinase kinase) inhibition (fig. S5). We investigated the role of distributed information processing by including the cAMP-GEF EPAC and the tyrosine phosphatase PTP-SL in model 2 (Fig. 2A) and then spatial compartmentalization of MKP in model 3 (Fig. 2A). These models (models 1 to 3) failed to account for transient CREB activity during MAPK inhibition with the MEK inhibitor U0126. To account for the transient CREB activity in the presence of the MEK inhibitor, which we attributed to the activity of a PKA inhibitor, we added the PKA inhibitor PKI (22) as an inhibitor of PKA catalytic activity in the nucleus to model 4 (Fig. 2B). We also used MAPK phosphorylation in response to the EPAC-specific agonist 8-pMeOPT-2′-O-Me-cAMP or the PKA-selective agonist 6-Bnz-cAMP in the presence or absence of U0126 and Rp-cAMPs to validate the network topology (Fig. 2C and fig. S4).
Fig. 2. Construction of sign-specified directed cAMP signaling network from receptor to CREB.
(A) Directed graphs of initial three models (models 1, 2, and 3) with increasing number of components used in the iterative model-building process. (B) The final right-sized model (model 4) with labels on individual reactions tested using a leave-one-out scheme for their effects on CREB and MAPK activity or MAPK and PKA translocation shown in (C). Deviation from experimental observations was quantified at each time point individually. Roman numerals represent the edges (reactions) that were tested. (C) Heatmap of RMS errors between individual computational models and the experimental observations. Each voxel represents the RMS error at an independent spatial or temporal data point of a given model. CREB (pCREB) and MAPK (pMAPK) activities were experimentally tested by immunoblotting; PKA activity in the nucleus (Nuc) and cytoplasm (Cyto) by PKA enzymatic assay (biochemistry); relative amount of pMAPK to total MAPK and the activity in the nucleus versus the cytoplasm were measured by immunofluorescence. Rows represent the different models, and the columns represent different data points. Triangles represent time course experiments (0 to 45 min). (D) Total error of all computational models from experimental observations as quantified by the sum of RMS errors in a given model divided by that of the right-sized model.
Simulation results of this last version best represented the experimental data (fig. S4B), so we called model 4 the “right-sized model” because it represents the minimum components and interactions needed to mathematically describe the experimental observations. Removal of components from this model resulted in simulation time courses that were substantially different from that observed experimentally. Overall, the deviation of simulated results from the experiments as quantified by the root mean square (RMS) error was the least for model 4 (Fig. 2, C and D). Although the RMS error analysis suggested that model 3 may also have represented the data, we did not consider this the correct model because it produced substantial deviations from the immunofluorescence experiments measuring MAPK translocation, indicating the importance of compartmentalization in our model. Model 4 had RMS errors approaching or above 0.5 for some of the observed data. These were differences in the simulation with data from individual time points and did not affect the overall shape of the time courses; thus, they were considered less important (fig. S4 and Fig. 2C). Although this was not a formal identifiability analysis, together, the experimental and computational data indicated that we achieved the minimum complexity necessary to describe the system.
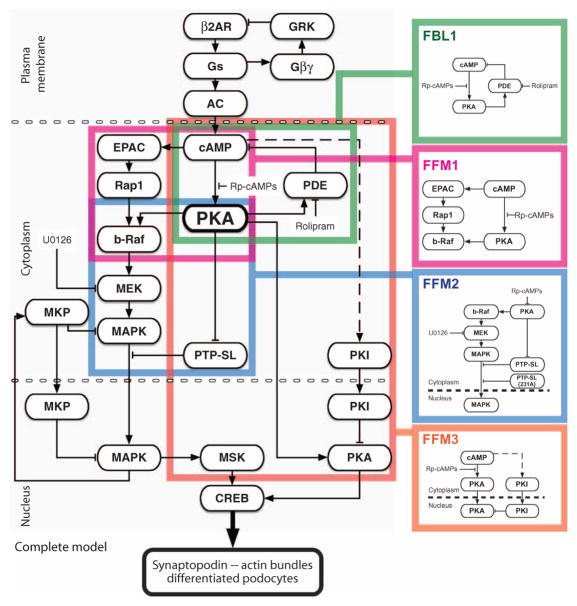
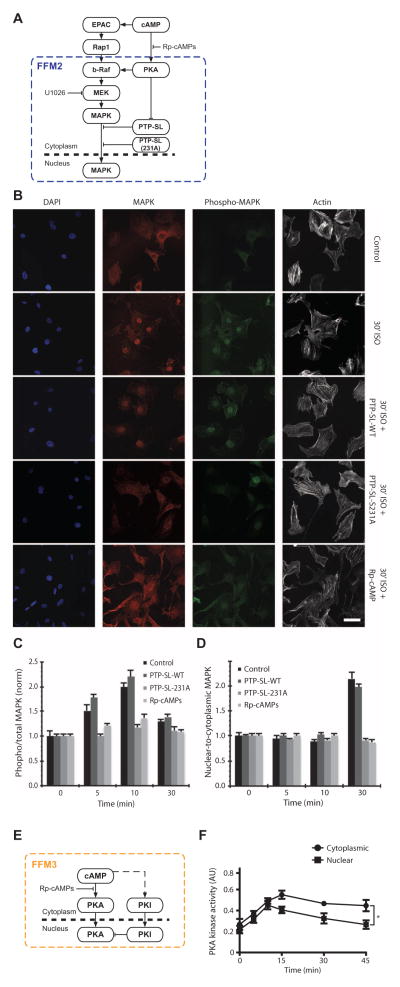
The right-sized model contained four nested and interconnected network motifs (Fig. 3). The first FFM represents cAMP-dependent activation of b-Raf and MEK through PKA and EPAC. The second FFM represents the activation of MAPK and the inhibition of its inhibitor PTP-SL (and other similar isoforms) by cAMP through PKA. FFM2 controls the translocation of MAPK and so can be predicted to contribute to the spatial information required to elicit an effect on gene expression associated with the change in podocyte cell fate as related to CREB activity. The third FFM controls the activity of PKA inside the nucleus: One path is the translocation of the PKA catalytic subunit from the cytoplasm, and the second path is the translocation of PKI from the cytoplasm into the nucleus (22). FFM3 is an incoherent motif with positive and negative effects on the output (the amount of activated PKA in the nucleus). The molecular mechanism for the cAMP-dependent activation of the PKA inhibitor is unknown. FBL1 reduces cAMP concentration through PKA-mediated activation of phosphodiesterases (PDEs) (23), thereby limiting activation of CREB through either the PKA or the EPAC paths.
Fig. 3. Minimally complex directed graph of the cAMP signaling network that controls CREB phosphorylation in the context of podocyte synaptopodin expression and actin bundling.
The right-sized model from Fig. 2B is marked with color-coded boxes to highlight the four key regulatory motifs; these motifs are shown separately on the right for clarity.
We performed simulations to determine the effects of the highlighted network motifs on the relationship between receptor activation and the duration of CREB activation. We analyzed the duration and amplitude of the activity of individual network motifs to understand how the signaling information from receptor to transcription factor is processed through the multiple interacting regulatory motifs to control the duration of CREB activity. To determine whether these motifs operated in a simultaneous or sequential fashion and to determine the relative contributions of the feed-forward loops to the amplitude and duration of CREB activity, we created network representations of the simulation results using a computational method based on Petri nets (fig. S6 and Supplementary Methods: Dynamic Graphs) (24). The resultant network representations (fig. S7), which we call “dynamic graphs,” enabled the analysis of the dynamics and characteristics of motifs, not just the individual signaling components, and provided an interactive display of the relative activity of the signaling components in the motif and the relative intensity of their interactions as outlined by the kinetic parameters.
Of the 106 parameters (74 reaction, 8 transport, and 20 initial concentration parameters), 68% were obtained from previously published studies that included biochemical data obtained from isolated proteins or in-cell assays, 17% were constrained using experimental data from podocytes, 9% were constrained using time courses from literature, and 6% were estimated on the basis of values of similar biochemical parameters (table S3). Because the pathways studied here are well conserved, it is likely that these parameters closely represent actual values in multiple cell types (25, 26). Nevertheless, we performed parameter sensitivity analysis (table S4), which indicated that the model was robust; the maximum variation that we observed was a difference of −3.192 and −2.0471 for the initial concentrations of PDE and PP2A, respectively, and a difference of 1.8309 and −1.831 for the forward and reverse rates of the interaction between Gαs and adenylyl cyclase (reaction 71, table S3) (27). These proteins were not identified as differentially expressed in our proteomics analysis (table S1).
Effects of inhibiting specific network motifs on indicators of podocyte differentiation
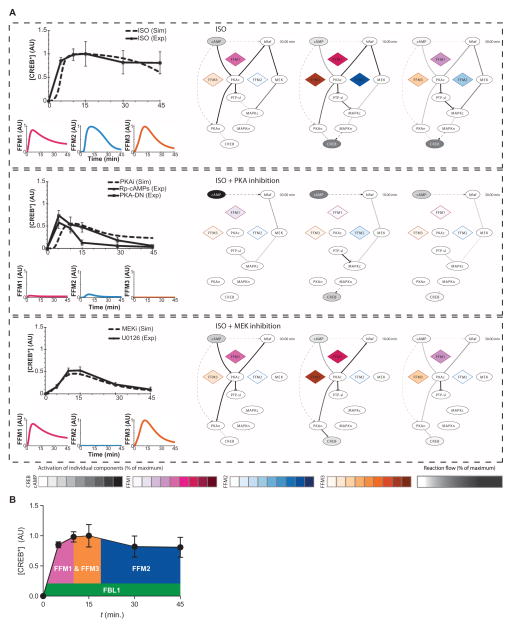
Simulating CREB phosphorylation (activity) in response to activation of the β-adrenergic receptor, dynamic graphs revealed the order in which the activity through FFM1, FFM2, and FFM3 occurred in the network (Fig. 4 and fig. S7). The simulations indicated that, upon receptor activation, PKA would be the first signal to phosphorylate CREB, and then MAPK would phosphorylate CREB (Fig. 4A, top). Once most of the nuclear PKA activities are inhibited, the signal from MAPK would sustain CREB phosphorylation. When PKA is inhibited (for example, by Rp-cAMPs), the simulations indicated that the input to CREB would come from cAMP through the EPAC pathway to MAPK (fig. S7), but CREB phosphorylation would be attenuated and shortened due to the loss of the PKA-mediated inhibition of the phosphatase PTP-SL (and its isoforms), thus resulting in transient CREB activation (Fig. 4A, middle). When MEK is inhibited (for example, by U0126), the simulations indicated that the only input to CREB would be from PKA (fig. S7). However, because cAMP stimulates both PKI and PKA activity, this is an incoherent FFM and would produce transient CREB activation (Fig. 4A, bottom). Because of the incoherent nature of this FFM and its dynamic features, the CREB activation would be transient in the absence of input from the MAPK pathway.
Fig. 4. Computational modeling and experiments show that interconnected feedforward loops control duration and extent of CREB activation.
(A) Top group represents the control condition of cells stimulated with isoproterenol (ISO), middle group represents the response in the presence of isoproterenol and PKA inhibition (Rp-cAMPs or PKA-DN), and bottom group represents the response in the presence of isoproterenol and MEK inhibition (MEKi or U0126). Upper line graphs on the left represent the computationally determined system response at the level of CREB (dashed black line) compared with the experimental observations (solid black line). Color-coded lower graphs represent the computationally determined activity of the indicated motifs over time. Network diagrams on the right show a schematic representation of the signal flow from cAMP to CREB upon isoproterenol stimulation at 3, 10, or 30 min. Activities of the simulated FFMs (diamonds) are depicted with color-coded scales; cAMP and CREB activities are depicted in grayscale. The intensity of the relationship lines represents the percent of the activity through that reaction, with dashed lines representing indirect and solid lines representing direct interactions (reaction flow). Interactive version of these dynamic graphs is available as fig. S7. (B) Schematic of the temporal contribution of the regulatory motifs in the maintenance of CREB activation as determined by motif lifetime analysis.
To verify these predictions from the simulations, we tested the response of differentiated mouse podocytes in culture to the β-adrenergic receptor agonist isoproterenol in the presence or absence of U0126, Rp-cAMPs, or dominant-negative PKA and monitored CREB phosphorylation (fig. S4 and Fig. 4A). The experimental inhibition of PKA activity either with Rp-cAMPs or by overexpression of the dominant-negative PKA did not temporally match the simulations completely; however, the overall transient profiles were similar.
To further understand how the network produced a transient CREB signal when PKA or MEK was inhibited and prolonged CREB activation when all parts of the network were operational, we used dynamic graph analysis. We measured the relative contribution of the separate motifs and the duration of upstream regulatory motifs in a nested FFM model (see Supplementary Methods: Computation of Motif Lifetime). We defined a parameter called “motif lifetime,” which represents the relationship between the output of the motif (for example, b-Raf activity for FFM1, or the amount of nuclear MAPK for FFM2) and the overall activity of the effector of the network (CREB phosphorylation). Motif lifetime was useful in estimating relative contributions of the various motifs to the amplitude and duration of activated CREB (Table 1). The simulations indicated that the FFMs operated with distinct time courses, and so contributed to the various temporal phases of CREB activation (fig. S8A). Using motif lifetime analysis, we quantified the relative contribution of paths within the network (Fig. 4B). This analysis indicated that a larger FFM arises from nesting of the three smaller motifs (fig. S8B). PKA was the major regulator of information flow through this network. However, information flowed through the MAPK pathway as well and not only through direct phosphorylation of CREB by PKA in the nucleus (fig. S8C). In the nucleus, MSK was predicted to mediate 86% of CREB phosphorylation and PKA only 14%; in the cytoplasm, the PKA–to–b-Raf interaction was predicted to stimulate 83% of MEK phosphorylation, and EPAC was predicted to stimulate 17%. Thus, the larger FFM from cAMP to PKA to the MAPK cascade (represented by MEK) in the cytosol and then from the MAPK cascade to CREB in the nucleus dominated the signal flow (fig. S8C). These predictions of the relative contributions are based on the rate constants used in the simulations (table S3). A study of the lifetime of the three FFMs showed that although all three FFMs were required to obtain maximal CREB activity, the two arms of FFM2, but not FFM1 or FFM3, must be operational to enable the prolonged maintenance of this activated state (Table 1 and Fig. 4B).
Table 1.
Description of the function and lifetime of the four motifs of the signaling network that were downstream of cAMP, activated CREB, and analyzed in this study.
| Regulatory motif | Motif type | Description of motif output | Lifetime of motif* represented by network activity (CREB phosphorylation) |
|---|---|---|---|
| FFM1 | Coherent OR gate feedforward | Activation of b-Raf by PKA and cAMP-GEF EPAC | Prolonged (>45 min) if PKA activates b-Raf, otherwise transient when PKA is inhibited (starting at 3 min and ending at 17 min, for a total duration of 14 min) |
| FFM2 | Coherent OR gate feedforward | cAMP regulation of the activation and translocation of MAPK | Prolonged (>45 min) if FFM1 produces a prolonged signal and PTP can be inhibited by PKA, otherwise transient when only PKA is inhibited (starting at 4 min and ending at 19 min, for a total duration of 15 min) |
| FFM3 | Incoherent OR gate feedforward | Delayed inactivation of PKA in nucleus after translocation | Transient (starting at 6 min and ending at 25 min, for a total duration of 19 min) |
| FBL1 | Negative feedback | Dynamic decrease of cAMP through PKA activation of PDE | Constantly operational after 3 min |
Lifetime values were calculated as described in the Supplementary Methods: Computational Assessment of Motif Lifetime.
Analysis of the contributions of the feedfoward motifs to differentiation in cultured podocyte
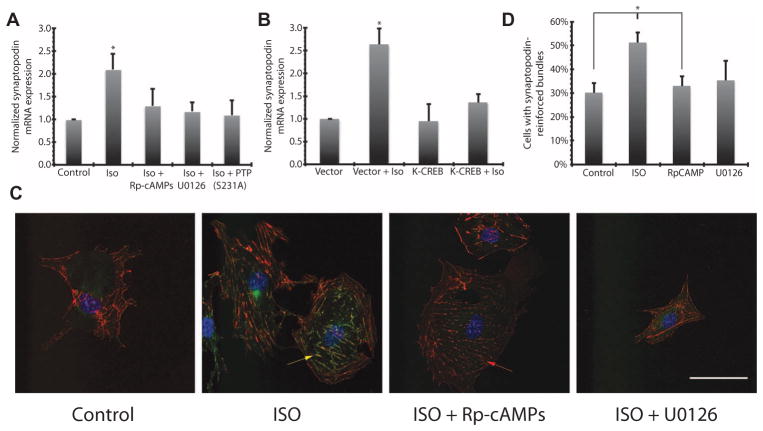
Podocytes proliferating in culture can be induced to adopt a differentiated state, and this change is accompanied by an increase in the expression of the gene encoding synaptopodin, a differentiation marker and a key regulator of podocyte structure (18). In differentiated podocytes, synaptopodin colocalizes with F-actin and stabilizes stress fibers, which are required for maintenance of the foot processes (18, 19). Hence, we quantified synaptopodin mRNA and the colocalization of the protein with stress fibers as morphological markers of differentiated podocytes. Activation of the β-adrenergic receptor with isoproterenol in cultured podocytes resulted in CREB activation for an extended duration (Fig. 4) and increased synaptopodin mRNA (Fig. 5A). Inhibition of FFM1 with Rp-cAMPs or FFM2 with U0126 or by overexpressing the PKA-insensitive mutant PTP-SL-S231A (28–30) abolished the isoproterenol-induced increase in synaptopodin mRNA (Fig. 5A). Cells overexpressing the dominant-negative K-CREB, which cannot bind to DNA due to a single amino acid mutation in its DNA binding domain (31), also failed to increase synaptopodin mRNA abundance in response to β-adrenergic receptor activation (Fig. 5B). Isoproterenol also stimulated increased formation of synaptopodin-associated actin stress fiber bundles (Fig. 5C), which was not observed in podocytes treated with Rp-cAMPs or U0126 (Fig. 5, C and D).
Fig. 5. Expression and functional localization of synaptopodin in cultured podocytes is controlled by PKA.
(A) Podocytes were treated with Rp-cAMPs or U0126, or transfected with PKA-insensitive PTP-SL-S231A mutant, and then stimulated with 10 μM isoproterenol (ISO). Synaptopodin mRNA abundance was quantified using RT-PCR and normalized against the amount of tubulin mRNA (all values are means ± SEM; *P < 0.001, one-way ANOVA; n = 3). (B) Cells transfected with dominant-negative K-CREB (or empty vector) and stimulated with 10 μM isoproterenol. Isoproterenol stimulation led to no change in synaptopodin expression in the K-CREB group, where significant increase was observed in vector-transfected cells (*P < 0.001, one-way ANOVA; n = 3). (C) Representative immunofluorescence images of cultured podocytes that have been treated with 10 μM isoproterenol. Colocalization of synaptopodin (green pseudocolor) with actin bundles (red pseudocolor) is indicated with a yellow arrow. Red arrow indicates loss of bundling in cells treated with isoproterenol and Rp-cAMPs. Scale bar, 50 μm. (D) Quantification of immunofluorescence images in (C) (*P < 0.05, one-way ANOVA; n = 18 cells over five slides in two experiments).
Spatial specification of information flow by FFMs
The network involves the spatial transfer of information from the plasma membrane [where the receptor, G proteins (heterotrimeric guanine nucleotide–binding proteins), and adenylyl cyclase are located and cAMP is produced] to the activation of PKA and EPAC by cAMP in the cytoplasm, and then the translocation of protein kinases into the nucleus to phosphorylate CREB. The translocation of MAPK into the nucleus is prevented by de-phosphorylation of MAPK at a tyrosine residue by PTP-SL (28–30). PKA phosphorylation of PTP-SL at the Ser231 inhibits its phosphatase activity; inhibition of PTP-SL promotes the movement of MAPK into the nucleus (28–30).
We tested if the inhibition of PTP-SL by PKA, which is part of FFM2, affected the spatial flow of information by blocking the activity of PKA using Rp-cAMPs or by overexpressing PTP-SL-S231A (Fig. 6A). This mutant form of PTP-SL cannot be phosphorylated by PKA, thus disrupting the downstream regulation of MAPK by PKA. To quantify the amount of MAPK in the cytoplasm and the nucleus, we performed immunofluorescence experiments (Fig. 6B), in which we quantified the amount of phosphorylated MAPK (Fig. 6C) and the nuclear-to-cytoplasmic ratio of MAPK (Fig. 6D) in cultured podocytes exposed to isoproterenol in the presence of various treatments to disrupt the effect of PTP-SL. Isoproterenol stimulated the translocation of MAPK to the nucleus in kidney podocytes; MAPK translocation to the nucleus was blocked by Rp-cAMPs or by expression of PTP-SL-S231A (Fig. 6D). These experiments indicated that the spatial specification of the predicted network topology was correct.
Fig. 6. Regulatory motifs within cAMP network are spatially specified.
(A) Schematic representation of FFM2. (B) Immunofluorescence images of cultured podocytes at baseline (Control) and 30 min of isoproterenol stimulation under basal (30′ ISO) conditions or with wild-type (WT) PTP-SL transfection, S231 mutant PTP-SL transfection, or preincubated with 100 μm Rp-cAMPs. Cells are stained for MAPK1,2 (red), phospho-MAPK (green), and actin (white), and nuclei-labeled with DAPI (4′,6-diamidino-2-phenylindole) (blue). Scale bar, 50 μm. (C) Time course of MAPK1,2 activation as measured by ratiometric quantification of phospho-MAPK and total MAPK immunofluorescence images. Values are means ± SEM; n = 24 cells over seven slides in three experiments. (D) Time course of MAPK1,2 translocation to the nucleus as measured by ratiometric quantification of immunofluorescence images. Values are means ± SEM; n = 24 cells over seven slides in three experiments. (E) Schematic representation of FFM3. (F) Time course of activated PKA in the cytoplasm and nucleus as measured by enzymatic assays after subcellular fractionation (*P < 0.05, two-way ANOVA; n = 3).
Another spatial component of the network is the translocation of PKA into the nucleus and the translocation of the PKA inhibitor (PKI) into the nucleus, which limits the duration of active PKA in the nucleus (22). The activation of both PKA and its inhibitor by cAMP and their translocation represent a multicompartmental, spatially specified incoherent FFM (Fig. 6E) (32). To test for the presence of FFM3 in podocytes, we measured PKA activity in the nuclear and cytoplasmic fractions from isoproterenol-treated podocytes. Consistent with the presence of an incoherent FFM, PKA activity in the nucleus reached a peak at 15 min and returned close to the basal amount 45 min after isoproterenol stimulation. In contrast, the amount of active PKA in the cytoplasm PKA remained high at 45 min (Fig. 6F).
We compared the results of the model simulations to the time course of CREB activation by isoproterenol when both PKA and MAPK were functional and when each of the kinases was individually inhibited (fig. S4B), which confirmed the topology in the multicompartmental model and accurately predicted the behavior of the system. The effect of FFM3 on CREB activity represents an example of the delayed inactivation feature of an incoherent FFM. The translocation of the PKA catalytic subunit in the nucleus starts after the stimulation of the β-adrenergic receptors by the agonist; PKA translocation is later matched with a nearly equal deactivation of PKA by PKI in the nucleus (fig. S7G). This activity time window enables a transient increase of PKA inside the nucleus, thus limiting the duration of CREB activation by phosphorylation by PKA. The spatial organization of this motif allows cytoplasmic PKA to stay active, and this cytoplasmic pool of PKA continues to stimulate MAPK signaling, which maintains CREB activation.
Predicting therapeutic interventions to restore podocyte function
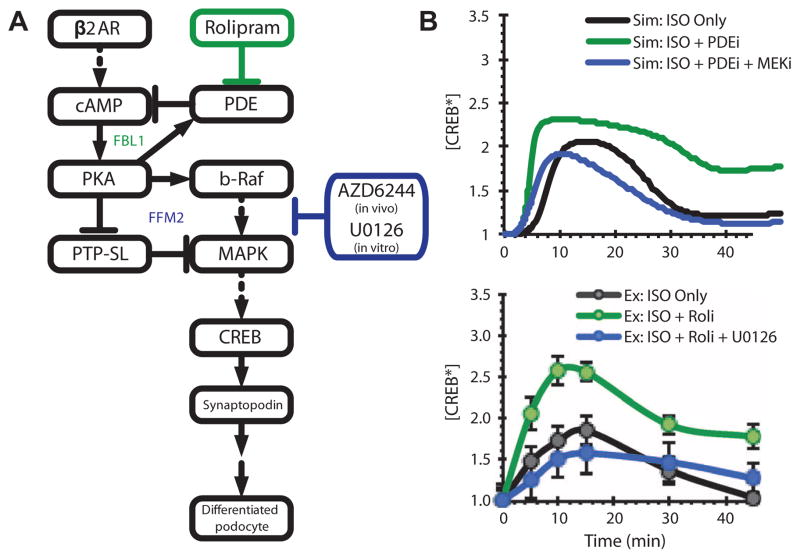
From the experimental cell culture and computational studies, we predicted interventions that would stimulate dedifferentiated podocytes with foot process effacement to revert to a differentiated state. The simplest approach to alter the system in vivo would be to use Food and Drug Administration (FDA)–approved drugs with therapeutic potential for treatment of kidney disease. Because the kidney is exposed to the low plasma concentrations of norepinephrine and epinephrine, which are the endogenous ligands of β-adrenergic receptors, one approach to increasing cAMP concentrations in podocytes in vivo would be to inhibit FBL1 by inhibiting PDEs, for example, with the clinically used PDE-4 inhibitor rolipram (Fig. 7A). To confirm that low-level stimulation of β-adrenergic receptors would produce transient activation of CREB, whereas inhibition of FBL1 by rolipram would produce prolonged activation, we exposed podocytes in culture to low-dose β-adrenergic stimulation (0.1 μM isoproterenol instead of 10 μM), which led to transient CREB activity (Fig. 7B). Inhibition of FBL1 by pretreatment with rolipram transformed the transient response into sustained activation, which was blocked with simultaneous pretreatment with U0126, a MEK inhibitor that blocks FFM2 (Fig. 7B). Simulations at 10% receptor occupancy with virtual ablation of FBL1 or simultaneous FBL1-FFM2 ablations showed similar changes in the dynamics of CREB activity (Fig. 7B).
Fig. 7. Overall network response to motif modulation predicted by the computational model can be experimentally validated in vitro using pharmacologic agents.
(A) Schematic diagram of FFM2, FBL1, and loci of pharmacological inhibition. (B) Effect of disruption of FBL1 with PDE-4 inhibitor rolipram (ISO + Roli) or disruption of FFM2 with the MEK inhibitor U0126 (ISO + Roli + U0126) during β-adrenergic stimulation with low-dose isoproterenol (0.1 μM; ISO Only) on CREB activity. Upper panel: Model predictions. Lower panel: Experimental observations.
From these studies, we predicted that in rats with puromycin-induced nephropathy, treatment with rolipram to block FBL1 would promote podocyte differentiation that would be associated with increased expression of synaptopodin and other podocyte differentiation markers and improved kidney filtration barrier function. These changes would be accompanied by morphological reorganization of the cells, leading to the formation of interdigitating foot processes and decreased proteinuria. Furthermore, coadministration of AZD6244, an FDA-approved MEK inhibitor, would disrupt FFM2 and thus block the rolipram-induced restoration of podocyte morphology and decrease in proteinuria.
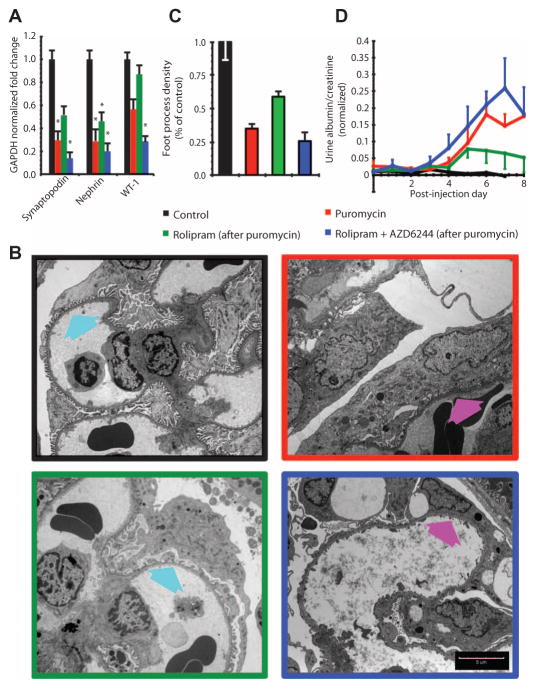
We initiated kidney dysfunction and morphological changes in podocytes with a single injection of puromycin into young rats. After the puromycin injection, the rats were injected with vehicle, rolipram, or rolipram plus AZD6244 twice a day for 7 days. Treatment with rolipram significantly increased mRNA abundance of the differentiation markers synaptopodin, nephrin, and WT-1 compared with their abundance in rats injected with puromycin followed by vehicle. In contrast, coadministration of AZD6244 with rolipram resulted in reduced mRNA abundance of these differentiation markers compared with their abundance in the vehicle-treated, puromycin-injected rats (Fig. 8A and fig. S9A).
Fig. 8. Compensatory modulation of a regulatory motif by pharmacologic agents restores podocyte morphology and function in vivo.
(A) Modulation of FBL1 and FFM2 in vivo recapitulates computationally predicted behavior in puromycin-induced nephropathy. Loss of expression of podocyte differentiation markers nephrin, synaptopodin, and WT-1, as measured by RT-PCR in rats subjected to the indicated treatments. Values are means ± SEM; *P < 0.05 versus control, one-way ANOVA; n = 4 rats for each condition. (B) Transmission electron microscopy images of glomerular podocytes from healthy animals (black border), puromycin-induced nephropathy (red border) animals, nephropathy animals with FBL1 inhibition by rolipram (green border), or nephropathy animals with simultaneous FBL1 and FFM2 inhibition by rolipram plus AZD6244 (blue border). Arrows point to interdigitating foot process morphology (cyan arrows) that was disrupted in untreated and rolipram plus AZD6244–treated puromycin animals (magenta arrows). Scale bar, 5 μm. (C) Blinded quantification of foot process morphology shown in (B). All groups are significantly different from each of the other groups (P < 0.05, one-way ANOVA; n = 60, 15 images from four rats for each condition). (D) Glomerular pathophysiology as measured by protein in urine. Glomerular function in animals treated with puromycin and animals treated with rolipram plus AZD6244 (after puromycin) was significantly different from control (P < 0.05, two-way ANOVA with post hoc Tukey test; n = 4 rats). There was no statistical difference between control animals and animals treated with rolipram (after puromycin).
Electron microscopy analysis of glomeruli from puromycin-injected rats showed that rolipram treatment increased the number of podocyte foot processes per unit circumference of the glomerular capillary (Fig. 8B). In comparison, podocytes exhibited flattened morphologies devoid of foot processes both in puromycin-injected animals treated with vehicle and in puromycin-injected animals treated with rolipram plus AZD6244 (Fig. 8B). Quantification of the foot processes revealed about 70% increase in rolipram-treated rats injected with puromycin compared to those treated with vehicle or rolipram plus AZD6244 (Fig. 8C and fig. S9B). At higher magnification, we observed actin bundles within the foot processes of podocytes from rolipram-treated rats (fig. S10). Consistent with the increase in foot processes, rolipram treatment significantly decreased proteinuria in puromycin-injected rats, and this amelioration in proteinuria was abolished by AZD6244 coadministration (Fig. 8D).
DISCUSSION
Here, we identified explicit relationships between regulatory motifs in signaling networks, differentiation states of cells, in situ morphology, and physiological function. This multiscale analysis provided insight into how the interaction of regulatory motifs enabled cells to change or maintain functional states in vivo. Proteomic analysis helped us identify the protein kinases and, thus, the signaling pathways associated with differentiated morphology and physiological function. From this initial comprehensive characterization, we chose one protein kinase, PKA, for further detailed study. The cAMP pathway is involved in podocyte differentiation (11, 17). Because the cAMP-to-CREB network contains multiple feedbacks and FFMs, we used this network as a model system to analyze how network motifs interact with one another to regulate biochemical and cellular activities.
The relationship between cellular components, morphology, and physiological function is complex, and no single pathway, network, or protein kinase is likely to be the sole determinant of such a relationship. Our choice of the cAMP network does not imply that other signaling pathways, regulatory networks, or protein kinases are not involved in podocyte differentiation; indeed, several protein kinases, such as Prkdc (also known as DNA-dependent protein kinase) (33) and the tyrosine kinase Fyn (34), have also been implicated as critical regulators of podocyte morphology and physiology.
We connected experiments on cultured cells with physiological experiments in the whole animal. For the cAMP network, use of rolipram, a clinically used PDE-4 inhibitor, enabled the connection of analyses of the network controlled by β-adrenergic receptor in cultured cells to experiments in the intact animal. Cell culture experiments were critical for obtaining a detailed understanding of the interactions between the network motifs in the cAMP network that controlled the sustained duration of β-adrenergic–dependent CREB activation needed to drive podocytes into a differentiated state. With the pharmacologic agents, rolipram (PDE inhibitor) and AZD6244 (MEK inhibitor), we recapitulated the motif relationships in vivo in the context of altered physiological function. The cAMP-CREB network is important for many other complex biological processes, such as learning and memory in the hippocampus and steroidogenesis in ovaries (35, 36). Although, by parsimonious reasoning, we expected that the signaling pathway from cAMP to CREB would be a short linear pathway (37, 38), we found that to represent the experimental data, the pathway required a network with at least four interconnected regulatory motifs that are stacked (operate sequentially) and, in the case of the FFMs, nested (where multiple small motifs are within one larger motif). Both motif arrangements can provide flexibility in temporal sequencing, which depends on the rate constants.
Our results suggested that one advantage of this complex network, involving two protein kinases that can target the same transcription factor, is the ability to tightly control the duration of activation of the common transcription factor, in this case CREB. The amplitude and duration of activation of transcription factors control decisions that change the differentiation state of the cell (39); however, the mechanisms by which such timing control is achieved through receptor regulation were unknown. In kidney podocytes, we showed how a set of three FFMs together with an upstream FBL can control the extent and duration of CREB activation. Although not experimentally studied here, there are two more negative FBLs: one for receptor desensitization involving the kinase GRK (G protein–coupled receptor kinase) (40) and the other involving MAPK-induced MKP for limiting MAPK activity (41). Because the network represented by model 4 included the effects of all of these motifs and produced the “right” duration and extent of CREB activity to increase synaptopodin expression and induce actin bundling, we did not experimentally analyze these two additional negative FBLs. Overall, we identified a minimum signaling network topology of multiple interacting regulatory motifs that enabled a key process required to control cell state. From this study, we can make two conclusions regarding molecular mechanisms that function at a systems level to control cell morphology and physiological function.
First, although in our animal study the pathophysiological phenomenon was triggered by one drug, computational modeling readily indicates that genomic or epigenomic changes (for example, single-nucleotide polymorphisms, DNA or histone methylation, or posttranslational modifications) that alter the function of any of the nodes within the motifs could produce an equivalent effect. Thus, understanding the network topology enables more effective targeting of key components and better prediction of the effects of perturbations, whether naturally occurring, such as mutations that are associated with altered or disease states, or pharmacological interventions. For example, knowing the activity of FFM2 can be more informative than knowing the activity of b-Raf or PTP-SL individually. Changes in the activity of b-Raf (a protein kinase that activates MAPK through MEK) or PTP-SL (a phosphatase that inhibits MAPK) can produce the same biological outcome. Such an effect by enzymes with two opposing activities can be readily understood when both are part of a regulatory motif. As more complex phenomena are studied, the size of regulatory networks will grow; thus, it will be more important to know the activity of such feedforward and feedback motifs and the relationships between their activities to understand how distal input and output relationships arise.
A second conclusion that we can draw is that compartmentalization enables regulatory motifs to function with the required amplitude and temporal characteristics to enable transcription factor activation for the appropriate duration. Both FFM2 (working through PTP-SL regulation in the cytoplasm and activated MAPK translocation to the nucleus) and FFM3 (working through the movement of PKI to the nucleus) require the cytoplasmic and nuclear compartments for the effects of the FFMs to be manifested. Thus, in understanding the effect of feedforward loops, spatial specification is as important as specificity in the interactions between components (connectivity based on mutual chemical specificity).
Dynamic graphs, which represent the integration of dynamic modeling (the ODE models) with network topology using Petri net formalisms, provide a facile computational approach for understanding the relationship between motifs in regulating signal flow through the network. Overall, the integrated experimental and computational approach revealed how distributed information processing within a regulatory network can control the amplitude and duration of output signals in vivo, with functional consequences that can have important clinical implications. Such distributed information processing is a consequence of the dynamics that arise from the underlying network topology; however, topological organization alone appears to be insufficient because spatial organization also plays a critical part. Thus, the spanning of regulatory motifs across multiple cellular compartments is a powerful feature of topological organization in signaling networks that regulate cell state. Despite our description of a reasonably complex system, we do not think that CREB is the sole driver of podocyte state or function. Podocytes are exposed to many other extracellular regulators, and we expect that many other networks and transcription factors are involved. Future experiments and simulations may help us determine how regulatory motifs for other transcription factors are configured, and how these different upstream signaling networks cooperate to produce cellular state changes that lead to physiological functions.
MATERIALS AND METHODS
Puromycin nephropathy model
All animal protocols in this study have been approved by the Institutional Animal Care and Use Committee of Icahn School of Medicine at Mount Sinai School. Young (5-week-old) male Sprague-Dawley rats were intravenously injected either with puromycin aminonucleoside (PAN; Sigma) at a concentration of 100 mg/kg or with an equivalent volume of PBS as negative control. Urine was collected each day, and the amount of excreted albumin (measured by Coomassie blue staining) and the amount of creatinine [measured by Jaffe’s method (42)] were determined. Protein in the urine was presented as the amount of excreted albumin normalized to creatinine. Animals were euthanized 7 days after PAN injection, kidneys were removed, and glomeruli were isolated by serial sieving in ice-cold PBS, after which they were split into two batches. One batch was used for proteomic analysis, whereas the other was used for quantitative RT-PCR. Purity of the glomerular preparation was confirmed by light microscopy and proteomic analysis (Supplementary Methods: Evaluation of Glomerular Purity and table S5).
Proteomics
Urea, octyl β-D-glucopyranoside, triethylammonium bicarbonate (TEAB) buffer, protease inhibitor cocktail, Phosphatase Inhibitor Cocktail I, and Phosphatase Inhibitor Cocktail II were from Sigma. Tris(2-carboxyethyl)-phosphine (TCEP), methyl methanethiosulfonate (MMTS), and iTRAQ reagents were from Ab Sciex. Sequencing grade–modified trypsin was from Promega Corp. PepClean C18 spin columns were from Pierce.
Samples were lysed by sonication in lysis buffer [8 M urea, 100 mM TEAB, 1.0% octyl β-D-glucopyranoside (pH 8.5)] containing protease and phosphatase inhibitors. The supernatant was cleared by centrifugation at 16,100g for 15 min at room temperature, and clarified samples were transferred to fresh microfuge tubes. A total of 100 μg of proteins was used for each sample. Proteins were reduced with TCEP, and subsequently, free thiols were blocked by MMTS. Proteins were first digested with Lys-C for 4 hours at 37°C. After diluting the urea concentration to 1 M, tryptic digestion was performed by addition of 2 μg of trypsin (Promega Corp.) to each of the samples at 37°C overnight. Peptides derived from each sample were labeled with iTRAQ tags as per the manufacturer’s instructions. After confirming the iTRAQ labeling efficiency by mass spectrometry (MS), peptides were mixed and dried overnight in a SpeedVac and desalted by Sep-Pak (Waters Corp.) C18 cartridges.
Resulting peptides were fractionated by strong cation exchange (SCX) chromatography as described (43) and collected in 2-ml fractions. Each SCX fraction was then desalted with PepClean C18 spin columns as described (44). After desalting, fractions with low amount of peptides (based on ultraviolet absorbance at 214 nm) were combined for further analysis, yielding 15 fractions with similar complexity of peptides. Liquid chromatography (LC)–MS/MS analyses were performed with an UltiMate 3000 nano LC system (Dionex) and LTQ Orbitrap Velos mass spectrometer. Desalted peptides from SCX fractions were first captured onto a reversed-phase (0.3-mm × 5-mm) trapping column and then resolved on a 75-μm × 150-mm capillary PepMap column (3 μm, 100 Å, C18, Dionex) with a 180-min gradient of solvent A [5% acetonitrile (ACN), 0.1% formic acid (FA)] and solvent B (85% ACN, 0.1% FA). Eluted peptides were introduced directly to LTQ Orbitrap Velos through a nanospray source (Proxeon) with a spray voltage of 2 kV and a capillary temperature of 275°C. Full-scan MS spectra were acquired in the positive ion mode with a scanning mass range of mass/charge ratio (m/z) 350 to 2000 and a resolution of 60,000 full-width at half maximum (FWHM). The higher-energy collision dissociation (HCD) MS/MS spectra were acquired in a data-dependent manner. The 10 most abundant ions were selected for HCD fragmentation per MS scan in the Orbitrap at a resolution of 7500 FWHM. The normalized collision energy was set to 45. The lock mass feature was engaged for accurate mass measurements.
The MS/MS spectra from the analyses were searched against rat protein sequences of SwissProt protein database with Mascot (version 2.3) and Sequest search engines via the Proteome Discoverer platform (version 1.3, Thermo Scientific). The precursor mass error window was set as 10 parts per million (ppm), and MS/MS error tolerance was set as 0.1 dalton for HCD spectra with up to two missed cuts. Methionine oxidation and 8plex iTRAQ labeling on tyrosine were set as variable modifications, whereas 8plex iTRAQ labeling on N terminus and lysine side chain and MMTS conjugation on cysteine were set as fixed modifications. The resulting *.dat files from Mascot and *.msf files from Sequest search were filtered with Scaffold (version 3.3.2, Proteome Software Inc.) for protein identification and quantification analyses. For additional validation of identification, X!Tandem search was engaged in Scaffold with same modification as described for Mascot and Sequest. All peptides were identified with at least 95% confidence interval value as specified by the PeptideProphet algorithm and less than 1% false discovery rate (FDR) based on forward/reverse database searches. Proteins were considered confidently identified with at least one unique peptide, and an experiment-wide FDR of no more than 1.0% at both the protein and the peptide levels. Proteins that share the same peptides and could not be differentiated on the basis of MS/MS analysis alone were grouped together to reduce the redundancy, using Scaffold (45). Relative quantification of proteins was determined with Scaffold Q+ module in a normalized log2-based relative iTRAQ ratio format, with iTRAQ 113 tag as the reference denominator.
Network enrichment analysis
All differentially expressed markers, in addition to down-regulated components alone, were used to identify highly enriched upstream kinases, pathways, and ontology terms with the EnrichR suite (http://amp.pharm.mssm.edu/Enrichr/). The following databases were mined for enriched terms: MGI Pathways from The Jackson Laboratories (13), KEA (14), human and mouse gene atlases (46), and the Gene Ontology “cellular components.” In addition, a protein-protein interaction network was constructed by mining the 18 curated databases with Genes2Networks (16) to include one intermediate that can connect the input proteins. The resultant network constructed with the X2K suite (15) was also used to identify enriched terms.
Podocyte cell culture
Conditionally immortalized murine podocytes are a gift from P. Mundel (Massachusetts General Hospital). Podocytes were cultivated in RPMI 1640 medium containing 10% fetal bovine serum, penicillin (100 U/ml), streptomycin (100 μg/ml), and 2 mM L-glutamine. To permit immortalized growth, the culture medium was supplemented with recombinant mouse interferon-γ (10 U/ml) to induce the expression of T antigen, and the cells were cultured at 33°C (permissive conditions). To induce differentiation, the cells were cultured on type I collagen at 37°C without interferon-γ for at least 5 days. We confirmed the degradation of the T antigen under nonpermissive condition (37°C) by immunoblot analysis. All podocytes used in the study were cultured in nonpermissive conditions for at least 5 days.
Quantitative real-time RT-PCR
Total RNA from cultured podocytes was extracted with the TRIzol reagent (Invitrogen). OneStep RT-PCR Kit (Qiagen) was used for quantitative, one-step, real-time PCR. Specific primers for synaptopodin (5′-GCCAGGGACCA-GCCAGATA-3′ and 3′-AGGAGCCCAGGCCTTCTCT-5′) and tubulin (5′-TGCCTTTGTGCACTGGTATG-3′ and 3′-CTGGAGCAGTTTGACGA-CAC-5′) were designed (MWG-Biotech).
Total RNA from isolated glomeruli was extracted with RNeasy Kit (Qiagen) according to the manufacturer’s instructions. SuperScript III (Invitrogen) and SYBR Green PCR (Qiagen) kits were used to perform quantitative, two-step, real-time RT-PCR analysis. Specific primers for rat GAPDH (5′-TGCACCACCAACTGCTTAGC-3′ and 3′-GGCATGGAC-TGTGGTCATGA-5′), synaptopodin (5′-GCCAGAGAATGGAGTCCT-GCGC-3′ and 3′-AGCACGAGGTGAGACAGTGCGT-5′), Wilms tumor 1 (5′-GAGAGCCAGCCCTCCATCC-3′ and 3′-GGGTCCTC-GTGTTTGAAGGAA-5′), and nephrin (5′-CCACAGCGAGGCACT-CCGTG-3′ and 3′-AGGATACGGTGCCGGGGACC-5′) were obtained from Sigma. The 2−ΔΔCT method was used for quantitative analysis of gene expression (47).
Immunoblotting
Podocytes were lysed with a buffer containing 1% NP-40, a protease inhibitor cocktail, and tyrosine and serine-threonine phosphorylation inhibitors. After determination of protein concentration, cell lysates were subjected to 8 to 12% SDS–polyacrylamide gel electrophoresis before transfer to polyvinylidene difluoride membranes. Immunoblotting was performed with the following specific antibodies for phospho-CREB (Ser133), phospho-MAPK1,2, CREB, and MAPK1,2 (Cell Signaling Laboratory), and β-actin (Sigma). The band intensity was quantified on a Kodak Image Station Imaging System.
Immunofluorescence studies in cultured podocytes
Podocytes cultured on coverslips were stimulated with isoproterenol (10 μM) at indicated time intervals and then fixed in 4% paraformaldehyde and 4% sucrose at room temperature for 15 min. After permeabilization in 0.5% Triton X, cells were incubated with antibodies recognizing phosphorylated or total MAPK1,2 (Cell Signaling Laboratory) or synaptopodin (Progen Biotechnik) overnight at 4°C. After washing, cells were incubated with fluorescence-labeled secondary antibodies from Invitrogen [Alexa Fluor 488 anti-rabbit immunoglobulin G (IgG) or Alexa Fluor 568 anti-mouse IgG]. Hoechst 33342 and Alexa Fluor 647 phalloidin (Invitrogen) were used to label nuclei or the F-actin cytoskeleton, respectively. After mounting with ProLong Gold (Invitrogen), slides were imaged with a Leica SP5 DMI laser scanning confocal microscope.
Transfection of podocytes
Podocytes were transiently transfected with the Nucleofector technology (Amaxa Biosystems), which used electroporation for gene transfer. Transfection efficiency was determined by transfecting podocytes with a vector (pMaxGFP) encoding the green fluorescent protein (GFP) and counting the percentage of GFP-positive cells. We tested several protocols based on the manufacturer’s instructions. After optimization, we achieved 80 to 90% transfection efficiency with Amaxa’s Nucleofector solution and Program T-20. Complementary DNA (1 μg/ml) was used for transfections. Wild-type PTP-SL and PTP-SL-S231A mutant vectors were obtained from R. Pulido. Expression of the transfected proteins was verified by immunoblot.
PKA kinase enzyme activity assay
Podocytes were pretreated with IBMX (3-isobutyl-1-methylxanthine) for 10 min and then treated with isoproterenol (10 μM) for 0, 5, 10, 15, 30, and 45 min. Both cytosolic and nuclear lysates were isolated in lysis buffer. PKA kinase activity was measured with nonradioactive PKA Kinase Activity Assay Kit (product # EKS-390A, Assay Designs Stressgen) according to the manufacturer’s instruction. The experiment was repeated three times.
In vivo modulation of regulatory motifs in puromycin nephropathy
Young (4-week-old) male Sprague-Dawley rats were randomly distributed to control, vehicle, rolipram, and rolipram plus AZD6244 groups and injected with PAN (or saline for control) as described above. Twenty-four hours after initial PAN injection, treatment animals started receiving twice-a-day intraperitoneal injections of rolipram (4.8 mg/kg; Sigma) dissolved sterilely in 10% Cremophor EL (Sigma) in PBS with or without daily intraperitoneal injections of AZD6244 (15 mg/kg; Selleck Chemicals) dissolved sterilely in 0.5% methyl cellulose and 0.4% Tween 80. The vehicle group received the equivalent volume of drug diluents. Proteinuria was quantified, as described above, for each day. Animals were euthanized with isoflurane overdose and rapid decapitation 7 days after PAN injection, kidneys were removed, and glomeruli were isolated by serial sieving in ice-cold PBS. RNA was isolated for quantitative analysis of gene expression, as described above.
Quantitative transmission electron microscopy
After dissection, ~8-mm3 cortical kidney tissue pieces were fixed with 2.5% glutaraldehyde in 0.2 M sodium cacodylate overnight, washed, and treated with 1% osmium tetroxide. After incremental ethanol dehydration and propylene oxide washes, the samples were embedded and thin-sectioned (70 to 90 nm). They were stained with uranyl acetate–lead citrate and imaged on a Hitachi H7000 transmission electron microscope. At least three glomeruli per animal were imaged at ×5000 resolution on 15 random sites, and the number of slit diaphragms per capillary circumferential distance was measured in a blinded fashion.
Mathematical modeling
The system was described as a multicompartment ODE model. This model uses the equation d[x]/dt = Σv, where [x] is the concentration of species x, and v is the sum of the rates v of the reactions affecting species x. The reversible reactions of the type x + A ↔ xA are described by mass action kinetics, v = − kon[X][A] + koff[AX], where v is the velocity, kon is the forward reaction rate constant, and koff is the reverse reaction rate constant. Enzyme-mediated reactions are approximated as irreversible, with Michaelis-Menten rates, v = kcat[E][x](Km + [x])−1, where [E] is the enzyme concentration, kcat is the catalytic efficiency constant, and Km is the Michaelis-Menten constant. Nuclear membrane flux densities are described: jx = kimport[x]cytosol − kexport[x]nucleus. The compartmental model was implemented in Virtual Cell (http://www.nrcam.uchc.edu) with the LSODA algorithm (variable order, variable time step). Each simulation was run for at least 3000 s. The kinetic parameters are described in table S3. Of 106 parameters for the entire model, 74 parameters were obtained from published biochemical and cell biological experimental data, 24 parameters were constrained with data published in this study, 8 parameters were constrained with time course data from the literature, and 6 parameters were given realistic estimates based on values of similar parameters. Computational constraints were achieved manually using values within an order of magnitude of values of similar parameters. Deviation between computational models and experimental observations was quantified with RMS error (absolute value of simulation datum − mean experimental datum).
The Virtual Cell model Podocyte Nested FFM Model is available in the public domain at http://www.vcell.org/ under the shared username sihar. The robustness of the model was evaluated with Virtual Cell local sensitivity analysis option. The analysis was performed by direct solution for variable sensitivities to a small variation of a single simulation parameter. The Virtual Cell model Podocyte Nested FFM Model (copy sensitivity) contains all the simulations performed for the sensitivity analysis and is available online under the same username.
Statistical analysis
All experiments were repeated at least three times, except for the in vivo assays, which were repeated twice. Means were obtained from three or more independent experiments. The data were presented as means ± SEM. For immunoblot and electrophoretic mobility shift assay (EMSA) experiments, bands are quantified by densitometric analyses with ImageJ. Statistical differences were determined by unpaired t test or one-way ANOVA followed by a post hoc Tukey test, where appropriate. Statistical differences in proteinuria between four in vivo groups and spatiotemporal PKA activity were assessed by two-way ANOVA with a post hoc Tukey test. Significance was defined as a P < 0.05.
Supplementary Material
Acknowledgments
We thank P. Mundel for the conditionally immortalized murine podocytes; R. Pulido for the PTP-SL constructs; H. Bell for technical assistance in electron microscopy; R. Huq for fluorescence microscopy; and R. D. Blitzer for critical comments on the manuscript.
Funding: This research was supported by NIH grants T-R01 DK087650 (R.I., J.C.H., and S.R.N.), GM54508 (R.I.), and DK78897 (J.C.H.) and by Systems Biology Center grant GM-071558 (R.I.). E.U.A. is a Howard Hughes Medical Institute Fellow of the Life Sciences Research Foundation; S.V.H. was a recipient of the Natural Sciences and Engineering Research Council of Canada fellowship BP-342902. Virtual Cell is supported by NIH grant P41RR013186 from the National Center for Research Resources. Confocal laser scanning microscopy was performed at the Mount Sinai School of Medicine–Microscopy Shared Resource Facility, supported with funding from NIH National Cancer Institute shared resources (5R24 CA095823-04), National Science Foundation Major Research Instrumentation (DBI-9724504), and NIH shared instrumentation (1 S10 RR0 9145-01) grants. The Orbitrap mass spectrometer was purchased in part with NIH grant P30NS046593.
Footnotes
www.sciencesignaling.org/cgi/content/full/7/311/ra12/DC1
Methods
Fig. S1. Results for enrichment analysis for down-regulated proteomic components in glomeruli from puromycin-induced nephropathy animals.
Fig. S2. Enrichment analysis for the second proteomics experiment (biological replicate) with puromycin-induced nephropathy rats.
Fig. S3. Protein-protein interaction network constructed with down-regulated components as seed nodes along with one intermediate.
Fig. S4. Multimodal experimental constraints for the multicompartmental dynamic model of cAMP/PKA signaling network topology.
Fig. S5. Analysis of CREB activity by EMSA to confirm CREB binding to DNA.
Fig. S6. Process for building dynamic graphs and visual summary of the associated computational methods.
Fig. S7. Dynamic graphs of individual component and motif activities.
Fig. S8. Relative contribution of FFM paths and their lifetimes.
Fig. S9. Spread of RT-PCR and morphometric data for animal studies.
Fig. S10. High-magnification transmission electron microscopy (TEM) images showing in vivo actin bundles.
Table S1. Proteins that were significantly up- or down-regulated according to differential proteomics of glomeruli from control (saline-injected) and puromycin-injected rats.
Table S2. Abundance of different PKA subunits as measured by the two independent proteomic experiments.
Table S3. Simulation parameters for the computational model.
Table S4. Parameter normalized local sensitivity analysis.
Table S5. Abundance of glomerular and tubular markers as quantified by proteomic analysis. References (48–93)
Data and materials availability: The raw proteomics data from two separate experiments are available at the Systems Biology Center New York Web site (http://sbcny.org/supplementary_materials.htm). Model 4 is accessible in the Virtual Cell database under the username sihar and the name Podocyte Nested FFM Model.
Author contributions: E.U.A. and S.V.H. designed and performed all experimental and computational studies, analyzed the data, and prepared the manuscript. N.J.E. designed and performed original computational modeling. Y.C. and G.J. performed animal, cell culture, and biochemistry experiments. P.Y.C., W.F., H.X., and S.R.N. provided support for cell culture and biochemistry experiments. M.R.J. and H.L. performed proteomics experiments and analysis. A.M. provided support for network analysis. R.E.G. provided support for electron microscopy experiments. J.C.H. and R.I. were responsible for the overall design and execution of the study and the final manuscript.
Competing interests: R.I. receives support for one postdoctoral position from GlaxoSmithKline (GSK). The person supported by GSK is not an author on this paper.
REFERENCES AND NOTES
- 1.Tryggvason K, Wartiovaara J. How does the kidney filter plasma? Physiology. 2005;20:96–101. doi: 10.1152/physiol.00045.2004. [DOI] [PubMed] [Google Scholar]
- 2.Haraldsson B, Nyström J, Deen WM. Properties of the glomerular barrier and mechanisms of proteinuria. Physiol Rev. 2008;88:451–487. doi: 10.1152/physrev.00055.2006. [DOI] [PubMed] [Google Scholar]
- 3.Lannigan R, Kark R, Pollak VE. The effect of a single intravenous injection of aminonucleoside of puromycin on the rat kidney: A light- and electron-microscope study. J Pathol Bacteriol. 1962;83:357–362. doi: 10.1002/path.1700830204. [DOI] [PubMed] [Google Scholar]
- 4.Bhalla US, Ram PT, Iyengar R. MAP kinase phosphatase as a locus of flexibility in a mitogen-activated protein kinase signaling network. Science. 2002;297:1018–1023. doi: 10.1126/science.1068873. [DOI] [PubMed] [Google Scholar]
- 5.Becskei A, Kaufmann BB, van Oudenaarden A. Contributions of low molecule number and chromosomal positioning to stochastic gene expression. Nat Genet. 2005;37:937–944. doi: 10.1038/ng1616. [DOI] [PubMed] [Google Scholar]
- 6.Isaacs FJ, Hasty J, Cantor CR, Collins JJ. Prediction and measurement of an autoregulatory genetic module. Proc Natl Acad Sci USA. 2003;100:7714–7719. doi: 10.1073/pnas.1332628100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alon U. Network motifs: Theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka K, Augustine GJ. A positive feedback signal transduction loop determines timing of cerebellar long-term depression. Neuron. 2008;59:608–620. doi: 10.1016/j.neuron.2008.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiong W, Ferrell JE., Jr A positive-feedback-based bistable ‘memory module’ that governs a cell fate decision. Nature. 2003;426:460–465. doi: 10.1038/nature02089. [DOI] [PubMed] [Google Scholar]
- 10.Yang Y, Atasoy D, Su HH, Sternson SM. Hunger states switch a flip-flop memory circuit via a synaptic AMPK-dependent positive feedback loop. Cell. 2011;146:992–1003. doi: 10.1016/j.cell.2011.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu TC, Wang Z, Feng X, Chuang P, Fang W, Chen Y, Neves S, Maayan A, Xiong H, Liu Y, Iyengar R, Klotman PE, He JC. Retinoic acid utilizes CREB and USF1 in a transcriptional feed-forward loop in order to stimulate MKP1 expression in human immunodeficiency virus-infected podocytes. Mol Cell Biol. 2008;28:5785–5794. doi: 10.1128/MCB.00245-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu J, Qian J, Borisov O, Pan S, Li Y, Liu T, Deng L, Wannemacher K, Kurnellas M, Patterson C, Elkabes S, Li H. Optimized proteomic analysis of a mouse model of cerebellar dysfunction using amine-specific isobaric tags. Proteomics. 2006;6:4321–4334. doi: 10.1002/pmic.200600026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaw DR. Searching the Mouse Genome Informatics (MGI) resources for information on mouse biology from genotype to phenotype. Curr Protoc Bioinformatics. 2009;Chapter 1(Unit 1.7) doi: 10.1002/0471250953.bi0107s05. [DOI] [PubMed] [Google Scholar]
- 14.Lachmann A, Ma’ayan A. KEA: Kinase enrichment analysis. Bioinformatics. 2009;25:684–686. doi: 10.1093/bioinformatics/btp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen EY, Xu H, Gordonov S, Lim MP, Perkins MH, Ma’ayan A. Expression2Kinases: mRNA profiling linked to multiple upstream regulatory layers. Bioinformatics. 2012;28:105–111. doi: 10.1093/bioinformatics/btr625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berger SI, Posner JM, Ma’ayan A. Genes2Networks: Connecting lists of gene symbols using mammalian protein interactions databases. BMC Bioinformatics. 2007;8:372. doi: 10.1186/1471-2105-8-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He JC, Lu TC, Fleet M, Sunamoto M, Husain M, Fang W, Neves S, Chen Y, Shankland S, Iyengar R, Klotman PE. Retinoic acid inhibits HIV-1-induced podocyte proliferation through the cAMP pathway. J Am Soc Nephrol. 2007;18:93–102. doi: 10.1681/ASN.2006070727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asanuma K, Yanagida-Asanuma E, Faul C, Tomino Y, Kim K, Mundel P. Synaptopodin orchestrates actin organization and cell motility via regulation of RhoA signalling. Nat Cell Biol. 2006;8:485–491. doi: 10.1038/ncb1400. [DOI] [PubMed] [Google Scholar]
- 19.Mundel P, Heid HW, Mundel TM, Krüger M, Reiser J, Kriz W. Synaptopodin: An actin-associated protein in telencephalic dendrites and renal podocytes. J Cell Biol. 1997;139:193–204. doi: 10.1083/jcb.139.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaywitz AJ, Greenberg ME. CREB: A stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- 21.Deak M, Clifton AD, Lucocq LM, Alessi DR. Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activation of CREB. EMBO J. 1998;17:4426–4441. doi: 10.1093/emboj/17.15.4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X, Dai JC, Orellana SA, Greenfield EM. Endogenous protein kinase inhibitor γ terminates immediate-early gene expression induced by cAMP-dependent protein kinase (PKA) signaling: Termination depends on PKA inactivation rather than PKA export from the nucleus. J Biol Chem. 2005;280:2700–2707. doi: 10.1074/jbc.M412558200. [DOI] [PubMed] [Google Scholar]
- 23.Ekholm D, Belfrage P, Manganiello V, Degerman E. Protein kinase A-dependent activation of PDE4 (cAMP-specific cyclic nucleotide phosphodiesterase) in cultured bovine vascular smooth muscle cells. Biochim Biophys Acta. 1997;1356:64–70. doi: 10.1016/s0167-4889(96)00159-0. [DOI] [PubMed] [Google Scholar]
- 24.Murata T. Petri nets: Properties, analysis and applications. Proc IEEE. 1989;77:541–580. [Google Scholar]
- 25.Castro LR, Gervasi N, Guiot E, Cavellini L, Nikolaev VO, Paupardin-Tritsch D, Vincent P. Type 4 phosphodiesterase plays different integrating roles in different cellular domains in pyramidal cortical neurons. J Neurosci. 2010;30:6143–6151. doi: 10.1523/JNEUROSCI.5851-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neves SR, Tsokas P, Sarkar A, Grace EA, Rangamani P, Taubenfeld SM, Alberini CM, Schaff JC, Blitzer RD, Moraru II, Iyengar R. Cell shape and negative links in regulatory motifs together control spatial information flow in signaling networks. Cell. 2008;133:666–680. doi: 10.1016/j.cell.2008.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ram PT. Overexpression of tightly regulated proteins: Protein phosphatase 2A overexpression in NIH 3T3 cells. Methods Enzymol. 2002;345:551–555. doi: 10.1016/s0076-6879(02)45046-x. [DOI] [PubMed] [Google Scholar]
- 28.Blanco-Aparicio C, Torres J, Pulido R. A novel regulatory mechanism of MAP kinases activation and nuclear translocation mediated by PKA and the PTP-SL tyrosine phosphatase. J Cell Biol. 1999;147:1129–1136. doi: 10.1083/jcb.147.6.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pulido R, Zúñiga A, Ullrich A. PTP-SL and STEP protein tyrosine phosphatases regulate the activation of the extracellular signal-regulated kinases ERK1 and ERK2 by association through a kinase interaction motif. EMBO J. 1998;17:7337–7350. doi: 10.1093/emboj/17.24.7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paul S, Nairn AC, Wang P, Lombroso PJ. NMDA-mediated activation of the tyrosine phosphatase STEP regulates the duration of ERK signaling. Nat Neurosci. 2003;6:34–42. doi: 10.1038/nn989. [DOI] [PubMed] [Google Scholar]
- 31.Walton KM, Rehfuss RP, Chrivia JC, Lochner JE, Goodman RH. A dominant repressor of cyclic adenosine 3′,5′-monophosphate (cAMP)-regulated enhancer-binding protein activity inhibits the cAMP-mediated induction of the somatostatin promoter in vivo. Mol Endocrinol. 1992;6:647–655. doi: 10.1210/mend.6.4.1350057. [DOI] [PubMed] [Google Scholar]
- 32.Mangan S, Itzkovitz S, Zaslaver A, Alon U. The incoherent feed-forward loop accelerates the response-time of the gal system of Escherichia coli. J Mol Biol. 2006;356:1073–1081. doi: 10.1016/j.jmb.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 33.Papeta N, Zheng Z, Schon EA, Brosel S, Altintas MM, Nasr SH, Reiser J, D’Agati VD, Gharavi AG. Prkdc participates in mitochondrial genome maintenance and prevents Adriamycin-induced nephropathy in mice. J Clin Invest. 2010;120:4055–4064. doi: 10.1172/JCI43721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verma R, Wharram B, Kovari I, Kunkel R, Nihalani D, Wary KK, Wiggins RC, Killen P, Holzman LB. Fyn binds to and phosphorylates the kidney slit diaphragm component Nephrin. J Biol Chem. 2003;278:20716–20723. doi: 10.1074/jbc.M301689200. [DOI] [PubMed] [Google Scholar]
- 35.Alberini CM. Transcription factors in long-term memory and synaptic plasticity. Physiol Rev. 2009;89:121–145. doi: 10.1152/physrev.00017.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altarejos JY, Montminy M. CREB and the CRTC co-activators: Sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol. 2011;12:141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhalla US, Iyengar R. Emergent properties of networks of biological signaling pathways. Science. 1999;283:381–387. doi: 10.1126/science.283.5400.381. [DOI] [PubMed] [Google Scholar]
- 38.Weng G, Bhalla US, Iyengar R. Complexity in biological signaling systems. Science. 1999;284:92–96. doi: 10.1126/science.284.5411.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wichterle H, Lieberam I, Porter JA, Jessell TM. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- 40.Premont RT, Inglese J, Lefkowitz RJ. Protein kinases that phosphorylate activated G protein-coupled receptors. FASEB J. 1995;9:175–182. doi: 10.1096/fasebj.9.2.7781920. [DOI] [PubMed] [Google Scholar]
- 41.Sun H, Charles CH, Lau LF, Tonks NK. MKP-1 (3CH134), an immediate early gene product, is a dual specificity phosphatase that dephosphorylates MAP kinase in vivo. Cell. 1993;75:487–493. doi: 10.1016/0092-8674(93)90383-2. [DOI] [PubMed] [Google Scholar]
- 42.Husdan H, Rapoport A. Estimation of creatinine by the Jaffe reaction. A comparison of three methods. Clin Chem. 1968;14:222–238. [PubMed] [Google Scholar]
- 43.Jain MR, Li Q, Liu T, Rinaggio J, Ketkar A, Tournier V, Madura K, Elkabes S, Li H. Proteomic identification of immunoproteasome accumulation in formalin-fixed rodent spinal cords with experimental autoimmune encephalomyelitis. J Proteome Res. 2012;11:1791–1803. doi: 10.1021/pr201043u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jain MR, Liu T, Hu J, Darfler M, Fitzhugh V, Rinaggio J, Li H. Quantitative proteomic analysis of formalin fixed paraffin embedded oral HPV lesions from HIV patients. Open Proteomics J. 2008;1:40–45. doi: 10.2174/1875039700801010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 46.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, Cooke MP, Walker JR, Hogenesch JB. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 48.Liu H, Sadygov RG, Yates JR., III A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 49.Rohr C, Marwan W, Heiner M. Snoopy—A unifying Petri net framework to investigate biomolecular networks. Bioinformatics. 2010;26:974–975. doi: 10.1093/bioinformatics/btq050. [DOI] [PubMed] [Google Scholar]
- 50.David R, Alla H. Discrete, Continuous, and Hybrid Petri Nets. 2. Springer; Heidelberg: 2010. p. 554. [Google Scholar]
- 51.Hardy S, Iyengar R. Modeling in Systems Biology. Vol. 16. Springer; London: 2011. pp. 225–251. [Google Scholar]
- 52.Heinrich R, Neel BG, Rapoport TA. Mathematical models of protein kinase signal transduction. Mol Cell. 2002;9:957–970. doi: 10.1016/s1097-2765(02)00528-2. [DOI] [PubMed] [Google Scholar]
- 53.Nemenoff RA, Winitz S, Qian NX, Van Putten V, Johnson GL, Heasley LE. Phosphorylation and activation of a high molecular weight form of phospholipase A2 by p42 microtubule-associated protein 2 kinase and protein kinase C. J Biol Chem. 1993;268:1960–1964. [PubMed] [Google Scholar]
- 54.Sanghera JS, Paddon HB, Bader SA, Pelech SL. Purification and characterization of a maturation-activated myelin basic protein kinase from sea star oocytes. J Biol Chem. 1990;265:52–57. [PubMed] [Google Scholar]
- 55.Takai A, Mieskes G. Inhibitory effect of okadaic acid on the p-nitrophenyl phosphate phosphatase activity of protein phosphatases. Biochem J. 1991;275:233–239. doi: 10.1042/bj2750233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pato MD, Sutherland C, Winder SJ, Walsh MP. Smooth-muscle caldesmon phosphatase is SMP-I, a type 2A protein phosphatase. Biochem J. 1993;293:35–41. doi: 10.1042/bj2930035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Foulkes JG, Strada SJ, Henderson PJF, Cohen P. A kinetic analysis of the effects of inhibitor-1 and inhibitor-2 on the activity of protein phosphatase-1. Eur J Biochem. 1983;132:309–313. doi: 10.1111/j.1432-1033.1983.tb07363.x. [DOI] [PubMed] [Google Scholar]
- 58.Charles CH, Sun H, Lau LF, Tonks NK. The growth factor-inducible immediate-early gene 3CH134 encodes a protein-tyrosine-phosphatase. Proc Natl Acad Sci USA. 1993;90:5292–5296. doi: 10.1073/pnas.90.11.5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brondello JM, Pouysségur J, McKenzie FR. Reduced MAP kinase phosphatase-1 degradation after p42/p44MAPK-dependent phosphorylation. Science. 1999;286:2514–2517. doi: 10.1126/science.286.5449.2514. [DOI] [PubMed] [Google Scholar]
- 60.Collins SP, Uhler MD. Characterization of PKIγ, a novel isoform of the protein kinase inhibitor of cAMP-dependent protein kinase. J Biol Chem. 1997;272:18169–18178. doi: 10.1074/jbc.272.29.18169. [DOI] [PubMed] [Google Scholar]
- 61.Force T, Bonventre JV, Heidecker G, Rapp U, Avruch J, Kyriakis JM. Enzymatic characteristics of the c-Raf-1 protein kinase. Proc Natl Acad Sci USA. 1994;91:1270–1274. doi: 10.1073/pnas.91.4.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seger R, Ahn NG, Posada J, Munar ES, Jensen AM, Cooper JA, Cobb MH, Krebs EG. Purification and characterization of mitogen-activated protein kinase activator(s) from epidermal growth factor-stimulated A431 cells. J Biol Chem. 1992;267:14373–14381. [PubMed] [Google Scholar]
- 63.Haystead TAJ, Dent P, Wu J, Haystead CMM, Sturgill TW. Ordered phosphorylation of p42mapk by MAP kinase kinase. FEBS Lett. 1992;306:17–22. doi: 10.1016/0014-5793(92)80828-5. [DOI] [PubMed] [Google Scholar]
- 64.Bolger GB, Erdogan S, Jones RE, Loughney K, Scotland G, Hoffmann R, Wilkinson I, Farrell C, Houslay MD. Characterization of five different proteins produced by alternatively spliced mRNAs from the human cAMP-specific phosphodiesterase PDE4D gene. Biochem J. 1997;328:539–548. doi: 10.1042/bj3280539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.MacKenzie SJ, Baillie GS, McPhee I, Bolger GB, Houslay MD. ERK2 mitogen-activated protein kinase binding, phosphorylation, and regulation of the PDE4D cAMP-specific phosphodiesterases. The involvement of COOH-terminal docking sites and NH2-terminal UCR regions. J Biol Chem. 2000;275:16609–16617. doi: 10.1074/jbc.275.22.16609. [DOI] [PubMed] [Google Scholar]
- 66.Paul S, Snyder GL, Yokakura H, Picciotto MR, Nairn AC, Lombroso PJ. The dopamine/D1 receptor mediates the phosphorylation and inactivation of the protein tyrosine phosphatase STEP via a PKA-dependent pathway. J Neurosci. 2000;20:5630–5638. doi: 10.1523/JNEUROSCI.20-15-05630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nika K, Hyunh H, Williams S, Paul S, Bottini N, Taskén K, Lombroso PJ, Mustelin T. Haematopoietic protein tyrosine phosphatase (HePTP) phosphorylation by cAMP-dependent protein kinase in T-cells: Dynamics and subcellular location. Biochem J. 2004;378:335–342. doi: 10.1042/BJ20031244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou B, Wang ZX, Zhao Y, Brautigan DL, Zhang ZY. The specificity of extra-cellular signal-regulated kinase 2 dephosphorylation by protein phosphatases. J Biol Chem. 2002;277:31818–31825. doi: 10.1074/jbc.M203969200. [DOI] [PubMed] [Google Scholar]
- 69.Sette C, Conti M. Phosphorylation and activation of a cAMP-specific phosphodiesterase by the cAMP-dependent protein kinase. Involvement of serine 54 in the enzyme activation. J Biol Chem. 1996;271:16526–16534. doi: 10.1074/jbc.271.28.16526. [DOI] [PubMed] [Google Scholar]
- 70.Hasler P, Moore JJ, Kammer GM. Human T lymphocyte cAMP-dependent protein kinase: Subcellular distributions and activity ranges of type I and type II isozymes. FASEB J. 1992;6:2735–2741. doi: 10.1096/fasebj.6.9.1319361. [DOI] [PubMed] [Google Scholar]
- 71.Smith SB, White HD, Siegel JB, Krebs EG. Cyclic AMP-dependent protein kinase I: Cyclic nucleotide binding, structural changes, and release of the catalytic subunits. Proc Natl Acad Sci USA. 1981;78:1591–1595. doi: 10.1073/pnas.78.3.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ogreid D, Døskeland SO. The kinetics of association of cyclic AMP to the two types of binding sites associated with protein kinase II from bovine myocardium. FEBS Lett. 1981;129:287–292. doi: 10.1016/0014-5793(81)80185-8. [DOI] [PubMed] [Google Scholar]
- 73.Rehmann H, Schwede F, Døskeland SO, Wittinghofer A, Bos JL. Ligand-mediated activation of the cAMP-responsive guanine nucleotide exchange factor Epac. J Biol Chem. 2003;278:38548–38556. doi: 10.1074/jbc.M306292200. [DOI] [PubMed] [Google Scholar]
- 74.Brinkmann T, Daumke O, Herbrand U, Kühlmann D, Stege P, Ahmadian MR, Wittinghofer A. Rap-specific GTPase activating protein follows an alternative mechanism. J Biol Chem. 2002;277:12525–12531. doi: 10.1074/jbc.M109176200. [DOI] [PubMed] [Google Scholar]
- 75.Kuiperij HB, de Rooij J, Rehmann H, van Triest M, Wittinghofer A, Bos JL, Zwartkruis FJ. Characterisation of PDZ-GEFs, a family of guanine nucleotide exchange factors specific for Rap1 and Rap2. Biochim Biophys Acta. 2003;1593:141–149. doi: 10.1016/s0167-4889(02)00365-8. [DOI] [PubMed] [Google Scholar]
- 76.Kraemer A, Brinkmann T, Plettner I, Goody R, Wittinghofer A. Fluorescently labelled guanine nucleotide binding proteins to analyse elementary steps of GAP-catalysed reactions. J Mol Biol. 2002;324:763–774. doi: 10.1016/s0022-2836(02)01136-1. [DOI] [PubMed] [Google Scholar]
- 77.Dessauer CW, Scully TT, Gilman AG. Interactions of forskolin and ATP with the cytosolic domains of mammalian adenylyl cyclase. J Biol Chem. 1997;272:22272–22277. doi: 10.1074/jbc.272.35.22272. [DOI] [PubMed] [Google Scholar]
- 78.Kim CM, Dion SB, Benovic JL. Mechanism of β-adrenergic receptor kinase activation by G proteins. J Biol Chem. 1993;268:15412–15418. [PubMed] [Google Scholar]
- 79.Gether U, Lin S, Kobilka BK. Fluorescent labeling of purified β2 adrenergic receptor. Evidence for ligand-specific conformational changes. J Biol Chem. 1995;270:28268–28275. doi: 10.1074/jbc.270.47.28268. [DOI] [PubMed] [Google Scholar]
- 80.Brandt DR, Ross EM. Catecholamine-stimulated GTPase cycle. Multiple sites of regulation by b-adrenergic receptor and Mg2+ studied in reconstituted receptor-Gs vesicles. J Biol Chem. 1986;261:1656–1664. [PubMed] [Google Scholar]
- 81.Mosser VA, Amana IJ, Schimerlik MI. Kinetic analysis of M2 muscarinic receptor activation of Gi in Sf9 insect cell membranes. J Biol Chem. 2002;277:922–931. doi: 10.1074/jbc.M104210200. [DOI] [PubMed] [Google Scholar]
- 82.Graziano MP, Freissmuth M, Gilman AG. Expression of Gsα in Escherichia coli. Purification and properties of two forms of the protein. J Biol Chem. 1989;264:409–418. [PubMed] [Google Scholar]
- 83.Seibold A, January BG, Friedman J, Hipkin RW, Clark RB. Desensitization of β2-adrenergic receptors with mutations of the proposed G protein-coupled receptor kinase phosphorylation sites. J Biol Chem. 1998;273:7637–7642. doi: 10.1074/jbc.273.13.7637. [DOI] [PubMed] [Google Scholar]
- 84.Costa M, Marchi M, Cardarelli F, Roy A, Beltram F, Maffei L, Ratto GM. Dynamic regulation of ERK2 nuclear translocation and mobility in living cells. J Cell Sci. 2006;119:4952–4963. doi: 10.1242/jcs.03272. [DOI] [PubMed] [Google Scholar]
- 85.Fujioka A, Terai K, Itoh RE, Aoki K, Nakamura T, Kuroda S, Nishida E, Matsuda M. Dynamics of the Ras/ERK MAPK cascade as monitored by fluorescent probes. J Biol Chem. 2006;281:8917–8926. doi: 10.1074/jbc.M509344200. [DOI] [PubMed] [Google Scholar]
- 86.Hagiwara M, Brindle P, Harootunian A, Armstrong R, Rivier J, Vale W, Tsien R, Montminy MR. Coupling of hormonal stimulation and transcription via the cyclic AMP-responsive factor CREB is rate limited by nuclear entry of protein kinase A. Mol Cell Biol. 1993;13:4852–4859. doi: 10.1128/mcb.13.8.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mayr BM, Canettieri G, Montminy MR. Distinct effects of cAMP and mitogenic signals on CREB-binding protein recruitment impart specificity to target gene activation via CREB. Proc Natl Acad Sci USA. 2001;98:10936–10941. doi: 10.1073/pnas.191152098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pang IH, Sternweis PC. Purification of unique α subunits of GTP-binding regulatory proteins (G proteins) by affinity chromatography with immobilized βγ subunits. J Biol Chem. 1990;265:18707–18712. [PubMed] [Google Scholar]
- 89.Post SR, Hilal-Dandan R, Urasawa K, Brunton LL, Insel PA. Quantification of signalling components and amplification in the β-adrenergic-receptor–adenylate cyclase pathway in isolated adult rat ventricular myocytes. Biochem J. 1995;311:75–80. doi: 10.1042/bj3110075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Taylor SS, Buechler JA, Yonemoto W. Camp-dependent protein kinase: Framework for a diverse family of regulatory enzymes. Annu Rev Biochem. 1990;59:971–1005. doi: 10.1146/annurev.bi.59.070190.004543. [DOI] [PubMed] [Google Scholar]
- 91.Storm S, Cleveland J, Rapp U. Expression of raf family proto-oncogenes in normal mouse tissues. Oncogene. 1990;5:345–351. [PubMed] [Google Scholar]
- 92.Carman CV, Som T, Kim CM, Benovic JL. Binding and phosphorylation of tubulin by G protein-coupled receptor kinases. J Biol Chem. 1998;273:20308–20316. doi: 10.1074/jbc.273.32.20308. [DOI] [PubMed] [Google Scholar]
- 93.Ohba Y, Mochizuki N, Matsuo K, Yamashita S, Nakaya M, Hashimoto Y, Hamaguchi M, Kurata T, Nagashima K, Matsuda M. Rap2 as a slowly responding molecular switch in the Rap1 signaling cascade. Mol Cell Biol. 2000;20:6074–6083. doi: 10.1128/mcb.20.16.6074-6083.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.