Abstract
The classical class I human leukocyte antigens (HLA-A, -B, and -C) present allele-specific self- or pathogenic peptides originated by intracellular processing to CD8+ immune effector cells. Even a single mismatch in the heavy chain (hc) of an HLA class I molecule can impact on the peptide binding profile. Since HLA class I molecules are highly polymorphic and most of their polymorphisms affect the peptide binding region (PBR), it becomes obvious that systematic HLA matching is crucial in determining the outcome of transplantation. The opposite holds true for the nonclassical HLA class I molecule HLA-E. HLA-E polymorphism is restricted to two functional versions and is thought to present a limited set of highly conserved peptides derived from class I leader sequences. However, HLA-E appears to be a ligand for the innate and adaptive immune system, where the immunological response to peptide-HLA-E complexes is dictated through the sequence of the bound peptide. Structural investigations clearly demonstrate how subtle amino acid differences impact the strength and response of the cognate CD94/NKG2 or T cell receptor.
1. Introduction
The few polymorphic HLA-E alleles are restricted to two functional variants HLA-E*01 : 01 and HLA-E*01 : 03. Officially, there are 13 HLA-E alleles recognized by the International Immunogenetics Database to date; however, only HLA-E*01 : 01 and *01 : 03 contribute to HLA-E function [1]. These two alleles are distributed almost equally among diverse populations. The maintenance of these two alleles is most likely based on a balancing selection, meaning that there is a heterozygote advantage for individuals that are heterozygous at the HLA-E gene locus [2]. This is in contrast to the classical HLA molecules that possess high frequencies of polymorphisms with crucial functional differences, maintained by overdominant selection [3].
The polymorphisms can be maintained by selection favouring the heterozygote genotype. In classical HLA class I molecules these polymorphisms impact on antigen presentation, such as alteration of peptide binding motifs [5–7] that result in modification of the whole peptide/HLA landscape [8]. HLA-E*01 : 01 and HLA-E*01 : 03 differ exclusively in one amino acid (AA) substitution at position 107, located on a loop between β-strands in the α2 domain of the heavy chain (hc), where an arginine for HLA-E*01 : 01 is substituted by a glycine for HLA-E*01 : 03. A substitution at this position is unlikely influencing peptide presentation, because it is not located in the eight stranded β-pleated sheet, or in the peptide binding groove's α-helix segments [9]; however, structural impact on the proximate AAs located within the peptide binding cleft cannot be excluded [10]. Nevertheless, the conservation of the two functional HLA-E alleles is equally among populations, concluding a likely functional difference [11, 12]. Evidence for this hypothesis is the difference in surface expression of HLA-E*01 : 01 and HLA-E*01 : 03. The surface expression of HLA-E*01 : 01 is reported to be significantly lower compared to HLA-E*01 : 03 on human lymphoblastic B cell lines (B-LCL) [10]. Consistent with the differences in surface expression thermal stability studies revealed that HLA-E*01 : 03 molecules have a higher thermal stability compared to HLA-E*01 : 01 molecules that were refolded in the presence of the same HLA class I signal peptide [10].
The binding of a peptide to HLA-E provides a stable trimeric complex that is presented on the cell surface. The main source of HLA-E peptide ligands is signal peptides derived from classical HLA molecules [13, 14]. The relative overall surface expression of both alleles is influenced by the combination of the present HLA-A, -B, and -C allotypes in individuals, since the cell surface expression and stability of HLA-E are based on the available HLA class I derived signal peptides [15].
As the HLA-E peptidome was first thought to be restricted to peptides derived from this source, in vitro studies with random peptide libraries have shown that HLA-E is capable of binding a range of different peptides and is not only restricted to peptides derived from classical HLA molecules [16], although a preference for hydrophobic residues at most positions of the peptide is evidenced. The range of HLA-E peptide selection includes the identified peptide ligand QMRPVSRVL derived from the HSP60 protein that upregulates HLA-E surface expression due to cellular stress response [17]. Additionally, peptide ligands with distinct differences in their AA sequence have been shown to bind to HLA-E. A peptide derived from the ATP-binding cassette transporter, multidrug resistance-associated protein 7 (MRP7) ALALVRMLI was identified to bind HLA-E during heat shock [18]; the peptide AISPRTLNA derived from the HIV Gag protein has been shown to upregulate HLA-E surface expression on HIV infected lymphocytes [19]. A peptide SQQPYLQLQ derived from gliadin, that is, the known antigen for priming the celiac disease pathogenesis, stabilizes HLA-E levels in celiac patients [20]. The HCV core35–44 peptide YLLPRRGPRL stabilized the HLA-E complex and conferred protection against NK cell mediated lysis through specific interaction with the CD94/NKG2A receptor [21]. Recent studies investigated the HLA-E derived peptide repertoire and confirmed striking differences in their anchor position and features [22]. In any case, the broadened peptide ligand varieties (Table 1) and functional potential of HLA-E gained more attention.
Table 1.
HLA-E peptide ligands.
| Peptide ligands (origin) |
HLA-E∗01:01 | HLA-E∗01:03 | TCR | CD94/NKG2A | Reference |
|---|---|---|---|---|---|
| Naturally presented ligands | |||||
| ALALVRMLI (ATP binding cassette transporter, MRP7) |
ND | ND | ND | + | [18] |
| VMAPRTLFL (HLA-G) |
+ | + | − | + | [33] |
| VMAPRTLIL (CMV UL40; HLA-C) |
+ | + | + | + | [26, 35, 38] |
| VMAPRTLVL (CMV; HLA-A) |
+ | + | + | + | [36, 57] |
|
| |||||
| Predicted ligands | |||||
| QMRPVSRVL (human HSP60) |
+ | + | ND | − | [17] |
| AISPRTLNA (HIV Gag protein) |
ND | ND | ND | + | [19] |
| SQQPYLQLQ (gliadin-wheat protein) | ND | ND | ND | + | [20] |
| SQAPLPCVL (EBV-BZLF1 protein) |
+ | + | + | + | [33] |
This table shows the diversity of naturally presented or predicted HLA-E peptide ligands and their interactions with immune receptors. Since CD94/NKG2C receptor interactions with HLA-E bound to noncanoncial peptide ligands are poorly reported in the literature, data is not included. +: positive for binding; −: no binding; ND: not determined.
2. Regulatory Interactions of pHLA-E Complexes
HLA-E is a mediator of NK cell activation and inhibition [23, 24]. Peptide-HLA-E (pHLA-E) complexes are ligands for either the inhibitory CD94/NKG2A or the stimulatory CD94/NKG2C NK cell receptor. The importance of the peptide sequence for the interaction with these receptors needs to be emphasized in this respect. Structural studies on the interaction between the CD94/NKG2 heterodimer and HLA-E are based on a pHLA-E complex bound to a peptide VMAPRTLFL derived from the HLA-G leader sequence (PDB: 3CDG). The affinity of HLA-EVMAPRTLFL to the CD94/NKG2A receptor is approximately 6 times higher than to the CD94/NKG2C receptor, as a result of differential interactions between distinct residues of either CD94 or NKG2 with particular residues on the HLA-E α1 helix and residues within the peptide [4, 25]. The distinct difference in affinity could also be verified with another nonameric peptide VMAPRTLIL derived from the UL40 protein of the human cytomegalovirus (HCMV), mimicking the signal peptide sequence present in most HLA-C allotypes [26]. Alterations of this peptide sequence have a strong impact on the recognition by CD94/NKG2A or NKG2C receptor and consequently have a high impact on NK cell cytotoxicity [27]. An example of peptide mediated cytotoxicity could be observed with the QMRPVSRVL peptide derived from HSP60, which showed total loss of recognition by the CD94/NKG2A inhibitory receptor [17], leading to cytotoxic NK cell responses due to the triggering by stimulatory receptors KIR2DS1 or NKG2D [28, 29]. These subtle alterations in the sequences of peptides bound to HLA-E impact extensively on the fine tuning of NK cell receptor binding and responses.
The structural analysis of HLA-EVMAPRTLFL in complex with the CD94/NKG2A receptor illustrates the impact of the peptide sequence, where an AA difference might result in the loss of the CD94/NKG2A receptor recognition [4] (Figure 1). The main contribution to direct interactions with the VMAPRTLFL peptide residues is mediated by the CD94 subunit through a hydrogen bond between CD94-Ser110 to the guanidinium group of the peptide's p5-Arg and a hydrogen bond of CD94-Gln112 to the main chain of the peptide's p6-Thr. The CD94-Gln112 additionally contacts p5-Arg and p8-Phe by van der Waals interactions. The p8-Phe is surrounded and contacted by the three polar CD94 residues Asn156, Asn158, and Asn160 and also interacts with Phe114. The NKG2A subunit of the CD94/NKG2A receptor complex exclusively contacts the peptide's p5-Arg with residue NKG2A-Pro171 through van der Waals interactions.
Figure 1.
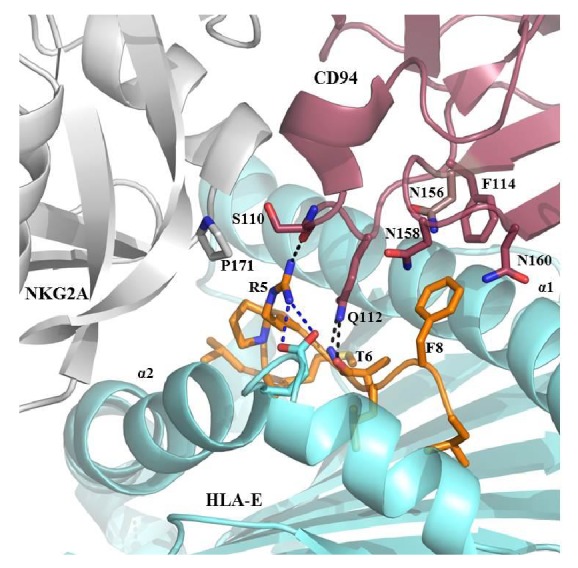
C-terminal peptide residues mediate contact between the HLA-EVMAPRTLFL complex and the CD94/NKG2A receptor. This structure represents the interactions between the CD94/NKG2A receptor and the HLA-EVMAPRTLFL complex (PDB: 3CDG) [4]. The CD94 subunit (raspberry) dominates the recognition of the peptide (orange sticks) with several contacts including hydrogen bonds of Ser110 to the peptide's p5-Arg and Gln112 to p6-Thr. The peptide's p8-Phe is surrounded by a polar pocket created by Asn156, Asn158, and Asn160 and van der Waals contacts with Phe114. p5-Arg also builds a salt bridge to Glu152 of the HLA-E heavy chain (pale teal) that may prevent more charged interactions with CD94/NKG2A. Residue Pro171 of NKG2A (pale grey) interacts with p5-Arg by van der Waals interactions. Hydrogen bonds are represented as black dashed lines, and salt bridges are given in blue.
The impact of the peptide sequence on CD94/NKG2A receptor recognition was also shown with the peptide VMAPRALLL derived from HLA-Cw*07 : 02 signal peptide that resulted in highly reduced recognition compared to the p6-Thr variant by the CD94/NKG2A receptor and could therefore not protect from NK cell lysis [30]. Although the peptide's p6-Thr (VMAPRTLFL) is substituted by p6-Ala (VMAPRALLL), this substitution did not change the conformation of the HLA-E heavy chain or the orientation of exposed side chains at p5 or p8 [31]. The subtle fine tuning of immune responses is explained as a result of peptide sequence alterations of HLA-E bound ligands.
HLA-E also plays a role in adaptive immunity, since certain pHLA-E complexes are recognized by subsets of CD8+ T cells [32]. Evidence for a function of HLA-E in T cell immunity could be observed with several pathogen derived ligands; in this case, certain pHLA-E complexes could not be recognized anymore by CD56+/CD94+/NKG2A+ NK cells. Here a nonameric peptide SQAPLPCVL derived from the Epstein-Barr virus BZLF1 protein bound to HLA-E could be recognized by the α β-T cell receptor (TCR) from a CD94+/NKG2A+/CD8+ T cell clone [33]. Furthermore, it has been demonstrated that CD4−/CD56−/CD8+ T cells, isolated from individuals that were immunized with a Salmonella enterica serovar Typhi vaccine, are specifically activated by B-LCLs that were expressing recombinant HLA-E and loaded with S. Typhi derived peptides [34]. Since it is known that HLA-E binds to a peptide VMAPRTLIL derived from the HCMV UL40 protein and serves as a ligand for the CD94/NKG2A and CD94/NKG2C NK cell receptor, this HLA-EVMAPRTLIL complex is also recognized by the TCR of CD45RA+/CD28−/CD27−CD56+ effector memory like T cells and leads to T cell mediated cytotoxicity [35].
The peptide-mediated fine tuning of immune responses within the innate immune system could be detected among subsets of the CD8+ T cell repertoire as well. The HCMV UL40 protein contains mutations among different HCMV strains, resulting in single AA exchanges within the peptide; consequently, a different subset of CD8+ T cells, specific for the complex HLA-EVMAPRTLVL [36], where the peptide's p8-Ile is exchanged for a p8-Val, could be identified.
The peptide specific recognition by its cognate TCR could be analyzed in the crystal structure of HLA-EVMAPRTLIL in complex with its cognate TCR, derived from a UL40 specific T cell clone. The affinity of this TCR to the HLA-EVMAPRTLIL complex is relatively lower compared to TCR interactions with classical pHLA complexes [37] that resulted in a lower on-rate of the interaction between the TCR and the HLA-E molecule. However, the half-life of this TCR/HLA-EVMAPRTLIL complex is comparable to classical TCR/HLA interactions [38]. Conformational changes of a pHLA-E molecule in complex with the cognate TCR showed comparable values to classical pHLA/TCR complexes. The mode of interaction between a TCR and a distinct pHLA-E complex could be explained by the importance of residue 8 within the VMAPRTLIL peptide. The contribution of p8-Ile to the binding of the TCR was shown to be crucial for a stable TCR/HLA-EVMAPRTLIL complex. Alterations at position 8 of the peptide to either Val or Leu resulted in a significant reduction of affinity to the TCR [39].
The importance of regulatory functions of HLA-E molecules and its peptide ligands for the balancing regulation of innate and adaptive immune response is highlighted and should be considered in situations where normal HLA class I expression is reduced or abrogated. Taken together, the source of peptide determines the role of the given pHLA-E complex in innate or adaptive immunity.
3. HLA-E and Malignancies
Since HLA-E is an effective inhibitory molecule whose main role is to prevent NK cell activation, it is a useful protective molecule for malignant cells in order to prevent their fate from NK cell killing. The role of HLA-E in controlling NK cell activity in the context of viral interference could be shown recently by demonstrating how miR-376a(e) regulates HLA-E expression during HCMV infection [40]. The downregulation of HLA class I molecules is a widespread escape mechanism of tumor cells to prevent the recognition by CD8+ T cells [41, 42]. The overexpression of HLA-E on tumor cells has recently been reported in colorectal cancer and was pointed out as a biomarker for tumor cell differentiation [43]. This overexpression of HLA-E is proposed to be associated with the inhibition of tumor tissue infiltrating NK or CD94+/NKG2A+/CD8+ T cells, resulting as a poor prognosis marker. Studies on colorectal cancer cell lines showed accordingly an overexpression of HLA-E that was correlated with the malignancy stage and furthermore the release of soluble HLA-E molecules from these cell lines [44]. A protective role of HLA-E in tumor cells was also underlined in patients with ovarian and cervical cancer, where tumor infiltrating CD8+ cytotoxic T cells (CTLs) showed an upregulation of the CD94/NKG2A inhibitory receptor, whereas NK cells were only found at very low numbers in the tumor tissues. Regarding HLA-E expression, the benefit of tumor infiltrating CTLs was abrogated presumably due to the inhibition of CD94+/NKG2A+ CTLs by HLA-E [45, 46]. Furthermore, HLA-E expression in early breast cancer patients was also proposed as a prognostic marker for the outcome on tumor progression [47]. Despite the downregulation of HLA class I molecules during the tumor immune escape, the surface expression of HLA-E is not affected. A similar immune escape mechanism has been reported for human fibroblasts infected with HCMV, where HLA-E is presented on the cell surface associated with downregulation of HLA class I [48]. Viral proteins specifically target and downregulate several components of the peptide loading complex, such as transporter associated with antigen processing (TAP) or tapasin (TPN) [49, 50]. Recently we found a TPN-independent peptide loading mechanism for HLA-E variants [51]. This observation explains the role of HLA-E and its differential surface expression levels in malignancies. Considering the HLA-E genotype and the differences in surface expression of HLA-E*01 : 01 and HLA-E*01 : 03, the lower surface expression of HLA-E*01 : 01 could be a benefit for antitumor immune responses resulting in less inhibitory ligands for CD94+/NKG2A+ immune cells.
4. Influence of HLA-E on HSCT Outcome
Even though the highly important properties of HLA-E regarding its antigen presentation and recognition by effector cells are clear, its clinical relevance for transplantation outcome is still controversy. It has been demonstrated that HLA-E polymorphisms in 10/10 HLA matched unrelated donor and recipients had no significant impact on the outcome of a hematopoietic stem cell transplantation (HSCT) [52]. In contrast to this finding an association between HLA-E*01 : 03 homozygosity and a significant improvement for HSCT outcome was published [53]. Both studies analyzed cohorts of intermediate size (N = 116; 83) but also with differences in patient treatment specifications, subsidiary diseases, and followup. Another clinical study provided data, where homozygous HLA-E*01 : 03 grafts revealed a significant lower risk for acute and chronic graft versus host disease (aGvHD; cGvHD) associated with a postulated impaired efficiency of minor histocompatibility antigen presentation by HLA-E*01 : 03, but also a significant increased risk for transplant related mortality (TRM) [54].
A general conclusion for the role of HLA-E in HSCT is not feasible, yet, since only a limited amount of clinical data considering cohort size and differences in patient treatment as well is available, this needs to be completed by comprehensive further studies. However, the fact that HLA-E is also a target for a subset of CD8+ T cells contributes to its posttransplant role. The recognition by HLA-E restricted CD8+ T cells emerges to the spotlight that was shown in HCMV-seropositive transplant patients that restore a subset of HLA-E-reactive CD8+ T cells [55]. These CD8+ T cells developed cytolytic activities when cultured with different HLA-E haplotyped endothelial cell cultures independently of the HLA-E allele. This activation was observed not only for HCMV seropositive but also for seronegative endothelial cells. This indeed suggests the direct recognition of allogeneic HLA-E on graft endothelial cells in solid organ transplantation. However, the activation could be inhibited due to the presence of specific HLA-C haplotypes that are ligands for the classical HLA class I NK receptor KIR2DL2 [56]. This receptor was highly expressed on the HLA-E restricted CD8+ T cell subsets. This data suggests that HCMV-associated HLA-E-restricted T cells could contribute to allograft rejection in the case of HLA-C haplotype/NK receptor mismatch on graft tissue.
5. Conclusion
The ongoing studies on HLA-E antigen presentation and functional effects revealed that HLA-E is a bridge between innate and adaptive immunity. Early research proposed a very restricted peptide repertoire capable of binding to HLA-E. Since HLA-E is not exclusively restricted to canonical peptides of HLA class I origin, its immunogenic features are probably more important for transplantation outcome due to its broader spectrum of presented peptides. For a certain estimation of the influence of HLA-E on transplantation outcome, the focus of clinical studies on HLA-E restricted HCMV-associated CD8+ T cell subsets needs to be extended and compared in greater cohorts.
The recognition of HLA-E molecules by the TCR of certain CTLs could be a promising option in anticancer therapies, since HLA-E surface expression has been shown on several types of tumors. However, the generation of CTLs with a specific TCR raised against antigenic peptides that are not alloreactive against nontumor HLA-E expressing cells is a crucial part that needs to be carefully considered. For this, the identification and functional assessment of tumor specific peptides presented by HLA-E and the presumable differences in the peptide repertoire of the two functional HLA-E alleles are of high importance.
Abbreviations
- HSCT:
Hematopoietic stem cell transplantation
- GvHD:
Graft versus host disease
- pHLA-E:
Peptide-HLA-E complexes
- AA:
Amino acid
- CTL:
Cytotoxic T lymphocytes.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Felício L. P., Porto I. O. P., Mendes-Junior C. T., Veiga-Castelli L. C., Santos K. E., Vianello-Brondani R. P., Sabbagh A., Moreau P., Donadi E. A., Castelli E. C. Worldwide HLA-E nucleotide and haplotype variability reveals a conserved gene for coding and 3′ untranslated regions. Tissue Antigens. 2014;83(2):82–93. doi: 10.1111/tan.12283. [DOI] [PubMed] [Google Scholar]
- 2.Grimsley C., Ober C. Population genetic studies of HLA-E: evidence for selection. Human Immunology. 1997;52(1):33–40. doi: 10.1016/S0198-8859(96)00241-8. [DOI] [PubMed] [Google Scholar]
- 3.Hughes A. L., Nei M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature. 1988;335(6186):167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- 4.Petrie E. J., Clements C. S., Lin J., Sullivan L. C., Johnson D., Huyton T., Heroux A., Hoare H. L., Beddoe T., Reid H. H., Wilce M. C. J., Brooks A. G., Rossjohn J. CD94-NKG2A recognition of human leukocyte antigen (HLA)-E bound to an HLA class I leader sequence. Journal of Experimental Medicine. 2008;205(3):725–735. doi: 10.1084/jem.20072525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bade-Doeding C., DeLuca D. S., Seltsam A., Blasczyk R., Eiz-Vesper B. Amino acid 95 causes strong alteration of peptide position P in HLA-B*41 variants. Immunogenetics. 2007;59(4):253–259. doi: 10.1007/s00251-007-0197-7. [DOI] [PubMed] [Google Scholar]
- 6.Huyton T., Ladas N., Schumacher H., Blasczyk R., Bade-Doeding C. Pocketcheck: updating the HLA class I peptide specificity roadmap. Tissue Antigens. 2012;80(3):239–248. doi: 10.1111/j.1399-0039.2012.01928.x. [DOI] [PubMed] [Google Scholar]
- 7.Huyton T., Schumacher H., Blasczyk R., Bade-Doeding C. Residue 81 confers a restricted C-terminal peptide binding motif in HLA-B*44:09. Immunogenetics. 2012;64(9):663–668. doi: 10.1007/s00251-012-0625-1. [DOI] [PubMed] [Google Scholar]
- 8.Bade-Döding C., Theodossis A., Gras S., Kjer-Nielsen L., Eiz-Vesper B., Seltsam A., Huyton T., Rossjohn J., McCluskey J., Blasczyk R. The impact of human leukocyte antigen (HLA) micropolymorphism on ligand specificity within the HLA-B∗41 allotypic family. Haematologica. 2011;96(1):110–118. doi: 10.3324/haematol.2010.030924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bjorkman P. J., Saper M. A., Samraoui B., Bennett W. S., Strominger J. L., Wiley D. C. The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature. 1987;329(6139):512–518. doi: 10.1038/329512a0. [DOI] [PubMed] [Google Scholar]
- 10.Strong R. K., Holmes M. A., Li P., Braun L., Lee N., Geraghty D. E. HLA-E allelic variants: correlating differential expression, peptide affinities, crystal structures, and thermal stabilities. Journal of Biological Chemistry. 2003;278(7):5082–5090. doi: 10.1074/jbc.M208268200. [DOI] [PubMed] [Google Scholar]
- 11.Zeng X., Chen H., Gupta R., Paz-Altschul O., Bowcock A. M., Liao W. Deletion of the activating NKG2C receptor and a functional polymorphism in its ligand HLA-E in psoriasis susceptibility. Experimental Dermatology. 2013;22(10):679–681. doi: 10.1111/exd.12233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J., Pan L., Chen L., Feng X., Zhou L., Zheng S. Non-classical MHC-Ι genes in chronic hepatitis B and hepatocellular carcinoma. Immunogenetics. 2012;64(3):251–258. doi: 10.1007/s00251-011-0580-2. [DOI] [PubMed] [Google Scholar]
- 13.Braud V. M., Allan D. S. J., Wilson D., McMichael A. J. TAP- and tapasin-dependent HLA-E surface expression correlates with the binding of an MHC class I leader peptide. Current Biology. 1998;8(1):1–10. doi: 10.1016/s0960-9822(98)70014-4. [DOI] [PubMed] [Google Scholar]
- 14.Braud V., Jones E. Y., McMichael A. The human major histocompatibility complex class Ib molecule HLA-E binds signal sequence-derived peptides with primary anchor residues at positions 2 and 9. European Journal of Immunology. 1997;27(5):1164–1169. doi: 10.1002/eji.1830270517. [DOI] [PubMed] [Google Scholar]
- 15.Merino A. M., Sabbaj S., Easlick J., Goepfert P., Kaslow R. A., Tang J. Dimorphic HLA-B signal peptides differentially influence HLA-E- and natural killer cell-mediated cytolysis of HIV-1-infected target cells. Clinical and Experimental Immunology. 2013;174(3):414–423. doi: 10.1111/cei.12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stevens J., Joly E., Trowsdale J., Butcher G. W. Peptide binding characteristics of the non-classical class Ib MHC molecule HLA-E assessed by a recombinant random peptide approach. BMC Immunology. 2001;2(1, article 5) doi: 10.1186/1471-2172-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michaëlsson J., Teixeira de Matos C., Achour A., Lanier L. L., Kärre K., Söderström K. A signal peptide derived from hsp60 binds HLA-E and interferes with CD94/NKG2A recognition. Journal of Experimental Medicine. 2002;196(11):1403–1414. doi: 10.1084/jem.20020797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wooden S. L., Kalb S. R., Cotter R. J., Soloski M. J. Cutting edge: HLA-E binds a peptide derived from the ATP-binding cassette transporter multidrug resistance-associated protein 7 and inhibits NK cell-mediated lysis. Journal of Immunology. 2005;175(3):1383–1387. doi: 10.4049/jimmunol.175.3.1383. [DOI] [PubMed] [Google Scholar]
- 19.Nattermann J., Nischalke H. D., Hofmeister V., Kupfer B., Ahlenstiel G., Feldmann G., Rockstroh J., Weiss E. H., Sauerbruch T., Spengler U. HIV-1 infection leads to increased HLA-E expression resulting in impaired function of natural killer cells. Antiviral Therapy. 2005;10(1):95–107. doi: 10.1177/135965350501000107. [DOI] [PubMed] [Google Scholar]
- 20.Terrazzano G., Sica M., Gianfrani C., Mazzarella G., Maurano F., De Giulio B., De Saint-Mezard S., Zanzi D., Maiuri L., Londei M., Jabri B., Troncone R., Auricchio S., Zappacosta S., Carbone E. Gliadin regulates the NK-dendritic cell cross-talk by HLA-E surface stabilization. Journal of Immunology. 2007;179(1):372–381. doi: 10.4049/jimmunol.179.1.372. [DOI] [PubMed] [Google Scholar]
- 21.Nattermann J., Nischalke H. D., Hofmeister V., et al. The HLA-A2 restricted T cell epitope HCV core35−44 stabilizes HLA-E expression and inhibits cytolysis mediated by natural killer cells. The American Journal of Pathology. 2005;166(2):443–453. doi: 10.1016/S0002-9440(10)62267-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lampen M. H., Hassan C., Sluijter M., Geluk A., Dijkman K., Tjon J. M., de Ru A. H., van der Burg S. H., van Veelen P. A., van Hall T. Alternative peptide repertoire of HLA-E reveals a binding motif that is strikingly similar to HLA-A2. Molecular Immunology. 2013;53(1-2):126–131. doi: 10.1016/j.molimm.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 23.Posch P. E., Borrego F., Brooks A. G., Coligan J. E. HLA-E is the ligand for the natural killer cell CD94/NKG2 receptors. Journal of Biomedical Science. 1998;5(5):321–331. doi: 10.1007/BF02253442. [DOI] [PubMed] [Google Scholar]
- 24.Braud V. M., Allan D. S. J., O'Callaghan C. A., Soderstrom K., D'Andrea A., Ogg G. S., Lazetic S., Young N. T., Bell J. I., Phillips J. H., Lanler L. L., McMichael A. J. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391(6669):795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 25.Kaiser B. K., Pizarro J. C., Kerns J., Strong R. K. Structural basis for NKG2A/CD94 recognition of HLA-E. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(18):6696–6701. doi: 10.1073/pnas.0802736105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomasec P. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science. 2000;287(5455):1031–1033. doi: 10.1126/science.287.5455.1031. [DOI] [PubMed] [Google Scholar]
- 27.Heatley S. L., Pietra G., Lin J., Widjaja J. M. L., Harpur C. M., Lester S., Rossjohn J., Szer J., Schwarer A., Bradstock K., Bardy P. G., Mingari M. C., Moretta L., Sullivan L. C., Brooks A. G. Polymorphism in human cytomegalovirus UL40 impacts on recognition of human leukocyte antigen-E (HLA-E) by natural killer cells. Journal of Biological Chemistry. 2013;288(12):8679–8690. doi: 10.1074/jbc.M112.409672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pegram H. J., Andrews D. M., Smyth M. J., Darcy P. K., Kershaw M. H. Activating and inhibitory receptors of natural killer cells. Immunology and Cell Biology. 2011;89(2):216–224. doi: 10.1038/icb.2010.78. [DOI] [PubMed] [Google Scholar]
- 29.Raulet D. H., Vance R. E. Self-tolerance of natural killer cells. Nature Reviews Immunology. 2006;6(7):520–531. doi: 10.1038/nri1863. [DOI] [PubMed] [Google Scholar]
- 30.Valés-Gómez M., Reyburn H. T., Erskine R. A., López-Botet M., Strominger J. L. Kinetics and peptide dependency of the binding of the inhibitory NK receptor CD94/NIKG2-A and the activating receptor CD94/NKG2-C to HLA-E. The EMBO Journal. 1999;18(15):4250–4260. doi: 10.1093/emboj/18.15.4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoare H. L., Sullivan L. C., Clements C. S., Ely L. K., Beddoe T., Henderson K. N., Lin J., Reid H. H., Brooks A. G., Rossjohn J. Subtle changes in peptide conformation profoundly affect recognition of the non-classical MHC class I molecule HLA-E by the CD94-NKG2 natural killer cell receptors. Journal of Molecular Biology. 2008;377(5):1297–1303. doi: 10.1016/j.jmb.2008.01.098. [DOI] [PubMed] [Google Scholar]
- 32.Pietra G., Romagnani C., Falco M., et al. The analysis of the natural killer-like activity of human cytolytic T lymphocytes revealed HLA-E as a novel target for TCR alpha/beta-mediated recognition. European Journal of Immunology. 2001;31(12):3687–3693. doi: 10.1002/1521-4141(200112)31:12<3687::AID-IMMU3687>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 33.Garcia P., Llano M., de Heredia A. B., Willberg C. B., Caparrós E., Aparicio P., Braud V. M., López-Botet M. Human T cell receptor-mediated recognition of HLA-E. European Journal of Immunology. 2002;32(4):936–944. doi: 10.1002/1521-4141(200204)32:4<936::AID-IMMU936>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 34.Salerno-Gonçalves R., Fernandez-Viña M., Lewinsohn D. M., Sztein M. B. Identification of a human HLA-E-restricted CD8+ T cell subset in volunteers immunized with Salmonella enterica serovar Typhi strain Ty21a typhoid vaccine. Journal of Immunology. 2004;173(9):5852–5862. doi: 10.4049/jimmunol.173.9.5852. [DOI] [PubMed] [Google Scholar]
- 35.Mazzarino P., Pietra G., Vacca P., Falco M., Colau D., Coulie P., Moretta L., Mingari M. C. Identification of effector-memory CMV-specific T lymphocytes that kill CMV-infected target cells in an HLA-E-restricted fashion. European Journal of Immunology. 2005;35(11):3240–3247. doi: 10.1002/eji.200535343. [DOI] [PubMed] [Google Scholar]
- 36.Pietra G., Romagnani C., Mazzarino P., Falco M., Millo E., Moretta A., Moretta L., Mingari M. C. HLA-E-restricted recognition of cytomegalovirus-derived peptides by human CD8+ cytolytic T lymphocytes. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(19):10896–10901. doi: 10.1073/pnas.1834449100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis M. M., Boniface J. J., Reich Z. Ligand recognition by alpha beta T cell receptors. Annual Review of Immunology. 1998;16:523–544. doi: 10.1146/annurev.immunol.16.1.523. [DOI] [PubMed] [Google Scholar]
- 38.Hoare H. L., Sullivan L. C., Pietra G., Clements C. S., Lee E. J., Ely L. K., Beddoe T., Falco M., Kjer-Nielsen L., Reid H. H., McCluskey J., Moretta L., Rossjohn J., Brooks A. G. Structural basis for a major histocompatibility complex class Ib-restricted T cell response. Nature Immunology. 2006;7(3):256–264. doi: 10.1038/ni1312. [DOI] [PubMed] [Google Scholar]
- 39.Sullivan L. C., Hoare H. L., McCluskey J., Rossjohn J., Brooks A. G. A structural perspective on MHC class Ib molecules in adaptive immunity. Trends in Immunology. 2006;27(9):413–420. doi: 10.1016/j.it.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 40.Nachmani D., Zimmermann A., Djian E. O., et al. MicroRNA editing facilitates immune elimination of HCMV infected cells. PLoS Pathogens. 2014;10(2) doi: 10.1371/journal.ppat.1003963.e1003963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garrido F., Algarra I. MHC antigens and tumor escape from immune surveillance. Advances in Cancer Research. 2001;83:117–158. doi: 10.1016/S0065-230X(01)83005-0. [DOI] [PubMed] [Google Scholar]
- 42.Ruiz-Cabello F., Cabrera T., Lopez-Nevot M.-A., Garrido F. Impaired surface antigen presentation in tumors: implications for T cell-based immunotherapy. Seminars in Cancer Biology. 2002;12(1):15–24. doi: 10.1006/scbi.2001.0406. [DOI] [PubMed] [Google Scholar]
- 43.Bossard C., Bézieau S., Matysiak-Budnik T., Volteau C., Laboisse C. L., Jotereau F., Mosnier J.-F. HLA-E/β2 microglobulin overexpression in colorectal cancer is associated with recruitment of inhibitory immune cells and tumor progression. International Journal of Cancer. 2012;131(4):855–863. doi: 10.1002/ijc.26453. [DOI] [PubMed] [Google Scholar]
- 44.Levy E. M., Bianchini M., Von Euw E. M., Barrio M. M., Bravo A. I., Furman D., Domenichini E., Macagno C., Pinsky V., Zucchini C., Valvassori L., Mordoh J. Human leukocyte antigen-E protein is overexpressed in primary human colorectal cancer. International Journal of Oncology. 2008;32(3):633–641. [PubMed] [Google Scholar]
- 45.Gooden M., Lampen M., Jordanova E. S., et al. HLA-E expression by gynecological cancers restrains tumor-infiltrating CD8+ T lymphocytes. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(26):10656–10661. doi: 10.1073/pnas.1100354108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sheu B.-C., Chiou S.-H., Lin H.-H., Chow S.-N., Huang S.-C., Ho H.-N., Hsu S.-M. Up-regulation of inhibitory natural killer receptors CD94/NKG2A with suppressed intracellular perforin expression of tumor-infiltrating CD8+ T lymphocytes in human cervical carcinoma. Cancer Research. 2005;65(7):2921–2929. doi: 10.1158/0008-5472.CAN-04-2108. [DOI] [PubMed] [Google Scholar]
- 47.de Kruijf E. M., Sajet A., van Nes J. G. H., Natanov R., Putter H., Smit V. T. H. B. M., Liefers G. J., van den Elsen P. J., van de Velde C. J. H., Kuppen P. J. K. HLA-E and HLA-G expression in classical HLA class I-negative tumors is of prognostic value for clinical outcome of early breast cancer patients. Journal of Immunology. 2010;185(12):7452–7459. doi: 10.4049/jimmunol.1002629. [DOI] [PubMed] [Google Scholar]
- 48.Wang E. C. Y., McSharry B., Retiere C., Tomasec P., Williams S., Borysiewicz L. K., Braud V. M., Wilkinson G. W. G. UL40-mediated NK evasion during productive infection with human cytomegalovirus. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(11):7570–7575. doi: 10.1073/pnas.112680099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mocarski E. S., Jr. Immunomodulation by cytomegaloviruses: manipulative strategies beyond evasion. Trends in Microbiology. 2002;10(7):332–339. doi: 10.1016/S0966-842X(02)02393-4. [DOI] [PubMed] [Google Scholar]
- 50.Halenius A., Hauka S., Dölken L., Stindt J., Reinhard H., Wiek C., Hanenberg H., Koszinowski U. H., Momburg F., Hengel H. Human cytomegalovirus disrupts the major histocompatibility complex class I peptide-loading complex and inhibits tapasin gene transcription. Journal of Virology. 2011;85(7):3473–3485. doi: 10.1128/JVI.01923-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kraemer T., Schumacher H., Huyton T., Badrinath S., Moneer S., Blasczyk R., Bade-Doeding C. Loading complex and presentation of non-canonical peptides by HLA-E impacts NKG2-mediated cellular function. Tissue Antigens. 2013;81(5):264–265. [Google Scholar]
- 52.Fürst D., Bindja J., Arnold R., Herr W., Schwerdtfeger R., Müller C. H., Recker K., Schrezenmeier H., Mytilineos J. HLA-E polymorphisms in hematopoietic stem cell transplantation. Tissue Antigens. 2012;79(4):287–290. doi: 10.1111/j.1399-0039.2011.01832.x. [DOI] [PubMed] [Google Scholar]
- 53.Danzer M., Polin H., Pröll J., Haunschmid R., Hofer K., Stabentheiner S., Hackl C., Kasparu H., König J., Hauser H., Binder M., Weiss R., Gabriel C., Krieger O. Clinical significance of HLA-E*0103 homozygosity on survival after allogeneic hematopoietic stem-cell transplantation. Transplantation. 2009;88(4):528–532. doi: 10.1097/TP.0b013e3181b0e79e. [DOI] [PubMed] [Google Scholar]
- 54.Ludajic K., Rosenmayr A., Faé I., Fischer G. F., Balavarca Y., Bickeböller H., Kalhs P., Greinix H. T. Association of HLA-E polymorphism with the outcome of hematopoietic stem-cell transplantation with unrelated donors. Transplantation. 2009;88(10):1227–1228. doi: 10.1097/TP.0b013e3181bbb8fe. [DOI] [PubMed] [Google Scholar]
- 55.Allard M., Tonnerre P., Nedellec S., Oger R., Morice A., Guilloux Y., Houssaint E., Charreau B., Gervois N. HLA-E-restricted cross-recognition of allogeneic endothelial cells by CMV-associated CD8 T cells: a potential risk factor following transplantation. PLoS ONE. 2012;7(11) doi: 10.1371/journal.pone.0050951.e50951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.David G., Djaoud Z., Willem C., Legrand N., Rettman P., Gagne K., Cesbron A., Retière C. Large spectrum of HLA-C recognition by killer Ig-like receptor (KIR)2DL2 and KIR2DL3 and restricted C1 specificity of KIR2DS2: dominant impact of KIR2DL2/KIR2DS2 on KIR2D NK cell repertoire formation. Journal of Immunology. 2013;191(9):4778–4788. doi: 10.4049/jimmunol.1301580. [DOI] [PubMed] [Google Scholar]
- 57.Sensi M., Pietra G., Molla A., Nicolini G., Vegetti C., Bersani I., Millo E., Weiss E., Moretta L., Mingari M. C., Anichini A. Peptides with dual binding specificity for HLA-A2 and HLA-E are encoded by alternatively spliced isoforms of the antioxidant enzyme peroxiredoxin 5. International Immunology. 2009;21(3):257–268. doi: 10.1093/intimm/dxn141. [DOI] [PubMed] [Google Scholar]


