Abstract
Human tyrosine hydroxylase activity is regulated by phosphorylation of its N-terminus and by an interaction with the modulator 14-3-3 proteins. We investigated the binding of singly or doubly phosphorylated and thiophosphorylated peptides, comprising the first 50 amino acids of human tyrosine hydroxylase, isoform 1 (hTH1), that contain the critical interaction domain, to 14-3-3ζ, by 31P NMR. Single phosphorylation at S19 generates a high affinity 14-3-3ζ binding epitope, whereas singly S40-phosphorylated peptide interacts with 14-3-3ζ one order-of-magnitude weaker than the S19-phosphorylated peptide. Analysis of the binding data revealed that the 14-3-3ζ dimer and the S19- and S40-doubly phosphorylated peptide interact in multiple ways, with three major complexes formed: 1), a single peptide bound to a 14-3-3ζ dimer via the S19 phosphate with the S40 phosphate occupying the other binding site; 2), a single peptide bound to a 14-3-3ζ dimer via the S19 phosphorous with the S40 free in solution; or 3), a 14-3-3ζ dimer with two peptides bound via the S19 phosphorous to each binding site. Our system and data provide information as to the possible mechanisms by which 14-3-3 can engage binding partners that possess two phosphorylation sites on flexible tails. Whether these will be realized in any particular interacting pair will naturally depend on the details of each system.
Introduction
Members of the 14-3-3 protein family are important modulators of several key signaling pathways that regulate critical biological activities, including progression through the cell cycle, growth, proliferation, and apoptosis (1–3). Extensive proteomics studies have revealed that the 14-3-3 family targets >200 distinct partners, ∼0.6% of the entire cellular proteome (4,5).
In humans, seven different 14-3-3 isoforms (β, γ, ε, η, σ, τ, and ζ) are found, each encoded by a distinct gene, that function as homo- or heterodimers of ∼30-kDa proteins (6,7). Crystal structures of all seven homodimeric mammalian proteins are available. The fold of the monomeric unit contains nine α-helices, arranged in an antiparallel fashion. Within the dimeric structure, two amphipathic grooves, one per monomer, constitute the binding pockets in which the conserved KRRY residues-Lys49, Arg56, Arg127, and Tyr128 in 14-3-3ζ-are positioned for the recognition of a phosphorylated serine or threonine target (8). Three different phosphoserine (pS)- or phosphothreonine-containing sequence motifs can be recognized by 14-3-3 proteins: RSXp(S/T)XP (motif I), RXXXp(S/T)XP (motif II), and a carboxyl-terminal p(S/T)XCOOH motif (motif III), where X is a nonproline amino acid (9–11). Some 14-3-3 family members have also been reported to bind nonphosphorylated peptides as well as motifs that do not strictly display the above canonical sequences (6,11).
Tyrosine hydroxylase (TH) and tryptophan hydroxylase were the first proteins that were identified as binding partners for 14-3-3 proteins (12). TH catalyzes the tetrahydrobiopterin-dependent hydroxylation of L-tyrosine to L-3,4-dihydroxyphenylalanine, which is the rate-limiting step in the biosynthesis of dopamine and other catecholamines (13). In humans, several isoforms of TH are expressed, with isoform 1 (hTH1; 497 residues, NP-000351.2) being the best characterized (14–16). hTH1 can be phosphorylated at T8 and/or three serine residues (S19, S31, and S40) by several protein kinases (17) on its highly dynamic 50-residue N-terminal region that precedes the regulatory ACT domain, characteristic of the aromatic amino-acid hydroxylase family. 14-3-3ζ and other isoforms have been reported to bind to S19 phosphorylated TH (18–20). Although not strictly exhibiting the canonical binding motifs, recent crystallographic analysis of the complex between hTH1 S19-phosphorylated N-terminal peptide (1–43 residues) and 14-3-3γ revealed that the phosphate interacts with the KRRY motif in the 14-3-3γ binding grove, and the region surrounding S19 engages in similar interactions as seen for peptides containing canonical motifs I and II (21). In addition, phosphorylation of hTH1 on S40 stimulates TH activity (17), and induces binding to the yeast 14-3-3 proteins BMH1 and BMH2 and to sheep brain 14-3-3 proteins, but not to 14-3-3ζ (18,19). On the other hand, 14-3-3ζ has been reported to interact with doubly S19- and S40-phosphorylated hTH1 (18). In a recent study (22) we have shown that the doubly phosphorylated hTH1 binds 14-3-3γ with similar stoichiometry and affinity as the singly phosphorylated enzyme on Ser19. However, 14-3-3γ interfered with the phosphorylation of TH1-pS19 on Ser40, supporting that Ser40 becomes inaccessible in the hTH1:14-3-3γ complex, although Ser40 does not significantly contribute to the binding of 14-3-3γ. Therefore the question of whether S40 phosphorylated hTH1 can be bound by 14-3-3 proteins has not been settled unequivocally.
The 14-3-3 proteins are thought to function by inducing a conformational change in the target proteins, thereby modulating the target proteins’ activities. In this way, 14-3-3 family members impart additional control onto key cellular enzymes, in addition to common enzymatic control mechanisms such as feedback loops or product inhibition. The contemporary prevalent model for the binding of doubly phosphorylated peptides by 14-3-3 suggests that one phosphorylated residue of the target protein engages 14-3-3 first, linking the two proteins together, followed by binding of the second phosphorylated residue, to effect the conformational change (23). The notion that binding of 14-3-3 to two phosphorylation sites is required to induce the activity altering conformational change, has led some authors to consider 14-3-3 as a binary computer (4,24).
We explored this model of 14-3-3 modulation using 31P NMR in a simplified peptide system. We investigated the binding of hTH1 peptides (region 1–50) with a single phosphorylation at S19 (pS19) or S40 (pS40) to 14-3-3ζ and determined individual binding affinities. We found that 14-3-3ζ can interact with the hTH1 peptide through either phosphorylated-S19 or -S40, although the binding affinity of 14-3-3ζ for the pS40 peptide is significantly lower than that for the pS19 peptide. Furthermore, binding of 14-3-3ζ to the doubly phosphorylated hTH1 peptide exhibits a complex binding profile involving three major complexes. Our results suggest that when considering 14-3-3 interactions with multiply phosphorylated targets in general, and hTH1 in particular, avidity considerations should be taken into account.
Methods
Cloning, expression, and purification of hTH1 peptides and 14-3-3ζ
The coding region for residues 1–50 of human tyrosine hydroxylase (hTH1–50) (Fig. 1 A) was inserted into a modified pET32a vector, in frame with N-terminal thioredoxin and 6×His tags, followed by a tobacco etch virus (TEV) cleavage site. A codon-optimized gene for 14-3-3ζ (GenScript, Piscataway Township, NJ) was inserted into pET15b with Cys25 and Cys189 codons converted to Ala using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). The DNA sequence for all constructs was confirmed by sequencing (Genewiz, South Plainfield, NJ).
Figure 1.
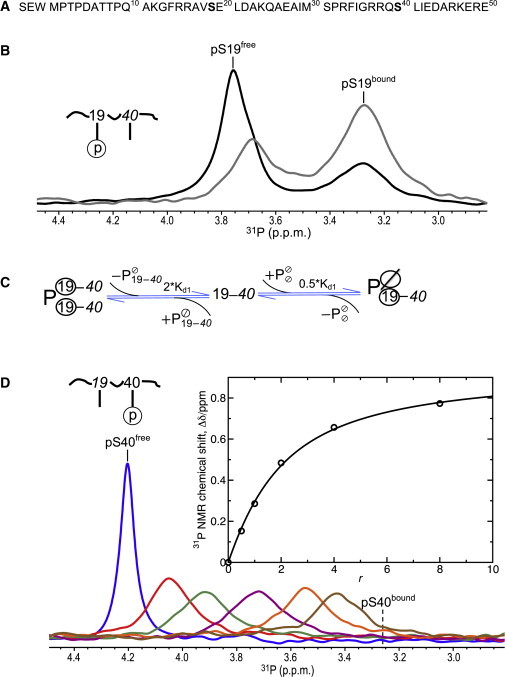
Binding of singly phosphorylated hTH1–50 peptides to 14-3-3ζ. (A) Amino-acid sequence of the hTH1–50 peptide with S19 and S40 highlighted (boldface). (B) 31P spectra of pS19-hTH1–50 at 0.83 mM peptide concentration and molar ratios of 14-3-3ζ to peptide (r) 0.5:1 (black) and 1:1 (gray). (C) Schematic depiction of pS19-hTH1–50 (denoted by 19–40) binding to the 14-3-3ζ homodimer (free protein dimer is depicted by , where Ø indicates an empty phosphoserine binding site). (Circled) Bound pS19 residues. (D) 31P spectra of pS40-hTH1–50 at 0.71 mM peptide concentration and molar ratios (r) of 14-3-3ζ to peptide of 0:1 (blue), 0.5:1 (red), 1:1 (dark green), 2:1 (purple), 4:1 (orange), and 8:1 (brown). (Inset) Chemical shift changes upon 14-3-3ζ binding (Δδ) are plotted as a function of molar ratio (r). The curve obtained by fitting Δδ to the quadratic equation shown in Eq. 2 is shown. To see this figure in color, go online.
Proteins were produced in Escherichia coli BL21(DE3) cells that were grown at 37°C in LB media to an OD600 of 0.8 and induced with 0.5 mM IPTG for 18 h at 18°C. Cells were harvested, resuspended in 50 mM TBS (50 mM Tris, 150 mM NaCl, pH 8.0) buffer, and lysed with a microfluidizer (Microfluidics, Westwood, MA). The cell lysate was clarified by centrifugation (21,000g for 60 min at 4°C), and the supernatant was loaded onto a HisTrap column (GE Healthcare, Piscataway, NJ). Proteins were eluted using a linear imidazole gradient (0.05–1 M in a total volume of 15 mL).
Fractions containing tagged hTH1 peptide were treated with TEV (1 mg/20 mg of peptide) at 4°C overnight, and applied to a Vydac reverse-phase C4 column (Grace Davison Discovery Sciences, Deerfield, IL). hTH1–50 was eluted from the reverse phase column using a linear gradient of 10–40% acetonitrile in 50 mL. Peptide-containing samples were further purified by gel filtration through a Superdex75 26/60 column (GE Healthcare) equilibrated in 20 mM Tris buffer, pH 7.5. The Trp-containing three amino-acid cloning artifact at the N-terminus of the hTH1–50 peptide (Fig. 1 A) allowed for detection during purification and concentration determination at 280 nm.
Fractions containing 14-3-3ζ were dialyzed into TBS buffer and treated with TEV overnight. Undigested protein, 6×His tag cleavage products, and TEV protease were removed by passing the sample over the HisTrap column for a second time and the flow-through containing untagged 14-3-3ζ was collected. The sample was further purified over anion exchange (HiTrap Q HP; GE Healthcare) followed by gel filtration on a Superdex75 26/60 column (GE Healthcare), equilibrated in 20 mM sodium phosphate buffer, pH 7.0.
The Cys25Ala/Cys189Ala 14-3-3ζ variant exhibited essentially the same stability as wild-type 14-3-3ζ protein, as evidenced by identical differential scanning calorimetry unfolding profiles.
Phosphorylation of hTH1–50
The serine residue at position 40 of the hTH1–50 peptide was phosphorylated using the catalytic subunit of cAMP-dependent protein kinase A (PKA; New England BioLabs, Ipswich, MA), whereas Ser19 was phosphorylated using p38 regulated/activated protein kinase (PRAK; obtained from the protein phosphorylation unit, University of Dundee, Dundee, Scotland). For phosphorylation at S19, the reaction mixture, containing hTH1–50 S40A peptide in 50 mM Tris buffer, pH 7.5, 1 mM ATP, 10 mM MgCl2, and PRAK kinase (50 μg for 10 mg of hTH1–50 S40A), was incubated for three days at 30°C. For phosphorylation at S40, hTH1–50 was incubated with PKA (0.5 μg for 15 mg hTH1–50) in 50 mM Tris buffer, pH 7.5,10 mM MgCl2, 200 μM ATP at 30°C for 2 h. Reaction products were purified by gel filtration using a Superdex30 16/60 column (GE Healthcare) followed by anion exchange (HiTrap Q HP; GE Healthcare).
Doubly phosphorylated peptide was prepared using S40 phosphorylated hTH1–50 peptide (pS40-hTH1–50) and PRAK kinase, as described above. Approximately 70% of pS40-hTH1–50 was converted to the doubly phosphorylated product (pS19pS40-hTH1–50), which was purified as described above. For thiophosphorylation, adenosine 5′-[γ-thio]triphosphate (ATPγS) was added to the PKA reaction mixture, instead of ATP. In all cases, the correct masses and purity of the final products were verified by matrix-assisted laser desorption/ionization-time of flight mass spectrometry.
31P NMR titrations
All 31P NMR spectra were recorded at 37°C on an AVANCE 600 MHz NMR spectrometer equipped with a broadband frequency probe (Bruker, Billerica, MA). Samples containing 14-3-3ζ and singly or doubly phosphorylated hTH1–50 peptides were prepared in 20 mM sodium phosphate, pH 7.0, at a constant peptide concentration (0.83 or 0.71 mM) with molar ratios of 14-3-3ζ to peptide of 0:1–8:1.
Modeling of binding scenarios
Binding schemes for the different complexes between 14-3-3ζ and singly or doubly phosphorylated peptides are presented in Fig. 1 C and Fig. 2 B, respectively. Equations for individual binding equilibria and equations describing mass conservation are provided in Fig. S1 and Fig. S2 in the Supporting Material. The equations were solved numerically using the software MATLAB (The MathWorks, Natick, MA) and results yield populations of the individual species as a function of the 14-3-3ζ to peptide molar ratios, r. Three different sets of solutions were obtained, only one of which was physically meaningful given that populations cannot be negative for any r value ≥0.
Figure 2.
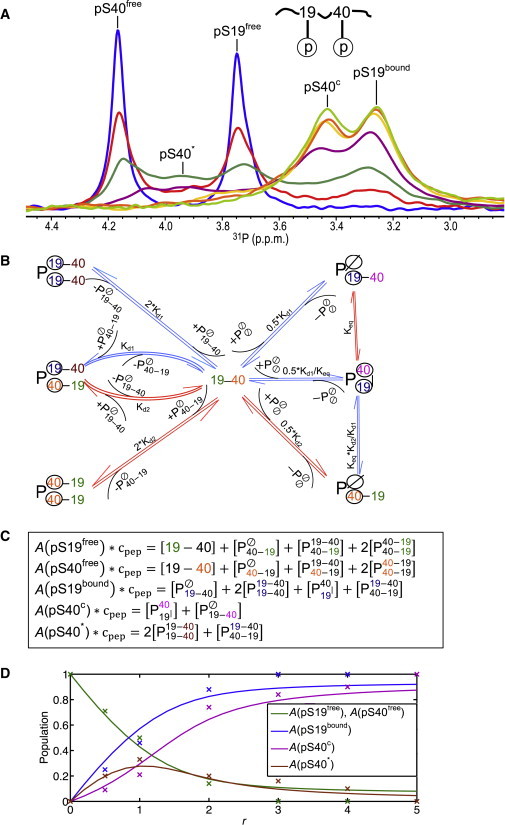
Binding of doubly phosphorylated peptide to 14-3-3ζ. (A) 31P spectra of pS19pS40-hTH1–50 at 0.83 mM and the molar ratio of 14-3-3ζ to the peptide (r) was 0:1 (blue), 0.5:1 (red), 1:1 (dark green), 2:1 (purple), 3:1 (yellow), 4:1 (orange), or 5:1 (light green). (B) Binding scheme of pS19pS40-hTH1–50 (denoted by 19–40) with respect to the 14-3-3ζ homodimer (). Dissociation constants Kd1 and Kd2 correspond to the values determined for the singly phosphorylated peptides. (Blue arrows) Equilibria for which slow exchange on the 31P chemical shift scale is observed; (red arrows) equilibria for which fast exchange on the 31P chemical shift scale is observed. (Circled) Bound phosphorous groups. Phosphorous groups contributing intensity to the same peak in the NMR spectra are shown in the same color. (C) Equations governing the relative peak intensities (denoted as “A”), based on the scheme depicted in panel B; each phosphopeptide contributes to one or more of the five peaks. The specific phosphoserine that contributes to intensity of a particular peak is colored in the same color as in panel B. (D) Populations of phosphoserine species that contribute to the experimentally observed peak intensities (x) for different 14-3-3ζ to peptide molar ratios, r. (Continuous lines) For comparison, the theoretically predicted populations, based on the binding model in panel B using Kd1 = 0.15 mM, Kd2 = 1.0 mM, and Keq = 1.5, are plotted. To see this figure in color, go online.
Results
Binding of singly phosphorylated hTH1 peptides to 14-3-3ζ
Binding between 14-3-3ζ and the phosphorylated N-terminus of hTH1 was probed by 31P NMR. hTH1 peptides, containing the first 50 residues (hTH1–50), were prepared and each serine in the peptide was phosphorylated individually, generating pS19-hTH1–50 and pS40-hTH1–50. PKA selectively and specifically phosphorylated only Ser40, but PRAK phosphorylated both Ser19 and Ser40 (J. Hritz and A.M. Gronenborn, unpublished results). Therefore, we introduced the S40A change, permitting the preparation of singly phosphorylated pS19-hTH1–50 peptide. Titration of 14-3-3ζ into a solution containing pS19-hTH1–50 revealed the system to be in slow exchange on the NMR chemical shift timescale. Thus, resonances for both free and the 14-3-3ζ-bound peptide can be observed separately (Fig. 1 B), and the relative fractions of the free and bound peptide can be determined from resonance intensities (25,26). A dissociation constant (Kd1) of 0.15 ± 0.03 mM for the 14-3-3ζ/pS19-hTH1–50 complex was derived, using
| (1) |
where cpep, A(pS19free), and A(pS19bound) comprise the total peptide concentration, and the free and bound fractions of the pS19-hTH1–50 peptide, respectively, with r the molar ratio of 14-3-3ζ to peptide. For both singly phosphorylated peptides, dissociation constants are reported for the 14-3-3ζ monomer, assuming that binding to a single site in the 14-3-3ζ homodimer is not influenced by binding to the second site.
To analyze our binding data, we generated a scheme for the interaction between pS19-hTH1–50 and the 14-3-3ζ dimer (Fig. 1 C). A pS19-hTH1–50 peptide (represented by 19–40 in Fig. 1 C) can bind to the free 14-3-3ζ dimer (depicted by in Fig. 1 C) at two different sites, with the dissociation constant being one-half of the observed Kd1; the binding of a second pS19-hTH1–50 peptide to the 14-3-3ζ dimer in which one site is already occupied (depicted by in Fig. 1 C) can only occur to the unoccupied site, and dissociation from a doubly occupied 14-3-3ζ dimer (depicted by in Fig. 1 C) can occur from either site, resulting in 2∗Kd1. The equations describing these two equilibria, taking mass conservation of the peptide and 14-3-3ζ into account, are presented in Fig. S1 in the Supporting Material. Based on this model, in a 1:1 mixture of pS19-hTH1–50 and 14-3-3ζ (monomer concentration) with a Kd1 of 0.15 mM, 34.4% of the peptide would be in the free state, 22.6% of peptide in the bound form, and 43% in the bound form.
In contrast to the result with the pS19 peptide, titration of 14-3-3ζ into a solution containing the pS40-hTH1–50 peptide revealed fast exchange on the NMR chemical shift scale, resulting in a single resonance at the weighted averaged chemical shifts of both the free and bound resonances (Fig. 1 D). By fitting the chemical shift change (Δδ) to Eq. 2, a Kd2 value of 1.0 ± 0.1 mM and a maximum chemical shift difference between free and bound peptide (Δδmax) of 0.94 p.p.m. were determined for the binding of 14-3-3ζ to pS40-hTH1–50:
| (2) |
These values allowed us to calculate the 31P bound chemical shift, δ(pS40bound), which was 3.26 p.p.m. (Fig. 1 D).
Interestingly, the bound 31P chemical shifts for both singly phosphorylated hTH1–50 peptides are essentially identical (3.26 p.p.m., Fig. 1, B and D), irrespective of the different neighboring amino acids around the phosphorylated Ser in the hTH1–50 peptide. This suggests that the binding of the phosphorylated residues to the identical binding pockets of 14-3-3ζ that are lined by residues Lys49, Arg56, Arg127, and Tyr128 is the major determinant of the chemical shift, irrespective of the local peptide sequence. We speculate that the side chain of Tyr128 most likely causes the upfield shift of the pS19 and pS40 31P resonances.
Binding of doubly phosphorylated hTH1 peptide to 14-3-3ζ
Binding between doubly phosphorylated proteins and 14-3-3 has previously been characterized using various methods, including isothermal titration calorimetry and fluorescence polarization (19,27,28). Although these methods provide overall binding properties, such as Kd or stoichiometry, important details for a system involving a homodimer and two binding motifs on the target are lacking. For example, answers to the question of how individual phosphorylated amino acids interact with 14-3-3, and what kind of complexes are possible, are still incomplete. 31P NMR is ideally suited to provide such information, since individual 31P signals can be monitored throughout the course of titration experiments between various interacting partners. We therefore prepared doubly phosphorylated hTH1–50 peptide (pS19pS40-hTH1–50) and monitored 14-3-3ζ binding to this peptide by 31P NMR. Our data reveal that a number of different bound species are present during the course of the titration (Fig. 2 A). The spectrum of free pS19pS40-hTH1–50 (blue spectrum in Fig. 2 A; r = 0) contains two well-separated resonances, at 3.76 p.p.m. and at 4.18 p.p.m. for the phosphorous atoms on pSer19 (pS19free) and pSer40 (pS40free), respectively. At the end of the titration (light green trace in Fig. 2 A; r = 5), the pS19pS40-hTH1–50 peptide is saturated with 14-3-3ζ, and resonances at 3.26 p.p.m. and 3.43 p.p.m. for the phosphorous atoms on pSer19 (pS19bound) and pSer40 (pS40c), respectively, are observed. The bound resonances are broader than those of the free peptide, reflecting the slower rotational correlation time of the peptide/14-3-3ζ complex (molecular mass ∼62-67 kDa).
The chemical shift of 3.43 p.p.m. is different from the completely saturated position (3.26 p.p.m.) obtained with a peptide that contains only phosphorylated Ser40 (pS40-hTH1–50, Fig. 1 D), indicating that this bound resonance position contains a contribution from the free pS40 resonance (due to fast exchange on the chemical shift scale). This signal, therefore, is designated pS40c, instead of pS40bound. During the titration, the intensities of both the pS19free and pS40free resonances decrease to the same degree with increasing 14-3-3ζ concentration, but only the bound pS19 31P resonance at 3.26 p.p.m. increases in intensity at the same rate (Fig. 2 A, e.g., red spectrum). The bound pS40 31P resonance at 3.43 p.p.m. is only visible for excess 14-3-3ζ over peptide (e.g., purple, yellow, orange, and light-green spectra). These data suggest that for excess peptide and little 14-3-3ζ (r < 1), the phosphorous group on Ser19 (strong binder) is the one predominantly bound by 14-3-3ζ, and two pS19pS40-hTH1–50 peptides are likely bound by one 14-3-3ζ dimer. For conditions where 14-3-3ζ is in excess (r > 1), both phosphorous groups on a single peptide can be bound by one 14-3-3ζ dimer.
The complete binding scenario, considering six possible complexes for the interaction between doubly phosphorylated peptide (free peptide is depicted as 19–40) and 14-3-3ζ (free 14-3-3ζ dimer is depicted as ), is presented in Fig. 2 B. Based on this interaction scenario and taking into account that resonances can be in slow or fast exchange on the chemical shift scale (marked by blue and red arrows, respectively), five different resonances are expected to be observable in the 31P NMR spectra (see below). Phosphorous groups that exhibit identical resonance frequencies, thus contributing intensity to the same peak, are labeled in the same colors in Fig. 2, B and C. The derived binding scheme is based on two assumptions:
-
1.
The bound 31P chemical shifts of pS19 and pS40 are identical when saturated with 14-3-3ζ, irrespective of whether the second binding site on the 14-3-3ζ dimer is occupied or not; and
-
2.
The individual phosphorous resonances in pS19pS40-hTH1–50 exhibit the same NMR exchange regime as those for the singly phosphorylated peptides (Fig. 1, B and D, i.e., the pS19 resonance is in slow exchange and that of pS40 is in fast exchange on the 31P NMR shift scale).
Note that the slow or fast exchange regimes apply not only to the interaction between free peptide (19–40) and the one site occupied 14-3-3ζ dimer complex but also to various additional 14-3-3ζ complexes (Fig. 2 B). For example, the pS40 phosphorous resonance of , the complex in which both sites on the protein dimer engage the two phosphate groups on 19‒40, is in fast exchange with that of , the complex in which one site on the dimer engages the pS19 phosphate group of the peptide, since very weak binding of the phosphate group of pS40 is involved. On the other hand, and are in slow exchange, since the exchange involves tight binding of the phosphate group of pS19. In agreement with the above binding scheme, five distinct peaks are observed in the 31P spectra during the titration (Fig. 2 A).
Multiple complexes contribute to the intensity of each phosphorous resonance. All peptides with the pS19 phosphate group free of 14-3-3ζ protein (19–40, ) contribute to the peak labeled “pS19free” (19 colored green in Fig. 2, B and C). Free peptide (19‒40) as well as any peptides bound to 14-3-3ζ via the pS40 phosphate group (40 colored orange in Fig. 2, B and C) will contribute to the peak labeled “pS40free” in Fig. 2 A. Given that the same peptide can contribute intensity to two different resonances (pS19free and pS40free), the total intensity of the two resonances is expected to be similar, as is indeed observed in the 31P spectra (Fig. 2 A).
Any complexes of 14-3-3ζ that contain peptide bound via the phosphate group on pS19 (, , , and ; 19 colored blue in Fig. 2, B and C) add intensity to the peak labeled “pS19bound.” The pS40c peak arises from the pS40 phosphate groups in the and complexes (40 colored purple in Fig. 2, B and C), which are in fast exchange. The contribution from the free pS40 phosphate group in causes the “pS40c” resonance to slightly shift toward the free position (4.18 p.p.m.) and, therefore, it is observed at 3.43 p.p.m., slightly downfield from the bound position (3.26 p.p.m.) of the singly phosphorylated pS40 peptide (Fig. 1 D). In contrast, the position of the pS19bound peak is at 3.27 p.p.m., identical to the bound position of the singly phosphorylated pS19 peptide (Fig. 1 B). The reason for this is that the phosphate groups that contribute to the pS19bound peak are in slow exchange, thus no weighted chemical shift averaging comes into play.
Finally, the free pS40 phosphate groups in the and complexes (40 colored brown in Fig. 2, B and C), denoted pS40∗, reside on the peptide that is bound to 14-3-3ζ via pS19, but they are not buried in the 14-3-3ζ binding site and are free in solution, surrounded by solvent. Note that pS40∗ is observed as a distinct peak at 3.93 p.p.m., in slow exchange with the pS40 phosphate resonance (4.18 p.p.m.) of the free peptide (Fig. 2 A), because the peptides in these complexes are bound to 14-3-3ζ via the pS19 phosphate group.
The relationships listed in Fig. 2 C imply that the total normalized area (denoted as “A”) of all peaks in Fig. 2 A corresponds to 2, because the peptide contains two phosphate groups:
| (3) |
Assignment of phosphorous resonances by selective thiophosphorylation
The 31P resonance assignment of the pS19 and pS40 in the pS19pS40-hTH1–50 peptide (Fig. 2 A) was initially obtained using the assignment of singly phosphorylated hTH1–50 peptides (Fig. 1, B and D). We confirmed the assignment using a hTH1–50 peptide that was phosphorylated on Ser19 and thiophosphorylated on Ser40 (pS19tpS40-hTH1–50). Thiophosphorylation causes a large downfield shift of the 31P resonance from ∼4.2 p.p.m. (blue spectra in Fig. 1 D and Fig. 2 A) to 43.95 p.p.m. (blue spectra in Fig. 3, A and B). Thus, in the spectrum of the pS19tpS40-hTH1–50 peptide (Fig. 3 B), both resonances are clearly separated.
Figure 3.
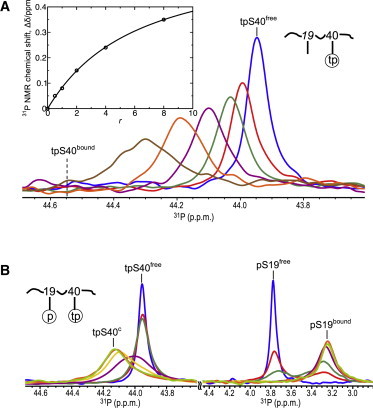
Binding of thiophosphorylated hTH1–50 peptides to 14-3-3ζ. (A) 31P spectra of tpS40-hTH1–50 peptide (0.71 mM) for varying molar ratios of 14-3-3ζ to the peptide (r); 0:1 (blue), 0.5:1 (red), 1:1 (dark green), 2:1 (purple), 4:1 (orange), and 8:1 (brown). (Inset) The chemical shift changes upon 14-3-3ζ binding (Δδ) are plotted as a function of molar ratio (r). (Continuous line) Predicted Δδ from fitting the experimental Δδ to the quadratic equation shown in Eq. 2. (B) 31P spectra of pS19tpS40-hTH1–50 (0.83 mM) for varying molar ratios of 14-3-3ζ to the peptide (r): 0:1 (blue), 0.5:1 (red), 1:1 (dark green), 2:1 (purple), 3:1 (yellow), 4:1 (orange), and 5:1 (light green). To see this figure in color, go online.
A thiophosphate group is assumed to functionally mimic a phosphate group, given its close chemical structure. To confirm that no gross changes to 14-3-3ζ binding were introduced into the system by thiophosphorylation, we carried out titration experiments with tpS40-hTH1–50 and 14-3-3ζ under the same conditions that were used for pS40-hTH1–50. We determined a Kd value of 3.8 mM for tpS40-hTH1–50 (Fig. 3 A), compared to 1.0 mM for the pS40-hTH1–50 (Fig. 1 D). This demonstrates that binding is slightly reduced by the replacement of an oxygen atom with a sulfur atom. Interestingly, binding of 14-3-3ζ to tpS40-hTH1–50 causes a downfield 31P chemical shift (Fig. 3 A), whereas binding to pS40-hTH1–50 induces an upfield shift (see above, and Fig. 1 D), illustrating that chemical shifts are extremely sensitive to any changes and hard to predict.
Whereas 14-3-3ζ binding to the pS19pS40-hTH1–50 peptide yields three distinct pS40 phosphorous signals that exhibit slow exchange, pS40free, pS40∗, and pS40c (Fig. 2 A), binding to the pS19tpS40-hTH1–50 peptide results in only two signals that are in fast-intermediate exchange (Fig. 3 B), caused in part by the smaller chemical shift difference between free (pS40free) and bound (pS40c) resonances (0.60 p.p.m) compared to that for pS19pS40-hTH1–50 (0.94 p.p.m.). However, even with this difference, it seems reasonable to assume that the binding scheme in Fig. 2 B can be applied to pS19tpS40-hTH1–50, with Kd2 = 3.8 mM instead of 1 mM and Keq = 0.26 (see below) rather than 1.5. Using the well-separated pS19bound 31P resonance in the thiophosphorylated peptide (Fig. 3 B) proved beneficial for deconvoluting the overlapping pS19 resonances in the 14-3-3ζ-bound pS19pS40-hTH1–50 spectra (see Fig. S3).
Analysis of the binding equilibrium between doubly phosphorylated hTH1 peptide to the 14-3-3ζ homodimer
Fig. 2 B illustrates a comprehensive equilibrium binding scenario for the interaction between the doubly phosphorylated hTH1 peptide and the 14-3-3ζ homodimer. Six possible complexes are considered. Because insufficient experimental data for the accurate determination of all individual dissociation constants are available, it is assumed that each subunit monomer in the 14-3-3ζ homodimer binds equivalent phosphate groups on pS19pS40-hTH1–50 with identical affinity, irrespective of the state of the other 14-3-3ζ monomer, i.e., no cooperativity is present. Therefore, three independent equilibrium constants, Kd1, Kd2, and Keq (Fig. 2 B), come into play. Kd1 (0.15 ± 0.03 mM) and Kd2 (1.0 ± 0.1 mM) are the experimentally determined dissociation constants for the interaction between singly phosphorylated hTH1–50 peptides, pS19-hTH1–50 and pS40-hTH1–50, respectively, and the 14-3-3ζ dimer (using the monomer at the concentration unit); Keq is the equilibrium constant defined by the concentration ratio of the complex to the complex. This is treated as the only variable parameter in this simplistic equilibrium binding model.
We used three approaches for determining Keq, as follows:
In the first approach, we derived a set of six equations for the individual, independent equilibria, along with two equations that are necessary to ensure mass conservation of the peptide and 14-3-3ζ (see Fig. S2 A). These equations were solved numerically in the software MATLAB. The resulting solutions provide the molar concentration of peptide complexes as a function of the 14-3-3ζ to doubly phosphorylated peptide ratio, r (see Fig. S2 B). The equations in Fig. 2 C link the theoretically predicted molar concentrations of individual complexes (see Fig. S2 B) with the intensities of five experimentally observable resonances. By fitting the theoretical profiles (continuous lines in Fig. 2 D) with the experimentally observed intensities (individual data points depicted by x in Fig. 2 D), for several 14-3-3ζ to peptide molar ratios, r, a Keq value of 1.5 was determined.
In the second approach, the value of Keq could, in principle, also be determined from the observed difference in the “pS40c” resonance position from that of “pS40bound,” if the resonance position of pS40 in the complex is known. Unfortunately, this is not known experimentally, but, assuming that it is identical to that of (pS40∗), then the following equation holds:
| (4) |
Considering that
then
| (5) |
If the values of δ(pS40∗), 3.93 p.p.m.; δ(pS40c), 3.43 p.p.m.; and δ(pS40bound), 3.26 p.p.m. are applied to Eq. 5, a Keq value of 3.0 is found.
In the third approach, the value of Keq can be estimated, based on Kd2 and the local concentration of the pS40 phosphate group in the complex. In an extended chain model of the doubly phosphorylated peptide, the distance between the Ser19 and Ser40 phosphate groups is estimated to be ∼6 nm. The local concentration of the Ser40 phosphate that resides within a sphere of 6 nm radius, if the Ser19 phosphate from the same peptide is fixed to the first binding site on the 14-3-3ζ dimer, would become cloc ∼ 1/(6.022∗1023∗4∗103∗(6.10−9)3)M, i.e., 1.9 mM. Thus, an estimated value of Keq is
The latter two estimated Keq values of 3.0 and 1.9 are very similar to the value determined from fitting (Keq =1.5), considering that Keq values of 3.0, 1.9, and 1.5 result in relative populations of 75, 66, and 60%, respectively, of the complex.
Using the simplified binding model (Fig. 2 B), with Kd1 = 0.15 mM, Kd2 = 1.0 mM, and Keq = 1.5, populations of , , and complexes were found to be significantly lower than other complexes (<5.5%; see Fig. S2 B). For example, at a 1:1 molar ratio of doubly phosphorylated peptide to 14-3-3ζ (r = 1), 37.0% of the peptide is unbound; 17.7% of the peptide is in the complex; 11.8% is in complex; and 24.1% (2 × 12.05%) is in the complex. Together, all other species account for 9.4% of the total population. At r = 5, the theoretical binding model predicts 52.5% of the peptide is in the complex and 35% is in the complex. Note that the 31P resonances of the pS40 phosphate groups in these two complexes are in fast exchange (Fig. 2 B), resulting in a single pS40c peak (Fig. 2 A).
Discussion
In the prevailing model of 14-3-3 binding to doubly phosphorylated partners, the partners contain a high affinity, sometimes called a “gatekeeper” residue, which initially interacts with 14-3-3, and a secondary, weaker epitope, which only binds when the gatekeeper site is already bound to 14-3-3 (23). hTH1 that is singly phosphorylated on Ser40 is thought to be unable to bind 14-3-3ζ, whereas the pS19 phosphate is the high affinity epitope (18,19). Our NMR results with the N-terminal 50 residue peptide, pS40-hTH1–50, indicate, however, that binding of pS40 to 14-3-3ζ is indeed possible, as evidenced by the change from the free peptide phosphorous resonance to the bound one. This interaction is clearly weaker (Kd = 1.0 ± 0.1 mM) than that between 14-3-3ζ and pS19-hTH1–50, containing the purported gatekeeper phosphate, which binds a factor-of-10 tighter (Kd = 0.15 ± 0.03 mM). Therefore, even under conditions where 14-3-3ζ is present in excess over the pS19pS40-hTH1–50 peptide, two complexes, and , form in a 60:40% (Keq = 1.5) ratio, irrespective of the 14-3-3ζ concentration. Our results show that it is possible that multiple complexes for doubly phosphorylated binding partners of 14-3-3ζ may exist, even if in the phosphorylated full-length hTH1 additional mechanisms may come into play and alter the relative binding affinities (18).
Indeed, for the full-length pS40-hTH1, the binding to 14-3-3γ was almost undetectable by native mass spectroscopy and surface plasmon resonance, while for the doubly phosphorylated enzyme Ser40 is involved in binding to 14-3-3γ (22). In addition, binding of one 14-3-3γ dimer to one of the monomers in tetrameric hTH1—which is organized as a dimer of dimers—appears to interfere with the binding of an additional 14-3-3γ dimer to the adjacent monomer in hTH1, but not to the dimer at the opposite end, as seen in electron micrographs of the complex (22). Thus, in the complex involving the full-length enzyme, steric factors appear to be responsible for moving the equilibrium toward a scenario where the pS19 site binds to a single monomer in the 14-3-3 dimer. Whether this is the case for all 14-3-3 interactions with phosphorylated regulatory regions is unclear. In general, the presence of additional interactions and their contributions to the overall regulatory mechanisms have to be ascertained and confirmed for each individual system.
At this point, it may be of interest to analyze the underlying thermodynamics that govern the observed interaction between 14-3-3ζ and pS19pS40-hTH1–50 (or other similar peptides, denoted as 1‒2, with two phosphorous sites), with the bound form as the dominant complex when 14-3-3ζ is in excess and dominant when peptide is in excess. For simplicity, we assume that the binding of site 1 (primary site) is independent of that of site 2 (secondary site) and that peptide binding to one binding site on the 14-3-3ζ dimer leads to a global entropy decrease (ΔSG < 0), whereas engagement of the second site on the same peptide only results in a local entropy decrease (ΔSL < 0). The enthalpy changes corresponding to the binding of the primary and secondary sites to 14-3-3 protein are denoted as ΔH1 and ΔH2. Given these assumptions, the condition where the amount of is larger than (when peptide is in excess over 14-3-3ζ; r < 1), i.e.,
results in
| (6) |
The opposite inequality
is expected for the case when the amount of is larger than (when r > 1). In the case that the two phosphorous groups exhibit comparable binding affinities (e.g., such is the case for the PKCε-V3 peptide that contains tandem repeat sequences (27)),
Because ΔH1 − ΔH2 ∼ 0 and T(ΔSG − ΔSL) < 0, the free energy difference
becomes negative, making the amount of complex larger than the amount of complex, irrespective of the r value.
Considering the large number of 14-3-3 binding partners and the above-described different binding modes, both cases are thermodynamically possible and may contribute to the regulation imparted by 14-3-3 on its targets.
Conclusions
A detailed analysis of the interaction between singly or doubly phosphorylated hTH1–50 peptides and 14-3-3ζ is presented. Using phosphorous NMR, dissociation constants for the binding of singly phosphorylated peptides, pS19-hTH1–50 and pS40-hTH1–50, to 14-3-3ζ were determined. They differ by a factor of 10, with the Ser19 phosphorylated peptide exhibiting a higher affinity (Kd = 0.15 ± 0.03 vs. 1.0 ± 0.1 mM). Furthermore, for the doubly phosphorylated peptide, pS19pS40-hTH1–50, a mixture of distinct complexes was observed, which permitted us to derive a comprehensive equilibrium binding scheme. All six possible different complexes are taken into account, and the system is described by three equilibrium constants: Kd1 = 0.15 mM, Kd2 = 1.0 mM, and Keq = 1.5. For excess pS19pS40-hTH1–50 peptide, the major complex is the 14-3-3ζ dimer with two peptides bound via the phosphate group on Ser19 , whereas, for excess 14-3-3ζ, two major complexes are seen, each one with a doubly phosphorylated peptide bound to one 14-3-3ζ dimer, with the Ser19 phosphate group bound to one 14-3-3ζ subunit, and the Ser40 phosphate group either bound to the second subunit or free in solution in a 60:40 ratio.
For other systems, the underlying thermodynamics will contribute to whether a doubly phosphorylated partner will bind with a 1:1 stoichiometry to the 14-3-3ζ dimer or if a mixture of complexes is formed. It is likely that most 14-3-3ζ binding partners will possess phosphorylated residues that exhibit different affinities for the binding sites on 14-3-3ζ. The presented methodology derived by monitoring individual 31P resonances has general applicability for any complex system involving 14-3-3 and doubly phosphorylated ligands, and it will be interesting to extend this approach to full-length hTH1 or other target proteins.
Author contributions
J.H., A.M., and A.M.G. conceived this study; J.H. and I.-J.L.B. acquired experimental data; J.H., T.K., I.-J.L.B., and A.M.G. performed data analysis and interpretation; J.H., I.-J.L.B., and A.M.G. drafted the article; all authors contributed to the final article.
Acknowledgments
We thank Jinwoo Ahn and Leonardus Koharudin for advice on protein purification, Doug Bevan and Phil Greer for computer technical support, and Michael J. Delk for NMR instrumental support. We also thank Teresa Brosenitsch for critical reading of the manuscript. We are grateful to Dr. Damodaran Krishnan, Department of Chemistry, University of Pittsburgh, for access to the Bruker 600 MHz NMR spectrometer with a BBF probe.
J.H. acknowledges financial support by an International Outgoing Fellowship of the European Community program “Support for Training and Career Development of Researchers (Marie Curie)”, under contract No. PIOF-GA-2009-235902. This work was supported in part by startup funds from the University of Pittsburgh School of Medicine.
Supporting Material
References
- 1.Morrison D.K. The 14-3-3 proteins: integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol. 2009;19:16–23. doi: 10.1016/j.tcb.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gardino A.K., Yaffe M.B. 14-3-3 proteins as signaling integration points for cell cycle control and apoptosis. Semin. Cell Dev. Biol. 2011;22:688–695. doi: 10.1016/j.semcdb.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freeman A.K., Morrison D.K. 14-3-3 proteins: diverse functions in cell proliferation and cancer progression. Semin. Cell Dev. Biol. 2011;22:681–687. doi: 10.1016/j.semcdb.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson C., Crowther S., MacKintosh C. Bioinformatic and experimental survey of 14-3-3-binding sites. Biochem. J. 2010;427:69–78. doi: 10.1042/BJ20091834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin J., Smith F.D., Pawson T. Proteomic, functional, and domain-based analysis of in vivo 14-3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr. Biol. 2004;14:1436–1450. doi: 10.1016/j.cub.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 6.Obsil T., Obsilova V. Structural basis of 14-3-3 protein functions. Semin. Cell Dev. Biol. 2011;22:663–672. doi: 10.1016/j.semcdb.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Yang X., Lee W.H., Elkins J.M. Structural basis for protein-protein interactions in the 14-3-3 protein family. Proc. Natl. Acad. Sci. USA. 2006;103:17237–17242. doi: 10.1073/pnas.0605779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardino A.K. Structural determinants of 14-3-3 binding specificities and regulation of subcellular localization of 14-3-3-ligand complexes: a comparison of the x-ray crystal structures of all human 14-3-3 isoforms. Semin. Cancer Biol. 2006;16:173–182. doi: 10.1016/j.semcancer.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Yaffe M.B., Rittinger K., Cantley L.C. The structural basis for 14-3-3: phosphopeptide binding specificity. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- 10.Ganguly S., Weller J.L., Klein D.C. Melatonin synthesis: 14-3-3-dependent activation and inhibition of arylalkylamine n-acetyltransferase mediated by phosphoserine-205. Proc. Natl. Acad. Sci. USA. 2005;102:1222–1227. doi: 10.1073/pnas.0406871102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aitken A. 14-3-3 proteins: a historical overview. Semin. Cancer Biol. 2006;16:162–172. doi: 10.1016/j.semcancer.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 12.Ichimura T., Isobe T., Fujisawa H. Brain 14-3-3 protein is an activator protein that activates tryptophan 5-monooxygenase and tyrosine 3-monooxygenase in the presence of Ca2+ calmodulin-dependent protein kinase II. FEBS Lett. 1987;219:79–82. doi: 10.1016/0014-5793(87)81194-8. [DOI] [PubMed] [Google Scholar]
- 13.Nagatsu T., Levitt M., Udenfriend S. Tyrosine hydroxylase: the initial step in norepinephrine biosynthesis. J. Biol. Chem. 1964;239:2910–2917. [PubMed] [Google Scholar]
- 14.Grima B., Lamouroux A., Mallet J. A single human gene encoding multiple tyrosine hydroxylases with different predicted functional characteristics. Nature. 1987;326:707–711. doi: 10.1038/326707a0. [DOI] [PubMed] [Google Scholar]
- 15.Kaneda N., Kobayashi K., Nagatsu T. Isolation of a novel cDNA clone for human tyrosine hydroxylase: alternative RNA splicing produces four kinds of mRNA from a single gene. Biochem. Biophys. Res. Commun. 1987;146:971–975. doi: 10.1016/0006-291x(87)90742-x. [DOI] [PubMed] [Google Scholar]
- 16.Fitzpatrick P.F. Tetrahydropterin-dependent amino acid hydroxylases. Annu. Rev. Biochem. 1999;68:355–381. doi: 10.1146/annurev.biochem.68.1.355. [DOI] [PubMed] [Google Scholar]
- 17.Daubner S.C., Le T., Wang S. Tyrosine hydroxylase and regulation of dopamine synthesis. Arch. Biochem. Biophys. 2011;508:1–12. doi: 10.1016/j.abb.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleppe R., Toska K., Haavik J. Interaction of phosphorylated tyrosine hydroxylase with 14-3-3 proteins: evidence for a phosphoserine 40-dependent association. J. Neurochem. 2001;77:1097–1107. doi: 10.1046/j.1471-4159.2001.00318.x. [DOI] [PubMed] [Google Scholar]
- 19.Obsilova V., Nedbalkova E., Obsil T. The 14-3-3 protein affects the conformation of the regulatory domain of human tyrosine hydroxylase. Biochemistry. 2008;47:1768–1777. doi: 10.1021/bi7019468. [DOI] [PubMed] [Google Scholar]
- 20.Itagaki C., Isobe T., Ichimura T. Stimulus-coupled interaction of tyrosine hydroxylase with 14-3-3 proteins. Biochemistry. 1999;38:15673–15680. doi: 10.1021/bi9914255. [DOI] [PubMed] [Google Scholar]
- 21.Skjevik A.A., Mileni M., Martinez A. The N-terminal sequence of tyrosine hydroxylase is a conformationally versatile motif that binds 14-3-3 proteins and membranes. J. Mol. Biol. 2013;426:150–168. doi: 10.1016/j.jmb.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleppe R., Rosati S., Martinez A. Phosphorylation dependence and stoichiometry of the complex formed by tyrosine hydroxylase and 14-3-3γ. Mol. Cell. Proteomics. 2014;13:2017–2030. doi: 10.1074/mcp.M113.035709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yaffe M.B. How do 14-3-3 proteins work? Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 2002;513:53–57. doi: 10.1016/s0014-5793(01)03288-4. [DOI] [PubMed] [Google Scholar]
- 24.Kleppe R., Ghorbani S., Haavik J. Modeling cellular signal communication mediated by phosphorylation dependent interaction with 14-3-3 proteins. FEBS Lett. 2014;588:92–98. doi: 10.1016/j.febslet.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 25.Lian L.Y., Roberts G.C.K. Effects of chemical exchange on NMR spectra. In: Robert R.C.K., editor. NMR of Macromolecules. A Practical Approach. IRL Press; Oxford, United Kingdom: 1993. pp. 153–182. [Google Scholar]
- 26.Bain A.D. Chemical exchange in NMR. Prog. Nucl. Magn. Reson. Spectrosc. 2003;43:63–103. [Google Scholar]
- 27.Kostelecky B., Saurin A.T., McDonald N.Q. Recognition of an intra-chain tandem 14-3-3 binding site within PKCε. EMBO Rep. 2009;10:983–989. doi: 10.1038/embor.2009.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Molzan M., Ottmann C. Synergistic binding of the phosphorylated S233-and S259-binding sites of C-RAF to one 14-3-3 ζ-dimer. J. Mol. Biol. 2012;423:486–495. doi: 10.1016/j.jmb.2012.08.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


