Abstract
Universal trees based on sequences of single gene homologs cannot be rooted. Iwabe et al. [Iwabe, N., Kuma, K.-I., Hasegawa, M., Osawa, S. & Miyata, T. (1989) Proc. Natl. Acad. Sci. USA 86, 9355-9359] circumvented this problem by using ancient gene duplications that predated the last common ancestor of all living things. Their separate, reciprocally rooted gene trees for elongation factors and ATPase subunits showed Bacteria (eubacteria) as branching first from the universal tree with Archaea (archaebacteria) and Eucarya (eukaryotes) as sister groups. Given its topical importance to evolutionary biology and concerns about the appropriateness of the ATPase data set, an evaluation of the universal tree root using other ancient gene duplications is essential. In this study, we derive a rooting for the universal tree using aminoacyl-tRNA synthetase genes, an extensive multigene family whose divergence likely preceded that of prokaryotes and eukaryotes. An approximately 1600-bp conserved region was sequenced from the isoleucyl-tRNA synthetases of several species representing deep evolutionary branches of eukaryotes (Nosema locustae), Bacteria (Aquifex pyrophilus and Thermotoga maritima) and Archaea (Pyrococcus furiosus and Sulfolobus acidocaldarius). In addition, a new valyl-tRNA synthetase was characterized from the protist Trichomonas vaginalis. Different phylogenetic methods were used to generate trees of isoleucyl-tRNA synthetases rooted by valyl- and leucyl-tRNA synthetases. All isoleucyl-tRNA synthetase trees showed Archaea and Eucarya as sister groups, providing strong confirmation for the universal tree rooting reported by Iwabe et al. As well, there was strong support for the monophyly (sensu Hennig) of Archaea. The valyl-tRNA synthetase gene from Tr. vaginalis clustered with other eukaryotic ValRS genes, which may have been transferred from the mitochondrial genome to the nuclear genome, suggesting that this amitochondrial trichomonad once harbored an endosymbiotic bacterium.
Full text
PDF

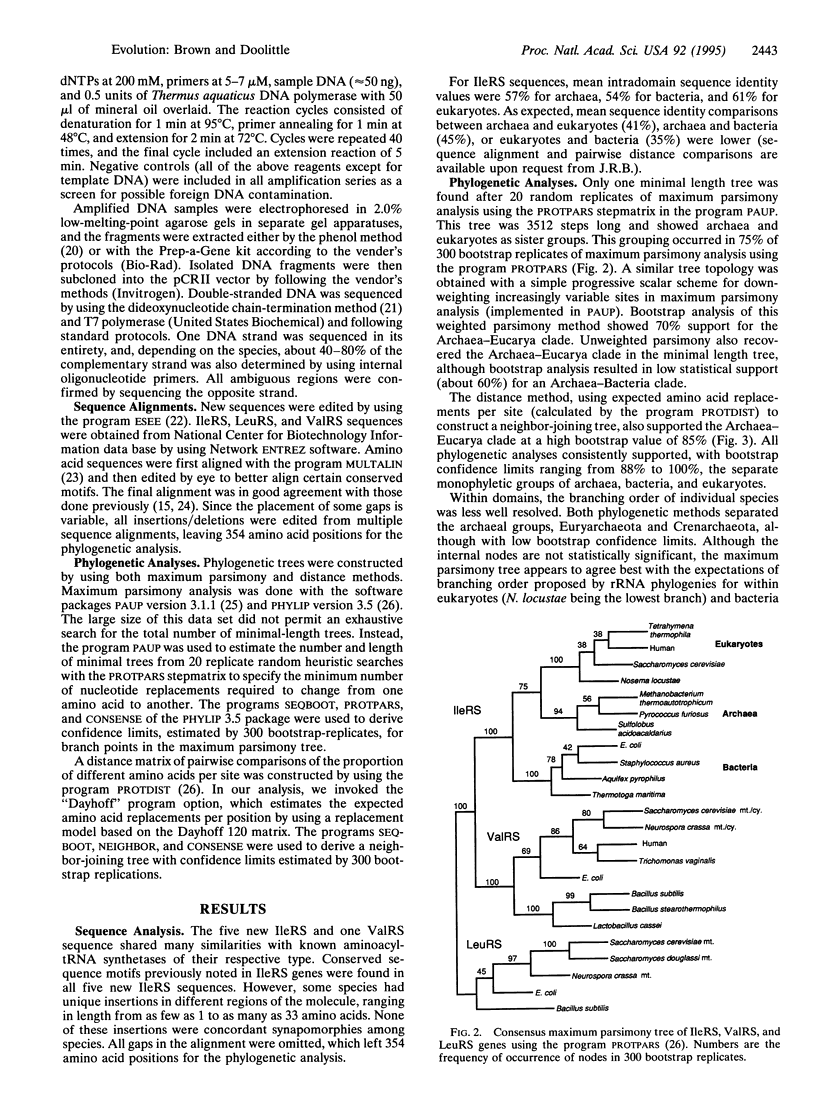
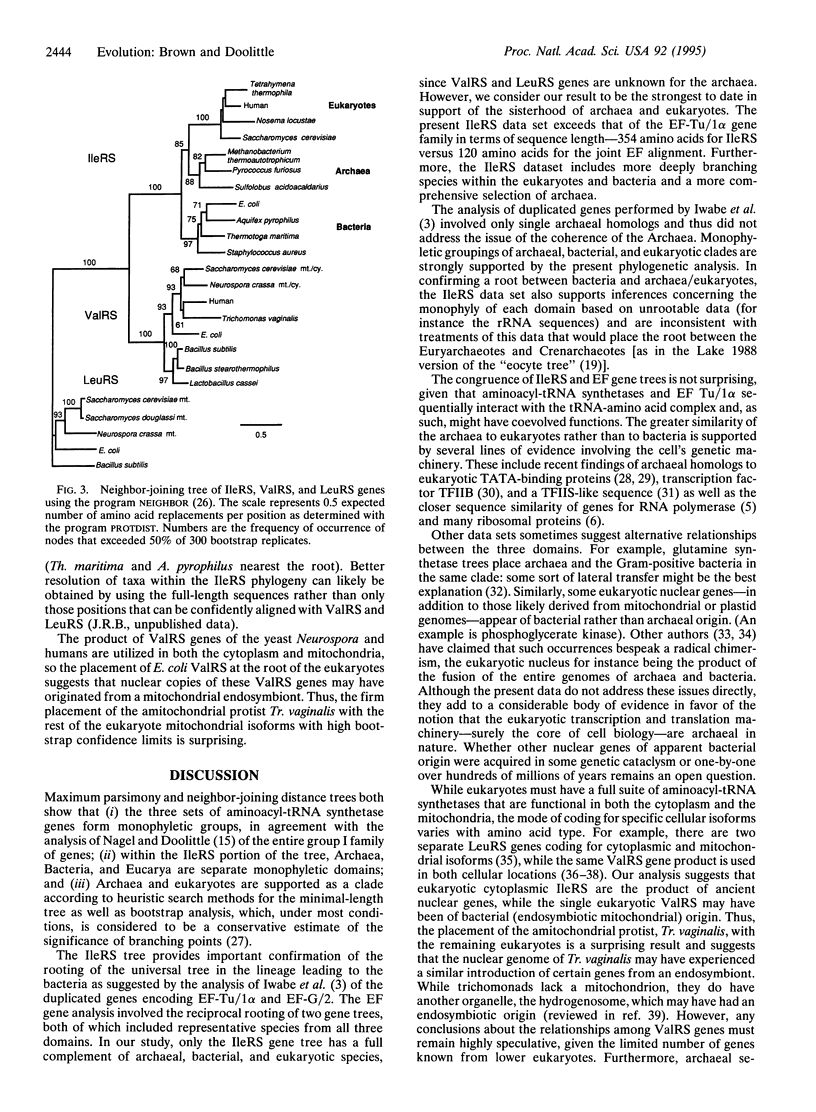

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Achenbach-Richter L., Gupta R., Stetter K. O., Woese C. R. Were the original eubacteria thermophiles? Syst Appl Microbiol. 1987;9:34–39. doi: 10.1016/s0723-2020(87)80053-x. [DOI] [PubMed] [Google Scholar]
- Brown J. R., Masuchi Y., Robb F. T., Doolittle W. F. Evolutionary relationships of bacterial and archaeal glutamine synthetase genes. J Mol Evol. 1994 Jun;38(6):566–576. doi: 10.1007/BF00175876. [DOI] [PubMed] [Google Scholar]
- Burggraf S., Olsen G. J., Stetter K. O., Woese C. R. A phylogenetic analysis of Aquifex pyrophilus. Syst Appl Microbiol. 1992 Aug;15(3):352–356. doi: 10.1016/S0723-2020(11)80207-9. [DOI] [PubMed] [Google Scholar]
- Cabot E. L., Beckenbach A. T. Simultaneous editing of multiple nucleic acid and protein sequences with ESEE. Comput Appl Biosci. 1989 Jul;5(3):233–234. doi: 10.1093/bioinformatics/5.3.233. [DOI] [PubMed] [Google Scholar]
- Carter C. W., Jr Cognition, mechanism, and evolutionary relationships in aminoacyl-tRNA synthetases. Annu Rev Biochem. 1993;62:715–748. doi: 10.1146/annurev.bi.62.070193.003435. [DOI] [PubMed] [Google Scholar]
- Cavarelli J., Moras D. Recognition of tRNAs by aminoacyl-tRNA synthetases. FASEB J. 1993 Jan;7(1):79–86. doi: 10.1096/fasebj.7.1.8422978. [DOI] [PubMed] [Google Scholar]
- Chow C. M., Rajbhandary U. L. Regulation of the nuclear genes encoding the cytoplasmic and mitochondrial leucyl-tRNA synthetases of Neurospora crassa. Mol Cell Biol. 1989 Nov;9(11):4645–4652. doi: 10.1128/mcb.9.11.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988 Nov 25;16(22):10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creti R., Ceccarelli E., Bocchetta M., Sanangelantoni A. M., Tiboni O., Palm P., Cammarano P. Evolution of translational elongation factor (EF) sequences: reliability of global phylogenies inferred from EF-1 alpha(Tu) and EF-2(G) proteins. Proc Natl Acad Sci U S A. 1994 Apr 12;91(8):3255–3259. doi: 10.1073/pnas.91.8.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriani G., Delarue M., Poch O., Gangloff J., Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990 Sep 13;347(6289):203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- Forterre P., Benachenhou-Lahfa N., Confalonieri F., Duguet M., Elie C., Labedan B. The nature of the last universal ancestor and the root of the tree of life, still open questions. Biosystems. 1992;28(1-3):15–32. doi: 10.1016/0303-2647(92)90004-i. [DOI] [PubMed] [Google Scholar]
- Gogarten J. P., Kibak H., Dittrich P., Taiz L., Bowman E. J., Bowman B. J., Manolson M. F., Poole R. J., Date T., Oshima T. Evolution of the vacuolar H+-ATPase: implications for the origin of eukaryotes. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6661–6665. doi: 10.1073/pnas.86.17.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilario E., Gogarten J. P. Horizontal transfer of ATPase genes--the tree of life becomes a net of life. Biosystems. 1993;31(2-3):111–119. doi: 10.1016/0303-2647(93)90038-e. [DOI] [PubMed] [Google Scholar]
- Hsieh S. L., Campbell R. D. Evidence that gene G7a in the human major histocompatibility complex encodes valyl-tRNA synthetase. Biochem J. 1991 Sep 15;278(Pt 3):809–816. doi: 10.1042/bj2780809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwabe N., Kuma K., Hasegawa M., Osawa S., Miyata T. Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9355–9359. doi: 10.1073/pnas.86.23.9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U., Rechsteiner T., Tan P. Y., Bühlmann E., Meile L., Leisinger T. Isoleucyl-tRNA synthetase of Methanobacterium thermoautotrophicum Marburg. Cloning of the gene, nucleotide sequence, and localization of a base change conferring resistance to pseudomonic acid. J Biol Chem. 1991 Jun 5;266(16):10570–10577. [PubMed] [Google Scholar]
- Jordana X., Chatton B., Paz-Weisshaar M., Buhler J. M., Cramer F., Ebel J. P., Fasiolo F. Structure of the yeast valyl-tRNA synthetase gene (VASI) and the homology of its translated amino acid sequence with Escherichia coli isoleucyl-tRNA synthetase. J Biol Chem. 1987 May 25;262(15):7189–7194. [PubMed] [Google Scholar]
- Kaine B. P., Mehr I. J., Woese C. R. The sequence, and its evolutionary implications, of a Thermococcus celer protein associated with transcription. Proc Natl Acad Sci U S A. 1994 Apr 26;91(9):3854–3856. doi: 10.1073/pnas.91.9.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakinuma Y., Igarashi K., Konishi K., Yamato I. Primary structure of the alpha-subunit of vacuolar-type Na(+)-ATPase in Enterococcus hirae. Amplification of a 1000-bp fragment by polymerase chain reaction. FEBS Lett. 1991 Nov 4;292(1-2):64–68. doi: 10.1016/0014-5793(91)80835-q. [DOI] [PubMed] [Google Scholar]
- Lake J. A. Origin of the eukaryotic nucleus determined by rate-invariant analysis of rRNA sequences. Nature. 1988 Jan 14;331(6152):184–186. doi: 10.1038/331184a0. [DOI] [PubMed] [Google Scholar]
- Marsh T. L., Reich C. I., Whitelock R. B., Olsen G. J. Transcription factor IID in the Archaea: sequences in the Thermococcus celer genome would encode a product closely related to the TATA-binding protein of eukaryotes. Proc Natl Acad Sci U S A. 1994 May 10;91(10):4180–4184. doi: 10.1073/pnas.91.10.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindale D. W., Gu Z. M., Csank C. Isolation and complete sequence of the yeast isoleucyl-tRNA synthetase gene (ILS1). Curr Genet. 1989 Feb;15(2):99–106. doi: 10.1007/BF00435455. [DOI] [PubMed] [Google Scholar]
- Müller M. The hydrogenosome. J Gen Microbiol. 1993 Dec;139(12):2879–2889. doi: 10.1099/00221287-139-12-2879. [DOI] [PubMed] [Google Scholar]
- Nagel G. M., Doolittle R. F. Evolution and relatedness in two aminoacyl-tRNA synthetase families. Proc Natl Acad Sci U S A. 1991 Sep 15;88(18):8121–8125. doi: 10.1073/pnas.88.18.8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouzounis C., Sander C. TFIIB, an evolutionary link between the transcription machineries of archaebacteria and eukaryotes. Cell. 1992 Oct 16;71(2):189–190. doi: 10.1016/0092-8674(92)90347-f. [DOI] [PubMed] [Google Scholar]
- Pühler G., Leffers H., Gropp F., Palm P., Klenk H. P., Lottspeich F., Garrett R. A., Zillig W. Archaebacterial DNA-dependent RNA polymerases testify to the evolution of the eukaryotic nuclear genome. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4569–4573. doi: 10.1073/pnas.86.12.4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlands T., Baumann P., Jackson S. P. The TATA-binding protein: a general transcription factor in eukaryotes and archaebacteria. Science. 1994 May 27;264(5163):1326–1329. doi: 10.1126/science.8191287. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba K., Schimmel P. Functional assembly of a randomly cleaved protein. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1880–1884. doi: 10.1073/pnas.89.5.1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin M. L. Early evolution and the origin of eukaryotes. Curr Opin Genet Dev. 1991 Dec;1(4):457–463. doi: 10.1016/s0959-437x(05)80192-3. [DOI] [PubMed] [Google Scholar]
- Sumi M., Sato M. H., Denda K., Date T., Yoshida M. A DNA fragment homologous to F1-ATPase beta subunit was amplified from genomic DNA of Methanosarcina barkeri. Indication of an archaebacterial F-type ATPase. FEBS Lett. 1992 Dec 21;314(3):207–210. doi: 10.1016/0014-5793(92)81472-x. [DOI] [PubMed] [Google Scholar]
- Tsutsumi S., Denda K., Yokoyama K., Oshima T., Date T., Yoshida M. Molecular cloning of genes encoding major two subunits of a eubacterial V-type ATPase from Thermus thermophilus. Biochim Biophys Acta. 1991 Dec 3;1098(1):13–20. [PubMed] [Google Scholar]
- Woese C. R., Fox G. E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci U S A. 1977 Nov;74(11):5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Kandler O., Wheelis M. L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillig W. Comparative biochemistry of Archaea and Bacteria. Curr Opin Genet Dev. 1991 Dec;1(4):544–551. doi: 10.1016/s0959-437x(05)80206-0. [DOI] [PubMed] [Google Scholar]




