Abstract
Age-related cognitive decline is a serious health concern in our aging society. Decreased cognitive function observed during healthy brain aging is most likely caused by changes in brain connectivity and synaptic dysfunction in particular brain regions. Here we show that aged C57BL/6J wild-type mice have hippocampus-dependent spatial memory impairments. To identify the molecular mechanisms that are relevant to these memory deficits, we investigated the temporal profile of mouse hippocampal synaptic proteome changes at 20, 40, 50, 60, 70, 80, 90, and 100 weeks of age. Extracellular matrix proteins were the only group of proteins that showed robust and progressive up-regulation over time. This was confirmed by immunoblotting and histochemical analysis, which indicated that the increased levels of hippocampal extracellular matrix might limit synaptic plasticity as a potential cause of age-related cognitive decline. In addition, we observed that stochasticity in synaptic protein expression increased with age, in particular for proteins that were previously linked with various neurodegenerative diseases, whereas low variance in expression was observed for proteins that play a basal role in neuronal function and synaptic neurotransmission. Together, our findings show that both specific changes and increased variance in synaptic protein expression are associated with aging and may underlie reduced synaptic plasticity and impaired cognitive performance in old age.
As the proportion of aged individuals in our population continues to grow, we are faced with an increase in age-related health problems. Brain aging invariably leads to functional decline and impairments in cognitive function and motor skills, which can seriously affect quality of life. A better understanding of the neurobiological mechanisms underlying age-related cognitive decline is crucial to facilitate maintenance of cognitive health in the elderly and to reveal potential causes of highly prevalent age-related forms of dementia, in particular Alzheimer disease, in which cognitive decline is severely impaired by yet unknown mechanisms.
Several studies showed that normal brain aging is associated with subtle morphological and functional alterations in specific neuronal circuits (1, 2) and that reduced cognitive function with increasing age is likely due to synaptic dysfunction (3). Increasing evidence supports the idea that alterations in hippocampal activity are correlated with deficits in learning and memory in healthy aging humans (4, 5). In addition, rodent models of healthy aging demonstrate strong correlations between impaired performance in learning and memory tests and disturbed hippocampal network activity (6, 7). Electrophysiological studies provide additional evidence that age-related disturbances in the hippocampus involve changes in the principal cellular features of learning and memory, synaptic long-term potentiation and long-term depression (8, 9). Together, these observations suggest that a decline in hippocampal synaptic efficacy and plasticity plays a critical role in age-dependent cognitive impairment.
Aging is also the primary risk factor for Alzheimer disease, which clinically manifests as severe and accelerated age-dependent cognitive decline (10). Genetic causes of familial early-onset Alzheimer disease all point to a key role in disease etiology for increased brain levels of the protein amyloid-β (11). Familial Alzheimer disease, however, is rare, and it is likely that increased amyloid-β levels in sporadic Alzheimer disease result from age-dependent and/or genetically determined alterations in the expression of other genes or proteins (12, 13). Thus, the identification of molecular mechanisms of normal brain aging might also contribute to our understanding of cognitive decline under pathological conditions, in particular in Alzheimer disease.
Although the exact mechanisms underlying brain aging remain to be fully determined, they likely include changes at the molecular, cellular, and neuronal-network levels. In particular, characterization of alterations in the molecular composition and dynamics of hippocampal synapses could potentially reveal important aspects of the underlying mechanisms of brain aging. Age-related changes in global hippocampal gene and protein expression have been investigated previously (14, 15), but these studies were not geared to identify the specific synaptic molecular substrates of brain aging. Here, we made use of iTRAQ1 technology and high-coverage mass spectrometry to study the effects of aging on the proteomic composition of mouse hippocampal synaptosomes. We investigated the synaptic proteomes of individual mice at 20, 40, 50, 60, 70, 80, 90, and 100 weeks of age. Our findings show that both specific changes and increased variance in synaptic protein expression are associated with age-related cognitive decline.
EXPERIMENTAL PROCEDURES
Animals
Male wild-type C57BL/6J mice were bred in-house and singly housed on sawdust in standard Makrolon type II cages (26.5 cm long, 20.5 cm wide, and 14.5 cm high) enriched with cardboard nesting material under a 12-h/12-h dark/light cycle with access to water and food ad libitum. All animal experiments were approved by the animal ethics committee of VU University.
Barnes Maze
The Barnes maze consisted of a circular gray platform (120-cm diameter) elevated 100 cm above the floor and containing 24 holes (4.5-cm diameter) spaced at equal distance from each other and 5 cm away from the edge of the platform. One hole was designated the escape hole and was equipped with a cylindrical entrance (4.5-cm diameter × 5-cm depth) mounted underneath the maze and providing access to an escape box (15.3 × 6.4 × 6.1 cm) containing a metal stairway for easy access that was not visible unless mice approached the hole closely. Other holes were equipped with identical cylindrical entrances, but without an escape box. Visual extra-maze cues (50 × 50 cm) composed of black and white patterns were mounted on the walls ∼70 cm from the maze. Three fans, surrounding the maze at a 60-cm distance and ∼120° apart, produced a variable airflow across the entire maze via a slow, 90° horizontal movement, both to provide an aversive environment and to disperse odor cues. Fluorescent tube lights mounted at the ceiling provided bright illumination (1000 lux), and a speaker mounted to the ceiling provided background sound.
Mice received two sessions per day, one in the morning and one in the afternoon, for five consecutive days. Mice were introduced in an opaque cylinder placed in the center of the maze, after which the experimenter left the room. The cylinder was pulled upward 30 s later, and mice were allowed to explore the maze and locate the escape hole. If the latency to enter the escape hole exceeded 300 s, mice were gently guided toward the escape hole. On day 1, mice received two habituation sessions, during which the escape box contained cage enrichment of each mouse's own home cage, and mice were left in the escape box for 60 s. On days 2–4, mice received two training sessions per day with the cage enrichment removed, and on day 5 they received one training session in the morning and a 300-s probe trial in the afternoon. During the probe trial, the escape hole was identical to all other holes. After each session, the platform and escape box were thoroughly cleaned with 70% ethanol, and the platform was rotated 90° to further prevent the use of odors cues.
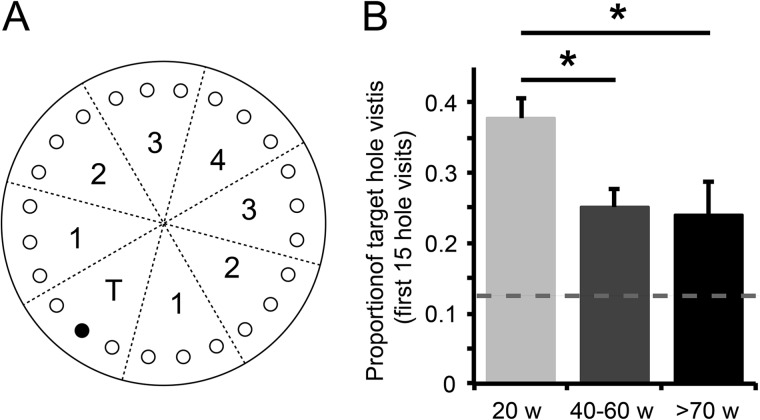
The path traveled by each mouse was video tracked by an overhead camera and analyzed using Viewer 2 software with the Barnes maze plugin (BIOBSERVE GmbH, Bonn, Germany). Multiple consecutive visits to the same hole were counted as a single hole visit. For data analysis, the Barnes maze was divided into eight octants. The octant containing the escape hole was designated the target octant, and the other octants were numbered 1–4 according to their distance from the target octant (see Fig. 1A). To assess learning, the proportion of hole visits in the target octant was calculated for the first 15 hole visits during the probe trial. Animals of different age categories (20 weeks, 40–60 weeks, and 70–100 weeks) were tested, and significance was determined via one-way ANOVA and Tukey's post-hoc testing. p values less than 0.05 were considered significant.
Fig. 1.
Aged mice are impaired in hippocampus-dependent spatial learning in a Barnes maze test. A, schematic drawing of the Barnes maze. The escape hole (black) is located in the target octant (T) of the maze and is removed during the probe trial; other octants are numbered 1–4 according to their distance from the target octant. B, the proportion of hole visits in the target octant during the first 15 hole visits revealed a significant reduction for mice aged 40 to 60 weeks (n = 9) and for mice aged 70 to 100 weeks (n = 13) relative to 20-week-old mice (n = 10) (one-way ANOVA; F2,29 = 4.43, p = 0.021). The dotted line indicates chance level (0.125). Mean ± S.E.; Tukey's post-hoc test; *p < 0.05.
Proteomics
Synaptosome Isolation and Sample Preparation
Animals were sacrificed by cervical dislocation, and hippocampi were dissected, frozen, and stored at −80 °C until protein isolation. Synaptosomes were isolated from hippocampi at eight different ages, 20, 40, 50, 60, 70, 80, 90, and 100 weeks, as described previously (16, 17). In brief, hippocampi were homogenized in ice-cold 0.32 m sucrose buffer with 5 mm HEPES at pH 7.4 and protease inhibitor (Roche) and centrifuged at 1000 × g for 10 min at 4 °C to remove debris. Supernatant was loaded on top of a discontinuous sucrose gradient consisting of 0.85 m and 1.2 m sucrose. After ultracentrifugation at 110,000 × g for 2 h at 4 °C, the synaptosome fraction was collected at the interface of 0.85 m and 1.2 m sucrose, resuspended, and pelleted by ultracentrifugation at 80,000 × g for 30 min at 4 °C. Pellets were redissolved in 5 mm HEPES, and protein concentrations were determined using a Bradford assay (Bio-Rad, Hercules, CA). For each sample, 50 μg of protein was transferred to a fresh tube and dried in a SpeedVac overnight.
Protein Digestion and iTRAQ Labeling
Per sample, 50 μg of synaptosome proteins were dissolved in 0.85% RapiGest (Waters Associates, Milford, MA), alkylated with methyl methanethiosulfonate, and digested with trypsin (sequencing grade; Promega, Madison, WI) as described elsewhere (16, 17). In each iTRAQ experiment, samples were labeled with iTRAQ reagents 113 = 20 weeks, 114 = 40 weeks, 115 = 50 weeks, 116 = 60 weeks, 117 = 70 weeks, 118 = 80 weeks, 119 = 90 weeks, and 121 = 100 weeks. To accommodate eight independent protein samples for each time point, a total of eight 8-plex iTRAQ experiments was performed (8 × 8).
Two-dimensional Liquid Chromatography
The lyophilized iTRAQ-labeled samples were separated in the first dimension on a strong cation exchange column (2.1 × 150 mm polysulfoethyl A column, PolyLC, Columbia, MD) and the in second dimension on an analytical capillary reverse phase C18 column (150 mm × 100 μm inner diameter) at 400 nL/min using the LC-Packing Ultimate system. The peptides were separated using a linear increase in concentration of acetonitrile from 4% to 28% in 75 min, to 36% in 7 min, and finally to 72% in 2 min. The eluent was mixed with matrix (7 mg of re-crystallized α-cyano-hydroxycinnaminic acid in 1 ml of 50% acetronitrile, 0.1% trifluoroacetic acid, 10 mm ammonium dicitrate) delivered at a flow rate of 1.5 μl/min and deposited onto an Applied Biosystems (Forster City, CA) matrix-assisted laser desorption ionization plate by means of a robot (Probot, Dionex, Sunnyvale, CA) once every 15 s for a total of 384 spots.
MALDI-MS/MS
Samples were analyzed on an ABI 5800 proteomics analyzer (Applied Biosystems). Peptide collision-induced dissociation was performed at 2 kV; the collision gas was air. MS/MS spectra were collected from 2000 laser shots. Peptides with signal-to-noise ratios greater than 50 at the MS mode were selected for MS/MS analysis, at a maximum of 30 MS/MS per spot. The precursor mass window was set to a relative resolution of 200. Peaklists were extracted from the instrument database using TS2Mascot software (Matrix Science, Boston, MA).
Protein Identification
Protein identification and quantification are described in detail in Ref. 18. To annotate spectra, Mascot (version 2.3.01, Matrix Science) searches were performed against Mus musculus sequences in the Swiss-Prot database (20/10/2010; 16,326 sequences searched) and in the larger but more redundant NCBI database (20/10/2010; 147,581 sequences searched). MS/MS spectra were searched with trypsin specificity and fixed iTRAQ modifications on lysine residues and N termini of the peptides and methylthio modifications on cysteine residues. Oxidation on methionine residues was allowed as a variable modification. The mass tolerance was 150 ppm for precursor ions and 0.4 Da for fragment ions, and a single site of miscleavage was allowed. For each spectrum the best scoring peptide sequence and protein accession were selected. The false discovery rate for peptide identification was calculated using a decoy database search (provided by the Mascot software), and the peptide list was limited to 5% false positives in order to improve protein identification confidence. Protein redundancy in the result files was removed by clustering the precursor protein sequences at a threshold of 90% sequence similarity over 85% of the sequence length. If present, a random Swiss-Prot protein entry was chosen as a representative for a protein cluster; if not, a random NCBInr sequence was chosen. Subsequently all peptides were matched against the protein clusters, and only those peptides that mapped uniquely to one protein were included. Proteins were considered for quantification if we identified at least three peptides in four replicate iTRAQ sets and at least one peptide in the four other sets.
Protein Quantification and Identification of Differentially Expressed Proteins
iTRAQ areas (m/z 113–121) were extracted from raw spectra and corrected for isotopic overlap. Peptides with iTRAQ signature peaks of less than 1500 were not considered for quantification. To compensate for the possible variations in the starting amounts of the samples, the individual peak areas of each iTRAQ signature peak were log transformed to yield a normal distribution and normalized to the mean peak area of every sample. No measures were taken to correct for iTRAQ compression. Next, iTRAQ reporter ions were standardized within each spectrum by subtracting the average intensities of all reporter ions in that spectrum. Protein abundances in every experiment were determined by taking the average normalized standardized iTRAQ peak area of all unique peptides annotated to a protein. To determine which proteins showed significant differential expression, three types of statistical analyses were performed. First, the “Two-Class Analysis” option implemented in the Significance Analysis of Microarrays (SAM) package (19) was used to determine significant regulation at individual time points. Two-class SAM analysis was used both as an unpaired analysis and as a paired analysis in which samples within iTRAQ sets were considered as paired with the 20-week time point as the baseline. Paired analysis was performed to reduce false positives resulting from technical variation between iTRAQ sets and resulted in more stringent selection of differentially regulated proteins than unpaired analysis. In addition, the “One-Class Time Course Analysis” option in SAM was used to determine significant regulation over time. In all SAM analyses a threshold q value of 10% was used to determine significant differential protein expression. The time course analysis was used to determine the set of proteins for which there was the strongest evidence of regulation over time. K-means clustering and Pearson correlation analysis were used to further analyze this set of proteins. The intersection of the paired and the unpaired two-class analysis was used to determine the set of proteins for which there was evidence of regulation at one or more time points.
Functional Protein Group Analysis
All proteins were assigned to one of 17 functional synaptic protein groups as previously defined (20), and overrepresentation of regulated proteins within functional groups was determined using Fisher's exact test. Enrichment was considered relevant only when overrepresented functional groups contained at least three proteins. In addition, functional enrichment was determined using the DAVID functional annotation tool (21, 22). The functional categories used were GO-term related to biological process, cellular component, and molecular function, as well as pathway annotations derived from KEGG. The entire set of detected proteins was used as the background set, and a modified Fisher's exact p value of <0.05 was considered significant. Enrichment was only considered relevant when enriched functional groups contained at least five proteins.
Protein Variance Analysis
For each detected protein, mean standardized peak areas and standard deviations of the mean standardized peak areas were calculated per time point for both the unpaired dataset and paired data set against the 20-week time point. Parametric one-way ANOVA and non-parametric Kruskal–Wallis (23) tests were used to determine whether the distribution of these sample means and deviations were dependent on time. Next, all standard deviations from all time points were collected and ranked, and the top 5% were selected and designated as high variable proteins, whereas the bottom 5% were selected and designated as low variable proteins. The intersection of the unpaired and paired data was used to determine the set of proteins with either high or low variability. The distribution of high and low variable proteins over time points was determined, and functional enrichment analysis was performed using DAVID (21, 22), using the whole set of detected proteins as a background and a modified Fisher's exact p value of <0.05 as significance threshold.
Immunoblotting
Immunoblot analysis was performed on six synaptosome protein extracts (six biological replicates) per time point. Samples were treated with chondroitinase ABC (Sigma Aldrich, Zwijndrecht, The Netherlands; 0.5 U/50 mg protein) for 90 min at 37 °C. Of each sample, 2.5 μg of protein was mixed with SDS sample buffer and heated to 90 °C for 5 min. Proteins were separated on a Criterion TGX Stain-Free Precast Gel (4–16% Tris-Glycine, Bio-Rad) in a Criterion Cell Electrophoresis System (Bio-Rad) and electroblotted onto PVDF membrane overnight at 4 °C. After blocking with 5% non-fat dry milk in TBS plus 0.5% Tween for 1 h, blots were incubated with primary antibodies followed by a horseradish-peroxidase-conjugated secondary antibody (Dako, Glostrup, Denmark; 1:10,000). The following antibodies were used: anti-brevican (gift from Dr. C. Seidenbecher, Magdeburg, Germany; 1:2000), anti-HAPLN1 (Abcam, Cambridge, UK; 1:1000), and anti-neurocan (Sigma; 1:1000). Blots were incubated with ECL substrate (GE Healthcare, Pollards Wood, UK), scanned with an Odyssey Imager (LI-COR, Lincoln, NE), and analyzed with Image Studio software (LI-COR, version 1.1.7) using background correction. To correct for differences in sample input, all gels were imaged before electroblotting, and the total protein densitometric values were used for sample normalization, which is more reliable then normalizing to the levels of a single protein (24, 25). Significance was determined using Student's t test (one-tailed, independent samples). p values less than 0.05 were considered significant.
Histology
Wisteria floribunda agglutinin (WFA) staining was performed on free-floating brain sections obtained from animals 3 and 18 months of age. Sections were quenched (10% methanol, 0.3% H2O2 in PBS) for 30 min, blocked with 0.2% Triton X-100 and 5% fetal bovine serum in PBS, and incubated overnight with fluorescein-labeled WFA (Vector Laboratories, Burlingame, CA; 1:400). Sections were then washed and coverslipped in Vectashield including DAPI as a nuclear dye (Vector Laboratories). WFA-positive cells in the CA1 region of the hippocampus were counted in 12 separate sections per animal using the “analyze particles” option in ImageJ (version 1.40g). The mean number of WFA-positive cells per section was calculated, and significance was determined using Student's t test (two-tailed, independent samples). p values less than 0.05 were considered significant.
RESULTS
Aged Mice Have Hippocampus-dependent Learning Deficits
To determine age-related hippocampal learning deficits, three age groups of C57BL/6J wild-type mice were tested in a Barnes maze. Mice were trained to find an escape hole among 24 holes that were located radially on the edge of a circular platform (Fig. 1A); the mice were guided by visual cues surrounding the platform. During a probe trial, the escape hole was not different from the other holes, and mice were tested for their ability to memorize its location. Taking the proportion of hole visits in the correct octant of the maze during first 15 hole visits as a measure of spatial memory, we observed a significant decrease in performance in both 40- to 60-week-old mice and 70- to 100-week-old mice relative to 20-week-old mice (one-way ANOVA; F2,29 = 4.43, p = 0.021) (Fig. 1B).
Proteomic Analysis Identifies 100 Synaptic Proteins That Are Significantly Regulated at Any Time Point
To identify changes in synaptic protein expression underlying age-related impairments in hippocampal learning, we next performed 8-plex iTRAQ proteomics on hippocampal synaptosomes isolated from C57BL/6J mice 20, 40, 50, 60, 70, 80, 90, and 100 weeks of age. For each time point, eight independent biological replicate samples of individual mice were measured. Each iTRAQ experiment contained samples from all eight time points, allowing comparison within and among age groups (supplemental Fig. S1A). Correct isolation of synaptosomes was confirmed by enrichment of the synaptic protein marker PSD-95 (supplemental Fig. S1B). Peptide and protein identification data for each iTRAQ experiment are summarized in supplemental Tables S1–S8. In total, 501 high-confidence proteins were detected (confidence interval ≥ 95%; number of quantifiable peptides = ≥3 in at least four experiments and ≥1 in all other experiments; supplemental Table S9). All mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (26) via the PRIDE partner repository with dataset identifier PXD001135 and DOI 10.6019/PXD001135.
To determine significant regulation at individual time points, paired and unpaired two-class analyses were performed using the SAM package (19). In the paired setup, samples within each iTRAQ set were considered as paired with respect to the 20-week time point, which reduced technical variation between iTRAQ sets. We considered proteins as regulated only when the false discovery rate was <10% in both the unpaired and the paired analyses. This approach resulted in 100 proteins being significantly differentially expressed at any time point (supplemental Table S9). As an initial indicator of the validity of these findings, we calculated the correlation coefficients for proteins that were part of multisubunit complexes and thus were expected to show similar regulation patterns. Indeed, we observed highly correlated expression for proteins within complexes, whereas expression between complexes was often very different (supplemental Fig. S2).
To allow functional characterization of these proteins, and to reduce the effect of potential false negatives at individual time points, we first combined differentially expressed proteins into three different age groups: early-aged (weeks 40 and 50), middle-aged (weeks 60, 70, and 80), and old-aged (weeks 90 and 100) (Fig. 2A; supplemental Tables S10–S12). For functional analysis, proteins were categorized in 17 functional synaptic protein groups as previously defined (20, 27), and regulated proteins at each age group were separately tested for overrepresentation of functional protein groups using all 501 detected proteins as the background set (Fig. 2B; Table I). In early-aged mice, we observed significant overrepresentation of up-regulated proteins involved in cell metabolism and structural plasticity (Figs. 2B and 2C). In middle-aged mice, overrepresentation was observed for proteins involved in cell adhesion (Figs. 2B and 2C). No significantly overrepresented functional protein groups were detected in old-aged mice (Fig. 2B). The same functional protein groups were found when functional enrichment analysis was performed on individual time points (supplemental Table S13), indicating that combining data from adjacent time points did not bias toward false positive findings.
Fig. 2.
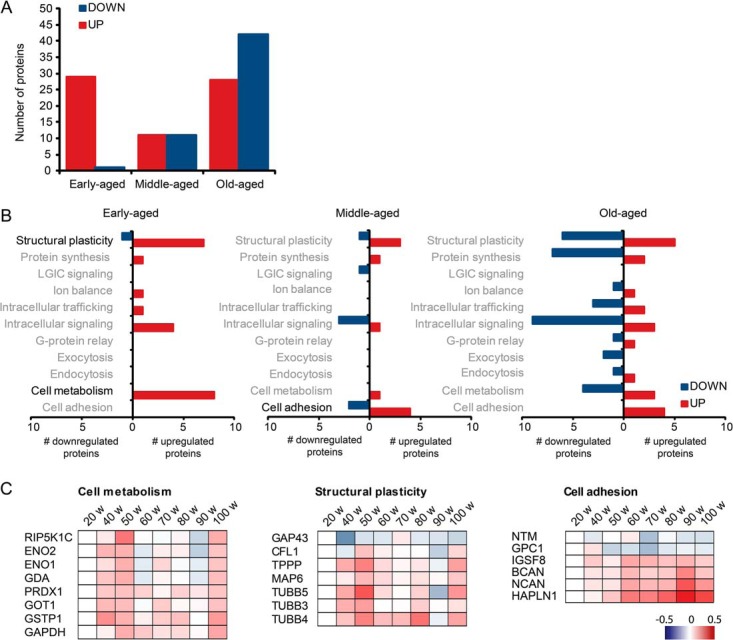
Proteomic analysis identifies 100 proteins that are significantly regulated at any time point. A, up- and down-regulated proteins were divided in three age groups: early-aged (40 to 50 weeks), middle-aged (60 to 80 weeks), and old-aged (90 to 100 weeks). B, all differentially expressed proteins were categorized in 17 functional synaptic protein groups as previously defined (20, 27). Only protein groups indicated in black print were significantly enriched in the corresponding age group (Fisher's exact test; p < 0.05; see also Table I). C, individual protein expression profiles of significantly enriched functional protein groups. Red indicates proteins that are up-regulated; blue indicates proteins that are down-regulated.
Table I. Functional enrichment of differentially expressed proteins. Significantly enriched (Fisher's exact p value < 0.05) functional protein groups (according to Ref. 20) are indicated per age group.
| Functional group | Proteins | Fold enrichment | Fisher's exact p value |
|---|---|---|---|
| Early-aged | |||
| Cell metabolism | GSTP1, GAPDH, GOT1, ENO1, ENO2, GDA, PIP5K1C, PRDX1 | 4.1 | 0.0003 |
| Structural plasticity | CFL1, TUBB4, TUBB5, DPYSL2, MTAP6, GAP43, TUBB3, TPPP | 2.2 | 0.0219 |
| Middle-aged | |||
| Cell adhesion/transsynaptic signaling | GPC1, NTM, HAPLN1, NCAN, IGSF8, BCAN | 4.9 | 0.0005 |
As a complementary approach, functional enrichment of differentially expressed proteins was also determined using GO and cellular pathway (KEGG) databases (supplemental Table S14). Early-aged mice showed an enrichment of several GO classes including cytoskeletal proteins, in particular tubulins. In middle-aged mice we observed a >15-times enrichment of GO classes related to the extracellular matrix (ECM). Old-aged mice showed significant enrichment for GO classes related to the cortical cytoskeleton (actins, actin-regulatory proteins, and neurofilament proteins), but enrichment was relatively low (<4 times) compared with early- and middle-aged animals.
Proteomic Analysis Identifies 25 Synaptic Proteins That Are Significantly Regulated Over Time
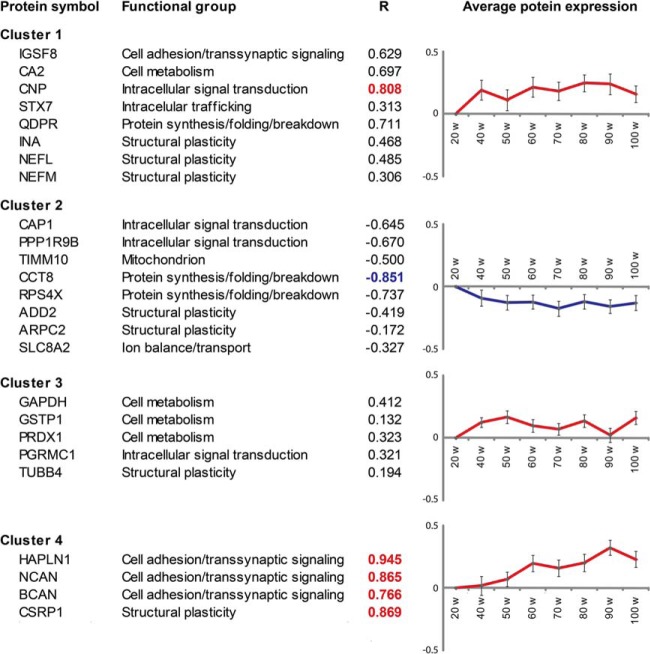
We next wanted to select differentially expressed proteins that were most consistently up- or down-regulated over time. One-class time course analysis in SAM using the eight individual time points as input revealed a total of 17 proteins that were significantly up-regulated over time and 8 proteins that were significantly down-regulated over time (Fig. 3). K-means clustering was used to separate these 25 proteins into four different expression clusters (Fig. 3). Clusters 1–3 contained proteins whose expression was increased or decreased already at early time points (40 to 50 weeks) and remained relatively constant over time thereafter. Notably, these three clusters included six cytoskeleton and associated proteins involved in structural plasticity. Only proteins in cluster 4 showed progressive up-regulation over time until week 90. These proteins also showed the strongest linear correlation between expression and time, as evidenced by R correlation coefficients (Fig. 3). Three out of the four proteins in cluster 4 represent protein constituents of the ECM (i.e. hyaluronan and proteoglycan link protein 1 (HAPLN1), neurocan (NCAN), and brevican (BCAN)). Hierarchical clustering of our data identified similar clusters of up- and down-regulated proteins (supplemental Fig. S3). In all, functional enrichment analysis of regulated proteins at individual time points and time course analyses both indicated (i) age-dependent dysregulation of the cytoskeleton and (ii) significant up-regulation of the ECM.
Fig. 3.
Proteomic analysis identifies 25 proteins that are significantly regulated over time. K-means clustering revealed four different expression clusters. Clusters 1–3 contained proteins whose expression was increased or decreased already early during aging and remained high or low over time. Only proteins in cluster 4 showed progressive up-regulation over time. These proteins also showed the strongest linear correlation between expression and age, as evidenced by linear regression R correlation coefficients. R-coefficients greater than 0.75 are indicated in red; R-coefficients less than or equal to 0.75 are indicated in blue. Graphs represent the average log2-fold change for all proteins in each cluster relative to the 20-week time point.
Extracellular Matrix Proteins Are Progressively Up-regulated Over Time
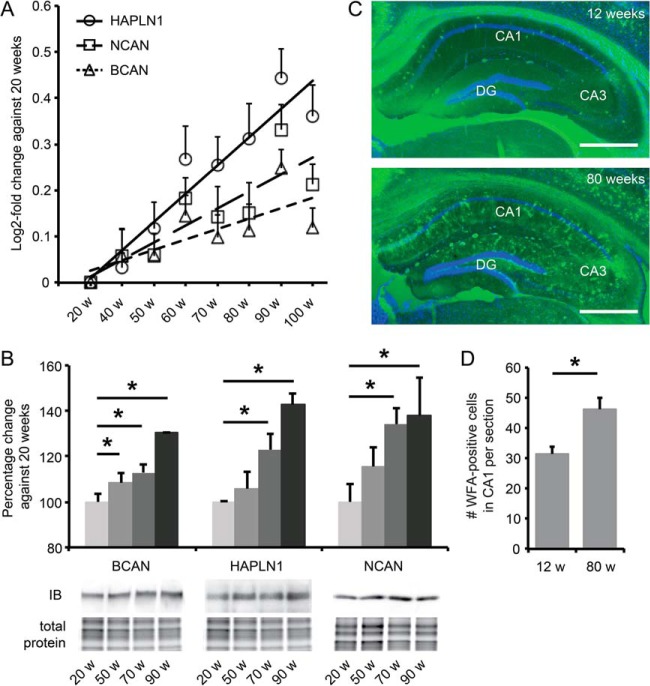
Three ECM proteins, HAPLN1, BCAN, and NCAN, showed a strong age-dependent increase in expression with maximum log2-fold changes of 0.25 to 0.45 at 90 weeks of age (Fig. 4A). Immunoblotting confirmed their age-dependent up-regulation (Fig. 4B). NCAN and BCAN are chondroitin sulfate proteoglycans that bind to the hyaluronic acid ECM backbone, whereas HAPLN1 is the principal link protein required for this binding. Increased expression of all three proteins is thus strongly indicative of an increase in ECM levels in the aged hippocampus. To test this, we next stained hippocampal sections of 3-month-old and 18-month-old mice with WFA, which specifically labels chondroitin sulfate proteoglycan glycosaminoglycan side chains. Indeed, we observed a strong increase in WFA staining in the aged hippocampus, both in the number of WFA-positive perineuronal nets (PNNs) and in diffuse ECM staining (Fig. 4C). Quantification of the number of PNNs in the CA1 region of the hippocampus revealed a 45% increase at old age relative to young control animals (n = 4 to 5; p = 0.02) (Fig. 4D). Together, these findings show that the most significant and characteristic age-dependent alteration in the hippocampal synaptic proteome is a progressive increase in ECM levels.
Fig. 4.
Hippocampal ECM levels increase progressively with age. A, temporal expression patterns of HAPLN1, BCAN, and NCAN as measured by iTRAQ proteomics (mean ± S.D.). B, immunoblotting (IB) confirmed the age-dependent up-regulation of HAPLN1, BCAN, and NCAN; n = 6 (Student's t test; mean ± S.E.; *p < 0.05). Representative blots are shown with the corresponding total amount of protein used for normalization. C, WFA staining (green) of coronal sections of the hippocampus confirms a robust increase in ECM levels in 18-month-old mice compared with 3-month-old mice. This was visible as an increase in the number of WFA-positive PNNs, as well as an increase in global WFA staining throughout the hippocampus. Blue, DAPI staining; CA, cornus ammonis; DG, dentate gyrus. Scale bars: 400 μm. D, quantification of WFA-positive PNNs in CA1 shows a significant increase in 18-month-old mice (n = 5; 12 sections per animal) relative to 3-month-old mice (n = 4; 12 sections per animal). Mean ± S.E.; Student's t test; *p < 0.05.
Variance in Synaptic Protein Expression Increases with Age
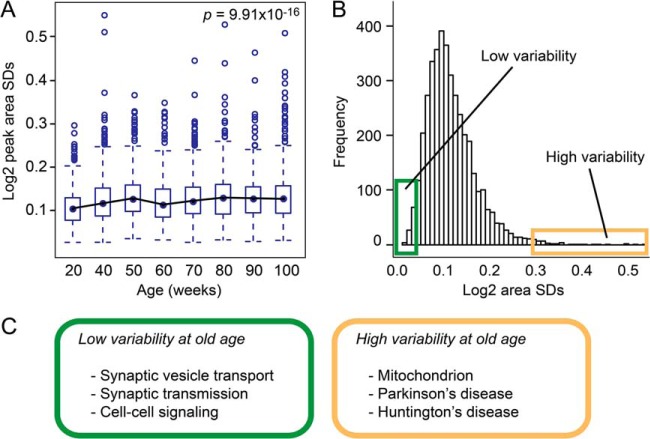
Recent studies show that heterogeneity and stochastic deregulation of gene expression increase with age (28–30) and possibly predict lifespan (31). To test whether synaptic protein expression is also subject to age-dependent loss of regulatory control, we measured stochasticity in protein levels as a function of age. We calculated the standard deviation (S.D.) of the average standardized peak areas per protein per time point (supplemental Table S9) and indeed observed a significant increase in S.D. with age (unpaired data: one-way ANOVA test, p = 9.91 × 10−16; Kruskal–Wallis test, p = 2.2 × 10−16; Fig. 5A). A similar age-dependent shift in S.D. distribution toward greater values was observed in the paired dataset (one-way ANOVA test, p = 1.07−11; Kruskal–Wallis test, p = 9.99−11). Peptide-based peak areas did not change with age (one-way ANOVA test, p = 0.134; Kruskal–Wallis test, p = 0.257), which excluded variation in absolute protein levels as an explanation for the observed increase in S.D. values. Next, we ranked the combined S.D. values derived from all time points and selected the 5% of proteins with the lowest S.D. values, which we designated as low-variability proteins, and the 5% of proteins with the highest S.D. values, which we referred to as high-variability proteins (Fig. 5B). The intersection of the unpaired and paired datasets revealed 102 proteins with extreme S.D. values, either low-variable or high-variable, at different time points.
Fig. 5.
Variance in synaptic protein expression increases with age. A, distributions of the standard deviations (S.D.) of the average standardized protein intensities (iTRAQ peak areas) per time point revealed a significant increase over time (unpaired data). B, distribution of all S.D. values from all time points indicating the highly constrained proteins as the top 5% of low-variability proteins (green box) and lowly constrained proteins as the top 5% of high-variability proteins (yellow box) (unpaired data). C, functional enrichment analysis for high-variability and low-variability proteins in the old-age group using GO term and KEGG pathway databases (see also supplemental Table S17).
We next selected the low-variability and high-variability proteins that were specifically associated with the two oldest age groups (weeks 90 and 100) (supplemental Tables S15 and S16) and performed a GO term and KEGG pathway functional enrichment analysis (Fig. 5C; supplemental Table S17). Low-variability proteins were specifically associated with synaptic vesicle transport, synaptic transmission, cell–cell signaling, and synaptic cell junctions. Proteins included in these groups were the presynaptic vesicle-associated proteins synapsin I, synaptic vesicle glycoprotein 2b, and syntaxin-binding protein 1 and the postsynaptic density proteins PSD-93, PSD-95, and β-catenin. High-variability proteins were found to be involved in cellular respiration and associated with Parkinson disease and Huntington disease. Proteins within these disease pathways are mainly mitochondrial complex I, complex IV, and inner membrane proteins. In addition, the neurodegeneration-related α-synuclein protein was found to be highly variable.
DISCUSSION
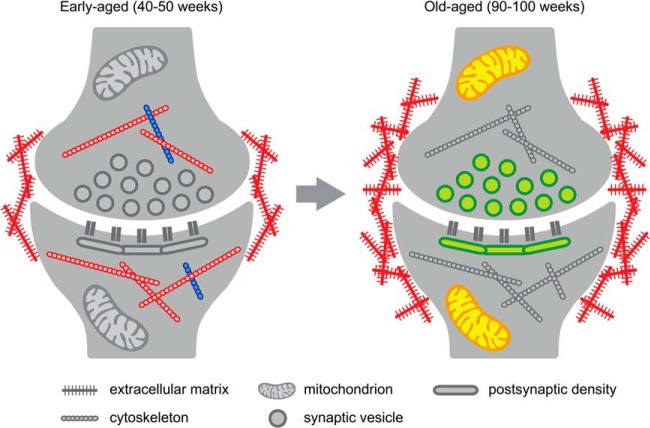
Age is the primary risk factor for cognitive decline and age-related neurodegenerative disorders such as Alzheimer disease. Age-related cognitive decline affects a significant proportion of the healthy aging population, but its causes remain to be determined. In mice, age-related cognitive decline can be observed as a decrease in hippocampus-dependent contextual fear memory and spatial learning (32). Here we confirmed that aged mice had spatial memory impairments in a Barnes maze task. We then identified proteomic alterations that correlate with these age-dependent memory deficits. Age-dependent hippocampal synapse loss is know to contribute to aging in rodents and humans (reviewed in Ref. 33), and to exclude the possibility that changes in relative protein abundances were due to synapse loss, we focused our proteomics approach on biochemically enriched synaptosome preparations and quantified relative protein levels within these preparations. Two types of proteomic alterations were identified. Firstly, we identified synaptic proteins that significantly changed in expression level during aging. Secondly, we found that some proteins showed increased stochasticity with aging, whereas others were more stably expressed. A schematic summary of these findings is provided in Fig. 6.
Fig. 6.
Schematic summary of protein expression and variance changes with age. The most consistent protein expression and protein variance changes observed in our study are indicated for early-aged and for old-aged hippocampal synapses. Only significantly enriched functional groups are shown for proteins that changed in abundance (up-regulated in red, down-regulated in blue) or in variance (high variance in yellow, low variance in green). In early-aged mice (40 to 50 weeks of age), we observed an increase in ECM protein expression and a dysregulation (both up and down) of the cytoskeleton. No functional enrichment was observed for proteins with extreme variance in early-aged mice. In old-aged mice (90 to 100 weeks of age), ECM protein expression continued to increase. In addition, mitochondrial protein expression became highly variable, whereas low variability was observed for proteins that are required to maintain basal pre- and postsynaptic functions (see text for details).
The most significant age-dependent change in synaptic protein levels was observed for a small group of four proteins (cluster 4 in Fig. 3), all of which were progressively up-regulated with age. Three out of these four proteins are ECM proteins (NCAN, BCAN, and HAPLN1), and their up-regulation was confirmed with immunoblotting. NCAN, BCAN, and HAPLN1 are typical components of ECM-containing PNNs (34–36), and our data indeed confirm a significant increase in the number of PNNs in the aged hippocampus. A developmental increase in PNNs corresponds with the ending of critical periods of maturation in various brain areas (37, 38), and PNNs are therefore believed to play an important role in limiting developmental plasticity. In the developing visual cortex, for instance, the absence of PNNs during the critical period allows ocular dominance plasticity to occur (39), and local enzymatic inactivation of PNNs restores ocular dominance plasticity in adult animals (40). More recent data indicate that PNNs also play a role in adult behavioral plasticity. Specifically, inactivation of PNNs in the basolateral amygdala enhances the extinction of fear memory (41), and in the perirhinal cortex PNN depletion enhances object recognition memory (42). These data strongly suggest that high ECM levels limit plasticity in the adult brain.
At the cell physiological level, ECM structures may limit synaptic plasticity in different ways. Firstly, ECM structures form physical barriers that restrict the lateral diffusion of AMPA receptors at postsynaptic sites (43, 44). Secondly, activity-dependent local degradation of the ECM by matrix metalloproteinases unmasks protein fragments that bind to synaptic integrin receptors and regulate long-term potentiation (45, 46). Finally, the ECM acts as a sink for growth factors and can either modulate or restrain their role in synaptic plasticity (47, 48). An age-dependent increase in hippocampal synaptic ECM levels as observed here may thus contribute to age-related learning and memory deficits by reducing molecular and cellular signaling mechanisms that are normally required in order for synaptic plasticity to occur. Several recent publications are in support of such a plasticity-limiting role of the ECM in the aging brain. Yamada and Jinno, for instance, reported age-dependent alterations in hippocampal ECM localization and composition in subjects up to one year of age (49). In addition, reducing cortical ECM levels in aged rats reactivates plasticity, both in sensorimotor cortex (50) and in visual cortex (51). Finally, increased expression of ECM proteins, in particular heparin sulfate proteoglycans, is associated with Alzheimer disease pathology (reviewed in Ref. 52), and we recently showed that reducing hippocampal ECM levels restores plasticity and memory deficits in a mouse model of Alzheimer disease (53).
In early-aged mice (40 to 50 weeks of age), we also observed alterations in the expression of proteins involved in structural plasticity. In particular, tubulins and microtubule-associated proteins (TUBB3, TUBB4, TUBB5, TPPP, and MAP6) and intermediate filament proteins (NEFL, NEFM) were up-regulated, whereas actin regulatory proteins (ADD2 and ARPC2) were down-regulated. Dysregulation of the cytoskeleton and of cytoskeleton-based transport is an important causative factor in age-related neurodegeneration. The microtubule stabilizing protein TAU, for instance, is causally linked with synaptic dysfunction and cognitive decline in Alzheimer disease and in frontotemporal lobe dementia (54, 55). Early-aged mice also showed an up-regulation of proteins involved in cell metabolism. Interestingly, several of these proteins were found to have neuroprotective properties (ENO2) (56), to prevent neurodegeneration (GSTP1) (57), or to play a role in eliminating harmful peroxides generated during metabolism (PRDX1) (58). Up-regulation of these proteins could thus protect neurons against the harmful consequences of aging.
In old-aged mice (90 to 100 weeks of age), we observed increased heterogeneity or stochasticity in synaptic protein expression. An age-dependent increase in variance has previously been reported for gene expression in the human brain (29). Moreover, increased variance in gene expression selectively affects cellular pathways that contribute to Parkinson's disease (59). These authors conclude that increased variance in expression changes the predictability or robustness of gene networks and thereby dysregulates cellular states, and that this might be an essential element that is shared among age-related disorders in general. A significant reduction in gene expression variance was observed in stem cells from schizophrenia patients, indicating that increased network robustness and reduced plasticity are equally detrimental to normal brain function (59). Our findings demonstrate that variance in protein expression levels also changes with age. Importantly, the increase in variance was specific for the old-age group, and did not the result from a decrease in protein abundance with aging, as the distributions of the standardized peak areas (i.e. protein levels) did not significantly change with age. Moreover, we found that distinct functional protein groups were associated with either high or low variability, suggesting that increased stochasticity specifically affects particular synaptic proteins, whereas others remain under tight control at old age.
Low-variability proteins in old-aged mice included proteins involved in synaptic vesicle transport and synaptic cell junction integrity, indicating high regulatory control with aging over mechanisms that play a basal role in synaptic neurotransmission. Highly variable proteins in old-aged mice were specifically involved in mitochondrial respiration and were frequently associated with neurodegenerative diseases, in particular Parkinson disease and Huntington disease. Disease conditions may arise when an increase in variance changes the predictability of the network outcome, resulting in dysregulation of the preferred state (59). Interestingly, many complex diseases lack genetic variants associated with the disease (60, 61), and in such cases variability in gene or protein expression may contribute to the development of diseases without obvious genetic heritability. Our data show that normal aging is associated with increased stochasticity in synaptic protein levels and dysregulation of protein networks that are known to be involved in neurodegenerative diseases, and thus they may explain to some extent the highly prevalent sporadic forms of these diseases among aging individuals.
Acknowledgments
We thank Ning Chen for assistance with immunoblotting, Jochem Cornelis for assisting with data analysis, and Rolinka van der Loo and Martha Peskens for assisting with behavioral studies.
Footnotes
Author contributions: M.J.V., K.L., A.d., A.B.S., S.S., and R.E.v. designed research; M.J.V., M.L., C.M.H., and I.P. performed research; M.J.V., A.R., M.L., C.M.H., W.J., P.v., and R.E.v. analyzed data; M.J.V., K.L., A.d., A.B.S., S.S., and R.E.v. wrote the paper.
* This work was supported by the Center of Medical Systems Biology (CMSB). A.B.S., K.W.L., S.S., P.v.N., and R.E.v.K. received funding from the EU-FP7 framework health program (SynSys, Grant No. 242167). M.J.V. received funding from the Top Institute Pharma.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- iTRAQ
- isobaric tag for relative and absolute quantification
- ANOVA
- analysis of variance
- ECM
- extracellular matrix
- GO
- gene ontology
- HAPLN1
- hyaluronan and proteoglycan link protein 1
- NCAN
- neurocan
- BCAN
- brevicn
- PNN
- perineuronal net
- SAM
- Significance Analysis of Microarrays
- S.D.
- standard deviation
- WFA
- Wisteria floribunda agglutinin.
REFERENCES
- 1. Rapp P. R., Gallagher M. (1996) Preserved neuron number in the hippocampus of aged rats with spatial learning deficits. Proc. Natl. Acad. Sci. U.S.A. 93, 9926–9930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rasmussen T., Schliemann T., Sorensen J. C., Zimmer J., West M. J. (1996) Memory impaired aged rats: no loss of principal hippocampal and subicular neurons. Neurobiol. Aging 17, 143–147 [DOI] [PubMed] [Google Scholar]
- 3. Burke S. N., Barnes C. A. (2006) Neural plasticity in the ageing brain. Nat. Rev. Neurosci. 7, 30–40 [DOI] [PubMed] [Google Scholar]
- 4. Beeri M. S., Lee H., Cheng H., Wollman D., Silverman J. M., Prohovnik I. (2009) Memory activation in healthy nonagenarians. Neurobiol. Aging 32, 515–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Daselaar S. M., Fleck M. S., Dobbins I. G., Madden D. J., Cabeza R. (2006) Effects of healthy aging on hippocampal and rhinal memory functions: an event-related fMRI study. Cereb. Cortex 16, 1771–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gerrard J. L., Burke S. N., McNaughton B. L., Barnes C. A. (2008) Sequence reactivation in the hippocampus is impaired in aged rats. J. Neurosci. 28, 7883–7890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wilson I. A., Ikonen S., Gureviciene I., McMahan R. W., Gallagher M., Eichenbaum H., Tanila H. (2004) Cognitive aging and the hippocampus: how old rats represent new environments. J. Neurosci. 24, 3870–3878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barnes C. A., Rao G., Houston F. P. (2000) LTP induction threshold change in old rats at the perforant path–granule cell synapse. Neurobiol. Aging 21, 613–620 [DOI] [PubMed] [Google Scholar]
- 9. Norris C. M., Korol D. L., Foster T. C. (1996) Increased susceptibility to induction of long-term depression and long-term potentiation reversal during aging. J. Neurosci. 16, 5382–5392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Forstl H., Kurz A. (1999) Clinical features of Alzheimer's disease. Eur. Arch. Psychiatry Clin. Neurosci. 249, 288–290 [DOI] [PubMed] [Google Scholar]
- 11. Querfurth H. W., LaFerla F. M. (2010) Alzheimer's disease. N. Engl. J. Med. 362, 329–344 [DOI] [PubMed] [Google Scholar]
- 12. Corder E. H., Saunders A. M., Strittmatter W. J., Schmechel D. E., Gaskell P. C., Small G. W., Roses A. D., Haines J. L., Pericak-Vance M. A. (1993) Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science 261, 921–923 [DOI] [PubMed] [Google Scholar]
- 13. Jonsson T., Stefansson H., Steinberg S., Jonsdottir I., Jonsson P. V., Snaedal J., Bjornsson S., Huttenlocher J., Levey A. I., Lah J. J., Rujescu D., Hampel H., Giegling I., Andreassen O. A., Engedal K., Ulstein I., Djurovic S., Ibrahim-Verbaas C., Hofman A., Ikram M. A., van Duijn C. M., Thorsteinsdottir U., Kong A., Stefansson K. (2013) Variant of TREM2 associated with the risk of Alzheimer's disease. N. Engl. J. Med. 368, 107–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Blalock E. M., Chen K. C., Sharrow K., Herman J. P., Porter N. M., Foster T. C., Landfield P. W. (2003) Gene microarrays in hippocampal aging: statistical profiling identifies novel processes correlated with cognitive impairment. J. Neurosci. 23, 3807–3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Walther D. M., Mann M. (2011) Accurate quantification of more than 4000 mouse tissue proteins reveals minimal proteome changes during aging. Mol. Cell. Proteomics 10, M110.004523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li K. W., Miller S., Klychnikov O., Loos M., Stahl-Zeng J., Spijker S., Mayford M., Smit A. B. (2007) Quantitative proteomics and protein network analysis of hippocampal synapses of CaMKIIalpha mutant mice. J. Prot. Res. 6, 3127–3133 [DOI] [PubMed] [Google Scholar]
- 17. Végh M. J., de Waard M. C., van der Pluijm I., Ridwan Y., Sassen M. J., van Nierop P., van der Schors R. C., Li K. W., Hoeijmakers J. H., Smit A. B., van Kesteren R. E. (2012) Synaptic proteome changes in a DNA repair deficient ercc1 mouse model of accelerated aging. J. Prot. Res. 11, 1855–1867 [DOI] [PubMed] [Google Scholar]
- 18. van Nierop P., Loos M. (2011) Bioinformatics procedures for analysis of quantitative proteomics experiments using iTRAQ. In Neuroproteomics (Li K. W., ed), pp. 275–296, Humana Press, New York [Google Scholar]
- 19. Tusher V. G., Tibshirani R., Chu G. (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U.S.A. 98, 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ruano D., Abecasis G. R., Glaser B., Lips E. S., Cornelisse L. N., de Jong A. P., Evans D. M., Davey Smith G., Timpson N. J., Smit A. B., Heutink P., Verhage M., Posthuma D. (2010) Functional gene group analysis reveals a role of synaptic heterotrimeric G proteins in cognitive ability. Am. J. Hum. Genet. 86, 113–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 22. Huang da W., Sherman B. T., Lempicki R. A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Siegel S., Castellan J. W. (1988) Nonparametric Statistics for the Behavioural Sciences, pp. 213–214, McGraw-Hill, New York [Google Scholar]
- 24. Van den Oever M. C., Goriounova N. A., Li K. W., Van der Schors R. C., Binnekade R., Schoffelmeer A. N., Mansvelder H. D., Smit A. B., Spijker S., De Vries T. J. (2008) Prefrontal cortex AMPA receptor plasticity is crucial for cue-induced relapse to heroin-seeking. Nat. Neurosci. 11, 1053–1058 [DOI] [PubMed] [Google Scholar]
- 25. Counotte D. S., Li K. W., Wortel J., Gouwenberg Y., Van Der Schors R. C., Smit A. B., Spijker S. (2010) Changes in molecular composition of rat medial prefrontal cortex synapses during adolescent development. Eur. J. Neurosci. 32, 1452–1460 [DOI] [PubMed] [Google Scholar]
- 26. Vizcaino J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolome S., Apweiler R., Omenn G. S., Martens L., Jones A. R., Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lips E. S., Cornelisse L. N., Toonen R. F., Min J. L., Hultman C. M., Holmans P. A., O'Donovan M. C., Purcell S. M., Smit A. B., Verhage M., Sullivan P. F., Visscher P. M., Posthuma D. (2012) Functional gene group analysis identifies synaptic gene groups as risk factor for schizophrenia. Mol. Psychiatry 17, 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bahar R., Hartmann C. H., Rodriguez K. A., Denny A. D., Busuttil R. A., Dolle M. E., Calder R. B., Chisholm G. B., Pollock B. H., Klein C. A., Vijg J. (2006) Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature 441, 1011–1014 [DOI] [PubMed] [Google Scholar]
- 29. Lu T., Pan Y., Kao S. Y., Li C., Kohane I., Chan J., Yankner B. A. (2004) Gene regulation and DNA damage in the ageing human brain. Nature 429, 883–891 [DOI] [PubMed] [Google Scholar]
- 30. Wang Q., Huang J., Zhang X., Wu B., Liu X., Shen Z. (2011) The spatial association of gene expression evolves from synchrony to asynchrony and stochasticity with age. PLoS One 6, e24076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rea S. L., Wu D., Cypser J. R., Vaupel J. W., Johnson T. E. (2005) A stress-sensitive reporter predicts longevity in isogenic populations of Caenorhabditis elegans. Nat. Genet. 37, 894–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Peleg S., Sananbenesi F., Zovoilis A., Burkhardt S., Bahari-Javan S., Agis-Balboa R. C., Cota P., Wittnam J. L., Gogol-Doering A., Opitz L., Salinas-Riester G., Dettenhofer M., Kang H., Farinelli L., Chen W., Fischer A. (2010) Altered histone acetylation is associated with age-dependent memory impairment in mice. Science 328, 753–756 [DOI] [PubMed] [Google Scholar]
- 33. Burke S. N., Barnes C. A. (2010) Senescent synapses and hippocampal circuit dynamics. Trends Neurosci. 33, 153–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Carulli D., Rhodes K. E., Brown D. J., Bonnert T. P., Pollack S. J., Oliver K., Strata P., Fawcett J. W. (2006) Composition of perineuronal nets in the adult rat cerebellum and the cellular origin of their components. J. Comp. Neurol. 494, 559–577 [DOI] [PubMed] [Google Scholar]
- 35. Carulli D., Rhodes K. E., Fawcett J. W. (2007) Upregulation of aggrecan, link protein 1, and hyaluronan synthases during formation of perineuronal nets in the rat cerebellum. J. Comp. Neurol. 501, 83–94 [DOI] [PubMed] [Google Scholar]
- 36. Yamaguchi Y. (2000) Lecticans: organizers of the brain extracellular matrix. Cell. Mol. Life Sci. 57, 276–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bruckner G., Grosche J., Schmidt S., Hartig W., Margolis R. U., Delpech B., Seidenbecher C. I., Czaniera R., Schachner M. (2000) Postnatal development of perineuronal nets in wild-type mice and in a mutant deficient in tenascin-R. J. Comp. Neurol. 428, 616–629 [DOI] [PubMed] [Google Scholar]
- 38. Galtrey C. M., Fawcett J. W. (2007) The role of chondroitin sulfate proteoglycans in regeneration and plasticity in the central nervous system. Brain Res. Rev. 54, 1–18 [DOI] [PubMed] [Google Scholar]
- 39. Hensch T. K. (2005) Critical period plasticity in local cortical circuits. Nat. Rev. Neurosci. 6, 877–888 [DOI] [PubMed] [Google Scholar]
- 40. Pizzorusso T., Medini P., Berardi N., Chierzi S., Fawcett J. W., Maffei L. (2002) Reactivation of ocular dominance plasticity in the adult visual cortex. Science 298, 1248–1251 [DOI] [PubMed] [Google Scholar]
- 41. Gogolla N., Caroni P., Luthi A., Herry C. (2009) Perineuronal nets protect fear memories from erasure. Science 325, 1258–1261 [DOI] [PubMed] [Google Scholar]
- 42. Romberg C., Yang S., Melani R., Andrews M. R., Horner A. E., Spillantini M. G., Bussey T. J., Fawcett J. W., Pizzorusso T., Saksida L. M. (2013) Depletion of perineuronal nets enhances recognition memory and long-term depression in the perirhinal cortex. J. Neurosci. 33, 7057–7065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Frischknecht R., Gundelfinger E. D. (2012) The brain's extracellular matrix and its role in synaptic plasticity. Adv. Exp. Med. Biol. 970, 153–171 [DOI] [PubMed] [Google Scholar]
- 44. Frischknecht R., Heine M., Perrais D., Seidenbecher C. I., Choquet D., Gundelfinger E. D. (2009) Brain extracellular matrix affects AMPA receptor lateral mobility and short-term synaptic plasticity. Nat. Neurosci. 12, 897–904 [DOI] [PubMed] [Google Scholar]
- 45. Michaluk P., Mikasova L., Groc L., Frischknecht R., Choquet D., Kaczmarek L. (2009) Matrix metalloproteinase-9 controls NMDA receptor surface diffusion through integrin beta1 signaling. J. Neurosci. 29, 6007–6012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nagy V., Bozdagi O., Matynia A., Balcerzyk M., Okulski P., Dzwonek J., Costa R. M., Silva A. J., Kaczmarek L., Huntley G. W. (2006) Matrix metalloproteinase-9 is required for hippocampal late-phase long-term potentiation and memory. J. Neurosci. 26, 1923–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Galko M. J., Tessier-Lavigne M. (2000) Function of an axonal chemoattractant modulated by metalloprotease activity. Science 289, 1365–1367 [DOI] [PubMed] [Google Scholar]
- 48. Kantor D. B., Chivatakarn O., Peer K. L., Oster S. F., Inatani M., Hansen M. J., Flanagan J. G., Yamaguchi Y., Sretavan D. W., Giger R. J., Kolodkin A. L. (2004) Semaphorin 5A is a bifunctional axon guidance cue regulated by heparan and chondroitin sulfate proteoglycans. Neuron 44, 961–975 [DOI] [PubMed] [Google Scholar]
- 49. Yamada J., Jinno S. (2013) Spatio-temporal differences in perineuronal net expression in the mouse hippocampus, with reference to parvalbumin. Neuroscience 253C, 368–379 [DOI] [PubMed] [Google Scholar]
- 50. Soleman S., Yip P. K., Duricki D. A., Moon L. D. (2012) Delayed treatment with chondroitinase ABC promotes sensorimotor recovery and plasticity after stroke in aged rats. Brain 135, 1210–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Scali M., Baroncelli L., Cenni M. C., Sale A., Maffei L. (2012) A rich environmental experience reactivates visual cortex plasticity in aged rats. Exp. Gerontol. 47, 337–341 [DOI] [PubMed] [Google Scholar]
- 52. Soleman S., Filippov M. A., Dityatev A., Fawcett J. W. (2013) Targeting the neural extracellular matrix in neurological disorders. Neuroscience 253C, 194–213 [DOI] [PubMed] [Google Scholar]
- 53. Végh M. J., Heldring C. M., Kamphuis W., Hijazi S., Timmerman A. J., Li K., van Nierop P., Mansvelder H. D., Hol E. M., Smit A. B., van Kesteren R. E. (2014) Reducing hippocampal extracellular matrix reverses early memory deficits in a mouse model of Alzheimer inverted question marks disease. Acta Neuropathol. Commun. 2, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ballatore C., Lee V. M., Trojanowski J. Q. (2007) Tau-mediated neurodegeneration in Alzheimer's disease and related disorders. Nat. Rev. Neurosci. 8, 663–672 [DOI] [PubMed] [Google Scholar]
- 55. Hoover B. R., Reed M. N., Su J., Penrod R. D., Kotilinek L. A., Grant M. K., Pitstick R., Carlson G. A., Lanier L. M., Yuan L. L., Ashe K. H., Liao D. (2010) Tau mislocalization to dendritic spines mediates synaptic dysfunction independently of neurodegeneration. Neuron 68, 1067–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hattori T., Takei N., Mizuno Y., Kato K., Kohsaka S. (1995) Neurotrophic and neuroprotective effects of neuron-specific enolase on cultured neurons from embryonic rat brain. Neurosci. Res. 21, 191–198 [DOI] [PubMed] [Google Scholar]
- 57. Sun K. H., Chang K. H., Clawson S., Ghosh S., Mirzaei H., Regnier F., Shah K. (2011) Glutathione-S-transferase P1 is a critical regulator of Cdk5 kinase activity. J. Neurochem. 118, 902–914 [DOI] [PubMed] [Google Scholar]
- 58. Rhee S. G., Yang K. S., Kang S. W., Woo H. A., Chang T. S. (2005) Controlled elimination of intracellular H(2)O(2): regulation of peroxiredoxin, catalase, and glutathione peroxidase via post-translational modification. Antioxid. Redox Signal. 7, 619–626 [DOI] [PubMed] [Google Scholar]
- 59. Mar J. C., Matigian N. A., Mackay-Sim A., Mellick G. D., Sue C. M., Silburn P. A., McGrath J. J., Quackenbush J., Wells C. A. (2011) Variance of gene expression identifies altered network constraints in neurological disease. PLoS Genet. 7, e1002207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Manolio T. A., Collins F. S., Cox N. J., Goldstein D. B., Hindorff L. A., Hunter D. J., McCarthy M. I., Ramos E. M., Cardon L. R., Chakravarti A., Cho J. H., Guttmacher A. E., Kong A., Kruglyak L., Mardis E., Rotimi C. N., Slatkin M., Valle D., Whittemore A. S., Boehnke M., Clark A. G., Eichler E. E., Gibson G., Haines J. L., Mackay T. F., McCarroll S. A., Visscher P. M. (2009) Finding the missing heritability of complex diseases. Nature 461, 747–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Eichler E. E., Flint J., Gibson G., Kong A., Leal S. M., Moore J. H., Nadeau J. H. (2010) Missing heritability and strategies for finding the underlying causes of complex disease. Nat. Rev. Genet. 11, 446–450 [DOI] [PMC free article] [PubMed] [Google Scholar]