Indaziflam is a cellulose biosynthesis inhibitor that has a different mechanism of action than previously characterized inhibitors and causes an increase in CELLULOSE SYNTHASE A particle density and reduced, but not paused, velocity at the plasma membrane focal plane.
Abstract
Cellulose biosynthesis is a common feature of land plants. Therefore, cellulose biosynthesis inhibitors (CBIs) have a potentially broad-acting herbicidal mode of action and are also useful tools in decoding fundamental aspects of cellulose biosynthesis. Here, we characterize the herbicide indaziflam as a CBI and provide insight into its inhibitory mechanism. Indaziflam-treated seedlings exhibited the CBI-like symptomologies of radial swelling and ectopic lignification. Furthermore, indaziflam inhibited the production of cellulose within <1 h of treatment and in a dose-dependent manner. Unlike the CBI isoxaben, indaziflam had strong CBI activity in both a monocotylonous plant (Poa annua) and a dicotyledonous plant (Arabidopsis [Arabidopsis thaliana]). Arabidopsis mutants resistant to known CBIs isoxaben or quinoxyphen were not cross resistant to indaziflam, suggesting a different molecular target for indaziflam. To explore this further, we monitored the distribution and mobility of fluorescently labeled CELLULOSE SYNTHASE A (CESA) proteins in living cells of Arabidopsis during indaziflam exposure. Indaziflam caused a reduction in the velocity of YELLOW FLUORESCENT PROTEIN:CESA6 particles at the plasma membrane focal plane compared with controls. Microtubule morphology and motility were not altered after indaziflam treatment. In the hypocotyl expansion zone, indaziflam caused an atypical increase in the density of plasma membrane-localized CESA particles. Interestingly, this was accompanied by a cellulose synthase interacting1-independent reduction in the normal coincidence rate between microtubules and CESA particles. As a CBI, for which there is little evidence of evolved weed resistance, indaziflam represents an important addition to the action mechanisms available for weed management.
Cellulose is a composite polymer of β-1,4-linked glucan chains and is the main load-bearing structure of plant cell walls (Jarvis, 2013). Although cellulose is a relatively simple polysaccharide molecule, its synthesis is quite complex. The principle catalytic unit is a plasma membrane (PM)-localized protein complex referred to as the cellulose synthase complex (CSC; Davis, 2012). In plants, the CSC, visualized with freeze fracture microscopy, is a solitary, hexagonal rosette-shaped complex (Herth and Weber, 1984; Delmer, 1999) and at least three of the catalytic CELLULOSE SYNTHASE A (CESA) proteins are required in each CSC for the production of cellulose (Desprez et al., 2007; Persson et al., 2007). In addition to CESAs, several accessory proteins have been discovered to be necessary for the production and deposition of cellulose, such as KORRIGAN (Lane et al., 2001), COBRA (Roudier et al., 2005) and CELLULOSE SYNTHASE INTERACTING1 (CSI1; Gu et al., 2010), as well as several others that are yet to be identified. The loss of function in any of the aforementioned proteins causes complete or partial loss of anisotropic growth in cells undergoing expansion, resulting in radial swelling. Severe radial swelling in rapidly expanding tissue is also a common symptomology observed in seedlings treated with cellulose biosynthesis inhibitors (CBIs). Therefore, numerous potential herbicidal targets exist (mechanisms of action) for the broad group of known CBIs.
Classification of an herbicide to the CBI designation was traditionally achieved by short-term [14C]radioisotope tracer studies focused on the incorporation of Glc into cellulose (Heim et al., 1990; Sabba and Vaughn, 1999). More recently, time-lapse confocal microscopy of reporter-tagged CESA proteins (Paredez et al., 2006) has been used to further classify CBIs. CBIs can be classified into at least three primary groups based on how treatment disrupts the normal tracking and localization of fluorescently labeled CESAs (for review, see Brabham and DeBolt, 2012). The disruption is, it can be assumed, the result of the inhibitory mechanism of the CBI. In the first group, isoxaben and numerous other compounds cause YELLOW FLUORESCENT PROTEIN YFP):CESAs to be depleted from the PM and concomitantly accumulate in cytosolic vesicles (called small CESA compartments or microtubule-associated cellulose synthase compartments; Paredez et al., 2006; Crowell et al., 2009; Gutierrez et al., 2009) The second group, consisting only of dichlobenil (DCB), causes YFP:CESAs to become immobilized and hyperaccumulated at distinct foci in the PM (Herth, 1987; DeBolt et al., 2007b). The third group influences CSC-microtubule (MT)-associated functions resulting in errant movement and localization of YFP:CESAs (DeBolt et al., 2007a; Yoneda et al., 2007). These different disruption processes suggest that each CBI group targets a different aspect of the complex cellulose biosynthetic process.
A lack of evolved weed resistance in the field suggests that CBIs are potentially underutilized tools for weed control (Sabba and Vaughn, 1999; Heap, 2014). CBIs have also been useful research tools in decoding fundamental aspects of cellulose biosynthesis. An exogenous application of a CBI provides spatial and temporal inhibition of cellulose. Resistance screens to CBIs have uncovered key genes in cellulose biosynthesis (Scheible et al., 2001; Desprez et al., 2002). Furthermore, CBIs such as isoxaben have also been effective in linking accessory proteins with CESAs in the CSC (Robert et al., 2005; Gu et al., 2010). Therefore, it is important to extend our range of CBI compounds. Indaziflam (Fig. 1A), an herbicide introduced by Bayer Crop Science, was recently proposed to be a CBI and was reported to have a photosystem II inhibition value of 9.4 (Meyer et al., 2009; Dietrich and Laber, 2012). Indaziflam is labeled for use in turf, for perennial crops, and for nonagricultural situations for preemergent control of grasses and broadleaf weeds (Meyer et al., 2009; Brosnan et al., 2011). The aim herein was to investigate indaziflam as a CBI and to characterize its inhibitory effect on cellulose biosynthesis.
Figure 1.
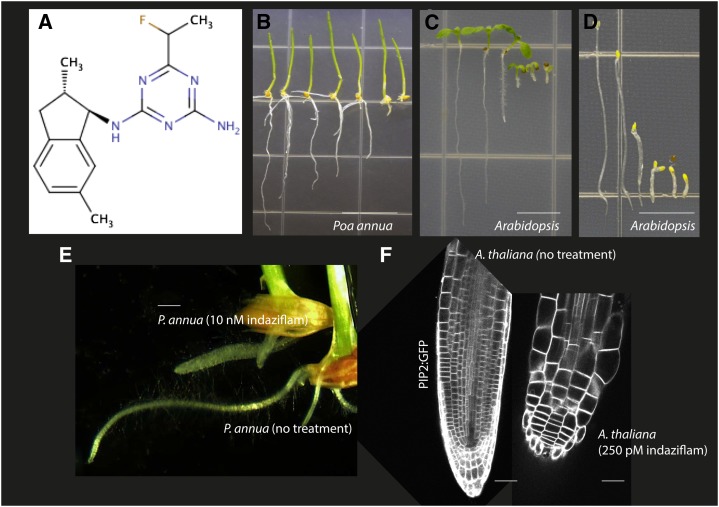
Indaziflam is a fluoroalkytriazine-containing compound that inhibits elongation in seedlings of P. annua and Arabidopsis. A, Chemical structure of indaziflam. B to D, Images of 7-d-old seedlings treated with increasing concentrations of indaziflam. B shows light-grown P. annua seedlings (indaziflam concentrations from left to right are 0, 100, 250, 500, 1,000, 5,000, and 10,000 pm). C and D show light-grown and dark-grown Arabidopsis seedlings, respectively (indaziflam concentrations from left to right are 0, 100, 250, 500, 1,000, and 2,500 pm). Indaziflam treatment induced swollen cells. E, Representative images of the primary root of P. annua grown in plates for 4 d with and without 10 nm indaziflam. F, Transgenic Arabidopsis seedlings expressing GFP:PIP2 were examined by laser scanning confocal microscopy and images represent visualization of the primary root grown vertically for 7-d plates without and with 250 pm indaziflam. PIP2, Plasma membrane intrinsic protein2. Bar = 10 mm in B, 5 mm in C and D, 2 mm in E, and 50 μm in F.
RESULTS
Indaziflam-Treated Seedlings Exhibit CBI Symptomologies
Dicotyledonous Arabidopsis (Arabidopsis thaliana) and monocotyledonous Poa annua were germinated and grown on plates for 7 d with various concentrations of indaziflam. Seedlings were grown using either a light (24-h light/0-h dark) or dark (0-h light/24-h dark) growth regimen to promote root or hypocotyl expansion, respectively. Both P. annua and Arabidopsis were susceptible to indaziflam and their growth was inhibited in a dose-dependent manner (Fig. 1, B–D). The growth reduced by 50% (GR50) values for light-grown P. annua, dark-grown Arabidopsis, and light-grown Arabidopsis were 671 ρm, 214 ρm, and 200 ρm of indaziflam, respectively (Supplemental Fig. S1). The similar GR50 values between the light- and dark-grown Arabidopsis seedlings suggests the phytotoxic effects of indaziflam do not require light. This eliminated several possible herbicidal modes of action for indaziflam that are dependent on light for toxicity (i.e. photosynthesis, chlorophyll, and pigment inhibitors). Visually, indaziflam-treated seedlings exhibited radial swelling (Fig. 1, E and F) and phloroglucinol staining revealed that indaziflam caused ectopic lignification, both of which are common characteristics of CBIs (Desprez et al., 2002; Supplemental Fig. S2).
Indaziflam Inhibits Cellulose Biosynthesis
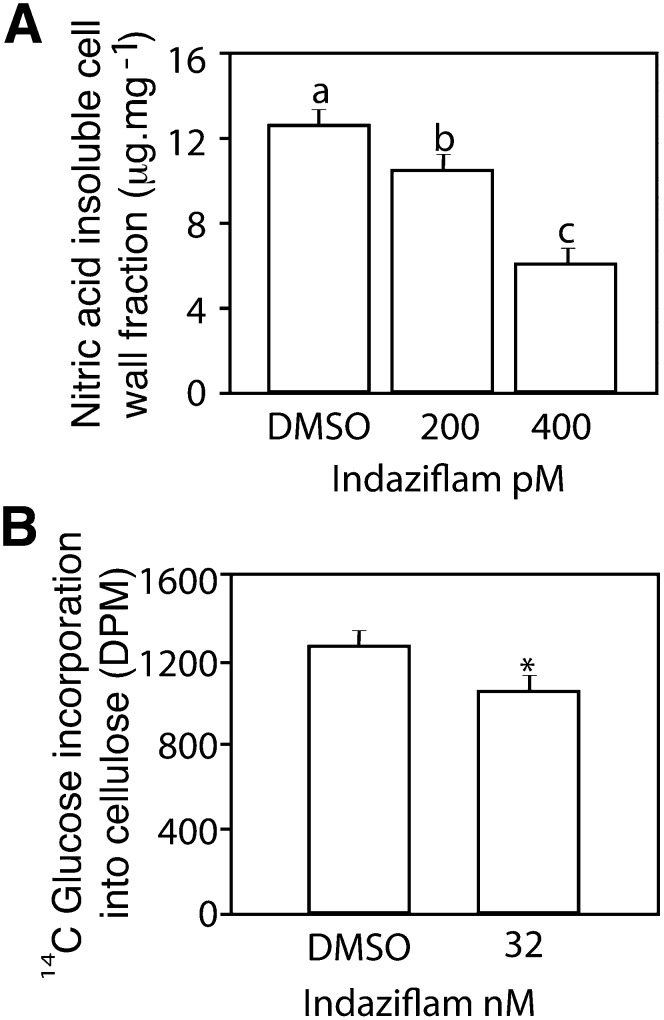
Classification of an herbicide as a CBI has traditionally been based on inhibition of cellulose synthesis in treated plants (Sabba and Vaughn, 1999). Cellulose is polymerized from the substrate UDP-Glc by glucosyltransferase CESA proteins (Delmer, 1999) and it can be partitioned from other polysaccharides by treatment with nitric acid. In crude cell wall extracts from the hypocotyl region of 5-d-old etiolated Arabidopsis seedlings, indaziflam reduced the amount of nitric acid-insoluble material (considered crystalline cellulose; Updegraff, 1969; Fig. 2A). This effect was dose dependent because indaziflam at 200 and 400 ρm reduced the Glc content of the acid-insoluble fraction by 18% and 51%, respectively, compared with the control (12.7 μg mg−1). Furthermore, indaziflam inhibited the incorporation of [14C]Glc into the acid-insoluble cellulose fraction within 1 h of treatment (Fig. 2B). Thus, indaziflam inhibited the production of cellulose soon after treatment (<1 h) and in a dose-dependent manner. This is consistent with inhibition of cellulose biosynthesis as the primary mode of action for indaziflam.
Figure 2.
Indaziflam treatment quantitatively inhibited the production of cellulose. A, The amount of acid-insoluble Glc content (crystalline cellulose) from pooled etiolated hypocotyl regions (5 mg of dry weight) of 5-d-old dark-grown Arabidopsis seedlings after treatment with indaziflam at 0 (0.01% DMSO mock), 200, or 400 pm. B, The inhibitory effects of indaziflam on the incorporation of [14C]Glc into the acid-insoluble cellulose fraction of 3-d-old etiolated dark-grown Arabidopsis seedlings after a 1-h treatment. The amount of radioactivity was determined by liquid scintillation spectrometry. In graphs, means were separated using Tukey’s test (A) or a Student’s t test (B) and different letters or asterisks indicate a significant difference at an α < 0.05. Error bars represent ±1 se (n = 5 for A and B). DPM, Disintegrations per minute.
Isoxaben- and Quinoxyphen-Resistant Plants Are Not Cross Resistant to Indaziflam
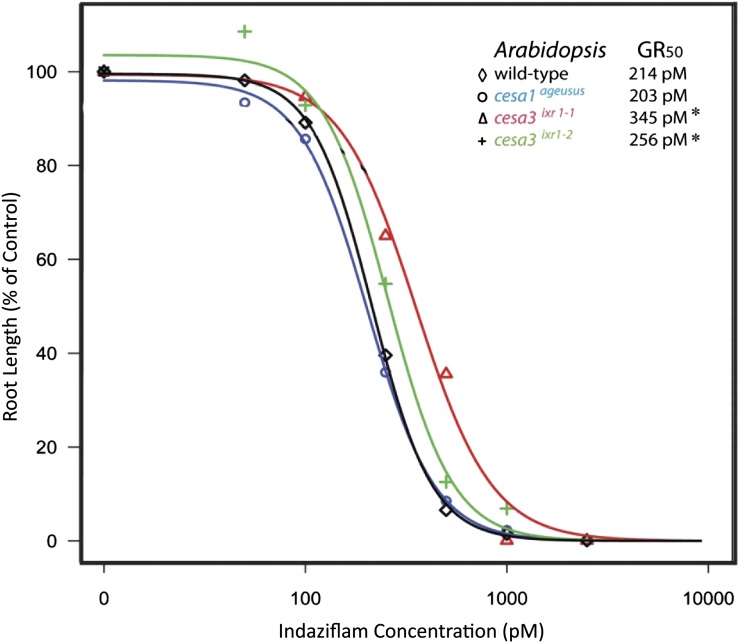
To determine whether indaziflam has the same mechanism of action as two other characterized CBIs, we tested whether known isoxaben- and quinoxyphen-resistant Arabidopsis mutants were cross resistant to indaziflam (Fig. 3). The mutants used were cesa3ixr1-1, cesa3ixr1-2, and cesa1ageusus. Isoxaben-resistant mutants cesa3ixr1-1 and cesa3ixr1-2 (Heim et al., 1989; Scheible et al., 2001) and the quinoxyphen-resistant mutant cesa1ageusus (Harris et al., 2012) have point mutations in the C terminus trans-spanning membrane domains and not in the cytosolic catalytic domain that confer resistance to their respective herbicide. The results were somewhat inconclusive as to whether the isoxaben- and quinoxyphen-resistant mutants were cross resistant to indaziflam. There were differences based upon GR50 values in the susceptibility of the wild type and mutants to indaziflam. The isoxaben-resistant mutants cesa3ixr1-1 (P < 0.0001) and cesa3ixr1-2 (P < 0.036) grown in the light both exhibited minor tolerance (<2-fold) to indaziflam compared with the wild type. However, these same mutants have a 300-fold and 90-fold level of resistance to isoxaben, respectively (Heim et al., 1989). In the dark, only cesa3ixr1-1 (P < 0.0001) exhibited any tolerance to indaziflam compared with the wild type (GR50 values of 275 versus 214 ρm). The cesa1ageusus mutant and an additional isoxaben-resistant mutant, cesa6ixr2-1 (Desprez et al., 2002; data not shown), were equally sensitive to indaziflam as wild-type plants whether grown in light or dark. Our results do not support indaziflam as having the same mechanism of action as quinoxyphen or isoxaben.
Figure 3.
Indaziflam dose response and GR50 values of light-grown Arabidopsis genotypes. To establish dose responses, seedlings were germinated in the light on agar plates containing indaziflam concentrations ranging from 0 to 10,000 pm. Seedling root length was measured and standardized as a percentage of the control. The Arabidopsis seedlings used in this assay were the Columbia ecotype as the wild type and mutants previously confirmed resistant to other CBIs. The cesa3ixr1-1 and cesa3ixr1-2 mutants are resistant to isoxaben and cesa1ageusus is resistant to quinoxyphen. The curves and GR50 values were generated by R software using the drc package. Asterisks indicate a significant difference (n = 60; P < 0.05) in the GR50 values between the mutant and the wild type. [See online article for color version of this figure.]
Indaziflam Caused Reduced Particle Velocity and Increased Accumulation of CESA Particles at the PM Focal Plane
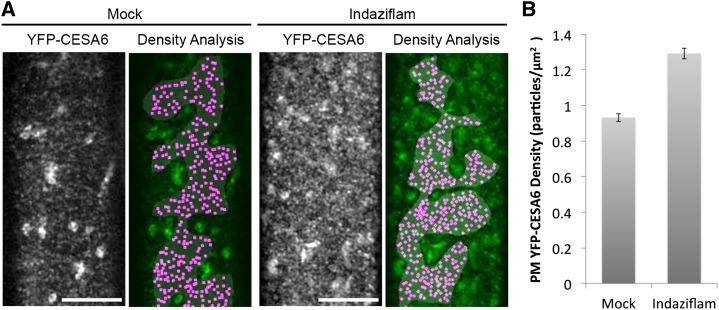
The question of how the PM-localized CSC population responds to indaziflam treatment in living cells is important to determine in order to understand the inhibitory mechanism of indaziflam. To explore this, we examined transgenic Arabidopsis plants expressing both YFP:CESA6 and RED FLUORESCENT PROTEIN (RFP):Tubulin α5 (TUA5; Gutierrez et al., 2009) during short-term exposure to indaziflam. Two questions were initially asked. First, does the entire organization of the CSC array change during indaziflam treatment or does the behavior of individual CESA particles change in response to indaziflam? Second, does indaziflam cause a similar or different inhibitory response on the PM-localized CSC population compared with previously described CBIs? To address the first question, we imaged the behavior of YFP:CESA6 and RFP:TUA5 in epidermal cells near the apical hook of etiolated Arabidopsis seedlings (Supplemental Movies S1 and S2). Analysis of time-lapse images from seedlings in the absence of indaziflam revealed a dynamic population of YFP:CESA6-labeled particles residing at the PM (Supplemental Movie S1). After indaziflam treatment (500 nm for 2 h), a greater population of YFP:CESA6 particles was observed at the PM focal plane (Fig. 4A). To quantify this, the number of distinct YFP:CESA6 particles displaying morphology and motility consistent with being membrane-localized particles was counted. In the absence of indaziflam, the density of discernable PM-localized YFP:CESA6 particles was 0.93 ± 0.02 μm−2 (Fig. 4B). By contrast, the density of YFP:CESA6 particles in indaziflam-treated cells was 30% greater (1.29 ± 0.02 μm−2; Fig. 4B). This response to indaziflam was consistent throughout the hypocotyl cells but was most prominent in expanding cells subtending the apical hook. Thus, indaziflam induced an atypical increase in the population density of CESA particles at the PM, consistent with broad disturbance of array organization.
Figure 4.
Indaziflam treatment induced a higher density of CESAs at the PM. Arabidopsis seedlings expressing YFP:CESA6 were grown in the dark for 3 d before imaging. A, Representative images and analysis of the PM-localized YFP:CESA6 particles in the prc1-1 background are shown. Single optical sections (monochrome) show the distribution of YFP:CESA6-labeled puncta upon 2-h 0.01% DMSO mock treatment (left) or 500 nm indaziflam treatment (right). The green/magenta overlay is a spatial count of the puncta that display morphology and motility consistent with PM YFP:CESA6 particles. A gray mask indicates the region of interest lacking underlying intracellular compartments, and magenta dots indicate local maxima of the fluorescence signal. B, Upon indaziflam treatment, the average density of YFP:CESA6 puncta at the PM increased. n = 15 cells from nine seedlings for mock and n = 18 cells from 12 seedlings for indaziflam. Error bars are ±1 se from the mean. Bar = 10 μm.
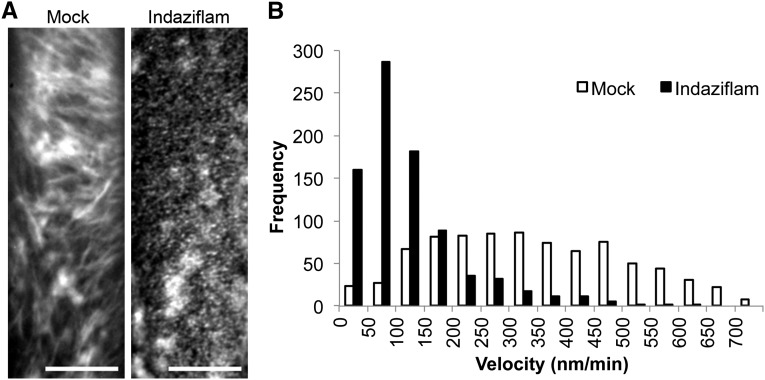
Individual CESA particles can also be tracked and some aspects of their behavior can be measured. One measurement is the velocity (positional movement) of PM-localized CESA particles. However, the actual movement of CESA particles at the PM is independent of MTs (Paredez et al., 2006; DeBolt et al., 2007a). Thus, an MT motor function in propelling CESA particles is unlikely. Rather, the movement of CESA particles was proposed to be a function of a polymerization force generated by the translocating glucan chain (Diotallevi and Mulder, 2007). The PM movement of CESA particles in untreated cells was bidirectional with an average velocity of 336 ± 167 nm min−1, which is consistent with numerous prior studies (Paredez et al., 2006; Bischoff et al., 2009; Crowell et al., 2009; Gutierrez et al., 2009; Gu et al., 2010; Li et al., 2012). After treatment with indaziflam, YFP:CESA6 velocity was reduced to 119 ± 95 nm min−1 (Fig. 5). Thus, indaziflam reduced CESA particle velocity by 65%, which is consistent with a role in inhibiting polymerization.
Figure 5.
Indaziflam reduced the velocity (particle movement rate) of YFP:CESA6. A, Representative time-lapse images of YFP:CESA6 particles in the prc1-1 background with and without indaziflam treatment (61 frames averaged). B, The histogram depicts the frequency of YFP:CESA6 particle velocities at the PM focal plane after a 2-h treatment with indaziflam or DMSO mock. Velocity was determined from images taken in the epidermal cells of 3-d-old dark-grown hypocotyls. The white bars are the recorded velocity from the mock and the black bars are indaziflam treatment (mean ±1 se). Bar = 10 μm
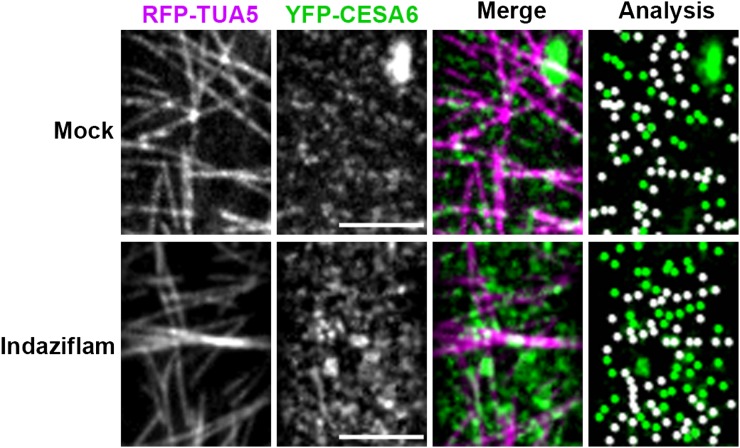
With the observed atypical increase in CESA density, we asked whether the rate of coincidence between MTs and CESA was altered by indaziflam. In the molecular rail hypothesis proposed by Giddings and Staehelin (1988), CESA particles are guided by the underlying cortical MTs. The coincidence between PM CESA particles and MTs is normally around 70% to 80% (Paredez et al., 2006; Li et al., 2012). The average colocalization rate over three experimental runs (total n = 544) between YFP:CESA6 particles and RFP:TUA5 after indaziflam treatment was 53% ± 4%. This was considerably less than the 71% ± 1% colocalization rate (total n = 303) observed in mock-treated cells (Fig. 6; Table I). This disruption in the colocalization between CESAs and MTs was prominent in expanding cells but was less apparent in cells that had undergone expansion further down the hypocotyl (Supplemental Fig. S3). Thus, the increased CESA density after indaziflam treatment appears to contribute to the decreased colocalization between MTs and CESA in the region close to the apical hook.
Figure 6.
Indaziflam treatment decreased the net colocalization between MTs and YFP:CESA6 at the PM. Arabidopsis seedlings expressing both RFP:TUA5 and YFP:CESA6 in prc1-1 were grown in the dark for 3 d before imaging. Representative single optical sections (monochrome) of cortical MTs labeled by RFP:TUA5 (magenta) and PM-localized YFP:CESA6 (green) were used for the colocalization analysis (Table I). After 2 h in 0.01% DMSO mock, 71% ± 1% of YFP:CESA6 particles were coaligned with MTs, which was not different from the ratio without any treatment (Li et al., 2012). After 2 h in 500 nm indaziflam, the colocalization ratio between YFP:CESA6 and RFP:TUA5 decreased to 53% ± 4%, which was not significantly different from the expected random ratio association of 47% ± 10%. Bar = 5 μm.
Table I. Quantification of colocalization between CESA and MTs.
Indaziflam treatment comprised 500 nm indaziflam for 2 h.
| YFP:CESA6 versus RFP:TUA5 | Mock Control | Indaziflam Treatment |
|---|---|---|
| No. of colocalized voxels | 303 | 544 |
| Percentage of material colocalized | 71 ± 1 | 53 ± 4 |
| P value | 0.013 | 0.398 |
| Percentage of expected random colocalized | 47 ± 10 | 47 ± 10 |
Reduced CESA Velocity after Indaziflam Treatment Is CSI1 Independent
CSI1 was identified as a primary linker protein between MTs and CSCs (Gu et al., 2010; Bringmann et al., 2012; Lei et al., 2012; Li et al., 2012). In csi1 mutants, CESA particles in the PM were found to display reduced velocity and their association with MTs was completely disrupted (Gu et al., 2010; Li et al., 2012). Because this cellular phenotype is similar to what we observed in wild-type seedlings treated with indaziflam, we explored the effect of indaziflam on the behavior of CESA particles in the csi1-3 mutant background.
The velocity of YFP:CESA6 at the PM focal plane in untreated csi1-3 was 236 ± 114 nm min−1 and, as expected, was slower than that observed in the untreated wild type (336 ± 167 nm min−1; Supplemental Fig. S4, A and B). However, upon treatment with indaziflam, YFP:CESA6 velocity in csi1-3 was further reduced from 236 ± 114 to 125 ± 102 nm min−1. Indaziflam also caused a significant increase in the number of PM-localized YFP:CESA6 particles on average to 1.25 particles per μm−2 in both csi1-3 and wild-type seedlings (Supplemental Fig. S5, A and B). These data suggest that the mechanism of action of indaziflam does not depend on a functional CSI1, otherwise the velocity of YFP:CESA6 in the csi1-3 background should not have been altered.
DISCUSSION
Indaziflam caused CBI symptomologies, including radial swelling and ectopic lignification, in both Arabidopsis and P. annua treated seedlings (Fig. 1). Furthermore, indaziflam inhibited the production of cellulose in Arabidopsis seedlings in a dose-dependent manner and within 1 h of treatment (Fig. 2). On the basis of these findings, the mode of action of indaziflam is consistent with its classification as a CBI. In characterizing the mechanisms of action of CBIs, it is important to understand the complexity of cellulose biosynthesis. In higher plants, a solitary, hexagonal rosette-shaped CSC synthesizes cellulose at the PM (Herth and Weber, 1984; Delmer, 1999). Recent data suggest that the CSC consists of 18 to 24 catalytic CESA proteins producing a microfibril with a cross sectional area of around 7 nm2 (Jarvis, 2013). Moreover, an incomplete but growing list of accessory proteins that are required for the functionality of CSCs may serve as potential CBI targets. Examples of such accessory proteins are KORRIGAN (endo-1,4-β-d-glucanase; Lane et al., 2001), COBRA (glycosyl-phosphatidyl inositol-anchored protein; Roudier et al., 2005), and CSI1 (Lei et al., 2012). Thus, there are many potential targets for CBIs and they may be further classified according to the specific mechanism of action. Traditional biochemical methodologies used to illustrate drug molecular mechanisms are not yet applicable to CBIs. To date, purification of functionally active cellulose-producing CSCs or CESAs has been challenging (Lai-Kee-Him et al., 2002) and insufficiently robust to enable in vitro drug affinity-binding assays. Furthermore, despite a crystallized bacterial CESA homolog (Morgan et al., 2013), both CESAs and CSCs have sufficiently diverged over time so that CBIs do not exhibit activity on bacteria (Tsekos, 1999; Morgan et al., 2013; Sethaphong et al., 2013). Therefore, determining how a given CBI disrupts cellulose biosynthesis has utilized live-cell imaging of CESA proteins in the presence of a CBI.
Through confocal microscopy, we demonstrated that indaziflam caused an atypical increase in CESA particle density and reduced, but not paused, velocity at the PM focal plane (Figs. 4 and 5). Indaziflam is clearly different from the CBIs quinoxyphen, isoxaben, and thaxtomin-A, which all induce a rapid clearance of CESA particles from the PM focal plane (Paredez et al., 2006; Bischoff et al., 2009; Harris et al., 2012). This corroborates our findings of a lack of cross resistance to indaziflam in isoxaben- or quinoxyphen-resistant mutants (Fig. 3). Similarly, morlin and cobteron (DeBolt et al., 2007a; Yoneda et al., 2007) affect both MT and CESA arrays, which was not the case for indaziflam. Indaziflam effects also share little similarity with those caused by DCB. DCB causes YFP:CESA6 particles to stop moving and hyperaccumulate at single foci in the PM focal plane (Herth and Weber, 1984; DeBolt et al., 2007b). Although both DCB and indaziflam caused CESA particles to accumulate in the PM, indaziflam induced CESA accumulation in both MT-rich and MT-poor regions and DCB appears to cause accumulation at distinct foci in MT-rich regions (DeBolt et al., 2007b). Furthermore, DeBolt et al. (2007b) found that the majority of the accumulated PM-localized YFP:CESA6 particles did not exhibit detectable movement 1 h after treatment (maximum velocity 34 nm min−1). However, in our study, the average particle velocity after indaziflam treatment was 119 ± 95 nm min−1. In all, the data suggest that indaziflam influences a different component of the complex cellulose biosynthetic process than other CBIs.
Interestingly, despite no obvious effect on the cortical MT morphology or motility, CESA-MT coincidence (Paredez et al., 2006) was uncoupled in indaziflam-treated cells (Fig. 6). Here, the behavior of YFP:CESA6 in indaziflam-treated cells resembled the behavior of CESAs in the CSC-MT linker protein, csi1, mutant background (Gu et al., 2010). Specifically, in the absence of CSI1, CESA particles at the PM were uncoupled from the MT array and exhibited reduced velocity (236 ± 114 nm min−1). Indaziflam also caused reduced CESA particle velocity and partial uncoupling from the MT array. Thus, utilizing the csi1-3 mutant, we asked whether indaziflam interacts with CSI1. Results for indaziflam-treated csi1-3 were comparable to indaziflam-treated wild-type cells, suggesting that the inhibitory mechanism of indaziflam was independent of CSI1 (Supplemental Figs. 2–4). Thus, the inhibitory mechanism of indaziflam does not mimic any prior characterized CBI or genetic lesion.
To date, there have yet to be any reported cases of weed species that have evolved field resistance to CBIs (Heap, 2014). The lack of CBI-resistant weeds could be attributable to several factors. First, CBIs may be used on a relatively small scale because they are mainly registered for use in perennial cropping systems (i.e. orchards and turf), for ornamentals, or for total vegetation control. Unlike some other herbicides such as glyphosate, CBIs are often used in combination with alternative modes of action and this can lower the probability of selecting for resistance to CBIs. Fitness of CBI-resistant weeds may be another factor. Although no field resistance has been reported, point mutations conferring resistance to isoxaben (Heim et al., 1990) and quinoxyphen (Harris et al., 2012) have been generated in Arabidopsis populations treated with the mutagen ethyl methane sulfonate. The mutations were mapped to CESA genes (Scheible et al., 2001; Desprez et al., 2002; Harris et al., 2012) and each point mutation was associated with a fitness penalty. Furthermore, plant cells can be habituated to a lethal dose of CBIs by significantly alternating their cell wall composition (Díaz-Cacho et al., 1999; Mélida et al., 2010). It is yet to be seen whether the mechanism for in vitro CBI habituation observed in the cell culture system could be mimicked in developmentally complex multicellular organisms, like a plant, to confer resistance. In lieu of these data, indaziflam is a potent herbicide used at low rates, has long soil residual activity, and has broad-spectrum activity on seedlings with type I (eudicots) or type II (Poaceae) cell walls, which is not the case for isoxaben. These properties could result in over-reliance on indaziflam alone, resulting in an increased selection pressure for indaziflam-resistant weeds. If resistance is managed, indaziflam has the potential to be a valuable alternative mode of action for weed management.
MATERIALS AND METHODS
Indaziflam Dose Response and Cross Resistance
All Arabidopsis (Arabidopsis thaliana) seedlings were grown vertically on one-half-strength Murashige and Skoog (MS) Basal Salt Mixture (PhytoTechnology Laboratories) agar plates under continuous light or dark conditions. The Arabidopsis Columbia ecotype was considered the wild type in all experiments. The CBI-resistant mutants used in conjunction with the dose-response assay were isoxaben-resistant cesa3ixr1-1, cesa3ixr1-2, and cesa6ixr2-1 (Heim et al., 1989; Scheible et al., 2001) and the quinoxyphen-resistant mutant cesa1ageusus (Harris et al., 2012) Poa annua were pregerminated and seedlings (n = 12) with a protruding radicle < 1.5 mm were placed in 9-cm-wide petri dishes and grown under constant light. The petri dishes contained two Whatman filter papers soaked with 4 mL of treatment. Appropriate indaziflam (Specticle 20 WSP 20% [w/w] active ingredient; Bayer Environmental Science) rates were predetermined prior to experiments. The compatibility and surfactant ingredients present as background in Specticle were not available and were replaced with 0.01% (v/v) dimethyl sulfoxide (DMSO) or deionized water. Treatments for Arabidopsis were indaziflam at 0, 50, 100, 250, 500, 1,000, and 10,000 pm and the DMSO concentration in agar media did not exceed more than 0.01% (v/v). P. annua treatments were indaziflam at 0, 100, 250, 500, 1,000, 5,000, and 10,000 pm in water. A total of 20 hypocotyl or root lengths from each Arabidopsis line and 12 P. annua roots were measured 7 d after treatment. Experiments were replicated in time, thrice. Length data were standardized to the percentage of the untreated control in each experiment. Percentage data were analyzed in R software using the drc package to determine and compare GR50 values (Knezevic et al., 2007).
Cellulose Assay and Lignin Staining
Cellulose content in the hypocotyl region of 5-d-old dark-grown Arabidopsis seedlings was determined by boiling 5 mg dry weight of the plant in nitric acid-acetic acid (Updegraff, 1969). Treatments were indaziflam at 0, 200, or 400 pm. The insoluble material was quantified colorimetrically for Glc content using the anthrone-sulphuric acid method and back calculated to cellulose (Scott and Melvin, 1953). For lignin staining, 7-d-old light-grown seedlings were incubated in ethanol (70%) for 24 h followed by 30 min in a 2% (w/v) phloroglucinol solution (20% hydrochloric acid). Images were taken with a bright-field stereomicroscope.
[14C]Glc Cell Wall Incorporation Assay
An adapted protocol similar to that of Heim et al. (1990) was used to measure the incorporation of radiolabeled Glc into the cellulose fraction of the cell wall. Dark-grown Arabidopsis seedlings were grown for 3 d in liquid MS media supplemented with 2% (w/v) Glc. After removal from the media, seedlings (20 mg fresh weight) were measured and placed in a 1.5-mL Eppendorf tube. This represents one replication. Seedlings were then washed twice with 0.5 mL of Glc-free MS media and centrifuged, and the supernatant was removed. Next, 0.5 mL of Glc-free MS media solution containing [14C]Glc at 1 μCi mL−1 was added to each tube followed by the addition of treatments. Seedlings were treated for 1 h in the dark with either DMSO (0.01% [v/v]) or indaziflam (32 nm). Samples were centrifuged and washed three times to remove unincorporated radioactivity. The material was then boiled in nitric acid-acetic acid for 30 min, cooled, and centrifuged for 5 min to pelletize insoluble material. A total of 400 μL of supernatant was removed and placed in a 10-mL liquid scintillation vial. The remaining liquid and insoluble material was washed with 0.5 mL of water and centrifuged for 5 min at 10,000 rpm. This was repeated thrice to remove any remaining [14C]Glc in solution. The pelletized material was resuspended in water and transferred to a liquid scintillation vial. Five mL of scintillation fluid cocktail (Bio-Safe II; Research Products International) was added to each vial with either soluble or insoluble fractions and radioactivity was determined by a liquid scintillation counter.
Confocal Microscopy
For live-cell imaging, 3-d-old dark-grown seedlings expressing YFP:CESA6 (Paredez et al., 2006) or YFP:CESA6- RFP:TUA5 (Gutierrez et al., 2009) were used. In addition, to visualize Arabidopsis expansion, we examined seedlings expressing the plasma membrane intrinsic protein2::GFP (Cutler et al., 2000). Seedlings were mounted in MS liquid medium for 2 h with or without indaziflam at 500 nm. Imaging was performed on a Yokogawa CSUX1 spinning disk system featuring a DMI6000 Leica motorized microscope, a Photometrics QuantEM:512SC CCD camera, and a Leica 100×/1.4 numerical aperture oil objective. An acousto-optic tunable filter laser with three laser lines (440, 491, and 561 nm) was used to enable faster shuttering and switching between different excitations. Band-pass filters (485/30 nm for CYAN FLUORESCENT PROTEIN, 520/50 nm for GFP, 535/30 nm for YFP, and 620/60 nm for RFP) were used for emission filtering. Image analysis was performed using Metamorph (Molecular Devices), ImageJ (version 1.36b; http://rsbweb.nih.gov/ij/), and Imaris (Bitplane) software.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Indaziflam dose response of P. annua and Arabidopsis.
Supplemental Figure S2. Indaziflam treatment-induced swollen cells in P. annua and Arabidopsis seedlings.
Supplemental Figure S3. Indaziflam treatment increased density of PM CSCs even in the regions where CESA was less expressed.
Supplemental Figure S4. Indaziflam reduced the velocity (particle movement rate) of YFP:CESA6 particles independent of CSI1.
Supplemental Figure S5. YFP:CESA6 particle density analysis in csi1 background.
Supplemental Movie S1. Time lapse imaging of YFP:CESA6 in the absence of indaziflam.
Supplemental Movie S2. Time lapse imaging of YFP:CESA6 in the presence of indaziflam.
Supplementary Material
Acknowledgments
We thank David W. Ehrhardt (Carnegie Institute for Science) for the mCherry:TUA5 YFP-CESA6 line.
Glossary
- CBI
cellulose biosynthesis inhibitor
- PM
plasma membrane
- CSC
cellulose synthase complex
- DCB
dichlobenil
- MT
microtubule
- GR50
growth reduced by 50%
- MS
Murashige and Skoog
- DMSO
dimethyl sulfoxide
Footnotes
This work was supported by the U.S. Department of Energy (grant no. DOE–FOA 10–0000368 to S.D. and C.B.), the U.S. Department of Agriculture (Hatch Act grant to S.D. and J.S.), and the National Science Foundation (grant no. 1121375 to Y.G. and L.L.).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Bischoff V, Cookson SJ, Wu S, Scheible WR. (2009) Thaxtomin A affects CESA-complex density, expression of cell wall genes, cell wall composition, and causes ectopic lignification in Arabidopsis thaliana seedlings. J Exp Bot 60: 955–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabham C, Debolt S. (2012) Chemical genetics to examine cellulose biosynthesis. Front Plant Sci 3: 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann M, Li E, Sampathkumar A, Kocabek T, Hauser MT, Persson S. (2012) POM-POM2/cellulose synthase interacting1 is essential for the functional association of cellulose synthase and microtubules in Arabidopsis. Plant Cell 24: 163–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosnan JT, McCullough PE, Breeden GK. (2011) Smooth crabgrass control with indaziflam at various springs timings. Weed Technol 25: 363–366 [Google Scholar]
- Crowell EF, Bischoff V, Desprez T, Rolland A, Stierhof YD, Schumacher K, Gonneau M, Höfte H, Vernhettes S. (2009) Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21: 1141–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR. (2000) Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA 97: 3718–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JK. (2012) Combining polysaccharide biosynthesis and transport in a single enzyme: dual-function cell wall glycan synthases. Front Plant Sci 3: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBolt S, Gutierrez R, Ehrhardt DW, Melo CV, Ross L, Cutler SR, Somerville C, Bonetta D. (2007a) Morlin, an inhibitor of cortical microtubule dynamics and cellulose synthase movement. Proc Natl Acad Sci USA 104: 5854–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBolt S, Gutierrez R, Ehrhardt DW, Somerville C. (2007b) Nonmotile cellulose synthase subunits repeatedly accumulate within localized regions at the plasma membrane in Arabidopsis hypocotyl cells following 2,6-dichlorobenzonitrile treatment. Plant Physiol 145: 334–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmer DP. (1999) Cellulose biosynthesis: exciting times for a difficult field of study. Annu Rev Plant Physiol Plant Mol Biol 50: 245–276 [DOI] [PubMed] [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Höfte H, Gonneau M, Vernhettes S. (2007) Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T, Vernhettes S, Fagard M, Refrégier G, Desnos T, Aletti E, Py N, Pelletier S, Höfte H. (2002) Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6. Plant Physiol 128: 482–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Cacho P, Moral R, Encina A, Acebes JL, Alvarez J. (1999) Cell wall modifaction in bean (Phaseolus vulgaris) callus cultures tolerant to isoxaben. Physiol Plant 107: 54–59 [Google Scholar]
- Dietrich H, Laber B. (2012) Inhibitors of cellulose biosynthesis. In Kramer W, Schirmer U, Jeschke P, Witschel M, eds, Modern Crop Protection Compounds, Ed 2, Vol 1 Wiley-VCH Verlag and Co. KGaA, Weinheim, Germany, p 356 [Google Scholar]
- Diotallevi F, Mulder B. (2007) The cellulose synthase complex: a polymerization driven supramolecular motor. Biophys J 92: 2666–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddings TH, Jr, Staehelin LA. (1988) Spatial relationship between microtubules and plasma-membrane rosettes during the deposition of primary wall microfibrils in Closterium sp. Planta 173: 22–30 [DOI] [PubMed] [Google Scholar]
- Gu Y, Kaplinsky N, Bringmann M, Cobb A, Carroll A, Sampathkumar A, Baskin TI, Persson S, Somerville CR. (2010) Identification of a cellulose synthase-associated protein required for cellulose biosynthesis. Proc Natl Acad Sci USA 107: 12866–12871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez R, Lindeboom JJ, Paredez AR, Emons AMC, Ehrhardt DW. (2009) Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat Cell Biol 11: 797–806 [DOI] [PubMed] [Google Scholar]
- Harris DM, Corbin K, Wang T, Gutierrez R, Bertolo AL, Petti C, Smilgies DM, Estevez JM, Bonetta D, Urbanowicz BR, et al. (2012) Cellulose microfibril crystallinity is reduced by mutating C-terminal transmembrane region residues CESA1A903V and CESA3T942I of cellulose synthase. Proc Natl Acad Sci USA 109: 4098–4103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heap I. (2014) The international survey of herbicide resistant weeds. http://www.weedscience.org (May 5, 2014)
- Heim DR, Roberts JL, Pike PD, Larrinua IM. (1989) Mutation of locus of Arabidopsis thaliana confers resistance to the herbicide isoxaben. Plant Physiol 90: 146–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim DR, Skomp JR, Tschabold EE, Larrinua IM. (1990) Isoxaben inhibits the synthesis of acid insoluble cell wall materials in Arabidopsis thaliana. Plant Physiol 93: 695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herth W. (1987) Effects of 2,6-DCB on plasma membrane rosettes of wheat root cells. Naturwissenschaften 74: 556–557 [Google Scholar]
- Herth W, Weber G. (1984) Occurrence of the putative cellulose-synthesizing “Rosettes” in the plasma membrane of Glycine max suspension culture cells. Naturwissenschaften 71: 153–154 [Google Scholar]
- Jarvis MC. (2013) Cellulose biosynthesis: counting the chains. Plant Physiol 163: 1485–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knezevic SZ, Streibig JC, Ritz C. (2007) Utilizing R software package for dose-response studies: the concept and data analysis. Weed Technol 21: 840–848 [Google Scholar]
- Lai-Kee-Him J, Chanzy H, Müller M, Putaux JL, Imai T, Bulone V. (2002) In vitro versus in vivo cellulose microfibrils from plant primary wall synthases: structural differences. J Biol Chem 277: 36931–36939 [DOI] [PubMed] [Google Scholar]
- Lane DR, Wiedemeier A, Peng L, Höfte H, Vernhettes S, Desprez T, Hocart CH, Birch RJ, Baskin TI, Burn JE, et al. (2001) Temperature-sensitive alleles of RSW2 link the KORRIGAN endo-1,4-β-glucanase to cellulose synthesis and cytokinesis in Arabidopsis. Plant Physiol 126: 278–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L, Li S, Gu Y. (2012) Cellulose synthase interactive protein 1 (CSI1) mediates the intimate relationship between cellulose microfibrils and cortical microtubules. Plant Signal Behav 7: 714–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Lei L, Somerville CR, Gu Y. (2012) Cellulose synthase interactive protein 1 (CSI1) links microtubules and cellulose synthase complexes. Proc Natl Acad Sci USA 109: 185–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mélida H, García-Angulo P, Alonso-Simón A, Alvarez JM, Acebes JL, Encina A. (2010) The phenolic profile of maize primary cell wall changes in cellulose-deficient cell cultures. Phytochemistry 71: 1684–1689 [DOI] [PubMed] [Google Scholar]
- Meyer DF, Hanrahan R, Michel J, Monke B, Mudge L, Norton L, Olsen C, Parker A, Smith J, Spak D. (2009) Indaziflam/BCS-AA10717-A new herbicide for pre-emergent control of grasses and broadleaf weeds for turf and ornamentals. WSSA Meeting Abstracts. http://wssa.net/meeting/meeting-abstracts (May 12, 2014)
- Morgan JLW, Strumillo J, Zimmer J. (2013) Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 493: 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez AR, Somerville CR, Ehrhardt DW. (2006) Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495 [DOI] [PubMed] [Google Scholar]
- Persson S, Paredez A, Carroll A, Palsdottir H, Doblin M, Poindexter P, Khitrov N, Auer M, Somerville CR. (2007) Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc Natl Acad Sci USA 104: 15566–15571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert S, Bichet A, Grandjean O, Kierzkowski D, Satiat-Jeunemaître B, Pelletier S, Hauser MT, Höfte H, Vernhettes S. (2005) An Arabidopsis endo-1,4-β- d-glucanase involved in cellulose synthesis undergoes regulated intracellular cycling. Plant Cell 17: 3378–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier F, Fernandez AG, Fujita M, Himmelspach R, Borner GHH, Schindelman G, Song S, Baskin TI, Dupree P, Wasteneys GO, et al. (2005) COBRA, an Arabidopsis extracellular glycosyl-phosphatidyl inositol-anchored protein, specifically controls highly anisotropic expansion through its involvement in cellulose microfibril orientation. Plant Cell 17: 1749–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabba RP, Vaughn KC. (1999) Herbicides that inhibit cellulose biosynthesis. Weed Sci 47: 757–763 [Google Scholar]
- Scheible WR, Eshed R, Richmond T, Delmer D, Somerville C. (2001) Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis Ixr1 mutants. Proc Natl Acad Sci USA 98: 10079–10084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott TA, Jr, Melvin EH. (1953) Determination of dextran with anthrone. Anal Chem 25: 1656–1661 [Google Scholar]
- Sethaphong L, Haigler CH, Kubicki JD, Zimmer J, Bonetta D, DeBolt S, Yingling YG. (2013) Tertiary model of a plant cellulose synthase. Proc Natl Acad Sci USA 110: 7512–7517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsekos I. (1999) The sites of cellulose synthesis in algae: diversity and evolution of cellulose-synthesizing enzyme complexes. J Phycol 35: 635–655 [Google Scholar]
- Updegraff DM. (1969) Semimicro determination of cellulose in biological materials. Anal Biochem 32: 420–424 [DOI] [PubMed] [Google Scholar]
- Yoneda A, Higaki T, Kutsuna N, Kondo Y, Osada H, Hasezawa S, Matsui M. (2007) Chemical genetic screening identifies a novel inhibitor of parallel alignment of cortical microtubules and cellulose microfibrils. Plant Cell Physiol 48: 1393–1403 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.