Two Arabidopsis membrane peroxins contribute to peroxisomal matrix protein import via distinct roles in recycling and degrading a matrix receptor protein.
Abstract
Peroxisomal matrix proteins carry peroxisomal targeting signals (PTSs), PTS1 or PTS2, and are imported into the organelle with the assistance of peroxin (PEX) proteins. From a microscopy-based screen to identify Arabidopsis (Arabidopsis thaliana) mutants defective in matrix protein degradation, we isolated unique mutations in PEX2 and PEX10, which encode ubiquitin-protein ligases anchored in the peroxisomal membrane. In yeast (Saccharomyces cerevisiae), PEX2, PEX10, and a third ligase, PEX12, ubiquitinate a peroxisome matrix protein receptor, PEX5, allowing the PEX1 and PEX6 ATP-hydrolyzing enzymes to retrotranslocate PEX5 out of the membrane after cargo delivery. We found that the pex2-1 and pex10-2 Arabidopsis mutants exhibited defects in peroxisomal physiology and matrix protein import. Moreover, the pex2-1 pex10-2 double mutant exhibited severely impaired growth and synergistic physiological defects, suggesting that PEX2 and PEX10 function cooperatively in the wild type. The pex2-1 lesion restored the unusually low PEX5 levels in the pex6-1 mutant, implicating PEX2 in PEX5 degradation when retrotranslocation is impaired. PEX5 overexpression altered pex10-2 but not pex2-1 defects, suggesting that PEX10 facilitates PEX5 retrotranslocation from the peroxisomal membrane. Although the pex2-1 pex10-2 double mutant displayed severe import defects of both PTS1 and PTS2 proteins into peroxisomes, both pex2-1 and pex10-2 single mutants exhibited clear import defects of PTS1 proteins but apparently normal PTS2 import. A similar PTS1-specific pattern was observed in the pex4-1 ubiquitin-conjugating enzyme mutant. Our results indicate that Arabidopsis PEX2 and PEX10 cooperate to support import of matrix proteins into plant peroxisomes and suggest that some PTS2 import can still occur when PEX5 retrotranslocation is slowed.
Peroxisomes are dynamic organelles housing critical oxidative metabolic reactions while sequestering harmful reactive oxygen species from the rest of the cell. In Arabidopsis (Arabidopsis thaliana), these single membrane-bound organelles are the sole site of β-oxidation and are essential for normal development (for review, see Hu et al., 2012). Triacylglycerols stored in seeds are cleaved by lipases during germination, and the released fatty acids are β-oxidized in peroxisomes to provide energy for early seedling development (for review, see Graham, 2008). Similarly, an auxin precursor, indole-3-butyric acid (IBA), is β-oxidized to active indole-3-acetic acid (IAA) in peroxisomes (Zolman et al., 2000, 2007, 2008; Strader et al., 2010; Strader and Bartel, 2011; Strader et al., 2011). IBA application enhances rooting in many plants (Woodward and Bartel, 2005a), and IAA produced from endogenous IBA promotes hypocotyl elongation, cotyledon expansion, root hair elongation, and lateral root proliferation in Arabidopsis seedlings (Zolman et al., 2001; Strader et al., 2010, 2011; De Rybel et al., 2012).
The enzymes needed for β-oxidation and other peroxisomal processes are imported into the peroxisome matrix from their site of synthesis in the cytosol using proteins known as peroxin (PEX) proteins. PEX5 and PEX7 are receptors that recognize peroxisomal targeting signals (PTSs) on proteins destined for the peroxisome matrix. PEX5 recognizes a three-amino acid C-terminal PTS1 (Keller et al., 1987), and PEX7 recognizes a nine-amino acid PTS2 located near the N terminus of the protein (Osumi et al., 1991; Swinkels et al., 1991). In plants, DEG15, a peroxisomal protease, cleaves the N-terminal PTS2 region after the protein enters the peroxisome (Helm et al., 2007; Schuhmann et al., 2008). Cargo-bound PEX5 and PEX7 associate with PEX13 and PEX14 on the peroxisomal membrane (for review, see Azevedo and Schliebs, 2006; Williams and Distel, 2006) and release their cargo into the peroxisome (Fig. 1A). In yeast (Saccharomyces cerevisiae), membrane-associated PEX5 is ubiquitinated, recognized by a complex of ATP-hydrolyzing enzymes comprised of PEX1 and PEX6, and retrotranslocated out of the peroxisome to be used in additional rounds of import (for review, see Fujiki et al., 2012; Grimm et al., 2012).
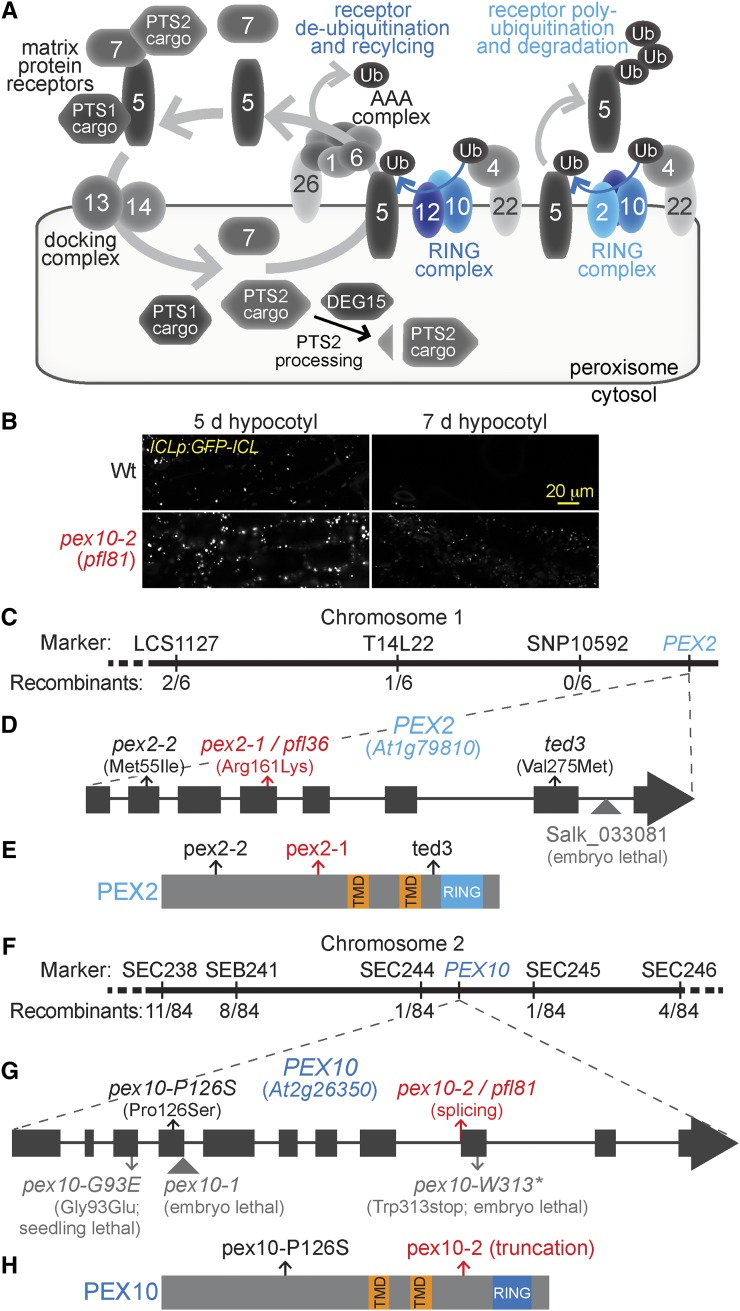
Figure 1.
Recombination mapping of pfl36 and pfl81 reveals mutations in PEX2 and PEX10. A, PEX proteins (numbered) implicated in matrix protein import serve as receptors (PEX5 and PEX7) recognizing PTS1 or PTS2 cargo proteins, dock receptors at the peroxisomal membrane (PEX13 and PEX14), or assist in PEX5 retrotranslocation (for review, see Hu et al., 2012). In yeast, the RING-finger proteins PEX2, PEX10, and PEX12 participate as heterooligomers in different modes of PEX5 ubiquitination (Ub; for review, see Platta et al., 2013). B, GFP-ICL fluorescence is detected in both 5- and 7-d-old pex10-2 (pfl81) seedlings carrying ICLp:GFP-ICL, whereas GFP-ICL is easily detected in 5- but not 7-d-old wild-type (Wt) ICLp:GFP-ICL seedlings. Hypocotyls of light-grown seedlings were imaged for GFP fluorescence using confocal microscopy. Bar = 20 µm. C, pfl36 was mapped to the bottom of chromosome 1 near the PEX2 gene using the phenotypes of prolonged GFP-ICL fluorescence accompanied by PMDH processing defects. The number of recombinants over the number of chromosomes scored is indicated for each marker assayed. D, A gene diagram of PEX2 depicting exons as rectangles and introns as lines. A missense mutation in the fourth exon of PEX2 in pfl36 (pex2-1) changes Arg161 to Lys. Three other pex2 alleles are indicated: pex2-2, ted3 (Hu et al., 2002), and the transfer DNA insertion allele Salk_033081 that confers embryo lethality (Hu et al., 2002). E, The locations of the lesions in viable pex2 alleles are indicated on a diagram depicting the PEX2 protein domains, which include two predicted transmembrane domains (TMDs) and a C-terminal RING domain. F, pfl81 was mapped using the IBA resistance phenotype to an interval on the lower arm of chromosome 2 that contained the PEX10 gene. The number of recombinants over the number of chromosomes scored is indicated for each marker assayed. G, pfl81 (pex10-2) carries a PEX10 splicing mutation in the last nucleotide of intron 8. Four other reported pex10 mutants are indicated on the gene diagram: the pex10-1 transfer DNA insertion allele (Schumann et al., 2003; Sparkes et al., 2003) and three Targeting Induced Local Lesions In Genomes (TILLING) alleles: pex10-G93E, pex10-P126S, and pex10-W313* (Prestele et al., 2010). H, The locations of the lesions in the two viable pex10 alleles are indicated on a diagram depicting the PEX10 protein domains, which include two predicted TMDs and a C-terminal RING domain.
Yeast PEX5 ubiquitination involves the peroxisomal membrane ubiquitin-protein ligases PEX2, PEX10, and PEX12 (for review, see Platta et al., 2013). The PEX12 ubiquitin-protein ligase monoubiquitinates PEX5 with the assistance of the ubiquitin-conjugating enzyme PEX4 (Platta et al., 2009), allowing PEX5 to be recycled back to the cytosol (Fig. 1A). When PEX4 is absent, yeast ubiquitin-conjugating enzyme4 (Ubc4) works with PEX2 to polyubiquitinate PEX5, marking PEX5 for proteasomal degradation (for review, see Thoms and Erdmann, 2006; Platta et al., 2007, 2013). In yeast, the Really Interesting New Gene (RING) domain of PEX10 binds both PEX2 and PEX12 RING domains to form a trimer (El Magraoui et al., 2012). PEX10 enhances in vitro ubiquitination activity of both PEX2-Ubc4 and PEX12-PEX4 (El Magraoui et al., 2012). Similarly, mammalian PEX12 enhances the in vitro ubiquitination activity of PEX10 (Okumoto et al., 2014). These findings suggest that these RING-finger proteins might act in heteromeric pairs to polyubiquitinate or monoubiquitinate PEX5 (Fig. 1A).
Arabidopsis PEX2, PEX10, and PEX12 each display zinc-dependent monoubiquitination activity in vitro (Kaur et al., 2013), but the comparative functions of the Arabidopsis RING-finger PEX proteins in PEX5 ubiquitination, recycling, and degradation have not been reported. This deficiency may, in part, reflect the fact that null alleles of the RING-finger PEX genes confer embryo lethality (Hu et al., 2002; Schumann et al., 2003; Sparkes et al., 2003; Fan et al., 2005; Prestele et al., 2010). RNA interference (RNAi) lines targeting PEX2, PEX10, or PEX12 inefficiently import matrix proteins, display the Suc dependence phenotype that typically accompanies fatty acid β-oxidation defects, and are resistant to 2,4-dichlorophenoxybutyric acid (Fan et al., 2005; Nito et al., 2007), a synthetic analog of IBA (Hayashi et al., 1998). Mutation of any one RING-finger PEX gene results in disassociation or reduced levels of the PEX2-PEX10-PEX12 complex in yeast (Hazra et al., 2002; Agne et al., 2003) and mammals (Okumoto et al., 2014). It is not known whether the defects of the Arabidopsis null and RNAi lines result directly from the loss of catalytic activity of the corresponding RING-finger protein or indirectly from destabilization of the complex and consequent loss of activity of one or both of the associated RING-finger PEX proteins.
Only one mutant defective in a RING-finger PEX gene has emerged from forward genetic screens for peroxisome defects in Arabidopsis. A partial loss-of-function pex12 missense allele, aberrant peroxisome morphology4 (apm4), was isolated from a GFP-PTS1 mislocalization screen (Mano et al., 2006). In addition to partially cytosolic GFP-PTS1, apm4 displays a PTS2 processing defect, Suc dependence, and 2,4-dichlorophenoxybutyric acid resistance (Mano et al., 2006), suggesting that PEX12 facilitates peroxisome protein import.
In addition to roles in matrix protein import suggested by analysis of RNAi lines (Nito et al., 2007), PEX2 and PEX10 may have plant-specific roles. A pex2 missense allele, ted3, was identified as a dominant suppressor of the photomorphogenic defects of the de-etiolated1 mutant and carries a mutation near the PEX2 RING-finger domain (Fig. 1, D and E; Supplemental Fig. S1; Hu et al., 2002). Moreover, PEX10 RNAi lines display pleiotropic phenotypes not commonly found in Arabidopsis pex mutants, including variegated leaves, reduced fertility (Nito et al., 2007), organ fusions, reduced cuticular wax deposition, and changes in endoplasmic reticulum structure (Kamigaki et al., 2009). Three pex10 alleles generated by TILLING have been reported (Fig. 1G; Supplemental Fig. S2): pex10-W313* truncates the RING-finger domain and is embryo lethal, pex10-G93E germinates but displays seedling lethality, and pex10-P126S displays reduced growth in both normal air and high CO2 conditions (Prestele et al., 2010). Although GFP-PTS1 is localized in peroxisomes in the pex10-P126S mutant, Suc dependence and IBA resistance were not reported (Prestele et al., 2010).
The consequences of overexpressing a mutant version of PEX10 carrying missense mutations in the RING-finger domain (ΔZn) in wild-type Arabidopsis also hint at plant-specific roles for PEX10. PEX10-ΔZn expression confers pleiotropic phenotypes, including smaller cells, serrated leaves, inefficient photorespiration, abnormal peroxisome size and shape, and reduced peroxisome-chloroplast association (Prestele et al., 2010). However, PEX10-ΔZn plants respond like the wild type to IBA and do not require Suc, suggesting that peroxisome metabolism is not dramatically impaired (Schumann et al., 2007; Prestele et al., 2010). In contrast, expressing PEX2-ΔZn in the wild type impairs GFP-PTS1 import without conferring morphological defects, and expressing PEX12-ΔZn confers no abnormal phenotypes (Prestele et al., 2010).
Here, we describe the identification and characterization of two unique mutants carrying lesions in Arabidopsis RING-finger PEX genes. We isolated pex2-1 and pex10-2 in a forward genetic screen for genes promoting peroxisomal matrix protein degradation and used these mutants to explore the roles of the corresponding proteins in peroxisome function. We found that PEX2 and PEX10 have independent but related functions that together support PEX5 recycling and matrix protein import into plant peroxisomes.
RESULTS
Identification of pex2 and pex10 Mutants
We isolated pex2 and pex10 mutants from a screen designed to identify proteins involved in peroxisomal matrix protein degradation (Burkhart et al., 2013). This microscopy-based screen exploits the peroxisome content remodeling that occurs during seedling photomorphogenesis. The glyoxylate cycle enzyme isocitrate lyase (ICL) is present during germination and early development but degraded as photosynthesis is established (Zolman et al., 2005; Lingard et al., 2009). We screened ethyl methanesulfonate-mutagenized seedlings expressing GFP-ICL driven by the ICL promoter for persistent GFP-ICL fluorescent (pfl) mutants, which retained GFP-ICL longer than the wild type (Fig. 1B), suggesting defects in peroxisomal matrix protein degradation (Burkhart et al., 2013).
We mapped the causal lesion in pfl36 to a region at the bottom of chromosome 1 that contained PEX2 (At1g79810; Fig. 1C). Sequencing PEX2 from pfl36 genomic DNA revealed a missense mutation in exon 4 that caused an amino acid change (Arg161Lys; Fig. 1D). The altered Arg residue is in a predicted cytosolic domain before the first predicted PEX2 transmembrane domain (Fig. 1E) and conserved among PEX2 isoforms in plants (Supplemental Fig. S1). We renamed pfl36 as pex2-1.
We mapped the pfl81 lesion to a region on the lower arm of chromosome 2 that contained PEX10 (At2g26350; Fig. 1F). Sequencing PEX10 from pfl81 genomic DNA revealed a G-to-A mutation of the last nucleotide of intron 8 that is predicted to disrupt PEX10 splicing (Fig. 1G). We isolated RNA from pfl81 seedlings and analyzed PEX10 splicing using reverse transcription (RT)-PCR followed by DNA sequencing. This analysis revealed two predominant PEX10 transcripts in pfl81 seedlings. One transcript retained intron 8, which if translated would result in 28 amino acids encoded by intron 8 followed by a stop codon. The second transcript was spliced one nucleotide after the wild-type splice acceptor site, which if translated would result in six out-of-frame amino acids followed by a stop codon. Either mutant transcript would encode a pex10 protein that was truncated before the RING-finger domain (Fig. 1H; Supplemental Fig. S2). pfl81 was renamed pex10-2.
Peroxisome-Related Defects in Peroxisomal RING-Finger Ubiquitin-Protein Ligase Mutants
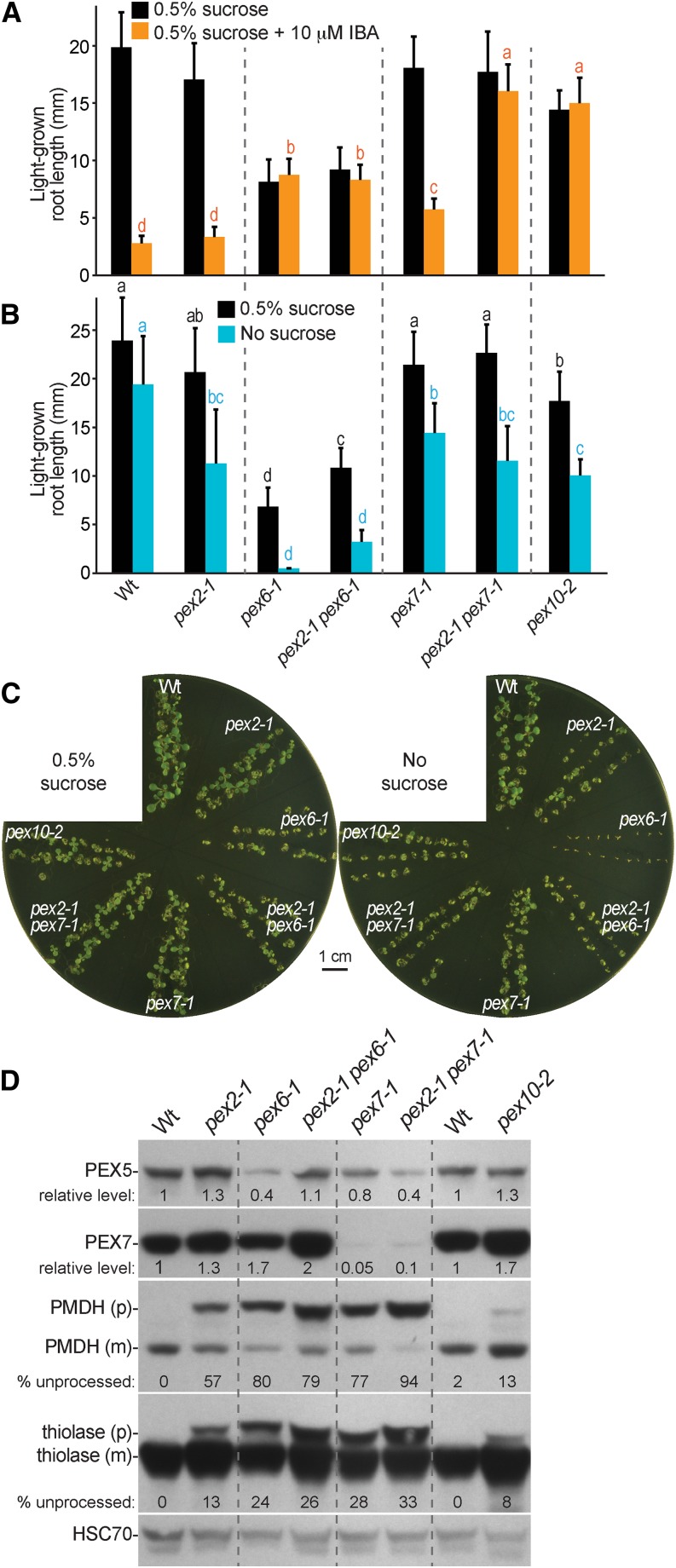
PEX2 and PEX10 are ubiquitin-protein ligases (Kaur et al., 2013) located in the peroxisomal membrane that are implicated in PEX5 recycling or degradation in yeast (Thoms and Erdmann, 2006; Platta et al., 2007, 2009; Williams et al., 2008; El Magraoui et al., 2012). To better understand the roles of Arabidopsis PEX2 and PEX10, we compared our pex2-1 and pex10-2 lines with ted3 (Hu et al., 2002), pex10-P126S (Prestele et al., 2010), and a viable pex2 TILLING allele carrying a Met55Ile missense mutation that we named pex2-2 (Fig. 1D). We compared the pex2 and pex10 alleles to previously characterized pex mutants: the PTS1 receptor mutant pex5-1 (Zolman et al., 2000) and the three receptor recycling mutants pex4-1 pex22-1 (Zolman et al., 2005), pex6-1 (Zolman and Bartel, 2004), and pex6-2 (Burkhart et al., 2013). Unlike typical peroxisome-defective mutants, pex2-1 and the other pex2 alleles were not Suc dependent in the light or the dark and responded to the inhibitory effects of IBA on light-grown root and dark-grown hypocotyl elongation similarly to the wild type (Fig. 2, A and B). However, pex2-1 and ted3 seemed to be slightly resistant to the promotive effect of IBA on lateral root production, similar to the weak pex6-2 allele (Fig. 2C), suggesting possibly impaired IBA-to-IAA β-oxidation in these two pex2 mutants. To examine whether these lateral rooting defects reflected β-oxidation defects or general auxin response defects, we examined lateral rooting in response to the synthetic auxin 1-naphthaleneacetic acid (NAA), which does not require β-oxidation for activity. We found that pex2-1 produced lateral roots similarly to the wild type in response to NAA, whereas the ted3 mutant had reduced lateral rooting in response to either IBA or NAA (Fig. 2C). This analysis suggested that pex2-1 was specifically impaired in peroxisome function, whereas pex2-2 resembled the wild type, and ted3 displayed slightly diminished general auxin responsiveness.
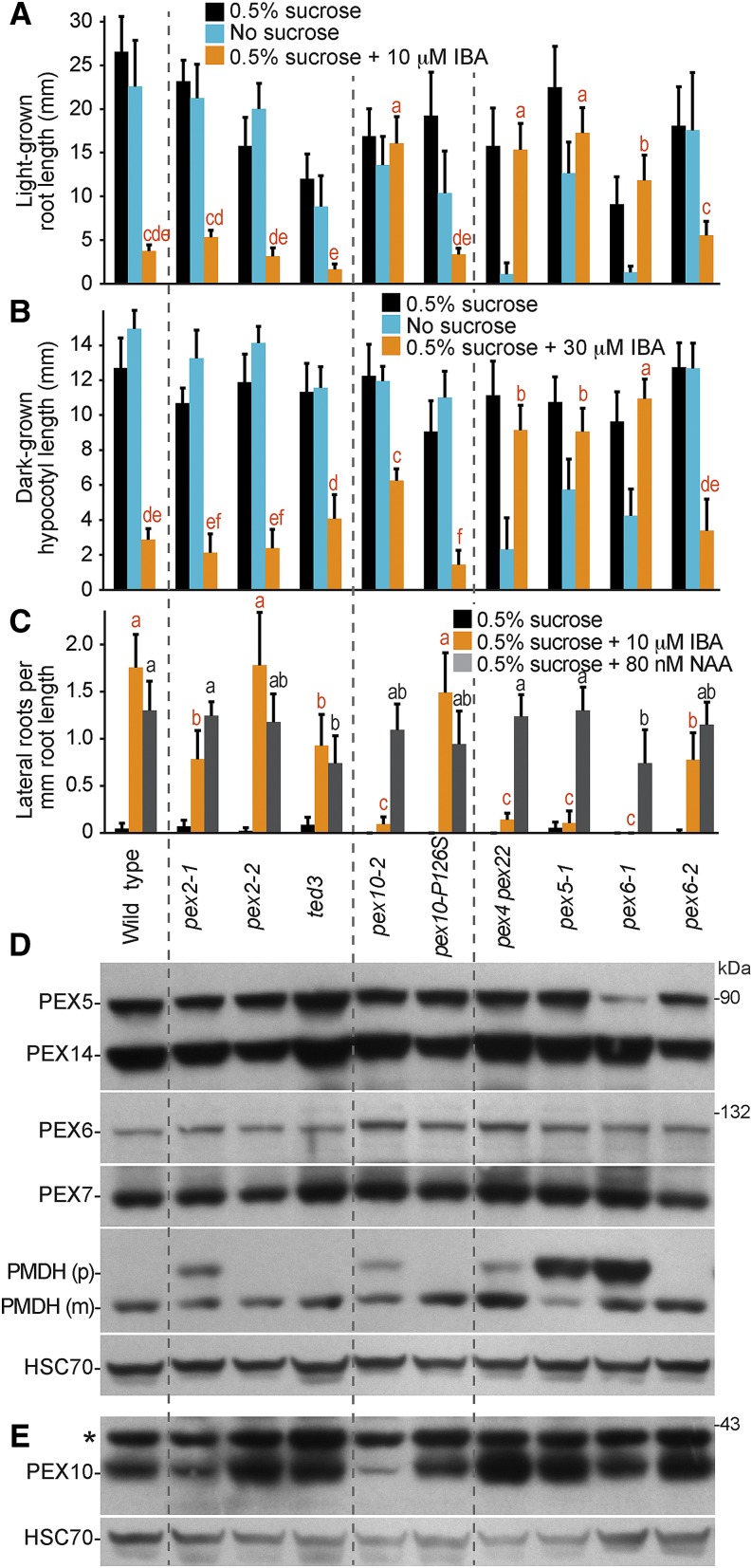
Figure 2.
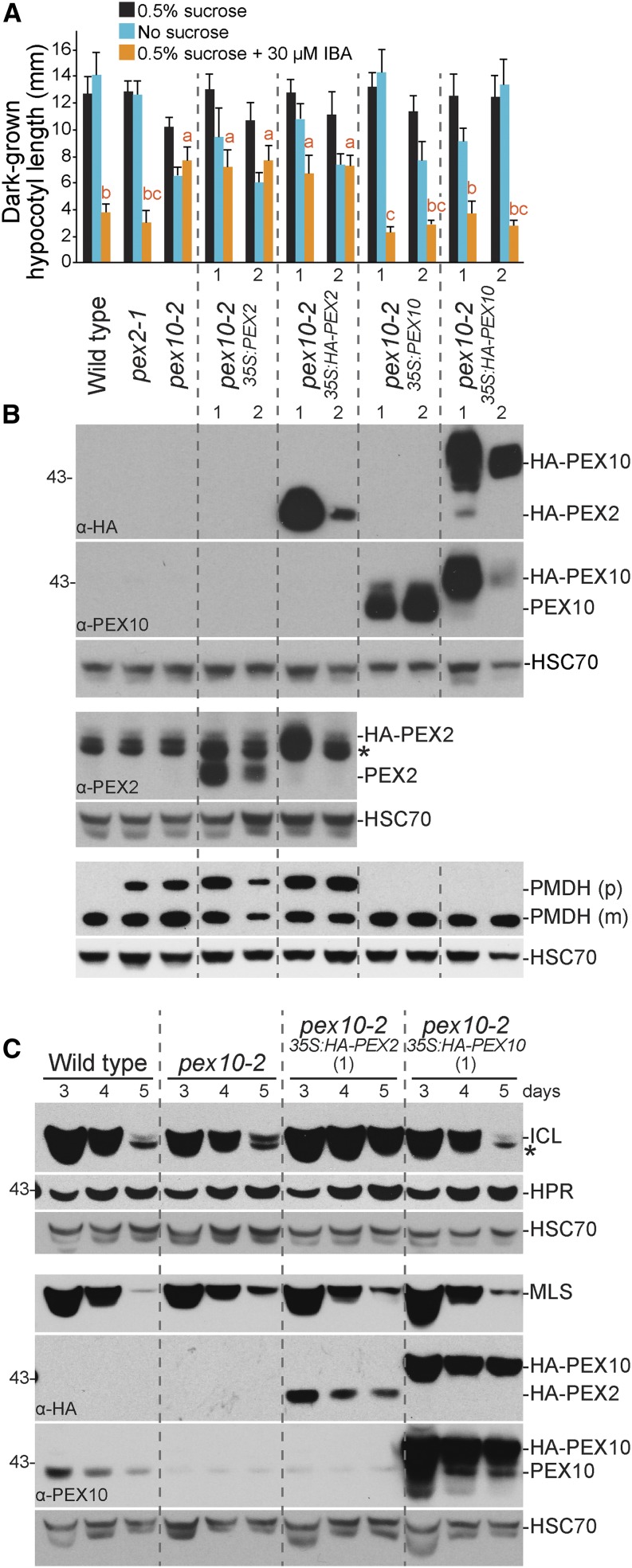
pex2-1 and pex10-2 mutants display IBA resistance and PTS2 processing defects. A, Root lengths of 8-d-old wild-type (Col-0) or pex seedlings grown in yellow-filtered light in the presence or absence of Suc or on Suc-supplemented medium containing 10 µm IBA are shown. Error bars show sds of the means (n ≥ 12). Different letters above bars represent significantly different means (one-way ANOVA, P < 0.005). B, Hypocotyl lengths of 6-d-old wild-type (Col-0) or pex seedlings grown in the dark in the presence or absence of Suc or on Suc-supplemented medium containing 30 µm IBA are shown. Error bars show sds of the means (n ≥ 13). Different letters above bars represent significantly different means (one-way ANOVA, P < 0.005). C, Lateral roots per millimeter of root length of 7-d-old wild-type (Col-0) or pex seedlings 3 d after transfer to Suc-containing medium with or without 10 µm IBA are shown. Error bars show sds of the means (n ≥ 8). Different letters above bars represent significantly different means (one-way ANOVA, P < 0.005). D, Protein extracts from the 8-d-old seedlings grown in the light on 0.5% Suc from A were processed for immunoblotting. The membrane was serially probed with antibodies to the indicated proteins. The positions of molecular mass markers (in kilodaltons) are indicated on the right. PMDH is synthesized as a precursor (p) containing the PTS2 signal that is processed into the mature (m) protein in the peroxisome. HSC70 was used to monitor loading. E, Protein extracts from 4-d-old seedlings grown in the light on 0.5% Suc were processed for immunoblotting and probed with the indicated antibodies. HSC70 was used to monitor loading. Experiments in A to E were repeated at least three times with similar results. *, A cross-reacting band that appears with the PEX10 antibody.
Like the pex2 alleles, the pex10-2 mutant did not exhibit Suc dependence in the light (Fig. 2A) or the dark (Fig. 2B). However, pex10-2 displayed robust IBA-resistant root or hypocotyl elongation in the light (Fig. 2A) or the dark (Fig. 2B), respectively. Moreover, pex10-2 was strongly resistant to the promotive effect of IBA on lateral root formation (Fig. 2C). In contrast, the PEX10 TILLING allele, pex10-P126S, resembled the wild type in these assays.
Because plant PTS2 proteins, such as peroxisomal malate dehydrogenase (PMDH), are cleaved upon peroxisomal entry by the peroxisomal protease DEG15 (Fig. 1A; Helm et al., 2007; Schuhmann et al., 2008), the molecular mass shift that accompanies this processing can be used as a proxy for matrix protein import. We found that pex2-1 and pex10-2 seedlings both displayed PMDH PTS2 processing defects that appeared to be less severe than those of pex5-1 or pex6-1 in 8-d-old seedlings (Fig. 2D). These PTS2 processing defects were apparent in younger seedlings as well (Fig. 3). In contrast, we did not detect PMDH processing defects in pex2-2, ted3, or pex10-P126S seedlings (Fig. 2D), consistent with the lack of peroxisome defects in the physiological assays of these mutants (Fig. 2, A–C).
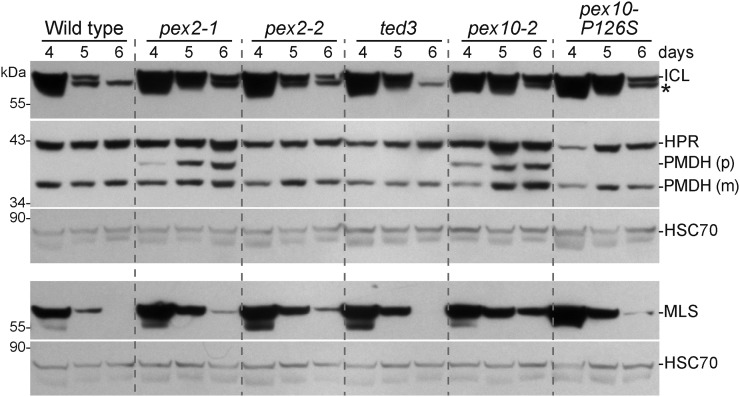
Figure 3.
Stabilization of glyoxylate cycle enzymes in pex2 and pex10 mutants. ICL and MLS are stabilized in several pex2 and pex10 mutants. Protein extracts from 4-, 5-, and 6-d-old light-grown seedlings of the wild type (Col-0) and pex2 and pex10 alleles were processed for immunoblotting. Membranes from duplicate gels were serially probed with antibodies to the indicated proteins. PMDH is synthesized as a precursor (p) with a cleavable PTS2 signal that is processed into mature (m) PMDH in the peroxisome. HSC70 was used to monitor loading. The positions of molecular mass markers (in kilodaltons) are indicated on the left. *, A cross-reacting band that appears with the ICL antibody.
Certain pex mutants display reduced steady-state levels of other PEX proteins. For example, the pex6-1 missense allele has low PEX5 levels (Zolman and Bartel, 2004), perhaps because PEX5 is polyubiquitinated and degraded when not efficiently retrotranslocated from the peroxisomal membrane. We used immunoblotting to examine PEX protein levels and found that 8-d-old pex2 and pex10 seedlings had wild-type levels of PEX5, PEX6, PEX7, and PEX14 proteins (Fig. 2D). We also examined PEX10 levels but were unable to reliably detect PEX10 by immunoblotting in 8-d-old wild-type seedling extracts (Figs. 4B and 5B). In extracts of 3- and 4-d-old seedlings, however, we could detect PEX10 in the wild type, and we found reduced PEX10 levels in pex10-2 seedlings (Figs. 2E and 5C). We did not detect the truncated pex10-2 protein(s) predicted from the RT-PCR analysis of pex10-2 complementary DNAs (cDNAs), suggesting that these truncation products may be unstable. We could not distinguish whether the reduced level of immunoreactivity migrating at the size of full-length PEX10 protein in the pex10-2 mutant (Figs. 2E and 5C) represented a protein that cross reacted with the PEX10 antibody or a small amount of pex10-2 protein resulting from a minor splice variant that we did not detect using RT-PCR. We also found that PEX10 was still present in pex2-1 seedlings (Fig. 2E), indicating that the pex2-1 missense mutation did not dramatically alter PEX10 protein levels.
Figure 4.
PEX2 (but not PEX10) rescues pex2-1 physiological and molecular defects. A, Overexpressing PEX2 (but not PEX10) restored IBA-responsive lateral rooting to pex2-1. Four-day-old seedlings were transferred to the indicated media for 4 additional d. Error bars show sds of the mean number of lateral roots per millimeter of root length (n = 8). Different letters above bars represent significantly different means (one-way ANOVA, P < 0.005). B, Expressing PEX2 (but not PEX10) rescued the PMDH PTS2 processing defect of pex2-1. Protein extracts from 8-d-old light-grown seedlings were processed for immunoblotting, and membranes from duplicate gels were serially probed with the indicated antibodies. The HA antibody detected both HA-PEX2 and HA-PEX10. The PEX2 (or PEX10) antibody detected untagged and HA-tagged PEX2 (or PEX10). HSC70 was used to monitor loading. *, A cross-reacting band that appears with the PEX2 antibody (Sparkes et al., 2005). C, The ICL and MLS stabilization of pex2-1 was rescued by transgenic HA-PEX2 (but not HA-PEX10). Extracts from 3-d-old to 5-d-old seedlings were processed for immunoblotting, and membranes from duplicate gels were probed with the indicated antibodies. *, A cross-reacting band that appears with the ICL antibody. In A and B, two independent transgenic lines (1 and 2) were examined for each transgene; C used the HA-PEX2 and HA-PEX10 lines showing higher steady-state protein levels. These experiments were repeated twice with similar results.
Figure 5.
PEX10 (but not PEX2) rescues pex10-2 physiological and molecular defects. A, Overexpressing PEX10 (but not PEX2) rescued the pex10-2 IBA resistance of dark-grown hypocotyls. Seedlings were grown on the indicated media in the dark for 5 d. Error bars show sds of the mean hypocotyl lengths (n = 12). Different letters above bars represent significantly different means (one-way ANOVA, P < 0.005). B and C, The PTS2 processing defect and ICL stabilization of pex10-2 were rescued by expressing HA-PEX10. Protein extracts from light-grown seedlings were processed for immunoblotting as described in Figure 3. PEX10 levels decreased from 3 to 5 d, and pex10-2 had lower endogenous PEX10 levels, even on day 3. *, Cross-reacting bands of the ICL or PEX2 antibodies. In A and B, two independent transgenic lines (1 and 2) were examined for each transgene; C used the HA-PEX2 and HA-PEX10 lines showing higher steady-state protein levels. These experiments were repeated twice with similar results.
Peroxisomal Matrix Proteins Are Partially Stabilized in pex2 and pex10 Mutants
pex2-1 and pex10-2 were originally isolated in a screen for GFP-ICL stabilization (Burkhart et al., 2013). To determine if this stabilization extended to endogenous peroxisomal proteins, we examined degradation of ICL and malate synthase (MLS) by using immunoblot analysis of developing seedlings. In the wild type, ICL and MLS were abundant in 4-d-old seedlings, present at reduced levels in 5-d-old seedlings, and not detected in 6-d-old seedlings (Fig. 3; Lingard et al., 2009; Burkhart et al., 2013). We detected both ICL and MLS longer in pex2-1, pex2-2, pex10-2, and pex10-P126S seedlings than in the wild type (Fig. 3), whereas ICL and MLS appeared to be degraded similarly to the wild type in ted3 (Fig. 3). We monitored levels of a photorespiration enzyme, hydroxypyruvate reductase (HPR), to determine if ICL and MLS stabilization was accompanied by developmental delays. In wild-type seedlings, HPR is already abundant in 4-d-old seedlings (Fig. 3; Burkhart et al., 2013). We found that HPR levels were similar in the wild type, pex2-1, pex2-2, and pex10-2 (Fig. 3), suggesting that ICL and MLS persistence in these mutants reflected slowed degradation rather than a developmental delay. In contrast, HPR protein levels were reduced at early time points in pex10-P126S (Fig. 3), indicating the ICL and MLS persistence observed in pex10-P126S could reflect a developmental delay rather than a degradation defect.
Complementation of pex2-1 and pex10-2 Mutants
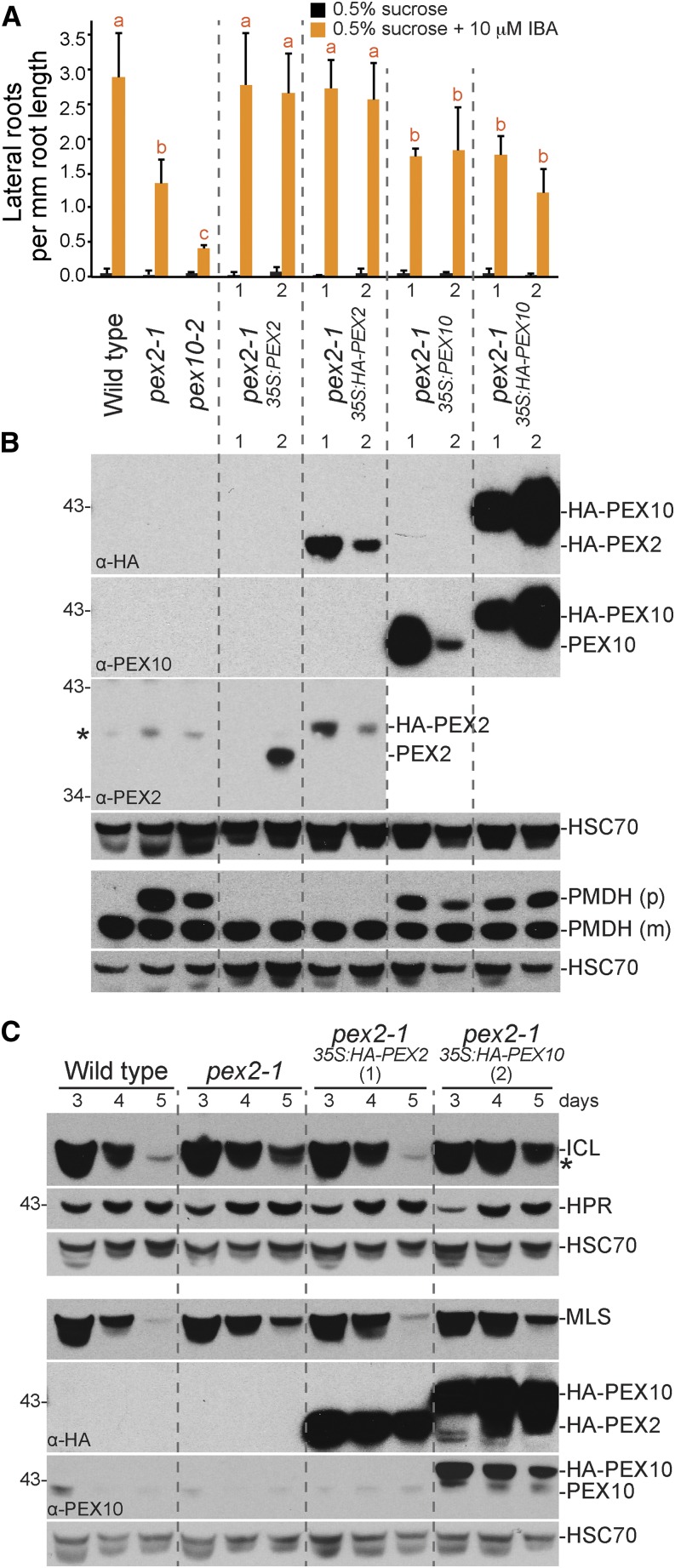
Because our physiological and molecular analyses revealed peroxisome-related defects in pex2-1 and pex10-2 but not in pex2-2, ted3 (Hu et al., 2002), or pex10-P126S (Prestele et al., 2010) mutants, we focused our detailed examination of PEX2 and PEX10 function on these two mutants. To determine if the phenotypic defects that we observed in pex2-1 were caused by the identified mutation, we expressed untagged and hemagglutinin (HA) epitope-tagged PEX2 cDNAs in pex2-1 from the strong cauliflower mosaic virus (CaMV) 35S promoter. We found that overexpressing either PEX2 or HA-tagged PEX2 in pex2-1 fully ameliorated the pex2-1 IBA resistance in lateral rooting (Fig. 4A) and the pex2-1 PMDH PTS2 processing defect (Fig. 4B). Additionally, when we expressed HA-PEX2 in pex2-1, ICL and MLS were no longer stabilized (Fig. 4C). We concluded that the observed peroxisomal defects were caused by the pex2-1 missense mutation and that the N-terminal HA tag did not interfere with PEX2 function.
We also found that overexpressing untagged or HA-tagged PEX10 in pex10-2 conferred wild-type IBA sensitivity to dark-grown (Fig. 5A) and light-grown (data not shown) pex10-2 seedlings and fully rescued the pex10-2 PMDH PTS2 processing defect (Fig. 5B). Moreover, HA-PEX10 expression in pex10-2 restored ICL degradation (Fig. 5C). We concluded that the peroxisomal defects observed in pex10-2 were caused by the identified splicing mutation and that the N-terminal HA tag did not interfere with PEX10 function.
PEX2 and PEX10 Specificity
Although PEX2 and PEX10 are not homologous at the level of primary sequence, both are peroxisomal transmembrane proteins with RING domains and ubiquitin-protein ligase activity (Kaur et al., 2013). In yeast, the PEX2-PEX10-PEX12 complex dissociates when PEX2 or PEX10 is deleted (Agne et al., 2003). Because PEX10 levels were not dramatically reduced in pex2-1 seedlings (Fig. 2E), the defects observed in pex2-1 did not seem to result from PEX10 degradation. To explore whether reduced PEX10 function contributed to pex2-1 phenotypic defects, we overexpressed PEX10 and HA-PEX10 in pex2-1. We found that the IBA-resistant lateral rooting (Fig. 4A) and the PMDH (Fig. 4B) PTS2 processing defects of pex2-1 were not ameliorated by PEX10 overexpression. In addition, ICL and MLS degradation was not restored to wild-type rates when HA-PEX10 was expressed in pex2-1 (Fig. 4C). These results suggest that increasing PEX10 is insufficient to compensate for pex2-1 defects.
In the reverse experiment, overexpression of PEX2 or HA-PEX2 in pex10-2 did not restore IBA responsiveness in hypocotyls (Fig. 5A), PTS2 processing of PMDH (Fig. 5B), or ICL and MLS degradation (Fig. 5C). These results suggest that the pex10-2 defects were not attributable to reduced PEX2 function. The observations that overexpressing PEX2 did not rescue pex10-2 defects and that overexpressing PEX10 did not rescue pex2-1 defects support previous indications that PEX2 and PEX10 have distinct roles in Arabidopsis (Schumann et al., 2007; Prestele et al., 2010) and other organisms (for review, see Platta et al., 2013).
PTS1-Targeted Matrix Proteins Are Mislocalized in pex2-1 and pex10-2 Mutants
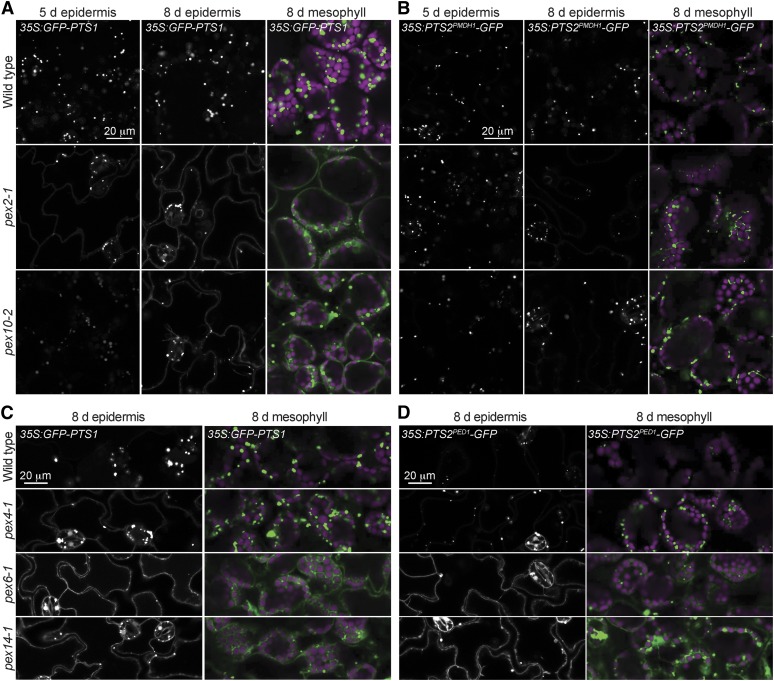
Because pex2-1 and pex10-2 inefficiently processed PTS2 proteins (Figs. 2–5), a defect that can result from reduced protein import into the peroxisome matrix (for review, see Hu et al., 2012), we directly examined peroxisomal matrix protein import in these mutants. We used confocal microscopy to examine seedlings expressing various peroxisomally targeted GFP derivatives driven by the CaMV 35S promoter; 35S:GFP-PTS1 expresses a GFP derivative that is C-terminally extended with a three-amino acid (Ser-Lys-Leu) PTS1 (Zolman and Bartel, 2004), and 35S:PST2PMDH1-GFP expresses the N-terminal PTS2 region from PMDH1 fused to GFP (Pracharoenwattana et al., 2007). GFP-PTS1 and PTS2PMDH1-GFP were each detected as punctate fluorescence in cotyledons of wild-type seedlings, suggesting efficient peroxisomal localization (Fig. 6). We found that GFP-PTS1 displayed a mixture of cytosolic and punctate fluorescence in epidermal cells of 5-d-old pex2-1 cotyledons and was primarily punctate in 5-d-old pex10-2 seedling cotyledons (Fig. 6A). In 8-d-old seedlings, pex2-1 and pex10-2 GFP-PTS1 mislocalization worsened; both mutants displayed mislocalized GFP-PTS1 in cotyledon epidermal and mesophyll cells (Fig. 6A), suggesting increased PTS1 import defects with age. In contrast to the PTS1 mislocalization, we were surprised to find that both 5- and 8-d-old pex2-1 and pex10-2 seedling cotyledon cells expressing PTS2PMDH1-GFP displayed punctate fluorescence that resembled the wild type (Fig. 6B), despite clear PMDH PTS2 processing defects in these mutants (Figs. 2–5). This result suggests that PEX2 and PEX10 are more important for PTS1 import than for PTS2 import and that the PTS2 processing defects observed in these mutants might result from defects in peroxisomal delivery of the DEG15 PTS2 processing protease, a PTS1 protein (Helm et al., 2007; Schuhmann et al., 2008).
Figure 6.
Peroxisomally targeted PTS1 proteins are mislocalized in pex2-1 and pex10-2 seedlings. Light-grown cotyledon epidermal and mesophyll cells were imaged in 5- or 8-d-old seedlings carrying the indicated reporters using confocal microscopy. GFP-PTS1 (A and C), PTS2PMDH1-GFP (B), and PTS2PED1-GFP (D) fluorescence is shown in white (epidermis) or green (mesophyll), and chlorophyll fluorescence is shown in magenta (mesophyll). These experiments were repeated at least twice with similar results. Bars = 20 µm.
To explore whether the apparent PTS1-specific defect observed in pex2-1 and pex10-2 extended to other peroxisome-defective mutants, we examined GFP-PTS1 and PTS2Peroxisome-defective1 (PED1)-GFP (Woodward and Bartel, 2005b) localization in pex4-1, pex6-1, and pex14-1. As previously observed (Monroe-Augustus et al., 2011; Woodward et al., 2014), both reporters displayed substantial cytosolic localization when the PEX14 receptor-docking protein was mutated (Fig. 6, C and D). Similarly, we detected cytosolic fluorescence in the pex6-1 mutant expressing either GFP-PTS1 or PTS2PED1-GFP (Fig. 6, C and D). Interestingly, however, the pex4-1 ubiquitin-conjugating enzyme mutant displayed a clear defect in GFP-PTS1 localization in the cotyledon epidermal and mesophyll cells while appearing to import PTS2PED1-GFP more efficiently (Fig. 6, C and D), suggesting that PEX4, like PEX2 and PEX10, is more important for PTS1 than PTS2 import.
The pex2-1 and pex10-2 PTS1 import defects (Fig. 6A) were consistent with PEX5 recycling defects. We therefore monitored PEX5 membrane association by using centrifugation to fractionate extracts prepared from 8-d-old seedlings. These experiments did not uncover clear differences from the wild type in PEX5 or PEX7 membrane association in either pex2-1 or pex10-2 (Supplemental Fig. S3), revealing that PEX5 retrotranslocation defects in these mutants, if present, were insufficient to be detected in this assay.
PEX5 Overexpression Exacerbates Most pex10-2 Defects
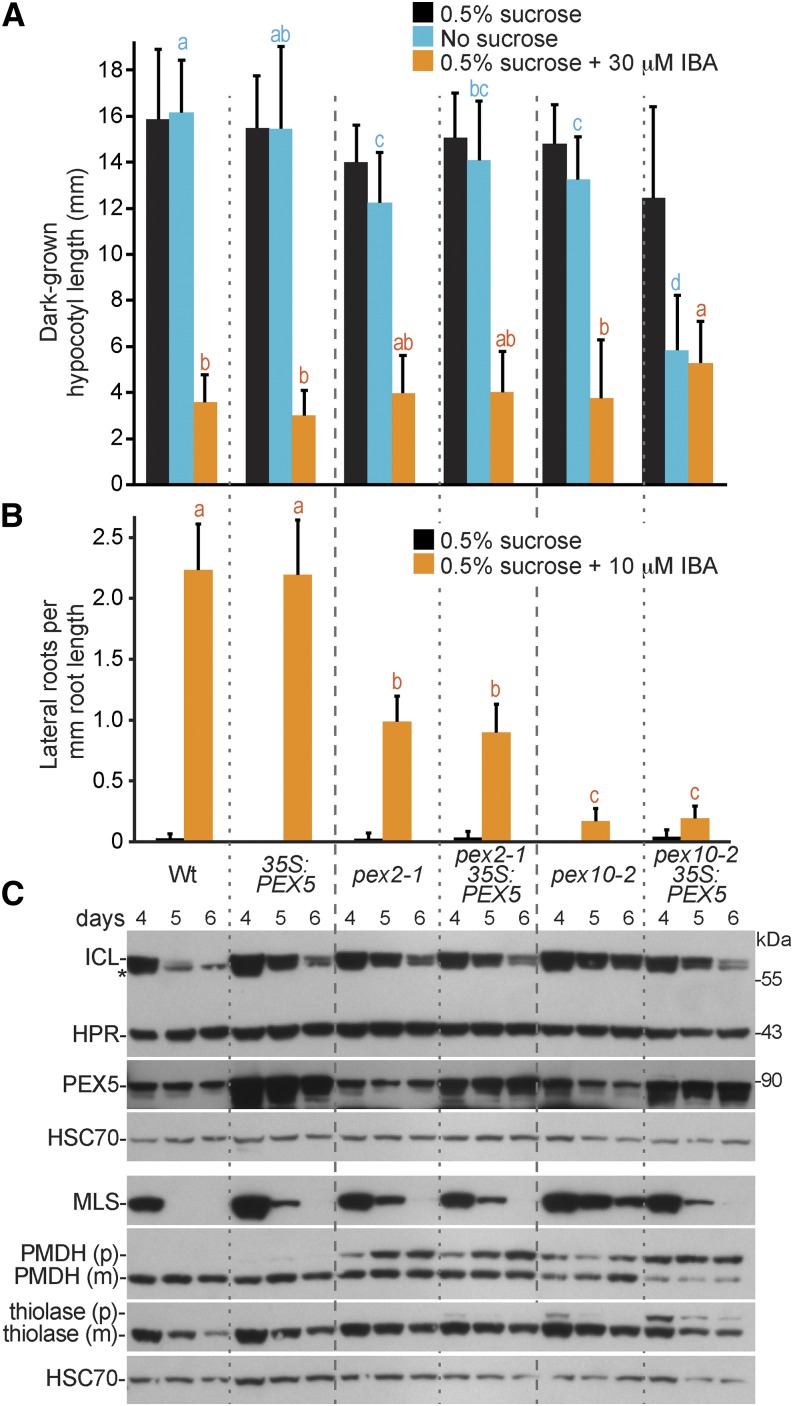
We overexpressed PEX5 in pex2-1 and pex10-2 to determine whether the physiological defects exhibited by these mutants were more likely to be caused by decreased cargo delivery, which might be rescued by PEX5 overexpression, or detrimental effects of PEX5 lingering in the peroxisomal membrane, which might be exacerbated by PEX5 overexpression. Although we generated pex2-1 35S:PEX5 and pex10-2 35S:PEX5 by crossing with the same wild-type plant carrying the 35S:PEX5 transgene (Woodward et al., 2014), PEX5 levels seemed somewhat lower in the resultant pex2-1 35S:PEX5 line than in the pex10-2 35S:PEX5 line (Fig. 7C). We found that overexpressing PEX5 in pex2-1 did not notably alter growth in the dark (Fig. 7A), IBA resistance in lateral roots (Fig. 7B), PMDH PTS2 processing defects (Fig. 7C), or ICL and MLS degradation (Fig. 7C). In contrast, PEX5 overexpression in pex10-2 seedlings conferred Suc dependence (Fig. 7A) and exacerbated the PTS2 processing defects (Fig. 7C). Interestingly, however, PEX5 overexpression partially restored the rates of ICL, MLS, and thiolase degradation in developing pex10-2 seedlings (Fig. 7C). The enhancement of pex10-2 physiological and PTS2-processing defects by excess PEX5 is consistent with a role for PEX10 in PEX5 retrotranslocation and suggests that excess PEX5 is detrimental if not promptly removed from the peroxisomal membrane.
Figure 7.
PEX5 overexpression alters pex10-2 defects but does not notably impact pex2-1. A, Hypocotyl lengths of 7-d-old wild-type (Wt; Col-0) or pex seedlings without or with PEX5 overexpression (35S:PEX5) grown in the dark in the presence or absence of Suc or on Suc-supplemented medium containing 30 µm IBA are shown. Error bars show sds of the means (n ≥ 22). Different letters above bars represent significantly different means (one-way ANOVA, P < 0.005). B, Lateral roots per millimeter of root length of 7-d-old seedlings 3 d after transfer to Suc-containing medium with or without 10 µm IBA are shown. Error bars show sds of the means (n ≥ 8). Different letters above bars represent significantly different means (one-way ANOVA, P < 0.005). C, Protein extracts from 4-, 5-, and 6-d-old seedlings grown in the light on 0.5% Suc were processed for immunoblotting. Membranes from duplicate gels were serially probed with antibodies to the indicated proteins. *, A cross-reacting band that appears with the ICL antibody. The positions of molecular mass markers (in kilodaltons) are indicated on the right. PMDH and thiolase are synthesized as precursor (p) containing the PTS2 signal that is processed into the mature (m) protein in the peroxisome. HSC70 was used to monitor loading. These experiments were repeated twice with similar results.
pex2-1 Restores PEX5 Levels in the pex6-1 Mutant
To test the role of PEX2 in PEX5 retrotranslocation, we crossed pex2-1 to pex mutants implicated in PEX5 degradation and monitored PEX5 levels and peroxisome defects in the double mutants. pex6-1 and pex7-1 both have reduced PEX5 protein levels (Zolman and Bartel, 2004; Ramón and Bartel, 2010). We found that pex2-1 restored the low PEX5 levels in the pex6-1 mutant to wild-type levels (Fig. 8D) and slightly suppressed pex6-1 Suc dependence (Fig. 8, B and C). However, the pex2-1 pex6-1 double mutant remained IBA resistant (Fig. 8A) and displayed similar PMDH and thiolase PTS2 processing defects as the pex6-1 parent (Fig. 8D), indicating that restoring PEX5 levels was insufficient to restore peroxisome function in the pex2-1 pex6-1 double mutant. In contrast, pex2-1 did not restore normal PEX5 levels to the pex7-1 mutant (Fig. 8D). Moreover, pex2-1 enhanced the IBA resistance (Fig. 8A) and PTS2 processing defects of pex7-1 (Fig. 8D), suggesting that the mechanism of PEX5 destabilization in pex7 mutants differs from the mechanism in pex6 mutants.
Figure 8.
pex2-1 restores PEX5 protein levels to pex6-1 but not pex7-1. A, Root lengths of 8-d-old wild-type (Wt; Col-0) or pex seedlings grown in yellow-filtered light in the presence of Suc or on Suc-supplemented medium containing 10 µm IBA are shown. Error bars show sds of the means (n ≥ 18). Different letters above bars represent significantly different means (one-way ANOVA, P < 0.005). B, Root lengths of 8-d-old seedlings grown in yellow-filtered light in the presence or absence of Suc. Error bars show sds of the means (n ≥ 16). Different letters above bars represent significantly different means (one-way ANOVA, P < 0.005). C, Photographs of 8-d-old seedlings from B. Bar = 1 cm. D, Protein extracts from 8-d-old seedlings grown in the light on 0.5% Suc from A were processed for immunoblotting. The membrane was serially probed with antibodies to the indicated proteins. PMDH and thiolase are synthesized as precursors (p) containing the PTS2 signal that is processed into the mature (m) protein in the peroxisome. Numbers below the PMDH or thiolase bands indicate the percentages remaining unprocessed in each lane. HSC70 was used to monitor loading, and numbers below PEX5 and PEX7 bands indicate ratios of the band to HSC70 that are normalized such that the ratio from the wild type was set to 1.0. These experiments were repeated three times with similar results.
We attempted to conduct similar analyses with pex10-2 but did not recover a pex10-2 pex6-1 double mutant, suggesting that combining the pex10-2 and pex6-1 mutations caused embryo lethality. Although we could not directly assay peroxisome-related phenotypes in the double mutant, this lethality is consistent with the possibility that pex10-2 enhances rather than suppresses pex6-1 defects.
The pex2-1 pex10-2 Double Mutant Shows Synergistic Defects
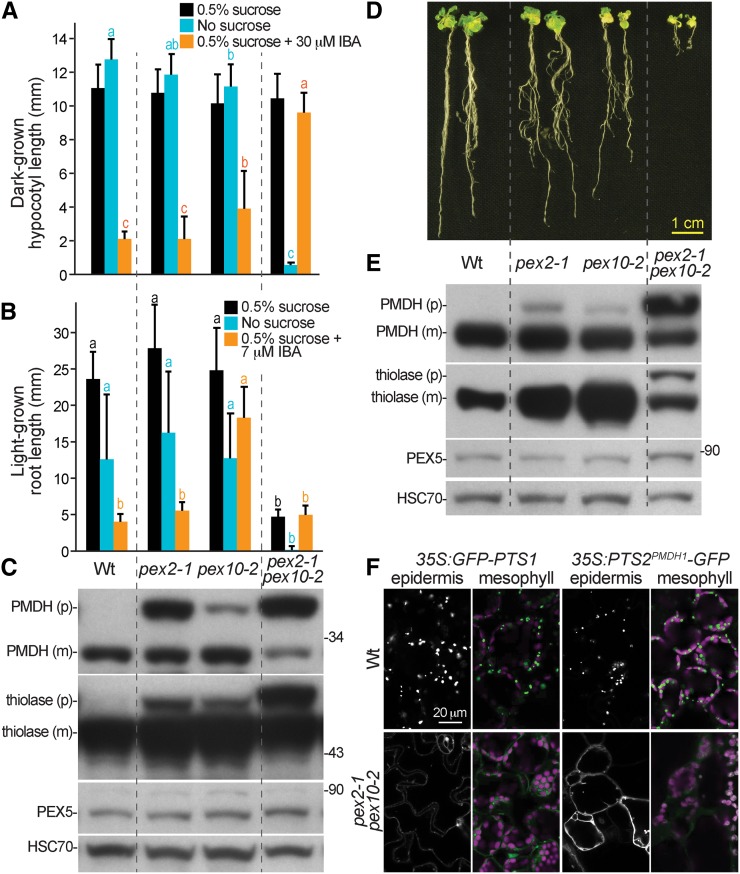
We isolated a pex2-1 pex10-2 double mutant to examine the effects of combining RING-finger protein lesions. Unlike either single mutant, the pex2-1 pex10-2 double mutant was significantly Suc dependent in both the dark (Fig. 9A) and the light (Fig. 9B) and strongly IBA resistant in the dark (Fig. 9A). Whereas the double mutant hypocotyl elongation in the dark was normal on Suc-supplemented medium (Fig. 9A), root elongation in the light was severely impaired (Fig. 9, B and D). Even at 17 d, double mutants had much shorter roots and smaller rosettes compared with the wild type or either parent (Fig. 9D), and the pex2-1 pex10-2 plants remained smaller than the wild type when grown to maturity in soil. In protein extracts from 8- or 17-d-old plants, we found that PEX5 levels were not notably altered in the double mutant (Fig. 9, C and E). However, the PMDH and thiolase PTS2 processing defects observed in the single mutants were dramatically enhanced in the double mutant (Fig. 9, C and E). We directly examined matrix protein import in the double mutant using confocal microscopy. As expected from the single-mutant phenotypes (Fig. 6), we found a severe defect in the import of GFP-PTS1 into peroxisomes in both epidermal and mesophyll cells of seedling cotyledons (Fig. 9F). However, in contrast to the single mutants, we also found that PTS2PMDH1-GFP was largely mislocalized in both the epidermis and mesophyll (Fig. 9F). The synergistic peroxisome-related defects observed in the pex2-1 pex10-2 double mutant are consistent with the possibility that either PEX2 or PEX10 can promote PEX5 retrotranslocation when function of the other RING-finger protein is reduced.
Figure 9.
The pex2-1 pex10-2 double mutant displays synergistic physiological and molecular defects. A, Hypocotyl lengths of 6-d-old seedlings grown in the dark in the presence or absence of Suc or on Suc-supplemented medium containing 30 µm IBA are shown. Error bars show sds of the means (n ≥ 16). Different letters above bars represent significantly different means (one-way ANOVA, P < 0.005). B, Root lengths of 8-d-old seedlings grown in yellow light in the presence or absence of Suc or on Suc-supplemented medium containing 7 µm IBA are shown. Error bars show sds of the means (n = 20). Different letters above bars represent significantly different means (one-way ANOVA, P < 0.005). C, Protein extracts from the 8-d-old seedlings grown in the light on 0.5% Suc from B were processed for immunoblotting. The membrane was serially probed with antibodies to the indicated proteins. The positions of molecular mass markers (in kilodaltons) are indicated on the right. PMDH and thiolase are synthesized as a precursor (p) containing the PTS2 signal that is processed into the mature (m) protein in the peroxisome. HSC70 was used to monitor loading. D, Photograph of the 17-d-old light-grown wild type (Wt; Col-0), pex2-1, pex10-2, and the pex2-1 pex10-2 double mutant. Bar = 1 cm. E, Protein extracts from 17-d-old seedlings grown in the light on 0.5% Suc were processed for immunoblotting. The membrane was serially probed with antibodies to the indicated proteins. The positions of molecular mass markers (in kilodaltons) are indicated on the right. PMDH and thiolase are synthesized as precursors containing the PTS2 signal that is processed into the mature proteins in the peroxisome. HSC70 was used to monitor loading. F, Light-grown cotyledon epidermal and mesophyll cells were imaged in 8-d-old seedlings using confocal microscopy. GFP-PTS1 and PTS2PMDH1-GFP fluorescence is shown in white (epidermis) or green (mesophyll); chlorophyll fluorescence is shown in magenta (mesophyll). These experiments were repeated at least twice with similar results. Bar = 20 µm.
DISCUSSION
Although a framework for understanding the roles of PEX proteins in matrix protein import is in place (Fig. 1A), the molecular roles of many PEX proteins are still unclear, and how functions vary or are conserved in different organisms is not well understood. We isolated the pex2-1 and pex10-2 peroxisomal ubiquitin-protein ligase mutants in a microscopy-based screen for matrix protein degradation defects (Burkhart et al., 2013). pex2-1 and pex10-2 are the first alleles of PEX2 and PEX10 to emerge from forward genetic screens for peroxisome defects in Arabidopsis. The isolation of pex2-1, which lacks dramatic physiological defects, adds to growing evidence that microscopy-based screens (Mano et al., 2006; Goto et al., 2011; Burkhart et al., 2013) can identify peroxisome-defective mutants with physiological defects too subtle to emerge from more common screens for reduced β-oxidation (for review, see Bartel et al., 2014a).
In addition to partial stabilization of endogenous matrix proteins (ICL and MLS; Fig. 3), pex2-1 and pex10-2 displayed IBA resistance and PTS2 processing defects (Fig. 2) that were rescued by overexpressing PEX2 or PEX10, respectively (Figs. 4 and 5). In contrast, overexpressing PEX2 in pex10-2 or PEX10 in pex2-1 did not complement the mutant defects (Figs. 4 and 5). These data suggest that the observed mutant defects were not primarily caused by reduced function of the other RING-finger PEX proteins and were consistent with our observation that PEX10 protein remained present in the pex2-1 mutant (Fig. 2E). However, we did not assess levels of the third RING-finger PEX protein, PEX12, in these experiments and cannot rule out the possibility that one or both lesions impair PEX12 function.
The pex10-2 mutation causes a splicing defect expected to prevent translation of the RING domain (Fig. 1, G and H; Supplemental Fig. S2). In Arabidopsis, the RING domains of both PEX2 and PEX10 possess ubiquitin-protein ligase activity in vitro (Kaur et al., 2013). The yeast PEX10 RING domain dimerizes with both PEX2 and PEX12 RING domains, enhancing the ubiquitin-protein ligase activity of the partner protein (El Magraoui et al., 2012). Because an Arabidopsis pex10 null allele is embryo lethal (Schumann et al., 2003; Sparkes et al., 2003), the viability of pex10-2 seems to suggest that PEX10 regions other than the RING domain (and by extension, PEX10 functions other than catalyzing ubiquitination) are important during embryogenesis. Arguing against this conclusion is the observation that the similarly positioned pex10-W313* truncation (Fig. 1G; Supplemental Fig. S2) confers embryo lethality (Prestele et al., 2010) and our detection of a small amount of apparently full-length PEX10 in the pex10-2 mutant (Figs. 2E and 5C). It is possible that a minor pex10-2 splice variant including the RING domain that escaped our detection accounts for the different viability of these two alleles.
The pex10-2 splicing allele generally displayed less severe PTS2 processing (Figs. 8D and 9C) and GFP-PTS1 import defects than the pex2-1 missense allele (Fig. 6A). We were, therefore, surprised to find that the pex10-2 mutant generally displayed stronger defects than pex2-1 in IBA responsiveness (Fig. 2, A–C) and matrix protein stabilization (Figs. 3 and 7C). This paradox may be resolved by the observation that pex10-2 displayed heightened sensitivity to the detrimental effects of PEX5 overexpression (Fig. 7), suggesting that the pex10-2 physiological and molecular defects result from not only matrix protein import defects but also negative consequences of PEX5 lingering in the peroxisomal membrane.
pex6-1 has low PEX5 levels (Figs. 2 and 8; Zolman and Bartel, 2004), presumably because PEX5 is polyubiquitinated and degraded when PEX6-driven retrotranslocation is compromised. Indeed, PEX5 levels in pex6-1 are restored when the PEX4 ubiquitin-conjugating enzyme is disabled (Ratzel et al., 2011). The ability of the pex2-1 lesion to similarly restore normal PEX5 levels to pex6-1 (Fig. 8) implies that PEX2 is necessary for efficient PEX5 degradation when retrotranslocation is slowed and implicates Arabidopsis PEX2 along with PEX4 (Ratzel et al., 2011) in PEX5 polyubiquitination. In contrast, pex2-1 enhanced the physiological and PTS2 processing defects of pex7-1 (Fig. 8), another mutant with low PEX5 levels (Ramón and Bartel, 2010). The failure of pex2-1 to restore PEX5 levels to pex7-1 is consistent with the idea that the reduced PEX5 levels observed in pex7-1 result primarily from PEX5 instability in the cytosol rather than impaired PEX5 receptor recycling.
We isolated pex2-1 and pex10-2 in a screen for GFP-ICL stabilization (Burkhart et al., 2013). We previously showed that peroxisomal matrix proteins, such as GFP-ICL, can be stabilized by slow development (lon2-6), delayed germination (lon2-6), or inefficient entry into the peroxisome (pex14 mutants; Burkhart et al., 2013). Moreover, the recovery of a weak pex6 allele (pex6-2) in this screen (Burkhart et al., 2013) and the stabilization of GFP-ICL in a ubiquitin-conjugating enzyme mutant (pex4-1 pex22-1; Lingard et al., 2009) suggest that GFP-ICL might be exported out of the peroxisome for cytosolic degradation. Similarly, the stabilization of ICL and MLS in pex2-1 and pex10-2 (Fig. 3) could reflect ubiquitination of matrix proteins by PEX2 or PEX10 analogously to the ubiquitin-protein ligases acting in endoplasmic reticulum-associated protein degradation (Hampton and Sommer, 2012). Such ubiquitination could promote PEX6-dependent retrotranslocation of substrates out of the peroxisome for cytosolic degradation. Supporting this idea is the recovery of ICL in a proteomic analysis of ubiquitinated Arabidopsis proteins (Kim et al., 2013a). Alternatively, matrix protein degradation could be indirectly reduced in pex2-1 or pex10-2, because impairing PEX5 recycling reduces protein import, which in turn, stabilizes matrix proteins (Lingard et al., 2009; Burkhart et al., 2013). Another possibility is suggested by the recent discovery of roles for autophagy of peroxisomes (pexophagy) and the peroxisomal protease LON2 in matrix protein degradation. Although lon2 single mutants do not stabilize ICL or MLS (Lingard and Bartel, 2009; Burkhart et al., 2013) and although ICL and MLS are only slightly stabilized in hypocotyls when autophagy is disrupted (Kim et al., 2013b), ICL and MLS are dramatically stabilized when autophagy is disrupted in an lon2 background (Farmer et al., 2013; Bartel et al., 2014b; Goto-Yamada et al., 2014). This stabilization suggests that LON2 promotes matrix protein degradation and that peroxisomes are targeted for autophagy when LON2 is dysfunctional. It remains to be determined whether LON2 directly degrades matrix proteins or whether LON2 dissociates matrix protein oligomers to enable their ubiquitin-dependent retrotranslocation from the peroxisome for cytosolic degradation. Our observation that PEX5 overexpression in pex10-2 accelerates ICL, MLS, and thiolase degradation while exacerbating PTS2 processing defects (Fig. 7C) suggests the intriguing possibility that pexophagy is enhanced when PEX5 accumulates in the pex10-2 mutant. It will be interesting to explore the roles of PEX2, PEX5, and PEX10 in Arabidopsis pexophagy.
In addition to defects in matrix protein degradation, we found that pex2-1 and pex10-2 partially mislocalized fluorescent PTS1 proteins to the cytosol (Fig. 6A). Because pex2-1 and pex10-2 both displayed PTS2 processing defects detectable through immunoblotting (Figs. 2–5), we were surprised to find that PTS2-GFP import defects were not apparent in these mutants (Fig. 6B). In Arabidopsis, PEX7 depends on PEX5 for cargo delivery (Hayashi et al., 2005; Woodward and Bartel, 2005b), and PEX5 levels depend on intact PEX7 (Ramón and Bartel, 2010). Consequently, many pex mutants, including pex5-10 and pex7 alleles, display both PTS1 and PTS2 import defects (Khan and Zolman, 2010; Ramón and Bartel, 2010). Indeed, combining pex2-1 and pex10-2 lesions confers Suc dependence and both PTS1 and PTS2 import defects in the double mutant (Fig. 9), and RNAi lines that reduce levels of PEX2 or PEX10 display Suc dependence and defects in importing both PTS1 and PTS2 proteins into peroxisomes (Nito et al., 2007). The Suc independence (Fig. 2, A and B) and continued PTS2 import (Fig. 6B) of the pex2-1 and pex10-2 single mutants indicate that our alleles are less completely impaired in PEX2 and PEX10 function than the RNAi lines. Like pex2-1 and pex10-2, we also observed a greater defect in PTS1 versus PTS2 import in pex4-1 (Fig. 6, C and D), which shows aberrant retention of PEX5 in the membrane (Ratzel et al., 2011). Intriguingly, the continued PTS2 import in the pex2-1, pex10-2, and pex4-1 partial loss-of-function alleles suggests that PTS2 import is less sensitive to slowed PEX5 recycling than PTS1 import. It will be interesting to learn whether PEX5 can still interact with PEX7 and facilitate some PTS2 import even when stuck in the peroxisomal membrane.
Despite the inability of PEX2 or PEX10 overexpression to compensate for pex10-2 or pex2-1 defects, respectively (Figs. 4 and 5), the synergistic physiological and molecular defects exhibited by the pex2-1 pex10-2 double mutant (Fig. 9) suggest that PEX2 and PEX10 function in related processes. Indeed, our data implicate both PEX2 and PEX10 in PEX5 regulation. The restoration by pex2-1 of PEX5 levels in pex6-1 (Fig. 8) suggests a role for PEX2 in PEX5 degradation when retrotranslocation is slowed. In addition, pex10-2 was sensitive to PEX5 overexpression (Fig. 7), consistent with a role for PEX10 in ubiquitinating PEX5 to promote retrotranslocation from the peroxisomal membrane. The apm4 mutant defective in the third RING-finger PEX protein, PEX12, accumulates PEX5 in the membrane (Mano et al., 2006), consistent with the possibility that PEX10 and PEX12 work together in PEX5 recycling in Arabidopsis. These related but distinct roles are consistent with our finding that PEX5 overexpression failed to enhance pex2-1 defects (Fig. 7). Future studies exploring PEX5 ubiquitination in these mutants along with mutants defective in PEX12 will be useful to further elucidate the roles of the RING-finger PEX proteins in Arabidopsis peroxisome biology.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) accession Columbia-0 (Col-0) was used as the wild type. pex4-1 (Zolman et al., 2005) carrying 35S:GFP-PTS1 (Zolman and Bartel, 2004) and 35S:PTS2PED1-GFP (Woodward and Bartel, 2005b), pex4-1 pex22-1 (Zolman et al., 2005), pex5-1 (Zolman et al., 2000), pex6-1 and pex6-1 35S:GFP-PTS1 (Zolman and Bartel, 2004), pex6-2 (Burkhart et al., 2013), pex7-1 (Woodward and Bartel, 2005b), pex10-P126S (Prestele et al., 2010), ted3 (Hu et al., 2002), and pex14-1 carrying 35S:GFP-PTS1 or 35S:PTS2PED1-GFP (Monroe-Augustus et al., 2011) were previously described. Before phenotypic analyses, pfl36/pex2-1, pex2-2, ted3, pfl81/pex10-2, and pex10-P126S were backcrossed to wild-type Col-0. Backcrossed pex2-1 and pex10-2 mutants were crossed to a wild-type (Col-0) plant carrying 35S:GFP-PTS1 (Zolman and Bartel, 2004), 35S:PTS2PMDH1-GFP (Pracharoenwattana et al., 2007), or 35S:PEX5 (Woodward et al., 2014). pex6-1 was crossed to a wild-type (Col-0) plant carrying 35S:PTS2PED1-GFP (Woodward and Bartel, 2005b). Homozygous mutants that were also homozygous for the transgene were selected from the progeny of the crosses using PCR-based genotyping.
Surface-sterilized seeds were plated on plant nutrient medium (Haughn and Somerville, 1986) supplemented with 0.5% (w/v) Suc (PNS) and solidified with 0.6% (w/v) agar. IBA stocks were dissolved in ethanol at 100 mm, and media normalized to the same ethanol content were used as controls. For assays of light-grown seedlings, seeds were stratified for 1 d, plated on the indicated IBA concentrations, and grown for 8 d at 22°C under continuous illumination through yellow long-pass filters that slow indolic compound breakdown (Stasinopoulos and Hangarter, 1990) unless otherwise indicated. For assays with dark-grown seedlings, seeds were stratified for 1 d, allowed to begin germinating in the absence of IBA under white light for 1 d, plated on the indicated media, placed under yellow light for 1 d, and placed in darkness for 4 d, after which hypocotyl lengths were measured. For lateral root assays, seeds were stratified for 1 to 3 d, plated on PNS, and grown in yellow light for 4 d. Four-day-old seedlings were then transferred to a mock or IBA-containing PNS plate and grown under yellow light for 3 additional d, after which the primary root length was measured and the number of lateral roots that had emerged from the primary root was counted.
Mutant Isolation and Recombination Mapping
pfl36 (pex2-1) and pfl81 (pex10-2; in the Col-0 ICLp:GFP-ICL background; Burkhart et al., 2013) were outcrossed to Landsberg erecta for recombination mapping. A mapping population for pfl36 was selected by screening the F2 population for light-grown seedlings with prolonged GFP-ICL fluorescence in cotyledons and hypocotyls and testing F3 progeny seedlings for PMDH PTS2 processing defects by immunoblotting. A mapping population for pfl81 was selected for IBA resistance in roots of light-grown seedlings. DNA isolated from progeny of selected F2 individuals was analyzed using published and newly developed PCR-based polymorphic markers (Supplemental Table S1).
Plasmid Construction and Plant Transformation
PEX2 (G18223) and PEX10 (G60512) cDNAs were obtained from the Arabidopsis Biological Resource Center at Ohio State University. Inserts of the pENTR223-PEX2 and pENTR223-PEX10 entry clones were recombined into the pEarleyGate 100 and pEarleyGate 201 plasmids (Earley et al., 2006) using LR Clonase (Invitrogen) to form pEG100-PEX2 (untagged), pEG100-PEX10 (untagged), pEG201-PEX2 (N-terminal HA tag), and pEG201-PEX10 (N-terminal HA tag), all of which are driven by the CaMV 35S promoter. Plasmids were transformed into Agrobacterium tumefaciens strain GV3101 (Koncz and Schell, 1986), which was used to transform pex2-1 carrying ICLp:GFP-ICL and pex10-2 using the floral dip method (Clough and Bent, 1998). Homozygous lines were selected in the progeny of transformants by following resistance to glufosinate ammonium (Basta).
Immunoblot Analysis
Protein was extracted from seedlings grown under continuous white light on PNS for the indicated number of days. To extract protein, frozen tissue was ground with a pestle, and two or three volumes of 2× loading buffer (Invitrogen) was added. Samples were centrifuged, and 15 μL of supernatant was transferred to a fresh tube with 1.6 μL of 0.5 m dithiothreitol and heated at 100°C for 5 min. Samples extracted from equal volumes of ground tissue were loaded onto NuPAGE 10% (w/v) BisTris gels (Invitrogen) alongside prestained protein markers (P7708S; New England Biolabs) and Cruz Markers (Santa Cruz Biotechnology) and electrophoresed using the MOPS-based buffer system suggested by the manufacturer (Invitrogen). After electrophoresis, proteins were transferred for 30 min at 24 V to a Hybond ECL nitrocellulose membrane (Amersham Pharmacia Biotech) using NuPAGE transfer buffer (Invitrogen). After transfer, membranes were rocked for 1 h at 4°C in blocking buffer (8% [w/v] nonfat dry milk, 20 mm Tris, pH 7.5, 150 mm NaCl, 0.1% [v/v] Tween 20) and incubated overnight at 4°C with primary antibodies diluted in blocking buffer: rabbit α-ICL (1:1,000 dilution; Maeshima et al., 1988), rabbit α-HPR (1:10,000 dilution; AS11 1797; Agrisera), rabbit α-MLS (1:25,000 dilution; Olsen et al., 1993), rabbit α-PEX2 (1:500 dilution; Sparkes et al., 2005), rabbit α-PEX5 (1:100 dilution; Zolman and Bartel, 2004), rabbit α-PEX6 (1:1,000 dilution; Ratzel et al., 2011), rabbit α-PEX7 (1:800 dilution; Ramón and Bartel, 2010), rabbit α-PEX10 (1:500; generated and affinity purified using a recombinant fusion protein including the N-terminal 273 amino acids of PEX10 by Proteintech Group, Inc.), rabbit α-PEX14 (1:10,000 dilution; AS08 372; Agrisera), rabbit α-PMDH2 (1:2,000 dilution; Pracharoenwattana et al., 2007), rabbit α-thiolase (PED1 isoform; 1:2,500 dilution; Lingard et al., 2009), mouse α-heat shock cognate70 (HSC70) (1:20,000–1:100,000 dilution; SPA-817; StressGen Bioreagents), mouse α-mitochondrial ATP synthase α-subunit (1:2,000; MS507; MitoScience), or rat α-HA (1:1,000 dilution; 3F10; Roche). After a 4-h incubation with horseradish peroxidase-linked goat α-rabbit, α-mouse, or α-rat secondary antibody (1:5,000 dilution in blocking buffer; sc-2030, sc-2031, or sc-2032, respectively; Santa Cruz Biotechnology), horseradish peroxidase was imaged using westernBright ECL reagent (Advansta) and autoradiography film. For serial probing of the same membrane with multiple antibodies, membranes were reblocked but not stripped between antibody incubations.
Films were photographed on a light box, and the resulting TIFF images were analyzed using ImageJ 64 Gel Analysis software. Areas under density curves from bands of interest were calculated after background subtraction and normalized to the appropriate control bands.
RNA Analysis
RNA was isolated from 4-d-old light-grown Col-0 and pex10-2 seedlings using the TRI Reagent RNA Extraction method according to the manufacturer’s instructions (Sigma) and dissolved in diethyl pyrocarbonate-treated water. cDNA was synthesized from RNA using a 3′ gene-specific primer (PEX10-3; Supplemental Table S2) and SuperScript III Reverse Transcriptase (Invitrogen). pex10-2 cDNAs were PCR amplified with the forward PEX10-6 primer located in exon 8 and the reverse PEX10-5 primer located in exon 10 (Supplemental Table S2). Amplicons were purified after agarose gel electrophoresis to separate splicing variants, and both observed variants were sequenced using the primers used for amplification.
Confocal Microscopy
PTS1 import was monitored using transgenic lines carrying 35S:GFP-PTS1, which encodes GFP extended with a C-terminal Ser-Lys-Leu PTS1 signal (Zolman and Bartel, 2004), or ICLp:GFP-ICL, which encodes ICL, a PTS1 protein, fused to the C terminus of GFP (Lingard et al., 2009). PTS2 import was monitored using 35S:PTS2PMDH1-GFP, which encodes GFP extended with the N-terminal 52-amino acid residues (including the PTS2 signal) of PMDH1 (Pracharoenwattana et al., 2007), or 35S:PTS2PED1-GFP, which encodes GFP extended with the N-terminal 49-amino acid residues (including the PTS2 signal) of the PED1 isozyme of thiolase (Woodward and Bartel, 2005b).
For confocal imaging, seedlings were mounted in water under a cover glass. Images were collected using a Carl Zeiss LSM 710 laser scanning confocal microscope equipped with a Meta detector. Samples were imaged through a 40× oil immersion objective after excitation with a 488-nm argon laser; GFP emission was collected between 493 and 530 nm, and chlorophyll emission was collected between 620 and 719 nm. Each image is an average of eight exposures using a 22.9-μm pinhole, corresponding to a 0.7-μm optical slice (epidermal cells), or a 70.4-μm pinhole, corresponding to a 1.8-μm optical slice (mesophyll cells).
Sequencing and Genotyping of Mutant Lesions
The PEX2 gene (At1g79810) was amplified from pfl36 genomic DNA using two oligonucleotide pairs (Supplemental Table S2). The resulting overlapping fragments covered the gene from 165 bp upstream of the translation start site to 90 bp downstream of the stop codon. Amplicons were purified using the Zymo PCR purification kit (Zymo Research) and sequenced directly (Lone Star Labs) with the primers used for amplification. The lesion identified in pfl36 changed PEX2 G703 (where one is the first nucleotide of the initiator codon) to an A, which created an amino acid change (Arg161Lys).
The PEX10 gene (At2g26350) was amplified from pfl81 genomic DNA using four oligonucleotide pairs (Supplemental Table S2). The resulting overlapping fragments covered the gene from 81 bp upstream of the translation start site to 275 bp downstream of the stop codon. Amplicons were purified using the Zymo PCR purification kit (Zymo Research) and sequenced directly (Lone Star Labs) with the primers used for amplification. The lesion identified in pfl81 changed G1709 (where one is the first nucleotide of the initiator codon) of PEX10 to an A, which alters a splice site.
pex2-1 and pex10-2 were followed in the progeny of crosses using PCR amplification with the primers listed in Supplemental Table S3 followed by digestion of the resultant amplicons with the restriction enzymes indicated in Supplemental Table S3.
Cell Fractionation
Eight-day-old light-grown seedlings (from 2 mg of dry seeds) were chopped with scissors in 1 mL of ice-cold fractionation buffer (150 mm Tris, pH 7.6, 10 mm KCl, 1 mm EDTA, 1 mm dithiothreitol, 100 mm Suc, 1× protease inhibitor cocktail; P9559; Sigma) followed by homogenization in a Dounce homogenizer (20 strokes) on ice. The homogenate was filtered through Miracloth (Calbiochem) and centrifuged at 44g at 4°C to remove cellular debris; 50 µL were removed as the homogenate fraction, and 300 µL of supernatant was centrifuged at 15,294g at 4°C and removed to give the soluble fraction. The pellet was washed one time with a volume of fractionation buffer equal to the homogenate volume, centrifuged for 20 min at 15,294g at 4°C to give a wash fraction, and suspended in a volume of fractionation buffer equal to the homogenate volume to give the pellet fraction. Soluble fractions contained cytosolic proteins as well as contents of lysed organelles, and pellet fractions contained organellar membranes and contents of intact organelles. NuPAGE 2× loading buffer (Invitrogen) was added to each sample to double the initial volume and processed for immunoblot analysis as described above.
Statistical Analysis
The SPSS Statistics software (version 21.0.0.1) was used to analyze statistical significance of measurements using one-way ANOVA followed by Duncan’s test. Mean values that are not significantly different (P < 0.005) from each other are marked with the same letter.
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL data libraries under accession numbers At1g79810 (PEX2) and At2g26350 (PEX10).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of PEX2 homologs from various organisms showing conserved transmembrane and RING domains and positions of pex2 mutations.
Supplemental Figure S2. Alignment of PEX10 homologs from various organisms showing conserved transmembrane and RING domains and positions of pex10 mutations.
Supplemental Figure S3. Membrane association of peroxisome matrix protein receptors in pex2-1, pex10-2, and pex2-1 pex10-2.
Supplemental Table S1. Markers used in recombination mapping of pex2-1 and pex10-2.
Supplemental Table S2. Primers used for amplification and sequencing of PEX2 and PEX10.
Supplemental Table S3. PCR-based markers for determining mutant genotypes.
Supplementary Material
Acknowledgments
We thank Alison Baker (University of Leeds), John Harada (University of California, Davis), and Masayoshi Maeshima (Nagoya University) for antibodies recognizing PEX2, MLS, and ICL, respectively; Steven Smith (University of Western Australia) for the anti-PMDH2 antibody and seeds from Col-0 transformed with 35S:PTS2PMDH1-GFP; Matthew Lingard (Rice University) for originating the pfl screen; Andrew Woodward (University of Mary Hardin-Baylor) for seeds from the pex2-2 and pex10-P126S TILLING alleles; Wendell Fleming (Rice University), Kim Gonzalez (Rice University), Mauro Rinaldi (Rice University), Pierce Young (Rice University), and Andrew Woodward (University of Mary Hardin-Baylor) for critical comments on the article; and the Arabidopsis Biological Resource Center at Ohio State University for cDNA clones and seeds from TILLING lines.
Glossary
- CaMV
cauliflower mosaic virus
- cDNA
complementary DNA
- Col-0
Columbia-0
- HPR
hydroxypyruvate reductase
- IAA
indole-3-acetic acid
- IBA
indole-3-butyric acid
- ICL
isocitrate lyase
- MLS
malate synthase
- NAA
1-naphthaleneacetic acid
- PMDH
peroxisomal malate dehydrogenase
- PNS
plant nutrient medium supplemented with 0.5% (w/v) Suc
- PTS
peroxisomal targeting signal
- RNAi
RNA interference
- RT
reverse transcription
Footnotes
This work was supported by the National Institutes of Health (grant no. R01GM079177 and Shared Instrumentation grant no. S10RR026399–01 for equipment on which confocal microscopy was performed) and by the Robert A. Welch Foundation (grant no. C–1309).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Agne B, Meindl NM, Niederhoff K, Einwächter H, Rehling P, Sickmann A, Meyer HE, Girzalsky W, Kunau WH. (2003) Pex8p: an intraperoxisomal organizer of the peroxisomal import machinery. Mol Cell 11: 635–646 [DOI] [PubMed] [Google Scholar]
- Azevedo JE, Schliebs W. (2006) Pex14p, more than just a docking protein. Biochim Biophys Acta 1763: 1574–1584 [DOI] [PubMed] [Google Scholar]
- Bartel B, Burkhart SE, Fleming WA. (2014a) Protein transport in and out of plant peroxisomes. In C Brocard, A Hartig, eds, Molecular Machines Involved in Peroxisome Biogenesis and Maintenance. Springer-Verlag, Vienna, pp 325–345 [Google Scholar]
- Bartel B, Farmer LM, Rinaldi MA, Young PG, Danan CH, Burkhart SE. (2014b) Mutation of the Arabidopsis LON2 peroxisomal protease enhances pexophagy. Autophagy 10: 518–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart SE, Lingard MJ, Bartel B. (2013) Genetic dissection of peroxisome-associated matrix protein degradation in Arabidopsis thaliana. Genetics 193: 125–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- De Rybel B, Audenaert D, Xuan W, Overvoorde P, Strader LC, Kepinski S, Hoye R, Brisbois R, Parizot B, Vanneste S, et al. (2012) A role for the root cap in root branching revealed by the non-auxin probe naxillin. Nat Chem Biol 8: 798–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- El Magraoui F, Bäumer BE, Platta HW, Baumann JS, Girzalsky W, Erdmann R. (2012) The RING-type ubiquitin ligases Pex2p, Pex10p and Pex12p form a heteromeric complex that displays enhanced activity in an ubiquitin conjugating enzyme-selective manner. FEBS J 279: 2060–2070 [DOI] [PubMed] [Google Scholar]
- Fan J, Quan S, Orth T, Awai C, Chory J, Hu J. (2005) The Arabidopsis PEX12 gene is required for peroxisome biogenesis and is essential for development. Plant Physiol 139: 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer LM, Rinaldi MA, Young PG, Danan CH, Burkhart SE, Bartel B. (2013) Disrupting autophagy restores peroxisome function to an Arabidopsis lon2 mutant and reveals a role for the LON2 protease in peroxisomal matrix protein degradation. Plant Cell 25: 4085–4100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y, Nashiro C, Miyata N, Tamura S, Okumoto K. (2012) New insights into dynamic and functional assembly of the AAA peroxins, Pex1p and Pex6p, and their membrane receptor Pex26p in shuttling of PTS1-receptor Pex5p during peroxisome biogenesis. Biochim Biophys Acta 1823: 145–149 [DOI] [PubMed] [Google Scholar]
- Goto S, Mano S, Nakamori C, Nishimura M. (2011) Arabidopsis ABERRANT PEROXISOME MORPHOLOGY9 is a peroxin that recruits the PEX1-PEX6 complex to peroxisomes. Plant Cell 23: 1573–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto-Yamada S, Mano S, Nakamori C, Kondo M, Yamawaki R, Kato A, Nishimura M. (2014) Chaperone and protease functions of LON protease 2 modulate the peroxisomal transition and degradation with autophagy. Plant Cell Physiol 55: 482–496 [DOI] [PubMed] [Google Scholar]
- Graham IA. (2008) Seed storage oil mobilization. Annu Rev Plant Biol 59: 115–142 [DOI] [PubMed] [Google Scholar]
- Grimm I, Saffian D, Platta HW, Erdmann R. (2012) The AAA-type ATPases Pex1p and Pex6p and their role in peroxisomal matrix protein import in Saccharomyces cerevisiae. Biochim Biophys Acta 1823: 150–158 [DOI] [PubMed] [Google Scholar]
- Hampton RY, Sommer T. (2012) Finding the will and the way of ERAD substrate retrotranslocation. Curr Opin Cell Biol 24: 460–466 [DOI] [PubMed] [Google Scholar]
- Haughn GW, Somerville C. (1986) Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol Gen Genet 204: 430–434 [Google Scholar]
- Hayashi M, Toriyama K, Kondo M, Nishimura M. (1998) 2,4-Dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid beta-oxidation. Plant Cell 10: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Yagi M, Nito K, Kamada T, Nishimura M. (2005) Differential contribution of two peroxisomal protein receptors to the maintenance of peroxisomal functions in Arabidopsis. J Biol Chem 280: 14829–14835 [DOI] [PubMed] [Google Scholar]
- Hazra PP, Suriapranata I, Snyder WB, Subramani S. (2002) Peroxisome remnants in pex3delta cells and the requirement of Pex3p for interactions between the peroxisomal docking and translocation subcomplexes. Traffic 3: 560–574 [DOI] [PubMed] [Google Scholar]
- Helm M, Lück C, Prestele J, Hierl G, Huesgen PF, Fröhlich T, Arnold GJ, Adamska I, Görg A, Lottspeich F, et al. (2007) Dual specificities of the glyoxysomal/peroxisomal processing protease Deg15 in higher plants. Proc Natl Acad Sci USA 104: 11501–11506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Aguirre M, Peto C, Alonso J, Ecker J, Chory J. (2002) A role for peroxisomes in photomorphogenesis and development of Arabidopsis. Science 297: 405–409 [DOI] [PubMed] [Google Scholar]
- Hu J, Baker A, Bartel B, Linka N, Mullen RT, Reumann S, Zolman BK. (2012) Plant peroxisomes: biogenesis and function. Plant Cell 24: 2279–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamigaki A, Kondo M, Mano S, Hayashi M, Nishimura M. (2009) Suppression of peroxisome biogenesis factor 10 reduces cuticular wax accumulation by disrupting the ER network in Arabidopsis thaliana. Plant Cell Physiol 50: 2034–2046 [DOI] [PubMed] [Google Scholar]
- Kaur N, Zhao Q, Xie Q, Hu J. (2013) Arabidopsis RING peroxins are E3 ubiquitin ligases that interact with two homologous ubiquitin receptor proteins(F). J Integr Plant Biol 55: 108–120 [DOI] [PubMed] [Google Scholar]
- Keller GA, Gould S, Deluca M, Subramani S. (1987) Firefly luciferase is targeted to peroxisomes in mammalian cells. Proc Natl Acad Sci USA 84: 3264–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan BR, Zolman BK. (2010) pex5 Mutants that differentially disrupt PTS1 and PTS2 peroxisomal matrix protein import in Arabidopsis. Plant Physiol 154: 1602–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DY, Scalf M, Smith LM, Vierstra RD. (2013a) Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell 25: 1523–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Lee H, Lee HN, Kim SH, Shin KD, Chung T. (2013b) Autophagy-related proteins are required for degradation of peroxisomes in Arabidopsis hypocotyls during seedling growth. Plant Cell 25: 4956–4966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J. (1986) The promoter of the TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204: 383–396 [Google Scholar]
- Lingard MJ, Bartel B. (2009) Arabidopsis LON2 is necessary for peroxisomal function and sustained matrix protein import. Plant Physiol 151: 1354–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingard MJ, Monroe-Augustus M, Bartel B. (2009) Peroxisome-associated matrix protein degradation in Arabidopsis. Proc Natl Acad Sci USA 106: 4561–4566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeshima M, Yokoi H, Asahi T. (1988) Evidence for no proteolytic processing during transport of isocitrate lyase into glyoxysomes in castor bean endosperm. Plant Cell Physiol 29: 381–384 [Google Scholar]
- Mano S, Nakamori C, Nito K, Kondo M, Nishimura M. (2006) The Arabidopsis pex12 and pex13 mutants are defective in both PTS1- and PTS2-dependent protein transport to peroxisomes. Plant J 47: 604–618 [DOI] [PubMed] [Google Scholar]
- Monroe-Augustus M, Ramón NM, Ratzel SE, Lingard MJ, Christensen SE, Murali C, Bartel B. (2011) Matrix proteins are inefficiently imported into Arabidopsis peroxisomes lacking the receptor-docking peroxin PEX14. Plant Mol Biol 77: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nito K, Kamigaki A, Kondo M, Hayashi M, Nishimura M. (2007) Functional classification of Arabidopsis peroxisome biogenesis factors proposed from analyses of knockdown mutants. Plant Cell Physiol 48: 763–774 [DOI] [PubMed] [Google Scholar]
- Okumoto K, Noda H, Fujiki Y. (2014) Distinct modes of ubiquitination of peroxisome-targeting signal type 1 (PTS1) receptor Pex5p regulate PTS1 protein import. J Biol Chem 289: 14089–14108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen LJ, Ettinger WF, Damsz B, Matsudaira K, Webb MA, Harada JJ. (1993) Targeting of glyoxysomal proteins to peroxisomes in leaves and roots of a higher plant. Plant Cell 5: 941–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osumi T, Tsukamoto T, Hata S, Yokota S, Miura S, Fujiki Y, Hijikata M, Miyazawa S, Hashimoto T. (1991) Amino-terminal presequence of the precursor of peroxisomal 3-ketoacyl-CoA thiolase is a cleavable signal peptide for peroxisomal targeting. Biochem Biophys Res Commun 181: 947–954 [DOI] [PubMed] [Google Scholar]
- Platta HW, El Magraoui F, Schlee D, Grunau S, Girzalsky W, Erdmann R. (2007) Ubiquitination of the peroxisomal import receptor Pex5p is required for its recycling. J Cell Biol 177: 197–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platta HW, Hagen S, Erdmann R. (2013) The exportomer: the peroxisomal receptor export machinery. Cell Mol Life Sci 70: 1393–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platta HW, El Magraoui F, Bäumer BE, Schlee D, Girzalsky W, Erdmann R. (2009) Pex2 and pex12 function as protein-ubiquitin ligases in peroxisomal protein import. Mol Cell Biol 29: 5505–5516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pracharoenwattana I, Cornah JE, Smith SM. (2007) Arabidopsis peroxisomal malate dehydrogenase functions in β-oxidation but not in the glyoxylate cycle. Plant J 50: 381–390 [DOI] [PubMed] [Google Scholar]
- Prestele J, Hierl G, Scherling C, Hetkamp S, Schwechheimer C, Isono E, Weckwerth W, Wanner G, Gietl C. (2010) Different functions of the C3HC4 zinc RING finger peroxins PEX10, PEX2, and PEX12 in peroxisome formation and matrix protein import. Proc Natl Acad Sci USA 107: 14915–14920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramón NM, Bartel B. (2010) Interdependence of the peroxisome-targeting receptors in Arabidopsis thaliana: PEX7 facilitates PEX5 accumulation and import of PTS1 cargo into peroxisomes. Mol Biol Cell 21: 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratzel SE, Lingard MJ, Woodward AW, Bartel B. (2011) Reducing PEX13 expression ameliorates physiological defects of late-acting peroxin mutants. Traffic 12: 121–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuhmann H, Huesgen PF, Gietl C, Adamska I. (2008) The DEG15 serine protease cleaves peroxisomal targeting signal 2-containing proteins in Arabidopsis. Plant Physiol 148: 1847–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann U, Prestele J, O’Geen H, Brueggeman R, Wanner G, Gietl C. (2007) Requirement of the C3HC4 zinc RING finger of the Arabidopsis PEX10 for photorespiration and leaf peroxisome contact with chloroplasts. Proc Natl Acad Sci USA 104: 1069–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann U, Wanner G, Veenhuis M, Schmid M, Gietl C. (2003) AthPEX10, a nuclear gene essential for peroxisome and storage organelle formation during Arabidopsis embryogenesis. Proc Natl Acad Sci USA 100: 9626–9631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes IA, Brandizzi F, Slocombe SP, El-Shami M, Hawes C, Baker A. (2003) An Arabidopsis pex10 null mutant is embryo lethal, implicating peroxisomes in an essential role during plant embryogenesis. Plant Physiol 133: 1809–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes IA, Hawes C, Baker A. (2005) AtPEX2 and AtPEX10 are targeted to peroxisomes independently of known endoplasmic reticulum trafficking routes. Plant Physiol 139: 690–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasinopoulos TC, Hangarter RP. (1990) Preventing photochemistry in culture media by long-pass light filters alters growth of cultured tissues. Plant Physiol 93: 1365–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Bartel B. (2011) Transport and metabolism of the endogenous auxin precursor indole-3-butyric acid. Mol Plant 4: 477–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Culler AH, Cohen JD, Bartel B. (2010) Conversion of endogenous indole-3-butyric acid to indole-3-acetic acid drives cell expansion in Arabidopsis seedlings. Plant Physiol 153: 1577–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Wheeler DL, Christensen SE, Berens JC, Cohen JD, Rampey RA, Bartel B. (2011) Multiple facets of Arabidopsis seedling development require indole-3-butyric acid-derived auxin. Plant Cell 23: 984–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinkels BW, Gould SJ, Bodnar AG, Rachubinski RA, Subramani S. (1991) A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J 10: 3255–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms S, Erdmann R. (2006) Peroxisomal matrix protein receptor ubiquitination and recycling. Biochim Biophys Acta 1763: 1620–1628 [DOI] [PubMed] [Google Scholar]
- Williams C, Distel B. (2006) Pex13p: docking or cargo handling protein? Biochim Biophys Acta 1763: 1585–1591 [DOI] [PubMed] [Google Scholar]
- Williams C, van den Berg M, Geers E, Distel B. (2008) Pex10p functions as an E3 ligase for the Ubc4p-dependent ubiquitination of Pex5p. Biochem Biophys Res Commun 374: 620–624 [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. (2005a) Auxin: regulation, action, and interaction. Ann Bot (Lond) 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. (2005b) The Arabidopsis peroxisomal targeting signal type 2 receptor PEX7 is necessary for peroxisome function and dependent on PEX5. Mol Biol Cell 16: 573–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Fleming WA, Burkhart SE, Ratzel SE, Bjornson M, Bartel B. (2014) A viable Arabidopsis pex13 missense allele confers severe peroxisomal defects and decreases PEX5 association with peroxisomes. Plant Mol Biol 86: 201–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Bartel B. (2004) An Arabidopsis indole-3-butyric acid-response mutant defective in PEROXIN6, an apparent ATPase implicated in peroxisomal function. Proc Natl Acad Sci USA 101: 1786–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Martinez N, Millius A, Adham AR, Bartel B. (2008) Identification and characterization of Arabidopsis indole-3-butyric acid response mutants defective in novel peroxisomal enzymes. Genetics 180: 237–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Monroe-Augustus M, Silva ID, Bartel B. (2005) Identification and functional characterization of Arabidopsis PEROXIN4 and the interacting protein PEROXIN22. Plant Cell 17: 3422–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Nyberg M, Bartel B. (2007) IBR3, a novel peroxisomal acyl-CoA dehydrogenase-like protein required for indole-3-butyric acid response. Plant Mol Biol 64: 59–72 [DOI] [PubMed] [Google Scholar]
- Zolman BK, Silva ID, Bartel B. (2001) The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid beta-oxidation. Plant Physiol 127: 1266–1278 [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Yoder A, Bartel B. (2000) Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156: 1323–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.