Significance
Genomic instability is a hallmark of human cancers. Common fragile sites (CFSs) pose nonrandom, preferential targets of genomic instability, such as chromosome breaks, in response to replicative stress. The tumor suppressor WW domain-containing oxidoreductase (WWOX) spans the CFS FRA16D and has been implicated in carcinogenesis by a currently unknown mechanism. In this study, we identify a direct role of WWOX in DNA damage response, which is a major antagonist of genomic instability. We show that WWOX maintains genomic stability and regulates DNA repair by modulating the activity of the DNA damage checkpoint kinase ataxia telangiectasia-mutated (ATM). Our findings provide evidence that a fragile gene product is directly involved in the response to DNA damage, suggesting a rationale for its preferential loss during carcinogenesis.
Keywords: genomic instability, common fragile sites, WW domain-containing oxidoreductase, ataxia telangiectasia-mutated, ITCH
Abstract
Genomic instability is a hallmark of cancer. The WW domain-containing oxidoreductase (WWOX) is a tumor suppressor spanning the common chromosomal fragile site FRA16D. Here, we report a direct role of WWOX in DNA damage response (DDR) and DNA repair. We show that Wwox deficiency results in reduced activation of the ataxia telangiectasia-mutated (ATM) checkpoint kinase, inefficient induction and maintenance of γ-H2AX foci, and impaired DNA repair. Mechanistically, we show that, upon DNA damage, WWOX accumulates in the cell nucleus, where it interacts with ATM and enhances its activation. Nuclear accumulation of WWOX is regulated by its K63-linked ubiquitination at lysine residue 274, which is mediated by the E3 ubiquitin ligase ITCH. These findings identify a novel role for the tumor suppressor WWOX and show that loss of WWOX expression may drive genomic instability and provide an advantage for clonal expansion of neoplastic cells.
Genomic instability is a common characteristic of human cancers. The DNA damage response (DDR) maintains the integrity of the genome in response to DNA damage. DDR is a complex signaling process that results in cell cycle arrest followed by either DNA repair or apoptosis if the DNA damage is too extensive to be repaired (1–3). Key mammalian damage response sensors are ataxia telangiectasia-mutated (ATM), ATM and Rad3-related, and DNA-dependent PKs (4, 5). Disruption of the DDR machinery in human cells leads to genomic instability and an increased risk of cancer progression (6, 7).
The WW domain-containing oxidoreductase (WWOX) gene spans the common fragile site (CFS) FRA16D (8, 9). Genomic alterations affecting the WWOX locus have been reported in several types of cancer and include homozygous and hemizygous deletions (10–13). Ectopic expression of WWOX in WWOX-negative cancer cells attenuates cell growth and suppresses tumor growth in immunocompromised mice (10, 11, 14). Importantly, targeted ablation of Wwox in mice results in higher incidence of spontaneous lesions resembling osteosarcomas and lung and mammary tumors (14–16). These findings suggest WWOX as a tumor suppressor. The WWOX protein contains two N-terminal WW domains mediating WWOX interaction with PP(proline)x(amino acid)Y(tyrosine)-containing proteins (11, 17) and a central short-chain deyhdrogenase/reductase domain that has been proposed to function in steroidogenesis (18). Recent characterization of WWOX domains revealed that they interact, mainly through the WW1 domain, with multiprotein networks (3). The mechanism by which WWOX suppresses tumorigenicity is, however, not well-known.
In vitro, CFSs are defined as gaps or breaks on metaphase chromosomes that occur in cells treated with inhibitors of DNA replication (19, 20). In vivo, CFSs are preferential targets of replication stress in preneoplastic lesions (21), and emerging evidence suggests that they represent early warning sensors for DNA damage (22–24). Both genetic and epigenetic factors are thought to regulate the fragility of CFS (25, 26). Recent profiling studies of CFS provide evidence that the functional fragility of CFS is tissue-specific (27–29). High-throughput genomic analyses of 3,131 cancer specimens (12) and 746 cancer cell lines (13) have recently identified large deletions in CFSs, including the FRA16D/WWOX locus. Although these deletions have been linked to the presence of DNA replication stress (30), the molecular function of gene products of CFSs, including the WWOX protein, is poorly understood. Here, we identify a direct role of WWOX in the DDR and show that the WWOX gene product functions as a modulator of the DNA damage checkpoint kinase ATM.
Results
Induction of WWOX Expression After DNA Damage.
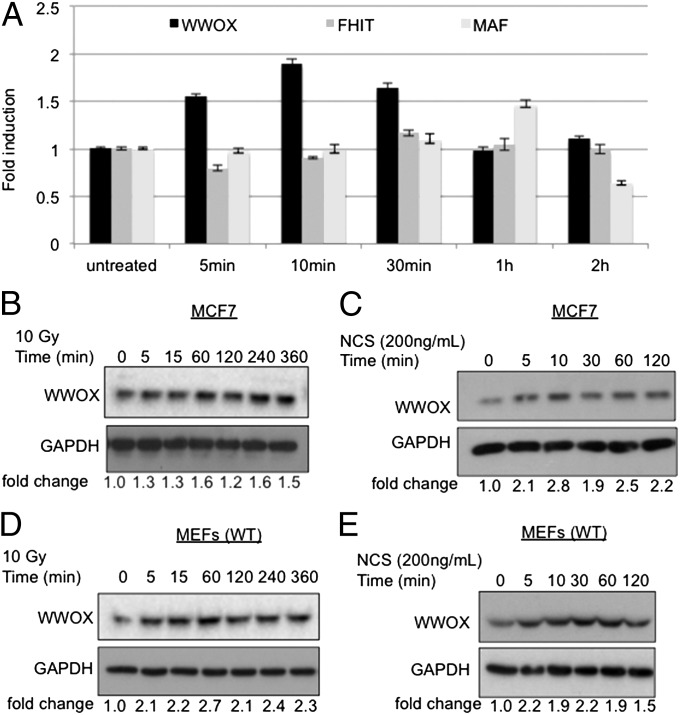
To determine whether WWOX plays a role in DDR, we examined the effect of induction of DNA double-strand breaks (DSBs) on WWOX mRNA levels using quantitative RT-PCR. DSBs were generated by using ionizing radiation or the well-established radiomimetic drug neocarzinostatin (NCS). Interestingly, 10 min after exposure of MCF7 cells to ionizing radiation, WWOX mRNA levels increased twofold (Fig. 1A). This increase was transient, and WWOX mRNA levels returned to baseline at 1–2 h (Fig. 1A). Of note, no altered expressions of the fragile site gene FHIT and the WWOX neighboring gene MAF after induction of DSBs were observed (Fig. 1A), suggesting that this effect is specific for WWOX.
Fig. 1.
Induction of WWOX expression early after DNA damage stimuli. (A) Real-time PCR analysis of WWOX, FHIT, and MAF in MCF7 after ionizing radiation treatment for the indicated time points. (B–E) Immunoblot blot analyses of WWOX levels in (B and C) MCF7 and (D and E) early-passage MEFs after treatment with ionizing radiation (10 Gy) or NCS (200 ng/mL) for different time points. Equal loading was confirmed by blotting with α-GAPDH–specific antibody. Fold change of WWOX expression upon DNA damage relative to untreated cells is shown below each blot.
Furthermore, immunoblot analysis revealed an increase of WWOX protein levels in response to induction of DSBs in MCF7 cells (Fig. 1 B and C). Of note, a comparable increase of WWOX protein expression was evident in primary mouse embryonic fibroblast (MEF) (Fig. 1 D and E), HEK293 (Fig. S1A), and LM7 cells (Fig. S1B), indicating that WWOX induction is not a cell line-specific phenomenon. These results indicate that WWOX protein expression is induced early after DNA damage, suggesting a potential role of WWOX in the DDR.
WWOX Depletion Decreases Genomic Stability Upon DNA Damage.
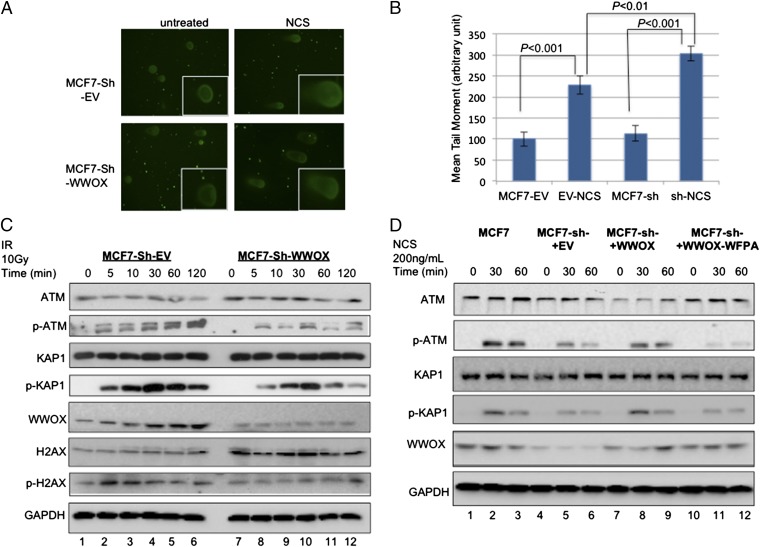
Because WWOX is induced by DNA damage, we set out to determine whether loss of WWOX expression affects genome stability. To this end, we performed comet assays with WWOX-depleted and control MCF7 cells and measured the comet tail moment of individual cells, which provides an indirect measurement of DNA DSBs. Interestingly, depletion of WWOX was associated with increased DSBs after induction of DNA damage (Fig. 2 A and B). These results suggest that loss of the WWOX gene product results in genomic instability upon DNA damage.
Fig. 2.
Depletion of WWOX renders cells more susceptible to DSBs and compromises DNA damage-induced ATM checkpoint activation. (A) Comet assay. Control- (EV-Sh) and WWOX-depleted MCF7 (WWOX-Sh) cells were treated or not treated with NCS (200 ng/mL) for 1 h. Single cells were then electrophoresed in agarose gel on a slide and stained with SyberGreen. Labeled DNA was visualized under a fluorescence microscope using 60× magnification. Representative images are shown. (B) Quantification of the comet assay in A. Bars show the comet tail as measured using ImageJ 1.47g software. Results show that MCF7-sh (WWOX-depleted) treated with NCS accumulates more DSBs relative to EV-control cells. (C) Control- and WWOX-depleted MCF7 cells were untreated or treated with ionizing radiation (10 Gy). Whole-cell lysates were prepared at the indicated time points, and immunoblot analysis was performed using specific antibodies against ATM, p-ATM (Ser1981), KAP1, p-KAP1 (pThr824), total H2A.X, γ-H2AX (pSer139), WWOX, and GAPDH. (D) WWOX-depleted MCF7 cells (MCF7-sh) were transduced with lentiviral vectors expressing scramble (EV), WT WWOX, or WT WWOX-WFPA mutant and selected with neomycin. The different cells were untreated or treated with NCS (200 ng/mL) for the indicated time points. Whole-cell lysates were prepared at the indicated time points, and immunoblot analysis was performed using specific antibodies against ATM, p-ATM, KAP1, p-KAP1, WWOX, and GAPDH. IR, ionizing radiation.
WWOX Regulates Activation of the ATM Checkpoint.
To evaluate the molecular nature of the defects observed in WWOX-depleted cells, we examined whether WWOX modulates the ATM signaling pathway, because ATM checkpoint kinase is the central coordinator of the DDR upon DSBs (31). After ionizing radiation treatment, MCF7 cells accumulated the activated Ser1981–phosphorylated- (p-) ATM. Accordingly, phosphorylation of the ATM substrates KAP1 and H2AX was evident (Fig. 2C, lanes 1–6). In contrast, WWOX-depleted MCF7 cells exhibited reduced p-ATM, p-H2AX (γ-H2AX), and p-KAP1 levels (Fig. 2C, lanes 7–12), suggesting blunted ATM function. In contrast, overexpression of WWOX but not WWOX-WFPA (a mutant form of WWOX, in which its WW1 domain has impaired interaction ability) (3, 32) in a WWOX-negative breast cancer cell line, MDA-MB231, was associated with improved DDR signaling (Fig. S2A). In addition, we also used WWOX+/+ (WT), WWOX+/− (HET), and WWOX−/− (KO) MEFs and analyzed p-KAP1 as an ATM substrate after induction of DSBs. Strikingly, genetic deletion of WWOX also resulted in reduced p-KAP1 levels (Fig. S2B), indicating that WWOX also regulates ATM signaling in nontumorigenic cells.
Next, we reconstituted WWOX expression in WWOX-depleted MCF7 cells using lentiviral transduction and analyzed the ATM checkpoint. WWOX-depleted MCF7 cells were transduced with scramble [Lenti–empty vector (EV)], Lenti-WWOX, or Lenti-WWOX-WFPA, and stable clones were generated. As expected, upon induction of DSBs, WWOX-depleted cells showed reduced p-ATM and p-KAP1 levels (Fig. 2D, lanes 4–6). Of note, reconstitution of WT WWOX expression (Fig. 2D, lanes 7–9) but not WWOX-WFPA (Fig. 2D, lanes 10–12) rescued ATM substrate phosphorylation upon DSB induction. Together, these results indicate that WWOX expression is necessary for proper ATM checkpoint function and that the WW1 domain of WWOX plays a critical role in this function.
WWOX Depletion Leads to Impaired γ-H2AX Foci Formation.
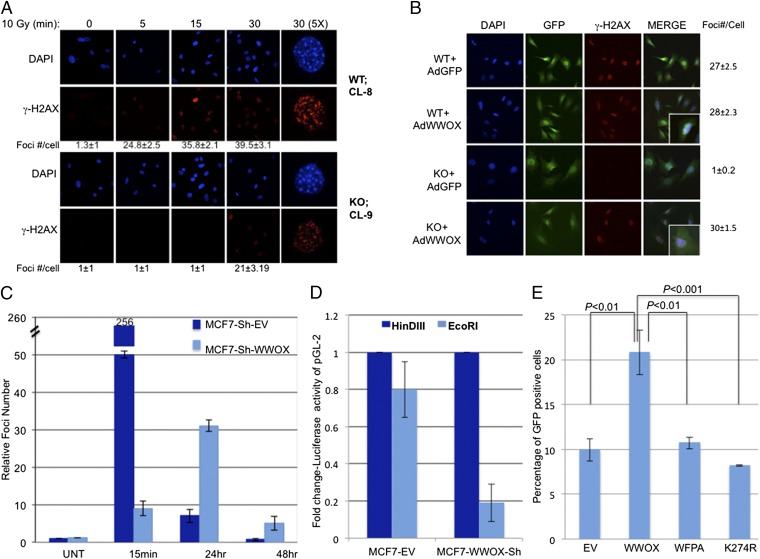
To further examine whether the checkpoint defect in WWOX-depleted and Wwox KO MEFs resulted from impaired checkpoint signaling, we examined γ-H2AX levels and foci formation (33–35). Consistent with our observations, immunoblotting indicated that WWOX knockdown in MCF7 cells was associated with attenuated histone H2AX phosphorylation (Fig. S3A). Confocal microscopy revealed that γ-H2AX foci formation was clearly delayed in KO MEFs compared with their WT counterparts (Fig. 3A).
Fig. 3.
Impaired γ-H2AX signals and DNA repair in WWOX-manipulated cells after DNA damage. (A) WT or KO MEFs were untreated or treated with ionizing radiation (10 Gy) and fixed at the indicated time points, and immunofluorescence analysis for γ-H2AX (red) was carried out. DAPI staining was performed to indicate the positions of nuclei. Cells were examined by confocal microscopy under 60× magnification. Close-up views of one cell are shown in column 5 (zoom = 5×). Quantification of γ-H2AX foci is shown under the panels as the average foci number per cell ± SEM; P < 0.002 in treatment of 30 min between WT and KO MEFs. (B) KO and WT MEFs were transduced with Ad-WWOX or Ad-GFP, treated with ionizing radiation (10 Gy), and fixed after 15 min, and immunofluorescence analysis for γ-H2AX (red) was carried out. A representative experiment is shown. Quantification of γ-H2AX foci is shown on the right side as the average foci number per cell ± SEM (P = 0.155 comparing KO-AdWWOX with WT-AdGFP; P = 0.118 comparing KO-AdWWOX with WT-AdWWOX). (C) Quantification of γ-H2AX foci in MCF7 control or WWOX-depleted cells after treatment with ionizing radiation (5 Gy) for different times. Representative figures are shown in Fig. S3B. The average of γ-H2AX foci in 10 fields is shown in the different treatments. UNT, untreated. (D and E) WWOX is important for HR. (D) WWOX-depleted MCF7 cells exhibit impaired micro-HR as revealed in cells transfected with pGL2-Luc vector that was linearized with either HindIII or EcoRI. Luciferase activity after EcoRI treatment was normalized to that of HindIII. (E) U2OS-DR-GFP cells were transduced with indicated Lenti-WWOX constructs (WWOX, WWOX-WFPA, or WWOX-K274R) (Fig. S3C). The newly generated cells were then transfected with I-SecI. Forty-eight hours later, cells were harvested and subjected to flow cytometric analysis to analyze the HR efficiency. The percentage of GFP-positive cells is shown. Data represent averages from three independent experiments.
To validate that the delay of γ-H2AX foci formation is caused by WWOX deficiency, we reconstituted Wwox KO MEFs using adenoviral transduction to reveal Ad-WWOX and Ad-GFP control MEFs. Strikingly, reconstitution of WWOX expression in KO MEFs rescued γ-H2AX foci formation to comparable levels as in WT cells (Fig. 3B). Together, these findings establish a role of WWOX in the early steps of the DDR.
To further examine the role of WWOX in DDR, we aimed to determine the consequences of WWOX depletion at early and late stages of the DDR. To this end, WWOX-depleted MCF7 cells were treated with ionizing radiation for 15 min, 24 h, and 48 h, and γ-H2AX foci were counted using confocal microscopy (Fig. 3C). We observed higher numbers of γ-H2AX foci in MCF7 control cells compared with WWOX-depleted cells at 15 min after NCS treatment (Fig. 3C and Fig. S3B). Intriguingly, the number of γ-H2AX foci, a well-established surrogate marker for DSBs, was also higher in WWOX-depleted MCF7 cells analyzed 24 and 48 h after treatment (Fig. 3C and Fig. S3B). Altogether, these results suggest that WWOX loss also affects DNA repair.
WWOX Regulates DNA Repair.
To address whether WWOX plays a role in DNA repair, we assessed DNA damage repair ability in WWOX-manipulated cells. Control or WWOX-depleted MCF7 cells were transfected with a microhomologous DNA damage repair reporter, pGL2-Luc, which has been linearized with either HindIII or EcoRI (36). Forty-eight hours posttransfection, the cells were collected, and luciferase activity was quantified. With HindIII digestion, no significant difference in Luc activity between control and WWOX-knockdown MCF7 cells was observed (Fig. 3D). In contrast, upon EcoRI digestion, control MCF7 cells were able to recover about 80% of Luc activity, whereas WWOX-depleted MCF7 cells recovered only 15% (Fig. 3D). Because EcoRI cuts within the coding sequence of the luciferase gene, restoration of luciferase activity requires precise rejoining of the short protruding ends, which involves microhomologous DNA damage repair. Reduced Luc activity in WWOX-depleted MCF7 cells indicates that WWOX is critical for microhomologous DNA damage repair. Because HindIII cuts within the linker region between the SV40 promoter and the Luc coding sequence, restoration of luciferase activity does not require precise end joining (36).
To further examine the role of WWOX in homologous recombination (HR), we used an I-SceI–dependent GFP reporter assay to measure HR in WWOX-expressing U2OS cells. This assay depends on reconstitution of GFP expression in consequence of repair of an induced I-SceI–induced DSB (37). We generated a stable U2OS-DR-GFP–expressing WWOX or control vector cells (Fig. S3C). Cells were transfected with I-SceI and harvested 48 h later, and the number of GFP-positive cells was assessed. The GFP-positive population increased approximately twofold from 10% to 21% after I-SceI expression in the cells expressing WWOX (Fig. 3E), suggesting a more efficient HR repair in the cells containing WWOX. In accordance, WWOX-WFPA or WWOX-K274R (see below) expression was not associated with improved HR (Fig. 3E). Taken together, these results suggest that WWOX affect DNA repair.
WWOX Physically Associates with ATM.
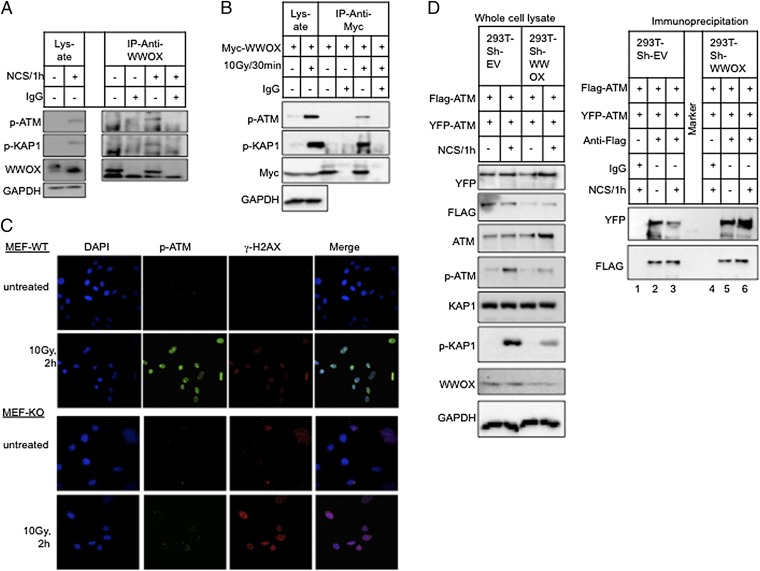
We next investigated the molecular mechanism by which WWOX contributes to the DDR and ATM checkpoint function. The observed defects in ATM activation and ATM substrate phosphorylation, including H2AX and KAP1 , prompted us to investigate potential physical interaction between WWOX and ATM. To this end, MCF7 cells were treated with NCS, and endogenous WWOX was immunoprecipitated from cell lysates with anti-WWOX antibody and analyzed by immunoblotting. Anti-IgG immunoprecipitation was used as the negative control. Strikingly, endogenous p-ATM and p-KAP1 specifically coimmunoprecipitated with WWOX after induction of DSBs, suggesting that both proteins interact after DNA damage (Fig. 4A). Comparable results were obtained in ionizing radiation-treated HEK293 cells ectopically expressing Myc-WWOX (Fig. 4B). These data indicate that WWOX forms a complex with ATM in response to DNA damage.
Fig. 4.
WWOX association with ATM. (A and B) WWOX–ATM physical association. (A) MCF7 cells were untreated or treated with NCS (200 ng/mL) for 1 h. Lysates were immunoprecipitated with anti-WWOX mAb. Anti-IgG was used as the negative control. The immunoprecipitates were analyzed by immunoblotting using antibodies against p-ATM, p-KAP1, and WWOX. (B) HEK293 cells were transfected with Myc-WWOX expression vector. Twenty-four hours later, cells were irradiated (10 Gy), and 30 min later, cells were lysed. Lysates were immunoprecipitated with anti-Myc antibody and blotted with anti–p-ATM, anti–p-KAP1, and Myc (WWOX). (C and D) WWOX–ATM functional association. (C) MEF-WT and MEF-KO cells were treated with ionizing radiation (10 Gy), and 2 h later, cells were fixed and stained for p-ATM (green) and p-H2AX (red). Nuclei were counterstained with DAPI (blue). Cells were visualized with confocal microscopy under 60× magnification. (D) WWOX depletion is associated with reduced ATM monomerization. HEK293-Sh-EV and HEK293-Sh-WWOX cells were cotransfected with Flag-ATM and YFP-ATM. At 48 h, cells were untreated or treated with NCS (200 ng/mL) for an additional 1 h. Whole-cell lysates were prepared and precleared with mouse anti-IgG followed by immunoprecipitation with anti-IgG (as negative control; lanes 1 and 4) or anti-Flag antibody overnight at 4 °C. Complexes were then washed, separated on SDS/PAGE, and analyzed by immunoblotting using antibodies against FLAG (self-immunoprecipitation) and YFP (coimmunoprecipitation). (Left) Whole-cell lysates were analyzed by immunoblotting using the indicated antibodies. GAPDH was used for normalization. IP, immunoprecipitation.
To map the WWOX–ATM interaction modules, HEK293 cells were transfected with a truncated form of GST-WWOX expressing intact or mutant WWOX, DSBs were induced, and GST pull-down assays were performed. ATM coprecipitated with the intact WW1 domain but not mutant GST-WW1-WFPA, GST-WW2, or GST-SDR, indicating that the WW1 domain of WWOX mediates the interaction of WWOX with ATM (Fig. S4A). To map the interaction interface on ATM, we synthesized truncated ATM proteins using in vitro translation and incubated them with bacterial purified GST-WWOX. The results suggest that WWOX interacts with the region spanning amino acids 1,530–2,100 (Fig. S4B). Together, these data indicate that WWOX physically interacts with the checkpoint kinase ATM.
WWOX Modulates ATM Activity.
Our findings so far suggest that WWOX interacts with ATM (Fig. 4 A and B), hence facilitating ATM proper kinase function (Figs. 2 and 3). These results prompted us to examine whether, in WWOX-deficient cells, ATM and other DDR proteins are properly recruited to DNA damage foci. Indeed, WWOX KO MEFs display reduced p-ATM recruitment to DNA damage foci upon ionizing radiation treatment compared with WT MEFs (Fig. 4C). Interestingly, the intensity of γ-H2AX is higher in KO cells, consistent with impaired DNA repair as in previous observations (Fig. 3C). Additionally, MCF7-shWWOX cells displayed reduced MDC1 and p-CHK2 recruitment to DNA damage foci (Fig. S5), which is consistent with impaired ATM function.
To analyze the molecular mechanisms by which WWOX affects ATM activation, we examined the effect of WWOX expression on ATM monomerization. ATM exists in vivo as an inactive dimer that dissociates into active monomers after DNA damage (38). To study ATM activation (monomerization), HEK293T control and WWOX-depleted cells expressing Flag-tagged ATM and YFP-tagged ATM were treated with NCS, and the ATM dimer was analyzed by immunoprecipitation using anti-Flag antibodies followed by immunoblotting with anti-YFP antibodies. As expected, NCS treatment was associated with reduced coimmunoprecipitation (dimerization) between Flag-ATM and YFP-ATM in HEK293-Sh-EV cells (Fig. 4D, lane 3 vs. lane 2), indicating proper activation (monomerization) of ATM. In contrast, WWOX-depleted cells (HEK293-Sh-WWOX) showed virtually no change in ATM dimer levels in response to NCS treatment (Fig. 4D, lane 6 vs. lane 5), suggesting impaired monomerization (activation) of ATM. Furthermore, consistent with impaired ATM activation, immunoblotting revealed reduced p-ATM and p-KAP1 levels in the WWOX-depleted cells. These findings further confirm our results showing inefficient ATM activation in WWOX-deficient cells after DNA damage and suggest a role of WWOX in ATM activation. Taken together, our results indicate that WWOX interacts with ATM and modulates its activation.
DNA Damage Triggers Nuclear Accumulation and Ubiquitination of WWOX.
WWOX has been reported to preferentially localize to the cytoplasm (39–41). Because WWOX binds ATM after DNA damage and ATM shows mainly nuclear distribution after DNA damage, we determined whether WWOX might accumulate in the nucleus in response to DNA damage. Indeed, confocal microscopy revealed time-dependent accumulation of GFP-WWOX in the cell nucleus after DSB induction (Fig. S6A).
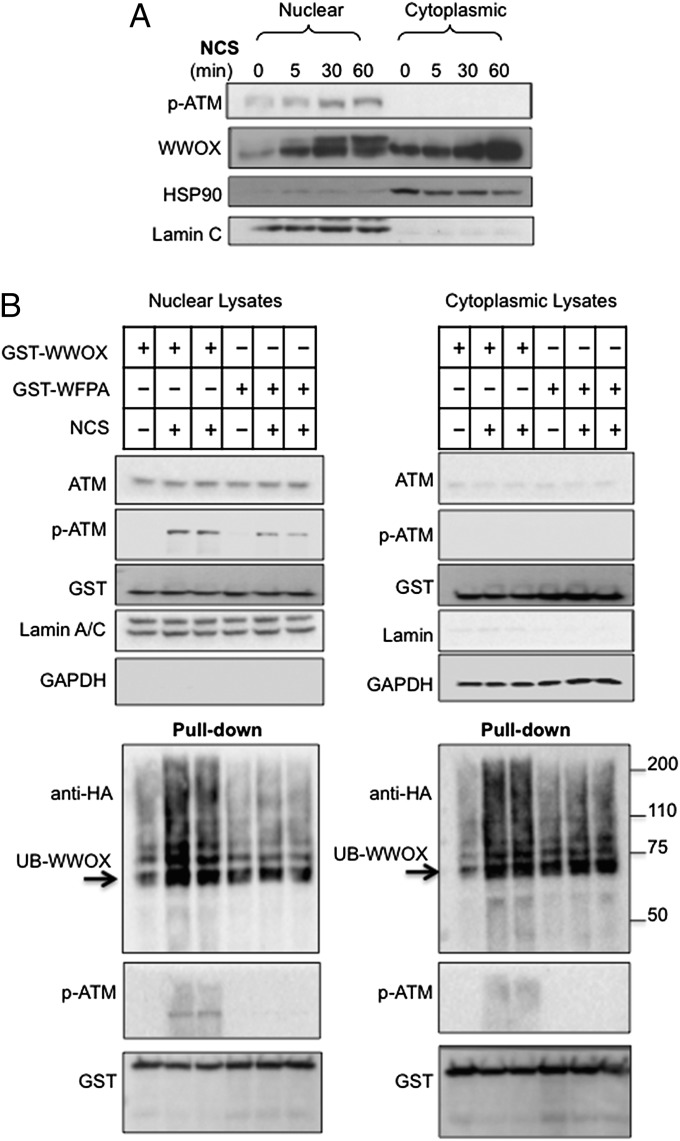
To confirm the subcellular localization of WWOX, nuclear and cytoplasmic fractions were purified from damaged and undamaged MCF7 cells and analyzed for WWOX expression. Subcellular fractionation was confirmed using HSP90 and Lamin expressions in the cytoplasmic and nuclear fractions, respectively. Upon DNA damage, as expected, general WWOX accumulation was observed. Interestingly, WWOX showed a pronounced accumulation in the nuclear fraction (Fig. 5A). Furthermore, immunoblotting indicated an additional higher molecular weight WWOX isoform in the nucleus (Fig. 5A), pointing to a specific posttranslational modification.
Fig. 5.
WWOX nuclear accumulation and ubiquitination. (A) MCF7 cells were untreated or treated with NCS (200 ng/mL). At 24 h after NCS treatment, the cells were fractionated into cytosolic and nuclear extracts. The fractionated cytosol and nuclear extracts were immunoblotted using antibodies against WWOX, HSP90, or Lamin. (B) WWOX ubiquitination after DNA damage. HEK293 cells transfected with HA-Ub and pEBG-WWOX or pEBG-WWOX-WFPA plasmids. At 24 h, cells were treated with NCS (200 ng/mL) for an additional 1 h. Cells were then subfractionated into nuclear and cytoplasmic fractions, and GST pull down was performed. Lysates were blotted against ATM, p-ATM, GST (WWOX), Lamin (nuclear fraction), GAPDH (cytoplasmic fraction), and anti-HA (Ub) antibodies. Arrows indicate ubiquitinated WWOX.
We recently reported that WWOX is the target of K63-linked polyubiquitination by the ubiquitin E3 ligase ITCH, which translocates WWOX to the nucleus (3). Thus, we tested whether the increased molecular weight is caused by WWOX ubiquitination. To this end, HEK293 cells expressing HA-Ubiquitin (HA-Ub), GST-WWOX, or GST-WWOX-WFPA were left untreated or treated with NCS to induce DSBs. GST pull down revealed that the higher molecular weight WWOX isoform is enriched and that WWOX, but not WWOX-WFPA, is modified by ubiquitin after DNA damage (Fig. S6B). In addition, treatment with UVC, a weaker inducer of DSBs, also enhanced WWOX polyubiquitination (Fig. S6B). To determine the Ub linkage of WWOX after DNA damage, HEK293 cells expressing GST-WWOX and HA-Ub, HA-Ub-K63 only, or HA-Ub-K48 only (ubiquitin mutants that are either K63 only or K48 only, whereas all other lysine residues are mutated to arginine) were treated with NCS, and lysates were subjected to GST pull down and immunoblotting analysis. The results indicate that WWOX undergoes specifically K63-linked polyubiquitination (Fig. S6C).
To determine whether WWOX ubiquitination is associated with nuclear WWOX, we performed subcellular fractionations. Nuclear GST-WWOX, but not WWOX-WFPA, was found to be polyubiquitinated (Fig. 5B). We also detected WWOX polyubiquitination in the cytoplasmic fraction, presumably because of the overexpression settings (Fig. 5B). Importantly, ATM activation (p-ATM) was reduced upon WWOX-WFPA expression compared with WT WWOX. Together, these data suggest that, after DNA damage, WWOX undergoes K63-linked polyubiquitination, facilitating its nuclear translocation.
WWOX Is Ubiquitinated at Lysine 274 on DNA Damage.
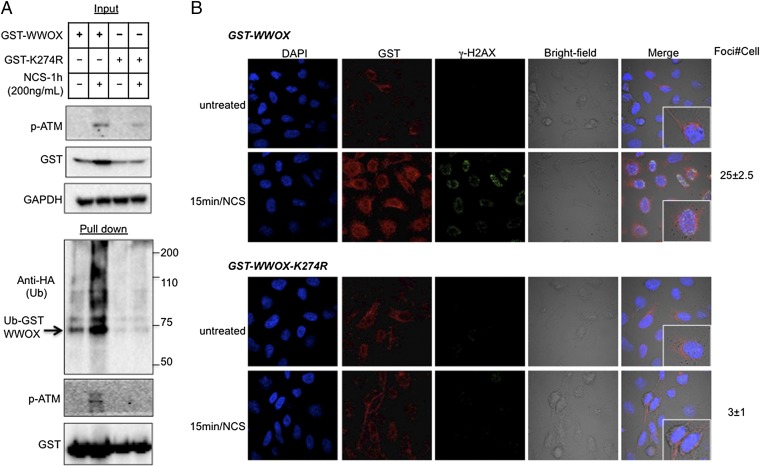
Our recent data revealed that ITCH mediates WWOX ubiquitination at K274 (3). To determine whether this lysine residue is critical for WWOX ubiquitination on DNA damage, we performed site-directed mutagenesis by replacing K274 by arginine (K274R) and examined ubiquitination after DNA damage. Indeed, K274R mutant WWOX lacked DNA damage-induced ubiquitination (Fig. 6A), identifying K274 as a main ubiquitin acceptor lysine residue on DSBs. Remarkably, expression of WWOX-K274R was associated with reduced activation of p-ATM (Fig. 6A, Upper), and reconstitution with WWOX-K274R in WWOX-depleted MCF7 cells failed to restore KAP1 activation (Fig. S7A), which is in contrast to WT WWOX (Fig. 2D). We next reasoned that the K274R mutant should be less stable compared with WT WWOX. To examine this assumption, we assessed the half-lives of WWOX and WWOX-K274R after treatment with cyclohexamide and induction of DSBs. We found, indeed, that half-life of WWOX-K274R is shorter compared with WWOX (Fig. S7B).
Fig. 6.
Ubiquitination of WWOX at Lys274 after DNA damage. (A) HEK293 cells were transfected with HA-Ub and GST-WWOX or GST-WWOX-K274R plasmids. At 24 h, cells were treated with NCS (200 ng/mL) for an additional 1 h. Cell lysates were blotted against p-ATM, GST (WWOX), and GAPDH. Pulled down complexes were blotted with anti-HA (Ub), anti-GST (WWOX), and anti–p-ATM antibodies. (B) HeLa cells were transfected with HA-Ub and GST-WWOX or GST-WWOX-K274R plasmids. At 24 h, cells were treated with NCS (200 ng/mL) for 15 min. Immunostaining was performed using anti-GST (red) and anti–p-H2AX (green). GST is also observed in control untreated cells, likely because anti-GST stains endogenous GST. DAPI was used as a marker for nuclei. Cells were examined by confocal microscopy under 60× magnification. Quantification of γ-H2AX foci is shown on the right side as the average foci number per cell ± SEM (P < 0.001).
Based on these results, we hypothesized that WWOX ubiquitination and stabilization might target it to the nucleus, where it interacts with ATM. To examine this possibility, we expressed GST-WWOX or GST-WWOX-K274R in HeLa cells, treated the cells with NCS, and analyzed WWOX localization by immunostaining. Untreated cells exclusively showed diffuse cytoplasmic WWOX staining (Fig. 6B, Upper). NCS treatment led to significant increased staining of γ-H2AX in cells expressing WT WWOX but not in WWOX-K274R–expressing cells. In addition, nuclear staining was significantly higher in cells expressing WT WWOX compared with WWOX-K274R (Fig. 6B).
We next sought to examine whether forcing WWOX-K274R to the nucleus is sufficient for ATM interaction and activation. Therefore, we fused WWOX and WWOX-K274R to a strong nuclear localization sequence to reveal WWOX-Nuc and WWOX-K274R-Nuc isoforms, respectively. Next, we determined their ubiquitination and ATM-activating functions. Remarkably, our results indicate that K274 is essential for not only ubiquitination of WWOX but also, interaction and activation of ATM (Fig. S7C).
Ub E3 Ligase ITCH Ubiquitinates WWOX After DNA Damage.
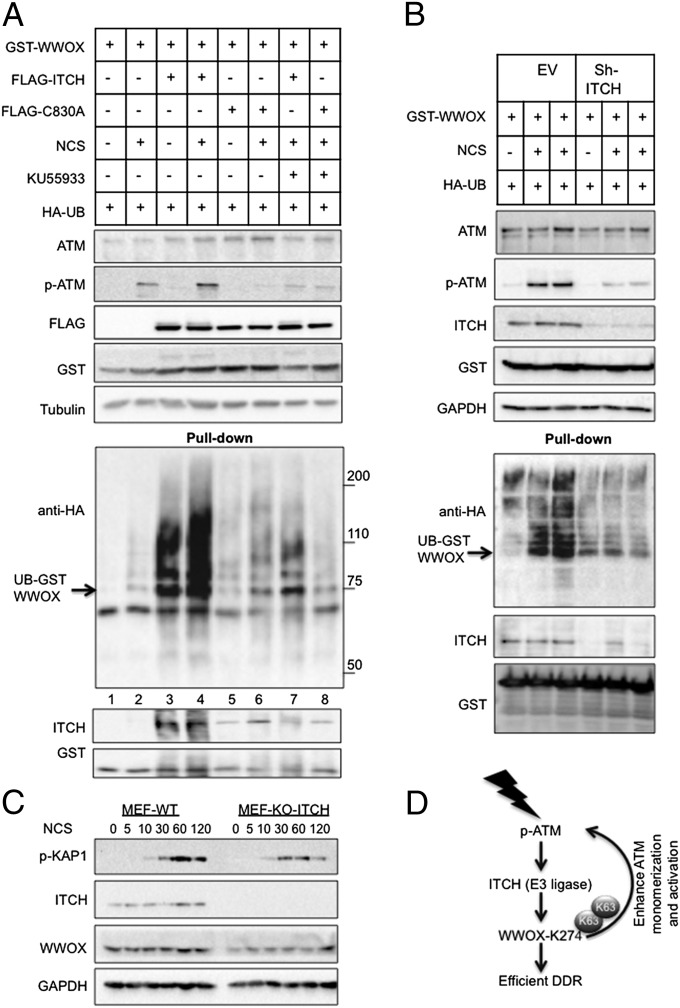
Next, we aimed to identify the Ub E3 ligase that mediates WWOX ubiquitination on DNA damage. Our recent work has identified ITCH as an E3 ligase that associates and mediates K63-linked polyubiquitination of WWOX, leading to its stabilization and nuclear translocation (3). However, the physiological context promoting this functional association remained unclear. These results prompted us to test whether ITCH ubiquitinates WWOX on DNA damage. Indeed, ectopic expression of ITCH but not a catalytic mutant mediated WWOX ubiquitination (Fig. 7A, lane 3 vs. lane 5). This effect was further enhanced after NCS treatment (Fig. 7A, lane 4). In addition, WWOX physically associated with ITCH under these conditions (Fig. 7A, Bottom), which is consistent with our recent observations (3).
Fig. 7.
ITCH ubiquitinates WWOX upon DNA damage. (A) HEK293 cells were transfected with HA-Ub, GST-WWOX, and FLAG-ITCH or FLAG-ITCHC830A plasmids. At 24 h, cells were mock or KU55933 treated overnight followed by NCS (200 ng/mL) treatment of 1 h. Cell lysates were blotted against the indicated antibodies. Pulled down complexes were blotted with anti-HA (Ub), anti-GST (WWOX), and anti-ITCH antibodies. (B) Control- and ITCH-depleted HEK293 cells were transfected with GST-WWOX and HA-Ub. At 24 h, cells were incubated with NCS (200 ng/mL) for an additional 1 h. Cell lysates and pulled down complexes were treated as in A. (C) Itch-KO and WT MEFs were treated with NCS (200 ng/mL) for the indicated times. Lysates were blotted using the indicated antibodies. (D) Hypothetical model of WWOX action in DDR. WWOX deficiency leads to an increased number of DNA strand breaks after DNA damage. After DNA damage, ATM positively enhances ITCH-mediated ubiquitination and translocation of WWOX into the nucleus. Nuclear WWOX physically interacts with ATM and mediates ATM monomerization and activation in a positive feed-forward loop manner. When WWOX is lost, ATM function is hampered.
Intriguingly, it has been recently suggested that ATM activates ITCH ubiquitin ligase activity (42). We, therefore, questioned whether ATM is required for WWOX ubiquitination. Indeed, we found that treatment with an ATM inhibitor (KU55933), attenuated WWOX-ITCH interaction and consequently, WWOX ubiquitination (Fig. 7A, lane 7). These results suggest that ATM activity is necessary to activate ITCH catalytic function.
To examine whether ITCH is required for WWOX ubiquitination after DNA damage, we analyzed the effect of ITCH depletion on WWOX ubiquitination. Remarkably, ITCH depletion resulted in decrease WWOX ubiquitination upon DNA damage (Fig. 7B).
In addition, we also used Itch-deficient and -proficient MEFs and determined the effect on the DDR. Our results revealed that Itch-KO cells display reduced p-KAP1 activation as well as reduced WWOX accumulation (Fig. 7C). Together, these results indicate an important role of ITCH in WWOX regulation on DNA damage.
Discussion
Targeted deletion of the Wwox gene in mice has previously shown that WWOX is a bona fide tumor suppressor (15, 16, 43). However, the molecular mechanism by which loss of WWOX facilitates tumorogenesis is poorly understood. In this study, we have identified an unforeseen direct function of WWOX in the DDR. Our results show that loss of WWOX results in delayed activation of the DNA damage checkpoint kinase ATM and impaired DNA repair. After DNA damage, ATM positively regulates the ligase activity of ITCH (42), which facilitates WWOX ubiquitination at Lys274 and thereby, promotes translocation of WWOX into the nucleus. Nuclear WWOX physically interacts with ATM and facilitates ATM monomerization and activation in a positive feed-forward loop manner (Fig. 7D). Although the detailed mechanism by which WWOX facilitates ATM activation remains to be elucidated, these findings argue for a direct function of WWOX in modulating DNA damage signaling.
Conflicting results were reported on WWOX expression upon exposure to DNA damage. In some reports, WWOX expression was down-regulated after ultraviolet (UV) exposure (44, 45) but not affected upon ionizing radiation (45). In contrast, Lai et al. (46) showed that WWOX levels are up-regulated upon UVB and that this induction is essential for UVB-induced apoptosis. It remained unknown whether these treatments induce genomic aberrations at the FRAD16 locus and/or loss of WWOX expression. It is possible that breaks in FRA16D, mostly affecting intron 8 in WWOX (9, 18, 47), do not affect WWOX transcription early after DNA damage. Although WWOX mRNA is up-regulated after DSBs, our findings show that posttranslational modification of the WWOX protein seems to be the predominant cause of WWOX accumulation.
Our results show that WWOX functions as an adaptor protein and is important for proper ATM activation upon DNA damage. Accordingly, WWOX depletion (hemizygous mutation) or loss (homozygous mutation) might result in reduced activation of ATM and substrate phosphorylation, including H2AX, CHK2, and KAP1. Furthermore, WWOX KO MEFs and WWOX-depleted MCF7 cells exhibited clear defects in p-ATM, γ-H2AX, MDC1, and p-CHK2 recruitment to DNA damage sites (38, 48). After longer time periods of DNA damage induction, these cells exhibited increased DSB numbers. These results are consistent with our finding that WWOX plays a role in DNA repair, suppression of genomic instability, and impaired activation of ATM. WWOX has also been shown to be essential for binding and stabilizing Ser46–p-p53, hence contributing to p53-mediated apoptosis (49) as well as enhancing p73 proapoptotic function (39). Intriguingly, the effects of WWOX on DDR signaling seem to be independent of p53, because the same consequences were observed in p53 WT (MCF7) and mutant (MDA-MB231) cells. Altogether, these results further indicate an essential role of WWOX in the DDR.
Our findings suggest that both the WWOX WW1 domain and K274 ubiquitination are important for ATM interaction and ATM activation. Interestingly, ITCH interacts with the WW1 domain of WWOX and mediates K63-linked polyubiquitination at Lys274, highlighting the significance of ITCH in the WWOX role in DDR. In fact, our results show that ITCH knockdown or genetic KO is associated with reduced WWOX ubiquitination and stabilization and thus, impaired DDR. ITCH ubiquitinates and regulates the stability of several substrates, including p73 (50), p63 (51), Gli (52), and others (53); therefore, we cannot exclude that other proteins could also contribute to its observed role in DDR. Although ITCH has been shown to function as a prosurvival factor (54), our results suggest that it can also contribute to other functions important for the neoplastic process. Future work shall be required to further delineate ITCH function in DDR and its role in tumorogenesis.
Our data show that the product of the WWOX fragile gene is directly involved in the DDR through regulating ATM checkpoint signaling and maintaining genomic stability. Similar results highlight the contribution of the FHIT gene, which encompasses FRA3B and encodes a 14-kDa protein with tumor suppressor functions (55). Deficiency of the FHIT gene as early as in preneoplastic lesions induced global genome instability and clonal expansion (56, 57). Likewise, the product of FRA15A (RORA) and FRA8I (SPIDR), which is inactivated in multiple tumors, has been shown to be involved in cellular stress response, DNA damage repair, and maintenance of chromosomal integrity (58, 59), suggesting that their impaired activity may contribute to genomic instability in cancer cells. These observations suggest that at least some products of CFSs might play a role in the DDR. Thus, one may speculate that they function as part of a highly conserved stress response network that is uniquely susceptible to genomic instability in cancer cells. Additional work will be required to decipher the exact function of other CFS gene products in cellular stress responses.
Our findings support the hypothesis that loss of WWOX provides a selective advantage in neoplastic transformation, because Wwox-deficient cells may preferentially acquire additional cancer-promoting mutations caused by an impaired DDR. In fact, Ras-mediated transformation of Wwox-KO MEFs results in an increased number of tumorigenic cells compared with WT cells, suggesting that WWOX deficiency facilitates transformation (60). Deletion or attenuation of WWOX is observed in early preneoplastic breast lesions (61, 62) and targeted ablation of murine Wwox-associated mammary tumor formation (16). It is, thus, likely that loss of WWOX at early stages of neoplasia acts as a driver of the transformation process through compromising genome stability.
Materials and Methods
Cell Culture and Transient Transfection.
HEK293, HeLa, LM7, MDA-MB231, and MEF cells were grown in DMEM, whereas MCF7 and U2OS cells were grown in RPMI. Both media were supplemented with 10% (vol/vol) FBS (Gibco), glutamine, and penicillin/streptomycin (Beit-Haemek). Cells were routinely authenticated, and cell aliquots from early passages were used. Transient transfections were achieved using Mirus TransLTi (Mirus Bio LLC) according to the manufacturer.
RNA Extraction, RT-PCR, and Real-Time PCR.
Total RNA was prepared using TRI reagent (Sigma Aldrich) as described by the manufacturer. One microgram RNA was used for cDNA synthesis using the First-Strand cDNA Synthesis Kit (Bio-Rad). Quantitative real-time PCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems). All measurements were performed in triplicate and standardized to the levels of GAPDH.
Comet Assay.
Comet assay was performed according to the manufacturer’s instructions (Trevigen Inc.). In brief, MCF7 and WWOX-depleted MCF7 cells were treated with NCS for 1 h, and then, cells were analyzed for DNA breaks by assessing the comet tail moment (product of tail length and relative tail content of DNA).
Cellular Fractionation.
Nuclear and cytoplasmic extracts were prepared as follows. First, cells were scraped in PBS, and after centrifugation, the cell pellet was reconstituted in a hypotonic lysis buffer (10 mmol/L Hepes, pH 7.9, 10 mmol/L KCl, 0.1 mmol/L EDTA) supplemented with 1 mmol/L DTT and a broad spectrum mixture of protease inhibitors (Sigma-Aldrich). The cells were allowed to swell on ice for 15 min; then, 0.5% Nonidet P-40 was added, and cells were lysed by vortex. After centrifugation, the cytoplasmic fraction was collected. Afterward, nuclear extracts were obtained by incubating nuclei in a hypertonic nuclear extraction buffer (20 mmol/L Hepes, pH 7.9, 0.42 mol/L KCl, 1 mmol/L EDTA) supplemented with 1 mmol/L DTT for 15 min at 4 °C. The nuclear fraction was collected after centrifugation.
Micro-HR Assay.
MCF7 cells were transfected with a microhomologous DNA damage repair reporter, pGL2-Luc vector, linearized with either HindIII or EcoRI. Forty-eight hours posttransfection, the cells were collected, and luciferase activity was quantified. Because EcoRI cuts within the coding sequence of the luciferase gene, the restoration of luciferase activity, therefore, requires the precise rejoining of the short protruding ends, which involves microhomologous DNA damage repair. However, because HindIII cuts within the linker region between the SV40 promoter and the Luc coding sequence, the restoration of luciferase activity does not require precise end joining. Luciferase activity after the cut with HindIII was used for normalization.
HR Assay.
The assay was established and modified by Weinstock et al. (37). U2OS cells stabilized with a single copy of DR-GFP were transduced with EV or Lenti-WWOX constructs. Stable clones were generated after G418 selection. Cells were transfected with I-SecI plasmid. Cells were harvested 2 or 3 d after infection and subjected to flow cytometric analysis. The GFP-positive cell population was measured.
Immunofluorescence.
Cells were seeded on round slide coverslips in 12-well plates. Twenty-four hours later, cells were transfected with the indicated expression plasmids. Twenty-four hours posttransfection, cells were treated with a specific DNA damaging agent for a specific time point; then, cells were fixed in 3.7% PBS-buffered formaldehyde and permeabilized with 0.05% Triton X-100 at room temperature. Cells were then incubated for 1 h in 10% goat serum (Invitrogen), primary antibody for 1 h, and secondary antibody for 30 min. Cells were examined by confocal microscopy (Olympus) under 60× magnification.
GST Pull Down.
Mammalian.
HEK293 cells were transfected with expression vectors encoding mammalian GST-WWOX (pEBG-WWOX), GST-WWOX-WFPA [where tryptophan (W44) and proline (P47) in the first WW domain of WWOX were replaced by phenylalanine (F) and alanine (A), respectively], or GST-WWOX-K274R [where lysine (K274) was replaced by arginine (R)]. Twenty-four hours later, cells were exposed to DNA damage and then lysed using 0.5% Nonidet P-40–containing buffer. After washing, cells were incubated with GST beads (Amersham) for 4 h at 4 °C, washed, and then, prepared for electrophoresis.
Bacterial.
Escherichia coli BL21 (DE3) was transformed with pGEX6.1-WWOX. After 4 h of induction with 1 mM isopropyl β-d-1-thiogalactopyranoside at 37 °C, bacteria were harvested. GST and GST-WWOX were purified with Fast Flow Glutathione-Sepharose (GE Healthcare) according to the manufacturer’s protocol. Then, bead-bound proteins were equilibrated in buffer of 20 mM Tris⋅Cl (pH 7.9), 5 mM MgCl2, 0.1 mM EDTA, 0.5 mM EGTA, 10% Glycerol, 250 mM KCl, and 0.2% Nonidet P-40 (AM250 buffer).
ATM deletion constructs were in vitro-transcribed and translated, including 35S-Methionine, according to the manufacturer’s protocol (TNT Coupled Reticulocyte Lysate System; Promega). TNT lysates and bead-bound GST-fusion proteins were incubated in AM250 buffer for 1 h at 4 °C and subsequently washed four times with AM250. Finally, the beads were boiled in 1× Laemmli buffer.
Statistical Analysis.
Results were expressed as means ± SDs. Student t test was used to compare values of test and control samples. P < 0.05 indicated significant difference.
Additional laboratory reagents used are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Eugenio Gaudio and Dr. Sonja Matt for technical help, Dr. Yossi Shiloh for the ataxia telangiectasia-mutated inhibitor, and Dr. Kay Huebner for the rabbit polyclonal WW domain-containing oxidoreductase antibody. This study was supported by a German Israeli Foundation Joint Grant (to T.G.H. and R.I.A.), Israeli Cancer Research Funds (to Z.S. and R.I.A.), Deutsche Forschungsgemeinschaft Grant SFB1036 (to T.G.H.), and the Deutsche Krebshilfe (T.G.H.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a Prearranged Editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1409252111/-/DCSupplemental.
References
- 1.Bartek J, Lukas J. DNA damage checkpoints: From initiation to recovery or adaptation. Curr Opin Cell Biol. 2007;19(2):238–245. doi: 10.1016/j.ceb.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Harper JW, Elledge SJ. The DNA damage response: Ten years after. Mol Cell. 2007;28(5):739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 3.Abu-Odeh M, et al. Characterizing WW domain interactions of tumor suppressor WWOX reveals its association with multiprotein networks. J Biol Chem. 2014;289(13):8865–8880. doi: 10.1074/jbc.M113.506790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shiloh Y. ATM and ATR: Networking cellular responses to DNA damage. Curr Opin Genet Dev. 2001;11(1):71–77. doi: 10.1016/s0959-437x(00)00159-3. [DOI] [PubMed] [Google Scholar]
- 5.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434(7033):605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 6.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability—an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11(3):220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 7.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 8.Bednarek AK, et al. WWOX, a novel WW domain-containing protein mapping to human chromosome 16q23.3-24.1, a region frequently affected in breast cancer. Cancer Res. 2000;60(8):2140–2145. [PubMed] [Google Scholar]
- 9.Ried K, et al. Common chromosomal fragile site FRA16D sequence: Identification of the FOR gene spanning FRA16D and homozygous deletions and translocation breakpoints in cancer cells. Hum Mol Genet. 2000;9(11):1651–1663. doi: 10.1093/hmg/9.11.1651. [DOI] [PubMed] [Google Scholar]
- 10.Del Mare S, Salah Z, Aqeilan RI. WWOX: Its genomics, partners, and functions. J Cell Biochem. 2009;108(4):737–745. doi: 10.1002/jcb.22298. [DOI] [PubMed] [Google Scholar]
- 11.Salah Z, Aqeilan R, Huebner K. WWOX gene and gene product: Tumor suppression through specific protein interactions. Future Oncol. 2010;6(2):249–259. doi: 10.2217/fon.09.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283):899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bignell GR, et al. Signatures of mutation and selection in the cancer genome. Nature. 2010;463(7283):893–898. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurek KC, et al. Frequent attenuation of the WWOX tumor suppressor in osteosarcoma is associated with increased tumorigenicity and aberrant RUNX2 expression. Cancer Res. 2010;70(13):5577–5586. doi: 10.1158/0008-5472.CAN-09-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aqeilan RI, et al. Targeted deletion of Wwox reveals a tumor suppressor function. Proc Natl Acad Sci USA. 2007;104(10):3949–3954. doi: 10.1073/pnas.0609783104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdeen SK, et al. Wwox inactivation enhances mammary tumorigenesis. Oncogene. 2011;30(36):3900–3906. doi: 10.1038/onc.2011.115. [DOI] [PubMed] [Google Scholar]
- 17.Chang NS, Hsu LJ, Lin YS, Lai FJ, Sheu HM. WW domain-containing oxidoreductase: A candidate tumor suppressor. Trends Mol Med. 2007;13(1):12–22. doi: 10.1016/j.molmed.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 18.Aqeilan RI, et al. Targeted ablation of the WW domain-containing oxidoreductase tumor suppressor leads to impaired steroidogenesis. Endocrinology. 2009;150(3):1530–1535. doi: 10.1210/en.2008-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durkin SG, Glover TW. Chromosome fragile sites. Annu Rev Genet. 2007;41:169–192. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- 20.Glover TW. Common fragile sites. Cancer Lett. 2006;232(1):4–12. doi: 10.1016/j.canlet.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 21.Tsantoulis PK, et al. Oncogene-induced replication stress preferentially targets common fragile sites in preneoplastic lesions. A genome-wide study. Oncogene. 2008;27(23):3256–3264. doi: 10.1038/sj.onc.1210989. [DOI] [PubMed] [Google Scholar]
- 22.Bartkova J, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434(7035):864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 23.Gorgoulis VG, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434(7035):907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 24.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319(5868):1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 25.Debatisse M, Le Tallec B, Letessier A, Dutrillaux B, Brison O. Common fragile sites: Mechanisms of instability revisited. Trends Genet. 2012;28(1):22–32. doi: 10.1016/j.tig.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 26.Ozeri-Galai E, Bester AC, Kerem B. The complex basis underlying common fragile site instability in cancer. Trends Genet. 2012;28(6):295–302. doi: 10.1016/j.tig.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 27.Le Tallec B, et al. Molecular profiling of common fragile sites in human fibroblasts. Nat Struct Mol Biol. 2011;18(12):1421–1423. doi: 10.1038/nsmb.2155. [DOI] [PubMed] [Google Scholar]
- 28.Le Tallec B, et al. Common fragile site profiling in epithelial and erythroid cells reveals that most recurrent cancer deletions lie in fragile sites hosting large genes. Cell Reports. 2013;4(3):420–428. doi: 10.1016/j.celrep.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 29.Hosseini SA, et al. Common chromosome fragile sites in human and murine epithelial cells and FHIT/FRA3B loss-induced global genome instability. Genes Chromosomes Cancer. 2013;52(11):1017–1029. doi: 10.1002/gcc.22097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dereli-Öz A, Versini G, Halazonetis TD. Studies of genomic copy number changes in human cancers reveal signatures of DNA replication stress. Mol Oncol. 2011;5(4):308–314. doi: 10.1016/j.molonc.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shiloh Y, Ziv Y. The ATM protein kinase: Regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14(4):197–210. [PubMed] [Google Scholar]
- 32.Salah Z, Alian A, Aqeilan RI. WW domain-containing proteins: Retrospectives and the future. Front Biosci (Landmark Ed) 2012;17:331–348. doi: 10.2741/3930. [DOI] [PubMed] [Google Scholar]
- 33.Su TT. Cellular responses to DNA damage: One signal, multiple choices. Annu Rev Genet. 2006;40:187–208. doi: 10.1146/annurev.genet.40.110405.090428. [DOI] [PubMed] [Google Scholar]
- 34.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273(10):5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 35.Kuo LJ, Yang LX. Gamma-H2AX - a novel biomarker for DNA double-strand breaks. In Vivo. 2008;22(3):305–309. [PubMed] [Google Scholar]
- 36.Wang RH, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14(4):312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weinstock DM, Nakanishi K, Helgadottir HR, Jasin M. Assaying double-strand break repair pathway choice in mammalian cells using a targeted endonuclease or the RAG recombinase. Methods Enzymol. 2006;409:524–540. doi: 10.1016/S0076-6879(05)09031-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bakkenist CJ, Kastan MB. Initiating cellular stress responses. Cell. 2004;118(1):9–17. doi: 10.1016/j.cell.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 39.Aqeilan RI, et al. Functional association between Wwox tumor suppressor protein and p73, a p53 homolog. Proc Natl Acad Sci USA. 2004;101(13):4401–4406. doi: 10.1073/pnas.0400805101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aqeilan RI, et al. WW domain-containing proteins, WWOX and YAP, compete for interaction with ErbB-4 and modulate its transcriptional function. Cancer Res. 2005;65(15):6764–6772. doi: 10.1158/0008-5472.CAN-05-1150. [DOI] [PubMed] [Google Scholar]
- 41.Aqeilan RI, et al. Physical and functional interactions between the Wwox tumor suppressor protein and the AP-2gamma transcription factor. Cancer Res. 2004;64(22):8256–8261. doi: 10.1158/0008-5472.CAN-04-2055. [DOI] [PubMed] [Google Scholar]
- 42.Santini S, et al. ATM kinase activity modulates ITCH E3-ubiquitin ligase activity. Oncogene. 2014;33(9):1113–1123. doi: 10.1038/onc.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aqeilan RI, et al. Inactivation of the Wwox gene accelerates forestomach tumor progression in vivo. Cancer Res. 2007;67(12):5606–5610. doi: 10.1158/0008-5472.CAN-07-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ishii H, et al. Components of DNA damage checkpoint pathway regulate UV exposure-dependent alterations of gene expression of FHIT and WWOX at chromosome fragile sites. Mol Cancer Res. 2005;3(3):130–138. doi: 10.1158/1541-7786.MCR-04-0209. [DOI] [PubMed] [Google Scholar]
- 45.Thavathiru E, Ludes-Meyers JH, MacLeod MC, Aldaz CM. Expression of common chromosomal fragile site genes, WWOX/FRA16D and FHIT/FRA3B is downregulated by exposure to environmental carcinogens, UV, and BPDE but not by IR. Mol Carcinog. 2005;44(3):174–182. doi: 10.1002/mc.20122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lai FJ, et al. WOX1 is essential for UVB irradiation-induced apoptosis and down-regulated via translational blockade in UVB-induced cutaneous squamous cell carcinoma in vivo. Clin Cancer Res. 2005;11(16):5769–5777. doi: 10.1158/1078-0432.CCR-04-2274. [DOI] [PubMed] [Google Scholar]
- 47.Paige AJ, et al. WWOX: A candidate tumor suppressor gene involved in multiple tumor types. Proc Natl Acad Sci USA. 2001;98(20):11417–11422. doi: 10.1073/pnas.191175898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stucki M, Jackson SP. gammaH2AX and MDC1: Anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair (Amst) 2006;5(5):534–543. doi: 10.1016/j.dnarep.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 49.Chang NS, et al. WOX1 is essential for tumor necrosis factor-, UV light-, staurosporine-, and p53-mediated cell death, and its tyrosine 33-phosphorylated form binds and stabilizes serine 46-phosphorylated p53. J Biol Chem. 2005;280(52):43100–43108. doi: 10.1074/jbc.M505590200. [DOI] [PubMed] [Google Scholar]
- 50.Rossi M, et al. The ubiquitin-protein ligase Itch regulates p73 stability. EMBO J. 2005;24(4):836–848. doi: 10.1038/sj.emboj.7600444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rossi M, et al. The E3 ubiquitin ligase Itch controls the protein stability of p63. Proc Natl Acad Sci USA. 2006;103(34):12753–12758. doi: 10.1073/pnas.0603449103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Di Marcotullio L, et al. Numb activates the E3 ligase Itch to control Gli1 function through a novel degradation signal. Oncogene. 2011;30(1):65–76. doi: 10.1038/onc.2010.394. [DOI] [PubMed] [Google Scholar]
- 53.Melino G, et al. Itch: A HECT-type E3 ligase regulating immunity, skin and cancer. Cell Death Differ. 2008;15(7):1103–1112. doi: 10.1038/cdd.2008.60. [DOI] [PubMed] [Google Scholar]
- 54.Salah Z, Melino G, Aqeilan RI. Negative regulation of the Hippo pathway by E3 ubiquitin ligase ITCH is sufficient to promote tumorigenicity. Cancer Res. 2011;71(5):2010–2020. doi: 10.1158/0008-5472.CAN-10-3516. [DOI] [PubMed] [Google Scholar]
- 55.Huebner K, Croce CM. FRA3B and other common fragile sites: The weakest links. Nat Rev Cancer. 2001;1(3):214–221. doi: 10.1038/35106058. [DOI] [PubMed] [Google Scholar]
- 56.Miuma S, et al. Fhit deficiency-induced global genome instability promotes mutation and clonal expansion. PLoS ONE. 2013;8(11):e80730. doi: 10.1371/journal.pone.0080730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saldivar JC, et al. Initiation of genome instability and preneoplastic processes through loss of Fhit expression. PLoS Genet. 2012;8(11):e1003077. doi: 10.1371/journal.pgen.1003077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33(3):275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 59.Wan L, et al. Scaffolding protein SPIDR/KIAA0146 connects the Bloom syndrome helicase with homologous recombination repair. Proc Natl Acad Sci USA. 2013;110(26):10646–10651. doi: 10.1073/pnas.1220921110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abu-Remaileh M, Aqeilan RI. Tumor suppressor WWOX regulates glucose metabolism via HIF1α modulation. Cell Death Differ. 2014;21(11):1805–1814. doi: 10.1038/cdd.2014.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guler G, et al. Concordant loss of fragile gene expression early in breast cancer development. Pathol Int. 2005;55(8):471–478. doi: 10.1111/j.1440-1827.2005.01855.x. [DOI] [PubMed] [Google Scholar]
- 62.Nunez MI, et al. Frequent loss of WWOX expression in breast cancer: Correlation with estrogen receptor status. Breast Cancer Res Treat. 2005;89(2):99–105. doi: 10.1007/s10549-004-1474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.