Abstract
We sought to explore the molecular mechanism of type 2 diabetes (T2D) and identify potential drug targets and candidate agents for T2D treatment. The differentially expressed genes (DEGs) were assessed between human pancreatic islets with T2D and normal islets. The dysfunctional pathways, the potential transcription factor, and microRNA targets were analyzed by bioinformatics methods. Moreover, a group of bioactive small molecules were identified based on the connectivity map database. The pathways of Eicosanoid Synthesis, TGF-beta signaling pathway, Prostaglandin Synthesis and Regulation, and Integrated Pancreatic Cancer Pathway were found to be significantly dysregulated in the progression of T2D. The genes of ZADH2 (zinc binding alcohol dehydrogenase domain containing 2), BTBD3 (BTB (POZ) domain containing 3), Cul3-based ligases, LTBP1 (latent-transforming growth factor beta binding protein 1), PDGFRA (alpha-type platelet-derived growth factor receptor), and FST (follistatin) were determined to be significant nodes regulated by potential transcription factors and microRNAs. Besides, two small molecules (sanguinarine and DL-thiorphan) were identified to be capable of reverse T2D. In the present study, a systematic understanding for the mechanism underlying T2D development was provided with biological informatics methods. The significant nodes and bioactive small molecules may be drug targets and candidate agents for T2D treatment.
1. Introduction
Type 2 diabetes (T2D) is a chronic metabolic disorder, which results from impaired insulin secretion and action in target tissues [1, 2]. Currently, the incidence of T2D is increasing worldwide [3]. And it is reported that there will be 280 million cases suffering from T2D in 2011 [4]. The prevalence trend is considered to be ascribed to genetic variants and environmental factors such as sedentary lifestyle, obesity [3, 5–7]. Despite the foundational evidence of the mechanism underlying T2D is far from being clear, great contributions have been made to address this health concern.
The variants of some critical genes are determined to contribute to T2D development. The TCF7L2 gene of transcription factor 7-like 2 commonly variant in individuals confers the risk of suffering from T2D [8]. Other genes that have expression variation in patients with T2D are indicated to be CAPN10 (calpain 10), KIR6. 2 (potassium inward-rectifier 6.2), PPAR γ (peroxisome proliferator-activated receptor γ), and IRS-1 (insulin receptor substrate-1) [9]. Another important understanding of the mechanism underlying T2D is associated with the dysfunction of β-cell in human pancreatic islets [10, 11]. The decreased β-cell mass and increased β-cell apoptosis resulted in T2D development and progression. The discovery of novel approaches for T2D treatment has concerned the uncharted area underlying mechanism.
In this work, we downloaded the microarray gene expression data of human pancreatic islets with or without T2D from GEO database. A comprehensive perspective was provided to understand the mechanism underlying T2D with the application of computational bioinformatics method. The dysfunction pathways, potential transcription factor targets, and microRNA targets were explored based on DEGs analysis. Besides, the candidate small molecules were identified, which were capable of ameliorating these genetic changes.
2. Data and Methods
2.1. Affymetrix Microarray Data and Differentially Expressed Genes Analysis
The cDNA microarray expression data (GSE38642) was downloaded from Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/), which was deposited by Taneera et al. [4]. The gene expression data were collected from human pancreatic islets including 54 nondiabetic samples and 9 T2D samples. As the progression of T2D is strongly associated with HbA1c expression [4], we only selected the 29 samples without T2D (HbA1c expression < 6.0) in control group and 8 samples with T2D (HbA1c expression > 6.0) in experimental group. We downloaded the raw data and annotation files for further analysis based on the platform of GPL6244 (Affymetrix Human Gene 1.0 ST Array).
Geoquery software is a tool for analysis and comprehension of microarray and genomics data directly from GEO database [12]. Limma statistics is commonly used for assessing differential expression genes [13, 14].
The microarray data was further performed by Geoquery in R statistical programming environment [15]. Then the differentially expressed genes between type 2 diabetic islets and nondiabetic islets were analyzed by limma package and were tested by modified t-test based on Empirical Bayes Methods [16].
2.2. Pathways Enrichment Analysis of Differentially Expressed Genes
WikiPathways is a public wiki for building research communities on biological pathways, which is characterized for pathway curation and pathway ontology annotations [17]. WebGestalt2 is a gene set analysis toolkit for functional enrichment analysis for large scale of genome [18].
We collected all the metabolic and nonmetabolic pathways from WikiPathways database and performed pathway enrichment analysis with the application of Gene Set Analysis Toolkit V2.
2.3. Prediction of Potential Transcription Factors Targets and MicroRNAs for Differential Expression Genes
Molecular Signatures Database (MSigDB) is freely available (http://www.broadinstitute.org/gsea/msigdb/index.jsp) collection of a large scale of well-annotated genomic data [19].
The entire set of transcription factor target gene signatures and microRNA data were obtained from the MSigDB. The gene set enrichment analysis was performed on hypergeometric algorithm. Finally, the potential transcription factors targets and microRNAs were obtained after testing by BH (Barnes-Hut) algorithm.
2.4. The Construction of Regulatory Network
We integrated the data of DEGs, potential transcription factor binding sites, and microRNAs obtained in our work and established the regulatory network. And we also constructed a regulatory motif with the DEGs regulated by multiple transcription factors and microRNAs for further analysis.
2.5. Identification of Candidate Small Molecules
The connectivity map (CMap) deposited genome-wide transcriptional expression data (7056 gene expression profiles) from 6100 small molecules treatment-control experiments [20].
We firstly divided the DEGs identified in our paper into two groups: upregulated DEGs and downregulated ones and selected the significantly differential expression genes (Top 500) in each group. The gene set enrichment analysis (GSEA) was performed between the significantly differential expressed genes and those from treatment-control pairs in CMap database. Then an enrichment score ranging from −1 to 1 was obtained, which represented the level of similarity. When the positive enrichment score was closed to 1, the corresponding bioactive small molecule (perturbagen) was considered to reversal the expression of query signature in the progression of disease, otherwise the perturbagen contributed to the development of disease.
3. Results
3.1. Identification of Differentially Expressed Genes
To assess the differentially expressed genes, we downloaded the GSE38642 gene expression profile from GEO database. After analyzed by limma package and t-test, we defined P < 0.0001 as the cutoff value. Total 225 genes were identified to be significantly differential expressed between T2D islets tissues and normal tissues.
3.2. Identification of Dysfunction Pathways
In order to investigate the DEGs in molecular functional level, we carried out pathway enrichment analysis based on WikiPathways database. Total of 15 pathways were revealed to be significantly dysregulated with P < 0.05 and at least 2 genes enriched.
As shown in Table 1, the enriched pathways terms relevant with cell surface function, signal transduction, hormone regulation, cellular metabolism, and immune response were determined to be dysregulated in the progression of T2D, such as focal adhesion, MAPK signaling pathway, Prostaglandin Synthesis and Regulation, Eicosanoid Synthesis, Mitochondrial LC-Fatty Acid, Beta-Oxidation, Selenium Pathway, Fatty Acid Biosynthesis, Tryptophan metabolism, IL-6 signaling pathway, IL-7 signaling pathway, IL-1 signaling pathway, Inflammatory Response Pathway, and Complement and Coagulation Cascades. Besides, the Integrated Pancreatic Cancer Pathway was also identified to be disturbed in T2D development.
Table 1.
Dysfunction pathways between human T2D islet cells and normal islet cells.
| Pathway | Count | P value |
|---|---|---|
| Eicosanoid synthesis | 3 | 0.0016 |
| MAPK signaling pathway | 7 | 0.0029 |
| IL-6 signaling pathway | 4 | 0.005 |
| Integrated Pancreatic Cancer Pathway | 7 | 0.0055 |
| Mitochondrial LC-Fatty Acid Beta-Oxidation | 2 | 0.0152 |
| Complement and Coagulation Cascades | 3 | 0.0205 |
| Focal adhesion | 6 | 0.021 |
| Selenium Pathway | 4 | 0.0316 |
| IL-7 signaling pathway | 2 | 0.0354 |
| TGF-beta signaling pathway | 3 | 0.0362 |
| IL-1 signaling pathway | 3 | 0.0377 |
| Fatty Acid Biosynthesis | 2 | 0.0436 |
| Tryptophan metabolism | 3 | 0.0441 |
| Inflammatory Response Pathway | 2 | 0.0464 |
| Prostaglandin Synthesis and Regulation | 2 | 0.0494 |
3.3. The Potential Transcription Factor Targets and MicroRNAs
The changes in the patterns of gene expression were affected by transcriptional regulation and posttranscriptional regulation; so we predicted the potential transcription factor targets and microRNA targets to further explore the mechanism underlying T2D progression.
After investigation by hypergeometric and BH algorithm, we defined P < 10−10 and P < 10−6 as threshold values in transcription factor targets analysis and microRNAs targets analysis, respectively.
As shown in Table 2, the enrichment transcription factor targets were explored based on the upstream sequences of DEGs. And the significant microRNAs and targets uncovered in this work were listed in Table 3.
Table 2.
Potential transcription factor targets.
| TF target | P value | TF target | P value |
|---|---|---|---|
| hsa_V$RP58_01 | 9.61E − 05 | hsa_V$SRF_Q6 | 0.0192 |
| hsa_GGARNTKYCCA_UNKNOWN | 0.0002 | hsa_V$PITX2_Q2 | 0.0201 |
| hsa_TTGTTT_V$FOXO4_01 | 0.0008 | hsa_V$IRF_Q6 | 0.0201 |
| hsa_V$CART1_01 | 0.0009 | hsa_V$HNF1_C | 0.0205 |
| hsa_RYTGCNWTGGNR_UNKNOWN | 0.0019 | hsa_V$CEBPA_01 | 0.0205 |
| hsa_TTAYRTAA_V$E4BP4_01 | 0.0021 | hsa_GGGNNTTTCC_V$NFKB_Q6_01 | 0.0209 |
| hsa_V$NFKB_Q6 | 0.0024 | hsa_V$TAL1BETAE47_01 | 0.0209 |
| hsa_RYAAAKNNNNNNTTGW_UNKNOWN | 0.0027 | hsa_V$CMYB_01 | 0.0214 |
| hsa_V$NFKB_C | 0.0027 | hsa_V$HLF_01 | 0.0214 |
| hsa_TATAAA_V$TATA_01 | 0.0041 | hsa_V$CDC5_01 | 0.0218 |
| hsa_V$CEBP_Q2 | 0.0052 | hsa_V$TAL1ALPHAE47_01 | 0.0223 |
| hsa_CTTTAAR_UNKNOWN | 0.0054 | hsa_V$RSRFC4_01 | 0.0223 |
| hsa_V$FOXJ2_02 | 0.0057 | hsa_V$CEBP_Q3 | 0.0232 |
| hsa_V$SMAD3_Q6 | 0.0058 | hsa_V$ICSBP_Q6 | 0.0252 |
| hsa_TGGAAA_V$NFAT_Q4_01 | 0.0063 | hsa_V$ZID_01 | 0.0267 |
| hsa_CAGGTA_V$AREB6_01 | 0.0064 | hsa_CCCNNNNNNAAGWT_UNKNOWN | 0.0269 |
| hsa_YTAAYNGCT_UNKNOWN | 0.0071 | hsa_V$RORA2_01 | 0.0291 |
| hsa_V$FREAC4_01 | 0.0079 | hsa_CAGGTG_V$E12_Q6 | 0.0293 |
| hsa_V$ER_Q6_02 | 0.0084 | hsa_V$GATA6_01 | 0.031 |
| hsa_V$CEBPB_02 | 0.0092 | hsa_V$E4BP4_01 | 0.0315 |
| hsa_AAAYRNCTG_UNKNOWN | 0.0096 | hsa_V$CREB_Q4_01 | 0.0321 |
| hsa_TAATTA_V$CHX10_01 | 0.0101 | hsa_V$IK2_01 | 0.0328 |
| hsa_TTANTCA_UNKNOWN | 0.0106 | hsa_V$CRX_Q4 | 0.0334 |
| hsa_V$HNF4ALPHA_Q6 | 0.0113 | hsa_GGATTA_V$PITX2_Q2 | 0.0347 |
| hsa_V$ER_Q6_01 | 0.0118 | hsa_V$ER_Q6 | 0.0352 |
| hsa_CTGCAGY_UNKNOWN | 0.0129 | hsa_V$RSRFC4_Q2 | 0.0364 |
| hsa_RNGTGGGC_UNKNOWN | 0.0133 | hsa_V$TATA_C | 0.0371 |
| hsa_V$HP1SITEFACTOR_Q6 | 0.0134 | hsa_V$FAC1_01 | 0.0378 |
| hsa_TGANTCA_V$AP1_C | 0.0141 | hsa_YKACATTT_UNKNOWN | 0.0384 |
| hsa_TGTYNNNNNRGCARM_UNKNOWN | 0.0153 | hsa_V$GATA1_05 | 0.0412 |
| hsa_V$HNF1_01 | 0.018 | hsa_WGTTNNNNNAAA_UNKNOWN | 0.0413 |
| hsa_CTTTGA_V$LEF1_Q2 | 0.0183 | hsa_GTGGGTGK_UNKNOWN | 0.0485 |
Table 3.
Potential microRNA targets.
| Target sequence | MicroRNAs | P value |
|---|---|---|
| hsa_TATTATA | MIR-374 | 0.0018 |
| hsa_TGAATGT | MIR-181A, MIR-181B, MIR-181C, MIR-181D | 0.0068 |
| hsa_TTGCACT | MIR-130A, MIR-301, MIR-130B | 0.0069 |
| hsa_GGGACCA | MIR-133A, MIR-133B | 0.0139 |
| hsa_ATGTCAC | MIR-489 | 0.016 |
| hsa_TGCTGCT | MIR-15A, MIR-16, MIR-15B, MIR-195, MIR-424, MIR-497 | 0.0174 |
| hsa_GTTTGTT | MIR-495 | 0.0209 |
| hsa_TACTTGA | MIR-26A, MIR-26B | 0.0245 |
| hsa_GACAATC | MIR-219 | 0.0262 |
| hsa_GTGTTGA | MIR-505 | 0.0269 |
| hsa_TCATCTC | MIR-143 | 0.0269 |
| hsa_ATACTGT | MIR-144 | 0.0283 |
| hsa_GTACTGT | MIR-101 | 0.0283 |
| hsa_CCCAGAG | MIR-326 | 0.0322 |
| hsa_CTACCTC | LET-7A, LET-7B, LET-7C, LET-7D, LET-7E, LET-7F, MIR-98, LET-7G, LET-7I | 0.0346 |
| hsa_ATAAGCT | MIR-21 | 0.0391 |
| hsa_CACCAGC | MIR-138 | 0.0393 |
| hsa_TTTGTAG | MIR-520D | 0.0394 |
| hsa_CACTGTG | MIR-128A, MIR-128B | 0.0419 |
| hsa_ACTGTGA | MIR-27A, MIR-27B | 0.0426 |
| hsa_AAGTCCA | MIR-422B, MIR-422A | 0.0458 |
3.4. The Regulatory Network Construction
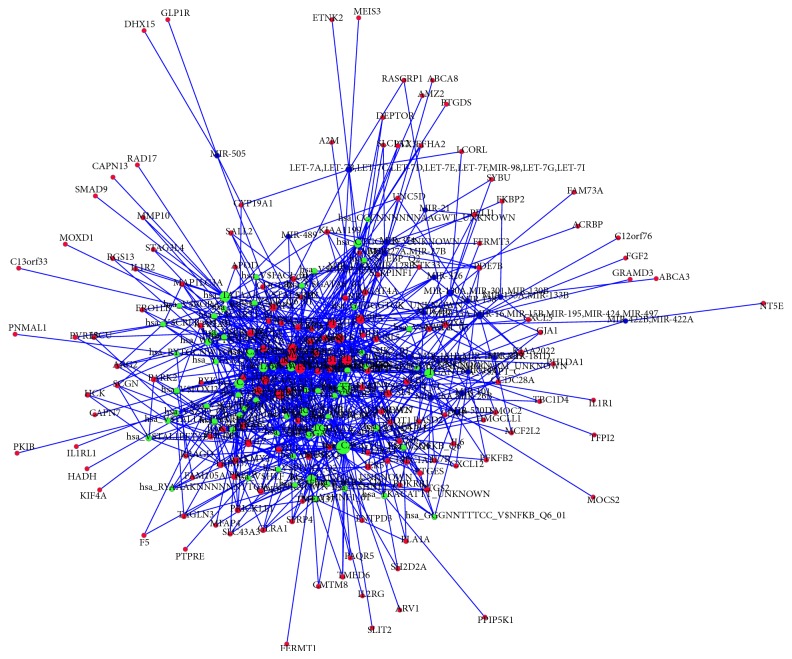
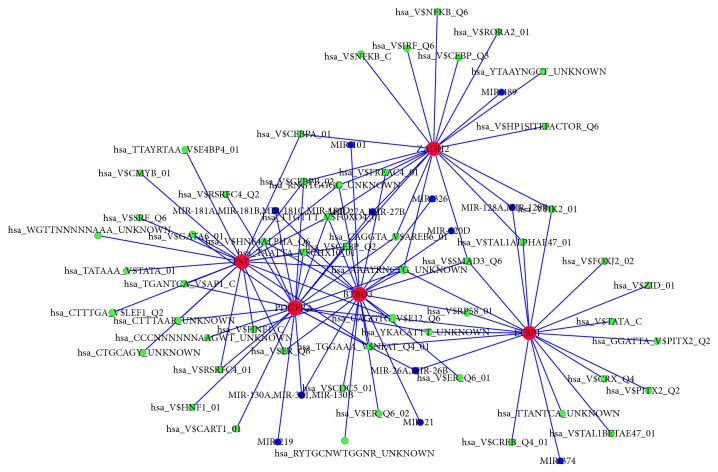
To investigate the associations between DEGs and microRNAs, transcription factors, we constructed the regulatory network. As shown in Figure 1, different DEGs were regulated by different microRNAs and transcription factors. The DEGs involved with multiple regulators might play key roles in the progression of T2D; therefore we selected the DEGs corresponding to multiple microRNAs and transcription factors (n ≥ 20) to establish the regulatory motif. Figure 2 showed that 5 genes played critical roles in the T2D development, including ZADH2, BTBD3, LTBP1, PDGFRA, and FST.
Figure 1.
The regulatory network of DEGs by transcription factors and microRNAs. Red notes: DEGs; green nodes: transcription factor targets; blue nodes: microRNA targets; edges: interactions between two nodes. The bigger nodes indicate to have more interactions with others.
Figure 2.
Regulatory motif of DEGs by transcription factors and microRNAs. Red notes: DEGs; green nodes: transcription factor targets; blue nodes: microRNA targets. The bigger nodes indicate to have more interactions with others.
3.5. Identification of Candidate Small Molecules
We performed computational bioinformatics analysis to identify the candidate drugs for T2D treatment. After comparing the query signatures induced by DEGs with data from CMap database, a large amount of small molecules was identified, which had positive or negative correlation to query signature. The top 20 small molecules closely relevant with T2D were listed in Table 4. The small molecules with higher positive enrichment scores were determined to be sanguinarine (enrichment score = 0.977) and DL-thiorphan (enrichment score = 0.956). In addition, small molecule of felbinac showed highly significant negative score (enrichment = −0.847).
Table 4.
Top 20 significant small molecules.
| CMap name | Enrichment | P |
|---|---|---|
| 8-Azaguanine | 0.932 | 0.00004 |
| Apigenin | 0.86 | 0.00052 |
| Chrysin | 0.931 | 0.00056 |
| Sulfametoxydiazine | 0.855 | 0.00056 |
| Lycorine | 0.803 | 0.00068 |
| Digoxin | 0.846 | 0.0008 |
| Prochlorperazine | 0.467 | 0.00086 |
| Sanguinarine | 0.977 | 0.00087 |
| Helveticoside | 0.733 | 0.00097 |
| Felbinac | −0.847 | 0.00101 |
| Adiphenine | −0.771 | 0.00118 |
| Diloxanide | −0.83 | 0.00157 |
| Etiocholanolone | −0.704 | 0.00157 |
| Heptaminol | −0.753 | 0.0017 |
| Acetylsalicylic acid | 0.499 | 0.00174 |
| Proscillaridin | 0.903 | 0.00182 |
| Cinchonine | −0.82 | 0.00197 |
| 0316684-0000 | −0.813 | 0.00229 |
| Proadifen | 0.806 | 0.00265 |
| DL-Thiorphan | 0.956 | 0.00342 |
4. Discussion
Nowadays, T2D is highlighted by its increasing epidemicity all over the word [3]. Although numerous studies have been conducted concerning the therapies for T2D, the effective approaches for T2D treatment are relatively rare. The current work provided the foundational evidences for T2D development with systematic informatics analysis. In this paper, we downloaded the microarray gene expression data (GSE38642) from GEO database and identified the DEGs between diabetic and nondiabetic human islets. Results showed that, using the cutoff value of P < 0.0001, total 225 genes were differentially expressed. By pathway enrichment analysis of the DEGs, 15 pathways were revealed to be significantly dysregulated such as Eicosanoid Synthesis, Prostaglandin Synthesis and Regulation, and Integrated Pancreatic Cancer Pathway.
Eicosanoid is a critical signaling molecules biological process and played diverse and complex roles in biological and pathological control [21]. Eicosanoids consist of multiple subfamilies including prostaglandins, thromboxanes, leukotrienes, and derivatives of arachidonate [22]. Many diseases such as cardiovascular disease [23], inflammatory bowel disease [24], and diarrhoeal diseases [25] were mediated by the secretion of eicosanoids. As outlined in previous study, eicosanoids played key roles in modulating platelet function of T2D patients. Thromboxane, served as a member of eicosanoid family, can induce platelet aggregation to vascular endothelium resulting in platelet dysfunction [26]. Platelet aggregation suppressed the normal interaction of intact healthy vascular endothelium with platelets, which might result in macrovascular and microvascular events T2D patients.
Prostaglandin is also a member of eicosanoids, deriving from unsaturated fatty acids [27]. The renal production of prostaglandins has been reported to be associated with nephropathy in T2D [28]. The expression of prostaglandins and their corresponding receptors induced in islets is revealed to be contributors of T2D development [29]. The expression of prostaglandin E2 (PGE2) was elevated, which was positively related with the activation of prostaglandin E receptor 3 (EP3). The activation of PGE2-to-EP3 signaling pathway resulted in the decline of the cAMP activation and insulin secretion induced by glucose. The accumulation of EP3 and PGE2 production contributed to T2D development and β-cell dysfunction. Thus, the pathways related with Eicosanoid Synthesis and Prostaglandin Synthesis and Regulation played crucial roles in T2D development and progression. Besides, Integrated Pancreatic Cancer Pathway was also indicated to be a significant pathway in T2D development. Although there were few evidences concerning the association between T2D and integrated pancreatic cancer, it implied that T2D might be a precipitating factor for patients suffering from integrated pancreatic cancer.
Our results also showed that the genes of LTBP1, PDGFRA, and FST were the most significant targets for potential transcription factors and microRNAs. Among these significant targets, LTBP1 encoded for latent-transforming growth factor beta binding protein 1 which is a member of carrier proteins [30]. LTBP1 has various interactions with extracellular matrix proteins and TGF-beta (TGF-β) [31]. TGF-β signaling pathway showed tightly association with diabetes development. It is reported that the level of glucose has a direct effect on TGF-β activation [32]. An elevated expression of TGF-β was observed in serum of patients with T2D and antidiabetic treatment was able to reverse this trend [33]. Another report suggested that the suppression of TGF-β-TGF-β receptor interaction is available for preventing diabetes progression by inhibiting the differentiation of islet-reactive CD8+ T cells in type 1 diabetes [34]. By pathway enrichment analysis, our results also showed that TGF-β signaling pathway was significant in the T2D progression.
In addition, PDGFRA encoded alpha-type platelet-derived growth factor receptor is one of the latent TGF-beta binding proteins [35]. The production of PDGFR is considered to be interacted with PI3K p85α and PI3Kp85αpY580 is activated by insulin receptor tyrosine kinase [36–38]. FST is the gene for follistatin which also served as activin-binding protein. Follistatin generally exists in blood and is considered to be involved in the inflammatory response stimulated by tissue injury or pathogenic incursion. Despite the clarification of mechanism underlying T2D concerning PDGFRA and FST was far from being clear, the significant nodes in regulatory networks may be potential drug targets for T2D treatment.
Besides, another important implication in our work was the identification of a group of small molecules. Data in Table 4 showed that the small molecules of sanguinarine (enrichment = 0.977) and DL-thiorphan (enrichment = 0.956) showed highly significant positive scores, suggesting that these small molecules are candidate agents targeting for T2D.
Sanguinarine is a benzophenanthridine alkaloid, which has been ascribed to a novel bioactive component extracted from plants [39]. And it has showed various properties including antimicrobial, antioxidant, and anti-inflammatory [40]. Previous researches proved that sanguinarine possessed potent anticancer activity against many different tumors, such as gastric osteosarcoma adenocarcinoma [41], osteosarcoma [42], prostate tumor [43], and oral cancers [44]. Sanguinarine prevented the development of cancers by inducing cancer cell apoptosis, suppressing tumor growth, migration, and invasion [45, 46]. A present study revealed that sanguinarine is involved in cell migration and angiogenesis suppression in cancer development by inhibiting the activity of vascular endothelial growth factor (VEGF) [39]. In spite of the increasing studies highlighting the anticancer property of sanguinarine, reviews also indicated the sanguinarine antidiabetic activity [47]. Sanguinarine derived from Fumaria parviflora plants has a hypoglycemic effect. In addition, sanguinarine has been used as an important drug against infections in one or more countries worldwide [48]. Moreover, DL-thiorphan is served as the specific neutral endopeptidase (NEP) inhibitor, which is widely used to differentiate NEP enzyme activity. NEP enzyme is a membrane-bound metallopeptidase that plays key roles in wound repair [49]. Fatty acids and glucose stimulated the expression of NEP. The activity of NEP was increased in the skin of objects with diabetic wound [50]. However, there are insufficient evidences indicating DL-thiorphan can be directly used in glucose control for patients with T2D. Therefore, sanguinarine and DL-thiorphan may be candidate agents for diabetes treatment in the near future.
In summary, the present study provides a systematic understanding for the mechanism underlying T2D development. The significant nodes such as LTBP1, PDGFRA, and FST assessed in regulatory network may be drug targets for T2D treatment. And sanguinarine and DL-thiorphan may be candidate agents targeting for T2D. However, more studies are required to confirm these discoveries in our work.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Defronzo R. A. From the triumvirate to the ominous octet: a new paradigm for the treatment of type 2 diabetes mellitus. Diabetes. 2009;58(4):773–795. doi: 10.2337/db09-9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeFronzo R. A. Pharmacologic therapy for type 2 diabetes mellitus. Annals of Internal Medicine. 1999;131(4):281–303. doi: 10.7326/0003-4819-131-4-199908170-00008. [DOI] [PubMed] [Google Scholar]
- 3.Tuomilehto J., Lindström J., Eriksson J. G., Valle T. T., Hamäläinen H., Ianne-Parikka P., Keinänen-Kiukaanniemi S., Laakso M., Louheranta A., Rastas M., Salminen V., Uusitupa M. Prevention of type 2 diabetes mellitus by changes in lifestyle among subjects with impaired glucose tolerance. The New England Journal of Medicine. 2001;344(18):1343–1350. doi: 10.1056/NEJM200105033441801. [DOI] [PubMed] [Google Scholar]
- 4.Taneera J., Lang S., Sharma A., Fadista J., Zhou Y., Ahlqvist E., Jonsson A., Lyssenko V., Vikman P., Hansson O., Parikh H., Korsgren O., Soni A., Krus U., Zhang E., Jing X.-J., Esguerra J. L. S., Wollheim C. B., Salehi A., Rosengren A., Renström E., Groop L. A systems genetics approach identifies genes and pathways for type 2 diabetes in human islets. Cell Metabolism. 2012;16(1):122–134. doi: 10.1016/j.cmet.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Tuomilehto J., Wolf E. Primary prevention of diabetes mellitus. Diabetes Care. 1987;10(2):238–248. doi: 10.2337/diacare.10.2.238. [DOI] [PubMed] [Google Scholar]
- 6.Hamman R. F. Genetic and environmental determinants of non-insulin-dependent diabetes mellitus (NIDDM) Diabetes/Metabolism Reviews. 1992;8(4):287–338. doi: 10.1002/dmr.5610080402. [DOI] [PubMed] [Google Scholar]
- 7.Manson J. E., Rimm E. B., Stampfer M. J., Colditz G. A., Willett W. C., Krolewski A. S., Rosner B., Hennekens C. H., Speizer F. E. Physical activity and incidence of non-insulin-dependent diabetes mellitus in women. The Lancet. 1991;338(8770):774–778. doi: 10.1016/0140-6736(91)90664-B. [DOI] [PubMed] [Google Scholar]
- 8.Grant S. F. A., Thorleifsson G., Reynisdottir I., Benediktsson R., Manolescu A., Sainz J., Helgason A., Stefansson H., Emilsson V., Helgadottir A., Styrkarsdottir U., Magnusson K. P., Walters G. B., Palsdottir E., Jonsdottir T., Gudmundsdottir T., Gylfason A., Saemundsdottir J., Wilensky R. L., Reilly M. P., Rader D. J., Bagger Y., Christiansen C., Gudnason V., Sigurdsson G., Thorsteinsdottir U., Gulcher J. R., Kong A., Stefansson K. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nature Genetics. 2006;38(3):320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 9.Stumvoll M., Goldstein B. J., van Haeften T. W. Type 2 diabetes: principles of pathogenesis and therapy. The Lancet. 2005;365(9467):1333–1346. doi: 10.1016/S0140-6736(05)61032-X. [DOI] [PubMed] [Google Scholar]
- 10.Butler A. E., Janson J., Bonner-Weir S., Ritzel R., Rizza R. A., Butler P. C. -cell deficit and increased -cell apoptosis in humans with type 2 diabetes. Diabetes. 2003;52(1):102–110. doi: 10.2337/diabetes.52.1.102. [DOI] [PubMed] [Google Scholar]
- 11.Meier J. J., Bonadonna R. C. Role of reduced β-cell mass versus impaired β-cell function in the pathogenesis of type 2 diabetes. Diabetes Care. 2013;36(2):S113–S119. doi: 10.2337/dcS13-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sean D., Meltzer P. S. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23(14):1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 13.Diboun I., Wernisch L., Orengo C. A., Koltzenburg M. Microarray analysis after RNA amplification can detect pronounced differences in gene expression using limma. BMC Genomics. 2006;7, article 252 doi: 10.1186/1471-2164-7-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smyth G. K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology. 2004;3(article 3) doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 15.Team R. D. C. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2005. http://www.R-project.org. [Google Scholar]
- 16.Yekutieli D., Benjamini Y. Resampling-based false discovery rate controlling multiple test procedures for correlated test statistics. Journal of Statistical Planning and Inference. 1999;82(1-2):171–196. doi: 10.1016/S0378-3758(99)00041-5. [DOI] [Google Scholar]
- 17.Kelder T., van Iersel M. P., Hanspers K., Kutmon M., Conklin B. R., Evelo C. T., Pico A. R. WikiPathways: building research communities on biological pathways. Nucleic Acids Research. 2012;40(1):D1301–D1307. doi: 10.1093/nar/gkr1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duncan D., Prodduturi N., Zhang B. WebGestalt2: an updated and expanded version of the Web-based Gene Set Analysis Toolkit. BMC Bioinformatics. 2010;11(supplement 4):P10. doi: 10.1186/1471-2105-11-S4-P10. [DOI] [Google Scholar]
- 19.Liberzon A., Subramanian A., Pinchback R., Thorvaldsdóttir H., Tamayo P., Mesirov J. P. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–1740. doi: 10.1093/bioinformatics/btr260.btr260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamb J., Crawford E. D., Peck D., Modell J. W., Blat I. C., Wrobel M. J., Lerner J., Brunet J.-P., Subramanian A., Ross K. N., Reich M., Hieronymus H., Wei G., Armstrong S. A., Haggarty S. J., Clemons P. A., Wei R., Carr S. A., Lander E. S., Golub T. R. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 21.Harizi H., Corcuff J.-B., Gualde N. Arachidonic-acid-derived eicosanoids: roles in biology and immunopathology. Trends in Molecular Medicine. 2008;14(10):461–469. doi: 10.1016/j.molmed.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Majerus P. W. Arachidonate metabolism in vascular disorders. The Journal of Clinical Investigation. 1983;72(5):1521–1525. doi: 10.1172/JCI111110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patrignani P. Aspirin insensitive eicosanoid biosynthesis in cardiovascular disease. Thrombosis Research. 2003;110(5-6):281–286. doi: 10.1016/S0049-3848(03)00382-7. [DOI] [PubMed] [Google Scholar]
- 24.Stenson W. F. Role of eicosanoids as mediators of inflammation in inflammatory bowel disease. Scandinavian Journal of Gastroenterology, Supplement. 1990;25(172):13–18. doi: 10.3109/00365529009091903. [DOI] [PubMed] [Google Scholar]
- 25.Rask-Madsen J. Eicosanoids and their role in the pathogenesis of diarrhoeal diseases. Clinics in Gastroenterology. 1986;15(3):545–566. [PubMed] [Google Scholar]
- 26.Vinik A. I., Erbas T., Sun Park T., Nolan R., Pittenger G. L. Platelet dysfunction in type 2 diabetes. Diabetes Care. 2001;24(8):1476–1485. doi: 10.2337/diacare.24.8.1476. [DOI] [PubMed] [Google Scholar]
- 27.Wolfe L. S. Eicosanoids: prostaglandins, thromboxanes, leukotrienes, and other derivatives of carbon-20 unsaturated fatty acids. Journal of Neurochemistry. 1982;38(1):1–14. doi: 10.1111/j.1471-4159.1982.tb10847.x. [DOI] [PubMed] [Google Scholar]
- 28.Okumura M., Imanishi M., Yamashita T., Yamamura Y., Kim S., Iwao H., Tanaka S., Fujii S. Renal production of thromboxane and prostaglandins in a rat model of type 2 diabetes. Life Sciences. 1999;66(5):371–377. doi: 10.1016/S0024-3205(99)00603-7. [DOI] [PubMed] [Google Scholar]
- 29.Kimple M. E., Keller M. P., Rabaglia M. R., Pasker R. L., Neuman J. C., Truchan N. A., Brar H. K., Attie A. D. Prostaglandin E2 receptor, EP3, is induced in diabetic islets and negatively regulates glucose- and hormone-stimulated insulin secretion. Diabetes. 2013;62(6):1904–1912. doi: 10.2337/db12-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Unsöld C., Hyytiäinen M., Bruckner-Tuderman L., Keski-Oja J. Latent TGF-β binding protein LTBP-1 contains three potential extracellular matrix interacting domains. Journal of Cell Science. 2001;114(1):187–197. doi: 10.1242/jcs.114.1.187. [DOI] [PubMed] [Google Scholar]
- 31.Jenkins G. The role of proteases in transforming growth factor-β activation. International Journal of Biochemistry and Cell Biology. 2008;40(6-7):1068–1078. doi: 10.1016/j.biocel.2007.11.026. [DOI] [PubMed] [Google Scholar]
- 32.Köppel H., Riedl E., Braunagel M., Sauerhoefer S., Ehnert S., Godoy P., Sternik P., Dooley S., Yard B. A. L-carnosine inhibits high-glucose-mediated matrix accumulation in human mesangial cells by interfering with TGF-β production and signalling. Nephrology Dialysis Transplantation. 2011;26(12):3852–3858. doi: 10.1093/ndt/gfr324. [DOI] [PubMed] [Google Scholar]
- 33.Pscherer S., Freude T., Forst T., Nussler A. K., Braun K. F., Ehnert S. Anti-diabetic treatment regulates pro-fibrotic TGF-β serum levels in type 2 diabetics. Diabetology and Metabolic Syndrome. 2013;5(1, article 48) doi: 10.1186/1758-5996-5-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Green E. A., Gorelik L., McGregor C. M., Tran E. H., Flavell R. A. CD4+CD25+ T regulatory cells control anti-islet CD8+ T cells through TGF-β-TGF-β receptor interactions in type 1 diabetes. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(19):10878–10883. doi: 10.1073/pnas.1834400100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schuit F., van Lommel L., Granvik M., Goyvaerts L., de Faudeur G., Schraenen A., Lemaire K. β-cell-specific gene repression: a mechanism to protect against inappropriate or maladjusted insulin secretion? Diabetes. 2012;61(5):969–975. doi: 10.2337/db11-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu P., Margolis B., Skolnik E. Y., Lammers R., Ullrich A., Schlessinger J. Interaction of phosphatidylinositol 3-kinase-associated p85 with epidermal growth factor and platelet-derived growth factor receptors. Molecular and Cellular Biology. 1992;12(3):981–990. doi: 10.1128/mcb.12.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hayashi H., Nishioka Y., Kamohara S., Kanai F., Ishii K., Fukui Y., Shibasaki F., Takenawa T., Kido H., Katsunuma N., Ebina Y. The α-type 85-kDa subunit of phosphatidylinositol 3-kinase is phosphorylated at tyrosines 368, 580, and 607 by the insulin receptor. Journal of Biological Chemistry. 1993;268(10):7107–7117. [PubMed] [Google Scholar]
- 38.Zhang Y., Wolf-Yadlin A., Ross P. L., Pappin D. J., Rush J., Lauffenburger D. A., White F. M. Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Molecular and Cellular Proteomics. 2005;4(9):1240–1250. doi: 10.1074/mcp.M500089-MCP200. [DOI] [PubMed] [Google Scholar]
- 39.Xu J.-Y., Meng Q.-H., Chong Y., et al. Sanguinarine is a novel VEGF inhibitor involved in the suppression of angiogenesis and cell migration. Molecular and Clinical Oncology. 2013;1(2):331–336. doi: 10.3892/mco.2012.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mackraj I., Govender T., Gathiram P. Sanguinarine. Cardiovascular Therapeutics. 2008;26(1):75–83. doi: 10.1111/j.1527-3466.2007.00037.x. [DOI] [PubMed] [Google Scholar]
- 41.Choi W. Y., Jin C.-Y., Han M. H., Kim G.-Y., Kim N. D., Lee W. H., Kim S.-K., Choi Y. H. Sanguinarine sensitizes human gastric adenocarcinoma AGS cells to TRAIL-mediated apoptosis via down-regulation of AKT and activation of caspase-3. Anticancer Research. 2009;29(11):4457–4465. [PubMed] [Google Scholar]
- 42.Park H., Bergeron E., Senta H., Guillemette K., Beauvais S., Blouin R., Sirois J., Faucheux N. Sanguinarine induces apoptosis of human osteosarcoma cells through the extrinsic and intrinsic pathways. Biochemical and Biophysical Research Communications. 2010;399(3):446–451. doi: 10.1016/j.bbrc.2010.07.114. [DOI] [PubMed] [Google Scholar]
- 43.Sun M., Lou W., Chun J. Y., Cho D. S., Nadiminty N., Evans C. P., Chen J., Yue J., Zhou Q., Gao A. C. Sanguinarine suppresses prostate tumor growth and inhibits survivin expression. Genes and Cancer. 2010;1(3):283–292. doi: 10.1177/1947601910368849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsukamoto H., Kondo S., Mukudai Y., Nagumo T., Yasuda A., Kurihara Y., Kamatani T., Shintani S. Evaluation of anticancer activities of benzo[c]phenanthridine alkaloid sanguinarine in oral squamous cell carcinoma cell line. Anticancer Research. 2011;31(9):2841–2846. [PubMed] [Google Scholar]
- 45.Jang B.-C., Park J.-G., Song D.-K., Baek W.-K., Yoo S. K., Jung K.-H., Park G.-Y., Lee T.-Y., Suh S.-I. Sanguinarine induces apoptosis in A549 human lung cancer cells primarily via cellular glutathione depletion. Toxicology in Vitro. 2009;23(2):281–287. doi: 10.1016/j.tiv.2008.12.013. [DOI] [PubMed] [Google Scholar]
- 46.Sun M., Liu C., Nadiminty N., Lou W., Zhu Y., Yang J., Evans C. P., Zhou Q., Gao A. C. Inhibition of Stat3 activation by sanguinarine suppresses prostate cancer cell growth and invasion. The Prostate. 2012;72(1):82–89. doi: 10.1002/pros.21409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chauhan A., Sharma P., Srivastava P., Kumar N., Dudhe R. Plants having potential antidiabetic activity: a review. Der Pharmacia Lettre. 2010;2(3):369–387. [Google Scholar]
- 48.Jerjomiceva N. Exploring Natural and Pharmocological Products for the Ability to Boost Neutrophils Against Bacterial Infections. Hannover, Germany: Tierärztliche Hochsch; 2013. [Google Scholar]
- 49.Muangman P., Tamura R. N., Gibran N. S. Antioxidants inhibit fatty acid and glucose-mediated induction of neutral endopeptidase gene expression in human microvascular endothelial cells. Journal of the American College of Surgeons. 2005;200(2):208–215. doi: 10.1016/j.jamcollsurg.2004.09.043. [DOI] [PubMed] [Google Scholar]
- 50.Antezana M. A., Sullivan S. R., Usui M. L., Gibran N. S., Spenny M. L., Larsen J. A., Ansel J. C., Bunnett N. W., Olerud J. E. Neutral endopeptidase activity is increased in the skin of subjects with diabetic ulcers. Journal of Investigative Dermatology. 2002;119(6):1400–1404. doi: 10.1046/j.1523-1747.2002.19618.x. [DOI] [PubMed] [Google Scholar]