Abstract
Selenoproteins are a distinct class of proteins that are characterized by the co-translational incorporation of selenium (Se) in the form of the 21st amino acid selenocysteine. Selenoproteins provide a key defense against oxidative stress, as many of these proteins participate in oxidation-reduction reactions neutralizing reactive oxygen species, where selenocysteine residues act as catalytic sites. Many selenoproteins are highly expressed in the brain and mouse knockout studies have determined that several are required for normal brain development. In parallel with these laboratory studies, recent reports of rare human cases with mutations in genes involved in selenoprotein biosynthesis have described individuals with an assortment of neurological problems that mirror those detailed in knockout mice. These deficits include impairments in cognition and motor function, seizures, hearing loss, and altered thyroid metabolism. Additionally, due to the fact that oxidative stress is a key feature of neurodegenerative disease, there is considerable interest in the therapeutic potential of selenium supplementation for human neurological disorders. Studies performed in cell culture and rodent models have demonstrated that selenium administration attenuates oxidative stress, prevents neurodegeneration, and counters cell signaling mechanisms known to be dysregulated in certain disease states. However, there is currently no definitive evidence in support of selenium supplementation to prevent and/or treat common neurological conditions in the general population. It appears likely, that in humans, supplementation with selenium may only benefit certain subpopulations, such as those that are either selenium-deficient or possess genetic variants that affect selenium metabolism.
Keywords: neurodegeneration, neurodevelopment, oxidative stress, selenium, selenocysteine, selenoprotein
INTRODUCTION
Selenium (Se) is an essential trace element that plays an important role in maintaining human health. It is obtained primarily through dietary sources and Se content differs depending upon the type of food consumed and, in the case of agricultural products, upon the Se bioavailability of the soil in which the crop was raised. Foods particularly rich in Se include Brazil nuts, organ meats, seafood, mushrooms, and onions. Se intake differs greatly across the world, with mean values of greater than 100 μg per day in the USA, Japan, and Venezuela, and daily intake of less than 40 μg in many European countries, such as the United Kingdom, Germany, Sweden, and the Czech Republic [1]. The potential importance of Se in biological processes was first recognized in 1954, when it was reported that trace amounts were needed to enable optimal activity of formic acid dehydrogenase in Escherichia coli [2]. Shortly thereafter, the significance of Se to mammalian life was described in a paper demonstrating liver necrosis in rats fed a Se-deficient diet [3]. Subsequent studies revealed that the antioxidant activity of glutathione peroxidase (GPx) is dependent upon Se [4], that Se is present in protein-bound selenocysteine (Sec) residues [5], and that Sec is located in the catalytic site of GPx [6]. It is now well established that the influence of Se on health is largely mediated by selenoproteins, a class of proteins characterized by the co-translational incorporation of Se in the form of the 21st amino acid Sec. Sec is structurally analogous to cysteine (Cys), with the sole difference being that the sulfur atom present in Cys is replaced with Se. In comparison to Cys, Sec is more nucleophilic and has a lower pKa (5.2 vs. 8.3), properties which enhance catalysis of redox reactions [7]. Selenoproteins have been demonstrated to play essential roles in a variety of functions, including thyroid hormone metabolism, brain development, male fertility, immune function, and energy metabolism. Many of these proteins partake in redox reactions neutralizing oxidative stress, where Sec residues act as catalytic sites. Oxidative stress is a key contributing factor to an array of disorders involving the central nervous system, including Alzheimer’s disease, epilepsy, Parkinson’s disease, and schizophrenia. This review will focus upon the role of selenoproteins in nervous system development and function.
Selenoproteins are encoded by 25 distinct genes in humans and includes the GPxs, thioredoxin reductases (Txnrds), and iodothyronine deiodinases (DIOs) [8]. The GPxs constitute an important class of antioxidant enzymes that utilize glutathione as a co-factor to catalyze redox reactions involving the reduction of hydrogen peroxide and/or phospholipid hydroperoxides. In humans, eight GPx genes have been discovered, with five of them encoding selenoproteins that contain Sec as the active site residue. The thioredoxin (Txn) system represents another key defense against oxidative stress. Txnrds are a family of homodimeric flavoenzymes that mediate reactions in which oxidized Txn is reduced at the expense of NADPH. In these proteins, Sec is incorporated as the penultimate C-terminal residue, where it is essential for enzymatic activity [9]. There are three distinct genes that code for Txnrds in mammals (Txnrd1, Txnrd2, Txnrd3), and constitutive knockout studies have demonstrated the indispensable function of Txnrd1 [10] and Txnrd2 [11] in supporting mammalian life. Another important class of selenoproteins is the DIOs, which play fundamental roles in thyroid hormone metabolism. These proteins are encoded by three distinct genes in mammals (DIO1, DIO2, DIO3) and mediate the activation and inactivation of thyroid hormones via reductive deiodination. Thyroid hormone activation is dependent upon conversion of the prohormone thyroxine (T4) to the biologically active form, 3,3′,5-triiodothyronine (T3). This process occurs via an outer ring monodeiodination reaction that can be catalyzed by either DIO1 or DIO2. Activated T3 interacts with thyroid hormone receptors in the nucleus, which in turn bind to specific DNA sequences and modulate gene expression. Conversely, DIO3 mediates the irreversible inactivation of T3 and T4 by means of inner ring monodeiodination reactions, yielding the inactive hormones T2 and rT3, respectively.
In addition to the selenoproteins in the aforementioned GPx, Txnrd, and DIO families, there are numerous other selenoproteins that have been characterized to varying degrees. Among these, selenoprotein P (SelP) is particularly distinctive due to the fact that it contains multiple Sec residues and has been postulated to act in selenium transport. First reported in 1982, SelP is a secreted glycoprotein, primarily produced in liver, which constitutes the majority of selenium found in plasma [12 – 14]. SelP can be divided into two functional domains, a large N-terminal domain containing one Sec residue (U) in a U-x-x-C redox motif and a smaller selenium-rich C-terminal domain that contains multiple Sec residues (up to nine in humans). This unique protein is believed to be multifunctional, in that the N-terminal redox motif is thought to contribute to the maintenance of extracellular redox balance [15], whereas the selenium-rich C-terminal domain has been found to mediate selenium transport to the brain and testes [16]. Selenium delivery is largely achieved by intracellular uptake of SelP upon binding to the apolipoprotein receptor, ApoER2 (also LRP8) [14]. ApoER2-mediated endocytosis of SelP has been shown to facilitate selenium delivery to brain [17], testes [18], muscle [19], placenta [20], and bone [21]. Additionally, a related lipoprotein receptor, megalin (also LRP2), has been demonstrated to mediate SelP uptake into the kidney and brain [22, 23].
Multiple selenoproteins (Sep15, SelK, SelM, SelN, SelS, SelT) reside in the endoplasmic reticulum (ER) and are thought to contribute to processes including calcium regulation, protein folding, and attenuation of ER stress [24]. SelS (also VIMP) is a component of the ER-associated degradation pathway that transports misfolded proteins from the ER to cytosol, where they are then tagged with ubiquitin and sent to the proteasome for degradation [25]. In this pathway, SelS is thought to mediate the formation of the retrotranslocation complex by recruiting cytosolic p97 ATPase and promoting its interaction with Derlin-1 in the ER membrane. Recent studies have also shown a functional role for SelK in receptor-mediated Ca2+ flux in immune cells [26], and demonstrated that SelK is a target of the calcium-activated protease, calpain, in resting macrophages [27]. SelN (also SEPN1) has been found to modulate calcium homeostasis in muscle cells via interactions with the ryanodine receptor, an intracellular calcium release channel [28]. Among ER-resident selenoproteins, Sep15, SelM, and SelT possess Txn-like folds containing Sec residues in the active site motif, suggesting that they function as thiol-disulfide oxidoreductases. Sep15 and SelM are structurally homologous proteins that have been proposed to form an evolutionary distinct selenoprotein family within the Txn superfamily [29]. Sep15 has been identified as a binding partner of ER-chaperone UDP-glucose: glycoprotein glucosyltransferase and is thought to promote protein folding [30], while cell culture studies have demonstrated that SelM has neuroprotective properties and contributes to intracellular calcium regulation [31]. SelT has been found to modulate neuroendocrine secretion and, like SelM, has been implicated in the regulation of calcium homeostasis [32].
Several other selenoproteins (SelH, SelO, SelV, SelW) that are not localized to the ER have also been observed to possess a Txn-like fold containing a C-x-x-U redox motif. Of these, SelH has been shown to be a redox-responsive DNA-binding protein that regulates the expression of genes involved in de novo glutathione synthesis and phase II detoxification [33]. SelW is widely expressed in a variety of tissues and has been documented to interact with glutathione [34] and the 14-3-3 proteins [35]. Both SelO and SelV are uncharacterized, with no published information available on their functional roles. SelI (also hEPT1) is another uncharacterized selenoprotein with an amino acid sequence containing a putative transmembrane region and a structural motif conserved among phospholipid synthases, suggestive of a potential role in phospholipid synthesis [36]. SelR (also MsrB1) is a member of the methionine sulfoxide reductase (Msr) family of enzymes, which also includes MsrA, MsrB2, and MsrB3 [37]. Among these proteins, only SelR contains Sec at the active site, whereas the other members of the Msr family utilize Cys residues. These enzymes mediate the reduction of oxidized methionine residues, as oxidative stress generates a mixture of S- and R-forms of methionine sulfoxide, which in turn are reduced by the MsrA and MsrB enzymes, respectively. Cell culture studies have revealed that SelR interacts with the magnesium channel TRPM6 and modulates its activity under conditions of oxidative stress [38]. Selenophosphate synthetase 2 (SPS2) is unique in that it is the only selenoprotein directly involved in selenoprotein biosynthesis. This protein catalyzes the generation of monoselenophosphate from selenide and ATP [39], which is then utilized for Sec synthesis as will be discussed next.
SELENOPROTEIN SYNTHESIS
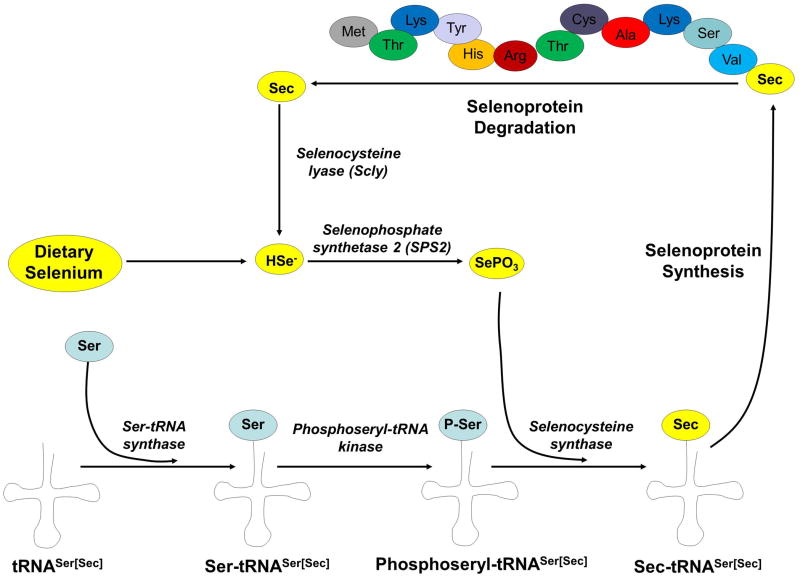
Upon the discovery in the 1960s that each individual amino acid in a protein is encoded by a corresponding 3 nucleotide sequence in the mRNA, amino acids were assigned to 61 of the 64 potential codons and the remaining three codons (UAA, UAG, UGA) were thought to act as stop codons. The notion that UGA functions solely as a stop codon remained unchallenged until the 1980s, when it was demonstrated that UGA codes for Sec in the formate dehydrogenase gene of E. coli [40] and in the mammalian glutathione peroxidase gene [41, 42]. In 1989, Sec was identified as the 21st amino acid, upon discovery of a novel tRNA that is coupled to Sec and recognizes UGA codons [43, 44]. It was also determined that in contrast to all other amino acids, the biosynthesis of Sec always occurs on its cognate tRNA, designated tRNA[Ser]Sec (Fig. 1). This process involves the initial aminoacylation of tRNA[Ser]Sec with serine by Ser-tRNA synthase, followed by serine phosphorylation by phosphoseryl-tRNA kinase to yield phosphoseryl-tRNA[Ser]Sec. The tRNA-bound phosphoseryl residue is then converted to Sec by the enzyme selenocysteine synthase (Sec synthase), a reaction in which selenium, in the form of monoselenophosphate, is transferred to tRNA[Ser]Sec to produce Sec-tRNA[Ser]Sec [45]. Also essential to this process is the selenoprotein SPS2, which generates the monoselenophosphate necessary for Sec synthesis. Another contributing factor to selenium metabolism is the enzyme selenocysteine lyase (Scly), which decomposes Sec into L-alanine and selenide, enabling selenium to be recycled for additional Sec biosynthesis [46].
Figure 1. Overview of selenocysteine biosynthesis and degradation.
tRNA[Ser]Sec is aminoacylated with serine by Ser-tRNA synthase, followed by serine phosphorylation by phosphoseryl-tRNA kinase to yield phosphoseryl-tRNA[Ser]Sec. The tRNA-bound phosphoseryl residue is then converted to Sec by the enzyme selenocysteine synthase (Sec synthase), a reaction in which Se, in the form of monoselenophosphate, is transferred to tRNA[Ser]Sec to produce Sec-tRNA[Ser]Sec. During selenoprotein synthesis, Sec-tRNA[Ser]Sec insertion occurs at UGA codons when directed by SECIS elements within selenoprotein mRNAs. Upon degradation, selenocysteine lyase decomposes Sec into L-alanine and selenide (represented as HSe−), enabling Se to be recycled for additional Sec biosynthesis.
In addition to the tRNA[Ser]Sec, several additional factors are required for successful incorporation of Sec at UGA codons rather than termination of protein synthesis. For eukaryotic selenoproteins, Sec incorporation is directed by a specific stem loop structural element, termed a selenocysteine insertion sequence (SECIS), that is located in the 3′-untranslated region of selenoprotein mRNAs [47]. The SECIS element forms the structural backbone for the assembly of several factors into a RNA-protein complex that directs Sec insertion [48]. Key components of this complex in eukaryotic organisms include the Sec-specific elongation factor (EFSec) and SECIS-binding protein 2 (SBP2) [49, 50]. EFSec is a GTP-dependent elongation factor that exclusively interacts with Sec-tRNA[Ser]Sec, whereas SBP2 is a transacting factor that coordinates Sec insertion by binding to SECIS elements located in selenoprotein mRNAs and recruiting EFSec. The mechanism of selenoprotein synthesis has been the subject of several excellent review articles where it is examined in much greater detail [51 – 54].
SELENIUM DISTRIBUTION IN THE BODY
Postmortem studies in humans have determined that the brain contains roughly 2.5% of total body selenium in comparison to 25–50% for skeletal muscle, 7% for liver, and 4% for kidney [55, 56]. When normalized for tissue weight, the highest Se concentrations are found in the kidney and liver, whereas Se levels in the brain are significantly lower. A similar distribution has been documented in rodents, with highest Se levels being observed in kidney, liver, and testes [57, 58]. Animal studies have also demonstrated that Se is preferentially retained in the brain and reproductive organs upon administration of a Se-deficient diet [58 – 61]. The preservation of Se in the brain under Se-deficient conditions is largely mediated by SelP and ApoER2. Mice lacking either SelP or ApoER2 have diminished Se levels in brain and quickly exhibit severe neurological dysfunction when fed a Se-deficient diet [17, 62]. Within the brain, Se content is highest in regions composed of gray matter and lesser in white matter [63]. Regional analysis of brain tissue from rats fed a Se-adequate diet found that Se concentrations are highest in cerebellum and lowest in brain stem, with intermediate levels observed in forebrain, hindbrain, and hippocampal regions [57]. This study also performed autoradiography on brain sections from Se-deficient rats administered a 75Se radiotracer. High levels of 75Se uptake were observed in the brain ventricles, superior olivary complex, inferior colliculus, dentate and CA3 regions of the hippocampus, olfactory bulb, and the cerebral and cerebellar cortices. Consistent with earlier observations that Se is more concentrated in gray than white matter, this study found that 75Se levels were lowest in regions comprised of white matter, such as the optic tract, cerebral peduncle, and corpus callosum.
SELENOPROTEIN EXPRESSION IN THE BRAIN
The most comprehensive investigation of selenoprotein expression in the adult mouse brain to date was performed by Zhang and colleagues [64]. Using the Allen Brain Atlas, along with supplemental immunohistochemical staining and mRNA analysis, they examined the relative expression of the complete adult murine selenoproteome (24 selenoproteins) in 159 brain regions. This study determined that selenoprotein expression is especially rich in the olfactory bulb, hippocampus, cerebral cortex, and cerebellar cortex, based upon the number of distinct selenoprotein mRNAs that were expressed at high levels. Their analyses also identified the following six selenoproteins as being highly expressed in the mouse brain: Sep15, SelM, SelK, SelP, GPx4 and SelW. The latter three proteins were considered the most abundant, as they were detectable at a high level in greater than 90% of the regions examined. For the sake of brevity, this review will focus upon discussing expression of these three selenoproteins (SelP, GPx4, SelW), supplementing the findings of Zhang and colleagues with both previous and recent work.
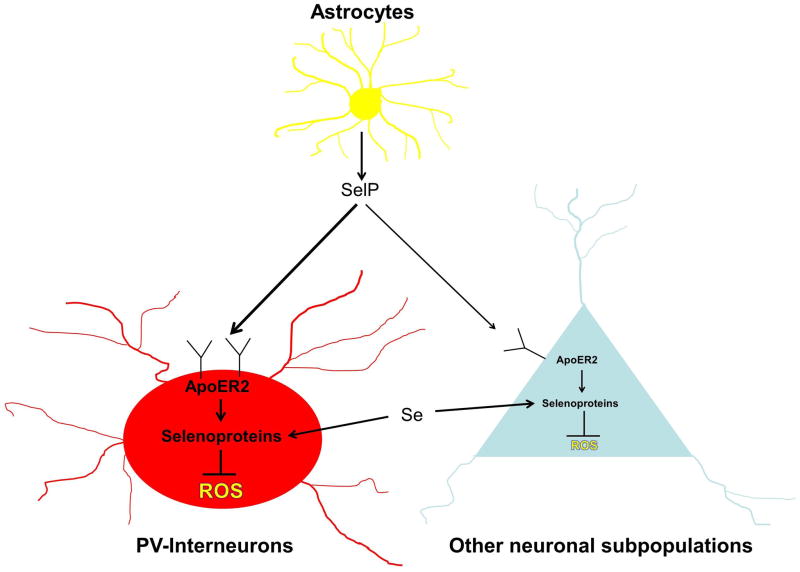
The unique role of SelP as the main supplier of selenium in the brain is reflected by its distinct expression pattern. Zhang and colleagues reported that SelP mRNA was homogenously expressed throughout the mouse brain, a distribution profile suggesting that it is primarily produced in glial cells [64]. This pattern was most distinct within the hippocampus, where SelP exhibited a uniform, diffuse expression pattern and appeared to be absent from neuronal layers where other selenoproteins were highly expressed. Given glial cells’ supportive role in maintaining neuronal function, it seems logical that SelP might be synthesized primarily by glia and later secreted for uptake by surrounding neuronal cells. Additional evidence that SelP is expressed in a manner complementary to that of other selenoproteins comes from analyses of the expression pattern of ApoER2, the primary receptor for SelP in the brain. ApoER2 mRNA was enriched in the four regions identified as having high selenoprotein expression [64], a finding consistent with the notion that ApoER2 facilitates selenium delivery via uptake of SelP. Another study reported that ApoER2 is highly expressed on parvalbumin (PV)-interneurons [65], a class of metabolically active GABAergic inhibitory interneurons that act to synchronize neural activity. PV-interneurons are particularly vulnerable to oxidative stress [66, 67] and selenoproteins are required for the normal development and function of these neurons [68]. Due to their intrinsic metabolic demands, PV-interneurons require greater redox capacity and elevated levels of ApoER2 expression may serve to preferentially sequester SelP to these highly active neurons (Fig. 2). There is also evidence from immunohistochemical studies on human brain tissue that SelP is expressed in specific subsets of neurons, including Purkinje cells of the cerebellum and pyramidal neurons of the cortex and hippocampus [69, 70]. These papers also found that SelP is produced in ependymal cells lining the brain ventricles and by astrocytes. Further studies are required to better clarify SelP expression in brain with respect to developmental time window, condition (baseline vs stress), brain region, and cell type.
Figure 2. Model of SelP-mediated selenium distribution in the brain.
Within the brain, SelP is synthesized and secreted primarily by astrocytes. ApoER2 mediates SelP uptake into neurons and the Se present is utilized in selenoprotein synthesis. Selenoproteins support normal cell function by neutralizing reactive oxygen species (ROS). PV-interneurons represent a class of metabolically active neurons in which ApoER2 is highly expressed. These neurons have been reported to be particularly affected in several mouse models of impaired selenium metabolism in the brain. It also important to note that SelP is not the sole source of Se for neuronal selenoprotein synthesis, as increasing dietary Se intake largely prevents neurological dysfunction and normalizes brain selenoprotein levels in SelP−/− mice.
The importance of GPx4 to neurological development and function has become increasingly apparent since its discovery in 1982 [71]. Unlike cellular GPx1, which is present in both neurons and glia, GPx4 seems to be exclusively expressed in neurons under normal conditions. In pathological states, its expression becomes upregulated in astrocytes [72], suggesting that GPx4 may also have a specialized role in glial function during brain injury. GPx4 exists in three isoforms, which are named according to their subcellular localization: mitochondrial GPx4 (m-GPx4), cytosolic GPx4 (c-GPx4) and nuclear GPx4 (n-GPx4). GPx4 mRNA is present through all stages of rat brain development and exhibits dynamic regional distribution during embryogenesis. In early development, m-GPx4 and c-GPx4 are highly expressed in forebrain, midbrain, and hindbrain regions, whereas n-GPx4 is weakly expressed [72]. The m-GPx4 isoform is critically important for normal brain development, as selective ablation of m-GPx4 with siRNA during in vitro embryogenesis induces apoptosis and impairs hindbrain formation [73]. Following birth, GPx4 is highly expressed in neurons, with most pronounced levels in the cortex, hippocampus, and cerebellum. Measurement of isoform-specific mRNA revealed that both m-GPx4 and c-GPx4 are the predominant transcripts in the adult brain [72].
SelW was originally identified due to its absence in muscle of myopathic lambs with White Muscle disease and was later purified and cloned [74]. It is the smallest mammalian selenoprotein (~10 kDa) and has been found to be highly expressed in the brains of rats [75], mice [64, 76, 77], and primates [78]. Expression of SelW within the brain is largely dependent upon SelP, as reduced levels of SelW mRNA and protein have been documented in mice lacking SelP [76, 77]. A recent immunohistochemical study demonstrated that SelW is highly expressed in neurons of the cortex, hippocampus, and cerebellum; and that within cells it is localized to both the soma and processes [77]. In this study, Western blot analysis of synaptosome fractions revealed that both SelW and GPx4 are present at synapses, as are several proteins involved in selenoprotein synthesis, including SBP2, SPS2, and EFSec. These results suggest that selenoproteins are not only present at synapses, but may be synthesized there as well.
CONSTITUTIVE KNOCKOUT MICE STUDIES
In recent years studies on genetically modified mice have provided major insights into the functional roles of various selenoproteins in the nervous system. Targeted deletion of the Sec tRNA gene was found to cause embryonic lethality in mice, demonstrating that selenoprotein synthesis is required to support mammalian life [79]. Subsequent studies revealed that genetic disruption of the genes encoding GPx4 [80], Txnrd1 [10], and Txnrd2 [11] all resulted in embryonic lethality, showing that these selenoproteins perform essential functions. The first viable selenoprotein knockout mouse targeted the GPx1 gene. Surprisingly, these mice manifested virtually no phenotype under standard laboratory conditions, as they were fertile and showed normal sensitivity to hyperoxia [81]. Subsequent studies revealed that upon challenge with the oxidative stress-inducing agents, paraquat and hydrogen peroxide, GPx1−/− mice were more sensitive than their wild type counterparts [82]. Further work showed that GPx1 promotes neuroprotection in response to oxidative challenge, as the brains of GPx1−/− mice were reported to be more vulnerable to mitochondrial toxin treatment [83], ischemia/reperfusion [84], and cold-induced brain injury [85]. Following the development of GPx1−/− mice, constitutive KO mice for several other selenoproteins were generated that exhibited neurological deficits to varying degrees.
In 2003, two independent groups simultaneously published the first papers describing mice lacking SelP [86, 87]. These initial publications documented neurological dysfunction in SelP−/− mice and showed that Se content and GPx activity were significantly reduced within both the brain and testes. The reported neurological deficits included ataxia, impaired motor coordination, reduced growth, and spontaneous death. Further investigation determined that neurological dysfunction was largely prevented when SelP−/− mice were fed a diet containing Se levels at or above 0.25 mg/kg [88]. Subsequent behavioral studies on SelP−/− mice fed a high Se diet (1 mg/kg) reported mild deficits in motor coordination and spatial learning, in conjunction with alterations in synaptic transmission and plasticity within the hippocampus [89]. Histological examination of SelP−/− mice fed a Se-deficient diet upon weaning revealed extensive neurodegeneration in brain regions associated with auditory and motor function, including the lateral lemniscus, inferior colliculus, red nucleus, and decussation of the cerebellar peduncle [90]. More recent studies on SelP−/− mice fed a Se-adequate diet (0.25 mg/kg) described behavioral impairments in contextual fear extinction, latent, inhibition, and sensorimotor gating [65]. In addition to these behavioral deficits, PV-interneuron density was reduced in the inferior collicus and this corresponded with a regional increase in oxidative stress. Altogether these studies demonstrate that: 1) SelP is particularly important for Se distribution and retention within the brain and 2) much of the neurological dysfunction derived from SelP deficiency can be circumvented by Se supplementation, indicating that Se transport can occur by means other than SelP.
Viable knockout mice for all three deiodinase genes (DIO1, DIO2, DIO3) have also been generated and characterized. DIO1−/− mice were observed to exhibit normal development, growth, and reproductive capacity [91]. These mice displayed elevated serum levels of T4, but levels of T3 and thyroid stimulating hormone (TSH) were normal. Additionally, beginning at 6 weeks of age, body weight was slightly elevated in DIO1−/− mice relative to age-matched controls. The initial characterization of DIO2−/− mice also reported normal development with no gross phenotypic abnormalities, but found elevated circulating levels of TSH and T4, providing evidence that DIO2 plays an important role in feedback regulation of TSH secretion [92]. Moreover, upon cold exposure these mice exhibited hypothermia due to impaired brown adipose tissue thermogenesis, as they survived by compensatory shivering accompanied by acute weight loss [93]. Later studies determined that DIO2−/− mice have hearing impairments and retarded cochlear development, demonstrating a critical function for DIO2 in the auditory system [94]. Transgenic mice lacking functional genes for both DIO1 and DIO2 have also been conceived and analyzed. These mice were observed to be relatively healthy, reproductively viable, and the phenotype largely reflected the sum of DIO1−/− and DIO2−/− mice [95]. However, the neurological impairments were greater than those previously reported for DIO2−/− mice, as double knockout mice exhibited altered brain gene expression and impaired spatial learning in the Morris water maze. The original publication on DIO3−/− mice reported that DIO3 deficiency results in central hypothyroidism, retarded growth, and impaired fertility [96]. These findings demonstrated that DIO3 is required for normal development and function of the thyroid axis. Later studies showed that DIO3 plays an important role in both the auditory and visual system. DIO3−/− mice were found to display auditory deficits and accelerated cochlear differentiation [97], in striking contrast to the cochlear retardation observed in DIO2−/− mice. In addition, survival of cone photoreceptors was reported to be greatly reduced in the retina of DIO3−/− mice, whereas rod photoreceptors remained intact [98]. Furthermore, altered cerebellar morphology and impaired locomotor performance on a vertical pole test were also recently documented in DIO3−/− mice [99]. In sum, these studies on mice lacking functional deiodinase genes illustrate the critical influence of thyroid hormone levels on brain development.
Constitutive knockout mice have also been developed for the structurally related ER-resident thioredoxin-like selenoproteins, Sep15 and SelM. Sep15−/− mice were reported to be viable and fertile with normal brain morphology and no apparent neurological deficits [100]. However, these mice developed cataracts at an early age, indicating an important role for Sep15 in glycoprotein folding in the lens. Mice lacking SelM were found to exhibit mild obesity without any apparent deficits in cognition or motor function [101]. In wild-type mice, SelM was observed to be highly expressed in hypothalamic nuclei involved in energy metabolism and SelM−/− mice displayed reduced leptin sensitivity within the hypothalamus. These findings allude to a functional role for SelM in modulating redox balance in brain regions involved in energy metabolism.
In addition to the papers mentioned above, there have also been published studies that reported no overt neurological deficits in constitutive knockout mice for several other selenoprotein genes. These include reports on mice lacking functional genes for SelK [26], GPx2 [102], GPx3 [103], SelN [104], and SelR [105]. We are currently not aware of any publications on constitutive knockout mice for genes encoding Txnrd3, SelH, SelI, SelO, SelS, SelT, SelV, SelW, or SPS2.
Knockout mice have also been generated for several genes involved in selenium metabolism that do not encode selenoproteins. These include the SelP receptors, ApoER2 and megalin, and the putative Se recycling enzyme, Scly. Knockout mice were produced for the lipoprotein receptors ApoER2 and megalin (Lrp2) prior to any knowledge of their involvement in selenium metabolism. In 1999, ApoER2 was identified as a receptor for reelin [106, 107] and the initial report on ApoER2−/− mice documented reduced fertility and malformation of cell layers within the cortex, hippocampus, and cerebellum of these mice [108]. Subsequent work showed that ApoER2−/− mice have impaired hippocampal synaptic plasticity and cognitive deficits [109]. It was later determined that brain selenium levels are significantly reduced in ApoER2−/− mice and that these mice develop neurological dysfunction and neurodegeneration analogous to SelP−/− mice when fed a Se-deficient diet [17, 62, 90]. Lrp2−/− mice were initially reported to exhibit developmental abnormalities in the lung, kidney, and brain, and to die perinatally from respiratory insufficiency [110]. Later studies provided evidence that megalin participates in SelP uptake, as mutant mice (Lrp2267/267) carrying a missense mutation in the extracellular domain of megalin were found to have reduced GPx activity in brain and kidney [23]. Mice lacking Scly were observed to exhibit mild obesity with no discernible neurological deficits when raised on a Se-adequate diet (0.25 mg/kg) [111, 112]. However, when challenged with a low Se diet (0.08 mg/kg), Scly−/− mice displayed cognitive deficits, had reduced brain GPx activity, and developed metabolic syndrome. These results suggest that Scly-mediated Sec recycling becomes physiologically important only when dietary Se supply is not adequate. To obtain insight into the relationship between Scly and SelP in selenium metabolism, double knockout mice lacking both of the aforementioned genes were generated and characterized [113]. Scly−/−SelP−/− mice required selenium supplementation to survive, and surviving mice exhibited impaired motor coordination, audiogenic seizures, and neurodegeneration in brainstem regions akin to those previously reported for ApoER2−/− and SelP−/− mice fed Se-deficient diets [90]. These results indicate that Scly and Sepp1 work cooperatively to maintain selenoprotein expression in the brain.
CONDITIONAL KNOCKOUT MICE STUDIES
As mentioned previously, constitutive deletion of several selenoprotein-related genes (Sec tRNA, GPx4, Txnrd1, Txnrd2) results in embryonic lethality in mice, demonstrating their required function for proper embryonic development. To further probe the functional role of these genes, conditional knockout mice have been developed utilizing Cre-Lox technology where targeted genetic ablation is restricted to specific cell types. Mice with conditional deletion of Sec tRNA within the liver (Alb-Cre/Trspfl/fl mice) were found to have reduced levels of selenium and GPx activity in kidney, but not brain [114]. Moreover, these mice exhibited no neurological deficits, suggesting that the neurological dysfunctional apparent in SelP−/− mice stems largely from a lack of SelP synthesis within the brain and not impaired synthesis in the liver. Additional studies in which Sec tRNA was conditionally inactivated in the forebrain (CamK-Cre/Trspfl/fl mice) reported that these mice quickly developed severe neurological dysfunction and frequent seizures, and rarely survived past 15 days [68]. Further analyses revealed dramatically reduced cerebral selenoprotein levels and impaired development of PV-interneurons in the hippocampus and cortex. These investigators generated another strain of mice with conditional inactivation of GPx4 (CamK-Cre/GPx4fl/fl mice) and observed a similar, albeit slightly milder, phenotype, as these mice quickly developed similar neurological problems. These mice were also found to exhibit a reduced density of PV-interneurons in the cerebral cortex on postnatal day 13. The density of GABAergic calretinin-expressing interneurons was observed to be normal, indicating that the reduction of PV-interneurons was subtype-specific. These results demonstrated that GPx4 plays an indispensable role in the normal development of PV-interneurons. Further work using conditional inactivation demonstrated that Txnrd1 also plays a major role in neurodevelopment [115]. For these studies, a Nestin-Cre transgenic line was used to create mice (Nestin-Cre/Txnrd1fl/fl) with targeted ablation of Txnrd1 in precursor cells of neuronal and glial lineages. These mice exhibited reduced growth, impaired motor coordination, ataxia, tremor, and cerebellar hypoplasia. When conditional inactivation was restricted to neurons (Tα1-Cre/Txnrd1fl/fl), the phenotype was largely rescued, suggesting an important role for Txnrd1 in glial and/or neuronal precursor cells. In contrast, mice with conditional inactivation of Txnrd2 (Nestin-Cre/Txnrd2fl/fl) in the nervous system did not display any apparent neurological deficits [115].
GENETIC DEFECTS OF SELENOPROTEIN BIOSYNTHESIS IN HUMANS
There have been several reported cases of human patients with certain mutations of the SBP2 gene that exhibit a unique clinical phenotype with multiple symptoms. As SBP2 encodes a factor that is required for selenoprotein synthesis, the reported mutations in humans are characterized by a global reduction in selenoprotein synthesis. In 2005, it was first reported that SBP2 mutations led to abnormal thyroid hormone metabolism, diminished type 2 iodothyronine activity levels, and reduced serum levels of SelP in four affected children from two separate families [116]. A subsequent study documented the case of a 12 year old Brazilian girl with nonsense mutations in the SBP2 gene that displayed altered thyroid metabolism in conjunction with reduced serum levels of Se and SelP [117]. This patient was also obese with a short stature and exhibited profound deficits in motor coordination and mental ability. Additionally, she also showed hearing loss and developed a progressive myopathy reminiscent of that reported in human patients with mutations in the SelN gene [118]. Another study provided an in depth examination of two male patients with SBP2 mutations and reported a similar phenotype, as patients displayed altered thyroid hormone metabolism, SelN-related myopathies, hearing loss, and reduced plasma levels of Se, SelP, and GPx3 [119]. Interestingly, both patients also exhibited the paradoxical metabolic symptoms of increased fat mass in combination with enhanced insulin sensitivity. Impaired spermatogenesis was observed in one male, whereas the other male in the study was prepubertal.
In addition to the reported human cases of SBP2 mutations, the syndrome of progressive cerebello-cerebral atrophy (PCCA) has been linked to mutations in the Sec synthase gene. PCCA is an autosomal recessive inherited disorder that is characterized by profound mental retardation, progressive microcephaly, and severe spasticity in the form of myoclonic and generalized tonic-clonic seizures [120]. Two different Sec synthase mutants were discovered in a cohort of PCCA-affected individuals, but not all PCCA patients carried Sec synthase mutations, suggesting that PCCA can be caused by a number of factors [121]. The observed human Sec synthase mutations were also unable to synthesize Sec in a bacterial complementation assay. Due to the fact that constitutive deletion of Sec tRNA is embryonic lethal in mice, it is likely that the reported individuals with Sec synthase mutations retained the ability to synthesize some selenoproteins at greatly reduced levels. With specific regard to the described PCCA patients who did not carry Sec synthase mutations, it was suggested that these individuals may carry mutations in other genes essential to selenium metabolism, such as tRNA[Ser]Sec, EFSec, SBP2, SPS2, GPx4, and SelP [122]. The authors of the aforementioned study also note the striking parallels between the neurological phenotype of PCCA patients and those reported for a number of transgenic mice strains with impaired selenium metabolism. These strains include constitutive knockout mice for Sepp1 and ApoER2 as well as mice with neuron-specific conditional deletion of Sec tRNA and GPx4.
SELENIUM, SELENOPROTEINS, AND NEUROLOGICAL DISEASE
Alzheimer’s Disease
Alzheimer’s Disease (AD) is a neurodegenerative disorder characterized by an accumulation of amyloid plaques and neurofibrillary tangles within the temporal lobe of the brain. Amyloid plaques consist primarily of aggregrates of β-amyloid (Aβ) proteins [123], whereas neurofibrillary tangles are composed of hyperphosphorylated tau, a protein whose normal function involves the stabilization of microtubules [124]. Oxidative stress is a key contributing factor to AD and, thus, there has been keen interest in the potential of Se supplementation for the prevention and treatment of AD.
Studies performed in cell culture have demonstrated that Se supplementation upregulates Se-dependent pathways and counteracts cell signaling mechanisms known to be dysregulated in AD. In particular, Se treatment has been reported to attenuate γ-secretase activity, reduce Aβ production, increase GPx activity, and prevent Aβ-induced neuronal death [125, 126]. Additional cell culture studies have also found that treatment with Se, in the form of sodium selenate (Na2SeO4), increased the activity of the serine-threonine phosphatase PP2A and reduced tau phosphorylation [127, 128]. Furthermore, increased Aβ-toxicity was observed in neuronal N2A cells when SelP levels were reduced by means of siRNA [129]. These findings are further supported by experiments using neurons derived from transgenic mice. Cultured neurons from GPx1−/− mice were reported to be more susceptible to Aβ-induced oxidative stress, and addition of ebselen, a Se-containing mimetic of GPx, reversed this effect [130]. Moreover, primary hippocampal neurons derived from mice overexpressing GPx4 were observed to be more resistant to Aβ-induced neurotoxicity [131].
In line with the aforementioned in vitro findings, Se supplementation has been found to attenuate AD-related pathology and cognitive deficits in several in vivo rodent models. Sodium selenate treatment reduced tau hyperphosphorylation, prevented neurodegeneration, and improved cognition in transgenic mice expressing several mutant forms of the human tau gene [127, 128]. In another study, of double knock-in transgenic mice expressing human mutations in the amyloid precursor protein and presenilin-1 genes, mice administered a Se-enriched diet were found to have increased total brain GPx activity, lower levels of Aβ plaque disposition, and decreased levels of RNA and DNA oxidation [132]. Also, gene expression analysis of transgenic mice expressing the human mutant presenilin-2 gene revealed significantly reduced expression of SelM, providing a link between SelM and a mutation associated with early-onset AD [133]. Follow-up studies on this observation, reported that Se-supplemented rats overexpressing SelM exhibited diminished β/γ-secretase activity, reduced tau phosphorylation, and increased activation of the ERK signaling pathway relative to Se-supplemented controls [134].
However, in contrast to the evidence presented above supporting the efficacy of Se administration in AD-related cell culture and animal models, there is currently no definitive evidence that Se supplementation is beneficial for the prevention and/or treatment of AD in humans. This topic was recently the subject of an extensive review article [135] in which the authors note the lack of consistency in both the methods and outcomes of the various human clinical trials conducted to date. Several factors likely contribute to the inconsistent results observed in humans. Among these, are significant differences between trials such as the administered form of Se (sodium selenite, sodium selenate, selenomethionine), Se dosage, whether Se was given alone or in combination with other vitamins, the duration of Se supplementation, the inclusion criteria for subjects at trial onset, and the measured outcomes. Currently underway in the United States is the Prevention of Alzheimer’s Disease with Vitamin E and Se (PREADVISE) trial (http://clinicaltrials.gov/show/NCT00040378). This is a double blind, placebo controlled study in which approximately 10,000 men aged 60 and above are supplemented daily with Vitamin E (400 IU) and Se (200 μg selenomethionine). The estimated completion date of this study is August 2014.
Likewise, the results of studies comparing Se levels and/or selenoprotein expression in postmortem brain tissue of AD patients to that of control subjects is extremely mixed [135]. For example, various studies have reported GPx activity to be diminished [136], increased [137, 138], or comparable [140, 141] between AD patients and controls. Of additional relevance, histological studies on human tissue from AD patients have found that SelP colocalizes with neurofibrillary tangles and Aβ plaques [70]. One potential effect of SelP binding to neurofibrillary tangles and Aβ plaques may be diminished Se delivery to neurons, as ApoER2-mediated endocytosis of SelP would be prevented. This would lead to diminished selenoprotein levels in neurons, and presumably less protection against oxidative stress.
Epilepsy
Epilepsy is a condition characterized by the recurrence of seizures in which there is a disruption in normal neuronal activity in the brain [141]. In the clinical literature, there is evidence for an association between low Se status and epilepsy in children [142–146]. Increased lipid peroxidation and reduced GPx activity was also reported in erythrocytes of adult patients with refractory epilepsy, and these parameters were found to be normalized in another cohort of epilepsy patients that received Se supplementation [147]. Similarly, in some cases Se supplementation has been found to attenuate pediatric epilepsy [142, 143]. These clinical findings parallel the observation that Se supplementation prevents the development of seizures in SelP−/− mice. Further studies in animals provide additional evidence for a relationship between Se status and epilepsy. Experiments in rodents have demonstrated that Se-deficiency increases susceptibility to kainate-induced seizures [148] and that sodium selenate supplementation suppresses seizures in several models [149].
Parkinson’s Disease
Parkinson’s Disease (PD) is a progressive neurodegenerative disorder associated with the malfunction and/or death of dopamine producing neurons in the substantia nigra (SN). A pathological hallmark of PD is the formation of Lewy bodies, which consist of aggregates of α-synuclein fibrils, within dopaminergic neurons of the SN. Oxidative stress is a contributing factor to PD, as dopamine metabolism produces hydrogen peroxide and superoxide as byproducts and also spontaneously generates highly reactive quinone molecules [150]. Studies on postmortem tissue showed that GPx4 is expressed in dopaminergic neurons of the SN in both control and PD subjects [151]. This study also found that overall GPx4 levels were significantly reduced in PD patients, but when normalized with respect to surviving cell density, GPx4 expression was higher per dopaminergic neuron in PD samples. Subsequent work showed that SelP is concentrated within the center of Lewy bodies in SN neurons and also expressed in dopaminergic axons and terminals of the putamen [152]. Another study demonstrated that GPx1 is expressed in microglia surrounding concentric Lewy bodies and postulated that GPx1 may be involved in the enzymatic degradation of Lewy bodies [153].
Studies in rodents provide evidence that selenoproteins play an important neuroprotective role in dopaminergic neurons. GPx1−/− mice exhibit increased vulnerability to the neurotoxin, 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP), which is widely used in rodent models to replicate the loss of dopaminergic neurons that is observed in PD [83]. Correspondingly, another publication reported that overexpression of GPx1 in mice attenuated the loss of dopaminergic neurons in response to administration of the neurotoxin 6-hydroxydopamine [154]. Several other rodent studies have also described the neuroprotective effect of Se supplementation against toxicity generated by administration of methamphetamine [155, 156], MPTP [157], and 6-hydroxydopamine [158]. As a whole, these studies demonstrate that selenoproteins mitigate dopaminergic neuron loss in multiple rodent models of acute toxicity thought to be of relevance to PD.
Schizophrenia
Schizophrenia is a neurodevelopmental disorder characterized by a heterogeneous mixture of positive, negative, and cognitive symptoms. A substantial body of work, from both human and animal studies, has established a link between dysfunction of cortical PV-interneurons and cognitive deficits in schizophrenia [159]. Another line of evidence has demonstrated that oxidative stress impairs PV-interneuron development and functionality [66, 67, 160]. Along a similar vein, several studies in selenoprotein knockout mice have reported deficits in PV-interneurons [65, 68, 113], presumably due to diminished antioxidant defense capabilities. Moreover, a microarray study examining gene expression in postmortem prefrontal cortex tissue found that SelP levels were significantly lower in subjects with schizophrenia [161]. A more recent publication reported that expression of selenium-binding protein 1 (SELENBP1) was significantly upregulated in both blood cells and postmortem brain tissue from patients with schizophrenia [162]. SELENBP1 does not encode a selenoprotein and its functional role is not well established. However, one study did find that SELENBP1 physically interacts with GPx1 and that the expression levels of SELENBP1 and GPx1 are inversely related [163]. This finding, in combination with the fact that it binds selenium, suggests that SELENBP1 may reduce levels of free selenium that are available for incorporation into selenoproteins. Thus, decreased neuronal selenoprotein synthesis may be a functional outcome of either SelP downregulation or SELENBP1 upregulation, which in turn may adversely affect PV-interneurons.
CONCLUDING REMARKS
Selenoproteins represent a particularly unique class of proteins that perform functions essential to life. Over the course of the past twenty years, evidence generated from targeted gene deletions in mice has demonstrated that several selenoproteins play important roles in normal brain development. Reports of rare human cases with defects in two genes integrally involved in selenoprotein biosynthesis, SBP2 and Sec synthase, have described individuals with an assortment of deficits that parallel those detailed in various selenoprotein knockout mice. Whereas the indispensable role of selenoproteins in brain development and neuroprotection is well established, there is limited evidence directly linking Se status or specific selenoprotein gene variants to common neurological diseases. For these conditions, Se supplementation may only be beneficial for patients with low Se status and/or compromised functionality of selenoprotein genes.
Table 1.
Summary of selenoproteins known to be involved in brain development (DIO2, DIO3, GPx4, SelP, Txnrd1) and/or highly expressed in the brain (GPx4, Sep15, SelK, SelM, SelP, SelW).
| Gene | Subcellular localization | Main sites of expression in brain | Dietary Se responsiveness | KO mouse phenotype |
|---|---|---|---|---|
| DIO2 | ER membrane | Cochlea, hypothalamus | Moderate | Hearing deficits; Impaired thermogenesis |
| DIO3 | Plasma membrane | Cochlea, cerebellum | Moderate | Hearing deficits; Altered cerebellar morphology and impaired locomotor function |
| GPx4 | Cytosol, Mitochondria, Nucleus | Cerebellum, hippocampus, hypothalamus | Resistant | Embryonic lethal; Seizures and ataxia in CamK- Cre/GPx4fl/fl mice |
| Sep15 | ER lumen | Cerebellum, hippocampus | Moderate | Early onset cataracts; No reported neurological deficits |
| SelK | ER, Plasma membrane | Cerebellum, hippocampus hypothalamus | Moderate | Impaired immune responsiveness; No reported neurological deficits |
| SelM | ER lumen | Olfactory bulb, cerebellum, hypothalamus, cochlea | Moderate | Mild obesity with no apparent cognitive or motor deficits |
| SelP | Secreted | Cerebellum, choroid plexus, olfactory bulb, hippocampus | Responsive | Cognitive and motor deficits; Seizures, ataxia, and neurodegeneration develop when fed Se-deficient diet |
| SelW | Cytosol | Hippocampus, cortex, cerebellum, olfactory bulb | Responsive | No published KO mouse |
| Txnrd1 | Cytosol, Nucleus | Cerebellum, hippocampus | Resistant | Embryonic lethal; Cerebellar hypoplasia and motor deficits in Nestin-Cre/Txnrd1fl/fl mice |
ABBREVIATIONS
- Aβ
amyloid beta
- AD
Alzheimer’s disease
- Cys
cysteine
- DIO
iodothyronine deiodinase
- EFSec
selenocysteine-specific elongation factor
- ER
endoplasmic reticulum
- GPx
glutathione peroxidase
- MPTP
1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine
- PD
Parkinson’s disease
- PV
parvalbumin
- SBP2
selenocysteine insertion sequence-binding protein 2
- Scly
selenocysteine lyase
- Se
selenium
- Sec
selenocysteine
- SECIS
selenocysteine insertion sequence
- SELENBP1
selenium binding protein 1
- SN
substantia nigra
- SPS2
selenophosphate synthetase 2
- T3
3,3′,5-triiodothyronine
- T4
thyroxine
- TSH
thyroid stimulating hormone
- Txn
thioredoxin
- Txnrd
thioredoxin reductase
References
- 1.Rayman MP. Food-chain selenium and human health: emphasis on intake. Br J Nutr. 2008;100:254–268. doi: 10.1017/S0007114508939830. [DOI] [PubMed] [Google Scholar]
- 2.Pinsent J. The need for selenite and molybdate in the formation of formic dehydrogenase by members of the coli-aerogenes group of bacteria. Biochem J. 1954;57:10–16. doi: 10.1042/bj0570010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwarz K, Foltz CM. Selenium as an integral part of factor 3 against dietary necrotic liver degeneration. J Am Chem Soc. 1957;79:3292–3293. [PubMed] [Google Scholar]
- 4.Rotruck JT, Pope AL, Ganther HE, Swanson AB, Hafeman DG, et al. Selenium: biochemical role as a component of glutathione peroxidase. Science. 1973;179:588–590. doi: 10.1126/science.179.4073.588. [DOI] [PubMed] [Google Scholar]
- 5.Cone JE, Del Rio RM, Davis JN, Stadtman TC. Chemical characterization of the selenoprotein component of clostridial glycine reductase: identification of selenocysteine as the organoselenium moiety. Proc Natl Acad Sci USA. 1976;73:2659–2663. doi: 10.1073/pnas.73.8.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forstrom JW, Zakowski JJ, Tappel AL. Identification of the catalytic site of rat liver glutathione peroxidase as selenocysteine. Biochemistry. 1978;17:2639–2644. doi: 10.1021/bi00606a028. [DOI] [PubMed] [Google Scholar]
- 7.Arnér ES. Selenoproteins – What unique properties can arise with selenocysteine in place of cysteine? Exp Cell Res. 2010;316:1296–1303. doi: 10.1016/j.yexcr.2010.02.032. [DOI] [PubMed] [Google Scholar]
- 8.Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, et al. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 9.Gladyshev VN, Jeang KT, Stadtman TC. Selenocysteine, identified as the penultimate C-terminal residue in human T-cell thioredoxin reductase, corresponds to TGA in the human placental gene. Proc Natl Acad Sci USA. 1996;93:6146–6151. doi: 10.1073/pnas.93.12.6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jakopoglu G, Przemeck GK, Schneider M, Moreno SG, Mayr N, et al. Cytoplasmic thioredoxin reductases is essential for embryogenesis but dispensable for cardiac development. Mol Cell Biol. 2005;25:1980–1988. doi: 10.1128/MCB.25.5.1980-1988.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conrad M, Jakupoglu C, Moreno SG, Lippl S, Banjac A, et al. Essential role for mitochondrial thioredoxin reductases in hematopoiesis, heart development, and heart function. Mol Cell Biol. 2004;24:9414–9423. doi: 10.1128/MCB.24.21.9414-9423.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burk RF, Gregory PE. Some characteristics of 75Se-P, a selenoprotein found in rat liver and plasma, and comparison of it with selenoglutathione peroxidases. Arch Biochem Biophys. 1982;213:73–80. doi: 10.1016/0003-9861(82)90441-6. [DOI] [PubMed] [Google Scholar]
- 13.Motsenbacher MA, Tappel AL. A selenocysteine-containing selenium-transport protein in rat plasma. Biochim Biophys Acta. 1982;719:147–153. doi: 10.1016/0304-4165(82)90318-x. [DOI] [PubMed] [Google Scholar]
- 14.Burk RF, Hill KE. Selenoprotein P – expression, functions, and roles in mammals. Biochim Biophys Acta. 2009;1790:1441–1447. doi: 10.1016/j.bbagen.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saito Y, Hayashi T, Tanaka A, Watanabe Y, Suzuki M, et al. Selenoprotein P in human plasma as an extracellular phospholipid hydroperoxide glutathione peroxidase. Isolation and enzymatic characterization of human selenoprotein P. J Biol Chem. 1999;274:2866–2871. doi: 10.1074/jbc.274.5.2866. [DOI] [PubMed] [Google Scholar]
- 16.Hill KE, Zhou J, Austin LM, Motley AK, Ham AJL, et al. The selenium-rich C-terminal domain of mouse selenoprotein P is necessary for the supply of selenium to brain and testis but not for the maintenance of whole body selenium. J Biol Chem. 2007;282:10972–10980. doi: 10.1074/jbc.M700436200. [DOI] [PubMed] [Google Scholar]
- 17.Burk RF, Hill KE, Olson GE, Weeber EJ, Motley AK, et al. Deletion of apolipoprotein E receptor-2 in mice lowers brain selenium and causes severe neurological dysfunction and death when a low selenium diet is fed. J Neurosci. 2007;27:6207–6211. doi: 10.1523/JNEUROSCI.1153-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olson GE, Winfrey VP, Nagdas SK, Hill KE, Burk RF. Apolipoprotein E receptor-2 (ApoER2) mediates selenium uptake from selenoprotein P by the mouse testis. J Biol Chem. 2007;282:12290–12297. doi: 10.1074/jbc.M611403200. [DOI] [PubMed] [Google Scholar]
- 19.Kurokawa S, Hill KE, McDonald WH, Burk RF. Long isoform mouse selenoprotein P (Sepp1) supplies rat myoblast L8 cells with selenium via endocytosis mediated by heparin binding properties and apolipoprotein E receptor-2. J Biol Chem. 2012;287:28717–28726. doi: 10.1074/jbc.M112.383521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burk RF, Hill KE, Winfrey VP, Kurokawa S, Mitchell SL, et al. Maternal-fetal transfer of selenium in the mouse. FASEB J. 2013;27:3249–3256. doi: 10.1096/fj.13-231852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pietschmann N, Rijntjes E, Hoeg A, Stoedter M, Schweizer U, et al. Selenoprotein P is the essential selenium transporter for bones. Metallomics. 2014;6:1043–1049. doi: 10.1039/c4mt00003j. [DOI] [PubMed] [Google Scholar]
- 22.Olson GE, Winfrey VP, Hill KE, Burk RF. Megalin mediates selenoprotein P uptake by kidney proximal tubule epithelial cells. J Biol Chem. 2008;283:6854–6860. doi: 10.1074/jbc.M709945200. [DOI] [PubMed] [Google Scholar]
- 23.Chiu-Ugalde J, Theilig F, Behrends T, Drebes J, Sieland C, et al. Mutation of megalin leads to urinary loss of selenoprotein P and selenium deficiency in serum, liver, kidneys, and brain. Biochem J. 2010;431:103–111. doi: 10.1042/BJ20100779. [DOI] [PubMed] [Google Scholar]
- 24.Shchedrina VA, Zhang Y, Labunskyy VM, Hatfield DL, Gladyshev VN. Structure- function relations, physiological roles, and evolution of mammalian ER-resident selenoproteins. Antioxid Redox Signal. 2010;12:839–849. doi: 10.1089/ars.2009.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429:841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- 26.Verma S, Hoffmann FW, Kumar M, Huang Z, Roe K, et al. Selenoprotein K knockout mice exhibit deficient calcium flux in immune cells and impaired immune responses. J Immunol. 2011;186:2127–2137. doi: 10.4049/jimmunol.1002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang Z, Hoffmann FW, Norton RL, Hashimoto AC, Hoffmann PR. Selenoprotein K is a novel target of m-calpain, and cleavage is regulated by Toll-like receptor-induced calpastatin in macrophages. J Biol Chem. 2011;286:34830–34838. doi: 10.1074/jbc.M111.265520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jurynec MJ, Xia R, Mackrill JJ, Gunther D, Crawford T, et al. Selenoprotein N is required for ryanodine receptor calcium release channel activity in human and zebrafish muscle. Proc Natl Acad Sci USA. 2008;105:12485–12490. doi: 10.1073/pnas.0806015105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferguson AD, Labunskyy VM, Fomenko DE, Arac D, Chelliah Y, et al. NMR structures of the selenoproteins Sep15 and SelM reveal redox activity of a new thioredoxin-like family. J Biol Chem. 2006;281:3536–3543. doi: 10.1074/jbc.M511386200. [DOI] [PubMed] [Google Scholar]
- 30.Korotkov KV, Kumaraswamy E, Zhou Y, Hatfield DL, Gladyshev VN. Association between the 15-kDa selenoprotein and UDP-glucose:glycoprotein glucosyltransferase in the endoplasmic reticulum of mammalian cells. J Biol Chem. 2001;276:15330–15336. doi: 10.1074/jbc.M009861200. [DOI] [PubMed] [Google Scholar]
- 31.Reeves MA, Bellinger FP, Berry MJ. The neuroprotective functions of selenoprotein M and its role in cystolic calcium regulation. Antioxid Redox Signal. 2010;12:809–819. doi: 10.1089/ars.2009.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grumolato L, Ghzili H, Montero-Hadjadje M, Gasman S, Lesage J, et al. Selenoprotein T is a PACAP-regulated gene involved in intracellular Ca2+ mobilization and neuroendocrine secretion. FASEB J. 2008;22:1756–1768. doi: 10.1096/fj.06-075820. [DOI] [PubMed] [Google Scholar]
- 33.Panee J, Stoytcheva ZR, Liu W, Berry MJ. Selenoprotein H is a redox-sensing high mobility group family DNA-binding protein that up-regulates genes involved in glutathione synthesis and phase II detoxification. J Biol Chem. 2007;282:23759–23765. doi: 10.1074/jbc.M702267200. [DOI] [PubMed] [Google Scholar]
- 34.Beilstein MA, Vendeland SC, Barofsky E, Jensen ON, Whanger PD. Selenoprotein W of rat muscle binds glutathione and an unknown small molecular weight moiety. J Inorg Biochem. 1996;61:117–124. doi: 10.1016/0162-0134(95)00045-3. [DOI] [PubMed] [Google Scholar]
- 35.Aachmann FL, Fomenko DE, Soragni A, Gladyshev VN, Dikiy A. Solution structure of selenoprotein W and NMR analysis of its interaction with 14-3-3 proteins. J Biol Chem. 2007;282:37036–37044. doi: 10.1074/jbc.M705410200. [DOI] [PubMed] [Google Scholar]
- 36.Horibata Y, Hirabayashi Y. Identification and characterization of human ethanolaminephosphotransferase1. J Lipid Res. 2007;48:503–508. doi: 10.1194/jlr.C600019-JLR200. [DOI] [PubMed] [Google Scholar]
- 37.Kim HY, Gladyshev VN. Methionine sulfoxide reductases: Selenoprotein forms and roles in antioxidant protein repair in mammals. Biochem J. 2007;407:321–329. doi: 10.1042/BJ20070929. [DOI] [PubMed] [Google Scholar]
- 38.Cao G, Lee KP, van der Wijst J, de Graaf M, van der Kemp A, et al. Methionine sulfoxide reductase B1 (MsrB1) recovers TRPM6 channel activity during oxidative stress. J Biol Chem. 2010;285:26081–26087. doi: 10.1074/jbc.M110.103655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu XM, Carlson BA, Irons R, Mix H, Zhong N, et al. Selenophosphate synthetase 2 is essential for selenoprotein biosynthesis. Biochem J. 2007;404:115–120. doi: 10.1042/BJ20070165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zinoni F, Birkmann F, Stadtman TC, Bock A. Nucleotide sequence and expression of the selenocysteine-containing polypeptide of formate dehydrogenase (formate-hydrogen-lyase-linked) from Escherichia coli. Proc Natl Acad Sci USA. 1986;83:4650–4654. doi: 10.1073/pnas.83.13.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chambers I, Frampton J, Goldfarb P, Affara N, McBain W, et al. The structure of the mouse glutathione peroxidase gene: the selenocysteine in the active site is encoded by the ‘termination codon’ TGA. EMBO J. 1986;5:1211–1227. doi: 10.1002/j.1460-2075.1986.tb04350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mullenbach GT, Tabrizi A, Irvine BD, Bell GI, Hallewell RA. Sequence of a cDNA coding for human glutathione peroxidase confirms TGA encodes the active site selenocysteine. Nucleic Acids Res. 1987;15:5484. doi: 10.1093/nar/15.13.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee BJ, Worland PJ, Davis JN, Stadtman TC, Hatfield DL. Identification of a selenocysteyl-tRNA(Ser) in mammalian cells that recognizes the nonsense codon, UGA. J Biol Chem. 1989;264:9724–9727. [PubMed] [Google Scholar]
- 44.Leinfelder W, Stadtman TC, Bock A. Occurrence in vivo of selenocysteyl-tRNA (SERUCA) in Escherichia coli. Effect of sel mutations. J Biol Chem. 1989;264:9720–9723. [PubMed] [Google Scholar]
- 45.Xu XM, Carlson BA, Irons R, Mix H, Zhong N, et al. Biosynthesis of selenocysteine on its tRNA in eukaryotes. PLoS Biol. 2007;5:e4. doi: 10.1371/journal.pbio.0050004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Esaki N, Nakamura T, Tanaka H, Soda K. Selenocysteine lyase, a novel enzyme that specifically acts on selenocysteine. Mammalian purification and distribution and properties of pig liver enzyme. J Biol Chem. 1982;257:4386–4391. [PubMed] [Google Scholar]
- 47.Berry MJ, Banu L, Chen YY, Mandel JD, Kieffer JD, et al. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature. 1991;353:273–276. doi: 10.1038/353273a0. [DOI] [PubMed] [Google Scholar]
- 48.Small-Howard A, Morozova N, Stoytcheva Z, Forry EP, Mansell JB, et al. Supramolecular complexes mediate selenocysteine incorporation in vivo. Mol Cell Biol. 2006;26:2337–2346. doi: 10.1128/MCB.26.6.2337-2346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Copeland PR, Fletcher JE, Carlson BA, Hatfield DL, Driscoll DM. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 2000;19:306–314. doi: 10.1093/emboj/19.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tujebajeva RM, Copeland PR, Xu XM, Carlson BA, Harney JW, et al. Decoding apparatus for eukaryotic selenocysteine incorporation. EMBO Rep. 2000;1:158–163. doi: 10.1093/embo-reports/kvd033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hatfield DL, Gladyshev VN. How selenium has altered our understanding of the genetic code. Mol Cell Biol. 2002;22:3565–3576. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Squires JE, Berry MJ. Eukaryotic selenoprotein synthesis: Mechanistic insight incorporating new factors and new functions for old factors. IUBMB Life. 2008;60:232–235. doi: 10.1002/iub.38. [DOI] [PubMed] [Google Scholar]
- 53.Donovan J, Copeland PR. Threading the needle: Getting selenocysteine into proteins. Antioxid Redox Signal. 2010;12:881–892. doi: 10.1089/ars.2009.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Flohé L. The labour pains of biochemical selenology: The history of selenoprotein biosynthesis. Biochim Biophys Acta. 2009;1790:1389–1403. doi: 10.1016/j.bbagen.2009.03.031. [DOI] [PubMed] [Google Scholar]
- 55.Oster O, Schmeidel G, Prellwitz W. The organ distribution of selenium in German adults. Biol Trace Elem Res. 1988;14:23–45. doi: 10.1007/BF02990125. [DOI] [PubMed] [Google Scholar]
- 56.Zachara BA, Pawluk H, Bloch-Boguslawska E, Sliwka KM, Korenkiewicz J, et al. Tissue level, distribution, and total body selenium content in healthy and diseased humans in Poland. Arch Environ Health. 2001;56:461–466. doi: 10.1080/00039890109604483. [DOI] [PubMed] [Google Scholar]
- 57.Behne D, Wolters W. Distribution of selenium and glutathione peroxidase in the rat. J Nutr. 1983;113:456–461. doi: 10.1093/jn/113.2.456. [DOI] [PubMed] [Google Scholar]
- 58.Kühbacher M, Bartel J, Hoppe B, Alber D, Bukalis G, et al. The brain selenoproteome: Priorities in the hierarchy and different levels of selenium homeostasis in the brain of selenium-deficient rats. J Neurochem. 2009;110:133–142. doi: 10.1111/j.1471-4159.2009.06109.x. [DOI] [PubMed] [Google Scholar]
- 59.Burk RF, Brown DG, Seely RJ, Scaief C. Influence of dietary and injected selenium on whole-body retention, route of excretion, and tissue retention of 75SeO32- in the rat. J Nutr. 1972;102:1049–1056. doi: 10.1093/jn/102.8.1049. [DOI] [PubMed] [Google Scholar]
- 60.Brown DG, Burk RF. Selenium retention in tissues and sperm of rats fed a torula yeast diet. J Nutr. 1972;102:102–108. doi: 10.1093/jn/103.1.102. [DOI] [PubMed] [Google Scholar]
- 61.Buckman TD, Sutphin MS, Eckhert CD. A comparison of the effects of dietary selenium on selenoprotein expression in rat brain and liver. Biochim Biophys Acta. 1993;1163:176–184. doi: 10.1016/0167-4838(93)90179-u. [DOI] [PubMed] [Google Scholar]
- 62.Burk RF, Hill KE, Motley AK, Winfrey VP, Kurokawa S, et al. Selenoprotein P and apolipoprotein E receptor-2 interact at the blood-brain barrier and also within the brain to maintain an essential selenium pool that protects against neurodegeneration. FASEB J. 2014 Apr 23; doi: 10.1096/fj.14-252874. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Höck A, Demmel U, Schicha H, Kasperek K, Feinendegen LE. Trace element concentration in the human brain. Brain. 1975;98:49–64. doi: 10.1093/brain/98.1.49. [DOI] [PubMed] [Google Scholar]
- 64.Zhang Y, Zhou Y, Schweizer U, Savaskan NE, Hua D, et al. Comparative analysis of selenocysteine machinery and selenoproteome gene expression in mouse brain identifies neurons as key functional sites of selenium in mammals. J Biol Chem. 2008;283:42427–42438. doi: 10.1074/jbc.M707951200. [DOI] [PubMed] [Google Scholar]
- 65.Pitts MW, Raman AV, Hashimoto AC, Todorovic C, Nichols RA, et al. Deletion of selenoprotein P results in impaired function of parvalbumin interneurons and alterations in fear learning and sensorimotor gating. Neuroscience. 2012;208:58–68. doi: 10.1016/j.neuroscience.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cabungcal JH, Nicolas D, Kraftsik R, Cuénod M, Do KQ, et al. Glutathione deficit during development induces anomalies in the rat anterior cingulate GABAergic neurons: relevance to schizophrenia. Neurobiol Dis. 2006;22:624–637. doi: 10.1016/j.nbd.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 67.Behrens MM, Ali SS, Dao DN, Lucero J, Shekhtman G, et al. Ketamine-induced loss of phenotype of fast-spiking interneurons is mediated by NADPH-oxidase. Science. 2007;318:1645–1647. doi: 10.1126/science.1148045. [DOI] [PubMed] [Google Scholar]
- 68.Wirth EK, Conrad M, Winterer J, Wozny C, Carlson BA, et al. Neuronal selenoprotein expression is required for interneuron development and prevents seizures and neurodegeneration. FASEB J. 2010;24:844–852. doi: 10.1096/fj.09-143974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Scharpf M, Schweizer U, Arzberger T, Roggendorf W, Schomburg L, et al. Neuronal and ependymal expression of selenoprotein P in the human brain. J Neural Transm. 2007;114:877–844. doi: 10.1007/s00702-006-0617-0. [DOI] [PubMed] [Google Scholar]
- 70.Bellinger FP, He QP, Bellinger MT, Lin Y, Raman AV, et al. Association of selenoprotein P with Alzheimer’s pathology in human cortex. J Alzheimers Dis. 2008;15:465–472. doi: 10.3233/jad-2008-15313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ursini F, Maiorino M, Valente M, Ferri L, Gregolin C. Purification from pig liver of a protein which protects liposomes and biomembranes from peroxidative degradation and exhibits glutathione peroxidase activity on phosphatidylcholine hydroperoxides. Biochim Biophys Acta. 1982;710:197–211. doi: 10.1016/0005-2760(82)90150-3. [DOI] [PubMed] [Google Scholar]
- 72.Savaskan NE, Borchert A, Bräuer A, Kuhn H. Role for glutathione peroxidase-4 in brain development and neuronal apoptosis: Specific induction of enzyme expression in reactive astrocytes following brain injury. Free Radic Biol Med. 2007;43:191–201. doi: 10.1016/j.freeradbiomed.2007.03.033. [DOI] [PubMed] [Google Scholar]
- 73.Borchert A, Wang CC, Ufer C, Schiebel H, Savaskan NE, et al. The role of phospholipid hydroperoxide glutathione peroxidase isoforms in murine embryogenesis. J Biol Chem. 2006;281:19655–19664. doi: 10.1074/jbc.M601195200. [DOI] [PubMed] [Google Scholar]
- 74.Whanger PD. Selenoprotein W: A review. Cell Mol Life Sci. 2000;57:1846–1852. doi: 10.1007/PL00000666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yeh JY, Beilstein MA, Andrews JS, Whanger PD. Tissue distribution and influence of selenium status on levels of selenoprotein W. FASEB J. 1995;9:392–396. doi: 10.1096/fasebj.9.5.7896009. [DOI] [PubMed] [Google Scholar]
- 76.Hoffmann PR, Hoge SC, Li PA, Hoffmann FW, Hashimoto AC, et al. The selenoproteome exhibits widely varying, tissue-specific dependence on selenoprotein P for selenium supply. Nucleic Acids Res. 2007;35:3963–3973. doi: 10.1093/nar/gkm355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Raman AV, Pitts MW, Hashimoto AC, Bellinger FP, Berry MJ. Selenoprotein W expression and regulation in mouse brain and neurons. Brain and Behavior. 2013;3:562–574. doi: 10.1002/brb3.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gu QP, Sun Y, Ream LW, Whanger PD. Selenoprotein W accumulates primarily in primate skeletal muscle, heart, brain and tongue. Mol Cell Biochem. 2000;204:49–56. doi: 10.1023/a:1007065829068. [DOI] [PubMed] [Google Scholar]
- 79.Bösl MR, Takaku K, Oshima M, Nishimura S, Taketo MM. Early embryonic lethality caused by targeted disruption of the mouse selenocysteine tRNA gene (Trsp) Proc Natl Acad Sci USA. 1997;94:5531–5534. doi: 10.1073/pnas.94.11.5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yant LJ, Ran Q, Rao L, Van Remmen H, Shibatani T, et al. The selenoprotein GPX4 is essential for mouse development and protects from radiation and oxidative damage insults. Free Radic Biol Med. 2003;34:496–502. doi: 10.1016/s0891-5849(02)01360-6. [DOI] [PubMed] [Google Scholar]
- 81.Ho YS, Magnenat JL, Bronson RT, Cao J, Gargano, et al. Mice deficient in cellular glutathione peroxidase develop normally and show no increased sensitivity to hyperoxia. J Biol Chem. 1997;272:16644–16651. doi: 10.1074/jbc.272.26.16644. [DOI] [PubMed] [Google Scholar]
- 82.De Haan JB, Bladier C, Griffiths P, Klener M, O’Shea RD, et al. Mice with a homozygous null mutation for the most abundant glutathione peroxidase, gpx1, show increased susceptibility to the oxidative stress-inducing agents paraquat and hydrogen peroxide. J Biol Chem. 1998;273:22528–22536. doi: 10.1074/jbc.273.35.22528. [DOI] [PubMed] [Google Scholar]
- 83.Klivenyi P, Andreassen OA, Ferrante RJ, Dedeoglu A, Mueller G, et al. Mice deficient in cellular glutathione peroxidase show increased vulnerability to malonate, 3-nitropropinoic acid, and 1-methyl-4-phenyl-1,2,5,6-tetrahydropyridine. J Neurosci. 2000;20:1–7. doi: 10.1523/JNEUROSCI.20-01-00001.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Crack PJ, Taylor JM, Flentjar NJ, de Haan J, Hertzog P, et al. Increased infarct size and exacerbated apoptosis in the glutathione peroxidase-1 (gpx-1) knockout mouse brain in response to ischemia/reperfusion injury. J Neurochem. 2001;78:1389–1399. doi: 10.1046/j.1471-4159.2001.00535.x. [DOI] [PubMed] [Google Scholar]
- 85.Flentjar NJ, Crack PJ, Boyd R, Malin M, de Haan JB, et al. Mice lacking glutathione peroxidase-1 activity show increased TUNEL staining and an accelerated inflammatory response in brain following a cold-induced injury. Exp Neurol. 2002;177:9–20. doi: 10.1006/exnr.2002.7927. [DOI] [PubMed] [Google Scholar]
- 86.Hill KE, Zhou J, McMahan WJ, Motley AK, Atkins JF, et al. Deletion of selenoprotein P alters distribution of selenium in the mouse. J Biol Chem. 2003;278:13640–13646. doi: 10.1074/jbc.M300755200. [DOI] [PubMed] [Google Scholar]
- 87.Schomburg L, Schweizer U, Holtmann B, Flohe L, Sendtner M, et al. Gene disruption discloses the role of selenoprotein P in selenium delivery to target tissues. Biochem J. 2003;370:397–402. doi: 10.1042/BJ20021853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hill KE, Zhou J, McMahan WJ, Motley AK, Burk RF. Neurological dysfunction occurs in mice with targeted deletion of the selenoprotein P gene. J Nutr. 2004;134:157–161. doi: 10.1093/jn/134.1.157. [DOI] [PubMed] [Google Scholar]
- 89.Peters MM, Hill KE, Burk RF, Weeber EJ. Altered hippocampus synaptic function in slenoprotein P deficient mice. Mol Neurodegener. 2006;1:12. doi: 10.1186/1750-1326-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Valentine WM, Abel YW, Hill KE, Austin L, Burk RF. Neurodegeneration in mice resulting from loss of functional selenoprotein P or its receptor apolipoprotein E receptor 2. J Neuropathol Exp Neurol. 2007;67:68–77. doi: 10.1097/NEN.0b013e318160f347. [DOI] [PubMed] [Google Scholar]
- 91.Schneider MJ, Fiering SN, Thai B, Wu SY, St Germain E, et al. Targeted disruption of the type 1 selenodeiodinase gene (Dio1) results in marked changes in thyroid hormone economy in mice. Endocrinology. 2006;147:580–589. doi: 10.1210/en.2005-0739. [DOI] [PubMed] [Google Scholar]
- 92.Schneider MJ, Fiering SN, Pallud SE, Parlow AF, St Germain DL, et al. Targeted disruption of the type 2 selenodeiodinase gene (DIO2) results in a phenotype of pituitary resistance to T4. Mol Endocrinol. 2001;15(12):2137–2148. doi: 10.1210/mend.15.12.0740. [DOI] [PubMed] [Google Scholar]
- 93.De Jesus LA, Carvalho SD, Riberio MO, Schneider M, Kim SW, et al. The type 2 iodothyronine deiodinase is essential for adaptive thermogenesis in brown adipose tissue. J Clin Invest. 2001;108:1379–1385. doi: 10.1172/JCI13803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ng L, Goodyear RJ, Woods CA, Schneider MJ, Diamond E, et al. Hearing loss and retarded cochlear development in mice lacking type 2 iodothyronine deiodinase. Proc Natl Acad Sci USA. 2004;101:3474–3479. doi: 10.1073/pnas.0307402101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Galton VA, Schneider MJ, Clark AS, St Germain DL. Life without T4 to T3 conversion: studies in mice devoid of the 5′ deiodinases. Endocrinology. 2009;150:2957–2963. doi: 10.1210/en.2008-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hernandez A, Martinez ME, Fiering S, Galton VA, St Germain D. Type 3 deiodinase is critical for the maturation and function of the thyroid axis. J Clin Invest. 2006;116:476–484. doi: 10.1172/JCI26240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ng L, Hernandez A, He W, Ren T, Srinivas M, et al. A protective role for type 3 deiodinase, a thyroid hormone-inactivating enzyme, in cochlear development and function. Endocrinology. 2009;150:1952–1960. doi: 10.1210/en.2008-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ng L, Lyubarsky A, Nikonov SS, Ma M, Srinivas M, et al. Type 3 deiodinase, a thyroid-hormone-inactivating enzyme, controls survival and maturation of cone photoreceptors. J Neurosci. 2010;30:3347–3357. doi: 10.1523/JNEUROSCI.5267-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Peeters RP, Hernandez A, Ng L, Ma M, Sharlin DS, et al. Cerebellar abnormalities in mice lacking type 3 deiodinase and partial reversal of phenotype by deletion of thyroid hormone receptor α1. Endocrinology. 2013;154:550–561. doi: 10.1210/en.2012-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kasaikina MV, Fomenko DE, Labunskyy VM, Lachke SA, Qiu W, et al. Roles of the 15-kDa selenoprotein (sep15) in redox homeostasis and cataract development revealed by the analysis of sep15 knockout mice. J Biol Chem. 2011;286:33203–33212. doi: 10.1074/jbc.M111.259218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pitts MW, Reeves MA, Hashimoto AC, Ogawa A, Kremer P, et al. Deletion of selenoprotein M leads to obesity without cognitive deficits. J Biol Chem. 2013;288:26121–26134. doi: 10.1074/jbc.M113.471235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Esworthy RS, Mann JR, Sam M, Chu FF. Low glutathione peroxidase activity in Gpx1 knockout mice protects jejunum crypts from gamma-irradation. Am J Physiol Gastrointest Liver Physiol. 2000;279:G426–G436. doi: 10.1152/ajpgi.2000.279.2.G426. [DOI] [PubMed] [Google Scholar]
- 103.Jin RC, Mahoney CE, Coleman Anderson L, Ottaviano F, Croce K, et al. Glutathione peroxidase-3 deficiency promotes platelet-dependent thrombosis in vivo. Circulation. 2011;123:1963–1973. doi: 10.1161/CIRCULATIONAHA.110.000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rederstorff M, Castets P, Arbogast S, Lainé J, Vassilopoulos S, et al. Increased muscle stress-sensitivity induced by selenoprotein N inactivation in mouse: a mammalian model for SEPN1-related myopathy. PLoS One. 2011;6:e23094. doi: 10.1371/journal.pone.0023094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fomenko DE, Novoselov SV, Natarajan SK, Lee BC, Koc A, et al. MsrB1 (methionine-R-sulfoxide reductase 1) knock-out mice: roles of MsrB1 in redox regulation and identification of a novel selenoprotein form. J Biol Chem. 2009;284:5986–5993. doi: 10.1074/jbc.M805770200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.D’Arcangelo G, Homayouni R, Keshvara L, Rice DS, Sheldon M, et al. Reelin is a ligand for lipoprotein receptors. Neuron. 1999;24:471–479. doi: 10.1016/s0896-6273(00)80860-0. [DOI] [PubMed] [Google Scholar]
- 107.Hiesberger T, Trommsdorff M, Howell BW, Goffinet A, Mumbry MC, et al. Direct binding of Reelin to VLDL receptor and ApoE receptor 2 induces tyrosine phosphorylation of disabled-1 and modulates tau phosphorylation. Neuron. 1999;24:481–489. doi: 10.1016/s0896-6273(00)80861-2. [DOI] [PubMed] [Google Scholar]
- 108.Trommsdorff M, Gotthardt M, Hiesberger T, Shelton J, Stockinger W, et al. Reeler/disabled-like disruption of neuronal migration in knock-out mice lacking the VLDL receptor and ApoE receptor 2. Cell. 1999;97:689–701. doi: 10.1016/s0092-8674(00)80782-5. [DOI] [PubMed] [Google Scholar]
- 109.Weeber EJ, Beffert U, Jones C, Christian JM, Forster E, et al. Reelin and ApoE receptors cooperate to enhance hippocampal synaptic plasticity and learning. J Biol Chem. 2002;277:39944–39952. doi: 10.1074/jbc.M205147200. [DOI] [PubMed] [Google Scholar]
- 110.Willnow TE, Hilpert J, Armstrong SA, Rohlmann A, Hammer RE, et al. Defective forebrain development in mice lacking gp330/megalin. Proc Natl Acad Sci USA. 1996;93:8460–8464. doi: 10.1073/pnas.93.16.8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Raman AV, Pitts MW, Seyedali A, Hashimoto AC, Seale LA, et al. Absence of selenoprotein P but not selenocysteine lyase results in severe neurological dysfunction. Genes Brain Behav. 2012;11:601–613. doi: 10.1111/j.1601-183X.2012.00794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Seale LA, Hashimoto AC, Kurokawa S, Gilman CL, Seyedali A, et al. Disruption of the selenocysteine lyase-mediated selenium recycling pathway leads to metabolic syndrome in mice. Mol Cell Biol. 2012;32:4141–4154. doi: 10.1128/MCB.00293-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Byrns CN, Pitts MW, Gilman CA, Hashimoto AC, Berry MJ. Mice lacking selenoprotein P and selenocysteine lyase exhibit severe neurological dysfunction, neurodegeneration, and audiogenic seizures. J Biol Chem. 2014;289:9662–9674. doi: 10.1074/jbc.M113.540682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schweizer U, Streckfu F, Pelt P, Carlson BA, Hatfield DA, et al. Hepatically derived selenoprotein P is a key factor for kidney but not brain selenium supply. Biochem J. 2005;386:221–226. doi: 10.1042/BJ20041973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Soerensen J, Jakupoglu C, Beck H, Forster H, Schmidt J, et al. The role of thioredoxin reductases in brain development. PLoS One. 2010;3:e1813. doi: 10.1371/journal.pone.0001813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dumitrescu AM, Liao XH, Abdullah MS, Lado-Abeal J, Majed FA, et al. Mutations in SECISBP2 result in abnormal thyroid metabolism. Nat Genet. 2005;37:1247–1252. doi: 10.1038/ng1654. [DOI] [PubMed] [Google Scholar]
- 117.Azevedo MF, Barra GB, Naves LA, Ribeiro Velasco LF, Godoy Garcia Castro P, et al. Selenoprotein-related disease in a young girl caused by nonsense mutations in the SBP2 gene. J Clin Endocrinol Metab. 2010;95:4066–4071. doi: 10.1210/jc.2009-2611. [DOI] [PubMed] [Google Scholar]
- 118.Moghadaszadeh B, Petit N, Jaillard C, Brockington M, Roy SQ, et al. Mutations in SEPN1 cause congenital muscular dystrophy with spinal rigidity and restrictive respiratory syndrome. Nat Genet. 2001;29:17–18. doi: 10.1038/ng713. [DOI] [PubMed] [Google Scholar]
- 119.Schoenmakers E, Agostini M, Mitchell C, Schoenmakers N, Papp L, et al. Mutations in the selenocysteine-binding protein 2 gene lead to a multisystem selenoprotein deficiency disorder in humans. J Clin Invest. 2010;120:4220–4235. doi: 10.1172/JCI43653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ben Zeev B, Hoffman C, Lev D, Watemberg N, Malinger G, et al. Progressive cerebello-cerebral atrophy: a new syndrome with microcephaly, mental retardation, and spastic quadriplegia. J Med Genet. 2003;40:e96. doi: 10.1136/jmg.40.8.e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Agamy O, Ben Zeev B, Lev D, Marcus B, Fine D, et al. Mutations disrupting selenocysteine formation cause progressive cerebello-cerebral atrophy. Am J Hum Genet. 2010;87:538–544. doi: 10.1016/j.ajhg.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schweizer U, Dehina N, Schomburg L. Disorders of selenium metabolism and selenoprotein function. Curr Opin Pediatr. 2011;23:429–435. doi: 10.1097/MOP.0b013e32834877da. [DOI] [PubMed] [Google Scholar]
- 123.Murpy MP, LeVine H., III Alzheimer’s Disease and the β-amyloid peptide. J Alzheimers Dis. 2010;19(1):311–323. doi: 10.3233/JAD-2010-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mandelkow EM, Mandelkow E. Tau in Alzheimer’s Disease. Trends Cell Biol. 1998;8:425–7. doi: 10.1016/s0962-8924(98)01368-3. [DOI] [PubMed] [Google Scholar]
- 125.Tung YT, Hsu WM, Wang BJ, Wu SY, Yen CT, et al. Sodium selenite inhibits gamma-secretase activity through activation of ERK. Neurosci Lett. 2008;440:38–43. doi: 10.1016/j.neulet.2008.05.048. [DOI] [PubMed] [Google Scholar]
- 126.Gwon AR, Park JS, Park JH, Baik SH, Jeong HY, et al. Selenium attenuates Aβ production and Aβ-induced neuronal death. Neurosci Lett. 2010;469:391–395. doi: 10.1016/j.neulet.2009.12.035. [DOI] [PubMed] [Google Scholar]
- 127.Corcoran NM, Martin D, Hutter-Paier B, Windisch M, Nguyen T, et al. Sodium selenate specifically activates PP2A phosphatase, dephosphorylates tau and reverses memory deficits in an Alzheimer’s disease model. J Clin Neurosci. 2010;17:1025–1033. doi: 10.1016/j.jocn.2010.04.020. [DOI] [PubMed] [Google Scholar]
- 128.Van Eersel J, Ke YD, Liu X, Delerue F, Kril JJ, et al. Sodium selenate mitigates tau pathology, neurodegeneration, and functional deficits in Alzheimer’s disease models. Proc Natl Acad Sci USA. 2010;107:13888–13893. doi: 10.1073/pnas.1009038107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Takemoto AS, Berry MJ, Bellinger FP. Role of selenprotein P in Alzheimer’s Disease. Ethn Dis. 2010;20 (1 Suppl 1):S1-92--5. [PMC free article] [PubMed] [Google Scholar]
- 130.Crack PJ, Cimdins K, Ali U, Hertzog PJ, Iannello RC. Lack of glutathione peroxidase-1 exacerbates Aβ-mediated neurotoxicity in cortical neurons. J Neural Transm. 2006;113:645–657. doi: 10.1007/s00702-005-0352-y. [DOI] [PubMed] [Google Scholar]
- 131.Ran Q, Gu M, Van Remmen H, Strong R, Roberts JL, et al. Glutathione peroxidase 4 protects cortical neurons from oxidative injury and amyloid toxicity. J Neurosci Res. 2006;84:202–208. doi: 10.1002/jnr.20868. [DOI] [PubMed] [Google Scholar]
- 132.Lovell MA, Xiong S, Lyubartseva G, Markesbery WR. Organoselenium (Sel-Plex diet) decreases amyloid burden and RNA and DNA oxidative damage in APP/PS1 mice. Free Radic Biol Med. 2009;46:1527–1533. doi: 10.1016/j.freeradbiomed.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hwang DY, Cho JS, Shim SB, Jee SW, Lee SH, et al. Differential expressed genes in transgenic mice carried human mutant presenilin-2 (N141I): Correlation of selenoprotein M with Alzheimer’s Disease. Neurochem Res. 2005;30:1009–1019. doi: 10.1007/s11064-005-6787-6. [DOI] [PubMed] [Google Scholar]
- 134.Yim SY, Chae KR, Shim SB, Hong JT, Park JY, et al. ERK activation induced by selenium treatment significantly downregulates β/γ secretase activity and tau phosphorylation in the trangenic rat overexpressing human selenoprotein M. Int J Mol Med. 2009;24:91–96. doi: 10.3892/ijmm_00000211. [DOI] [PubMed] [Google Scholar]
- 135.Loef M, Schrauzer GN, Walach H. Selenium and Alzheimer’s disease: a systematic review. 2011;26:81–104. doi: 10.3233/JAD-2011-110414. [DOI] [PubMed] [Google Scholar]
- 136.Ansari MA, Scheff SW. Oxidative stress in the progression of Alzheimer’s disease in the frontal cortex. J Neuropathol Exp Neurol. 2010;69:155–167. doi: 10.1097/NEN.0b013e3181cb5af4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lovell MA, Ehmann WD, Butler SM, Markesbery WR. Elevated thiobarbituric acid-reactive substances and antioxidant activity in the brain in Alzheimer’s disease. Neurology. 1995;45:1594–1601. doi: 10.1212/wnl.45.8.1594. [DOI] [PubMed] [Google Scholar]
- 138.Schuessel K, Leutner S, Cairns NJ, Muller WE, Eckert A. Impact of gender on upregulation of antioxidant defense mechanisms in Alzheimer’s disease brain. J Neural Transm. 2004;111:1167–1182. doi: 10.1007/s00702-004-0156-5. [DOI] [PubMed] [Google Scholar]
- 139.Kish SJ, Morito CL, Hornykiewicz O. Brain glutathione peroxidase in neurodegenerative disorders. Neurochem Pathol. 1986;4:23–28. doi: 10.1007/BF02834296. [DOI] [PubMed] [Google Scholar]
- 140.Marcus DL, Thomas C, Rodriguez C, Simberkoff K, Tsai JS, et al. Increased peroxidation and reduced antioxidant enzyme activity in Alzheimer’s disease. Exp Neurol. 1998;150:40–44. doi: 10.1006/exnr.1997.6750. [DOI] [PubMed] [Google Scholar]
- 141.Fisher RS, van Emde Boas W, Blume W, Elger C, Genton P, et al. Epileptic seizures and epilepsy: definitions proposed by the International League Against Epilepsy (ILAE) and the International Bureau for Epilepsy (IBE) Epilepsia. 2005;46:470–472. doi: 10.1111/j.0013-9580.2005.66104.x. [DOI] [PubMed] [Google Scholar]
- 142.Weber GF, Maertens P, Meng XZ, Pippenger CE. Glutathione peroxidase deficiency and childhood seizures. Lancet. 1991;337:1443–1444. doi: 10.1016/0140-6736(91)93130-2. [DOI] [PubMed] [Google Scholar]
- 143.Ramaekers VT, Calomme M, Vanden Berghe D, Makropoulos W. Selenium deficiency triggering intractable seizures. Neuropediatrics. 1994;25:217–223. doi: 10.1055/s-2008-1073025. [DOI] [PubMed] [Google Scholar]
- 144.Ashrafi MR, Shams S, Nouri M, Mohseni M, Shabanian R, et al. A probable causative factor for an old problem: selenium and glutathione peroxidase appear to play important roles in epilepsy pathogenesis. Epilepsia. 2007;48:1750–1755. doi: 10.1111/j.1528-1167.2007.01143.x. [DOI] [PubMed] [Google Scholar]
- 145.Amiri M, Farzin L, Moassesi ME, Sajadi F. Serum trace element levels in febrile convulsion. Biol Trace Elem Res. 2010;135:38–44. doi: 10.1007/s12011-009-8487-6. [DOI] [PubMed] [Google Scholar]
- 146.Seven M, Basaran SY, Cengiz M, Unal S, Yuksel A. Deficiency of selenium and zinc as a causative factor for idiopathic intractable epilepsy. Epilepsy Res. 2013;104:35–39. doi: 10.1016/j.eplepsyres.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 147.Yürekli VA, Nazıroğlu M. Selenium and topiramate attenuates blood oxidative toxicity in patients with epilepsy: a clinical pilot study. Biol Trace Elem Res. 2013;152:180–186. doi: 10.1007/s12011-013-9616-9. [DOI] [PubMed] [Google Scholar]
- 148.Savaskan NE, Bräuer AU, Kühbacher M, Eyüpoglu IY, Kyriakopoulos A, et al. Selenium deficiency increases susceptibility to glutamate-induced excitotoxicity. FASEB J. 2003;17:112–114. doi: 10.1096/fj.02-0067fje. [DOI] [PubMed] [Google Scholar]
- 149.Jones NC, Nguyen T, Corcoran NM, Velakoulis D, Chen T, et al. Targeting hyperphosphorylated tau with sodium selenate suppresses seizures in rodent models. Neurobiol Dis. 2012;45:897–901. doi: 10.1016/j.nbd.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 150.Stokes AH, Hastings TG, Vrana KE. Cytotoxic and genotoxic potential of dopamine. J Neurosci Res. 1999;55:659–665. doi: 10.1002/(SICI)1097-4547(19990315)55:6<659::AID-JNR1>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 151.Bellinger FP, Bellinger MT, Seale LA, Takemoto AS, Raman AV, et al. Glutathione peroxidase 4 is associated with neuromelanin in substantia nigra and dystrophic axons in putamen of Parkinson’s brain. Mol Neurodegener. 2011;6:8. doi: 10.1186/1750-1326-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bellinger FP, Raman AV, Rueli RH, Bellinger MT, Dewing AS, et al. Changes in selenoprotein P in substantia nigra and putamen in Parkinson’s disease. J Parkinsons Dis. 2012;2:115–126. doi: 10.3233/JPD-2012-11052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Power JH, Blumbergs PC. Cellular glutathione peroxidase in human brain: cellular distribution, and its potential role in the degradation of Lewy bodies in Parkinson’s disease and dementia with Lewy bodies. Acta Neuropathol. 2009;117:63–73. doi: 10.1007/s00401-008-0438-3. [DOI] [PubMed] [Google Scholar]
- 154.Bensadoun JC, Mirochnitchenko O, Inouye M, Aebischer P, Zurn AD. Attenuation of 6-OHDA-induced neurotoxicity in glutathione peroxidase transgenic mice. Eur J Neurosci. 1998;10:3231–3236. doi: 10.1046/j.1460-9568.1998.00345.x. [DOI] [PubMed] [Google Scholar]
- 155.Kim HC, Jhoo WK, Choi DY, Im DH, Shin EJ, et al. Protection of methamphetamine nigrostriatal toxicity by dietary selenium. Brain Res. 1999;851:76–86. doi: 10.1016/s0006-8993(99)02122-8. [DOI] [PubMed] [Google Scholar]
- 156.Imam SZ, Newport GD, Islam F, Slikker W, Jr, Ali SF. Selenium, an antioxidant, protects against methamphetamine-induced dopaminergic neurotoxicity. Brain Res. 1999;818:575–578. doi: 10.1016/s0006-8993(98)01311-0. [DOI] [PubMed] [Google Scholar]
- 157.Khan HA. Selenium partially reverses the depletion of striatal dopamine and its metabolites in MPTP-treated C57BL mice. Neurochem Int. 2010;57:489–491. doi: 10.1016/j.neuint.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 158.Zafar KS, Siddiqui A, Sayeed I, Ahmad M, Salim S, et al. Dose-dependent protective effect of selenium in rat model of Parkinson’s disease: neurobehavioral and neurochemical evidences. J Neurochem. 2003;84:438–446. doi: 10.1046/j.1471-4159.2003.01531.x. [DOI] [PubMed] [Google Scholar]
- 159.Lewis DA, Curley AA, Glausier JR, Volk DW. Cortical parvalbumin interneurons and cognitive dysfunction in schizophrenia. Trends Neurosci. 2012;35:57–67. doi: 10.1016/j.tins.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Cabungcal JH, Steullet P, Morishita H, Kraftsik R, Cuenod M, et al. Perineuronal nets protect fast-spiking interneurons against oxidative stress. Proc Natl Acad Sci USA. 2013;110:9130–9135. doi: 10.1073/pnas.1300454110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hakak Y, Walker JR, Li C, Wong WH, Davis KL, et al. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proc Natl Acad Sci USA. 2001;98:4746–4751. doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Glatt SJ, Everall IP, Kremen WS, Corbeil J, Sasik R, et al. Comparative gene expression analysis of blood and brain provides concurrent validation of SELENBP1 up-regulation in schizophrenia. Proc Natl Acad Sci USA. 2005;102:15533–15538. doi: 10.1073/pnas.0507666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Fang W, Goldberg ML, Pohl NM, Bi X, Tong C, et al. Functional and physical interaction between the selenium-binding protein 1 (SBP1) and glutathione peroxidase 1 selenoprotein. Carcinogenesis. 2010;31:1360–1366. doi: 10.1093/carcin/bgq114. [DOI] [PMC free article] [PubMed] [Google Scholar]