Abstract
Many mosquito species serve as vectors of diseases such as malaria and yellow fever, wherein pathogen transmission is tightly associated with the reproductive requirement of taking vertebrate blood meals. Toxorhynchites is one of only three known mosquito genera that does not host-seek and initiates egg development in the absence of a blood-derived protein bolus. These remarkable differences make Toxorhynchites an attractive comparative reference for understanding mosquito chemosensation as it pertains to host-seeking. We performed deep transcriptome profiling of adult female Toxorhynchites amboinensis bodies, antennae and maxillary palps, and identified 25,084 protein-coding “genes” in the de novo assembly. Phylogenomic analysis of 4,266 single-copy “genes” from T. amboinensis, Aedes aegypti, Anopheles gambiae, and Culex quinquefasciatus robustly supported Ae. aegypti as the closest relative of T. amboinensis, with the two species diverged approximately 40 Ma. We identified a large number of T. amboinensis chemosensory “genes,” the majority of which have orthologs in other mosquitoes. Finally, cross-species expression analyses indicated that patterns of chemoreceptor transcript abundance were very similar for chemoreceptors that are conserved between T. amboinensis and Ae. aegypti, whereas T. amboinensis appeared deficient in the variety of expressed, lineage-specific chemoreceptors. Our transcriptome assembly of T. amboinensis represents the first comprehensive genomic resource for a nonblood-feeding mosquito and establishes a foundation for future comparative studies of blood-feeding and nonblood-feeding mosquitoes. We hypothesize that chemosensory genes that display discrete patterns of evolution and abundance between T. amboinensis and blood-feeding mosquitoes are likely to play critical roles in host-seeking and hence the vectorial capacity.
Keywords: Toxorhynchites, mosquito, chemosensation, chemosensory receptor, transcriptome, RNA-seq, host-seeking, blood feeding, disease vector, chemical ecology
Introduction
The notorious reputation garnered by many mosquito species as human disease vectors is exclusively attributable to the concomitant behaviors of host-seeking and blood feeding (hematophagy) exhibited by adult female mosquitoes. These blood meals provide the female with a concentrated nutritive source that initiates egg development and allows the completion of the reproductive cycle. Notably, there are three nonhematophagous mosquito genera (Malaya, Topomyia, and Toxorhynchites) that lack a reproductive requirement for blood meals and do not exhibit host-seeking behavior(s). Of these, Toxorhynchites is the most widely distributed and comprised over 90 species found throughout the tropical areas of Africa, Asia, the South Pacific, and the Americas. Adult Toxorhynchites are physically much larger than other blood-feeding mosquitoes and feed exclusively on sugar sources. Moreover, the mouthparts (labium) of adult Toxorhynchites are noticeably elongated and curved downward, a presumed morphological refinement toward nectar feeding that also might render them less amenable to piercing skin (Clements 1999).
Although Toxorhynchites is a morphologically distinctive genus within the family Culicidae, its precise placement among other genera is uncertain. Previous studies that included Toxorhynchites have produced several incongruent phylogenies. For example, early phylogenetic studies based on morphologic characters positioned Toxorhynchites as its own subfamily within Culicidae, whereas subsequent molecular studies placed Toxorhynchites at various locations within the subfamily Culicinae (Miller et al. 1997; Harbach and Kitching 1998; Mitchell et al. 2002; Shepard et al. 2006; Reidenbach et al. 2009). The precise phylogenetic position of Toxorhynchites within Culicidae is important as it would allow inference as to whether the blood-feeding phenotype represented a trait that was lost in Toxorhynchites or a novel adaptation that occurred only in certain Culicidae lineages, after their divergence from Toxorhynchites. Given that hematophagy is thought to have arisen and been lost independently multiple times in dipterans, the order of events within mosquito taxa is not immediately clear (Wiegmann et al. 2011).
Toxorhynchites mosquitoes are also interesting from a chemoreception perspective because, unlike the vast majority of hematophagous mosquitoes, adults have no reproductive requirement to engage in host-seeking behaviors. Host-seeking in mosquitoes is facilitated through a range of sensory cues and is largely mediated through the chemoreception of host-associated kariomones. Consequently, a characterization of the chemosensory gene repertoire of Toxorhynchites and its comparison to transcriptome profiles from blood-feeding mosquitoes is expected to yield insights into the molecular aspects of the chemoreceptive requirements that encompass host-seeking or blood-feeding behaviors.
Thus far, studies of chemoreceptor expression and function in mosquitoes have been limited to vector species where the identification of large gene families and the fundamental properties of chemosensory signaling continue to be elucidated. Odorant receptors (ORs), variant ionotropic receptors (IRs), and gustatory receptors (GRs) comprise the major families of chemoreceptors defined to date and no studies of chemoreceptors in nonblood-feeding mosquitoes have been published. To address these open questions regarding the ecology, evolution, and comparative genomics of Toxorhynchites, we have sequenced total RNA from the bodies of female Toxorhynchites amboinensis along with each of the two major chemosensory organs of mosquitoes, the antennae and the maxillary palps. Our data facilitated the construction of a transcriptome of the most abundant transcripts present within the adult female T. amboinensis as a whole, along with the characterization of chemosensory-associated transcripts that are enriched within the head appendages. To do so, we leveraged extant genomic resources of disease vector mosquitoes Anopheles gambiae (vector of malaria [Holt et al. 2002]), Aedes aegypti (vector of yellow fever and dengue [Nene et al. 2007]), and Culex quinquefasciatus (vector of West Nile virus [Arensburger et al. 2010]) to annotate the assembled T. amboinensis transcriptome as well as place T. amboinensis into a well-defined phylogenetic context. Finally, we have compared the transcript abundance patterns of the major chemoreceptor gene families in the olfactory tissues of three mosquito species to further refine our understanding of the molecular bases for host-seeking behaviors. This study brings together a de novo assembly of the T. amboinensis transcriptome and a novel comparative analysis of chemoreceptor evolution and expression between hematophageous and nonhematophageous mosquitoes.
Materials and Methods
Mosquitos and Mosquito Rearing
Toxorhynchites amboinensis eggs were obtained from the US Centers for Disease Control and Prevention Dengue Laboratory in San Juan, Puerto Rico. Eggs were hatched in distilled water in an upright incubator at 26 °C and 75% relative humidity under a 12:12 light dark cycle. Toxorhynchites amboinensis larvae were reared on a diet of Ae. aegypti or An. gambiae larvae, with prey developmental stage (L1–L4) approximating that of predator. Pupae were transferred to small cups of clean distilled water and allowed to eclose inside a 100 × 50 × 50 cm cage. Adults were provided apple slices plus cotton balls soaked with a solution of 10% sucrose, 10% honey ad libitum. Females were given access to a small, black melamine bowl, approximately 50 mm in diameter, partially filled with 100 ml distilled water plus 1% oak leaf infusion for oviposition.
RNA Isolation and RNA Sequencing
Approximately 200 antennae or maxillary palps were removed using surgical forceps from randomly selected 7- to 14-day-old adult females and placed into Trizol reagent for total RNA extraction. Additionally, 20 bodies from the same females, minus antennae and palps, were treated similarly. Messenger RNA (mRNA) isolation and cDNA library preparation were carried out using the Illumina mRNA sequencing kit (Illumina Inc., San Diego, CA). Libraries were sequenced by the HudsonAlpha Institute for Biotechnology (Huntsville, AL) using an Illumina HiSeq2000.
Data Preprocessing and De Novo Transcriptome Assembly
Initial quality assessment of raw RNA sequencing (RNA-seq) data was performed using FastQC (version 0.10.1; http://www.bioinformatics.babraham.ac.uk/projects/fastqc/, last accessed October 22, 2014) and several issues were identified, including adapter contamination and low quality reads. To ensure the quality of transcriptome analyses, the following preprocessing procedures were applied to RNA-seq data from each tissue: 1) Reads that failed the Illumina “chastity” filter (a measurement of the overall signal purity of reads) were first removed, 2) reads with adapter contamination were trimmed using Trimmomatic (version 0.22; Lohse et al. 2012), 3) reads were mapped against known T. amboinensis ribosomal RNA sequences and sequenced mitochondrial genomes of Culicidae species (downloaded from National Center for Biotechnology Information [NCBI] at http://www.ncbi.nlm.nih.gov/genomes/OrganelleResource.cgi?taxid=7157, accessed July, 2012) using Bowtie 2 (version 2.0.0; Langmead and Salzberg 2012) to remove reads that are derived from rRNA/mitochondrial genes, and 4) a final quality-based trimming was performed using Trimmomatic (version 0.22) with the following parameters: LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:25. All abovementioned data filtrations were conducted in paired-end mode. Cleaned data from all three tissues were combined together and assembled using Trinity (version 2012-06-08; Grabherr et al. 2011) to generate a de novo transcriptome assembly. Trinity was run with default settings and only reported transcripts of 200 bp or longer. No filtering of low-abundance transcripts was performed in order to maximize the detection of chemosensory genes as some of them may have low transcript abundances.
Gene Identification
To annotate protein-coding genes in the transcriptome assembly, open reading frames (ORFs) of at least 300 bp long were first extracted from all assembled transcripts in all six frames, allowing for overlap. For each ORF, the translated protein sequence was compared against the NCBI nonredundant (NR) protein database using BLASTP (version 2.2.26+; Altschul et al. 1997) with an e value cutoff of 1e-5 and the bit score of the best match was recorded. Results of this similarity search were later used to generate the taxonomic distribution of best matches for annotated genes. ORFs with significant coding potential were also identified from the complete set of ORFs using the TransDecoder utility of Trinity.
This initial set of annotation contains a high level of redundancy as more than half of the annotated transcripts are alternative splicing isoforms (i.e., transcripts reconstructed from the same Trinity component which ideally represent one gene). To avoid this redundancy in subsequent sequence and expression analyses, a reference transcriptome assembly was created by selecting one representative transcript for each Trinity subcomponent. The representative transcripts were determined as following: 1) For each transcript, the highest bit score sum was calculated by comparing all possible combinations of nonoverlapping ORFs identified on that transcript, including both the ORFs with homologs in the NR database and the ORFs with coding potential; 2) for each Trinity component, the transcript with the highest bit score sum was selected as the representative; and 3) the longest one was chosen if multiple isoforms have the same bit score sum. For each representative transcript, the ORFs in the combination that gave rise to the highest bit score sum were considered annotated genes; the one with the longest total ORF length was selected if multiple combinations have the same bit score sum.
Functional Annotation
Two approaches were employed to obtain functional information about annotated T. amboinensis genes. First, translated protein sequences of all genes were compared against the UniProtKB/Swiss-Prot database (release 2013_11) using BLASTP with an e value cutoff of 1e-5; each T. amboinensis genes was assigned the gene ontology (GO) terms associated with its best match in the database. Second, as a more conservative strategy, InterProScan 5 (data version 38.0; Quevillon et al. 2005; Hunter et al. 2012) was used to identify conserved protein domains encoded by T. amboinensis genes and the GO terms associated with protein domains were transferred to corresponding genes. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotations of T. amboinensis genes were generated using the KEGG Automatic Annotation Server. Genes encoding (retro-)transposons were identified using TransposonPSI (version 08222010; http://transposonpsi.sourceforge.net/, last accessed October 22, 2014).
Phylogenetic Analysis and Divergence Time Estimation
OrthoMCL (version 2.0.3; Li et al. 2003) was used to identify ortholog groups between T. amboinensis and four other dipteran species, including An. gambiae (version AgamP3.6), Ae. aegypti (version AeagL1.3), C. quinquefasciatus (version CpipJ1.3), and Drosophila melanogaster (version 5.48). Gene annotations of the three mosquitoes and the fruit fly were downloaded from the websites of VectorBase (https://www.vectorbase.org, last accessed October 22, 2014) and FlyBase (http://flybase.org/, last accessed October 22, 2014), respectively, and only the longest isoform of each gene was used in this study. The ortholog groups that contain a single gene from each of the five species were used to infer the phylogenetic relationship between species. For each of these groups, a codon-based alignment of the coding sequences was created using MAFFT (version 7.037b; Katoh and Standley 2013) with the “auto” option and poorly aligned regions in the alignment were removed using trimAl (version 1.4; Capella-Gutierrez et al. 2009) with the “automated1” option. Maximum-likelihood (ML) trees were constructed from both individual alignments and a concatenated alignment of all single-copy ortholog groups using RAxML (version 7.4.2; Stamatakis 2006) with the “GTRGAMMA” model and 100 bootstrap replicates were carried out to evaluate the reliability of tree topologies.
The 144 orthologous groups that recovered the same ML tree topology as the concatenated analysis with full support were used for the divergence time estimation using BEAST2 (version 2.0.2; Bouckaert et al. 2014). The analysis was performed with the following settings: Partitions, data set was partitioned by codon positions; site model, GTR+GAMMA (four categories); clock model, relaxed clock log normal; and tree priors, birth–death model. The separation of mosquitoes and Drosophila at 227 Ma (95% confidence interval 210–244 Ma), as suggested by a recent study (Wheat and Wahlberg 2013), was used as a calibration point. Four independent runs were carried out, each with 100 million generations and one tree being sampled every 10,000 generations. Convergence check and tree summarization were performed using Tracer (version 1.5) and TreeAnnotator (version 2.0.2), respectively.
Chemosensory Gene Family Analysis
The annotation of chemosensory genes was performed following a previous protocol (Zhou et al. 2012). In brief, previously reported chemosensory genes from An. gambiae, Ae. aegypti, C. quinquefasciatus, and D. melanogaster were used as queries in TBLASTN searches against the complete T. amboinensis transcriptome assembly. Putative chemosensory gene coding regions were identified after filtering out low-scoring blast hits. For each region, the query sequence that yields the highest bit score was selected as reference to perform homology-based gene prediction using GeneWise (version 2.2.0; Birney et al. 2004) to avoid fragmented gene models due to unspliced introns. All gene models were manually inspected and modified if needed, and redundant models derived from isoforms were removed.
For each of the OR/GR/IR families, protein sequences of genes in the four mosquitoes were aligned using MAFFT with a high accuracy option (–maxiterate 1000 –genafpair). The multiple sequence alignments were manually curated and poorly aligned regions were removed using trimAl with the “automated1” option. ML trees were constructed using RAxML with the PROTGAMMALG model and 100 bootstrap replicates. Trees were annotated and displayed using the website of iTOL (Letunic and Bork 2011). Toxorhynchites amboinensis OR/GR/IR genes were named (supplementary table S7, Supplementary Material online) according to their (co-)orthologs in Ae. aegypti (or An. gambiae if the former is absent; e.g., TaOr7 is the ortholog of AaOr7 and AgOr7). Lineage-specific duplicates were further distinguished by appending a unique number identifier following the orthologs number (e.g., TaOr3.1 and TaOr3.2 are co-orthologs to AaOr3)
Transcript Abundance Calculation
Gene expression values for An. gambiae samples were taken directly from our previous study on nonblood-fed female chemosensory appendages (Pitts et al. 2011). Gene expression values for Ae. aegypti were generated by mapping read sets from nonblood-fed female antenna (Sequence Read Archive [SRA] depositions: SRR1103960, SRR1103962, and SRR1103964) and palp (courtesy of Dr Joseph Dickens [USDA, ARS]) to the current version of the genome using Bowtie2/Tophat2 (Langmead et al. 2009) and then counting the reads mapped to each gene from the resulting alignment file to the Ae. aegypti genome features (Vectorbase; version 2.2 with misassigned transcript IDs removed). Gene expression values for T. amboinesis were generated by mapping the reads used in the assembly to the Trinity transcriptome assembly (representative isoforms) using Bowtie2. In all cases, normalized gene expression levels were calculated for each tissue in terms of reads per kilobase per million reads mapped (RPKM) (Mortazavi et al. 2008).
Data Availability
The raw sequence reads data generated in this study are available through the NCBI SRA database under the BioProject accession number: Pending. The complete transcriptome assembly of T. amboinesis will be available through the NCBI Transcriptome Shotgun Assembly database with the accession number: Pending. The protein and coding sequences as well as functional annotations of all annotated T. amboinensis genes are available at http://dx.doi.org/10.6084/m9.figshare.1092617 (last accessed October 22, 2014).
Results and Discussion
A Comprehensive Transcriptome Assembly of T. amboinensis
In-depth RNA-seq profiling of antennae, maxillary palps, and whole bodies of female T. amboinensis generated more than 90 million pairs of raw reads from each sample, most of which passed rigorous quality control procedures (supplementary table S1, Supplementary Material online). Collectively, we obtained approximately 300 million pairs of high-quality sequence reads and created a de novo transcriptome assembly that was 267.6 Mb in size and consisted of 303,383 transcripts belonging to 199,064 Trinity components (table 1), which ideally each represents one gene. These numbers are an order of magnitude higher than that of annotated transcripts and genes in other available mosquito genomes. However, alternative assembly strategies and programs consistently generated comparable numbers of transcripts and components (supplementary table S2, Supplementary Material online), suggesting that these surprisingly large numbers were unlikely due to artifacts/redundancies in the assembly. Our presumption is that the vast majority of these assembled transcripts is noncoding.
Table 1.
Summary of Toxorhynchites amboinensis Transcriptome Assembly
| Complete Assembly | Protein-Coding Components |
||
|---|---|---|---|
| All Isoforms | Representative Isoforms | ||
| Total size | 267.6 Mb | 171.21 Mb | 41.0 Mb |
| Number of components | 199,064 | 21,605 | |
| Number of transcripts | 303,383 | 80,552 | 21,605 |
| Longest transcript | 28,920 bp | 28,920 bp | 28,908 bp |
| N50 transcript length | 2,082 bp | 3,575 bp | 3,041 bp |
| GC content | 41.51% | 43.42% | 43.86% |
| CEGMA completeness | 248/248 | 241/248 | 240/248 |
Note.—The completeness of T. amboinensis transcriptome assembly was evaluated using CEGMA based on the presence/absence of 248 conserved eukaryotic genes. All the seven genes missed in annotated transcripts (all isoforms) and seven of the eight genes missed in annotated transcripts (representative isoforms) are less than 300 bp long, which is the lower limit of gene length to be considered in our annotation.
We then annotated protein-coding regions (hereafter referred to as “genes” unless otherwise indicated) from the transcriptome assembly. All genes represent continuous ORFs supported by either sequence homology to known protein-coding genes or significant protein-coding potential as determined by a likelihood-based approach. All subsequent analyses in this study were based on genes rather than transcripts because a nonnegligible portion (approximately 13.5%) of the transcripts contained more than one gene (supplementary fig. S1A, Supplementary Material online), which may confound the investigation of gene expression and evolution. In total, we identified 21,605 components each of which contains at least one annotated gene (table 1) about half of which (10,195) contain multiple transcripts (supplementary fig. S1, Supplementary Material online). We used homology criteria (see Materials and Methods) to select the most representative transcript for each component, resulting in a final annotation set composed of 25,084 genes (table 2).
Table 2.
Summary of Toxorhynchites amboinensis Transcriptome Annotation
| Total Gene Count | 25,084 |
| Genes with homolog in NR | 20,278 |
| Genes with coding potential | 4,770 |
| Genes coding (retro-)transposon | 4,888 |
| Total gene length | 26.7 Mb |
| Longest gene length | 24,453 bp (8,151 amino acids) |
| Median gene length | 663 bp (221 amino acids) |
| GC content | 46.79% |
Approximately 80% of the annotated T. amboinensis genes had at least one homolog in the NCBI NR database (conserved genes). Not surprisingly, the best match for more than 90% of T. amboinensis genes was with sequences from insects, with Ae. aegypti, C. quinquefasciatus, and An. gambiae being the top three source species in descending order (fig. 1A). Previous studies have revealed significant contributions of transposable elements (TEs) to the three sequenced mosquito genomes (Holt et al. 2002; Nene et al. 2007; Arensburger et al. 2010); strikingly, almost half of the Ae. aegypti genome assembly was composed of various types of TEs. Similarly, we found 4,888 T. amboinensis genes (approximately 20%) likely encode (retro-) transposon-related proteins, which cover many major types of TEs (table 2 and supplementary table S3, Supplementary Material online).
Fig. 1.—
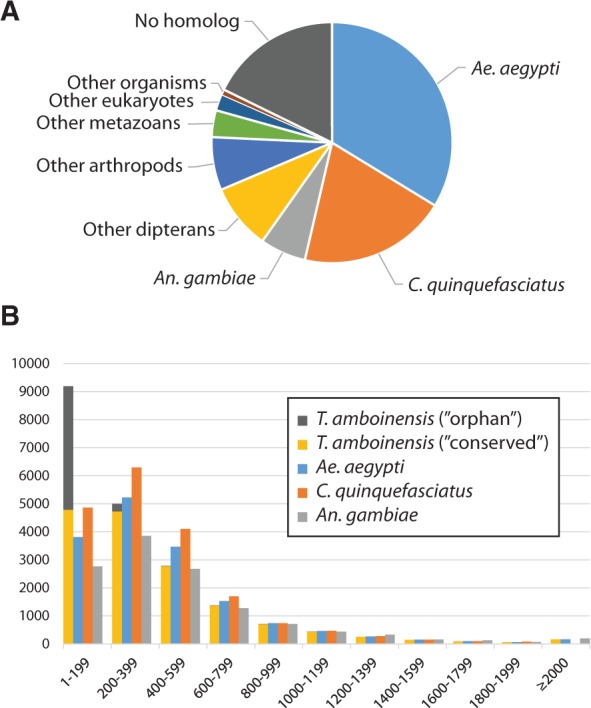
Summary characteristics of T. amboinensis proteins identified in the transcriptome assembly. (A) Taxonomic distribution of best hits of annotated T. amboinensis proteins in the NCBI NR protein database. (B) Length distribution of proteins identified in the transcriptome assembly of T. amboinensis and sequenced genomes of three blood-feeding mosquitoes, including Ae. aegypti, C. quinquefasciatus, and An. gambiae. For T. amboinensis, the “conserved” and “orphan” proteins refer to proteins with and without homolog in the NR database, respectively.
The remaining 20% of genes lacked detectable homologs in the database, but are likely protein coding according to their sequence signatures. These types of genes are also termed “orphan” and have been recently documented in a wide range of organisms (reviewed in Tautz and Domazet-Loso 2011), including insects (Wissler et al. 2013). For example, orphans comprise approximately 5% of the annotated genes in each of the three mosquito genomes (Wissler et al. 2013). Such genes may be fast evolving and thus too divergent from their homologs in other species to be recognized solely based on sequence similarity. Alternatively, orphans may represent novel genes that recently evolved in the T. amboinensis lineage, after its divergence from other mosquitoes. In general, transcripts of orphan genes were less abundant than those of conserved genes in T. amboinensis (supplementary fig. S2, Supplementary Material online). That said, more than 1,500 of them had transcript abundances above the transcriptome median level in at least one of the three samples, and over 2,000 showed significant differential enrichment among samples. In addition, approximately 300 orphan genes encode hypothetical protein products that are longer than 200 amino acids, with the largest one comprising 1,002 residues (fig. 1B). Altogether, the data suggest that some of these orphan genes may indeed be functional and have contributed to the morphological and physiological differences between T. amboinensis and its relatives.
Completeness of Transcriptome Assembly of T. amboinensis
The transcriptome assembly and gene annotations generated here constitute valuable resources for the study of T. amboinensis biology, with the caveat that, to some extent, the assembly is likely incomplete. Indeed, although we have performed in-depth transcriptome profiling on whole bodies and two chemosensory appendages of female T. amboinensis, genes that have low abundance levels or show restricted expression in specific tissues and/or developmental stages beyond our sample set will be missed or at the very least, underrepresented. However, these limitations aside, it is reasonable to posit that the transcriptome assembly generated here is relatively comprehensive based on the following lines of evidence (see supplementary note, Supplementary Material online, for details).
An evaluation of the assembly completeness using Core Eukaryotic Gene Mapping Approach (CEGMA) (Parra et al. 2007) showed that all of the 248 conserved eukaryotic genes can be found in the complete transcriptome assembly (table 1). In addition, the number and length distribution of genes annotated in the T. amboinensis transcriptome assembly closely resemble those in the three sequenced mosquito genomes analyzed here (fig. 1B). More importantly, our parallel de novo transcriptome assembly analysis in An. gambiae (using much smaller transcriptome data sets but following the same assembly and annotation protocol) was able to recover more than 70% of An. gambiae genes, more than half of which was recovered at full length. Taken together, these considerations indicate that the T. amboinensis transcriptome assembly generated here likely represents a comprehensive genomic resource. Sequences and transcript abundances of all annotated T. amboinensis genes are available along with detailed functional annotations, including GO, protein domains, and metabolic pathways (see Data Availability).
Compared with other mosquitoes, T. amboinensis larvae are predatory whereas adults are very large and do not take blood meals, which suggests that salient aspects of its development, immunity, metabolism, and neuronal systems would likely be unique. In this regard, a comprehensive transcriptome assembly also enabled a genome-wide survey of gene families of interest. For example, we have identified 354 immunity-related genes in T. amboinensis, which covered all major pathways currently known in mosquitoes and this number is similar to the total number of immunity genes reported in the three mosquito genomes (Waterhouse et al. 2007; Bartholomay et al. 2010) (supplementary table S4, Supplementary Material online; also see later section on chemosensory gene families). In conclusion, the sequence and functional information generated here is expected to significantly facilitate future studies of this nonblood-feeding mosquito as well as comparative analyses with its blood-feeding relatives.
Resolving the Phylogenetic Placement of T. amboinensis within Culicidae
The transcriptome assembly of T. amboinensis provides a unique opportunity to examine its precise placement within Culicidae using genome-scale data. We constructed orthologous gene groups from the four mosquitoes plus D. melanogaster, which was included as outgroup, and identified 4,266 genes that are single-copy in all five species. The concatenated maximum-likelihood analysis of these single-copy genes (6,807,855 nucleotide positions in total) resulted in a fully resolved tree placing T. amboinensis sister to Ae. aegypti (fig. 2A) relative to the other two mosquitoes. This grouping of T. amboinensis and Ae. aegypti was also strongly supported by most of the single gene phylogenies (supplementary fig. S3, Supplementary Material online). Furthermore, using the 144 genes that fully supported the concatenated ML tree, we recovered the same topology in a Bayesian analysis and estimated that the ancestors of T. amboinensis and Ae. aegypti separated about 39.6 Ma, soon after the estimated divergence of the ancestors of Ae. aegypti and C. quinquefasciatus at about 52.5 Ma (fig. 2A).
Fig. 2.—
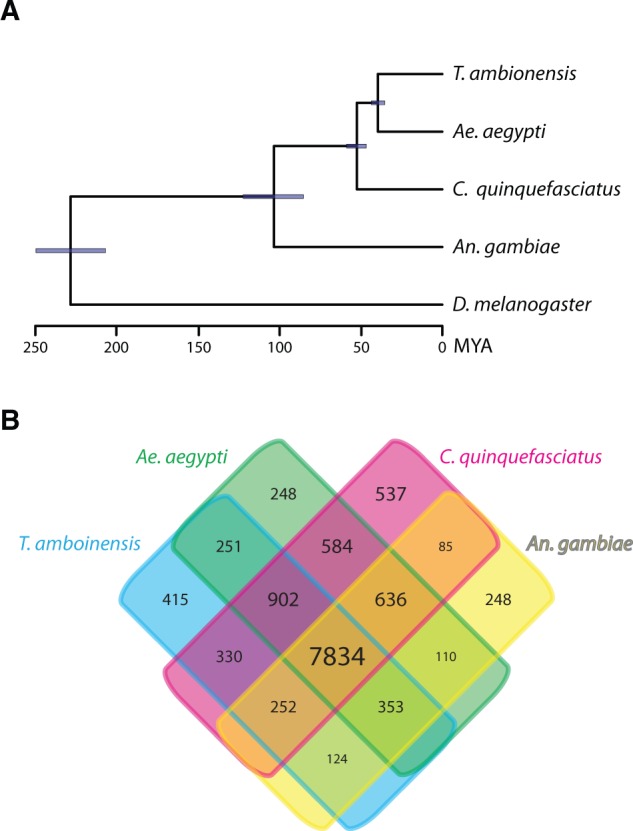
Phylogenetic analyses of orthologous genes supporting a sister relationship between T. amboinensis and Ae. aegypti. (A) Chronogram showing the phylogenetic relationships and estimated divergence times between T. amboinensis and other mosquitoes. Drosophila melanogaster was included as an outgroup, and its separation with Culicidae at 227 Ma (95% confidence interval: 210–244 Ma) was used as a calibration point in the divergence time estimation. The purple bar on each node indicates the 95% confidence interval of estimated divergence time. The same tree topology was recovered in a concatenated ML analysis of 4,266 single-copy orthologous genes with maximum supports for all nodes, and also received overwhelming supports from ML analyses of individual genes. (B) A Venn diagram showing the number of orthologous gene groups shared between four mosquito species.
The genome-scale data presented here display overwhelming support for a sister relationship between T. amboinensis and Ae. aegypti. Additional phylogenomic studies with broader taxon sampling will be helpful to further evaluate this relationship, as only four mosquito species were included here. Nevertheless, the phylogeny proposed here provides a solid evolutionary context for comparative studies between T. amboinensis and other mosquitoes. When one considers that most mosquito genera are populated with species that are capable of taking blood meals for reproduction, the more derived position of T. amboinensis suggested here indicates that blood feeding is an ancestral trait that has been lost in the lineage leading to T. amboinensis.
Characterization of the Chemosensory Gene Repertoire of T. amboinensis
The transition from the ancestral hematophagous state to the derived nonblood-feeding state in T. amboinensis is expected to be accompanied by changes in lifestyle requirements, most notably the loss of a need to engage in host-seeking behavior(s). Inasmuch as these behaviors are strongly mediated by chemosensory cues in hematophagous mosquitoes, it is reasonable to ask whether or not the repertoire of peripherally expressed, chemosensory genes in T. amboinensis may have changed concomitantly with the shift in feeding behavior. With a high quality transcriptome assembly of T. amboinensis available and its phylogenetic affiliation resolved, we now have the opportunity to carry out comparative analyses of chemosensory genes between blood-feeding and nonblood-feeding mosquitoes.
A large number of chemosensory genes, including 87 ORs, 21 GRs, and 38 IRs were identified in the transcriptome assembly of T. amboinensis (also see supplementary table S5, Supplementary Material online, for information on additional chemosensory genes). In comparison to its closest relative, Ae. aegypti, we identified fewer chemosensory genes in T. amboinensis, which may reflect excessive loss in T. amboinensis (table 3) or may be attributed to genes that are present in the genome but whose transcripts were absent in the samples included in this study. In fact, a previous transcriptome profile in An. gambiae indicated that many chemosensory genes have very low expression levels in adult chemosensory appendages and whole bodies of both sexes (Pitts et al. 2011). Moreover, several An. gambiae OR (AgOr) genes are specifically expressed during larval stages or other tissues (Xia et al. 2008; Pitts et al. 2011). Indeed, 31 of the 38 IR genes identified in the T. amboinensis transcriptome assembly belong to a subfamily (“antennal” IRs) whose members in other insects have been shown to be expressed in antennae (Croset et al. 2010); this number is close to that in Ae. aegypti which has many more IR genes (supplementary table S5, Supplementary Material online).
Table 3.
Number of Chemosensory Genes in Mosquitoes
| OR | GR | IR | |
|---|---|---|---|
| Anopheles gambiae | 75 | 61 | 46 (22) |
| Culex quinquefasciatus | 178 | 65 | 69 (45) |
| Aedes aegypti | 127 | 70 | 95 (37) |
| Toxorhynchites amboinensis | 87 | 21 | 38 (31) |
Note.—Numbers in parenthesis indicate antennal IRs.
To better understand the changes in the chemosensory gene repertoire of T. amboinensis, we reconstructed the evolutionary histories of mosquito genes in the three major chemosensory receptor gene families, namely ORs, GRs, and IRs. We focused on these gene families, which together comprise the chemosensory receptome, because their functional roles in chemosensation are much better characterized compared with other gene families such as odorant-binding proteins. This analysis reveals that most chemosensory receptor genes in T. amboinensis have clear (co-)orthologs in Ae. aegypti. In the OR gene family phylogeny (fig. 3), we identified 31 well-supported clades (bootstrap support ≥ 70%) that include both T. amboinensis and Ae. aegypti ORs (AaOrs), each of which likely represents one single gene in the common ancestor of the two mosquitoes; 27 of them also contain ORs in C. quinquefasciatus and/or An. gambiae. In total, the 31 clades encompass 58 of the 87 T. amboinensis OR genes (TaOrs). Among these clades, T. amboinensis and Ae. aegypti have the same number of genes in 16 clades, including 13 one-to-one orthologous pairs, and they differ by only one gene in nine clades (fig. 3 and supplementary fig. S5, Supplementary Material online). Similar results were obtained from the phylogenetic analyses of GR and IR families (supplementary figs. S6 and S7, Supplementary Material online); 19 of 21 T. amboinensis GR genes (TaGrs) and 35 of 38 T. amboinensis IR genes (TaIrs) have well-supported (co-) orthologs in Ae. aegypti, many of which are one-to-one orthologous pairs (13 TaGrs and 15 TaIrs).
Fig. 3.—
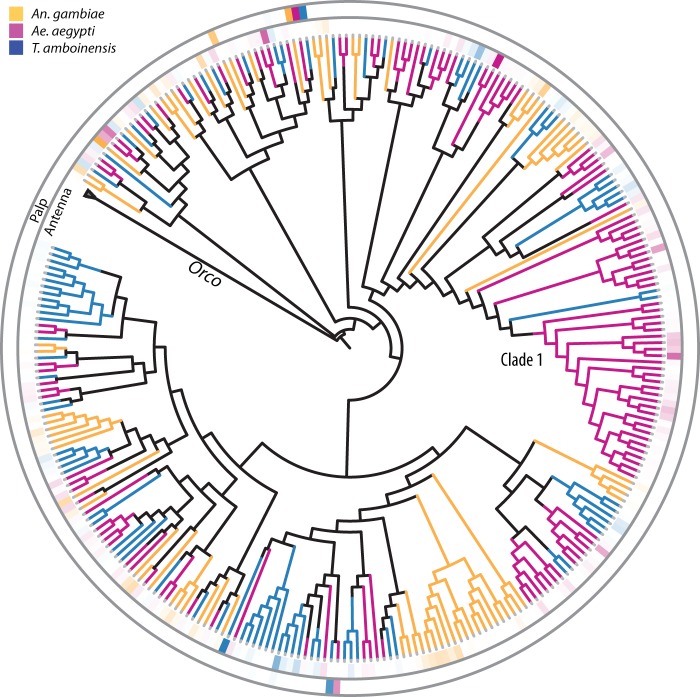
OR gene tree and their relative expression in the antennae and palp of T. amboinensis, Ae. aegypti, and An. gambiae. Gene phylogeny of the OR-family of chemoreceptors. Branches are color coded by the respective species and the heat maps at the branch tips represent the RPKM-percent composition of each family member within the entirety of that family’s expression within the tissue of each species. The outer heat map ring displays the tissue expression profile in maxillary palps and the inner heat map ring displays the tissue expression profile in antennae. Orco was used to root the tree and the Orco clade was collapsed to emphasize differences between the tuning ORs.
These data suggest that the chemosensory receptor gene repertoire of T. amboinensis (as recovered from its transcriptome assembly) is relatively stable compared with the other three mosquito genomes. In particular, six OR clades, eight GR clades, and nine IR clades are well supported and each is comprised by a single gene in each of the four mosquitoes. Among these single-copy clades are several genes with well-characterized or potentially important chemosensory functions, such as AaOrco (for convenience, here we use the name of Ae. aegypti gene in each clade) which encodes the highly conserved OR coreceptor, AaGr1-3 which encodes receptors for carbon dioxide (Kent et al. 2008), AaGr7 and AaGr11 which encode candidate sugar receptors (Kent et al. 2008), AaGr14 which is orthologous to a D. melanogaster Gr (DmGr66a) required for caffeine response (Moon et al. 2006; Kent et al. 2008), as well as a number of aforementioned conserved “antennal” IR genes, including AaIr8a, AaIr21a, AaIr25a, AaIr31a, AaIr40a, AaIr64a, AaIr75i, AaIr76b, and AaIr93a.
There are also several apparent differences between the chemosensory gene repertoires of T. amboinensis and Ae. aegypti. There are eight TaOrs, two TaGrs, and three TaIrs whose (co-)orthologs are missing in the Ae. aegypti genome, but can be found in the other two, more distant mosquito genomes, indicating lineage-specific loss in Ae. aegypti. In addition, 21 TaOrs do not have clear orthologs in any of the three blood-feeding mosquitoes, which may suggest gene loss in these species; alternatively, it is possible that these genes have experienced relatively rapid sequence divergence and thus their relationship with other genes has become more difficult to resolve. Furthermore, we found a relatively modest expansion of genes in T. amboinensis relative to Ae. aegypti in two OR clades (six TaOrs vs. two AaOrs in one clade, and five TaOrs vs. one AaOrs in the other) and one IR clade (six TaIrs vs. two AaIrs). Conversely, more clades in the three families have higher numbers of Ae. aegypti genes than observed in T. amboinensis. Most strikingly, one such clade in the OR family included 40 AaOrs and only 2 TaOrs. Importantly, a large number of Ae. aegypti chemosensory genes lack orthologs in the transcriptome assembly of T. amboinensis, including 30 AaOrs and 43 AaGrs.
Chemoreceptive Transcript Abundance in T. amboinensis
To contextualize these newly annotated ORs, IRs, and GRs within the olfactory tissues of T. amboinensis, we next calculated the relative transcript abundance levels in terms of RPKM. We then compared the chemoreceptor transcriptome profile of T. amboinensis’ with both that of its close relative Ae. aegypti as well as An. gambiae. To accommodate the fact that de novo assembly is unable to account for T. amboinensis chemoreceptors that yield little to no transcriptional signal (i.e., those that may be present in the genome but are otherwise not discernable), we have limited our quantitative analysis to transcripts that showed a minimum abundance of two RPKM. This level is designated as being “detectable” and is applied to all data sets analyzed here.
Odorant Receptors
ORs appear to have evolved exclusively within insects and form heteromeric ion channel complexes consisting of at least two subunits: One a highly conserved coreceptor (Orco) whereas the other (ORx) belongs to a class of highly divergent ligand-specifying partners (Clyne et al. 1999; Vosshall et al. 1999; Larsson et al. 2004; Sato et al. 2008; Missbach et al. 2014). Orco is required for membrane localization of OR complexes, whereas ORx confers the odor tuning properties (Benton et al. 2006). Within this paradigm, ligands for numerous AgOrs and a more limited number of ORs from AaOrs and C. quinquefasciatus ORs (CqOrs) have been identified (Carey et al. 2010; Pelletier et al. 2010; Wang et al. 2010).
Antennae
The tuning OR composition of the T. amboinensis antenna is complex with 67 of the 86 annotated TaOrs showing abundance at reliably detectable levels, with the majority of those occurring above the transcriptome wide median level of abundance. Forty-seven of the detectable tuning TaOrs (70%) showed relative abundances above the median level of all antennal transcripts (5.39 RPKM), a very similar number to the 52 AaOrs (52% of the 91 detectable AaOrs) above the transcriptome median in Ae. aegypti antennae (6.02 RPKM; supplementary table S6, Supplementary Material online). Therefore, although the vast majority of the antennal OR population in both T. amboinensis and Ae. aegypti appears comprised nearly identical numbers of ORs, there are twice as many AaOrs detectable below the transcriptome median. This is potentially important as the presence of this additional, low abundance AaOr complexity may represent a salient difference in the effective odor receptivity between these two species.
Next, we compared the antennal expression profile of ORs of the three mosquito species within the context of their evolutionary history to investigate the degree to which species-specific ORs contribute to the transcriptional profile of their respective species (fig. 3). Analyzed in this way, the OR abundance profiles within each species’ antennae show a nearly equal compositional division between those ORs that are conserved among all three species and ORs that are species-specific. In An. gambiae, Ae. aegypti, and T. amboinensis, approximately 50% of the summed expression levels of tuning OR abundances are attributable to conserved ORs whose orthologs show expression in the antennae of all three species. Clades containing orthologs to such pan-Culcidae conserved ORs as AgOr11, AgOr35, AgOr39, and AgOr80 account for over one-third of the total OR abundance in their respective species alone. Perhaps, the best-characterized example of functionally conserved ORs is the orthologous group containing AgOr2 (AaOr2, CqOr2, TaOr2) and AgOr10 (AaOr10, CqOr10, TaOr10) and that have retained similar odorant sensitivities in their respective species (Hughes et al. 2010; Pelletier et al. 2010; Bohbot et al. 2011). In the antennae, Or2/Or10 comprises 2%, 3 and 4% of the total abundance of tuning ORs in Ae. aegypti, An. gambiae and T. amboinensis, respectively. This high degree of interspecific sequential, functional and transcriptional conservation suggests a common role for ORs in these mosquito species, and their persistence in T. amboinensis further suggests that this role ranges beyond host-seeking.
The remaining half of antennal tuning OR abundance is attributable to subfamily or lineage-specific ORs within each species. For example, a large, species-specific expansion (fig. 3, Clade1) accounts for approximately 20% of the total level of tuning ORs in Ae. aegypti. Similarly, the Culicinae-specific clade containing an expansion of 17 TaOrs and just 7 AaOrs accounts for over 30% of the total level of tuning ORs in T. amboinensis. This expansion includes such TaOrs as TaOr13.3, TaOr3.2, and TaOr71.1, the latter being the single, most highly abundant TaOr in the antennae. Although TaOr71.1 alone accounts for 17.8% of all tuning ORs (57 times that of the transcriptome median), the sum of all seven AaOr orthologs in this expansion account for just over 3% of tuning ORs in Ae. aegypti.
The Orco coreceptor represented the most highly abundant OR family member in each of the three species. As reported previously, levels of Orco in the chemosensory tissues of two Anophelinae mosquitoes were consistently close to one-to-one as compared with the sum total of all tuning ORs in anopheline antennae, suggesting a possible mechanism of Orco-OR complex regulation within olfactory receptor neurons (ORNs) (Pitts et al. 2011; Rinker, Pitts, et al. 2013; Rinker, Zhou, et al. 2013). In the two Culicinae analyzed here this ratio remained close to one-to-one, although interestingly, Orco abundance in the antennae appears proportionally higher than the summed levels of tuning OR in both Ae. aegypti (1.40) and in T. amboinensis (1.30). This is in contrast to the relationship observed in the Anophelinae and suggests that Orco may be regulated differently between the two mosquito subfamilies.
Palp
OR expression in the maxillary palps of mosquitoes is largely restricted to basiconic sensilla present on the most distal segment. Each of these sensilla is innervated by three ORNs, two of which stereotypically express one of a pair of turning ORs (Lu et al. 2007); one ORN expresses a conserved ortholog of Or8 (e.g., AgOr8, AaOr8, CqOr8) whereas the second contains a tuning OR that appears specific to either the Anophelinae subfamily (e.g., AgOr28) or Culicinae subfamily (e.g., AaOr49, CqOr49).
Similarly, the palps of T. amboinensis displayed only two detectable tuning TaOrs: TaOr8 and TaOr49 (fig. 3). In terms of levels of OR abundance, it is notable that the ratio of TaOrco to the summed RPKM of the two tuning TaOrs (2.02) is nearly identical to that seen in Ae. aegypti palp (1.90) but is reciprocal to the relationship in An. gambiae palp (0.89). This again recapitulates the quantitative relationships we observed between Orco and the tuning ORs in the antennae of these species, and its presence in a tissue that is well characterized as having only two tuning ORs further suggests a difference in regulatory mechanisms, rather than the existence of unannotated ORs in Culicinae.
Ionotropic Receptors
The IRs are the most ancient of the three chemoreceptive gene families, yet are the most recently discovered (Benton et al. 2009; Croset et al. 2010; Liu et al. 2010) and the least well characterized in terms of their localization and function. IRs as a class appear particularly tuned to detection of ammonia, amines, acids and alcohols, and are expressed in ORNs, which may or may not also express ORs (Benton et al. 2009). In Drosophila, IRs appear to be expressed in specific ORNs of the antenna and proboscis, with the distribution of IR transcripts in the mosquito suggesting similar enrichments in the antenna (Pitts et al. 2011; Leal et al. 2013; Rinker, Pitts, et al. 2013) and labellum (Sparks et al. 2014).
Functionally, the IR gene family resembles ORs in that it is split into expansive groups of IRs that seem to confer odorant specificity, and several putative IR coreceptors (orthologs to DmIr25a, DmIr8a, and DmIr76b) that are highly conserved and are likely to be necessary for proper IR function (Croset et al. 2010).
Antennae
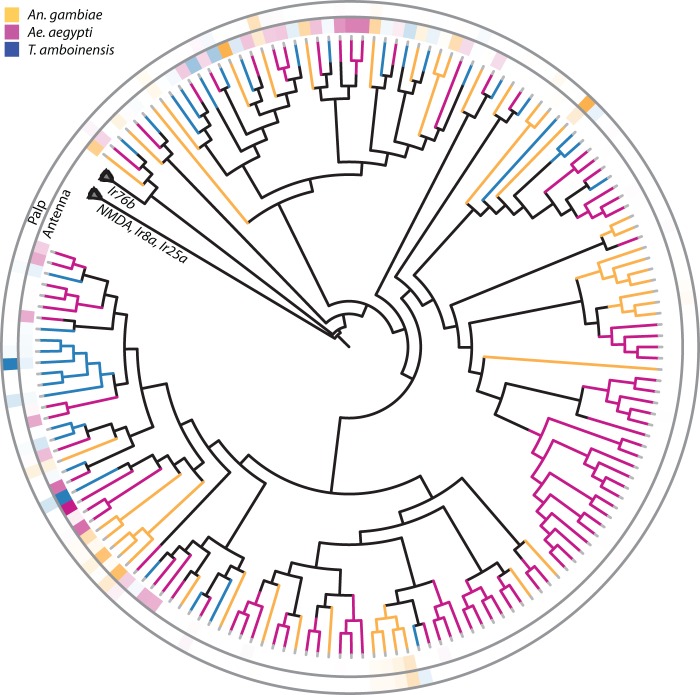
The IR composition of the antennae of all three species was very similar. Twenty-eight ligand specifying TaIr genes were reliably detected in T. amboinensis antennae, nearly equal to the numbers seen in An. gambiae (27) and Ae. aegypti (26). Orthologs to all three putative, IR coreceptors were found to have very high transcript abundances, consistent with transcriptomic studies in the antenna of other insects (Pitts et al. 2011; Zhou et al. 2014). Indeed the most highly abundant IR transcripts in the antennae of each of the three species were the homologs to Ir25a and Ir76b, with Ir8a consistently showing an abundance level of approximately one-third that of Ir25a. Moreover in both of the Culicinae, the ratio of total RPKM for the three IR coreceptors to the RPKM total of the remaining IRs was remarkably consistent (0.998 in Ae. aegypti and 0.997 in T. amboinensis), a correlation reminiscent of the conserved Orco/tuning OR relationship in Anopheles and implying an interdependency between these two IR subcategories.
The IRs appear to differ markedly from the ORs with regard to their evolution and transcriptome profiles. When the abundance levels of antennal IRs are viewed within the context of the evolutionary history of the IR gene family, it appears that greater than 90% of the antennal abundance of ligand specifying IRs is derived from those residing in single-copy, three species clades (fig. 4). For example, the most highly abundant IRs in An. gambiae (AgIr75l), Ae. aegypti (AAEL008587), and T. amboinensis (TaIr41r) each have orthologs in the other two species that are also moderately or highly expressed within those species. Furthermore, there are few notable instances of detectable IRs that appear to be species-specific. Only An. gambiae shows a modest level of some members of an IR clade (AgIr41n, AgIr41c, AgIr41t.2) that has no discernable orthologs in Culicinae. Overall, the underrepresentation in terms of abundance of lineage-specific IRs within mosquito species appears to confirm the observation that even though members of the IR gene family rapidly duplicate and diversify within species, most of those IRs that are detectable are highly conserved (Benton et al. 2009).
Fig. 4.—
IR gene tree and their relative expression in the antennae and palp of T. amboinensis, Ae. aegypti, and An. gambiae. Gene phylogeny of the IR-family of chemoreceptors. Branches are color coded by the respective species and the heat maps at the branch tips represent the RPKM-percent composition of each family member within the entirety of that family’s expression within the tissue of each species. The outer heat map ring displays the tissue expression profile in maxillary palps and the inner heat map ring displays the tissue expression profile in antennae. NMDA receptors were used to root the tree and the NMDA clades were collapsed to emphasize differences between the ligand specifying IRs.
Palp
Although primarily concentrated in the antennae, IR transcripts have also been reported in the maxillary palps of both An. gambiae (Pitts et al. 2011) and Ae. aegypti (Bohbot et al. 2014). Toxorhynchites amboinensis palps show a similar profile with a very limited number of IRs being detectable along with what may be the stereotypical absence (Pitts et al. 2011; Guo et al. 2013) of the IR8a coreceptor in insect palps. More strikingly, the Ir76b ortholog—the second putative IR coreceptor expressed in the palps of both An. gambiae and Ae. aegypti—is also not detectable within the palps of T. amboinensis and this absence may indicate a significant shift in the chemoreceptive capacity in the palp of this particular lineage.
Gustatory Receptors
GRs, along with ORs, are the other class of seven transmembrane chemoreceptors that have been identified in insects (Hill et al. 2002; Bohbot et al. 2007; Kent et al. 2008; Kent and Robertson 2009). As their name implies, they are most associated with contact chemosensation although several of the family members play a role in olfaction and host-seeking (Kent and Robertson 2009; McMeniman et al. 2014).
Antennae
In mosquitoes, GR transcripts are detectable in the antennae but always at very low levels (Pitts et al. 2011; Rinker, Pitts, et al. 2013). In T. amboinensis we observed nine detectable TaGrs in the antennae, seven of which were present at abundance levels greater than the transcriptome wide median. In contrast, only two AaGrs and five AgGrs were detectable, and in both cases none reached an RPKM level above their respective transcriptome median values.
All of the detectable TaGrs had orthologs in both Ae. aegypti and Culex. Of the most abundant antennal TaGrs, the top three, TaGr2, TaGr23 and TaGr21 (RPKMS of 15.0, 13.6, and 10.3, respectively), had detectable orthologs in Ae. aegypti antennae (AaGr14, AaGr2, and AaGr7, respectively). Notably, none of the detectable TaGrs had any clear orthologs within An. gambiae.
Palp
GR expression in the maxillary palps of mosquitoes has been localized to the non-OR expressing ORNs innervating the basiconic sensilla in An. gambiae and Ae. aegypti (Lu et al. 2007; Erdelyan et al. 2012). Orthologs of two of the three GRs expressed here (AaGr1/AaGr3, AgGr22/AgGr24) are necessary for CO2 reception in D. melanogaster (Dm21a/Dm63a) (Dahanukar et al. 2007; Jones et al. 2007; Kwon et al. 2007), and the presence of a third GR in the basiconic sensilla of mosquitoes has been postulated to confer enhanced sensitivity of CO2 in An. gambiae (Lu et al. 2007). Each of the three GRs (AaGr1/AaGr2/AaGr3, AgGr22/AgGr23/AgGr24, and TaGr1/TaGr2/TaGr3) shows high sequence conservation between species and appears to function together to confer sensitivity to CO2 as well as other semiochemicals in basiconic sensilla of the palp (Kellogg 1970; Grant et al. 1995; Tauxe et al. 2013). This is significant inasmuch as CO2 appears to be an important long-range activator of mosquito host-seeking (Gillies 1980; Takken and Knols 1999).
All three GRs are expressed at very high levels in the palp in each of the three species (supplementary fig. S4, Supplementary Material online). Moreover, the pattern of expression between orthologous GRs appears conserved for TaGr1 and TaGr2 with each detected at nearly equal ratios to one another in all species. In contrast, the abundance of TaGr3 relative to the level of the most abundant palpal Gr was markedly lower in T. amboinensis (40%) than in either An. gambiae (56%) or Ae. aegypti (68%). Because Gr3 is essential for CO2 detection in flies and confounds host-seeking in Ae. aegypti (McMeniman et al. 2014), the lower relative abundance of TaGr3 could hint at an alteration in the sensitivity of this particular mode of GR-mediated chemoreception.
The expression analysis of chemoreceptive genes present on the antennae and palps shows that the peripheral chemoreceptive repertoire of female T. amboinensis mosquitoes displays two broad tendencies that are also observed in the other two Culicidae analyzed here. First, we see that the overwhelming majority of detectable GRs and IRs, along with at least half of the detectable ORs consist of genes that have clear orthologs in the transcriptomes of all three taxa. Second, we see that the remaining half of the OR composition of the antennae in each of the three taxa consist of lineage-specific OR genes. Together, these observations suggest a generalizable model of peripheral chemoreceptor variation that exists between mosquito species. One rationale for this is that the expression of conserved GRs, IRs, and ORs provides a Culicidae-wide chemoreceptive foundation upon which lineage-specific expansions of ORs can then dynamically and independently explore sequence and odor space within each lineage.
Beyond the generalities described above, the transcriptome profile of T. amboinensis also displays subtle characteristics that appear more specific to the Culicinae as well as some which distinguish it from Ae. aegypti. First, in all chemosensory tissues, T. amboinensis and Ae. aegypti show a modest yet inverted relationship, between the amounts of Orco transcript and total tuning OR transcripts relative to An. gambiae, suggesting alternative modes of OR regulation within the two subfamilies. Second, T. amboinensis displays the conserved presence of an ortholog for AaOr49/CqOr49 in the palp along with the absence of any AgOr28 ortholog. These trends may represent two, broad divergences in stereotypic OR-expression that distinguish Anophelinae from Culicinae.
Such examples of chemoreceptive similarities between T. amboinensis and other mosquito species hint at conserved ranges of olfactory tuning that reflect the constraints of chemical ecology common to all Culicidae. In this light, behaviorally salient chemosensory signals such as plant, host, and oviposition site cues likely share many similarities in their odor profiles and may underlie the prevalence of the observed set of ancient and abundant chemoreceptors. Indeed, such a shared chemoreceptive repertoire of interspecific oviposition cues would be of particular importance to T. amboinensis whose life cycle depends heavily on depositing eggs in water sources that contain the larvae of other mosquitoes.
Where T. amboinensis appears to show some distinction from Ae. aegypti is in the number and relative abundance of certain chemoreceptive genes. First, the palp of T. amboinensis is lacking the detectable presence of an ortholog to Ag/AaIr76b which, as a putative IR coreceptor, could indicate an overall shift in functional TaIr diversity in the palp. Second, T. amboinensis females appear to have a markedly reduced antennal OR diversity than Ae. aegypti, and this diversity is lacking specifically in the numbers of lineage-derived OR genes. Such examples of chemoreceptive “simplification” in T. amboinensis could be the result of the loss of blood feeding in this lineage, with the reduction in the variety of tuning ORs supporting the role ORs play in conferring host specificity in mosquitoes (DeGennaro et al. 2013).
Conclusions
In this study we have performed a comparative analysis of the transcriptome profiles between T. amboinensis and three disease vector mosquitoes, with a focus on chemosensory gene families. Through in-depth RNA-seq of chemosensory appendages and whole bodies of adult female T. amboinensis, we obtained a comprehensive de novo transcriptome assembly, and confidently resolved for the first time the phylogenetic position of T. amboinensis thereby providing an important context for subsequent comparative studies. The transcriptome assembly, together with the detailed functional annotations we have generated (e.g., protein domains and GO), represents a valuable genomic resource that will facilitate future studies of T. amboinensis. In the transcriptome assembly of T. amboinensis, we identified a large chemosensory gene repertoire that is comparable to other mosquitoes. Phylogenetic analyses of the three major chemosensory receptor gene families revealed both clades that have maintained stable copy numbers in the four mosquitoes, as well as those with differential gain-and-loss among these blood-feeding and nonblood-feeding mosquitoes.
Subsequent transcript abundance analysis revealed that T. amboinensis is generally similar to other mosquito species while lacking certain chemoreceptive components present in the other taxa analyzed. Specifically, we observed a reduction in the numeric variety of antennal ORs as compared with Ae. aegypti. This marked depletion in OR complexity co-occurs with the retention of a large number of conserved mosquito ORs, indicating that T. amboinensis retains a core repertoire of expressed ORs that complement sets of conserved GRs and IRs. In the palp, we also observed the absence of one of the putative IR coreceptors along with indications of a potential decrease in CO2 receptivity.
Taken together, our analyses show that in its divergence from Ae. aegypti, T. amboinensis not only lost its reproductive requirement for the taking of blood meals but also experienced a measurable, diminution in the qualitative complexity of peripheral chemoreceptive coding. Chemoreceptors that appear to have been conserved in both copy number and abundance may be centrally involved in such common phenotypic behaviors as nectar feeding and ovipositing that reflect the constraints of chemical ecology.
Supplementary Material
Supplementary note, tables S1–S7, and figures S1–S7 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Dr Joseph Dickens (USDA, ARS) for palpal RNA-seq reads from Ae. aegypti, and Manuel Amador (CDC, Puerto Rico) for providing T. amboinensis eggs. They are also indebted to the members of the Rokas/Zwiebel labs for helpful discussions and critical readings. This work was conducted in part using the resources of the Advanced Computing Center for Research and Education at Vanderbilt University and was supported by funds provided by the National Science Foundation (DEB-0844968) and the National Institutes of Health (NIAID, AI105619) to A.R., as well as grants from the Innovation and Discovery in Engineering and Science (IDEAS) program of Vanderbilt University and from the National Institutes of Health (NIAID, AI056402) to L.J.Z. This work was also supported by the NIDCD through an NRSA (F31 DC012991) to D.C.R.
Literature Cited
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arensburger P, et al. Sequencing of Culex quinquefasciatus establishes a platform for mosquito comparative genomics. Science. 2010;330:86–88. doi: 10.1126/science.1191864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomay LC, et al. Pathogenomics of Culex quinquefasciatus and meta-analysis of infection responses to diverse pathogens. Science. 2010;330:88–90. doi: 10.1126/science.1193162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4:e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Vannice K, Gomezdiaz C, Vosshall L. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136:149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Clamp M, Durbin R. GeneWise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohbot J, et al. Molecular characterization of the Aedes aegypti odorant receptor gene family. Insect Mol Biol. 2007;16:525–537. doi: 10.1111/j.1365-2583.2007.00748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohbot JD, et al. Conservation of indole responsive odorant receptors in mosquitoes reveals an ancient olfactory trait. Chem Senses. 2011;36:149–160. doi: 10.1093/chemse/bjq105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohbot JD, Sparks JT, Dickens JC. The maxillary palp of Aedes aegypti, a model of multisensory integration. Insect Biochem Mol Biol. 2014;48:29–39. doi: 10.1016/j.ibmb.2014.02.007. [DOI] [PubMed] [Google Scholar]
- Bouckaert R, et al. BEAST2: A software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 2014;10:e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey AF, Wang G, Su C-Y, Zwiebel LJ, Carlson JR. Odorant reception in the malaria mosquito Anopheles gambiae. Nature. 2010;464:66–71. doi: 10.1038/nature08834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements AN. The biology of mosquitoes: sensory reception and behaviour. Wallinford (United Kingdom): CABI; 1999. [Google Scholar]
- Clyne PJ, et al. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron. 1999;22:327–338. doi: 10.1016/s0896-6273(00)81093-4. [DOI] [PubMed] [Google Scholar]
- Croset V, et al. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 2010;6:e1001064. doi: 10.1371/journal.pgen.1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahanukar A, Lei Y-T, Kwon JY, Carlson JR. Two gr genes underlie sugar reception in Drosophila. Neuron. 2007;56:503–516. doi: 10.1016/j.neuron.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGennaro M, et al. orco mutant mosquitoes lose strong preference for humans and are not repelled by volatile DEET. Nature. 2013;498:487–491. doi: 10.1038/nature12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdelyan CN, Mahood TH, Bader TS, Whyard S. Functional validation of the carbon dioxide receptor genes in Aedes aegypti mosquitoes using RNA interference. Insect Mol Biol. 2012;21:119–127. doi: 10.1111/j.1365-2583.2011.01120.x. [DOI] [PubMed] [Google Scholar]
- Gillies M. The role of carbon dioxide in host-finding by mosquitoes (Diptera: Culicidae): a review. Bull Entomol Res. 1980;70:525–532. [Google Scholar]
- Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant A, Wigton B, Aghajanian J, O’Connell R. Electrophysiological responses of receptor neurons in mosquito maxillary palp sensilla to carbon dioxide. J Comp Physiol A. 1995;177:389–396. doi: 10.1007/BF00187475. [DOI] [PubMed] [Google Scholar]
- Guo M, et al. Variant ionotropic receptors are expressed in olfactory sensory neurons of coeloconic sensilla on the antenna of the desert locust (Schistocerca gregaria) Int J Biol Sci. 2013;10:1–14. doi: 10.7150/ijbs.7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbach R, Kitching I. Phylogeny and classification of the Culicidae (Diptera) Syst Entomol. 1998;23:327–370. [Google Scholar]
- Hill CA, et al. G protein-coupled receptors in Anopheles gambiae. Science. 2002;298:176–178. doi: 10.1126/science.1076196. [DOI] [PubMed] [Google Scholar]
- Holt RA, et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- Hughes DT, Pelletier J, Luetje CW, Leal WS. Odorant receptor from the southern house mosquito narrowly tuned to the oviposition attractant skatole. J Chem Ecol. 2010;36:797–800. doi: 10.1007/s10886-010-9828-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S, et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AK, Brown LA, Sattelle DB. Insect nicotinic acetylcholine receptor gene families: from genetic model organism to vector, pest and beneficial species. Invert Neurosci. 2007;7:67–73. doi: 10.1007/s10158-006-0039-6. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg FE. Water vapour and carbon dioxide receptors in Aedes aegypti. J Insect Physiol. 1970;16:99–108. doi: 10.1016/0022-1910(70)90117-4. [DOI] [PubMed] [Google Scholar]
- Kent L, Robertson H. Evolution of the sugar receptors in insects. BMC Evol Biol. 2009;9:50. doi: 10.1186/1471-2148-9-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent LB, Walden KR, Robertson HM. The Gr family of candidate gustatory and olfactory receptors in the yellow-fever mosquito Aedes aegypti. Chem Senses. 2008;33:79–95. doi: 10.1093/chemse/bjm067. [DOI] [PubMed] [Google Scholar]
- Kwon JY, Dahanukar A, Weiss LA, Carlson JR. The molecular basis of CO2 reception in Drosophila. Proc Natl Acad Sci U S A. 2007;104:3574–3578. doi: 10.1073/pnas.0700079104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson MC, et al. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43:703–714. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Leal WS, Choo YM, Xu P, da Silva CS, Ueira-Vieira C. Differential expression of olfactory genes in the southern house mosquito and insights into unique odorant receptor gene isoforms. Proc Natl Acad Sci U S A. 2013;110:18704–18709. doi: 10.1073/pnas.1316059110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011;39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, et al. Distinct olfactory signaling mechanisms in the malaria vector mosquito Anopheles gambiae. PLoS Biol. 2010;8:e1000467. doi: 10.1371/journal.pbio.1000467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, et al. RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res. 2012;40:W622–W627. doi: 10.1093/nar/gks540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, et al. Odor coding in the maxillary palp of the malaria vector mosquito Anopheles gambiae. Curr Biol. 2007;17:1533–1544. doi: 10.1016/j.cub.2007.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMeniman CJ, Corfas RA, Matthews BJ, Ritchie SA, Vosshall LB. Multimodal integration of carbon dioxide and other sensory cues drives mosquito attraction to humans. Cell. 2014;156:1060–1071. doi: 10.1016/j.cell.2013.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BR, Crabtree MB, Savage HM. Phylogenetic relationships of the Culicomorpha inferred from 18S and 5.8S ribosomal DNA sequences (Diptera:Nematocera) Insect Mol Biol. 1997;6:105–114. doi: 10.1111/j.1365-2583.1997.tb00078.x. [DOI] [PubMed] [Google Scholar]
- Missbach C, et al. Evolution of insect olfactory receptors. Elife. 2014;3:e02115. doi: 10.7554/eLife.02115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A, Sperling FAH, Hickey DA. Higher-level phylogeny of mosquitoes (Diptera: Culicidae): mtDNA data support a derived placement for Toxorhynchites. Insect Syst Evol. 2002;33:163–174. [Google Scholar]
- Moon SJ, Kottgen M, Jiao Y, Xu H, Montell C. A taste receptor required for the caffeine response in vivo. Curr Biol. 2006;16:1812–1817. doi: 10.1016/j.cub.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Nene V, et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- Pelletier J, Hughes DT, Luetje CW, Leal WS. An odorant receptor from the southern house mosquito Culex pipiens quinquefasciatus sensitive to oviposition attractants. PLoS One. 2010;5:e10090. doi: 10.1371/journal.pone.0010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts RJ, Rinker DC, Jones PL, Rokas A, Zwiebel LJ. Transcriptome profiling of chemosensory appendages in the malaria vector Anopheles gambiae reveals tissue- and sex-specific signatures of odor coding. BMC Genomics. 2011;12:271. doi: 10.1186/1471-2164-12-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevillon E, et al. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidenbach KR, et al. Phylogenetic analysis and temporal diversification of mosquitoes (Diptera: Culicidae) based on nuclear genes and morphology. BMC Evol Biol. 2009;9:298. doi: 10.1186/1471-2148-9-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinker DC, Pitts RJ, et al. Blood meal-induced changes to antennal transcriptome profiles reveal shifts in odor sensitivities in Anopheles gambiae. Proc Natl Acad Sci U S A. 2013;110:8260–8265. doi: 10.1073/pnas.1302562110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinker DC, Zhou X, et al. Antennal transcriptome profiles of anopheline mosquitoes reveal human host olfactory specialization in Anopheles gambiae. BMC Genomics. 2013;14:749. doi: 10.1186/1471-2164-14-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Pellegrino M, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452:1002–1006. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- Shepard JJ, Andreadis TG, Vossbrinck CR. Molecular phylogeny and evolutionary relationships among mosquitoes (Diptera: Culicidae) from the northeastern United States based on small subunit ribosomal DNA (18S rDNA) sequences. J Med Entomol. 2006;43:443–454. doi: 10.1603/0022-2585(2006)43[443:mpaera]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Sparks JT, Bohbot JD, Dickens JC. The genetics of chemoreception in the labella and tarsi of Aedes aegypti. Insect Biochem Mol Biol. 2014;48:8–16. doi: 10.1016/j.ibmb.2014.02.004. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Takken W, Knols BG. Odor-mediated behavior of Afrotropical malaria mosquitoes. Annu Rev Entomol. 1999;44:131–157. doi: 10.1146/annurev.ento.44.1.131. [DOI] [PubMed] [Google Scholar]
- Tautz D, Domazet-Loso T. The evolutionary origin of orphan genes. Nat Rev Genet. 2011;12:692–702. doi: 10.1038/nrg3053. [DOI] [PubMed] [Google Scholar]
- Tauxe GM, MacWilliam D, Boyle SM, Guda T, Ray A. Targeting a dual detector of skin and CO2 to modify mosquito host seeking. Cell. 2013;155:1365–1379. doi: 10.1016/j.cell.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725–736. doi: 10.1016/s0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- Wang G, Carey AF, Carlson JR, Zwiebel LJ. Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proc Natl Acad Sci U S A. 2010;107:4418–4423. doi: 10.1073/pnas.0913392107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RM, et al. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science. 2007;316:1738–1743. doi: 10.1126/science.1139862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheat CW, Wahlberg N. Critiquing blind dating: the dangers of over-confident date estimates in comparative genomics. Trends Ecol Evol. 2013;28:636–642. doi: 10.1016/j.tree.2013.07.007. [DOI] [PubMed] [Google Scholar]
- Wiegmann BM, et al. Episodic radiations in the fly tree of life. Proc Natl Acad Sci U S A. 2011;108:5690–5695. doi: 10.1073/pnas.1012675108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissler L, Gadau J, Simola DF, Helmkampf M, Bornberg-Bauer E. Mechanisms and dynamics of orphan gene emergence in insect genomes. Genome Biol Evol. 2013;5:439–455. doi: 10.1093/gbe/evt009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, et al. The molecular and cellular basis of olfactory-driven behavior in Anopheles gambiae larvae. Proc Natl Acad Sci U S A. 2008;105:6433–6438. doi: 10.1073/pnas.0801007105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou SS, Sun Z, Ma W, Chen W, Wang MQ. De novo analysis of the Nilaparvata lugens (Stal) antenna transcriptome and expression patterns of olfactory genes. Comp Biochem Physiol Part D Genomics Proteomics. 2014;9:31–39. doi: 10.1016/j.cbd.2013.12.002. [DOI] [PubMed] [Google Scholar]
- Zhou X, et al. Phylogenetic and transcriptomic analysis of chemosensory receptors in a pair of divergent ant species reveals sex-specific signatures of odor coding. PLoS Genet. 2012;8:e1002930. doi: 10.1371/journal.pgen.1002930. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence reads data generated in this study are available through the NCBI SRA database under the BioProject accession number: Pending. The complete transcriptome assembly of T. amboinesis will be available through the NCBI Transcriptome Shotgun Assembly database with the accession number: Pending. The protein and coding sequences as well as functional annotations of all annotated T. amboinensis genes are available at http://dx.doi.org/10.6084/m9.figshare.1092617 (last accessed October 22, 2014).