Abstract
Cyclins are essential for cell proliferation, the cell cycle and tumorigenesis in all eukaryotes. UbcH10 regulates the degradation of cyclins in a ubiquitin-dependent manner. Here, we report that UbcH10 is likely involved in tumorigenesis. We found that cancer cells exposed to n-acetyl-leu-leu-norleucinal (ALLN) treatment and UbcH10 depletion exhibit a synergistic therapeutic effect. Abundant expression of UbcH10 drives resistance to ALLN-induced cell death, while cells deficient in UbcH10 were susceptible to ALLN-induced cell death. The depletion of UbcH10 hindered tumorigenesis both in vitro and in vivo, as assessed by colony formation, growth curve, soft agar and xenograft assays. These phenotypes were efficiently rescued through the introduction of recombinant UbcH10. In the UbcH10-deficient cells, alterations in the expression of cyclins led to cell cycle changes and subsequently decreases in tumorigenesis. The tumorigenesis of xenograft tumors from UbcH10-deficient cells treated with ALLN was decreased relative to wild-type cells treated with ALLN in nude mice. On the molecular level, we observed that UbcH10 deficiency enhances the activation of caspase 8 and caspase 3 but not caspase 9 to impair cell viability upon ALLN treatment. Collectively, our results suggest that, as an oncogene, UbcH10 is a potential drug target for the treatment of colorectal cancer.
Malignant neoplasm is one of the most common lethal diseases in the world. Colorectal carcinoma is the 3rd most common malignancy and the 4th most common cause of cancer mortality worldwide, and more than 1 million new cases are diagnosed globally in each year1.
Carcinogenesis occurs when mutations accumulate in critical oncogenes and tumor suppressor genes, leading to the dysregulation of cellular proliferation and the cell cycle and changes in protein expression2. Proper cell cycle progression is controlled by a series of cell cycle checkpoint events3. The dysregulation of cell cycle control often gives rise to uncontrollable cell proliferation and chromosomal instability, which results in tumorigenesis3.
The dynamic regulation of cell cycle proteins, such as the proteasome-dependent proteolysis of cyclin-dependent kinases and cyclin-dependent kinase inhibitors, plays an important role in cell cycle progression4,5,6. In eukaryotes, the degradation of a specific protein is initiated by adding a polyubiquitin chain to a lysine residue in the target protein, and this chain is recognized by the 26S proteasome. This system requires three enzymes, including ubiquitin-activating enzyme E1, which transfers ubiquitin to the ubiquitin-conjugating enzyme E2. The E3 ubiquitin ligase then recruits ubiquitin-charged E2 and the substrate to aid in the transfer of ubiquitin to the target protein via E27,8.
UbcH10, also known as UBE2C, belongs to the ubiquitin-conjugating enzymes family. This E2 protein catalyzes the ubiquitination and degradation of cyclins A and B and acts together with the E3 ligase of the anaphase-promoting complex (APC) to participate in the regulation of the spindle assembly checkpoint9,10,11. A high UbcH10 protein expression level has been reported in many human tumor types, including esophageal adenocarcinomas12, anaplastic thyroid carcinomas13, malignant breast carcinomas14,15, hepatocellular carcinomas16, astrocytic tumors17, lung cancer18, lymphoma19 and colorectal cancer20, suggesting that UbcH10 is closely associated with tumor onset and progression. The UbcH10 mRNA and protein levels are high in several primary cancerous tissues such as those of the lung, stomach, uterus and bladder21. The mRNA level is even higher than that of the other 16 E2 genes21. In colorectal cancer, knockdown of UbcH10 by RNA interference has been shown to block the proliferation of cancer cells, indicating that UbcH10 could be a potential target for cancer therapy22. Although previous studies identified UbcH10 as the E2 of the APC, which is responsible for the degradation of cyclin A and B in cell cycle control, the precise mechanisms by which UbcH10 is related to accelerated tumorigenesis are not yet understood.
ALLN (N-acetyl-Leu-Leu-Norleu-al), also known as calpain inhibitor 1, inhibits calpain I and II and blocks the proteasome pathway23. ALLN strongly activates p53-dependent apoptosis, and our previous studies have shown that BAX is translocated in cells exposed to ALLN24,25. Bartus RT et al. reported that calpain inhibitors may provide a unique and potentially powerful means for treating stroke and other ischemic cerebral events26, and the inhibition of calpain prevented neuronal and behavioral deficits in an N-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) mouse model of Parkinson's disease27; this strongly supports the important role of ALLN.
Here, we report that UbcH10 is highly expressed in colorectal cancer tissues. UbcH10 deficiency was found to significantly impede cell proliferation, colony formation and tumor formation ability in vitro and in vivo. The reintroduction of UbcH10 notably rescued these phenotypes. We also determined that the expression levels of UbcH10 played an important role in sensitizing the cells to ALLN. This is the first report to investigate mechanisms underlying the involvement of UbcH10 in colorectal cancer and the first to propose UbcH10 as a potential drug target. It is also the first to propose the use of ALLN to treat cancer cells with low levels of UbcH10 expression to obtain an improved therapeutic effect.
Results
UbcH10 is highly expressed in colorectal cancer
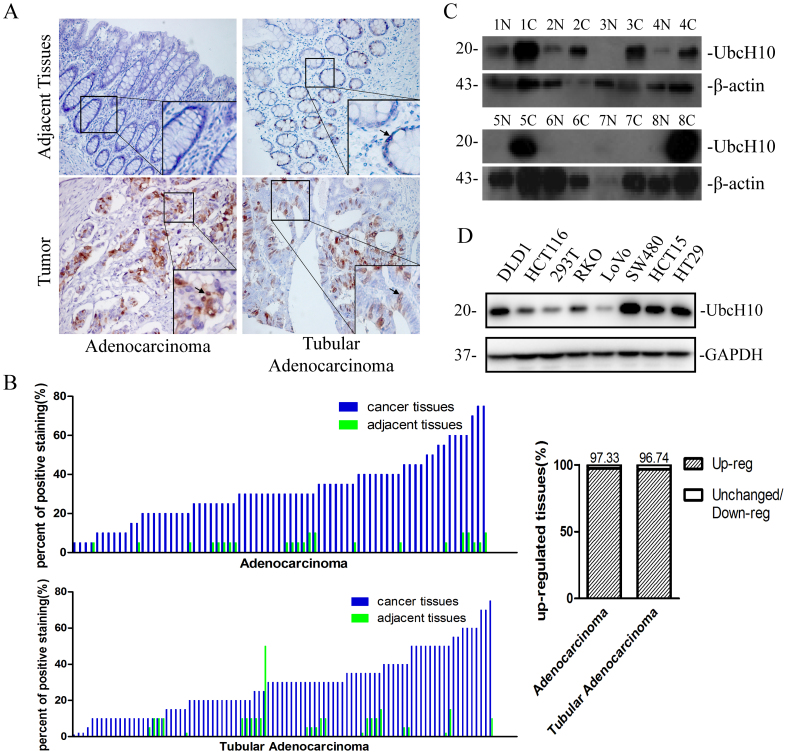
To analyze the expression level of UbcH10 in colorectal cancer, we performed an immunohistochemical analysis of UbcH10 using tissue microarrays containing 75 colon cancer (adenocarcinoma) and 92 rectal cancer (tubular adenocarcinoma) tissue samples, as well as the corresponding adjacent tissues. We estimated the percent of positive stained cells. Most of the cancer tissues showed a higher rate of positive staining for UbcH10 than adjacent tissues (Figure 1A, 1B). The immunoblotting analysis using clinical colorectal samples from Wuhan Union Hospital showed similar results (Figure 1C). Moreover, 5 out of 7 colorectal cancer-derived cell lines showed UbcH10 up-regulation (Figure 1D). Taken together, these results imply that aberrant expression of UbcH10 is related to colorectal cancer.
Figure 1. Expression of UbcH10 in colorectal cancer.
(A) Immunohistochemical staining of UbcH10 on tissue microarrays containing colorectal cancer tissues and adjacent normal tissues. Representative stainings of colon cancer tissues (bottom, left) and rectal cancer tissues (bottom, right) are shown. Arrows indicate the cells that stained positively. (B) Tissue microarray data analysis of UbcH10 expression in tumors and adjacent normal tissues from 75 patients with colon cancer and 92 patients with rectal cancer. Here shows the percent of positive staining cells in the corresponding tissue sample. (C) The UbcH10 protein expression levels of colorectal tumors and adjacent normal tissues were examined using Western blot. β-actin was used as a loading control. N represents normal tissue. C represents tumor tissue. (D) The protein expression levels of UbcH10 in 7 colorectal cancer cell lines and the 293T cell line were examined. GAPDH was used as a loading control.
Targeted depletion of UbcH10 represses tumor formation in vivo and in vitro
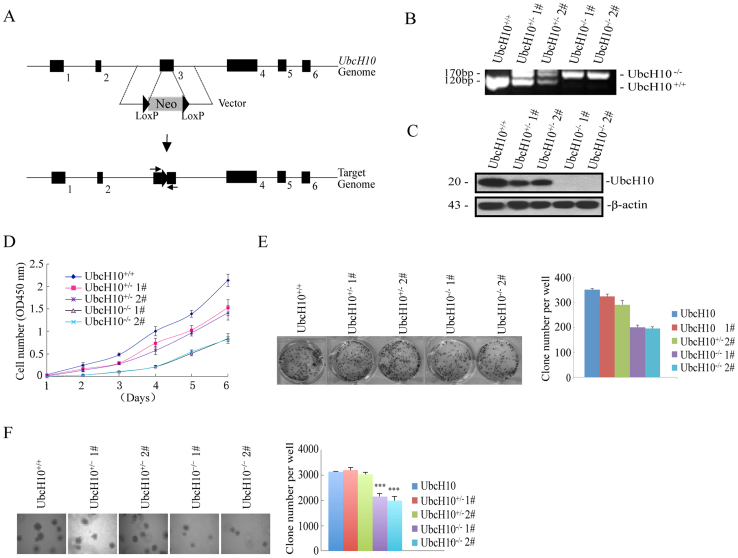
Although UbcH10 is highly expressed in colorectal cancer, it is unclear whether the depletion of UbcH10 suppresses tumorigenesis. As shown in Figure 1D, UbcH10 was abundantly expressed in the DLD1 cell line. Therefore, we inactivated endogenous UbcH10 expression in this cell line using an AAV(Adeno-Associated Virus)-mediated somatic cell knock-out method. The human UbcH10 gene consists of 7 exons; the conserved ubiquitin-conjugating and catalytic (UBCc) domain is located in C terminus. A stop codon added in exon 3 will cause in-frame translation and hence delete the UBCc of UbcH10. Therefore, both alleles of exon 3 were targeted by two rounds of rAAV-mediated homologous recombination (Figure 2A). The targeted cell line was validated via genomic PCR and Western blot analyses. The genomic PCR resulted in a band shift of exon 3, and the immunoblotting analysis detected no trace of the UbcH10 protein in the targeted knockout cells (Figure 2B and C).
Figure 2. The depletion of UbcH10 represses tumor formation in vitro.
(A) Targeting strategy of the UbcH10 genomic locus. Homologous arms (left: 1.1 kb; right: 0.92 kb) were constructed in an AAV vector containing the neomycin-resistance gene (Neo). The homologous recombination resulted in the frame shift and expression silencing of the UbcH10 gene. The genotyping primers are shown with two arrows. (B and C) UbcH10-deficient cells were identified via genomic PCR and immunoblotting. For the DNA analysis, the lower band represents the normal UbcH10 allele and the upper band represents the disrupted allele. For protein analysis, the cell lysates were analyzed via immunoblotting using an anti-UbcH10 antibody. β-actin was used as a loading control. (D) Cell proliferation assay. Approximately 1 × 103 cells of the indicated cell lines were seeded in 96-well plates and analyzed using the CCK8 method each day for seven consecutive days. (E) The indicated cells (3 × 103) were seeded in 6-well plates and then cultured for approximately 10 days. The cell colonies were stained with crystal violet and photographed. The number of clones was counted and plotted. (F) Soft agar colony formation assay. The indicated cells (1 × 104) were seeded in 6-well plates with 0.35% upper agar and 0.7% lower agar for 14 days. The cell colonies were photographed, counted and plotted. The numbers of colonies are expressed as the means ± S.E. from three assays. ***P< 0.001 compared with controls.
Next, we examined the effects of UbcH10 on cellular proliferation and colony formation. Proliferation assays and growth curve analyses were performed to determine the growth properties of these cells. As shown in Figure 2D, the targeted UbcH10 DLD1 cells showed significant growth inhibition, compared with the wild-type cells. To detect the viability and tumorigenicity of the cells in the presence or absence of UbcH10 and/or ALLN, we carried out colony formation and soft agar assays. In the colony formation experiment, the wild-type DLD1 cells form a significant number of colonies, whereas the colony number was decreased when the UbcH10 was depleted (Figure 2E). The soft agar colony formation assays showed that UbcH10 deficiency resulted in a notable decrease in colony size and number, demonstrating that UbcH10 affects anchorage-independent growth in DLD1 cells (Figure 2F).
UbcH10-/- DLD1 cell phenotypes are rescued by the reintroduction of UbcH10
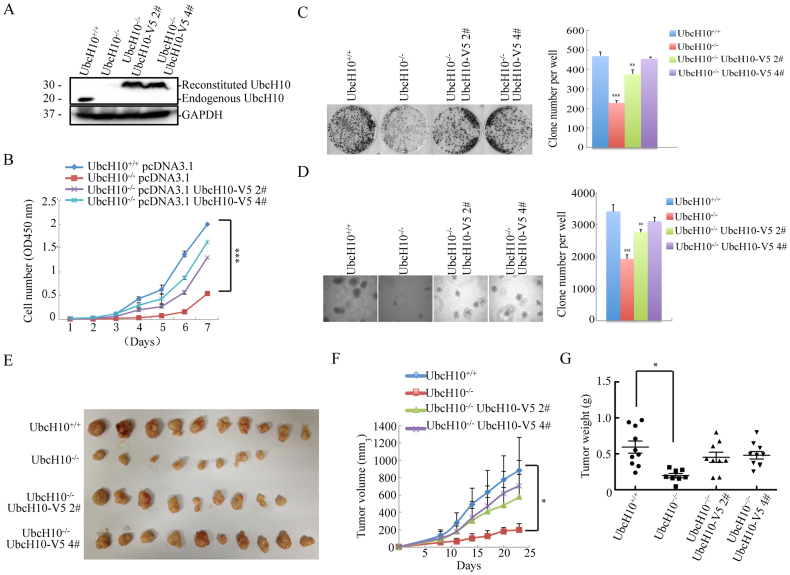
To further confirm the role of UbcH10 in driving tumor growth rather than promoting other nonspecific effects, we performed rescue experiments by stably expressing UbcH10 in the UbcH10-/- DLD1 cell line (Figure 3A). The cell growth assays showed that wild-type DLD1 cells exhibited a higher growth rate than the UbcH10-depleted cells, whereas expression of the UbcH10 protein significantly rescued the cell growth inhibition phenotype of the UbcH10-deficient DLD1 cells (Figure 3B). Both the clone formation and soft agar assays demonstrated that the exogenous expression of UbcH10 in UbcH10-deficient DLD1 cells potently increased the colony number (Figure 3C and D).
Figure 3. Rescue of repressed tumor formation via exogenous UbcH10 expression in vivo and in vitro.
(A) For the exogenous expression of wild-type recombinant UbcH10 in UbcH10-/- DLD1 cells, stable cell lines were generated via plasmid-mediated transfection. The expression of UbcH10 in the DLD1 cells was examined using Western blotting. GAPDH was used as a loading control. (B) Cell proliferation assay. Approximately 1 × 103 cells of the indicated cell lines were seeded in 96-well plates and analyzed using the CCK8 method each day for seven consecutive days. Three repeats were performed and representative results are shown. (C) The indicated cells (3 × 103) were seeded in 6-well plates and then cultured for approximately 10 days. The cell colonies were stained with crystal violet and photographed. The clones were counted and plotted. (D) Soft agar colony formation assay. The indicated cells (1 × 104) were seeded in 6-well plates with 0.35% upper agar and 0.7% lower agar for 14 days. The cell colonies were photographed, counted, and plotted. (E–G) Xenograft experiments were performed by injecting the indicated cells (5 × 106) into the flanks of 6-week-old nude mice to form tumors. The tumors were measured every 3 days, and the tumor volume was calculated using the formula ([length × width2] × 0.5). The tumors were removed and weighed 23 days after injection. The numbers of colonies are expressed as the means ± S.E. from six assays. *P< 0.05, **P< 0.01, ***P< 0.001 compared with controls.
For the in vivo analysis, we injected wild-type, UbcH10-/- and two rescued DLD1 cell lines into nude mice to observe whether tumor growth is specifically affected by UbcH10. This assay demonstrated that the UbcH0 recombinant protein significantly rescued the tumor growth inhibition to a wild-type level. The sizes of the tumors originating from the wild-type cells and rescued cells were only slightly different. Additionally, both of these types of tumors were up to 2-fold larger than the tumors originating from the UbcH10-/- DLD1 cells on day 23 (Figure 3E). Accordingly, notable differences in tumor weight were also observed in the presence and absence of UbcH10. The weight of the tumors from the UbcH10-/- DLD1 cells was more than 2-fold lower than that of the tumors from the rescued cells (Figure 3F and G). These results suggest that UbcH10 plays an important role in the regulation of colorectal cancer tumorigenesis.
Genetic inactivation of UbcH10 stabilizes cyclin A and cyclin B1
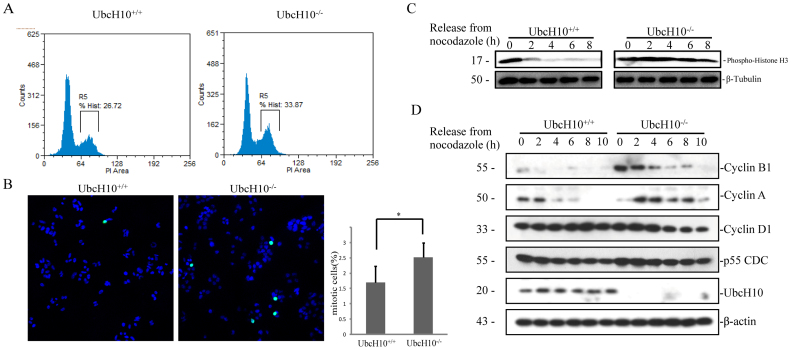
To determine whether UbcH10 affects tumor growth by regulating the rate of proliferation, we assessed the cell cycle profiles and the expression of cyclins over the cell cycle in UbcH10-deficient cells. As shown in Figure 4A and B, the number of UbcH10-/- cells in G2/M phase was higher than that of the wild-type cells. So we examined the G2/M marker histone H3.1(phosphor-Ser10) level after releasing the cells from nocodazole blocking. As expected, the amount of phospho-H3.1 decreases rapidly in UbcH10 normal cells, while in UbcH10-/- cells, phospho-H3.1 remains high level even after releasing for 8 hours. Then we examined the effect of UbcH10 disruption on the expression levels of cyclin proteins. We found that degradation of cyclin B1 was blocked in the UbcH10-/- cells more than in the wild-type cells over the entire cell cycle. Interestingly, cyclin A was modestly decreased at the beginning of the cell cycle and then accumulated over the rest of the cycle in the UbcH10-/- cells. However, the protein levels of cyclin D1, p55CDC and UbcH10 did not notably change under this condition. These experiments demonstrated that UbcH10 affects colon cancer cell growth in vitro and in vivo by regulating the expression of cell cycle proteins, mainly cyclin A and cyclin B1.
Figure 4. The cell cycle distribution is altered in UbcH10-deficient cells.
(A) The proportion of cells in each phase of the cell cycle was analyzed using flow cytometry in UbcH10-deficient cells. The DNA contents of the treated cells were detected using flow cytometry. The number of cells in each phase of the cell cycle was counted and plotted via R5. (B) The cells in G2/M phase were stained via immunofluorescence of Histone H3.1 (phospho Ser10). The blue and green dots represent DAPI and phosphor-H3.1 positive cells, respectively. (mean ± S.E.; *P< 0.05) (C) Histone H3.1 (phospho Ser10) level change after nocodazole blocking. (D) Protein expression levels of cyclins in the indicated cell lines. The cells were synchronized with thymidine and nocodazole, and collected at the indicated times after release from nocodazole treatment. The cell lysates were analyzed via immunoblotting using the indicated antibodies. β-Tubulin and β-actin were used as loading control.
UbcH10-/- DLD1 cells are more sensitive to ALLN than wild-type cells
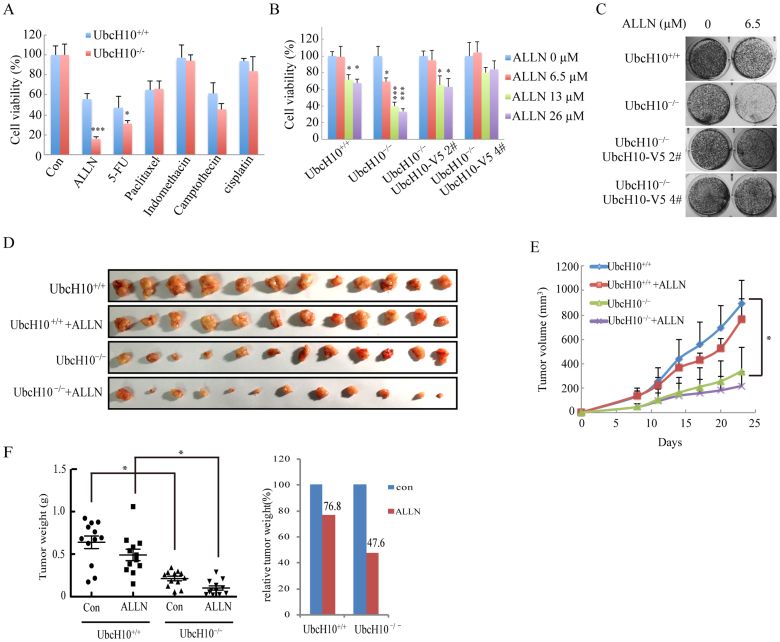
We next determined the response of UbcH10-/- cells and their parental cells to a variety of anticancer agents. A cell viability analysis in different conditions revealed a dramatic increase in ALLN-induced cell death and a moderate increase in 5-fluorouracil and camptothecin-induced cell death in the UbcH10-/- cells compared with their parental cells (Figure 5A and B). The long-term viability of the UbcH10-/- cells, as measured using colony formation assays, was decreased after ALLN treatment and was significantly different from that of the parental cells after the same treatment (Figure 5C). However, the rescued cells displayed restored phenotypes (Figure 5B and C).
Figure 5. UbcH10-deficient DLD1 cells were more susceptible to ALLN.
(A) ALLN was identified as an effective drug for UbcH10-/- DLD1 cells. Approximately 1 × 104 cells of the indicated cell lines were seeded in 96-well plates and treated with various drugs for 24 hours. The viability of the cells was measured 12 hours later using the CCK8 method. The concentration of ALLN was 10 μg/mL (26 μM). (B) ALLN was effective at decreasing the viability of the UbcH10-/- DLD1 cells at a concentration of 26 μM. Approximately 1 × 104 cells of the indicated cell lines were seeded in 96-well plates and treated with various concentrations of ALLN for 24 hours. The viability of the cells was measured immediately using the CCK8 method. (B) Colony formation assay to determine the sensitivity of the indicated cell lines to ALLN. Approximately 1 × 103 cells of the indicated cell lines were seeded in 24-well plates and treated with the indicated concentrations of ALLN for 24 hours. The drug was then removed, and the cells were cultured for an additional two weeks. The cell colonies were stained with crystal violet and photographed. (D–F) Xenograft experiments were performed by injecting wild-type DLD1 or UbcH10-/- DLD1 cells (5 × 106) into the flanks of 6-week-old nude mice to form tumors. The mice were then randomly assigned to the vehicle control group or the ALLN treatment group. The mice in the vehicle control group were intraperitoneally injected with vehicle (saline and ethanol), and the mice in the treatment group were intraperitoneally injected with 10 mg/kg/day of ALLN. The injections were performed once each day for two weeks. The tumor volumes were measured every 3 days. At the end of the experiment, the mice were euthanized, and the tumors were weighed on an electronic balance. The numbers of colonies are expressed as the means ± S.E. from three assays. *P< 0.05, **P< 0.01, ***P< 0.001 compared with controls.
To determine whether the sensitivity of UbcH10-/- cells to ALLN can be effectively used to treat tumors in nude mice, we injected UbcH10-/- or wild-type DLD1 cells into the flanks of nude mice. As shown in Figure 5D and 5E, for the wild-type group, the tumor volume of the treated mice was reduced to 85.7% on day 23 compared with the control mice. However, for the UbcH10-/- group, the tumor volume of the treated mice was reduced to 64.5% compared with the control mice. The tumor weight was also measured and is presented in Figure 5F. Consistent with the tumor volume, the tumor weights of the treated groups were decreased to 76.8% in the wild-type group and to 47.6% in the UbcH10-/- group compared with the control groups. These findings suggest that ALLN treatment is more effective in tumors with lower UbcH10 expression.
UbcH10 deficiency promotes the apoptotic effect of ALLN
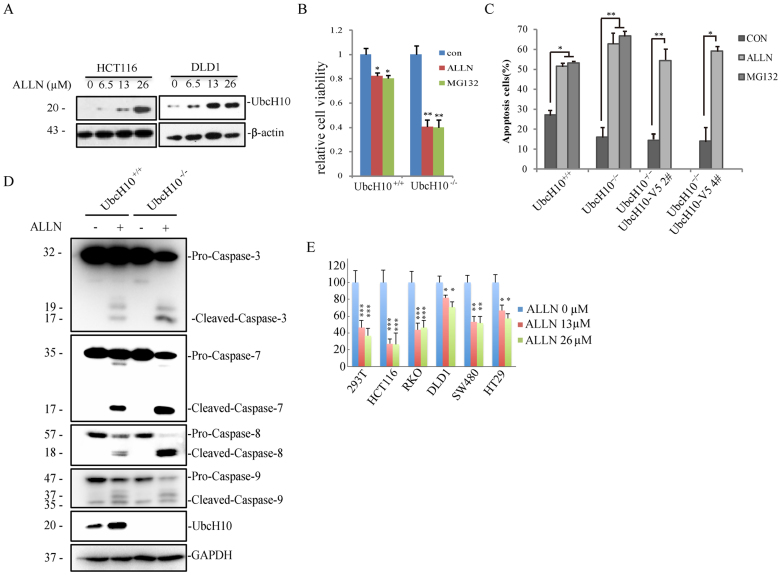
As an up-regulated protein in tumor cells, we assumed that UbcH10 is responsible for resistance to apoptosis drugs. The protein level of UbcH10 as detected using western blotting partly confirms this hypothesis because Ubch10 increased in the DLD1 and HCT116 cells as the ALLN concentration increased (Figure 6A). Because ALLN itself is a proteasome inhibitor and causes ER stress, we introduced MG132 as a positive control. Interestingly, the phenotypes caused by ALLN are the same as those of MG132. In the cell viability experiment, the UbcH10-deficient cells showed more sensitivity to MG132, as well as ALLN (Figure 6B). An analysis of nuclear morphology and apoptosis via Annexin V/PI staining confirmed that ALLN-induced apoptosis was notably reinforced in the UbcH10-/- cells compared with the parental cells, while re-introduction of UbcH10 slightly rescued the phenotype (Figure 6C and S1). The relative apoptosis rates of ALLN and MG132 also supported the cell viability results. To further investigate the mechanism of apoptosis in this process, we examined the expression of apoptosis markers using western blotting. We found that cleaved caspase 3, caspase 7 and caspase 8 were clearly increased upon ALLN treatment in the UbcH10-/- cells compared with the wild-type cells, whereas cleaved caspase 9 was not notably changed upon ALLN treatment between these two cell types (Figure 6D). These experiments suggest that UbcH10 is closely involved in ALLN-induced cell apoptosis.
Figure 6. UbcH10 deficiency aggravates ALLN-induced apoptosis.
(A) HCT116 and DLD1 cells were exposed to the indicated concentrations of ALLN for 24 hours. The cell lysates were analyzed via immunoblotting using anti-UbcH10 antibody. β-actin was used as a loading control. (B) DLD1-UbcH10–/– cells are sensitive to both ALLN- and MG132-induced apoptosis. Approximately 1 × 104 cells of the indicated cell lines were seeded in 96-well plates and treated with 26 μM ALLN or 3.3 μM MG132 for 24 hours. The viability of the cells was measured immediately using the CCK8 method. (C) Annexin-V/PI staining followed by flow cytometry. DLD1 and DLD1-UbcH10–/– cells were exposed to ALLN (26 μM) or MG132(3.3 μM) for 24 hours. The cells were then stained with Annexin-V/PI for 15 min, and apoptosis was analyzed via flow cytometry. The early and late apoptosis rates of the cells are shown. (D) DLD1 and DLD1-UbcH10–/– cells were treated with ALLN (0 or 26 μM) for 24 hours. The cell lysates were analyzed via immunoblot using the indicated antibodies. GAPDH was used as a loading control. (E) Cell susceptibility to ALLN is related to UbcH10 level. Approximately 1 × 104 cells of the indicated cell lines were seeded in 96-well plates and treated with various concentrations of ALLN for 24 hours. The viability of the cells was measured immediately using the CCK8 method. The cell numbers are expressed as the means ± S.E. from six assays. *P< 0.05, **P< 0.01, ***P< 0.001 compared with controls.
Nevertheless, we were interested in whether UbcH10 overexpression in other cells has similar effects on cell death. As expected, the cell lines that abundantly expressed UbcH10 were significantly resistant to ALLN-induced cell death, but the cell lines with relatively lower UbcH10 expression were susceptible to ALLN-induced cell death (Figure 6E).
Discussion
Tumorigenesis usually results from cell cycle defects and blocking of the apoptosis pathway28,29. UbcH10 has been widely reported to be highly expressed in various cancer types and to be significantly involved in tumorigenesis30. Townsley et al. cloned UbcH10, a human homolog of E2-C, and reported that dominant-negative human UbcH10 proteins inhibit the destruction of both cyclin A and B in mammalian cells9. Recent work has revealed that UbcH10 is E2 in APC/C and is involved in a specific E2-E3 interface and regulates APC activity via its N-terminal extension for substrate selection and checkpoint control11,31,32. However, there is little evidence indicating that UbcH10 could be a drug target, and little is known about how UbcH10 promotes tumorigenesis. In this report, we demonstrate that inhibition of the expression of UbcH10 could be a valid method to mitigate tumor growth in colon cancer and that ALLN could be an effective drug to treat tumors by lowering UbcH10 expression.
We are interested in exploring the function of UbcH10 due to substantial reports that UbcH10 is ectopically expressed in various cancers. To verify this result, we analyzed the expression of UbcH10 in a number of colorectal cancer cell lines and patient samples. Consistent with previous studies, we found that UbcH10 is highly expressed in colorectal cancer. After confirming this phenomenon, we knocked out UbcH10 by disrupting the genomic DNA in DLD1 cells. Using these cells, we demonstrated that the inhibition of UbcH10 potently retarded the proliferation of tumor cells. Chen et al. obtained similar results via RNA interference-mediated silencing of UbcH1022. Importantly, cell cycle changes and the accumulation of cyclin A and cyclin B1 were directly tested in endogenous conditions. We first studied the protein expression profiles of cyclin A and cyclin B1 throughout the entire cell cycle in the absence of UbcH10. Interestingly, after being released from nocodazole, cyclin A was modestly down regulated in the knock-out cells compared with the wild-type cells. In contrast, cyclin B1 was highly expressed throughout the entire cell cycle in the knock-out cells; the up-regulation of cyclin B1 is consistent with previous studies that demonstrated that cyclin B1 is down regulated when UbcH10 is overexpressed in MEFs from UbcH10 transgenic mice33. The blockage of cyclin B1 degradation is always considered a pivotal event when cells encounter problems in exiting mitosis. The flow cytometry and H3S10 staining results strongly indicated that UbcH10-deficient cells are arrested in the G2/M phase. The UbcH10 deficiency-induced phenotypes were significantly rescued through the reintroduction of UbcH10. UbcH10 was found to be predominantly localized in the nucleus, which also indicates a close relationship between UbcH10 and cyclins. These findings strongly indicate that UbcH10 is involved in cell cycle regulation during tumorigenesis.
We also examined the different effects of anti-cancer drugs on UbcH10-/- cells and wild-type cells. ALLN possessed a more powerful lethality when UbcH10 was absent in a screen of various anti-cancer drugs. The results of our rescue experiments corroborated the increased susceptibility of UbcH10-/- cells to ALLN, which was very encouraging. Therefore, we attempted to confirm this result under physiological conditions. Various cell lines with different UbcH10 expression levels were used to measure ALLN sensitivity. As expected, the cells with low UbcH10 expression were more susceptible to ALLN-induced cell death. Strikingly, when the cells were treated with ALLN, the expression of UbcH10 became markedly increased in both the DLD1 and HCT116 cells. This finding appeared to be paradoxical, but this may be a compensation mechanism in which the ALLN-induced inhibition of the proteasome-dependent degradation pathway gives rise to the increase in E2/UbcH10. Our results strongly indicate that UbcH10 is closely related to ALLN-induced cell death. In the in vivo experiments, we found that in ALLN-treated mice, the volume and weight of tumors from UbcH10-/- cells were potently suppressed compared with the volume and weight of tumors from wild-type cells; however, this suppression was apparently not associated with ALLN-induced cell death in the UbcH10-/- cells.
We showed that a complete knockout of UbcH10 could reduce tumorigenesis. However, it is likely that down-regulating UbcH10 to different extents results in a range of therapeutic effectiveness. The lack of currently available therapies that can target UbcH10 also limits the applicability of the results of this study; the development of potential small molecule drugs that can inhibit UbcH10 is necessary for making advances in this field. Calpain inhibitors are widely used for therapeutic purposes in mouse models of neurological disease34 as well as for suppressing cell cycle progression35. The limited effectiveness of tumor drugs is thought to be partly caused by the DNA damage checkpoint escape of tumor cells. Previous research by Reddy et al showed that the APC-specific protein UbcH10 promotes cell mitosis36, corresponding with our results that ALLN functions more effectively in UbcH10-deleted cells. Furthermore, we also observed that ALLN sensitizes UbcH10-/- cells to the same extent as MG132 (a proteasomal inhibitor that induces apoptosis) does. This serves as an explanation for the intrinsic mechanism of UbcH10.
In summary, our study demonstrates that UbcH10 is an important oncogene involved in colorectal cancer. UbcH10 is essential for tumorigenesis and functions by changing the cell cycle profile of tumor cells. Cells with lower UbcH10 expression are more susceptible to ALLN treatment. Therefore, UbcH10 could be a target for cancer therapy, and the combination of lowering UbcH10 expression and administering ALLN or other drugs may be more effective in the treatment of colorectal tumor patients.
Methods
Ethics Statement
All studies involving human cells were approved by the Committee on the Use of Human Subjects in Research of Wuhan University. All of the animal studies were conducted in accordance with the Guidelines of the China Animal Welfare Legislation, as approved by the Committee on Ethics in the Care and Use of Laboratory Animals of Wuhan University (Permit Number: 11100E). All efforts were made to minimize suffering.
Xenografts
Four-week-old female athymic nude mice were obtained from HFK Bio (Peking, China). When the mice were 6 weeks old, UbcH10-/- cells and parental cells were collected and washed twice with PBS. A total of 5 × 106 cells were resuspended in 0.2 mL PBS and inoculated into the flanks of 10 mice. On the 8th day following inoculation, the mice were divided into two groups: the control group was intraperitoneally injected with vehicle (saline and ethanol), and the treatment group was intraperitoneally injected with 200 μL ethanol containing 10 mg/kg (26 μM) ALLN. The tumors were measured every 3 days, and the tumor volumes were calculated using the formula ([length × width2] × 0.5). The tumors were removed and weighed 23 days after injection.
Cell culture
Human colorectal carcinoma DLD1, HCT116, RKO, LoVo, HCT15, HT29, and SW480 cells were obtained from the American Type Culture Collection and maintained in McCoy's 5A Medium (AppliChem, Darmstadt, Germany, A1324, 9050) with 10% (v/v) fetal bovine serum (FBS; HyClone, Logan, UT) and 100 U penicillin-streptomycin (Gibco, Gaithersburg, MD) at 37°C in 5% CO2. Additionally, HEK293T cells were maintained in DMEM Medium (HyClone) with 10% (v/v) FBS (HyClone) and 100U penicillin-streptomycin (Gibco) at 37°C in 5% CO2.
Immunohistochemical analysis
The tissue microarray included tumors and adjacent normal tissues from 75 cases of colon cancer and 92 cases of rectal cancer (Shanghai Biochip Co., Ltd). Paraffin-embedded tissue sections (4 μm) were prepared according to standard methods, and the expression of UbcH10 was detected using immune peroxidase. The expression of UbcH10 was evaluated and scored according to the staining intensity and area, estimated jointly by three observers under a multi-head microscope, clinical pathological information was previously masked to the observers.
Genetic knock-out and knock-in of UbcH10 in human somatic cells
The genetic inactivation of UbcH10 in DLD1 human colorectal cancer cells was performed using AAV-mediated recombination following a previously described method (Kohli et al, 2004). Briefly, two groups of homologous arms (1 and 1 kb, 1.1 and 0.92 kb) were constructed in an AAV vector containing the neomycin resistance gene (Neo). HEK293T cells were transfected with a target vector containing two homologous arms and two packaging vectors (AAV-Helper and AAV-RC). After 3 days, the cells were collected with a cell scraper and subjected to three freeze-thaw cycles using liquid nitrogen. The supernatant was harvested as the rAAV stock. A total of 1 × 105 DLD1 cells were infected with rAAV virus for 2 days and then split into six 96-well plates to obtain as many single clones as possible. After 2 weeks of selection in 0.5 mg/mL G418, the single clones were screened using PCR, and the positive clones were amplified. Then, the clones were passaged into one well of a 24-well plate, and GFP-Cre adenovirus was added into the well to excise the resistant gene. The clones with one disrupted allele were screened via PCR using the appropriate primers. The second allele was then disrupted using the same method.
Chemicals and antibodies
ALLN (208719) was purchased from Cal Bio Tech (Spring Valley, CA, USA). DAPI (4′,6-diamidino-2-phenylindole) (D3571) was obtained from Invitrogen (Grand Island, NY, USA). Thymidine was purchased from Sigma (St. Louis, MO, USA). Rabbit monoclonal anti-caspase 3 antibody (9665), mouse monoclonal anti-caspase 7 antibody (9494S), mouse monoclonal anti-caspase 8 antibody (9746) and rabbit polyclonal anti-caspase 9 antibody (9502) were obtained from Cell Signaling Technology (Beverly, MA, USA). Rabbit polyclonal anti-UbcH10 antibody (AB3861) was purchased from Millipore (Billerica, MA, USA). Mouse monoclonal anti-Flag antibody (AC004) was obtained from Sigma (St. Louis, MO, USA). Mouse polyclonal anti-GAPDH antibody (CW0100A) was purchased from the CWBIO (Peking, China). Rabbit histone H3.1 (phospho-ser10) antibody (11184-1) was purchased from Signalway Antibody (Baltimore, MD, USA). Horseradish peroxidase- conjugated secondary antibody was obtained from Jackson Immuno Research. Alexa Fluor® 488 goat anti-rabbit IgG (H+L) antibody (A-11070) was purchased from Invitrogen (Grand Island, NY, USA).
Transient and stable overexpression
Transient transfections were performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions (Life Technologies, Inc.). The transfection reagent and DNA were mixed in Opti-MEM (Invitrogen), and the complex was added to cells grown to 40–80% confluence. After 4 hours, the medium was replaced with fresh medium. For stable transfection with pcDNA3.1 UbcH10-V5, 1 mg/mL G418 was added to the medium 48 hours after transient transfection, and the cells were selected for 2 weeks. The stable transfected cells were maintained in 0.5 mg/mL G418-containing medium.
Colony formation and soft agar assay
For the colony formation assay, DLD1 cells (3 × 103) were exposed to 6.5 μM ALLN for 24 hours. The medium and drugs were then replaced with fresh medium. Ten days later, the cell colonies were stained with crystal violet and photographed. For the soft agar assay, Sea Plaque Agarose (Cambrex Bio Science Rockland, 50101) and 2 mL 0.7% lower agar-McCoy's 5A Medium was plated into each well of 6-well plates. Next, 1 mL DLD1 cells (1 × 104) were mixed with 1 mL 0.7% agar-McCoy's 5A mix and added on top of the solidified lower agar, 2 mL McCoy's 5A medium was added to the upper agar, and the plates were incubated at 37°C in 5% CO2 for approximately 3 weeks. Finally, the clones were counted and photographed.
Fluorescence confocal microscopy
The UbcH10 knock-out and wild-type DLD1 cells were cultured on cover slips. After 24 hours, the cells were fixed with 4% paraformaldehyde for 10 min. The fixed cells were incubated with the indicated antibodies or DAPI and observed via Olympus (FV 1000, Tokyo, Japan) confocal microscope under a 10× objective.
Cell cycle synchronization and cell cycle assay
Cells were plated in standard growth medium to achieve approximately 40% confluence on the following day, the standard growth medium was replaced with medium containing 0.4 mM thymidine(Sigma) for 18 hours, washed with PBS and grown in fresh medium for 4 hrs. Followed by grown in culture medium contain 100 ng/mL nocodazole for 12 hours, then the release of the cells. The cells were collected at various time points after nocodazole was removed, allowing for the collection of cells at specific cell cycle phases. For flow cytometric analysis, the collected cells were fixed in 1 mL 70% ice-cold ethanol, incubated at 4°C for 30 minutes, and then centrifuged at 1,000 g for 5 minutes to remove the ethanol. The cell pellet was washed with PBS and suspended in 0.5 mL PBS containing 50 μg/mL RNase A at 37°C for 30 minutes. Then, 50 μg/mL PI staining solution was added and incubated for 30 min at room temperature in the dark. The samples were analyzed via flow cytometry to determine the cell cycle distribution (Harper, 2005).
Cell viability and apoptosis assay
Cell viability was assayed using Cell Counting Kit-8 (CCK-8, Dojindo Laboratories, Kumamoto, Japan) following the manufacturer's protocol. All cell types (1 × 104) were seeded into each well of a 96-well plate, cultured to 80% confluence and treated with different doses of ALLN and other anticancer drugs for 24 hours. The medium was then replaced with 100 μl fresh McCoy's 5A complete medium with 10% CCK-8 reagent, and the cells were incubated for 1 hour at 37°C. The absorbance was measured at 450 nm using an ELx800 microplate reader (BioTek, USA). The results are shown as cell viability percentages. For the apoptosis assay, all cell types (1 × 105) were plated in 6-well plates and cultured to 80% confluence. After treatment with ALLN for 24 hours, the attached and floating cells were harvested at various time points, washed twice with phosphate-buffered saline (PBS), and resuspended in 1x binding buffer at a concentration of 1 × 106 cells/mL. The cells were stained with PI and Annexin V for 15 minutes at room temperature. The samples were analyzed via flow cytometry (Beckman Coulter, Fullerton, CA, USA).
Immunoblotting analysis
The immunoblotting analysis was performed as described previously25. Briefly, cells were collected at various time points and lysed with SDS-sample buffer. Protein concentrations were determined with a BCA protein assay kit (Thermo). The 30-μg samples were loaded onto a gel for SDS-PAGE, transferred and incubated with primary and HRP-conjugated secondary antibodies. The membranes were then developed with the Immobilon Western Chemiluminescent HRP Substrate kit (Merck Millipore).
Data statistical analysis
Each experiment was performed in triplicate. We show the most representative experiments, and the values are presented as the means ±S.D. Student's t-test was used to determine the difference between the experimental and control groups. p<0.05 indicated statistical significance.
Author Contributions
S.-Z.L., R.-L.D. and X.-D.Z. designed the experiments; S.-Z.L., Y.S., H.-H.Z., B.-X.J. and Y.L. performed the experiments; S.-Z.L.,Y.S. and H.-H.Z. analyzed the data; S.-Z.L., R.-L.D., X.-D.Z., W.-B.L. and Y.L. prepared the manuscript. All authors reviewed the paper.
Supplementary Material
Supplementary Materials
Acknowledgments
This work was supported by grants from the National Basic Research Program of China (2011CB944404), the National Natural Science Foundation of China (81270306), the National Science and Technology Support Project (2014BAI02B01, 2012BAI39B02, 2012BAI39B03), the Trans-Century Training Programme Foundation for the Talents by the State Education Commission (NCET-10-0655), and Fundamental Research Funds for the Central Universities (204275771).
References
- Tenesa A. & Dunlop M. G. New insights into the aetiology of colorectal cancer from genome-wide association studies. Nat Rev Genet 10, 353–358 (2009). [DOI] [PubMed] [Google Scholar]
- Fearon E. R. & Vogelstein B. A genetic model for colorectal tumorigenesis. Cell 61, 759–767 (1990). [DOI] [PubMed] [Google Scholar]
- Hartwell L. H. & Kastan M. B. Cell cycle control and cancer. Science 266, 1821–1828 (1994). [DOI] [PubMed] [Google Scholar]
- Glotzer M., Murray A. W. & Kirschner M. W. Cyclin is degraded by the ubiquitin pathway. Nature 349, 132–138 (1991). [DOI] [PubMed] [Google Scholar]
- King R. W., Deshaies R. J., Peters J. M. & Kirschner M. W. How proteolysis drives the cell cycle. Science 274, 1652–1659 (1996). [DOI] [PubMed] [Google Scholar]
- Nakayama K. I. & Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer 6, 369–381 (2006). [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet 30, 405–439 (1996). [DOI] [PubMed] [Google Scholar]
- Glickman M. H. & Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 82, 373–428 (2002). [DOI] [PubMed] [Google Scholar]
- Townsley F. M., Aristarkhov A., Beck S., Hershko A. & Ruderman J. V. Dominant-negative cyclin-selective ubiquitin carrier protein E2-C/UbcH10 blocks cells in metaphase. Proc Natl Acad Sci U S A 94, 2362–2367 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastians H., Topper L. M., Gorbsky G. L. & Ruderman J. V. Cell cycle-regulated proteolysis of mitotic target proteins. Mol Biol Cell 10, 3927–3941 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers M. K., Pan B., Mukhyala K. & Jackson P. K. The unique N terminus of the UbcH10 E2 enzyme controls the threshold for APC activation and enhances checkpoint regulation of the APC. Mol Cell 31, 544–556 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. et al. Expression and effect of inhibition of the ubiquitin-conjugating enzyme E2C on esophageal adenocarcinoma. Neoplasia 8, 1062–1071 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallante P. et al. UbcH10 overexpression may represent a marker of anaplastic thyroid carcinomas. Brit J Cancer 93, 464–471 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlingieri M. T. et al. UbcH10 is overexpressed in malignant breast carcinomas. Eur J Cancer 43, 2729–2735 (2007). [DOI] [PubMed] [Google Scholar]
- Fujita T. et al. Clinicopathological relevance of UbcH10 in breast cancer. Cancer Sci 100, 238–248 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieta K. et al. Identification of overexpressed genes in hepatocellular carcinoma, with special reference to ubiquitin-conjugating enzyme E2C gene expression. Int J Cancer 121, 33–38 (2007). [DOI] [PubMed] [Google Scholar]
- Jiang L. et al. Expression of ubiquitin-conjugating enzyme E2C/UbcH10 in astrocytic tumors. Brain Res 1201, 161–166 (2008). [DOI] [PubMed] [Google Scholar]
- Kadara H. et al. Identification of gene signatures and molecular markers for human lung cancer prognosis using an in vitro lung carcinogenesis system. Cancer Rrev Res 2, 702–711 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troncone G. et al. UbcH10 expression in human lymphomas. Histopathology 54, 731–740 (2009). [DOI] [PubMed] [Google Scholar]
- Chen S. et al. Association of clinicopathological features with UbcH10 expression in colorectal cancer. J Cancer Res Clin 136, 419–426 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto Y. et al. UbcH10 is the cancer-related E2 ubiquitin-conjugating enzyme. Cancer Res 63, 4167–4173 (2003). [PubMed] [Google Scholar]
- Chen S. M. et al. RNA interference-mediated silencing of UBCH10 gene inhibits colorectal cancer cell growth in vitro and in vivo. Clinical Exp Pharmacol P 37, 525–529 (2010). [DOI] [PubMed] [Google Scholar]
- Lee D. H. & Goldberg A. L. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Bio 8, 397–403 (1998). [DOI] [PubMed] [Google Scholar]
- Atencio I. A., Ramachandra M., Shabram P. & Demers G. W. Calpain inhibitor 1 activates p53-dependent apoptosis in tumor cell lines. Cell Growth Differ 11, 247–253 (2000). [PubMed] [Google Scholar]
- Li S. Z. et al. ALLN hinders HCT116 tumor growth through Bax-dependent apoptosis. Biochem Bioph Res Co 437, 325–330 (2013). [DOI] [PubMed] [Google Scholar]
- Bartus R. T. et al. Postischemic administration of AK275, a calpain inhibitor, provides substantial protection against focal ischemic brain damage. J Cereb Blood Flow Metab 14, 537–544 (1994). [DOI] [PubMed] [Google Scholar]
- Crocker S. J. et al. Inhibition of calpains prevents neuronal and behavioral deficits in an MPTP mouse model of Parkinson's disease. J Neurosci 23, 4081–4091 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin P. J., Vogelstein B. & Kinzler K. W. Apoptosis and APC in colorectal tumorigenesis. Proc Natl Acad Sci U S A 93, 7950–7954 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zornig M., Hueber A., Baum W. & Evan G. Apoptosis regulators and their role in tumorigenesis. Biochim Biophys Acta 1551, F1–37 (2001). [DOI] [PubMed] [Google Scholar]
- Kapoor S. UbcH10 and its emerging role in systemic carcinogenesis. Int J Surg Pathol 21, 538 (2013). [DOI] [PubMed] [Google Scholar]
- Rape M., Reddy S. K. & Kirschner M. W. The processivity of multiubiquitination by the APC determines the order of substrate degradation. Cell 124, 89–103 (2006). [DOI] [PubMed] [Google Scholar]
- Williamson A. et al. Identification of a physiological E2 module for the human anaphase-promoting complex. Proc Natl Acad Sci U S A 106, 18213–18218 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ree J. H., Jeganathan K. B., Malureanu L. & van Deursen J. M. Overexpression of the E2 ubiquitin-conjugating enzyme UbcH10 causes chromosome missegregation and tumor formation. J Cell Biol 188, 83–100 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinchese F. et al. Inhibition of calpains improves memory and synaptic transmission in a mouse model of Alzheimer disease. J Clin Invest 118, 2796–2807 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storr S. J., Carragher N. O., Frame M. C., Parr T. & Martin S. G. The calpain system and cancer. Nat Rev Cancer 11, 364–374 (2011). [DOI] [PubMed] [Google Scholar]
- Reddy S. K., Rape M., Margansky W. A. & Kirschner M. W. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature 446, 921–925 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials