ABSTRACT
In the context of deciphering the metabolic strategies of the obligate pathogenic fungi in the genus Pneumocystis, the genomes of three species (P. carinii, P. murina, and P. jirovecii) were compared among themselves and with the free-living, phylogenetically related fission yeast (Schizosaccharomyces pombe). The underrepresentation of amino acid metabolism pathways compared to those in S. pombe, as well as the incomplete steroid biosynthesis pathway, were confirmed for P. carinii and P. jirovecii and extended to P. murina. All three Pneumocystis species showed overrepresentation of the inositol phosphate metabolism pathway compared to that in the fission yeast. In addition to those known in S. pombe, four genes, encoding inositol-polyphosphate multikinase (EC 2.7.1.151), inositol-pentakisphosphate 2-kinase (EC 2.7.1.158), phosphoinositide 5-phosphatase (EC 3.1.3.36), and inositol-1,4-bisphosphate 1-phosphatase (EC 3.1.3.57), were identified in the two rodent Pneumocystis genomes, P. carinii and P. murina. The P. jirovecii genome appeared to contain three of these genes but lacked phosphoinositide 5-phosphatase. Notably, two genes encoding enzymes essential for myo-inositol synthesis, inositol-1-phosphate synthase (INO1) and inositol monophosphatase (INM1), were absent from all three genomes, suggesting that Pneumocystis species are inositol auxotrophs. In keeping with the need to acquire exogenous inositol, two genes with products homologous to fungal inositol transporters, ITR1 and ITR2, were identified in P. carinii and P. murina, while P. jirovecii contained only the ITR1 homolog. The ITR and inositol metabolism genes in P. murina and P. carinii were expressed during fulminant infection as determined by reverse transcriptase real-time PCR of cDNA from infected lung tissue. Supplementation of in vitro culture with inositol yielded significant improvement of the viability of P. carinii for days 7 through 14.
IMPORTANCE
Microbes in the genus Pneumocystis are obligate pathogenic fungi that reside in mammalian lungs and cause Pneumocystis pneumonia in hosts with weakened immune systems. These fungal infections are not responsive to standard antifungal therapy. A long-term in vitro culture system is not available for these fungi, impeding the study of their biology and genetics and new drug development. Given that all genomes of the Pneumocystis species analyzed lack the genes for inositol synthesis and contain inositol transporters, Pneumocystis fungi, like S. pombe, appear to be inositol auxotrophs. Inositol is important for the pathogenesis, virulence, and mating processes in Candida albicans and Cryptococcus neoformans, suggesting similar importance within the Pneumocystis species as well. This is the first report to (i) characterize genes in the inositol phosphate metabolism and transport pathways in Pneumocystis species and (ii) identify inositol as a supplement that improved the viability of P. carinii in in vitro culture.
INTRODUCTION
The fungal pathogen Pneumocystis jirovecii causes a potentially lethal pneumonia, Pneumocystis pneumonia (PCP), in humans. PCP is an opportunistic infection and primarily occurs in patients with a compromised immune system, including those infected with the human immunodeficiency virus (HIV/AIDS), with chronic obstructive pulmonary disease (COPD) (1), and in other immunosuppressed populations, e.g., those receiving anti-tumor necrosis factor alpha (TNF) therapy for autoimmune diseases or immunosuppressive therapies after organ transplantation (2). Currently, the therapeutic options available for PCP are limited (3). These fungi are not susceptible to the commonly used antifungal agents, such as the azoles or amphotericin B, and despite concerted efforts to identify additional chemotherapeutic agents, the combination of trimethoprim and sulfamethoxazole (TMP-SMX) continues to be the standard prophylactic and therapeutic modality in use today, as it has been for the last few decades (4). However, TMP-SMX treatment can cause serious side effects, while other, second-line therapies are not as efficacious or are more toxic, like pentamidine isethionate (5).
The development of new drug therapies for treatment of PCP is lagging because an in vitro cultivation system for any species of Pneumocystis that permits continuous passage does not exist. In vitro evaluation of new drugs for activity against P. jirovecii must be performed using surrogate Pneumocystis species obtained from laboratory immunosuppressed rodents, such as Pneumocystis carinii, which infects rats, and Pneumocystis murina, which infects mice, as there are no animal models for P. jirovecii and thus no culture stocks. The current in vitro systems are limited to short-term maintenance of the fungi in primary cultures, which does not allow determination of cidal or static effects. In addition, the use of Pneumocystis species from rodents leaves questions about how these drugs will perform in patients infected by P. jirovecii.
The Pneumocystis genus is comprised of obligate fungal pathogens that live only in the lungs of their mammalian hosts. The lack of a long-term in vitro culture indicates that some crucial nutrients are missing or undersupplemented in the culture medium. Previous metabolic studies indicated that the pathogen does not synthesize ergosterol and has to scavenge cholesterol from its host to thrive (6). More recent genome-sequencing studies pointed out that the fungus has dramatic underrepresentation of amino acid synthesis pathways (7). Despite the years of trials to supplement Pneumocystis culture with various sterols, amino acids, and other nutrients, the goal of obtaining a long-term in vitro culture of the fungus has remained beyond reach (8).
In this work, we conducted a comparative genome analysis of the three currently sequenced species of the Pneumocystis genus, P. jirovecii, P. carinii, and P. murina, and a free-living but phylogenetically related fungus, the fission yeast Schizosaccharomyces pombe (9). The analysis was focused on enzymes and metabolic pathways to better understand the metabolic strategies adopted by the obligate pathogen in the mammalian host milieu and, therefore, to identify nutrients that it may require from exogenous sources. The analysis identified an overrepresentation of the inositol phosphate metabolism pathway in the Pneumocystis genomes compared to that in the fission yeast. Genomic sequences and gene expression of the identified enzymes, as well as inositol transporters, were experimentally confirmed in P. carinii and P. murina, as P. jirovecii is not readily available. Supplementation with inositol in a primary in vitro culture system significantly improved the viability of P. carinii (as indicated by ATP levels) throughout a 14-day period, a time frame when ATP levels declined dramatically without this additive. These new data suggest that Pneumocystis species are inositol auxotrophs and that inositol is one of the critical nutrients needed for in vitro culture but in and of itself is not sufficient for continuous cultivation. Importantly, the lack of synthesis and reliance on exogenous sources of inositol provide a new attractive drug target for the pneumonia caused by these fungi.
RESULTS
Choice of the genome annotation tool.
With the goal of unveiling the metabolic biological pathways encoded in the fungal genomes, two genome annotation tools, Blast2GO (10) and SHARKhunt (11), were validated for their ability to identify enzyme-coding genes within a well annotated fungal organism, S. pombe, which is phylogenetically close to Pneumocystis (9). Definitions of enzymes and metabolic pathways in S. pombe were retrieved from the KEGG Pathway database. The results in Fig. 1 illustrate the performance of both methods using various sequence homology cutoffs. Details of the distribution of the identified enzymes per pathway can be found in Table S1 in the supplemental material or at the Pneumocystis Genome Project website (http://pgp.cchmc.org/).
FIG 1 .
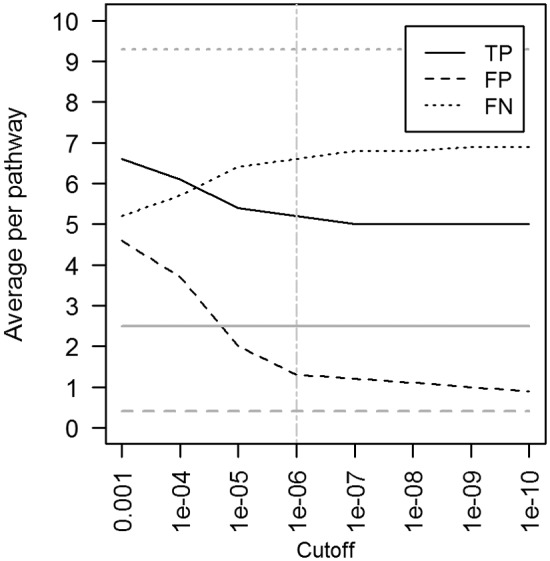
Identification of enzymes, as defined in the KEGG Pathway database, by Blast2GO (gray horizontal lines) and SHARKhunt (black curves) in S. pombe used as a gold standard organism. TP, true positives (enzymes that are known in S. pombe and which were identified by the tool); FP, false positives (not known in the fungus but were mistakenly found); FN, false negatives (known but were not identified). The numbers of found or missed enzymes are averages per metabolic pathway present in S. pombe. The vertical line indicates the sequence homology cutoff used in this work in the SHARKhunt search for enzymes within Pneumocystis genomes.
The Blast2GO results (Fig. 1, horizontal gray lines) appeared to be independent of the homology cutoff used. Blast2GO showed very low sensitivity, finding only 2.5 enzymes on average per pathway, and yielded a high level of missing enzymes (9.3 enzymes on average). The low level of performance of Blast2GO and its independence from the homology cutoff used indicate either an outdated table of enzymes or a highly conservative heuristic used in the identification of enzymes based on the matched gene ontology (GO) categories. For example, it did not find any enzymes in steroid biosynthesis (KEGG Pathway identifier [ID] 00100) and found very few enzymes in glycerolipid metabolism (2 out of 15 known in S. pombe) or glycerophospholipid metabolism (4 out of 29) pathways (KEGG Pathway IDs 00561 and 00564, respectively; see Table S1 in the supplemental material). Of note, BlastX used in Blast2GO did show strong hits to genomic sequences of the enzymes of the steroid biosynthesis pathway of S. pombe. However, its subsequent enzyme annotation module failed to assign EC codes to them.
SHARKhunt showed a better performance than Blast2GO in recognizing known enzymes from the genomic sequences. At a homology cutoff with an E value of ≤10−6 (Fig. 1, vertical line), it reached an optimal balance between true- and false-positive instances, yielding 5.2, 1.3, and 6.6 enzymes on average for true positive, false positive, and false negative, respectively. Since SHARKhunt was more sensitive in finding enzymes, while sustaining a low level of false positives similar to that obtained with Blast2GO, we have chosen SHARKhunt as a search tool for subsequent annotation of Pneumocystis genomic sequences with respect to enzymes and metabolic pathways.
Comparative genome analysis of the Pneumocystis genus.
There were 481, 490, and 460 total enzymes identified by SHARKhunt as being encoded in the P. carinii, P. murina, and P. jirovecii genomes, respectively. Of these, 328, 333, and 315 enzymes were mapped to metabolic pathways. The majority of enzymes identified (274 mapped and 124 not mapped) were shared across all three species of Pneumocystis (Fig. 2). The entire list of identified enzymes and the corresponding pathways can be found in Table S2 in the supplemental material.
FIG 2 .
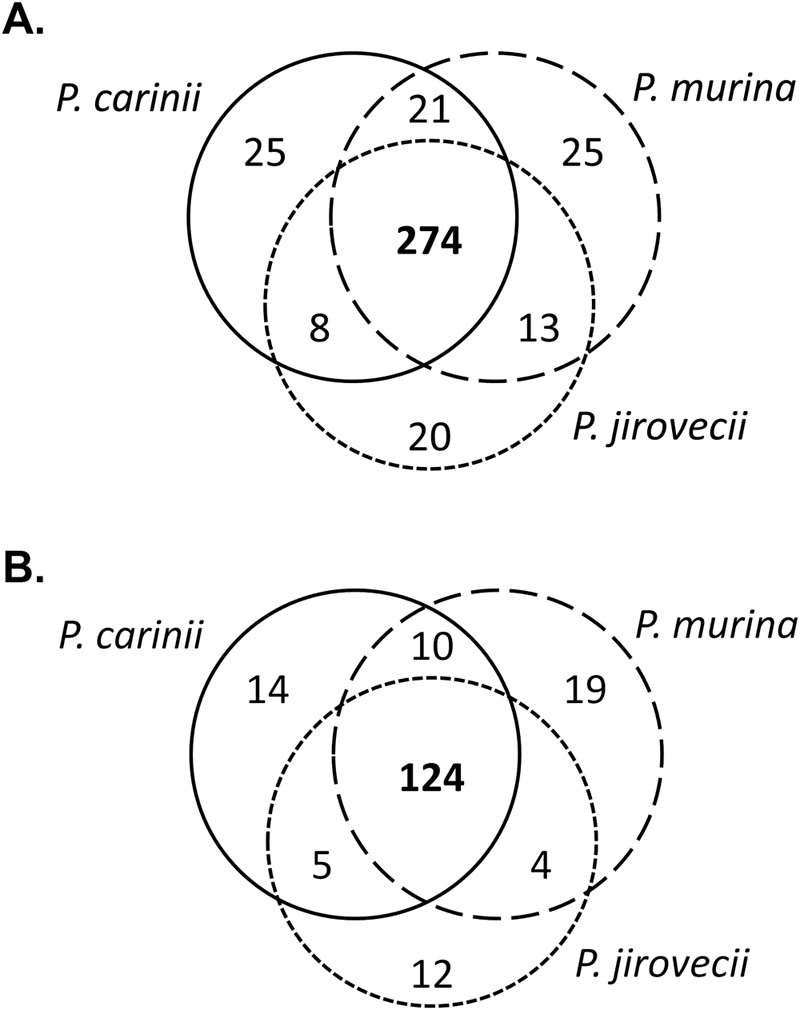
Overlap of identified enzymes between three Pneumocystis species. (A) Enzymes mapped into metabolic pathways defined in the KEGG Pathway database. (B) Enzymes not mapped into metabolic pathways.
Among the enzymes not shared between all three species, two observations are worth mentioning. The Pneumocystis species that inhabit rats and mice (P. carinii and P. murina) have five additional enzymes in the tryptophan metabolism pathway (KEGG Pathway ID 00380) compared to P. jirovecii residing in humans. These are indoleamine 2,3-dioxygenase (EC 1.13.11.52), 3-hydroxyanthranilate 3,4-dioxygenase (EC 1.13.11.6), kynurenine 3-monooxygenase (EC 1.14.13.9), arylformamidase (EC 3.5.1.9), and kynureninase (EC 3.7.1.3) that constitute the functional module tryptophan → kynurenine → 2-aminomuconate (KEGG Module ID M00038). Moreover, P. carinii has an additional enzyme in this pathway, alpha-ketoglutarate dehydrogenase (EC 1.2.4.2). On the other hand, P. jirovecii has three enzymes in the steroid biosynthesis pathway (KEGG Pathway ID 00100) that are specific to this species, namely, lathosterol 5-desaturase (EC 1.14.21.6; Erg3) and 7-dehydrocholesterol reductase (EC 1.3.1.21), which are the two final steps in cholesterol biosynthesis (KEGG Module ID M00101), and cycloeucalenol cycloisomerase (EC 5.5.1.9), which is an enzyme in the phytosterol branch of the steroid biosynthesis pathway and may represent a false-positive artifact of SHARKhunt predictions. The actual presence of these enzymes, as well as their expression, has yet to be confirmed. Their functional significance and biological implications, such as relevance to host specificity, will be the subject of future studies and are beyond the scope of this work.
Pneumocystis metabolic pathways and their comparison with those of S. pombe.
To reveal the metabolic strategies employed by the obligate Pneumocystis species in contrast to those of a free-living fungus, all identified enzymes have been mapped to metabolic pathways in KEGG Pathways and compared to the known pathways in fission yeast. Table 1 represents metabolic pathways defined for S. pombe in KEGG; for the complete list refer to Table S3 in the supplemental material.
TABLE 1 .
Representation of metabolic pathways of the three Pneumocystis species compared to those in S. pombe by the numbers of enzymes identified
| KEGG Pathway and ID | Metabolic pathway | No. of enzymes present in: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
S. pombe |
P. cariniic |
P. murina |
P. jirovecii |
|||||||||
| Per KPDa |
Per SHb |
Per SH |
Also in S. pombe |
Not in S. pombe |
Per SH |
Also in S. pombe |
Not in S. pombe |
Per SH |
Also in S. pombe |
Not in S. pombe |
||
| Amino acid metabolism | ||||||||||||
| 00250 | Alanine, aspartate, and glutamate metabolism | 21 | 11 | 11 | 10 | 1 | 12 | 11 | 1 | 12 | 11 | 1 |
| 00260 | Glycine, serine, and threonine metabolism | 22 | 16 | 10 | 7 | 3 | 9 | 7 | 2 | 9 | 7 | 2 |
| 00270 | Cysteine and methionine metabolism | 27 | 13 | 10 | 6 | 4 | 9 | 5 | 4 | 8 | 4 | 4 |
| 00280 | Valine, leucine and isoleucine degradation | 8 | 5 | 5 | 4 | 1 | 8 | 4 | 4 | 7 | 5 | 2 |
| 00290 | Valine, leucine and isoleucine biosynthesis | 7 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
| 00300 | Lysine biosynthesis | 13 | 6 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 00310 | Lysine degradation | 8 | 5 | 4 | 4 | 0 | 4 | 3 | 1 | 3 | 3 | 0 |
| 00330 | Arginine and proline metabolism | 29 | 21 | 8 | 4 | 4 | 8 | 5 | 3 | 8 | 4 | 4 |
| 00340 | Histidine metabolism | 12 | 3 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 |
| 00350 | Tyrosine metabolism | 13 | 6 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 |
| 00360 | Phenylalanine metabolism | 8 | 5 | 2 | 2 | 0 | 3 | 2 | 1 | 2 | 2 | 0 |
| 00380 | Tryptophan metabolism | 9 | 7 | 8 | 3 | 5 | 7 | 2 | 5 | 2 | 2 | 0 |
| 00400 | Phenylalanine, tyrosine and tryptophan biosynthesis | 20 | 12 | 10 | 10 | 0 | 8 | 8 | 0 | 10 | 10 | 0 |
| Carbohydrate metabolism | ||||||||||||
| 00010 | Glycolysis/gluconeogenesis | 24 | 10 | 16 | 14 | 2 | 15 | 14 | 1 | 14 | 13 | 1 |
| 00020 | Citrate cycle (TCA cycle) | 16 | 9 | 16 | 15 | 1 | 15 | 13 | 2 | 14 | 12 | 2 |
| 00030 | Pentose phosphate pathway | 15 | 7 | 11 | 10 | 1 | 10 | 10 | 0 | 11 | 11 | 0 |
| 00040 | Pentose and glucuronate interconversions | 4 | 3 | 3 | 2 | 1 | 3 | 2 | 1 | 3 | 2 | 1 |
| 00051 | Fructose and mannose metabolism | 13 | 6 | 10 | 8 | 2 | 10 | 8 | 2 | 11 | 8 | 3 |
| 00052 | Galactose metabolism | 11 | 5 | 8 | 5 | 3 | 5 | 4 | 1 | 6 | 4 | 2 |
| 00500 | Starch and sucrose metabolism | 14 | 12 | 16 | 10 | 6 | 16 | 9 | 7 | 14 | 9 | 5 |
| 00520 | Amino sugar and nucleotide sugar metabolism | 17 | 10 | 15 | 11 | 4 | 12 | 11 | 1 | 14 | 11 | 3 |
| 00562 | Inositol phosphate metabolism | 10 | 8 | 12 | 8 | 4 | 12 | 8 | 4 | 11 | 8 | 3 |
| 00620 | Pyruvate metabolism | 18 | 11 | 11 | 9 | 2 | 14 | 10 | 4 | 11 | 8 | 3 |
| 00630 | Glyoxylate and dicarboxylate metabolism | 10 | 9 | 9 | 6 | 3 | 9 | 6 | 3 | 8 | 6 | 2 |
| 00640 | Propanoate metabolism | 12 | 6 | 6 | 5 | 1 | 5 | 4 | 1 | 5 | 4 | 1 |
| 00650 | Butanoate metabolism | 8 | 3 | 2 | 2 | 0 | 3 | 2 | 1 | 3 | 2 | 1 |
| Energy metabolism | ||||||||||||
| 00190 | Oxidative phosphorylation | 9 | 7 | 9 | 8 | 1 | 10 | 8 | 2 | 10 | 8 | 2 |
| 00680 | Methane metabolism | 13 | 4 | 8 | 6 | 2 | 9 | 7 | 2 | 7 | 5 | 2 |
| 00910 | Nitrogen metabolism | 11 | 6 | 4 | 2 | 2 | 5 | 3 | 2 | 4 | 2 | 2 |
| 00920 | Sulfur metabolism | 14 | 8 | 5 | 2 | 3 | 3 | 1 | 2 | 4 | 2 | 2 |
| Glycan biosynthesis and metabolism | ||||||||||||
| 00510 | N-Glycan biosynthesis | 21 | 7 | 11 | 11 | 0 | 10 | 10 | 0 | 10 | 10 | 0 |
| 00511 | Other glycan degradation | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 |
| 00513 | Various types of N-glycan biosynthesis | 13 | 5 | 5 | 5 | 0 | 4 | 4 | 0 | 4 | 3 | 1 |
| 00514 | Other types of O-glycan biosynthesis | 2 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 |
| 00563 | Glycosylphosphatidylinositol anchor biosynthesis | 5 | 2 | 2 | 2 | 0 | 2 | 2 | 0 | 2 | 2 | 0 |
| Lipid metabolism | ||||||||||||
| 00061 | Fatty acid biosynthesis | 5 | 4 | 4 | 2 | 2 | 4 | 2 | 2 | 3 | 1 | 2 |
| 00062 | Fatty acid elongation | 6 | 1 | 2 | 1 | 1 | 3 | 1 | 2 | 2 | 1 | 1 |
| 00071 | Fatty acid degradation | 5 | 4 | 2 | 2 | 0 | 2 | 2 | 0 | 2 | 2 | 0 |
| 00072 | Synthesis and degradation of ketone bodies | 2 | 1 | 2 | 2 | 0 | 2 | 2 | 0 | 2 | 2 | 0 |
| 00100 | Steroid biosynthesis | 16 | 6 | 9 | 7 | 2 | 9 | 7 | 2 | 11 | 7 | 4 |
| 00561 | Glycerolipid metabolism | 15 | 9 | 6 | 3 | 3 | 7 | 3 | 4 | 6 | 3 | 3 |
| 00564 | Glycerophospholipid metabolism | 29 | 13 | 11 | 11 | 0 | 13 | 12 | 1 | 11 | 11 | 0 |
| 00565 | Ether lipid metabolism | 10 | 4 | 4 | 3 | 1 | 5 | 3 | 2 | 4 | 2 | 2 |
| 00590 | Arachidonic acid metabolism | 7 | 6 | 2 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 |
| 00591 | Linoleic acid metabolism | 5 | 2 | 1 | 1 | 0 | 2 | 1 | 1 | 1 | 0 | 1 |
| 00600 | Sphingolipid metabolism | 8 | 5 | 6 | 2 | 4 | 5 | 2 | 3 | 7 | 2 | 5 |
| 01040 | Biosynthesis of unsaturated fatty acids | 6 | 2 | 3 | 1 | 2 | 4 | 1 | 3 | 3 | 1 | 2 |
| Metabolism of cofactors and vitamins | ||||||||||||
| 00130 | Ubiquinone and other terpenoid-quinone biosynthesis | 6 | 2 | 3 | 0 | 3 | 4 | 0 | 4 | 1 | 0 | 1 |
| 00670 | One carbon pool by folate | 13 | 5 | 9 | 8 | 1 | 8 | 7 | 1 | 9 | 8 | 1 |
| 00730 | Thiamine metabolism | 6 | 4 | 4 | 3 | 1 | 2 | 2 | 0 | 3 | 2 | 1 |
| 00740 | Riboflavin metabolism | 10 | 2 | 8 | 6 | 2 | 8 | 6 | 2 | 7 | 6 | 1 |
| 00750 | Vitamin B6 metabolism | 7 | 5 | 2 | 2 | 0 | 2 | 2 | 0 | 2 | 2 | 0 |
| 00760 | Nicotinate and nicotinamide metabolism | 10 | 6 | 5 | 3 | 2 | 6 | 4 | 2 | 5 | 3 | 2 |
| 00770 | Pantothenate and coenzyme A biosynthesis | 12 | 7 | 7 | 5 | 2 | 7 | 5 | 2 | 8 | 6 | 2 |
| 00780 | Biotin metabolism | 6 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 |
| 00785 | Lipoic acid metabolism | 3 | 1 | 3 | 3 | 0 | 3 | 3 | 0 | 1 | 1 | 0 |
| 00790 | Folate biosynthesis | 8 | 2 | 7 | 6 | 1 | 8 | 6 | 2 | 7 | 6 | 1 |
| 00860 | Porphyrin and chlorophyll metabolism | 15 | 12 | 14 | 10 | 4 | 16 | 10 | 6 | 14 | 10 | 4 |
| Metabolism of other amino acids | ||||||||||||
| 00410 | Beta-alanine metabolism | 8 | 4 | 2 | 1 | 1 | 3 | 2 | 1 | 2 | 1 | 1 |
| 00430 | Taurine and hypotaurine metabolism | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 |
| 00450 | Selenocompound metabolism | 6 | 6 | 4 | 3 | 1 | 4 | 3 | 1 | 3 | 2 | 1 |
| 00460 | Cyanoamino acid metabolism | 4 | 3 | 2 | 2 | 0 | 2 | 2 | 0 | 2 | 2 | 0 |
| 00480 | Glutathione metabolism | 16 | 11 | 8 | 6 | 2 | 9 | 7 | 2 | 7 | 6 | 1 |
| Metabolism of terpenoids and polyketides | ||||||||||||
| 00900 | Terpenoid backbone biosynthesis | 17 | 10 | 13 | 12 | 1 | 13 | 11 | 2 | 11 | 10 | 1 |
| 00909 | Sesquiterpenoid and triterpenoid biosynthesis | 2 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 2 | 1 | 1 |
| Nucleotide metabolism | ||||||||||||
| 00230 | Purine metabolism | 42 | 29 | 34 | 27 | 7 | 32 | 28 | 4 | 34 | 28 | 6 |
| 00240 | Pyrimidine metabolism | 26 | 18 | 20 | 16 | 4 | 19 | 16 | 3 | 18 | 15 | 3 |
| Translation | ||||||||||||
| 00970 | Aminoacyl-tRNA biosynthesis | 23 | 13 | 21 | 21 | 0 | 21 | 21 | 0 | 21 | 21 | 0 |
| Xenobiotic biodegradation and metabolism | ||||||||||||
| 00627 | Aminobenzoate degradation | 0 | 3 | 0 | 3 | 4 | 0 | 4 | 4 | 0 | 4 | |
| 00983 | Drug metabolism: other enzymes | 0 | 5 | 0 | 5 | 5 | 0 | 5 | 5 | 0 | 5 | |
KPD, defined in the KEGG Pathway database.
SH, identified by SHARKhunt.
For Pneumocystis species, first, the total number of enzymes identified by SHARKhunt; second, the number of these enzymes overlapping with S. pombe genes defined in the KEGG Pathway database; and third, the number of enzymes not known in S. pombe.
Besides the remarkable representation of the tryptophan metabolism pathway in the two rodent Pneumocystis species compared to that in the human Pneumocystis species, S. pombe is also missing this functional module. Otherwise, metabolic pathways constituting amino acid metabolism lack many enzymes in the Pneumocystis genomes compared to these pathways in the fission yeast. This has recently been noted for P. carinii (7) and P. jirovecii (12), and the present work extends these observations to P. murina. While amino acids appear to be an important ingredient of the culture medium, they do not suffice to support the long-term in vitro viability of Pneumocystis. Most growth media previously tested have been serum based and contained amino acids as essential components in their formulations.
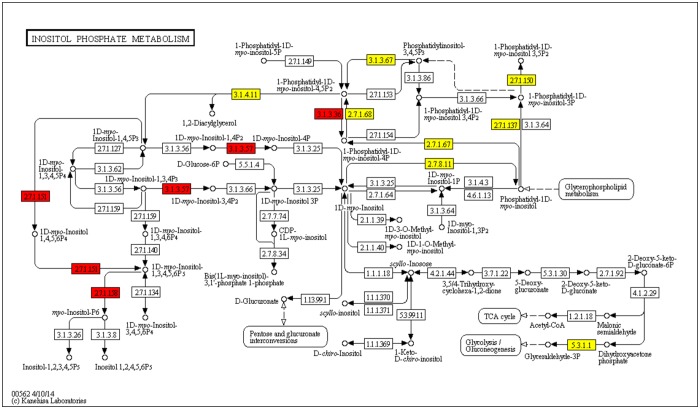
Metabolic pathways of carbohydrate metabolism are well represented in all Pneumocystis genomes, and the expression of the genes from this category has been previously confirmed for P. carinii using an expressed sequence tag (EST) library (13). In contrast to this previous study, more specific metabolic pathways in this category are now identified, along with the larger number of enzymes comprising them. Three pathways are worth mentioning here: starch and sucrose metabolism (KEGG Pathway ID 00500), amino sugar and nucleotide sugar metabolism (KEGG Pathway ID 00520), and inositol phosphate metabolism (KEGG Pathway ID 00562), These pathways, in addition to enzymes shared with the fission yeast, have 6, 4, and 4 additional enzymes in P. carinii, 7, 1, and 4 in P. murina, 5, 3, and 3 in P. jirovecii, respectively. Since inositol is an essential nutrient in eukaryotes and a critical proliferative factor for many pathogens, the increased number of Pneumocystis-specific genes in the inositol phosphate metabolism pathway in comparison to those in fission yeast was of great interest to us (Fig. 3). The enzymes highlighted in yellow in Fig. 3 are those shared between all fungi and those in red are Pneumocystis specific, whereas inositol polyphosphate 1-phosphatase (EC 3.1.3.57) is specific to P. carinii and P. murina. In addition to a large set of genes for inositol-metabolizing enzymes, Pneumocystis genomes contain sequences homologous to inositol transporter genes, including ITR1_SCHPO, ITR2_SCHPO, and putative inositol transporters in other fungi of the Aspergillus genus. These findings are highly significant in light of the fact that genes encoding inositol-1-phosphate synthase (INO1, EC 5.5.1.4), an enzyme essential for the synthesis of inositol from glucose-6-phosphate to myo-inositol 1-phosphate, were absent from each of the 3 genomes, suggesting that Pneumocystis fungi, like S. pombe, require an external source of inositol and are therefore inositol auxotrophs. The enzyme that catalyzes the next step in the synthesis, inositol monophosphatase (INM1, EC 3.1.3.25), which dephosphorylates myo-inositol 1-phosphate to myo-inositol, was also absent from the Pneumocystis and S. pombe genomes.
FIG 3 .
Inositol phosphate metabolism pathway as defined in the KEGG Pathway database. Highlighted are the enzymes found in the fungi: yellow, enzymes shared between S. pombe and Pneumocystis; red, Pneumocystis-specific enzymes.
In the effort to reduce the possibility that the apparent lack of genes for myo-inositol-synthesizing enzymes in the Pneumocystis genomes is not a false-negative finding, a genome of another closely related fungus was retrieved that might have these two enzymes. Taphrina deformans belongs to the same subphylum (Taphrinomycotina) as S. pombe and Pneumocystis spp. and, according to the genome annotation project maintained by the Joint Genome Institute, is predicted to have both these enzymes (see EC 5.5.1.4 and 3.1.3.25; http://genome.jgi.doe.gov/cgi-bin/metapathways?db=Tapde1_1&map=MAP00562&models=0&col=1&batchidTapde1_1%3A1). Sequence homology searches, both on the DNA and protein levels, for T. deformans EC 5.5.1.4 and 3.1.3.25 did not yield homologous hits to any of the three Pneumocystis genomes.
Given the significance of inositol and its metabolites in the proliferation and virulence of other pathogenic fungi, as well as the lack of synthetic enzymes in Pneumocystis, we focused on further investigation of the inositol phosphate metabolism pathway. The computationally identified genes relevant to this pathway were subsequently validated using the experimental approaches described below.
Reverse transcriptase real-time PCR of the inositol phosphate metabolism pathway.
Reverse transcriptase real-time PCR was used to confirm the expression of the four Pneumocystis-specific genes in the inositol phosphate metabolism (IPM) pathway. Since P. jirovecii samples were not available for this study, the experimental results pertain to rodent Pneumocystis species only. When possible, the primers were designed to span an intron junction so that they would amplify cDNA exclusively. The threshold cycle (CT) value was determined by averaging the threshold cycles for the triplicate reactions. The ΔCT value between the IPM pathway gene and the thymidylate synthase (TS) gene was calculated by subtracting the average CT value of the single-copy reference gene, encoding TS, from the average CT value of the IPM gene (Table 2). This comparison was made to evaluate the expression levels of the four genes as they related to this single-copy gene/enzyme from Pneumocystis. No change in the ΔCT value would indicate that the level of expression of the gene of interest was similar to that of TS. A positive ΔCT value would indicate that the gene was expressed at a lower level than the reference gene. Conversely, a negative ΔCT value would indicate that the gene was expressed at a higher level than the reference gene. All genes except the inositol-1,4-bisphosphate 1-phosphatase gene were found to be expressed at higher levels than the TS gene in both P. murina and P. carinii. Inositol-1,4-bisphosphate 1-phosphatase, while still expressed, was expressed at a lower level than TS in both P. murina and P. carinii. Bands of the expected size were observed when representative products were run on an agarose gel, verifying that all genes were expressed (Fig. 4).
TABLE 2 .
ΔCT values for genes in the inositol phosphate metabolism pathway versus the thymidylate synthase genea
| Species | Enzyme | EC | ΔCTb |
|---|---|---|---|
| P. murina | Inositol-polyphosphate multikinase | 2.7.1.151 | −1.54 |
| Inositol-pentakisphosphate 2-kinase | 2.7.1.158 | −3.42 | |
| Phosphoinositide 5-phosphatase | 3.1.3.36 | −4.99 | |
| Inositol-1,4-bisphosphate 1-phosphatase | 3.1.3.57 | 5.78 | |
| P. carinii | Inositol-polyphosphate multikinase | 2.7.1.151 | −2.94 |
| Inositol-pentakisphosphate 2-kinase | 2.7.1.158 | −3.21 | |
| Phosphoinositide 5-phosphatase | 3.1.3.36 | −2.26 | |
| Inositol-1,4-bisphosphate 1-phosphatase | 3.1.3.57 | 0.63 |
Thymidylate synthase is an enzyme essential for Pneumocystis viability.
Reactions were performed in triplicate, and the average threshold cycle (CT) value of the three replicates was determined. ΔCT values were calculated by subtracting the average CT value of the thymidylate synthase gene from the average CT value of the gene from the inositol phosphate metabolism pathway.
FIG 4 .
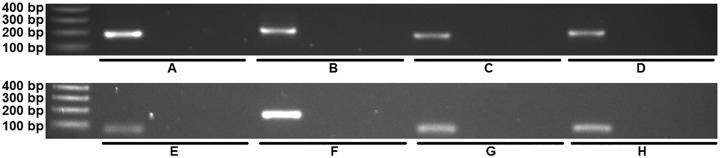
Representative products from reverse transcriptase real-time PCR of genes in the inositol phosphate metabolism pathway that were found to be conserved between P. murina and P. carinii but were not present in S. pombe. All products were found to be of the expected size, and all negative controls (mouse/rat cDNA or no cDNA) were negative. (A) P. murina inositol-polyphosphate multikinase, mouse cDNA negative control, and no-cDNA negative control. (B) P. murina inositol-pentakisphosphate 2-kinase, mouse cDNA negative control, and no-cDNA negative control. (C) P. murina phosphoinositide 5-phosphatase, mouse cDNA negative control, and no-cDNA negative control. (D) P. murina inositol-1,4-bisphosphate 1-phosphatase, mouse cDNA negative control, and no-cDNA negative control. (E) P. carinii inositol-polyphosphate multikinase, rat cDNA negative control, and no-cDNA negative control. (F) P. carinii inositol-pentakisphosphate 2-kinase, rat cDNA negative control, and no-cDNA negative control. (G) P. carinii phosphoinositide 5-phosphatase, rat cDNA negative control, and no-cDNA negative control. (H) P. carinii inositol-1,4-bisphosphate 1-phosphatase, rat cDNA negative control, and no-cDNA negative control.
Genomic analysis of ITR1 and ITR2 in Pneumocystis.
Genes with homology to fungal inositol transporter genes were identified in the Pneumocystis species. Homologs to ITR1 were identified in P. murina (PmITR1), P. carinii (PcITR1), and P. jirovecii (PjITR1) (Table 3). The lengths of the genes were quite similar, and the locations of the single intron were identical in the rodent Pneumocystis species and quite similar in P. jirovecii. P. carinii and P. murina homologs were more similar to one another than they were to homologs in the species infecting humans, which is in keeping with their phylogenetic relatedness.
TABLE 3 .
Structural analysis of inositol transporter genes in Pneumocystis species
| Gene product and fungal species | gDNA size (bp) | No. of introns (position[s]) | % gDNA identity |
No. of amino acids | % Amino acid identity |
||||
|---|---|---|---|---|---|---|---|---|---|
| P. carinii | P. murina | P. jirovecii | P. carinii | P. murina | P. jirovecii | ||||
| ITR1 | |||||||||
| P. carinii | 1,568 | 1 (94–140) | 90.26 | 79.87 | 507 | 89.35 | 75.83 | ||
| P. murina | 1,571 | 1 (96–142) | 90.26 | 78.81 | 507 | 89.35 | 75.24 | ||
| P. jirovecii | 1,582 | 1 (97–136) | 79.87 | 78.81 | 513 | 75.83 | 75.24 | ||
| ITR2 | |||||||||
| P. carinii | 1,968 | 6 (95–143, 225–272, 742–786, 1166–1217, 1504–1549, 1593–1637) | 89.28 | 558 | 89.29 | ||||
| P. murina | 1,960 | 6 (98–145, 224–267, 741–785, 1161–1210, 1497–1540, 1586–1630) | 89.28 | 560 | 89.29 | ||||
DNAMAN version 5.2.9 was used to analyze the putative Pneumocystis ITR genes.
ITR2 was identified in P. murina (PmITR2) and P. carinii (PcITR2) but not in P. jirovecii (Table 3). The PmITR2 gene had 1,960 bp of genomic sequence, and the PcITR2 gene had 1,968 bp. Both had six introns, ranging from 44 to 50 bp in length, which were located in almost exactly the same places in each genome. The putative amino acid sequences and the genomic DNA (gDNA) were about 90% homologous.
Reverse transcriptase real-time PCR of Pneumocystis inositol transporter genes.
Reverse transcriptase real-time PCR was utilized to confirm the expression of the two transporter genes involved in IPM that were found to be conserved between S. pombe, P. murina, and P. carinii. The results of the real-time PCR on the inositol transporter genes were analyzed in the same fashion as the results for the inositol metabolism pathway, and the ΔCT value between the inositol transporter gene and TS was calculated (Table 4). All of the ITR genes were expressed at higher levels than the TS gene in both P. murina and P. carinii. When representative products were run on an agarose gel, bands of the expected size were observed, indicating that all genes were expressed (Fig. 5).
TABLE 4 .
ΔCT values for inositol transporter genes versus the thymidylate synthase genea
| Species | Gene product | ΔCT |
|---|---|---|
| P. murina | ITR1 | −0.30 |
| P. carinii | ITR1 | −2.76 |
| P. murina | ITR2 | −4.56 |
| P. carinii | ITR2 | −1.13 |
Reactions were performed in triplicate, and the average threshold cycle (CT) value of the three replicates was determined. ΔCT values were calculated by subtracting the average CT value of the thymidylate synthase gene from the average CT value of the gene from the inositol phosphate metabolism pathway.
FIG 5 .

Representative products from reverse transcriptase real-time PCR of Pneumocystis ITR genes. All products were found to be of the expected size, and all negative controls (mouse/rat cDNA or no cDNA) were negative. (A) P. murina ITR1, mouse cDNA negative control, and no-cDNA negative control. (B) P. carinii ITR1, rat cDNA negative control, and no-cDNA negative control. (C) P. murina ITR2, mouse cDNA negative control, and no-cDNA negative control. (D) P. carinii ITR2, rat cDNA negative control, and no-cDNA negative control.
Culture supplementation with inositol.
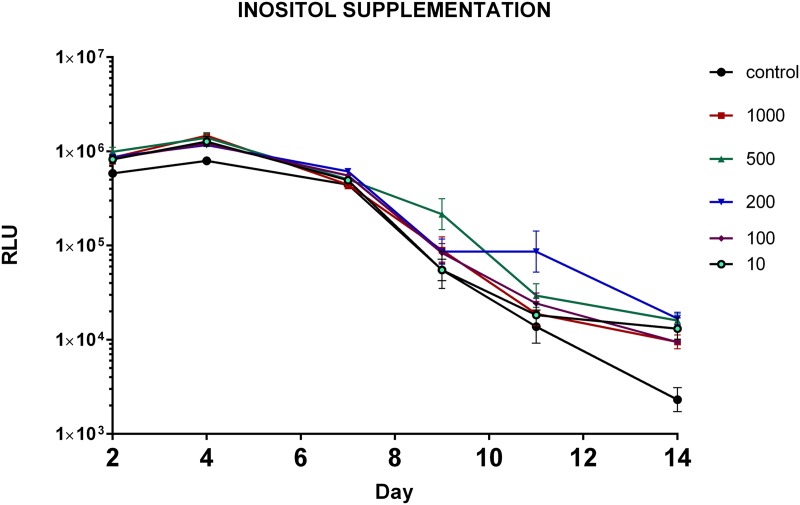
The ATP bioluminescence assay is used to assess the viability (total ATP content) of P. carinii in inositol-supplemented medium versus unsupplemented medium. This method has been routinely used to assess the effects of anti-Pneumocystis agents with the anticipation that effective drugs will decrease the viability of treated versus untreated organisms (4). In the case of supplements, the desired outcome is an increase in the ATP levels versus the results for untreated organisms. In this set of experiments, a wide range of myo-inositol concentrations, from 10,000 to 0.01 µg/ml, were assessed for their effects on the ATP content. The most consistent results were achieved with concentrations ranging between 1,000 and 10 µg/ml (Fig. 6). All readings of the assays were performed in triplicate wells and analyzed using analysis of variance (ANOVA) with Tukey’s posttest and significance set to a P value of <0.05.
FIG 6 .
ATP content of myo-inositol-supplemented versus unsupplemented P. carinii in vitro. The ATP content of P. carinii in an RPMI 1640-based axenic culture that was supplemented with myo-inositol at concentrations ranging from 1,000 to 10 µg/ml (or not supplemented, control) over a 14-day time period were measured using a bioluminescence-based assay which emits light (relative light units [RLU]) in a linear relationship with the amount of ATP present. Cultures were run in triplicate, and significance (P < 0.05) of the results for supplemented versus unsupplemented medium was found at all time points (note the log scale).
As observed by the results in Fig. 6, the ATP content as measured by the output of relative light units (RLU) was significantly increased at all concentrations of the myo-inositol supplementation (note the log scale). Greater effects were observed after 7 days in culture, when the viability of the unsupplemented control P. carinii dramatically declined in ATP content. A clear dose response was not obvious with the different concentrations of myo-inositol. This was likely due to the presence of myo-inositol in both the medium and serum, which was not controlled for in these studies and may have interfered with the ability to evaluate the optimal concentration in that context. Although the inositol clearly improved the viability of the P. carinii extracted from the rat lung, it was insufficient as the sole supplement to sustain a high level of viability throughout the 14-day assay period.
DISCUSSION
myo-inositol is an essential nutrient in eukaryotes, where it is involved in at least 3 vital functions (14). As a constituent of phosphatidylinositol, it is a component of biological membranes and, when phosphorylated, participates in exocytosis, cytoskeletal restructuring, and intracellular membrane trafficking; phosphatidylinositol-4,5-bisphosphate is critical in signal transduction pathways that control many cellular function; and finally, it is a precursor of glycosylphosphatidylinositol membrane anchors which affix glycolipids to cell surfaces (15).
myo-inositol is also a critical proliferative factor for many pathogens, including mycobacteria, fungi, and protozoa, which adopt different strategies of acquiring inositol either from the host or by synthesizing it (16). In infections with Candida albicans and Cryptococcus neoformans, inositol (17) and inositol transporters (18) have been associated with pathogenesis and virulence. Inositol has also been implicated in the mating processes of S. pombe and C. neoformans (19). In the case of C. neoformans, inositol secreted by plants stimulates mating, as does exogenous inositol imported by transporters in S. pombe (20). The connection of inositol with mating in S. pombe and C. neoformans is additionally intriguing, as Pneumocystis species reproduce sexually within the lung (9). myo-inositol may then play more than a nutritional role in supporting the growth of Pneumocystis and could potentially function as a signal or stimulant for initiation of the sexual cycle.
The lack of inositol-1-phosphate synthase and inositol monophosphatase, two enzymes critical to the synthesis of myo-inositol, and the presence of inositol transporters and enzymes involved in the metabolism of inositol strongly suggest that Pneumocystis species cannot synthesize their own inositol and must rely on the host for its exogenous sources.
Saccharomyces cerevisiae can both synthesize inositol and import it via two distinct transporter genes, ITR1 and ITR2 (21). C. albicans also has the ability to synthesize inositol but only has a single high-affinity inositol transporter (17). Remarkably, C. neoformans possesses 10 or 11 inositol transporters, depending on the variety (19), which is thought to result from the requirement for inositol from plants in its native environment. C. albicans has a number of opportunities to salvage inositol from its human host. Dietary intake of myo-inositol is about 1 g/day, making for a reliable source for C. albicans as an intestinal commensal (17). In the disseminated state, C. albicans can be exposed to 15 to 70 µM in human serum and up to 200 to 270 µM in liver and lymphatic tissue as ready sources. Within the lung, Pneumocystis would be exposed to myo-inositol from serum due to alveolar damage induced by the pneumonia, with subsequent leakage of serum into the alveolar compartments, and also from catabolism of phosphatidylinositol, which comprises approximately 1.5% of pulmonary surfactant (22).
Conclusions.
A comparative genome analysis has provided critical new insights into the Pneumocystis genus with respect to enzymes and metabolic pathways. The genomes of three species of Pneumocystis (P. carinii, P. murina, and P. jirovecii) were compared among themselves and with the free-living, phylogenetically related fungus S. pombe. Pneumocystis species and S. pombe are both members of the monophyletic Taphrinomycotina (Archiascomycota) (23, 24).
The majority of the computationally identified enzymes were shared across Pneumocystis species. We chose to focus on the inositol phosphate metabolism pathway as it appeared that Pneumocystis species could not synthesize myo-inositol due to a lack of INO1 and INM1 genes in their genomes. Four enzymes and two inositol transporters were found in the rodent Pneumocystis genomes but not in S. pombe. At the same time, P. jirovecii lacked both the second inositol transporter (ITR2) and phosphoinositide 5-phosphatase (EC 3.1.3.36) that are found in P. murina and P. carinii. These differences support the genetic literature that contends the rodent species are more closely related to each other on a phylogenetic level than they are to the species inhabiting humans.
Supplementation of in vitro culture with inositol statistically improved the viability of P. carinii during the later days of culture, 7 through 14, compared to the viability of the fungus in unsupplemented medium. However, it also appears that inositol, together with amino acids and the additional cofactors and ingredients in the RPMI 1640 medium and serum, are still not sufficient to maintain a long-term culture. Nonetheless, the strategy of comparative genomics employed here holds the greatest promise to date to identify all of the essential nutrients that Pneumocystis fungi require to live and thrive both in the mammalian lung and in an artificial environment. Further biochemical characterization of the nutrient acquisition mechanisms identified should lay the groundwork for rational drug design of a completely new class of anti-Pneumocystis agents.
MATERIALS AND METHODS
Genome annotation.
Genomic sequences were retrieved from the following sources: P. carinii sequences from the Pneumocystis Genome Project (http://pgp.cchmc.org/), P. murina sequences from the Pneumocystis murina Database (http://www.broadinstitute.org/annotation/genome/Pneumocystis_group/MultiHome.html), and P. jirovecii and S. pombe sequences from the NCBI Genome database (http://www.ncbi.nlm.nih.gov/genome?LinkName=pubmed_genome&from_uid=23269827 and http://www.ncbi.nlm.nih.gov/genome/14, respectively).
P. murina genomic sequences used in this study are revealed and reported here with permission of the genome holders. Genome-wide analysis and identification of putative enzymes is in concordance with the National Institutes of Health (NIH) policy of genomic data sharing.
Annotation of genomic sequences was conducted using sequence homology-based approaches implemented in BlastX (25), Blast2GO (10), and SHARKhunt (11). Sequence homology search was performed across the NCBI nr database and using the PRIAM enzyme-specific sequence profiles (26). Identified enzymes were mapped to metabolic pathways defined in the KEGG Pathway database (27) and compared with a reference genome, that of S. pombe, using the EC2KEGG tool (28).
Reverse transcriptase real-time PCR of the inositol phosphate metabolism pathway.
RNA was isolated from cryogenically preserved P. murina and P. carinii organisms with TRIzol reagent (Life Technologies, Grand Island, NY) according to the manufacturer’s protocol. cDNA was immediately synthesized from the extracted RNA using SuperScript II reverse transcriptase (Life Technologies, Grand Island, NY) according to the manufacturer’s protocol. The remaining RNA and the freshly synthesized cDNA were stored at −70° C.
Oligonucleotide primers, based on sequences from the Pneumocystis murina Database (PmD, http://www.broadinstitute.org/annotation/genome/Pneumocystis_group.2/MultiHome.html) and Pneumocystis Genome Project (PGP, http://pgp.cchmc.org/), were designed to amplify genes in the inositol phosphate metabolism pathway that were found to be conserved between P. murina and P. carinii but not present in S. pombe. When possible, primers were designed to span an intron junction to prevent amplification of genomic DNA. In addition to the primers designed to amplify these genes, reactions were also performed with sets of primers designed to amplify the Pneumocystis thymidylate synthase (TS) gene, which is present as a single copy in the genome, as a comparison for expression of the inositol phosphate metabolism pathway genes, and the Pneumocystis mitochondrial large ribosomal subunit (mtLSU), as a positive control. Each set of primers also had a negative control that contained no cDNA template.
Real-time PCR was performed in an Applied Biosystems 7500 fast real-time PCR system (Life Technologies, Grand Island, NY). The reactions were performed in triplicate in a final volume of 22 µl containing 1× SYBR green PCR master mix (Life Technologies, Grand Island, NY) and 0.8 mM of the appropriate primer pair (Table 5). Undiluted cDNA from either P. murina or P. carinii was used as the template. The reactions were initially incubated at 50°C for 2 min and then at 95°C for 10 min, followed by 40 to 45 cycles of 95°C for 15 s and 50 to 60°C (Table 5) for 1 min. Fluorescence data were captured during the 50 to 60°C step. Representative reaction products were run on a 1.0% agarose gel prestained with ethidium bromide and visualized with UV light to confirm the presence of products of the predicted size.
TABLE 5 .
Primers used for real-time PCR analysis of genes in the inositol phosphate metabolism pathway that were found in the P. murina, P. carinii, and P. jirovecii genomes but not in the S. pombe genome
| Species and target | Primer |
Annealing temp (°C) | |
|---|---|---|---|
| Left | Right | ||
| P. murina | |||
| Inositol-polyphosphate multikinase (EC 2.7.1.151) | AGAGCCTGATTTTGACATCGA | AGCCTCTTCCCATATCTTTTGCA | 55 |
| Inositol-pentakisphosphate 2-kinase (EC 2.7.1.158) | GGCATATTTCTCAGAAGGAAACCG | ATGAAAGAGGGGAGTCACAACC | 55 |
| Phosphoinositide 5-phosphatase (EC 3.1.3.36) | TTGATAGGGCCAATGAATTTTC | TCTGACTTATATTTTCGCCCTGA | 55 |
| Inositol-1,4-bisphosphate 1-phosphatase (EC 3.1.3.57) | TTTGTCAAAGCCATGCACCG | ACCTGCAGCATGATCCCAAG | 56 |
| P. carinii | |||
| Inositol-polyphosphate multikinase (EC 2.7.1.151) | TGGCTATATAAAGGGTTTGCGA | AGCCTCTTCCCATATCTTTTGCA | 60 |
| Inositol-pentakisphosphate 2-kinase (EC 2.7.1.158) | AGTCGTAACTCCCCTCTTTCAT | CGCATGTGTTTCGTTAATGTCT | 55 |
| Phosphoinositide 5-phosphatase (EC 3.1.3.36) | AAAGGCTCTAATTACGTCAACT | TGATCAGAAAATGTCAATGGTGA | 50 |
| Inositol-1,4-bisphosphate 1-phosphatase (EC 3.1.3.57) | TGTTAGCACGTGGGGAATGT | ACCTGCTGCATGATCCCAAA | 60 |
Assembly and completion of the putative P. carinii ITR1/2 sequence.
Two putative sequences for the P. murina ITR1 and ITR2 (PmITR1 and PmITR2), as determined by the Broad Institute and their collaborators, were retrieved from the PmD (Pm B123: PNEG_01850.1 and PNEG_00943.1). These sequences were used in homology searches against the sequence database of the PGP using the Basic Local Alignment Search Tool (BLAST) to identify sequences with homology to PmITR1 and PmITR2. The retrieved sequences were assembled and trimmed in DNAMAN version 5.2.9 (Lynnon Corporation, Quebec, Canada) to produce two putative P. carinii ITR contigs, designated PcITR1 and PcITR2. The contigs were analyzed by BLAST against the GenBank Fungal database to ensure that they were homologous to other fungal ITRs. This analysis also identified the sequence for a putative P. jirovecii ITR from GenBank (accession number CCJ30095).
After assembly from the PGP sequence database, the putative PcITR1 sequence was found to have a sequencing gap. To complete the sequence, oligonucleotide primers were designed that flanked the sequence gap starting at base pair 49 (PcITR1-L, AAG GAA GCA GTA GAA AGT GA, and PcITR1-R, CAT AAT GAC CTC CAA TAG CCA). PCR was performed in a 20-µl reaction mixture containing 1× Jump-Start REDTaq ReadyMix (Sigma, St. Louis, MO), 0.8 mM of the PcITR1-L and PcITR1-R oligonucleotide primers, and genomic P. carinii DNA as the template. The PCR conditions were as follows: 95°C hot start for 3 min, a 95°C denaturing step for 30 s, 50°C annealing step for 30 s, and 72°C extension step for 1 min for 45 cycles, and a final extension of 5 min at 72°C. Reaction products were run on a 0.7% agarose gel prestained with ethidium bromide and visualized with UV light for the presence of a product of the predicted size. PCR products were ligated into the pCR2.1-TOPO vector (Life Technologies, Grand Island, NY) and transformed into TOP10F′ cells (Life Technologies, Grand Island, NY) according to the manufacturer’s protocol. Plasmids were purified, screened for inserts of the proper size, and then sequenced on an ABI Prism 3730xl DNA analyzer (Life Technologies, Grand Island, NY) at a local facility.
Reverse transcriptase real-time PCR of inositol transporter genes.
Primers for real-time PCR were designed as described above based on sequences of the PmITR1, PmITR2, PcITR1, and PcITR2 genes (Table 6). Real-time PCR was performed on the inositol transporter genes as described above. Representative reaction products were run on a 1.0% agarose gel prestained with ethidium bromide and visualized with UV light to confirm the presence of products of the predicted size.
TABLE 6 .
Primers used for real-time PCR analysis of the putative inositol transporter genes in P. murina and P. carinii
| Target | Primer |
Annealing temp (°C) | |
|---|---|---|---|
| Left | Right | ||
| PmITR1 | CAGCATTTGTTACTGCACCACA | AATCCAAACCCAAGCACCCA | 50 |
| PcITR1 | TGCATTTGTTACTGCACCACA | ACAATCCAAATCCAAGCACCT | 60 |
| PmITR2 | GACCTGTTCCGTGGGCATTA | GGCTACTCCCTTTGCTCGAA | 55 |
| PcITR2 | GGAACAAGGTTTATCGATTCTAC | ACCGGGTACAATTTGAATCAA | 50 |
Culture media and supplementation. (i) Compound preparation.
Serial dilutions of myo-inositol (Sigma-Aldrich, St. Louis, MO) ranging from 10,000 to 0.01 µg/ml were made in RPMI 1640 containing 20% calf or horse serum, 1% MEM (minimal essential medium) vitamin solution, 1% minimal essential medium with nonessential amino acids (MEM-NEAA), 2,000 units/ml penicillin-streptomycin, and 50 µg/ml vancomycin. Negative controls included medium without added inositol and with 10 µg/ml ampicillin. There was no vehicle control as the myo-inositol was miscible in the medium.
(ii) P. carinii ATP assay.
Cryopreserved and characterized (ATP content, no microbial contaminants) P. carinii cells isolated from rat lung tissue were distributed into triplicate wells of 48-well plates with a final volume of 1,000 µl of RPMI 1640 supplemented with 20% calf serum and other additives and a P. carinii cell total of 5 × 107/ml as previously described (29). Plates were incubated at 37°C with 5% CO2 in a water-jacketed incubator.
At the assigned time points, 5% of the well volume (50 µl) was removed and washed, and the ATP content was measured using the ATP-liteM luciferin-luciferase assay (PerkinElmer) according to the vendor’s instructions. The luminescence generated by the ATP content of the samples was measured by a BMG Polarstar optima spectrophotometer. Samples of each group were examined microscopically to rule out the presence of bacteria or other microbial contaminants that may have been introduced during the culture process. Cultures were refed every other day by removing 50% of the medium and replacing it with fresh medium.
Statistical analysis.
Background luminescence was subtracted, and triplicate well readings of the assays were analyzed using ANOVA with a Tukey posttest. Significance was accepted at a P value of <0.05.
Quench control test.
Inositol levels were checked for any direct effect on the ATP bioluminescence assay. The ATP standard was diluted and added to the medium with the inositol dilutions used in the assays. No quenching of the enzyme reaction was observed.
Genomic and protein sequences.
Sequences of genes, transcripts, and proteins of the Pneumocystis-specific enzymes from the inositol phosphate metabolism pathway and inositol transporters are available in Text S1 and S2, respectively, in the supplemental material. New sequences of P. carinii inositol transporters are published in the NCBI GenBank with the following accession numbers: PcITR1 (KM263199) and PcITR2 (KM263200).
SUPPLEMENTAL MATERIAL
Genomic sequences of identified Pneumocystis genes and their transcripts: enzymes. gDNA, cDNA, and amino acid sequences of genes and their transcripts described in the text, in the FASTA format. Download
Genomic sequences of identified Pneumocystis genes and their transcripts: transporters. gDNA, cDNA, and amino acid sequences of genes and their transcripts described in the text, in the FASTA format. Download
Performance of Blast2GO and SHARKhunt tools in annotating S. pombe genome with respect to enzymes and corresponding metabolic pathways. The numbers represent counts of the enzymes known in the genome and identified by one of the tools using different sequence homology cutoffs.
Full list of enzymes (using EC numbers) identified in the three Pneumocystis genomes and their overlaps among the analyzed species.
Overlaps of the Pneumocystis enzymes with the known enzymes in S. pombe in the corresponding metabolic pathways.
ACKNOWLEDGMENTS
The work was supported by NHLBI grant 5R01HL119190 (A.P. and M.T.C.) and a Merit Review grant from the Veterans Affairs program in Biomedical Laboratory Research & Development (M.T.C.).
Footnotes
Citation Porollo A, Sesterhenn TM, Collins MS, Welge JA, Cushion MT. 2014. Comparative genomics of Pneumocystis species suggests the absence of genes for myo-inositol synthesis and reliance on inositol transport and metabolism. mBio 5(6):e01834-14. doi:10.1128/mBio.01834-14.
REFERENCES
- 1. Morris A, Sciurba FC, Lebedeva IP, Githaiga A, Elliott WM, Hogg JC, Huang L, Norris KA. 2004. Association of chronic obstructive pulmonary disease severity and Pneumocystis colonization. Am. J. Respir. Crit. Care Med. 170:408–413. 10.1164/rccm.200401-094OC. [DOI] [PubMed] [Google Scholar]
- 2. Kaur N, Mahl TC. 2007. Pneumocystis jiroveci (carinii) pneumonia after infliximab therapy: a review of 84 cases. Dig. Dis. Sci. 52:1481–1484. 10.1007/s10620-006-9250-x. [DOI] [PubMed] [Google Scholar]
- 3. Porollo A, Meller J, Joshi Y, Jaiswal V, Smulian AG, Cushion MT. 2012. Analysis of current antifungal agents and their targets within the Pneumocystis carinii genome. Curr. Drug Targets 13:1575–1585. 10.2174/138945012803530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cushion MT, Walzer PD. 2009. Preclinical drug discovery for new anti-pneumocystis compounds. Curr. Med. Chem. 16:2514–2530. 10.2174/092986709788682038. [DOI] [PubMed] [Google Scholar]
- 5. Monnet X, Vidal-Petiot E, Osman D, Hamzaoui O, Durrbach A, Goujard C, Miceli C, Bourée P, Richard C. 2008. Critical care management and outcome of severe Pneumocystis pneumonia in patients with and without HIV infection. Crit. Care 12:R28. 10.1186/cc6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kaneshiro ES. 2004. Sterol metabolism in the opportunistic pathogen Pneumocystis: advances and new insights. Lipids 39:753–761. 10.1007/s11745-004-1292-5. [DOI] [PubMed] [Google Scholar]
- 7. Hauser PM, Burdet FX, Cissé OH, Keller L, Taffé P, Sanglard D, Pagni M. 2010. Comparative genomics suggests that the fungal pathogen pneumocystis is an obligate parasite scavenging amino acids from its host’s lungs. PLoS One 5:e15152. 10.1371/journal.pone.0015152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cushion MT, Ebbets D. 1990. Growth and metabolism of Pneumocystis carinii in axenic culture. J. Clin. Microbiol. 28:1385–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cushion MT, Stringer JR. 2010. Stealth and opportunism: alternative lifestyles of species in the fungal genus Pneumocystis. Annu. Rev. Microbiol. 64:431–452. 10.1146/annurev.micro.112408.134335. [DOI] [PubMed] [Google Scholar]
- 10. Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 11. Pinney JW, Shirley MW, McConkey GA, Westhead DR. 2005. metaSHARK: software for automated metabolic network prediction from DNA sequence and its application to the genomes of Plasmodium falciparum and Eimeria tenella. Nucleic Acids Res. 33:1399–1409. 10.1093/nar/gki285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cissé OH, Pagni M, Hauser PM. 2012. De novo assembly of the Pneumocystis jirovecii genome from a single bronchoalveolar lavage fluid specimen from a patient. mBio 4(1):e00428-12. 10.1128/mBio.00428-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cushion MT, Smulian AG, Slaven BE, Sesterhenn T, Arnold J, Staben C, Porollo A, Adamczak R, Meller J. 2007. Transcriptome of Pneumocystis carinii during fulminate infection: carbohydrate metabolism and the concept of a compatible parasite. PLoS One 2:e423. 10.1371/journal.pone.0000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Michell RH. 2008. Inositol derivatives: evolution and functions. Nat. Rev. Mol. Cell Biol. 9:151–161. 10.1038/nrm2334. [DOI] [PubMed] [Google Scholar]
- 15. Simonsen A, Wurmser AE, Emr SD, Stenmark H. 2001. The role of phosphoinositides in membrane transport. Curr. Opin. Cell Biol. 13:485–492. 10.1016/S0955-0674(00)00240-4. [DOI] [PubMed] [Google Scholar]
- 16. Reynolds TB. 2009. Strategies for acquiring the phospholipid metabolite inositol in pathogenic bacteria, fungi and protozoa: making it and taking it. Microbiology 155:1386–1396. 10.1099/mic.0.025718-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jin JH, Seyfang A. 2003. High-affinity myo-inositol transport in Candida albicans: substrate specificity and pharmacology. Microbiology 149:3371–3381. 10.1099/mic.0.26644-0. [DOI] [PubMed] [Google Scholar]
- 18. Wang Y, Liu TB, Delmas G, Park S, Perlin D, Xue C. 2011. Two major inositol transporters and their role in cryptococcal virulence. Eukaryot. Cell 10:618–628. 10.1128/EC.00327-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xue C. 2012. Cryptococcus and beyond—inositol utilization and its implications for the emergence of fungal virulence. PLoS Pathog. 8:e1002869. 10.1371/journal.ppat.1002869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Niederberger C, Gräub R, Schweingruber AM, Fankhauser H, Rusu M, Poitelea M, Edenharter L, Schweingruber ME. 1998. Exogenous inositol and genes responsible for inositol transport are required for mating and sporulation in Shizosaccharomyces pombe. Curr. Genet. 33:255–261. 10.1007/s002940050334. [DOI] [PubMed] [Google Scholar]
- 21. Nikawa J, Tsukagoshi Y, Yamashita S. 1991. Isolation and characterization of two distinct myo-inositol transporter genes of Saccharomyces cerevisiae. J. Biol. Chem. 266:11184–11191. [PubMed] [Google Scholar]
- 22. Zuo YY, Veldhuizen RA, Neumann AW, Petersen NO, Possmayer F. 2008. Current perspectives in pulmonary surfactant—inhibition, enhancement and evaluation. Biochim. Biophys. Acta 1778:1947–1977. 10.1016/j.bbamem.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 23. Zytkiewicz A, Czarkowska D, Pietron K, Papierkowski A. 1965. Pneumopathies in the newborn period. Ann.Univ. Mariae Curie Sklodowska Med. 20:239–248 (In Polish.) [PubMed] [Google Scholar]
- 24. Liu Y, Leigh JW, Brinkmann H, Cushion MT, Rodriguez-Ezpeleta N, Philippe H, Lang BF. 2009. Phylogenomic analyses support the monophyly of Taphrinomycotina, including Schizosaccharomyces fission yeasts. Mol. Biol. Evol. 26:27–34. 10.1093/molbev/msn221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 26. Claudel-Renard C, Chevalet C, Faraut T, Kahn D. 2003. Enzyme-specific profiles for genome annotation: PRIAM. Nucleic Acids Res. 31:6633–6639. 10.1093/nar/gkg847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. 2012. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 40:D109–D114. 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Porollo A. 2014. EC2KEGG: a command line tool for comparison of metabolic pathways. Source Code Biol. Med. 9:19. 10.1186/1751-0473-9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cushion MT, Collins MS. 2011. Susceptibility of pneumocystis to echinocandins in suspension and biofilm cultures. Antimicrob. Agents Chemother. 55:4513–4518. 10.1128/AAC.00017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genomic sequences of identified Pneumocystis genes and their transcripts: enzymes. gDNA, cDNA, and amino acid sequences of genes and their transcripts described in the text, in the FASTA format. Download
Genomic sequences of identified Pneumocystis genes and their transcripts: transporters. gDNA, cDNA, and amino acid sequences of genes and their transcripts described in the text, in the FASTA format. Download
Performance of Blast2GO and SHARKhunt tools in annotating S. pombe genome with respect to enzymes and corresponding metabolic pathways. The numbers represent counts of the enzymes known in the genome and identified by one of the tools using different sequence homology cutoffs.
Full list of enzymes (using EC numbers) identified in the three Pneumocystis genomes and their overlaps among the analyzed species.
Overlaps of the Pneumocystis enzymes with the known enzymes in S. pombe in the corresponding metabolic pathways.