Abstract
17β-estradiol (E2) plays critical roles in a number of target tissues including the mammary gland, reproductive tract, bone, and brain. Although it is clear that E2 reduces inflammation and ischemia-induced damage in the cerebral cortex, the molecular mechanisms mediating the effects of E2 in this brain region are lacking. Thus, we examined the cortical transcriptome using a mouse model system. Female adult mice were ovariectomized and implanted with silastic tubing containing oil or E2. After 7 days, the cerebral cortices were dissected and RNA was isolated and analyzed using RNA-sequencing. Analysis of the transcriptomes of control and E2-treated animals revealed that E2 treatment significantly altered the transcript levels of 88 genes. These genes were associated with long term synaptic potentiation, myelination, phosphoprotein phosphatase activity, mitogen activated protein kinase, and phosphatidylinositol 3-kinase signaling. E2 also altered the expression of genes linked to lipid synthesis and metabolism, vasoconstriction and vasodilation, cell-cell communication, and histone modification. These results demonstrate the far-reaching and diverse effects of E2 in the cerebral cortex and provide valuable insight to begin to understand cortical processes that may fluctuate in a dynamic hormonal environment.
Introduction
The effects of 17β-estradiol (E2) have been extensively studied in the female reproductive tract where it is required for reproductive competency. E2 also targets a variety of other tissues, including the mammary gland [1], bone [2], [3], cardiovasculature [4], and brain [5]. E2 plays several critical roles in brain development, such as influencing sexual dimorphism [6] and forming synapses [7]. In the cycling female, E2 is an important regulator of ovulation through its communication with the hypothalamus and pituitary [8], [9]. E2 can also act on brain regions not associated with reproduction and can influence pain perception, locomotion, and mood [10].
Numerous experiments have demonstrated that E2 protects the brain from a variety of insults [11]–[13]. For example, E2 protects neuroblastoma cells from H2O2 [14] and beta amyloid [15], [16] toxcicity. Additionally, E2 decreases cellular damage in neurons that have been treated with excitotoxic levels of glutamate [17] and hippocampal slice cultures that have been exposed to oxygen and glucose deprivation [18]. In vivo, E2 reduces inflammation [19], [20] and ischemia-induced damage [21], [22] and this protection is most evident in the cerebral cortex.
In addition to its neuroprotective effects, E2 modulates synaptic plasticity [23], influences neurotransmission [24], [25], and acts as a neurotrophin [26] to support brain homeostasis. These cumulative reports suggest that critical changes in gene expression in the brain are induced by E2. Although the cerebral cortex receives input from several brain regions and is essential for cognitive and executive functions [27], the mechanism by which E2 mediates its effects in the cerebral cortex are unclear. To better understand the molecular consequences of E2 in the cerebral cortex, we analyzed RNA sequencing (RNA-Seq) data from the cortices of oil- and E2- treated, ovariectomized female mice. This unbiased approach identified E2-regulated genes that provide insight into the multiple biological processes influenced by E2 treatment.
Materials and Methods
Animals and surgery
14 week old female C57BL/6J mice were obtained from Jackson Laboratory (Bar Harbor, ME) and maintained on a 12 hr light/dark schedule with access to water and food ad libitum. After 7 days, mice were anesthetized by inhalation of 4% isoflurane, bilaterally ovariectomized and then implanted subcutaneously with silastic tubing (0.062 in/0.125 in, inner/outer diameter, 1 in length; Dow Corning, Midland, MI) plugged at both ends with medical adhesive (Dow Corning). The silastic tubing, which remained in the mice for 7 days, contained either 35 µl of cottonseed oil or 35 µl of cottonseed oil with 180 µg/ml E2 and produced a low, physiological level of circulating E2 (∼25 pg/ml) [21], [28] that is equivalent to estrus levels in mice [29]. Ovariectomized mice were fed phytoestrogen-free chow and after 7 days, the mice were sacrificed, the brains were dissected, and cerebral cortices were harvested. This method of E2 treatment has been extensively used to demonstrate the anti-inflammatory and neuroprotective actions of E2 in the cerebral cortex [19], [21], [30], [31]. The protocol (#12014) for this study was approved and carried out in strict accordance with guidelines from the University of Illinois at Urbana-Champaign Institutional Animal Care and Use Committee and Division of Animal Resources. Analgesics were administered after surgery and all efforts were made to minimize suffering.
RNA isolation
Total RNA was isolated from each cerebral cortex using Ambion RNAqueous according to the manufacturer’s protocol (Life Technologies, Grand Island, NY) and treated with Turbo DNA-free reagent (Ambion, Life Technologies, Austin, TX) to remove genomic DNA. RNA purity was assessed with native agarose gel electrophoresis and analysis of the 28S and 18S rRNA bands. RNA was of high purity, showed no degradation, and was free of DNA (Fig. S1).
RNA-Sequencing
RNA-seq was completed at the W.M. Keck Center for Comparative and Functional Genomics at the University of Illinois Urbana-Champaign. The TruSeq RNA sample prep kit (Illumina, San Diego, CA) and 1 µg of total RNA were used to make poly-A selected and barcoded RNA-Seq libraries for each cortical sample. cDNA libraries were pooled and quantified using real-time PCR with the Library Quantification kit (Kapa Biosystems, Woburn, MA). The libraries were sequenced using 3 lanes for 101 cycles with 7 additional cycles for the index read on the Illumina HiSeq2000 according to the manufacturer’s instructions. The RNA-Seq libraries produced over 255 million reads with each individual sample having more than 29 million reads. The data was then used to generate Fastq files using Casava 1.8.2.
RNA-Seq alignment and statistics
Sequences were aligned using TopHat v. 1.4.1 [32] and Bowtie 1.0 [33]. The genome sequence index was mm10 from UCSC (http://hgdownload.soe.ucsc.edu/downloads.html#mouse). Raw read counts were tabulated for each sample at the gene level using the GTF gene model file for mm10 from UCSC and htseq-count, from HTSeq v0.5.3p9 (http://www-huber.embl.de/users/anders/HTSeq/doc/index.html) using the default “exon” feature type and “gene_id” attribute.
The raw read counts were used in R 3.0.0 [34] for data pre-processing and statistical analysis using packages from Bioconductor [35] as indicated below. Data are available in the Array Express database under accession number E-MTAB-2762. Genes without 1 count per million (CPM) mapped reads in at least 4 samples, irrespective of group, were filtered out and 14,908 of the 37,482 genes passed this filter and were analyzed using edgeR 3.2.3 [36]. The raw count values were used in a negative binomial statistical model that accounted for the total library size for each sample and an extra TMM normalization factor for any biases due to changes in total RNA composition of the samples [37], [38]. Tests for the pairwise comparisons were pulled from the model and separately adjusted for multiple testing using the False Discovery Rate (FDR) method [39].
Comparable expression values were generated from read counts using voom normalized values [40]. The voom values were scaled to the standard deviation of the mean, hierarchically clustered, and displayed as heatmaps. Additional annotation information (gene names, descriptions) was obtained from Ensembl Genes 71, Mus musculus genes (GRCm38.p1) database using the Ensembl gene IDs provided in the GTF gene model file.
Cytoscape (Version 3.0.1) was used in conjunction with the plug-in ClueGO (Version 2.0.7) for network creation [41], [42]. KEGG [43], Reactome [44], and Gene Ontology (biological process) [45] databases were used within the program for network categorization. Over-representation (enrichment) was calculated in the program using a right-sided hypergeometric test and Bonferroni step-down method for multiple test correction.
Transcriptomine from the Nuclear Receptor Signaling Atlas website was used to determine previously identified E2-regulated genes. 17β-estradiol was selected as the ligand and >1.1 fold change in either direction with p<0.05 significance was selected for ‘CNS, all tissues and cell lines’ and ‘all tissues, all cell lines’ RNA sources.
Real-Time PCR (RT-PCR)
RNA concentrations were measured and cDNA was synthesized using the iScript kit (Bio-Rad, Hercules, CA) as described by the manufacturer. cDNA was combined with iQ SYBR Green Supermix (Bio-Rad, Hercules, CA), and forward and reverse primers (0.9 µM) for receptor transporter protein 1, Rtp1, (5′-CTGCCCTGCCTTACACTTAC -3′ and 5′-TCACCTCCTCCATCTTCTCG -3′), macrophage galactose N-acetyl-galactosamine specific lectin 2, Mgl2, (5′- GTGACAAGAAAGGAGGAATG -3′ and 5′- GAGATGACCACCAGTAGC -3′), NLR-pyrin domain containing 3, Nlrp3, (5′- CCAAGGAGGAAGAAGAAGAG -3′ and 5′- AAGAGACCACGGCAGAAG -3′), fatty acid binding protein 7, Fabp7, (5′- GTGACCAAACCAACTGTGATTATC -3′ and 5′- TGTCTCCATCCAACCGAACC-3′), lysozyme 2, Lyz2, (5′- TGAAGACTCTCCTGACTC-3′ and 5′- ACGGTTGTAGTTTGTAGC -3′), succinate dehydrogenase complex, subunit A, flavoprotein, Sdha, (5′- GCTCATCGGTGTTGCTGTG-3′ and 5′-TTGCTCTTATTCGGTGTATGGAC -3′), aldolase A fructose-bisphosphate, Aldoa, (5′- GAGAACACCGAGGAGAAC-3′ and 5′-CCTTGGACTTGATAACTTGG -3′), or ribosomal protein L7, Rpl7, (5′- CGCACTGAGATTCGGATG-3′ and 5′-TTAATTGAAGCCTTGTTGAGC-3′). RT-PCR was carried out using a Bio-Rad iQ5 multicolor Real-Time PCR Detection System. Samples were run in triplicate with each primer set along with a standard curve. Ct values were normalized to Rpl7 using the delta-delta Ct method. Combined data are expressed as the mean ± SEM and Student’s t-test was used to detect significant (p<0.05) differences.
Western Blotting
Extracts from cortical tissue were prepared using RIPA buffer (Thermo Scientific, Rockford, IL), protease inhibitors (Complete Mini, Roche, Mannheim, Germany) and phosphatase inhibitors (Phosphatase Inhibitor Cocktail Set III, Calbiochem, San Diego, CA). Samples were homogenized using a Pro Homogenizer (ProScientific Inc., Oxford, CT) and protein concentration was determined using the bicinchoninic acid (BCA) assay (ThermoScientific) with bovine serum albumin as a standard. 30 µg of protein was loaded per lane on a 4–12% gradient acrylamide gel. Proteins were transferred to a nitrocellulose membrane and probed with an anti-phosphorylated ERK (p44/p42 MAPK, #9101, Cell Signaling Technology, Danvers, MA) or an anti-ERK (p44/p42 MAPK, #4695, Cell Signaling Technologies) specific antibody. Western blots were imaged and quantitated using the Licor Odyssey Infrared Imaging System and pERK was normalized to total ERK. Combined data are expressed as the mean ± SEM and Student’s t-test was used to detect significant (p<0.05) differences in the levels of pERK.
Results and Discussion
To identify E2-mediated alterations in the female cerebral cortical transcriptome, a comprehensive study was carried out using a mouse model system. Female mice were ovariectomized and implanted with silastic tubing containing oil alone or oil with E2, which produced a low, physiological level of E2 [21]. This method of E2 treatment has been used in several studies to demonstrate the anti-inflammatory and neuroprotective actions of E2 in the mouse cerebral cortex [19], [21], [30], [31]. After 7 days, brains were dissected and total RNA was isolated from the cerebral cortices. The isolated RNA was poly-A selected, converted to cDNA, and analyzed using RNA-Seq.
E2 significantly altered gene expression in cerebral cortex
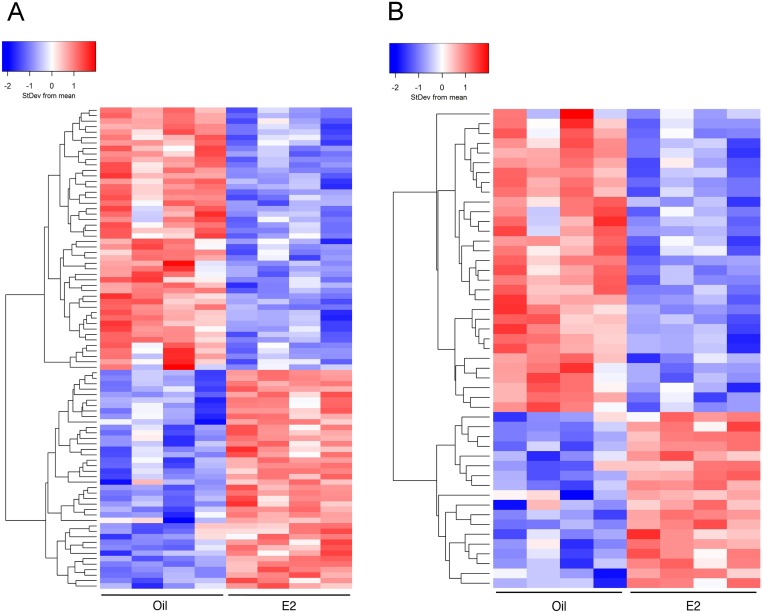
E2 significantly (FDR p<0.05) altered the expression of 88 genes in the cerebral cortex (Table 1, Table S1). The expression of these genes is displayed on a heat map (Fig. 1A), where red indicates a significant increase and blue indicates a significant decrease in transcript levels. Interestingly, the number of genes decreased (48) by E2 treatment was slightly greater than the number of genes increased (40) by E2 treatment.
Table 1. 88 E2-responsive genes in the cerebral cortex.
| Gene symbol | Description | FDR p-value | Fold change |
| 2410137F16Rik | RIKEN cDNA 2410137F16 gene | 0.020 | 1.7 |
| Adcy9 | adenylate cyclase 9 | 0.019 | 1.2 |
| Agxt2l1 | alanine-glyoxylate aminotransferase 2-like 1 | 3.1E-05 | 1.8 |
| Ankrd33b | ankyrin repeat domain 33B | 0.0071 | 1.2 |
| Apln | apelin | 0.0043 | −1.3 |
| Aqp4 | aquaporin 4 | 0.00059 | −1.3 |
| Bcas1 | breast carcinoma amplified sequence 1 | 3.1E-05 | −1.3 |
| Bhlhe40 | basic helix-loop-helix family, member e40 | 0.0040 | 1.2 |
| Btbd17 | BTB (POZ) domain containing 17 | 0.020 | −1.2 |
| Cd82 | CD82 antigen | 0.0058 | −1.3 |
| Cdhr1 | cadherin-related family member 1 | 0.038 | −2.8 |
| Cmtm5 | CKLF-like MARVEL transmembrane domain containing 5 | 0.0038 | −1.2 |
| Cnp | 2′,3′-cyclic nucleotide 3′ phosphodiesterase | 0.010 | −1.2 |
| Col19a1 | collagen, type XIX, alpha 1 | 0.048 | 1.3 |
| Cpeb1 | cytoplasmic polyadenylation element binding protein 1 | 0.030 | 1.1 |
| Cryab | crystallin, alpha B | 0.0017 | −1.2 |
| Dusp4 | dual specificity phosphatase 4 | 0.038 | 1.3 |
| Ednrb | endothelin receptor type B | 0.011 | −1.3 |
| Elfn2 | leucine rich repeat and fibronectin type III, extracellular 2 | 0.019 | 1.1 |
| Elovl5 | ELOVL family member 5, elongation of long chain fatty acids (yeast) | 0.011 | −1.2 |
| Erbb4 | v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) | 0.044 | 1.1 |
| Fa2h | fatty acid 2-hydroxylase | 0.041 | −1.2 |
| Fabp7 | fatty acid binding protein 7, brain | 0.001 | −1.6 |
| Fam107a | family with sequence similarity 107, member A | 0.016 | 1.2 |
| Fbxo33 | F-box protein 33 | 0.026 | 1.1 |
| Fcrls | Fc receptor-like S, scavenger receptor | 0.030 | 1.2 |
| Flnb | filamin, beta | 0.046 | 1.1 |
| Gdpd5 | glycerophosphodiester phosphodiesterase domain containing 5 | 0.029 | −1.2 |
| Gfap | glial fibrillary acidic protein | 0.0013 | −1.3 |
| Gja1 | gap junction protein, alpha 1 | 0.044 | −1.1 |
| Gjc2 | gap junction protein, gamma 2 | 0.00059 | −1.3 |
| Gltp | glycolipid transfer protein | 0.023 | −1.2 |
| Gm20634 | predicted gene 20634 | 0.019 | −1.3 |
| Gsn | gelsolin | 0.0071 | −1.2 |
| Hadha | hydroxyacyl-Coenzyme A dehydrogenase, alpha subunit | 0.019 | −1.1 |
| Herc1 | hect domain and RCC1-like domain 1 | 0.039 | 1.1 |
| Hist1h2bc | histone cluster 1, H2bc | 0.029 | −1.2 |
| Hivep3 | human immunodeficiency virus type I enhancer binding protein 3 | 0.010 | 1.2 |
| Igfbp2 | insulin-like growth factor binding protein 2 | 0.00090 | −1.5 |
| Igfbpl1 | insulin-like growth factor binding protein-like 1 | 0.023 | −2.3 |
| Irs2 | insulin receptor substrate 2 | 0.0022 | 1.3 |
| Jam2 | junction adhesion molecule 2 | 0.011 | −1.2 |
| Lyz2 | lysozyme 2 | 0.0043 | −1.7 |
| Mag | myelin-associated glycoprotein | 0.0017 | −1.3 |
| Mgl2 | macrophage galactose N-acetyl-galactosamine specific lectin 2 | 2.6E-06 | 3.5 |
| Mid1ip1 | Mid1 interacting protein 1 (gastrulation specific G12-like (zebrafish)) | 0.038 | −1.1 |
| Mll1 | myeloid/lymphoid or mixed-lineage leukemia 1 | 0.0020 | 1.2 |
| Mvd | mevalonate (diphospho) decarboxylase | 0.048 | −1.2 |
| Myoc | myocilin | 0.0017 | −1.4 |
| Ndufa3 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3 | 0.022 | −1.2 |
| Nlrp3 | NLR family, pyrin domain containing 3 | 0.0020 | 2.0 |
| Nov | nephroblastoma overexpressed gene | 0.010 | 1.3 |
| Nuak1 | NUAK family, SNF1-like kinase, 1 | 0.0013 | 1.2 |
| Olfml3 | olfactomedin-like 3 | 7.0E-06 | −1.4 |
| Pcnt | pericentrin (kendrin) | 0.038 | 1.1 |
| Pdgfb | platelet derived growth factor, B polypeptide | 8.8E-08 | 1.3 |
| Phf15 | PHD finger protein 15 | 0.0013 | 1.2 |
| Phf21b | PHD finger protein 21B | 0.021 | 1.2 |
| Plk2 | polo-like kinase 2 | 0.022 | 1.1 |
| Pllp | plasma membrane proteolipid | 0.033 | −1.2 |
| Prdx6 | peroxiredoxin 6 | 0.043 | −1.1 |
| Ptn | pleiotrophin | 0.016 | −1.2 |
| Ptpn7 | protein tyrosine phosphatase, non-receptor type 7 | 0.046 | −1.7 |
| Rtp1 | receptor transporter protein 1 | 0.0043 | 2.8 |
| Serpinb1a | serine (or cysteine) peptidase inhibitor, clade B, member 1a | 0.011 | −1.5 |
| Siglec1 | sialic acid binding Ig-like lectin 1, sialoadhesin | 0.048 | 1.9 |
| Slc13a3 | solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 | 0.014 | −1.4 |
| Slc25a1 | solute carrier family 25 (mitochondrial carrier, citrate transporter), member 1 | 0.019 | −1.2 |
| Slc38a3 | solute carrier family 38, member 3 | 0.048 | −1.2 |
| Sntb2 | syntrophin, basic 2 | 0.012 | 1.3 |
| Sowahb | sosondowah ankyrin repeat domain family member B | 0.039 | 1.2 |
| Spred2 | sprouty-related, EVH1 domain containing 2 | 0.023 | 1.1 |
| Srrm4 | serine/arginine repetitive matrix 4 | 0.0017 | 1.2 |
| Tcn2 | transcobalamin 2 | 0.022 | −1.2 |
| Tet3 | tet methylcytosine dioxygenase 3 | 0.016 | 1.1 |
| Tfrc | transferrin receptor | 0.0017 | 1.2 |
| Tgfbr1 | transforming growth factor, beta receptor I | 7.1E-07 | 1.3 |
| Tmem116 | transmembrane protein 116 | 0.038 | 1.4 |
| Top2a | topoisomerase (DNA) II alpha | 0.022 | −1.7 |
| Trib2 | tribbles homolog 2 (Drosophila) | 0.017 | 1.2 |
| Trp53inp2 | transformation related protein 53 inducible nuclear protein 2 | 0.038 | -1.1 |
| Tsc1 | tuberous sclerosis 1 | 0.017 | 1.1 |
| Tst | thiosulfate sulfurtransferase, mitochondrial | 0.0052 | −1.2 |
| Ugt8a | UDP galactosyltransferase 8A | 0.038 | −1.3 |
| Unc5b | unc-5 homolog B (C. elegans) | 0.021 | −1.2 |
| Vcam1 | vascular cell adhesion molecule 1 | 0.016 | −1.3 |
| Vstm4 | V-set and transmembrane domain containing 4 | 0.038 | −1.4 |
| Vwf | Von Willebrand factor homolog | 3.1E-05 | 1.6 |
Figure 1. Heatmaps comparing cerebral cortices from oil- and E2-treated mice.
Hierarchical clustering was used to visualize the transcript levels of (A) 88 genes that were significantly altered (FDR p<0.05) by E2 treatment or (B) 49 genes that were altered 1.2 fold or more by E2 treatment (FDR p<0.05). Each column represents cortical tissue from one mouse (8 mice total) and rows indicate genes. Colors symbolize increased (red) or decreased (blue) transcript levels.
Of the 88 genes that responded to E2 treatment, 49 were altered 1.2 fold or more (FDR p<0.05, Fig. 1B). Again, the number of genes decreased (blue) by E2 treatment was greater than the number of genes increased (red) by E2 treatment suggesting that E2 is a more potent repressor than activator of gene expression in the cerebral cortex. However, both heatmaps demonstrated that E2 differentially regulates gene expression in the mouse cerebral cortex.
Validation of E2 regulated genes
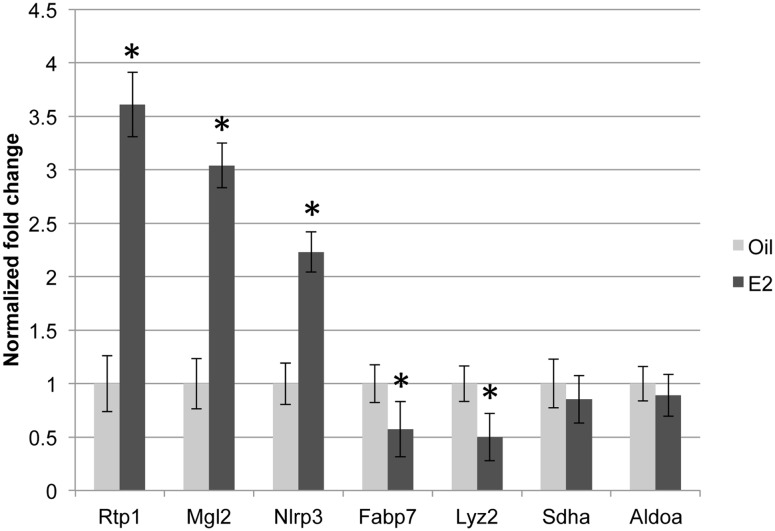
To validate the RNA-Seq data, we examined a subset of E2-regulated genes using RT-PCR analysis. In agreement with the RNA-Seq data, E2 increased Rtp1, Mgl2, and Nlrp3 expression, decreased Fabp7 and Lyz2 expression, and did not alter the expression of Sdha and Aldoa (Fig. 2, *p<0.05). Thus, RT-PCR analysis provided evidence of the accuracy of the RNA-Seq dataset.
Figure 2. Validation of transcripts altered by E2 treatment.
(A) Quantitative real-time PCR was conducted with gene-specific primers. The normalized fold change ± SEM was calculated using the delta-delta Ct method with Rpl7 as a control gene. The Student’s t-test was used to detect significant differences in oil- and E2- treated animals (4 animals/treatment, *p<0.05).
E2-regulated genes
We were interested in determining if any of the 88 genes that were significantly altered by E2 in the cerebral cortex have previously been reported as E2-responsive genes. Using the genome-wide expression profiling database Transcriptomine in the Nuclear Receptor Signaling Atlas, we found that each of the 88 genes except Rtp1, Gm20634, and 2410137FRik was listed as an E2-responsive gene in a variety of tissues or cultured cells [46], but only 5 genes, Aqp4, Bhlhe40, Ednrb, Erbb4 and Igfbp2, were designated as E2-responsive in the central nervous system. Two additional studies have reported that Gfap [47] and Slc13a3 [20] are E2-responsive in the brain. Thus, based on literature and database searches, the majority of the 88 genes identified in our dataset are novel, E2-regulated genes in the mouse cerebral cortex. Interestingly, earlier reports suggest that the gene expression profile of the hippocampus differs substantially from the gene expression profile of the cerebral cortex and that acute and chronic E2 treatments may differentially alter gene expression [48], [49].
The 10 genes that declined most significantly in response to E2 treatment are shown in Table 2. The largest E2-mediated decrease was observed in cadherin-related family member 1 (Cdhr1), which is a protocadherin in the cadherin superfamily, and functions as a calcium-dependent cell adhesion and signaling molecule [50]. The greatest E2-mediated increases in transcript levels are listed in Table 3. The expression of macrophage galactose N-acetyl-galactosamine specific lectin 2 (Mgl2), also referred to as CD301, was most significantly increased. The function of this gene in the cerebral cortex has not been described. However, microglia, the resident immune cells in the brain, often express multiple cluster of differentiation (CD) cell surface proteins [51]. Siglec1, another CD gene (CD169), is expressed on macrophages associated with the perivasculature in the rat brain [52]. These results suggest that E2 may be altering gene expression in immune cells.
Table 2. E2-regulated genes with the most decreased expression.
| Gene symbol | Description | Fold decrease | FDR p value |
| Cdhr1 | cadherin-related family member 1 | −2.8 | 0.038 |
| Igfbpl1 | insulin-like growth factor binding protein-like 1 | −2.3 | 0.023 |
| Ptpn7 | protein tyrosine phosphatase, non-receptor type 7 | −1.7 | 0.046 |
| Top2a | topoisomerase (DNA) II alpha | −1.7 | 0.022 |
| Lyz2 | lysozyme 2 | −1.7 | 0.0043 |
| Fabp7 | fatty acid binding protein 7, brain | −1.6 | 0.0013 |
| Serpinb1a | serine (or cysteine) peptidase inhibitor, clade B, member 1a | −1.5 | 0.011 |
| Igfbp2 | insulin-like growth factor binding protein 2 | −1.5 | 0.0009 |
| Olfml3 | olfactomedin-like 3 | −1.4 | 7.00E-06 |
| Myoc | myocilin | −1.4 | 0.0017 |
Table 3. E2-regulated genes with the most increased expression.
| Gene symbol | Description | Fold increase | FDR p value |
| Mgl2 | macrophage galactose N-acetyl-galactosamine specific lectin 2 | 3.5 | 2.60E-06 |
| Rtp1 | receptor transporter protein 1 | 2.8 | 0.0043 |
| Nlrp3 | NLR family, pyrin domain containing 3 | 2 | 0.002 |
| Siglec1 | sialic acid binding Ig-like lectin 1, sialoadhesin | 1.9 | 0.048 |
| Agxt2l1 | alanine-glyoxylate aminotransferase 2-like 1 | 1.8 | 3.10E-05 |
| 2410137F16Rik | RIKEN cDNA 2410137F16 gene | 1.7 | 0.02 |
| Vwf | Von Willebrand factor homolog | 1.6 | 3.10E-05 |
| Tmem116 | transmembrane protein 116 | 1.4 | 0.038 |
| Pdgfb | platelet derived growth factor, B polypeptide | 1.3 | 8.80E-08 |
| Sntb2 | syntrophin, basic 2 | 1.3 | 0.012 |
Networks of E2-responsive biological processes and pathways
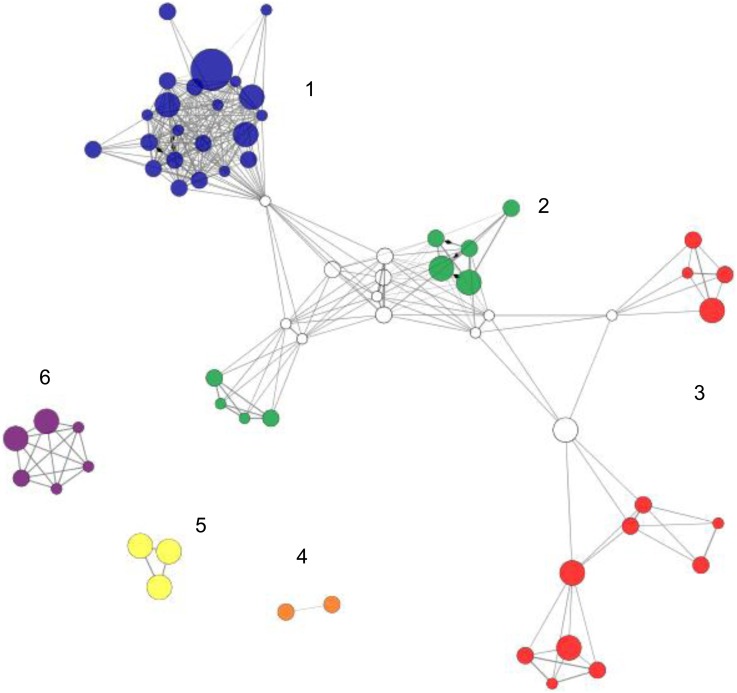
To begin to understand how E2 regulates the cerebral cortical transcriptome, the 88 E2-responsive genes were uploaded to ClueGO [42] to identify networks of biological processes and pathways that are altered by E2 treatment (Fig. 3 and Table 4). The nodes (filled circles) represent biological processes or pathways associated with the E2-regulated genes based on gene ontology terms [45], Reactome [44] and KEGG [43] databases. Related nodes are clustered together in color-coded networks and all of the nodes in a network are the same color. However, a node can participate in two networks and those nodes are white. The size of the node reflects the level of statistical significance of each of the E2-regulated biological processes or pathways. Thus larger nodes have increased statistical significance.
Figure 3. Networks of E2-regulated genes.
ClueGO analysis classified the 88 E2-regulated genes into 6 networks. White nodes indicate that a biological process is associated with two networks. Node size indicates the statistical significance of the biological process represented. Thus, larger nodes indicate greater statistical significance.
Table 4. E2-responsive networks and associated genes.
| Biological process GO term or Pathway | Genes |
| Network 1 | |
| Regulation of centrosome cycle | Gja1, Plk2 |
| Inactivation of MAPK activity | Dusp4, Spred2 |
| Cell-cell junction assembly | Gja1, Ugt8a |
| Regulation of tissue remodeling | Gja1, Tfrc |
| Regulation of mRNA splicing, via spliceosome | Gja1, Srrm4 |
| Carbohydrate derivative transport | Gja1, Gltp |
| Regulation of signal transduction by p53 class mediator | Gja1, Spred2 |
| Long-term potentiation | Gfap, Plk2 |
| Regulation of cell junction assembly | Gja1, Tsc1 |
| Lens development in camera-type eye | Cryab, Gja1, Tgfrb1 |
| Regulation of phosphoprotein phosphatase activity | Nuak1, Tsc1 |
| rRNA transport | Gja1, Tsc1, Tst |
| Network 2 | |
| Cerebellum morphogenesis | Herc1, Pcnt |
| Vasoconstriction | Apln, Ednrb, Pdgfb |
| Vasodilation | Apln, Cnp, Pdgfb |
| Network 3 | |
| Fatty acid elongation | Elovl5, Hadha |
| Triglyceride biosynthesis | Elovl5, Slc25a1 |
| Histone H4 acetylation | Mll1, Phf15 |
| Histone H3-K4 methylation | Mll1, Tet3 |
| Regulation of ligase activity | Mid1ip1, Trib2 |
| Lysine degradation | Hadha, Mll1 |
| Negative regulation of protein complex disassembly | Gsn, Mid1ip1 |
| Network 4 | |
| Response to estradiol stimulus | Aqp4, Cryab, Igfbp2 |
| Vasopressin-regulated water reabsorption | Adcy9, Aqp4 |
| Network 5 | |
| Myelination | Fa2h, Pllp, Tsc1, Ugt8a |
| Network 6 | |
| Meiotic chromosome separation | Bhlhe40, Top2a |
| Network 1 and 2 | |
| Olfactory lobe development | Erbb4, Pcnt |
| Regulation of phosphatidylinositol 3-kinase activity | Erbb4, Pdgfb |
| Tissue regeneration | Erbb4, Gja1 |
| Regulation of lipid kinase activity | Erbb4, Pdgfb |
| Network 2 and 3 | |
| Positive regulation of fatty acid metabolic process | Irs2, Mid1ip1 |
| Fatty acid beta oxidation | Hadha, Irs2 |
| Regulation of polysaccharide metabolic process | Irs2, Pdgfb |
The largest network is comprised of 22 nodes (Fig. 3, blue nodes) and the 14 genes associated with these nodes are listed in Table 4. Genes associated with this network include glial fibrillary acidic protein (Gfap) and polo-like kinase 1 (Plk2), which play a role in long term synaptic potentiation. Synaptic plasticity, learning, and memory are linked to long term synaptic potentiation. Dual specificity phosphatase 4 (Dusp4) and sprouty-related, EVH1 domain containing 2 (Spred2) are associated with the MAPK pathway, which is important in synaptic plasticity and also plays a role in cell signaling [53]. NUAK family, SNF 1-like kinase (Nuak1) and tuberous sclerosis 1 (Tsc1) are associated with phosphoprotein phosphatase regulation and may be contributing to modulation of protein phosphorylation. Taken together, these results suggest that E2 is affecting important signal integration pathways in the cerebral cortex.
Unique to network 2 (green) were biological processes involved in vasoconstriction and vasodilation which included platelet derived growth factor B (Pdgfb) and endothelin receptor type B (Ednrb). Networks 1 and 2 shared genes involved in PI3K activity such as v-erb-a erythroblastic leukemia viral oncogene homolog 4 (Erbb4) and Pdgfb. The PI3K pathway is important for E2 signaling and inhibition of this pathway blocks downstream ERK activation by E2 in cortical neuron cultures [54]. Fatty acid synthesis is a critical function in the brain, which contains the second highest level of lipids in the body after adipose tissue [55]. Fatty acid metabolic processes were associated with insulin receptor substrate 2 (Irs2) and Mid1 interacting protein 1 (Mid1ip1) in network 3. Mid1ip1 enhances fatty acid synthesis and its overexpression in the liver causes triglyceride accumulation [56]. Irs2 is critical in regulating brain size, since the brains of Irs2 null mice are reduced by ∼50% due to decreased neuronal proliferation [57].
Pathways involved in lipid synthesis were also present in network 3 (red) and included genes ELOVL family member 5, elongation of long chain fatty acids (Elovl5) and hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-coenzyme A thiolase/enoyl-Coenzyme A hydratase alpha subunit (Hadha) and the solute carrier 25 (Slc25a1). Regulation of ligase activity was associated with Mid1ip1 and tribbles homolog 2 (Trib2). In addition to its role in fatty acid synthesis, Mid1ip1 interacts with Mid1, a ubiquitin ligase and microtubule associated protein [58], [59]. Trib2 functions as an adaptor for protein degradation through interactions with the E3-ubiquitin ligase Cop1 [60]. Gelsolin (Gsn), which encodes an actin binding protein involved in signaling and cytoskeletal remodeling [61] is associated with negative regulation of protein complex disassembly.
Three networks (4–6) were not connected to any of the other networks. Genes previously reported to be estrogen responsive in various cell types were included in network 4 (orange). The genes in this group included crystallin alpha b (Cryab), aquaporin 4 (Aqp4), and insulin-like growth factor binding protein 2 (Igfbp2), which have been reported as E2-responsive genes in the mouse uterus [62], cultured rat cortical neurons [63], and rat hippocampal tissue, respectively [64], [65]. Myelin is essential for proper nerve conduction [66] and network 5 (yellow) contained several genes associated with myelination. Fatty acid 2-hydroxylase (Fa2h), plasma membrane proteolipid (Pllp), UDP galactosyltransferase 8A (Ugt8a), and tuberous sclerosis 1 (Tst1) have been associated with oligodendrocytes, which produce myelin. Network 6 (purple) included DNA topoisomerase 2A (Top2a) and basic helix loop helix family, member 40 (Bhlhe40) which are both involved in chromosome separation. However, Top2a has also been reported to be expressed in cortical neurons [67] and Bhlhe40, also known as Stra13, has been implicated in neuronal differentiation [68]. Thus the roles of these genes extend beyond chromosomal separation.
The network analysis provided insight into the diverse array of functions that were affected by E2 treatment. However, this analysis is constrained by existing information in the databases used, which consequently did not include all of the 88 genes we identified. Thus we utilized literature searches to provide a more comprehensive picture of the pathways and processes that were affected by E2 treatment.
Signaling Pathways
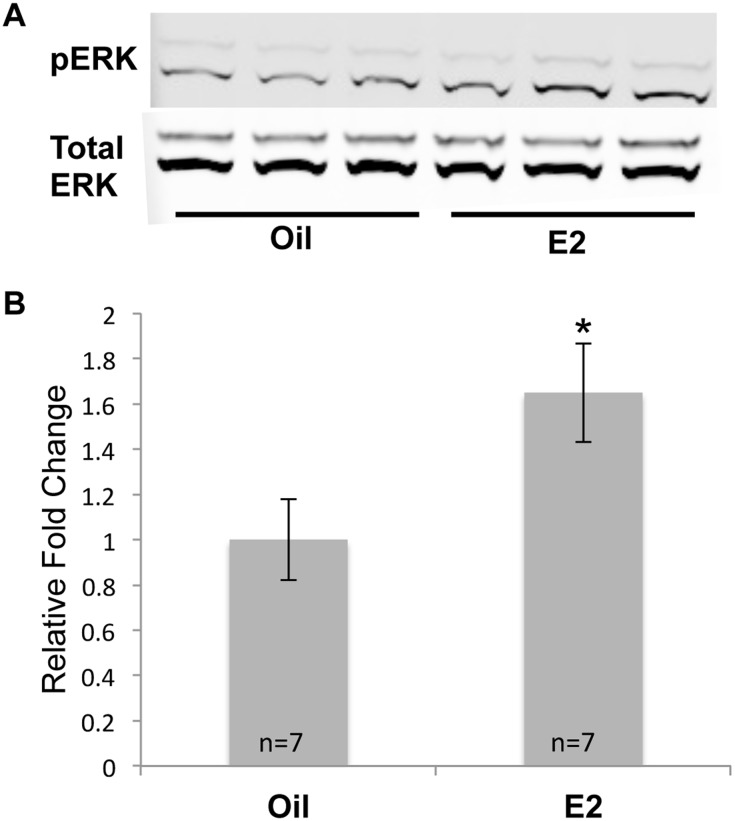
The MAP kinase pathway plays a critical role in neuronal plasticity and survival [53] and E2 has been implicated in inducing rapid signaling through this pathway in neuroblastoma cells, primary cortical neurons, cortical explants, and the cerebral cortex in vivo [54], [69]–[71]. To determine whether MAP kinase signaling was activated after longer E2 treatment, we examined the level of phosphorylated extracellular regulated kinase (pERK) in the cortices of mice that had been treated with oil or E2 for 7 days. In fact, the level of pERK was significantly increased in the E2-treated animals (Fig. 4) demonstrating that E2 modulation of pERK and MAP kinase signaling is not limited to acute exposures (5–30 min), but is still enhanced after a longer treatment time.
Figure 4. E2 increases pERK protein levels.
(A)Western blot analysis was used to monitor pERK and total ERK levels in the cortices of mice that had been treated with oil or E2 for 7 days. (B) pERK values were normalized to total ERK and are displayed as the normalized fold change ± SEM. The Student’s t-test was used to detect significant differences in oil- and E2- treated animals (*p<0.05). The number of animals in each treatment group is indicated at the base of each bar.
We identified several E2-regulated genes in the cerebral cortex that modulate the MAP kinase signaling pathway. Dual specificity phosphatase 4 (Dusp4) dephosphorylates ERK [72] and is increased by E2 in breast cancer cells [73]. Thus the E2-mediated increase in Dusp4 expression could lead to decreased ERK phosphorylation. Interestingly, expression of protein tyrosine phosphatase, non-receptor type 7 (Ptpn7) was decreased with E2 treatment and may also be involved in ERK dephosphorylation [74]. In addition, expression of a repressor of MAP kinase activity, Spred2, [75] was modestly increased with E2 treatment. The fine-tuned expression of these genes by E2 highlights the balance that is needed between phosphorylation and dephosphorylation in the MAP kinase pathway [74]. Dusp4, Ptpn7 and Spred2 are novel, E2-responsive modulators of MAP kinase in the cerebral cortex.
Cerebral cortex microvasculature
The brain is one of the most highly perfused organs in the body [76]. Proper brain function relies on maintenance of an extensive network of capillaries that form the cerebral microvasculature, which supplies oxygen and nutrients to meet the demands of this highly metabolic tissue. The microvasculature is comprised of endothelial cells surrounded by an extracellular matrix and a variety of cell types including neurons, astrocytes, microglia, and pericytes [51]. This complex network is referred to as the “neurovascular unit” since cooperation between these cells is necessary to maintain microvascular function [77]. Tight junctions between endothelial cells, together with the surrounding astrocyte end feet and pericytes form the blood-brain barrier (BBB), which carefully regulates the exchange of nutrients, water and other molecules. A dysfunctional BBB can lead to neurodegeneration and is the hallmark of several brain injuries [78].
Previous studies have shown that E2 decreases BBB permeability and thereby limits ischemic damage [79]. We identified several genes involved in BBB regulation that were altered by E2 treatment. Pdgfb transcript levels were increased by E2 treatment. Since Pdgfb binds to the Pdgfb receptor on pericytes [80] and mice with low Pdgfb levels have a dysfunctional BBB [81], Pdgfb is necessary for pericyte proliferation and maintenance [82], [83] and a functional neurovascular unit. Therefore, E2 may be acting to stimulate synthesis of Pdgfb in pericytes and endothelial cells, which could enhance autocrine and paracrine signaling to support BBB function.
E2 decreased the expression of Aqp4, a water transporter present on astrocyte end feet. These findings are in agreement with a previous report which indicated that E2 decreases Aqp4 expression and reduces hypoxia-induced swelling of rat cortical astrocytes in vitro [63]. E2 also decreased the expression of two solute carriers, Slc13a3, a sodium decarboxylate cotransporter and Slc38a3, an amino acid transporter, that have been associated with the BBB [84]. Together, the E2 mediated reduction in Aqp4, Slc13a3, and Slc38a3 could alter the exchange of water and solutes at the BBB and help to maintain fluid balance and homeostasis in the brain.
Von Willebrand Factor (Vwf) was increased by E2 treatment. Vwf is highly expressed in endothelial cells of brain microvasculature [85] and Vwf-null mice have increased damage compared to their wild-type counterparts after exposure to hypoxia and reoxygenation [86], suggesting that this factor is necessary for BBB adaptability and may help the brain to recover from an hypoxic event.
E2 has antinflammatory effects on the vasculature [87]. We found that E2 treatment decreased Vcam1 expression in the cerebral cortex. Vcam1 attracts leukocytes and monocytes to inflamed endothelial cells [88]. It has been proposed that E2 may decrease inflammation of endothelial cell cultures that have been subjected to an inflammatory agent by decreasing Vcam1 expression [89]. In addition, the E2 induced increase of Tgfb receptor 1 (Tgfbr1) may increase the sensitivity of Tgfb signaling, thus reducing inflammation [90]. The combined effects of E2 on maintaining the BBB (Pdgfb, Aqp4, Slc13a3, Slc38a3, Vwf) and reducing inflammation (Vcam1, Tgfbr1) could help to protect the cerebral cortex from injury.
Oligodendrocytes and Myelin
Oligodendrocytes insulate neuronal axons by extending processes that produce lipid rich myelin. Myelin ensheathment of axons is important for nerve conduction and the loss of myelin leads to neurodegeneration. We were surprised at the number of oligodendrocyte-associated genes that were E2 responsive. Expression of myocillin (Myoc), myelin-associated glycoprotein (Mag), UDP galactosyltransferase 8a (Ugt8a), fatty acid 2-hydrolase (Fa2h), 2′, 3′-cyclic nucleotide 3′ phosphodiesterase (Cnp), CKLF-like MARVEL transmembrane domain containing 5 (Cmtm5) and plasma membrane proteolipid (Pllp) were all decreased by E2 treatment. A previous study found that turnover of oligodendrocytes in female rodents was increased and that Cnp protein expression was less than in males [91]. The decreased expression of these genes could indicate that E2 increases oligodendrocyte turnover rates in the cerebral cortex as well. However, much remains to be learned about the molecular consequences of decreased expression in this subset of oligodendrocyte- associated genes.
Neurite extension
Neurite outgrowth is important for neuronal development, communication and function [92]. Impairment of neurite extension is associated with aging and neurodegeneration [93]. E2 can increase neurite extension in a variety of brain regions through several pathways including growth factor signaling, PI3K, and MAP kinase pathways [94]. We identified several genes involved in neurite extension that were altered by E2 treatment. E2 treatment modestly increased expression of Erbb4, which encodes a transmembrane protein that binds to neuregulin 1 (Nrg1). Erbb4-Nrg1signaling enhances neurite outgrowth through activation of the PI3K and MAP kinase pathways [95]. The E2-mediated increase in Erbb4 in the cerebral cortex could enhance neuritogenesis.
The expression of Igfbpl1 and Igfbp2 was decreased by E2 treatment. Igfbpl1 and Igfbp2 bind and sequester growth factors such as Igf1 [96]. Although Igfbpl1 is expressed in the developing mouse forebrain [97], the role of Igfbpl1 in the cerebral cortex has not been examined. In breast cancer cells, a decrease in Igfbpl1 has been associated with an increase in Igf1 levels [98]. E2 decreases Igfbp2 in the hippocampus [64] which can modulate Igf1 signaling pathways [99]. Moreover, Igf1 and E2 act synergistically to promote neurite outgrowth [100]. The E2-mediated decrease in expression of both Igfbpl1 and Igfbp2 could potentially allow growth factors such as Igf1 to circulate and promote neurite extension.
Glial fibrillary acidic protein (Gfap), an intermediate filament protein specifically expressed by astrocytes, can inhibit neurite outgrowth [47], [101]. The E2-mediated decrease in expression of Gfap in our studies suggests that E2 supports neurite extension, and could prevent an age-related increase in Gfap expression.
Overall Implications
E2 alters gene expression through classical pathways that involve binding of the E2-occupied receptor to DNA. E2 can also act through non-classical pathways, by activation of membrane-associated proteins and rapid signaling pathways such as MAP kinase and PI3K, both of which have been shown to be important in the brain [102], [103]. It has been suggested that cross-talk occurs amongst the various E2 signaling pathways [104], [105] and that the cumulative E2-activation of several pathways may be required for effective E2-mediated neuroprotection [102].
Our study reflects the complex nature of E2 action and suggests that multiple signaling pathways in the cerebral cortex converge to orchestrate a diverse array of molecular events including those related to cerebrovascular function, neurite outgrowth, and brain homeostasis. The molecular impact of E2 treatment has particular relevance when considering the physiological consequences of menopause and estrogen replacement therapy. Further understanding of these events may provide insight into mechanisms responsible for estrogen-mediated gene expression and promote development of targeted treatments that support brain homeostasis.
Supporting Information
RNA gel demonstrating intact RNA samples. Native agarose gel electrophoresis was used to resolve the intact 28s and 18s rRNA bands. 2 µg of RNA were run per lane.
(PDF)
88 E2-responsive genes in the cerebral cortex.
(XLSX)
Acknowledgments
We thank Alicia Dietrich and Lisi Yuan for comments and help with mouse surgeries. We also thank Samantha Pisani and Claire Scavuzzo for thought-provoking discussions.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. Sequencing files are available from EMBL-EBI Array Express under accession number E-MTAB-2762.
Funding Statement
This work was supported by a grant (R01 DK053884) from the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (www.niddk.nih.gov) to AN. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Brisken C, O'Malley B (2010) Hormone action in the mammary gland. Cold Spring Harb Perspect Biol 2: a003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Richelson LS, Wahner HW, Melton LJ 3rd, Riggs BL (1984) Relative contributions of aging and estrogen deficiency to postmenopausal bone loss. N Engl J Med 311: 1273–1275. [DOI] [PubMed] [Google Scholar]
- 3. Nieves JW, Komar L, Cosman F, Lindsay R (1998) Calcium potentiates the effect of estrogen and calcitonin on bone mass: Review and analysis. Am J Clin Nutr 67: 18–24. [DOI] [PubMed] [Google Scholar]
- 4. Mendelsohn ME (2000) Mechanisms of estrogen action in the cardiovascular system. J Steroid Biochem Mol Biol 74: 337–343. [DOI] [PubMed] [Google Scholar]
- 5. McEwen B (2002) Estrogen actions throughout the brain. Recent Prog Horm Res 57: 357–384. [DOI] [PubMed] [Google Scholar]
- 6. McCarthy MM (2008) Estradiol and the developing brain. Physiol Rev 88: 91–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McEwen BS, Woolley CS (1994) Estradiol and progesterone regulate neuronal structure and synaptic connectivity in adult as well as developing brain. Exp Gerontol 29: 431–436. [DOI] [PubMed] [Google Scholar]
- 8. Nakai Y, Plant TM, Hess DL, Keogh EJ, Knobil E (1978) On the sites of the negative and positive feedback actions of estradiol in the control of gonadotropin secretion in the rhesus monkey. Endocrinology 102: 1008–1014. [DOI] [PubMed] [Google Scholar]
- 9. Harlan RE (1988) Regulation of neuropeptide gene expression by steroid hormones. Mol Neurobiol 2: 183–200. [DOI] [PubMed] [Google Scholar]
- 10. McEwen BS, Alves SE (1999) Estrogen actions in the central nervous system. Endocr Rev 20: 279–307. [DOI] [PubMed] [Google Scholar]
- 11. Garcia-Segura LM, Azcoitia I, DonCarlos LL (2001) Neuroprotection by estradiol. Prog Neurobiol 63: 29–60. [DOI] [PubMed] [Google Scholar]
- 12. Behl C (2002) Oestrogen as a neuroprotective hormone. Nat Rev Neurosci 3: 433–442. [DOI] [PubMed] [Google Scholar]
- 13. Green PS, Simpkins JW (2000) Neuroprotective effects of estrogens: Potential mechanisms of action. Int J Dev Neurosci 18: 347–358. [DOI] [PubMed] [Google Scholar]
- 14. Wang X, Dykens JA, Perez E, Liu R, Yang S, et al. (2006) Neuroprotective effects of 17beta-estradiol and nonfeminizing estrogens against H2O2 toxicity in human neuroblastoma SK-N-SH cells. Mol Pharmacol 70: 395–404. [DOI] [PubMed] [Google Scholar]
- 15. Sortino MA, Chisari M, Merlo S, Vancheri C, Caruso M, et al. (2004) Glia mediates the neuroprotective action of estradiol on beta-amyloid-induced neuronal death. Endocrinology 145: 5080–5086. [DOI] [PubMed] [Google Scholar]
- 16. Liang K, Yang L, Yin C, Xiao Z, Zhang J, et al. (2010) Estrogen stimulates degradation of beta-amyloid peptide by up-regulating neprilysin. J Biol Chem 285: 935–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Singer CA, Figueroa-Masot XA, Batchelor RH, Dorsa DM (1999) The mitogen-activated protein kinase pathway mediates estrogen neuroprotection after glutamate toxicity in primary cortical neurons. J Neurosci 19: 2455–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Raval AP, Bramlett H, Perez-Pinzon M (2006) Estrogen preconditioning protects the hippocampal CA1 against ischemia. Neuroscience 141: 1721–1730. [DOI] [PubMed] [Google Scholar]
- 19. Suzuki S, Brown CM, Dela Cruz CD, Yang E, Bridwell DA, et al. (2007) Timing of estrogen therapy after ovariectomy dictates the efficacy of its neuroprotective and antiinflammatory actions. Proc Natl Acad Sci U S A 104: 6013–6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sarvari M, Kallo I, Hrabovszky E, Solymosi N, Toth K, et al. (2010) Estradiol replacement alters expression of genes related to neurotransmission and immune surveillance in the frontal cortex of middle-aged, ovariectomized rats. Endocrinology 151: 3847–3862. [DOI] [PubMed] [Google Scholar]
- 21. Dubal DB, Zhu H, Yu J, Rau SW, Shughrue PJ, et al. (2001) Estrogen receptor alpha, not beta, is a critical link in estradiol-mediated protection against brain injury. Proc Natl Acad Sci U S A 98: 1952–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Simpkins JW, Rajakumar G, Zhang Y, Simpkins CE, Greenwald D, et al. (1997) Estrogens may reduce mortality and ischemic damage caused by middle cerebral artery occlusion in the female rat. J Neurosurg 87: 724–730. [DOI] [PubMed] [Google Scholar]
- 23. Adams MM, Shah RA, Janssen WGM, Morrison JH (2001) Different modes of hippocampal plasticity in response to estrogen in young and aged female rats. Proceedings of the National Academy of Sciences of the United States of America 98: 8071–8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Balthazart J, Ball GF (2006) Is brain estradiol a hormone or a neurotransmitter? Trends Neurosci 29: 241–249. [DOI] [PubMed] [Google Scholar]
- 25. Fink G, Sumner BE, Rosie R, Grace O, Quinn JP (1996) Estrogen control of central neurotransmission: Effect on mood, mental state, and memory. Cell Mol Neurobiol 16: 325–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Toran-Allerand CD (1996) The estrogen/neurotrophin connection during neural development: Is co-localization of estrogen receptors with the neurotrophins and their receptors biologically relevant? Dev Neurosci 18: 36–48. [DOI] [PubMed] [Google Scholar]
- 27. Keenan PA, Ezzat WH, Ginsburg K, Moore GJ (2001) Prefrontal cortex as the site of estrogen's effect on cognition. Psychoneuroendocrinology 26: 577–590. [DOI] [PubMed] [Google Scholar]
- 28. Wise PM, Camp-Grossman P, Barraclough CA (1981) Effects of estradiol and progesterone on plasma gonadotropins, prolactin, and LHRH in specific brain areas of ovariectomized rats. Biol Reprod 24: 820–830. [DOI] [PubMed] [Google Scholar]
- 29. Nelson JF, Felicio LS, Osterburg HH, Finch CE (1992) Differential contributions of ovarian and extraovarian factors to age-related reductions in plasma estradiol and progesterone during the estrous cycle of C57BL/6J mice. Endocrinology 130: 805–810. [DOI] [PubMed] [Google Scholar]
- 30. Dubal DB, Shughrue PJ, Wilson ME, Merchenthaler I, Wise PM (1999) Estradiol modulates bcl-2 in cerebral ischemia: A potential role for estrogen receptors. J Neurosci 19: 6385–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dubal DB, Kashon ML, Pettigrew LC, Ren JM, Finklestein SP, et al. (1998) Estradiol protects against ischemic injury. J Cereb Blood Flow Metab 18: 1253–1258. [DOI] [PubMed] [Google Scholar]
- 32. Trapnell C, Pachter L, Salzberg SL (2009) TopHat: Discovering splice junctions with RNA-seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langmead B (2010) Aligning short sequencing reads with bowtie. Curr Protoc Bioinformatics Chapter 11: Unit 11.7. [DOI] [PMC free article] [PubMed]
- 34.R Core Team (2013) R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statisical Computing.
- 35. Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, et al. (2004) Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson MD, Oshlack A (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11: R25-2010-11-3-r25. Epub 2010 Mar 2. [DOI] [PMC free article] [PubMed]
- 38. McCarthy DJ, Chen Y, Smyth GK (2012) Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Res 40: 4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 57: 289–300. [Google Scholar]
- 40.Law C (2013) Melbourne, Australia: PhD Thesis. University of Melbourne.
- 41. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, et al. (2003) Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, et al. (2009) ClueGO: A cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25: 1091–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kanehisa M, Goto S, Kawashima S, Nakaya A (2002) The KEGG databases at GenomeNet. Nucleic Acids Res 30: 42–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Joshi-Tope G, Gillespie M, Vastrik I, D'Eustachio P, Schmidt E, et al. (2005) Reactome: A knowledgebase of biological pathways. Nucleic Acids Res 33: D428–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. (2000) Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat Genet 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ochsner SA, Watkins CM, McOwiti A, Xu X, Darlington YF, et al. (2012) Transcriptomine, a web resource for nuclear receptor signaling transcriptomes. Physiol Genomics 44: 853–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rozovsky I, Wei M, Stone DJ, Zanjani H, Anderson CP, et al. (2002) Estradiol (E2) enhances neurite outgrowth by repressing glial fibrillary acidic protein expression and reorganizing laminin. Endocrinology 143: 636–646. [DOI] [PubMed] [Google Scholar]
- 48. Aenlle KK, Kumar A, Cui L, Jackson TC, Foster TC (2009) Estrogen effects on cognition and hippocampal transcription in middle-aged mice. Neurobiol Aging 30: 932–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Aenlle KK, Foster TC (2010) Aging alters the expression of genes for neuroprotection and synaptic function following acute estradiol treatment. Hippocampus 20: 1047–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Morishita H, Umitsu M, Murata Y, Shibata N, Udaka K, et al. (2006) Structure of the cadherin-related neuronal receptor/protocadherin-alpha first extracellular cadherin domain reveals diversity across cadherin families. J Biol Chem 281: 33650–33663. [DOI] [PubMed] [Google Scholar]
- 51. Guillemin GJ, Brew BJ (2004) Microglia, macrophages, perivascular macrophages, and pericytes: A review of function and identification. J Leukoc Biol 75: 388–397. [DOI] [PubMed] [Google Scholar]
- 52. Willis CL, Camire RB, Brule SA, Ray DE (2013) Partial recovery of the damaged rat blood–brain barrier is mediated by adherens junction complexes, extracellular matrix remodeling and macrophage infiltration following focal astrocyte loss. Neuroscience 250: 773–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Thomas GM, Huganir RL (2004) MAPK cascade signalling and synaptic plasticity. Nat Rev Neurosci 5: 173–183. [DOI] [PubMed] [Google Scholar]
- 54. Mannella P, Brinton RD (2006) Estrogen receptor protein interaction with phosphatidylinositol 3-kinase leads to activation of phosphorylated akt and extracellular signal-regulated kinase 1/2 in the same population of cortical neurons: A unified mechanism of estrogen action. J Neurosci 26: 9439–9447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marin R, Casanas V, Perez JA, Fabelo N, Fernandez C, et al.. (2013) Oestrogens as modulators of neuronal signalosomes and brain lipid homeostasis related to protection against neurodegeneration. J Neuroendocrinol. [DOI] [PubMed]
- 56. Kim CW, Moon YA, Park SW, Cheng D, Kwon HJ, et al. (2010) Induced polymerization of mammalian acetyl-CoA carboxylase by MIG12 provides a tertiary level of regulation of fatty acid synthesis. Proc Natl Acad Sci U S A 107: 9626–9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schubert M, Brazil DP, Burks DJ, Kushner JA, Ye J, et al. (2003) Insulin receptor substrate-2 deficiency impairs brain growth and promotes tau phosphorylation. J Neurosci 23: 7084–7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Berti C, Fontanella B, Ferrentino R, Meroni G (2004) Mig12, a novel opitz syndrome gene product partner, is expressed in the embryonic ventral midline and co-operates with Mid1 to bundle and stabilize microtubules. BMC Cell Biol 5: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Trockenbacher A, Suckow V, Foerster J, Winter J, Krauss S, et al. (2001) MID1, mutated in opitz syndrome, encodes an ubiquitin ligase that targets phosphatase 2A for degradation. Nat Genet 29: 287–294. [DOI] [PubMed] [Google Scholar]
- 60. Keeshan K, Bailis W, Dedhia PH, Vega ME, Shestova O, et al. (2010) Transformation by tribbles homolog 2 (Trib2) requires both the Trib2 kinase domain and COP1 binding. Blood 116: 4948–4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sun HQ, Yamamoto M, Mejillano M, Yin HL (1999) Gelsolin, a multifunctional actin regulatory protein. J Biol Chem 274: 33179–33182. [DOI] [PubMed] [Google Scholar]
- 62. Tian XC, Wang QY, Li DD, Wang ST, Yang ZQ, et al. (2013) Differential expression and regulation of cryab in mouse uterus during preimplantation period. Reproduction 145: 577–585. [DOI] [PubMed] [Google Scholar]
- 63. Rutkowsky JM, Wallace BK, Wise PM, O'Donnell ME (2011) Effects of estradiol on ischemic factor-induced astrocyte swelling and AQP4 protein abundance. Am J Physiol Cell Physiol 301: C204–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pechenino AS, Frick KM (2009) The effects of acute 17beta-estradiol treatment on gene expression in the young female mouse hippocampus. Neurobiol Learn Mem 91: 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Takeo C, Ikeda K, Horie-Inoue K, Inoue S (2009) Identification of Igf2, Igfbp2 and Enpp2 as estrogen-responsive genes in rat hippocampus. Endocr J 56: 113–120. [DOI] [PubMed] [Google Scholar]
- 66. Sherman DL, Brophy PJ (2005) Mechanisms of axon ensheathment and myelin growth. Nat Rev Neurosci 6: 683–690. [DOI] [PubMed] [Google Scholar]
- 67. Peng ZF, Koh CH, Li QT, Manikandan J, Melendez AJ, et al. (2007) Deciphering the mechanism of HNE-induced apoptosis in cultured murine cortical neurons: Transcriptional responses and cellular pathways. Neuropharmacology 53: 687–698. [DOI] [PubMed] [Google Scholar]
- 68. Boudjelal M, Taneja R, Matsubara S, Bouillet P, Dolle P, et al. (1997) Overexpression of Stra13, a novel retinoic acid-inducible gene of the basic helix-loop-helix family, inhibits mesodermal and promotes neuronal differentiation of P19 cells. Genes Dev 11: 2052–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Watters JJ, Campbell JS, Cunningham MJ, Krebs EG, Dorsa DM (1997) Rapid membrane effects of steroids in neuroblastoma cells: Effects of estrogen on mitogen activated protein kinase signalling cascade and c-fos immediate early gene transcription. Endocrinology 138: 4030–4033. [DOI] [PubMed] [Google Scholar]
- 70. Singh M, Setalo G, Jr, Guan X, Warren M, Toran-Allerand CD (1999) Estrogen-induced activation of mitogen-activated protein kinase in cerebral cortical explants: Convergence of estrogen and neurotrophin signaling pathways. J Neurosci 19: 1179–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bryant D, Bosch M, Rønnekleiv O, Dorsa D (2005) 17-beta estradiol rapidly enhances extracellular signal-regulated kinase 2 phosphorylation in the rat brain. Neuroscience 133: 343–352. [DOI] [PubMed] [Google Scholar]
- 72. Haneda M, Sugimoto T, Kikkawa R (1999) Mitogen-activated protein kinase phosphatase: A negative regulator of the mitogen-activated protein kinase cascade. Eur J Pharmacol 365: 1–7. [DOI] [PubMed] [Google Scholar]
- 73. Deroo BJ, Hewitt SC, Collins JB, Grissom SF, Hamilton KJ, et al. (2009) Profile of estrogen-responsive genes in an estrogen-specific mammary gland outgrowth model. Mol Reprod Dev 76: 733–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Keyse SM (2000) Protein phosphatases and the regulation of mitogen-activated protein kinase signalling. Curr Opin Cell Biol 12: 186–192. [DOI] [PubMed] [Google Scholar]
- 75. Ullrich M, Bundschu K, Benz PM, Abesser M, Freudinger R, et al. (2011) Identification of SPRED2 (sprouty-related protein with EVH1 domain 2) as a negative regulator of the hypothalamic-pituitary-adrenal axis. J Biol Chem 286: 9477–9488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cipolla M (2009) The cerebral circulation. San Rafael, CA: Morgan & Claypool Life Sciences. [PubMed]
- 77. Iadecola C (2004) Neurovascular regulation in the normal brain and in alzheimer's disease. Nat Rev Neurosci 5: 347–360. [DOI] [PubMed] [Google Scholar]
- 78. Hawkins BT, Davis TP (2005) The blood-brain barrier/neurovascular unit in health and disease. Pharmacol Rev 57: 173–185. [DOI] [PubMed] [Google Scholar]
- 79. Liu R, Wen Y, Perez E, Wang X, Day AL, et al. (2005) 17beta-estradiol attenuates blood-brain barrier disruption induced by cerebral ischemia-reperfusion injury in female rats. Brain Res 1060: 55–61. [DOI] [PubMed] [Google Scholar]
- 80. Ribatti D, Nico B, Crivellato E (2011) The role of pericytes in angiogenesis. Int J Dev Biol 55: 261–268. [DOI] [PubMed] [Google Scholar]
- 81. Armulik A, Genove G, Mae M, Nisancioglu MH, Wallgard E, et al. (2010) Pericytes regulate the blood-brain barrier. Nature 468: 557–561. [DOI] [PubMed] [Google Scholar]
- 82. Lindblom P, Gerhardt H, Liebner S, Abramsson A, Enge M, et al. (2003) Endothelial PDGF-B retention is required for proper investment of pericytes in the microvessel wall. Genes & Development 17: 1835–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Enge M, Bjarnegard M, Gerhardt H, Gustafsson E, Kalen M, et al. (2002) Endothelium-specific platelet-derived growth factor-B ablation mimics diabetic retinopathy. EMBO J 21: 4307–4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Dahlin A, Royall J, Hohmann JG, Wang J (2009) Expression profiling of the solute carrier gene family in the mouse brain. J Pharmacol Exp Ther 329: 558–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yamamoto K, de Waard V, Fearns C, Loskutoff DJ (1998) Tissue distribution and regulation of murine von willebrand factor gene expression in vivo. Blood 92: 2791–2801. [PubMed] [Google Scholar]
- 86. Suidan GL, Brill A, De Meyer SF, Voorhees JR, Cifuni SM, et al. (2013) Endothelial von willebrand factor promotes blood-brain barrier flexibility and provides protection from hypoxia and seizures in mice. Arterioscler Thromb Vasc Biol 33: 2112–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Krause DN, Duckles SP, Pelligrino DA (2006) Influence of sex steroid hormones on cerebrovascular function. J Appl Physiol (1985) 101: 1252–1261. [DOI] [PubMed] [Google Scholar]
- 88. Collins T, Read MA, Neish AS, Whitley MZ, Thanos D, et al. (1995) Transcriptional regulation of endothelial cell adhesion molecules: NF-kappa B and cytokine-inducible enhancers. FASEB J 9: 899–909. [PubMed] [Google Scholar]
- 89. Simoncini T, Maffei S, Basta G, Barsacchi G, Genazzani AR, et al. (2000) Estrogens and glucocorticoids inhibit endothelial vascular cell adhesion molecule-1 expression by different transcriptional mechanisms. Circ Res 87: 19–25. [DOI] [PubMed] [Google Scholar]
- 90. Spittau B, Wullkopf L, Zhou X, Rilka J, Pfeifer D, et al. (2013) Endogenous transforming growth factor-beta promotes quiescence of primary microglia in vitro. Glia 61: 287–300. [DOI] [PubMed] [Google Scholar]
- 91. Cerghet M, Skoff RP, Bessert D, Zhang Z, Mullins C, et al. (2006) Proliferation and death of oligodendrocytes and myelin proteins are differentially regulated in male and female rodents. J Neurosci 26: 1439–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kwiatkowski AV, Rubinson DA, Dent EW, Edward van Veen J, Leslie JD, et al. (2007) Ena/VASP is required for neuritogenesis in the developing cortex. Neuron 56: 441–455. [DOI] [PubMed] [Google Scholar]
- 93. Wong TP (2002) Aging of the cerebral cortex. MJM 6: 104–114. [Google Scholar]
- 94. Arevalo MA, Ruiz-Palmero I, Scerbo MJ, Acaz-Fonseca E, Cambiasso MJ, et al. (2012) Molecular mechanisms involved in the regulation of neuritogenesis by estradiol: Recent advances. J Steroid Biochem Mol Biol 131: 52–56. [DOI] [PubMed] [Google Scholar]
- 95. Krivosheya D, Tapia L, Levinson JN, Huang K, Kang Y, et al. (2008) ErbB4-neuregulin signaling modulates synapse development and dendritic arborization through distinct mechanisms. J Biol Chem 283: 32944–32956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Firth SM, Baxter RC (2002) Cellular actions of the insulin-like growth factor binding proteins. Endocr Rev 23: 824–854. [DOI] [PubMed] [Google Scholar]
- 97. Gonda Y, Sakurai H, Hirata Y, Tabata H, Ajioka I, et al. (2007) Expression profiles of insulin-like growth factor binding protein-like 1 in the developing mouse forebrain. Gene Expr Patterns 7: 431–440. [DOI] [PubMed] [Google Scholar]
- 98. Smith P, Nicholson LJ, Syed N, Payne A, Hiller L, et al. (2007) Epigenetic inactivation implies independent functions for insulin-like growth factor binding protein (IGFBP)-related protein 1 and the related IGFBPL1 in inhibiting breast cancer phenotypes. Clin Cancer Res 13: 4061–4068. [DOI] [PubMed] [Google Scholar]
- 99. Chesik D, De Keyser J, Wilczak N (2007) Insulin-like growth factor binding protein-2 as a regulator of IGF actions in CNS: Implications in multiple sclerosis. Cytokine Growth Factor Rev 18: 267–278. [DOI] [PubMed] [Google Scholar]
- 100. Topalli I, Etgen AM (2004) Insulin-like growth factor-I receptor and estrogen receptor crosstalk mediates hormone-induced neurite outgrowth in PC12 cells. Brain Res 1030: 116–124. [DOI] [PubMed] [Google Scholar]
- 101. Rozovsky I, Wei M, Morgan TE, Finch CE (2005) Reversible age impairments in neurite outgrowth by manipulations of astrocytic GFAP. Neurobiol Aging 26: 705–715. [DOI] [PubMed] [Google Scholar]
- 102. Mhyre AJ, Dorsa DM (2006) Estrogen activates rapid signaling in the brain: Role of estrogen receptor alpha and estrogen receptor beta in neurons and glia. Neuroscience 138: 851–858. [DOI] [PubMed] [Google Scholar]
- 103. Raz L, Khan MM, Mahesh VB, Vadlamudi RK, Brann DW (2008) Rapid estrogen signaling in the brain. Neurosignals 16: 140–153. [DOI] [PubMed] [Google Scholar]
- 104. Vasudevan N, Pfaff DW (2008) Non-genomic actions of estrogens and their interaction with genomic actions in the brain. Front Neuroendocrinol 29: 238–257. [DOI] [PubMed] [Google Scholar]
- 105. Belcher SM, Zsarnovszky A (2001) Estrogenic actions in the brain: Estrogen, phytoestrogens, and rapid intracellular signaling mechanisms. J Pharmacol Exp Ther 299: 408–414. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RNA gel demonstrating intact RNA samples. Native agarose gel electrophoresis was used to resolve the intact 28s and 18s rRNA bands. 2 µg of RNA were run per lane.
(PDF)
88 E2-responsive genes in the cerebral cortex.
(XLSX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. Sequencing files are available from EMBL-EBI Array Express under accession number E-MTAB-2762.