Abstract
Background
Salmonella enterica serovar Typhimurium is the most important serovar associated with human salmonellosis worldwide. Here we aimed to explore the molecular epidemiology and genetic characteristics of this serovar in Guangdong, China.
Methodology
We evaluated the molecular epidemiology and genetic characteristics of 294 endemic Salmonella Typhimurium clinical isolates which were collected from 1977 to 2011 in Guangdong, China, and compared them with a global set of isolates of this serovar using epidemiological data and Multilocus Sequence Typing (MLST) analysis.
Principal Finding
The 294 isolates were assigned to 13 Sequencing types (STs) by MLST, of which ST34 and ST19 were the most common in Guangdong. All the STs were further assigned to two eBurst Groups, eBG1 and eBG138. The eBG1 was the major group endemic in Guangdong. Nucleotide and amino acid variability were comparable for all seven MLST loci. Tajima’s D test suggested positive selection in hisD and thrA genes (p<0.01), but positive selection was rejected for the five other genes (p>0.05). In addition, The Tajima’s D test within each eBG using the global set of isolates showed positive selection in eBG1 and eBG138 (p<0.05), but was rejected in eBG243 (p>0.05). We also analyzed the phylogenetic structure of Salmonella Typhimurium from worldwide sources and found that certain STs are geographically restricted. ACSSuT was the predominant multidrug resistance pattern for this serovar. The resistant profiles ACSSuTTmNaG, ACSSuTTmNa and ACSuTTmNaG seem to be specific for ST34, and ASSuTNa for ST19.
Conclusion
Here we presented a genotypic characterization of Salmonella Typhimurium isolates using MLST and found two major STs are endemic in Guangdong. Our analyses indicate that genetic selection may have shaped the Salmonella Typhimurium populations. However, further evaluation with additional isolates from various sources will be essential to reveal the scope of the epidemiological characteristics of Salmonella Typhimurium in Guangdong, China.
Introduction
Salmonella enterica is one of the leading causes of zoonotic food-borne disease worldwide [1], [2]. The global burden of disease caused by Salmonella infections is substantial, with more than 90 million human cases per year and costs of over $7 billion annually in Europe and the USA [2], [3]. Salmonella Typhimurium, one of ∼2500 serovars of Salmonella enterica, has been isolated in many geographically diverse regions and has caused sporadic outbreaks [3], [4], [5]. Salmonella Typhimurium is the most common cause of salmonellosis in Africa and North America [1], whereas in Europe, it is the second most common endemic serovar isolated from humans [5], [6].
In Asia, multiple epidemiological studies have addressed the characteristics of Salmonella Typhimurium in Thailand, Japan, Hongkong and Taiwan [7], [8], [9]. However, the incidence of Salmonella infections on the mainland of China has not been well documented. Laboratory-based surveillance of nontyphoidal Salmonella infections was initiated in China in 2008 [10], and the results showed that serotypes Enteritidis (12.5%) and Typhimurium (45.2%) were frequently isolated. Studies in Henan province in China also showed that Salmonella Typhimurium was predominant [11]. Our previous study of 1,764 Salmonella enterica isolates from 128 serovars in Guangdong province demonstrated that during 2007 to 2012, Salmonella Typhimurium (n = 523, 29.65%) was one of the most common serovars causing infant salmonellosis [12]. In addition, we found a major serovar shift from Salmonella Enteritidis to Salmonella Typhimurium in Guangdong after 2008. Meanwhile, a worrying percentage of multidrug resistant strains, including those resistant to quinolones and cephalosporin, were also observed in Salmonella Typhimurium [12]. However, the available routine surveillance methods that were used, including serotyping and pulsed-field gel electrophoresis (PFGE), are not suitable for reconstructing evolutionary paths of globally distributed clonal lineages in Salmonella Typhimurium. We therefore used a DNA sequence-based approach to investigate the population structure and potential evolutionary trends within Salmonella Typhimurium in Guangdong, China.
MLST is a DNA sequence-based method using multiple housekeeping genes which can reveal the genetic relatedness of bacterial strains and reconstruct their evolutionary paths. MLST was first developed by Maiden et al. in Neisseria meningitidis [13], and it has since been applied to many other bacterial species [14], [15], [16]. MLST has several advantages over non-sequence based methods, including PFGE, which rely on gel banding patterns. In particular, the DNA sequence is unambiguous, and data from different labs can easily be compared. MLST also allows the reconstruction of phylogenetic relationships within a population using a single combined character matrix with no loss of sequence information.
Several different MLST strategies have been examined in Salmonella isolated from the environment, animals or humans [17], [18], [19], [20]. We employed one of the schemes based on seven housekeeping genes that has been widely used and provides access to global data from a publicly accessible database (http://mlst.warwick.ac.uk/mlst/dbs/Senterica). We used this scheme to differentiate closely related Salmonella Typhimurium isolates into phylogenetically relevant clusters, to analyze the characters of nucleotide sequence data and genetic variation, and to explore the potential population structure and association with antimicrobial susceptibility profiles of Salmonella Typhimurium isolates in Guangdong, China.
Materials and Methods
Ethical standards
Ethical approval was granted by the Ethical Committee of Guangdong Provincial Center for Disease Control and Prevention. The present study complies with the World Health Organization and international guidelines on global surveillance. The procedure of sampling from outpatients is part of the standard diagnostic work-up of patients.
Sampling, bacterial culture and identification
The sampling plan was previously described [12]. According to the Global Salmonella Surveillance program, the recruitment of patients followed specific guidelines. Patients who had two of the following three symptoms were chosen for the study: 1) diarrhea more than three times within 24 hours with watery stools. 2) fever >38°C, headache, chills and malaise. 3) diarrhea with vomiting, abdominal pain and watery stools. Stool samples were collected from clinical diarrhea outpatients of hospitals in fifteen cities from 2007 to 2011. All of the samples were cultured in local hospitals using solid broth agar media (Huankai, Guangzhou, China) for the initial evaluation of Salmonella growth, and then suspicious colonies were picked into vials containing semi-solid Rappaport Vassiliadis Medium (OXOID, France), and shipped to Guangdong Provincial Center for Disease Control and Prevention (Guangzhou, China) at room temperature, where they were incubated daily and checked for growth. The cultures that grew were Gram stained, subcultured and identified at the species level by standard biochemical methods, including the characteristic growth on Kligler Iron Agar (Huankai, Guangzhou, China), tests for urease, oxidase, β-galactosidase and indole production, and positive tests for lysine decarboxylase. The serovar of all the isolates was determined with commercial antiserum (Remel, Lenexa, Kansas, USA) following the Kauffmann-White scheme [21]. The outbreak isolates were collected separately, and all the strains collected as described above were considered epidemiologically unrelated. Age, gender andgeographic origin were recorded as part of the standard information present on the laboratory request forms. In addition, nineteen Salmonella Typhimurium strains isolated from 1977 to 2002 were included in this study (Table S1).
Genomic DNA isolation, PCR amplification, sequencing and MLST
Genomic DNA was isolated using the InstaGene Matrix (BioRad, Hercules, USA) according the manufacturer’s protocol. In brief, colonies from overnight cultures were washed in water. InstaGene Matrix was added to the cells, and the mixture was incubated at 56°C for 30 min. After being vortexed vigorously for 10 s, the cells-matrix mixture was incubated at 100°C for 8 min and then centrifuged at 13,000 g for 10 min. The supernatants were collected and used for PCR templates. PCR reactions were prepared by combining 2 µl of isolated DNA with PCR buffer containing a final concentration of 1.5 mM MgCl2 (Applied BioSystems, Foster, USA), 0.2 mM of each dNTPs (Promega, Madison, USA), 0.2 mM of each of the appropriate forward and reverse primers, and 1.25 U of GoTaqDNA polymerase (Promega, Madison, USA). All the primer sequences for amplification and sequencing were obtained from the MLST Databases at the University of Warwick (www.mlst.warwick.ac.uk/mlst/dbs/Senterica). The PCR cycling conditions followed the online instructions. The PCR products were purified with Sephadex G-50 fine (GE Healthcare Bio-Sciences AB, Uppsala, Sweden). Nucleotide cycle-sequencing was performed directly on purified PCR templates using automated Sanger dideoxychain termination methods and the primers described on the MLST website. The sequences of seven housekeeping genes, aroC, dnaN, hemD, hisD, purE, sucA and thrA, were compared with the available MLST database (http://mlst.warwick.ac.uk/mlst/dbs/Senterica) to get the allele number and sequence typing (ST) number for each isolate. Sequence information of newly assigned alleles and STs were deposited in the MLST database.
DNA polymorphic analysis and neutrality test
DNA polymorphism analysis, including the number of mutations, the number of alleles, the nucleotide diversity, the number of synonymous and nonsynonymous mutations, was carried out using DnaSPv5.10.00 software [22]. Tajima’s D test was used to compare the population mutation rate, which is estimated from the number of segregating sites, to the average pairwise nucleotide distance to determine whether the observed frequency of segregating mutations agreed with the expected frequency under the standard neutral model [23]. Allelic types and STs were assigned according to the MLST database of Salmonella.
Phylogenetic reconstruction and clustering analyses
All of the sequences were edited using SeqMan software from the Lasergene software package (DNASTAR, USA). Iterative alignment was performed automatically with manual adjustment in BioNumerics 6.5 (Applied Maths, Belgium). A minimal spanning tree was generated from the allelic profiles of isolates using the predefined template in BioNumerics 6.5 designated as MST for categorical data, which preferentially joins single and double locus variants with the largest number of isolates per ST. All the reliable STs that belong to Salmonella Typhimurium on the website (http://mlst.warwick.ac.uk) were used for this tree construction (Table S2).
A dendrogram was built using MEGA software 5.0 [24] to determine the relationships among the different STs. Phylogenetic neighbor joining trees were inferred for concatenated sequences to determine the variable sites in seven loci using MEGA 5.0 software [24]. The percentage of replicate trees in which the associated taxa clustered together was estimated by a bootstrap test inferred from 1000 replicates. ClonalFrame [25] was used to infer their phylogenetic relatedness based on sequence alignments of gene fragments for one representative from each of these STs. ClonalFrame identifies regions that are likely to have arisen by homologous recombination and accounts for them when reconstructing the clonal genealogy. The lengths of the branches in the evolutionary tree are measured in coalescent time units, which are equal to the effective population size multiplied by the average duration of a generation. The nucleotide sequence alignments for all gene fragments from unique STs were used to infer clonal relationships with ClonalFrame from the 50% consensus of 10 runs, each with 100,000 iterations following a burn-in phase of 100,000 iterations.
Antimicrobial susceptibility
The antimicrobial susceptibility test was performed according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) [26]. Agar diffusion assays were performed on Muller-Hinton agar with disks containing streptomycin, 10 µg (S), gentamicin, 10 µg (G), ceftazidime, 30 µg (Caz), cefotaxime, 30 µg (Ctx), cefepime, 30 µg (Fep), ampicillin, 10 µg (A), nalidixic acid, 30 µg (Nal), ciprofloxacin, 5 µg (Cp), tetracycline, 30 µg (T), chloramphenicol, 30 µg (C), sulfamethoxazole, 300 µg (Su), and trimethoprim, 5 µg (Tm), respectively. The results were interpreted according to the CLSI guidelines [26]. Escherichia coli ATCC 25922 was used as the quality control organism. Multi-resistance was identified as resistance to more than three classes of antimicrobials. The antimicrobial resistance pattern as well as the source of isolation of each strain is shown in Table S1.
Results
Representative assessment of Salmonella Typhimurium and MLST determination
According to the latest data, the frequency of human salmonelosis in China was 549 per 100,000 people in 2013 (China CDC surveillance system, unpublished data), over 33 times as high as human infections in the USA in 2012 (16.4 per 100,000) [27]. Based on this frequency, 549,000 people in Guangdong Province (total population size 100 million) are infected by Salmonella annually. Thirty percent of Salmonella isolates in Guangdong Province are of serovar Typhimurium [12], which corresponds to approximately 164,700 human infections per year. The 275 isolates from 2007–2011 that are described here represent 0.0334% (275/164,700*5) of the estimated number of infections over five years.
We performed MLST sequencing of seven housekeeping genes on all 275 isolates as well as on 19 other Salmonella Typhimurium strains which were isolated between 1977 and 2002, for a total of 294 isolates (Table S1). We found only one hemD nucleotide sequence (allele) but multiple alleles in aroC (2 alleles), dnaN (4), hisD (3), purE (3), sucA (3) and thrA (6), of which one in each of purE, sucA and thrA was novel. The aligned sequences of the concatenated loci were 3,336 base pairs long, with 59 polymorphic sites (58 parsimony informative and 1 singleton sites). The seven loci yielded 13 Sequence types (STs), of which ten (11 isolates) were novel (Table 1). Most of the isolates were assigned to the previously defined ST34 (209, 71.1%) and ST19 (69, 23.5%) (Table 1). ST36 and ST1782 contained 5 and 2 other isolates, respectively, and one isolate was identified for each of the other nine STs.
Table 1. Polymorphism summary and tests for neutral evolution in each locus of the Guangdong isolates and the global set of strains.
| Parameters | Phylogenetic Marker (a/b) | ||||||||
| aroC | dnaN | hemD | hisD | purE | sucA | thrA | Concatenated loci | ||
| No. ofsequences | 294/1186 | 294/1186 | 294/1186 | 294/1186 | 294/1186 | 294/1186 | 294/1186 | 294/1186 | |
| No. ofcharacters | 501/501 | 501/501 | 432/432 | 501/501 | 399/399 | 501/501 | 501/501 | 3336/3336 | |
| No. ofcodons | 167/167 | 167/167 | 144/144 | 167/167 | 133/133 | 167/167 | 167/167 | 1112/1112 | |
| DNA polymorphism analysis (a/b) | |||||||||
| No. ofmutations (η) | 5/14 | 9/10 | 0/9 | 15/23 | 4/11 | 5/14 | 21/29 | 59/110 | |
| synonymouschanges | 5/13 | 7/7 | 0/3 | 13/17 | 3/8 | 4/11 | 21/28 | 53/87 | |
| nonsynonymouschanges | 0/1 | 2/3 | 0/6 | 2/6 | 1/3 | 1/3 | 0/1 | 6/23 | |
| nucleotideVariability | 0.998%/2.794% | 1.796%/1.996% | 0/2.083% | 2.994%/4.591% | 1.002%/2.757% | 0.998%/2.794% | 4.191%/5.788% | 0.177%/3.297% | |
| amino acidVariability | 0/0.599% | 1.197%/1.796% | 0/4.166% | 1.197%/3.593% | 0.752%/2.256% | 0.598%/1.796% | 0/0.598% | 0.539%/2.068% | |
| nucleotidediversity (Pi) | 0.00047/0.00113 | 0.00125/0.00145 | 0/0.00033 | 0.00026/0.00013 | 0.00012/0.00070 | 0.00039/0.00076 | 0.00087/0.00130 | 0.0005/0.00084 | |
| allele | 2/6 | 4/5 | 1/5 | 3/9 | 3 (1)/9 | 3 (1)/11 | 6 (1)/15 | 13 (10)/47 | |
| Neutrality test (a/b) | |||||||||
| Tajima’s D | −1.29146 (p>0.10)/−1.47225 (p>0.10) | −1.22590 (p>0.10)/−0.86813 (p>0.10) | na/−1.67213(0.10>p>0.05) | −2.31354(p<0.01)/−2.30983 (p<0.05) | −1.56342 (0.10>p>0.05)/−1.62052(0.10>p>0.05) | −1.38243(p>0.10)/−1.69198 (0.10>p>0.05) | −2.26094 (p<0.01)/−2.05595 (p<0.05) | −2.40175 (p<0.01)/−2.23945 (p<0.01) | |
a/b: the Guangdong isolates/the Global set of isolates.
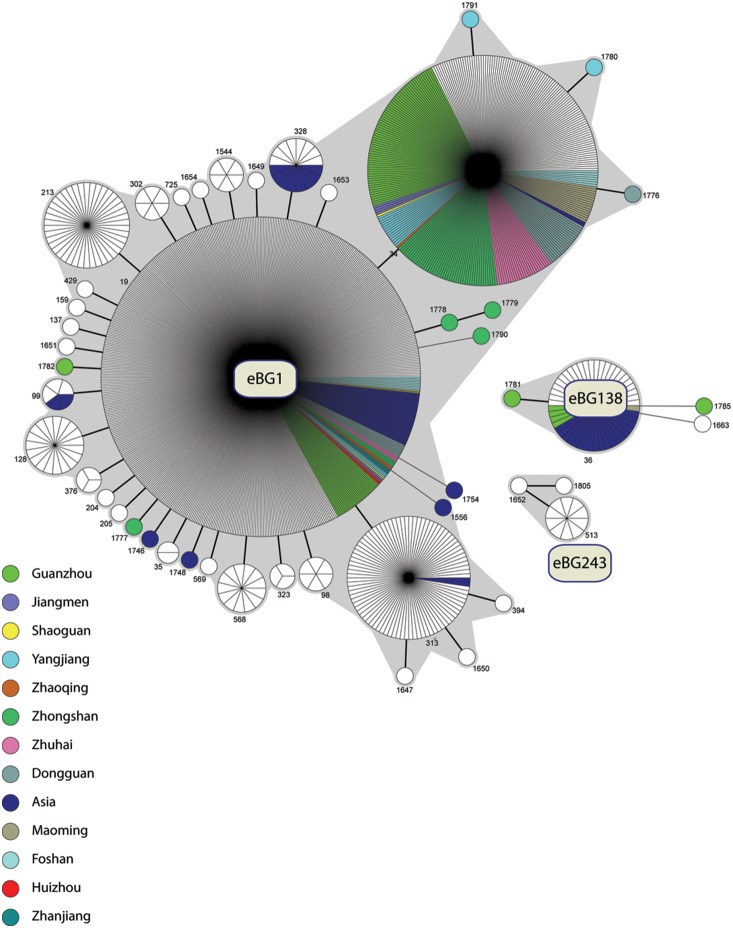
Achtman et al. [17] grouped Salmonella enterica STs into discrete eBGs (eBurstGroups), each of which includes two or more STs that are connected by pair-wise identities for six of the seven gene fragments, i.e., they share six of the seven alleles that define the ST. In other bacterial species, these groups are also referred to as “Clonal Complex” or “ST Complex”. We extended these groupings to include the ten novel STs described here as well as novel Typhimurium STs that have recently been uploaded to the MLST website by other scientists (Fig. 1). All Typhimurium STs fall into three eBURST groups (eBG1, eBG138 and eBG243), except for STs 1665 and 1783, which are double locus variants of eBG138. Most of the novel STs described in this study belong to eBG1 (ST1791, 1780, 1776, 1782, 1777), but ST1781 was assigned to eBG138. We did not find any isolates in eBG243, which is generally rare, and has not yet been isolated from Asia. STs 19 and 34, the two most common STs among the isolates from Guangdong province, are the two most commonly isolated STs at the global level and belong to eBG1. The most common ST in eBG138 is ST36 (Fig. 1).
Figure 1. minimal spanning tree (MSTree) of the MLST data of S. enterica serovar Typhimurium.
Each circle corresponds to each STs with the size proportional to the number of isolates. The topological arrangement within the MSTree is dictated by its graphic algorithm, which uses an iterative network approach to identify sequential links of increasing distance beginning with central STs that contain the largest numbers of isolates. As a result, singleton STs are scattered throughout the MSTree proximal to the first node that was encountered with shared alleles, even if equal levels of identity to other nodes that are distant within the MSTree exist. The figure only shows the links of six identical gene fragments (SLVs, thick black line) and five identical gene fragments (DLVs, thin black line) because these correlate with eBGs, which are indicated by grey shading. The key shows the isolates from Guangdong or Asia.
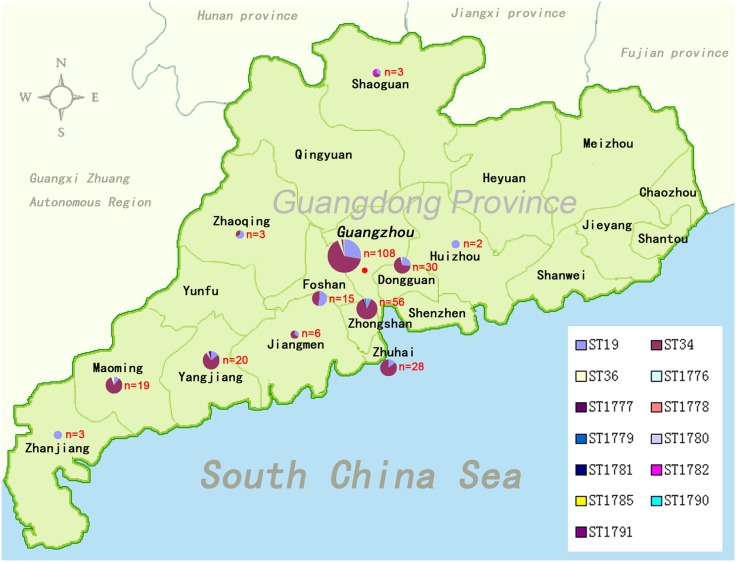
Comparative Geographical analysis showed that most isolates from Guangdong belong to ST34 within eBG1, rather than ST19, which is more common outside Asia (Fig. 2). The novel STs in eBG1 were ST 1776–1780, 1782, and 1790–1791. ST1781 belongs to eBG138, and ST1785 is a double locus variant of that eBG. All 19 isolates collected between 1977 and 2002 were assigned to ST19, and all 11 isolates assigned to novel STs were isolated after 2007 (Fig. 2).
Figure 2. The locations where clinical diarrhea outpatients were collected and the STs assigned in each location in this study.
Field sites are shown in relation to the ranges of the 12 proposed cities in Guangdong Province, China. The STs in each city are marked with red.
Descriptive analysis of nucleotide sequence data
The nucleotide diversity of DNA sequences in the isolates from Guangdong ranged from 0 for hemD to 0.00125 for dnaN, corresponding to between 0 and 4.191% of the nucleotides being variable in each locus. The amino acid variability ranged from 0–1.197% due to two variable amino acids in dnaN and hisD and one each in purE and sucA (Table 1). Both the nucleotide diversity and the amino acid variability in the global set of isolates showed the same trend, but with higher values than in Guangdong isolate (Table 1). The nonsynonymous and synonymous base substitution and amino acid variability were summarized in Table 2.
Table 2. Synonymous and non-synonymous changes in DNA and amino acid sequence in seven genes of Salmonella serovar Typhimurium in this study.
| Gene | Total codons | 1st base | 2nd base | 3rd base | Amino acid change | Strains |
| aroC | 167 | 14 | ATG→ATA/M→I | ST328 | ||
| dnaN | 167 | 3 | 7 | GGC→AGC/G→S | ST1653 | |
| TCG→GCG/S→A | ST36, ST1663 | |||||
| GTG→ATG/V→M | ST34 | |||||
| hemD | 144 | 1 | 2 | 3 | GCG→ACG/S→T | ST1754 |
| GTG→GCG/V→S | ST513, ST1652, ST1754, ST1805 | |||||
| GCG→TCG/S→A | ST513, ST1652, ST1754, ST1805 | |||||
| GCG→GTG/S→V | ST98 | |||||
| CGC→CAC/R→H | ST513, ST1652, ST1805 | |||||
| CGC→CAC/R→H | ST1663 | |||||
| hisD | 167 | 2 | 3 | 18 | AAG→AAC/K→N | ST1754, 1785, 1790 |
| GGC→GAC/G→D | ST1790 | |||||
| GCG→GAG/S→E | ST1663 | |||||
| CCG→CTG/P→L | ST394 | |||||
| GAT→AAT/D→N | ST1649 | |||||
| GTC→ATC/V→I | ST1544 | |||||
| purE | 133 | 1 | 3 | GTA→ATA/V→I | ST213 | |
| GCC→GAC/A→D | ST1777 | |||||
| CAG→GAG/Q→E | ST302, 313 | |||||
| sucA | 167 | 4 | 1 | 9 | ACC→ATC/T→I | ST1780 |
| GGC→TGC/G→C | ST137 | |||||
| GTG→CTG/V→L | ST128 | |||||
| thrA | 167 | 2 | 27 | ACG→TCG/T→A | ST99 | |
| Total | 787 | 12 | 7 | 81 |
A, Alanine; C, Cysteine; D, Ariginine; E, Glutamic acid; G, Glycine; H, Histidine; I, Isoleucine; K, Lysine; L, Leucine; M, Methionine; N, Asparagine; P, Proline; Q, Glutamine; R, Argnine; S, Serine; T, Threonine; V,Valine.
Tajima’s D test for the data set showed positive selection in hisD and thrA genes (p<0.05), but was rejected in the other genes in the isolates from Guangdong as well as in the global set of isolates (p>0.05) (Table 1). The neutrality evolution test within each eBG using the global set of isolates showed positive selection in aroC, hemD, hisD, sucA and thrA genes (p<0.05) in eBG1 as well as in hisD and thrA (p<0.05) in eBG138, but was rejected in eBG243 (p>0.05) (Table 3).
Table 3. A neutrality test of three eBGs on seven loci in the global strains from MLST database.
| eBG1 (ST19, 34, 1776–80,1782, 1790–91) (n = 1123) | eBG138 (ST36,1781, 1785) (n = 51) | eBG243 (ST513, 1805, 1652) (n = 12) | |
| aroC (501 bp) | −1.86501 (p<0.05) | 0 | 0 |
| dnaN (501 bp) | −0.97769 (p>0.10) | 0 | 0 |
| hemD (432 bp) | −1.72921 (p<0.05) | −1.10080 (p>0.10) | 0 |
| hisD (501 bp) | −2.23874 (p<0.01) | −2.29677 (p<0.01) | 0 |
| purE (399 bp) | −1.53932 (0.10>p>0.05) | 0 | 0 |
| sucA (501 bp) | −1.88897 (p<0.05) | 0 | 0 |
| thrA (501 bp) | −2.31093 (p<0.01) | −2.12375 (p<0.05) | −1.02689 (p>0.10) |
| Concatenated loci | −2.51802 (p<0.001) | −2.51435 (p<0.001) | −1.02689 (p>0.10) |
Phylogenetic analysis of the Salmonella serovar Typhimurium population
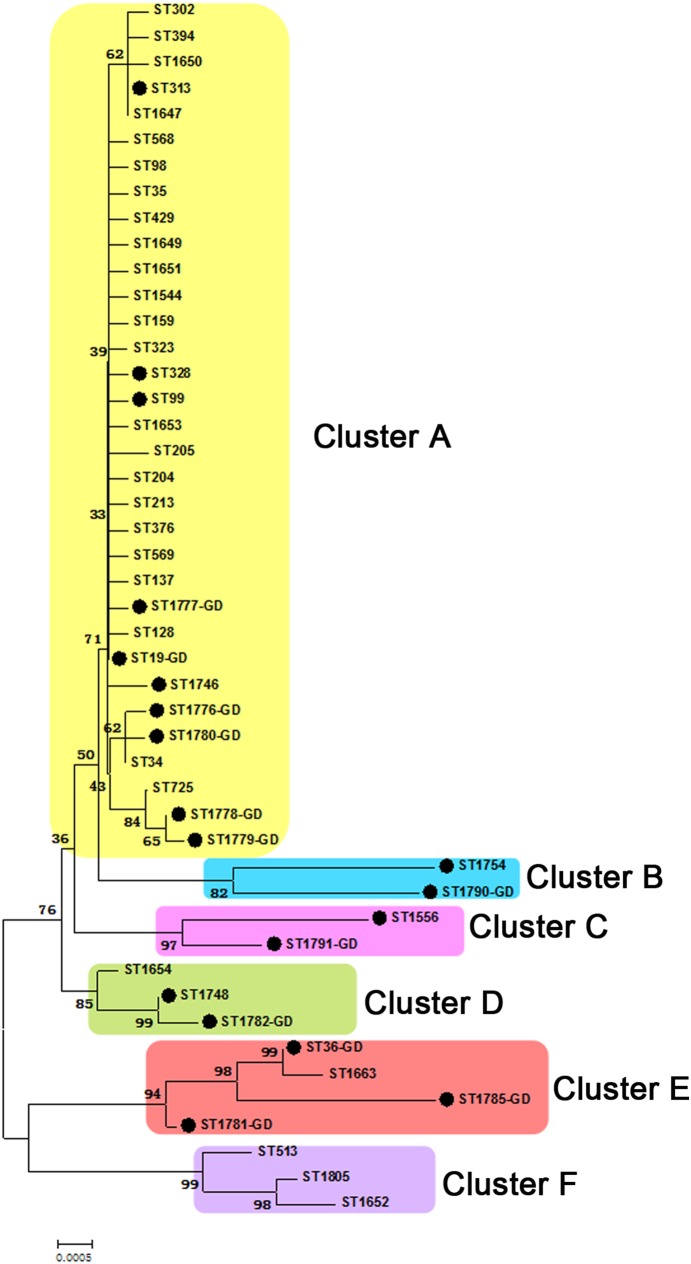
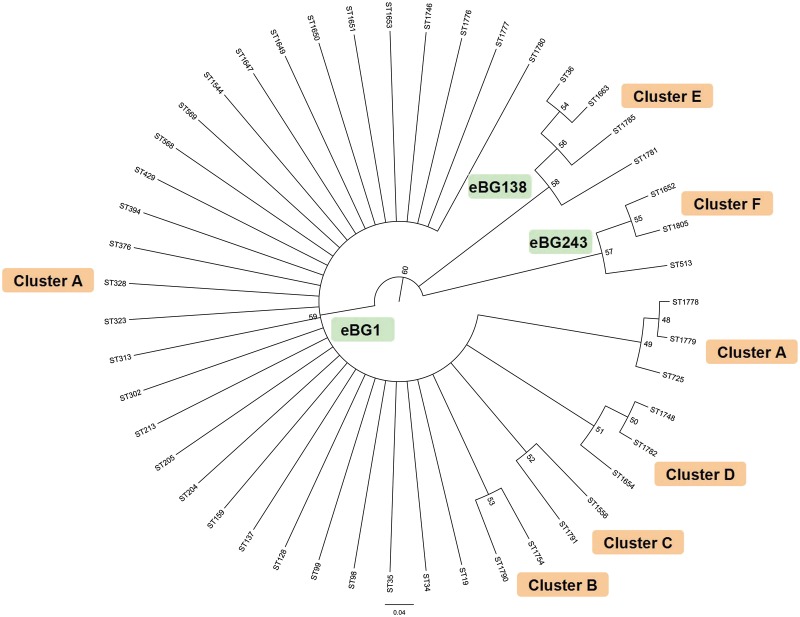
The evolutionary relationships of the 294 Salmonella isolates tested in this study were determined with sequence information from the MLST database. A neighbor joining tree was generated for this purpose by concatenating sequences (3336 bp) from the unique STs (n = 47) (Fig. 3). The tree demonstrated that STs were clustered into six clusters (bootstrap ≥70), represented by A (yellow), B (blue), C (pink), D (green), E (red) and F (purple). Cluster A is composed of 7 STs from Guangdong, which covered most of the isolates from 12 cities. Clusters B–D are composed of 3 STs, including 4 isolates from Guangzhou, Zhongshan, Yangjiang and Shaoguan, whereas cluster E (3 STs, 7 isolates) was mostly found in Guangzhou (n = 6) and Maoming (n = 1). No STs from Guangdong were detected in cluster F (3 STs). However, we found that all the STs from Guangdong were closed to the STs originally observed in Asia (Fig. 3, black dot). According to the ClonalFrame analysis, Salmonella Typhimurium contains three groups, which correspond to eBG1, eBG138 and eBG243 on MStree (Fig. 4). The clusters B–D branch from the same root position within eBG1 as single STs in that eBG and a small cluster not recognized visually within the phylogenetic tree. These results indicate that much of the apparent phylogenetic clustering within eBG1 reflects recombination events in single locus. The estimated recombination ratio was 0.1691+/−0.0039 (rho/theta; frequency of recombination events per mutation event) resulting in 0.517+/−0.0098 (r/m) nucleotide replacements due to recombination rather than mutation.
Figure 3. A neighbor joining tree showing the phylogenetic relationships of the Salmonella Typhimurium STs included in this study (n = 47).
All STs were delineated into five major groups (bootstrap ≥70). The STs from Asia were labeled with a black dot in the phylogenetic tree, whereas STs from Guangdong were labeled GD. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replications) are indicated. The evolutionary distances were computed using the neighbor joining method and are presented in units of the number of base substitutions per site.
Figure 4. A consensus ClonalFrame tree constructed using ClonalFrame 1.1 in this study (ST = 47).
The groups were marked using assigned clusters from the neighbor joining and minimal spanning trees.
Antimicrobial susceptibility
A large burden of antibiotics resistance was found for ampicillin (87.75%), nalidixic acid (82.65%), sulfamethoxazole (89.79%), trimethoprim (71.43%), tetracycline (89.12%), gentamicin (65.31%), streptomycin (78.57%) and chloramphenicol (74.49%) in the isolates from Guangdong (Table 4), while less resistance in four antimicrobial agents consist of ceftazidime, cefotaxime, cefepime and ciprofloxacin (5.74%–13.39%). In particular, the resistance to cephalosporins is not infrequent observed. The isolates from ST 34 showed higher lever of resistance to all eleven antibiotics (except ciprofloxacin) than the isolates from ST 19. A few novel STs, including ST1778 and ST1790, showed high level of resistance to first or partial third or fourth generation antibiotics.
Table 4. Antimicrobial susceptibility and MDR among 294 Salmonella enterica isolates obtained from humans with infections in Guangdong province, China, between 2007 and 2011.
| No. ofisolates | ampicillin | ceftazidime | cefotaxime | cefepime | ciprofloxacin | nalidixicacid | Sulfamethoxazole | trimethoprim | tetracycline | gentamicin | streptomycin | chloramphenicol | MDR | |
| ST34 | 209 | 202(96.65) | 21(10.05) | 28(13.39) | 25(11.97) | 12(5.74) | 197(94.26) | 204 (98.06) | 163(77.99) | 202(96.65) | 157(75.12) | 172(82.30) | 178 (85.17) | 206 (98.56) |
| ST19 | 69 | 44(63.77) | 2(2.89) | 6(8.69) | 4(5.79) | 10(14.49) | 36(52.17) | 46 (66.67) | 37(53.62) | 46(66.66) | 28(40.58) | 47(68.12) | 31(30.55) | 49(71.01) |
| ST36 | 5 | 3(60.00) | 1 (20.00) | 2 (40.00) | 0 | 1 (20.00) | 2(40.00) | 4 (80.00) | 2(40.00) | 4(80.00) | 1(20.00) | 4(80.00) | 3(60.00) | 4 (80.00) |
| ST1776 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ST1777 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ST1778 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 |
| ST1779 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ST1780 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ST1781 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ST1782 | 2 | 2 | 0 | 0 | 0 | 1 | 2 | 2 | 2 | 2 | 1 | 2 | 1 | 1 |
| ST1785 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 |
| ST1790 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| ST1791 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 |
| Total(% resistance)b | 294 | 258(87.75) | 25(8.50) | 38(12.93) | 32(10.88) | 24(8.16) | 243(82.65) | 264(89.79) | 210(71.43) | 262(89.12) | 192(65.31) | 231(78.57) | 219(74.49) | 268(91.16) |
Number of isolates resistant to indicated agent at the indicated breakpoint (% resistance)a.
The percentage means the proportion of resistant isolates for each antibiotics.
The percentage means the proportion of MDR isolates among the total isolates.
Multidrug resistance occurred in 91.16% (268/294) of the isolates. A large proportion of the MDR isolates were observed in ST 34 (98.56%) and in most of the novel STs (Table 4). The antimicrobial resistance patterns are shown in Table 4. In total, 57 resistant profiles were observed, of which 9 resistant profiles (more than 5 strains in each profile, corresponding to 5 STs including ST34, 19 and 36) accounted for 65.64% (n = 193) of all isolates (Table 5). The resistant profiles, including ACSSuTTmNaG, ACSSuTTmNa, ACSuTTmNaG, and ASSuTNa were found to be specific for ST34 and ST19. ACSSuT, the typical resistance pattern for Salmonella Typhimurium, was found in 179 isolates (66.79%). Three Salmonella Typhimurium isolates resistant to all twelve antimicrobial agents were obtained (ESS221, ESS463 and ESS797). Only four isolates were fully susceptible to all the agents tested (Table S1).
Table 5. Salmonella Typhimurium resistance type and the corresponding MLST type in this study.
| Mainly MDR Type (N, %)a | MLST Type (n/N, %)b | |
| Typical pattern | ACSSuT (179, 66.79%) | ST34 (146, 81.56%) |
| ST19 (25, 13.97%) | ||
| ST36 (3, 1.68) | ||
| 1 | ACSSuTTmNaG (107, 39.93%) | ST34 (97, 90.65%) |
| ST19 (6, 5.61%) | ||
| 2 | ACSSuTTmNa (17, 6.34%) | ST34 (14, 82.35%) |
| ST19 (3, 17.65%) | ||
| 3 | ACSuTTmNaG (13, 4.85%) | ST34 (13, 100%) |
| 4 | ASSuTNa (13, 4.85%) | ST34 (11, 84.62%) |
| ST19 (1, 7.69%) | ||
| 5 | ACSSuTTmNaGCp (12, 4.48%) | ST34 (6, 50%) |
| ST19 (6, 50%) | ||
| 6 | ACSSuTNaG (11, 4.11%) | ST34 (10, 90.90%) |
| ST19 (1, 9.10%) | ||
| 7 | ACSSuTTmNaGCazCtxFep (8, 2.99%) | ST34 (8, 100%) |
| 8 | ACSSuTTm (7, 2.61%) | ST34 (5, 71.43%) |
| ST19 (1, 14.29%) | ||
| ST36 (1, 14.29%) | ||
| 9 | ASSuT (5, 1.87%) | ST34 (5, 100%) |
| Totalc | 193/294, 65.64% | ST34 (169/268, 63.06%) |
| ST19 (18/268, 6.72%) | ||
| ST36 (1/268, 0.37%) |
Resistant pattern of Salmonella Typhimurium occurring in at least 5 strains. N = total number of isolates with the specific resistant pattern, % = hundred percentage in all isolates.
MLST type that includes more than 2 strains. n = number of isolates occurring in the present STs, N = total number of isolates involving the present resistant pattern, % = hundred percentage in this ST.
The proportion of MDR profile isolates among the total isolates, as well as in each ST.
Discussion
Sequence analysis of all the isolates found that the most common STs observed were ST19 and ST34 in 12 cities of Guangdong province. The comparison of these data with the MLST database indicated that these two STs are relatively common in a range of hosts, including poultry, soya, fishmeal, lizards and humans (http://www.mlst.net). ST19 and ST34 were previously defined as a unique eBG1 by Achtman et al. [17], thus eBG1 was the predominant group in Guangdong, China. The archived information from the MLST database showed that the most prevalent ST in eBG1 is ST19. Unusually, more of the isolates from Guangdong were ST34 rather than ST19. Marcus et al. [8] showed that ST34 was also the major MDR ST of Salmonella Typhimurium in Hongkong, China. Therefore, one possibility is that ST34 is mainly endemic in Asia, whereas ST19 seems to be globally distributed. Within eBG1, there are common STs found elsewhere but not in Asia, including ST213, 128, 568, and ST313 which was the predominant ST in Africa but has spread globally. However, we did not find any isolates in ST313 and its descendants, ST1647, 1650, 394, or in ST328 and ST99, which are common in Taiwan and elsewhere. These observations were unexpected given the increasing cargo trade and frequent human travel between Guangdong and other countries. However, we did find STs in eBG138 that have been previously isolated in all continents, particularly in Taiwan, Japan, Malaysia and Thailand of Asia, suggesting that there are geographic specificities for the prevalence of different STs, possibly reflecting different intensities of the trade of food products with countries within Asia and elsewhere, or different dynamics in the spread of individual STs. Food associated with domestic animals (e.g., pork, chicken, egg and milk) are considered as the primary source of infection in Guangdong [12]. However, recent results have indicated that Typhimurium infection in humans is associated with clades distinct from those common in domesticated animals [3]. Therefore, it can be speculated that the transmission of the major eBGs or STs is a mixture spreading due to human travel and international trade in food products between different continents [28], [29], [30].
Searching the allele profiles of the MLST international database for Salmonella, we found that most of the novel STs are SLVs of either ST19 or ST34, which reflect a mixture of recombination and mutation, consistent with previous conclusions for the serovar Newport [20], [31]. Similar trends were observed for ST36 and its descendent STs. These data indicated that these strains probably displayed divergent evolution. ST19 and ST34 were the predominant clone lineage over time, and the novel STs were expanded from this lineage by a recent genetic event (mutation or recombination). Tajima’s D test, an effective tool for making inferences about population demographics [23], supports the hypothesis that ecological adaptation or slight geographic expansion occurred within some of the STs (Table 1). That is because the test values were significantly different from zero for all seven loci (p<0.01) and for two individual loci in the isolates from Guangdong (hisD and thrA, p<0.05). In addition, Tajima’s test within each eBG that potentially share a recent common ancestor, also showed positive selection in hisD and thrA in a global set of isolates (p<0.05). However, Zhou et al. claim that in Salmonella Agona [32] most genetic changes are due to changes of bacterial phages and other accessory genes, which result in changing the PFGE pattern but not the phenotype. The similar study by Zhou et al. in Salmonella Paratyphi A [33] showed that evolution is not adaptive but neutral due to transient Darwinian evolution; thus adaptations for particular ecological niches at the level of STs are rare. Indeed, we found a high estimated recombination ratio in the ClonalFrame tree, which means that the nucleotide replacements are due to recombination rather than mutation. However, greater genomic resolution will be needed to address such adaptation within Salmonella Typhimurium in a future study. Phylogenetic trees constructed by concatenating sequences from all the STs of Salmonella Typhimurium in MLST database (n = 47) showed that all STs were separated into six clusters, which correspond to three clades in the Clonalframe tree, whereas previous phylogenetic analyses by Sangal et al. [31] indicated that all Typhimurium belonged to a single clade with the exception of ST36, which was closely related to that clade. In this study, a much larger number of bacterial isolates and many more STs contributed to the result of three related clades, corresponding to both neighbor joining (Fig. 3) and the ClonalFrame tree (Fig. 4).
Synonymous and nonsynonymous substitutions were found in coding regions. There are 95 synonymous and 6 nonsynonymous substitutions in the isolates from Guangdong, whereas many more substitutions were found in a global set of isolates. One coding frame shift mutation was detected in hisD. Nonsynonymous substitutions was also detected in previous studies of Salmonella enterica [34], and is attributed to the threshold for slightly deleterious mutations within clones of a species [35]. However, a more traditional explanation of nonsynonymous substitutions is that the genes concerned were changing under selection pressure. A similar finding reported by Feil et al. [36] in a study of Staphylococcus aureus showed that a higher proportion of nonsynonymous substitutions occurred in closely related isolates that at the species or higher taxon levels. Nevertheless, the nonsynonymous substitutions within the one serovar of Salmonella enterica examined in this study will be worthy of further investigation.
The previous studies of salmonellosis in Guangdong illustrated the emergence of antimicrobial resistance in local populations; however, the genotype could not account for the observed phenotype [37], [38], [39]. Using MLST in this study, we demonstrated a higher proportion of MDR in ST19 and ST34, which were the most common STs in Guangdong. In particular, ST34 has been proved to be the predominant ST carrying multiple resistance determinants in Hongkong, China [8]. In addition, a few novel STs, including ST1778 and ST1790, showed high level of resistance to all the antibiotics tested. Therefore, further studies to determine the nature of resistance to related antibiotics are ongoing, whereas mobile genetic elements, including integrons, plasmids, genomic islands and transposons are possibly responsible for the observed resistance.
In conclusion, we presented the genotypic characterization of 294 Salmonella isolates from Guangdong, China, and used MLST to compare these isolates with a global set of isolates of this serovar. The 294 isolates were assigned to 13 STs, of which 10 were novel. All STs were further assigned to two eBurst Groups, eBG1 and eBG138. The genetic diversity analysis suggested a likely slight genetic selection within Salmonella Typhimurium population. Resistant profiles specific for particular STs were observed. Further evaluations with additional isolates obtained from various sources and regions are needed to reveal the scope of the epidemiological characteristics of Salmonella Typhimurium in Guangdong, China.
Supporting Information
The Salmonella Typhimurium strain information isolated from 1977 to 2002 in this study (Table S1).
(XLS)
The information of all the reliable STs of Salmonella Typhimurium on the website ( http://mlst.warwick.ac.uk ).
(XLS)
Acknowledgments
We acknowledge Mark Achtman and Zhemin Zhou from the Warwick Medical School, University of Warwick, United Kingdom for their great contributions to the MNtree and the ClonalFrame tree reconstruction, manuscript revision, comprehensive communication and scientific comments for the present study.
Funding Statement
This project was supported by the China–U.S. Collaborative Program on Emerging and Re-Emerging Infectious Diseases and the Medical Scientific Research Foundation in Guangdong (1U2GGH000018-01), the 12th five-year-major-projects of China’s Ministry of Public Health (2012zx10004-213), and Science and Technology Planning Project of Guangdong Province Program (2012B060400012; 2013B060400012). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Herikstad H, Motarjemi Y, Tauxe RV (2002) Salmonella surveillance: a global survey of public health serotyping. Epidemiol Infect 129: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, et al. (2010) The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis 50: 882–889. [DOI] [PubMed] [Google Scholar]
- 3. Mather AE, Reid SW, Maskell DJ, Parkhill J, Fookes MC, et al. (2013) Distinguishable Epidemics of Multidrug-Resistant Salmonella Typhimurium DT104 in Different Hosts. Science 341: 1514–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cui S, Li J, Sun Z, Hu C, Jin S, et al. (2008) Ciprofloxacin-resistant Salmonella enterica serotype Typhimurium, China. Emerg Infect Dis 14: 493–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Galanis E, Lo Fo Wong DM, Patrick ME, Binsztein N, Cieslik A, et al. (2006) Web-based surveillance and global Salmonella distribution, 2000–2002. Emerg Infect Dis 12: 381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Olsen SJ, Bishop R, Brenner FW, Roels TH, Bean N, et al. (2001) The changing epidemiology of Salmonella: trends in serotypes isolated from humans in the United States, 1987–1997. J Infect Dis 183: 753–761. [DOI] [PubMed] [Google Scholar]
- 7. Lee HY, Su LH, Tsai MH, Kim SW, Chang HH, et al. (2009) High rate of reduced susceptibility to ciprofloxacin and ceftriaxone among nontyphoid Salmonella clinical isolates in Asia. Antimicrob Agents Chemother 53: 2696–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wong MH, Yan M, Chan EW, Liu LZ, Kan B, et al. (2013) Expansion of Salmonella enterica serovar Typhimurium ST34 clone carrying multiple resistance determinants in China. Antimicrob Agents Chemother 57: 4599–4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Torpdahl M, Lauderdale TL, Liang SY, Li I, Wei SH, et al. (2013) Human isolates of Salmonella enterica serovar Typhimurium from Taiwan displayed significantly higher levels of antimicrobial resistance than those from Denmark. Int J Food Microbiol 161: 69–75. [DOI] [PubMed] [Google Scholar]
- 10. Deng X, Ran L, Wu S, Ke B, He D, et al. (2012) Laboratory-based surveillance of non-typhoidal Salmonella infections in Guangdong Province, China. Foodborne Pathog Dis 9: 305–312. [DOI] [PubMed] [Google Scholar]
- 11. Xia S, Hendriksen RS, Xie Z, Huang L, Zhang J, et al. (2009) Molecular characterization and antimicrobial susceptibility of Salmonella isolates from infections in humans in Henan Province, China. J Clin Microbiol 47: 401–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ke B, Sun J, He D, Li X, Liang Z, et al. (2014) Serovar distribution, antimicrobial resistance profiles, and PFGE typing of Salmonella enterica strains isolated from 2007–2012 in Guangdong, China. BMC Infectious Diseases 14: 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, et al. (1998) Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A 95: 3140–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lacher DW, Steinsland H, Blank TE, Donnenberg MS, Whittam TS (2007) Molecular evolution of typical enteropathogenic Escherichia coli: clonal analysis by multilocus sequence typing and virulence gene allelic profiling. J Bacteriol 189: 342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gonzalez-Escalona N, Martinez-Urtaza J, Romero J, Espejo RT, Jaykus LA, et al. (2008) Determination of molecular phylogenetics of Vibrio parahaemolyticus strains by multilocus sequence typing. J Bacteriol 190: 2831–2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Godoy D, Randle G, Simpson AJ, Aanensen DM, Pitt TL, et al. (2003) Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei . J Clin Microbiol 41: 2068–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Achtman M, Wain J, Weill FX, Nair S, Zhou Z, et al. (2012) Multilocus sequence typing as a replacement for serotyping in Salmonella enterica . PLoS Pathog 8: e1002776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tankouo-Sandjong B, Sessitsch A, Liebana E, Kornschober C, Allerberger F, et al. (2007) MLST-v, multilocus sequence typing based on virulence genes, for molecular typing of Salmonella enterica subsp. enterica serovars. J Microbiol Methods 69: 23–36. [DOI] [PubMed] [Google Scholar]
- 19. Sukhnanand S, Alcaine S, Warnick LD, Su WL, Hof J, et al. (2005) DNA sequence-based subtyping and evolutionary analysis of selected Salmonella enterica serotypes. J Clin Microbiol 43: 3688–3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Harbottle H, White DG, McDermott PF, Walker RD, Zhao S (2006) Comparison of multilocus sequence typing, pulsed-field gel electrophoresis, and antimicrobial susceptibility typing for characterization of Salmonella enterica serotype Newport isolates. J Clin Microbiol 44: 2449–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grimont PAD, Weill F-X (2007) Antigenic formulae of the Salmonella Serovars. Paris: World Health Organization, Institut Pasteur: 166p.
- 22. Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- 23. Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Didelot X, Falush D (2007) Inference of bacterial microevolution using multilocus sequence data. Genetics 175: 1251–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.CLSI (2012) Performance standards for antimicrobial susceptibility testing;. Twenty-second informational supplement Wayne, PA: Clinical and Laboratory Standards Institute: 188p.
- 27.Centers for Disease Control and Prevention (2012) Trends in foodborne illness in the United States. Atlanta: Centers for Disease Control and Prevention April 18, 2013 (www.cdc.gov/features/dsfoodnet2012/reportcard.html).
- 28. Hakanen A, Kotilainen P, Huovinen P, Helenius H, Siitonen A (2001) Reduced fluoroquinolone susceptibility in Salmonella enterica serotypes in travelers returning from Southeast Asia. Emerg Infect Dis 7: 996–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lunguya O, Lejon V, Phoba MF, Bertrand S, Vanhoof R, et al. (2013) Antimicrobial resistance in invasive non-typhoid Salmonella from the Democratic Republic of the Congo: emergence of decreased fluoroquinolone susceptibility and extended-spectrum beta lactamases. PLoS Negl Trop Dis 7: e2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Feasey NA, Dougan G, Kingsley RA, Heyderman RS, Gordon MA (2012) Invasive non-typhoidal Salmonella disease: an emerging and neglected tropical disease in Africa. Lancet 379: 2489–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sangal V, Harbottle H, Mazzoni CJ, Helmuth R, Guerra B, et al. (2010) Evolution and population structure of Salmonella enterica serovar Newport. J Bacteriol 192: 6465–6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhou Z, McCann A, Litrup E, Murphy R, Cormican M, et al. (2013) Neutral genomic microevolution of a recently emerged pathogen, Salmonella enterica serovar Agona. PLoS Genet 9: e1003471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou Z, McCann A, Weill FX, Blin C, Nair S, et al. (2014) Transient Darwinian selection in Salmonella enterica serovar Paratyphi A during 450 years of global spread of enteric fever. Proc Natl Acad Sci U S A 111: 12199–12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thampapillai G, Lan R, Reeves PR (1994) Molecular evolution in the gnd locus of Salmonella enterica . Mol Biol Evol 11: 813–828. [DOI] [PubMed] [Google Scholar]
- 35. Hu H, Lan R, Reeves PR (2006) Adaptation of multilocus sequencing for studying variation within a major clone: evolutionary relationships of Salmonella enterica serovar Typhimurium. Genetics 172: 743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Feil EJ, Cooper JE, Grundmann H, Robinson DA, Enright MC, et al. (2003) How clonal is Staphylococcus aureus? J Bacteriol 185: 3307–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ke BX, Deng XL, Li BS, Tan HL, He DM, et al. (2011) Surveillance on Salmonella infection in Guangdong province, 2008–2009. Zhonghua Liu Xing Bing Xue Za Zhi 32: 789–792. [PubMed] [Google Scholar]
- 38. Ke BX, Deng XL, Zhang LH, Chen JL, Ke CW, et al. (2008) Surveillance and pathogenic analysis on non-typhoidal Salmonella in Guangdong province, 2007. Zhonghua Liu Xing Bing Xue Za Zhi 29: 1199–1203. [PubMed] [Google Scholar]
- 39. He DM, Ke BX, Deng XL, Ke CW, Liang ZM, et al. (2012) Surveillance and analysis on the pathogenic features of Salmonella in Guangdong province in 2010. Zhonghua Yu Fang Yi Xue Za Zhi 46: 424–429. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Salmonella Typhimurium strain information isolated from 1977 to 2002 in this study (Table S1).
(XLS)
The information of all the reliable STs of Salmonella Typhimurium on the website ( http://mlst.warwick.ac.uk ).
(XLS)