The authors sought to assess the impact of recommendations with regard to identifying patients eligible for anti-HER2 agents by fluorescence in situ hybridization and to elucidate whether multiplex ligation-dependent probe amplification (MLPA) may be of help in assessing HER2 gene status. Recent guidelines seem to improve the identification of HER2-positive carcinomas. MLPA can be of help if heterogeneous amplification has been ruled out.
Keywords: Breast cancer, HER2 amplification, Equivocal HER2 status, Guidelines, Heterogeneity
Abstract
Background.
The primary objectives of this study on carcinomas with equivocal HER2 expression were to assess the impact of distinct recommendations with regard to identifying patients eligible for anti-HER2 agents by fluorescence in situ hybridization (FISH) and to elucidate whether multiplex ligation-dependent probe amplification (MLPA) may be of support in assessing HER2 gene status.
Methods.
A cohort of 957 immunohistochemistry-evaluated HER2-equivocal cases was analyzed by dual-color FISH. The results were assessed according to U.S. Food and Drug Administration (FDA) and European Medicines Agency (EMA) guidelines and American Society of Clinical Oncology (ASCO) and College of American Pathologists (CAP) 2007 and 2013 guidelines for dual- and single-signal in situ hybridization (ISH) assays. A subgroup of 112 cases was subjected to MLPA.
Results.
HER2 amplification varied from 15% (ASCO/CAP 2007 HER2/CEP17 ratio) to 29.5% (FDA/EMA HER2 copy number). According to the ASCO/CAP 2013 interpretation of the dual-signal HER2 assay, ISH-positive carcinomas accounted for 19.7%. In contrast with the ASCO/CAP 2007 ratio, this approach labeled as positive all 32 cases (3.34%) with a HER2/CEP17 ratio <2 and an average HER2 copy number ≥6.0 signals per cell. In contrast, only one case showing a HER2 copy number <4 but a ratio ≥2 was diagnosed as positive. MLPA data correlated poorly with FISH results because of the presence of heterogeneous HER2 amplification in 33.9% of all amplified carcinomas; however, MLPA ruled out HER2 amplification in 75% of ISH-evaluated HER2-equivocal carcinomas.
Conclusion.
The ASCO/CAP 2013 guidelines seem to improve the identification of HER2-positive carcinomas. Polymerase chain reaction-based methods such as MLPA can be of help, provided that heterogeneous amplification has been ruled out by ISH.
Implications for Practice:
Our comprehensive portrait of breast carcinomas exhibiting equivocal HER2 expression demonstrates the urgent need for uniform interpretation of HER2 gene status for this controversial category. The recently updated American Society of Clinical Oncology and College of American Pathologists 2013 guidelines have led to significant improvement in the detection of HER2-positive breast cancer. Carcinomas remaining equivocal following in situ hybridization display predominantly low levels of HER2 at both protein and gene levels but represent a subgroup of high-grade estrogen receptor-positive breast cancers with higher proliferative activity than HER2 not amplified carcinomas.
Introduction
Detecting HER2 positivity in breast cancer is crucial for identification of patients who can be offered a potentially life-saving treatment, such as trastuzumab and lapatinib. With the advent of an antibody-drug conjugate consisting of trastuzumab linked to the cytotoxin emtansine (i.e., T-DM1), which is currently being used in clinical trials [1–5], administration of chemotherapy and anti-HER2 agents will rely on pathological assessment of HER2 positivity only [1].
By using immunohistochemistry (IHC) to evaluate protein overexpression, breast carcinomas can be classified into two main groups with completely different clinicotherapeutic implications: patients with a positive (score 3+) breast cancer are suitable candidates for anti-HER2 agents, whereas those harboring a HER2-negative result (score 0 and 1+) are not.
In daily practice we encounter a significant proportion of breast carcinomas with equivocal HER2 expression (score 2+, evaluated by immunohistochemistry [IHC; IHC-HER2-equivocal]). As defined by the American Society of Clinical Oncology (ASCO) and College of American Pathologists (CAP) 2007 recommendations [6] or by the U.S. Food and Drug Administration (FDA) and European Medicines Agency (EMA) guidelines [7, 8], this type accounts for up to 18% of all newly diagnosed breast carcinomas [6, 9–11]. The new 2013 guidelines from ASCO/CAP [12] also include incomplete membrane positivity in the definition of score 2+ breast carcinomas, and that is likely to lead to higher prevalence of IHC-HER2-equivocal carcinomas in the near future.
In score 2+ breast carcinomas, HER2 positivity must be confirmed by in situ hybridization (ISH); however, several factors may affect ISH results. First, distinct recommendations are available in Europe [7] and in the U.S. for eligibility for trastuzumab treatment [6, 8, 12]; these recommendations adopt different cutoffs for both the HER2/CEP17 ratio and the absolute HER2 copy number evaluation. In addition, the new ASCO/CAP guidelines [12] introduced an algorithm that integrates the HER2/CEP17 ratio and the HER2 copy number, but the influence of these recommendations on the final rate of patients eligible for HER2-targeted therapy has not been determined. Second, heterogeneity of HER2 gene amplification has been described in up to 41% of IHC-HER2-equivocal carcinomas [13]. Third, we [14] and others [15–17] have provided direct evidence of CEP17 gain and amplification causing misleading interpretation of ISH results. Fourth, even after ISH testing, a variable proportion of tumors remain equivocal.
We designed the present study to provide a definitive portrait of this controversial category of breast cancers. The specific aims were to analyze the impact of different scoring methods (dual- vs. single-signal ISH) and cutoffs for defining HER2 amplification and to determine whether an alternative technique to ISH may be of use with equivocal cases.
Materials and Methods
Cohort
Because our center (Pathology Unit, Azienda Ospedaliera Citta' della Salute e della Scienza di Torino) is the referral center for fluorescence ISH (FISH; dual-color) testing, we routinely receive IHC-HER2-equivocal breast cancer tissue samples that require gene testing. Between 2009 and 2012, we collected 1,084 consecutive IHC-HER2-equivocal breast carcinomas from 8 centers. All centers successfully participated in the quality control program for HER2 assessment in IHC (regional program of the Italian Society of Pathology [18], technical procedures according to ASCO/CAP guidelines 2007 [6]). For purposes of the study, histological features (histological type and grade) were reassessed, and original HER2 slides provided by each center were reviewed to confirm HER2-equivocal expression. A final set of 957 cases was included in the study.
IHC
All centers used the HercepTest preparation kit (Dako, Glostrup, Denmark, http://www.dako.com) and scored the results according to FDA/EMA recommendations [7, 8]; therefore, score 2+ cases were those exhibiting mild to moderate complete membranous staining in at least 10% of cells. IHC data on estrogen receptor (ER) and progesterone receptor (PR) expression and on proliferation index (Ki-67) were obtained from the original pathology reports.
FISH
FISH was performed according to the manufacturer’s instructions with probes for HER2 and CEP17 (Abbott Molecular Diagnostics, Abbott Park, IL, https://www.abbottmolecular.com), as described previously [14]. For analysis, 10 invasive areas on each slide were selected and automatically acquired at ×40 with the motorized Metafer scanning system (Carl Zeiss MetaSystems GmbH, Jena, Germany, http://www.zeiss.com) and Axio Imager epifluorescence microscope (one focus plane for DAPI (4′,6-diamidino-2-phenylindole) and 13 focus planes for green and red spots). PathVysion V2 software (MetaSystems Hard & Software GmbH, Altlußheim, Germany) (FDA approved) was used to automatically analyze HER2 and CEP17 probes. A range of 300–800 cells was examined.
Results were reviewed by three of the authors (L. Verdun di Cantogno, A. Sapino, C. Marchiò) and scored according to both FDA/EMA recommendations [7, 8] and ASCO/CAP guidelines from 2007 [6] and 2013 [12] for dual-signal assay analyses. In addition, we also simulated the scoring for the single-signal assay by taking into account HER2 gene counting only. In the latest ASCO/CAP 2013 guidelines, HER2 gene counting is the only parameter evaluated for single-probe ISH and is also integrated into the evaluation of the results of the HER2/CEP17 ratio for dual-signal ISH. The cutoffs for the different categories of HER2 gene status are reported in Table 1. Heterogeneity was defined according to the supplemental material shown in the ASCO/CAP 2013 guidelines [12].
Table 1.
Summary of the different scoring methods for HER2 gene testing by in situ hybridization (single- and dual-signal assays) with corresponding cutoffs
MLPA
Two 4 μm-thick paraffin sections were manually mesodissected by scraping off relevant areas of infiltrating carcinomas (foci of ductal carcinoma in situ discarded) with a pipet tip into DNase-free tubes [19]. Genomic DNA was extracted using the PureLink Genomic DNA extraction kit (Invitrogen, Carlsbad, CA, http://www.invitrogen.com), following the manufacturers’ instructions.
MLPA reactions were performed with 100–200 ng of purified genomic DNA using the P004-C1 ERBB2 probe mix (MRC-Holland, Amsterdam, The Netherlands, http://www.mlpa.com). All tests were performed using an MJ Thermal Cycler (MJ Research, Waltham, MA). Polymerase chain reaction (PCR) products were separated on an ABI 3130 capillary sequencer (Applied Biosystems, Foster City, CA, http://www.appliedbiosystems.com), and gene copy numbers were analyzed using GeneMapper 4.0 (Applied Biosystems) and Coffalyser (version 8.0; MRC-Holland) software. Because four probes for HER2 are included in the kit (17-035.1 ERBB2 exon 07, 17-035.1 ERBB2 exon 22, 17-035.1 ERBB2 exon 28, 17-035.1 ERBB2 exon 29), the mean of each probe-specific normalized ratio of the HER2 gene was calculated. By using the mean value, cases were defined as follows: normal: HER2 <1.3; gain: HER2 >1.3 but <2; amplified: HER2 >2 [20–22]. These thresholds were also validated in breast cancer cell lines showing HER2 amplification (BT474: HER2 copy number >6 by FISH, MLPA ratio >5), HER2 gain (T47D: HER2 copy number of 4 by FISH, MLPA ratio of 1.32) and lacking HER2 gain or amplification (MCF7: HER2 copy number of 2 by FISH, MLPA ratio of 0.61).
In Situ Proximity Ligation Assay
To study the protein levels in IHC-HER2-equivocal carcinomas, we performed a proximity ligation assay (PLA) in a subgroup of 26 cases with score 2+. For comparison, we also analyzed 10 cases with score 0 or 1+ and HER2 not amplified and 10 cases with score 3+ and HER2 amplified breast carcinomas. All reagents used for PLA analysis were from Olink Bioscience (Uppsala, Sweden, http://www.olink.com). HER2 assessment was performed using the Duolink II detection kit according to the manufacturer’s instructions (Olink Bioscience) and the polyclonal rabbit anti-human c-erbB-2 (A0485; Dako) primary antibody directed against HER2. Images were acquired at ×40 magnification using the Metafer scanning system and Axio Imager epifluorescence microscope (Carl Zeiss MetaSystems GmbH). We consistently acquired an image stack consisting of 9 image planes spaced 0.3 μm apart per region of interest (in total 10 regions of interest per case), and images were finally analyzed by using the Duolink Image Tool. In cases showing heterogeneity, the areas to be analyzed were selected based on the IHC staining for HER2.
Data Analysis
The following data were recorded in a dedicated database: histological type and histological grade; percentage of positivity for ER, PR, and Ki-67; HER2 gene copy number; CEP17 copy number; HER2/CEP17 ratio; and percentage of amplified cells. Thresholds for positivity were as follows: ≥1% for ER [23], >20% for PR [23, 24], ≥20 for Ki-67 [23]. In cases with genetic heterogeneity subjected to MLPA, FISH results were analyzed in two distinct ways: by considering distinct populations of amplified and not amplified cells, thus performing separate counts for the two populations, and by performing a mean between amplified and not amplified cells indistinctly throughout the overall population.
Statistical analysis was performed using SPSS StatViewer (version 19; IBM Corp., Armonk, NY, http://www-01.ibm.com/software/analytics/spss/), correlations were performed by chi-square test and Fisher’s exact test. A p value <.05 was considered statistically significant.
Results
Histopathological and Immunophenotypical Features of the Cohort
According to the World Health Organization classification [25], 78.8% were invasive carcinomas of no special type and 21.2% were of special histological type (9.3% lobular carcinomas, 7.7% mixed ductal-lobular carcinomas, and 4.2% other special histological types).
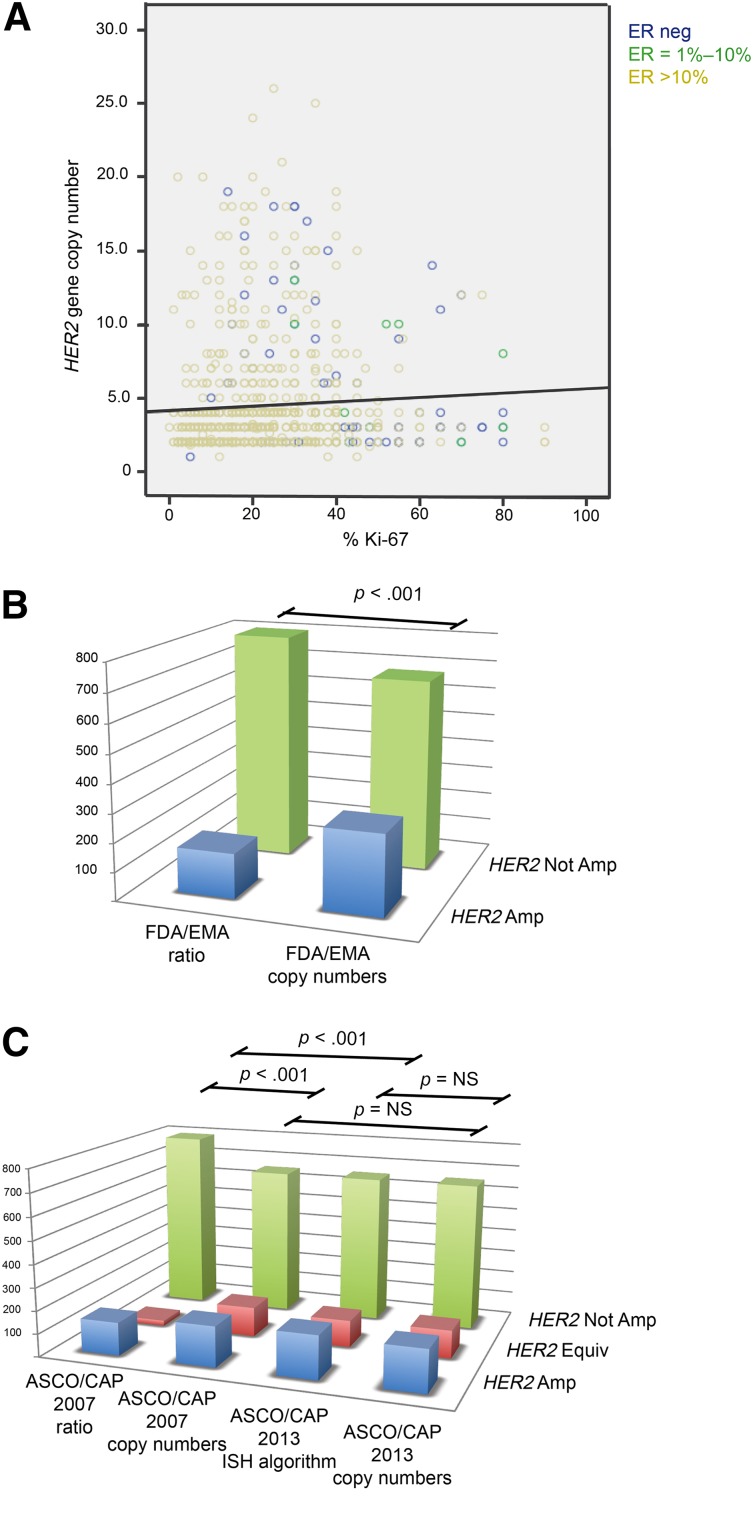
Analysis of the immunophenotype revealed that IHC-HER2-equivocal carcinomas were mostly ER and PR positive (76%) (Fig. 1A). Of the ER-positive carcinomas, 83% had ≥50% positive cells.
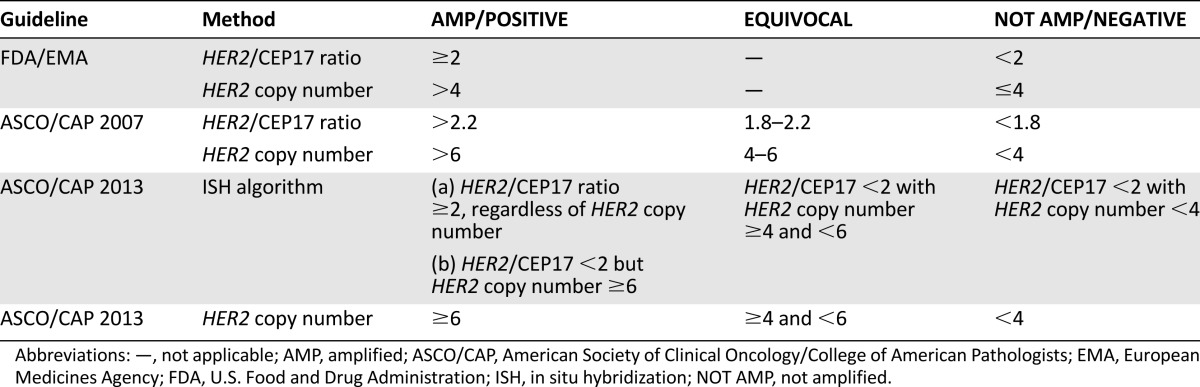
Figure 1.
Distribution of immunophenotypical variables throughout the cohort and prevalence of amplification according to distinct guidelines. (A): Relationship of ER, Ki-67, and HER2 gene copy number values in immunohistochemistry-evaluated HER2-equivocal carcinomas. These carcinomas are largely ER positive (green circles between 1%–10%, yellow circles when >10% of positive cells) with high proliferative activity; however, presence of amplification is associated with ER-negative carcinomas (blue circles exhibiting high HER2 copy numbers). A great proportion of ER-positive cases with low proliferation indexes also harbor a low HER2 gene copy number. (B, C): Statistically significant differences in the distribution of HER2 amplified, not amplified, and equivocal cases (the latter for ASCO/CAP only) are observed with the different scoring methods of the FDA/EMA and ASCO/CAP recommendations, as shown by p values. Notably, no statistically significant differences were observed between the scoring of dual- and single-signal assays according to ASCO/CAP 2013 and between HER2 copy number by ASCO/CAP 2007 and both ISH algorithm and HER2 copy number by ASCO/CAP 2013.
Abbreviations: Amp, amplified; ASCO/CAP, American Society of Clinical Oncology/College of American Pathologists; EMA, European Medicines Agency; Equiv, equivocal; ER, estrogen receptor; FDA, U.S. Food and Drug Administration; ISH, in situ hybridization; neg, negative; Not Amp, not amplified; NS: not statistically significant.
The mean proliferation index was 23.8% (SD ±15.7%), with a median of 20% (Fig. 1A). A high proliferation index (≥20%) was observed in 55.2% of carcinomas.
HER2 and CEP17 Status According to Distinct Guidelines
The frequency of HER2 amplification varied from 15% using the ASCO/CAP 2007 ratio to 29.5% using the FDA/EMA copy number (Table 2; Fig. 1B, 1C). In general, the HER2 copy number identified a significantly higher number of amplified cases compared with the HER2/CEP17 ratio (p values <.001) (Table 2; Fig. 1B, 1C). Although the occurrence of amplification differed slightly between FDA/EMA and ASCO/CAP 2007 recommendations by using the HER2/CEP17 ratio (Table 2), a greater difference was observed by using the gene copy number as the single parameter of amplification (Table 2).
Table 2.
Prevalence of HER2 amplification in the cohort according to distinct guidelines
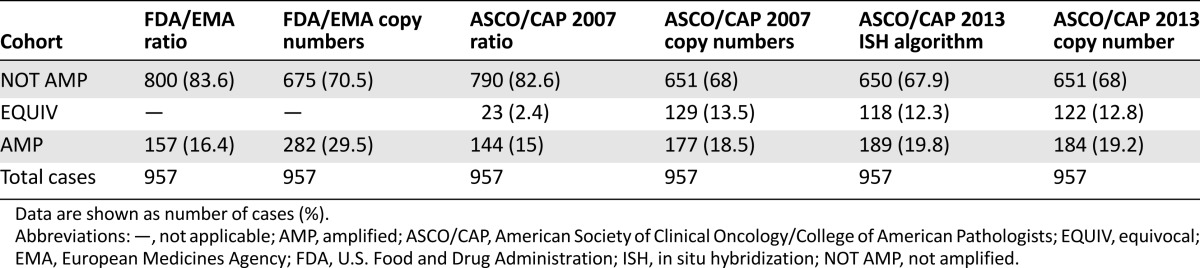
According to the algorithm proposed by the ASCO/CAP 2013 guidelines, which takes into account the ratio and then the copy number values, 68% of cases were ISH negative, 12.2% were ISH equivocal, and 19.8% were ISH positive (Table 2), exhibiting high similarity with the results obtained by using the HER2 copy number as the single parameter in both 2007 and 2013 guidelines (Table 2; Fig. 1C). In general, the new dual-signal ISH algorithm labeled a significantly greater number of equivocal cases by reducing the number of not amplified cases when compared with the ratio of ASCO/CAP 2007 (p < .001) (Fig. 1C; Table 2). However, all 32 cases (3.34%) with a ratio <2 and an average HER2 copy number ≥6.0 signals per cell were labeled as positive.
With respect to the copy number parameter of the previous edition, the ASCO/CAP 2013 recommendations labeled a greater number of cases as amplified by lowering the number of equivocal cases; however, this difference was not statistically significant (Fig. 1C; Table 2). Finally, by comparing the dual-signal ISH with the single-signal ISH results of the ASCO/CAP 2013 guidelines, only 5 cases (0.5%) were differently classified: 1 case from amplified switched to not amplified (HER2 was 3.9, CEP17 was 1.6; HER2/CEP17 ratio of 2.3) and four cases from amplified to equivocal (HER2 ranging from 4.2 to 5.9; CEP17 ranging from 1.6 to 2.72; HER2/CEP17 ratio ranging from 2 to 3.6).
When considering CEP17 status, approximately one third of the whole cohort (257 of 957, 28.1%) had a mean CEP17 copy number >3 (range: 3.1–9.3) and 12.7% (122 of 957) showed a mean CEP17 copy number <2 (range: 1–1.9). Finally, a CEP17 copy number >3 was more frequent (mean value: 76.5%) in ISH-equivocal cases (both ASCO/CAP 2007 copy number and ASCO/CAP 2013) than in HER2 amplified carcinomas (mean: 45.3%) (p ≤ .01) and not amplified carcinomas (mean: 14.5%) (p < .001).
Correlations of FISH Results and Histopathological and Immunophenotypical Features
HER2 amplification, as defined by any of the guidelines, was more frequent in ER- and PR- negative carcinomas (Tables 3, 4). HER2 amplification was significantly associated with a high proliferative index only when defined by gene copy numbers (p values ≤.001) (Tables 3, 4).
Table 3.
Correlations between histopathological and immunophenotypical features and HER2 gene amplification defined according to FDA/EMA recommendations
Table 4.
Correlations between histopathological and immunophenotypical features and HER2 gene amplification defined according to ASCO/CAP guidelines

In contrast, ISH-HER2-equivocal carcinomas, as defined by both ASCO/CAP guideline editions, were enriched with ER-positive cases (mean: 94.8%) and displayed a higher proliferation index (mean: 63.4%) than HER2 not amplified carcinomas (ER mean: 84%; Ki-67 mean: 51.5%) (all p values <.05).
A mean CEP17 copy number >3 was more represented in the ER- and PR-positive subgroups (p = .001), was strictly associated with high proliferating activity (p = .007), and was more likely to be associated with G2 or G3 carcinomas (p = .003) (supplemental online Table 1).
Heterogeneity According to ASCO/CAP 2013
Of the 189 ISH-positive cases, we identified 64 cases (33.9%) that harbored HER2 genetic heterogeneity. Of these, 81.3% (n = 52) could be defined as a diffuse intermingling of amplified and not amplified cells across the tumor, whereas 18.7% (n = 12) displayed two discrete populations (clones) of amplified and not amplified tumor cells [12]. Up to 48.4% (n = 31) of heterogeneous cases also exhibited a mean CEP17 copy number >3 (supplemental online Table 2).
MLPA in IHC-HER2-Equivocal Carcinomas
A subgroup of 112 cases was subjected to MLPA. The analysis was performed using the mean value of the four HER2 probes. Because MLPA results are scored according to a three-tiered scoring system, data were analyzed by categorizing cases according to the ASCO/CAP 2013 categories for HER2 copy number.
MLPA confirmed a normal HER2 gene status in 93% of the 71 ISH-negative and 75% of the 12 ISH-equivocal cases examined, with the remaining cases displaying HER2 gain (Fig. 2). Of the 29 ISH-positive cases harboring ≥6 HER2 copies (range: 6–24; of which 58% had a HER2/CEP17 ratio ≥2), 14 (48.2%) were confirmed as amplified, 7 (24.1%) had HER2 gain, and 8 (27.5%) had normal HER2 status by MLPA. No significant differences were observed between cases with CEP17 copy numbers >3 or ≤3 (Fig. 2).
Figure 2.
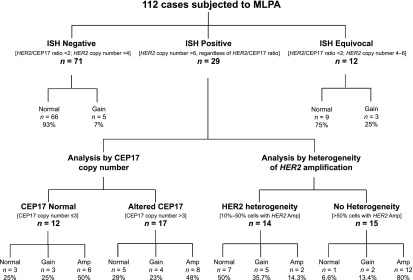
Schematic representation of MLPA results in 112 immunohistochemistry-evaluated HER2-equivocal carcinomas and comparison with fluorescence ISH (FISH) data. Cases are subdivided as ISH negative, ISH equivocal, and ISH positive, according to American Society of Clinical Oncology/College of American Pathologists 2013 guidelines. ISH-positive cases are categorized by CEP17 copy numbers and by the presence of heterogeneity of HER2 amplification. For MLPA results, cases were defined as follows: normal: HER2 <1.3; gain: HER2 >1.3 but <2; amplified: HER2 >2. Concordance between FISH and MLPA results was observed regardless of CEP17 status but was affected by the presence of heterogeneity of HER2 amplification (50% in heterogeneous cases vs. 80% in carcinomas with HER2 amplification in >50% of tumor cells).
Abbreviations: Amp, amplified; ISH, in situ hybridization; MLPA, multiplex ligation-dependent probe amplification.
Taken together, these results indicate a disagreement between FISH and MLPA results (FISH amplification vs. MLPA gain or normal) for 15 ISH-positive cases: 3 cases had a mean HER2 copy number of 6–6.6, whereas the remaining 12 cases (80%) had heterogeneous HER2 amplification (range: 10%–50%). Of the 12 tumors with heterogeneous HER2 amplification, 6 were defined as discrete populations of amplified and not amplified tumor cells and 6 as diffuse intermingling of amplified and not amplified cells across the tumor. For these heterogeneous cases, we further calculated the FISH results of HER2 copy number for the whole population: Nine cases were scored as equivocal and three as not amplified.
By analyzing ISH-positive cases without HER2 genetic heterogeneity (n = 15), FISH and MLPA displayed 80% agreement (Fig. 2).
HER2 Protein Variability in IHC-HER2-Equivocal Carcinomas
We assessed the mean value and range of HER2 signals by PLA in a series of 10 cases with score 0 or 1+ and HER2 not amplified and 10 cases with score 3+ and HER2 amplified carcinomas. In the first group of cases, the number of signals ranged between 4.58 and 5.83 (mean: 5.51), whereas in the second group, the PLA signals ranged between 10.37 and 29.22 (mean: 20.9) (Table 5).
Table 5.
Quantification of HER2 protein by PLA in different in situ hybridization categories as defined by the guidelines of American Society of Clinical Oncology and College of American Pathologists—2013
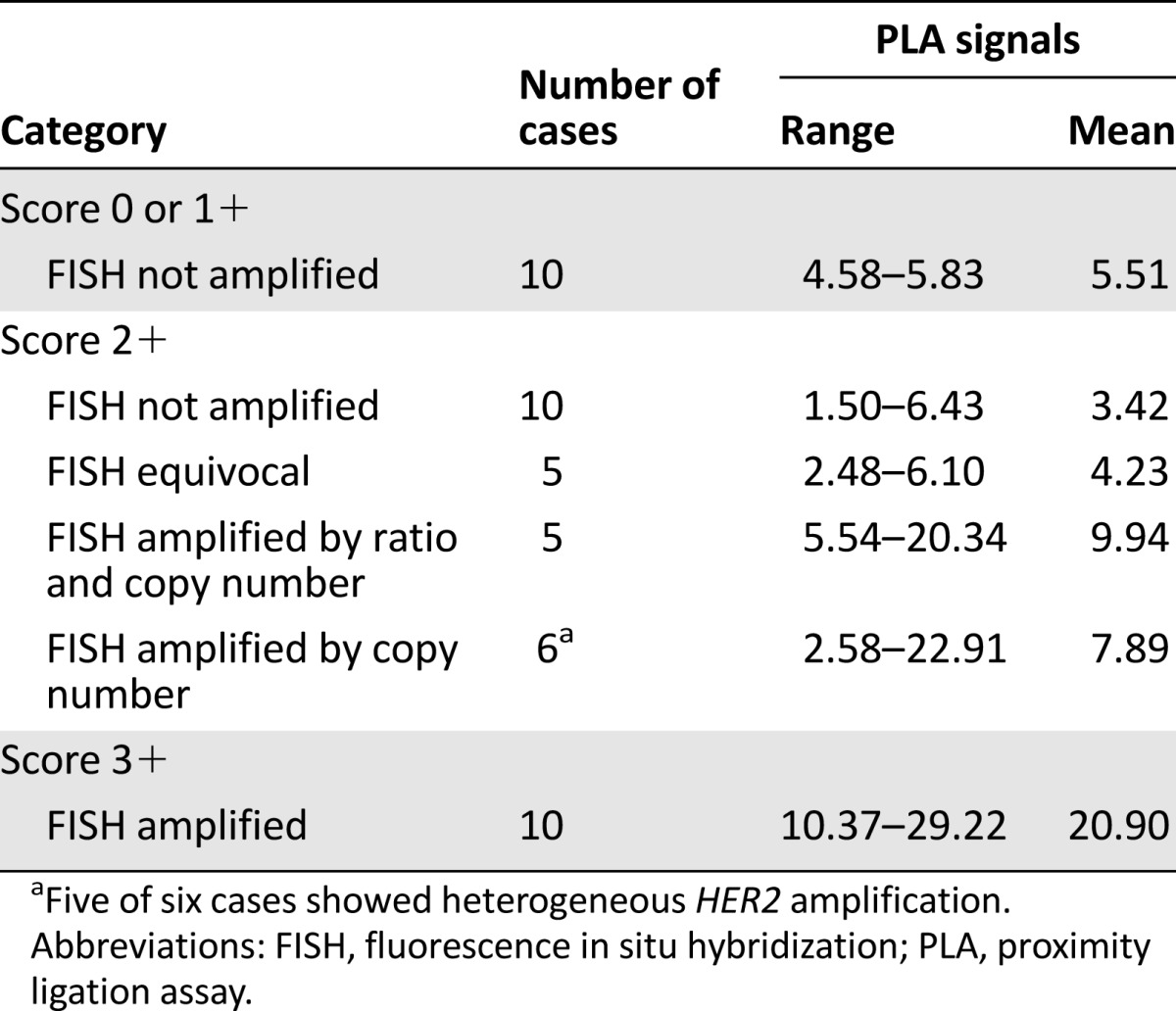
Score 2+ cases showed a wide variability in terms of protein levels assessed by PLA signals (range: 1.50–22.91; mean: 6.28), as detailed in Table 5 and illustrated in supplemental online Figure 1. When subdivided into different categories according to the FISH results [12], those carcinomas lacking HER2 amplification and those defined as ISH-HER2-equivocal showed values that recapitulated the score 0 or 1+ category (i.e., no significant difference between categories, p values >.05), whereas carcinomas proven to harbor HER2 amplification displayed a number of PLA signals much closer to score 3+ breast cancers (Table 5). Cases showing ≥6 HER2 signals but a HER2/CEP17 ratio <2 displayed a mean value of PLA signals that was significantly lower than score 3+ cases (p = .001) but not significantly different from score 2+ cases harboring HER2 amplification, as defined by both ratio and copy numbers (p = .55). Of note, five of six carcinomas showing HER2 amplification only defined by copy numbers displayed heterogeneous amplification. Although the PLA analysis was carried out based on heterogeneous HER2 expression, thus reporting the results of the population showing the highest number of signals, we cannot rule out the possibility that better concordance would have been achieved by performing a parallelism with the FISH pattern, given the type of heterogeneity of these cases, which showed diffuse intermingling of amplified and not amplified cells across the tumor.
Discussion
This study reports data from the largest cohort of breast carcinomas showing IHC-HER2-equivocal status. These carcinomas are a specific subset of ER-positive breast cancers with high proliferation activity, consistent with previous data from smaller series [11, 26]. Although the IHC-HER2-equivocal carcinomas were largely ER positive, HER2 amplification was more pervasive in ER-negative carcinomas.
The frequency of HER2 amplification in the whole cohort varied from 15% (ASCO/CAP 2007 ratio) to 29.5% (FDA/EMA HER2 copy number). The cutoffs of ratio ≥2 and >6 HER2 copies identified amplification for 16.4% and 18.5%, respectively. Consequently, depending on the recommendation or parameter of choice, there is important variability in the selection of patients who can be targeted with anti-HER2 agents. The frequencies of HER2 amplification reported above are slightly lower than those reported in previous series of IHC-HER2-equivocal carcinomas [26, 27]. This could be attributable mainly to technical causes, such as the different cutoff used for IHC scoring (10% vs. 30%), the large number of cases included in the present study, and the high number of fields (10 tumor areas) and of cells (300–800) counted with an automated instrument. However, a selection bias for IHC-HER2-equivocal cases depending on the immunohistochemical procedures cannot be ruled out, although all centers successfully participated in quality control for HER2 assessment in IHC. A significantly higher frequency of amplification was observed based on the HER2 gene copy number compared with the HER2/CEP17 ratio. This suggests that the HER2/CEP17 ratio is a more conservative method of calculating gene amplification; however, it is important to note that only cases categorized as amplified because of the increase of the HER2 gene copy number (both >4 and >6 thresholds) exhibited significantly greater proliferation compared with those not amplified. This association may reflect the effect of HER2 as a driver of proliferation [28, 29] or may represent the epiphenomenon of what is normally observed in the S phase of the cell cycle. In addition, the HER2/CEP17 ratio can underestimate the presence of HER2 amplification, particularly in cases with high CEP17 copy numbers.
The ASCO/CAP 2013 guidelines [12] have proposed an algorithm to report dual-signal ISH results that takes into account both HER2/CEP17 ratios and HER2 copy numbers. We demonstrated that such an algorithm rescued all cases with high CEP17 copy numbers and HER2 copy numbers ≥6 (3.34%) that could be labeled as negative based on the ratio alone. Indeed, as quantified by PLA, these carcinomas show HER2 protein levels that, although variable, reach values similar to score 3+ carcinomas.
The main caveat with the new recommendations regards a potential pitfall during dual-signal ISH interpretation. Although rare (4 of 957 cases, 0.4%), there are cases in which the HER2/CEP17 ratio is above the cutoff (≥2) due to a mean number of CEP17 signals <2. In such cases, the evaluation of HER2 copy number is imperative; however, the new guidelines suggest labeling these cases harboring fewer than four copies as ISH positive. This change could lead to overestimation of amplification, which does not make biological sense, as recently pointed out by Bhargava and Dabbs [30]. In fact, a lack of response to trastuzumab treatment in cases with HER2 amplification associated with monosomy of chromosome 17 has been described [31].
The ISH algorithm [12] still identifies a category of carcinomas with an equivocal HER2 result, and the guidelines stress the need for data defining their profile. We demonstrated that this category exhibits distinct features when compared with HER2 not amplified and HER2 amplified carcinomas and represents a subgroup of ER-positive carcinomas with high proliferative activity, aberrant CEP17 copy numbers, and low levels of HER2 protein, as quantified by PLA. In a small series of ISH-HER2-equivocal cases, MLPA revealed mainly lack of amplification; however, 25% of samples still showed HER2 gain. Consequently, analyses of larger series by alternative techniques are warranted to confirm this observation.
Our analysis has also provided insight into the heterogeneity of HER2 amplification, which was observed in 33.9% of amplified cases, similar to data reported for other series of IHC-HER2-equivocal carcinomas [13, 32]. Of note, HER2 heterogeneity was also frequently observed in cases with mean CEP17 copy numbers >3 (approximately 37%), as reported previously [13]. In diagnostic terms, this association may lead exponentially to troublesome interpretation of ISH results. HER2 genetic heterogeneity has recently been considered in the ASCO/CAP 2013 guidelines [12], in which panelists suggested reporting separate calculations of ISH results whenever two discrete populations are encountered. In our cohort, most (81.3%) of the heterogeneous cases exhibited diffuse intermingling of amplified and not amplified cells across the tumor. The potential relevance of subpopulations of HER2 amplified cells within otherwise HER2 not amplified tumors has been a topic of debate [33]. Although some have questioned the recommendation for routine reporting on a histopathological phenomenon for which clinical significance remains poorly defined [33], subpopulations of HER2 amplified cells may be responsible for worse outcomes [33–35]. The sensitivity of these cells to trastuzumab remains to be fully determined; although some data are available for focal amplified clones [36, 37], studies on the biological significance of the most prevalent type of heterogeneity are missing. If counted as a homogeneous population, these tumors would not reach the threshold for amplification and would not be eligible for trastuzumab. This phenomenon may well account for the observation of trastuzumab responses in HER2-negative tumors [33, 38], and, as also noted by Hanna et al. [33], the results of the ongoing National Surgical Adjuvant Breast and Bowel Project B-47 trial of adjuvant trastuzumab in low-HER2 breast cancer may shed light on this controversy.
From a technical standpoint, HER2 genetic heterogeneity provided insight into the reasons underlying the poor correlation between FISH and MLPA data for ISH-defined HER2 amplified breast carcinomas. Within the latter subgroup, discordant results between the two techniques were observed in up to 50% of cases. This is important in diagnostic practice because some have advocated the use of alternative methods [1] because of the occurrence of altered CEP17. MLPA is a PCR-based method that performs well on fragmented DNA and may be reliably implemented in routine practice; however, because this technique is based on DNA extraction, tissue morphology is lost, as is the genetic heterogeneity, potentially leading to underestimation of HER2 amplification. Our data suggest that, at least in IHC-HER2-equivocal carcinomas, the best practice would be to rule out the presence of heterogeneous HER2 amplification and subsequently to apply MLPA in ISH-HER2-equivocal carcinomas, for which up to 75% could be definitely labeled as not amplified.
Conclusion
Our data on a large cohort of score 2+ invasive breast carcinomas show that, depending on the recommendation/parameter of choice to assess HER2 gene status, there is an important variability in the selection of patients targetable with anti-HER2 agents. The recently updated ASCO/CAP guidelines appear to improve the detection of HER2 positive breast cancer, as the algorithm for dual-signal ISH allows to rescue all cases with high CEP17 copy numbers and HER2 copy number ≥6 that could be labeled as negative based on the ratio alone. In addition, we demonstrate that PCR based methods such as MLPA may be of assistance in those cases defined as equivocal after ISH testing, provided that heterogeneity of amplification is first ruled out by in situ techniques.
See http://www.TheOncologist.com for supplemental material available online.
Supplementary Material
Acknowledgments
This work was supported by AIRC (MFAG13310 to Caterina Marchiò) and by Ricerca Sanitaria Finalizzata (RF-2010-2310674 to Anna Sapino).
Author Contributions
Conception/Design: Anna Sapino, Caterina Marchiò
Provision of study material or patients: Anna Sapino, Francesca Pietribiasi, Paolo Bernardi, Riccardo Arisio, Laura Viberti, Stefano Guzzetti, Renzo Orlassino
Collection and/or assembly of data: Francesca Maletta, Ludovica Verdun di Cantogno, Luigia Macrì, Cristina Botta, Patrizia Gugliotta, Maria Stella Scalzo, Caterina Marchiò, Davide Balmativola
Data analysis and interpretation: Anna Sapino, Francesca Maletta, Ludovica Verdun di Cantogno, Luigia Macrì, Cristina Botta, Patrizia Gugliotta, Cristiana Ercolani, Marcella Mottolese, Giuseppe Viale, Caterina Marchiò, Laura Annaratone
Manuscript writing: Anna Sapino, Marcella Mottolese, Giuseppe Viale, Caterina Marchiò
Final approval of manuscript: Anna Sapino, Francesca Maletta, Ludovica Verdun di Cantogno, Luigia Macrì, Cristina Botta, Patrizia Gugliotta, Maria Stella Scalzo, Francesca Pietribiasi, Paolo Bernardi, Riccardo Arisio, Laura Viberti, Stefano Guzzetti, Renzo Orlassino, Cristiana Ercolani, Marcella Mottolese, Giuseppe Viale, Caterina Marchiò
Disclosures
Giuseppe Viale: Dako, Roche, Novartis (C/A); Dako, Roche (H). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
References
- 1.Sapino A, Goia M, Recupero D, et al. Current challenges for HER2 testing in diagnostic pathology: State of the art and controversial issues. Front Oncol. 2013;3:129. doi: 10.3389/fonc.2013.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peddi PF, Hurvitz SA. Trastuzumab emtansine: The first targeted chemotherapy for treatment of breast cancer. Future Oncol. 2013;9:319–326. doi: 10.2217/fon.13.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verma S, Miles D, Gianni L, et al. Trastuzumab emtansine for HER2-positive advanced breast cancer. N Engl J Med. 2012;367:1783–1791. doi: 10.1056/NEJMoa1209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krop IE, Beeram M, Modi S, et al. Phase I study of trastuzumab-DM1, an HER2 antibody-drug conjugate, given every 3 weeks to patients with HER2-positive metastatic breast cancer. J Clin Oncol. 2010;28:2698–2704. doi: 10.1200/JCO.2009.26.2071. [DOI] [PubMed] [Google Scholar]
- 5.Krop IE, LoRusso P, Miller KD, et al. A phase II study of trastuzumab emtansine in patients with human epidermal growth factor receptor 2-positive metastatic breast cancer who were previously treated with trastuzumab, lapatinib, an anthracycline, a taxane, and capecitabine. J Clin Oncol. 2012;30:3234–3241. doi: 10.1200/JCO.2011.40.5902. [DOI] [PubMed] [Google Scholar]
- 6.Wolff AC, Hammond ME, Schwartz JN, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25:118–145. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 7.European Medicines Agency: Latest news. Available at http://www.ema.europa.eu/ema/. Accessed September 26, 2014.
- 8.U.S. Food and Drug Administration: Drugs. Available at http://www.fda.gov/Drugs/default.htm. Accessed September 25, 2014.
- 9.Atkinson R, Mollerup J, Laenkholm AV, et al. Effects of the change in cutoff values for human epidermal growth factor receptor 2 status by immunohistochemistry and fluorescence in situ hybridization: A study comparing conventional brightfield microscopy, image analysis-assisted microscopy, and interobserver variation. Arch Pathol Lab Med. 2011;135:1010–1016. doi: 10.5858/2010-0462-OAR. [DOI] [PubMed] [Google Scholar]
- 10.Meijer SL, Wesseling J, Smit VT, et al. HER2 gene amplification in patients with breast cancer with equivocal IHC results. J Clin Pathol. 2011;64:1069–1072. doi: 10.1136/jclinpath-2011-200019. [DOI] [PubMed] [Google Scholar]
- 11.Rossi V, Sarotto I, Maggiorotto F, et al. Moderate immunohistochemical expression of HER-2 (2+) without HER-2 gene amplification is a negative prognostic factor in early breast cancer. The Oncologist. 2012;17:1418–1425. doi: 10.1634/theoncologist.2012-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolff AC, Hammond ME, Hicks DG, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31:3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 13.Ohlschlegel C, Zahel K, Kradolfer D, et al. HER2 genetic heterogeneity in breast carcinoma. J Clin Pathol. 2011;64:1112–1116. doi: 10.1136/jclinpath-2011-200265. [DOI] [PubMed] [Google Scholar]
- 14.Marchiò C, Lambros MB, Gugliotta P, et al. Does chromosome 17 centromere copy number predict polysomy in breast cancer? A fluorescence in situ hybridization and microarray-based CGH analysis. J Pathol. 2009;219:16–24. doi: 10.1002/path.2574. [DOI] [PubMed] [Google Scholar]
- 15.Moelans CB, de Weger RA, van Diest PJ. Absence of chromosome 17 polysomy in breast cancer: Analysis by CEP17 chromogenic in situ hybridization and multiplex ligation-dependent probe amplification. Breast Cancer Res Treat. 2010;120:1–7. doi: 10.1007/s10549-009-0539-2. [DOI] [PubMed] [Google Scholar]
- 16.Varga Z, Tubbs RR, Wang Z, et al. Co-amplification of the HER2 gene and chromosome 17 centromere: A potential diagnostic pitfall in HER2 testing in breast cancer. Breast Cancer Res Treat. 2012;132:925–935. doi: 10.1007/s10549-011-1642-8. [DOI] [PubMed] [Google Scholar]
- 17.Yeh IT, Martin MA, Robetorye RS, et al. Clinical validation of an array CGH test for HER2 status in breast cancer reveals that polysomy 17 is a rare event. Mod Pathol. 2009;22:1169–1175. doi: 10.1038/modpathol.2009.78. [DOI] [PubMed] [Google Scholar]
- 18.Rete Oncologica del Piemonte e della Valle d'Aosta. Available at http://www.reteoncologica.it/index.php?option=com_content&view=article&id=449:raccomandazioni-di-retetumori-del-pancreas-&catid=34:news&Itemid=78. Accessed September 30, 2014.
- 19.Farshid G, Cheetham G, Davies R, et al. Validation of the multiplex ligation-dependent probe amplification (MLPA) technique for the determination of HER2 gene amplification in breast cancer. Diagn Mol Pathol. 2011;20:11–17. doi: 10.1097/PDM.0b013e3181ed7832. [DOI] [PubMed] [Google Scholar]
- 20.Bunyan DJ, Eccles DM, Sillibourne J, et al. Dosage analysis of cancer predisposition genes by multiplex ligation-dependent probe amplification. Br J Cancer. 2004;91:1155–1159. doi: 10.1038/sj.bjc.6602121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coffa J, van de Wiel MA, Diosdado B, et al. MLPAnalyzer: Data analysis tool for reliable automated normalization of MLPA fragment data. Cell Oncol. 2008;30:323–335. doi: 10.3233/CLO-2008-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moelans CB, de Wegers RA, Monsuurs HN, et al. Molecular differences between ductal carcinoma in situ and adjacent invasive breast carcinoma: A multiplex ligation-dependent probe amplification study. Cell Oncol (Dordr) 2011;34:475–482. doi: 10.1007/s13402-011-0043-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldhirsch A, Winer EP, Coates AS, et al. Personalizing the treatment of women with early breast cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol. 2013;24:2206–2223. doi: 10.1093/annonc/mdt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prat A, Cheang MC, Martín M, et al. Prognostic significance of progesterone receptor-positive tumor cells within immunohistochemically defined luminal A breast cancer. J Clin Oncol. 2013;31:203–209. doi: 10.1200/JCO.2012.43.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lakhani SR, Ellis I, Schnitt SJ, et al. WHO Classification of Tumours of the Breast (IARC WHO Classification of Tumours) Lyon, France: International Agency for Research on Cancer; 2012. [Google Scholar]
- 26.Dieci MV, Barbieri E, Bettelli S, et al. Predictors of human epidermal growth factor receptor 2 fluorescence in-situ hybridisation amplification in immunohistochemistry score 2+ infiltrating breast cancer: A single institution analysis. J Clin Pathol. 2012;65:503–506. doi: 10.1136/jclinpath-2011-200643. [DOI] [PubMed] [Google Scholar]
- 27.Chibon F, de Mascarel I, Sierankowski G, et al. Prediction of HER2 gene status in Her2 2+ invasive breast cancer: A study of 108 cases comparing ASCO/CAP and FDA recommendations. Mod Pathol. 2009;22:403–409. doi: 10.1038/modpathol.2008.195. [DOI] [PubMed] [Google Scholar]
- 28.Hudis CA. Trastuzumab—mechanism of action and use in clinical practice. N Engl J Med. 2007;357:39–51. doi: 10.1056/NEJMra043186. [DOI] [PubMed] [Google Scholar]
- 29.Yarden Y. The EGFR family and its ligands in human cancer. Signalling mechanisms and therapeutic opportunities. Eur J Cancer. 2001;37(suppl 4):S3–S8. doi: 10.1016/s0959-8049(01)00230-1. [DOI] [PubMed] [Google Scholar]
- 30.Bhargava R, Dabbs DJ. Interpretation of human epidermal growth factor receptor 2 (HER2) in situ hybridization assays using 2013 update of American Society of Clinical Oncology/College of American Pathologists HER2 guidelines. J Clin Oncol. 2014;32:1855. doi: 10.1200/JCO.2013.53.9213. [DOI] [PubMed] [Google Scholar]
- 31.Risio M, Casorzo L, Redana S, et al. HER2 gene-amplified breast cancers with monosomy of chromosome 17 are poorly responsive to trastuzumab-based treatment. Oncol Rep. 2005;13:305–309. [PubMed] [Google Scholar]
- 32.Murthy SS, Sandhya DG, Ahmed F, et al. Assessment of HER2/Neu status by fluorescence in situ hybridization in immunohistochemistry-equivocal cases of invasive ductal carcinoma and aberrant signal patterns: A study at a tertiary cancer center. Indian J Pathol Microbiol. 2011;54:532–538. doi: 10.4103/0377-4929.85087. [DOI] [PubMed] [Google Scholar]
- 33.Hanna WM, Rüschoff J, Bilous M, et al. HER2 in situ hybridization in breast cancer: Clinical implications of polysomy 17 and genetic heterogeneity. Mod Pathol. 2014;27:4–18. doi: 10.1038/modpathol.2013.103. [DOI] [PubMed] [Google Scholar]
- 34.Bartlett AI, Starcyznski J, Robson T, et al. Heterogeneous HER2 gene amplification: Impact on patient outcome and a clinically relevant definition. Am J Clin Pathol. 2011;136:266–274. doi: 10.1309/AJCP0EN6AQMWETZZ. [DOI] [PubMed] [Google Scholar]
- 35.Seol H, Lee HJ, Choi Y, et al. Intratumoral heterogeneity of HER2 gene amplification in breast cancer: Its clinicopathological significance. Mod Pathol. 2012;25:938–948. doi: 10.1038/modpathol.2012.36. [DOI] [PubMed] [Google Scholar]
- 36.Miller DV, Jenkins RB, Lingle WL, et al. Focal HER2/neu amplified clones partially account for discordance between immunohistochemistry and fluorescence in-situ hybridization results: Data from NCCTG N9831 intergroup adjuvant trial. J Clin Oncol. 2004;22(suppl):568a. [Google Scholar]
- 37.Oakman C, Sapino A, Marchiò C, et al. Chemotherapy with or without trastuzumab. Ann Oncol. 2010;21(suppl 7):vii112–vii119. doi: 10.1093/annonc/mdq283. [DOI] [PubMed] [Google Scholar]
- 38.Allison KH, Dintzis SM, Schmidt RA. Frequency of HER2 heterogeneity by fluorescence in situ hybridization according to CAP expert panel recommendations: Time for a new look at how to report heterogeneity. Am J Clin Pathol. 2011;136:864–871. doi: 10.1309/AJCPXTZSKBRIP07W. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.