Abstract
The C-terminal fragment of the c-Met receptor tyrosine kinase is present in the nuclei of some cells irrespective of ligand stimulation, but the responsible nuclear localization signal (NLS) has not been previously reported. Here, we report that two histidine residues separated by a 10-amino-acid spacer (H1068–H1079) located in the juxtamembrane region of c-Met function as a putative novel NLS. Deletion of these sequences significantly abolished the nuclear translocation of c-Met, as did substitution of the histidines with alanines. This substitution also decreased the association of c-Met fragment with importin β. The putative NLS of c-Met is unique in that it relies on histidines, whose positive charge changes depending on pH, rather than the lysines or arginines, commonly found in classical bipartite NLSs, suggesting the possible ‘pH-dependency' of this NLS. Indeed, decreasing the cytosolic pH either with nigericin, an Na+/H+ exchanger or pH 6.5 KRB buffer significantly increased the level of nuclear c-Met and the interaction of the c-Met fragment with importin β, indicating that low pH itself enhanced nuclear translocation. Consistent with this, nigericin treatment also increased the nuclear level of endogenous c-Met in HeLa cells. The putative aberrant bipartite NLS of c-Met seems to be the first example of what we call a ‘pH-dependent' NLS.
Introduction
Met is a proto-oncogene initially identified as a transforming gene from a chemically induced human sarcoma cell line.1 The product of this gene, c-Met, is a cell surface receptor tyrosine kinase (RTK) with a single transmembrane domain. It is synthesized as a 170-kDa single-chain precursor, and undergoes intracellular proteolytic cleavage to the mature form. This mature c-Met is a heterodimer composed of an extracellular α chain (50 kDa) and a membrane-spanning β chain (145 kDa) that are linked together by disulfide bonds. The extracellular portion of c-Met is responsible for binding with the c-Met ligand, hepatocyte growth factor (HGF). The intracellular portion of c-Met consists of a juxtamembrane domain that harbors regulatory motifs,2, 3, 4 a tyrosine kinase domain that has tyrosine kinase activity and a C-terminal domain that contains a multifunctional docking site for various signaling molecules. Upon binding with HGF, c-Met becomes dimerized and activated, and then undergoes autophosphorylation in the intracellular domain; thereafter, substrate phosphorylation leads to changes in various cellular responses, including cell proliferation, motility, survival, adhesion and angiogenesis. Dysregulation of the HGF-c-Met pathway via either autocrine or paracrine means can lead to malignant cell transformation. Interestingly, the expression level of c-Met is associated with the progression of many different types of tumors, and upregulation of c-Met is observed in metastatic tumors.5
Various RTKs, such as epidermal growth factor receptor (EGFR), ErbB-2, ErbB-4 and fibroblast growth factor receptor 1, reportedly translocate into the nuclei of cells as either intact or truncated forms.6,7 A cytosolic fragment may be generated from a transmembrane protein in two different ways: cleavage within the transmembrane domain by γ-secretase8 or cleavage within the cytoplasmic domain by proteases such as the proteasome9 or caspases.10 Nuclear EGFR is able to induce the transcription of various genes that are essential for cell proliferation,11 cell cycle regulation, DNA repair and cell survival,12, 13, 14 and has been correlated with treatment resistance and poor prognosis in cancer.15, 16, 17 Furthermore, a truncated fragment of ErbB-4 found in the nucleus has been shown to be involved in activating or regulating the transcription of target genes.6,7 In the case of c-Met, a C-terminal fragment of~60 kDa has been identified in the nuclear fractions of several cancerous and noncancerous cell lines.18 Interestingly, nuclear c-Met was observed even in the absence of its ligand in MDA-MB231 breastcarcinoma cells.19 However, we do not yet fully understand the mechanism(s) underlying the nuclear translocation of c-Met.
The nuclear C-terminal fragment of c-Met is thought to play a substantial role as a transcription factor, activating certain genes associated with either cell proliferation20 or cell migration which is associated with an invasive phenotype.19
The translocation of protein molecules larger than 30–40 kDa21,22 from the cytoplasm to the nucleus requires a specific amino acid sequence called a nuclear localization signal (NLS).23,24 In general, the nuclear import of proteins occurs via the formation of a ternary complex composed of the carrier proteins, that is, importin α and importin β, and the NLS-containing cargo protein. Importin α recognizes and binds the NLS of the cargo protein, whereas importin β enables the formation of a trimeric complex by mediating the binding of importin α with the NLS.
Many classical and non-classical NLSs have been identified to date. The classical monopartite NLS generally consists of 4–6 predominantly basic amino acids, such as lysine and arginine; a characteristic example is found in the Simian Virus 40 Large T-antigen.25 A second type of classical NLS is the bipartite NLS, which is characterized by the presence of two basic amino acid clusters separated by a 10- to 12-amino-acid spacer, as seen in nucleoplasmin (KRPAATKKAGQAKKKK).26,27 Importin α binds with the NLS-containing cargo protein through ionic interactions that form between the basic amino acids of the NLS and acidic amino acids of importin α.28 Nevertheless, there are some previous reports that some of the proteins are imported to the nucleus by direct binding with importin β without the involvement of importin α as in Rex protein of HTLV-1,29 Smad 3,30 ribosomal protein L23a31 and histones.32 The nuclear localization of the c-Met C-terminal fragment should require the presence of an NLS, but neither relevant sequence was found by the available nuclear localization search engine nor had been reported in previous studies.
Here, we report that c-Met harbors a novel functional NLS in its juxtamembrane domain, consisting of two histidines separated by a 10-amino-acid linker. Although this is similar to the known bipartite NLSs, the NLS found in c-Met contains histidines rather than lysines or arginines, making the efficiency of nuclear localization by this NLS uniquely sensitive to changes in cytosolic pH.
Materials and methods
Materials
F12 (Dulbecco's Modified Eagle's Medium/Nutrient Mixture F-12 Ham), PEI (Polyethylenimine), 2-deoxyglucose, NaN3, leptomycin B and nigericin were purchased from Sigma-Aldrich Chemical Co. (St Louis, MO, USA). DMEM (Dulbecco's Modified Eagle's Medium) and Lipofectamine 2000 were obtained from Invitrogen Corporation (Carlsbad, CA, USA). BCECF-AM [2',7'-Bis-(2-Carboxyethyl)-5-(and-6)-Carboxyfluorescein, Acetoxymethyl Ester] was from Molecular Probes (Eugene, OR, USA). Mouse monoclonal anti-c-Met antibody (25H2) was from Cell Signaling Technology (Beverly, MA, USA). Rabbit polyclonal anti-c-Met (C-28) (sc-161), mouse monoclonal anti-GFP (sc-9996), anti-α-tubulin (sc-23948) and anti-lamin B (sc-374015) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti-importin β antibody (3E9) was from Abcam (Cambridge, UK).
Plasmid construction
Plasmids for Jxtm1,-2,-3 green fluorescent protein (GFP) were from Dr Rimm (Yale University, New Haven, CT, USA). For the cloning of GFP-Juxta-kinase domain (JKD), fragment number-2 (F-2) and fragment number-3 (F-3), truncating in the juxtamembrane region at K957 and G1090, D972 and Y1026, P1027 and I1084 of c-Met were respectively cloned into the pEGFP-C3 fusion vector. We amplified using the primers containing XhoI and BamHI sites, respectively, in a PCR using the template DNA of human c-Met. The DNA products were cloned into pEGFP-C3 fusion vector in the similar manner. The primer sequences used are in the Supplementary Table 2A. A series of mutant plasmids were generated by PCR mutagenesis using either QuickChange Multi Site-Directed Mutagenesis kit (Stratagene, #200531, Agilent Technologies, Santa Clara, CA, USA) for amino acid substitution according to manufacturer's instructions. The primers used in the PCR reactions are listed in the Supplementary Table 2B. All PCR products were checked by sequencing to confirm the presence of mutation and to verify the absence of secondary point mutations.
Cell culture and transfection
HeLa and Chang cell lines have been purchased from ATCC (Manassas, VA, USA), and passaged and used in our laboratory for fewer than 6 months after resuscitation. ATCC performs cell line authentication by using STR profiling. HeLa cells were grown in F12 and Chang cells were grown in DMEM, respectively supplemented with 10% fetal bovine serum (FBS) and 1% antibiotic-antimycotic (Life Technologies, Carlsbad, CA, USA) at 37 °C under 5% CO2 in air and 95% humidity. Cells were transiently transfected with each GFP-fusion plasmids or si-RNA using PEI or Lipofectamine 2000 (Life Technologies), respectively, according to the manufacturer's instructions.
Cellular fractionation
Cells pretreated with MG132 (10 μM) for 2 h were harvested and washed with phosphate-buffered saline (PBS), resuspended in 200 μl of TM-2 buffer (20 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 0.3 M sucrose containing freshly added protease inhibitors) and incubated on ice for 5 min, subsequently added with 10 μl 10% Triton X-100 and again incubated on ice for 5 min. Cells were sheared by needle (22-guage) and then centrifuged (1,200g, 4 °C, 15 min). The supernatant was used as the cytoplasmic fraction. The pellet was resuspended in the TM-2 buffer, sheared again using a needle, and then centrifuged (1200g, 4 °C, 15 min). Pellet was washed three times, and resuspended in the mixture of 85 μl of TM-2 buffer containing 10 μl of 3 M KCl, 5 μl of 10% Triton X-100 and incubated for 10 min, and then centrifuged at 12 000g for 2 min. Supernatant was used as the nuclear fraction.
Immunocytochemistry
Cells (1 × 105 in number) grown on coverslips were transfected with plasmids or si-RNAs. At 24 h after transfection, cells were supplied with complete media. At 48 h after transfection, cells were washed with PBS and fixed with 1:1 ice-cold methanol-acetone for 15 min. Cells were then permeabilized with 0.2% (V/V) Triton X-100 in PBS for 5 min, and then washed three times with PBS and then blocked with 1% (W/V) BSA for 1 h. Cells were then incubated with primary antibodies at 4 °C for overnight and then washed with PBS. Cells were incubated with goat anti-mouse or goat anti-rabbit Alexa 448 for 1 h at room temperature. After washing three times with PBS, cells were stained with diamidino-2-phenylindole (DAPI) for 10 min and washed three times with PBS. Coverslips were then mounted with mounting medium (Biomeda, Foster city, CA, USA). Images were taken under fluorescence or confocal microscope (LSM 710, Carl Zeiss, Jena, Germany).
Decrease of cytosolic pH
HeLa cells transfected with GFP-fusion constructs were treated either with nigericin at different concentrations as indicated in the text or with pH 6.5 Krebs-Ringer bicarbonate (KRB) solution (140 mM NaCl, 3.6 mM KCl, 0.5 mM NaH2PO4, 0.5 mM MgSO4, 1.5 mM CaCl2, 10 mM HEPES, 2 mM NaHCO3, 5.5 mM glucose). Cells were then incubated at 37 °C under a humidified atmosphere of 5% CO2 for different durations. Cells were then harvested for further assays.
Cytosolic pH measurements
HeLa cells seeded on poly-L-lysine-coated coverslips were loaded with a pH-sensitive fluorescence dye, BCECF-AM (2.5 μM) for 30 min at room temperature. After the wash-out, nigericin (1 μM) or acidic pH (6.5) was applied for the indicated time (0–60 min) in KRB solution containing sulfinpyrazone (100 μM). Fluorescence imaging of BCECF was performed by using an inverted microscope (IX-70, Olympus, Tokyo, Japan) equipped with a cooled charge-coupled device camera (Cascade 512B, Photometrics, Tucson, AZ, USA). Cells were alternately excited at 440 nm and 490 nm by a light source (DG-4, Sutter, Novato, CA, USA) and acquired images at 540 nm were analysed using Metafluor 6.3 software (Universal Imaging, Molecular Devices, Sunnyvale, CA, USA). Titration of pH was carried out by clamping the cytosolic pH with high K+ buffers (125 mM KCl, 5 mM NaCl, 1 mM NaH2PO4, 1 mM MgSO4 and 10 mM HEPES) of defined pH containing nigericin (5 μM) and monensin (5 μM).
Importin-β binding assay
Lysate (500 μg) of HeLa cells transfected with various constructs was incubated with 1 μg of anti-importin β (3E9) antibody for 16 h at 4 °C with gentle rotation. Twenty-five microliter of 50% slurry of rProtein G Agarose were added and the mixture was incubated for additional 1 h to pull down the immune complex. After three successive wash with RIPA buffer containing freshly added protease inhibitors (l mg ml−1 aprotinin and l mg ml−1 leupeptin) and phosphatase inhibitors (1 mM sodium fluoride, 1 mM NaVO4), bound proteins were subjected to western blotting.
Time-lapse analysis
HeLa cells were seeded onto a six-well plate and transfected with GFP-fusion constructs. Twenty-four hours after transfection, two-channel time-lapse video microscopy was performed using a fully motorized Axiovert 200 M microscope (Carl Zeiss), equipped with AxiocamHRm. Temperature and CO2 control was maintained using the Incubator S-M and heating insert M06 controlled by Tempcontrol 37-2 and CTI-Controller 3700. Both phase contrast and GFP fluorescence images were acquired for 48 h with a lapse time of 5 min using AxioVision 4.3 software (Carl Zeiss). For routine and quantitative analyses, images were acquired using a × 20 objective lens (LD Plan-Neofluar × 20/0.4 Corr Ph2, Carl Zeiss).
Statistical analyses
All data were presented as mean±s.d. Each experiment was performed in triplicates. Statistical differences were analyzed by Student's t-test or analysis of variance test, and P-values <0.05 were regarded as statistically significant.
Results
The c-Met fragment localizes to the nuclei of HeLa cells
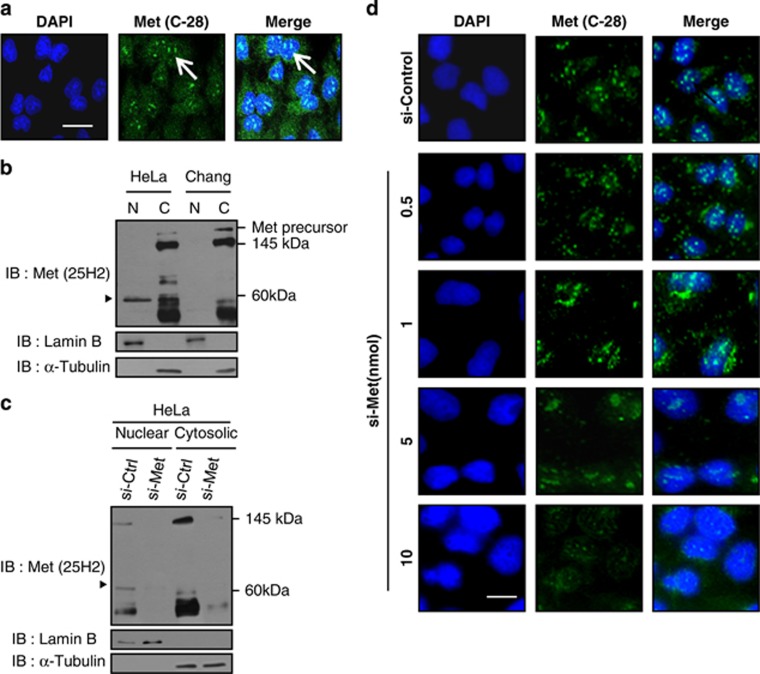
To search for the NLS of c-Met, we first needed to identify a suitable cell line that contained all the molecular machineries required for nuclear translocation of the c-Met fragment. To this end, we investigated the presence of the c-Met fragment in the nuclei of HeLa cells. Cells were stained with an antibody specific for the C terminus of c-Met (C-28) and observed by confocal microscopy, which revealed a fine and distinct granular pattern of staining in the nuclei (Figure 1a). We next performed cellular fractionation to confirm the immunocytochemical data. Immunoblotting using another antibody specific to the C terminus of c-Met (25H2) clearly detected the ~60 kDa C-terminal fragment of c-Met in the nuclear fraction. This is consistent with previous reports on the nuclear C-terminal fragment of c-Met in various cells.18,19 In contrast, the nuclear fraction from Chang liver cells did not show the nuclear c-Met band despite the presence of wild-type c-Met in the non-nuclear fraction (Figure 1b), indicating that the nuclear translocation of the c-Met fragment is cell-type-specific. The observed expression of the c-Met fragment in the nuclear fraction of HeLa cells was clearly abolished by the c-Met-small interfering RNA (siRNA) (Figure 1c), confirming the presence of the nuclear c-Met fragment in the nuclei of HeLa cells. To confirm the specificity of this signal, we carried out RNAi-mediated gene knockdown experiments, and found that the distinct punctate pattern was significantly and dose-dependently abrogated by a c-Met-specific siRNA (Figure 1d) supporting the specificity of antibody. Based on these findings, we used the HeLa cell line as the model for our investigations.
Figure 1.
The C-terminal fragment of c-Met localizes in the nuclei of HeLa cells. (a) Sub-confluent HeLa cells were fixed and stained with C-terminal antibody for c-Met (C-28, green) and DAPI for nucleus (blue). Representative images show the nuclear patches of c-Met. Size bar,10 μm. (b) Cell lysates from the indicated cells were fractionated as described in Materials and methods, and then subjected to western blot analysis by using antibody against C terminus of c-Met (25H2). Notice the c-Met RTK (145 kDa) in cytosolic fraction and a 60 kDa fragment of c-Met (triangle) in nuclear fraction only in HeLa cells. Purity of cellular fractionation was checked by Lamin B (nuclear marker) and tubulin (cytoplasmic marker). (c) Cell lysates from HeLa cells transfected with indicated siRNAs were fractionated and subjected to western blot analysis as in (b), triangle; 60kDa nuclear c-Met fragment. (d) HeLa cells were transfected with control or c-Met siRNAs at the indicated concentrations and fixed at 24 h after transfection. Immunocytochemistry was performed using C-28 antibody, and DAPI staining was used to visualize nuclei. Representative images show disappearance of nuclear c-Met signal in the presence of c-Met siRNA. Met (green) and nuclei (blue). Size bar; 10 μm. Each experiment was performed in triplicate with similar results.
Serial deletion of the juxtamembrane domain shows that c-Met contains an NLS at H1068-H1079
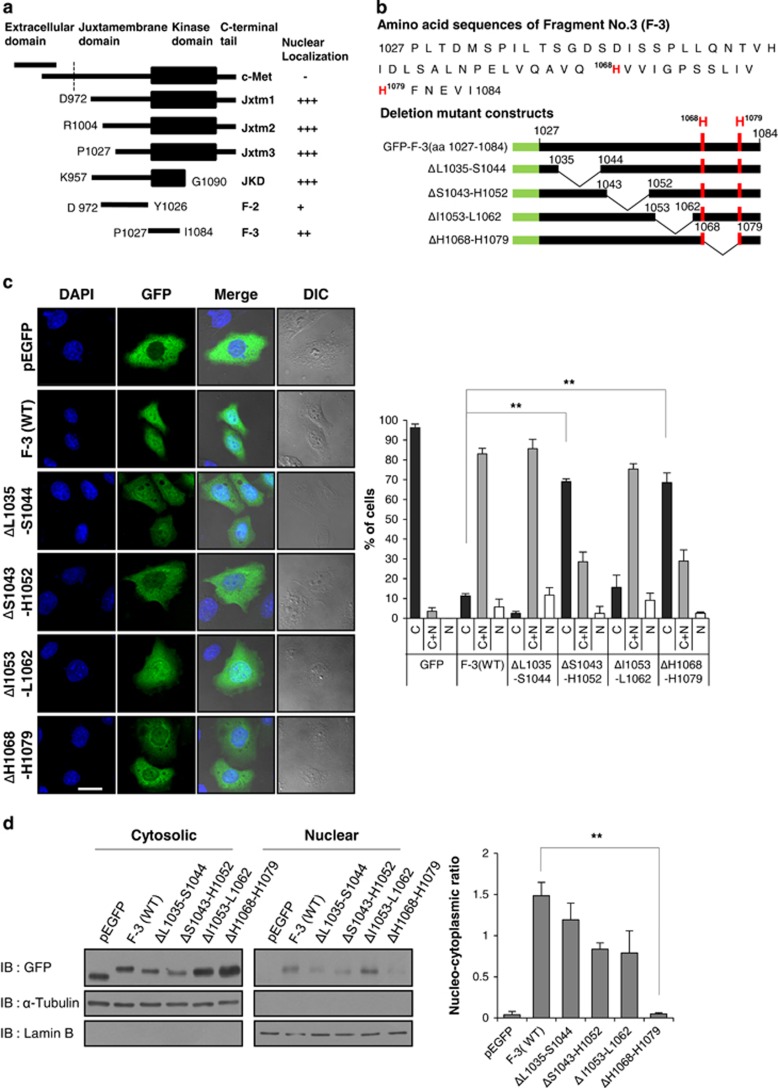
Although the nuclear c-Met fragment clearly originates from the cytosolic portion of full-size Met, no previous study had clearly identified the precise mechanism of translocation or the specific NLS involved. A previous report found that c-Met does not contain a classical NLS.18 Therefore, our main goal was to identify one or more motifs essential for the nuclear translocation of the c-Met fragment. We generated various GFP fusion constructs of the cytosolic portion of c-Met (Figure 2a), and transfected these constructs into HeLa cells. Immunocytochemistry, and western blotting after subcellular fractionation enabled us to localize the putative NLS to between P1027 and I1084 in the juxtamembrane domain of c-Met (data not shown); this is the F-3 fragment shown in Figure 2a.
Figure 2.
Serial deletion of juxtamembrane domain shows that c-Met harbors NLS motif at aaH1068-H1079. (a) Schematic representation of full and different truncated forms of Met with different domains fused to C terminus of pEGFP plasmid as described in Materials and methods. dotted line; the boundary between extracellular domain and transmembrane domain. (b) Amino acid sequences of fragment No.3 (F-3) and schematic diagrams of deletion mutant constructs of F-3 fragment. GFP (green), vertical lines (red) denote the position of H1068 and H1079. (c) HeLa cells were transfected with indicated deletion mutant constructs of GFP-F3, and subjected to immunocytochemistry using GFP antibody. Confocal images were taken at × 40 objective. Size bar; 10 μm. The percentages of cells depicting subcellular distribution of deletion mutant constructs are shown in the bar graph on the right. Error bars; standard deviations. Three independent experiments were performed in triplicates. **P<0.01 by Student's t-test, n=200–270. (d) Cellular fractionation and western blot analysis by using anti-GFP was performed after transfection as in (c). Purity of cellular fractionation was checked by Lamin B (nuclear marker) and tubulin (cytoplasmic marker). Each experiment was performed in triplicates with equivalent results. The densitometry graph on the right shows the nucleo-cytoplasmic ratio (nuclear GFP intensity divided by lamin intensity/cytosolic GFP intensity divided by tubulin intensity).**P<0.01 by Student's t-test.
To uncover the exact NLS, we generated a series of deletion mutants of GFP-(F-3) fusion constructs (Figure 2b) and investigated their intracellular distributions using Immunocytochemistry, and western blotting after subcellular fractionation. Two of the deletion mutants (ΔL1035–S1044 and ΔI1053–L1062) showed nuclear localization patterns similar to that of the wild-type F-3 fragment, indicating that the putative NLS was not located in these fragments. In contrast, the ΔH1068–H1079 deletion robustly abolished nuclear localization, as observed by immunocytochemistry (Figure 2c, left panel). The percentage of cells showing ubiquitous (cytosolic and nuclear) localization was significantly decreased by this deletion, whereas the proportion showing cytosol-only localization was significantly increased compared to the results obtained with the wild-type F-3 fragment (Figure 2c, right panel). Similar results were obtained from a subcellular fractionation assay, wherein the protein level of the ΔH1068–H1079 mutant was substantially increased in the cytosolic fraction but decreased in the nuclear fraction (Figure 2d). Although the ΔS1043–H1052 deletion mutant also showed decreased nuclear localization in our immunocytochemical analysis (Figure 2c), our subcellular fractionation assay clearly indicated that ΔH1068–H1079 but not ΔS1043–H1052 harbored the major NLS in the juxtamembrane fragment of c-Met (Figure 2d).
Two histidine residues are important for the nuclear translocation of the c-Met fragment
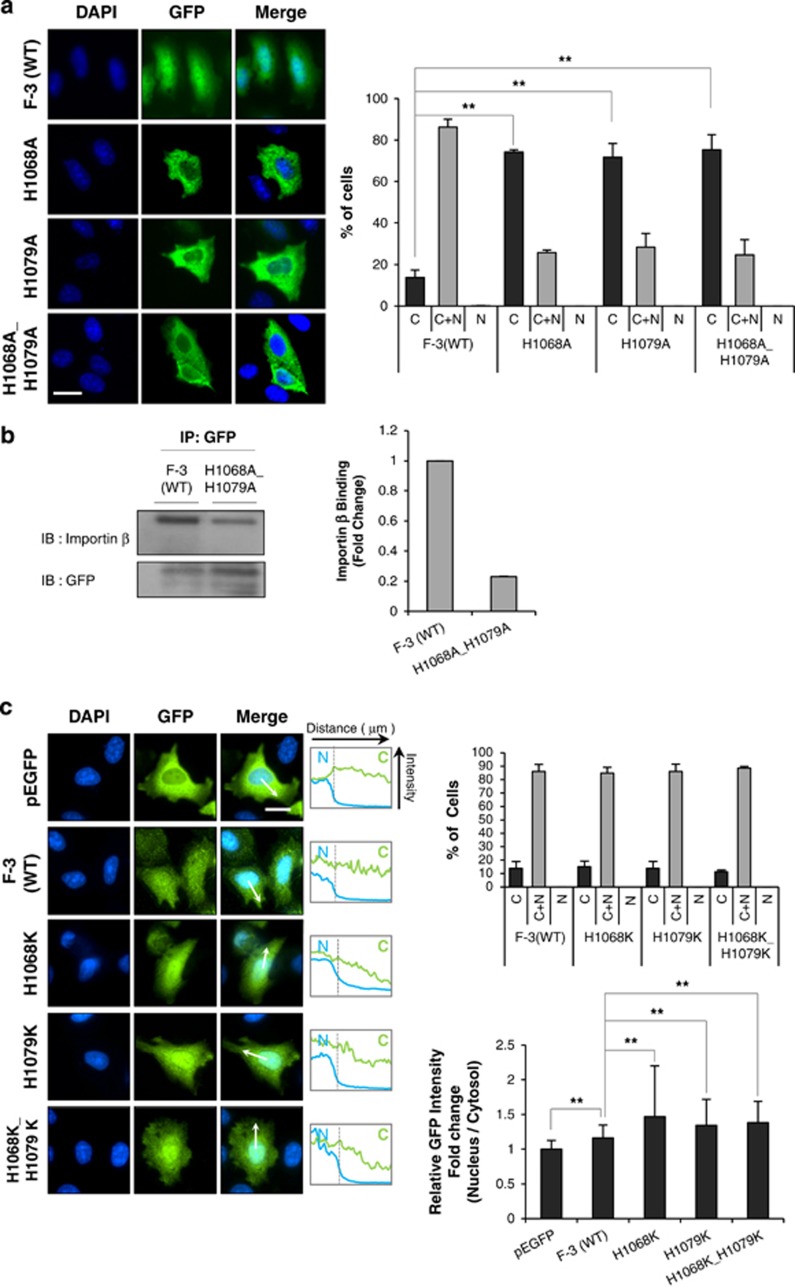
Classical bipartite NLSs comprise two stretches of basic amino acids (lysines or arginines) separated by a 10- to 12-amino-acid spacer, as previously described for many nuclear proteins, including nucleoplasmin26,27 (Supplementary Table 1). In bipartite NLSs, the positive charges provided by lysines or arginines are essential because they enable the electrostatic interaction with negatively charged amino acid residues on importin α, resulting in binding between the importin and the cargo protein.33, 34, 35 We hypothesized that the putative NLS found in c-Met (H1068-H1079) might be a variant of this classical bipartite NLS, with two histidines replacing the lysines or arginines found in classical bipartite NLSs. To test this, we created point mutants of the F-3 protein wherein one or both histidines of the putative NLS motif were substituted with alanine (H1068A, H1079A and H1068A_H1079A), and examined the importance of the histidine residues for the nuclear localization of c-Met. Indeed, the mutant constructs all yielded significantly higher fractions of cells showing cytoplasm-only localization (from ~10% to 70–80%), indicating that nuclear translocation was blocked in the mutant-expressing cells (Figure 3a). Next, we performed biochemical analysis to confirm the essential role of these histidine residues for nuclear translocation. Since mammalian cells may express six different forms of importin α but only one form of importin β,36,37 and association with importin β is an essential process for the cargo proteins to translocate into nucleus, we examined whether the association of cargo with importin β changes depending on the histidine residues. Clearly, the wild-type F-3 fragment co-immunoprecipitated with importin β, and substitution of both histidines with alanine significantly decreased this protein–protein interaction (Figure 3b). These results are consistent with our hypothesis that these two histidines are essential for the function of the NLS of c-Met. Moreover, considering that positive charges of lysine or arginine residues in classical bipartite NLS enable electrostatic interaction with importin α, the histidine residues might also confer positivity needed for the interaction between cargo and importin α, which subsequently enhance the formation of cargo-importin α-importin β ternary complex.
Figure 3.
Two histidine residues are crucial for the nuclear translocation of c-Met fragment. (a) HeLa cells were transfected with wild-type F-3, indicated histidine-to-alanine mutant constructs or pEGFP as a vector control. Immunocytochemistry by using anti-GFP antibody was performed as in Figure 2. Representative images were taken from fluorescence microscopy at × 40 objective. Bar graph on right panel shows the percentage of subcellular distribution of cells (n=120–225). ***P<0.001 by Student's t-test. (b) Cell lysates from HeLa cells transfected with F-3(WT) or H1068A_H1079A mutant were subjected to immunoprecipitation by using anti-GFP antibody, which was followed by immunoblotting with importin β antibody. Bar graph in right shows the relative binding tendency of importin β with cargo protein compared to F-3(WT). (c) HeLa cells were transfected with wild-type F-3 or indicated histidine-to-lysine mutant constructs, and subjected to immunocytochemistry by using anti-GFP antibodies. Bar graph displaying on right upper panel shows the percentage of subcellular distribution of cells (n=102–156) and on the right lower panel shows the relative nucleo-cytoplasmic ratio of green fluorescence intensity. Middle panel shows the intensity changes revealed by line scan analysis performed following the arrow. Error bars; standard deviations of three independent experiments. *P<0.05, **P<0.01 by Student's t-test, n=25.
When we replaced either or both of the histidine residues with lysine (which is a more basic amino acid), the nucleo-cytoplasmic ratios of the lysine-substituted mutants were higher than that of the wild-type F-3 construct (Figure 3c, lower right panel). In this case, we measured nucleo-cytoplasmic intensity ratios. Simple counting of the cells showing different subcellular localizations failed to reveal any difference (Figure 3c, right upper panel) because the category of cells showing both nuclear and cytosolic localization (Figure 3c, right upper panel) contained cells with varied proportions of nuclear and cytosolic signals. A representative line scan analysis also revealed a similar tendency (Figure 3c, middle panel). Together, these findings strengthen our assumption that the two histidine residues in the NLS of c-Met might confer the positive charges needed for the complex formation with importin molecules and the subsequent nuclear translocation of the c-Met fragment.
The NLS-directed nuclear accumulation of the c-Met fragment is pH-dependent
Because the imidazole group of histidine has a pKa of 6.0–6.2, which is fairly close to physiological pH, it can be protonated or deprotonated depending on the pH inside the cell.38, 39, 40 Thus, under physiological conditions, a relatively small shift in the environmental pH (especially to the acidic side) will readily change the average positive charge of a histidine residue. Thus, we speculated that a small change in intracellular pH could change the strength of the interaction between importin and the histidine-containing NLS of the c-Met fragment, thereby affecting its nuclear translocation.
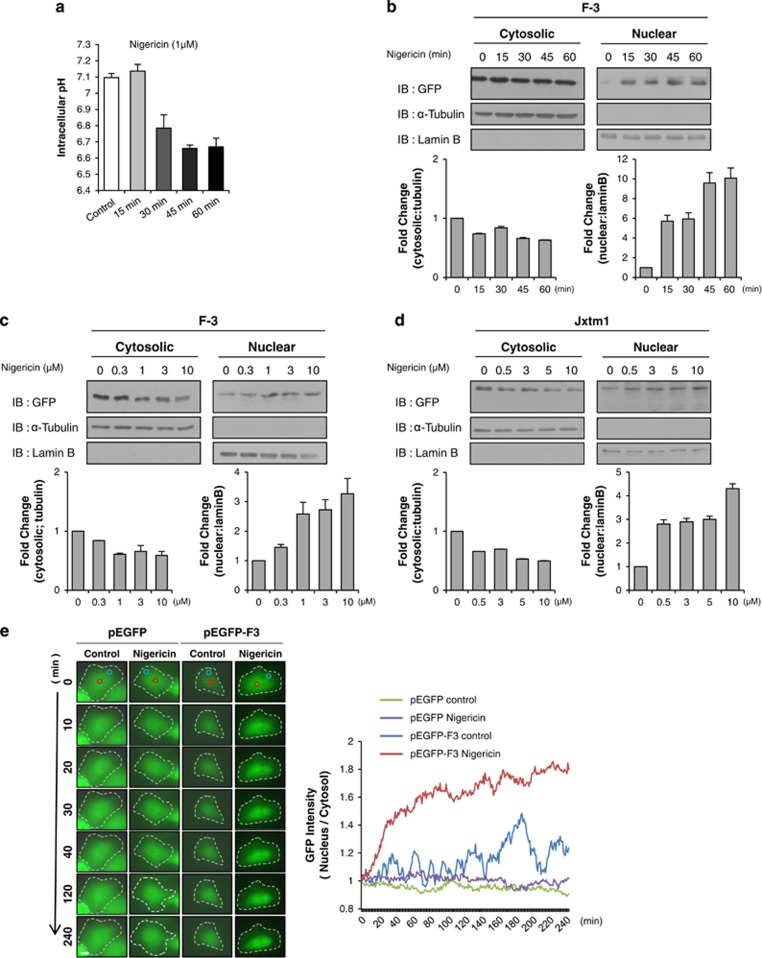
To explore the effect of decreased cytosolic pH on the nuclear translocation of the c-Met fragment, we used nigericin, which reduces cytosolic pH via the exchange of Na+ and H+ from vesicular organelles.41,42 We measured the actual cytosolic pH change with the pH-sensitive dye, BCECF-AM.43,44 As shown in Figure 4a, treatment of HeLa cells with 1 uM nigericin significantly decreased the cytosolic pH as early as 30 min posttreatment, and this decrease continued time-dependently thereafter. We then explored the effect of nigericin on the nuclear translocation of the c-Met fragment. As expected, decreasing the cytosolic pH significantly increased the nuclear accumulation of the fusion protein both time- and dose-dependently (Figures 4b and c). The increase in the nuclear protein level was observed as early as 15 min after nigericin treatment and reached up to 10-fold at 1 h posttreatment; the protein level in the cytosolic fraction decreased only slightly because most of the fusion proteins were localized in the cytosol (Supplementary Figure 1). We obtained essentially the same result using the Jxtm1 fragment of c-Met protein (Figure 2a), which has similar size to the~60 kDa C-terminal fragment of c-Met, ruling out the possibility that this result was an artifact due to our use of a small fragment of c-Met (Figure 4d). Since there was some discrepancy in the timings of the pH decrease and the nuclear translocation, we used time-lapse microscopy for real-time measurement of nuclear translocation (Figure 4e). When the fluorescence intensities of two circular areas inside the cytosol and nucleus, respectively, were measured from the time-lapse images (Figure 4e, right panel), we clearly saw that the nucleo-cytoplasmic ratio of the fusion protein increased significantly at ~30 min after nigericin treatment, which was consistent with the cytosolic pH change measured using BCECF-AM (Figure 4a). These data collectively indicated that the novel NLS of c-Met appears to be sensitive to changes in pH (hereinafter called ‘pH-dependent').
Figure 4.
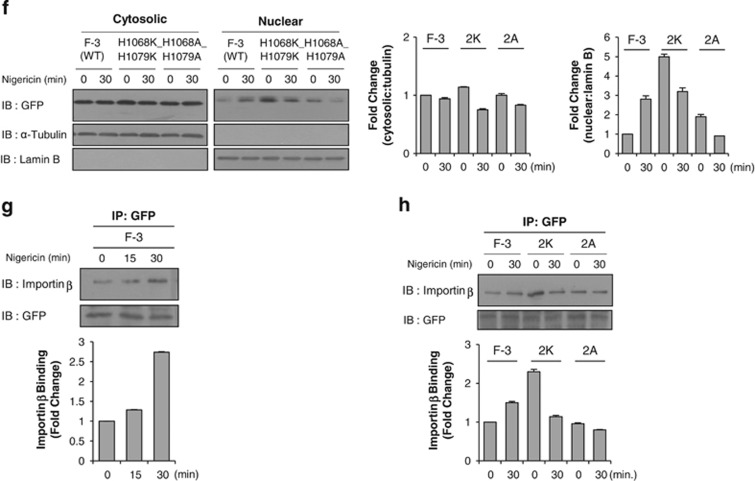
Nuclear accumulation of c-Met fragment is cytosolic pH-dependent. (a) Bar graph shows the intracellular pH change measured by BCECF dye as described in Materials and methods with 1 μM nigericin treatment in a time-dependent manner. Representative result from three independent experiments. pH value of each experiment is the average of data from more than 15 cells on a cover-slip. (b) HeLa cells transiently transfected with F-3 construct were treated with or without 1 μM nigericin at indicated time intervals. Western blot analysis was performed using anti-GFP antibody after cellular fractionation. Tubulin and lamin B were used as cytoplasmic and nuclear markers respectively. Bar graphs below each panel show their respective subcellular distribution of the protein in fold change. (c, d) Western blot analysis using anti-GFP antibody was performed after subcellular fractionation of F-3 (in c) or Jxtm1 (in d)-transfected HeLa cells treated with nigericin at indicated concentration for 30 min. Bar graphs below show the subcellular distribution of the protein in fold change. (e) Representative time-lapse microscopy images of either pEGFP or F-3 transfected HeLa cells treated with 1 μM nigericin at indicated time intervals. The graph displaying on right shows nucleo-cytoplasmic ratio of GFP intensity over time. Red circle; nuclear area, white circle; cytoplasmic area. Margin of the cells are indicated by white broken line, n=10. (f) Western blot analysis using indicated antibodies were performed after subcellular fractionation of F-3 (WT), histidine-to-lysine (2K), or histidine-to-alanine (2A) mutants-transfected HeLa cells treated with or without 1 μM nigericin for 30 min. Bar graphs on right panel show the subcellular distribution of the protein in fold change. (g) Importin β binding assay (as described in Figure 3b) was performed in F-3-transfected HeLa cells after the treatment with nigericin at indicated time points. Bar graph below shows the amount of cargo-binding importin β in fold change. (h) Immunoprecipitation was performed with anti-GFP antibody with the cell lysate of F-3 (WT), histidine-to-lysine (2K) or histidine-to-alanine (2A) mutants-transfected HeLa cells treated with nigericin for 30 min followed by immunoblotting with importin β antibody. Bar graph below shows amount of cargo-binding importin β in fold change.
Next, we examined whether this pH dependency could be altered by replacing the critical histidines with other amino acids. Indeed, the increase of nuclear localization by nigericin treatment was abolished when histidine residues were substituted with either lysines or alanines (Figure 4f). Unexpectedly, nigericin treatment decreased the protein levels of these mutants in the nuclear fraction, which we don't understand the reason yet. Collectively, however, these data provide additional evidence that the putative bipartite NLS is pH-dependent, and this pH dependency stems from the critical histidine residues.
We then examined whether a decrease in cytosolic pH could enhance the binding of the c-Met fragment to importin α and β. Indeed, we found that a decrease in cytosolic pH enhanced the interaction between importin β and the F-3 fragment in immunoprecipitation assays run after 30 min of nigericin treatment (Figure 4g). This is consistent with the time point at which the decrease in cytosolic pH became significant (Figure 4a). In lysine- or alanine-mutant-transfected cells, in contrast, a decrease in cytosolic pH did not alter the level of cargo-associated importin β (Figure 4h), indicating that the pH-dependent change in the association of the c-Met fragment with importin is governed by these histidine residues.
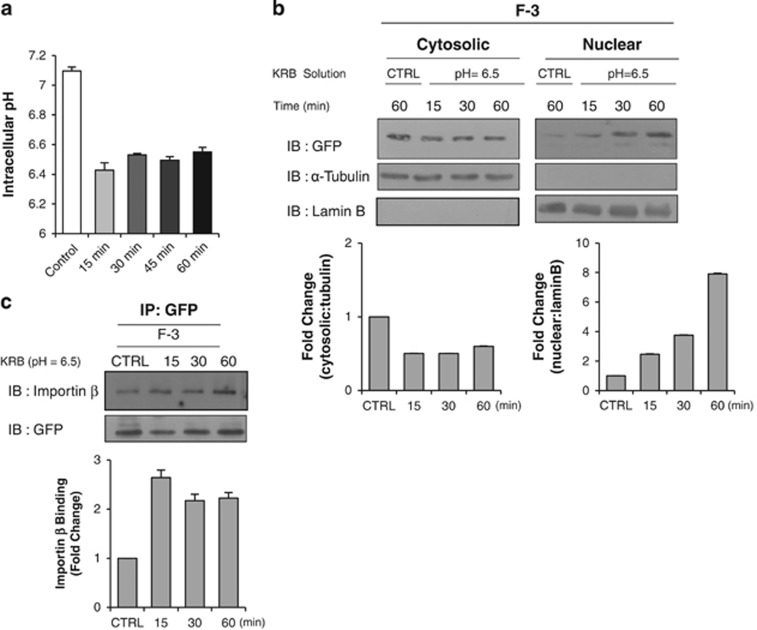
To further confirm the effect of altered cytosolic pH on the nuclear translocation of the c-Met fragment, we next used a low pH buffer. The actual cytosolic pH in cells significantly decreased to 6.5 after as little as 15 min of incubation with a pH 6.5 KRB buffer solution (Figure 5a). This was associated with a significant and time-dependent increase in the nuclear accumulation of the F-3 fusion protein (Figure 5b). This increase was observed after as little as 15 min of the low pH buffer treatment, and the nuclear fraction of the F-3 fusion protein was increased up to seven-fold after 1 h of treatment; meanwhile, the cytosolic fraction showed a slight decrease. The associations between the F-3 fragment and the importin proteins were also significantly and time-dependently when the cells were incubated with the low pH buffer (Figure 5c).
Figure 5.
Low pH KRB buffer solution enhances the nuclear accumulation of c-Met fragment. (a) Bar graph shows the intracellular pH change measured by BCECF dye as described in Materials and Methods after the treatment with low pH-KRB (pH 6.5) at indicated time intervals. Representative result from three independent experiments. pH value of each experiment is the average of data from more than 15 cells on a coverslip. (b) HeLa cells transiently transfected with F-3 construct were treated with KRB (pH=6.5) solution for indicated durations. Immunoblotting of the cell lysates after subcellular fractionation was performed using anti-GFP antibody. Bar graphs below show the changes in fold of densitometric intensity compared to control with neutral pH KRB solution (CTRL). (c) Importin binding assay was performed in F-3 – transfected HeLa cells treated with low pH buffer for indicated durations. For this, immunoprecipitation was carried out in the same way as mentioned above. Densitometric bar graph shows the binding amount of importin β (in fold change) with cargo.
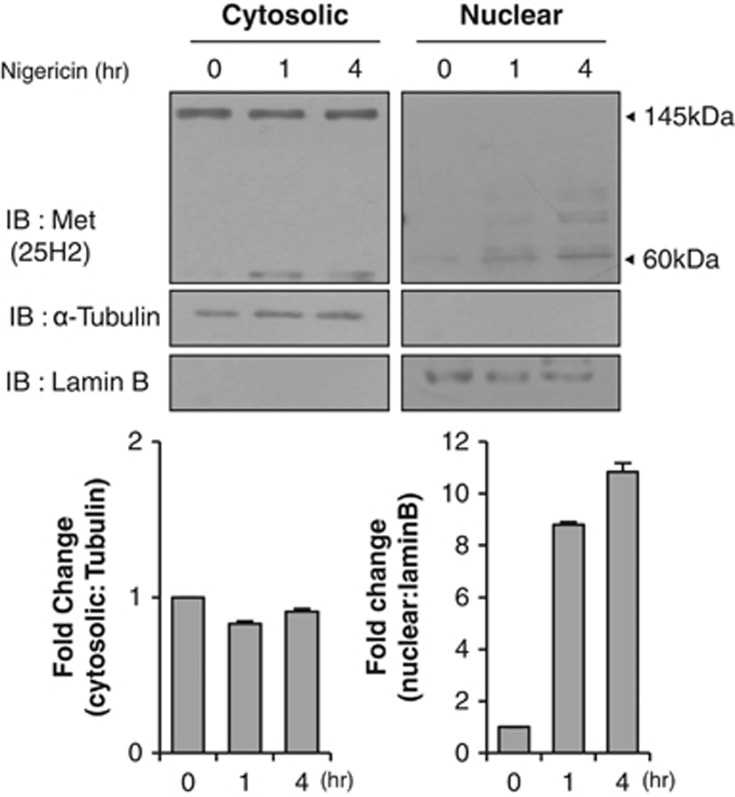
Finally, we explored whether a decrease in cytosolic pH could also enhance the nuclear translocation of the endogenous 60-kDa c-Met fragment. As expected, we found that the amount of the nuclear c-Met fragment increased significantly and time-dependently increased in nigericin-treated cells compared to untreated controls (Figure 6). This indicates that the pH-dependent nuclear localization observed with the ectopically expressed c-Met fragment could also be observed with endogenous c-Met in HeLa cells.
Figure 6.
Decrease of cytosolic pH enhances nuclear translocation of~60 kDa endogenous c-Met fragment. HeLa cells treated with 1 μM nigericin for indicated durations were subjected to subcellular fractionation followed by immunoblotting with an antibody against C terminus of c-Met (25H2). Protein densitometry graph depicts either 145 kDa Met (cytoplasm) or 60 kDa C-terminal fragment of c-Met level (nucleus) in fold change.
In sum, we herein used two different methods to change the cytosolic pH of HeLa cells, and our results verified the hypothesis that the putative novel NLS found in c-Met is pH-dependent, most probably due to the histidine residues that provide the positive charges needed to bind with the importin molecules. To our knowledge, the putative non-classical bipartite NLS found in c-Met can be the first reported example of a pH-dependent NLS.
Discussion
We herein show for the first time that a variant of the classical bipartite NLS, in which two histidines are separated by a 10-amino-acid spacer in the juxtamembrane region, is necessary for the nuclear translocation of the C-terminal fragment of c-Met. We also demonstrate that this variant is a pH-dependent NLS, providing what we believe to be the first example of such an NLS.
Regarding pH-dependent changes in the subcellular localization of proteins, the p26 protein was shown to undergo nuclear-cytoplasmic shuttling during aerobic-anoxic transition in embryos of the brine shrimp, Artemia franciscana.45 However, this previous study did not examine the amino acid sequences that might be responsible for this phenomenon. A recent study found that von Hippel–Lindau (VHL) tumor suppressor protein is confined to the nucleoli when the extracellular pH drops below normal physiological conditions.46 However, this prior study did not clearly elucidate the responsible motif or the underlying mechanism for the subnuclear confinement of VHL in an acidic environment. To the best of our knowledge, the present study is the first to provide an example of an NLS that shows pH dependency.
The classical bipartite NLSs consists of two stretches of basic amino acid residues separated by a 10- to 12-amino-acid linker, as seen in nucleoplasmin.26,27 Numerous bipartite NLSs have been found in various proteins, as shown in Supplementary Table 1. Some optimal consensus sequences have been proposed, including (K/R)(K/R)X10-12(K/R),26 KRX10–12KRRK47 and KRX10–12K(K/R).48 Studies on classical bipartite NLS recognitions and interactions have shown that the upstream basic residues bind to the minor groove on importin α, whereas a downstream monopartite-like sequence combines with the major binding pocket on importin α.48, 49, 50, 51 The latter interaction occurs via salt bridges and H-bonding interactions between positive charges on the NLS and negative charges in the binding pockets of importin α. With two separate binding pockets, importin α can accommodate the 10-residue spacers linking the two basic amino acid clusters in classical bipartite NLSs.26,27 Although we could not directly show the association between cargo and importin α, the necessity of positive charges for the cargo-importin β interaction is consistent with the widely accepted role of basic aminoacid residues in the formation of cargo-importin α – importin β ternary comlex
The putative novel bipartite NLS reported herein differs from the classical bipartite NLSs by having critical histidine residues rather than lysines or arginines. Our results clearly show that these histidine residues are crucial for the nuclear translocation and pH dependency of this putative NLS. However, the positivity of these individual histidine residues are weaker than those of the two or three lysine/arginine residues commonly present in classical bipartite NLSs. The positivity of histidine residues was expected to be increased with the decrease of intracellular pH either by nigericin treatment or by low pH buffer at the physiological pH range and it might increase the binding potential of the cargo with the carrier proteins via electrostatic interaction forming the trimer complex. Some previous reports have indicated that the linker sequences of an NLS, particularly hydrophobic amino acids, are involved in importin-cargo binding.35,47,52 But the deletion of hydrophobic amino acids (1069VVI1071 and 1076LIV1078) in the H1068-H1079 linker of c-Met did not abolish the nuclear import (data not shown) indicating that only the histidine residues provide interacting site for the cargo-importin binding. In addition, this experiment strongly suggest a specific role for the histidines and not a general fold or domain for this region of the protein. The mode of regulatory mechanism or nuclear import pathway of c-Met is still unclear and it requires the further extensive study.
Most of our experiments were performed on both the F-3 fragment and the Jxtm1 fragment (Figure 4d; Supplementary Figures 2A–D), which is similar to the ~60 kDa C-terminal fragment of c-Met in terms of its molecular size and intramolecular location. We consistently obtained similar results with the F-3 fragment and Jxtm1. In addition, we found that nigericin treatment up-regulated the endogenous nuclear c-Met fragment in HeLa cells (Figure 6). We are therefore convinced that this putative pH-dependent NLS exists and functions in the nuclear translocation of the c-Met fragment.
What would be the biological relevance of the pH-dependent nuclear accumulation of c-Met fragment? In the first place, the functional role and biological relevance of nuclear c-Met is not well understood. However, some authors suggested that C-terminal fragment of nuclear c-Met may play a substantial role as a transcription factor in enhancing the c-Met signaling pathway by activating certain genes which may be associated with either cell proliferation20 or cell migration and motility which leads to invasive phenomenon in MDA-MB231 breast carcinoma cells.19 pH-dependency of nuclear translocation of c-terminal fragment of c-Met gives us an insight that it may confer proliferative advantage and/or migratory capacity for cancer cells in conditions when cytosolic pH is lowered; that is, hypoxia and/or high lactic acid condition which are frequently encountered in cancers.
In conclusion, we herein show for the first time that the H1068-H1079 sequence in the juxtamembrane region of c-Met may play a functional role in its trafficking to the nucleus. Our unique finding of a putative pH-dependent NLS in c-Met may provide important new insights into the nuclear translocation of RTKs in mammalian cells. Moreover, if histidine-governed bipartite NLSs are found in other proteins, this work might lead to the discovery a new group of proteins whose nuclear localizations are regulated by cytosolic pH.
Acknowledgments
We thank Dr Rimm (Yale University, New Haven, CT, USA) for providing Jxtm1,-2,-3 GFP constructs, Dr Hyeseong Cho (Ajou University, Korea) for the critical comments on the experimental design. This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (No. 2011-0030043).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Experimental & Molecular Medicine website (http://www.nature.com/emm)
Supplementary Material
References
- Park M, Dean M, Kaul K, Braun MJ, Gonda MA, Vande Woude G. Sequence of MET protooncogene cDNA has features characteristic of the tyrosine kinase family of growth-factor receptors. Proc. Natl Acad. Sci. 1987;84:6379–6383. doi: 10.1073/pnas.84.18.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigna E, Gramaglia D, Longati P, Bardelli A, Comoglio PM. Loss of the exon encoding the juxtamembrane domain is essential for the oncogenic activation of TPR-MET. Oncogene. 1999;18:4275–4281. doi: 10.1038/sj.onc.1202791. [DOI] [PubMed] [Google Scholar]
- Lee JH, Han SU, Cho H, Jennings B, Gerrard B, Dean M, et al. A novel germ line juxtamembrane Met mutation in human gastric cancer. Oncogene. 2000;19:4947–4953. doi: 10.1038/sj.onc.1203874. [DOI] [PubMed] [Google Scholar]
- Ma PC, Kijima T, Maulik G, Fox EA, Sattler M, Griffin JD, et al. c-MET mutational analysis in small cell lung cancer: novel juxtamembrane domain mutations regulating cytoskeletal functions. Cancer Res. 2003;63:6272–6281. [PubMed] [Google Scholar]
- Jiang W, Hiscox S, Matsumoto K, Nakamura T. Hepatocyte growth factor/scatter factor, its molecular, cellular and clinical implications in cancer. Crit. Rev. Oncol. Hematol. 1999;29:209–248. doi: 10.1016/s1040-8428(98)00019-5. [DOI] [PubMed] [Google Scholar]
- Maher PA. Nuclear translocation of fibroblast growth factor (FGF) receptors in response to FGF-2. J. Cell Biol. 1996;134:529–536. doi: 10.1083/jcb.134.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuro A, Nagai M, Navin NE, Sudol M. WW Domain-containing protein YAP associates with ErbB-4 and Acts as a Co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J. Biol. Chem. 2003;278:33334–33341. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- Hoppe T, Matuschewski K, Rape M, Schlenker S, Ulrich HD, Jentsch S. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell. 2000;102:577–586. doi: 10.1016/s0092-8674(00)00080-5. [DOI] [PubMed] [Google Scholar]
- Bae SS, Choi JH, Oh YS, Perry DK, Ryu SH, Suh PG. Proteolytic cleavage of epidermal growth factor receptor by caspases. FEBS Lett. 2001;491:16–20. doi: 10.1016/s0014-5793(01)02167-6. [DOI] [PubMed] [Google Scholar]
- Lo HW, Xia W, Wei Y, Ali-Seyed M, Huang SF, Hung MC. Novel prognostic value of nuclear epidermal growth factor receptor in breast cancer. Cancer Res. 2005;65:338–348. [PubMed] [Google Scholar]
- Dittmann K, Mayer C, Rodemann HP. Nuclear EGFR as novel therapeutic target: insights into nuclear translocation and function. Strahlentherapie und Onkologie: Organ der Deutschen Rontgengesellschaft... [et al] 2010;186:1–6. doi: 10.1007/s00066-009-2026-4. [DOI] [PubMed] [Google Scholar]
- Wang SC, Nakajima Y, Yu YL, Xia W, Chen CT, Yang CC, et al. Tyrosine phosphorylation controls PCNA function through protein stability. Nat. Cell Biol. 2006;8:1359–1368. doi: 10.1038/ncb1501. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay D, Mandal M, Adam L, Mendelsohn J, Kumar R. Physical interaction between epidermal growth factor receptor and DNA-dependent protein kinase in mammalian cells. J. Biol. Chem. 1998;273:1568–1573. doi: 10.1074/jbc.273.3.1568. [DOI] [PubMed] [Google Scholar]
- Hoshino M, Fukui H, Ono Y, Sekikawa A, Ichikawa K, Tomita S, et al. Nuclear expression of phosphorylated EGFR is associated with poor prognosis of patients with esophageal squamous cell carcinoma. Pathobiology. 2007;74:15–21. doi: 10.1159/000101047. [DOI] [PubMed] [Google Scholar]
- Laimer K, Spizzo G, Gastl G, Obrist P, Brunhuber T, Fong D, et al. High EGFR expression predicts poor prognosis in patients with squamous cell carcinoma of the oral cavity and oropharynx: a TMA-based immunohistochemical analysis. Oral Oncol. 2007;43:193–198. doi: 10.1016/j.oraloncology.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Hung LY, Tseng JT, Lee YC, Xia W, Wang YN, Wu ML, et al. Nuclear epidermal growth factor receptor (EGFR) interacts with signal transducer and activator of transcription 5 (STAT5) in activating Aurora-A gene expression. Nucleic Acids Res. 2008;36:4337–4351. doi: 10.1093/nar/gkn417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozner-Moulis S, Pappas DJ, Rimm DL. Met, the hepatocyte growth factor receptor, localizes to the nucleus in cells at low density. Cancer Res. 2006;66:7976–7982. doi: 10.1158/0008-5472.CAN-05-4335. [DOI] [PubMed] [Google Scholar]
- Matteucci E, Bendinelli P, Desiderio MA. Nuclear localization of active HGF receptor Met in aggressive MDA-MB231 breast carcinoma cells. Carcinogenesis. 2009;30:937–945. doi: 10.1093/carcin/bgp080. [DOI] [PubMed] [Google Scholar]
- Rodrigues MA, Gomes DA, Leite MF, Grant W, Zhang L, Lam W, et al. Nucleoplasmic calcium is required for cell proliferation. J. Biol. Chem. 2007;282:17061–17068. doi: 10.1074/jbc.M700490200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda Y. Nucleocytoplasmic protein traffic and its significance to cell function. Genes Cells. 2000;5:777–787. doi: 10.1046/j.1365-2443.2000.00366.x. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- Kiseleva EM, Goldberg MW, Cronshaw J, Allen TD. The Nuclear Pore Complex: Structure. Function and Dynamics. 2000;10:12. [PubMed] [Google Scholar]
- Sorokin AV, Kim ER, Ovchinnikov LP. Nucleocytoplasmic transport of proteins. Biochemistry. Biokhimiia. 2007;72:1439–1457. doi: 10.1134/s0006297907130032. [DOI] [PubMed] [Google Scholar]
- Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Robbins J, Dilwortht SM, Laskey RA, Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- Dingwall C, Sharnick SV, Laskey RA. A polypeptide domain that specifies migration of nucleoplasmin into the nucleus. Cell. 1982;30:449–458. doi: 10.1016/0092-8674(82)90242-2. [DOI] [PubMed] [Google Scholar]
- Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J. Biol. Chem. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmeri D, Malim MH. Importin beta can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin alpha. Mol. Cell. Biol. 1999;19:1218–1225. doi: 10.1128/mcb.19.2.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Liu X, Lodish HF. Importin beta mediates nuclear translocation of Smad 3. J. Biol. Chem. 2000;275:23425–23428. doi: 10.1074/jbc.C000345200. [DOI] [PubMed] [Google Scholar]
- Jakel S, Gorlich D. Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 1998;17:4491–4502. doi: 10.1093/emboj/17.15.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlhausser P, Muller EC, Otto A, Kutay U. Multiple pathways contribute to nuclear import of core histones. EMBO Rep. 2001;2:690–696. doi: 10.1093/embo-reports/kve168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C, Laskey RA. Nuclear import: a tale of two sites. Curr. Biol. 1998;8:R922–R924. doi: 10.1016/s0960-9822(98)00010-4. [DOI] [PubMed] [Google Scholar]
- Chook YM, Blobel G. Karyopherins and nuclear import. Curr. Opin. Struct. Biol. 2001;11:703–715. doi: 10.1016/s0959-440x(01)00264-0. [DOI] [PubMed] [Google Scholar]
- Lange A, McLane LM, Mills RE, Devine SE, Corbett AH. Expanding the definition of the classical bipartite nuclear localization signal. Traffic (Copenhagen, Denmark) 2010;11:311–323. doi: 10.1111/j.1600-0854.2009.01028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler M, Ansieau S, Prehn S, Leutz A, Haller H, Hartmann E. Cloning of two novel human importin-alpha subunits and analysis of the expression pattern of the importin-alpha protein family. FEBS Lett. 1997;417:104–108. doi: 10.1016/s0014-5793(97)01265-9. [DOI] [PubMed] [Google Scholar]
- Gabriel G, Klingel K, Otte A, Thiele S, Hudjetz B, Arman-Kalcek G, et al. Differential use of importin-[alpha] isoforms governs cell tropism and host adaptation of influenza virus. Nat. Commun. 2011;2:156. doi: 10.1038/ncomms1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Hong M. Protonation, tautomerization, and rotameric structure of histidine: a comprehensive study by magic-angle-spinning solid-state NMR. J. Am. Chem. Soc. 2011;133:1534–1544. doi: 10.1021/ja108943n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rötzschke O, Lau JM, Hofstätter M, Falk K, Strominger JL. A pH-sensitive histidine residue as control element for ligand release from HLA-DR molecules. Proc. Natl Acad. Sci. 2002;99:16946–16950. doi: 10.1073/pnas.212643999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampmann T, Mueller DS, Mark AE, Young PR, Kobe B. The role of histidine residues in low-pH-mediated viral membrane fusion. Structure. 2006;14:1481–1487. doi: 10.1016/j.str.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Nilsson C, Kagedal K, Johansson U, Ollinger K. Analysis of cytosolic and lysosomal pH in apoptotic cells by flow cytometry. Methods Cell Sci. 2003;25:185–194. doi: 10.1007/s11022-004-8228-3. [DOI] [PubMed] [Google Scholar]
- Lin HJ, Herman P, Lakowicz JR. Fluorescence lifetime-resolved pH imaging of living cells. Cytometry. 2003;52:77–89. doi: 10.1002/cyto.a.10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corvini PFX, Gautier H, Rondags E, Vivier H, Goergen JL, Germain P. Intracellular pH determination of pristinamycin-producing Streptomyces pristinaespiralis by image analysis. Microbiology. 2000;146:2671–2678. doi: 10.1099/00221287-146-10-2671. [DOI] [PubMed] [Google Scholar]
- Weinlich M, Theiss C, Lin CT, Kinne RK. BCECF in single cultured cells: inhomogeneous distribution but homogeneous response. J. Exp. Biol. 1998;201:57–62. doi: 10.1242/jeb.201.1.57. [DOI] [PubMed] [Google Scholar]
- Clegg JS, Jackson SA, Liang P, MacRae TH. Nuclear-cytoplasmic translocations of protein p26 during aerobic-anoxic transitions in embryos of Artemia franciscana. Exp. Cell Res. 1995;219:1–7. doi: 10.1006/excr.1995.1197. [DOI] [PubMed] [Google Scholar]
- Mekhail K, Gunaratnam L, Bonicalzi ME, Lee S. HIF. activation by pH-dependent nucleolar sequestration of VHL. Nat. Cell Biol. 2004;6:642–647. doi: 10.1038/ncb1144. [DOI] [PubMed] [Google Scholar]
- Fontes MR, Teh T, Jans D, Brinkworth RI, Kobe B. Structural basis for the specificity of bipartite nuclear localization sequence binding by importin-alpha. J. Biol. Chem. 2003;278:27981–27987. doi: 10.1074/jbc.M303275200. [DOI] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Matsumura N, Takashima H, Miyamoto-Sato E, Tomita M, et al. Six classes of nuclear localization signals specific to different binding grooves of importin alpha. J. Biol. Chem. 2009;284:478–485. doi: 10.1074/jbc.M807017200. [DOI] [PubMed] [Google Scholar]
- Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- Conti E, Kuriyan J. Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin alpha. Structure. 2000;8:329–338. doi: 10.1016/s0969-2126(00)00107-6. [DOI] [PubMed] [Google Scholar]
- Fontes MR, Teh T, Kobe B. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-alpha. J. Mol. Biol. 2000;297:1183–1194. doi: 10.1006/jmbi.2000.3642. [DOI] [PubMed] [Google Scholar]
- Jang YH, Min do S. The hydrophobic amino acids involved in the interdomain association of phospholipase D1 regulate the shuttling of phospholipase D1 from vesicular organelles into the nucleus. Exp. Mol. Med. 2012;44:571–577. doi: 10.3858/emm.2012.44.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.