Abstract
Fast neutron radiation has been used as a mutagen to develop extensive mutant collections. However, the genome-wide structural consequences of fast neutron radiation are not well understood. Here, we examine the genome-wide structural variants observed among 264 soybean [Glycine max (L.) Merrill] plants sampled from a large fast neutron-mutagenized population. While deletion rates were similar to previous reports, surprisingly high rates of segmental duplication were also found throughout the genome. Duplication coverage extended across entire chromosomes and often prevailed at chromosome ends. High-throughput resequencing analysis of selected mutants resolved specific chromosomal events, including the rearrangement junctions for a large deletion, a tandem duplication, and a translocation. Genetic mapping associated a large deletion on chromosome 10 with a quantitative change in seed composition for one mutant. A tandem duplication event, located on chromosome 17 in a second mutant, was found to cosegregate with a short petiole mutant phenotype, and thus may serve as an example of a morphological change attributable to a DNA copy number gain. Overall, this study provides insight into the resilience of the soybean genome, the patterns of structural variation resulting from fast neutron mutagenesis, and the utility of fast neutron-irradiated mutants as a source of novel genetic losses and gains.
Keywords: soybean, fast neutron, structural variation, deletion, duplication
DNA damage from ionizing radiation occurs in both natural and induced settings. Structural characterization of this DNA damage is useful for understanding environmental consequences and for targeted research objectives. Many mutant populations have been created from ionizing radiation for forward and reverse genetics purposes. These resources have been utilized to select for phenotypes of interest and to enable connections between gene and function. Mutants created from fast neutron (FN) irradiation have been utilized in many species, including Arabidopsis (Li et al. 2001), Medicago truncatula (Oldroyd and Long 2003; Rogers et al. 2009), Glycine soja (Searle et al. 2003), Citrus clementina (Rios et al. 2008), Hordeum vulgare (Zhang et al. 2006), Pisum sativum (Domoney et al. 2013), and Lotus japonicus (Hoffmann et al. 2007). These studies were conducted for the primary purpose of identifying gene–phenotype associations.
Whole-genome experimental platforms provide the technologies to examine the impact of radiation damage. The stability and functionality of whole genomes can be evaluated, in addition to the influence of single genes or loci. To date, observations on a handful of mutants derived from FN irradiation have identified genomic deletions in the range of a single base pair to a few base pairs to several megabases (Bruggemann et al. 1996; Li et al. 2001; Men et al. 2002). With the increased capabilities of whole-genome technologies, we now have the ability to screen for structural variation (SV) in more individuals with greater genome coverage than ever before.
Soybean [Glycine max (L.) Merrill] is a paleopolyploid species with a sequenced genome size of ∼1.1 Gb that contains ancient homeologous regions and other paralogous genes (Schlueter et al. 2004; Doyle and Egan 2010; Schmutz et al. 2010; Severin et al. 2011). Due to multiple rounds of genome duplication, up to 12 duplicated copies of a region may occur in the soybean genome (Cannon and Shoemaker 2012). The duplicative nature of the genome indicates that there are regions of the genome that may be dispensable (reviewed in Marroni et al. 2014). In addition, the majority of the soybean genome (57%) is composed of heterochromatic pericentromeric regions that are low in gene density and rich in repeat sequences (Du et al. 2010; Schmutz et al. 2010). Therefore, the natural composition of the genome may act as a buffer, preventing phenotypes that would otherwise result from mutagenesis-induced genomic variation.
A soybean FN mutant population (Bolon et al. 2011) was previously developed to serve as a phenotypic and genomic resource for genetic screens and functional genomics research. FN radiation was used to induce lesions in the soybean genome and generate a library of >27,000 unique soybean mutants. Forward genetic screening resulted in the identification of phenotypes that deviated from wild-type for ∼2% of the population. Genomic analyses on a preliminary subset of 30 mutants using array-comparative genomic hybridizations (aCGHs) uncovered genomic regions that exhibited significant SV, including deletions and duplications that ranged in size from 986 bp to almost 3 Mb (Bolon et al. 2011).
In this study, we report the largest set of independent genome-wide structural observations from a FN-irradiated mutant population to date. Array-comparative genomic hybridization and next-generation sequencing (NGS) technologies were applied to soybean FN-irradiated mutants from the population resource. Data from 264 mutants were analyzed from three selection classes: forward genetic screen mutants, no-phenotype mutants, and reverse genetic screen mutants. The objectives were to catalog genome-wide patterns of induced genomic SV and discover SV regions associated with specific mutant phenotypes. The results define the distribution of induced deletions and reveal the prevalence of segmental duplications in genomes following FN irradiation. These findings provide insight toward the sustainable damage and recoverable variations in the soybean genome and indicate that novel deletions and duplications arising in the population hold promise for generating useful traits.
Materials and Methods
Soybean fast neutron mutant population development and forward genetic screens
Soybean FN mutants were created as previously described (Bolon et al. 2011). Seeds (60,000) from 2006 Crop Improvement Association stocks of mid-maturity group I cv. ‘M92-220’ were shipped to the McClellan Nuclear Radiation Center at the University of California, Davis and exposed to FN radiation doses of 4, 8, 16, and 32 Gy. An additional 60,000 seeds of the same germplasm stock were irradiated at 16 and 32 Gy. M1 seed was planted and propagated by single seed descent. M2 seed was planted in a row by column grid format, and M2 plants were individually tagged with an assigned barcode as a unique identifier. Young leaf tissue was collected for each M2 plant, and M3 seed was single-plant harvested from 27,000 unique M2 plants. Twenty M3 seeds derived from each unique M2 plant were subsequently planted in rows, and individual plants were tagged and harvested in subsequent generations such that each plant could be tracked back to its unique M2 source. Additionally, a small sample of M3 seed derived from each of the 27,000 unique M2 plants was collected, labeled, and frozen for long-term storage.
For the forward genetic screen on seed composition phenotypes, seeds were initially analyzed for seed composition by near infrared (NIR) spectroscopy using a Perten DA7200 diode array instrument (Huddinge, Sweden) equipped with calibration equations developed by Perten in cooperation with the University of Minnesota. For the forward genetic screen on above-ground phenotypes, M2 and later generation plants were evaluated for the morphology and pigmentation of visible plant tissues and for days to maturity. For the forward genetic screen on below-ground phenotypes, plants were grown for 3 weeks in quartz sand inoculated with US Department of Agriculture Bradyrhizobium and watered with 50% Hoagland’s solution. Roots were evaluated for architecture, size, depth, color, and nodulation.
Reverse genetic screen on the fast neutron mutants
Sequenom MassARRAY technology was used for high-throughput quantification of homeologous gene proportions within soybean FN mutants. Primers were designed for extension coverage across known SNPs identified from the soybean reference genome sequence (Schmutz et al. 2010) in 29 homeologous genes between chromosomes 8 and 15, and 30 homeologous genes between chromosomes 3 and 19. DNA was isolated from the leaf tissue of 760 randomly selected unique M2 individuals and used as a template for PCR and extension reactions across the SNP regions. Quantification of mass spectrometry results on the PCR and extension products was performed at the University of Minnesota Genomics facility. Results were confirmed by aCGH for mutants showing putative copy number differences between the homeologous intervals.
Structural variant detection by comparative genomic hybridization microarrays
Labeling and aCGH methodologies for the initial 30 hybridizations are described in detail in Bolon et al. (2011). All subsequent aCGH experiments were performed using similar methods, with some minor modifications. Briefly, labeling reactions for mutant (Cy3 dye) and reference (Cy5 dye) samples were performed with 1 μg each of genomic DNA from mutant and reference leaf tissue samples. DNA from the cv. ‘M92-220’ wild-type background was used for the reference sample. The mutant samples each consisted of DNA from one FN individual, with no bulking or pooling of mutant samples. Genomic DNA was isolated using the Qiagen DNeasy kit and the 3′–5′ exo-Klenow fragment from DNA polymerase I was used to incorporate the label. After quantification of labeled DNA, the labeled DNA was hybridized for 72 hr at 42° on the 700K feature or the 1.4M feature NimbleGen aCGH arrays designed for G. max. Each array consisted of unique probe sequences (50–60mers) based on the reference ‘Williams 82’ soybean sequence Glyma.Wm82.a1.v1.1 (www.phytozome.net) (Schmutz et al. 2010) and spaced at ∼0.5- to 1.1-kb intervals on average. The 700K and 1.4M array platforms have been described in previous publications (Haun et al. 2011; Anderson et al. 2014).
Comparative genome hybridization data were analyzed using the Roche NimbleGen NimbleScan v2.5 segMNT algorithm and by Roche NimbleGen DEVA software. Corrected log2 ratios were obtained for each probe datapoint. Minimum segment lengths of two probes and segment log2 ratio differences of 0.1 between segments at 0.999 acceptance percentile were required to define genomic segment borders. Significant copy number changes were determined by retrieving segments with an average corrected log2 ratio greater than the probe average plus three standard deviations (increase) or less than the average minus three standard deviations (decrease). SV segment increases were considered to be duplication segments. SV segment decreases of 3 standard deviations or more were called homozygous deletions unless the magnitude of the deviation was less than or equal to half of the maximum deviation found for the sample. If SV segment decreases were >1.5 standard deviations but less than or equal to half of the maximum deviation within the sample, these segments were considered to be hemizygous deletions. In addition, if potential segment gaps were less than half the size of the total distance covered by neighboring segments and showed the same direction of change, then the entire region was considered a single SV event as described (Bolon et al. 2011). Visual inspection of aCGH profiles was performed to identify any obvious SVs that were not detected by the automated pipeline.
Final mutant deletion and duplication regions were determined after filtering against regions of natural heterogeneity (Haun et al. 2011) in the population. To determine candidate regions of genetic heterogeneity or natural variation within the population, the standard deviation from the average corrected log2 ratio of Cy3 (sample) to Cy5 (control) intensities was calculated at each probe position. After the average absolute value of the above was calculated for each probe position as described (Bolon et al. 2011), the 95th percentile border was calculated across the unique probe positions, and the median value across each 11-probe sliding window was determined. Regions with median values that peaked above the 95th percentile border or with SV in both directions for >5% of samples without shared traits were removed from the analysis.
Genes from the December 2012 annotation download (Glyma.Wm82.a1.v1.1, Glyma189) that overlapped SV events were determined using a custom Perl script. The genes in the soybean genome that have retained paralogs within syntenic blocks were identified using BLAST (Altschul et al. 1990), DAGChainer (Haas et al. 2004), and selection of gene pairs from syntenic blocks with average Ks values between 0.03 and 0.60; the full list of such genes has been previously published (Anderson et al. 2014). Chromosome diagrams were visualized using CViT (Cannon and Cannon 2011).
Development and screening of mapping populations with fast neutron soybean mutants
Soybean FN mutant FN0172932 was crossed to lines ‘Anoka’ (PI 548508) and ‘Minsoy’ (PI 548389) and backcrossed to cv. ‘M92-220’ to create mapping populations. In subsequent generations, soybean seeds were analyzed for seed protein and oil composition by NIR using a Perten DA7200 diode array instrument equipped with calibration equations developed by Perten in cooperation with the University of Minnesota. Genomic DNA samples from nine individuals at tail ends of the seed composition phenotype were isolated using the Qiagen DNeasy kit protocol and assayed on the Illumina Goldengate platform for genotyping using 1536 soybean Universal SNP BARC markers (Hyten et al. 2010). SNP markers were subject to BLAST (Altschul et al. 1990) analysis to identify the corresponding physical SNP position along the reference genome sequence, and polymorphic SNP locations were examined for FN-induced structural variations. PCR confirmation of the deletion in FN0172932 was performed on DNA from 81 segregating backcrossed progeny at each end of the seed composition phenotype distribution using flanking primer sequences 5′-GCTGGAGCTAGCTGAGGAACCTG-3′ and 5′-GGGGGAGATTGTGGAAGCAATGACT-3′ and interior primer sequences 5′-TGGCCGATGTGATGGACTTCTTCC-3′ and 5′-GGGGGAGATTGTGGAAGCAATGACT-3′.
Soybean FN mutant FN0163764 was crossed to cv. ‘MN1410’ to create a mapping population for a short petiole mutant phenotype. Eighty F3 progeny derived from 80 random F2 plants were phenotyped. Flower color was recorded, and petiole lengths were measured from the lowest three nodes at maturity. Genomic DNA was extracted from young leaf tissue of these F3 progeny for PCR detection and confirmation of structural variation events. PCR confirmation of the tandem duplication in FN0163764 was performed using primer sequences 5′-TGCCTTATTTCTTCAATTGCCC-3′, 5′-TAGAATCCACGCCCTTTACG-3′, and 5′-CGTCATTTTCAGGGACCAGC-3′ with an expected product size of 145 bp across the tandem duplication junction and 339 bp for the control.
Structural variant detection by high-throughput DNA sequencing
Genomic DNA from mutant plants FN0172932, FN0163764, and the wild-type background ‘M92-220’ were extracted using the Qiagen Plant DNeasy system. Barcoded libraries for each sample were prepared by the TruSeq DNA method, pooled, and sequenced across eight lanes. For each mutant sample, sequencing results were generated for the equivalent of one lane of sequencing reads, with a minimum of 125 million pass filter reads. For the wild-type sample, sequencing results were generated for the equivalent of two lanes of sequencing reads. The average library size was 400–500 bp with an insert of ∼280–380 bp. Libraries were sequenced on 100-bp paired-end runs by Illumina HiSeq, and the average quality score was >Q30. Paired-end 100-bp sequencing reads were filtered and aligned to the soybean genome sequence (Phytozome v9.0, ftp://ftp.jgi-psf.org/pub/compgen/phytozome/v9.0/Gmax/) (Schmutz et al. 2010) using BWA-mem (Li and Durbin 2010), and structural variation was analyzed using a combination of CREST (Wang et al. 2012), Breakdancer (Chen et al. 2009), and SAMtools (Li et al. 2009). All resequencing data are available through the Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra/) under accession no. SRP036841, with specific genotypes as accession nos. SRX467195, SRX467189, and SRX467197.
Results
Differential screening of a soybean fast neutron mutant population resource
The soybean FN mutant population was developed by exposing >120,000 soybean seeds of cv ‘M92-220’ to fast neutron radiation doses of 4, 8, 16, or 32 Gy (Bolon et al. 2011). Approximately 27,000 M1 individuals were propagated by single seed descent to the M2 generation. As such, the population can be viewed as 27,000 lineages each derived from a unique M2 individual. Propagation of subsequent generations beyond the M3 has been variable among the lineages.
The M2 individuals and M3 seed were respectively evaluated by forward genetic screens for visual above-ground phenotypes and seed composition alterations (Figure 1). In addition, progeny from >5000 M2-derived lineages were evaluated for visual below-ground phenotypes on roots and nodules. To date, ∼500 mutants have been validated based on identification of a phenotype and confirmation of the observed phenotype over an additional generation. These mutants were propagated for further evaluation over multiple locations and years.
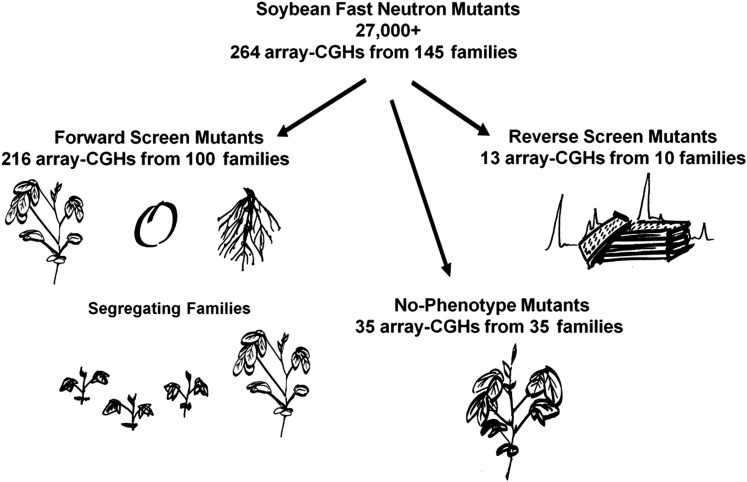
Figure 1.
Fast neutron-irradiated mutants analyzed by aCGH for genomic SV. The soybean fast neutron mutant population resource consists of >27,000 M2-derived lineages. Three categories (forward screen, no-phenotype, and reverse screen) of mutants were assayed based on distinct selection criteria. aCGHs on 216 forward screen mutants representing 100 unique M2-derived lineages were chosen from among mutants with documented aberrations in visual above-ground appearance, seed composition, or root and nodule screening. In some cases, multiple sibling plants from segregating families were assayed. No-phenotype plants showing no observed differences from the wild-type background were represented by 35 unique M2-derived individuals. Ten M2 plants with large deletions/duplications in specific homeologous intervals were identified from the Sequenom MassARRAY reverse screen. Thirteen plants representing these 10 lineages were analyzed by aCGH.
Forward genetic screens based on seed composition, above-ground, or below-ground visual phenotypes identified 216 mutant plants that were selected for genomic analyses. Herein, these plants will be described as “forward screen” mutants. Some M2-derived lineages are represented by more than one plant, as sibling plants were sometimes sampled from segregating rows in subsequent generations. The 216 forward screen mutant plants thus represent 100 unique M2-derived lineages (Figure 1). Furthermore, 35 FN-derived plants with no observed phenotype were randomly selected from the population to represent individuals with no phenotype, and are herein referred to as the “no-phenotype” class. Phenotypic information derived from both the forward screen and no-phenotype plants was deposited at the updated mutant database: http://www.soybase.org/mutants.
A reverse genetic screen using Sequenom MassARRAY technology was also conducted on a subset of the soybean FN-irradiated mutant population. Sequenom MassARRAY technology provides a quantification of the ratio between SNPs at any given locus (Springer and Stupar 2007). As a reverse genetic screen in soybean, the method relies on the presence of SNPs between otherwise conserved homeologous genes within the sequenced soybean genome. The genomic DNA for 760 randomly selected M2 individuals was screened for unbalanced SNP proportions, as indications of deletion or duplication events within the homeologous regions relative to one another. SNPs at 29 homeologous gene pairs between syntenic regions on chromosomes 8 and 15 (Lin et al. 2010) and 10 homeologous gene pairs between syntenic regions on chromosome 3 and 19 (Supporting Information, Table S1) were assayed for unbalanced DNA ratios. Therefore, the Sequenom MassARRAY technology served as a reverse genetic screen for deletions and duplications in 760 M2 plants at large homeologous regions located between chromosomes 8 and 15 and 3 and 19, respectively.
Ten M2 individuals exhibited clear copy number differences among homeologous groups. Six of these M2 individuals were directly advanced to aCGH analyses. Sufficient DNA was not available to perform aCGH on the remaining four M2 individuals; therefore, one or two individuals from more advanced generations (M3 or M5) were chosen to investigate the putative deletions/duplications identified in these lineages. Altogether, 13 plants representing the original 10 M2 individuals (Figure 1) were selected for aCGH analyses and are herein referred to as the “reverse screen” mutants.
Genomic analyses of fast neutron mutants by aCGH
Previously, genomic analysis was carried out on 30 soybean FN mutants using a custom NimbleGen soybean tiling aCGH platform with nearly 700,000 features (Bolon et al. 2011). For a more comprehensive survey of the soybean FN-irradiated mutant population, genome-wide analysis through aCGH was carried out on 90 additional mutants on the ∼700,000 feature platform. To add greater resolution to the analyses, 144 additional mutants were genotyped on an updated ∼1.4 million feature platform. Altogether, a total of 264 soybean FN-irradiated mutants were structurally genotyped between the two aCGH platforms (Table S2). Each of the 264 aCGH experiments were used to profile the genomic SV of individual plants (i.e., no bulking of multiple individuals was done for these experiments). Furthermore, the 264 plants represented a wide range of FN generations, including 13 M2, 127 M3, 69 M4, 23 M5, eight M6, six M7, and 18 M8 individuals (Table S2).
As mentioned above, several individuals genotyped by aCGH were descended from the same M2 individual. After accounting for multigenerational and sibling individuals, including those from advanced families with segregating phenotypes, a total of 145 unique M2-derived lineages were represented in the panel of 264 plants (Figure 1). Analysis by aCGH was also performed on two additional arrays, representing controls for the cultivar ‘M92-220’ wild-type progenitor background. Categorization of all 264 mutants analyzed by aCGH is documented in Table S2.
Analysis of SV within the 264 FN plants assayed by aCGH uncovered segmental duplications and deletions, down to ∼0.5 kb (the smallest segment was 493 bp). Chromosomal regions with frequently reoccurring SV among the 264 plants were inferred to be intervals of natural genomic heterogeneity among the cv. ‘M92-220’ seeds that were mutagenized (Table S3). Because these SV events were not caused by FN irradiation, these regions were filtered out of the downstream analysis. In total, 1216 duplications and deletions inferred to be induced by FN irradiation were detected within the genomes of the 264 FN mutants (Table S4). On average, approximately one segmental duplication was observed in any single mutant (Table 1). Furthermore, homozygous deletions were observed two to three times per mutant, and hemizygous deletions were observed approximately once per mutant (Table 1). Detected SV events were deposited at SoyBase (Grant et al. 2009) under www.soybase.org/mutants and genome-wide views of duplication and deletion coverage are displayed with online browse and search features via the relational database and at the “fast neutron mutant” gbrowse track at www.soybase.org (example screenshot, Figure S1).
Table 1. Summary of genome-wide structural variant mean values from fast neutron-irradiated mutants.
| Event | Mean event number per plant | Mean event size (kb) | Mean gene number per event |
|---|---|---|---|
| Segmental duplication | 1.02 | 2189 | 150 |
| Homozygous deletion | 2.50 | 420 | 13 |
| Hemizygous deletion | 1.09 | 1773 | 58 |
Prevalence of chromosome end duplications following fast neutron irradiation
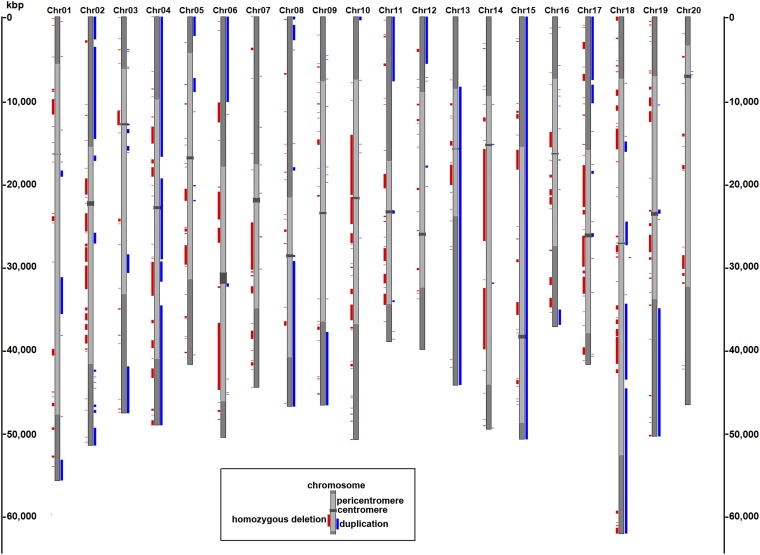
A tremendous range of SV events were identified in the FN mutant population. The genome-wide coverage of duplication and homozygous deletion events in the genomes of the 264 mutants is summarized in Figure 2. Segmental duplication events were detected on every chromosome and revealed a total of 268 segmental duplications among the 264 mutants. It is noteworthy that duplication events represented a total coverage of >586 Mb and a tiling coverage of over a quarter of the soybean genome (∼289 Mb) (Figure 2, Figure S2). Duplications ranged in size from 499 bp to >50 Mb, with a mean size of >2 Mb and a median size of ∼51 kb (Table 1 and Table 2). Approximately 15% (40/268) of segmental duplications included regions at or near the telomere (defined as intervals within 10 kb of a chromosome end). Therefore, while the majority of duplications are interstitial, there is a clear enrichment for duplications at or near chromosome ends (Figure 3A).
Figure 2.
Genome-wide view of segmental duplication and deletion event coverage detected in fast neutron-irradiated soybean mutants. Each bar represents 1 of 20 soybean chromosomes. Light gray lines indicate pericentromeric regions while euchromatic and centromeric regions are shown in dark gray. Homozygous deletion event coverage is represented by red bars to the left of the chromosome, and segmental duplication event coverage is represented by blue bars to the right of the chromosome. The scale across the vertical chromosomes is represented in kilobase pairs (kbp).
Table 2. Summary of genome-wide structural variant median values from fast neutron-irradiated mutants.
| Event | Median event number per plant | Median event size (kb) | Median gene number per event |
|---|---|---|---|
| Segmental duplication | 0 | 51 | 0 |
| Homozygous deletion | 2 | 38 | 3 |
| Hemizygous deletion | 0 | 938 | 27 |
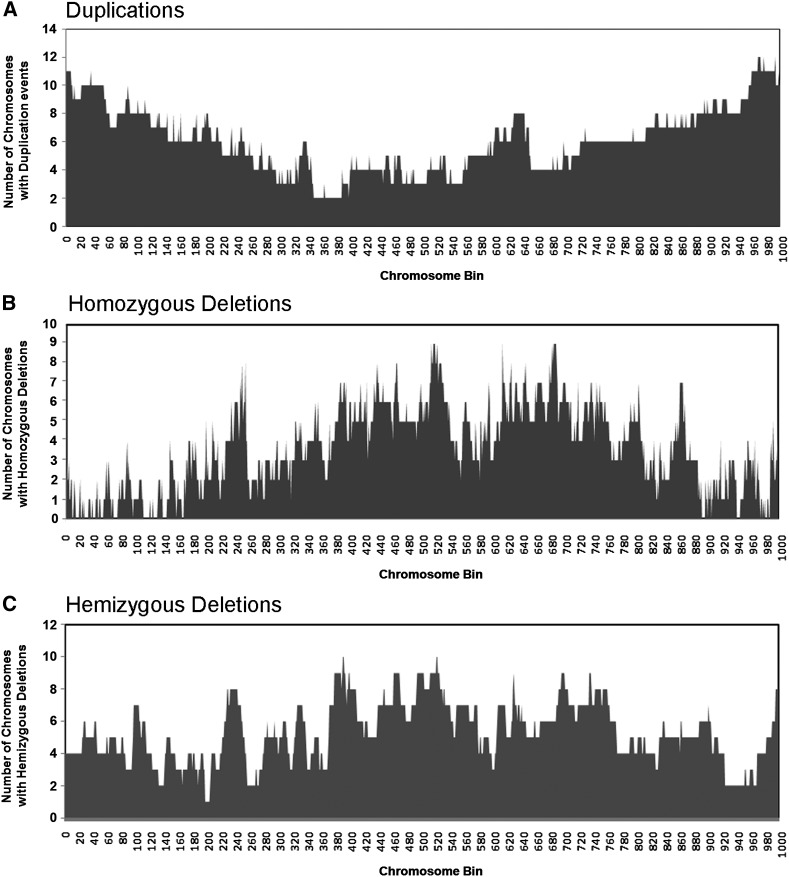
Figure 3.
Frequency of SV across chromosome regions in fast neutron-irradiated soybean mutants. Each chromosome was divided into 1000 bins of equal size in numerical order. The number of chromosomes (out of 20) with at least one SV event for each representative chromosome bin was calculated. (A) Segmental duplication events are most prevalent at chromosome ends. (B) Homozygous deletions are most prevalent in the pericentromeric regions. (C) Hemizygous deletions are found across the entire chromosome.
Deletion events occurred more frequently in the heterochromatic and pericentromeric chromosomal regions than in the gene-rich chromosomal arms (Figure 2, Figure S3, and Figure S4). Homozygous deletion sizes ranged from 493 bp to 8.1 Mb, and hemizygous deletion sizes ranged from 4.9 kb to 9.3 Mb. Homozygous deletions (∼38 kb, median size) were substantially smaller than hemizygous deletions (∼938 kb, median size) (Table 2), perhaps due to increased lethality associated with large homozygous deletions. Tiling coverage of homozygous deletion events (∼157 Mb) within these mutants encompassed approximately one-seventh of the soybean genome.
The chromosomal distribution of SV events were further examined to reveal distinct trends for segmental duplications vs. deletions. Composite representations of these events and their prevalence among chromosomes are shown in Figure 3. The highest frequency of segmental duplications involved chromosome ends compared to other regions of the chromosome (Figure 3A). In contrast, deletion events, particularly homozygous deletions, occurred more frequently away from chromosome ends (Figure 3, B and C). We estimated that only 0.9% (6/661) of homozygous deletions and 5.2% (15/287) of hemizygous deletions occur at or near the telomeres, in contrast to the 15% observed for duplications (see above).
Validation and segregation of large duplications and deletions
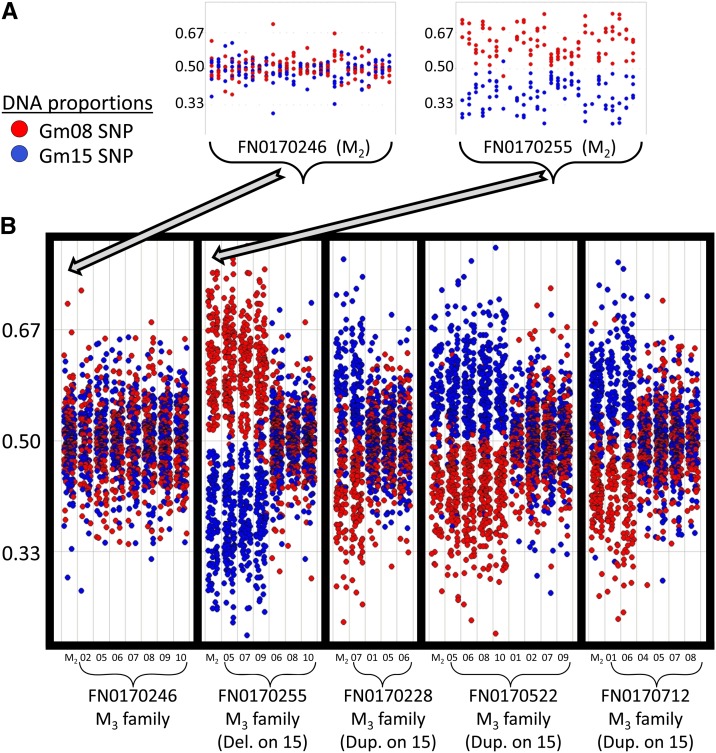
We sought to validate a subset of large SV events, as the size and frequency of such events (particularly the duplications) revealed by aCGH was surprising. Individual M2 plants harboring putative hemizygous deletions or duplications within the ∼1-Mb syntenic intervals on chromosomes 8 and 15 were identified based on reverse screens (explained above) and aCGH analysis. Following one generation of self-pollination, a subset of M3 individuals was subjected to Sequenom MassARRAY analysis to quantify the relative SNP ratios between the chromosome 8 and 15 regions (Lin et al. 2010). Biased SNP ratios indicated individuals that inherited the deletion or duplication, while balanced SNP ratios indicated individuals that segregated back to diploid status in these regions. Four segregating families are shown in Figure 4. aCGH data were used to determine the approximate size and chromosomal location of each event and whether each event was a deletion or duplication (Figure S5). Segregation patterns indicated that approximately half of the M3 progeny inherited the duplications/deletions, while the other half segregated back to wild type. These data confirm that the large deletions and duplications are present in the M2 generation, and hemizygous events are subject to segregation in subsequent generations. These results indicate that large SVs may be generated by FN mutagenesis; however, more advanced generations will likely show fewer events as these loci may segregate back to the wild-type status.
Figure 4.
Validation of large segmental deletions and duplications in soybean fast neutron plants. Sequenom MassARRAY analysis was used to provide a relative quantification of DNA copy number between the ∼1-Mb syntenic intervals on chromosomes 8 and 15. SNPs differentiating a series of homeologous genes on chromosomes 8 and 15 were identified and quantified relative to one another. Red spots indicate the proportion of SNPs at each position that match the chromosome 8 homeolog, while blue spots indicate the proportion of SNPs at each position that match the chromosome 15 homeolog. Twenty-seven SNPs located throughout the ∼1-Mb intervals are shown. Plants with wild-type chromosomes would be expected to exhibit approximately equal proportions (e.g., 0.50 and 0.50, respectively) of blue and red spots throughout the interval, while plants harboring large duplications or deletions in these intervals would exhibit biased proportions (e.g., 0.33 blue and 0.67 red proportions when a deletion spans the chromosome 15 interval on one of the homologous chromosomes). (A) Results for two M2 individuals are shown, representing a wild-type proportion (plant FN0170246) and a biased proportion (plant FN0170255). FN0170255 shows a strong bias favoring the chromosome 8 SNPs throughout the syntenic interval, indicating the presence of either a large duplication in the chromosome 8 interval or a large deletion in the chromosome 15 interval. (B) The plots are tightened to display the data for more individuals. For each segregating family, the M2 parent is shown on the far left. The M3 individuals (numbered on the x-axis) are grouped according to whether they maintained the large deletion/duplication (left side) or whether they segregated back to wild-type proportions (right side). The aCGH data (shown in Figure S5) indicated that the FN0170255 M2 plant harbored a large deletion that included the syntenic interval on chromosome 15. Therefore, the FN0170255 M3 family showed segregation of this large deletion. Meanwhile, the aCGH data (Figure S5) indicated that the FN0170228, FN0170522, and FN0170712 M2 plants, respectively, harbored large duplications, each including the syntenic interval on chromosome 15. Therefore, these families showed segregation of the large duplications.
SV patterns specific to mutation selection category
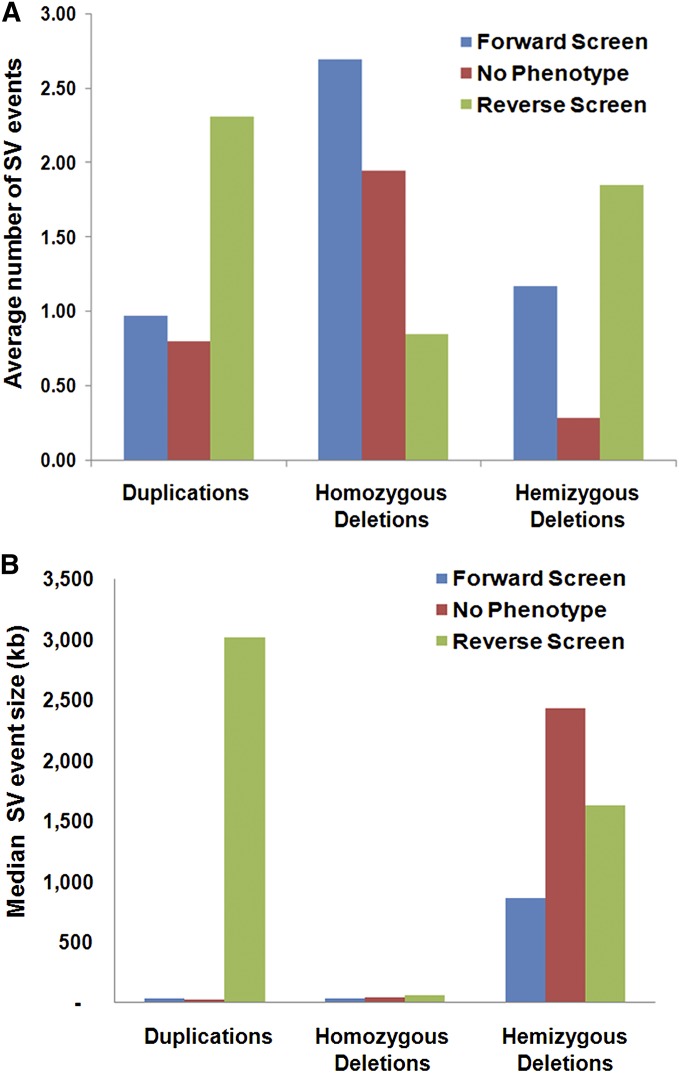
Characteristic size and frequency patterns were observed for SV events detected in forward screen, no-phenotype, and reverse screen mutant categories (Figure 5, Figure S2, Figure S3, and Figure S4). The reverse screen subpopulation recovered the highest average number of duplication events (Figure 5A). Furthermore, the reverse screen duplication events exhibited a remarkably large size compared to the forward screen and no-phenotype subpopulations (Figure 5B). This included the striking example of reverse screen mutant FN0170228, where duplication events spanned nearly all of chromosome 15 (Figure 2, Figure S2C, and Table S4).
Figure 5.
Summary of observed SV number and size within fast neutron mutant selection classes. (A) Bar graph of average number of SVs in forward screen, no-phenotype, and reverse screen mutants for duplications, homozygous deletions, and hemizygous deletions. (B) Bar graph of the median size of SVs in forward screen, no-phenotype, and reverse screen mutants for duplications, homozygous deletions, and hemizygous deletions.
A comparison of SV event frequency by class showed that the highest average number of homozygous deletion events was found in mutants that belonged to the forward screen subpopulation (Figure 5A). This interesting observation may indicate that mutants exhibiting clear phenotypes may be more likely to harbor conspicuous homozygous deletions. For the no-phenotype class, an average of almost two homozygous deletion events per plant was observed at a median deletion size of ∼41 kb (Figure 5, A and B). Notably, a homozygous deletion event of >1.6 Mb in size was detected in a no-phenotype FN plant (Table S4).
Genic coverage of soybean fast neutron mutant SV events
Since the functional effects of gene mutations (knockouts, duplications, etc.) are of great interest, the number of soybean genes that mapped within SV segments detected by aCGH was assessed. The mean number of genes per genotype associated with segmental duplications, homozygous deletions, and hemizygous deletions was 150, 13, and 58, respectively (Table 1). Of the 53,927 unique soybean genes (www.phytozome.net, Glyma.Wm82.a1.v1.1, Glyma189) annotated within the sequenced soybean genome (Schmutz et al. 2010), >20,000 genes (40.5%) were found within duplication regions, ∼5000 genes (9.1%) were found within homozygous deletion regions, and >10,000 genes (19.4%) were found within hemizygous deletion regions detected by aCGH among these 264 mutants (Table 3).
Table 3. Summary of the genes found within detected SV events among fast neutron-irradiated mutants.
| Category | Whole genome | Segmental duplication | Homozygous deletion | Hemizygous deletion |
|---|---|---|---|---|
| Total genes | 53,927 | 21,588 | 4924 | 10,455 |
| Genes with syntenic paralog | 32,579 | 8265 | 1313 | 3769 |
| Genes without syntenic paralog | 21,349 | 13,323 | 3611 | 6686 |
| % with syntenic paralog | 60.41 | 38.29 | 26.67 | 36.05 |
| % without syntenic paralog | 39.59 | 61.71 | 73.33 | 63.95 |
The relative rates for genes with a retained syntenic paralog are compared to genes that have not retained a syntenic paralog.
One of the underlying questions in conducting a genome-wide analysis of hundreds of FN mutants is whether the presence of certain genes is essential or dispensable. Of the 53,927 unique genes annotated in the G. max v1.1 reference soybean genome, 32,579 have maintained a syntenic paralog since whole-genome duplication, while 21,349 do not maintain a syntenic paralog (though they may maintain a paralog(s) elsewhere in the genome) (Anderson et al. 2014). Notably, the rate of induced SV in the FN population was found to be considerably higher for genes that have not maintained a syntenic paralog (Table 3). While it may be hypothesized that genes retaining a syntenic paralog may be more susceptible to mutagenic disturbances because of the buffer provided by the redundant paralogous gene, this does not appear to be the case in our dataset, particularly regarding homozygous deletion events (3611 were observed in genes without syntenic paralogs, while only 1313 were observed in genes with syntenic paralogs). However, these results are consistent with the observation of higher rates of SV in the pericentromeric regions, where genes that have not retained a syntenic paralog are at a higher relative density (Du et al. 2012).
Association of a fast neutron mutant deletion with a quantitative change in seed composition
Due to the nature of FN mutagenesis, mutants are likely to possess more than one deletion or duplication. The above observations on the frequency of deletions or duplications in mutants confirmed this premise. To demarcate the genomic region linked to a quantitative mutant phenotype, several outcross and backcross populations were created using soybean FN mutants. One particular seed composition mutant of interest was from the mutant family FN0172932. This mutant displayed higher seed oil and lower seed protein levels compared to the wild-type background. Outcrossed mapping populations were developed by crossing FN0172932 with soybean lines ‘Anoka’ and ‘Minsoy.’ In addition, a backcross population was created between the FN0172932 mutant and the wild-type background ‘M92-220.’ The subsequent segregating populations were evaluated for both seed composition phenotypes (NIR for oil/protein and combustion protocol for seed protein concentrations) and genotypes (high-throughput SNP assays and PCR-based markers).
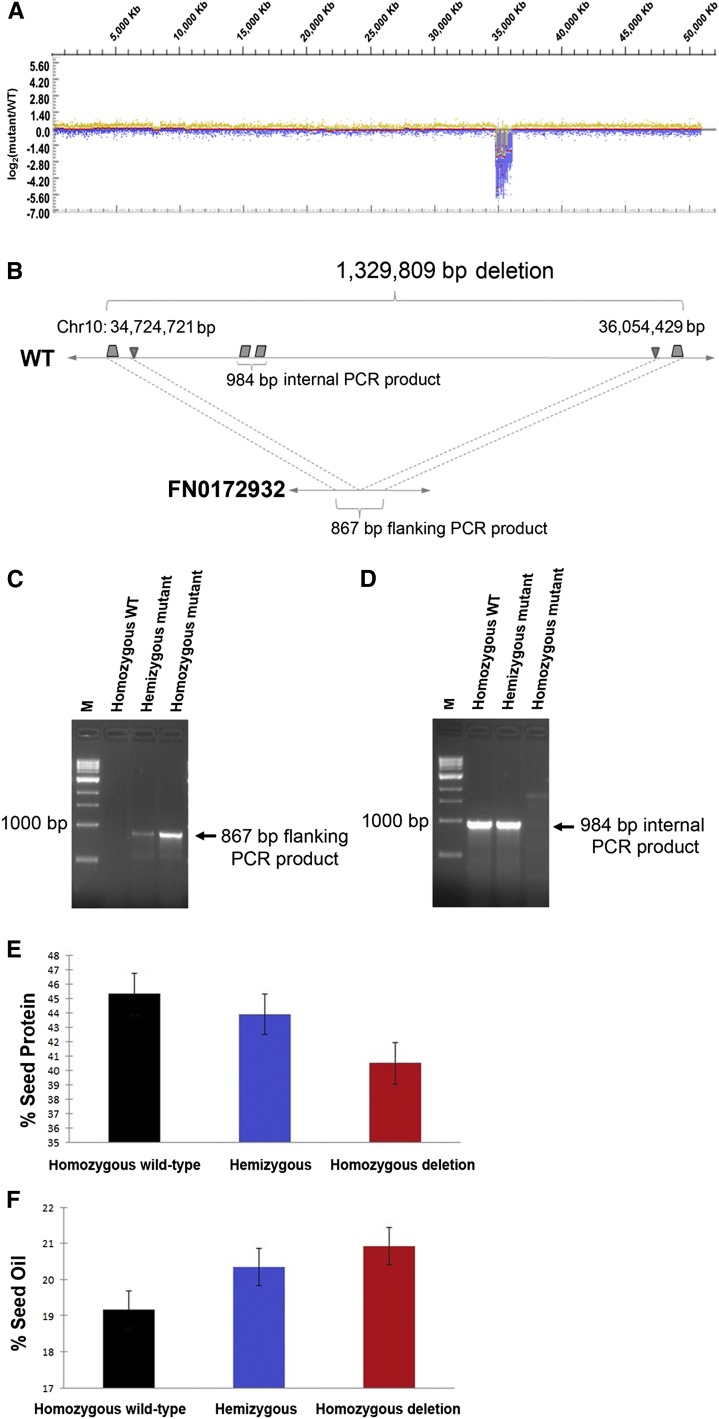
Multiple SV events were identified in FN0172932 by aCGH. Therefore, bulk segregation analysis (Michelmore et al. 1991) was performed on the outcrossed populations to determine associations between the seed composition phenotype and a chromosomal region. The bulk segregation analysis identified a region on chromosome 10 cosegregating with the mutant phenotype. This chromosomal region was coincident with a large deletion detected by aCGH, spanning the probe coordinates 34,724,962 to 36,053,334 bp on chromosome 10 (Figure 6A). DNA from FN0172932 was evaluated by high-throughput paired-end sequencing to determine the exact breakpoints on chromosome 10 for this ∼1.3 Mb deletion. The sequence data revealed that the deletion occurs between positions 34,724,721 and 36,054,429 bp; this was further confirmed by PCR and Sanger sequencing (Figure 6, B–D). Subsequent PCR analysis and phenotyping of 81 individuals from a segregating BC1F3 population confirmed this locus–trait association, wherein the presence of the deletion on chromosome 10 corresponded to an additive increase in percent oil and decrease in percent protein composition of the soybean seed (Figure 6, E and F, Figure S6). Seeds from individuals that segregated as homozygous present (i.e., wild-type) on chromosome 10 averaged 19.2% oil and 45.3% protein, while seeds from individuals homozygous for the chromosome 10 deletion averaged 20.9% oil and 40.5% protein.
Figure 6.
Association of a fast neutron mutant deletion with a quantitative change in seed composition. (A) The aCGH log2 ratios of mutant FN0172932 to wild-type (WT) signals are shown for chromosome 10. A deletion is visible from ∼34 to 36 Mb. (B) The exact coordinates of the deletion were resolved by high-throughput paired-end sequencing followed by PCR. Deletion junctions are represented by inverted triangles from 34,724,721 to 36,054,429 bp, framing a 1,329,809-bp deletion. (C) PCR across the deletion region with flanking primers (shaded trapezoids in B) confirmed the presence of the deletion allele. (D) PCR within the deletion region using internal primers (shaded parallelograms in B) confirmed the presence of the wild-type allele. (E and F) Bar graphs show that the presence of the deletion allele on chromosome 10 corresponds to a simultaneous decrease in percent protein (E) and additive increase in percent oil (F) composition of the soybean seed.
These data suggest that the chromosome 10 deletion is directly associated or in close linkage with the high oil/low protein seed composition phenotype in FN0172932. In addition, these methods demonstrate the ability to detect and trace associated loci and to develop markers for quantitative traits of interest using soybean FN-irradiated mutants.
Lastly, the genotyping of the BC1F3 population exhibited a lower than expected frequency of the deletion haplotype. The homozygous deletion was detected in 24 plants and the hemizygous form was found in 12 plants, whereas the homozygous wild-type appeared in 43 plants. The structure of this population would have predicted an equal frequency of the deletion and wild-type haplotypes, indicating that this population exhibited a fitness differential or segregation distortion favoring the wild-type version of chromosome 10.
Association of a fast neutron mutant genomic copy number gain with a short petiole phenotype
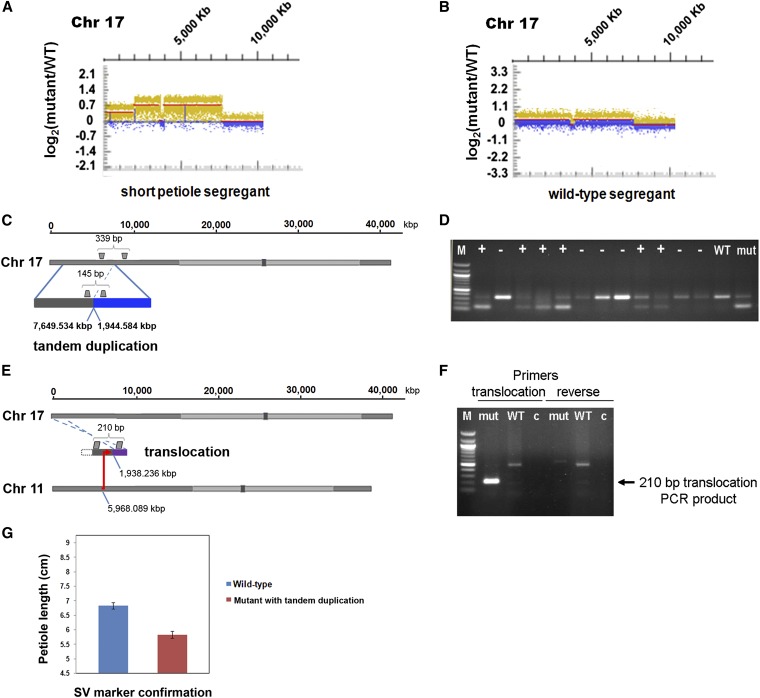
Five M4 descendants of the mutant family FN0163764 segregated for a short petiole and lanceolate leaf phenotype (3:2, mutant:wild-type) (Figure S7A). Four of these individuals were assayed by aCGH, and three displayed the mutant phenotype while one displayed the wild-type phenotype. Figure 7, A and B shows that a duplication event on chromosome 17 from the end of the chromosome to position 7,649,252 bp was shared among the short petiole and wild-type siblings. However, a higher order segmental duplication from 1,944,742 bp to 7,649,252 bp on chromosome 17 was found only in the short petiole mutant individuals (Figure 7A) and not in the wild-type segregant (Figure 7B).
Figure 7.
Association of a copy number gain with the short petiole phenotype. (A) The aCGH normalized log2 ratios of mutant FN0163764 to the ‘M92-220’ progenitor are shown for the first 10 Mb of chromosome 17. The short petiole segregant in the FN0163764 family showed segmental duplication coverage from the start of the chromosome end to ∼7 Mb and also showed an additional level of genomic copy number gain above this region from ∼2 to 7 Mb. (B) The wild-type petiole segregant of the FN0163764 mutant showed a single level of segmental duplication coverage from the chromosome end to ∼7 Mb. (C) NGS-derived results provided evidence for a tandem duplication on chromosome 17. (D) The presence of the tandem duplication was confirmed by the presence of the 145-bp PCR product (lower band) in the short petiole mutant (mut) but not in the wild-type (WT) ‘M92-220’ background. Evidence for the inheritance of the tandem duplication (designated by the “+” symbol) was also found in 50% of FN0163764 × ‘MN1410’ progeny. (E) NGS results also provided evidence for a translocation between chromosome 17 and chromosome 11. (F) The presence of the translocation was confirmed with the expected 210-bp PCR product in the mutant and not the WT ‘M92-220’ background. No-template control lanes (labeled as “c”) are also shown. (G) Association of the presence of the tandem duplication (PCR confirmation) with a decrease (P-value 2.22E-05) in petiole length in F3 segregants of the FN0163764 × ‘MN1410’ outcross, shown with standard error bars.
Additional analyses were required to map the location of the duplications on chromosome 17. Resequencing of the short petiole FN0163764 mutant using paired-end high-throughput sequencing revealed the presence of at least two rearranged sequence junctions that map to the duplicated interval (Figure 7, C and E). An insertion was detected by mapping 22 short-clipped sequence reads that aligned position 7,649,534 bp next to position 1,944,584 bp of chromosome 17, providing evidence for a tandem duplication event on this chromosome. This rearranged junction was confirmed by PCR and Sanger sequencing (Figure 7, C and D, Figure S7B). In addition, a translocation was mapped between position 1,944,476 bp on chromosome 17 and position 5,958,801 bp on chromosome 11, which was also confirmed by PCR and sequencing (Figure 7E). The tandem duplication on chromosome 17 and the translocation between chromosome 17 and chromosome 11 were confirmed structural variations detected only in the mutant individuals (Figure 7, C–F).
To test for association of SV events with the short petiole phenotype, a segregating population was produced by mating FN0163764 to cv. ‘MN1410.’ PCR of the tandem duplication and translocation junctions were conducted on 80 F3 progeny derived from 80 random F2 plants. The tandem duplication event on chromosome 17 was associated with the short petiole mutant phenotype, as F3 individuals harboring the tandem repeat exhibited petioles that were 13% shorter on average (Figure 7, D and G, t-test P-value 2.22E-05). Flower color in the F3 progeny segregated at the expected 5:3 ratio (51 purple: 29 white) for this dominant trait. However, the presence of the tandem duplication event (scored with a dominant PCR marker) was detected in only 50% of the F3 progeny (the tandem duplication was present in 40 plants and absent in 40 plants), indicating that this locus was transmitted at a lower frequency than expected.
Discussion
This study provides a global view of the resilience of the soybean genome to structural changes. Genome-wide segmental duplications and deletions across the 20 soybean chromosomes were mapped for 264 FN soybean mutants representing 145 M2-derived lineages. This effort produced a catalog of FN-induced genomic variation on an unprecedented scale. The prevalence of segmental duplications in addition to deletions within FN-irradiated mutants was described. Moreover, we demonstrated the value of these mutants for functional studies through the recovery of specific genomic copy number changes linked to interesting (and potentially beneficial) phenotypes.
Fast neutron-irradiated mutants as a source of induced genomic copy number variation
This study represents a relatively deep survey of large deletion and duplication mapping in a soybean FN-irradiated mutant population. Previous studies have shown the effectiveness of using FN mutants for functional studies and genome-wide analyses. These studies provided valuable insight into the contribution of individual genes and the detection of large deletions or single-base deletions and substitutions (Bruggemann et al. 1996; Li et al. 2001; Men et al. 2002; Belfield et al. 2012; O’Rourke et al. 2013). In this study, the importance of both large deletions and segmental duplications generated within the FN mutant genome was demonstrated. Furthermore, more complex rearrangements were also observed in this dataset. For example, the short petiole mutant FN0163764 provided a striking illustration of a multitiered copy number region, and a putative chromosomal translocation, that were generated following FN mutagenesis. These findings were in contrast to prior reports of FN mutant studies that focused on gene presence/absence as opposed to quantitative copy number variation. The discovery that the FN population harbors a wide range of segmental duplications stands in contrast to the common perception that FN mutagenesis is solely a system for inducing deletions.
The ability to induce and sustain genomic copy number changes following FN mutagenesis expands our understanding of the potential mechanisms that underlie the recovery of unique traits in mutant populations. The promise of recovering advantageous phenotypes from a FN deletion event is primarily based on the concept of knocking out genes that result in recovery of novel plants with nondeleterious qualities. While this may be the primary source of novel phenotypes in a population, the high frequency of large duplications reported in this study also supports the possibility that plants with unique properties may be recovered from segmental duplications. This further implies that novel plant traits generated in a FN mutant population are not necessarily associated with gene losses or loss-of-function phenotypes, and that beneficial gain-of-function phenotypes may be recovered without loss of genetic background material. As has been demonstrated in several recent studies of natural genomic variation, copy number gains can often result in phenotypic plasticity and novel traits that are advantageous in plants (Cook et al. 2012; Maron et al. 2013; Zmienko et al. 2014).
There are additional DNA alterations that are generated by FN irradiation that we could not assess in this analysis, as they require a deep resequencing coverage that was unaffordable across hundreds of individuals. For example, a recent study reported on the genome-wide discovery of induced single base substitutions and the prominence of single base deletions in six FN-irradiated Arabidopsis mutants (Belfield et al. 2012). PCR-based screens have also identified small deletions (Waugh et al. 2006) below the detection level of the aCGH platform. Furthermore, the prevalence of duplications located on chromosomal ends (∼15% of duplications) suggests that many of these events may be translocations, perhaps the result of adjacent segregation of reciprocal translocations. Such translocations may be more efficiently resolved by whole-genome sequencing (Korbel et al. 2007).
Sequencing-based genotyping may also identify more complex rearrangements, such as inversions and multichromosomal translocations. Prior studies have inferred that double strand breaks and their repair mechanisms, including nonhomologous end joining, microhomology-mediated end joining, and possibly single strand annealing, play important roles in genome structure following exposure to radiation (Cornforth 2006; Povirk 2006; Sankaranarayanan et al. 2013). Analysis of breakpoint signatures and microhomologies at SV junctions through NGS may be used to provide insight on the formation mechanisms of these complex structures (Gribble et al. 2014). While more expensive and time consuming than aCGH analysis, sequencing-based approaches offer the potential to deeply resolve the range of DNA polymorphisms and SV events harbored by a subset of high-value mutants.
Genome resilience and gene essentiality in soybean
We examined the coverage and distribution of large deletions and duplications and noted the genes and regions that were associated with the different types of polymorphisms in the soybean FN mutants. In the soybean genome, chromosomes are composed of 57% pericentromeric regions (Du et al. 2010; Schmutz et al. 2010). In the soybean FN mutant population, homozygous deletions were predominately found within pericentromeric space, which may be expected as this region has a lower density of genes and would be expected to have a reduced rate of lethality associated with such deletions. Subdividing the mutants by class also revealed some expected results. For example, mutants exhibiting phenotypes (the forward screen group) were found to have a greater portion of the genome deleted (including more total genes) than no-phenotype plants. In contrast, the high rate of segmental duplications within the mutant population was unexpected. The prevalence of copy number variation at regions of heterochromatin near telomeres and centromeres has been documented as a natural phenomenon, for example, in the human genome (She et al. 2004; Zhang et al. 2005; Nguyen et al. 2006; Shao et al. 2008). However, the retention of segmental duplications, particularly at chromosome ends, in genomes with induced mutations, is a novel observation.
The likelihood of identifying a gene of interest affected by a homozygous deletion event among these 145 mutant families is approximately 1 in 10. In addition, 4 out of 10 genes were found within at least one duplication event from this dataset. However, the subset of mutants examined in this study may not be representative of the population, as they were predominately composed of visible phenotype mutants and included 10 reverse screen mutant families with large duplications. While previous studies have examined smaller plant genomes (Ibarra-Laclette et al. 2013), the large-scale examination of genome reduction potential may provide a foundation for understanding the minimal soybean genome.
Viable seed is evidence of “recovery” from FN radiation in the generations following exposure. All soybean FN mutants that are cataloged as part of the mutant resource have reached at least the M2 generation and produced M3 seed. Therefore, the detected SV events uncovered for each mutant were not lethal to the plant. The high rate and large size of the hemizygous deletions detected in this study indicated that many of these deletions would be lethal if homozygous. Still, mutant genomes were able to survive despite losing up to 8 Mb of total genomic space due to homozygous deletions. Furthermore, nearly complete chromosomal duplications were observed in a subset of surviving plants. Such phenomena are a testament to the resilience of the soybean genome.
SV event transmission and marker–trait associations
Two mutants were analyzed to identify marker–trait associations between specific SV events and phenotypes. FN0172932 contained a large (∼1.3 Mb) deletion on chromosome 10 that was associated with a shift in the seed composition profile toward higher oil and lower protein. This was the first quantitative trait genetically mapped in this population. In addition, this association provides evidence that a simple molecular marker (in this case, a PCR-based marker) can be developed to predict the phenotypic outcomes caused by SV for traits that are difficult to phenotype. For example, this marker could be used to introgress the high oil/low protein seed composition quantitative trait locus into elite materials. In addition, the experimental template (utilizing a combination of aCGH, bulk segregant analysis and genome resequencing) establishes the development of such marker–trait associations in this population.
The second mutant FN0163764 represents the first discovery of a trait linked to a segmental duplication in this population. This short petiole mutant is linked with a large (∼7 Mb) duplication near the end of the long arm of chromosome 17. While the molecular basis for the short petiole phenotype cannot be confirmed at present, it is an intriguing possibility that the disrupted dosage caused by the large (putatively tandem) duplication may be partially or completely responsible for this phenotype. A previously reported short petiole mutant in this FN population was associated with a large deletion that mapped to an interval that overlaps the FN0163764 chromosome 17 duplication (Bolon et al. 2011). Since duplicated regions have at times been linked to negative regulation and gene silencing (Tuteja et al. 2004; Lacombe et al. 2009), it is possible that duplication of a gene or locus may result in a similar phenotype as deletion of the same region.
The two SV events associated with phenotypes in this study exhibited lower transmission than wild-type chromosomes in segregating populations. Whether caused by fitness penalties or meiotic mechanisms, recognition of reduced transmission is fundamental for utilization of this population, both as a functional genomics tool and as a potential germplasm resource. Nearly all M1 SV events were likely generated as hemizygous events. Therefore, while some SV events were fixed in the homozygous state in early generations, many events with reduced transmission were lost during early rounds of selfing. Furthermore, it is probable that many hemizygous SV events in the population were homozygous lethal, consequently leading to the recovery of homozygous wild-type chromosomes in successive generations. Therefore, the suite of SV events and phenotypes observed in early generations following mutagenesis is likely more extensive than those observed in later generations (which are the generations that would be used as parents for introgressing loci of interest into elite backgrounds). It is possible that more advanced generations (e.g., M10) may be essentially devoid of large segmental deletions and duplications, unless these events can be fixed in the homozygous state in early generations or are artificially selected for in later generations.
Supplementary Material
Acknowledgments
We are grateful to Dimitri Von Ruckert, William Haun, Dinesha Walek, Jill Miller-Garvin, and Aaron Becker for project support. We thank Ethalinda Cannon for prerelease CViT software updates. We also acknowledge use of resources at the Minnesota Supercomputing Institute at the University of Minnesota. We thank the Minnesota Soybean Research and Promotion Council (no. 18-12C and no. 21-13C), the United Soybean Board (no. 1320-532-5688), the National Science Foundation (no. IOS-1127083), and the US Department of Agriculture-Agricultural Research Service for funding and supporting this work.
Footnotes
Available freely online through the author-supported open access option.
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.170340/-/DC1.
The sequence read data from the fast neutron plants FN0172932 and FN0163764, along with the parent line of the population (cv. ‘M92-220’), are deposited in the Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra/) under accession no. SRP036841. The SRX accession nos. for these genotypes are SRX467185, SRX467191, and SRX467183, respectively. All comparative genomic hybridization microarray data can be found as accession nos. GSE31111, GSE31038, and GSE58172 in the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo).
Communicating editor: S. Poethig
Literature Cited
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 494–498. [DOI] [PubMed] [Google Scholar]
- Anderson, J. E., M. B. Kantar, T. Y. Kono, F. Fu, A. O. Stec et al., 2014 A roadmap for functional structural variants in the soybean genome. G3 (Bethesda) 4: 1307–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfield E. J., Gan X., Mithani A., Brown C., Jiang C., et al. , 2012. Genome-wide analysis of mutations in mutant lineages selected following fast-neutron irradiation mutagenesis of Arabidopsis thaliana. Genome Res. 22: 1306–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolon Y. T., Haun W. J., Xu W. W., Grant D., Stacey M. G., et al. , 2011. Phenotypic and genomic analyses of a fast neutron mutant population resource in soybean. Plant Physiol. 156: 240–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggemann E., Handwerger K., Essex C., Storz G., 1996. Analysis of fast neutron-generated mutants at the Arabidopsis thaliana HY4 locus. Plant J. 10: 755–760. [DOI] [PubMed] [Google Scholar]
- Cannon E. K., Cannon S. B., 2011. Chromosome visualization tool: a whole genome viewer. Int. J. Plant Genomics 2011: 373875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon S. B., Shoemaker R. C., 2012. Evolutionary and comparative analyses of the soybean genome. Breed. Sci. 61: 437–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Wallis J. W., McLellan M. D., Larson D. E., Kalicki J. M., et al. , 2009. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat. Methods 6: 677–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D. E., Lee T. G., Guo X., Melito S., Wang K., et al. , 2012. Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science 338: 1206–1209. [DOI] [PubMed] [Google Scholar]
- Cornforth M. N., 2006. Perspectives on the formation of radiation-induced exchange aberrations. DNA Repair (Amst.) 5: 1182–1191. [DOI] [PubMed] [Google Scholar]
- Domoney C., Knox M., Moreau C., Ambrose M., Palmer S., et al. , 2013. Exploiting a fast neutron mutant genetic resource in Pisum sativum (pea) for functional genomics. Funct. Plant Biol. 40: 1261–1270. [DOI] [PubMed] [Google Scholar]
- Doyle J. J., Egan A. N., 2010. Dating the origins of polyploidy events. New Phytol. 186: 73–85. [DOI] [PubMed] [Google Scholar]
- Du J., Tian Z., Hans C. S., Laten H. M., Cannon S. B., et al. , 2010. Evolutionary conservation, diversity and specificity of LTR-retrotransposons in flowering plants: insights from genome-wide analysis and multi-specific comparison. Plant J. 63: 584–598. [DOI] [PubMed] [Google Scholar]
- Du J., Tian Z., Sui Y., Zhao M., Song Q., et al. , 2012. Pericentromeric effects shape the patterns of divergence, retention, and expression of duplicated genes in the paleopolyploid soybean. Plant Cell 24: 21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant D., Nelson R. T., Cannon S. B., Shoemaker R. C., 2009. SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res. 38: D843–D846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribble S. M., Wiseman F. K., Clayton S., Prigmore E., Langley E., et al. , 2014. Massively parallel sequencing reveals the complex structure of an irradiated human chromosome on a mouse background in the Tc1 model of Down syndrome. PLoS ONE 8: e60482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B. J., Delcher A. L., Wortman J. R., Salzberg S. L., 2004. DAGchainer: a tool for mining segmental genome duplications and synteny. Bioinformatics 20: 3643–3646. [DOI] [PubMed] [Google Scholar]
- Haun W. J., Hyten D. L., Xu W. W., Gerhardt D. J., Albert T. J., et al. , 2011. The Composition and origins of genomic variation among individuals of the soybean reference cultivar Williams 82. Plant Physiol. 155: 645–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann D., Jiang Q., Men A., Kinkema M., Gresshoff P. M., 2007. Nodulation deficiency caused by fast neutron mutagenesis of the model legume Lotus japonicus. J. Plant Physiol. 164: 460–469. [DOI] [PubMed] [Google Scholar]
- Hyten D. L., Cannon S. B., Song Q., Weeks N., Fickus E. W., et al. , 2010. High-throughput SNP discovery through deep resequencing of a reduced representation library to anchor and orient scaffolds in the soybean whole genome sequence. BMC Genomics 11: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra-Laclette E., Lyons E., Hernandez-Guzman G., Perez-Torres C. A., Carretero-Paulet L., et al. , 2013. Architecture and evolution of a minute plant genome. Nature 498: 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel J. O., Urban A. E., Affourtit J. P., Godwin B., Grubert F., et al. , 2007. Paired-end mapping reveals extensive structural variation in the human genome. Science 318: 420–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe S., Souyris I., Berville A. J., 2009. An insertion of oleate desaturase homologous sequence silences via siRNA the functional gene leading to high oleic acid content in sunflower seed oil. Mol. Genet. Genomics 281: 43–54. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R., 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Song Y., Century K., Straight S., Ronald P., et al. , 2001. A fast neutron deletion mutagenesis-based reverse genetics system for plants. Plant J. 27: 235–242. [DOI] [PubMed] [Google Scholar]
- Lin J. Y., Stupar R. M., Hans C., Hyten D. L., Jackson S. A., 2010. Structural and functional divergence of a 1-Mb duplicated region in the soybean (Glycine max) genome and comparison to an orthologous region from Phaseolus vulgaris. Plant Cell 22: 2545–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maron L. G., Guimaraes C. T., Kirst M., Albert P. S., Birchler J. A., et al. , 2013. Aluminum tolerance in maize is associated with higher MATE1 gene copy number. Proc. Natl. Acad. Sci. USA 110: 5241–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marroni F., Pinosio S., Morgante M., 2014. Structural variation and genome complexity: Is dispensable really dispensable? Curr. Opin. Plant Biol. 18C: 31–36. [DOI] [PubMed] [Google Scholar]
- Men A. E., Laniya S. T., Searle I. R., Iturbe-Ormaetxe I., Gresshoff I., et al. , 2002. Fast neutron mutagenesis of soybean (Glycine soja L.) produces a supernodulating mutant containing a large deletion in linkage group H. Genome Lett. 3: 147–155. [Google Scholar]
- Michelmore R. W., Paran I., Kesseli R. V., 1991. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 88: 9828–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D. Q., Webber C., Ponting C. P., 2006. Bias of selection on human copy-number variants. PLoS Genet. 2: e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke J. A., Iniguez L. P., Bucciarelli B., Roessler J., Schmutz J., et al. , 2013. A re-sequencing based assessment of genomic heterogeneity and fast neutron-induced deletions in a common bean cultivar. Front. Plant Sci. 4: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd G. E., Long S. R., 2003. Identification and characterization of nodulation-signaling pathway 2, a gene of Medicago truncatula involved in Nod actor signaling. Plant Physiol. 131: 1027–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povirk L. F., 2006. Biochemical mechanisms of chromosomal translocations resulting from DNA double-strand breaks. DNA Repair (Amst.) 5: 1199–1212. [DOI] [PubMed] [Google Scholar]
- Rios G., Naranjo M. A., Iglesias D. J., Ruiz-Rivero O., Geraud M., et al. , 2008. Characterization of hemizygous deletions in citrus using array-comparative genomic hybridization and microsynteny comparisons with the poplar genome. BMC Genomics 9: 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers C., Wen J., Chen R., Oldroyd G G., 2009. Deletion-based reverse genetics in Medicago truncatula. Plant Physiol. 151: 1077–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaranarayanan K., Taleei R., Rahmanian S., Nikjoo H., 2013. Ionizing radiation and genetic risks. XVII. Formation mechanisms underlying naturally occurring DNA deletions in the human genome and their potential relevance for bridging the gap between induced DNA double-strand breaks and deletions in irradiated germ cells. Mutat. Res. 753: 114–130. [DOI] [PubMed] [Google Scholar]
- Schlueter J. A., Dixon P., Granger C., Grant D., Clark L., et al. , 2004. Mining EST databases to resolve evolutionary events in major crop species. Genome 47: 868–876. [DOI] [PubMed] [Google Scholar]
- Schmutz J., Cannon S. B., Schlueter J., Ma J., Mitros T., et al. , 2010. Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183. [DOI] [PubMed] [Google Scholar]
- Searle I. R., Men A. E., Laniya T. S., Buzas D. M., Iturbe-Ormaetxe I., et al. , 2003. Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 299: 109–112. [DOI] [PubMed] [Google Scholar]
- Severin A. J., Cannon S. B., Graham M. M., Grant D., Shoemaker R. C., 2011. Changes in twelve homoeologous genomic regions in soybean following three rounds of polyploidy. Plant Cell 23: 3129–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L., Shaw C. A., Lu X. Y., Sahoo T., Bacino C. A., et al. , 2008. Identification of chromosome abnormalities in subtelomeric regions by microarray analysis: a study of 5,380 cases. Am. J. Med. Genet. A. 146A: 2242–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X., Horvath J. E., Jiang Z., Liu G., Furey T. S., et al. , 2004. The structure and evolution of centromeric transition regions within the human genome. Nature 430: 857–864. [DOI] [PubMed] [Google Scholar]
- Springer N. M., Stupar R. M., 2007. Allele-specific expression patterns reveal biases and embryo-specific parent-of-origin effects in hybrid maize. Plant Cell 19: 2391–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja J. H., Clough S. J., Chan W. C., Vodkin L. O., 2004. Tissue-specific gene silencing mediated by a naturally occurring chalcone synthase gene cluster in Glycine max. Plant Cell 16: 819–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Mullighan C. G., Easton J., Roberts S., Heatley S. L., et al. , 2012. CREST maps somatic structural variation in cancer genomes with base-pair resolution. Nat. Methods 8: 652–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waugh R., Leader D. J., McCallum N., Caldwell D., 2006. Harvesting the potential of induced biological diversity. Trends Plant Sci. 11: 71–79. [DOI] [PubMed] [Google Scholar]
- Zhang L., Lu H. H., Chung W. Y., Yang J., Li W. H., 2005. Patterns of segmental duplication in the human genome. Mol. Biol. Evol. 22: 135–141. [DOI] [PubMed] [Google Scholar]
- Zhang L., Fetch T., Nirmala J., Schmierer D., Brueggeman R., et al. , 2006. Rpr1, a gene required for Rpg1-dependent resistance to stem rust in barley. Theor. Appl. Genet. 113: 847–855. [DOI] [PubMed] [Google Scholar]
- Zmienko A., Samelak A., Kozlowski P., Figlerowicz M., 2014. Copy number polymorphism in plant genomes. Theor. Appl. Genet. 127: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.