Abstract
Most complex diseases have well-established genetic and non-genetic risk factors. In some instances, these risk factors are likely to interact, whereby their joint effects convey a level of risk that is either significantly more or less than the sum of these risks. Characterizing these gene-environment interactions may help elucidate the biology of complex diseases, as well as to guide strategies for their targeted prevention. In most cases, the detection of gene-environment interactions will require sample sizes in excess of those needed to detect the marginal effects of the genetic and environmental risk factors. Although many consortia have been formed, comprising multiple diverse cohorts to detect gene-environment interactions, few robust examples of such interactions have been discovered. This may be because combining data across studies, usually through meta-analysis of summary data from the contributing cohorts, is often a statistically inefficient approach for the detection of gene-environment interactions. Ideally, single, very large and well-genotyped prospective cohorts, with validated measures of environmental risk factor and disease outcomes should be used to study interactions. The presence of strong founder effects within those cohorts might further strengthen the capacity to detect novel genetic effects and gene-environment interactions. Access to accurate genealogical data would also aid in studying the diploid nature of the human genome, such as genomic imprinting (parent-of-origin effects). Here we describe two studies from northern Sweden (the GLACIER and VIKING studies) that fulfill these characteristics.
Keywords: Lifestyle, Genetics, Genealogy, Biobank, Complex disease, Scandinavia
Introduction
Most common complex diseases share risk factors and may be caused by interactions between genetic susceptibility and environmental exposures (such as diet, physical activity, smoking, sunlight, pollutants, and toxins). Moreover, diseases that had previously been considered etiologically independent, such as type 2 diabetes and prostate cancer or dyslipidemia and cognitive decline, are now known to involve some of the same biological pathways. Gaining a deeper understanding of the risk factors and pathways that cause common diseases will facilitate the prediction, prevention, and management of those diseases.
Variation in the nuclear genome is at the very basis of human biology, acting as a conduit through which environmental exposures, including diet, physical activity, and smoking, affect the many phenotypes of health and disease. Recent large-scale population genetic studies have revealed novel human pathobiology by highlighting pathways within which common genetic variants influence disease predisposition [1•], with the majority of these variants conveying similar risk regardless of variation in environmental exposures. Much less is known of genetic variants whose risk is conditional on the environment (gene-environment interactions), with few adequately replicated examples published to date [2]. Aside from providing information about human biology, genetic studies can also aid in disease prediction and treatment stratification, although most instances where this is true have involved rare, monogenic diseases such as maturity-onset diabetes of the young [3], phenylketonuria [4], and non-small-cell lung carcinoma [5]. The objective to achieve similar successes for complex diseases like type 2 diabetes, cardiovascular disease (CVD), and common cancers is being aggressively pursued by investigators around the globe.
To discover and confirm joint risk factors, pathways, and gene-environment interactions will likely require single, large prospective cohort studies within which exposures and phenotypes have been well characterized. Moreover, it is quite plausible that gene-environment interactions that convey the largest effects will be those that involve genetic variants that are relatively rare (i.e. effect allele frequencies<0.05 %), as gene-environment interactions that are common and of large effect magnitudes are likely to have already been detected in previous studies. Identifying rare gene variants and their interactions will likely require very large sample sizes, but such effects may be more detectable in low-admixture populations where strong founder effects are present [6•], several of which reside in Scandinavia.
Scandinavia hosts some of the world's premier medical biobanks and databases, amongst which those in northern Sweden are some of the largest and most extensively characterized (http://bbmri.se/). The current generation of investigators owe much to the foresight, diligence and hard work of scientists several decades ago who began the process of building up these biobanks; no less important has been the commitment of the public to donate their time, blood, and other biosamples, and to communicate information about their personal characteristics, lifestyle, and social circumstances for the sake of biomedical research.
Meta-analyzing results from multiple diverse and relatively small cohorts has facilitated huge progress in population genetics, yet the heterogeneous characteristics of these cohorts and the consequent need to work with the lowest common denominators when defining traits and exposures of interest is a considerable limitation of the approach, not least because statistical power is often diminished and the range of research questions that can be addressed is restricted. Hence, focusing on single (or a small number of comparable), large prospective cohorts may be a fruitful strategy for future studies of genes, environments, and complex disease [7•].
To address some of these issues, we conceived two studies: GLACIER (Gene-Lifestyle Interactions and Complex Traits Involved in Elevated Disease Risk) and VIKING (Västerbotten Imputation Databank of Near-Complete Genomes). The GLACIER Study has already contributed to many publications focused on gene-lifestyle interactions and genetic association studies in complex cardiometabolic traits (described below), whereas the VIKING Study is underway, building on GLACIER by including genealogical data that will be used to undertake heritability studies and long-range phasing of the genome, which will enable the relationships between the diploid nature of human genome and diseases to be investigated. Both studies capitalize on existing resources and characteristics, which include i) a large, well-characterized prospective cohort study (the Northern Sweden Health and Disease Study Cohort); ii) a low-admixture population within which several major complex diseases are common; iii) expertly curated registries (of disease, prescription medications, and vital statistics, etc.); iv) access to a 12-digit code, unique to each and every Swedish citizen, that can be used to link together these bioresources at an individual level.
New Perspectives on Complex Diseases
Understanding connections between different complex diseases is an important and emerging challenge in the field of biomedicine, not least because researching these connections will help illustrate the integrated nature of multiple aging-related diseases; more importantly though, by better understanding these links, simultaneous disease prevention or treatment may be possible.
Since 80 % of type 2 diabetics are obese at diagnosis and 65 % of them progress to CVD, the shared risk factors and biologic pathways for these diseases is widely recognized [8]. More recently, connections between these and other complex diseases (e.g., infection, cognitive decline, osteoporosis, and malignancies) have emerged. For example, herpes simplex virus 1 (HSV-1) causes cold sores, but can also promote beta-amyloid deposition, a component of amyloid plaques, tau phosphorylation, which destabilizes CNS microtubules, and demyelination of neuronal myelin sheaths, each being key features of Alzheimer's pathophysiology [9, 10]. HSV-1 has also been linked with CVD [11].
The major genetic risk marker for Alzheimer's is the apolipoprotein E (APOE) e4 allele [12]. APOE encodes a major component of triglyceride-rich lipoproteins (chylomicrons and chylomicron remnants), VLDL (very low density lipoproteins), and IDL (intermediate density lipoproteins), and transports cholesterol, lipoproteins, and fat-soluble vitamins to the blood via the lymphatic system. APOE mutations are associated with common hypercholesterolemia [13], renal cell carcinoma [14], and familial type III hyperlipoproteinemia. Thus, infection, cognitive decline, cancer, and CVD are all biologically linked diseases.
Bone-derived proteins such as osteocalcin (encoded by gamma-carboxyglutamic acid-containing protein (BGLAP) and secreted by osteoblasts) are bone formation markers. Osteoblast/clast-secreted proteins also play an important role in insulin secretion and action, glucose metabolism [15], cancer [16], and vascular calcification [17], thereby linking bone formation, diabetes, malignancies, and atherosclerosis.
A major determinant of bone formation, and an important health problem in Sweden as a result of the long and dark winter months, is vitamin D deficiency [18]. The health implications of vitamin D deficiency include osteoporosis, diabetes, and a range of cancers, amongst other morbidities [19]. Whether causal or not [20, 21], these relationships indicate an underlying unifying component to these diseases that is at least correlated with vitamin D deficiency.
A large European collection of prospective cohort studies, called Me-Can, has reported associations between cardiometabolic traits (body mass index [BMI], blood pressure, fasting glucose, total cholesterol, and triglycerides) and numerous cancer sites [22–25, 26•, 27, 28]. Generally, the metabolic risk factors are associated with cancers in a manner consistent with their risk for CVD endpoints, although not always. For example, total cholesterol is inversely associated with multiple cancers [22], and elevated fasting glucose and triglyceride concentrations conveyed lower risk of prostate cancer [26•], as does type 2 diabetes [29]. Genetic association studies, which are less prone to reverse-causality and confounding than conventional observational studies, support these findings. For example, variation at TCF7L2 impacts type 2 diabetes [30], colorectal cancer [31], and obesity [32•]; the JAZF1 locus influences endometrial cancer [33] risk; JAZF1 and HNF1B influence type 2 diabetes and prostate cancer [34] risk; and the FTO locus associates with obesity, type 2 diabetes, and breast cancer [35, 36]. In addition, chromosomal abnormalities such as aneuploidy or copy-neutral loss of heterozygosity (clonal mosaic events) are abundant in cancer [37] and type 2 diabetes [38].
Materials & Methods
Participants
The GLACIER and VIKING Studies draw upon a wealth of existing biomedical data and survey materials available in a prospective biobank collated under the umbrella of the NSHDS (Northern Sweden Health and Disease Study) at the Department of Biobank Research (also called the Medical Biobank) at Umeå University (http://www.biobank.umu.se/biobank/). The NSHDS comprises three ongoing studies [39••], the largest of which is the population-based Västerbottens Hälsoundersökning (Västerbottens Health Survey, VHU), also known as the Västerbottens Intervention Program (VIP). Other cohorts within the NSHDS include the population-based Northern Sweden MONICA (Multinational Monitoring of Trends and Determinants in Cardiovascular Disease) project and the Northern Sweden Mammography Screening Project. Currently, these three studies account for roughly 95,000, 29,000, and 11,000 unique biobank participants, respectively, with 135,000, 54,000, and 14,000 registered sampling occasions to date. The GLACIER and VIKING studies primarily focus on the VHU data, although as outlined below in the Summary of Key Findings, the use of disease registry data requires the inclusion of participants outside the VHU.
Established in 2008, the GLACIER Study comprises a cohort suitable for studies on the genetics of complex cardio-metabolic traits and gene-environment interactions, nested within the NSHDS. The GLACIER Study includes roughly 20,000 participants who underwent the baseline health examination between 1985 and 2004, of whom around 7,000 have undergone a 10-year follow-up exam (with ongoing data collection). All participants whose data are collated in the NSHDS have provided written, informed consent for their data and biomaterials to be used for biomedical research, and the regional ethical review board in Umeå has approved all aspects of the GLACIER Study as well as a pilot project for the VIKING Study. Further ethics permissions will be sought as the VIKING Study evolves and new projects emerge.
Physical Measurements
Since 1985, all residents within the county of Västerbotten (total population ~260,000) have been invited to visit their primary health care center for a clinical health examination in the year of their 40th, 50th, and 60th birthdays. The protocol is standardized across study centers and conducted by trained research nurses. The extensive health examination in VHU includes basic anthropometry: weight (to the nearest 0.1 kg) was measured with a calibrated balance-beam scale with the participants wearing light clothes without shoes. Height was measured to the nearest 1 cm using a wall-mounted stadiometer. BMI was calculated as weight (kg) divided by height (m) squared. Waist circumference was measured in a subgroup using a non-stretchable nylon tape at the midpoint between the 12th rib and the iliac crest. Systolic and diastolic blood pressure were measured once using a mercury sphygmomanometer following a 5 min rest with the participant in the supine position.
Biochemistry
Fasting whole blood samples were drawn from an antecubital vein, processed (into plasma, buffy coat, and erythrocytes), and stored at −80 °C at the Northern Sweden Biobank pending biochemical analyses. Capillary blood was drawn prior to and 2 h after administration of a 75-g oral glucose load, and plasma glucose, serum total cholesterol, and triglycerides were measured with a Reflotron bench-top analyzer (Roche Diagnostics Scandinavia AB). High-density lipoprotein (HDL) cholesterol was measured in a subgroup of participants. Information on participants’ fasting status was also collected: in GLACIER more than 80 % report being fasted>8 h, ~3 % between 4 to 8 h, ~1 %<4 h, and information on fasting is missing for ~10 % of GLACIER participants. DNA was extracted from peripheral white blood cells using the Plasmid Maxi Kit (Qiagen AB, Solna, Sweden) and genomic DNA samples were diluted to 4 ng/μl.
Lifestyle Data
During their visits to the primary health care centers participants complete a self-administered questionnaire that collects information about socioeconomy, psychosocial conditions, and diet via a validated semi-quantitative food-frequency questionnaire (FFQ) [40, 41••]. A modified version of the International Physical Activity Questionnaire, the Short Form-36 (SF-36) querying self-rated health, and additional questions about family history of CVD and diabetes, quality of life [42], social support, working conditions [43, 44], tobacco use, and alcohol consumption [45, 46] are also included.
The FF Q is designed to capture the habitual diet consumed by people living in northern Sweden and initially consisted of 84 food items, but in 1996 was reduced to 66 items by combining questions on some foods. Participants are asked to indicate intake of each food on a nine-point frequency scale ranging from never to four or more times per day, and also indicate general portion size for three main food groups (potatoes/rice/pasta, meat/fish, and vegetables). The data from the FFQ (used in both VHU and MONICA) are collated, processed, and managed in a separate database, the Northern Sweden Diet Database (NSDD) at the Department of Biobank Research (http://www.biobank.umu.se/biobank/biobank-for-researchers/northern-sweden-diet-database/).
Genotyping
All GLACIER participants have been genotyped for approximately 150 gene variants, the majority of which were selected from published GWAS meta-analyses of cardiometabolic traits. In 6,000 GLACIER participants, CardioMetaboChip (Illumina Inc.) genotyping was performed (CardioMetaboChip is a fine-mapping tool containing around 200,000 SNPs from regions of the genome known to harbor cardiometabolic trait-regulating loci [47]). Further genotyping with the Illumina Human ExomeChip array has been undertaken in roughly 2,500 GLACIER participants, which characterizes around 245,000 gene variants restricted to the coding DNA regions (http://genome. sph.umich.edu/wiki/Exome_Chip_Design). Additionally, 3,000 genome-wide Illumina scans have been arrayed on Biobank participants’ samples as part of case-control studies for glioma (PI: B. Melin/Umeå University) and osteoporosis (PIs: C. Ohlsson and U. Pettersson Kymmer, Gothenburg/ Umeå University). About 2,000 of these participants overlap with the GLACIER cohort.
Genealogies
The VIKING Study focuses on a target population in the northern and western parts of the county of Västerbotten in northern Sweden, which to a large extent is mapped in a genealogical database with individual-level multigenerational data, administrated by the Demographic Database (DDB) at Umeå University. Data cover the period 1700-1950 and the database is built from the detailed Swedish population registers that, until 1990, constituted the key component of the national system of registration. In more recent years, genealogical data have been collected and managed by Statistics Sweden (http://www.scb.se/en_/). These registers are characterized by their completeness, detail, and wealth of information, including basic demographic information as well as a variety of social variables and different kinds of exposure data. Via linkage to registry data at Statistics Sweden, data can be extended to include the extant population.
The population of Västerbotten has experienced little immigration and migration for many centuries, with >50 % of the extant population stemming from at least seven ancestral Västerbotten generations [48]; this information is derived primarily from church-book records by the DDB. The church-books are stored on microfiche and are gradually being digitized by trained personnel at the DDB. The genealogical database in the VIKING Study involves complete registration (i.e., all individuals present in a selected geographic region during a selected period are included); this makes the data particularly suitable for genotype imputation and heritability studies. The target population for VIKING has been specifically selected to include geographical areas where a large part of the population was previously included in medical research registers or in prospective cohorts in the Northern Sweden Biobank. However, in order to gain optimal coverage and correspondence, the database is being updated with population data from other major regions of the county. Adding data to the database is of strategic value to VIKING, as there is an extraordinarily high density of biomedical data in this population. For example, biospecimens are available for almost 100 % of age groups 45 – 64 and 65 – 79 years, as well as for a substantial portion in older and younger age groups across the region. The participation rate in the longitudinal VHU is very high and a large part of the population is also included in the MONICA Study, the Northern Sweden Mam-mary Screening Cohort, and in the Northern Sweden Maternity Cohort [39••]. All cohorts’ data and materials are stored at the Northern Sweden Biobank. A strategic addition of data from new geographic areas also improves the scientific value of the database, by providing bridges between existing regions. Building longitudinal population databases over larger regions, including several neighboring parishes and geographically connected population clusters, will help account for regional migration, thereby increasing the prospects of finding complete life courses and long genealogies for a large majority of the Västerbotten population. This expansion of the VIKING Study to include population data at present missing in the DDB databases will significantly increase the number of genetically informative individuals, and thus also make an important contribution to the completeness and geographic coverage of the VIKING Study.
Data entry and linkage is executed by the DDB and Statistics Sweden, according to consistent principles and international standards, ensuring consistent data and reliable genealogies [49, 50]. The quality and consistency of the VIKING Study will distinguish it from other resources including genealogical data such as the Icelandic deCODE database and the Utah Resource for Genetic and Epidemiologic Research.
The digitization process involves several phases, all of which are necessary to create a comprehensive database useful for large-scale studies. Data entry from the original sources is followed by different steps of linkage, where numerous person-level records are brought together and genealogies are created. This includes automatic record linkage according to established principles followed by a semi-automated process where residual notations are examined and, if possible, linked. Particular attention is directed towards distinguishing between biological vs. non-biological relationships, as data will be used for issues where information on heritability is needed.
Methods for secure linkage to biobank data and other civil registration-based registry resources are developed and evaluated in close cooperation with Statistics Sweden, and have recently been implemented and evaluated on a large-scale basis by the DDB. All procedures have been established in accordance with the Swedish legal framework on confidentiality and protection of privacy, allowing a high level of data security as well as high fidelity. The Swedish Data Inspection Board has been consulted during the process. Linkage to modern registry data will be executed by Statistics Sweden by way of a separate, permanently maintained key table. Data retrievals will, as a rule, be coded. The quality of the linkage between longitudinal population data and civil registration number-based registry resources, using this method, has been validated.
Clinical Registries
The possibility to merge biologic (including genetic), genealogic, socio-demographic, and lifestyle data with data from regional disease registries and the national prescription medication encashment registry creates an exciting opportunity for large-scale studies on gene-lifestyle and gene-drug interactions for a wide-range of disease traits. These registries include myocardial infarction (FIA, N~3,100), stroke (CAST RO, N~2,370), Alzheimer's (N~500), and several cancer sites including breast (N~2,670), prostate (N~2,200), melanoma (N~670), and colorectal (N~1,600). Data from each of these clinical outcome registries are available within the Northern Sweden Biobank, thus access to these databases is accessible to both the VIKING and GLACIER studies.
Overview of Published Findings from the GLACIER Study
We have used the GLACIER Study to undertake study-specific and consortia-based projects on gene-environment interactions and on the genetic determinants of cardiometabolic disease traits. Below is a summary of key works.
Gene-Lifestyle Interactions in Glycemic Traits
In 2009 we helped established a nutritional genetics working group as part of the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium. Here, the GLACIER Study, in collaboration with >10 smaller international cohorts, was used to study interactions between gene variants previously associated with glucose or insulin concentrations and dietary exposures in relation to fasting glucose or insulin levels. In the initial study, which reported interactions between these variants and dietary whole grain intake, the strongest interaction effect was for a variant at the GCKR locus and fasting insulin concentrations [51]; we went on to report interactions between variants at the SLC30A8 locus and total (dietary and supplementary) zinc intake on fasting insulin levels [52]. Additional analyses of interactions between these variants and dietary patterns [53] and magnesium intake [54] yielded predominantly negative findings.
In analyses conducted as part of the MAGIC consortium, the GLACIER Study also contributed to two rare genome-wide discovery analyses of genetic interactions with physical activity [55] and BMI [56] in relation to fasting and post-challenge glucose concentrations respectively. Whilst both sets of analyses yielded predominantly negative results, these attempts to conduct such analyses on a genome-wide scale will no doubt represent important stepping stones for subsequent successful gene-environment interaction discovery studies.
Gene-Lifestyle Interactions in Obesity
One of the most widely replicated examples of a gene-lifestyle interaction is between variants at the FTO locus and physical activity in obesity-related traits. The initial study, emanating from a Danish cohort, was soon followed by several replication attempts, which yielded mixed results. Because of this equipoise, we organized a large consortium of cohorts comprising around 240,000 participants to test the hypothesis [57]. Overall, we were able to replicate the interaction effect, but this was driven primarily by the inclusion of cohorts from North America, raising further questions about the nature of this interaction effect that are yet to be answered. Elsewhere, we focused on gene-smoking interactions [58], seeking replication of earlier findings reported by Freathy et al. [59] and extending these to examine also gene interactions with a type of chewing tobacco (snus) that is common in Sweden and some other Scandinavian countries. In this study we confirmed the original interaction effect, but showed that despite approximately comparable quantities of nicotine (the presumed molecular agent for the interaction) in cigarettes and snus, no such interactions for the latter were evident. We have also studied interactions between a polygenic risk score and physical activity in obesity [60]. Our analysis, which included the GLACIER cohort and other Scandinavian and European-ancestry cohorts, sought in part to replicate a finding reported previously in a cohort from Norfolk in the UK [61]. Our study included around 110,000 adults and aside from demonstrating that genetic risk of obesity is marginally less in physically active people, we also described a range of methodological issues that should be considered when studying gene-environment interactions.
Genetic Associations with Nutrients and Dietary Factors
In studies on macronutrient intake [62], genetic variation close to FGF21 (rs838145) was associated at genome-wide levels of statistical significance with higher dietary carbohydrate and lower dietary fat intake. The same variants were also associated with circulating FGF21 protein levels (p<0.05), but not with FGF21 expression in blood or brain. We also identified an association between the BMI-associated allele at FTO (rs1421085) and higher protein intake, which was genome-wide significant before adjustment for BMI, and marginally less so after BMI adjustment. In subsequent analyses [63], we confirmed that the BMI-increasing allele of the FTO variant is associated with higher dietary protein intake, and also reported its association with lower dietary carbohydrate and total energy intake. Of these, the associations between FTO and protein and total energy intakes remained statistically significant after adjustment for BMI. In separate analyses involving the GLACIER Study (Cornelis et al. Mol Psych. In press), we identified eight loci that were associated at a genome-wide level of significance with coffee intake (0.03 – 0.14 cups/day per effect allele), explaining ~1.3 % of the variance in coffee intake. Six variants are proximal to genes that are candidates for caffeine pharmacokinetics (ABCG2, AHR, POR, CYP1A2) and pharmacodynamics (BDNF, SLC6A4).
Genetic Associations with Blood Lipid Concentrations and Coronary Artery Disease (CAD)
The GLACIER Study and its investigators have played central roles in large-scale consortia designed to discover genetic risk variants for dyslipidemia and CAD, which have since led to several high profile publications [13, 60, 64–66]. Using the GLACIER Study, we proceeded to examine whether the variants associated cross-sectionally with blood lipid concentrations also predict the long-term deterioration in blood lipids [67••], the latter of which is informative as an intermediate risk factor for CAD. Through those analyses in the GLACIER cohort, which we replicated in a cohort from southern Sweden (the Malmö Diet and Cancer Study), we were able to determine that whilst some genetic variants appear to be associated with chronically elevated blood lipid levels, others are associated with change in lipid levels over time. The 10-year repeated measures data for lipids (and most other variables) render the GLACIER dataset especially powerful for the analysis of temporal effects (Fig. 1). Although the factors that underlie this effect are unknown, it is likely that it reflects interactions between these gene variants and aging-related factors (i.e., gene-environment interactions).
Fig. 1.
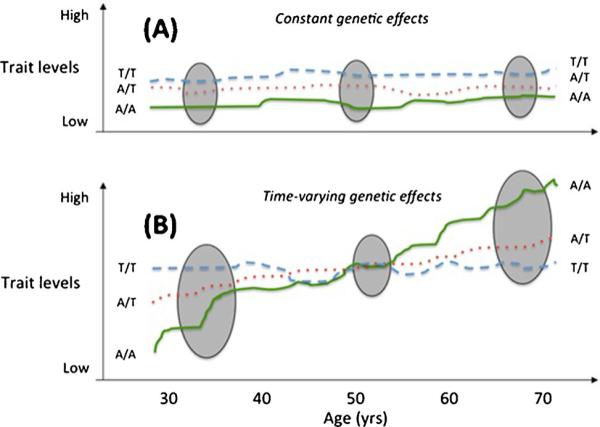
Hypothetical genetic effects that are either constant (Panel A) or time-varying (Panel B). The grey ellipses are sampling occasions for cross-sectional studies. Cross-sectional studies are most likely to detect loci with constant effects, as the sampling time (age) has no impact on genetic effects. By contrast, time-varying genetic effects may be undetectable in a cross-sectional analysis if the age-range of the participating cohorts is broad, or if the sampling occasion is at the cross-over point. Moreover, cross-sectional studies in younger adults would conclude that genotype A/A is protective, even though the genotype conveys high risk later in life
Genetic Associations with Glycemic Traits and Type 2 Diabetes
Our initial publication in the Västerbotten materials focused on replicating and extending associations between common variants at the wolframin-encoding gene (WFS1) and type 2 diabetes and glycemic traits [68]. Defects in WFS1 (4 Chr 4p16.1) cause a rare, childhood-onset autosomal-recessive neurological disorder called Wolfram syndrome (OMIM: #222300). The disease is also called DIDMOAD, referring to its key pathophysiological features: diabetes insipidus, (non-autoimmune) diabetes mellitus with optic atrophy, and deafness. In 2007, several common WFS1 variants were reportedly associated with adult-onset type 2 diabetes [69]. Our study in the Västerbotten population [68] replicated those findings and reported novel associations with fasting and post-challenge blood glucose concentrations. We combined these data with those from several other cohorts using meta-analysis to support our findings. We later used data from the Västerbotten cohort along with these from elsewhere to fine-map the WFS1 locus [70].
In a subsequent analysis [71], we examined the extent to which loci previously associated with blood concentrations of glucose, insulin, lipids, and with BMI increase the discriminative ability of type 2 diabetes prediction models. Model 1, including only SNPs associated with type 2 diabetes, had a discriminative power of 0.591 (p<1.00×10−20 vs. null model) as estimated by the area under the receiver operator characteristic curve (ROC AUC). Model 2, including only fasting glucose/insulin SNPs, had a significantly higher discriminative power than the null model (ROC AUC 0.543; p=9.38× 10−6 vs. null model), but lower discriminative power than model 1 (p=5.92×10−5). Model 3, with only lipid-associated SNPs, had significantly higher discriminative power than the null model (ROC AUC 0.565; p=1.44×10−9) and was not statistically different from model 1 (p=0.083). The ROC AUC of model 4, which included only obesity SNPs, was 0.557 (p= 2.30×10−7 vs. null model) and smaller than model 1 (p= 0.025). Finally, the model including all SNPs yielded a significant improvement in discriminative power compared with the null model (p<1.0×10−20) and model 1 (p=1.32×10−5); its ROC AUC was 0.626, which is, nevertheless, likely too low to be of use in clinical practice.
The GLACIER Study and its investigators have also helped lead efforts to discover genetic variants associated with blood glucose concentrations as part of the MAGIC consortium [72]. We carried forward some of those discoveries in the prospective sub-cohort of the GLACIER Study to assess whether they predict long-term changes in fasting and post-oral glucose challenge blood glucose levels [73••], and whether their predictive ability complimented that of conventional risk prediction algorithms including non-genetic data. In those analyses, we were able to distinguish genetic variants that are associated with chronically elevated blood glucose levels from those that are associated with a loss of glucose control over a decade of follow-up; however, we also showed that these variants, when modeled jointly, yield only marginal improvements in predictive accuracy compared with conventional risk prediction algorithms.
A recent trend in complex traits genetics research has been towards the fine mapping of specific regions of the genome, such as those that code functional elements. Through collaboration with the Danish LuCamp consortium, we used our data to replicate a number of quantitative trait loci, although no striking evidence of association with type 2 diabetes was found [74].
Genetic Associations with Obesity-Associated Traits
Our initial obesity genetics paper focused on associations of nine loci discovered by the GIANT consortium. Our study extended those findings by demonstrating associations of these variants with adipose fat mass and type 2 diabetes [75]. Here we determined for the first time that a variant in the MC4R region was associated with type 2 diabetes, a result that was later confirmed by the DIAGRAM+ consortium [76]. The GLACIER Study has since contributed to a number of meta-analyses that have sought to discover novel associations for anthropometric traits [77–79] or to confirm existing purported associations (Stijnen et al. Am J Epid. In press).
Overview of Ongoing Work Within the VIKING Study
The VIKING Study's overarching objectives are i) to build on the GLACIER Study and the genealogical data curated by the DDB and Statistics Sweden to obtain near-complete genome sequences in a large proportion of the Västerbotten population, ii) to combine these data with information from the Northern Sweden Biobank and regional/national disease registries, and iii) to discover gene variants, which, owing to interactions with exposures and treatments, might be used to guide stratified treatment decisions for complex diseases.
Specific aims
Estimate heritability for a range of phenotypes, whilst also accounting for the presence of gene-environment interactions
Use genetic data in index persons from the Skellefteå and Norsjö regions of northern Sweden to impute genotypes into the Västerbotten population at large. Analyses using data from the Umeå region are also planned, as those data will soon be available
Use imputed genotype data to study genetic susceptibility to environmental risk factors for complex diseases (e.g., type 2 diabetes and CVD)
Combine genetic and genealogical data to study genetic imprinting and parent-of-origin effects in relation to complex disease predisposition
Long-Range Phasing and Haplotype Imputation
Determining the parental origin of a pair of alleles at a given genotype can prove useful for numerous genetic analyses (see below), but this requires genetic data to be ‘phased’. Knowing a population's genealogical structure, and combining this with targeted direct genotyping in selected individuals has previously been used successfully to impute large amounts of genetic data in Iceland [80, 81] and in Finland as part of the SiSu project (Sequencing Initiative Suomi) [82]. In VIKING this can be achieved using high quality genealogical data for northern Sweden, digitized by the DDB. An example is given below:
In a pilot study we mapped the genealogies of people living in the Skellefteå region between 1700 and 1950, (see Table 1) taking GLACIER participants with existing genetic data as the index individuals. In total there are 467 families comprising 4,235 genotyped individuals, with each family ranging in size from one (175 genotyped participants have no record of a relative in the database) to 151,823 (one family with extended genealogy) (see Table 1). The family with the extended genealogy spans >12 generations, descending from 4,155 founders and containing 3,747 individuals with existing genotype data, and constitutes 80 – 95 % of the total population during the lifetime of this pedigree (see Table 1). The longest ancestral path includes 12 ancestors (43 individuals have 12 ancestors in the pedigree). Most of the individuals in this pedigree have ancestral paths including between six and 10 individuals. Assuming all founders are unrelated, the minimum inbreeding coefficient is 0.000019. The average inbreeding coefficient is 0.0028 and the average inbreeding coefficient in individuals born from consanguineous unions is 0.0077. However, in the complex family structures, different breeding schemes can result in similar inbreeding coefficient and making conclusions about mating solely based on the inbreeding coefficients can sometimes be misleading. Among those in this extended pedigree, ~10,000 have detailed phenotype data and blood samples stored in the Northern Sweden Biobank (with about 2,000 having detailed genetic data). Of these 10,000, there are 133 trios, 55 father-offspring pairs, 274 mother-offspring pairs, and 3,556 sib-pairs.
Table 1.
Population statistics for the Skellefteå region found in the Demographic Database for the period 1719 – 1950; total population and Family 1 with the extended genealogy
| Year | Total DDB Skellefteå population | Family 1 | % Family 1 |
|---|---|---|---|
| 1719 | 1105 | 884 | 80 % |
| 1750 | 3841 | 3418 | 89 % |
| 1800 | 6705 | 6309 | 94 % |
| 1850 | 16947 | 16107 | 95 % |
| 1900 | 39864 | 36999 | 93 % |
| 1950 | 60321 | 48825 | 81 % |
Parent-of-Origin Studies
Knowing from which parent an allele has been inherited is important when maternally and paternally inherited alleles convey different effects on disease predisposition. The impact of these so-called parent-of-origin effects on complex traits is unclear. Although in murine models, many studies have reported parent-of-origin effects for a range of traits [83]. Evidence in humans is far less extensive, in part because the data needed to study such effects is often unavailable. However, recent methodological developments using variance estimates extracted from genome-wide genotype data in unrelated individuals [84] will likely facilitate parent-of-origin effect studies in human populations. Although this approach shows promise, examining differential transcription at an allelic level, which is an important feature of parent-of-origin effect studies, requires the knowledge of the maternal and paternal alleles. In the VIKING Study we have a unique opportunity to study parent-of-origin effects for a number of phenotypes owing to the availability of detailed biomedical and demographic data.
Summary & Conclusions
The first whole genome sequences published in 2001 cost around 70 million euros to obtain, [85, 86] which dramatically contrasts today's ~2,000 euros price tag (for 30X coverage, which is at least sufficient to identify all genomic variants down to 0.5 – 1 % allele frequency). Even with whole genome sequencing set to fall below 1,000 euros within the next few years, [87, 88] it remains infeasible to directly sequence entire regional populations the size of Västerbotten (~200 million euros). However, in low-admixed populations where detailed, accurate genealogies are available, near-complete genomes can also be obtained by imputing genome sequences, providing genetic information is available for a suitable subsample, i.e. in the Icelandic population imputation into 296,256 ungenotyped relatives of 95,085 genotyped individuals having GWAS data and additional 2,230 having sequence data [81]. As shown in Table 2, roughly one half of the Västerbotten population is estimated to have ancestral lineages extending back between seven and 12 generations (1700s and earlier); this, combined with low immigration and emigration, means that many Västerbotten residents are direct decedents of the population's founders, making Västerbotten ideal for the whole-genome phasing and imputation.
Table 2.
Generational structure of people in the Umeå Demographic Database. Number of people (and proportion of the total cohort) are shown by generational depth
| Approximate birth year of known ancestor | Generational depth (N generations) | Individuals (N) | Proportion of total cohort (%) |
|---|---|---|---|
| 1875-1900 | 2 | 15,290 | 15.0 |
| 1850-1875 | 3 | 10,699 | 10.5 |
| 1825-1850 | 4 | 7,526 | 7.4 |
| 1800-1825 | 5 | 9,020 | 8.8 |
| 1780-1800 | 6 | 7,922 | 7.8 |
| 1760-1780 | 7 | 7,970 | 7.8 |
| 1740-1760 | 8 | 11,204 | 11.0 |
| 1720-1740 | 9 | 16,474 | 16.1 |
| 1700-1720 | 10 | 14,531 | 14.2 |
| 1680-1700 | 11 | 1,540 | 1.5 |
| 1660-1680 | 12 | 1 | 0 |
The generational depths shown above assume on average two generations for the population since 1950 and that all lineages from 1950 are represented in the extant population. The exact number of generations and size of the extant population will be known when the database is complete.
The optimal scenario is thus one where a genome sequence databank is accessible for an entire regional population in which detailed information on non-genetic risk factors and disease incidence is also available. Although this idea was inconceivable only a few years ago, major advances in computational methods and high-throughput genotyping technologies now permit high-density genotyping arrays to be undertaken in sample collections of several thousand individuals at affordable costs. With the VIKING Study, it is plausible that near-complete genome sequences will be obtainable in the majority of the Västerbotten population, and owing to the dense exposure and phenotype data available within the Northern Sweden Biobank, studies of the nature outlined above may be possible.
Acknowledgments
We are grateful to the study participants whose data have contributed to the GLACIER and VIKING Studies through the studies that comprise the Northern Sweden Health and Disease Study. We are also thankful for the work and expertise of the many investigators and support staff who collected and managed data and biomaterials within those studies. The VIKING and GLACIER Studies have been primarily funded by the Swedish Research Council, Novo Nordisk Foundation, Swedish Heart Lung Foundation, Albert Påhlsson Foundation and the Swedish Diabetes Association (all grants to PWF).
Footnotes
Compliance with Ethics Guidelines
Conflict of Interest Azra Kurbasic declares that she has no conflict of interest.
Alaitz Poveda has received research support through a grant from the Basque Government.
Yan Chen declares that he has no conflict of interest.
Åsa Ågren declares that she has no conflict of interest.
Elisabeth Engberg declares that she has no conflict of interest.
Frank B. Hu declares that he has no conflict of interest.
Ingegerd Johansson declares that she has no conflict of interest.
Ines Barroso, along with her spouse, owns stock in GlaxoSmithKline and Incyte Corporation.
Anders Brändström declares that he has no conflict of interest.
Göran Hallmans declares that he has no conflict of interest.
Frida Renström declares that she has no conflict of interest.
Paul W. Franks declares that he has no conflict of interest.
Human and Animal Rights and Informed Consent This article does not contain any studies with human or animal subjects performed by any of the authors.
Contributor Information
Azra Kurbasic, Department of Clinical Sciences, Genetic and Molecular Epidemiology Unit, Lund University, Skåne University Hospital Malmö, CRC, Building 91, Level 10, Jan Waldenströms gata 35, SE-205 02 Malmö, Sweden.
Alaitz Poveda, Department of Clinical Sciences, Genetic and Molecular Epidemiology Unit, Lund University, Skåne University Hospital Malmö, CRC, Building 91, Level 10, Jan Waldenströms gata 35, SE-205 02 Malmö, Sweden; Department of Genetics, Physical Anthropology and Animal Physiology, Faculty of Science and Technology, University of the Basque Country, Bilbao, Spain.
Yan Chen, Department of Clinical Sciences, Genetic and Molecular Epidemiology Unit, Lund University, Skåne University Hospital Malmö, CRC, Building 91, Level 10, Jan Waldenströms gata 35, SE-205 02 Malmö, Sweden.
Åsa Ågren, Department of Biobank Research, Umeå University, Umeå, Sweden.
Elisabeth Engberg, Umeå Demographic Database, Umeå University, Umeå, Sweden.
Frank B. Hu, Department of Nutrition, Harvard School of Public Health, Boston, MA, USA
Ingegerd Johansson, Department of Odontology, Umeå University, Umeå, Sweden.
Ines Barroso, Wellcome Trust Sanger Institute, Hinxton, Cambridge, UK; Metabolic Research Laboratories Institute of Metabolic Science, Addenbrooke's Hospital, University of Cambridge, Cambridge, UK; NIHR Cambridge Biomedical Research Centre, Institute of Metabolic Science, Addenbrooke's Hospital, Cambridge, UK.
Anders Brändström, Umeå Demographic Database, Umeå University, Umeå, Sweden.
Göran Hallmans, Department of Biobank Research, Umeå University, Umeå, Sweden; Department of Public Health & Clinical Medicine, Umeå University, Umeå, Sweden.
Frida Renström, Department of Clinical Sciences, Genetic and Molecular Epidemiology Unit, Lund University, Skåne University Hospital Malmö, CRC, Building 91, Level 10, Jan Waldenströms gata 35, SE-205 02 Malmö, Sweden; Department of Biobank Research, Umeå University, Umeå, Sweden.
Paul W. Franks, Department of Clinical Sciences, Genetic and Molecular Epidemiology Unit, Lund University, Skåne University Hospital Malmö, CRC, Building 91, Level 10, Jan Waldenströms gata 35, SE-205 02 Malmö, Sweden Department of Nutrition, Harvard School of Public Health, Boston, MA, USA; Department of Public Health & Clinical Medicine, Umeå University, Umeå, Sweden.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of outstanding importance
- 1•.Langenberg C, Sharp SJ, Franks PW, Scott RA, Deloukas P, Forouhi NG, et al. Gene-lifestyle interaction and type 2 diabetes: the EPIC interact case-cohort study. PLoS Med. 2014;11(5) doi: 10.1371/journal.pmed.1001647. [This paperdescribes the largest study of gene-lifestyle interactions in incident type 2 diabetes to date. The study found that genetic effects on diabetes are greater at a younger age and in leaner participants. No evidence of interaction with Mediterranean diet or physical activity were observed.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmad S, Varga TV, Franks PW. Gene × environment interactions in obesity: the state of the evidence. Hum Hered. 2013;75(2–4):106–15. doi: 10.1159/000351070. [DOI] [PubMed] [Google Scholar]
- 3.Pearson ER, Starkey BJ, Powell RJ, Gribble FM, Clark PM, Hattersley AT. Genetic cause of hyperglycaemia and response to treatment in diabetes. Lancet. 2003;362(9392):1275–81. doi: 10.1016/S0140-6736(03)14571-0. [DOI] [PubMed] [Google Scholar]
- 4.Horner FA, Streamer CW. Effect of a phenylalanine-restricted diet on patients with phenylketonuria; clinical observations in three cases. J Am Med Assoc. 1956;161(17):1628–30. doi: 10.1001/jama.1956.62970170004005b. [DOI] [PubMed] [Google Scholar]
- 5.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 6•.Hatzikotoulas K, Gilly A, Zeggini E. Using population isolates in genetic association studies. Brief Funct Genomics. 2014 doi: 10.1093/bfgp/elu022. [This paper describes statistical approaches to genetic analysis in heavily admixed populations.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7•.Tenesa A, Haley CS. The heritability of human disease: estimation, uses and abuses. Nat Rev Genet. 2013;14(2):139–49. doi: 10.1038/nrg3377. [This paper outlines appropriate ways of interpreting heritability estimates, and explains how heritability estimates are frequently misinterpreted.] [DOI] [PubMed] [Google Scholar]
- 8.Mannucci E, Dicembrini I, Lauria A, Pozzilli P. Is glucose control important for prevention of cardiovascular disease in diabetes? Diabetes Care. 2013;36(Suppl 2):S259–63. doi: 10.2337/dcS13-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santana S, Recuero M, Bullido MJ, Valdivieso F, Aldudo J. Herpes simplex virus type I induces the accumulation of intracellular beta-amyloid in autophagic compartments and the inhibition of the non-amyloidogenic pathway in human neuroblastoma cells. Neurobiol Aging. 2012;33(2):430.e19, 33. doi: 10.1016/j.neurobiolaging.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 10.Ball MJ, Lukiw WJ, Kammerman EM, Hill JM. Intracerebral propagation of Alzheimer's disease: strengthening evidence of a herpes simplex virus etiology. Alzheimers Dement. 2013;9(2):169–75. doi: 10.1016/j.jalz.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siscovick DS, Schwartz SM, Corey L, Grayston JT, Ashley R, Wang SP, et al. Chlamydia pneumoniae, herpes simplex virus type 1, and cytomegalovirus and incident myocardial infarction and coronary heart disease death in older adults : the Cardiovascular Health Study. Circulation. 2000;102(19):2335–40. doi: 10.1161/01.cir.102.19.2335. [DOI] [PubMed] [Google Scholar]
- 12.Caselli RJ, Dueck AC, Osborne D, Sabbagh MN, Connor DJ, Ahern GL, et al. Longitudinal modeling of age-related memory decline and the APOE epsilon4 effect. N Engl J Med. 2009;361(3):255–63. doi: 10.1056/NEJMoa0809437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Global Lipids Genetics Consortium. Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45(11):1274–83. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moore LE, Brennan P, Karami S, Menashe I, Berndt SI, Dong LM, et al. Apolipoprotein E/C1 locus variants modify renal cell carcinoma risk. Cancer Res. 2009;69(20):8001–8. doi: 10.1158/0008-5472.CAN-09-1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawai M, Devlin MJ, Rosen CJ. Fat targets for skeletal health. Nat Rev Rheumatol. 2009;5(7):365–72. doi: 10.1038/nrrheum.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roodman GD. Osteoblast function in myeloma. Bone. 2011;48(1):135–40. doi: 10.1016/j.bone.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 17.Cho HJ, Cho HJ, Kim HS. Osteopontin: a multifunctional protein at the crossroads of inflammation, atherosclerosis, and vascular calcification. Curr Atheroscler Rep. 2009;11(3):206–13. doi: 10.1007/s11883-009-0032-8. [DOI] [PubMed] [Google Scholar]
- 18.Nilson F, Moniruzzaman S, Andersson R. A comparison of hip fracture incidence rates among elderly in Sweden by latitude and sunlight exposure. Scand J Public Health. 2013 doi: 10.1177/1403494813510794. [DOI] [PubMed] [Google Scholar]
- 19.Hossein-nezhad A, Holick MF. Vitamin D for health: a global perspective. Mayo Clin Proc. 2013;88(7):720–55. doi: 10.1016/j.mayocp.2013.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wactawski-Wende J, Kotchen JM, Anderson GL, Assaf AR, Brunner RL, O'Sullivan MJ, et al. Calcium plus vitamin D supplementation and the risk of colorectal cancer. N Engl J Med. 2006;354(7):684–96. doi: 10.1056/NEJMoa055222. [DOI] [PubMed] [Google Scholar]
- 21.Shapses SA, Manson JE. Vitamin D and prevention of cardiovascular disease and diabetes: why the evidence falls short. JAMA. 2011;305(24):2565–6. doi: 10.1001/jama.2011.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strohmaier S, Edlinger M, Manjer J, Stocks T, Bjorge T, Borena W, et al. Total serum cholesterol and cancer incidence in the Metabolic syndrome and Cancer Project (Me-Can). PLoS One. 2013;8(1):e54242. doi: 10.1371/journal.pone.0054242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ulmer H, Bjorge T, Concin H, Lukanova A, Manjer J, Hallmans G, et al. Metabolic risk factors and cervical cancer in the metabolic syndrome and cancer project (Me-Can). Gynecol Oncol. 2012;125(2):330–5. doi: 10.1016/j.ygyno.2012.01.052. [DOI] [PubMed] [Google Scholar]
- 24.Haggstrom C, Rapp K, Stocks T, Manjer J, Bjorge T, Ulmer H, et al. Metabolic factors associated with risk of renal cell carcinoma. PLoS One. 2013;8(2):e57475. doi: 10.1371/journal.pone.0057475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindkvist B, Almquist M, Bjorge T, Stocks T, Borena W, Johansen D, et al. Prospective cohort study of metabolic risk factors and gastric adenocarcinoma risk in the Metabolic Syndrome and Cancer Project (Me-Can). Cancer Causes Control. 2013;24(1):107–16. doi: 10.1007/s10552-012-0096-6. [DOI] [PubMed] [Google Scholar]
- 26•.Haggstrom C, Stocks T, Ulmert D, Bjorge T, Ulmer H, Hallmans G, et al. Prospective study on metabolic factors and risk of prostate cancer. Cancer. 2012;118(24):6199–206. doi: 10.1002/cncr.27677. [This study describes observational analyses linking features of the metabolic syndrome and prostate cancer in European adults. The study includes a cohort from the Northern Sweden Biobank, where the GLACIER and VIKING Studies are set.] [DOI] [PubMed] [Google Scholar]
- 27.Nagel G, Stocks T, Spath D, Hjartaker A, Lindkvist B, Hallmans G, et al. Metabolic factors and blood cancers among 578,000 adults in the metabolic syndrome and cancer project (Me-Can). Ann Hematol. 2012;91(10):1519–31. doi: 10.1007/s00277-012-1489-z. [DOI] [PubMed] [Google Scholar]
- 28.Stocks T, Van Hemelrijck M, Manjer J, Bjorge T, Ulmer H, Hallmans G, et al. Blood pressure and risk of cancer incidence and mortality in the Metabolic Syndrome and Cancer Project. Hypertension. 2012;59(4):802–10. doi: 10.1161/HYPERTENSIONAHA.111.189258. [DOI] [PubMed] [Google Scholar]
- 29.Pierce BL. Why are diabetics at reduced risk for prostate cancer? A review of the epidemiologic evidence. Urol Oncol. 2012;30(5):735–43. doi: 10.1016/j.urolonc.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 30.Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38(3):320–3. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 31.Seshagiri S, Stawiski EW, Durinck S, Modrusan Z, Storm EE, Conboy CB, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488(7413):660–4. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32•.Helgason A, Palsson S, Thorleifsson G, Grant SF, Emilsson V, Gunnarsdottir S, et al. Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat Genet. 2007;39(2):218–25. doi: 10.1038/ng1960. [This paper provides an eloquent example of how genetic association signals can be refined using pedigree-based data in a population isolate.] [DOI] [PubMed] [Google Scholar]
- 33.Koontz JI, Soreng AL, Nucci M, Kuo FC, Pauwels P, van Den Berghe H, et al. Frequent fusion of the JAZF1 and JJAZ1 genes in endometrial stromal tumors. Proc Natl Acad Sci U S A. 2001;98(11):6348–53. doi: 10.1073/pnas.101132598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stevens VL, Ahn J, Sun J, Jacobs EJ, Moore SC, Patel AV, et al. HNF1B and JAZF1 genes, diabetes, and prostate cancer risk. Prostate. 2010;70(6):601–7. doi: 10.1002/pros.21094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3(7):e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45(4):353–61. 361e1–2. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacobs KB, Yeager M, Zhou W, Wacholder S, Wang Z, Rodriguez-Santiago B, et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet. 2012;44(6):651–8. doi: 10.1038/ng.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonnefond A, Skrobek B, Lobbens S, Eury E, Thuillier D, Cauchi S, et al. Association between large detectable clonal mosaicism and type 2 diabetes with vascular complications. Nat Genet. 2013;45(9):1040–3. doi: 10.1038/ng.2700. [DOI] [PubMed] [Google Scholar]
- 39••.Hallmans G, Agren A, Johansson G, Johansson A, Stegmayr B, Jansson JH, et al. Cardiovascular disease and diabetes in the Northern Sweden Health and Disease Study Cohort - evaluation of risk factors and their interactions. Scand J Public Health Suppl. 2003;61:18–24. doi: 10.1080/14034950310001432. [This paper describes the Northern Sweden Health and Disease Study, the collection of cohorts making up the Northern Sweden Biobank, within which the VIKING and GLACIER Studies are set.] [DOI] [PubMed] [Google Scholar]
- 40.Johansson G, Wikman A, Ahren AM, Hallmans G, Johansson I. Underreporting of energy intake in repeated 24-hour recalls related to gender, age, weight status, day of interview, educational level, reported food intake, smoking habits and area of living. Public Health Nutr. 2001;4(4):919–27. doi: 10.1079/phn2001124. [DOI] [PubMed] [Google Scholar]
- 41••.Johansson I, Hallmans G, Wikman A, Biessy C, Riboli E, Kaaks R. Validation and calibration of food-frequency questionnaire measurements in the Northern Sweden Health and Disease cohort. Public Health Nutr. 2002;5(3):487–96. doi: 10.1079/phn2001315. [This paper describes some of the dietary methods and available data in the GLACIER and VIKING Studies.] [DOI] [PubMed] [Google Scholar]
- 42.Sullivan M, Karlsson J, Bengtsson C, Furunes B, Lapidus L, Lissner L. “The Goteborg Quality of Life Instrument”–a psychometric evaluation of assessments of symptoms and well-being among women in a general population. Scand J Prim Health Care. 1993;11(4):267–75. doi: 10.3109/02813439308994842. [DOI] [PubMed] [Google Scholar]
- 43.Karasek R, Theorell T. Healthy work : stress, productivity, and the reconstruction of working life. Basic Books; New York: 1990. [Google Scholar]
- 44.Theorell T, Perski A, Akerstedt T, Sigala F, Ahlberg-Hulten G, Svensson J, et al. Changes in job strain in relation to changes in physiological state. A longitudinal study. Scand J Work Environ Health. 1988;14(3):189–96. doi: 10.5271/sjweh.1932. [DOI] [PubMed] [Google Scholar]
- 45.Babor TF, Higgins-Biddle JC, Saunders JB, Monteiro MG, World Health Organization. Dept. of Mental Health and Substance Dependence . AUDIT : the Alcohol Use Disorders Identification Test : guidelines for use in primary health care. 2nd ed. World Health Organization; Geneva: 2001. p. 38. [Google Scholar]
- 46.Aertgeerts B, Buntinx F, Kester A. The value of the CAGE in screening for alcohol abuse and alcohol dependence in general clinical populations: a diagnostic meta-analysis. J Clin Epidemiol. 2004;57(1):30–9. doi: 10.1016/S0895-4356(03)00254-3. [DOI] [PubMed] [Google Scholar]
- 47.Voight BF, Kang HM, Ding J, Palmer CD, Sidore C, Chines PS, et al. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet. 2012;8(8):e1002793. doi: 10.1371/journal.pgen.1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Einarsdottir E, Egerbladh I, Beckman L, Holmberg D, Escher SA. The genetic population structure of northern Sweden and its implications for mapping genetic diseases. Hereditas. 2007;144(5):171–80. doi: 10.1111/j.2007.0018-0661.02007.x. [DOI] [PubMed] [Google Scholar]
- 49.Vikström P, Edvinsson S, Brändström A. Longitudinal databases – sources for analyzing the life course: characteristics, difficulties and possibilities. Hist Comput. 2004;14:1–2. [Google Scholar]
- 50.Mandemakers K, Dillon L. Best Practices with Large Databases on Historical Populations. Historical Methods. 2004;37(1) [Google Scholar]
- 51.Nettleton JA, McKeown NM, Kanoni S, Lemaitre RN, Hivert MF, Ngwa J, et al. Interactions of dietary whole-grain intake with fasting glucose- and insulin-related genetic loci in individuals of European descent: a meta-analysis of 14 cohort studies. Diabetes Care. 2010;33(12):2684–91. doi: 10.2337/dc10-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanoni S, Nettleton JA, Hivert MF, Ye Z, van Rooij FJ, Shungin D, et al. Total zinc intake may modify the glucose-raising effect of a zinc transporter (SLC30A8) variant: a 14-cohort meta-analysis. Diabetes. 2011;60(9):2407–16. doi: 10.2337/db11-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nettleton JA, Hivert MF, Lemaitre RN, McKeown NM, Mozaffarian D, Tanaka T, et al. Meta-analysis investigating associations between healthy diet and fasting glucose and insulin levels and modification by loci associated with glucose homeostasis in data from 15 cohorts. Am J Epidemiol. 2013;177(2):103–15. doi: 10.1093/aje/kws297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hruby A, Ngwa JS, Renstrom F, Wojczynski MK, Ganna A, Hallmans G, et al. Higher magnesium intake is associated with lower fasting glucose and insulin, with no evidence of interaction with select genetic loci, in a meta-analysis of 15 CHARGE Consortium Studies. J Nutr. 2013;143(3):345–53. doi: 10.3945/jn.112.172049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scott RA, Chu AY, Grarup N, Manning AK, Hivert MF, Shungin D, et al. No interactions between previously associated 2-hour glucose gene variants and physical activity or BMI on 2-hour glucose levels. Diabetes. 2012;61(5):1291–6. doi: 10.2337/db11-0973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Manning AK, Hivert MF, Scott RA, Grimsby JL, Bouatia-Naji N, Chen H, et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet. 2012;44(6):659–69. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kilpelainen TO, Qi L, Brage S, Sharp SJ, Sonestedt E, Demerath E, et al. Physical activity attenuates the influence of FTO variants on obesity risk: a meta-analysis of 218,166 adults and 19,268 children. PLoS Med. 2011;8(11):e1001116. doi: 10.1371/journal.pmed.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Varga TV, Hallmans G, Hu FB, Renstrom F, Franks PW. Smoking status, snus use, and variation at the CHRNA5-CHRNA3-CHRNB4 locus in relation to obesity: the GLACIER study. Am J Epidemiol. 2013;178(1):31–7. doi: 10.1093/aje/kws413. [DOI] [PubMed] [Google Scholar]
- 59.Freathy RM, Kazeem GR, Morris RW, Johnson PC, Paternoster L, Ebrahim S, et al. Genetic variation at CHRNA5-CHRNA3-CHRNB4 interacts with smoking status to influence body mass index. Int J Epidemiol. 2011;40(6):1617–28. doi: 10.1093/ije/dyr077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ahmad S, Rukh G, Varga TV, Ali A, Kurbasic A, Shungin D, et al. Gene × physical activity interactions in obesity: combined analysis of 111,421 individuals of European ancestry. PLoS Genet. 2013;9(7):e1003607. doi: 10.1371/journal.pgen.1003607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li S, Zhao JH, Luan J, Ekelund U, Luben RN, Khaw KT, et al. Physical activity attenuates the genetic predisposition to obesity in 20,000 men and women from EPIC-Norfolk prospective population study. PLoS Med. 2010;7(8) doi: 10.1371/journal.pmed.1000332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tanaka T, Ngwa JS, van Rooij FJA, Zillikens MC, Wojczynski MK, Frazier-Wood AC, et al. Genome-wide meta-analysis of observational studies shows common genetic variants associated with macronutrient intake. Am J Clin Nutr. 2013;97(6):1395–402. doi: 10.3945/ajcn.112.052183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qi Q, Kilpelainen TO, Downer MK, Tanaka T, Smith CE, Sluijs I, et al. FTO genetic variants, dietary intake and body mass index: insights from 177 330 individuals. Hum Mol Genet. 2014 doi: 10.1093/hmg/ddu411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Do R, Willer CJ, Schmidt EM, Sengupta S, Gao C, Peloso GM, et al. Common variants associated with plasma triglycerides and risk for coronary artery disease. Nat Genet. 2013;45(11):1345–52. doi: 10.1038/ng.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Voight BF, Peloso GM, Orho-Melander M, Frikke-Schmidt R, Barbalic M, Jensen MK, et al. Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. Lancet. 2012;380(9841):572–80. doi: 10.1016/S0140-6736(12)60312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Consortium CAD, Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45(1):25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67••.Varga TV, Sonestedt E, Shungin D, Koivula RW, Hallmans G, Escher SA, et al. Genetic determinants of long-term changes in blood lipid concentrations: 10-year follow-up of the GLACIER study. PLoS Genet. 2014;10(6):e1004388. doi: 10.1371/journal.pgen.1004388. [This paper provides a detailed description of the GLACIER Study and reports data on the association of gene variants at 157 loci and long-term deteriorations in blood lipid concentrations in people from Northern Sweden.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Franks PW, Rolandsson O, Debenham SL, Fawcett KA, Payne F, Dina C, et al. Replication of the association between variants in WFS1 and risk of type 2 diabetes in European populations. Diabetologia. 2008;51(3):458–63. doi: 10.1007/s00125-007-0887-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sandhu MS, Weedon MN, Fawcett KA, Wasson J, Debenham SL, Daly A, et al. Common variants in WFS1 confer risk of type 2 diabetes. Nat Genet. 2007;39(8):951–3. doi: 10.1038/ng2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fawcett KA, Wheeler E, Morris AP, Ricketts SL, Hallmans G, Rolandsson O, et al. Detailed investigation of the role of common and low-frequency WFS1 variants in type 2 diabetes risk. Diabetes. 2010;59(3):741–6. doi: 10.2337/db09-0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fontaine-Bisson B, Renstrom F, Rolandsson O, Magic, Payne F, Hallmans G, et al. Evaluating the discriminative power of multi-trait genetic risk scores for type 2 diabetes in a northern Swedish population. Diabetologia. 2010;53(10):2155–62. doi: 10.1007/s00125-010-1792-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42(2):105–16. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73••.Renstrom F, Shungin D, Johansson I, Investigators M, Florez JC, Hallmans G, et al. Genetic predisposition to long-term nondiabetic deteriorations in glucose homeostasis: Ten-year follow-up of the GLACIER study. Diabetes. 2011;60(1):345–54. doi: 10.2337/db10-0933. [This paper provides a detailed description of the GLACIER Study and reports data on the association of gene variants at 16 loci and long-term deteriorations in blood glucose concentrations in people from Northern Sweden.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Albrechtsen A, Grarup N, Li Y, Sparso T, Tian G, Cao H, et al. Exome sequencing-driven discovery of coding polymorphisms associated with common metabolic phenotypes. Diabetologia. 2013;56(2):298–310. doi: 10.1007/s00125-012-2756-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Renstrom F, Payne F, Nordstrom A, Brito EC, Rolandsson O, Hallmans G, et al. Replication and extension of genome-wide association study results for obesity in 4923 adults from northern Sweden. Hum Mol Genet. 2009;18(8):1489–96. doi: 10.1093/hmg/ddp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Morris AP, Voight BF, Teslovich TM, Ferreira T, Segre AV, Steinthorsdottir V, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44(9):981–90. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lindgren CM, Heid IM, Randall JC, Lamina C, Steinthorsdottir V, Qi L, et al. Genome-wide association scan meta-analysis identifies three Loci influencing adiposity and fat distribution. PLoS Genet. 2009;5(6):e1000508. doi: 10.1371/journal.pgen.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Berndt SI, Skibola CF, Joseph V, Camp NJ, Nieters A, Wang Z, et al. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat Genet. 2013;45(5):501–12. doi: 10.1038/ng.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Randall JC, Winkler TW, Kutalik Z, Berndt SI, Jackson AU, Monda KL, et al. Sex-stratified genome-wide association studies including 270,000 individuals show sexual dimorphism in genetic loci for anthropometric traits. PLoS Genet. 2013;9(6):e1003500. doi: 10.1371/journal.pgen.1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kong A, Masson G, Frigge ML, Gylfason A, Zusmanovich P, Thorleifsson G, et al. Detection of sharing by descent, long-range phasing and haplotype imputation. Nat Genet. 2008;40(9):1068–75. doi: 10.1038/ng.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Styrkarsdottir U, Thorleifsson G, Sulem P, Gudbjartsson DF, Sigurdsson A, Jonasdottir A, et al. Nonsense mutation in the LGR4 gene is associated with several human diseases and other traits. Nature. 2013;497(7450):517–20. doi: 10.1038/nature12124. [DOI] [PubMed] [Google Scholar]
- 82.Palotie A, Widen E, Ripatti S. From genetic discovery to future personalized health research. N Biotechnol. 2012 doi: 10.1016/j.nbt.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mott R, Yuan W, Kaisaki P, Gan X, Cleak J, Edwards A, et al. The architecture of parent-of-origin effects in mice. Cell. 2014;156(1–2):332–42. doi: 10.1016/j.cell.2013.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hoggart CJ, Venturini G, Mangino M, Gomez F, Ascari G, Zhao JH, et al. Novel Approach Identifies SNPs in SLC2A10 and KCNK9 with Evidence for Parent-of-Origin Effect on Body Mass Index. PLoS Genet. 2014;10(7):e1004508. doi: 10.1371/journal.pgen.1004508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291(5507):1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 86.Lander ES, Consortium IHGS. Linton LM, Birren B, Nusbaum C, Zody MC, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 87.Bennett ST, Barnes C, Cox A, Davies L, Brown C. Toward the 1,000 dollars human genome. Pharmacogenomics. 2005;6(4):373–82. doi: 10.1517/14622416.6.4.373. [DOI] [PubMed] [Google Scholar]
- 88.Mardis ER. Anticipating the 1,000 dollar genome. Genome Biol. 2006;7(7):112. doi: 10.1186/gb-2006-7-7-112. [DOI] [PMC free article] [PubMed] [Google Scholar]


